Coordinating the balance between haematopoietic stem cell (HSC) quiescence and self-renewal is crucial for maintaining haematopoiesis lifelong. Equally important for haematopoietic function is modulating their localisation within their bone marrow niches, as maintenance of HSC function is tightly controlled by a complex network of intrinsic molecular mechanisms and extrinsic signalling interactions with their surrounding microenvironment 1. In this study we demonstrate that nuclear factor erythroid 2-related factor 2 (Nfe2l2, or Nrf2), well established as a global regulator of oxidative stress response, plays a regulatory role in all these aspects of HSC homeostasis. Nrf2-deficiency results in an expansion of haematopoietic stem and progenitor cells (HSPCs) due to cell-intrinsic hyperproliferation, which was accomplished at the expense of HSC quiescence and self-renewal. We further show that Nrf2 mediates both migration and retention of HSCs in their niche. Moreover, we identify a previously unrecognized link between Nrf2 and CXCR4, contributing, at least partially, to the maintenance of the main modalities of HSC function.
HSCs are characterised by their ability to self-renew and differentiate into all blood cell lineages. Consequently, regulating HSC function is critical in maintaining haematopoiesis continuously for the lifespan of the organism. Postnatally, the most primitive, quiescent HSCs reside in a relatively hypoxic microenvironmental niche in the bone marrow (BM) 2, 3 and are capable of sustaining lifelong haematopoiesis. In fact, previous studies indicate that low oxygen concentration is critical for maintaining stem cell quiescence in various tissues 4-7, including the bone marrow niche 8. Conversely, reactive oxygen species (ROS) have recently been shown to prime HSPCs for differentiation in Drosophila 9, while disruption of oxidative stress pathways leads to premature HSC senescence due to loss of HSC quiescence and decline in the capacity for self-renewal in mouse models 10-12. Taken together, these findings indicate that modulation of the redox balance plays a pivotal role in maintaining HSC functions.
Nrf2 belongs to the “cap-n-collar” subfamily of the basic leucine zipper (b-Zip) transcription factors 13, and is ubiquitously expressed as a master regulator in the antioxidant response pathway 14-17. Although Nrf2 was found to be dispensable for murine development, growth and erythropoiesis 18, due to the continual requirement for HSC protection from oxidative stress, along with recent findings that Nrf2 maintains homeostatic quiescence of intestinal stem cells in Drosophila 19, we sought to assess the role of Nrf2 in regulating HSC functions.
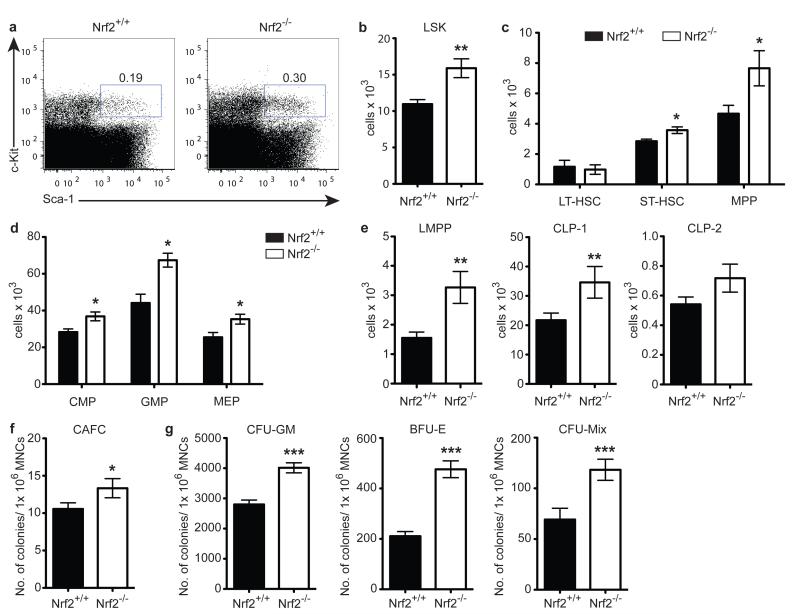
To determine whether Nrf2 plays a role in haematopoiesis, we first quantified the primitive HSPCs in Nrf2−/− mice in steady-state conditions. In young adult Nrf2-deficient animals, we observed an increase in both percentage and absolute number of LSK (Lineage−Sca-1+c-Kit+) cells compared to age-matched Nrf2+/+ wild-type (WT) controls (Fig. 1a, b), although overall BM cellularity was unchanged (data not shown). More comprehensive analysis of the LSK fraction revealed significant increases at the short-term HSC (ST-HSC, CD34+Flt3−) and continued through multipotent progenitor (MPP, CD34+Flt3+) stages, but did not impact on long-term HSCs (LT-HSC, CD150+CD48−CD34−Flt3−) (Fig. 1c). We also found increases in the number of committed downstream progenitors of both the myeloid and lymphoid lineages (Fig. 1d, e). Consistent with these findings, we also observed significantly increased colony formation from Nrf2−/− BM in cobblestone area-forming cell (CAFC) and colony-forming cell (CFC) assays (Fig. 1f, g). Together, these results suggest that loss of functional Nrf2 leads to expansion of the haematopoietic progenitor pool but spares the most primitive LT-HSCs.
Figure 1. Nrf2−/− Mice Exhibit an Expanded Haematopoietic Stem and Progenitor Pool.
WT and Nrf2−/− bone marrow (BM) under steady-state were examined from 8-10 week old female mice. (a) Representative flow cytometric analysis comparing WT to Nrf2−/− LSK frequency, gated on lineage− (Lin−) cells. (b - e) Absolute number of LSK cells (b), HSC subsets (c), myeloid progenitors (d) or lymphoid progenitors (e), mean + SEM of data from 8 animals per strain over 2 independent experiments. CMP, Common myeloid progenitor (Lin−c-kit+Sca-1− CD34+CD16/32mid); GMP, Granulocyte-macrophage progenitor (Lin−c-kit+Sca-1− CD34+CD16/32high); MEP, Megakaryocyte-erythroid progenitor (Lin−c-kit+Sca-1−CD34− CD16/32low); LMPP, Lymphoid-primed multipotent progenitor (Lin−c-kit+Sca-1 +CD34+Flt3+CD62Lhigh); CLP, Common lymphoid progenitor (CLP-1: Lin−IL-7Ra+c-kit+; CLP-2: Lin−IL-7Ra+B220+). (f) Number of colonies formed from WT or Nrf2−/− BM in cobblestone area-forming cells assay (CAFC), mean + SEM of data from 11 animals per strain over 3 independent experiments. MNCs, Mononucleated cells. (g) Number of colonies formed from WT or Nrf2−/− BM in myeloid (CFU-GM), erythroid (BFU-E), and mixed (CFU-Mix) lineages in colony-forming cell assay (CFC), data represent mean + SEM from 13 WT and 12 Nrf2−/− mice over 3 independent experiments.
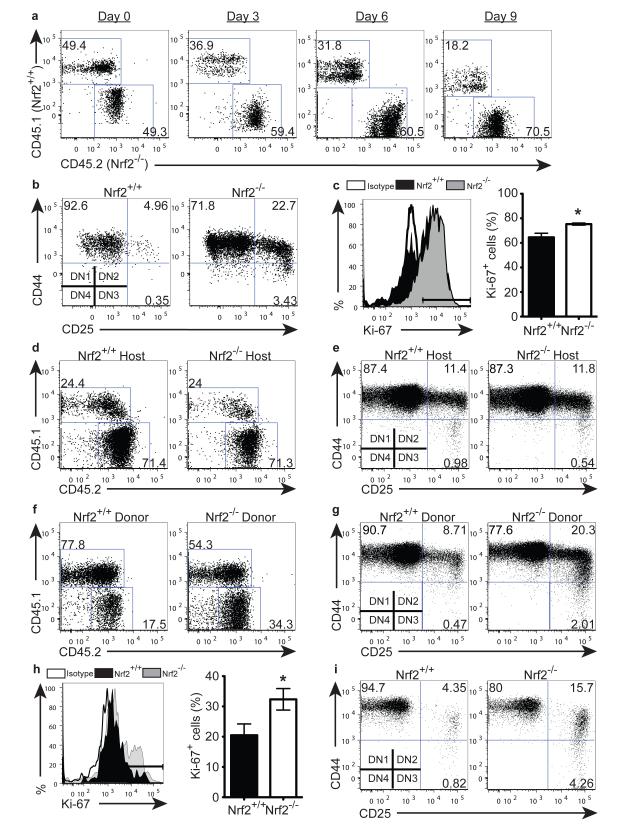
To test the impact of Nrf2 on HSPC function, we co-cultured sorted LSK cells from Nrf2−/− (CD45.2) and WT (CD45.1) BM in a competitive setting on OP9-DL1 culture, which allows the in vitro analysis of T cell development from HSPCs. As expected given their increased number in vivo (Fig. 1), even when seeded in equal numbers, Nrf2−/− LSKs dominated the culture as early as day 3 (Fig. 2a, S1a), indicating a competitive advantage in proliferation as a result of Nrf2 deficiency. T cell development progresses through a series of distinct stages that can be identified based on their differential expression of CD4, CD8, CD25 and CD44 20. In addition to their enhanced proliferation, we also observed that Nrf2−/− LSKs had enhanced differentiation kinetics compared to WT controls (Fig. 2b, S1b), rapidly progressing to the DN2 (CD4−CD8− CD44+CD25+) and DN3 (CD4−CD8−CD44−CD25+) stages. Moreover, we found significantly increased expression of Ki-67 on LSKs from Nrf2−/− compared with WT BM in vivo (Fig 2c), in concordance with the enhanced proliferation observed in vitro.
Figure 2. Nrf2 Regulates HSPC Proliferation and Differentiation Intrinsically.
(a - b) Sorted BM LSK cells were seeded onto OP9-DL1 stromal cells. (a) Representative flow cytometric analysis showing the relative contribution of Nrf2−/− (CD45.2) and WT (CD45.1) cells when co-cultured at a 1:1 ratio on day 0; reproducible in 4 independent experiments. (b) Differentiation of WT or Nrf2−/− LSKs cultured for 5 days, gated on double-negative (DN, CD4− CD8−) population; reproducible in 5 independent experiments. (c) Expression of Ki-67 in WT and Nrf2−/− BM LSKs, representative plot (left) and bar graph (right) showing mean + SEM of 4 animals per strain over 2 independent experiments. (d - e) CD45.1+ WT LSKs were preconditioned in CD45.2+ WT or Nrf2−/− animals in vivo then seeded onto OP9-DL1. (d) Co-culture of preconditioned CD45.1+ WT LSKs with untransplanted CD45.2+ WT LSKs at a 1:1 ratio on day 0. Representative flow cytometric analysis showing relative contribution of cells cultured for 12 days, reproducible in 2 independent transplant experiments. (e) Representative flow cytometric analysis showing T-cell differentiation kinetics cultured for 6 days, reproducible in 2 independent transplant experiments. (f - g) CD45.2+ WT or Nrf2−/− LSKs were preconditioned in CD45.1+ WT animals in vivo then seeded onto OP9-DL1. (f) Co-culture of untransplanted CD45.1+ WT LSKs with preconditioned CD45.2+ WT or Nrf2−/− LSKs at a 1:1 ratio at day 0. Representative flow cytometric analysis showing contribution of cells cultured for 12 days, reproducible in 2 independent transplant experiments. (g) Representative flow cytometric analysis showing T cell differentiation cultured for 6 days, reproducible in 2 independent transplant experiments. (h) Expression of Ki-67 in WT and Nrf2−/− BM LT-HSCs, representative plot (left) and bar graph (right) showing mean + SEM of 4 animals per strain over 2 independent experiments. (i) Differentiation of WT or Nrf2−/− LT-HSCs co-cultured with OP9-DL1 for 18 days, reproducible in 2 independent experiments.
Given the ubiquitous expression of Nrf2, it is essential to clarify whether these differences in HSPC proliferation and differentiation kinetics are caused extrinsically by the BM niche or intrinsically by the HSPC itself. To assess the role of the BM niche, we generated chimeras using CD45.1+ WT LSKs transferred into either CD45.2+ WT or CD45.2+ Nrf2−/− hosts. After 8 weeks, which allowed for effective primary reconstitution, we sorted LSKs from these chimeric mice and co-cultured them with OP9-DL1 stromal cells. Transplanted CD45.1+ WT LSKs behaved in a similar manner irrespective of their pre-conditioning microenvironment (Fig. 2d, e, S1c, d). In contrast, when we performed the reciprocal chimeric experiment where CD45.2+ WT or CD45.2+ Nrf2−/− LSKs, previously primed in CD45.1+ WT hosts, were co-cultured on OP9-DL1 stromal cells, we discovered once again that transplanted CD45.2+ Nrf2−/− LSKs proliferated more and differentiated faster than transplanted Nrf2+/+ WT LSKs (Fig. 2f, g, S1e, f). Taken together, these results suggest that Nrf2 regulates proliferation and differentiation of HSPCs in a cell-intrinsic manner.
Nrf2 could be found at the transcript level in both LT-HSCs as well as their downstream MPPs (Fig. S1g), suggesting that despite their unaltered frequency in deficient animals (Fig. 1c), Nrf2 could act on this earliest haematopoietic population. Consistent with our findings on the bulk LSK cells, we found a greater proportion of Nrf2−/− LT-HSCs expressed Ki-67 (Fig. 2h), indicating that more HSCs were undergoing proliferation. Functionally, Nrf2-deficient LT-HSCs differentiated more rapidly on OP9-DL1 culture than WT controls (Fig. 2i, S1h). These data demonstrate that Nrf2 plays an important role in HSPC proliferation and differentiation starting at the LT-HSC stage.
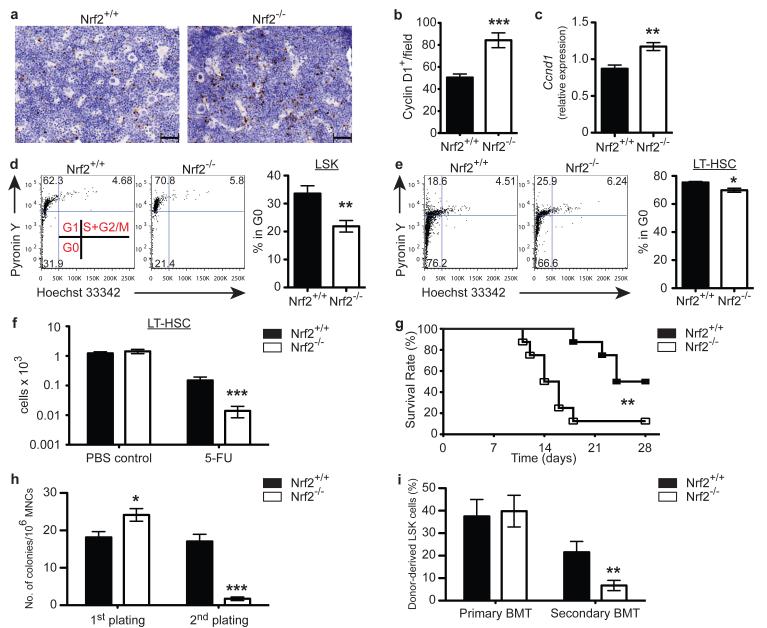
Based on these findings, we hypothesised that Nrf2 functions as a negative regulator of cell cycle entry in HSCs, actively maintaining the balance between HSC quiescence and self-renewal. As the synthesis of cyclin D is initiated during G1 phase, and its presence is required for the transition to S phase, we first assessed the level of cyclin D expression as an indirect measure of quiescence in the bone marrow. We found a 2-fold increase in cyclin D1+ cells in the Nrf2−/− BM (Fig. 3a, b), and higher levels of cyclin D at the transcript level in purified Nrf2−/− LSKs (Fig 3c). Furthermore, by analyzing RNA and DNA contents using Pyronin Y and Hoeschst 33342, we found fewer LSKs from Nrf2−/− BM in the quiescent G0 state (Fig. 3d), and importantly also reduced proportion of the most primitive LT-HSCs in G0 stage (Fig. 3e). These findings support our hypothesis that Nrf2 acts as a negative regulator of cell cycle progression, thereby maintaining HSC quiescence. To further explore this phenomenon in vivo, we exploited the characteristic of 5-Fluorouracil (5-FU), which kills cycling cells and spares quiescent stem cells. Following a single dose of 5-FU, we observed a profound reduction in the number of LT-HSCs in Nrf2−/− mice compared to WT controls (Fig. 3f). These findings suggest that the HSC compartment of Nrf2 deficient mice is considerably less quiescent than WT controls. Given the hyperproliferation of Nrf2−/− HSCs, coupled with the reduction of HSC quiescence, continued insult should lead to accelerated haematopoietic exhaustion. Consistent with our hypothesis, Nrf2−/− mice challenged with sequential 5-FU treatment, died significantly earlier than WT controls (Fig. 3g). To directly assess whether Nrf2 controls the self-renewal ability of HSCs, in addition to maintaining HSPC quiescence, we performed sequential CAFC assay in vitro and serial competitive transplantation in vivo. In both the secondary plating and transplantation, we saw a dramatic reduction in the contribution of Nrf2−/− HSCs (Fig. 3h, i). Taken together, our data demonstrate that Nrf2 plays an important role in maintaining the balance between HSC quiescence and self-renewal.
Figure 3. Nrf2 Maintains HSC Quiescence and Self-renewal.
(a) Representative immunohistochemistry images and (b) Quantification of Cyclin D+ cells in WT and Nrf2−/− BM sections. Data represent mean + SEM of 8 animals per strain. (c) Expression of Ccnd1 mRNA transcripts in WT and Nrf2−/− LSKs, relative to Actb expression. Data represent mean + SEM of 4 WT and 3 Nrf2−/− animals. (d) Representative flow cytometric analysis (Left panel) showing cell cycle status of WT and Nrf2−/− LSKs and bar graph (Right panel) showing the percentage of cells in the quiescent G0 phase. Data represent mean + SEM of 5 WT and 6 Nrf2−/− animals. (e) Representative flow cytometric analysis (Left panel) showing cell cycle status of WT and Nrf2−/− LT-HSCs and bar graph (Right panel) showing the percentage of cells in the quiescent G0 phase. Data represent mean + SEM of 4 animals per strain. (f) Absolute number of BM LT-HSCs 3.5 days after a single dose of PBS or 5-FU. Data represent the mean + SEM of 5 WT and 6 Nrf2−/− animals in PBS group and 8 WT and 9 Nrf2−/− animals in 5-FU group, reproducible in 3 independent experiments. (g) Survival of WT and Nrf2−/− mice following sequential 5-FU treatment, n = 8 animals per strain over 2 independent experiments. (h) Bar graphs showing colony formation in serial CAFC assays, mean + SEM of 8 animals per strain over 3 independent experiments. (i) Equal numbers of donor CD45.2+ (WT or Nrf2−/−) and competitor CD45.1+ (WT) LSK cells from BM were serially transplanted into lethally irradiated CD45.1+ (WT) recipients. Contribution of donor-derived (CD45.2+) BM LSKs was assessed 4 months after primary and secondary bone marrow transplants (BMT). Data represent the mean + SEM of 12 recipients of WT and 14 recipients of Nrf2−/− donor cells in primary transplant and 9 recipients of WT and 10 recipients of Nrf2−/− donor cells in secondary transplant over 2 independent experiments.
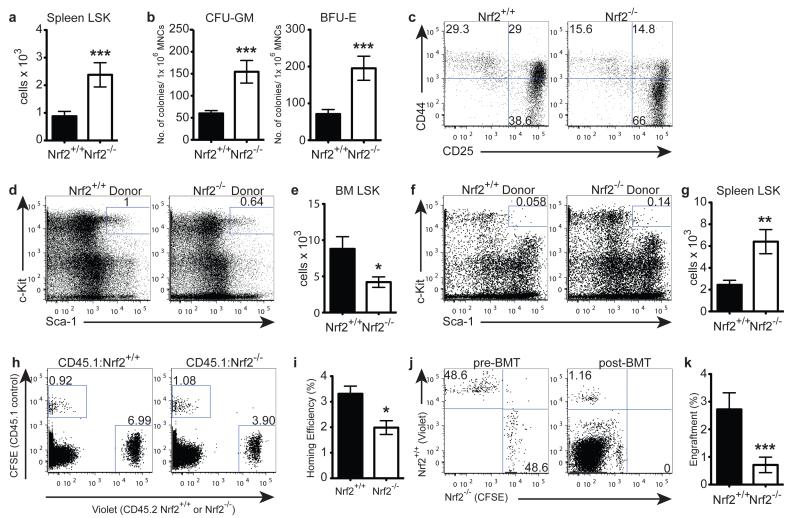
Surprisingly, despite their hyperproliferative potential in vitro, Nrf2−/− LSKs did not possess a competitive advantage in vivo in primary recipients (Fig. 3i), suggesting a potential role for Nrf2 in the homing and retention of HSPCs in the BM niche. Interestingly, in the steady-state, Nrf2−/− mice displayed a significantly higher frequency and absolute number of HSPCs in the spleen (Fig. 4a). Consistent with this finding, Nrf2−/− splenocytes gave rise to significantly more CFU-GM and BFU-E colonies in CFC assays, and sorted Nrf2−/− splenic LSKs mimicked their BM counterparts and once again differentiated more rapidly when co-cultured with OP9-DL1 stromal cells (Fig. 4b, c). To address whether this increase in peripheral LSKs was due to a cell-intrinsic defect in HSPC retention, we generated chimeras with sorted CD45.2+ WT or Nrf2−/− LSKs transplanted into CD45.1+ WT hosts. After 8 weeks following transplant, we found significantly fewer Nrf2−/− LSKs in the BM and drastically more in the periphery (Fig. 4d-g). To explore the homing capacity, we labelled purified Lineage-depleted (Lin−) BM cells from CD45.1+ WT and CD45.2+ WT or Nrf2−/− mice with fluorescent dyes, injected them into lethally irradiated WT recipients, and quantified the contribution of cells present in the recipient BM 18 hours after transplant by flow cytometry. We observed that Nrf2−/− Lin− BM cells trafficked to the bone marrow niche less efficiently than CD45.2+ WT controls (Fig. 4h, i), implying that Nrf2 is required for efficient homing of BM cells. To specifically examine the impact of Nrf2 on the homing of HSCs, we labelled and mixed WT and Nrf2−/− BM at 1:1 ratio, and compared the engraftment efficiency of the LT-HSCs 16 hours after transplant. Although present at equal numbers in the inoculum, Nrf2−/− LT-HSCs showed a significant impairment in BM engraftment, indicating a defect in their homing ability (Fig. 4j, k). Collectively, these findings suggest that Nrf2 governs the retention of HSCs and their homing to the BM niche.
Figure 4. Nrf2 Governs HSPC Retention and Migration to Bone Marrow Niche.
(a) Absolute number of LSK cells in untreated spleen; mean + SEM of 13 WT and 14 Nrf2−/− animals over 4 independent experiments. (b) Splenocytes were cultured in CFC conditions and colonies were scored for myeloid and erythroid lineages; mean + SEM of 9 WT and 8 Nrf2−/− animals over 2 independent experiments. (c) LSK cells were isolated from spleen and co-cultured for 11 days with OP9-DL1 stromal cells, representative plots of 2 independent experiments. (d - g) CD45.2+ WT or Nrf2−/− LSKs were transplanted into lethally irradiated CD45.1+ WT recipients. Retention in the BM niche was assessed 8 weeks post-BMT. (d) Representative flow cytometric analysis and (e) quantification of WT or Nrf2−/− LSKs retained in the WT BM; mean + SEM of 10 recipients of WT and 8 recipients of Nrf2−/− LSKs over 2 independent experiments. (f) Representative flow cytometric analysis and (g) quantification of WT or Nrf2−/− LSKs mobilised to the WT spleen; mean + SEM of 10 recipients of WT and 8 recipients of Nrf2−/− LSKs over 2 independent experiments. (h - i) Equal numbers of CFSE+ (CD45.1+ WT) and Violet+ (CD45.2+ WT or Nrf2−/−) Lin− BM cells were transplanted into lethally irradiated CD45.1+ WT recipients and BM content was assessed 18 hours post-transplant. Representative flow cytometric analysis (h) and bar graphs (i) of WT or Nrf2−/− Lin− BM cells. Homing efficiency = absolute number of dye+ Lin− cells found in the BM 18 hours post-transplant ÷ absolute number of dye+ Lin− cells transplanted; mean + SEM of 10 recipients per strain over 2 independent experiments. (j - k) Equal numbers of Violet+ (CD45.2+ WT) and CFSE+ (CD45.2+ Nrf2−/−) BM cells were transplanted into lethally irradiated CD45.2+ WT recipients, and content of LT-HSCs was assessed 16 hours post-transplant. Representative flow cytometric analysis (j) and bar graphs (k) showing the proportion of dye+ LT-HSCs in the pre-transplant mix (Left panel, pre-BMT) and in recipients 16 hours post-transplant (Right panel, post-BMT); mean + SEM of 16 recipients per strain over 3 independent experiments.
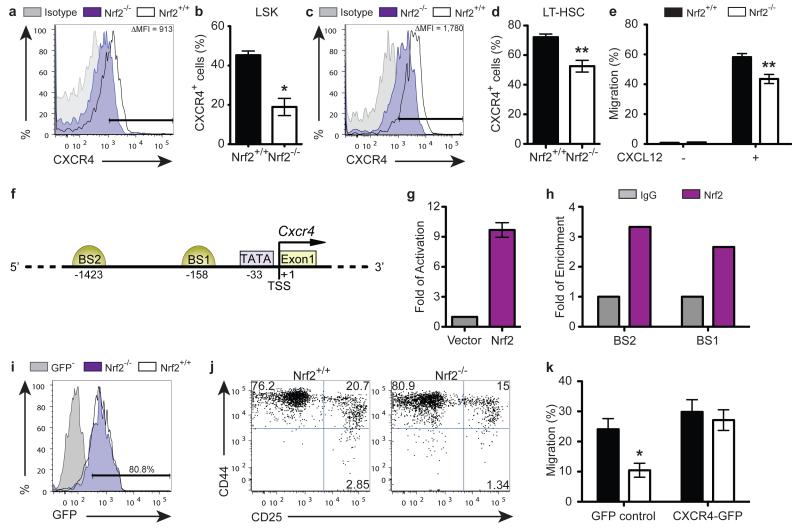
In addition to its well-described role as a crucial chemokine receptor for HSPC homing and retention, there has been considerable interest recently in the role of CXCR4 signalling for its role in maintaining HSC quiescence 21-23. Strikingly, our findings on the role of Nrf2 on HSC homing and quiescence closely mirrored the observations in Cxcr4−/− adult animals. This directed us to investigate the interaction between Nrf2 and CXCR4. Although there was no change in the expression of VCAM-1 (Fig. S2), another key molecule for HSPC homing, we found a 2-fold reduction in CXCR4 expression in Nrf2−/− LSKs compared to WT controls (Fig. 5a, b), which again was observed as early as in LT-HSC stage (Fig. 5c, d). This finding was functionally confirmed by performing in vitro transwell migration assays, where we found that Nrf2−/− LSKs migrated less efficiently towards CXCL12 at 6 hours (Fig. 5e), consistent with the reduced expression of CXCR4 caused by Nrf2-deficiency in HSPCs.
Figure 5. Nrf2 Mediates HSPC Functions via CXCR4.
(a) Representative flow cytometric analysis and (b) bar graphs showing the proportion of sorted WT and Nrf2−/− BM LSK cells expressing CXCR4; mean + SEM of 4 animals per strain; Δ MFI (Mean fluorescence intensity) = (MFI of WT LSKs) − (MFI of Nrf2−/− LSKs). (c) Representative flow cytometric analysis and (d) bar graphs showing the proportion of sorted WT and Nrf2−/− BM LT-HSCs expressing CXCR4; mean + SEM of 6 animals per strain. (e) Percentage of LSK cells that migrated toward unconditioned media with (+) or without (−) supplementation of CXCL12 at 6 hours of assays; mean + SEM of 6 observations per strain over 2 independent experiments. (f) Schematic diagram representing the regulatory region of Cxcr4 promoter. The positions of potential transcription factor binding sites are noted (bp). BS, putative Nrf2 Binding Site; TSS, Transcription Start Site. (g) HEK293T cells were transfected with Cxcr4 promoter-driven luciferase reporter plasmids (covering −0.5 kb from TSS), and luciferase activity was assessed 24 hours after transfection. Data represent the mean + SEM of 3 independent experiments. (h) Chromatin bound DNA from WT BM was immunoprecipitated with a Nrf2-specific antibody or IgG control. Quantitative PCR was performed using primers amplifying potential Nrf2 binding sites on Cxcr4 promoter. Data represent the mean of 2 independent experiments. (i - k) Lentiviral overexpression of CXCR4 in Nrf2−/− HSPCs. (i) Transduction efficiency of WT or Nrf2−/− LSKs with CXCR4-expressing vector (marked by expression of GFP) assessed 48 hours after transduction. (j) Representative flow cytometric analysis showing differentiation of WT or Nrf2−/− LSKs transduced to overexpress with CXCR4 then co-cultured with OP9-DL1 for 11 days, reproducible in 3 independent experiments. (k) Percentage of GFP-transduced or CXCR4-overexpressing WT and Nrf2−/− LSK cells migrated toward unconditioned media supplemented with CXCL12 at 6 hours of assays; mean + SEM of 5 independent experiments.
To determine whether CXCR4 is a direct target of Nrf2, we scrutinised the promoter region of the mouse and human CXCR4 gene, and identified two putative conserved Nrf2 binding sites in the Cxcr4 promoter (Fig. 5f). We cloned the Cxcr4 promoter from mouse genomic DNA into a dual luciferase reporter vector, and demonstrated that exogenous Nrf2 transactivated the minimal promoter region in vitro (Fig. 5g). In addition, to assess whether endogenous Nrf2 binds to the Cxcr4 promoter in BM, we conducted chromatin immunoprecipitation assays. We noted enrichment of both putative binding sites immunoprecipitated by Nrf2, confirming the physical interaction of endogenous Nrf2 and the Cxcr4 promoter in vivo (Fig. 5h). Thus, Nrf2 directly binds to Cxcr4 promoter and activates its expression.
Finally, we sought to examine whether dysregulation of CXCR4 expression contributed to the dual quiescence and migration defects observed in Nrf2−/− HSPCs. Lentiviral overexpression of CXCR4 in Nrf2−/− LSK cells rescued the phenotype of Nrf2−/− HSPCs in their differentiation kinetics on OP9-DL1 co-culture to a comparable pace with WT LSKs, as well as restored the homing ability of Nrf2−/− HSPCs in transwell migration assays (Fig. 5i-k, S3). Taken together, these results suggest that Nrf2 controls multiple aspects of haematopoiesis at least partially through regulation of CXCR4 signalling.
Although Nrf2 has previously been reported to be dispensable for erythropoiesis and megakaryocyte differentiation 18, 24, 25, here we saw that it has a critical role in maintaining HSC function through its impacts on quiescence and self-renewal and, by extension, differentiation. Consistent with previous observations, we found that Nrf2 did not appear to control lineage specification, as Nrf2−/− HSPCs displayed typical myeloid and lymphoid commitment. Rather, our data argue that Nrf2 functions as a master regulator of the main modalities of HSC function. Not only do we show that Nrf2 can regulate the balance between quiescence and proliferation, but also between self-renewal and differentiation, as well as homing and retention of HSCs in the BM niche. This firmly establishes an important role for Nrf2 in the most primitive haematopoietic compartment. Moreover, our studies suggest that Nrf2 exerts its influence, at least in part, through direct regulation of CXCR4 expression. Importantly, previous studies have shown that among the cell cycle regulators investigated, cyclin D1 was singularly upregulated in Cxcr4−/− LSKs23, consistent with our findings that cyclin D1 was upregulated in Nrf2−/− HSPCs.
There is increasing evidence that maintaining intracellular ROS at its basal level is necessary for HSC quiescence 9-11, 26, 27. The most widely studied transcription factor linking the ROS pathway to HSC quiescence and self-renewal are the FOXO (Forkhead O) proteins, which are important downstream effectors of the insulin/IGF-1 like signalling (ISS) pathway. Intriguingly, recent studies suggest that ISS not only inhibits DAF-16, the C. elegans homologue of FOXO, but also directly suppresses SKN-1, the C. elegans homologue of Nrf2, in aging 28. There is also mounting evidence that the ISS and ROS signalling pathways cross-talk in the regulation of aging 29. Interestingly, groups studying FOXO-deficient HSCs have found a similar phenotype to the one we describe here, alluding to the possibility that Nrf2 functions in parallel to the FOXO proteins as a downstream target of the PI3K-Akt pathway. Future research to validate the involvement of Nrf2 in the ISS pathway in mammalian models could be of considerable interest in understanding HSC aging 30.
Finally, a recent study has demonstrated a similar negative regulatory role for Nrf2 in intestinal stem cell proliferation 19. Considering that Nrf2 is ubiquitously expressed, and stem cells and their progenitors are critical to the maintenance and function of various tissues, these findings indicate that Nrf2 serves as a master regulator of stem cell integrity and longevity in adult tissues. Future research into understanding and manipulating Nrf2 in tissue-specific stem cells and their niches may provide insights and innovative approaches to the field of regenerative medicine.
Methods
Mice
Nrf2−/− mice were a kind gift from Dr. Jefferson Chan (University of California, Irvine), and were backcrossed to C57BL/6 background for 7-8 generations. Wild-type (Nrf2+/+) C57BL/6 (CD45.2) or B6.SJL-PtprcaPepcb/BoyJ (CD45.1) mice were purchased from the Jackson Laboratory (Bar Harbor, ME). All mice were age-matched, female Nrf2+/+ and Nrf2−/− mice (8 to 12 weeks old). All mice were maintained in the MSKCC Animal Facility in Thorensten units with filtered germ-free air. Experiments were conducted in compliance with institutional guidelines at MSKCC.
Cell Isolation
BM cells were flushed from intact femurs and tibia, and spleens were mashed with glass slides to generate single cell suspension. Collection of the cells was performed in RPMI media with 10% FBS or PBS with 0.5% BSA, and filtered through a 70-μm strainer. Unless otherwise stated, all cell numbers in this study were standardized as total counts per leg or per spleen.
Flow Cytometry
Monoclonal antibodies (mAbs) recognizing the following markers were used for flow cytometric analyses and cell sorting (LSR II or FACSAria III, BD Biosciences, NJ): (from BD Pharmingen, NJ) c-kit (2B8), Sca-1 (D7), CD11b (M1/70), CD11c (HL3), CD19 (ID3), CD3ε (145-2C11), CD34 (RAM34), CD4 (RM4-5), CD45 (30-F11), CD45.1 (A20), CD45.2 (104), CD48 (HM48-1), CD8α (53-6.7), CD62L (MEL-14), CD135 (A2F10.1), Gr-1 (RB6-8C5), NK1.1 (PK136), TER-119 (TER-119), CXCR4 (2B11), VCAM-1 (429), Rat IgG2aκ isotype (B39-4); (from eBioscience, CA) CD127 (A7R34), CD150 (mShad150), CD16/CD32 (93); (from Invitrogen, CA) B220 (RA3-6B2), and streptavidin (N/A). The lineage antibody cocktail included anti-CD3, anti-CD4, anti-CD8α, anti-CD19, anti-CD11b, anti-CD11c, anti-Gr-1, anti-NK1.1, and anti-TER119. Nuclear staining of Ki-67 was done using anti-human Ki-67 antibody (MOPC-21) (BD Pharmingen, NJ) and fixation/permeabilisation solutions (eBiosciences, CA).
Colony-Forming Cell Assays
BM cells (5 × 104) or splenocytes (1 × 105) were plated in triplicate in 35-mm tissue culture dishes (Nalge Nunc, NY) containing 1 mL assay medium consisting of IMDM, 1.2% methylcellulose (Fisher Scientific, NJ), 30% FBS, 5 × 10−5 M 2-mercaptoethanol, 2 mM L-glutamine, and 0.5 mM hemin, and supplemented with rmSCF (20 ng/mL), rmIL-3 (20 ng/mL), and human erythropoietin (EPO, 6 U/mL) (Amgen Inc, CA). After 10 days of incubation at 37°C in 5% CO2 in air, CFU-GM, BFU-E, and CFU-Mix were scored under an inverted microscope.
Cobblestone Area Forming Cell Assay
MS-5 cells were seeded in 12.5 cm2 tissue culture flasks in α-MEM with 10% FBS. When the cells reached confluence, medium was replaced with long-term culture (LTC) medium: MEM-α, 12.5% FBS, 12.5% horse serum (HyClone Lab. Inc., UT), 10−6 M hydrocortisone (Sigma Chemical Co., MO) and 5 × 10−5 M 2-mercaptoethanol. BM cells (5 × 105) or Lin− splenocytes (1.5 × 104) were added per flask in triplicates. The cultures were demidepopulated weekly and fed with fresh LTC medium. After 3 weeks of culture, CAFCs were scored as phase-dark hematopoietic clones of at least 5 cells beneath the stromal layer using an inverted microscope. After scoring CAFCs at week 3, suspension cells and adherent cells that were detached by treatment with 0.05% trypsin/0.53 mM EDTA were assayed in methylcellulose for CFU-GM or replated in fresh MS-5 cells for secondary CAFC assay. The co-cultures of secondary CAFC assay were again scored at week 3.
Cell Culture
Co-culture of HSPCs and OP9-DL1 stromal cell lines was previously described 31. Briefly, isolated LSK cells were seeded on a 60-80% confluent monolayer of OP9-DL1 cells into six-well tissue culture plates in α-MEM, supplemented with 20% FBS, 100U/mL of penicillin, 100μg/mL of streptomycin, 5ng/mL of rmIL-7 (Miltenyi Biotec, CA), and rm5ng/mL of Flt3L (Miltenyi Biotec, CA). Cells were maintained at 37°C in a humidified atmosphere containing 5% CO2, and were collected at indicated intervals.
Bone Marrow Transplant
In the serial transplantation assay, we purified BM LSK cells from CD45.2+ (WT or Nrf2−/−) donors and CD45.1+ (WT) competitors, mixed them at 1:1 ratio, and transplanted 2 × 103 cells into lethally irradiated CD45.1+ (WT) mice (1,100 cGy total body irradiation, split dose) via tail vein injection. After 4 months of reconstitution, we assessed chimerism of donor-derived LSK cells (CD45.2+) in the recipient BM, as well as sort purified these cells, mixed them with new CD45.1+ (WT) competitors at 1:1 ratio, and transplanted them into a second set of lethally irradiated CD45.1+ (WT) mice. Chimerism in the BM was again assessed 4 months after secondary transplant. In the chimeric repopulation study, we isolated LSK cells from donor BM and transplanted 2 × 103 cells into lethally irradiated mice, and analysed reconstitution in BM after 8 weeks. In the in vivo homing assays, we isolated Lin− BM cells from CD45.1+ (WT) and CD45.2+ (WT or Nrf2−/−) donors using a lineage cell depletion kit (Miltenyi Biotec, CA) per manufacturer’s instructions. We labelled the isolated cells with 5μM CellTrace™ Violet Cell Proliferation Kit for flow cytometry (Invitrogen, CA) or CFSE (Invitrogen, CA) per manufacturer’s recommendation, mixed the labelled CD45.1+ and CD45.2+ Lin− BM cells at 1:1 ratio, then transplanted 2 × 106 cells into lethally irradiated recipients for analysis 18 hours post-transplant. We also isolated WBM cells from CD45.2+ WT and Nrf2−/− donors, labelled them with CellTrace™ Violet Cell Proliferation Kit and CFSE respectively, mixed them at 1:1 ratio, then transplanted 16 × 106 cells into lethally irradiated recipients for analysis of LT-HSC engraftment 16 hours following the transplant.
Cell Cycle Analysis
Purified BM cells (LSKs or LT-HSCs) were fixed with ice-cold 4% paraformaldehyde at 4°C overnight, washed with PBS twice, then labeled with 20μg/mL Hoechst 33342 (Invitrogen, CA) and 1μg/mL pyronin Y (Sigma-Aldrich, MO) for 20 minutes before flow cytometric analysis.
Immunohistochemistry
Fresh tibiae were collected and fixed in 10% formalin at room temperature overnight, decalcified for 24 hours using Decalcifier I (Surgipath, IL), washed with water, and embedded in wax. 5 μm paraffin sections were prepared using a microtome (Leica Microsystems, Germany). The sections were dewaxed in xylene, hydrated in graded alcohols, blocked in 1% hydrogen peroxide, and treated in pH6.0 10mM citrate buffer. The sections were then incubated with primary antibody Cyclin D1 (Thermo Scientific, MA) overnight, followed by appropriate secondary antibodies and avidin-biotin complexes (Vector Laboratories, CA). Antibody reaction was visualized with 3-3′ Diaminobenzidine (Sigma-Aldrich, MO) and counterstained with haematoxylin. Tissue sections were dehydrated in graded alcohols, cleared in xylene and mounted.
5-FU Treatment
5-FU (InvivoGen, CA) was administered to mice intraperitoneally at a dose of 150mg/kg either as a single dose or once every 7 days consecutively for 3 times. BM cellularity was examined at day 3.5 in the former experiment, and the survival of individual mice was monitored daily in the latter.
Transwell Cell Migration Assay
Purified LSK cells were incubated in IMDM media containing 20% FBS, 20ng/mL murine IL-3 (PeproTech Inc., NJ), 20ng/mL SCF (PeproTech Inc., NJ), and 20ng/mL Flt3L (Amgen Inc., CA) 37°C in a humidified atmosphere containing 5% CO2 for 18 hours. LSK cells were then washed with IMEM media twice, resuspended in QBSF media (Quality Biologicals Onc., MD) in 0.1 × 106/mL. 100μL of cell suspension was loaded to the upper chamber of the transwell, and 600μL of QBSF media with or without CXCL12 (200ng/mL, R&D systems, MN) was added to the lower chamber of the transwell plate (pore size 5μm, Corning, NY). Cells were allowed to migrate across the membrane at 37°C with 5% CO2 incubator, and their migration efficiency was assessed at 6 hours.
Luciferase Reporter Assay
CXCR4 promoter regions were identified within 2kb upstream from the transcription start site of mouse CXCR4 sequence (Genebank accession number NC_000067) as previously described32. Putative Nrf2 binding sites were predicted using PROMO (http://www.lsi.upc.es/~alggen). 500bp upstream from transcription start site of mouse CXCR4 was amplified by PCR from WT C57BL/6 mouse genomic DNA and subsequently subcloned into the pGL4.23 luciferase reporter vector (Promega, WI), and named pCXCR4. pCXCR4 construct was co-transfected into HEK293T cells using lipid-based 293T TransIT Reagent (Mirus Bio, WI) with either pUC19 empty vector or Nrf2-pCMV vector, as well as phRL-TK vector as an internal control. Cells were collected 24 hours after transfection and both luciferase activity was determined using Dual-Luciferase Reporter Assay System (Promega, WI) as per manufacturer’s instructions.
Chromatin Immunoprecipitation Assay (ChIP)
ChIP assays were performed using the ChIP Assay Kit (Upstate Biotechnology, EMD Millipore, MA) as per manufacturer’s instruction. Mouse WBM cells were used to assess the binding of Nrf2 to potential binding sites in Cxcr4 promoter. Chromatin was incubated with normal mouse IgG or an anti-Nrf2 antibody (C-20) (Santa Cruz, CA). Input and immunoprecipitated DNA were analyzed by quantitative PCR with primer pairs spanning the Nrf2 binding sites identified in the Cxcr4 promoter (TBS2: AACCGAAAGCCTTCCTTAGC and TGATGATCCCGTTTGTCACC; TBS3: ATCCACGTGGGTAAGGATGG and AGAAGTCCAAGAGCCACTGC).
Lentiviral Transduction
CXCR4 cDNA from pORF-mCXCR4 plasmid (Invivogen, CA) was subcloned into a plasmid encoding recombinant lentiviral vector with eGFP reporter (a kind gift from Dr. Michel Sadelain). Viral particles were produced in HEK293T cells using lipid-based 293T TransIT Reagent (Mirus Bio, WI) as previously described33. Freshly purified LSK cells were resuspended in 0.2 × 106/mL of QBSF media (Quality Biologicals Onc., MD), supplemented with 10ng/mL rmSCF (Miltenyi Biotec, CA), 20ng/mL rhIGF-2 (Miltenyi Biotec, CA), 10ng/mL rhFGF II (Miltenyi Biotec, CA), 100ng/mL rhTPO (Miltenyi Biotec, CA). Concentrated viral particles were added to cell suspension with 0.8 μg/mL polybrene and spinoculated at 2 × 103RPM for 90min at 22°C. Cells were collected for subsequent assays and eGFP fluorescence determined using flow cytometric analysis 48 hours after transduction.
Statistical Analysis
Data were processed in GraphPad Prism 5.0 software. Statistical analysis for comparisons between two groups was performed with nonparametric unpaired Mann-Whitney U test. Survival data were analyzed with the Mantel-Cox log-rank test. * p < 0.05; ** p < 0.01; and *** p < 0.005 were considered as statistically significant.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Jefferson Chan (University of California, Irvine) for providing the Nrf2−/− mice; the staff of the Memorial Sloan-Kettering Cancer Center Research Animal Resources Center for excellent animal care; the staff of the Flow Cytometry Core Facility and Maria S. Jiao of the Comparative Pathology Laboratories for assistance with sample preparation. This research was supported by National Institutes of Health award numbers RO1-HL069929 (MvdB), R01-AI100288 (MvdB), R01-AI080455 (MvdB), R01-AI101406 (MvdB), and P01-CA023766 (ROR). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Support was also received from the Radiation Effects Research Foundation (RERF-NIAID) (MvdB), The Experimental Therapeutics Center of Memorial Sloan-Kettering Cancer Center funded by Mr. William H. Goodwin and Mrs. Alice Goodwin, The Lymphoma Foundation, Alex’s Lemonade Stand, The Geoffrey Beene Cancer Research Center at Memorial Sloan-Kettering Cancer Center, and The Peter Solomon Fund. JAD was supported by an Australian National Health and Medical Research Council Biomedical Training Fellowship and a Research Fellowship from the Leukemia and Lymphoma Society. EV was supported by a fellowship from Italian Foundation for Cancer Research.
Footnotes
AUTHOR CONTRIBUTIONS JJT designed and performed the experiments. JAD, KT, JH, EV assisted with experiments and provided substantial intellectual inputs. NVS, MLW, OMS, and LFY assisted with experiments. MAM and MvdB guided the research. JJT, JAD, KT, AMH, YS, and MvdB wrote the manuscript.
REFERENCES
- 1.Kiel MJ, Morrison SJ. Uncertainty in the niches that maintain haematopoietic stem cells. Nat Rev Immunol. 2008;8:290–301. doi: 10.1038/nri2279. [DOI] [PubMed] [Google Scholar]
- 2.Zhang J, Li L. Stem cell niche: microenvironment and beyond. J Biol Chem. 2008;283:9499–9503. doi: 10.1074/jbc.R700043200. [DOI] [PubMed] [Google Scholar]
- 3.Schofield R. The relationship between the spleen colony-forming cell and the haemopoietic stem cell. Blood Cells. 1978;4:7–25. [PubMed] [Google Scholar]
- 4.Yun Z, Maecker HL, Johnson RS, Giaccia AJ. Inhibition of PPAR gamma 2 gene expression by the HIF-1-regulated gene DEC1/Stra13: a mechanism for regulation of adipogenesis by hypoxia. Dev Cell. 2002;2:331–341. doi: 10.1016/s1534-5807(02)00131-4. [DOI] [PubMed] [Google Scholar]
- 5.Yun Z, Lin Q, Giaccia AJ. Adaptive myogenesis under hypoxia. Mol Cell Biol. 2005;25:3040–3055. doi: 10.1128/MCB.25.8.3040-3055.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ezashi T, Das P, Roberts RM. Low O2 tensions and the prevention of differentiation of hES cells. Proc Natl Acad Sci U S A. 2005;102:4783–4788. doi: 10.1073/pnas.0501283102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mazumdar J, et al. O2 regulates stem cells through Wnt/beta-catenin signalling. Nat Cell Biol. 2010;12:1007–1013. doi: 10.1038/ncb2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takubo K, et al. Regulation of the HIF-1alpha level is essential for hematopoietic stem cells. Cell Stem Cell. 2010;7:391–402. doi: 10.1016/j.stem.2010.06.020. [DOI] [PubMed] [Google Scholar]
- 9.Owusu-Ansah E, Banerjee U. Reactive oxygen species prime Drosophila haematopoietic progenitors for differentiation. Nature. 2009;461:537–541. doi: 10.1038/nature08313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tothova Z, et al. FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell. 2007;128:325–339. doi: 10.1016/j.cell.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 11.Ito K, et al. Reactive oxygen species act through p38 MAPK to limit the lifespan of hematopoietic stem cells. Nat Med. 2006;12:446–451. doi: 10.1038/nm1388. [DOI] [PubMed] [Google Scholar]
- 12.Ito K, et al. Regulation of oxidative stress by ATM is required for self-renewal of haematopoietic stem cells. Nature. 2004;431:997–1002. doi: 10.1038/nature02989. [DOI] [PubMed] [Google Scholar]
- 13.Moi P, Chan K, Asunis I, Cao A, Kan YW. Isolation of NF-E2-related factor 2 (Nrf2), a NF-E2-like basic leucine zipper transcriptional activator that binds to the tandem NF-E2/AP1 repeat of the beta-globin locus control region. Proc Natl Acad Sci U S A. 1994;91:9926–9930. doi: 10.1073/pnas.91.21.9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li W, Kong AN. Molecular mechanisms of Nrf2-mediated antioxidant response. Mol Carcinog. 2009;48:91–104. doi: 10.1002/mc.20465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nguyen T, Nioi P, Pickett CB. The Nrf2-antioxidant response element signaling pathway and its activation by oxidative stress. J Biol Chem. 2009;284:13291–13295. doi: 10.1074/jbc.R900010200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Itoh K, et al. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem Biophys Res Commun. 1997;236:313–322. doi: 10.1006/bbrc.1997.6943. [DOI] [PubMed] [Google Scholar]
- 17.Ishii T, et al. Transcription factor Nrf2 coordinately regulates a group of oxidative stressinducible genes in macrophages. J Biol Chem. 2000;275:16023–16029. doi: 10.1074/jbc.275.21.16023. [DOI] [PubMed] [Google Scholar]
- 18.Chan K, Lu R, Chang JC, Kan YW. NRF2, a member of the NFE2 family of transcription factors, is not essential for murine erythropoiesis, growth, and development. Proc Natl Acad Sci U S A. 1996;93:13943–13948. doi: 10.1073/pnas.93.24.13943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hochmuth CE, Biteau B, Bohmann D, Jasper H. Redox regulation by Keap1 and Nrf2 controls intestinal stem cell proliferation in Drosophila. Cell Stem Cell. 2011;8:188–199. doi: 10.1016/j.stem.2010.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zuniga-Pflucker JC. T-cell development made simple. Nat Rev Immunol. 2004;4:67–72. doi: 10.1038/nri1257. [DOI] [PubMed] [Google Scholar]
- 21.Tzeng YS, et al. Loss of Cxcl12/Sdf-1 in adult mice decreases the quiescent state of hematopoietic stem/progenitor cells and alters the pattern of hematopoietic regeneration after myelosuppression. Blood. 2011;117:429–439. doi: 10.1182/blood-2010-01-266833. [DOI] [PubMed] [Google Scholar]
- 22.Sugiyama T, Kohara H, Noda M, Nagasawa T. Maintenance of the hematopoietic stem cell pool by CXCL12-CXCR4 chemokine signaling in bone marrow stromal cell niches. Immunity. 2006;25:977–988. doi: 10.1016/j.immuni.2006.10.016. [DOI] [PubMed] [Google Scholar]
- 23.Nie Y, Han YC, Zou YR. CXCR4 is required for the quiescence of primitive hematopoietic cells. J Exp Med. 2008;205:777–783. doi: 10.1084/jem.20072513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Motohashi H, et al. NF-E2 domination over Nrf2 promotes ROS accumulation and megakaryocytic maturation. Blood. 2010;115:677–686. doi: 10.1182/blood-2009-05-223107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuroha T, et al. Ablation of Nrf2 function does not increase the erythroid or megakaryocytic cell lineage dysfunction caused by p45 NF-E2 gene disruption. J Biochem. 1998;123:376–379. doi: 10.1093/oxfordjournals.jbchem.a021947. [DOI] [PubMed] [Google Scholar]
- 26.Juntilla MM, et al. AKT1 and AKT2 maintain hematopoietic stem cell function by regulating reactive oxygen species. Blood. 2010;115:4030–4038. doi: 10.1182/blood-2009-09-241000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Banning A, Deubel S, Kluth D, Zhou Z, Brigelius-Flohe R. The GI-GPx gene is a target for Nrf2. Mol Cell Biol. 2005;25:4914–4923. doi: 10.1128/MCB.25.12.4914-4923.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tullet JM, et al. Direct inhibition of the longevity-promoting factor SKN-1 by insulinlike signaling in C. elegans. Cell. 2008;132:1025–1038. doi: 10.1016/j.cell.2008.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Papaconstantinou J. Insulin/IGF-1 and ROS signaling pathway cross-talk in aging and longevity determination. Mol Cell Endocrinol. 2009;299:89–100. doi: 10.1016/j.mce.2008.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Geiger H, Rudolph KL. Aging in the lympho-hematopoietic stem cell compartment. Trends Immunol. 2009;30:360–365. doi: 10.1016/j.it.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 31.Zakrzewski JL, et al. Adoptive transfer of T-cell precursors enhances T-cell reconstitution after allogeneic hematopoietic stem cell transplantation. Nat Med. 2006;12:1039–1047. doi: 10.1038/nm1463. [DOI] [PubMed] [Google Scholar]
- 32.Hayashi H, Kume T. Forkhead transcription factors regulate expression of the chemokine receptor CXCR4 in endothelial cells and CXCL12-induced cell migration. Biochem Biophys Res Commun. 2008;367:584–589. doi: 10.1016/j.bbrc.2007.12.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Na IK, et al. Concurrent visualization of trafficking, expansion, and activation of T lymphocytes and T-cell precursors in vivo. Blood. 2010;116:e18–25. doi: 10.1182/blood-2009-12-259432. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.