Abstract
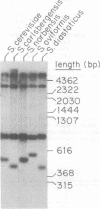
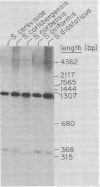
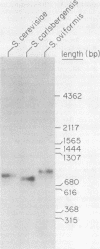
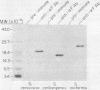
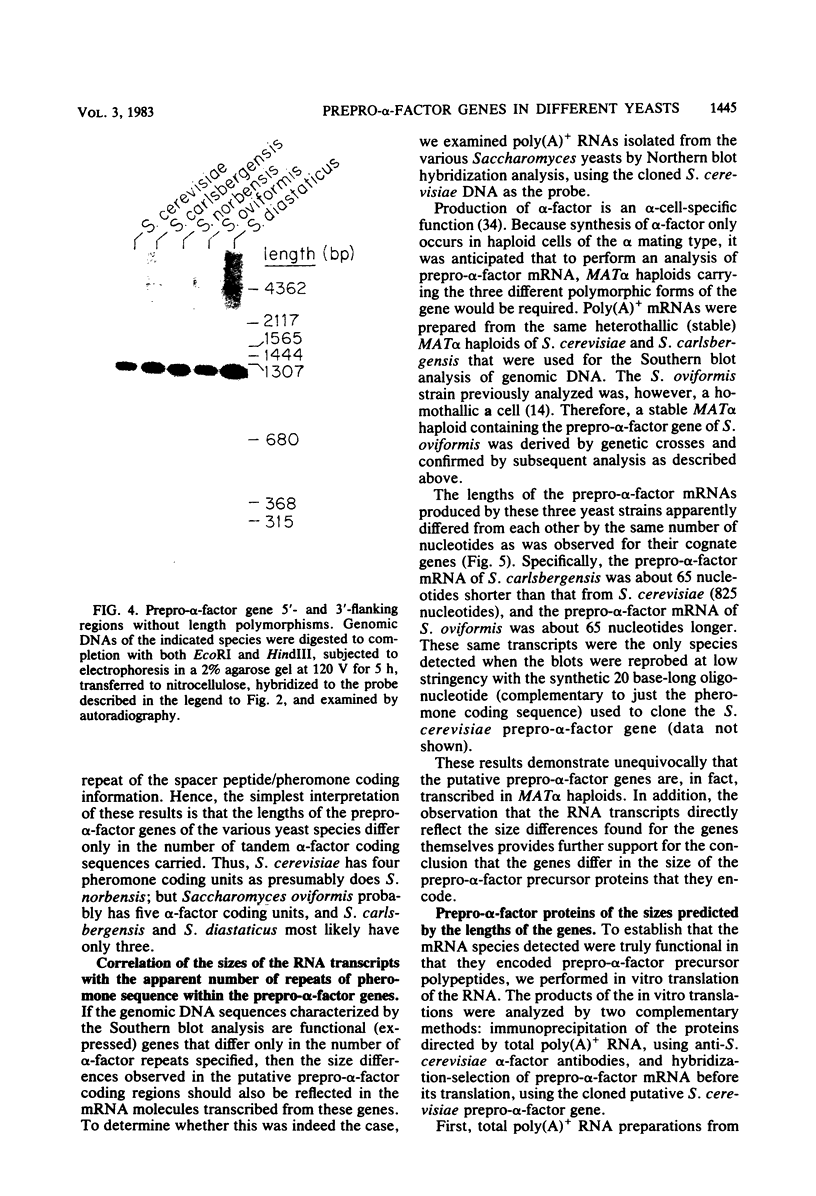
The chromosomal region containing a structural gene for the mating pheromone precursor prepro-alpha-factor was examined in a variety of Saccharomyces yeasts by using a cloned putative prepro-alpha-factor gene of Saccharomyces cerevisiae as the probe. Analysis by restriction endonuclease digestion and Southern blot hybridization indicated that the physical arrangement of this region is highly conserved in all the Saccharomyces species analyzed, but displays length polymorphisms of limited size (50 to 60 base pairs). The observed polymorphisms were shown to be due solely to differences in the number of tandemly arranged spacer peptide/pheromone units within the coding sequence of these genes. Analysis of polyadenylated RNA indicated that these genes specified RNA transcripts and that these RNA molecules could be translated in vitro into prepro-alpha-factor polypeptides immunoprecipitable with anti-alpha-factor antibodies. The sizes of both the mRNAs and the proteins synthesized from them reflected exactly the differences observed in the lengths of the genes. These findings demonstrate conclusively that the putative prepro-alpha-factor DNA cloned from S. cerevisiae, as well as the sequences detected in the other Saccharomyces species, are indeed expressed and functional genes, and suggest that proper proteolytic processing of prepro-alpha-factor is unaffected by the number of pheromone repeats encoded within this precursor protein.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bennetzen J. L., Hall B. D. The primary structure of the Saccharomyces cerevisiae gene for alcohol dehydrogenase. J Biol Chem. 1982 Mar 25;257(6):3018–3025. [PubMed] [Google Scholar]
- Bonner W. M., Laskey R. A. A film detection method for tritium-labelled proteins and nucleic acids in polyacrylamide gels. Eur J Biochem. 1974 Jul 1;46(1):83–88. doi: 10.1111/j.1432-1033.1974.tb03599.x. [DOI] [PubMed] [Google Scholar]
- Botstein D., Falco S. C., Stewart S. E., Brennan M., Scherer S., Stinchcomb D. T., Struhl K., Davis R. W. Sterile host yeasts (SHY): a eukaryotic system of biological containment for recombinant DNA experiments. Gene. 1979 Dec;8(1):17–24. doi: 10.1016/0378-1119(79)90004-0. [DOI] [PubMed] [Google Scholar]
- Broach J. R., Strathern J. N., Hicks J. B. Transformation in yeast: development of a hybrid cloning vector and isolation of the CAN1 gene. Gene. 1979 Dec;8(1):121–133. doi: 10.1016/0378-1119(79)90012-x. [DOI] [PubMed] [Google Scholar]
- Burke D., Mendonça-Previato L., Ballou C. E. Cell-cell recognition in yeast: purification of Hansenula wingei 21-cell sexual agglutination factor and comparison of the factors from three genera. Proc Natl Acad Sci U S A. 1980 Jan;77(1):318–322. doi: 10.1073/pnas.77.1.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson M., Botstein D. Two differentially regulated mRNAs with different 5' ends encode secreted with intracellular forms of yeast invertase. Cell. 1982 Jan;28(1):145–154. doi: 10.1016/0092-8674(82)90384-1. [DOI] [PubMed] [Google Scholar]
- Chamberlain J. P. Fluorographic detection of radioactivity in polyacrylamide gels with the water-soluble fluor, sodium salicylate. Anal Biochem. 1979 Sep 15;98(1):132–135. doi: 10.1016/0003-2697(79)90716-4. [DOI] [PubMed] [Google Scholar]
- Chevallier M. R., Bloch J. C., Lacroute F. Transcriptional and translational expression of a chimeric bacterial-yeast plasmid in yeasts. Gene. 1980 Oct;11(1-2):11–19. doi: 10.1016/0378-1119(80)90082-7. [DOI] [PubMed] [Google Scholar]
- Comb M., Seeburg P. H., Adelman J., Eiden L., Herbert E. Primary structure of the human Met- and Leu-enkephalin precursor and its mRNA. Nature. 1982 Feb 25;295(5851):663–666. doi: 10.1038/295663a0. [DOI] [PubMed] [Google Scholar]
- Faye G., Leung D. W., Tatchell K., Hall B. D., Smith M. Deletion mapping of sequences essential for in vivo transcription of the iso-1-cytochrome c gene. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2258–2262. doi: 10.1073/pnas.78.4.2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazer A. N. Phycobilisomes: structure and dynamics. Annu Rev Microbiol. 1982;36:173–198. doi: 10.1146/annurev.mi.36.100182.001133. [DOI] [PubMed] [Google Scholar]
- Hanahan D., Meselson M. Plasmid screening at high colony density. Gene. 1980 Jun;10(1):63–67. doi: 10.1016/0378-1119(80)90144-4. [DOI] [PubMed] [Google Scholar]
- Heidecker G., Messing J., Gronenborn B. A versatile primer for DNA sequencing in the M13mp2 cloning system. Gene. 1980 Jun;10(1):69–73. doi: 10.1016/0378-1119(80)90145-6. [DOI] [PubMed] [Google Scholar]
- Julius D., Blair L., Brake A., Sprague G., Thorner J. Yeast alpha factor is processed from a larger precursor polypeptide: the essential role of a membrane-bound dipeptidyl aminopeptidase. Cell. 1983 Mar;32(3):839–852. doi: 10.1016/0092-8674(83)90070-3. [DOI] [PubMed] [Google Scholar]
- Kessler S. W. Rapid isolation of antigens from cells with a staphylococcal protein A-antibody adsorbent: parameters of the interaction of antibody-antigen complexes with protein A. J Immunol. 1975 Dec;115(6):1617–1624. [PubMed] [Google Scholar]
- Kurjan J., Herskowitz I. Structure of a yeast pheromone gene (MF alpha): a putative alpha-factor precursor contains four tandem copies of mature alpha-factor. Cell. 1982 Oct;30(3):933–943. doi: 10.1016/0092-8674(82)90298-7. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Laskey R. A., Mills A. D. Enhanced autoradiographic detection of 32P and 125I using intensifying screens and hypersensitized film. FEBS Lett. 1977 Oct 15;82(2):314–316. doi: 10.1016/0014-5793(77)80609-1. [DOI] [PubMed] [Google Scholar]
- McCullough J., Herskowitz I. Mating pheromones of Saccharomyces kluyveri: pheromone interactions between Saccharomyces kluyveri and Saccharomyces cerevisiae. J Bacteriol. 1979 Apr;138(1):146–154. doi: 10.1128/jb.138.1.146-154.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonell M. W., Simon M. N., Studier F. W. Analysis of restriction fragments of T7 DNA and determination of molecular weights by electrophoresis in neutral and alkaline gels. J Mol Biol. 1977 Feb 15;110(1):119–146. doi: 10.1016/s0022-2836(77)80102-2. [DOI] [PubMed] [Google Scholar]
- Nasmyth K. A., Tatchell K. The structure of transposable yeast mating type loci. Cell. 1980 Mar;19(3):753–764. doi: 10.1016/s0092-8674(80)80051-1. [DOI] [PubMed] [Google Scholar]
- Paterson B. M., Roberts B. E. Structural gene identification utilizing eukaryotic cell-free translational systems. Gene Amplif Anal. 1981;2:417–437. [PubMed] [Google Scholar]
- Phillips S. L., Tse C., Serventi I., Hynes N. Structure of polyadenylic acid in the ribonucleic acid of Saccharomyces cerevisiae. J Bacteriol. 1979 May;138(2):542–551. doi: 10.1128/jb.138.2.542-551.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Sprague G. F., Jr, Rine J., Herskowitz I. Control of yeast cell type by the mating type locus. II. Genetic interactions between MAT alpha and unlinked alpha-specific STE genes. J Mol Biol. 1981 Dec 5;153(2):323–335. doi: 10.1016/0022-2836(81)90281-3. [DOI] [PubMed] [Google Scholar]
- Sripati C. E., Warner J. R. Isolation, characterization, and translation of mRNA from yeast. Methods Cell Biol. 1978;20:61–81. doi: 10.1016/s0091-679x(08)62009-9. [DOI] [PubMed] [Google Scholar]
- Strathern J., Hicks J., Herskowitz I. Control of cell type in yeast by the mating type locus. The alpha 1-alpha 2 hypothesis. J Mol Biol. 1981 Apr 15;147(3):357–372. doi: 10.1016/0022-2836(81)90488-5. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G. pBR322 restriction map derived from the DNA sequence: accurate DNA size markers up to 4361 nucleotide pairs long. Nucleic Acids Res. 1978 Aug;5(8):2721–2728. doi: 10.1093/nar/5.8.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taussig R., Carlson M. Nucleotide sequence of the yeast SUC2 gene for invertase. Nucleic Acids Res. 1983 Mar 25;11(6):1943–1954. doi: 10.1093/nar/11.6.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas P. S. Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5201–5205. doi: 10.1073/pnas.77.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl G. M., Stern M., Stark G. R. Efficient transfer of large DNA fragments from agarose gels to diazobenzyloxymethyl-paper and rapid hybridization by using dextran sulfate. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3683–3687. doi: 10.1073/pnas.76.8.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret K. S., Sherman F. DNA sequence required for efficient transcription termination in yeast. Cell. 1982 Mar;28(3):563–573. doi: 10.1016/0092-8674(82)90211-2. [DOI] [PubMed] [Google Scholar]