Abstract
Interleukin-1β (IL-1β) is a potent proinflammatory and immunoregulatory cytokine playing an important role in the progression of rheumatoid arthritis (RA). However, the signaling network of IL-1β in synoviocytes from RA patients is still poorly understood. Here, we show for the first time that phospholipase D1 (PLD1), but not PLD2, is selectively upregulated in IL-1β-stimulated synoviocytes, as well as synovium, from RA patients. IL-1β enhanced the binding of NF-κB and ATF-2 to the PLD1 promoter, thereby enhancing PLD1 expression. PLD1 inhibition abolished the IL-1β-induced expression of proinflammatory mediators and angiogenic factors by suppressing the binding of NF-κB or hypoxia-inducible factor 1α to the promoter of its target genes, as well as IL-1β-induced proliferation or migration. However, suppression of PLD1 activity promoted cell cycle arrest via transactivation of FoxO3a. Furthermore, PLD1 inhibitor significantly suppressed joint inflammation and destruction in IL-1 receptor antagonist-deficient (IL-1Ra−/−) mice, a model of spontaneous arthritis. Taken together, these results suggest that the abnormal upregulation of PLD1 may contribute to the pathogenesis of IL-1β-induced chronic arthritis and that a selective PLD1 inhibitor might provide a potential therapeutic molecule for the treatment of chronic inflammatory autoimmune disorders.
INTRODUCTION
Rheumatoid arthritis (RA) is a chronic, disabling, systemic, inflammatory autoimmune joint disease characterized by an impaired cell-mediated vigorous immune response within the joint space, leading to the invasive growth of synovial tissue and the destruction of cartilage and bone (1). In the synovium, synoviocytes actively participate in the chronic inflammatory responses of RA as a major cell population of the invasive pannus (2). Synovial fibroblasts isolated from RA patients have the potential to produce proinflammatory cytokines, chemokines, angiogenic and proinflammatory factors, and matrix metalloproteinases (MMPs) that degrade the extracellular matrix and cartilage (3).
Interleukin-1β (IL-1β) is one of the most potent and pleiotropic proinflammatory cytokines and has been associated with a variety of inflammatory and autoimmune diseases. IL-1β and tumor necrosis factor α (TNF-α) are key cytokines in the inflammatory cascade of RA and have been intensively studied for therapeutic interventions (4, 5). In contrast to tumor necrosis factor alpha (TNF-α), which is detected predominantly in the early stages of RA, IL-1β is detected long after the onset of RA (6), thus justifying the use of IL-1-blocking therapy at all stages. Inflammatory arthritis in experimental animal models can be prevented or significantly ameliorated by the blockade of IL-1 (6). Therefore, inhibition of IL-1β-mediated signaling might be a promising form of therapy. Although blockade of these cytokines is beneficial for RA treatment, it is not curative and the effect is only partial, with failures to respond very common (7).
Incidentally, despite a large body of literature on the inflammatory pathways triggered by IL-1β in various cell types, no significant validation of their potential signaling targets has been documented. Therefore, it seems possible that a novel IL-1β-mediated signaling target molecule(s) contributes to the pathophysiology of inflammation in RA patients.
Angiogenesis is one of the key mechanisms for maintaining and perpetuating chronic inflammation in RA, and some angiogenic mediators have been described in rheumatoid synovial joints (8). Aberrant angiogenesis is associated with a variety of diseases, including tumor growth and metastasis, as well as fibrovascular disorders such as RA (8).
Phospholipase D (PLD) is considered a promising drug target for the treatment of inflammation and cancer (9, 10). However, a lack of effective small-molecule pharmaceutical inhibitors of PLD has so far hampered rigorous disease-relevant target validation. Recently, selective inhibitors of PLD with considerable pharmacological characterization have been developed by using a diversity-oriented synthetic approach (11), thus providing an important new set of tools to better define the physiological role of PLD. PLD catalyzes the hydrolysis of phospholipids to phosphatidic acid (PA), which has emerged as a new class of potent bioactive molecules that are implicated in a variety of cellular processes, such as cell proliferation, differentiation, migration, survival, membrane trafficking, vesicular transport, and cytoskeletal reorganization (12). Two mammalian isoforms of phosphatidylcholine (PC)-specific PLD, PLD1 and PLD2, have been identified and characterized. Although PLD1 and PLD2 show structural similarities, their modes of activation and functional roles are distinct.
It has been reported that the expression of PLD1 is significantly increased in the heart during experimental autoimmune myocarditis (13). Moreover, microarray analysis in the transcriptional profiling of peripheral blood mononuclear cells during acute pancreatitis showed that PLD1 is the top upregulated gene (14). However, the functional role of PLD in a pathological lesion of chronic inflammatory diseases, such as RA, remains unknown.
Thus, in this study, we investigated the role of PLD in IL-1β-activated rheumatoid synoviocytes, the actual sites of chronic inflammation, as well as in IL-1 receptor antagonist-deficient (IL-1Ra−/−) mice, which exhibit excess IL-1 signaling and are an animal model of spontaneous arthritis. Our results demonstrate that PLD1, as a new target of IL-1β, plays a critical role in synoviocyte activation and is tightly linked to the production of various cytokines/chemokines, adhesion molecules, and angiogenic and matrix-degrading factors, as well as to the modulation of cell cycle arrest and proliferation involving transcription factors such as NF-κB, hypoxia-inducible factor 1α (HIF-1α), and forkhead box class O3a (FoxO3a) to form an interactive molecular mechanism responsible for the amplification and perpetuation of RA. Moreover, a selective inhibitor of PLD1 significantly hampered IL-1β-induced synoviocyte activation and pathogenesis in IL-1Ra−/− mice, thereby providing a novel therapeutic strategy for the treatment of chronic inflammatory autoimmune diseases such as RA.
MATERIALS AND METHODS
Reagents.
Recombinant human IL-1β, TNF-α, and IL-6 were obtained from R&D Systems Inc. (Minneapolis, MN). MG132 and 1- or 3-butanol were purchased from Sigma-Aldrich (St. Louis, MO). Various inhibitors were purchased from Calbiochem. The concentrations of the inhibitors used were as follows: farnesyltransferase I (FTase I), 5 μM; U0126, 20 μM; AG126, 10 μM; pyrrolidine dithiocarbamate (PDTC), 50 μM; SP600125, 20 μM; SB203580, 20 μM; wortmannin, 10 μM. PLD1 selective inhibitor VU155069 was purchased from Cayman Chemical. Dual-luciferase assay kits were purchased from Promega.
Synovial cell and mouse embryo fibroblast (MEF) cell cultures.
Synovial tissue samples were obtained from 10 patients with active RA and 10 patients with osteoarthritis (OA) who were currently undergoing synovectomy or joint replacement. RA was diagnosed according to the 1987 revised criteria of the American College of Rheumatology (formerly the American Rheumatism Association). To isolate fibroblasts, synovial tissues were diced into small pieces and digested overnight with 1 mg/ml of type I collagenase (Worthington, Lakewood, NJ). Primary cultured synovial cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% heat-inactivated fetal bovine serum (FBS) and 1% penicillin-streptomycin (all from Invitrogen, Carlsbad, CA) as described previously (15). Cells from passages 3 to 7 were used in the experiments. This work was approved by the Institutional Review Committee of the Catholic Medical Center, Catholic University of Korea. Informed consent for the use of human synovial tissues was obtained from all of the study subjects. Akt1+/+/Akt2+/+ (wild-type [WT]) and Akt1−/−/Akt2−/− (double-knockout) mice were kindly provided by Sun Sik Bae (Pusan National University) (16). Embryos were dissected from a pregnant double heterozygote Akt1/Akt2 (Akt1+/−/Akt2+/−) female that had been bred with a double heterozygote Akt1/Akt2 (Akt1+/−/Akt2+/−) male. The yolk sacs, heads, and internal organs were isolated and used for genotyping by reverse transcription (RT)-PCR. Carcasses were treated with trypsin-EDTA for 30 min at 37°C, and clumps of cells were disrupted by chopping with scissors. After centrifugation, the cells were resuspended in culture medium (Dulbecco's modified Eagle's medium [DMEM] supplemented with 10% FBS and antibiotics) and maintained at 37°C in 5% CO2.
Animal experiments.
IL-1Ra−/− mice on the BALB/c background were kindly provided by Y. Iwakura (University of Tokyo), maintained under specific-pathogen-free conditions at the Institute of Medical Science, Catholic University of Korea, and fed standard mouse chow (Ralston Purina, St. Louis, MO) and water ad libitum. All experimental procedures were examined and approved by the Animal Research Ethics Committee at the Catholic University of Korea. IL-1Ra−/− mice were maintained until 8 weeks of age. Exactly 10 mg/kg of body weight PLD1 inhibitor was injected intraperitoneally three times per week for 4 weeks. Untreated mice were injected intraperitoneally with the same volume of phosphate-buffered saline containing 10% dimethyl sulfoxide three times per week during the inhibitor treatment period.
Plasmids and small interfering RNA (siRNA).
Dominant-negative TRAF6 (dn-TRAF6), dn extracellular signal-regulated kinase 2 (dn-ERK2), dn-p38, and dn-IκBα (S32A, S36A IκBα), along with pGL2-3X NF-κB (NF-κB–Luc), pGL2-HRE (HIF-1α-responsive element [HRE]–Luc), 3× AP1 (AP1-Luc), 3× STAT (STAT-Luc), 3× C/EBP (C/EBP-Luc), pGL2-3× NF-AT (NF-AT–Luc), and 3× FoxO (FoxO-Luc) containing repeating DNA-binding motifs of the indicated transcription factors, were used. Several on the NF-κB or ATF-2 binding site mutant constructs in the human PLD1 (pGL4-PLD1 Luc; 1.9 kb) promoter reporter plasmids have been described elsewhere (see Table S1 in the supplemental material) (17). siRNAs for control and PLD1 were purchased from Dharmacon Research Inc. (Lafayette, CO). siRNAs for NF-κB, ATF-2, and FoxO3a were obtained from Santa Cruz. Two kinds of siRNA sequences for PLDs were previously described (18).
Transient transfection and reporter gene assay.
Following the manufacturer's instructions, expression plasmids or siRNAs were transiently transfected into cells with Lipofectamine 2000, Lipofectamine Plus (Invitrogen), or Polyfect (Qiagen) reagent. Transfection and luciferase assays were performed as previously described (17). Relative luciferase activity was obtained by normalization of the activity of firefly luciferase against the activity of the internal control, Renilla luciferase.
RNA isolation and real-time quantitative PCR (qPCR).
cDNA was synthesized from total RNA extracted with TRIzol (Invitrogen). qPCR and RT-PCR were performed as previously described (19). Two independent experiments were performed in triplicate for each reaction. All data were normalized with β-actin gene expression values. For sequences of the target-specific primers used in real-time qPCR and RT-PCR, see Tables S2 and S3 in the supplemental material.
IHC analysis.
Immunohistochemical (IHC) staining of sections of synovium samples obtained from RA and OA patients was performed. The tissues were embedded in optimum cutting temperature compound (Tissue-Tek TT 4583; Sakura Finetech, Torrance, CA), snap-frozen in liquid nitrogen, and stored at −80°C. Tissue sections (7 μm) were fixed in 4% paraformaldehyde solution overnight at 4°C. The sections were depleted of endogenous peroxidase activity by adding methanolic H2O2 and then blocked with normal serum for 30 min. The tissues were first incubated with primary antibodies (Abs) overnight at 4°C, a biotinylated secondary linking Ab for 20 min, and then a streptavidin-peroxidase complex for 1 h. The final color product was developed with 3,3′-diaminobenzidine chromogen (Dako, Carpinteria, CA). The sections were counterstained with hematoxylin. The IL-1Ra−/− mouse joint tissues were then fixed in 4% paraformaldehyde, decalcified in EDTA bone decalcifier, and embedded in paraffin. Next, 7-μm sections were prepared and stained with hematoxylin and eosin (H&E), safranin O, and toluidine blue to detect proteoglycans. The sections were dewaxed with xylene, after which they were dehydrated in a graded alcohol series. Endogenous peroxidase activity was quenched with methanol and 3% H2O2. IHC was performed with the Vectastain ABC kit (Vector Laboratories, Burlingame, CA). Images were captured with a DP71 digital camera (Olympus, Center Valley, PA) attached to an Olympus BX41 microscope at a magnification of ×400. For histological evaluation of collagen-induced arthritis, sections were evaluated in a blind manner as described previously. The scores were evaluated as previously described (20).
IHC analysis and quantification.
IHC analysis and quantification were performed as described previously (18). IHC analysis was performed with Abs specific to p65 and FoxO3a. The positive cells in 10 different fields were quantitated by fluorescence.
ChIP assay.
Chromatin immunoprecipitation (ChIP) experiments were performed as previously described (21). RA fibroblast-like synoviocytes (RAFLS) were used for cross-linking with 1% paraformaldehyde in phosphate-buffered saline (PBS) for 10 min. Cells were scraped and collected by centrifugation. Cells were lysed in lysis buffer (50 mM HEPES, pH 7.5, 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% deoxycholate, 1.0 mM protease inhibitor cocktail) and sonicated for 20 s three times. Immunoprecipitation was carried out at 4°C overnight. Immunocomplexes were extracted three times with 1% SDS and 0.1 M NaHCO3, and cross-linking was reversed by incubation at 65°C overnight. The saved chromatin input fraction was also processed in the same manner. Samples were then digested with DNase- and RNase-free proteinase K at 50°C for 4 h, followed by extraction with phenol-chloroform-isoamyl alcohol. About 1/20 of the immunoprecipitated DNA was used in each qPCR. For the sequences of the promoter-specific primers used in qPCR, see Table S4 in the supplemental material.
PLD activity assay.
PLD activity was assessed by measuring the formation of [3H]phosphatidylbutanol, the product of PLD-mediated transphosphatidylation, in the presence of 1-butanol by a previously described procedure with a slight modification (22). RAFLS and OAFLS in six-well plates were serum starved in the presence of 1 μCi of [3H]myristic acid/ml. After overnight starvation, the cells were washed three times with 5 ml of PBS and preequilibrated in serum-free DMEM for 1 h. For the final 10 min of preincubation, 0.3% butan-1-ol was included. At the end of the preincubation period, cells were treated with or without agonists for the indicated times. Incubations were terminated by removing the medium, washing the cells on ice with 5 ml of ice-cold PBS, and adding 1.5 ml of ice-cold methanol. Cells were scraped off the plates with a rubber policeman, and the lipids were extracted and separated with methanol–chloroform–0.1 N HCl (1:1:1). The lower phase was dried under N2, resuspended in 30 μl of chloroform-methanol (2:1), and spotted onto silica gel 60A thin-layer chromatography plates (Whatman). The plates were developed in the upper phase of the solvent system of ethyl acetate–iso-octane–H2O–acetic acid (55:25:50:10) and stained with iodine. A phosphatidylbutanol standard (Avanti Polar Lipids) was used to locate the bands, which were scraped into a scintillation mixture. The radioactivity incorporated into the total phospholipids was measured, and the result is presented as a percentage of the total lipid counts per minute incorporated into phosphatidylbutanol.
Western blotting.
Protein samples were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) on 8 to 15% gels and transferred to a nitrocellulose membrane. The blots were then blocked with 5% nonfat milk and incubated with the appropriate primary Abs, followed by incubation with a horseradish peroxidase-conjugated secondary Ab. Immunoreactive bands were detected by enhanced chemiluminescence. The primary Abs used were anti-α-tubulin, antihemagglutinin, anti-lamin B, anti-NF-κB (p65), anti-cyclooxygenase 2 (anti-COX-2), anti-phospho-FoxO3a, and anti-FoxO3a from Santa Cruz; anti-vascular endothelial growth factor (anti-VEGF) and anti-HIF-1α Abs from BD; anti-phospho-ERK, anti-ERK, anti-phospho-ATF-2, anti-ATF-2, anti-phospho-Jun N-terminal protein kinase (anti-phospho-JNK), anti-JNK, anti-phospho-p38, anti-p38, anti-phospho-IκBα, anti-IκBα, anti-phospho-Akt, anti-Akt, anti-phospho-FoxO3a, and anti-FoxO3a Abs from Cell Signaling; and anti-Akt1 and anti-Akt2 Abs from Upstate Biotech. A PLD1- or PLD2-selective Ab was generated as described previously (23). A polyclonal anti-PLD Ab that recognizes both PLD1 and PLD2 was generated as described previously (4).
Separation of nuclear and cytosolic fractions.
With a commercially available kit, nuclear and cytosolic fractions were separated according to the manufacturer's protocol (Pierce). The purity of the nuclear and cytosolic fractions was ensured by immunoblotting with anti-lamin B and anti-α-tubulin Abs.
Zymography.
The zymography assay for MMP-2 activity was performed as previously described, with a slight modification (24). Samples were applied to an SDS-polyacrylamide gel copolymerized with 0.5% gelatin. Gels were rinsed twice after electrophoresis in 2.5% Triton X-100 and then incubated for 18 h at 37°C in incubation buffer (50 mM Tris-HCl, 5 mM CaCl2, 1 μM ZnCl2, 0.05% NaN3). The gelatin gels were stained with Coomassie brilliant blue and destained with 10% acetic acid and 10% isopropanol. Zones of gelatinolytic activity were detected as clear bands against a black background.
Cell migration.
RAFLS migration was assayed with the CytoSelect 24-well cell migration assay in a colorimetric format (12-μm pore size; Cell Biolabs). Before the addition of cells, the upper chambers were coated with type 1 collagen (BD Biosciences). RAFLS were seeded into the upper chambers, incubated for 36 h, transfected with siRNA specific to PLD1 or a PLD1 inhibitor, and then stimulated with IL-1β. The assay was performed exactly as specified by the manufacturer's protocol. Cells that had migrated to the bottom of the membrane were stained, extracted, and quantified at 560 nm. Human umbilical vein endothelial cell (HUVEC) migration was assayed as previously described (25). A total of 2 × 104 HUVECs per well were seeded onto a layer of previously polymerized Matrigel in a 24-well plate, and conditioned medium of RAFLS was added to each well. Following 24 h of incubation, the morphology of the cells was observed under a phase-contrast microscope and photographed at ×100 magnification.
ELISA.
The expression levels of human VEGF, IL-6, IL-8, IL-15, and MCP-1 secreted by RAFLS were measured with an enzyme-linked immunosorbent assay (ELISA) development kit (eBioscience) according to the manufacturer's instructions.
Tube formation.
A total of 2 × 104 HUVECs were seeded onto a layer of previously polymerized Matrigel in a 24-well plate, and conditioned medium of RAFLS was added to each well. Following 12 h of incubation, the morphology of the cells was observed under a phase-contrast microscope and photographed at ×100 magnification.
Cell proliferation and cell cycle analysis.
Cell proliferation was assayed with a cell proliferation ELISA (colorimetric) bromodeoxyuridine (BrdU) incorporation assay (Roche Diagnostics, Indianapolis, IN) according to the manufacturer's protocol. Phases of the cell cycle were analyzed by flow cytometry. Cells were trypsinized, washed with 1× PBS, fixed and permeabilized with cold 70% ethanol, and finally incubated for 30 min with 1 ml of propidium iodide (contains NP-40; Biosure, CA) in the dark. The DNA content of these cells was measured on the basis of the presence of propidium iodide-stained cells. Flow cytometric analysis of at least 10,000 cells from each sample was done, and cell cycle data were analyzed with a FACSCalibur flow cytometer (BD Biosciences, San Jose, CA) with an excitation wavelength of 488 nm and an emission wavelength of 530 nm.
Statistical analysis.
Data are presented as means ± standard deviations (SDs). Data were analyzed by Student's t test, and P < 0.05 was considered to be statistically significant.
RESULTS
PLD1 is highly expressed in rheumatoid synovium and selectively induced by inflammatory cytokines in rheumatoid synoviocytes.
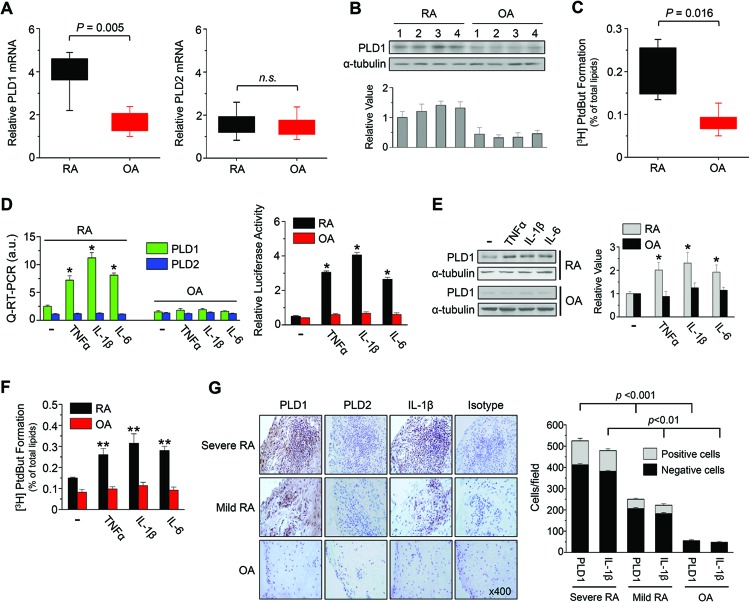
We examined the expression of PLD in RAFLS and OAFLS. We found that the basal expression of PLD1 but not PLD2 in RAFLS (n = 10) was statistically significantly higher than that in OAFLS (n = 10), as analyzed by qPCR (Fig. 1A). Moreover, the PLD1 protein level was significantly upregulated in RAFLS (n = 4), as analyzed by immunoblot assay with an Ab specific to PLD that recognizes both PLD1 and PLD2 (Fig. 1B). The basal expression of PLD2 protein could hardly be detected by Ab specific to PLD in both RAFLS and OAFLS. These results suggest different magnitudes of PLD1 expression in RAFLS and OAFLS. The enzymatic activity of PLD in RAFLS was also significantly greater than that in OAFLS (Fig. 1C).
Fig 1.
PLD1 is upregulated in the RA synovium and is selectively induced by inflammatory responses in RAFLS. (A and B) The expression of PLD was analyzed by qPCR (A) and immunoblotting with an Ab specific to PLD (B) in RAFLS and OAFLS. (C) Cells were labeled with [3H]myristate for 12 h, after which PLD activity was measured. (D) RAFLS and OAFLS were transfected with or without the PLD1 promoter and stimulated with TNF-α (1 ng/ml), IL-1β (10 ng/ml), or IL-6 (10 ng/ml) for 36 h, after which the expression of PLD isozymes was analyzed by qPCR (left) and promoter assay (right). (E) Cells were treated with cytokines for 36 h, after which the lysates were immunoblotted with an anti-PLD Ab. (B and E) Relative PLD1 protein levels were quantitated by densitometer analysis. The data shown are representative of at least three independent experiments. (F) Cells were treated with the cytokines for 30 min, and the enzymatic activity of PLD was examined. (G) IHC staining of PLD and IL-1β (×400, left) and quantification of PLD1- or IL-1β-positive cells in the synovium from mild or severe RA and OA patients (right). *, P < 0.01; **, P < 0.05. The data presented are the means ± SDs of four independent experiments.
Next, we investigated whether or not proinflammatory cytokines are responsible for the induction of PLD expression. IL-1β, TNF-α, and IL-6 are recognized as important cytokines in developing synovial inflammation and play predominant roles in the pathogenesis of RA. These inflammatory cytokines selectively induced PLD1 expression in RAFLS but not in OAFLS, as analyzed by qPCR and promoter assay (Fig. 1D). However, the expression of PLD2 was not affected by cytokines. The PLD1, but not the PLD2, protein level was also significantly increased by these cytokines in RAFLS but not in OAFLS (Fig. 1E). Moreover, IL-1β, TNF-α, and IL-6 enhanced the enzymatic activity of PLD in RAFLS but not in OAFLS (Fig. 1F). This result is of interest and potential significance in relation to the search for possible markers of RA. To further investigate the expression of PLD in joint tissues, we performed IHC staining of the synovium of RA patients with Abs selective for the PLD isoforms. Positive staining was observed mainly in the lining layer of the hyperplastic synovium (n = 10). Expression of PLD1 and IL-1β was significantly correlated with disease severity in the RA synovium (Fig. 1G, left). Moreover, the number of PLD1- or IL-1β-positive cells in the synovium was significantly correlated with RA disease severity (Fig. 1G, right). However, the expression of PLD1 and IL-1β was very low in the OA synovium. Further, the expression of PLD2 was not detectable in the synovium of RA and OA patients. As a negative control, isotype Ab did not show any immunoreactivity with PLD Ab in the RA synovium. Taken together, these results indicate that expression of PLD1 is involved in the progression of chronic arthritic inflammation during RA and is selectively induced by inflammatory cytokines important to RA pathogenesis, suggesting that PLD1 as a new target of chronic inflammation in RAFLS.
TRAF6/ERK/NF-κB and TRAF6/p38/ATF-2 signaling pathways are required for IL-1β-induced PLD1 expression.
IL-1 is a central cytokine playing an important role in the progression of RA (26). Consistent with our findings (Fig. 1G), it has been reported that IL-1β is elevated in serum during RA, and this is correlated with higher disease activity (27). However, the signaling network of IL-1β in RAFLS is still poorly understood.
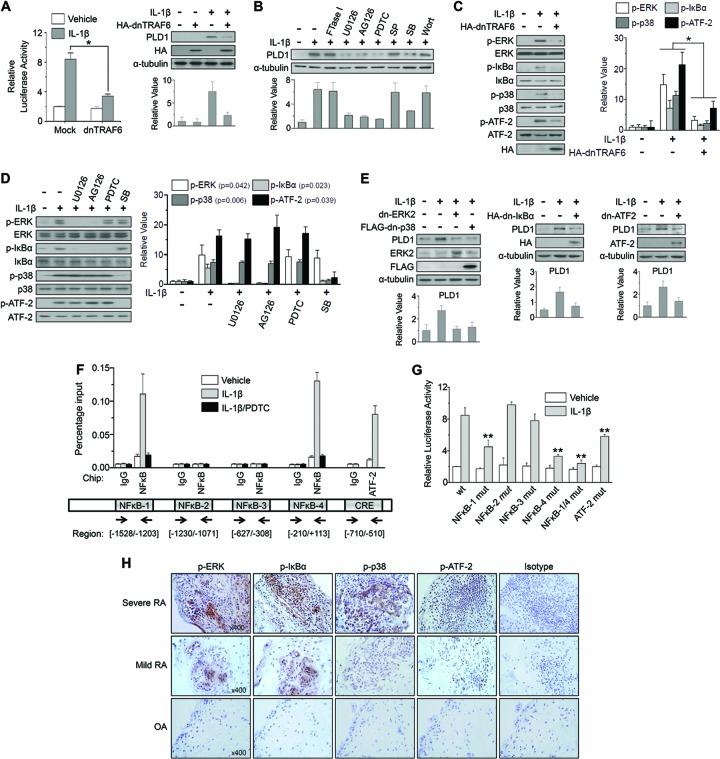
As our in vitro studies showed that IL-1 was the most potent inducer of PLD1 among the proinflammatory cytokines examined in RAFLS, we investigated the signaling pathways of IL-1β-induced PLD1 expression. TRAF6 is known to be a signal transducer for IL-1, as an IL-1 receptor adaptor molecule (28). As shown in Fig. 2A, the expression of dn-TRAF6 significantly suppressed the PLD1 promoter activity and protein expression induced by IL-1β in RAFLS. dn-TRAF6 also inhibited PLD1 mRNA expression, as analyzed by qPCR (see Fig. S1A in the supplemental material). Next, we investigated whether or not IL-1β might increase PLD1 expression via a downstream signaling pathway of TRAF6. IL-1β has been shown to activate all three mitogen-activated protein kinase (MAPK) pathways, as well as IκB kinases (IKKα and IKKβ), through activation of TRAF6 in several cell types (29). IL-1β-induced PLD1 expression was significantly suppressed by inhibitors specific to MEK (U0126), ERK (AG126), NF-κB (PDTC), and p38 (SB203580) but not by those specific to Ras (FTase I), JNK (SP600125), and phosphatidylinositol 3-kinase (PI3K) (wortmannin), as analyzed by Western blotting (Fig. 2B) and qPCR (see Fig. S1B in the supplemental material). Expression of dn-TRAF6 significantly suppressed the IL-1β-induced phosphorylation of ERK, IκBα, p38, and ATF-2 in RAFLS (Fig. 2C). We further observed that IL-1β-induced phosphorylation of IκBα but not ATF-2 was specifically inhibited by inhibitors of MEK/ERK and NF-κB, suggesting that ERK is an upstream kinase for the activation of NF-κB signaling (Fig. 2D). However, inhibition of NF-κB activation by ERK inhibitors does not necessarily put ERKs upstream of IKKs. The effect of the inhibitor could be very indirect, e.g., by blocking the synthesis of an NF-κB-activating cytokine. Moreover, IL-1β-induced phosphorylation of ATF-2 but not IκBα was specifically suppressed by p38 MAPK inhibitor (Fig. 2D). ATF-2, a transcriptional factor, is known to be phosphorylated and activated by JNK or p38 MAPK (19). Furthermore, expression of dn-ERK2, dn-p38, dn-IκBα, or dn-ATF-2 abolished IL-1β-induced PLD1 expression, as analyzed by Western blotting (Fig. 2E). Additionally, transfection of dn-ERK, dn-IκBα, dn-p38, or dn-ATF2, as well as depletion of NF-κB and ATF-2, abolished IL-1β-induced PLD1 mRNA expression, promoter activity, and protein expression (see Fig. S1C to F in the supplemental material). Taken together, these results suggest that IL-1β-stimulated PLD1 induction is mediated via both the TRAF6/ERK/NF-κB and TRAF6/p38/ATF-2 pathways.
Fig 2.
IL-1β-induced PLD1 expression is mediated by the TRAF6/ERK/NF-κB and TRAF6/p38/ATF-2 signaling pathways. (A) RAFLS were cotransfected with pGL4-PLD1 and dn-TRAF6 and treated with IL-1β (10 ng/ml) for 36 h. A luciferase activity assay was performed (left). PLD1 expression was analyzed by Western blotting (right). (B) RAFLS were pretreated with FTase I, U0126, AG126, PDTC, SP600125 (SP), SB203580 (SB), and wortmannin (Wort) for 30 min and treated with IL-1β for 36 h, after which the lysates were analyzed by Western blotting with an anti-PLD Ab. (C) RAFLS were transfected with dn-TRAF6 and treated with IL-1β. HA, hemagglutinin. (D) RAFLS were pretreated with the indicated inhibitors and treated with IL-1β. (E) RAFLS were transfected with the indicated dominant negative constructs and then stimulated with IL-1β for 36 h. (C to E) Lysates were immunoblotted with the indicated Abs. (A to E) Relative PLD1 protein levels were quantitated by densitometer analysis. The data shown are representative of three independent experiments. (F) RAFLS were pretreated with PDTC for 30 min and then treated with or without IL-1β for 12 h. A ChIP assay was performed with preimmune IgG, anti-NF-κB, or anti-ATF-2 Ab, and the product was analyzed by qPCR. Arrows indicate the positions of the primers used in the ChIP experiment. (G) RAFLS were cotransfected with WT pGL4-PLD1, one or two NF-κB or ATF-2 binding site mutant forms (mut) of pGL4-PLD1 and then treated with or without IL-1β. A luciferase activity assay was then performed. (H) IHC staining of the indicated proteins in synovium from mild or severe RA and OA patients (×400). *, P < 0.01; **, P < 0.05. The data presented are the means ± SDs of four independent experiments.
NF-κB positively regulates the expression of inflammation-responsive genes, as well as oncogenes, through direct binding to their promoters (30). Recently, we reported that mitogens and growth factors selectively induce PLD1 but not PLD2 expression in various cancer cells via binding of NF-κB to the PLD1 promoter (17, 18, 24). ATF-2 is a member of the CREB family of transcription factors that can bind to both AP-1- and CRE-responsive elements. ATF-2 plays a role in the expression of genes central to rheumatic diseases such as those for TNF-α and E-selectin (31). However, it remains unknown whether or not ATF-2 is involved in IL-1β-dependent TRAF6 signaling. Four putative NF-κB binding sites and one CRE are present in the PLD1 promoter (see Fig. S2 in the supplemental material). As shown in Fig. 2F, IL-1β increased the activity of p65 NF-κB binding to the two NF-κB binding sites (NF-κB-1 and NF-κB-4), as well as the activity of ATF-2 binding to the CRE, on the PLD1 promoter, as analyzed by ChIP assay (Fig. 2F). When RAFLS was treated with PDTC, binding of p65 to the PLD1 promoter was dramatically decreased (Fig. 2F). We further observed that the two NF-κB-binding sites and one ATF-2-binding site were functionally critical for the transcriptional activation of IL-1β-induced PLD1 by site-directed mutagenesis (Fig. 2G). Furthermore, expression of the phosphorylated forms of the IkBα, ERK, p38, and ATF-2 proteins strongly increased in the RA synovium, in contrast to that in the OA synovium (Fig. 2H), as analyzed by IHC staining, suggesting the in vivo relevance of PLD1 expression signaling networks. Taken together, these data implicate PLD1 as a direct transcriptional target of the NF-κB and ATF-2 signaling pathways mediated by IL-1β in RAFLS.
PLD1 activity is required for IL-1β-induced expression of proinflammatory mediators, matrix-degrading enzymes, and adhesion molecules via binding of NF-κB to promoters of its target genes.
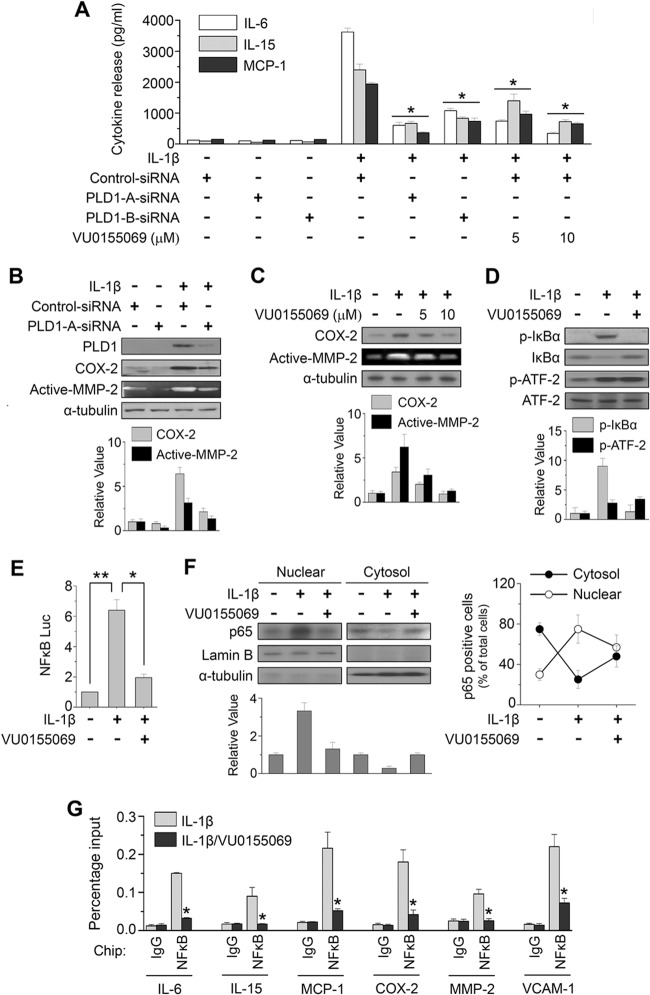
IL-1β is a potent inducer of inflammatory cytokine and chemokine production in RAFLS. Stimulation of RAFLS with IL-1β for 24 h resulted in a dramatic induction of proinflammatory cytokines or chemokines, such as IL-6, IL-15, and monocytic chemoattractant protein 1 (MCP-1)/CCL2, as analyzed by ELISA (Fig. 3A). Depletion of PLD1 with two kinds of siRNA and a PLD1-selective inhibitor (VU0155069) significantly abolished IL-1β-induced IL-6, IL-15, and MCP1 production. The effect of PLD1 siRNAs on PLD1 expression is shown in Fig. S3 in the supplemental material. Moreover, PLD1 inhibitor suppressed the IL-1β-induced expression of COX-2, an inflammatory mediator, as well as the gelatinolytic activity of MMP-2, a matrix-degrading enzyme (Fig. 3B and C). The destruction of articular cartilage and bone is mediated by the release of matrix-degrading enzymes. RAFLS are a major source of MMPs in the synovium and actively drive joint destruction via these enzymes (32). Additionally, PLD1 inhibitor significantly suppressed both basal and IL-1β-induced PLD activities (see Fig. S4A in the supplemental material). Furthermore, the involvement of PLD activity in the production of inflammatory mediators was further examined by using 1-butanol to block PA production through PLD by virtue of the formation of phosphatidylbutanol through the transphosphatidylation reaction. 1-Butanol essentially abrogated the release or expression of inflammatory factors, whereas an identical concentration of 3-butanol, an inactive analogue for PLD-mediated PA formation, did not have a significant effect on the release or expression of factors in IL-1β-stimulated RAFLS, as analyzed by ELISA, RT-PCR, and Western blotting (see Fig. S4B to D). Moreover, depletion of PLD1 significantly abrogated the expression of inflammatory mediator mRNAs in RAFLS (see Fig. S4E and F). The transcription factor NF-κB is recognized as a pivotal regulator of inflammation in RA. PLD1 inhibitor suppressed the IL-1β-induced phosphorylation of IκBα (Fig. 3D), as well as the transactivation (Fig. 3E) and nuclear localization of NF-κB (Fig. 3F), in RAFLS. Although AP-1, STAT3, C/EBPs, and NF-AT are known to play major roles in the regulation of inflammatory genes, their transactivation was not significantly affected by PLD1 inhibitor under IL-1β-treated conditions (see Fig. S5 in the supplemental material), suggesting that off-target effects of the PLD inhibitor can be ruled out. Analyses of nuclear extracts from synovial explants have revealed the increased DNA binding activity of NF-κB in RA patients but not in OA patients (33). Thus, we examined whether or not PLD1 inhibitor affects the binding of NF-κB to the promoters of its target genes. As shown in Fig. 3G, PLD1 inhibitor significantly suppressed the binding of IL-1β-induced NF-κB to the promoters of its target genes (the genes for IL-6, IL-8, IL-15, MCP-1, COX-2, MMP-2, and VCAM-1), as analyzed by ChIP assay. Moreover, selective inhibition of PLD1 abrogated the IL-1β-induced expression of proinflammatory mediators, matrix-degrading enzyme, and cell adhesion molecules, as analyzed by qPCR (see Fig. S6 in the supplemental material). Taken together, these results suggest that PLD1 activity is required for the IL-1β-induced expression of proinflammatory mediators via the binding of NF-κB to promoters of its target genes in RAFLS.
Fig 3.
PLD1 activity is required for IL-1β-induced expression of proinflammatory mediators, matrix-degrading enzymes, and adhesion molecules via inhibition of NF-κB. (A) RAFLS were pretreated with the PLD1 inhibitor VU0155069 or transfected with two kinds of siRNAs specific to PLD1 and then stimulated with IL-1β for 24 h. Secretion of IL-6, IL-15, and MCP-1 was quantified by ELISA. (B, C) RAFLS were transfected with siRNA specific to PLD1 and then treated with IL-1β for 36 h. Lysates were immunoblotted by the indicated Abs, and the activity of MMP-2 was analyzed by gelatin zymography of the conditioned medium. (D) RAFLS were pretreated with VU0155069 and stimulated with IL-1β. Lysates were immunoblotted with the indicated Abs. (E) Luciferase assay for NF-κB in RAFLS. (F) RAFLS were pretreated with VU0155069 for 30 min and then stimulated with IL-1β for 36 h. The lysate was fractionated into the cytosol and nucleus and then analyzed by immunoblotting with the indicated Abs (left). NF-κB-positive cells identified in different fields by IHC analysis were quantitated as described in Materials and Methods (right). Mean scores ± SDs (error bars) are shown. (B to D, F) The relative levels of the indicated proteins were quantitated by densitometer analysis. The data shown are representative of four independent experiments. (G) ChIP assay of the binding of NF-κB to the promoters of its target genes. RAFLS were pretreated with a PLD1 inhibitor and stimulated with IL-1β for 12 h. A ChIP assay was then performed with preimmune IgG or an anti-NF-κB Ab, followed by qPCR. *, P < 0.05; **, P < 0.01. The data presented are the means ± SDs of four independent experiments.
Selective inhibition of PLD1 abrogates IL-1β-induced expression of angiogenic factors by inhibiting HIF-1α binding to promoters of its target genes.
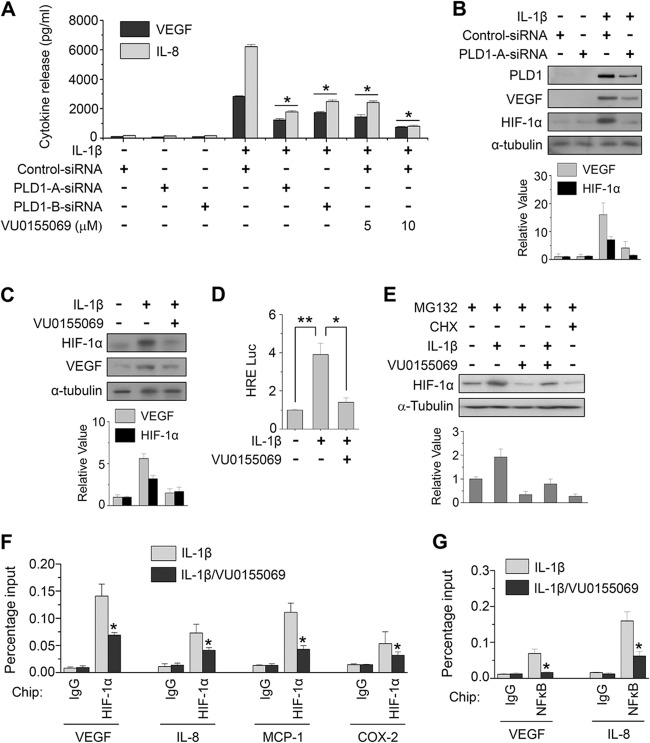
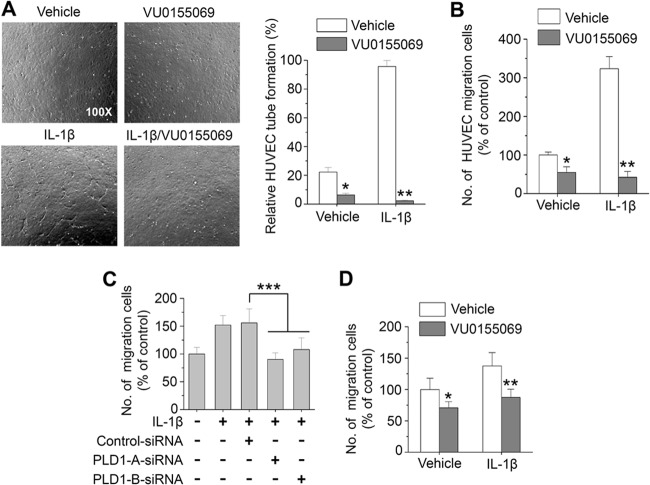
Angiogenesis is an early and critical event in the pathogenesis of RA that increases the number of synovial capillaries within the activated endothelium. Newly formed vessels can maintain a chronic inflammatory state by transporting inflammatory cells to the sites of synovitis, as well as by supplying nutrients and oxygen to the pannus. IL-1β profoundly modulates synovial angiogenesis through angiogenic factors such as VEGF and IL-8, which are produced mostly by RAFLS (34, 35). Depletion or inhibition of PLD1 abolished the production of VEGF and IL-8 in IL-1β-stimulated RAFLS, as analyzed by ELISA and immunoblotting (Fig. 4A to C). The RA synovium is under hypoxic conditions, and a variety of cytokines such as IL-1β and TNF-α are able to stabilize and activate HIF-1α, a transcription factor that increases the transcriptional activation of VEGF. PLD1 inhibitor suppressed the IL-1β-induced HIF-1α transactivation and protein expression in RAFLS (Fig. 4D and E). HIF-1α regulates transcription levels of angiogenic and proinflammatory genes. Thus, we examined whether or not PLD1 inhibitor affects the binding of HIF-1α to the promoters of its target genes. As shown in Fig. 4F, PLD1 inhibitor significantly suppressed the binding of IL-1β-induced HIF-1α to the promoters of its targets (the genes for VEGF, IL-8, MCP-1, and COX-2), as analyzed by ChIP assay. These genes are also known to be transcriptional targets of NF-κB. PLD1 inhibitor also abolished the binding of IL-1β-induced NF-κB to the promoters of VEGF and IL-8 (Fig. 4G). Knockdown and inhibition of PLD1 suppressed the IL-1β-induced expression of these genes, as analyzed by RT-PCR (see Fig. S4C to F in the supplemental material). Moreover, exogenous PLD1 completely rescued the inhibitory effect of PLD1 inhibitor on the IL-1β-induced expression of these genes (see Fig. S7 in the supplemental material). To further examine the antiangiogenic effect of PLD1 inhibitor, RAFLS were stimulated with IL-1β in the presence of PLD1 inhibitor for 36 h, after which conditioned media were applied to HUVECs for an angiogenesis assay. PLD1 inhibitor significantly decreased the IL-1β-induced tube formation of HUVECs (Fig. 5A). Moreover, PLD1 inhibitor significantly abrogated thes IL-1β-stimulated migration of HUVECs, an important feature of angiogenesis (Fig. 5B). Additionally, depletion of PLD1 also suppressed the IL-1β-induced migration of RAFLS (Fig. 5C and D). Taken together, these data suggest that PLD1 activity is required for the IL-1β-induced expression of angiogenic factors via the binding of HIF-1α to the promoters its target genes, angiogenesis, and migration.
Fig 4.
PLD1 activity is required for IL-1β-induced expression of angiogenic factors via binding of HIF-1α to the promoters of its target genes. (A) RAFLS were pretreated with VU0155069 or transfected with two kinds of siRNAs specific to PLD1, followed by stimulation with IL-1β for 24 h. Secretion of VEGF and IL-8 was quantified by ELISA. (B and C) RAFLS were transfected with siRNA specific to PLD1, pretreated with or without VU0155069, and stimulated with IL-1β for 36 h. Cell lysates were immunoblotted with the indicated Abs. The relative levels of the indicated proteins were quantitated by densitometer analysis. The data shown are representative of four independent experiments. (D) RAFLS were transfected with HRE-Luc, pretreated with VU0155069, and stimulated with IL-1β for 36 h, and then luciferase activity was determined. (E) RAFLS were pretreated with VU0155069 or cycloheximide (CHX, 5 μM) for 30 min, treated with IL-1β for 36 h, and then treated with MG132 (20 μM) for 6 h. The HIF-1α protein level was determined by Western blotting. Relative HIF-1α protein levels were quantitated by densitometer analysis. The data shown are representative of four independent experiments. (F and G) RAFLS were pretreated with VU0155069 for 30 min and treated with IL-1β for 12 h, and a ChIP assay was performed with preimmune IgG, anti-HIF-1α, and anti-NF-κB Abs, followed by qPCR. *, P < 0.01; **, P < 0.05. The data presented are the means ± SDs of four independent experiments.
Fig 5.
PLD1 activity is involved in IL-1β-induced angiogenesis and migration. (A, B) RAFLS were preincubated with 10 μM VU0155069 and treated with IL-1β for 36 h. Conditioned medium was collected and applied to HUVECs, and then tube formation (A) and migration (B) were measured. RAFLS were transfected with siRNAs specific to PLD1 (C) or pretreated with or without 10 μM VU0155069 (D), seeded into collagen type I (0.1% solution)-coated migration chambers, and then stimulated with IL-1β for 36 h. The extent of migration is expressed as the average number of cells per microscopic field. *, P < 0.01; **, P < 0.01; ***, P < 0.05. The data presented are the means ± SDs of four independent experiments.
PLD1 activity is required for IL-1β-induced phosphorylation of FoxO3a and suppression of cell cycle arrest via transactivation of FoxO3a.
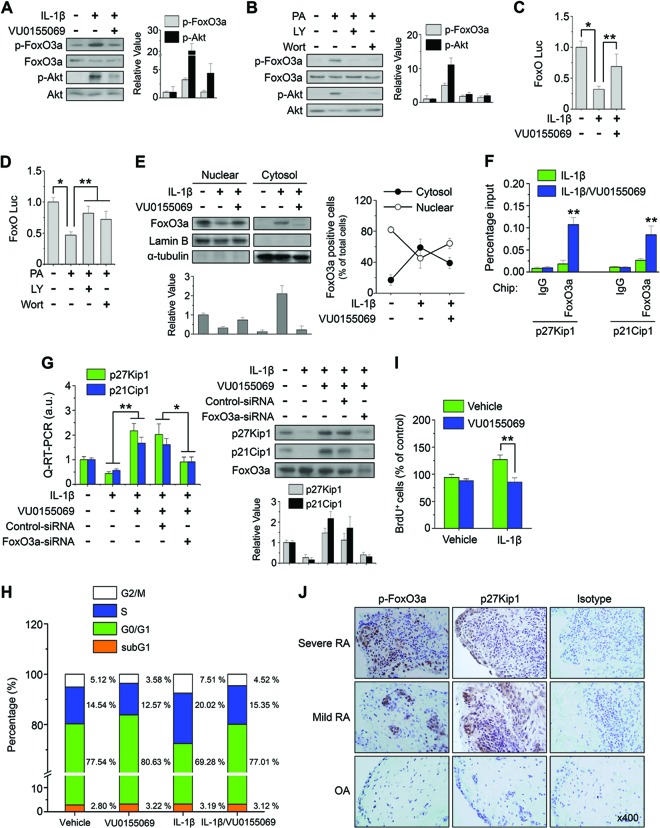
PI3K-dependent activation of Akt has been observed in RA patient synovial tissue, and mechanisms that interfere with this process are protective in animal models of arthritis. Akt can regulate cell survival and proliferation via phosphorylation-dependent inactivation of FoxO transcription factors. Akt-mediated phosphorylation of FoxO family members occurs in the cytoplasm and interferes with the binding of FoxO to target genes involved in cell cycle arrest (36). FoxO3a is highly expressed and phosphorylated in synovial tissues from RA patients (37). Thus, we examined whether or not PLD1 is involved in the regulation of FoxO3a. PLD1 inhibitor abolished the IL-1β-induced phosphorylation of Akt and FoxO3a in RAFLS (Fig. 6A). Ectopic expression of WT PLD1 but not a catalytically inactive mutant form of PLD1 increased the phosphorylation of Akt and FoxO3a (see Fig. S8A in the supplemental material). Moreover, PA, a product of PLD activity, enhanced the phosphorylation of these proteins, whereas inhibitors of PI3K suppressed PA-induced phosphorylation (Fig. 6B). These results suggest that PLD activity is required for the phosphorylation of Akt and FoxO3a. Moreover, PLD1 inhibitor restored the IL-1β-induced suppression of FoxO3a transactivation (Fig. 6C), whereas inhibitors of PI3K restored the PA-induced suppression of FoxO3a transactivation (Fig. 6D). However, IL-1β and PLD1 inhibitors did not affect the transactivation or phosphorylation of FoxO3a in Akt1/2 double-knockout MEFs, suggesting that Akt is required for the phosphorylation and transactivation of FoxO3a (see Fig. S8B). Additionally, PLD1 inhibitor suppressed the IL-1β-induced cytosolic localization of FoxO3a and increased the nuclear localization of FoxO3a (Fig. 6E). Unphosphorylated active FoxO family proteins promote the transcription of genes that induce cell cycle arrest, such as the genes for p27Kip1 and p21Cip1 (36). PLD1 inhibitor promoted the binding of FoxO3a to the promoters of its targets, the genes for p27Kip1 and p21Cip1 (Fig. 6F). Moreover, PLD1 inhibitor-induced expression of p27Kip1 or p21Cip1 was abolished by the depletion of FoxO3a in IL-1β-treated RAFLS, as analyzed by qPCR and Western blotting (Fig. 6G).
Fig 6.
Inhibition of PLD1 suppresses IL-1β-induced phosphorylation of FoxO3a and enhances cell cycle arrest via transactivation of FoxO3a. (A) RAFLS were pretreated with VU0155069 (10 μM) and stimulated with IL-1β for 30 min. (B) RAFLS were pretreated with a PI3K inhibitor (wortmannin [Wort], LY294002 [LY]) and stimulated with PA (50 μM) for 30 min. (A and B) Lysates were immunoblotted with the indicated Abs. The relative levels of the indicated proteins were quantitated by densitometer analysis. The data shown are representative of four independent experiments. (C and D) RAFLS were transfected with FoxO-Luc and treated with the indicated drugs, and a luciferase activity assay was performed. (E) RAFLS were pretreated with VU0155069 and treated with IL-1β for 30 min. Lysate was fractionated into the cytosol and nucleus and then immunoblotted with the indicated Abs (left). Relative FoxO3a protein levels were quantitated by densitometer analysis. The data shown are representative of four independent experiments. The FoxO3a-positive cells identified in different fields by IHC were quantitated (right). (F) RAFLS were pretreated with VU0155069 and treated with IL-1β for 12 h, and a ChIP assay was performed with the indicated Abs. (G) RAFLS were transfected with or without siRNA specific to FoxO3a and subjected to real-time qPCR (left) and immunoblotting (right). a.u., arbitrary units. The relative levels of the indicated proteins were quantitated by densitometer analysis. The data shown are representative of four independent experiments. RAFLS were treated with or without VU0155069 and IL-1β for 36 h and then analyzed by FACS (H) and BrdU incorporation assay (I). (J) IHC staining of p-FoxO3a and p27Kip1 in the synovium from mild/severe RA and OA patients. *, P < 0.01; **, P < 0.05. The data presented are the means ± SDs of four independent experiments.
Since the increased proliferation of FLS contributes to the pathogenesis of RA and PLD regulates the phosphorylation and transactivation of FoxO, we examined whether or not PLD1 inhibitor affects the cell cycle and proliferation of RAFLS. PLD1 inhibitor increased the population of cells in the G0/G1 phase and decreased the population of cells in the S and G2/M phases, in comparison with those of IL-1β-treated RAFLS, as analyzed by fluorescence-activated cell sorting (FACS) (Fig. 6H). Moreover, the IL-1β-stimulated proliferation of RAFLS was significantly suppressed by PLD1 inhibitor, as analyzed by BrdU incorporation assay (Fig. 6I). p27Kip1 and p21Cip1 participate in the maintenance of RAFLS in the quiescent G0/G1 state both in synovial tissue and in culture (38). Therefore, the PLD1 inhibitor-mediated suppression of RAFLS proliferation may be due to cell cycle arrest via the induction of p27Kip1/p21Cip1 caused by the transactivation of FoxO3a.
PLD1 inhibitor suppresses the development of spontaneous arthritis, decreases the expression of NF-κB and HIF-1α target genes, and promotes the expression of FoxO3a target genes in IL-1Ra-deficient mice.
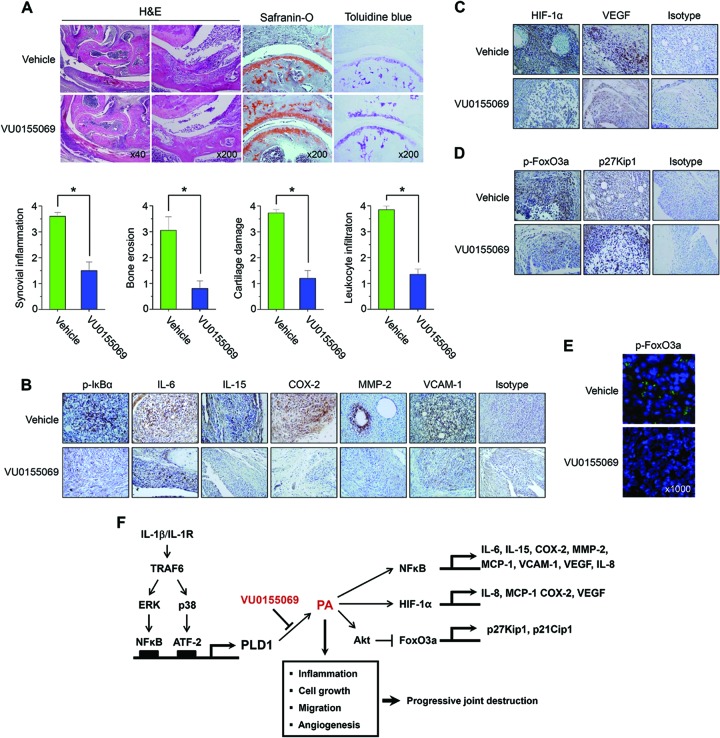
IL-1Ra is an endogenous inhibitor of IL-1 and supposedly regulates IL-1 activity. Polyarthritis develops in IL-1Ra−/− mice on the BALB/c background at age 5 weeks, and by 12 weeks of age, almost all mice become affected (39). The histopathology of the lesions from excess IL-1 closely resembles that of RA in humans, with marked synovial and periarticular inflammation, as well as articular erosion caused by the invasion of granulation tissue (39). In this model, excess IL-1 signaling because of a deficiency in IL-1Ra results in joint-specific inflammation and bone destruction. To further verify the pathogenic role of PLD1, IL-1Ra−/− mice, an animal model of spontaneous arthritis, were injected with PLD1 inhibitor and analyzed by several approaches. Histochemical analysis revealed a severe pathology, including articular cartilage erosion, synovial inflammation, pannus formation, and leukocyte infiltration, which was attenuated in IL-1Ra−/− mice injected with PLD1 inhibitor compared with that in the vehicle-treated groups (Fig. 7A). Moreover, PLD1 inhibitor suppressed the phosphorylation of IκBα, as well as the expression of proinflammatory mediators (IL-6, IL-15, and COX-2), matrix-degrading enzyme (MMP-2), and adhesion molecules (VCAM-1) in the synovium of IL-1Ra−/− mice, as analyzed by IHC staining (Fig. 7B). Treatment with PLD1 inhibitor also decreased the expression of angiogenic factors (HIF-1α and VEGF) in the synovium of IL-1Ra−/− mice (Fig. 7C). Furthermore, PLD1 inhibitor suppressed the phosphorylation of FoxO3a and enhanced the expression of its target, the gene for p27Kip1, in the synovium of IL-1Ra−/− mice (Fig. 7D). Additionally, PLD1 inhibitor suppressed the expression of p-FoxO3a in the cytosol of spleen cells from IL-1Ra−/− mice (Fig. 7E). These results are correlated with those obtained from RAFLS and imply that the PLD1-mediated signaling network may play a very important role in the pathogenesis of spontaneous arthritis in IL-1Ra−/− mice.
Fig 7.
PLD1 inhibitor suppresses the pathogenesis of spontaneous arthritis, decreases the expression of NF-κB and HIF-1α target genes, and promotes the expression of FoxO3a target genes in IL-1Ra-deficient mice. (A) IL-1Ra−/− mice were treated with intraperitoneal injections of PLD1 inhibitor (10 mg/kg; n = 8) or vehicle (n = 8). Tissue sections from joints of each mouse were stained with H&E, toluidine blue, or safranin O. Representative photographs from each group are shown. The histological scores of mice treated with PLD1 inhibitor or vehicle are shown at the bottom. The data represent individual values and the average value of five individual mice in each group. *, P < 0.05 compared with the vehicle-treated group. (B to D) Tissue sections from the joints of mice treated with PLD1 inhibitor or vehicle were stained with the indicated Abs. The cells stained with each Ab are shown in brown (×400). (E) Tissue sections from the spleens of IL-1Ra-deficient mice were stained with 4′,6-diamidino-2-phenylindole (DAPI; blue) or anti-p-FoxO3a Ab (green). Tissues were monitored with a Zeiss LSM 510 confocal microscope. Anti-p-FoxO3a- and DAPI-stained images were overlaid to visualize the nuclear and cytoplasmic localization of p-FoxO3a. The data shown are representative of three independent experiments. (F) Model illustrating the roles of PLD1 in RAFLS. IL-1β binds to its receptor and stimulates the TRAF6-ERK/NF-κB and TRAF6/p38/ATF-2 signaling pathways, leading to selective PLD1 expression. PLD1-derived PA is involved in the production of proinflammatory mediators and angiogenic factors. PLD1 inhibitor abolishes the expression of molecules involved in RA pathogenesis by suppressing the binding of NF-κB/HIF-1α to the promoters of IL-1β target genes and enhances the expression of cell cycle arrest genes (p27Kip1 and p21Cip1 genes) via transactivation of FoxO3a. The abnormal upregulation of PLD1 in RAFLS may contribute to the pathogenesis of chronic arthritis and thus provide a potential target for the control of inflammatory arthritis.
DISCUSSION
In the present study, we demonstrate that IL-1β is functionally coupled to PLD1 but not PLD2 in RA synoviocytes. Since IL-1β is associated with various inflammatory diseases, understanding the intracellular signal transduction mechanisms that regulate IL-1β-mediated responses has profound implications, not the least of which is to identify novel molecules as potential therapeutic targets. Regulation of molecules involved in the IL-1β-triggered signaling pathways has gained attention in this regard, as a variety of signaling molecules, such as PDE4, p38 MAPK, and NF-κB inhibitors, are being studied in clinical trials (40) but have produced undesired side effects so far. More efficient therapies may become available upon the elucidation of the molecular mechanisms and the roles of key molecules involved in the IL-1β-triggered signaling events. Thus, we investigated the role of PLD in the IL-1β-mediated intracellular signaling mechanism in human RA synoviocytes and validated the in vitro and in vivo relevance of PLD1 in the process of chronic autoimmune inflammatory arthritis. In this study, we demonstrated for the first time that PLD1 plays a critical role in IL-1β-induced synoviocyte activation, as well as in the progression of chronic inflammatory arthritis, in IL-1 receptor antagonist-deficient mice, a model of spontaneous arthritis.
Our study showed that PLD1 expression and activity in the synovium or FLS were higher in RA patients than in OA patients. PLD1 expression correlated well with the severity of RA.
The increase in the expression of PLD1 but not PLD2 was triggered by proinflammatory cytokines such as IL-1β, TNF-α, and IL-6 in RAFLS but not in OAFLS. A great deal of evidence indicates that in activated rheumatoid synoviocytes, many pathological processes, including inflammatory cytokine production, are regulated by intracellular signaling. IL-1β, TNF-α, and IL-6 are all critical cytokines involved in the pathogenesis of RA. These cytokines are abundantly produced in RA patient synovium and are highly concentrated in the synovium and serum of RA patients.
IL-1β-induced PLD1 expression was mediated via the TRAF6/ERK/NF-κB and TRAF6/p38/ATF-2 signaling pathways. Recently, isoform-selective small-molecule PLD inhibitors have been developed by using a diversity-oriented synthetic approach with considerable pharmacological characterization (11). In our study, PLD1 inhibitor significantly abolished IL-1β-induced proinflammatory cytokines or chemokines such as IL-6, IL-15, and MCP-1 by suppressing the binding of NF-κB to the PLD1 promoter. Activation of the NF-κB transcription factor plays a central role in inflammatory responses based on the ability of NF-κB to regulate proinflammatory gene transcription. Moreover, excessive NF-κB activation has been implicated in diverse chronic diseases, including RA. Thus, PLD1 inhibitor may effectively induce an anti-inflammatory response in RA by modulating inflammatory gene activation via inhibition of NF-κB activity. Although AP-1, STAT3, C/EBPs, and NF-AT are also known to play major roles in the regulation of inflammatory genes, their transactivation was not affected by PLD1 inhibitor under IL-1β-stimulated conditions, suggesting that off-target PLD inhibitor effects can be ruled out.
In fact, activation of STAT3 is mediated mainly by IL-6 or IL-10 (41) and the activation of NF-AT is mediated mainly by Ca2+ signaling (42). As IL-1β is not a strong inducer of transactivation of STAT3 and NF-AT, the ability of IL-1β to induce Stat3 and NFAT activity is likely to be indirect.
Abnormally reduced oxygen concentrations leading to dysfunctional cell metabolism are present in RA. HIF-1α is an important molecular switch that guides cellular responsiveness in synovial tissue exhibiting the chronic inflammation associated with RA. This factor controls the outgrowth of new vasculature that supports the on-going demand for nutrients, oxygen, and leukocyte recruitment by facilitating the expression of many proangiogenic and proinflammatory genes. PLD1 inhibitor suppressed IL-1β-induced HIF-1α expression and significantly decreased the binding of IL-1β-induced HIF-1α to the promoters of the genes for VEGF, IL-8, MCP-1, and COX-2, thereby inhibiting their expression. Angiogenesis is an early and critical event in the pathogenesis of RA. VEGF is a prototypic angiogenic factor that induces endothelial cell proliferation, angiogenesis, and capillary permeability. Neovascularization is dependent on endothelial cell activation, migration, and proliferation of angiogenesis and may provide a novel approach to RA therapy. In this study, we documented that PLD1 inhibitor suppressed the IL-1β-induced angiogenesis, migration, and proliferation of HUVECs or RAFLS. Collectively, these data indicate that PLD1 is a critical mediator of synovial inflammation. Our findings further suggest that specific inhibition of PLD1 activation may be considered a promising anti-inflammatory approach with therapeutic potential for RA.
PI3K-dependent activation of Akt has been observed in RA synovial tissue, and mechanisms that interfere with this process have been shown to be protective in an animal model of arthritis. Akt can regulate cell survival and proliferation via phosphorylation-dependent inactivation of FoxO.
Further analysis of the activation status of the intracellular signaling pathways regulating FoxO transcriptional activity in RA synovial tissues may therefore be helpful in identifying the molecular networks that differentially support inflammation and joint destruction in RA patients. IL-1β mediates the proliferation of synoviocytes through the coupling of PLD1 to FoxO3a; so far, this is the first example of this pathway to be shown in cytokine signaling. Taken together, IL-1β enhances proinflammatory mediators, angiogenic factors, and proliferation and migration via PLD1-mediated NF-κB or HIF-1α activation and the FoxO3a inactivation signaling pathway in RA synoviocytes. Moreover, injection of PLD1 inhibitor into IL-1Ra−/− mice significantly suppressed the pathogenesis of spontaneous chronic arthritis. The PLD1 inhibitor effect in IL-1Ra−/− mice is well correlated with that in RAFLS. Thus, it is suggested that the biological activity of PLD1 apparently exacerbates the joint inflammation of IL-1Ra−/− mice. Our results suggest that IL-1β-induced PLD1 expression plays an essential role in the pathogenesis of spontaneous arthritis (Fig. 7F). A variety of chronic inflammatory diseases are associated with altered cellular signaling. Understanding the intracellular pathways might lead to new approaches to the treatment of inflammatory diseases, including the use of bioavailable small molecules that regulate cytokine function and production.
In summary, our present study demonstrates that a selective PLD1 inhibitor has therapeutic potential for improving chronic autoimmune inflammatory diseases such as RA but also other inflammatory diseases where IL-1β plays a role.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by a grant from the Translational Research Center for Protein Function Control, NSF (2009-0092960), South Korea, and a National Research Foundation of Korea grant funded by the South Korean Government (MEST) (2012002009).
Footnotes
Published ahead of print 20 May 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.01519-12.
REFERENCES
- 1. Feldmann M. 2001. Pathogenesis of arthritis: recent research progression. Nat. Immunol. 2:771–773 [DOI] [PubMed] [Google Scholar]
- 2. Firestein GS. 1996. Invasive fibroblast-like synoviocytes in rheumatoid arthritis: passive responders or transformed aggressors? Arthritis Rheum. 39:1781–1790 [DOI] [PubMed] [Google Scholar]
- 3. Mor A, Abramson SB, Pillinger MH. 2005. The fibroblast-like synovial cell in rheumatoid arthritis: a key player in inflammation and joint destruction. Clin. Immunol. 115:118–128 [DOI] [PubMed] [Google Scholar]
- 4. Min DS, Kwon TK, Park WS, Chang JS, Park SK, Ahn BH, Ryoo ZY, Lee YH, Lee YS, Rhie DJ, Yoon SH, Hahn SJ, Kim MS, Jo YH. 2001. Neoplastic transformation and tumorigenesis associated with overexpression of phospholipase D isozymes in cultured murine fibroblasts. Carcinogenesis 22:1641–1647 [DOI] [PubMed] [Google Scholar]
- 5. van den Berg WB, Miossec P. 2009. IL-17 as a future therapeutic target for rheumatoid arthritis. Nat. Rev. Rheumatol. 5:549–553 [DOI] [PubMed] [Google Scholar]
- 6. Burger D, Dayer JM, Palmer G, Gabay C. 2006. Is IL-1 a good therapeutic target in the treatment of arthritis? Best Pract. Res. Clin. Rheumatol. 20:879–896 [DOI] [PubMed] [Google Scholar]
- 7. Brennan FM, McInnes IB. 2008. Evidence that cytokines play a role in rheumatoid arthritis. J. Clin. Invest. 118:3537–3545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Folkman J. 1995. Angiogenesis in cancer, vascular, rheumatoid and other disease. Nat. Med. 1:27–31 [DOI] [PubMed] [Google Scholar]
- 9. Steed PM, Chow AH. 2001. Intracellular signaling by phospholipase D as a therapeutic target. Curr. Pharm. Biotechnol. 2:241–256 [DOI] [PubMed] [Google Scholar]
- 10. Su W, Chen Q, Frohman MA. 2009. Targeting phospholipase D with small-molecule inhibitors as a potential therapeutic approach for cancer metastasis. Future Oncol. 5:1477–1486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Scott SA, Selvy PE, Buck JR, Cho HP, Criswell TL, Thomas AL, Armstrong MD, Arteaga CL, Lindsley CW, Brown HA. 2009. Design of isoform-selective phospholipase D inhibitors that modulate cancer cell invasiveness. Nat. Chem. Biol. 5:108–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Peng X, Frohman MA. 2012. Mammalian phospholipase D physiological and pathological roles. Acta Physiol. (Oxf.) 204(2):219–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ahn M, Lee Y, Sim KB, Min DS, Matsumoto Y, Wie MB, Shin YG, Shin T. 2004. Increased expression of phospholipase D in the heart with experimental autoimmune myocarditis in Lewis rats. Immunol. Invest. 33:95–105 [DOI] [PubMed] [Google Scholar]
- 14. Bluth M, Lin YY, Zhang H, Viterbo D, Zenilman M. 2008. Use of gene expression profiles in cells of peripheral blood to identify new molecular markers of acute pancreatitis. Arch. Surg. 143:227–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kim HR, Cho ML, Kim KW, Ju JH, Park MK, Oh HJ, Kim JS, Park SH, Lee SH, Kim HY. 2007. Up-regulation of IL-23p19 expression in rheumatoid arthritis synovial fibroblasts by IL-17 through PI3-kinase-, NF-kappaB- and p38 MAPK-dependent signalling pathways. Rheumatology (Oxford) 46:57–64 [DOI] [PubMed] [Google Scholar]
- 16. Kim EK, Tucker DF, Yun SJ, Do KH, Kim MS, Kim JH, Kim CD, Birnbaum MJ, Bae SS. 2008. Linker region of Akt1/protein kinase B-alpha mediates platelet-derived growth factor-induced translocation and cell migration. Cell. Signal. 20:2030–2037 [DOI] [PubMed] [Google Scholar]
- 17. Kang DW, Park MH, Lee YJ, Kim HS, Kwon TK, Park WS, Min DS. 2008. Phorbol ester up-regulates phospholipase D1 but not phospholipase D2 expression through a PKC/Ras/ERK/NFκB-dependent pathway and enhances matrix metalloproteinase-9 secretion in colon cancer cells. J. Biol. Chem. 283:4094–4104 [DOI] [PubMed] [Google Scholar]
- 18. Kang DW, Park MH, Lee YJ, Kim HS, Lindsley CW, Brown HA, Min DS. 2011. Autoregulation of phospholipase D activity is coupled to selective induction of phospholipase D1 expression to promote invasion of breast cancer cells. Int. J. Cancer 128:805–816 [DOI] [PubMed] [Google Scholar]
- 19. Breitwieser W, Lyons S, Flenniken AM, Ashton G, Bruder G, Willington M, Lacaud G, Kouskoff V, Jones N. 2007. Feedback regulation of p38 activity via ATF-2 is essential for survival of embryonic liver cells. Genes Dev. 21:2069–2082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Camps M, Rückle T, Ji H, Ardissone V, Rintelen F, Shaw J, Ferrandi C, Chabert C, Gillieron C, Françon B, Martin T, Gretener D, Perrin D, Leroy D, Vitte PA, Hirsch E, Wymann MP, Cirillo R, Schwarz MK, Rommel C. 2005. Blockade of PI3Kgamma suppresses joint inflammation and damage in mouse models of rheumatoid arthritis. Nat. Med. 11:936–943 [DOI] [PubMed] [Google Scholar]
- 21. Kang DW, Lee SH, Yoon JW, Park WS, Choi KY, Min DS. 2010. Phospholipase D1 drives a positive feedback loop to reinforce the Wnt/beta-catenin/TCF signaling axis. Cancer Res. 70:4233–4242 [DOI] [PubMed] [Google Scholar]
- 22. Ahn BH, Kim SY, Kim EH, Choi KS, Kwon TK, Lee YH, Chang JS, Kim MS, Jo YH, Min DS. 2003. Transmodulation between PLD and c-Src enhances cell proliferation. Mol. Cell. Biol. 23:3103–3115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kim SY, Min DS, Choi JS, Choi YS, Park HJ, Sung KW, Kim JM, Lee MY. 2004. Differential expression of phospholipase D isozymes in the hippocampus following kainic acid-induced seizures. J. Neuropathol. Exp. Neurol. 63:812–820 [DOI] [PubMed] [Google Scholar]
- 24. Kang DW, Min DS. 2010. Platelet derived growth factor increases phospholipase D1 but not phospholipase D2 expression via NFκB signaling pathway and enhances invasion of breast cancer cells. Cancer Lett. 294:125–133 [DOI] [PubMed] [Google Scholar]
- 25. Pickens SR, Volin MV, Mandelin AM, Kolls JK, Pope RM, Shahrara S. 2010. IL-17 Contributes to angiogenesis in rheumatoid arthritis. J. Immunol. 184:3233–3241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schiff MH. 2000. Role of interleukin 1 and interleukin 1 receptor antagonist in the mediation of rheumatoid arthritis. Ann. Rheum. Dis. 59(Suppl 1):103–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chen DY, Hsieh TY, Chen YM, Hsieh CW, Lan JL, Lin FJ. 2009. Proinflammatory cytokine profiles of patients with elderly-onset rheumatoid arthritis: a comparison with younger-onset disease. Gerontology 55:250–258 [DOI] [PubMed] [Google Scholar]
- 28. Cao Z, Xiong J, Takeuchi M, Kurama T, Goeddel DV. 1996. TRAF6 is a signal transducer for interleukin-1. Nature 383:443–446 [DOI] [PubMed] [Google Scholar]
- 29. Deng L, Wang C, Spencer E, Yang L, Braun A, You J, Slaughter C, Pickart C, Chen ZJ. 2000. Activation of the kappaB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell 103:351–361 [DOI] [PubMed] [Google Scholar]
- 30. Lin A, Karin M. 2003. NF-kappaB in cancer: a marked target. Semin. Cancer Biol. 13:107–114 [DOI] [PubMed] [Google Scholar]
- 31. Firestein GS, Manning AM. 1999. Signal transduction and transcription factors in rheumatic disease. Arthritis Rheum. 42:609–621 [DOI] [PubMed] [Google Scholar]
- 32. Neumann E, Lefèvre S, Zimmermann B, Gay S, Müller-Ladner U. 2010. Rheumatoid arthritis progression mediated by activated synovial fibroblasts. Trends Mol. Med. 16:458–468 [DOI] [PubMed] [Google Scholar]
- 33. Asahara H, Asanuma M, Ogawa N, Nishibayashi S, Inoue H. 1995. High DNA-binding activity of transcription factor NF-kappaB in synovial membranes of patients with rheumatoid arthritis. Biochem. Mol. Biol. Int. 37:827–832 [PubMed] [Google Scholar]
- 34. Larsen H, Akhavani MA, Raatz Y, Paleolog EM. 2007. Gene expression studies to investigate disease mechanisms in rheumatoid arthritis: does angiogenesis play a role? Curr. Rheumatol. Rev. 3:243–251 [Google Scholar]
- 35. Szekanecz Z, Koch AE. 2009. Angiogenesis and its targeting in rheumatoid arthritis. Vascul. Pharmacol. 51:1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Huang H, Tindall DJ. 2007. Dynamic FoxO transcription factors. J. Cell Sci. 120(Pt 15):2479–2487 [DOI] [PubMed] [Google Scholar]
- 37. Turrel-Davin F, Tournadre A, Pachot A, Arnaud B, Cazalis MA, Mougin B, Miossec P. 2010. FoxO3a involved in neutrophil and T cell survival is overexpressed in rheumatoid blood and synovial tissue. Ann. Rheum. Dis. 69:755–760 [DOI] [PubMed] [Google Scholar]
- 38. Helmchen B, Weckauf H, Ehemann V, Wittmann I, Meyer-Scholten C, Berger I. 2005. Expression pattern of cell cycle-related gene products in synovial stroma and synovial lining in active and quiescent stages of rheumatoid arthritis. Histol. Histopathol. 20:365–372 [DOI] [PubMed] [Google Scholar]
- 39. Horai R, Saijo S, Tanioka H, Nakae S, Sudo K, Okahara A, Ikuse T, Asano M, Iwakura Y. 2000. Development of chronic inflammatory arthropathy resembling rheumatoid arthritis in interleukin 1 receptor antagonist-deficient mice. J. Exp. Med. 191:313–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Palladino MA, Bahjat FR, Theodorakis EA, Moldawer LL. 2003. Anti-TNF-alpha therapies: the next generation. Nat. Rev. Drug Discov. 2:736–746 [DOI] [PubMed] [Google Scholar]
- 41. Murray PJ. 2007. The JAK-STAT signaling pathway: input and output integration. J. Immunol. 178:2623–2629 [DOI] [PubMed] [Google Scholar]
- 42. Im SH, Rao A. 2004. Activation and deactivation of gene expression by Ca2+/calcineurin-NFAT-mediated signaling. Mol. Cells 18:1–9 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.