Abstract
Hypoxia-inducible factor 1α (HIF-1α) plays a crucial role in the progression of glioblastoma multiforme tumors, which are characterized by their effective immune escape mechanisms. As major histocompatibility complex class I (MHC-I) is involved in glioma immune evasion and since HIF-1α is a pivotal link between inflammation and glioma progression, the role of tumor necrosis factor alpha (TNF-α)-induced inflammation in MHC-I gene regulation was investigated. A TNF-α-induced increase in MHC-I expression and transcriptional activation was concurrent with increased HIF-1α, ΝF-κΒ, and β-catenin activities. While knockdown of HIF-1α and β-catenin abrogated TNF-α-induced MHC-I activation, NF-κB had no effect. β-Catenin inhibition abrogated HIF-1α activation and vice versa, and this HIF-1α–β-catenin axis positively regulated CREB phosphorylation. Increased CREB activation was accompanied by its increased association with β-catenin and CBP. Chromatin immunoprecipitation revealed increased CREB enrichment at CRE/site α on the MHC-I promoter in a β-catenin-dependent manner. β-Catenin replaced human Brahma (hBrm) with Brg1 as the binding partner for CREB at the CRE site. The hBrm-to-Brg1 switch is crucial for MHC-I regulation, as ATPase-deficient Brg1 abolished TNF-α-induced MHC-I expression. β-Catenin also increased the association of MHC-I enhanceosome components RFX5 and NF-YB at the SXY module. CREB acts as a platform for assembling coactivators and chromatin remodelers required for MHC-I activation in a HIF-1α/β-catenin-dependent manner.
INTRODUCTION
The oxygen-sensing transcription factor hypoxia-inducible factor (HIF) is an important contributor to tumor growth and function (1, 2). HIF-1 is a heterodimer of the regulatory subunit HIF-1α and the constitutive subunit HIF-1β/ARNT (aryl hydrocarbon receptor nuclear translocator) (3). Though hypoxia stabilizes HIF-1α through inhibition of prolyl-hydroxylases and a subsequent decrease in HIF-1α ubiquitination and degradation (4), HIF-1α can also be activated under normoxia in response to proinflammatory cytokines (5). Glioblastoma multiforme (GBM) tumors, the most aggressive of brain tumors, contain hypoxic areas (6), and HIF-1 plays a key role in glioma invasion, metastasis, and angiogenesis, among other cellular functions (7).
Gliomas are characterized by their effective immune escape mechanisms, which evade immunosurveillance (8). The immune response to tumors is dependent predominantly on the activation of cytotoxic T lymphocytes that recognize tumor-specific antigens in the context of classical major histocompatibility complex class I (MHC-I) molecules and eliminate them (9). Downregulation of MHC-I, which characterizes many cancers, decreases their susceptibility to lysis by cytotoxic T cells (10). In contrast, glioma cells expressing high levels of MHC-I resist natural killer (NK) cell-mediated cytolysis (11). Glioma cells are therefore paradigmatic for tumor-mediated immune suppression, with high MHC-I levels contributing to the immune escape mechanisms through interaction with NK cell receptors. Moreover, inhibition of the highly elevated HLA levels in multiple myeloma triggers cell death without effector function (12).
Interestingly, the hypoxic tumor microenvironment contributes to the immune escape of cancers (13). Hypoxia contributes to tumor cell shedding of MHC-I chain-related molecule A (MICA), which plays an important role in tumor immune surveillance through its interaction with the NKG2D receptor on NK cells and cytotoxic T cells (14). Tumor necrosis factor alpha (TNF-α) increases MHC-I expression in neuroblastoma cells (15) and stabilizes MHC-I complexes (16). We have previously reported TNF-α-induced HIF-1α expression under normoxia (17) and the existence of the NF-κB–HIF-1α axis in interleukin-1β-treated glioma cells (18). As NF-κB transactivates MHC-I (19), we investigated whether TNF-α-induced HIF-1α and NF-κB affect MHC-I expression in glioma cells.
Canonical Wnt signaling requires the association of β-catenin with nuclear T-cell factor (TCF)–leukocyte enhancer factor (LEF) and results in the transcriptional activation of target genes (20). Wnt signaling is also known to regulate HIF-1α activity (21). Additionally, β-catenin interacts with HIF-1α to promote cellular adaptation to hypoxia (22). We have shown the involvement of Wnt-1 in inflammation-induced HIF-1α activation in glioma cells (18). Glioma cells possess elevated β-catenin activation (23), and TNF-α induces β-catenin activation (24). Though β-catenin plays a role in the expression of MICA (25), the role of canonical glycogen synthase kinase 3β (GSK-3β)/β-catenin signaling in MHC-I regulation is largely unknown.
Besides HIF-1α, the transcription factor cyclic AMP-responsive element-binding protein (CREB) is also involved in cellular responses to hypoxia (26). CREB regulates the expression of several genes, including the hypoxia-responsive ones (27). CREB, with its intrinsic oxygen-sensing mechanism, binds more efficiently to its cognate binding site, CREB response elements (CREs), to increase gene expression under hypoxia compared to that which occurs under normoxia (28). Moreover, CREB has been shown to regulate both the constitutive and cytokine-induced promoter activity of the MHC gene (29). CREB-binding protein (CBP)/p300, which is a transcriptional coactivator of CREB (30), interacts with HIF-1α under hypoxia (31). Also, β-catenin binds CBP (32) and the histone acetyltransferase activity of CBP facilitates the recruitment of chromatin remodelers SWI/SNF, which enable remodeling of local chromatin structures to facilitate transcriptional activation (33, 34). The chromatin-modifying SWI/SNF family, consisting of two closely related ATPases, human Brahma (hBrm) and Brahma-related gene 1 (Brg1), is required for HIF-1α expression and transcriptional activity under hypoxia (35).
Three conserved regulatory elements in the MHC-I promoter, enhancer A, the interferon (IFN)-stimulated response element, and site α, play an important role in the constitutive and cytokine-induced transactivation of MHC-I genes (36). Of these cis-regulatory elements, site α, which recruits CREB and regulates IFN-γ and class II transactivator-induced MHC-I transactivation, is particularly important (29). The enhanceosome complex that assembles on the SXY module of the MHC-I promoter consists of CREB, regulatory factor X (RFX), and nuclear factor Y (NF-Y), and the cooperative binding of these factors to their cognate site regulates MHC gene expression (37). Expression of MHC-I can be induced by immune regulatory cytokines to meet the local requirements of the immune response. As different stimuli affect MHC-I expression differentially, tight regulation at the transcriptional level is maintained (38). Hypoxic tumor microenvironment plays an important role in the enhancement of tumor escape from immune surveillance (39). Besides, inflammation, which is recognized as an indispensable participant in tumor progression, induces HIF-1α activation in gliomas (17). As MHC-I expression, either constitutive or inducible, is dependent on microenvironmental cues, we investigated whether TNF-α-induced HIF-1α regulates MHC-I expression through the modulation of a distinct repertoire of transcription factors, coactivators, and chromatin remodelers.
MATERIALS AND METHODS
Cell culture and treatment.
Human glioma cell lines T98G (CRL-1690; ATCC) and U87MG (CRL-11268; ATCC) were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10%heat-inactivated fetal bovine serum (Gibco) and penicillin (100 U/ml)-streptomycin (100 μg/ml; Gibco). Glioma cell lines were transiently transfected with Lipofectamine 2000 or Lipofectamine RNAiMAX (Invitrogen) in serum-free medium (SFM) according to the manufacturer's protocol. On attaining semiconfluence, cells were switched to SFM and after 4 h, cells were pretreated with quercetin or the phosphoinositide 3-kinase inhibitor LY294002 for 2 h and subsequently cotreated in the presence or absence of TNF-α (50 ng/ml) in SFM. Dimethyl sulfoxide-treated cells were used as controls. For hypoxia treatment, cells were exposed to 2%hypoxia (2%O2, 5%CO2, 93%N2) in a preprogrammed Anoxomat Mark II chamber (MART Microbiology, Drachten, Netherlands) for 24 h according to the manufacturer's instructions. All reagents were purchased from Sigma unless otherwise stated. Recombinant human TNF-α was purchased from R&D Systems. LY294002 was purchased from Calbiochem.
Small interfering RNA (siRNA) transfection.
Glioma cells were seeded into the wells of 24-well plates or onto 90-mm tissue culture dishes in DMEM without antibiotics, and transfection with 50 nmol/liter duplex HIF-1α, β-catenin, or a nonspecific siRNA (Thermo Fischer Scientific) was carried out with Lipofectamine RNAiMAX reagent (Life Technologies-Invitrogen) as described previously (18). Cells were stimulated at 18 h posttransfection with 50 ng/ml TNF-α (R&D Systems). After 12 or 24 h of stimulation, cells were harvested and analyzed by flow cytometry, Western blot analysis, chromatin immunoprecipitation (ChIP), and semiquantitative reverse transcriptase PCR (RT-PCR). The control nontargeting siRNA (catalog no. D-001210-03-20) and siRNAs targeting β-catenin (catalog no. L-003482-00-0005) and HIF-1α (catalog no. H-00765-00-0023) were obtained from Dharmacon (Thermo Scientific).
Western blot analysis and immunoprecipitation (IP).
Western blot analysis was performed with protein from whole lysates and nuclear extracts of cells transfected with different constructs or siRNA and treated with different inhibitors in the presence or absence of TNF-α as described previously (18), with antibodies against MHC-I, β2-microglobulin (β2m), β-catenin, HIF-1α, CREB, pCREB, CBP, Brg1, Brm, RFX5, and NF-Y subunit beta (NF-YB). Antibodies were purchased from Cell Signaling Inc. unless otherwise mentioned. HIF-1α, NF-YB, and RFX5 antibodies were purchased from Santa Cruz Inc. Secondary antibodies were purchased from Vector Laboratories. After the addition of a chemiluminescent reagent (Millipore), blots were exposed to the Chemigenius bioimaging system (Syngene) for development and images were captured with GeneSnap software (Syngene). The blots were stripped and reprobed with anti-β-actin (Sigma) or anti-C-23 (Santa Cruz) antibody to determine equivalent loading as described previously (18). For endogenous IP, nuclear extracts from glioma cells were incubated with 4 μg of the indicated antibodies for 16 h. Lysate was then incubated for 4 h with a mixture of protein G-Sepharose (Amersham Pharmacia). Beads were washed five times in IP buffer, and immunoprecipitated proteins were resolved by 8 to 10%SDS-PAGE. Control IP was performed with an anti-IgG antibody (Abcam). For Brg1 and hBrm acetylation studies, cells were lysed and the endogenous Brg1 and hBrm proteins were immunoprecipitated with anti-Brg1 and anti-hBrm antibodies (Abcam). Western blot analysis was performed with the immunoprecipitates with a specific anti-acetylated-lysine antibody (Cell Signaling Technology) as described previously (40).
Semiquantitative RT-PCR analysis.
Semiquantitative RT-PCR was performed for HLA-A, HLA-B, TAP1, LMP2, and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) or 18S rRNA with a one-step RT-PCR kit (Qiagen) as described previously (41). The PCR products were separated on a 1.7 to 2%agarose gels, stained with ethidium bromide, and photographed with Gene-Sys software provided with the Chemigenius bioimaging system (Syngene). The PCR primers used for RT-PCR are listed as follows: HLA-A forward and reverse, 5′-AAAAGGAGGGAGTTACACTCAGG-3′ and 5′-GCTGTGAGGGACACATCAGAG-3′, respectively; TAP1 forward and reverse, 5′-AGGGCTGGCTGGCTGCTTTGA-3′ and 5′-ACGTGGCCCATGGTGTTGTTAT-3′, respectively; LMP2 forward and reverse, 5′-CGTTGTGATGGGTTCTGATTCC-3′ and 5′-GACAGCTTGTCAAACACTCGGTT-3′, respectively; GAPDH forward and reverse, 5′-AGAAGGCTGGGGCTCATTTG-3′ and 5′-AGGGGCCATCCACAGTCTTC-3′, respectively; 18S rRNA forward and reverse, 5′-CAGCCACCCGAGATTGAGCA-3′ and 5′-TAGTAGCGACGGGCGGTGTG-3′, respectively.
Surface labeling and flow cytometric analysis.
Cells were treated for 24 h with 50 nM TNF-α, harvested by trypsinization, and fixed for 10 min in 1%paraformaldehyde in phosphate-buffered saline (PBS). To detect surface MHC-I (HLA-A, -B, and -C), cells were stained with an anti-MHC I antibody (Abcam) for 2 h at room temperature and then with a fluorescein-conjugated donkey anti-rabbit secondary antibody for 30 min under nonpermeabilizing conditions. Cells were washed twice and resuspended in fluorescence-activated cell sorting (FACS) buffer (1%bovine serum albumin in PBS with 0.1%NaN3). At least 20,000 events per sample were collected on a FACSCalibur flow cytometer (Becton Dickinson) and analyzed with CellQuest Pro software (Becton Dickinson) as described previously (17).
Luciferase reporter gene assays.
Reporter gene assays were performed with cells transfected with different combinations of MHC-I reporter, HIF-1α–luciferase construct, β-catenin overexpression construct or siRNA, wild-type (WT) HIF-1α, and WT CREB constructs and TCF4 TOP Flash and treated with TNF-α in the presence or absence of quercetin or Akt inhibitor as described previously (18). Briefly, glioma cells were transfected with 300 ng of expression plasmids and 100 ng of the indicated luciferase reporter constructs. A Renilla luciferase vector (pRL-TK; Promega) at 20 ng/well was included for normalization of transfection efficiency. Cells were harvested at the indicated time points, and cell lysates were analyzed with the Dual-Luciferase reporter assay system (Promega) as described previously (17). The pGL3-HLA-B7 and pGL3-HLA-B7Δα luciferase reporters were a kind gift from Peter J. van den Elsen, Leiden University Medical Center, Leiden, Netherlands (29). The HIF-1α-responsive element luciferase construct used was a gift from Chinmay Mukhopadhyay, Jawaharlal Nehru University, New Delhi, India (42). The pEGFP-C1 HIF-1α WT construct was a gift from Georgios Simos (43), University of Thessaly, Thessaly, Greece. Plasmids pBJ5-BRG1 (Addgene plasmid 17873) and pBJ5-BRG1-K-R (Addgene plasmid 17874; Crabtree lab) were obtained from Addgene and described previously (44). M50 Super 8X TOP Flash TCF4 reporter plasmid (Addgene plasmid 12456; Moon lab) was also obtained from Addgene. Wild-type (WT) and constitutively active (S37A) β-catenin expression plasmids were a kind gift from Mien-Chie Hung, University of Texas, Houston (45). The WT CREB expression plasmid was a kind gift from Marc R. Montminy, Salk Institute, La Jolla, CA (46). NF-κB luciferase and IκB-α mutant constructs were purchased from Clontech Inc.
ChIP and ChIP-qPCR assays.
ChIP was performed by enzymatic DNA shearing (Chip-IT Enzymatic; Active Motif) as previously described (18). Cells treated with or without TNF-α for 12 h were fixed in 1%formaldehyde at room temperature for 10 min. Isolated nuclei were lysed and then enzymatically sheared with the Enzymatic Shearing kit (Active Motif). Anti-CREB, -Brg1, -hBrm, -RFX5, and -NF-YB antibodies were used for IP, and a nonspecific IgG antibody (Abcam) was used as a control. Following reverse cross-linking and DNA purification, DNA from input (1:10 diluted) or immunoprecipitated samples was assayed by PCR. The SXY region of the MHC-I promoter was PCR amplified for 36 cycles from the immunoprecipitated chromatin. The PCR product was resolved on a 3%agarose gel and visualized by ethidium bromide staining and UV illumination. DNA recovered by ChIP was also analyzed by quantitative PCR (qPCR) with Power SYBR green PCR Master Mix (Applied Biosystems Inc.), with a ViiA7 real-time thermocycler (Applied Biosystems Inc.) for 40 cycles. The cycle thresholds (CTs) of immunoprecipitated material were normalized to their corresponding input DNA (1%input) and corrected by using nonspecific IgG CTs to allow direct comparison of different conditions. Relative (n-fold) enrichment was calculated with respect to the control levels. Nontarget sequences were amplified from the same material as a control for the IP in each sample, and results were corrected accordingly. The sequences of the primers used for endpoint and qPCR analyses of the amplified regions were as follows: HLA-A (ChIP) forward and reverse, 5′-TCCGCAGTTTCTTTTCTCCC-3′ and 5′-GGAGAATCTGAGTCCCGGTGG-3′, respectively; nontarget region 1 forward and reverse, 5′-AGGAATTCTCAAGAAACTGGTCG-3′ and 5′-CCTGGATGGTCTCATGTATCTCA-3′, respectively; nontarget region 2 forward and reverse, 5′-TGATCACTCAGTGCCCCTGAGCTC-3′ and 5′-GCCCAAAGCCCTCACTCACCT-3′, respectively.
Statistical analysis.
All comparisons between groups were performed with the two-tailed paired Student t test. All P values of less than 0.05 were taken as significant.
RESULTS
TNF-α-induced HIF-1α regulates MHC-I promoter activity and expression.
High levels of MHC-I in glioma cells facilitate the immune escape mechanism (11), and HIF-1α confers tumor cell resistance to lysis mediated by immune effectors (13). As we have previously reported that TNF-α induces HIF-1α activation in glioma cells under normoxia (17), we investigated the role of TNF-α-induced HIF-1α in MHC-I gene regulation. Western blot analysis indicated an increase in MHC-I expression upon TNF-α treatment in a time-dependent manner (Fig. 1A).
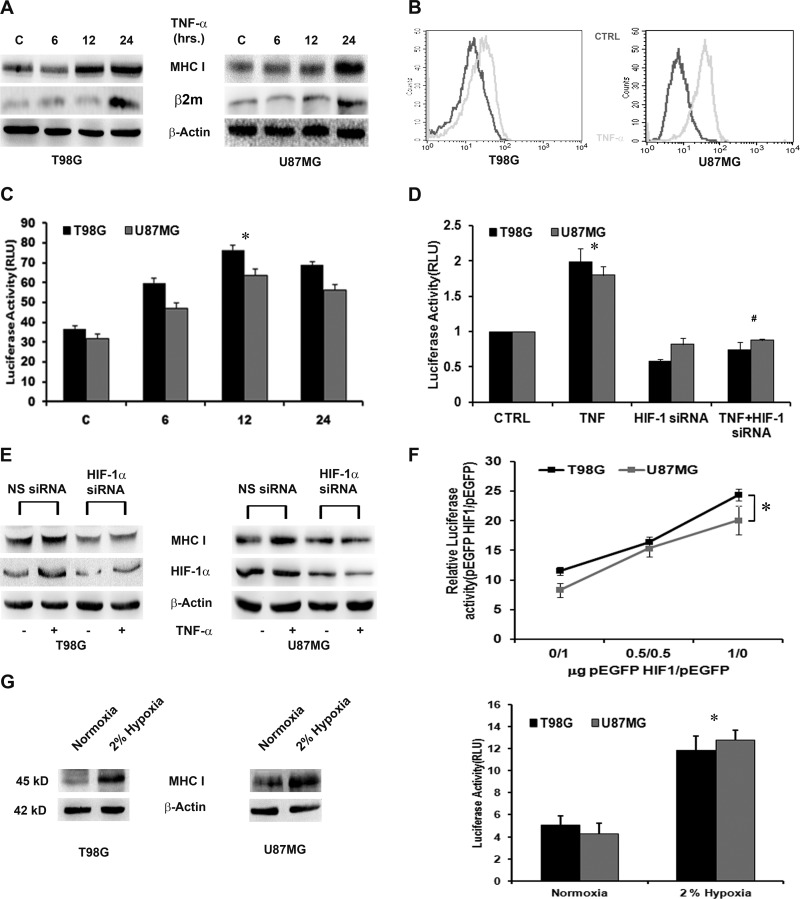
Fig 1.
TNF-α increases MHC class I expression and transcriptional activation in a HIF-1α-dependent manner. (A) Western blot analysis depicting MHC-I and β2m expression in TNF-α-treated glioma cells. Increased MHC-I heavy chain and β2m expression was also evident by Western blotting. The data are representative of three independent experiments. Blots were reprobed for β-actin to establish equivalent loading. C, control. (B) Surface expression of MHC-I in TNF-α-treated glioma cells as determined by FACS analysis. Shown is a histogram representative of three independent experiments showing the same trend. CTRL, control. (C) TNF-α increases MHC-I transcriptional activity in glioma cells. Luciferase reporter assay of MHC-I transcriptional activity in cells transfected with HLA-B7 reporter constructs and treated with TNF-α for 12 h. RLU, relative light units. (D) TNF-α-induced HIF-1α regulates MHC-I expression. Shown is a Western blot analysis depicting MHC-I levels in glioma cells transfected with HIF-1α siRNA in the presence or absence of TNF-α. The data are representative of three independent experiments. Blots were reprobed for β-actin to establish equivalent loading. NS, nonspecific. (F) TNF-α-induced MHC-I transcriptional activation is HIF-1α dependent. Glioma cells cotransfected either with WT HIF-1α or HIF-1α siRNA and HLA-B7 reporter constructs were treated with TNF-α for 12 h, and a luciferase reporter assay was performed to determine MHC-I promoter activity. Overexpression of WT HIF increases while that of HIF-1α siRNA decreases TNF-α-induced MHC-I promoter activity. (G) Western blot analysis and luciferase reporter assay demonstrating MHC-I expression and activity in cells exposed to 2%hypoxia for 24 h. In panels C, F, and G, the graphs represent normalized firefly luciferase activity. Values represent the means ± the standard errors of the means from three independent experiments. Symbols: *, significant increase over the untreated control; #, significant decrease from TNF-α-treated cells (P < 0.05).
As the formation of an MHC-I heavy chain–β2m heterodimer necessary to render MHC-I molecules functionally active is regulated by β2m levels (47), we investigated the status of β2m upon TNF-α stimulation. The increase in MHC-I was accompanied by an increase in β2m expression (Fig. 1A). Flow cytometric analysis also indicated an increase in MHC-I membrane expression upon TNF-α treatment (Fig. 1B). A luciferase reporter assay was performed to determine whether TNF-α also modulates MHC-I at the transcriptional level. Transfection of T98G cells with a pGL3-HLA-B7 luciferase reporter construct, followed by TNF-α treatment for 6, 12, and 24 h, resulted in ∼1.8-, 2.3-, and 2-fold increases in luciferase activity over control levels, respectively (Fig. 1C). A similar trend in MHC-I promoter activity was observed in TNF-α-treated U87MG cells (Fig. 1C). While elevated MHC-I promoter activity was observed at 6, 12, and 24 h, the activity peaked significantly at 12 h after TNF-α treatment (P = 0.033, T98G; P = 0.039, U87MG). As TNF-α-induced MHC-I promoter activity was maximal at 12 h, subsequent experiments to dissect the effect of TNF-α on MHC-1 promoter activity were performed at 12 h. We also determined the status of the MHC-I antigen-processing components TAP and LMP in TNF-α-treated cells. An increase in both TAP1 and LMP2, as evidenced by RT-PCR (see Fig. S1A in the supplemental material), was observed upon TNF-α treatment. Since we have previously reported TNF-α-induced increased HIF-1α activation in glioma cells (17), we questioned its role in MHC-I gene regulation. TNF-α-induced increased MHC-I transcriptional activity (Fig. 1D) and expression (Fig. 1E) were abrogated in cells transfected with HIF-1α siRNA. To further ascertain the role of HIF-1α, MHC-I promoter activity was determined in cells transfected with graded doses of the WT HIF-1α construct. Importantly, introduction of HIF-1α enhanced MHC-I transcriptional activity in a dose-dependent manner, as evidenced by its ability to transactivate the MHC-I promoter (P = 0.0136) (Fig. 1F). As inflammation-induced HIF-1α regulated MHC-I expression, we determined whether hypoxia also regulates MHC-I in glioma cells. An increase in both MHC-I expression and promoter activity was observed in glioma cell lines exposed to 2%hypoxia (Fig. 1G). Thus, HIF-1α independently of the stimulus of its induction regulates MHC-I expression and activity in glioma cells.
TNF-α induces β-catenin activation in glioma cells.
β-Catenin activation requires its dephosphorylation at serine residues 33, 37, and 41 (48). As TNF-α induces β-catenin activation in human breast epithelial cells (24), we determined the status of GSK3β/β-catenin signaling and activation in TNF-α-treated glioma cells. TNF-α increased active β-catenin, as evidenced by the increase in nonphosphorylated β-catenin levels with a concomitant increase in phosphoserine 9 GSK3β levels, without affecting the steady-state level of either protein (Fig. 2A, top). TNF-α also increased the accumulation of β-catenin in the nucleus in a time-dependent manner (Fig. 2A, middle). Immunocytochemistry analysis also revealed the increased nuclear accumulation of active β-catenin in glioma cells (see Fig. S2 in the supplemental material). Since hypoxia elevated MHC-I levels in glioma cells, we also investigated its effect on β-catenin activation. An increase in active β-catenin was also observed in glioma cells exposed to 2%hypoxia (Fig. 2A, bottom).
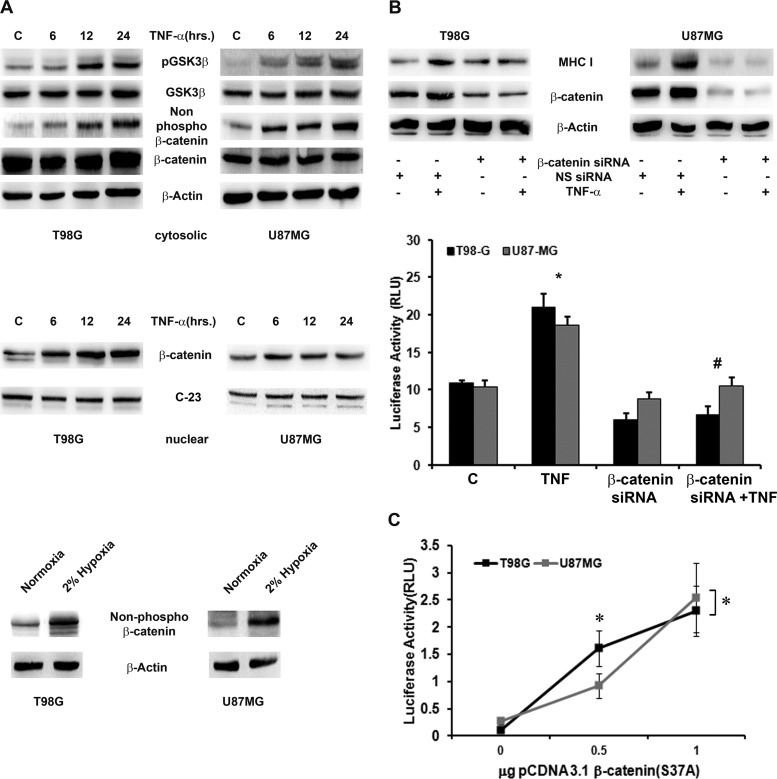
Fig 2.
MHC-I expression and transcriptional activation are dependent on β-catenin activation. (A) Increased GSK-3β/β-catenin signaling and nuclear β-catenin expression in TNF-α-treated glioma cells. Shown are Western blot analyses indicating nonphosphorylated and total β-catenin levels in TNF-α-treated cells. The data are representative of three independent experiments. β-Actin and C-23 (nucleolin) levels are shown as loading controls. Western blot analyses also demonstrate nonphosphorylated β-catenin levels in glioma cells treated with 2%hypoxia for 24 h. C, control. (B) TNF-α-induced β-catenin regulates MHC-I expression and activity. Western blot analysis and a luciferase reporter assay were performed with glioma cells either transfected with β-catenin siRNA alone or cotransfected with an HLA-B7 reporter construct in the presence or absence of TNF-α. Blots were reprobed for β-actin to establish equivalent loading. (C) MHC-I transcriptional activity is β-catenin dependent. Glioma cells were cotransfected with an HLA-B7 reporter construct and the indicated amounts of a constitutively active (S37A) β-catenin expression construct. The graph represents relative MHC-I luciferase activity normalized to Renilla luciferase activity. RLU, relative light units. The data shown are representative of three independent experiments. Symbols: *, significant increase over the untreated control; #, significant decrease from TNF-α-treated cells (P < 0.05).
TNF-α-induced increase in MHC-I expression and activation is β-catenin dependent.
As increased MHC-I expression was concomitant with increased β-catenin activation, we investigated the role of β-catenin in TNF-α-induced MHC-I expression. siRNA-mediated knockdown of β-catenin prevented a TNF-α-induced increase in MHC-I expression (Fig. 2B, top) and its promoter activity (P = 0.018) (Fig. 2B, bottom). To further validate the role of β-catenin in MHC-I transcriptional activity, TNF-α-induced β-catenin activation was mimicked by the expression of constitutively active β-catenin (S37A) in glioma cells in graded doses. The increase in MHC-I promoter activity positively correlated (Pearson's correlation coefficient = 0.94; P = 0.0035) with the expression of β-catenin in a dose-dependent manner (Fig. 2C). Thus, increased β-catenin activation regulates a TNF-α-mediated elevation in MHC-I expression and promoter activity. However, a TNF-α-induced increase in TAP1 and LMP2 is independent of β-catenin, as evidenced by RT-PCR (see Fig. S1B in the supplemental material).
β-Catenin-mediated MHC-I promoter activation and expression is TCF4 independent.
β-Catenin activation requires its association with nuclear TCF4 to trigger the transcriptional activation of its target genes (49). As hypoxia-induced HIF-1α is known to inhibit the β-catenin–TCF4 complex (22), we investigated the role of TNF-α-induced HIF-1α in the β-catenin–TCF4 interaction and its role in MHC-I expression. T98G glioma cells were treated with TNF-α in the presence or absence of HIF-1α siRNA, and the interaction between β-catenin and TCF4 was determined by IP. An increased interaction of β-catenin and TCF4 was observed upon TNF-α treatment and was abrogated upon HIF-1α knockdown (Fig. 3A). To further investigate the role of the β-catenin–TCF4 interaction in MHC-I expression and promoter activity, glioma cells were treated with TNF-α in the presence or absence of quercetin, which inhibits β-catenin–TCF4 signaling by interfering with β-catenin–TCF complex formation (50). Surprisingly, blockage of the transactivation function of β-catenin by quercetin failed to abrogate TNF-α-induced MHC-I promoter activity, P = 0.048 (Fig. 3B) and expression (Fig. 3C). This suggested that β-catenin regulates MHC-I independently of its association with TCF4.
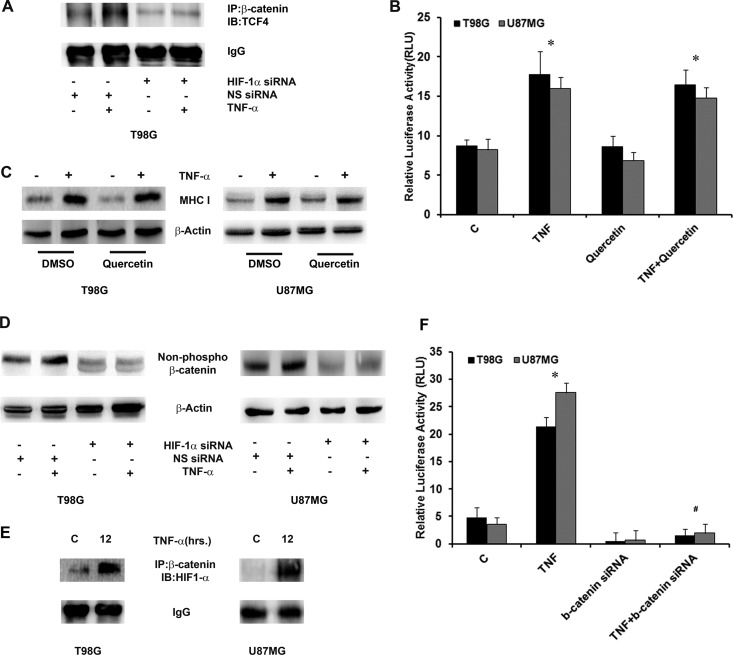
Fig 3.
β-Catenin–HIF-1α axis regulates MHC-I expression and transcriptional activation. (A) The TNF-α-induced β-catenin–TCF4 interaction is regulated by HIF-1α. β-Catenin was immunoprecipitated from nuclear lysates as indicated and probed for TCF4. IgG bands were used to normalize levels. IB, immunoblot; NS, nonspecific. (B) MHC I promoter activity and expression are independent of the β-catenin–TCF4 interaction. Following transfection with an HLA-B7 reporter construct, cells were treated with TNF-α in the presence or absence of 10 μM quercetin and a reporter assay was performed. The graph represents relative luciferase activities normalized to control Renilla reporter activity. RLU, relative light units. (C) Western blot analysis was performed to determine MHC I levels in glioma cells treated with different combinations of quercetin and TNF-α. DMSO, dimethyl sulfoxide. (D) Western blot analyses were performed to determine the levels of nonphosphorylated β-catenin in cells transfected with either HIF-1α siRNA or nontargeting siRNA in the presence or absence of TNF-α. The data are representative of three independent experiments. Blots were reprobed for β-actin to establish equivalent loading. (E) TNF-α increases β-catenin–HIF-1α interaction in glioma cells. Nuclear lysates from TNF-α-treated cells were immunoprecipitated with β-catenin and probed with antibody against HIF-1α. IgG levels are shown to establish equivalent loading. A representative blot from two independent experiments with identical results is shown. C, control. (F) Inhibition of β-catenin prevents TNF-α-mediated HIF-1α activation. Following cotransfection with HIF-1α reporter construct and β-catenin siRNA, cells were treated with TNF-α and reporter assay was performed to determine HIF-1α activity. The graph represents relative luciferase activity normalized to Renilla control reporter activity. Data bars represent the means ± the standard errors of the means from three independent experiments. Symbols: *, significant increase over the untreated control, #, significant decrease from TNF-α-treated cells (P < 0.05).
HIF-1α stabilizes TNF-α-induced β-catenin activation.
Though HIF-1α inhibits β-catenin-dependent transcription (22), a concomitant increase in β-catenin and HIF-1α activation had no negative effect on MHC-I transcriptional activation. As HIF-1α positively regulates MHC-I expression and because of the noninvolvement of TCF4-driven β-catenin function, we further investigated the mode of regulation of β-catenin owing to the possibility of a rather direct action of HIF-1α on β-catenin than on the β-catenin–TCF4 interaction. TNF-α-treated glioma cells showed reduced active levels of β-catenin under conditions where HIF-1α was knocked down with siRNA (Fig. 3D). As HIF-1α is known to bind β-catenin to regulate its function during hypoxic adaptation (22), we determined whether TNF-α increases the association of HIF-1α with β-catenin. Coimmunoprecipitation revealed an increased physical interaction of β-catenin with HIF-1α in TNF-α-treated cells (Fig. 3E).
TNF-α induces a HIF-1α–β-catenin axis in glioma cells.
As β-catenin regulates HIF-1α activity under hypoxic conditions (22), we investigated whether similar regulation occurs under inflammatory conditions under normoxia. TNF-α-induced HIF-1α promoter activity was abrogated in cells transfected with β-catenin siRNA (Fig. 3F). This, together with the ability of HIF-1α to regulate β-catenin activity, suggests the existence of a β-catenin–HIF-1α axis in TNF-α-treated cells under normoxia. We have reported that TNF-α-induced HIF-1α is Akt dependent (17). As Akt is involved in Wnt signaling (51), we investigated whether β-catenin activation is Akt dependent. Selective inhibition of Akt by LY294002 abrogated TNF-α-induced β-catenin activation (see Fig. S3 in the supplemental material).
Noninvolvement of NF-κB in TNF-α-mediated induction of MHC-I.
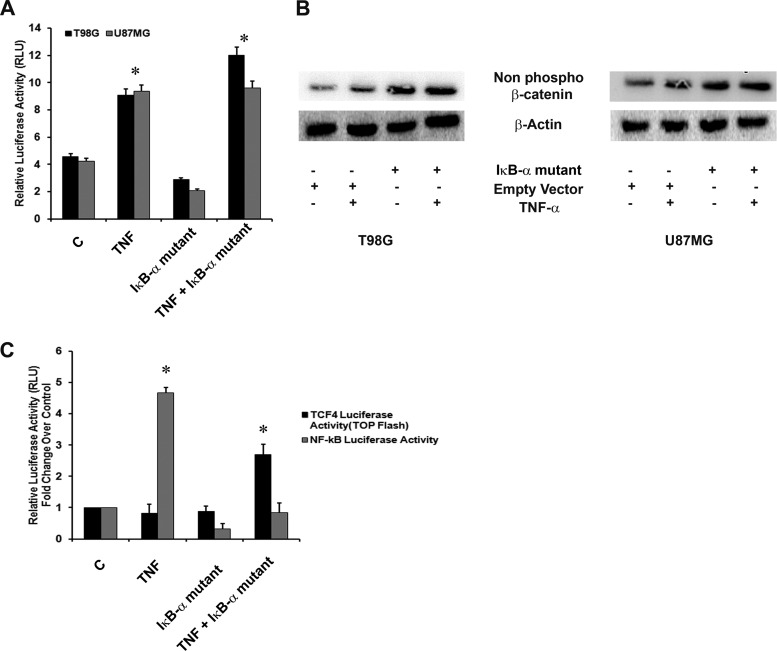
NF-κB regulates MHC-I expression (19), and we have shown the presence of the NF-κB–HIF-1α axis in glioma cells (18). While NF-κB overexpression inhibits β-catenin–TCF4-mediated transcription, GSK-3β represses TNF-α-induced NF-κB activity (52). As we observed the involvement of β-catenin–HIF-1α in MHC-I regulation, we questioned the importance of NF-κB in the regulation of MHC-I. Transfection with the IκB-α mutant construct had no effect on TNF-α-induced MHC promoter activity in cells (Fig. 4A). Although abrogation of NF-κB activation downregulated basal levels MHC-I promoter activity, the changes were not statistically significant (Fig. 4A). As β-catenin interacts with and inhibits NF-κB (45) and since NF-κB negatively regulates β-catenin–TCF4–p300 signaling loops (52), we investigated the role of NF-κB in β-catenin signaling. Western blot analysis revealed an increase in β-catenin activation, as evidenced by an increase in nonphosphorylated β-catenin levels in cells transfected with the IκB-α mutant construct (Fig. 4B). To further evaluate the effect of NF-κB on β-catenin-mediated transcription, TCF4 luciferase reporter activity was determined in TNF-α-treated cells transfected with the IκB-α mutant construct (Fig. 4C). Abrogation of NF-κB activation enhanced TNF-α-induced TCF4 activity by ∼2.5-fold (P = 0.012). TCF4 promoter activity in cells treated with TNF-α alone were comparable to that in untreated control cells and cells transfected with the IκB-α mutant construct (Fig. 4C).
Fig 4.
Noninvolvement of NF-κB in TNF-α-induced MHC class I expression and transcriptional activation. (A) TNF-α regulates MHC-I activation in an NF-κB-independent manner. Luciferase reporter assay of MHC-I transcriptional activity in cells cotransfected with HLA-B7 luciferase reporter and IκB-α mutant constructs and treated with TNF-α. C, control; RLU, relative light units. (B) TNF-α-induced NF-κB regulates β-catenin expression. Western blot analyses indicate nonphosphorylated β-catenin levels in TNF-α-treated cells transfected with the IκB-α mutant construct. β-Actin levels are shown as a loading control. (C) TNF-α-induced TCF4 transcriptional activation is NF-κB dependent. Glioma cells cotransfected with the TOP Flash TCF4 reporter construct and the IκB-α mutant construct were treated with TNF-α for 12 h, and a luciferase reporter assay was performed to determine TCF4 promoter activity. The graphs in panels A and C represent normalized luciferase activity over Renilla luciferase values. Values represent the means ± the standard errors of the means from three independent experiments. An asterisk indicates a significant increase over the untreated control.
β-Catenin-mediated regulation of MHC-I involves CREB.
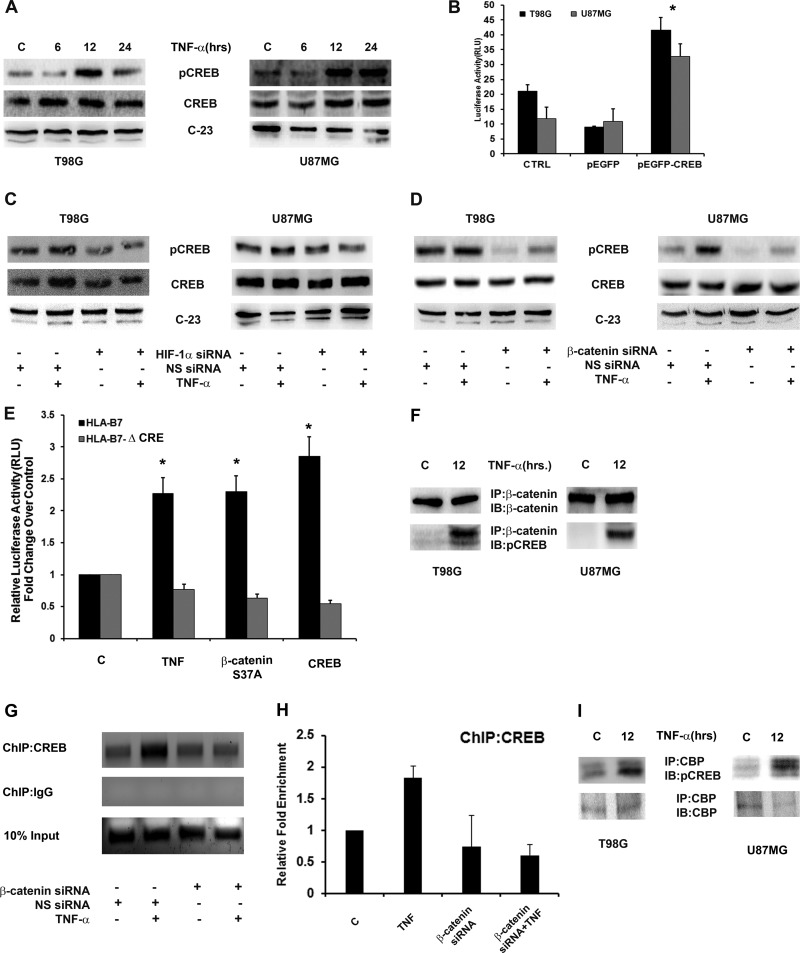
β-Catenin-mediated transcriptional activation of promoters requires sequences within the promoter related to consensus TCF/LEF-binding sites (53). The absence of putative TCF-binding sites and the inability of quercetin to affect TNF-α-induced MHC-I suggested a TCF-independent transcriptional program. Importantly, CREB is involved in MHC-I gene transcription through its binding to a cognate site within the SXY module of the proximal MHC-I promoter (37). To investigate the role of CREB in β-catenin-mediated MHC-I regulation, we determined the status of CREB in TNF-α-treated cells. TNF-α increased CREB phosphorylation at serine 133 in a time-dependent manner (Fig. 5A). As CREB is involved in MHC-I promoter activity (37), we determined its role in TNF-α-induced MHC-I transcription. Overexpression of WT CREB increased MHC-I transcriptional activity (P = 0.038) (Fig. 5B), confirming previous findings that CREB regulates MHC-I promoter activity (37). This increase in CREB is dependent on the β-catenin–HIF-1α axis, as a decrease in TNF-α-induced CREB phosphorylation was observed upon the siRNA-mediated knockdown of HIF-1α (Fig. 5C) and β-catenin (Fig. 5D). We next investigated the involvement of TNF-α-induced CREB in the regulation of MHC-I through site α by treating cells transfected with the HLA-B7Δα luciferase reporter. TNF-α had no effect on MHC-I promoter activity in the absence of functional CRE/site α, as evidenced by luciferase reporter assay (Fig. 5E). The importance of HIF-1α–β-catenin axis-induced CREB in the regulation of MHC-I activation through the CRE site was validated in cells cotransfected with HLA-B7Δα luciferase reporter and β-catenin or CREB overexpression plasmids. Overexpression of either β-catenin or CREB failed to induce MHC promoter activity in the absence of a functional CRE site (Fig. 5E). This confirmed that HIF-1α–β-catenin-mediated regulation of MHC-I involves CREB.
Fig 5.
Involvement of CREB in β-catenin-mediated transactivation of the MHC-I promoter. (A) TNF-α increases CREB phosphorylation in glioma cells. Shown is a Western blot analysis demonstrating CREB levels in cells treated with TNF-α for different intervals of time. C, control. (B) MHC-I transcriptional activation is CREB dependent. Luciferase reporter assay indicating MHC-I activity in glioma cells cotransfected with an HLA-B7 reporter and a pEGFP CREB-WT or an empty pEGFP vector construct. CTRL, control. TNF-α increases CREB phosphorylation in a HIF-1α (C)- and β-catenin (D)-dependent manner. Shown is a Western blot analysis demonstrating CREB phosphorylation in cells transfected with either HIF-1α or β-catenin and nonspecific siRNA and treated with TNF-α. (E) MHC I promoter activity is CRE site dependent. Graphs represent relative luciferase activities normalized to Renilla control reporter activity. Data bars represent the means ± the standard errors of the means of three independent experiments. An asterisk indicates a significant increase over the untreated control (P < 0.05). (F) TNF-α increases β-catenin–CREB interaction in glioma cells. Nuclear extracts from TNF-α-treated cells were immunoprecipitated with β-catenin antibody and then immunoblotted (IB) with CREB antibody. (G and H) ChIP and ChIP-qPCR assays indicating enhanced CREB interaction with CRE in the SXY module of the MHC-I promoter in TNF-α-treated glioma cells. DNA isolated from control and TNF-α-treated cells before and after IP with anti-CREB antibody was amplified with specific primer sets. A representative result from pre-IP (Input) and anti-CREB antibody-immunoprecipitated samples after PCR amplification is shown. ChIP-qPCR data bars represent change in fold enrichment ± SD from 2 independent experiments. NS, nonspecific. (I) IP demonstrating increased interaction between CREB and CBP upon TNF-α treatment. Nuclear extracts from TNF-α-treated cells were immunoprecipitated with CBP antibody and then immunoblotted with pCREB antibody. Panels A, C, and D are representative of three independent experiments. Blots were reprobed for C-23 to establish equivalent loading. Panels F and I are representative blots from two independent experiments with identical results.
TNF-α increases the association of CREB and β-catenin.
As CREB interacts with β-catenin (54), we investigated whether TNF-α affects the interaction between β-catenin and CREB in addition to their expression. IP revealed an increased association between β-catenin and CREB upon TNF-α treatment (Fig. 5F).
TNF-α induces binding of CREB to the CRE site of the MHC-I promoter.
To determine whether increased CREB activation and its interaction with β-catenin are accompanied by its enhanced binding to CRE/site α on the MHC-I promoter in a β-catenin manner, ChIP was performed with a CREB antibody in TNF-α-treated cells transfected with β-catenin siRNA. Endpoint ChIP-PCR and ChIP-qPCR assays revealed an increased enrichment of CREB at its cognate site on the MHC-I promoter in TNF-α-treated cells (Fig. 5G and H). This increased association of CREB was β-catenin dependent (Fig. 5G and H). Though β-catenin interacts with CREB (55), we failed to detect an interaction of β-catenin with CRE of MHC-I (data not shown). Since CREB binds to CRE directly and β-catenin interacts with CREB, it is likely that β-catenin regulates MHC-I via its interaction with CREB. The transcriptional coactivator CBP is essential not only for CREB (56) and β-catenin (32) but also for HIF-1α-dependent transcriptional activation (31). Activation of CREB through phosphorylation at serine 133 results in the recruitment of CBP and the activation of CRE (27). As increased CREB phosphorylation facilitates the recruitment of CBP, the interaction between CREB and CBP was determined in TNF-α-treated cells. IP revealed increased CREB-CBP interaction upon TNF-α treatment (Fig. 5I). Thus, a transcriptionally active MHC-I promoter requires HIF-1α–β-catenin axis-driven activation of CREB, its interaction with CBP, and its recruitment to the CRE site.
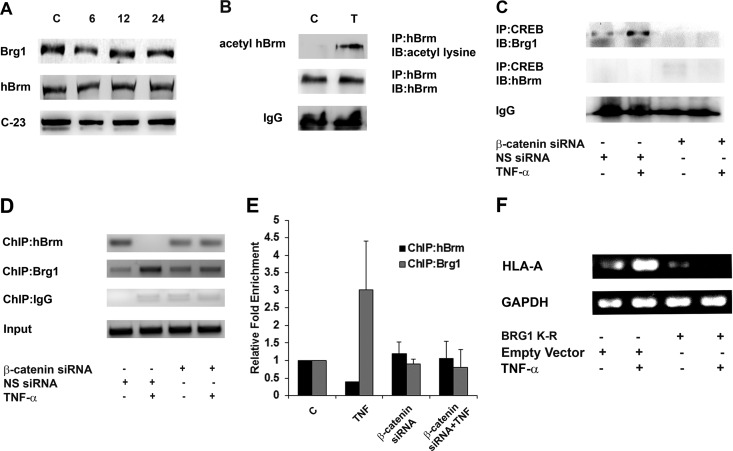
β-Catenin recruits Brg1 at the CRE site of the MHC-I promoter by replacing hBrm.
As CBP facilitates SWI/SNF recruitment to remodel chromatin structures (57) and since the SWI/SNF component Brg1, but not Brm, is involved in the regulation of enhancer A of the MHC-I promoter (58), the role of SWI/SNF in MHC-I activation through the SXY module was determined. TNF-α had no effect on the level of either hBrm or Brg1 expression, as demonstrated by Western blotting (Fig. 6A). However, an increase in the acetylated form of hBrm, but not Brg1 (data not shown), was observed upon exposure to TNF-α (Fig. 6B). The acetylated form of hBrm signifies its inactivation (59). Besides, β-catenin interacts with Brg1 to activate target gene promoters (60). Since the acetylated form of Brg1 was not detectable in TNF-α-treated cells (data not shown), we further determined the status of CREB-bound Brg1 by coimmunoprecipitation as a measure of Brg1 activation. IP with CREB antibody revealed the presence of Brg1 in the complex in TNF-α-treated cells, suggesting increased involvement of Brg1 (Fig. 6C). However, Brm was not associated with CREB in either control or TNF-α-treated cells (Fig. 6C). The TNF-α-induced increased CREB/Brg1 interaction was abrogated in cells transfected with β-catenin siRNA (Fig. 6C). An hBrm-to-Brg1 switch in the IFN-γ-activated sequence is associated with increased gene activity (59). Also, Brg1 and hBrm can compensate each other functionally to potentiate hypoxia-driven gene regulation (61). As CREB showed Brg1 binding specificity, the specific roles of these SWI/SNF factors in the regulation of MHC through the CRE site was evaluated by ChIP assay. While both Brg1 and hBrm occupy the SXY region in untreated cells, TNF-α treatment resulted in the eviction of Brm and enhanced enrichment of Brg1 at the CRE-binding site in the SXY module (Fig. 6D). This increased interaction of Brg1 at the CRE site in TNF-α-treated cells was abolished upon siRNA-mediated knockdown of β-catenin (Fig. 6D). Quantitative assessment of the change in the relative enrichment of Brg1/hBrm promoter occupancy also indicated a similar trend (Fig. 6E). Moreover, the decreased recruitment of hBrm to the CRE site upon TNF-α treatment was reversed upon the inhibition of β-catenin, as evidence by ChIP and ChIP-qPCR assays, respectively (Fig. 6D and E).
Fig 6.
Increased association of CREB with Brg1 but not hBrm. (A) TNF-α has no effect on the levels of hBrm and Brg1. Shown is a Western blot analysis indicating nuclear hBrm and Brg1 levels in TNF-α-treated cells. C-23 levels are shown as a loading control (lane C). (B) TNF-α induces acetylation of hBrm. Nuclear extracts from TNF-α-treated cells were immunoprecipitated with anti-hBrm antibody and analyzed for the levels of acetylated hBrm with a pan-acetylated-lysine antibody. IB, immunoblot. (C) TNF-α increases the association between CREB and Brg1 but not hBrm in a β-catenin-dependent manner. Nuclear extracts from cells transfected with β-catenin siRNA and treated with TNF-α were immunoprecipitated with CREB antibody, and an immunoblot analysis was done with Brg1 and hBrm antibodies. Band density was normalized against IgG levels under the same conditions. NS, nonspecific. (D, E) ChIP and ChIP-qPCR analyses indicating the relative changes in hBrm and Brg1 binding levels at the SXY module of the MHC-I promoter upon TNF-α treatment in a β-catenin-dependent manner. A representative gel image of the precleared fraction (Input) and the anti-Brg1- or anti-hBrm antibody-immunoprecipitated sample after PCR amplification is shown. DNA samples immunoprecipitated with the indicated antibodies were also subjected to qPCR along with the diluted input (1%), and the n-fold enrichment was calculated relative to control levels after correction for background signals. qPCR data bars indicate relative changes (n-fold) in enrichment over the control levels ± the standard deviations from two independent sets. (F) Brg1 activity is crucial for MHC-I expression. RT-PCR for MHC-I expression in TNF-α-treated cells transfected with ATPase-deficient Brg1 with a mutation in the ATPase subunit (K-R). GAPDH levels were used as internal controls. All experiments were performed with glioma cell line T98G.
TNF-α fails to activate MHC-I gene transcription in glioma cells lacking the ATPase subunit of Brg1.
ATP-dependent multiprotein SWI/SNF complexes disrupt the nucleosomal barrier and allow transcription to initiate and proceed (62). To investigate the direct effect of this hBrm-to-Brg1 switch on MHC-I transcription, we performed RT-PCR for MHC-I expression in TNF-α-treated cells transfected with a Brg1 expression plasmid with a mutation in the ATPase subunit (K-R). Expression of ATPase-deficient Brg1 abrogated the TNF-α-induced increase in MHC-I expression, confirming the direct involvement of Brg1-mediated chromatin remodeling in MHC-I gene expression (Fig. 6F). Thus, the increased recruitment of Brg1 to the CRE site is involved in MHC-I gene regulation.
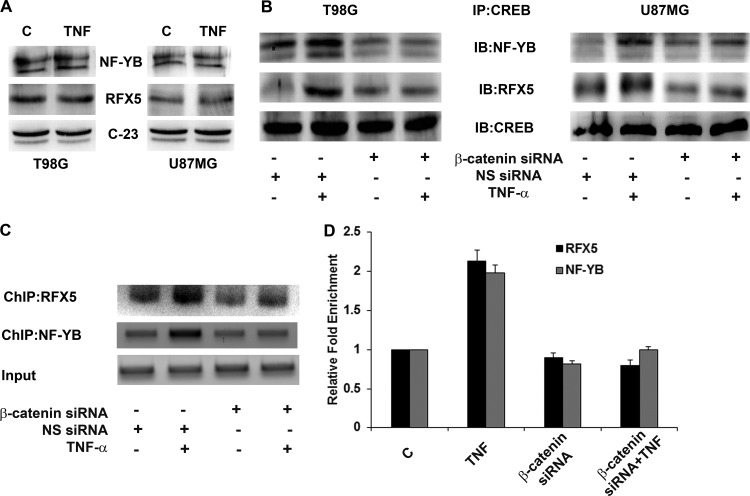
β-Catenin facilitates recruitment of the MHC enhanceosome complex.
In addition to CREB, the simultaneous recruitment of regulatory factor RFX5 and nuclear factor NF-YB to the MHC promoter regulates the formation of an enhanceosome complex, an essential component of the regulation of MHC (37). No change in the expression of either RFX5 or NF-YB was observed upon TNF-α treatment (Fig. 7A). However, coimmunoprecipitation revealed TNF-α-induced increased interaction of both RFX5 and NF-YB with CREB in a β-catenin-dependent manner (Fig. 7B). To determine whether this increased association was accompanied by increased occupancy of the SXY module, ChIP was performed with RFX5 and NF-YB antibodies. TNF-α increased the recruitment of both RFX5 and NF-YB at the SXY module (Fig. 7C). While RFX5 recruitment was abrogated upon β-catenin inhibition, NF-YB occupancy was reduced to control levels (Fig. 7C). β-Catenin thus regulates the recruitment of CREB, RFX5, and NF-YB within the SXY regulatory module to facilitate enhanceosome formation.
Fig 7.
β-Catenin regulates MHC-I enhanceosome formation. (A) TNF-α has no effect on the expression of RFX5 and NF-YB in glioma cells. Shown is a Western blot analysis indicating nuclear RFX5 and NF-YB levels in cells treated with TNF-α for 12 h. C-23 levels are shown as a loading control. (B) TNF-α increases the association of CREB with RFX5 and NF-YB, and this increased association of enhanceosome components is β-catenin dependent. Nuclear extracts from cells transfected with either β-catenin or nontargeting siRNA and treated with TNF-α for 12 h were immunoprecipitated with CREB antibody, and an immunoblot analysis was performed with RFX5 and NF-YB antibodies. Immunoprecipitated CREB levels were used to normalize bands. (C and D) TNF-α regulates the promoter occupancy of RFX5 and NF-YB at the SXY module in a β-catenin-dependent manner in T98G cells. DNA isolated from control and TNF-α-treated cells before and after IP with antibodies as indicated was amplified with specific primer sets. Representative results obtained with pre-IP (Input) and immunoprecipitated samples after PCR amplification are shown. DNA samples immunoprecipitated with the indicated antibodies were also subjected to a qPCR assay along with diluted input (1%), and the relative enrichment was calculated with respect to control levels after correction for background signals. qPCR data bars indicate relative changes in enrichment over control levels ± the standard deviations from two independent sets. The DNA gels and Western blot analyses shown are representative of three independent experiments with similar results. NS, nonspecific.
DISCUSSION
Targeting of the aberrant MHC-I level that is associated with evasion of immune responses is considered an important antimyeloma strategy (63). As high MHC-I levels also facilitate the evasion of immune responses in glioma tumors (11), the identification of mechanisms of its regulation might be useful for the development of anti-immune evasion strategies for the treatment of GBM. The hypoxic tumor microenvironment contributes to immune evasion (39). Although HIF-1α has been established as an important contributor to glioma progression, its role in the regulation of immunomodulatory HLA is not well understood. Besides hypoxia, inflammation-induced HIF-1α also plays an important role in glioma progression under normoxia (17). Our finding that both inflammation and hypoxia increase MHC-I expression suggests that HIF-1α, independently of its stimulus of induction, is an important contributor to MHC-I expression in glioma.
Increased GSK3 activity and decreased β-catenin activation induce MICA and MICB in cancer cells, which facilitate NK cell lysis via an NKG2D-restricted mechanism (25). NK cells reject tumors expressing MHC-I if the tumors express a ligand for NKG2D (64). This suggests that NK2GD stimulation could counteract the inhibitory signals triggered by MHC-I recognition. High levels of MHC-I in glioma cells mask the activating signals of NKG2DL and resist NK cell-mediated cytolysis (11). Since β-catenin inhibition induces MICA to facilitate NK cell lysis in cancer cells (25), it is possible that β-catenin activation increases MHC-I while decreasing MICA to subsequently abolish the activating potential of NKG2DL.
β-Catenin inhibits NF-κB (45), and constitutive NF-κB activation in GBM tumors regulates the expression of genes that promote their growth and survival (65). Although NF-κB transactivates MHC-I (19), TNF-α-induced NF-κB activation has no effect on MHC-I expression in glioma cells. Inhibition of NF-κB activation enhanced TNF-α-induced β-catenin activation and TCF4-driven transcription in glioma cells. Moreover, β-catenin induced MHC-I promoter activation in a TCF4-independent manner. This could possibly account for no apparent change in MHC-I promoter activity despite an increase in NF-κB-dependent, TCF4-driven transcription. However, inhibition of NF-κB activity resulted in a slight reduction in the basal MHC-I promoter activity in the absence of TNF-α. It is likely that elevated β-catenin activation in IκB-α mutant construct-transfected, TNF-α-treated cells overrides the NF-κB deficiency.
To better understand the β-catenin regulatory mechanisms that activate MHC-I transcriptional machinery independently of TCF, we used STRING, a search tool for incorporating known and predicted protein interaction information, to predict putative regulatory factors. This search led to the identification of CREB, which is known to regulate MHC-I gene transcription through its binding to the CRE site within the SXY module of the proximal MHC-I promoter (37). As hypoxia increases the efficiency of CREB binding to its cognate elements (28), it is possible that inflammation-induced HIF-1α enriches CREB at the CRE site of the MHC-I promoter.
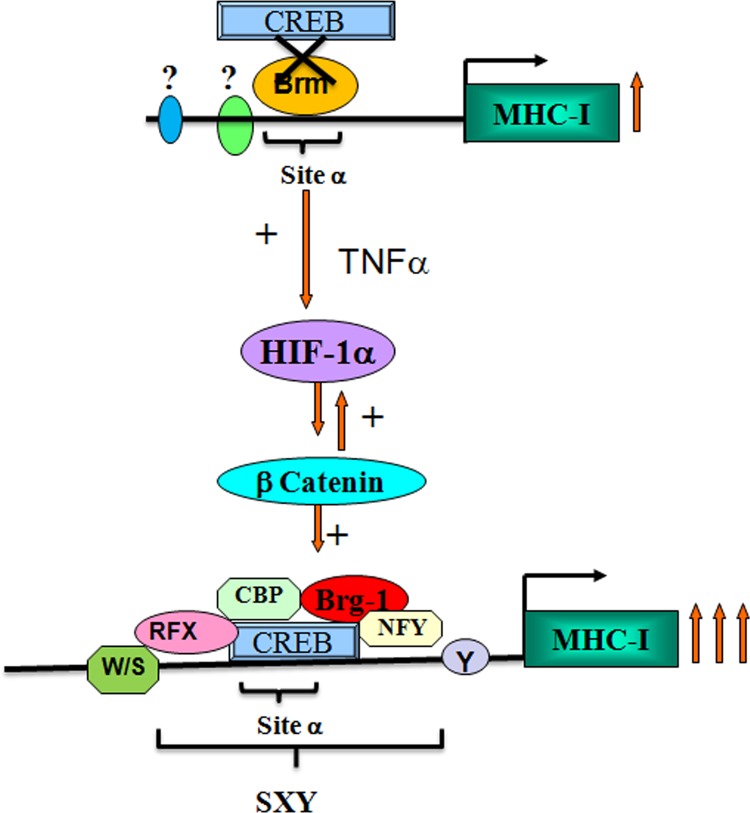
SWI/SNF components are required for HIF-1α expression and transcriptional activity under hypoxia (35). Though Brg1 and hBrm are recruited to the erythropoietin gene promoter in a hypoxia-dependent manner, hBrm is more potent than Brg1 with regard to HIF-1-mediated transcription of the erythropoietin gene (61). However, an hBrm-to-Brg1 switch in the IFN-γ-activated sequence is associated with increased gene activity (59). It is possible that Brg1 is crucial for β-catenin-driven MHC-I expression, since recruitment of Brg1 by β-catenin to target gene promoters facilitates transcriptional activation through chromatin remodeling (60). Besides, Brg1 is involved in higher-order chromatin remodeling of MHC upon IFN-γ treatment (66). Also, increased CREB binding at CRE/site α was accompanied by its increased association with CBP, which is an important coactivator in HIF-1α-dependent activation of transcription (31). Brg1 and CBP synergistically activate MHC-I genes through enhancer A (58), and CBP recruits Brg1 to the IFN-β promoter (33). It is likely that HIF-1α-driven recruitment of Brg1 at the CRE site at the expense of hBrm allows greater transcriptionally permissive chromatin architecture, which is conducive to greater MHC-I activation than in the constitutive state. Without cytokine stimulation, CREB cannot associate with its cognate elements on the SXY module, possibly because of the inhibitory effect of hBrm on the promoter (Fig. 8). Constitutive MHC-I expression could have resulted from the participation of regulatory elements other than site α, such as κB sites. As RFX controls the constitutive expression of MHC-I (67), it is possible that RFX levels are sufficient to maintain a high level of MHC-I, even in the absence of inflammation. By coordinating the dynamic interactions of CREB with the chromatin modifier Brg1 and components of the MHC enhanceosome, the HIF-1α–β-catenin axis controls MHC-I transcription under inflammatory conditions. Our findings suggest that the HIF-1α–β-catenin axis exclusively transactivates the MHC-I promoter through the SXY module in TNF-α-treated glioma cells.
Fig 8.
Model depicting the roles of β-catenin and HIF-1α in TNF-α-mediated MHC-I gene activation. Shown is a schematic representation of the hBrm-to-Brg1 switch at CRE/site α of the MHC-I promoter by HIF-1α. In the absence of inflammation, hBrm occupies the CRE site and prevents the access of CREB to its cognate element. Phosphorylated CREB induced by the TNF-α-triggered HIF-1α–β-catenin axis binds to the CRE site, which is followed by recruitment of CBP and Brg1. TNF-α promotes the formation of the MHC-I enhanceosome by recruiting RFX5 and NF-YB at the CREB/CBP/Brg1 complex. Brg1 mediates an open conformation around the CRE promoter and enhances the HIF-1α-mediated increased induction of MHC-I (arrows). Sites other than CRE/site α drive constitutive MHC-I expression in glioma cells.
As the biological function of MHC-I involves effective antigen presentation, modulation of its expression will affect the recognition of the presented peptides. Targeting of elevated MHC-I is considered therapy for multiple myeloma (68). As elevated MHC-I levels have a direct bearing on the immune escape mechanism in glioma cells and as HIF-1α is considered a potential antiglioma target (69), future endeavors to inhibit MHC-I by targeting the HIF-1α–β-catenin axis may provide a novel therapeutic approach to the modulation of immune surveillance in these tumors.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by a research grant from the Department of Biotechnology, Government of India (BT/PR/12924/Med/30/235/2009), to E.S.
Footnotes
Published ahead of print 13 May 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.01254-12.
REFERENCES
- 1.Semenza GL. 2010. Defining the role of hypoxia-inducible factor 1 in cancer biology and therapeutics. Oncogene 29:625–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Semenza GL. 2003. Targeting HIF-1 for cancer therapy. Nat. Rev. Cancer 3:721–732 [DOI] [PubMed] [Google Scholar]
- 3.Wang GL, Semenza GL. 1995. Purification and characterization of hypoxia-inducible factor 1. J. Biol. Chem. 270:1230–1237 [DOI] [PubMed] [Google Scholar]
- 4.Wang GL, Jiang BH, Rue EA, Semenza GL. 1995. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc. Natl. Acad. Sci. U. S. A. 92:5510–5514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jung Y, Isaacs JS, Lee S, Trepel J, Liu ZG, Neckers L. 2003. Hypoxia-inducible factor induction by tumour necrosis factor in normoxic cells requires receptor-interacting protein-dependent nuclear factor kappa B activation. Biochem. J. 370:1011–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaur B, Khwaja FW, Severson EA, Matheny SL, Brat DJ, Van Meir EG. 2005. Hypoxia and the hypoxia-inducible-factor pathway in glioma growth and angiogenesis. Neuro Oncol. 7:134–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zagzag D, Zhong H, Scalzitti JM, Laughner E, Simons JW, Semenza GL. 2000. Expression of hypoxia-inducible factor 1alpha in brain tumors: association with angiogenesis, invasion, and progression. Cancer 88:2606–2618 [PubMed] [Google Scholar]
- 8.Walker PR, Calzascia T, Dietrich PY. 2002. All in the head: obstacles for immune rejection of brain tumours. Immunology 107:28–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dunn GP, Old LJ, Schreiber RD. 2004. The immunobiology of cancer immunosurveillance and immunoediting. Immunity 21:137–148 [DOI] [PubMed] [Google Scholar]
- 10.Garrido F, Ruiz-Cabello F, Cabrera T, Perez-Villar JJ, Lopez-Botet M, Duggan-Keen M, Stern PL. 1997. Implications for immunosurveillance of altered HLA class I phenotypes in human tumours. Immunol. Today 18:89–95 [DOI] [PubMed] [Google Scholar]
- 11.Friese MA, Platten M, Lutz SZ, Naumann U, Aulwurm S, Bischof F, Buhring HJ, Dichgans J, Rammensee HG, Steinle A, Weller M. 2003. MICA/NKG2D-mediated immunogene therapy of experimental gliomas. Cancer Res. 63:8996–9006 [PubMed] [Google Scholar]
- 12.Sekimoto E, Ozaki S, Ohshima T, Shibata H, Hashimoto T, Abe M, Kimura N, Hattori K, Kawai S, Kinoshita Y, Yamada-Okabe H, Tsuchiya M, Matsumoto T. 2007. A single-chain Fv diabody against human leukocyte antigen-A molecules specifically induces myeloma cell death in the bone marrow environment. Cancer Res. 67:1184–1192 [DOI] [PubMed] [Google Scholar]
- 13.Barsoum IB, Hamilton TK, Li X, Cotechini T, Miles EA, Siemens DR, Graham CH. 2011. Hypoxia induces escape from innate immunity in cancer cells via increased expression of ADAM10: role of nitric oxide. Cancer Res. 71:7433–7441 [DOI] [PubMed] [Google Scholar]
- 14.Siemens DR, Hu N, Sheikhi AK, Chung E, Frederiksen LJ, Pross H, Graham CH. 2008. Hypoxia increases tumor cell shedding of MHC class I chain-related molecule: role of nitric oxide. Cancer Res. 68:4746–4753 [DOI] [PubMed] [Google Scholar]
- 15.Drew PD, Lonergan M, Goldstein ME, Lampson LA, Ozato K, McFarlin DE. 1993. Regulation of MHC class I and beta 2-microglobulin gene expression in human neuronal cells. Factor binding to conserved cis-acting regulatory sequences correlates with expression of the genes. J. Immunol. 150:3300–3310 [PubMed] [Google Scholar]
- 16.Hallermalm K, Seki K, Wei C, Castelli C, Rivoltini L, Kiessling R, Levitskaya J. 2001. Tumor necrosis factor-alpha induces coordinated changes in major histocompatibility class I presentation pathway, resulting in increased stability of class I complexes at the cell surface. Blood 98:1108–1115 [DOI] [PubMed] [Google Scholar]
- 17.Tewari R, Choudhury SR, Ghosh S, Mehta VS, Sen E. 2012. Involvement of TNFalpha-induced TLR4-NF-kappaB and TLR4-HIF-1alpha feed-forward loops in the regulation of inflammatory responses in glioma. J. Mol. Med. (Berl.) 90:67–80 [DOI] [PubMed] [Google Scholar]
- 18.Sharma V, Dixit D, Koul N, Mehta VS, Sen E. 2011. Ras regulates interleukin-1beta-induced HIF-1alpha transcriptional activity in glioblastoma. J. Mol. Med. (Berl.) 89:123–136 [DOI] [PubMed] [Google Scholar]
- 19.Mansky P, Brown WM, Park JH, Choi JW, Yang SY. 1994. The second kappa B element, kappa B2, of the HLA-A class I regulatory complex is an essential part of the promoter. J. Immunol. 153:5082–5090 [PubMed] [Google Scholar]
- 20.Arce L, Yokoyama NN, Waterman ML. 2006. Diversity of LEF/TCF action in development and disease. Oncogene 25:7492–7504 [DOI] [PubMed] [Google Scholar]
- 21.Lee SH, Kim MH, Han HJ. 2009. Arachidonic acid potentiates hypoxia-induced VEGF expression in mouse embryonic stem cells: involvement of Notch, Wnt, and HIF-1alpha. Am. J. Physiol. Cell Physiol. 297:C207–C216 [DOI] [PubMed] [Google Scholar]
- 22.Kaidi A, Williams AC, Paraskeva C. 2007. Interaction between beta-catenin and HIF-1 promotes cellular adaptation to hypoxia. Nat. Cell Biol. 9:210–217 [DOI] [PubMed] [Google Scholar]
- 23.Zheng H, Ying H, Wiedemeyer R, Yan H, Quayle SN, Ivanova EV, Paik JH, Zhang H, Xiao Y, Perry SR, Hu J, Vinjamoori A, Gan B, Sahin E, Chheda MG, Brennan C, Wang YA, Hahn WC, Chin L, DePinho RA. 2010. PLAGL2 regulates Wnt signaling to impede differentiation in neural stem cells and gliomas. Cancer Cell 17:497–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim MJ, Kim DH, Na HK, Surh YJ. 2010. TNF-alpha induces expression of urokinase-type plasminogen activator and beta-catenin activation through generation of ROS in human breast epithelial cells. Biochem. Pharmacol. 80:2092–2100 [DOI] [PubMed] [Google Scholar]
- 25.Skov S, Pedersen MT, Andresen L, Straten PT, Woetmann A, Odum N. 2005. Cancer cells become susceptible to natural killer cell killing after exposure to histone deacetylase inhibitors due to glycogen synthase kinase-3-dependent expression of MHC class I-related chain A and B. Cancer Res. 65:11136–11145 [DOI] [PubMed] [Google Scholar]
- 26.Abramovitch R, Tavor E, Jacob-Hirsch J, Zeira E, Amariglio N, Pappo O, Rechavi G, Galun E, Honigman A. 2004. A pivotal role of cyclic AMP-responsive element binding protein in tumor progression. Cancer Res. 64:1338–1346 [DOI] [PubMed] [Google Scholar]
- 27.Mayr B, Montminy M. 2001. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat. Rev. Mol. Cell Biol. 2:599–609 [DOI] [PubMed] [Google Scholar]
- 28.Goren I, Tavor E, Goldblum A, Honigman A. 2001. Two cysteine residues in the DNA-binding domain of CREB control binding to CRE and CREB-mediated gene expression. J. Mol. Biol. 313:695–709 [DOI] [PubMed] [Google Scholar]
- 29.Gobin SJ, Peijnenburg A, Keijsers V, van den Elsen PJ. 1997. Site alpha is crucial for two routes of IFN gamma-induced MHC class I transactivation: the ISRE-mediated route and a novel pathway involving CIITA. Immunity 6:601–611 [DOI] [PubMed] [Google Scholar]
- 30.Kwok RP, Lundblad JR, Chrivia JC, Richards JP, Bachinger HP, Brennan RG, Roberts SG, Green MR, Goodman RH. 1994. Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature 370:223–226 [DOI] [PubMed] [Google Scholar]
- 31.Arany Z, Huang LE, Eckner R, Bhattacharya S, Jiang C, Goldberg MA, Bunn HF, Livingston DM. 1996. An essential role for p300/CBP in the cellular response to hypoxia. Proc. Natl. Acad. Sci. U. S. A. 93:12969–12973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takemaru KI, Moon RT. 2000. The transcriptional coactivator CBP interacts with beta-catenin to activate gene expression. J. Cell Biol. 149:249–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Agalioti T, Lomvardas S, Parekh B, Yie J, Maniatis T, Thanos D. 2000. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-beta promoter. Cell 103:667–678 [DOI] [PubMed] [Google Scholar]
- 34.Bannister AJ, Kouzarides T. 1996. The CBP co-activator is a histone acetyltransferase. Nature 384:641–643 [DOI] [PubMed] [Google Scholar]
- 35.Kenneth NS, Mudie S, van Uden P, Rocha S. 2009. SWI/SNF regulates the cellular response to hypoxia. J. Biol. Chem. 284:4123–4131 [DOI] [PubMed] [Google Scholar]
- 36.van den Elsen PJ, Gobin SJ, van Eggermond MC, Peijnenburg A. 1998. Regulation of MHC class I and II gene transcription: differences and similarities. Immunogenetics 48:208–221 [DOI] [PubMed] [Google Scholar]
- 37.Gobin SJ, van Zutphen M, Westerheide SD, Boss JM, van den Elsen PJ. 2001. The MHC-specific enhanceosome and its role in MHC class I and beta(2)-microglobulin gene transactivation. J. Immunol. 167:5175–5184 [DOI] [PubMed] [Google Scholar]
- 38.Ting JP, Baldwin AS. 1993. Regulation of MHC gene expression. Curr. Opin. Immunol. 5:8–16 [DOI] [PubMed] [Google Scholar]
- 39.Noman MZ, Messai Y, Carre T, Akalay I, Meron M, Janji B, Hasmim M, Chouaib S. 2011. Microenvironmental hypoxia orchestrating the cell stroma cross talk, tumor progression and antitumor response. Crit. Rev. Immunol. 31:357–377 [DOI] [PubMed] [Google Scholar]
- 40.Wolf D, Rodova M, Miska EA, Calvet JP, Kouzarides T. 2002. Acetylation of beta-catenin by CREB-binding protein (CBP). J. Biol. Chem. 277:25562–25567 [DOI] [PubMed] [Google Scholar]
- 41.Dixit D, Sharma V, Ghosh S, Mehta VS, Sen E. 2012. Inhibition of Casein kinase-2 induces p53-dependent cell cycle arrest and sensitizes glioblastoma cells to tumor necrosis factor (TNFalpha)-induced apoptosis through SIRT1 inhibition. Cell Death Dis. 3:e271. 10.1038/cddis.2012.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Biswas S, Gupta MK, Chattopadhyay D, Mukhopadhyay CK. 2007. Insulin-induced activation of hypoxia-inducible factor-1 requires generation of reactive oxygen species by NADPH oxidase. Am. J. Physiol. Heart Circ. Physiol. 292:H758–H766 [DOI] [PubMed] [Google Scholar]
- 43.Mylonis I, Chachami G, Paraskeva E, Simos G. 2008. Atypical CRM1-dependent nuclear export signal mediates regulation of hypoxia-inducible factor-1alpha by MAPK. J. Biol. Chem. 283:27620–27627 [DOI] [PubMed] [Google Scholar]
- 44.Khavari PA, Peterson CL, Tamkun JW, Mendel DB, Crabtree GR. 1993. BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature 366:170–174 [DOI] [PubMed] [Google Scholar]
- 45.Deng J, Miller SA, Wang HY, Xia W, Wen Y, Zhou BP, Li Y, Lin SY, Hung MC. 2002. Beta-catenin interacts with and inhibits NF-kappa B in human colon and breast cancer. Cancer Cell 2:323–334 [DOI] [PubMed] [Google Scholar]
- 46.Mayr BM, Canettieri G, Montminy MR. 2001. Distinct effects of cAMP and mitogenic signals on CREB-binding protein recruitment impart specificity to target gene activation via CREB. Proc. Natl. Acad. Sci. U. S. A. 98:10936–10941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rock KL, Gamble S, Rothstein L, Gramm C, Benacerraf B. 1991. Dissociation of beta 2-microglobulin leads to the accumulation of a substantial pool of inactive class I MHC heavy chains on the cell surface. Cell 65:611–620 [DOI] [PubMed] [Google Scholar]
- 48.Staal FJ, van Noort M, Strous GJ, Clevers HC. 2002. Wnt signals are transmitted through N-terminally dephosphorylated beta-catenin. EMBO Rep. 3:63–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van de Wetering M, Cavallo R, Dooijes D, van Beest M, van Es J, Loureiro J, Ypma A, Hursh D, Jones T, Bejsovec A, Peifer M, Mortin M, Clevers H. 1997. Armadillo coactivates transcription driven by the product of the Drosophila segment polarity gene dTCF. Cell 88:789–799 [DOI] [PubMed] [Google Scholar]
- 50.Park CH, Chang JY, Hahm ER, Park S, Kim HK, Yang CH. 2005. Quercetin, a potent inhibitor against beta-catenin/Tcf signaling in SW480 colon cancer cells. Biochem. Biophys. Res. Commun. 328:227–234 [DOI] [PubMed] [Google Scholar]
- 51.Naito AT, Akazawa H, Takano H, Minamino T, Nagai T, Aburatani H, Komuro I. 2005. Phosphatidylinositol 3-kinase-Akt pathway plays a critical role in early cardiomyogenesis by regulating canonical Wnt signaling. Circ. Res. 97:144–151 [DOI] [PubMed] [Google Scholar]
- 52.Saegusa M, Hashimura M, Kuwata T, Hamano M, Okayasu I. 2007. Crosstalk between NF-kappaB/p65 and beta-catenin/TCF4/p300 signalling pathways through alterations in GSK-3beta expression during trans-differentiation of endometrial carcinoma cells. J. Pathol. 213:35–45 [DOI] [PubMed] [Google Scholar]
- 53.Tetsu O, McCormick F. 1999. Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature 398:422–426 [DOI] [PubMed] [Google Scholar]
- 54.Wang H, Wen S, Bunnett NW, Leduc R, Hollenberg MD, MacNaughton WK. 2008. Proteinase-activated receptor-2 induces cyclooxygenase-2 expression through beta-catenin and cyclic AMP-response element-binding protein. J. Biol. Chem. 283:809–815 [DOI] [PubMed] [Google Scholar]
- 55.Pradeep A, Sharma C, Sathyanarayana P, Albanese C, Fleming JV, Wang TC, Wolfe MM, Baker KM, Pestell RG, Rana B. 2004. Gastrin-mediated activation of cyclin D1 transcription involves beta-catenin and CREB pathways in gastric cancer cells. Oncogene 23:3689–3699 [DOI] [PubMed] [Google Scholar]
- 56.Parker D, Ferreri K, Nakajima T, LaMorte VJ, Evans R, Koerber SC, Hoeger C, Montminy MR. 1996. Phosphorylation of CREB at Ser-133 induces complex formation with CREB-binding protein via a direct mechanism. Mol. Cell. Biol. 16:694–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ogiwara H, Ui A, Otsuka A, Satoh H, Yokomi I, Nakajima S, Yasui A, Yokota J, Kohno T. 2011. Histone acetylation by CBP and p300 at double-strand break sites facilitates SWI/SNF chromatin remodeling and the recruitment of non-homologous end joining factors. Oncogene 30:2135–2146 [DOI] [PubMed] [Google Scholar]
- 58.Brockmann D, Lehmkuhler O, Schmucker U, Esche H. 2001. The histone acetyltransferase activity of PCAF cooperates with the brahma/SWI2-related protein BRG-1 in the activation of the enhancer A of the MHC class I promoter. Gene 277:111–120 [DOI] [PubMed] [Google Scholar]
- 59.Zhang Y, Cheng MB, Zhang YJ, Zhong X, Dai H, Yan L, Wu NH, Shen YF. 2010. A switch from hBrm to Brg1 at IFNgamma-activated sequences mediates the activation of human genes. Cell Res. 20:1345–1360 [DOI] [PubMed] [Google Scholar]
- 60.Barker N, Hurlstone A, Musisi H, Miles A, Bienz M, Clevers H. 2001. The chromatin remodelling factor Brg-1 interacts with beta-catenin to promote target gene activation. EMBO J. 20:4935–4943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang F, Zhang R, Beischlag TV, Muchardt C, Yaniv M, Hankinson O. 2004. Roles of Brahma and Brahma/SWI2-related gene 1 in hypoxic induction of the erythropoietin gene. J. Biol. Chem. 279:46733–46741 [DOI] [PubMed] [Google Scholar]
- 62.Naidu SR, Love IM, Imbalzano AN, Grossman SR, Androphy EJ. 2009. The SWI/SNF chromatin remodeling subunit BRG1 is a critical regulator of p53 necessary for proliferation of malignant cells. Oncogene 28:2492–2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ikegame A, Ozaki S, Tsuji D, Harada T, Fujii S, Nakamura S, Miki H, Nakano A, Kagawa K, Takeuchi K, Abe M, Watanabe K, Hiasa M, Kimura N, Kikuchi Y, Sakamoto A, Habu K, Endo M, Itoh K, Yamada-Okabe H, Matsumoto T. 2012. Small molecule antibody targeting HLA class I inhibits myeloma cancer stem cells by repressing pluripotency-associated transcription factors. Leukemia 26:2124–2134 [DOI] [PubMed] [Google Scholar]
- 64.Cerwenka A, Baron JL, Lanier LL. 2001. Ectopic expression of retinoic acid early inducible-1 gene (RAE-1) permits natural killer cell-mediated rejection of a MHC class I-bearing tumor in vivo. Proc. Natl. Acad. Sci. U. S. A. 98:11521–11526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nozell S, Laver T, Moseley D, Nowoslawski L, De Vos M, Atkinson GP, Harrison K, Nabors LB, Benveniste EN. 2008. The ING4 tumor suppressor attenuates NF-kappaB activity at the promoters of target genes. Mol. Cell. Biol. 28:6632–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Christova R, Jones T, Wu PJ, Bolzer A, Costa-Pereira AP, Watling D, Kerr IM, Sheer D. 2007. P-STAT1 mediates higher-order chromatin remodelling of the human MHC in response to IFNgamma. J. Cell Sci. 120:3262–3270 [DOI] [PubMed] [Google Scholar]
- 67.Gobin SJ, Peijnenburg A, van Eggermond M, van Zutphen M, van den Berg R, van den Elsen PJ. 1998. The RFX complex is crucial for the constitutive and CIITA-mediated transactivation of MHC class I and beta2-microglobulin genes. Immunity 9:531–541 [DOI] [PubMed] [Google Scholar]
- 68.Yang J, Yi Q. 2010. Killing tumor cells through their surface beta(2)-microglobulin or major histocompatibility complex class I molecules. Cancer 116:1638–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sen E. 2011. Targeting inflammation-induced transcription factor activation: an open frontier for glioma therapy. Drug Discov. Today 16:1044–1051 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.