Abstract
Genomic variation and related evolutionary dynamics of human respiratory syncytial virus (RSV), a common causative agent of severe lower respiratory tract infections, may affect its transmission behavior. RSV evolutionary patterns are likely to be influenced by a precarious interplay between selection favoring variants with higher replicative fitness and variants that evade host immune responses. Studying RSV genetic variation can reveal both the genes and the individual codons within these genes that are most crucial for RSV survival. In this study, we conducted genetic diversity and evolutionary rate analyses on 36 RSV subgroup B (RSV-B) whole-genome sequences. The attachment protein, G, was the most variable protein; accordingly, the G gene had a higher substitution rate than other RSV-B genes. Overall, less genetic variability was found among the available RSV-B genome sequences than among RSV-A genome sequences in a comparable sample. The mean substitution rates of the two subgroups were, however, similar (for subgroup A, 6.47 × 10−4 substitutions/site/year [95% credible interval {CI 95%}, 5.56 × 10−4 to 7.38 × 10−4]; for subgroup B, 7.76 × 10−4 substitutions/site/year [CI 95%, 6.89 × 10−4 to 8.58 × 10−4]), with the time to their most recent common ancestors (TMRCAs) being much lower for RSV-B (19 years) than for RSV-A (46.8 years). The more recent RSV-B TMRCA is apparently the result of a genetic bottleneck that, over longer time scales, is still compatible with neutral population dynamics. Whereas the immunogenic G protein seems to require high substitution rates to ensure immune evasion, strong purifying selection in conserved proteins such as the fusion protein and nucleocapsid protein is likely essential to preserve RSV viability.
INTRODUCTION
Human respiratory syncytial virus (RSV) is the leading cause of severe respiratory tract infections in infants under the age of 2 years (1). Premature neonates and children with chronic pulmonary or congenital heart diseases are especially at risk, but immunocompromised adults and the elderly have also been reported as high-risk groups (2–4). Individuals infected by RSV mostly suffer from rhinitis and common cold-like symptoms but can also develop serious pneumonia or bronchiolitis, thereby requiring hospitalization (5). According to the Centers for Disease Control (CDC), RSV infections annually account for the hospitalization of up to 126,000 children and 62,000 elderly people in the United States alone (http://www.cdc.gov/rsv/about/faq.html). Seasonal epidemic outbreaks of RSV disease in Europe and North America occur during the winter and early spring months, with a peak incidence in December (6). In tropical countries, there is a correlation between increased RSV infection rates and rainy seasons (7). Relative humidity is an important factor for RSV activity, but the exact influence on the course of infection or on virus transmission dynamics is unknown (8).
RSV is an enveloped, nonsegmented, negative-sense, single-stranded RNA virus of approximately 15,000 nucleotides that is classified in the genus Pneumovirus belonging to the family of Paramyxoviridae. The viral genome, which is surrounded by a nucleocapsid protein complex, encodes 11 proteins that have roles in three different stages of the RSV life cycle. The nucleocapsid protein (N), phosphoprotein (P), and large polymerase (L) and the transcription regulatory proteins M2-1 and M2-2 are essential mediators of the RSV transcription and replication processes (9). Virus entry and assembly are mediated by the structural matrix protein (M), attachment glycoprotein (G), and fusion glycoprotein (F) (10, 11).
Virus attachment occurs via binding of the immunogenic RSV G to the glycosaminoglycans (GAGs) of the host target cell (12, 13). RSV entry into host cells is then mediated via the fusion protein, which requires enzymatic cleavage by furin-like proteases for RSV F activation (14). RSV F alone can initiate infection, as proven with RSV G deletion mutants (15). Recently, the structural small hydrophobic protein (SH) has been identified as a viroporin which permeabilizes the host membrane, suggesting facilitation of viral entry (16, 17). This accessory structural protein is not pivotal for RSV infection and mainly accumulates in the lipid raft structures of the Golgi complex (18, 19). Finally, RSV nonstructural proteins 1 and 2 (NS1 and -2) are host interference proteins that impair the antiviral state by impeding interferon activity (20). In addition, the soluble truncated form of RSV G has been described as an antigen decoy in antibody-mediated neutralization, thereby impairing the antibody-mediated antiviral effects of Fc receptor-bearing leukocytes (21).
For RSV, two antigenic subgroups (A and B) have been identified via antibody cross-reactivity patterns and these were further classified into genotypes according to genetic divergence within the highly variable G gene (22–24). The two subgroups can cocirculate, and RSV reinfections occur frequently throughout life, which indicates that there is only partial cross-immunity against different strains (25). Initial infection with RSV-A is frequently followed by infection with RSV-B, but this order is not consistently observed (26). It has been suggested that the antigenic variability of the G protein, both within and between the antigenic subgroups, particularly facilitates evasion of the preexisting host immune responses (27, 28). Multiple genotypes can be present in one population, although new genotypes may replace older predominating genotypes in time over successive epidemic seasons (24, 29). The heterogeneity in the genotypic distribution patterns observed among different populations is most likely influenced by variations in the herd immunity and also by community-specific cultural and behavioral patterns (30, 31).
The pathogenesis and disease outcome of RSV infection are most likely determined by the course of the induced immune response (32) and the capacity of RSV to modify this response for its own benefits (e.g., through Toll-like receptor-signaling interference [33]). This has been hypothesized on the basis of several vaccine studies, where the critical immune balance was negatively affected in numerous ways. Early vaccination attempts in children in the 1960s with a formalin-inactivated RSV vaccine resulted in enhanced disease severity upon subsequent virus exposure (34–37). It is thought that inadequate serum neutralizing-antibody levels, low CD8+ memory T cell response, and enhanced CD4+ memory T cell response were the consequences of this intensified disease profile (35, 38–41). Later vaccination attempts with live, attenuated RSV vaccines did not show similar respiratory disease exacerbation upon natural infection but nevertheless failed to provide appreciable protection from infection. From clinical-immunological data and the epidemiological profile of RSV, it is clear that this virus, which lacks clear-cut virulence factors, causes disease by modifying the host immune response in a way that affects pathogenesis and that it has typical seasonal transmission behavior. However, the association between RSV genetic diversity, RSV-induced immune responses, and the formation of specific epidemiological patterns remains poorly understood.
Understanding fluctuations in the genetic diversity of viral populations that are driven by ecological and evolutionary processes could elucidate the roles of specific genes and proteins in RSV survival. In addition, knowledge of evolutionarily conserved domains or substitution hot spots could highlight potentially useful therapeutic targets. Phylodynamic analyses are increasingly used to infer viral population dynamics from phylogenetic patterns (42). These analyses reveal the impact of both short- and long-term genomic evolutionary changes that enable viruses to escape from host immune responses and how these changes affect viral epidemic behavior. Although the molecular epidemiology and evolutionary dynamics of RSV have been extensively studied, these analyses have primarily focused on single genes—in particular, the G gene (43, 44). Little is known about either the diversity or evolution rates of other genes or the RSV genome as a whole. Importantly, as has recently been demonstrated in many other virus groups, full-genome analyses can provide valuable insights into the processes that shape viral epidemiology and evolution (45). Complete genome sequences from RSV clinical isolates have, however, been generated in sufficiently large numbers to facilitate such analyses only recently (46–48), and that work is further extended with this study.
Here, we conducted phylodynamic analyses on a collection of RSV-B strains and present the first calculations made within the phylodynamic framework of RSV-B genomic diversity through time. In addition to detailed genome-wide genetic variability analyses, we inferred genome-wide evolution rates and estimate the time since the most recent common ancestor (TMRCA) of all the currently available RSV-B genome sequences. Furthermore, we compare these estimates with those previously made for RSV-A (48). We show that whereas RSV-B genomes evolved with rates similar to RSV-A genomes, there is substantially less protein variability among currently sampled RSV-B strains, suggesting that these circulating RSV-B strains have a more recent ancestry due either to a chance fixation of a particular variant or to fixation of a variant harboring advantageous mutations. The observed differences in G, SH, and M2-2 gene variability between RSV-A and -B have the highest impact on this dissimilarity. As has been previously shown for RSV-A, the RSV G gene is evolving considerably faster than the remainder of the RSV genome—a finding that is consistent with evidence of both persistent and episodic diversifying selection in this gene. Whereas positive selection in the G gene likely reflects the impact of host immune responses, more pervasive evidence of purifying selection within the more conserved genes likely reflects selection favoring the optimization of viral replication and transmission.
MATERIALS AND METHODS
Clinical virus isolates and ethics statement.
From 2002 to 2012, nasopharyngeal aspirates and nose-throat swabs were collected from 34 patients who were hospitalized in the Wilhelmina Children's Hospital of Utrecht, the Netherlands, or in the Gasthuisberg University Medical Hospital of Leuven, Belgium (see Table S1 in the supplemental material). At the time of sampling, 28 patients were under the age of 2 and 6 were adults ranging from 27 to 79 years of age. As part of the routine diagnostic process, collected samples were cultured and checked for the formation of syncytia in HEp2 cell cultures, a typical RSV phenotype. The anonymized clinical strains were used in this study according to the guidelines of the institutions' ethical committees (Medisch Ethische Toetsingsingscommissie [METC] for Dutch samples; Continuing Medical Education [CME] for Belgian samples), and the study was performed in concordance with Dutch privacy legislation. The institutional review board (IRB) confirmed (protocol 12/320) that viral strains are not regarded as patient owned and that the use of these strains is not restricted in the applicable Dutch law (Law Medical Scientific Research with People, Wet Maatschappelijke Ondersteuning [WMO]; article 1b).
Viral RNA extraction, cDNA synthesis, and real-time TaqMan PCR.
Viral genomic RNA was directly isolated from patient material using MagnaPure LC total nucleic acid kits (Roche Diagnostics, Mannheim, Germany). Multiscribe reverse transcriptase (RT) kits and random hexamers (Applied Biosystems, Foster City, CA) were used for reverse transcription of isolated viral RNA. Subtyping of the RSV patient strains was achieved using real-time PCR on viral cDNA with primers and probes designed on the basis of highly conserved genomic regions of the N gene for RSV subgroups A (RSV-A) and B (RSV-B) as described by Tan et al. (48).
Synthesis and sequencing of genomic RSV PCR fragments.
Human RSV-B PCR fragments were obtained via fractional amplification of MagNAPure LC genomic RNA isolates using a Superscript III one-step RT-PCR system with a Platinum Taq High Fidelity kit (Invitrogen) according to the manufacturer's protocol and a 9800 Fast thermal cycler (Applied Biosystems). PCR products were purified from 1% agarose gels by the use of a GeneJET gel extraction kit (Fermentas) and were sequenced according the conventional Sanger technique. Fragments ranging between 650 and 1,400 nucleotides in length were sequenced on an ABI 3730 48-capillary DNA analyzer using BigDye Terminator 3.1 (ABI) and sequence-specific primers (see Table S2 in the supplemental material). Whole-genome sequences were assembled with these strain-specific PCR fragments by aligning them to the reference RSV strain, B1 (AF013254.1), using the computer program Seqman Pro (Lasergene 10 software; DNASTAR, Inc.).
Substitution analysis on the whole-genome and individual protein levels.
Individual gene sequences were extracted from the whole-genome sequences of all individual RSV-B strains (34 RSV-B isolates and 2 reference RSV-B strains) in silico, using Seqman. Seaview4 was used to translate genes into protein coding sequences that were subsequently aligned with the EMBL-EBI ClustalW2-Multiple Sequence Alignment tool (49). The sequence variability scores per gene and coding protein were calculated relative to the gene and protein consensus sequences, respectively, obtained from the data set of aligned clinical and reference strains. Genomic and protein substitutions per site were mapped using the Plot0.997 program (http://plot.micw.eu/). The NetNGlyc 1.0 server (shown to achieve correct prediction of 86% glycosylated and 61% nonglycosylated sites with an accuracy of 76%) (50) and NetOGlyc 3.1 server (shown to achieve correct prediction of 76% glycosylated and 93% nonglycosylated sites with an accuracy of 21%) (51) were used to predict the gain and loss of N-glycosylation and O-glycosylation sites, respectively. Previously described RSV-A genome sequences (48) were included in all these analyses for comparison with RSV-B-specific genetic variations. In addition, we analyzed previously obtained RSV-A data for evidence of O-glycosylation.
Data set assembly and recombination analysis.
Analysis of RSV complete genome evolutionary dynamics was conducted on a data set containing a combination of newly obtained RSV-B isolates and strains from a recent study by Rebuffo-Scheer et al. (47). The latter five full-genome sequences (JN032115 to JN032117 and JN032119 and JN032120) were derived from nasopharyngeal and nasal swabs collected from patients in the Milwaukee metropolitan area. A total of 41 RSV-B complete genomes were aligned using Mafft. Manual editing of the alignment was performed using Se-Al (available at http://tree.bio.ed.ac.uk/software/seal). Strain recombination was evaluated using the RDP, GENECONV, RECSCAN, MAXCHI, CHIMAERA, SISCAN, and 3SEQ recombination detection methods implemented in RDP3 (52). Evidence of likely recombination events was considered robust only when these were detected by two or more different recombination detection methods together with evidence that different genome fragments apparently derived from different parental strains clearly fell within different clades of RSV-B phylogenetic trees. Two Dutch-Belgian RSV-B strains showed significant evidence of recombination. Since the analyses carried out in this study relied on accurate phylogenetic inference and a single phylogenetic tree cannot adequately describe the evolutionary history of recombinant sequences, we opted to remove the minor recombinant parts of these two genomes from the data set and replaced them by gaps in the alignment. These two strains were incorporated only in the data set for Bayesian inference and were not included in the substitution analysis.
The evolutionary dynamics of the G gene were investigated based on an alignment of the G gene partitions from our complete RSV-B genomes together with two other data sets previously evaluated by Zlateva et al. in 2005 (43) and Baek et al. in 2012 (53) (see overview of strains in Table S3 in the supplemental material). Zlateva et al. aligned 204 sequences encompassing the region of amino acid positions 58 to 299 of the G protein (according the AF013254 reference strain). Baek et al. aligned full-length G proteins of Korean origin. All RSV-B estimates obtained by the molecular evolutionary analyses were compared with recent published RSV-A estimates of molecular evolutionary analyses (48).
Bayesian inference of the RSV-B evolutionary history.
In order to determine whether there existed a clear temporal signal of nucleotide divergence within our complete genome data set, exploratory analyses were performed with the computer program Path-O-Gen (available at http://tree.bio.ed.ac.uk/software/pathogen/) (54), which performs a linear regression analysis of root-to-tip divergence of individual samples against their sample dates. For this purpose, we constructed a maximum-likelihood tree using PhyML (55) with a general time-reversible (GTR) substitution model and a discretized gamma distribution to model rate variation among sites. These exploratory analyses were performed with the exact sampling date (month and year) for our novel RSV-B genomes. Collection months were not available for the five sequences from Rebuffo-Scheer et al. (47), so we set the sampling date at the midpoint of the reported sampling year.
The RSV evolutionary and demographic histories were reconstructed for both the whole-genome sequences and the G gene data set using Bayesian genealogical inference implemented in BEAST v1.7 (56). Using Markov Chain Monte Carlo (MCMC) analyses, we estimated a posterior distribution of time-measured genealogies based on a full probabilistic model that included a nucleotide substitution model, a molecular clock model, and a coalescent model as prior distribution for the phylogenetic tree. We used the GTR substitution model with a discretized gamma distribution to model rate variation among sites. Substitution rate variability across the RSV genome was studied using a partition model that allowed for separate substitution rates for all individual genes and a single concatenated noncoding region. The best molecular clock model fit was assessed on the basis of marginal-likelihood estimates using path sampling (PS) estimators and stepping-stone (SS) estimators (57). We compared strict and relaxed clock models that assume homogenous and heterogeneous substitutions rates across phylogenetic branches, respectively (58). Tip ages of the five sequences with unknown exact sample date were estimated within a 1-year time interval with a uniform prior distribution (59). With the Bayesian skyline plot model as a flexible demographic prior distribution, we analyzed the changes in the effective population size through time (60). MCMC analyses were performed until convergence could be convincingly assumed according to evaluation with Tracer (available at http://tree.bio.ed.ac.uk/software/tracer/). Marginal posterior distributions for evolutionary rates and times to the most common ancestors (TMRCA) of various sequence groups were summarized using means and 95% highest posterior density intervals (HPDs). A maximum clade credibility (MCC) tree annotated with divergence time and evolutionary rate summaries was used as a representation of the evolutionary history and visualized using FigTree (available at http://tree.bio.ed.ac.uk/software/figtree) (58).
A genealogical test based on posterior predictive simulation was conducted in order to determine whether the posterior phylogenetic tree shapes deviated from neutral expectations (61). In short, the tree shapes estimated by the Bayesian coalescent analysis were summarized with the genealogic Fu and Li statistic (DF), which compares the length of the terminal branches to the total length of the coalescent genealogy and returns negative values for long terminal branch lengths. This in turn indicates an excess of slightly deleterious mutations on these branches and consequently a deviation from neutrality. Trees with the same tip numbers and the same tip ages and under the same neutral coalescent model as applied to the real data were randomly simulated by the posterior predictive simulation analysis. In this study, we used the genealogical neutrality test extension that allowed simulation of trees under the Bayesian skyline plot model (62). On the basis of the frequency with which the DF of the inferred trees is more extreme than the DF of the simulated trees, we derived P values to test departures from neutrality.
Diversifying selection analyses.
We identified diversifying selection in the G gene data using a combination of a fixed-effect-likelihood (FEL) approach (63), a recently developed renaissance counting (RC) procedure (64), and a random-effect-likelihood (REL) approach (65). The FEL method fits codon models to each site independently and uses a likelihood ratio test to assess whether a model assuming equal nonsynonymous and synonymous rates (dN = dS) can be rejected in favor of a model with different dN and dS rates. We used a P value < 0.1 to consider sites as diversifying in our consensus approach (see below). Estimates of site-specific dN/dS ratios via the RC approach were obtained by combining stochastic mapping under nucleotide substitution models and empirical Bayes regularization. Neutrality was rejected and sites were considered to be under positive diversifying selection only when the 97.5% lower limit of the dN/dS cumulative density interval exceeded a value of 1. The REL approach fits a codon model to the entire alignment but allows the dN/dS ratio to vary among sites. We used the Akaike's information criterion to select the best-fitting codon substitution model (comparing the nonsynonymous and dual and lineage-dual models [65]), and we identified positively selected sites using an empirical Bayes method with a log Bayes factor (ln BF) cutoff value of 3. The cutoff values for the three different approaches (FEL, RC, and dual REL) are fairly liberal, but we use a consensus approach in considering evidence for diversifying selection as advised by Kosakovsky Pond and Frost (63), and in analogy to the method described in reference 48, we list only sites that are identified as under diversifying selection by at least two of these methods.
Whether diversifying selection in the G gene was pervasive or episodic was evaluated using the recently developed mixed-effects model of evolution (MEME) (66). This model allows the dN/dS distribution to vary from site to site and, importantly, also from branch to branch at a specific site, thereby relaxing the assumption that the strength of natural selection is constant among all lineages. MEME extends the FEL approach by modeling two categories of lineage-specific dN rates for each site; one category has a dN ≤ dS (ß−), and the other category is characterized by an unrestricted dN rate (ß+). The codon substitution model that includes these categories of sites is tested against the same model in which also ß+ is constrained to values below or equal to 1. We reported only sites identified as evolving under diversifying selection with an associated P value ≤ 0.05. For these sites, we list the ß+ estimate and the proportion of branches estimated to be part of this dN rate class (p+).
Nucleotide sequence accession numbers.
The nucleotide sequences from the Dutch and Belgian RSV-B isolates were deposited in the GenBank database (http://www.ncbi.nlm.nih.gov/GenBank/index.html) under accession numbers JX576729 to JX576762. Previously documented prototype RSV-B reference strains B1 (AF013254) and 9320 (AY353550) plus RSV-A strains JQ901447 to JQ901458, JX015479 to JX015499, A2 (M74568), Long (AY911262), Line19 (FJ614813), and RSS-2 (NC_001803) were included in genomic and protein substitution analysis studies. All sequence alignments used in this study are available upon request.
RESULTS
Protein substitution mapping of RSV-A and RSV-B strains.
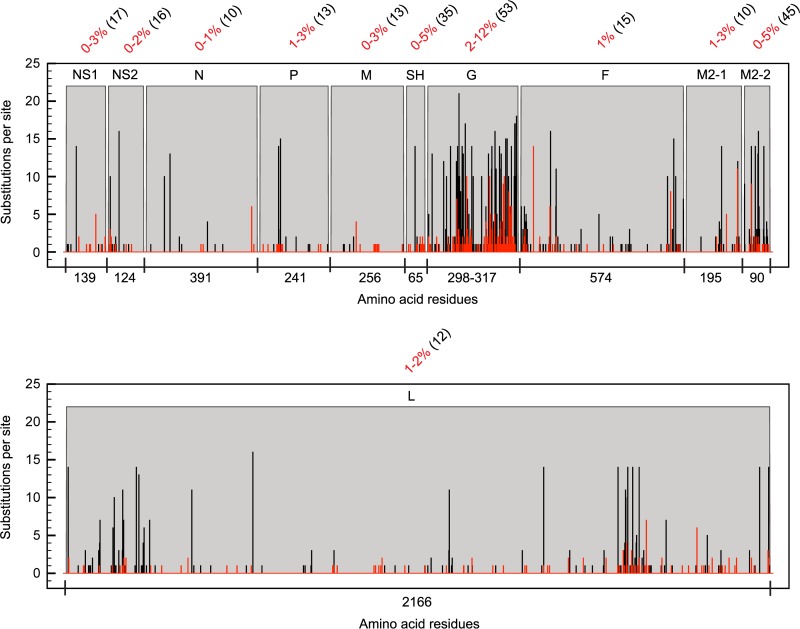
For each RSV protein, the amino acid residue changes within 34 Dutch-Belgian RSV-B clinical strains (see Table S1 in the supplemental material) and two reference laboratory strains were mapped to determine the protein substitution density (Fig. 1). Changes were relative to the consensus of all aligned RSV-B strains, and the degree of variation with respect to the consensus was indicated as the percentage of sequence variability per protein. The overall variation within RSV-B proteins is much lower than that previously described for RSV-A (48). This is clearly reflected by the greater amount of sites that are substituted among the RSV-A subgroup and the greater number of RSV-A strains having a substitution at one unique site. The G protein is the most variable RSV-B protein, with 2% to 12% variation compared to less than 5% variation in the other 10 proteins. In comparisons of variation between the RSV subgroup A and B consensus sequences, the structural proteins SH and G and the accessory transcription regulation protein M2-2 displayed the highest sequence variability (35%, 53%, and 45%, respectively).
Fig 1.
RSV protein sequence variability. The number of substitutions per site for RSV-A (black bars) and RSV-B (red bars) and the protein sequence variability (percent numbers in red) in each RSV-B protein were calculated per strain relative to the consensus. The numbers within parentheses indicate percentages of protein sequence variability between the consensus sequences of type A and type B RSV strains.
Since the SH and G proteins can be glycosylated, a process that might affect their folding and function, variations in glycosylation were studied both for these proteins and for the more conserved RSV F glycoprotein that is essential for initializing RSV infection. Predicted glycosylation patterns show remarkable differences between the G, F, and SH proteins derived from subgroup A and B strains.
Insertions and deletions alter N- and O-glycosylation predictions in the RSV-B G protein.
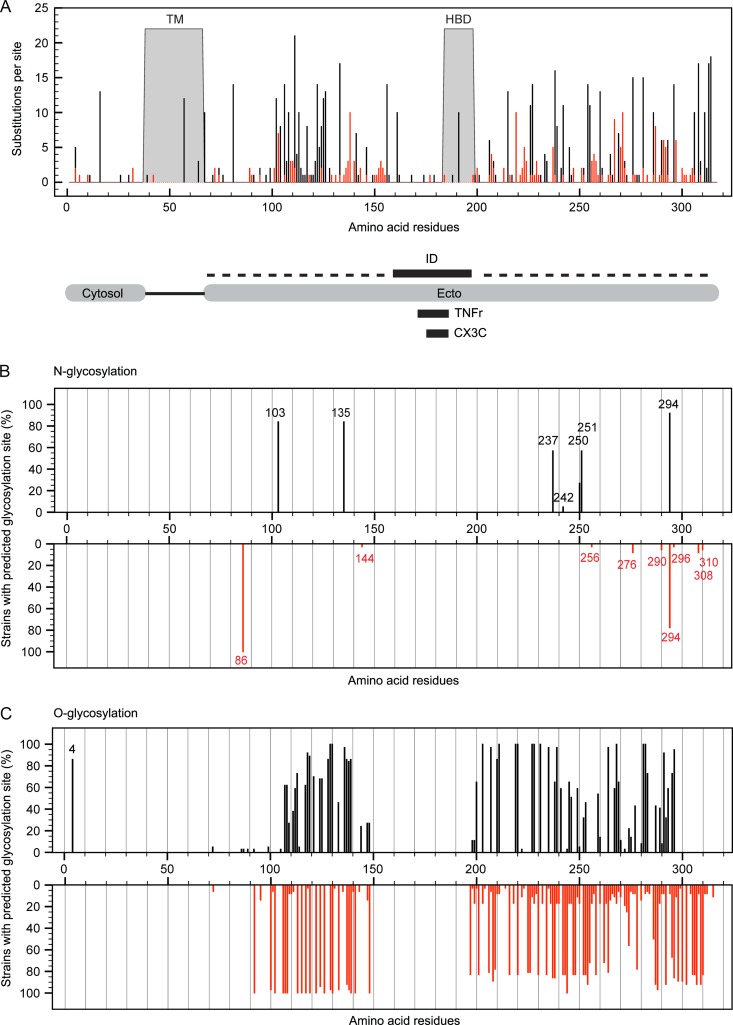
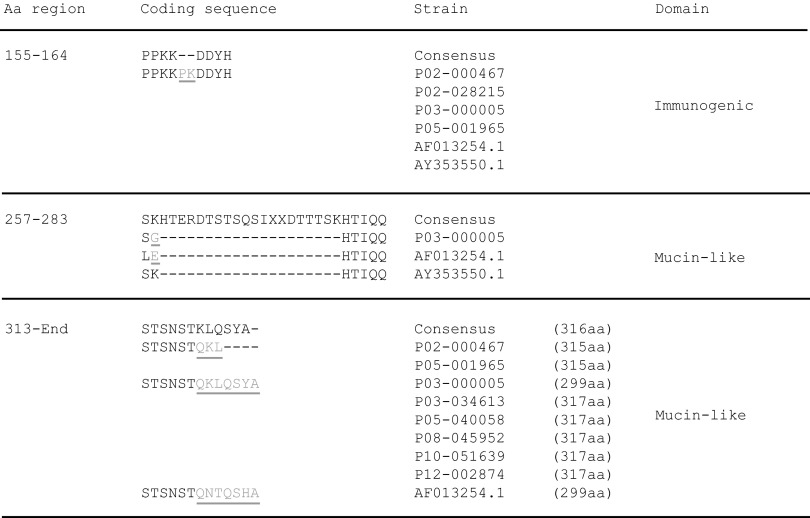
The highly immunogenic RSV G protein binds to glycosaminoglycans (GAGs) of the host target cell to facilitate subsequent virus-host fusion (12, 13). Amino acid residues at sites located in the highly variable domains present in the ectodomain of the G protein are most prone to substitutions in both RSV-A and RSV-B strains (Fig. 2A). The high degree of amino acid sequence variation found in these RSV G domains, also designated the mucin-like regions, indicates that there are more relaxed selective constraints operating on these regions, which may allow molecular adaptation without loss of protein function. These regions act as pathogenicity factors and are characterized by the high prevalence of serine and threonine residues, which allow the binding of O-linked glycans (67). The central conserved domain, which contains the immunogenic domain, the tumor necrosis factor receptor (TNFr) homologous region, and the CX3C chemokine motif, displays only substitutions at individual sites that are unique to single RSV-B sequences. This conserved part of the ectodomain is also marked by the absence of N-glycosylation (Fig. 2B) or O-glycosylation (Fig. 2C) sites. Specifically, N-glycan binding sites were predicted for RSV G asparagine residues 86, 144, 256, 276, 290, 294, 296, 308, and 310 within the RSV-B data set. The first N residue (N86) is predicted to be glycosylated in all RSV-B strains, while only a single strain was predicted to have glycosylation potential at N144 or N256 due to NKP-to-NKS sequon changes for N144 or a K256N substitution, respectively. The N-glycan binding residues at positions 276, 290, 294, and 296 all contain an asparagine residue (underlined) within the same PENTPNXXQTPASE acceptor sequence context. Variations in the sequence coordinates of this asparagine residue are the consequence of deletions and insertions located upstream of this acceptor sequence in various sequences (Fig. 3). The majority (33 of 36) of the studied RSV-B strains have the duplication regions of 240 to 259 (ERDTSTPQSTVLDTTTSKHT) and 260 to 280 (ERDTSTSQSIXXDTTTSKHT), but in the two reference strains AF013254 and AY353550 and in the clinical strain P03-000005, the second 260-to-280 sequence is missing. On the other hand, the sequences from strains 02-000467, 02-028215, 03-000005, 05-001965, AF013254, and AY353550 have a PK insertion in the 155-to-164 (PPKKPKDDYH) sequence. Asparagine residues with similar acceptor sequences were found for N308 and N310 as well, which correspond to the N-glycosylation site in the STSNST C-terminal ectodomain sequence. This acceptor sequence needs to be followed by additional amino acids, which give alternative RSV G sequence ends (Fig. 3) and are necessary for glycosylation potential, because strains lacking these extra residues are not predicted to be N-glycosylated at STSNST.
Fig 2.
Substitution hot spots in specific RSV G protein domains and consequences for glycosylation. (A) Schematic representation of the RSV G protein and its specific domains: the transmembrane domain (TM), heparin binding domain (HBD), N-terminal cytosolic domain (Cytosol), C-terminal ectodomain (Ecto), immunogenic domain (ID; aa 159 to 198), CX3C chemokine motif (CX3C; aa 182 to 186), and the region homologous to the fourth subdomain of TNFr (TNFr; aa 171–186). Dashed lines represent mucin-like regions. (B and C) The percentage of strains with predicted sites for N-glycosylation (B) and O-glycosylation (C) within the G protein at a certain amino acid position for both RSV-A (black; 37 strains) and RSV-B (red; 36 strains).
Fig 3.
Amino acid sequence divergence in the G protein of RSV-B strains. Insertions (gray, underlined), deletions (−), and variable C-terminal ends are indicated.
The total number of predicted O-glycosylation sites in the G protein is 36% higher for RSV-B strains than for RSV-A strains, and these sites are all present in the ectodomain of this protein. The O-glycan binding site at residue T4 is found in more than 80% of known RSV-A strains and is exceptional in that it lies within the cytosol domain of the attachment protein. However, this site is not present in the reference strains.
N-Glycan binding sites are exclusively predicted in the F2 domain of the highly conserved fusion protein.
Virus-host membrane fusion mediated via the enzymatic activated fusion protein occurs directly after host attachment via RSV G (14). RSV F is highly conserved among the RSV-B strains and, as with the other proteins, contains fewer substitutions than its RSV-A counterpart. Variability is particularly restricted to the signal peptide sequence of the F2 domain. In the RSV-B sequences, the asparagine residues at sites 27, 70, 116, 120, and 126 in the F2 domain are predicted to have N-glycosylation potential in all strains sequenced in this study. This is different within the RSV A strain data set, where some strains lack glycosylation potential at N120 (48). Despite the fact that none of the RSV-B strains have a predicted N-glycosylation site at N500 (N-glycosylation potential < 0.5), this specific site, with sequon NQS in both RSV-A and –B, has previously been shown to be glycosylated according to in vitro experiments (68).
Although rich in threonine and serine residues, the fusion protein is the only protein among the three RSV structural glycoproteins for which no O-glycosylation sites were predicted. This is in strong contrast with the attachment protein of RSV and may contribute to the low level of molecular variation in RSV F.
Comparison of N- and O-glycosylation in the SH protein of subgroup A and B strains.
SH is a small hydrophobic type II integral membrane protein that acts as a viroporin in ion channel formation (16, 17), thereby enhancing host membrane permeability. It is not pivotal for RSV infection (18), and the amino acid substitutions observed within this protein are mainly located in the C-terminal ectodomain. The O-glycosylation predicted for the SH protein at threonine position T64 of RSV-A strains is missing in the RSV-B subgroup strains. The difference of one amino acid in protein length, between subgroup A (64 amino acids [aa]) and subgroup B (65 aa), is most likely the cause of either the O-glycosylation potential loss at the last C-terminally located threonine in RSV-B strains or the gain of an O-glycan binding site in RSV-A strains. The asparagine residues at positions 3 and 52 in the SH proteins of RSV-A and RSV-B are predicted to be N-glycan binding sites. However, the reference RSV-B strain, B1, lacks N-glycosylation at N52, which is most likely caused by a K53N substitution at the second position of the NKT glycosylation sequon. The T4 residue is a predicted O-linked glycan site for all RSV strains. For RSV-B strain 08-045420, the three additional O-glycosylation sites, S5, T7, and T8, were predicted to have sugar binding potential as well. It is unclear why the S5 and T7 sites, which are present in all RSV-B strains, are specific potential glycosylation sites in 08-045420. The I8T substitution in this specific strain, however, is predicted to have created a new O-linked glycan site, which could also influence the sugar acceptor potential for the other two sites.
M2-2 protein transcription variants differ between RSV subgroups A and B.
The transcription regulatory protein, M2-2, acts as a switch from RNA transcription to genome replication during the RSV infection cycle and is tightly regulated by the M2-1 protein (69, 70). M2-2 can be transcribed in different truncated forms via a ribosomal termination-dependent reinitiation mechanism (71). Of the alternative start codons at positions 1, 3, and 7 described for RSV-A strains, only those at positions 1 and 7 are present in all RSV-B strains included in this study. Nevertheless, the first methionine is, in some RSV-A strains, also substituted by a threonine or proline residue, which is predicted to result in the transcription of only the truncated M2-2 forms.
The phylodynamic history of RSV-B.
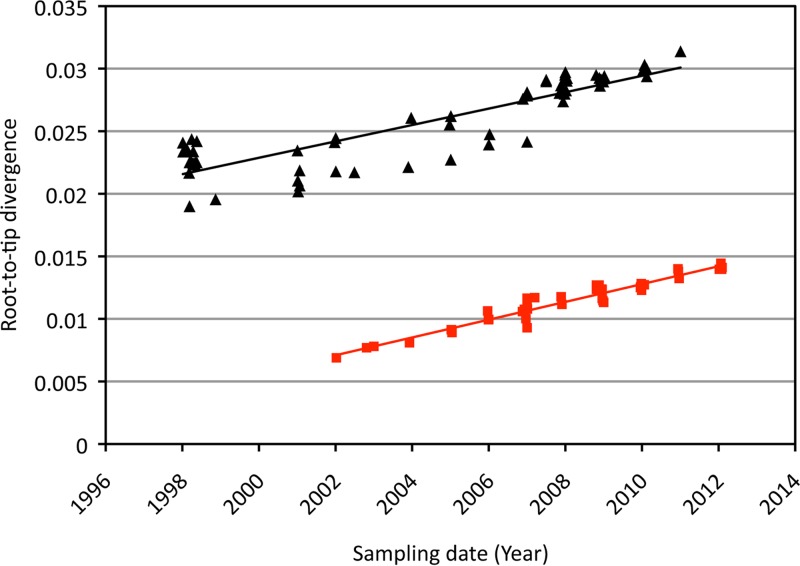
Plotting the root-to-tip divergence as a function of sampling time for each RSV-B genome clearly showed a temporal nucleotide divergence signal for the RSV-B complete genomes sampled over the last 10 years (Fig. 4). Although this analysis is exploratory in nature, a comparison with RSV-A already suggests a number of interesting aspects. First, the regression lines are roughly parallel, which indicates similar rates of evolution. Second, the levels of divergence from the root are considerably higher for RSV-A than for RSV-B, implying a shorter TMRCA for the latter, which offers an explanation for the overall lower degrees of genomic variability among the analyzed RSV-B genomes. Finally, the regression analyses also suggest lower variability in evolutionary rates among lineages for the RSV-B genomes.
Fig 4.
Plot of the root-to-tip divergence as a function of sampling time for the RSV-A and RSV-B genomes. RSV-A is indicated by black triangles, and RSV-B is indicated by red squares; the corresponding regression lines are plotted in the same colors.
The root-to-tip divergence analysis clearly justified the application of a dated tip molecular clock model during a Bayesian evolutionary reconstruction. Strict and relaxed clock models were compared using two different approaches to compare model fit in a Bayesian framework (Table 1). The best model fit was indicated by the highest log marginal-likelihood estimate (in bold in Table 1). Both the path sampling and stepping-stone sampling approaches, which have been shown to be the most accurate Bayesian phylogenetic model selection methods (57), agreed with the assumption of a constant substitution rate across phylogenetic tree branches based on genomic sequences. This is in line with the root-to-tip regression exploration and argues for the use of the strict clock model for further analysis of these data. However, for the G gene data set that includes many more RSV-B strains (43, 47, 53), this assumption had to be rejected and a relaxed clock model was required to accommodate rate variation among lineages.
Table 1.
Model-fit of molecular clock models based on log likelihood estimates
| Model | Log marginal-likelihood estimate |
|||
|---|---|---|---|---|
| Whole genome |
G gene |
|||
| Stricta | Relaxed | Strict | Relaxeda | |
| Path sampling | −32,304.88367 | −32,307.19482 | −7,706.969731 | −7,682.638227 |
| Stepping-stone sampling | −32,305.06152 | −32,307.41622 | −7,718.684767 | −7,696.065831 |
Highest log marginal-likelihood estimates are marked in bold.
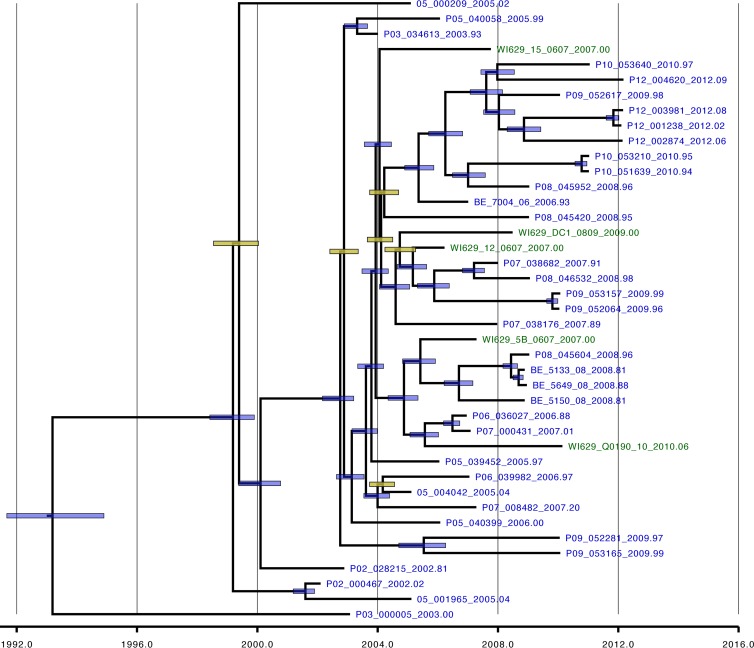
The estimated posterior tree distribution for the genome-wide RSV-B data set represented by a maximum clade credibility tree did not provide evidence for geographical or strong temporal clustering of the RSV-B genomes (Fig. 5). Strains from several different epidemic seasons are present in the same clusters, and the Milwaukee strains are phylogenetically interspersed with the Dutch-Belgian genomes. The MRCA of the RSV-B genome phylogeny dated back to approximately 1993 (95% credibility interval [CI 95%], 1991 to 1995; Table 2), which is, as suggested by the linear regression analyses, considerably later than estimates for a similar sample of RSV-A genomes (1964; CI 95%, 1956 to 1973 [48]). The shorter TMRCA for RSV-B explains why we observed lower genetic diversity over the complete genome, but this does not appear to be the result of a lower rate of evolution because the latter was estimated to be 7.76 × 10−4 substitutions per site per year (CI 95%, 6.89 × 10−4 to 8.58 × 10−4), which is very similar to that of RSV-A (6.47 × 10−4; CI 95%, 5.56 × 10−4 to 7.38 × 10−4) (48).
Fig 5.
RSV-B whole-genome-based phylogeny. The distribution of Dutch-Belgian strains (blue) and Milwaukee strains (green) is indicated. The node bars depict the credibility intervals for nodes showing a posterior probability support > 95% (blue) or < 95% (yellow).
Table 2.
Evolutionary rate and TMRCA estimates for RSV-A and RSV-B
| Input | Mean evolutionary rate × 10−4 (no. of substitutions/site/yr) |
TMRCA (yr) |
||||
|---|---|---|---|---|---|---|
| Mean | 95% HPD lower | 95% HPD upper | Mean | 95% HPD lower | 95% HPD upper | |
| RSV-A (genomic) | 6.47 | 5.56 | 7.38 | 1964 | 1957 | 1972 |
| RSV-B (genomic) | 7.76 | 6.89 | 8.58 | 1993 | 1991 | 1995 |
| RSV-A (G gene) | 22.2 | 19.3 | 25.6 | 1942 | 1929 | 1953 |
| RSV-B (G gene) | 27.8 | 23.5 | 32.3 | 1955 | 1946 | 1960 |
In light of the short TMRCA, we studied the clustering of the strains for which complete genomes are available in the G gene data set (data not shown). This revealed that these strains also coalesce into a single internal node that dates back to 1994 (CI 95%, 1992 to 1995) whereas the root of the G gene tree is dated back to 1955 (CI 95%, 1946 to 1960) (Table 2). So, whereas a recently obtained RSV-A full-genome sample appeared to be representative of the strain diversity in G gene phylogeny (48), the RVS-B sample represents the progeny of a more recent ancestor within the complete strain diversity. This could represent a chance fixation in a largely neutral epidemiological scenario, or it may reflect the fixation of a variant with advantageous mutations. This also impacts the classification of recent RSV-B strains; according to the G gene tree, the GB13 lineage is predominant among the Dutch-Belgian strains, and only strain 03-000005 belongs to another lineage (GB12).
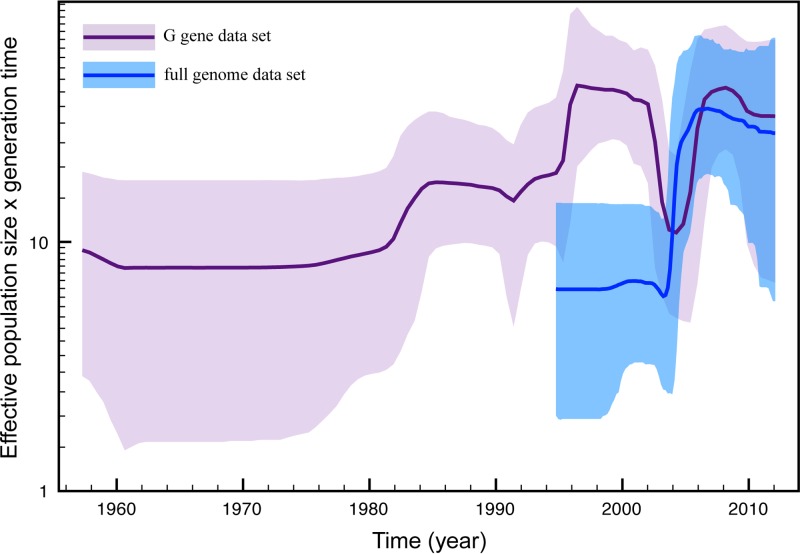
To further investigate the impact of this fixation event on RSV-B population dynamics, we reconstructed the change in relative genetic diversity through time using a Bayesian skyline plot model in our Bayesian genealogical inference for both the G gene and full-genome data sets (superimposed in Fig. 6). This reveals a noticeable degree of variation in relative genetic diversity through time, including a contraction and expansion between 2002 and 2008. From the full-genome sample, we reconstructed only the recovery dynamics due to the absence of samples prior to 2002. The increase in relative genetic diversity from the early 1990s in the G gene skyline might also reflect such a bottleneck event because the paucity in samples before this time hampers the reconstruction of the complete ancestral diversity. The timing of such a potential bottleneck event coincides with the MRCA of the strains for which complete genomes are available in the G gene data set.
Fig 6.
Bayesian skyline plot. The estimated change in effective population size over time for both the full-genome (blue) and G gene (purple) data sets is indicated. The thick lines represent the mean estimate, whereas the transparent areas represent the 95% highest-posterior-density intervals.
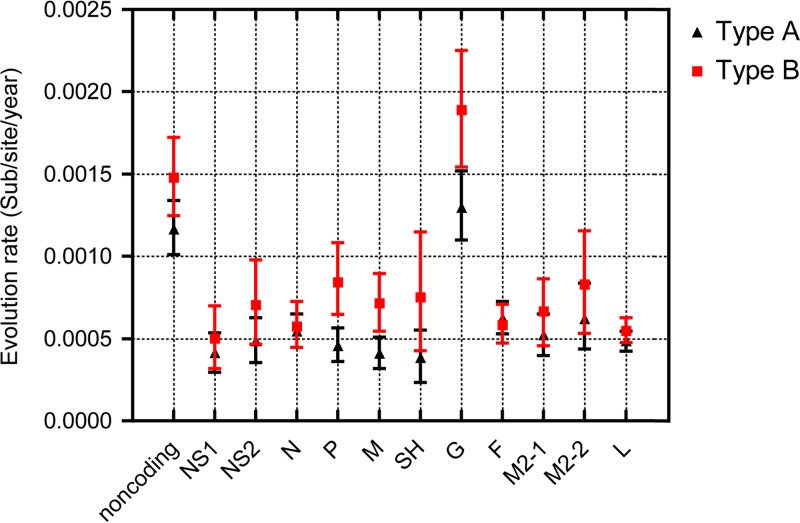
It is important to point out that, as is the case with RSV-A, the RSV-B attachment protein gene appears to have a significantly higher substitution rate than the RSV-B full genome (Table 2) (43, 44). The variability in evolutionary rates across RSV genomes was further determined for all the various gene partitions and the combined noncoding regions via the Bayesian estimation approach in order to individually identify the impacts of different genes on the genome-wide substitution rate (Fig. 7). In analogy to the previous RSV-A results, considerably elevated substitution rates were observed in the noncoding regions and G gene, while the other genes were more conserved in both RSV subgroups. Relatively high G gene variability (1% to 8% variability; Fig. 8) and the associated relatively high substitution rate estimate (almost 3-fold higher than that estimated for other genes), which was in line with previous estimates (44), indicate that either there is a general relaxation of negative selection (i.e., selection disfavoring change) acting on this gene or there are various codons within this gene that are evolving under positive selection (i.e., selection favoring change).
Fig 7.
Comparison of RSV-A and RSV-B evolutionary rate partitions. The mean rate of evolutionary changes indicated by the numbers of substitutions per site for each year was estimated per gene and for the combined noncoding sequence parts for the RSV-A (black triangles) and RSV-B (red squares) data sets.
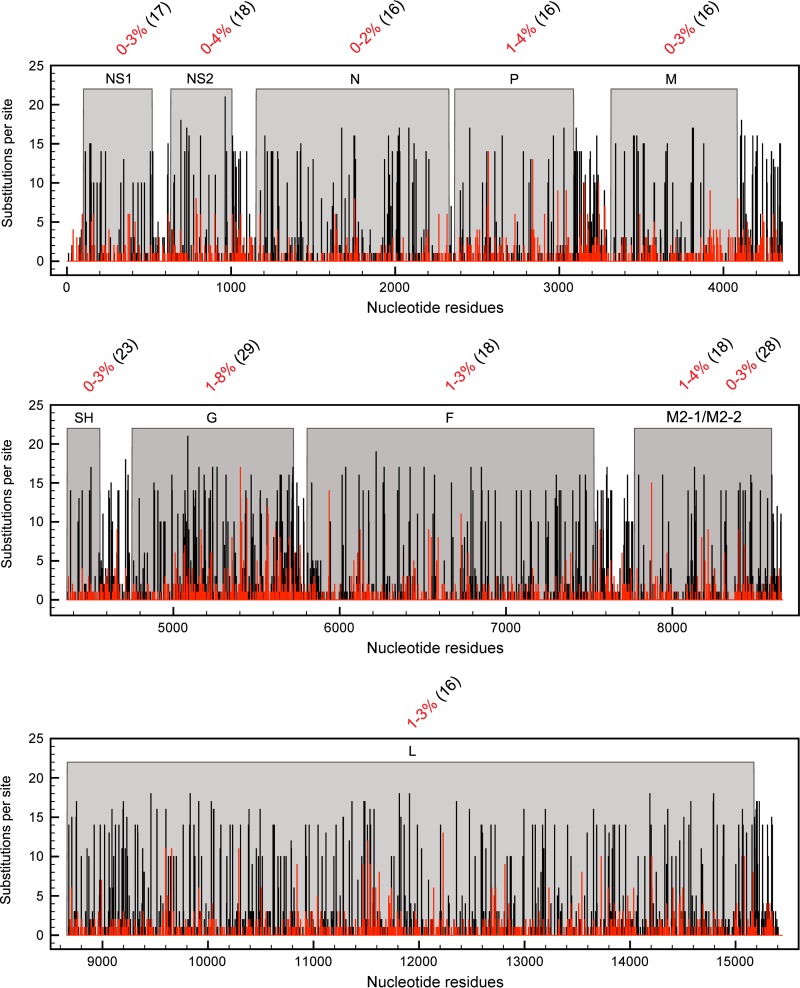
Fig 8.
RSV nucleotide sequence variability in the whole genome. The number of substitutions per site for the RSV-A (black bars) and RSV-B (red bars) genomes and the nucleotide sequence variability (percent numbers in red) in each RSV-B gene were calculated per strain relative to the consensus. The numbers within parentheses indicate percentages of nucleotide sequence variability between the consensus of type A and type B RSV strains.
Patterns of natural selection acting at individual sites.
Sites within the RSV G protein that are potentially evolving under conditions of diversifying selection were identified by querying nonsynonymous/synonymous substitution rate ratios (dN/dS) at each codon position of the G-protein gene using the FEL, RC, and REL methods (Table 3). Of the 11 sites identified by at least two of these three methods as displaying evidence of diversifying selection, only sites 224 and 230 (marked in bold) were significantly supported by all three of the methods. As has been previously found with RSV-A, most of the 11 sites were within the two hypervariable regions of the G protein ectodomain. However, sites 50 and 53 are part of the transmembrane domain. In general, the methods applied here to study site-specific selective patterns identify sites that are under pervasive diversifying selection (i.e., diversifying selection in most lineages of the phylogeny). However, the RC, FEL, and REL methods cannot efficiently detect episodic diversifying selection, which involves diversifying selection of specific sites in a restricted number of branches in the phylogeny with either neutral or negative selection occurring at these sites along the remaining branches. Therefore, MEME analysis was conducted to discriminate between these two kinds of diversifying selection. In total, 10 sites were identified to be under diversifying selection by MEME (with an associated P value ≤ 0.05). Five of these sites, which are apparently evolving under diversifying selection across a considerable proportion of branches in the phylogeny (p+), were also identified by two of the three other selection detection methods (marked in bold in Table 4) and can therefore be considered sites under pervasive diversifying selection. All other sites detected via MEME are evolving under diversifying selection across only small numbers of subsets of branches in the phylogeny (ß+) and are thus identified as evolving under episodic diversifying selection.
Table 3.
Sites under diversifying selection in the G gene data set
| Sitea | RC (dN/dS) | FEL |
REL |
||
|---|---|---|---|---|---|
| dN − dS | P value | E (dN/dS) | Log (BF) | ||
| 50 | 8.00 (3.59, 15.47) | 3.60 | 0.05 | 3.21 | 2.54 |
| 53 | 4.74 (1.50, 9.46) | 1.88 | 0.26 | 3.67 | 3.49 |
| 81 | 7.74 (3.62, 15.23) | 3.58 | 0.05 | 3.20 | 2.53 |
| 85 | 3.77 (1.20, 7.56) | 1.41 | 0.37 | 2.53 | 3.15 |
| 160 | 6.80 (3.17, 13.35) | 10.15 | 0.00 | 2.90 | 2.10 |
| 187 | 4.82 (2.12, 9.35) | 2.00 | 0.20 | 3.79 | 3.91 |
| 210 | 7.82 (1.70, 18.98) | 10.39 | 0.06 | 2.15 | 1.20 |
| 224 | 7.62 (3.29, 15.19) | 3.50 | 0.06 | 3.84 | 4.15 |
| 226 | 2.99 (1.40, 5.90) | 1.67 | 0.31 | 3.52 | 3.12 |
| 230 | 9.79 (3.90, 19.48) | 4.76 | 0.03 | 3.75 | 3.78 |
| 239 | 4.97 (2.14, 9.88) | 1.86 | 0.26 | 3.72 | 3.66 |
Sites marked in bold are positive selected sites that meet the criteria of all applied methods.
Table 4.
Sites under episodic diversifying selection in the G gene data seta
| Sitea | β+ | p+ | P value |
|---|---|---|---|
| 18 | 58.53 | 0.01 | 0.00 |
| 138 | 135.60 | 0.01 | 0.04 |
| 145 | 10,000.00 | 0.01 | 0.04 |
| 153 | 600.31 | 0.01 | 0.02 |
| 160 | 3.12 | 1.00 | 0.02 |
| 187 | 3.34 | 0.42 | 0.04 |
| 226 | 6.35 | 0.23 | 0.03 |
| 230 | 4.65 | 0.61 | 0.01 |
| 238 | 768.59 | 0.01 | 0.04 |
| 239 | 1.84 | 0.97 | 0.04 |
Sites marked in bold are positive selected sites that meet the criteria of one or more of the applied methods. Data in columns 2 and 3 represent lineage-specific unrestricted dN rate estimates for indicated sites (ß+) and proportions of branches estimated to be part of the indicated dN-rate class (p+), respectively.
Testing genealogical departures from neutrality.
To assess the impact of diversifying selection in the G gene on RSV-B circulation patterns, we performed a genealogical test of neutrality using posterior predictive simulation. Phylogenetic tree shapes deduced from complete genome Bayesian inference were compared to those simulated with the same population dynamics and under a model of neutrality. This test indicated that neutrality could be rejected at the population level for the complete genome RSV-B data set (P = 0.01). This is in contrast with the RSV-A population, which did not show departures from neutrality (P = 0.24) (48). Although the data approach statistical significance, neutrality cannot be convincingly rejected when the large data set of the highly variable G gene is considered in the predictive simulation (RSV-B P = 0.06; RSV-A P = 0.08).
DISCUSSION
Here we have used 34 RSV-B full-genome sequences sampled between 2002 and 2012 to infer genome-wide mutation density maps and nucleotide substitution rates and compared these with those determined previously for RSV-A (48). Although the identified substitution hot spots closely resemble those detected for RSV-A, particular differences may reflect RSV subtype-specific variation in replication and/or immune evasion strategies and could therefore be helpful in explaining RSV subtype-specific differences in transmission dynamics and epidemiology.
As with RSV-A, most sequence variability within RSV-B strains was detected in the intergenic noncoding regions and the G gene; in the G gene, high nucleotide sequence variability correlated with an elevated amino acid substitution rate. In contrast with the RSV-A strains analyzed to date, the G genes from RSV-B strains showed a high frequency of nucleotide insertions and deletions within the G ectodomain. The highly variable and immunogenic G protein has been previously suggested to be susceptible to neutralizing antibodies; this might drive the selection of strains carrying immune escape mutations within the ectodomain (72).
In contrast to the results seen with RSV-A strains, we did not observe high variability in the M2-2 gene in the RSV-B strains studied. Comparing RSV-A and RSV-B gene products, the highest protein sequence variation between these subtypes was found between the G, SH, and M2-2 proteins. For the SH protein, variation is mainly caused by C-terminal polymorphisms in the ectodomain, which were more common among RSV-A strains. Apparently, the impact of immunogenic pressure on the evolution of other RSV genes is far less substantial since these genes show high degrees of conservation. However, even low substitution rates in these genes could still have a large effect on viral fitness and the course of RSV transmission dynamics. It is therefore entirely relevant to map substitutions in conserved regions to both support future research into the biological functions of the various RSV proteins and inform the development of preventive and therapeutic approaches. In the latter case, it is particularly relevant to compare RSV-A and RSV-B variations in efforts to develop protective strategies that are effective against all RSV infections.
Amino acid site alterations that are correlated with either changes in N- and O-glycosylation potential or susceptibility to antibody neutralization can trigger RSV phenotypic differences. Such differences might include increased RSV infectiousness through adaptation to host defenses and altered transmission dynamics. Although we explicitly recognize the urge to experimentally confirm the true nature of glycosylation predictions, we also feel that the in silico assessment of N- and O-glycosylation potential on the extended whole-genome database presented here yields an excellent starting point for further in-depth analyses throughout the RSV field. We show that RSV-A and -B strains contain similar numbers of predicted N-glycan binding sites, although the exact glycosylation sites vary greatly due to substantial sequence variability within and between the two subgroups. Only two sites, N86 and N294, were found predominately in the G protein of RSV-B strains. The latter asparagine position was indicated as N276, N290, and N296 within other strains of the RSV-B subgroup due to deletions or insertions in upstream sequences. Binding sites for N-glycans on the fusion protein of RSV-B strains were largely similar to those predicted for RSV-A strains, with binding potential being predicted for all of the analyzed RSV-B strains. In contrast to the findings of Zimmer et al. (68), neither the asparagine residue at position N500 in the RSV-B strains nor the homologous asparagine in RSV-A strains had any predicted glycosylation potential. Zimmer et al. showed that glycosylation at this site was required for efficient syncytium formation. This repeatedly emphasizes the requirement for in vitro analysis to verify predicted glycosylation sites. O-glycosylation was predicted only for the RSV attachment protein where it potentially occurs mainly in the mucin-like regions of this protein. Surprisingly, the T4 O-glycan binding site in the cytosolic domain of the G protein was predicted for 80% of the RSV-A strains, but it was not detected in either the reference strains or any of the analyzed RSV-B strains. Relative to the RSV-B strains, the RSV-A strains contained fewer sites with O-glycan binding potential. Furthermore, the RSV F protein, which is rich in threonine and serine residues, also appears to lack the potential to be O-glycosylated. Since the extensive glycosylation of the G protein may shield these viruses from immune recognition, the conserved F protein, with its minimal degree of glycosylation, is possibly more suitable as a target for intervention strategies.
RSV-B evolutionary rates and demography were analyzed using Bayesian phylogenetic techniques based on a recombination-free whole-genome sequence data set. Two of the RSV genomes in our initial data set showed some evidence of potential recombination. Since genomic recombination in RSV is believed to be extremely rare (73), it is most likely that these recombinants arose as a result of PCR or sequencing artifacts. We removed the genomic parts that appeared to be inherited from minor parental strains from these mosaic genomes and treated them as unobserved sequences for further phylogenetic, population genetic, molecular clock, and selection analyses.
For both the RSV-B strains analyzed here and the RSV-A strains analyzed previously, it is evident that strains isolated from different geographical regions tend to have a phylogenetically mixed distribution within the RSV phylogeny. This indicates that there is no strong long-term geographical or temporal structuring of RSV populations.
Nevertheless, the RSV-B strains we sampled coalesced into a relatively recent root of the complete genome phylogeny (dated back to 1993 [CI 95%, 1991 to 1995]) as well as into an internal node with similar age in the G gene phylogeny. As a consequence of the relatively recent TMRCA, all except one of the RSV-B strains (03-000005) in this study belong to the GB13 clade. The relatively recent ancestry was not the case for the RSV-A genome sample, which appeared to be representative of the complete diversity in the corresponding G gene tree. This recent TMRCA may be the result of a chance fixation of a variant or a variant driven to fixation by some advantageous mutation in its genome. The Bayesian skyline plot reconstructions indicated that such fixations could impose population genetic bottlenecks, which can result in relatively constant levels of genetic diversity over longer time scales. The fact that we were able to detect these dynamics for RSV-B and not for RSV-A may be due to differences in sampling. Whereas the RVS-B complete-genome sample was restricted to 2002 to 2012, the RSV-A data set contained a considerable amount of strains sampled in 1998, which increases the probability of capturing more ancestral diversity. Also, despite the fact that there does not seem to be strong geographical clustering of RSV-B diversity, additional sampling from different locations may be useful to further elucidate RSV population dynamics.
As with the RSV-A strain data set, the majority of RSV-B genes are apparently evolving under pervasive negative selection against a background of neutrally evolving sites. Both pervasive positive selection and episodic diversifying selection were, however, readily detectable within the G gene. Of the 11 positively selected sites identified by the RC, FEL, and REL methods, only five sites were confirmed by the MEME method to be evolving under pervasive positive selection. In addition, MEME identified episodic selection at four other sites. This novel method for selective pressure analyses is less sensitive to the influences of sequence sampling and to false identification of diversifying selection. Surprisingly, besides the fact that most diversifying selection was detected in the highly variable mucin-like regions of the RSV-B G gene, some positively selected sites were also found in the cytosolic domain (site 18), transmembrane domain (sites 50 and 53), and the more conserved region of the ectodomain (sites 160 and 187). This contrasts with the patterns of positively selected sites previously observed for RSV-A strains for which positive selection was primarily found in the mucin-like regions. Also, in contrast to RSV-A strains, we did not detect any positively selected sites in RSV-B that might directly influence N-glycosylation.
According to the genealogical tests of neutrality, the genetic bottlenecks are still compatible with population turnover corresponding to neutral expectations for the G gene evolutionary history even though the test approaches significance (P = 0.06). This was similar to the situation observed with the RSV-A data set (48). Although it would be useful to explore other genealogical test statistics in the posterior predictive simulation, such as a recently proposed temporal clustering statistic (74), which may prove more sensitive in detection of departures from neutrality, it is clear that population turnover is not as pervasive and selection driven as that observed for human influenza A virus (62) and norovirus GII.4 (61). Neutrality was, however, rejected for the complete-genome RSV-B data set, but since this focuses on a smaller time scale that accommodates one of bottleneck events observed over longer time scales (Fig. 6), it remains difficult to make strong conclusions based on this. The exclusive accumulation of neutral substitutions in the G gene and the diversifying selection detected in multiple RSV G domains suggest that there are relaxed evolutionary constraints on this gene. Therefore, the RSV G protein may not be the best target for effective vaccines as it is not expected to induce functional B cell-mediated immunity.
For the determination of the evolutionary rate, as well as for constructing a phylogenetic tree for the genotyping of RSV strains, the whole-genomic approach followed both in this paper and in that of Tan et al. (48) yielded results that were broadly consistent with those obtained from analyses of the G gene partitions. From that perspective, a conclusion that G gene analysis is an appropriate choice for molecular epidemiology studies might be justified. This is due to the fact that the G gene accounts for the majority of genomic variation, which in turn is mainly situated in regions of the gene encoding the highly variable mucin-like regions of the G protein. Most likely, the only structural and functional requirement for these regions is the presence of heavily glycosylated amino acids. However, the whole-genome analyses presented in this paper and that of Tan et al. (48) revealed potentially important aspects of M2-2, F, and SH gene diversity, information that would have remained undisclosed when focusing solely on the G gene.
In summary, RSV-A and RSV-B whole-genome evolutionary analyses have revealed that these subgroups have similar evolutionary rates and display comparable degrees of conservation. In both subgroups, the G gene is exceptionally variable and displays elevated substitution rates compared to other genes. In both subgroups, the high substitution rates observed in this gene are at least in part due to the fact that a substantial number of residues are evolving under diversifying selection. Despite sporadic instances of positive selection, the majority of accumulating substitutions are likely neutral in both subgroups. This implies that the high degrees of RSV G gene diversity are not entirely driven by the evasion of adaptive host immunity. Individuals attempting to acquire immunological memory only weaken their immunological balance, ironically decreasing their tolerance to RSV and increasing the probability of experiencing more severe disease. Overall, the high degrees of G protein variability, its likely extensive glycosylation, and evidence that various of its amino acids are evolving under diversifying selection imply that the G protein might not be as good a target for prevention/intervention strategies as the more conserved F protein, which displays far less evidence of glycosylation and diversifying selection.
At present, this report represents the broadest conceivable RSV genome analysis, which provides us with unique information on the degree of genetic conservation and the existence of natural genomic variants. These data constitute a key source for further extensive research in the field of RSV protein structure-function analyses and immune regulatory onset studies and are highly valuable for specific peptide selection in therapeutic development studies.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Anton M. van Loon and Marc van Ranst for allowing access to their collections of Dutch and Belgian RSV-B isolates, respectively.
L.T., P.L., M.C.V., E.J.H.J.W., and F.E.J.C. conceived and designed the experiments. L.T. and M.C.V. performed the experiments. L.T., P.L., M.C.V., and D.P.M. analyzed the data. L.H. and G.M.V.B. contributed reagents/materials/analysis tools. L.T., F.E.J.C., D.P.M., and P.L. wrote the paper.
Footnotes
Published ahead of print 22 May 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.03278-12.
REFERENCES
- 1. Henrickson KJ, Hoover S, Kehl KS, Hua W. 2004. National disease burden of respiratory viruses detected in children by polymerase chain reaction. Pediatr. Infect. Dis. J. 23:S11–S18 [DOI] [PubMed] [Google Scholar]
- 2. Falsey AR, Hennessey PA, Formica MA, Cox C, Walsh EE. 2005. Respiratory syncytial virus infection in elderly and high-risk adults. N. Engl. J. Med. 352:1749–1759 [DOI] [PubMed] [Google Scholar]
- 3. Falsey AR, Walsh EE. 2000. Respiratory syncytial virus infection in adults. Clin. Microbiol. Rev. 13:371–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Han LL, Alexander JP, Anderson LJ. 1999. Respiratory syncytial virus pneumonia among the elderly: an assessment of disease burden. J. Infect. Dis. 179:25–30 [DOI] [PubMed] [Google Scholar]
- 5. Nair H, Nokes DJ, Gessner BD, Dherani M, Madhi SA, Singleton RJ, O'Brien KL, Roca A, Wright PF, Bruce N, Chandran A, Theodoratou E, Sutanto A, Sedyaningsih ER, Ngama M, Munywoki PK, Kartasasmita C, Simoes EA, Rudan I, Weber MW, Campbell H. 2010. Global burden of acute lower respiratory infections due to respiratory syncytial virus in young children: a systematic review and meta-analysis. Lancet 375:1545–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zlateva KT, Vijgen L, Dekeersmaeker N, Naranjo C, Van Ranst M. 2007. Subgroup prevalence and genotype circulation patterns of human respiratory syncytial virus in Belgium during ten successive epidemic seasons. J. Clin. Microbiol. 45:3022–3030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. White LJ, Mandl JN, Gomes MG, Bodley-Tickell AT, Cane PA, Perez-Brena P, Aguilar JC, Siqueira MM, Portes SA, Straliotto SM, Waris M, Nokes DJ, Medley GF. 2007. Understanding the transmission dynamics of respiratory syncytial virus using multiple time series and nested models. Math Biosci. 209:222–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Meerhoff TJ, Paget JW, Kimpen JL, Schellevis F. 2009. Variation of respiratory syncytial virus and the relation with meteorological factors in different winter seasons. Pediatr. Infect. Dis. J. 28:860–866 [DOI] [PubMed] [Google Scholar]
- 9. Cowton VM, McGivern DR, Fearns R. 2006. Unravelling the complexities of respiratory syncytial virus RNA synthesis. J. Gen. Virol. 87:1805–1821 [DOI] [PubMed] [Google Scholar]
- 10. Batonick M, Wertz GW. 2011. Requirements for human respiratory syncytial virus glycoproteins in assembly and egress from infected cells. Adv. Virol. 2011:343408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mitra R, Baviskar P, Duncan-Decocq RR, Patel D, Oomens AG. 2012. The human respiratory syncytial virus matrix protein is required for maturation of viral filaments. J. Virol. 86:4432–4443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hallak LK, Kwilas SA, Peeples ME. 2007. Interaction between respiratory syncytial virus and glycosaminoglycans, including heparan sulfate. Methods Mol. Biol. 379:15–34 [DOI] [PubMed] [Google Scholar]
- 13. Hallak LK, Spillmann D, Collins PL, Peeples ME. 2000. Glycosaminoglycan sulfation requirements for respiratory syncytial virus infection. J. Virol. 74:10508–10513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. González-Reyes L, Ruiz-Arguello MB, Garcia-Barreno B, Calder L, Lopez JA, Albar JP, Skehel JJ, Wiley DC, Melero JA. 2001. Cleavage of the human respiratory syncytial virus fusion protein at two distinct sites is required for activation of membrane fusion. Proc. Natl. Acad. Sci. U. S. A. 98:9859–9864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Techaarpornkul S, Barretto N, Peeples ME. 2001. Functional analysis of recombinant respiratory syncytial virus deletion mutants lacking the small hydrophobic and/or attachment glycoprotein gene. J. Virol. 75:6825–6834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gan SW, Ng L, Lin X, Gong X, Torres J. 2008. Structure and ion channel activity of the human respiratory syncytial virus (hRSV) small hydrophobic protein transmembrane domain. Protein Sci. 17:813–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gan SW, Tan E, Lin X, Yu D, Wang J, Tan GM, Vararattanavech A, Yeo CY, Soon CH, Soong TW, Pervushin K, Torres J. 2012. The small hydrophobic protein of the human respiratory syncytial virus forms pentameric ion channels. J. Biol. Chem. 287:24671–24689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Collins PL, Mottet G. 1993. Membrane orientation and oligomerization of the small hydrophobic protein of human respiratory syncytial virus. J. Gen. Virol. 74(Pt 7):1445–1450 [DOI] [PubMed] [Google Scholar]
- 19. Rixon HW, Brown G, Aitken J, McDonald T, Graham S, Sugrue RJ. 2004. The small hydrophobic (SH) protein accumulates within lipid-raft structures of the Golgi complex during respiratory syncytial virus infection. J. Gen. Virol. 85:1153–1165 [DOI] [PubMed] [Google Scholar]
- 20. Lo MS, Brazas RM, Holtzman MJ. 2005. Respiratory syncytial virus nonstructural proteins NS1 and NS2 mediate inhibition of Stat2 expression and alpha/beta interferon responsiveness. J. Virol. 79:9315–9319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bukreyev A, Yang L, Fricke J, Cheng L, Ward JM, Murphy BR, Collins PL. 2008. The secreted form of respiratory syncytial virus G glycoprotein helps the virus evade antibody-mediated restriction of replication by acting as an antigen decoy and through effects on Fc receptor-bearing leukocytes. J. Virol. 82:12191–12204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Anderson LJ, Hierholzer JC, Tsou C, Hendry RM, Fernie BF, Stone Y, McIntosh K. 1985. Antigenic characterization of respiratory syncytial virus strains with monoclonal antibodies. J. Infect. Dis. 151:626–633 [DOI] [PubMed] [Google Scholar]
- 23. Mufson MA, Orvell C, Rafnar B, Norrby E. 1985. Two distinct subtypes of human respiratory syncytial virus. J. Gen. Virol. 66(Pt 10):2111–2124 [DOI] [PubMed] [Google Scholar]
- 24. Peret TC, Hall CB, Schnabel KC, Golub JA, Anderson LJ. 1998. Circulation patterns of genetically distinct group A and B strains of human respiratory syncytial virus in a community. J. Gen. Virol. 79(Pt 9):2221–2229 [DOI] [PubMed] [Google Scholar]
- 25. Hall CB, Walsh EE, Long CE, Schnabel KC. 1991. Immunity to and frequency of reinfection with respiratory syncytial virus. J. Infect. Dis. 163:693–698 [DOI] [PubMed] [Google Scholar]
- 26. Mufson MA, Belshe RB, Orvell C, Norrby E. 1987. Subgroup characteristics of respiratory syncytial virus strains recovered from children with two consecutive infections. J. Clin. Microbiol. 25:1535–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Johnson PR, Spriggs MK, Olmsted RA, Collins PL. 1987. The G glycoprotein of human respiratory syncytial viruses of subgroups A and B: extensive sequence divergence between antigenically related proteins. Proc. Natl. Acad. Sci. U. S. A. 84:5625–5629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sullender WM. 2000. Respiratory syncytial virus genetic and antigenic diversity. Clin. Microbiol. Rev. 13:1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cane PA, Matthews DA, Pringle CR. 1994. Analysis of respiratory syncytial virus strain variation in successive epidemics in one city. J. Clin. Microbiol. 32:1–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Anderson LJ, Hendry RM, Pierik LT, Tsou C, McIntosh K. 1991. Multicenter study of strains of respiratory syncytial virus. J. Infect. Dis. 163:687–692 [DOI] [PubMed] [Google Scholar]
- 31. Peret TC, Hall CB, Hammond GW, Piedra PA, Storch GA, Sullender WM, Tsou C, Anderson LJ. 2000. Circulation patterns of group A and B human respiratory syncytial virus genotypes in 5 communities in North America. J. Infect. Dis. 181:1891–1896 [DOI] [PubMed] [Google Scholar]
- 32. Tang YW, Graham BS. 1997. T cell source of type 1 cytokines determines illness patterns in respiratory syncytial virus-infected mice. J. Clin. Invest. 99:2183–2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Klein Klouwenberg P, Tan L, Werkman W, van Bleek GM, Coenjaerts F. 2009. The role of Toll-like receptors in regulating the immune response against respiratory syncytial virus. Crit. Rev. Immunol. 29:531–550 [DOI] [PubMed] [Google Scholar]
- 34. Chin J, Magoffin RL, Shearer LA, Schieble JH, Lennette EH. 1969. Field evaluation of a respiratory syncytial virus vaccine and a trivalent parainfluenza virus vaccine in a pediatric population. Am. J. Epidemiol. 89:449–463 [DOI] [PubMed] [Google Scholar]
- 35. Kapikian AZ, Mitchell RH, Chanock RM, Shvedoff RA, Stewart CE. 1969. An epidemiologic study of altered clinical reactivity to respiratory syncytial (RS) virus infection in children previously vaccinated with an inactivated RS virus vaccine. Am. J. Epidemiol. 89:405–421 [DOI] [PubMed] [Google Scholar]
- 36. Kim HW, Canchola JG, Brandt CD, Pyles G, Chanock RM, Jensen K, Parrott RH. 1969. Respiratory syncytial virus disease in infants despite prior administration of antigenic inactivated vaccine. Am. J. Epidemiol. 89:422–434 [DOI] [PubMed] [Google Scholar]
- 37. Murphy BR, Walsh EE. 1988. Formalin-inactivated respiratory syncytial virus vaccine induces antibodies to the fusion glycoprotein that are deficient in fusion-inhibiting activity. J. Clin. Microbiol. 26:1595–1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Castilow EM, Olson MR, Meyerholz DK, Varga SM. 2008. Differential role of gamma interferon in inhibiting pulmonary eosinophilia and exacerbating systemic disease in fusion protein-immunized mice undergoing challenge infection with respiratory syncytial virus. J. Virol. 82:2196–2207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kruijsen D, Bakkers MJ, van Uden NO, Viveen MC, van der Sluis TC, Kimpen JL, Leusen JH, Coenjaerts FE, van Bleek GM. 2010. Serum antibodies critically affect virus-specific CD4+/CD8+ T cell balance during respiratory syncytial virus infections. J. Immunol. 185:6489–6498 [DOI] [PubMed] [Google Scholar]
- 40. Kruijsen D, Schijf MA, Lukens MV, van Uden NO, Kimpen JL, Coenjaerts FE, van Bleek GM. 2011. Local innate and adaptive immune responses regulate inflammatory cell influx into the lungs after vaccination with formalin inactivated RSV. Vaccine 29:2730–2741 [DOI] [PubMed] [Google Scholar]
- 41. Olson MR, Varga SM. 2007. CD8 T cells inhibit respiratory syncytial virus (RSV) vaccine-enhanced disease. J. Immunol. 179:5415–5424 [DOI] [PubMed] [Google Scholar]
- 42. Grenfell BT, Pybus OG, Gog JR, Wood JL, Daly JM, Mumford JA, Holmes EC. 2004. Unifying the epidemiological and evolutionary dynamics of pathogens. Science 303:327–332 [DOI] [PubMed] [Google Scholar]
- 43. Zlateva KT, Lemey P, Moes E, Vandamme AM, Van Ranst M. 2005. Genetic variability and molecular evolution of the human respiratory syncytial virus subgroup B attachment G protein. J. Virol. 79:9157–9167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zlateva KT, Lemey P, Vandamme AM, Van Ranst M. 2004. Molecular evolution and circulation patterns of human respiratory syncytial virus subgroup a: positively selected sites in the attachment G glycoprotein. J. Virol. 78:4675–4683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Holmes EC, Grenfell BT. 2009. Discovering the phylodynamics of RNA viruses. PLoS Comput. Biol. 5:e1000505. 10.1371/journal.pcbi.1000505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kumaria R, Iyer LR, Hibberd ML, Simoes EA, Sugrue RJ. 2011. Whole genome characterization of non-tissue culture adapted HRSV strains in severely infected children. Virol. J. 8:372. 10.1186/1743-422X-8-372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rebuffo-Scheer C, Bose M, He J, Khaja S, Ulatowski M, Beck ET, Fan J, Kumar S, Nelson MI, Henrickson KJ. 2011. Whole genome sequencing and evolutionary analysis of human respiratory syncytial virus A and B from Milwaukee, WI 1998–2010. PLoS One 6:e25468. 10.1371/journal.pone.0025468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tan L, Lemey P, Houspie L, Viveen MC, Jansen NJG, van Loon AM, Wiertz E, van Bleek GM, Martin DP, Coenjaerts FE. 2012. Genetic variability among complete human respiratory syncytial virus subgroup A genomes: bridging molecular evolutionary dynamics and epidemiology. PLoS One 7:e51439. 10.1371/journal.pone.0051439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948 [DOI] [PubMed] [Google Scholar]
- 50. Gupta R, Brunak S. 2002. Prediction of glycosylation across the human proteome and the correlation to protein function. Pac. Symp. Biocomput. 2002:310–322 [PubMed] [Google Scholar]
- 51. Julenius K, Molgaard A, Gupta R, Brunak S. 2005. Prediction, conservation analysis, and structural characterization of mammalian mucin-type O-glycosylation sites. Glycobiology 15:153–164 [DOI] [PubMed] [Google Scholar]
- 52. Martin DP, Lemey P, Lott M, Moulton V, Posada D, Lefeuvre P. 2010. RDP3: a flexible and fast computer program for analyzing recombination. Bioinformatics 26:2462–2463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Baek YH, Choi EH, Song MS, Pascua PN, Kwon HI, Park SJ, Lee JH, Woo SI, Ahn BH, Han HS, Hahn YS, Shin KS, Jang HL, Kim SY, Choi YK. 2012. Prevalence and genetic characterization of respiratory syncytial virus (RSV) in hospitalized children in Korea. Arch. Virol. 157:1039–1050 [DOI] [PubMed] [Google Scholar]
- 54. Drummond A, Pybus OG, Rambaut A. 2003. Inference of viral evolutionary rates from molecular sequences. Adv. Parasitol. 54:331–358 [DOI] [PubMed] [Google Scholar]
- 55. Guindon S, Delsuc F, Dufayard JF, Gascuel O. 2009. Estimating maximum likelihood phylogenies with PhyML. Methods Mol. Biol. 537:113–137 [DOI] [PubMed] [Google Scholar]
- 56. Drummond AJ, Suchard MA, Xie D, Rambaut A. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 29:1969–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Baele G, Lemey P, Bedford T, Rambaut A, Suchard MA, Alekseyenko AV. 2012. Improving the accuracy of demographic and molecular clock model comparison while accommodating phylogenetic uncertainty. Mol. Biol. Evol. 29:2157–2167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Drummond AJ, Ho SY, Phillips MJ, Rambaut A. 2006. Relaxed phylogenetics and dating with confidence. PLoS Biol. 4:e88. 10.1371/journal.pbio.0040088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Shapiro B, Ho SY, Drummond AJ, Suchard MA, Pybus OG, Rambaut A. 2011. A Bayesian phylogenetic method to estimate unknown sequence ages. Mol. Biol. Evol. 28:879–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ho SY, Phillips MJ, Cooper A, Drummond AJ. 2005. Time dependency of molecular rate estimates and systematic overestimation of recent divergence times. Mol. Biol. Evol. 22:1561–1568 [DOI] [PubMed] [Google Scholar]
- 61. Drummond AJ, Suchard MA. 2008. Fully Bayesian tests of neutrality using genealogical summary statistics. BMC Genet. 9:68. 10.1186/1471-2156-9-68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Siebenga JJ, Lemey P, Kosakovsky Pond SL, Rambaut A, Vennema H, Koopmans M. 2010. Phylodynamic reconstruction reveals norovirus GII. 4 epidemic expansions and their molecular determinants. PLoS Pathog. 6:e1000884. 10.1371/journal.ppat.1000884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kosakovsky Pond SL, Frost SD. 2005. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol. Biol. Evol. 22:1208–1222 [DOI] [PubMed] [Google Scholar]
- 64. Lemey P, Minin VN, Bielejec F, Kosakovsky Pond SL, Suchard MA. 2012. A counting renaissance: combining stochastic mapping and empirical Bayes to quickly detect amino acid sites under positive selection. Bioinformatics 28:3248–3256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Pond SK, Muse SV. 2005. Site-to-site variation of synonymous substitution rates. Mol. Biol. Evol. 22:2375–2385 [DOI] [PubMed] [Google Scholar]
- 66. Murrell B, Wertheim JO, Moola S, Weighill T, Scheffler K, Kosakovsky Pond SL. 2012. Detecting individual sites subject to episodic diversifying selection. PLoS Genet. 8:e1002764. 10.1371/journal.pgen.1002764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wertheim JO, Worobey M. 2009. Relaxed selection and the evolution of RNA virus mucin-like pathogenicity factors. J. Virol. 83:4690–4694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zimmer G, Trotz I, Herrler G. 2001. N-glycans of F protein differentially affect fusion activity of human respiratory syncytial virus. J. Virol. 75:4744–4751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Bermingham A, Collins PL. 1999. The M2-2 protein of human respiratory syncytial virus is a regulatory factor involved in the balance between RNA replication and transcription. Proc. Natl. Acad. Sci. U. S. A. 96:11259–11264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Cheng X, Park H, Zhou H, Jin H. 2005. Overexpression of the M2-2 protein of respiratory syncytial virus inhibits viral replication. J. Virol. 79:13943–13952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ahmadian G, Randhawa JS, Easton AJ. 2000. Expression of the ORF-2 protein of the human respiratory syncytial virus M2 gene is initiated by a ribosomal termination-dependent reinitiation mechanism. EMBO J. 19:2681–2689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Woelk CH, Holmes EC. 2001. Variable immune-driven natural selection in the attachment (G) glycoprotein of respiratory syncytial virus (RSV). J. Mol. Evol. 52:182–192 [DOI] [PubMed] [Google Scholar]
- 73. Spann KM, Collins PL, Teng MN. 2003. Genetic recombination during coinfection of two mutants of human respiratory syncytial virus. J. Virol. 77:11201–11211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Gray RR, Pybus OG, Salemi M. 2011. Measuring the temporal structure in serially-sampled phylogenies. Methods Ecol. Evol. 2:437–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.