Abstract
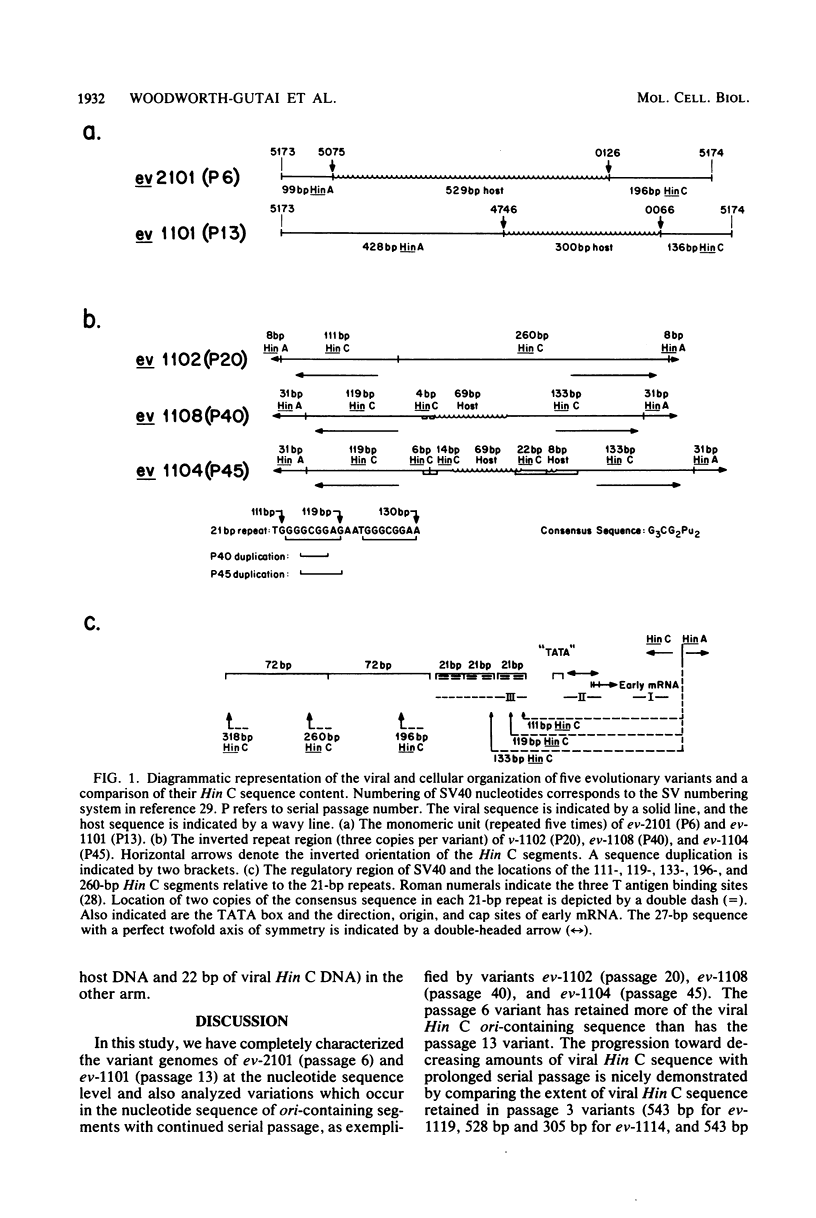
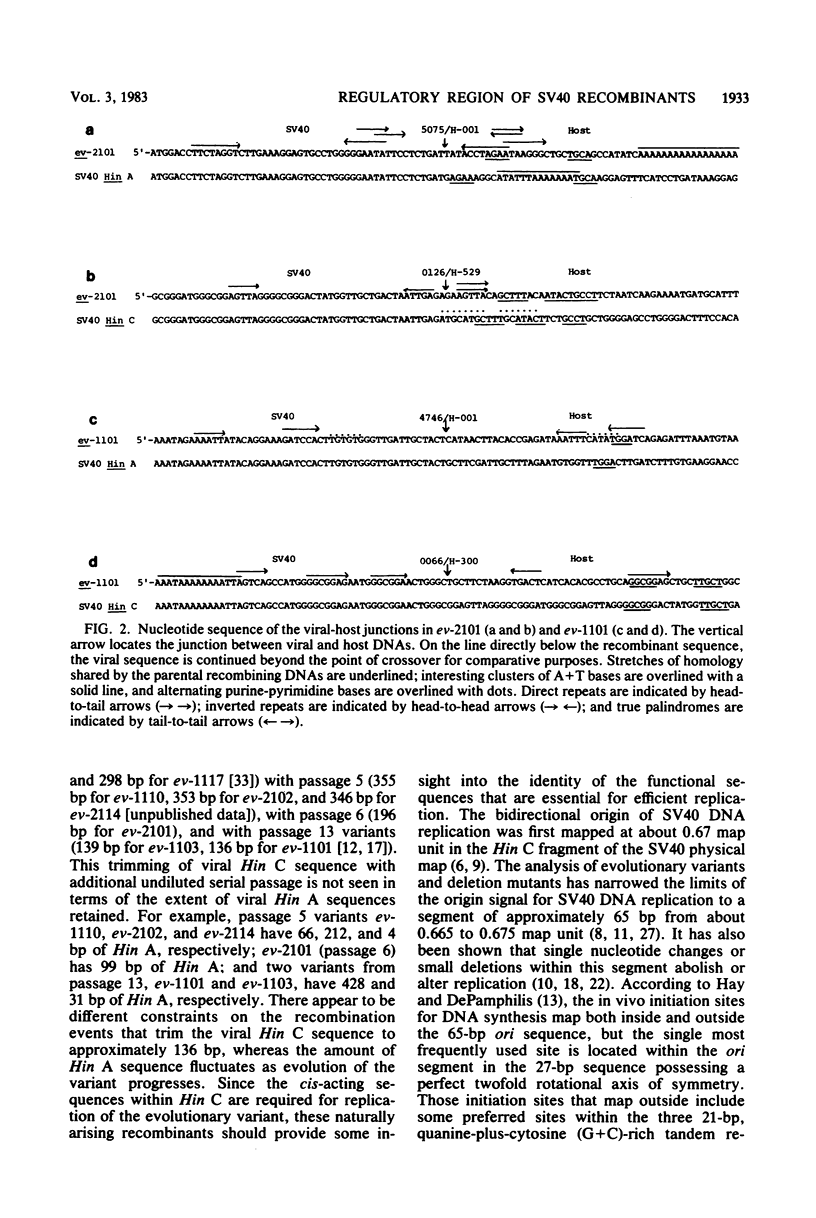
When simian virus 40 (SV40) is serially passaged at high multiplicity, a heterogeneous collection of naturally arising variants is generated. Those which are the most abundant presumably have a selective replicative advantage over other defective and wild-type helper SV40s. Two such naturally arising host-substituted variants of SV40 have been characterized in terms of complete nucleotide sequence determination. Evolutionary variant ev-1101 (previously isolated by Lee et al., Virology 66:53-69, 1975) is from undiluted serial passage 13, whereas ev-2101 is newly isolated from undiluted serial passage 6 of an independently-derived evolutionary series. Both variants contain a five-times tandemly repeated segment of DNA consisting of viral Hin C and Hin A sequences that have recombined with a segment of host DNA that is not highly reiterated in the monkey genome. The monkey segment differs in the two variants as does the size of the viral segment retained. In two additional host-substituted variants, ev-1102 (previously isolated from serial passage 20 by Brockman et al., Virology 54:384-397, 1973) and ev-1108 (newly isolated from serial passage 40), the SV40 sequences derived from the replication origin are present as inverted repetitions. The inverted repeat regions of these two variants have been analyzed at the nucleotide sequence level and are compared with SV40 variant ev-1104 from passage 45 (previously characterized by Gutai and Nathans, J. Mol. Biol. 126:259-274, 1978). The viral segment containing the regulatory signals for replication and viral gene expression is considerably shortened in later serial passages as demonstrated by these five variants. It is of interest that the variants presumably arose due to their enhanced replication efficiency, yet are missing some of the sequence elements implicated in the regulation of replication. Furthermore, a comparison of the structure of the replication origin regions indicates that additional changes occur in the SV40 regulatory region with continued undiluted serial passage.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bergsma D. J., Olive D. M., Hartzell S. W., Subramanian K. N. Territorial limits and functional anatomy of the simian virus 40 replication origin. Proc Natl Acad Sci U S A. 1982 Jan;79(2):381–385. doi: 10.1073/pnas.79.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botchan M., Stringer J., Mitchison T., Sambrook J. Integration and excision of SV40 DNA from the chromosome of a transformed cell. Cell. 1980 May;20(1):143–152. doi: 10.1016/0092-8674(80)90242-1. [DOI] [PubMed] [Google Scholar]
- Brockman W. W. Evolutionary variants of simian virus 40. Prog Med Virol. 1977;23:69–95. [PubMed] [Google Scholar]
- Brockman W. W., Lee T. N., Nathans D. The evolution of new species of viral DNA during serial passage of simian virus 40 at high multiplicity. Virology. 1973 Aug;54(2):384–397. doi: 10.1016/0042-6822(73)90151-7. [DOI] [PubMed] [Google Scholar]
- Brockman W. W., Nathans D. The isolation of simian virus 40 variants with specifically altered genomes. Proc Natl Acad Sci U S A. 1974 Mar;71(3):942–946. doi: 10.1073/pnas.71.3.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danna K. J., Nathans D. Bidirectional replication of Simian Virus 40 DNA. Proc Natl Acad Sci U S A. 1972 Nov;69(11):3097–3100. doi: 10.1073/pnas.69.11.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhruva B. R., Shenk T., Subramanian K. N. Integration in vivo into simian virus 40 DNA of a sequence that resembles a certain family of genomic interspersed repeated sequences. Proc Natl Acad Sci U S A. 1980 Aug;77(8):4514–4518. doi: 10.1073/pnas.77.8.4514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMaio D., Nathans D. Cold-sensitive regulatory mutants of simian virus 40. J Mol Biol. 1980 Jun 15;140(1):129–142. doi: 10.1016/0022-2836(80)90359-9. [DOI] [PubMed] [Google Scholar]
- Fareed G. C., Garon G. F., Salzman N. P. Origin and direction of simian virus 40 deoxyribonucleic acid replication. J Virol. 1972 Sep;10(3):484–491. doi: 10.1128/jvi.10.3.484-491.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gluzman Y., Sambrook J. F., Frisque R. J. Expression of early genes of origin-defective mutants of simian virus 40. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3898–3902. doi: 10.1073/pnas.77.7.3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutai M. W., Nathans D. Evolutionary variants of simian virus 40: Cellular DNA sequences and sequences at recombinant joints of substituted variants. J Mol Biol. 1978 Dec 5;126(2):275–288. doi: 10.1016/0022-2836(78)90363-7. [DOI] [PubMed] [Google Scholar]
- Gutai M. W., Nathans D. Evolutionary variants of simian virus 40: Nucleotide sequence of a conserved SV40 DNA segment containing the origin of viral DNA replication as an inverted repetition. J Mol Biol. 1978 Dec 5;126(2):259–274. doi: 10.1016/0022-2836(78)90362-5. [DOI] [PubMed] [Google Scholar]
- Hay R. T., DePamphilis M. L. Initiation of SV40 DNA replication in vivo: location and structure of 5' ends of DNA synthesized in the ori region. Cell. 1982 Apr;28(4):767–779. doi: 10.1016/0092-8674(82)90056-3. [DOI] [PubMed] [Google Scholar]
- Jelinek W. R., Schmid C. W. Repetitive sequences in eukaryotic DNA and their expression. Annu Rev Biochem. 1982;51:813–844. doi: 10.1146/annurev.bi.51.070182.004121. [DOI] [PubMed] [Google Scholar]
- Lee T. N., Brockman W. W., Nathans D. Evolutionary variants of simian virus 40: cloned substituted variants containing multiple initiation sites for DNA replication. Virology. 1975 Jul;66(1):53–69. doi: 10.1016/0042-6822(75)90178-6. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McCutchan T., Singer M., Rosenberg M. Structure of simian virus 40 recombinants that contain both host and viral DNA sequences. II. The structure of variant 1103 and its comparison to variant CVPS/1P2 (EcoRI res). J Biol Chem. 1979 May 10;254(9):3592–3597. [PubMed] [Google Scholar]
- Myers R. M., Tjian R. Construction and analysis of simian virus 40 origins defective in tumor antigen binding and DNA replication. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6491–6495. doi: 10.1073/pnas.77.11.6491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordheim A., Rich A. The sequence (dC-dA)n X (dG-dT)n forms left-handed Z-DNA in negatively supercoiled plasmids. Proc Natl Acad Sci U S A. 1983 Apr;80(7):1821–1825. doi: 10.1073/pnas.80.7.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg H., Singer M., Rosenberg M. Highly reiterated sequences of SIMIANSIMIANSIMIANSIMIANSIMIAN. Science. 1978 Apr 28;200(4340):394–402. doi: 10.1126/science.205944. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R. The use of thin acrylamide gels for DNA sequencing. FEBS Lett. 1978 Mar 1;87(1):107–110. doi: 10.1016/0014-5793(78)80145-8. [DOI] [PubMed] [Google Scholar]
- Shortle D., Nathans D. Regulatory mutants of simian virus 40: constructed mutants with base substitutions at the origin of DNA replication. J Mol Biol. 1979 Jul 15;131(4):801–817. doi: 10.1016/0022-2836(79)90202-x. [DOI] [PubMed] [Google Scholar]
- Sol C. J., Hassing I., Maris W., Walig C., van der Noordaa J. Evolutionary variants of simian virus 40 which are impaired in early lytic functions but transform nonpermissive cells. J Virol. 1981 Jan;37(1):395–410. doi: 10.1128/jvi.37.1.395-410.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sompayrac L., Danna K. J. Simian virus 40 deletion mutants that transform with reduced efficiency. Mol Cell Biol. 1983 Mar;3(3):484–489. doi: 10.1128/mcb.3.3.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringer J. R. DNA sequence homology and chromosomal deletion at a site of SV40 DNA integration. Nature. 1982 Mar 25;296(5855):363–366. doi: 10.1038/296363a0. [DOI] [PubMed] [Google Scholar]
- Stringer J. R. Integrated simian virus 40 DNA: nucleotide sequences at cell-virus recombinant junctions. J Virol. 1981 May;38(2):671–679. doi: 10.1128/jvi.38.2.671-679.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian K. N., Shenk T. Definition of the boundaries of the origin of DNA replication in simian virus 40. Nucleic Acids Res. 1978 Oct;5(10):3635–3642. doi: 10.1093/nar/5.10.3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjian R. Protein-DNA interactions at the origin of simian virus 40 DNA replication. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 2):655–661. doi: 10.1101/sqb.1979.043.01.073. [DOI] [PubMed] [Google Scholar]
- Wakamiya T., McCutchan T., Rosenberg M., Singer M. Structure of simian virus 40 recombinants that contain both host and viral DNA sequences. I. The structure of variant CVPS/1/P2 (EcoRI res). J Biol Chem. 1979 May 10;254(9):3584–3591. [PubMed] [Google Scholar]
- Wang A. H., Quigley G. J., Kolpak F. J., Crawford J. L., van Boom J. H., van der Marel G., Rich A. Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature. 1979 Dec 13;282(5740):680–686. doi: 10.1038/282680a0. [DOI] [PubMed] [Google Scholar]
- Wilson J. H., Berget P. B., Pipas J. M. Somatic cells efficiently join unrelated DNA segments end-to-end. Mol Cell Biol. 1982 Oct;2(10):1258–1269. doi: 10.1128/mcb.2.10.1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodworth-Gutai M. Recombination in SV40-infected cells: nucleotide sequences at viral-viral recombinant joints in naturally arising variants. Virology. 1981 Mar;109(2):344–352. doi: 10.1016/0042-6822(81)90505-5. [DOI] [PubMed] [Google Scholar]
- Woodworth-Gutai M. Recombination in SV40-infected cells: viral DNA sequences at sites of circularization of transfecting linear DNA. Virology. 1981 Mar;109(2):353–365. doi: 10.1016/0042-6822(81)90506-7. [DOI] [PubMed] [Google Scholar]


