Abstract
A critical component of vaccine design is to generate and maintain antigen-specific memory lymphocytes of sufficient quantity and quality to give the host life-long protection against re-infection. Therefore, it is important to understand how memory T cells acquire the ability for self-renewal while retaining a potential for heightened recall of effector functions. During acute viral infection or following vaccination, antigen-specific T cells undergo extensive phenotypic and functional changes during differentiation to the effector and memory phases of the immune response. The changes in cell phenotype that accompany memory T-cell differentiation are predominantly mediated through acquired transcriptional regulatory mechanisms, in part achieved through epigenetic modifications of DNA and histones. Here we review our current understanding of epigenetic mechanisms regulating the off-on-off expression of CD8 and CD4 T-cell effector molecules at naive, effector and memory stages of differentiation, respectively, and how covalent modifications to the genome may serve as a mechanism to preserve ‘poised’ transcriptional states in homeostatically dividing memory cells. We discuss the potential of such mechanisms to control genes that undergo on-off-on patterns of expression including homing and pro-survival genes, and the implications on the development of effector-memory and central-memory T-cell differentiation. Lastly, we review recent studies demonstrating epigenetic modifications as a mechanism for the progressive loss of transcriptional adaptation in antigen-specific T cells that undergo sustained high levels of T-cell receptor signalling.
Keywords: epigenetic, exhaustion, memory T cell, transcription, viral infection
OTHER ARTICLES PUBLISHED IN THIS SERIES
The MYSTerious MOZ, a histone acetyltransferase with a key role in haematopoiesis. Immunology 2013;139:161–165.
Epigenetic aspects of lymphocyte antigen receptor gene rearrangement or ‘when stochasticity completes randomness’. Immunology 2013;139:141–150.
Why detailed model gene studies in higher eukaryotes are still necessary. Immunology 2013;139:158–160.
Roles of repressive epigenetic machinery in lineage decision of T cells. Immunology 2013;139:151–157.
Epigenetic regulation of inducible gene expression in the immune system. Immunology 2013;139:285–293.
Transcription factors and CD4 T cells seeking identity: masters, minions, setters and spikers. Immunology 2013;139:294–298.
Regulators of chromatin state and transcription in CD4 T cell polarization. Immunology 2013;139:299–308.
CD8 T-cell memory
The ability of functional memory CD8 T cells to directly target and kill infected cells provides a vital component in a vaccine's arsenal against viral infections. To achieve the maximal benefit from this component of cellular immunity it is important to understand when and how T-cell memory is generated. During acute viral infection, antigen-driven differentiation of naive CD8 T cells results in expression of cytolytic molecules and cytokines at the effector stage of the response that facilitate control of the infection. Following pathogen clearance, a subset of antigen-specific CD8 T cells survive to the memory stage of the immune response1 (Fig. 1a). Antigen-specific CD8 T cells that survive the contraction phase of the response have obtained the unique properties of self-renewal in lymphoid and non-lymphoid tissues, and a heightened ability to recall effector functions relative to their naive precursors.2–5 Extensive molecular and cellular studies of CD8 T-cell differentiation during acute viral infection have revealed that cells destined to survive into the memory phase of the response can be identified at the effector stage, referred to as memory precursors.6–9 The initial identification of a memory precursor subset came from gene expression studies broadly demonstrating that the acquired functions of virus-specific CD8 T cells were coupled to changes in the corresponding gene's transcriptional regulation. Kinetic analysis of the gene expression profile of the antigen-specific CD8 T cells during acute viral infection revealed that gene expression programmes could be divided into distinct patterns. Particularly informative was the subset of genes that appeared to have an on-off-on gene expression profile at naive, effector and memory stages of the immune response, respectively (Fig. 1b,c).10–12 Such genes include those that encode pro-survival and homing molecules such as interleukin-7 receptor α (IL-7Rα), Bcl-2, CD62L (L-selectin) and others that are predictive of either the ability to homeostatically proliferate following the clearance of antigen or enhanced recall capacity following re-encounter with antigen. Within this category of genes, expression of the transcript for IL7Ra is a key determinant of cell survival and homeostasis at the memory stage.7,13 Identification of memory precursor cells was born out of using IL7Ra expression as a marker for a subset of effector cells with the ability to survive in the absence of antigen. Identification of memory cell precursors at the effector stage of the response was further refined by including the down-regulated expression of CD25 and Klrg1 for subsetting.8,9
Figure 1.
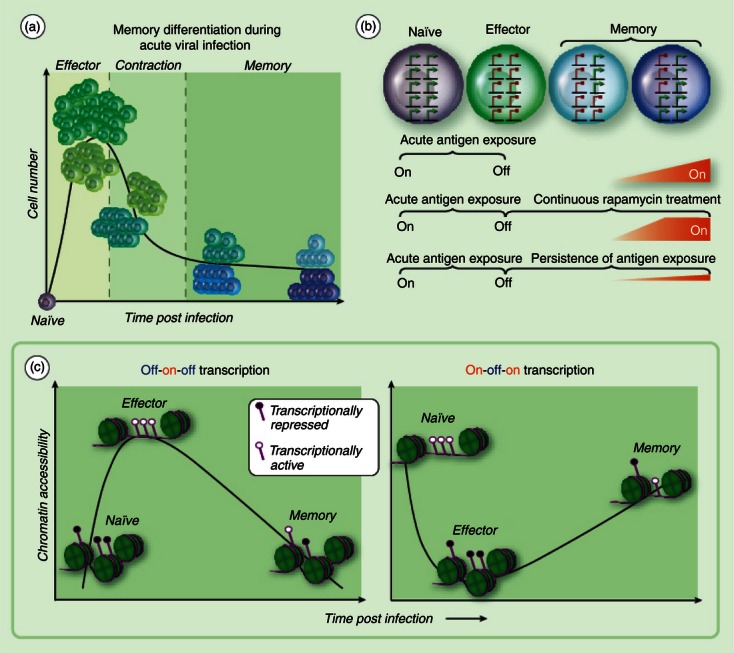
Adaptations in gene regulation during memory T-cell differentiation. (a) Naive antigen-specific CD8 T cells undergo clonal expansion upon presentation with the cognate antigen. The quantity of antigen-specific CD8 T cells at peak of clonal expansion is ∼ 1 × 105 greater than the naive population. Approximately 90–95% of antigen-specific CD8 T cells die during the contraction stage of the immune response. A subset of effector cells progressively modify the transcriptional programme, acquiring homing and pro-survival properties before and into the contraction phase of the response. (b) Longitudinal gene expression profiling studies of total antigen-specific CD8 T cells during acute viral infection have revealed a group of genes that are expressed (green arrows) in naive cells, repressed (red circle) in effector cells, and re-expressed at different memory time points, i.e. on-off-on gene expression. Continuous treatment of mice with rapamycin at the effector stage of an acute immune response enhances the kinetics of gene re-expression in CD8 T cells. Persistence of antigen presentation during chronic viral infection retains the ‘off’ state for several genes in the category of on-off-on expression. (c) Off-on-off gene expression is coupled to the loss of repressive epigenetic transcriptional regulatory programmes at the effector stage of the immune response followed by reacquisition of repressive and poised epigenetic programmes. A hypothetical model for epigenetic regulation of on-off-on gene expression would involve reduction in chromatin accessibility and acquisition of repressive epigenetic programming at the effector stage of the immune response followed by permissive epigenetic programming and chromatin accessibility during the transition to central memory T cells. Acquisition of repressive epigenetic modifications are indicated by a filled circle marker on the purple line (DNA) at transcriptional regulatory regions. Chromatin accessibility is indicated by the absence of green histone octamers.
The ability to identify memory precursor effector cells has generated much interest in delineating when and how memory CD8 T cells are generated and the role of T-cell receptor (TCR) signalling strength and duration in the functional outcome of the ensuing memory T cells.6 Significant efforts are now focused on determining the mechanism(s) that mediate the progressive changes in phenotype and function of antigen-specific T cells as they develop in response to both acute and chronic pathogens. Here we review our current understanding of transcriptional regulatory mechanisms of genes directly related to effector and memory functions and highlight potential mechanisms for the generation of phenotypically distinct memory T-cell subsets.
Memory CD8 T-cell heterogeneity
It is believed that memory T cell heterogeneity has evolved as a mechanism for partitioning memory-associated functions into specialized cells to protect against a range of pathogens and routes of exposure. Memory CD8 T cells that populate non-lymphoid tissues and provide immediate recall of effector functions are loosely categorized as effector-memory (Tem) cells. Tem cells maintain down-regulation of the molecules CD62L and CCR7 and serve as the first line of defence against pathogen re-exposure. In contrast, memory CD8 T cells that express CD62L and CCR7 and preferentially home to lymphoid tissues are referred to as central-memory cells (Tcm). The preferential lymphoid homing of Tcm cells is believed to facilitate their encounter with antigen-presenting dendritic cells, thereby generating a self-renewing source of cells with effector functions, which can then migrate to the site of infection.14–17 Importantly, many of the differentially acquired traits of Tem versus Tcm cells, including CD62L- and CCR7-mediated lymphoid homing, are the result of differential transcriptional regulation of gene products from the ‘on-off-on’ subset of genes (Fig. 1b). A current challenge for the field is to determine how acquired transcriptional programmes, those common among all memory cells as well as the transcriptional programmes that are unique to memory subsets, are maintained during cell division of memory T cells.
Drawing upon insights from other developmental systems, epigenetic modifications may provide a transcriptional regulatory mechanism that can be propagated during homeostatic cell division of memory cells.18,19 Recently several laboratories have demonstrated that epigenetic modifications, namely histone modifications and DNA methylation, modulate transcriptional activation of effector molecules via the restriction of access to chromatin by transcription factors and polymerase. Our current understanding of epigenetic regulation of memory cell function has come from studies that have focused on the mechanisms controlling expression of effector molecules such as the genes for interferon-γ (IFNg), interleukin 2 (IL-2) granzyme b and perforin.20–25 As these genes become transcriptionally up-regulated, the proximal promoter region loses repressive epigenetic marks (DNA and histone modifications). Following control of the viral infection, these loci become epigenetically ‘poised’ for polymerase accessibility and transcriptional activation in memory cells (Fig. 1c). In the case of IFNg, Kersh et al.22 determined that the promoter re-acquires a repressive DNA methylation, but can demethylate this region within 6 hr of TCR stimulation. Additionally the laboratories of both Turner and Shen revealed that the IFNg promoter obtained permissive histone modifications at the effector stage of differentiation which were maintained into the memory stage.21,26 These data demonstrate that the acquired ability of memory cells to rapidly recall cytokine production is coupled to modification of the epigenetic programme at these loci by establishing a poised transcriptional state. Moreover, these studies firmly establish epigenetic programming as a mechanism that adapts to TCR signalling. In addition to these important studies on transcriptional regulation of effector molecules, our laboratory has recently demonstrated that the promoter of the immuno-inhibitory molecule programmed death 1 (PD-1) undergoes dynamic epigenetic modifications during acute versus chronic viral infection.27 Our data demonstrated that epigenetic modification of the PD-1 promoter was tuned to the duration and or strength of the TCR signal.27 A commonality among the effector molecules and immuno-inhibitory receptor is that their off-on-off pattern of gene expression during naive to effector to memory differentiation is regulated in part through epigenetic modifications at their promoters (Fig. 1c). Taken together, these studies demonstrate that epigenetic modifications are used to control immune function by not only directly regulating the expression of cytolytic molecules, but also by controlling the sensitivity of the cell to activating inhibitory signals.
Indeed, the rapid recall of effector molecules is a defining feature of memory CD8 T cells, yet equally important is the ability of memory CD8 T cells to persist at a higher quantity relative to their naive counterparts in the absence of antigen. This acquired function is critical to the design of vaccines that generate life-long T-cell immunity. Importantly the dramatic increase in quantity of antigen-specific CD8 T cells at the memory stage of the response over the naive stage is in part achieved through up-regulation of pro-survival molecules in a subset of effector cells. Therapeutic strategies designed to enhance the quantity of effector cells that survive to the memory stage of the response following acute infection or vaccination through manipulation of pro-survival gene expression programmes in antigen-specific CD8 T cells is now the focus of intense investigation.28 Support for this strategy has recently come from studies using rapamycin therapy. It was demonstrated that mice treated daily with rapamycin, the inhibitor of mammalian target of rapamycin (mTOR), during the course of acute lymphocytic choriomeningitis virus infection developed a greater quantity and quality of memory CD8 T cells.29 Importantly, this effect was observed when rapamycin treatment was initiated at the effector stage of the immune response, after extensive proliferation had already occurred (Fig. 1b). Of particular interest, rapamycin treatment resulted in faster re-expression kinetics for several molecules within the ‘on-off-on’ subset of genes including CD62L and IL-7Ra (Fig. 1b).29 These studies using rapamycin demonstrate that antigen-specific CD8 T-cell gene expression programmes can be modified after the initial encounter with antigen and that the modification of the gene expression programme can translate into changes in the quantity of memory T cells. Taken together, these data suggest that the elevated quantity of antigen-specific CD8 T cells at the memory stage of the response is the result of progressive changes in gene regulation at the effector stage. Additionally, these studies highlight a need for further investigation into the transcription factors or epigenetic mechanisms that may be downstream of the mTOR pathway.
Extrapolating from our understanding of off-on-off gene regulatory mechanisms, it may be reasoned that the acquired epigenetic modifications at the transcriptional regulatory regions of on-off-on genes initiates with the acquisition of repressive epigenetic modifications during the progression of an antigen-specific T cell into the effector stage of the response. This hypothetical repressive epigenetic programme may then undergo erasure during contraction and enter the memory phase of the response (Fig. 1c). Additionally, this would indicate that kinetics of ‘off to on’ gene expression at the antigen-independent stage of the memory response could be controlled by the manipulation of epigenetic enzymes or interpreting proteins. Future efforts focused on on-off-on epigenetic regulatory mechanisms will undoubtedly be informative regarding the adaptation of transcriptional programmes during memory CD8 T-cell differentiation.
Effector and memory CD4 T-cell differentiation
Similar to CD8 T-cell memory differentiation, dramatic changes in gene expression and function accompany the differentiation of CD4 effector and memory T cells. The full significance of such gene regulation remains unresolved. The dissection of CD4 memory differentiation becomes more complicated by the extensive T helper lineage diversity that exists within the effector CD4 T-cell population. Following activation with antigen, naive CD4 T cells undergo extensive proliferation and differentiation toward different T helper lineages, including Th1, Th2, Th17, regulatory T and T follicular helper lineages.30,31 Lineage differentiation of CD4 T helper cells is regulated by extrinsic factors such as the cytokine milieu provided by antigen-presenting cells during priming, as well as intrinsic factors including the lineage-associated transcription factors Tbet, Gata3, RORg, Foxp3 and Bcl6.31,32 Lineage-specific gene expression programmes confer specialized effector functions of each individual T helper subset to generate immune responses that are tailored to express specific effector molecules and cytokines for the effective control of varying types of pathogens.30,31,33,34 Differentiation of one particular T helper lineage may be accompanied by the suppression of gene expression programmes that inhibit genes commonly expressed by other T helper lineages.32 The occurrence of lineage commitment during proliferation has prompted a focus to understand the maintenance of acquired transcrip-tional programmes through epigenetic mechanisms. It is believed that a specific set of epigenetic modifications may accompany the differentiation of a particular T helper lineage that permit the expression of genes associated with that lineage, including demethylation of DNA and the acquisition of permissive histone modifications, while maintenance or de novo generation of inhibitory marks may occur at loci associated with other T helper lineages.32,35–37
One method that has aided the biochemical analysis of such gene regulation following CD4 T-cell activation is the ability to polarize naive CD4 T cells toward these T helper lineages through in vitro culturing conditions.30,38,39 The polarized cells that are products of such conditions can then be exposed to alternative polarizing conditions to measure their ‘plasticity’, or capacity to convert to alternate T helper lineages and express the specific gene expression programmes of the associated T helper fates. Epigenetic regulation plays an important role in regulating the expression of T helper lineage-specific genes, with the classic example being differential regulation of the IFNg and IL4 loci during the differentiation of Th1 and Th2 cells. Th1 cells produce large amounts of IFN-γ and do not express IL4, whereas Th2 cells produce the signature cytokine IL-4, as well as IL-5 and IL-13, but do not express IFNg.33 Analysis of the IFNg expression in Th1 cells is accompanied by permissive histone modifications and demethylation of conserved non-coding sequences at the IFNg locus, while these same regions maintain repressive histone marks and methylated DNA in Th2 cells.37 In contrast, the IFNg locus remains in a repressed state in differentiating Th2 cells,37 whereas the IL4 locus undergoes chromatin remodelling and DNA demethylation.40 Further evidence that epigenetics influence the gene expression programmes of T helper lineages is demonstrated by deletion of genes that encode enzymes necessary for DNA methylation. The maintenance methyltransferase Dnmt1 plays an important role in the repression of the IL4 and Foxp3 loci, and deficiency of Dnmt1 results in inappropriate expression of these genes.41–43 Likewise, CD4 T cells lacking the de novo methyltransferase Dnmt3a can simultaneously express IFNg and IL4 under non-skewing activation conditions, and hypomethylation of both of these loci allows for the development of Th2 cells with a propensity to express IFNg when re-stimulated under Th1 conditions.44 Furthermore, re-stimulation of Th2, Th17 and iTreg Dnmt3a knockout cells in the presence of IL-12 results in IFN-γ production, indicating that de novo methylation restricts plasticity of T helper lineages.45
The majority of studies regarding the T helper lineage gene expression and epigenetic programmes of CD4 T cells have been conducted using in vitro generated effector subsets. Whereas such experiments may be useful for looking at the potential of polarized cells to express genes that have been programmed under certain skewing conditions, they may not fully represent what happens to memory T cells generated in vivo following the clearance of antigen. Hence, an important question that emerges is whether the cells that comprise the memory CD4 T-cell pool maintain their potential to recall a T helper lineage-specific gene expression programme. In other words, are epigenetic programmes maintained, such that memory CD4 T cells ‘remember’ the gene expression programme associated with cells at the effector stage (Fig. 1c)? This question highlights the need for epigenetic analysis of antigen-specific memory CD4 T-cell subsets to provide insight into T helper lineage maintenance and plasticity upon boosting or re-exposure to pathogen.
It is unclear to what extent memory CD4 T cells are derived from committed effector cells of each of these lineages. To this end, several studies have investigated the recall potential of Th1 memory cells. It has been shown that Th1 memory cells exist in vivo following infection, and are derived from Tbet and IFN-γ-expressing Th1 effector cells.46 Th1 memory cells exhibit minimal (or possibly delayed) re-expression of CD62L and CCR7, suggesting that these cells are Th1 effector-memory cells.46,47 Besides Th1 memory cells, other studies have demonstrated the generation of and recall by Th2 committed memory cells,48–50 whereas it is currently unclear whether long-lived Th17 cells can be generated following infection.51 In addition, there may be central-memory cells that do not have commitment toward any of the T helper lineages, and following reactivation with antigen, can potentially generate secondary effector cells of several different T helper lineages.47 Given the complexity and extensive heterogeneity that exists within the memory CD4 T-cell pool, an important question is whether memory CD4 T cells transition through an effector stage. Again, interrogation of epigenetic modifications may prove particularly useful when focused on loci such as IFNg, IL4, IL17, and others that are associated with T helper lineage-specific functions.
Further work is needed to determine the extent to which T helper lineages are maintained in the memory pool, and to further define memory differentiation at both the cellular and epigenetic levels. Although it is clear that there are indeed subsets of Th1 memory cells within the memory pool, there may be varying degrees of plasticity between different T helper lineages depending on the degree of terminal differentiation and polarity toward each lineage for each individual memory cell. In addition, there may be subsets of CD4 memory cells that are not biased toward any lineage, and so display the highest degree of lineage potential following reactivation with antigen. In the case of such non-committed cells, the prediction would be that lineage-associated transcription factor and/or effector genes (i.e Tbx21/Tbet, Gata3, Rorg, Bcl6, IFNg, IL4, etc.) have not yet acquired epigenetic modifications consistent with expression of these genes that would skew their response toward any particular lineage. In depth gene expression and epigenetic analysis of memory subsets will be useful in determining whether ‘Th uncommitted’ memory CD4 T cells contribute significantly to the pool of memory T cells. Further, analysis of on-off-on gene regulation for genes such as CD62L, CCR7 and Bcl2 in memory cells will be useful for understanding factors that govern homing and survival during homeostasis in the absence of their antigen, and possibly be predictive of the fate of memory cells following re-encounter with antigen.
It is essential to understand how antigen-specific CD4 memory T cells behave in response to repeated exposure to pathogen, or throughout the course of vaccination, where priming and repeated boosting to antigen, results in reactivation of memory cells. Determining whether memory CD4 T cells ‘remember’ and efficiently recall lineage-specific gene expression programmes that were acquired during their progenitors at the effector stage will provide an important framework for predicting the capacity of memory CD4 T-cell subsets to provide cellular immune responses and provide help for humoral immune responses upon boosting or challenge with pathogen.
T-cell exhaustion: selection or progression?
A shared feature of CD4 and CD8 T-cell memory differentiation is that the strength and duration of TCR signalling determines the function and phenotype of the cells. At the extreme end of the TCR strength/duration of the signal spectrum are cells differentiated during chronic viral infections. Therefore, additional insights into the mechanism for differentiation of functional memory T cells may be gained from interrogating the mechanism for development of non-functional memory cells during conditions of antigen persistence.
Failure to control viral infection results in a diminished ability of antigen-specific CD8 T cells to rapidly up-regulate cytokine expression and to kill antigen-presenting cells, often regarded as T-cell exhaustion.52,53 It is now well accepted that these functionally impaired exhausted T cells can be rejuvenated through manipulation of their inhibitory receptor signalling, and therapeutic strategies that target these inhibitory mechanisms play an important role in clearance of chronic viral infections such as HIV or hepatitis C virus, as well as control of several types of cancer.54–57 Discovery of the reversible quality of T-cell exhaustion through blockade of inhibitory receptor signalling has focused our attention on the temporal relationship of the exhaustion phenotype with duration of antigen exposure. Longitudinal studies of chronically infected mice indeed reveal that the development of the exhausted phenotype of antigen-specific CD8 T cells occurs during a gradual progression of changes to the gene expression programme.52,58 Specifically, the reduction in cytokine production and killing potential is coupled to persistence of high viral load and is exacerbated in the absence of CD4 T-cell help.59–61 What is not definitively demonstrated by these longitudinal studies is whether development of an exhaustion transcriptional programme is solely accomplished through survival of a subset of cells that were prone to exhaustion or if the resulting phenotype is an acquired property obtained through progressive modification of transcriptional programmes in antigen-specific cells. To address the issue of selection versus progression, the Walker laboratory recently investigated clonal selection of HIV-specific CD8 T cells from HIV controllers versus progressors. Their data indicate that the different functions of HIV-specific CD8 T cells from HIV progressors versus HIV controllers is a result of the different chronic environments (high versus low viral load) promoting survival of distinct antigen-specific CD8 T-cell clones.62 Further analysis is needed to completely resolve the contribution of clonal selection of virus-specific cells as the majority of the functional data came from cells following ex vivo expansion. It is important to note that these data do not rule out the progression of transcriptional regulation.
The apparent gross difference in gene expression profiles between functional memory and exhausted antigen-specific T cells as well as the recent report by the Walker laboratory on distinct clonal selection during differing severities of HIV infection raise the question as to whether the state of exhaustion is obtained through progressive changes in gene regulation. An initial examination of this complex issue has been performed using mouse model systems. West et al.63 controlled for clonal selection by adoptively transferring clonal naive and functional memory CD8 T cells (generated from P14 TCR transgenic mice) into naive recipient mice, which were then challenged with the chronic strain of lymphocytic choriomeningitis virus. Surprisingly, naive cells were better suited than functional memory cells for generating cells that persisted during chronic infection. These data demonstrate that naive cells contain a cell intrinsic mechanism that allows them to adapt to the chronic antigen whereas this mechanism is absent in memory CD8 T cells. In a different set of experiments, Shin et al.64 showed that exhausted CD8 T cells that were adoptively transferred into naive mice or epitope variant chronic infection-matched mice decline over the course of several weeks in the absence of TCR ligation. This is in contrast to the rapid contraction that occurs following the effector phase of the immune response of an acute infection.1,4,10 Taken together these data indicate that the duration/strength of TCR ligation results in a progressive reinforcement of expression programmes that are downstream of the TCR signal.
Fixation of epigenetic modifications and or the expression of unique transcription factors are a likely mechanism for preserving the exhausted state in the absence of antigen (Fig. 1b). Indeed, gene expression profiling studies demonstrate the preservation of many effector transcriptional programmes including persistent down-regulation of several on-off-on genes (Fig. 1b). Consistent with this idea, we have recently reported on preservation of acquired epigenetic modifications at the PD-1 locus regulatory regions in virus-specific CD8 T cells during chronic viral infection.27 Our data demonstrated that the transient up-regulation of PD-1 expression in functional virus-specific CD8 T cells was coupled to chromatin accessibility, permissive histone modifications, and acquisition of an unmethylated transcriptional regulatory region at the peak of acute viraemia. Following clearance of the acute viral infection, the PD-1 transcriptional regulatory region regained the DNA methylation programme and became less sensitive to DNase challenge. Importantly, the repressive transcriptional programme was not reacquired in virus-specific CD8 T cells during chronic infection of mice and humans.27 To our surprise, the permissive epigenetic transcriptional programme at the PD-1 locus was retained in PD-1lo cells following reduction in chronic viral load. Preservation of the permissive transcriptional programme facilitated enhanced re-expression of PD-1 relative to functional memory cells that contained the repressive programme at the PD-1 locus.27 The kinetic analysis of epigenetic regulation of PD-1 during acute and chronic infections as well as analysis of effector molecule regulation during CD4 and CD8 T-cell memory cell differentiation have set the stage for further analysis of the enzymes that catalyse the epigenetic modifications and their specificity determinates.
Summary
Further scrutiny of gene regulatory mechanisms related to the identification and function of phenotypically distinct effector and memory T-cell subsets is necessary. Undoubtedly such studies will further clarify when memory cells are generated and how progressive changes in phenotype and function are obtained. Specifically, analysis of epigenetic modifications will provide a snapshot of the differentiation status of effector and memory T cells. Epigenetic profiling of antigen-specific CD4 and CD8 memory T cells will immediately benefit vaccine development as it will provide a novel parameter for identifying poised expression programmes aiding in the assessment of T-cell memory quality. As we better understand the intricacies of the mechanisms involved in acquiring and adapting transcriptional programmes, it may be possible to rationally modify established programmes, so providing a novel strategy for improving T-cell-based therapies against chronic infections.
Acknowledgments
This work was supported by the National Institutes of Health (NIH) grant P01 AI080192-01 (to R.A.), grant R37 AI30048-17 (to R.A.), grant AHMED05GCGH0 (to R.A.), Center for HIV/AIDS Vaccine Immunology and Immunogen Discovery UM1AI100663 (to R.A.), and post-doctoral fellowship F32 A1096709-01A1 (to J.S.H.).
Disclosures
The authors have no conflicts of interest to disclose.
References
- 1.Lau LL, Jamieson BD, Somasundaram T, Ahmed R. Cytotoxic T-cell memory without antigen. Nature. 1994;369:648–52. doi: 10.1038/369648a0. [DOI] [PubMed] [Google Scholar]
- 2.Ahmed R, Gray D. Immunological memory and protective immunity: understanding their relation. Science. 1996;272:54–60. doi: 10.1126/science.272.5258.54. (New York, NY) [DOI] [PubMed] [Google Scholar]
- 3.Bevan MJ, Goldrath AW. T-cell memory: you must remember this. Curr Biol. 2000;10:R338–40. doi: 10.1016/s0960-9822(00)00461-9. [DOI] [PubMed] [Google Scholar]
- 4.Doherty PC, Topham DJ, Tripp RA. Establishment and persistence of virus-specific CD4+ and CD8+ T cell memory. Immunol Rev. 1996;150:23–44. doi: 10.1111/j.1600-065x.1996.tb00694.x. [DOI] [PubMed] [Google Scholar]
- 5.Wherry EJ, Ahmed R. Memory CD8 T-cell differentiation during viral infection. J Virol. 2004;78:5535–45. doi: 10.1128/JVI.78.11.5535-5545.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ahmed R, Bevan MJ, Reiner SL, Fearon DT. The precursors of memory: models and controversies. Nat Rev Immunol. 2009;9:662–8. doi: 10.1038/nri2619. [DOI] [PubMed] [Google Scholar]
- 7.Kaech SM, Tan JT, Wherry EJ, Konieczny BT, Surh CD, Ahmed R. Selective expression of the interleukin 7 receptor identifies effector CD8 T cells that give rise to long-lived memory cells. Nat Immunol. 2003;4:1191–8. doi: 10.1038/ni1009. [DOI] [PubMed] [Google Scholar]
- 8.Kalia V, Sarkar S, Subramaniam S, Haining WN, Smith KA, Ahmed R. Prolonged interleukin-2Rα expression on virus-specific CD8+ T cells favors terminal-effector differentiation in vivo. Immunity. 2010;32:91–103. doi: 10.1016/j.immuni.2009.11.010. [DOI] [PubMed] [Google Scholar]
- 9.Sarkar S, Kalia V, Haining WN, Konieczny BT, Subramaniam S, Ahmed R. Functional and genomic profiling of effector CD8 T cell subsets with distinct memory fates. J Exp Med. 2008;205:625–40. doi: 10.1084/jem.20071641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaech SM, Hemby S, Kersh E, Ahmed R. Molecular and functional profiling of memory CD8 T cell differentiation. Cell. 2002;111:837–51. doi: 10.1016/s0092-8674(02)01139-x. [DOI] [PubMed] [Google Scholar]
- 11.Wherry EJ, Ha SJ, Kaech SM, et al. Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity. 2007;27:670–84. doi: 10.1016/j.immuni.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 12.Wirth TC, Xue HH, Rai D, Sabel JT, Bair T, Harty JT, Badovinac VP. Repetitive antigen stimulation induces stepwise transcriptome diversification but preserves a core signature of memory CD8+ T cell differentiation. Immunity. 2010;33:128–40. doi: 10.1016/j.immuni.2010.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goldrath AW, Sivakumar PV, Glaccum M, Kennedy MK, Bevan MJ, Benoist C, Mathis D, Butz EA. Cytokine requirements for acute and basal homeostatic proliferation of naive and memory CD8+ T cells. J Exp Med. 2002;195:1515–22. doi: 10.1084/jem.20020033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lefrancois L, Marzo AL. The descent of memory T-cell subsets. Nat Rev Immunol. 2006;6:618–23. doi: 10.1038/nri1866. [DOI] [PubMed] [Google Scholar]
- 15.Lefrancois L, Masopust D. T cell immunity in lymphoid and non-lymphoid tissues. Curr Opin Immunol. 2002;14:503–8. doi: 10.1016/s0952-7915(02)00360-6. [DOI] [PubMed] [Google Scholar]
- 16.Masopust D, Vezys V, Marzo AL, Lefrancois L. Preferential localization of effector memory cells in nonlymphoid tissue. Science. 2001;291:2413–7. doi: 10.1126/science.1058867. (New York, NY) [DOI] [PubMed] [Google Scholar]
- 17.Bevan MJ. Memory T cells as an occupying force. Eur J Immunol. 2011;41:1192–5. doi: 10.1002/eji.201041377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chappell C, Beard C, Altman J, Jaenisch R, Jacob J. DNA methylation by DNA methyltransferase 1 is critical for effector CD8 T cell expansion. J Immunol. 2006;176:4562–72. doi: 10.4049/jimmunol.176.8.4562. [DOI] [PubMed] [Google Scholar]
- 19.Youngblood B, Davis CW, Ahmed R. Making memories that last a lifetime: heritable functions of self-renewing memory CD8 T cells. Int Immunol. 2010;22:797–803. doi: 10.1093/intimm/dxq437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bachmann MF, Barner M, Viola A, Kopf M. Distinct kinetics of cytokine production and cytolysis in effector and memory T cells after viral infection. Eur J Immunol. 1999;29:291–9. doi: 10.1002/(SICI)1521-4141(199901)29:01<291::AID-IMMU291>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 21.Northrop JK, Thomas RM, Wells AD, Shen H. Epigenetic remodeling of the IL-2 and IFN-γ loci in memory CD8 T cells is influenced by CD4 T cells. J Immunol. 2006;177:1062–9. doi: 10.4049/jimmunol.177.2.1062. [DOI] [PubMed] [Google Scholar]
- 22.Kersh EN, Fitzpatrick DR, Murali-Krishna K, Shires J, Speck SH, Boss JM, Ahmed R. Rapid demethylation of the IFN-γ gene occurs in memory but not naive CD8 T cells. J Immunol. 2006;176:4083–93. doi: 10.4049/jimmunol.176.7.4083. [DOI] [PubMed] [Google Scholar]
- 23.Araki Y, Fann M, Wersto R, Weng NP. Histone acetylation facilitates rapid and robust memory CD8 T cell response through differential expression of effector molecules (eomesodermin and its targets: perforin and granzyme B) J Immunol. 2008;180:8102–8. doi: 10.4049/jimmunol.180.12.8102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Juelich T, Sutcliffe EL, Denton A, et al. Interplay between chromatin remodeling and epigenetic changes during lineage-specific commitment to granzyme B expression. J Immunol. 2009;183:7063–72. doi: 10.4049/jimmunol.0901522. [DOI] [PubMed] [Google Scholar]
- 25.Zediak VP, Johnnidis JB, Wherry EJ, Berger SL. Cutting edge: persistently open chromatin at effector gene loci in resting memory CD8+ T cells independent of transcriptional status. J Immunol. 2011;186:2705–9. doi: 10.4049/jimmunol.1003741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Denton AE, Russ BE, Doherty PC, Rao S, Turner SJ. Differentiation-dependent functional and epigenetic landscapes for cytokine genes in virus-specific CD8+ T cells. Proc Natl Acad Sci USA. 2011;108:15306–11. doi: 10.1073/pnas.1112520108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Youngblood B, Oestreich KJ, Ha SJ, et al. Chronic virus infection enforces demethylation of the locus that encodes PD-1 in antigen-specific CD8+ T cells. Immunity. 2011;35:13. doi: 10.1016/j.immuni.2011.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Araki K, Youngblood B, Ahmed R. The role of mTOR in memory CD8 T-cell differentiation. Immunol Rev. 2010;235:234–43. doi: 10.1111/j.0105-2896.2010.00898.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Araki K, Turner AP, Shaffer VO, Gangappa S, Keller SA, Bachmann MF, Larsen CP, Ahmed R. mTOR regulates memory CD8 T-cell differentiation. Nature. 2009;460:108–12. doi: 10.1038/nature08155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu J, Paul WE. CD4 T cells: fates, functions, and faults. Blood. 2008;112:1557–69. doi: 10.1182/blood-2008-05-078154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crotty S. Follicular helper CD4 T cells (TFH) Annu Rev Immunol. 2011;29:621–63. doi: 10.1146/annurev-immunol-031210-101400. [DOI] [PubMed] [Google Scholar]
- 32.Wilson CB, Rowell E, Sekimata M. Epigenetic control of T-helper-cell differentiation. Nat Rev Immunol. 2009;9:91–105. doi: 10.1038/nri2487. [DOI] [PubMed] [Google Scholar]
- 33.Heinzel FP, Sadick MD, Holaday BJ, Coffman RL, Locksley RM. Reciprocal expression of interferon-γ or interleukin 4 during the resolution or progression of murine leishmaniasis. Evidence for expansion of distinct helper T cell subsets. J Exp Med. 1989;169:59–72. doi: 10.1084/jem.169.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ouyang W, Kolls JK, Zheng Y. The biological functions of T helper 17 cell effector cytokines in inflammation. Immunity. 2008;28:454–67. doi: 10.1016/j.immuni.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Avni O, Lee D, Macian F, Szabo SJ, Glimcher LH, Rao A. TH cell differentiation is accompanied by dynamic changes in histone acetylation of cytokine genes. Nat Immunol. 2002;3:643–51. doi: 10.1038/ni808. [DOI] [PubMed] [Google Scholar]
- 36.Lee DU, Agarwal S, Rao A. Th2 lineage commitment and efficient IL-4 production involves extended demethylation of the IL-4 gene. Immunity. 2002;16:649–60. doi: 10.1016/s1074-7613(02)00314-x. [DOI] [PubMed] [Google Scholar]
- 37.Schoenborn JR, Dorschner MO, Sekimata M, Santer DM, Shnyreva M, Fitzpatrick DR, Stamatoyonnapoulos JA, Wilson CB. Comprehensive epigenetic profiling identifies multiple distal regulatory elements directing transcription of the gene encoding interferon-γ. Nat Immunol. 2007;8:732–42. doi: 10.1038/ni1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nurieva RI, Chung Y, Hwang D, et al. Generation of T follicular helper cells is mediated by interleukin-21 but independent of T helper 1, 2, or 17 cell lineages. Immunity. 2008;29:138–49. doi: 10.1016/j.immuni.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu KT, Kanno Y, Cannons JL, et al. Functional and epigenetic studies reveal multistep differentiation and plasticity of in vitro-generated and in vivo-derived follicular T helper cells. Immunity. 2011;35:622–32. doi: 10.1016/j.immuni.2011.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Agarwal S, Rao A. Modulation of chromatin structure regulates cytokine gene expression during T cell differentiation. Immunity. 1998;9:765–75. doi: 10.1016/s1074-7613(00)80642-1. [DOI] [PubMed] [Google Scholar]
- 41.Makar KW, Perez-Melgosa M, Shnyreva M, Weaver WM, Fitzpatrick DR, Wilson CB. Active recruitment of DNA methyltransferases regulates interleukin 4 in thymocytes and T cells. Nat Immunol. 2003;4:1183–90. doi: 10.1038/ni1004. [DOI] [PubMed] [Google Scholar]
- 42.Makar KW, Wilson CB. DNA methylation is a nonredundant repressor of the Th2 effector program. J Immunol. 2004;173:4402–6. doi: 10.4049/jimmunol.173.7.4402. [DOI] [PubMed] [Google Scholar]
- 43.Josefowicz SZ, Wilson CB, Rudensky AY. Cutting edge: TCR stimulation is sufficient for induction of Foxp3 expression in the absence of DNA methyltransferase 1. J Immunol. 2009;182:6648–52. doi: 10.4049/jimmunol.0803320. [DOI] [PubMed] [Google Scholar]
- 44.Gamper CJ, Agoston AT, Nelson WG, Powell JD. Identification of DNA methyltransferase 3a as a T cell receptor-induced regulator of Th1 and Th2 differentiation. J Immunol. 2009;183:2267–76. doi: 10.4049/jimmunol.0802960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thomas RM, Gamper CJ, Ladle BH, Powell JD, Wells AD. De novo DNA methylation is required to restrict T helper lineage plasticity. J Biol Chem. 2012;287:22900–9. doi: 10.1074/jbc.M111.312785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harrington LE, Janowski KM, Oliver JR, Zajac AJ, Weaver CT. Memory CD4 T cells emerge from effector T-cell progenitors. Nature. 2008;452:356–60. doi: 10.1038/nature06672. [DOI] [PubMed] [Google Scholar]
- 47.Pepper M, Pagan AJ, Igyarto BZ, Taylor JJ, Jenkins MK. Opposing signals from the Bcl6 transcription factor and the interleukin-2 receptor generate T helper 1 central and effector memory cells. Immunity. 2011;35:583–95. doi: 10.1016/j.immuni.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mohrs M, Shinkai K, Mohrs K, Locksley RM. Analysis of type 2 immunity in vivo with a bicistronic IL-4 reporter. Immunity. 2001;15:303–11. doi: 10.1016/s1074-7613(01)00186-8. [DOI] [PubMed] [Google Scholar]
- 49.Stetson DB, Mohrs M, Mallet-Designe V, Teyton L, Locksley RM. Rapid expansion and IL-4 expression by Leishmania-specific naive helper T cells in vivo. Immunity. 2002;17:191–200. doi: 10.1016/s1074-7613(02)00363-1. [DOI] [PubMed] [Google Scholar]
- 50.Zaph C, Rook KA, Goldschmidt M, Mohrs M, Scott P, Artis D. Persistence and function of central and effector memory CD4+ T cells following infection with a gastrointestinal helminth. J Immunol. 2006;177:511–8. doi: 10.4049/jimmunol.177.1.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pepper M, Linehan JL, Pagan AJ, Zell T, Dileepan T, Cleary PP, Jenkins MK. Different routes of bacterial infection induce long-lived TH1 memory cells and short-lived TH17 cells. Nat Immunol. 2010;11:83–9. doi: 10.1038/ni.1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wherry EJ, Blattman JN, Murali-Krishna K, van der Most R, Ahmed R. Viral persistence alters CD8 T-cell immunodominance and tissue distribution and results in distinct stages of functional impairment. J Virol. 2003;77:4911–27. doi: 10.1128/JVI.77.8.4911-4927.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xiong Y, Luscher MA, Altman JD, Hulsey M, Robinson HL, Ostrowski M, Barber BH, MacDonald KS. Simian immunodeficiency virus (SIV) infection of a rhesus macaque induces SIV-specific CD8+ T cells with a defect in effector function that is reversible on extended interleukin-2 incubation. J Virol. 2001;75:3028–33. doi: 10.1128/JVI.75.6.3028-3033.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brahmer JR, Tykodi SS, Chow LQ, et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med. 2012;366:2455–65. doi: 10.1056/NEJMoa1200694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Topalian SL, Hodi FS, Brahmer JR, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012;366:2443–54. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Barber DL, Wherry EJ, Masopust D, Zhu B, Allison JP, Sharpe AH, Freeman GJ, Ahmed R. Restoring function in exhausted CD8 T cells during chronic viral infection. Nature. 2006;439:682–7. doi: 10.1038/nature04444. [DOI] [PubMed] [Google Scholar]
- 57.Sharpe AH, Wherry EJ, Ahmed R, Freeman GJ. The function of programmed cell death 1 and its ligands in regulating autoimmunity and infection. Nat Immunol. 2007;8:239–45. doi: 10.1038/ni1443. [DOI] [PubMed] [Google Scholar]
- 58.Doering TA, Crawford A, Angelosanto JM, Paley MA, Ziegler CG, Wherry EJ. Network analysis reveals centrally connected genes and pathways involved in CD8+ T cell exhaustion versus memory. Immunity. 2012;37:1130–44. doi: 10.1016/j.immuni.2012.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Battegay M, Moskophidis D, Rahemtulla A, Hengartner H, Mak TW, Zinkernagel RM. Enhanced establishment of a virus carrier state in adult CD4+ T-cell-deficient mice. J Virol. 1994;68:4700–4. doi: 10.1128/jvi.68.7.4700-4704.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Matloubian M, Concepcion RJ, Ahmed R. CD4+ T cells are required to sustain CD8+ cytotoxic T-cell responses during chronic viral infection. J Virol. 1994;68:8056–63. doi: 10.1128/jvi.68.12.8056-8063.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zajac AJ, Blattman JN, Murali-Krishna K, Sourdive DJ, Suresh M, Altman JD, Ahmed R. Viral immune evasion due to persistence of activated T cells without effector function. J Exp Med. 1998;188:2205–13. doi: 10.1084/jem.188.12.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen H, Ndhlovu ZM, Liu D, et al. TCR clonotypes modulate the protective effect of HLA class I molecules in HIV-1 infection. Nat Immunol. 2012;13:691–700. doi: 10.1038/ni.2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.West EE, Youngblood B, Tan WG, et al. Tight regulation of memory CD8+ T cells limits their effectiveness during sustained high viral load. Immunity. 2011;35:285–98. doi: 10.1016/j.immuni.2011.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shin H, Blackburn SD, Blattman JN, Wherry EJ. Viral antigen and extensive division maintain virus-specific CD8 T cells during chronic infection. J Exp Med. 2007;204:941–9. doi: 10.1084/jem.20061937. [DOI] [PMC free article] [PubMed] [Google Scholar]


