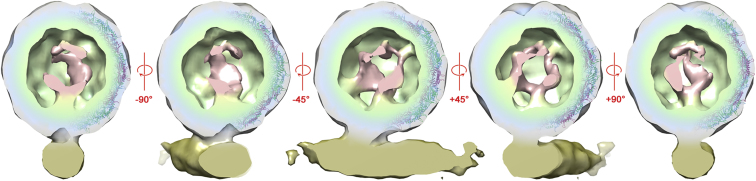
Summary
Simple, spherical RNA viruses have well-understood, symmetric protein capsids, but little structural information is available for their asymmetric components, such as minor proteins and their genomes, which are vital for infection. Here, we report an asymmetric structure of bacteriophage MS2, attached to its receptor, the F-pilus. Cryo-electron tomography and subtomographic averaging of such complexes result in a structure containing clear density for the packaged genome, implying that the conformation of the genome is the same in each virus particle. The data also suggest that the single-copy viral maturation protein breaks the symmetry of the capsid, occupying a position that would be filled by a coat protein dimer in an icosahedral shell. This capsomere can thus fulfill its known biological roles in receptor and genome binding and suggests an exit route for the genome during infection.
Highlights
-
•
The asymmetric structure of a virus receptor complex is described
-
•
The density for ordered genomic RNA was observed in the structure
-
•
Viral maturation protein was visualized
Dent et al. describe how the asymmetric structure of a single-stranded RNA virus bound to its receptor shows a unique feature in the otherwise highly symmetric protein capsid that protects its genome. This suggests a model for how the genome could leave its protective container during infection.
Introduction
Many simple viruses exploit high symmetry to build protective capsids from the minimum number of genes (Crick and Watson, 1956; Hodgkin, 1950). Selective evolutionary pressures maximize both genetic economy and packaging capacity, resulting in capsids with the highest symmetry that can be built from a single, or very few, component(s): icosahedral symmetry. Even larger containers can be built if the viral coat protein (CP) is able to adopt different conformations, as described in the theory of quasi-equivalence (Caspar and Klug, 1962).
Structural biology has taught us much about how icosahedral symmetry is realized in three-dimensional (3D) space in viruses (for a review, see Abrescia et al., 2012; Rossmann and Johnson, 1989). In almost all such studies, using both X-ray diffraction and cryo-electron microscopy (cryo-EM), icosahedral symmetry averaging was applied to produce the final structure. This might not appear to be a problem until we recall that at least one component of all viruses is asymmetric: their genome. Many viruses also incorporate single-copy proteins that function as movement, maturation, or infectivity proteins. Applying icosahedral symmetry averages away details of such asymmetric features and thus obscures vital aspects of virus biology. This is perhaps most clearly shown in the structures of large double-stranded DNA (dsDNA) viruses (Cherrier et al., 2009; Xiao et al., 2009), which were at first thought to be icosahedral but which are now known to have unique vertices (Xiao and Rossmann, 2011). However, it is extremely difficult to avoid such averaging. In cryo-EM data, asymmetric features contribute to the images of virions and could be used to determine a structure without symmetry averaging. So far, however, this has only been regularly achieved for dsDNA viruses where the asymmetry is very large, overcoming the effects of the poor contrast in EM images. This has been particularly successful for tailed bacteriophages (Jiang et al., 2006; Lander et al., 2006; Morais et al., 2001). Such viruses incorporate a special vertex that is occupied by the genome packaging motor used during assembly and subsequently by the portal and tail complexes that allow attachment to hosts and genome release during early events in the next infection. This feature is shared with herpes viruses (Cardone et al., 2007). The tails, or large special vertices, can be used as fiduciary markers allowing asymmetric structure determination using single-particle methods. This has yielded unique insights into the biology of these organisms (reviewed in Johnson and Chiu, 2007).
In contrast, positive-sense, single-stranded (+ss)RNA viruses, one of the largest classes of viral pathogens, are often described as lacking such special vertices and forming complete protein shells with icosahedral surface lattices (Harrison et al., 1978; Rossmann and Johnson, 1989). Here we report the asymmetric structure of such a virus, bacteriophage MS2, bound to its receptor the E. coli F-pilus (Brinton et al., 1964; Valentine and Strand, 1965). MS2 has a T = 3 morphology, an architecture that requires the single CP to adopt three quasi-equivalent conformations (Caspar and Klug, 1962). The building block of the MS2 capsid is a CP dimer, and two distinct types of this dimer are found in the capsid; an asymmetric (A/B) dimer, and a symmetric (C/C) dimer (Figure 1A) (Valegård et al., 1990). The A/B dimers extend from the 5-fold axes to the 3-fold axes, while the C/C-type dimers sit on the 2-fold axes. MS2 encapsidates an ∼3.6 kb, +ssRNA genome that encodes four gene products: maturation protein (MP), CP, and lysis and replicase proteins (Figure 1B) (Fiers et al., 1976). The genomic RNA is not resolved in the crystal structure, a result that was thought to arise from a lack of order in its packing. More recently, however, cryo-EM reconstruction using icosahedral symmetry averaging has shown that the genome is extensively ordered at intermediate resolution (Koning et al., 2003; Toropova et al., 2008), a discrepancy explained by the fact that, in early X-ray diffraction studies, the low-resolution data needed to resolve imperfectly ordered material were rarely recorded (Tsuruta et al., 1998). Cryo-EM shows the genomic RNA as two concentric shells of density (Toropova et al., 2008). The first (at higher radius) is intimately associated with the inner surface of the CP shell, which presents a lattice of RNA binding sites (Valegård et al., 1994). This is important because RNA binding promotes an allosteric shift in conformation from a symmetric, C/C-like dimer to an asymmetric A/B-like conformation; i.e., the genome specifies the quasi-equivalent conformation of its own CP (Stockley et al., 2007; Dykeman et al., 2010). The second shell (at lower radius) corresponds to RNA not bound to the CP layer (Rolfsson et al., 2010) and appears to be ordered by steric and/or electrostatic interactions, including contacts to the MP.
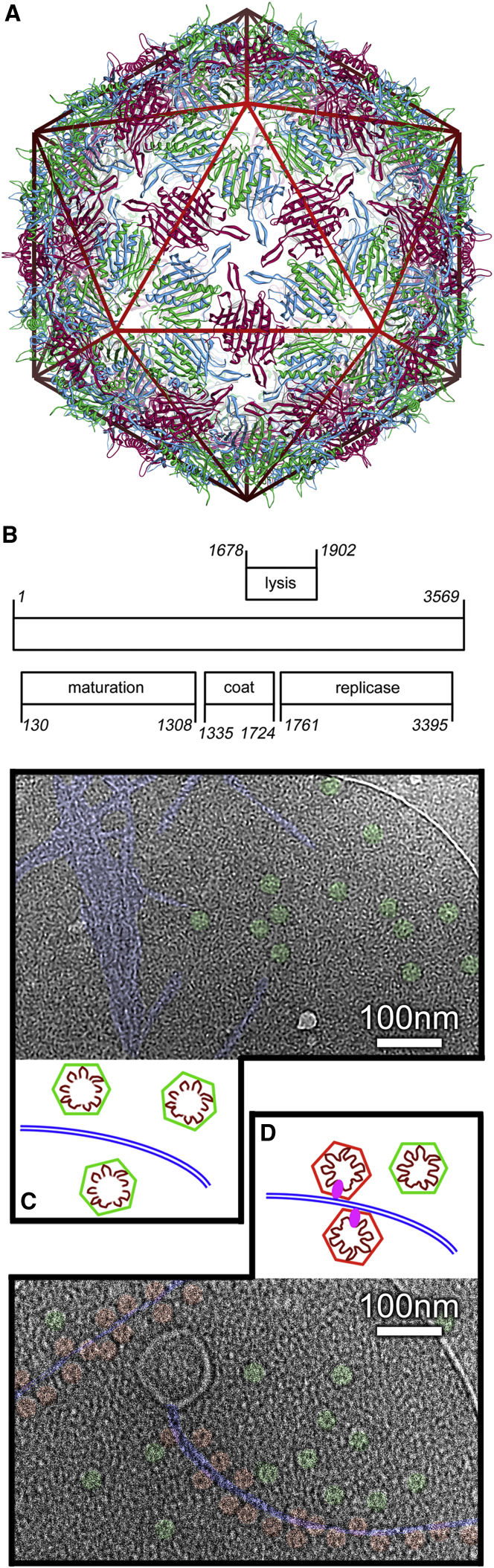
Figure 1.
Bacteriophage MS2
(A) The X-ray structure of MS2 at 3.0 Å resolution in cartoon representation (Golmohammadi et al., 1993). The T = 3 capsid contains a single type of CP subunit found in three quasi-equivalent conformations: A, B, and C (blue, green, and red respectively). The virion has a diameter of ∼285 Å, and no density for the ∼3.6 kb RNA genome is resolved.
(B) The genomic RNA encodes four gene products: MPs, CPs, and lysis and replicase proteins.
(C) A cryo-EM image of the isolated pili incubated with recombinant MS2 VLPs that lack MP. No binding of the VLPs is seen, and isolated F-pili clump into bundles.
(D) A cryo-EM image of the same pilus preparation incubated with wild-type MS2. Heavy decoration of the pili is observed. (C) and (D) are colored to show the pili (blue), bound MS2 (pink), and unbound MS2 (green). All structural figures were prepared using UCSF Chimera (Pettersen et al., 2004).
Despite our understanding of the structure and assembly of MS2, the molecular details of how it infects its host remain poorly understood. A single copy of the MP is present in wild-type (wt) MS2 virions, which bind to F-pili, while particles lacking MP lack infectivity and do not bind to pili (Krahn et al., 1972; Roberts and Steitz, 1967). Indeed, mutations in MP abolish infectivity and prevent interaction with the pilus (Lodish et al., 1965). The MP is thus thought to mediate the interaction between the virus and pilus. To test this, we imaged the same preparation of isolated F-pili with either recombinant MS2 virus-like particles (VLPs), which lack MP (Figure 1C), or wt MS2 virions, most of which contain it (Roberts and Steitz, 1967) (Figure 1D), using cryo-EM. Although these experiments were performed in identical solution conditions, it was extremely difficult to image F-pili in the presence of recombinant MS2 VLPs, as the pili aggregate into large bundles, to which the MS2 VLPs do not bind and are thus excluded. By contrast, wt MS2 particles bind the F-pili readily, helping to solubilize them from the bundles. This experiment therefore reinforces the key role of MP in receptor recognition. Infection proceeds by release of the complex between the genome and MP from the capsid (Kozak and Nathans, 1971; Krahn et al., 1972) but only when the pilus is attached to a living cell (Danziger and Paranchych, 1970). This complex enters the cell by a currently unknown pathway requiring divalent cations (Paranchych, 1966), leaving empty but essentially intact capsids outside the bacterium. During internalization, MP is proteolytically cleaved into two fragments (Krahn et al., 1972) allowing the viral RNA to begin its programmed expression of phage proteins. The complexes imaged here therefore represent the earliest point in an infection cycle.
All structural studies to date used icosahedral symmetry averaging and so describe MS2 as a perfect icosahedral assembly. However, this must be an oversimplification because, as discussed earlier, infectivity requires the incorporation of two asymmetric features: a single copy of the MP and the genome. As well as binding pilus, the MP binds to sequences close to the 5′ and 3′ ends of the genomic RNA (Nathans et al., 1966; Shiba and Suzuki, 1981), hence circularizing it. These are very different roles necessitating very different locations within the virion. Binding the genome requires that part of the MP be inside the capsid, while binding pilus requires a part of it to be surface exposed. Since it is a small (44 kDa) asymmetric feature in a large structure (∼2.5 MDa) to which icosahedral symmetry averaging has been applied, it has not been seen in any previous structural study.
Imaging the MP is complicated owing to its extreme insolubility in isolation (Roberts and Steitz, 1967), which has made working with MP challenging. To address this lack of structural information, we sought to determine the 3D structure of the MS2-pilus complex, reasoning that this would allow us to generate an asymmetric density distribution in which details of the MP’s location and organization would be apparent. Using cryo-electron tomography and subtomographic averaging, we show that the RNA genome is packaged in a defined orientation within the receptor-bound virus capsid, in stark contrast to prevailing ideas that RNA encapsidation is often nonspecific (Belyi and Muthukumar, 2006). In addition, we show that the contact point between the virus and pilus suggests that the MP is an integral structural component of the capsid. We propose that the MP replaces a CP dimer in the capsid, spanning the thickness of the protein shell, consistent with its known roles in receptor and genome binding. The MS2 capsid is thus asymmetric, in contrast to previous structural studies in which symmetry averaging was applied.
Results
The starting point for our study was purified F-pili and MS2 virions. Since the pili are not attached to E. coli, having been sheared off the cells that expressed them during purification, this enables us to dissect the initial binding of virus to receptor from any downstream structural rearrangement. Multiple MS2 particles bind to the sides of each pilus, allowing us to image large numbers of attached virions simultaneously. We incubated MS2 with purified pili, together with 10 nm colloidal gold, and vitrified the resulting complexes. Tomographic tilt series were then collected in the electron microscope. Owing to the need to minimize radiation damage, electron doses must be strictly limited, and the resulting data have an extremely poor signal-to-noise (S/N) ratio (Figure 2A). The contrast is thus extremely low, essentially making tilt series alignment using features within the images impossible. All tilt series were therefore aligned based on the positions of the fiducial gold markers. A 3D tomographic reconstruction was then calculated from the aligned tilt series by back projection. Shown in Figure 2B (see also Movie S1 available online) is a slice through the 3D reconstruction calculated from the tilt series in Figure 2A, together with a 3D model of all the pilus-attached virus particles in the 3D volume (Figures 2C and 2D).
Figure 2.
Electron Tomography of MS2-Decorated F-Pili
(A) The untilted (0°) image from a tilt series of an F-pilus heavily decorated with MS2. To prevent radiation damage, very low doses are used (∼1 e−/Å2 per tilt); thus, contrast is very poor. The dark black dots are 10 nm gold particles used for alignment.
(B) A section through the tomographic reconstruction of the sample in (A).
(C and D) 3D models of the pili (tubes) and MS2 particles (spheres) in (A) and (B).
See also Movie S1.
Although contrast is much improved in the tomograms, their resolution is not sufficient to allow us to interpret the molecular details of the virus-receptor interaction directly. This is especially so because our limited ability to tilt EM specimens leads to a substantial “missing wedge” of information in the resulting 3D reconstruction, and structural distortion. However, each of our tomographic reconstructions in principle contains many copies of identically bound virus particles that can be averaged together (see Frank, 1992). This brings two enormous benefits. First, because the virus decorates the pilus in a full range of azimuthal orientations, the missing wedge in each is in a different position. Averaging particles together therefore fills in this missing wedge. Second, it increases the S/N ratio. Together, these two factors dramatically increase the resolution of our map, allowing us to validate our structure against other structural information and increase its biological interpretability. It is important to note that the averaging we apply is of repeating examples of a single asymmetric virus-receptor complex rather than the symmetry averaging usually used in virus structure determination.
We performed such a subtomographic averaging experiment on the pilus-attached MS2 particles, using a starting model derived from the data by averaging particles along a single pilus (see Experimental Procedures). It was apparent during subtomographic averaging that an alignment of the particles based on the entire structure (virus and pilus) resulted in a poor map, presumably owing to flexibility in the interaction between the two (Toropova et al., 2011). We therefore applied a spherical mask to force the alignment to ignore the pilus (other than as a constraint on orientation) and focus on features in the capsid itself. The resulting asymmetric reconstruction of the pilus-bound MS2 virion at ∼39 Å resolution is shown in Figure 3 (see also Figure S1).
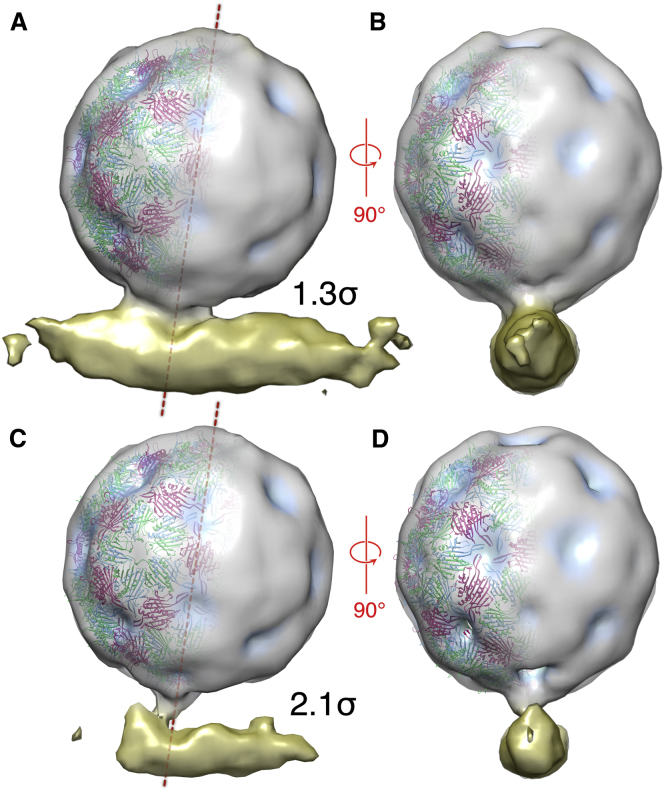
Figure 3.
Asymmetric Structure of a Virus-Receptor Complex
(A) A view perpendicular to the long axis of the pilus. MS2 is shown as a radially colored density at 1.3 σ; the surface of the virion is gray, but depressions in that surface are indicated by a blue coloration. The pilus is gold. The fitted X-ray structure for MS2, colored as in Figure 1, is shown on the left of the virion. MS2 binds to pilus at a 9° angle indicated by the dashed line through the center of the particle.
(B) A view rotated by 90° to look down the long axis of the pilus.
(C and D) The same views as in (A) and (B) but at a higher contour level (2.1 σ), at which the connection between virion and pilus is only just maintained.
See also Figure S1.
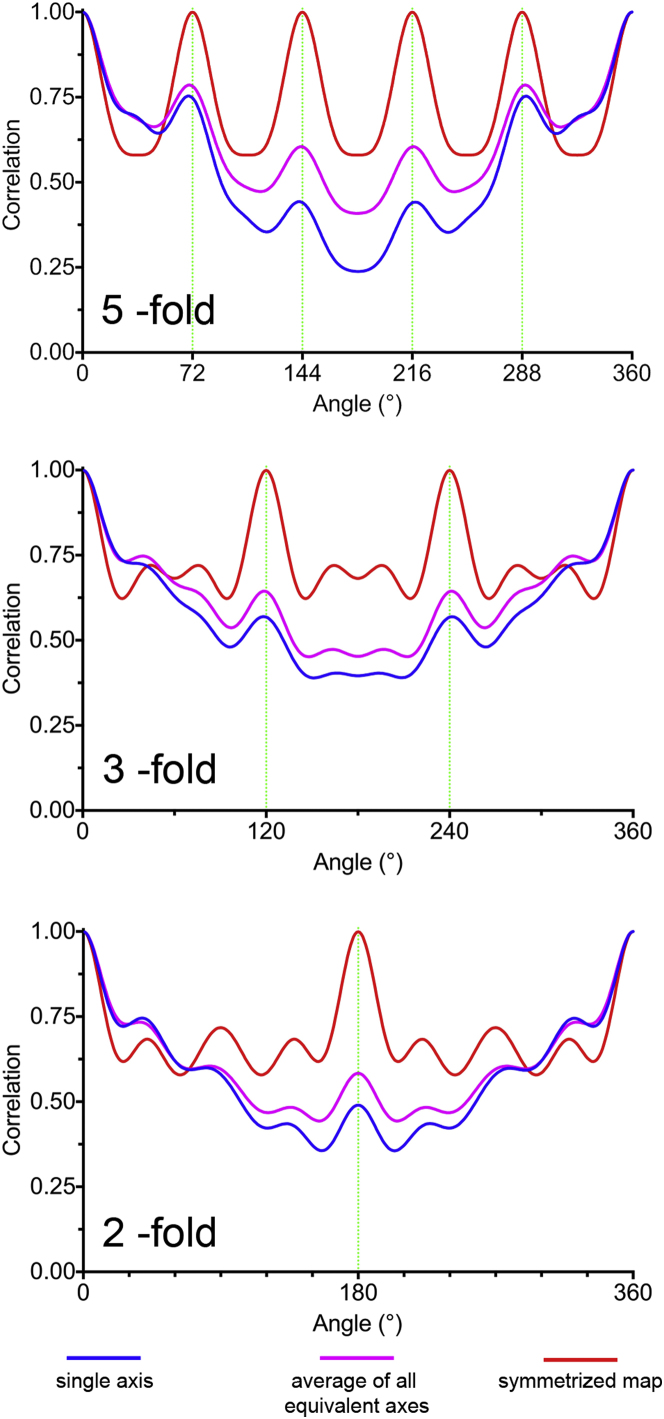
The validation of the asymmetric structure of MS2 depends critically on the many icosahedrally averaged structures available for this virus. Despite the fact that no symmetry averaging has been applied, the receptor-bound virus has the faceted, symmetric appearance of an icosahedral object. This implies that receptor-bound MS2 is predominantly a symmetric structure that is accurately described by previous X-ray structures. To confirm this, we calculated self-rotation functions for our electron density map at the positions where we would expect to find icosahedral symmetry axes in a symmetrized map. These confirm that the particle has approximate 5-fold, 3-fold, and 2-fold symmetry despite the lack of such symmetry being imposed (Figure 4). It is also confirmed visually by the very small change in appearance when the density is icosahedrally averaged (see Figure S1). Given this, we fitted such a structure (2ms2; Golmohammadi et al., 1993) into our map. Visually the X-ray model fits the electron density of the capsid well. The virus capsid is bound with a small tilt (of ∼9°, as indicated by the dashed line in Figures 3A and 3C), consistent with our previous observations (Toropova et al., 2011). The structure of MS2 has distinctive pores (∼15 Å in diameter) at both the icosahedral 5-fold and 3-fold axes. At the resolution of this study, these are seen as depressions in the surface of the map rather than as pores. The depressions and the facets that contain them are located correctly. The asymmetric electron density thus reveals the symmetric features of the capsid when bound to its receptor. Given this, in the following discussion, we follow the convention of describing structural features with respect to a generalized position in an icosahedrally averaged lattice, i.e., “at a 2-fold axis” or “around a 5-fold axis.” However, no such averaging has been applied, and each position is different.
Figure 4.
The Asymmetric Structure Has Icosahedral Symmetry
Rotation functions showing the symmetry around what would be 5-fold, 3-fold, and 2-fold positions. For each type of icosahedral symmetry axis, a representative rotation function for one axis (in blue), the average of all such symmetry axes in the asymmetric reconstruction (in magenta), and the symmetrized results (in red) are shown.
The idea that MS2 is a symmetric structure is profoundly misleading. When we look inside the virus capsid, by clipping away the front half of the structure, the asymmetry of the receptor-bound virion is revealed (Figure 5; Movies S2–S5). Given the good agreement with the symmetrical model, we can assign the asymmetric data to a protein shell surrounding density corresponding to the genome and the MP. Note that the MS2 CP does not contain extended polypeptide arms and so is entirely restricted to this outer protein shell. There is substantial non-CP density that must correspond mostly to viral RNA (∼1.2 MDa) rather than MP (∼44 kDa). Part of this density forms a shell located immediately beneath the CP layer, presumably corresponding to the CP-bound RNA seen in structures of the virus, and of VLPs lacking MP (green in Figure 5). The density at lower radii (pink in Figure 5) is discontinuous but, when symmetry averaging is applied, corresponds well with the second shell seen in previous symmetry-averaged reconstructions (data not shown) (Dykeman et al., 2011; Toropova et al., 2008, 2011). These results therefore provide further validation for our interpretation of the experimental density.
Figure 5.
Internal Density Suggests that the Genome Is Packaged in a Defined Conformation
In the center is the same view shown in Figure 3A (at 1.3 σ) but with the front half of the structure clipped away to show internal density. The MS2-pilus complex and the MS2 X-ray structure (right-hand side of each view) are shown and colored as in Figure 3. A layer of density (in green) in close association with the CP shell corresponds to CP-bound genomic RNA, while the discontinuous density (in pink) corresponds to the inner shell of RNA seen in previous icosahedrally averaged cryo-EM structures (Rolfsson et al., 2010; Toropova et al., 2008).
This density for the genome has been reinforced by the averaging of 1,500 individual 3D reconstructions of the MS2-receptor complex. We suggest that this is consistent with the genomic RNA being packaged within MS2 virions in a single conformation (or more conservatively, an ensemble of closely related conformations that are indistinguishable at this resolution) with respect to the MP. The bridges that link the inner (pink) to outer (green) density all arrive at positions beneath the A/B dimers, consistent with the locations of the density linking shells in the icosahedrally averaged cryo-EM reconstruction (Figure 5) (Dykeman et al., 2011; Toropova et al., 2008). It is thus consistent with our current models of capsid assembly where the RNA follows a path around the outer shell, presenting stem-loop structures (packaging signals) to CP dimers, thus facilitating the switching of quasi-equivalent conformations necessary to build a T = 3 capsid (Basnak et al., 2010; Morton et al., 2010a; Rolfsson et al., 2010; Stockley et al., 2007; Dykeman et al., 2013a).
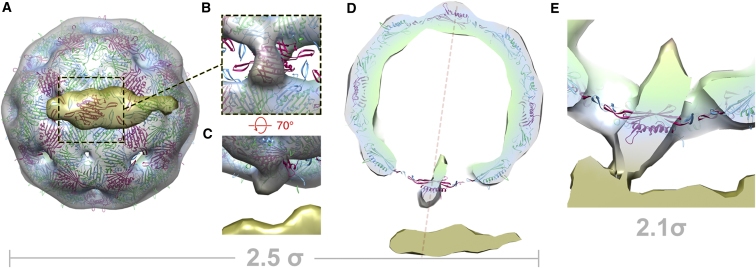
The most obvious example of asymmetry in the virus-receptor complex is of course the interaction with the receptor itself. Our previous studies suggested that this interaction might occur close to one of the 5-fold vertices of the virion (Hill et al., 1997; Toropova et al., 2011). However, the current work overturns this hypothesis (Figure 6; Movie S6). Shown in Figure 6A is a view through the pilus on to the viral capsid, which locates the interaction as occurring at what would be an icosahedral 2-fold axis, i.e., at the position normally associated with a dimer in the C/C quasi-conformation. This discrepancy is explained by the small size of the MS2 particle and the resulting close proximity of the symmetry axes in our previous analysis, together with the flexibility in the interaction between pilus and virus described earlier. The location of MP at a 2-fold axis is surprising because there is no pore in the capsid at this position, and it is difficult to understand how MP could span the capsid layer. If it replaces a CP dimer (or CP subunit), it could easily fulfill its known biological functions. The presence of MP in the CP lattice would thus break the icosahedral symmetry of the capsid. The asymmetric structure presented here provides direct structural evidence to support such a model. Shown in Figure 6B is a close-up of the same view of the virus-receptor complex as in Figure 6A but with the front clipping plane altered to exclude the pilus and reveal the structure of the capsid at the point of interaction. This reveals that the density at the point of attachment is very different from all the other 2-fold axes. The electron density at this point fits a C/C dimer poorly (Figure 6B), the only such point in the capsid where this is so (i.e., one out of 30 equivalent positions). This density extends beyond the CP layer, projecting both toward the pilus and the center of the viral particle (Figure 6C). In the figure, the C/C dimer shown is positioned by virtue of a global fit of an icosahedrally averaged X-ray structure. The discrepancy between this model and the density is even more apparent when viewed from the side (Figures 6D and 6E): The long axis of the C/C dimer (magenta) is aligned to complete the symmetry-averaged protein shell, but the electron density is both larger than a C/C dimer and oriented in an almost orthogonal direction. Furthermore, the pores in the density map on either side of the attachment site are much larger than those found elsewhere, implying a general perturbation of the structure in that region. We suggest that the most plausible explanation for this density is that MP occupies the position normally associated with a CP dimer in the C/C conformation.
Figure 6.
The MP Replaces a CP Dimer
(A) A view through the pilus on to the capsid beneath at 2.5 σ.
(B) A close-up from an identical viewpoint, but with the pilus density clipped away, showing the point of interaction. This position is occupied by a C/C-type dimer on a 2-fold axis in the icosahedrally averaged structure.
(C) An oblique view of the same point from the side, showing that density at this point is different from all other 29 equivalent positions.
(D) A 25-Å-thick section through the complex (the view in Figure 3A) showing the coordinates for a C/C dimer and their poor fit to density that we ascribe to MP. The disruption of the capsid structure around this point is indicated by the large pores at either side.
(E) A close-up view of the interaction between virus and pilus. The section is thicker (35 Å) and at a lower contour level (2.1 σ) than in (D) to show connected density between pilus and virus.
See also Movie S6.
Discussion
The results presented here fundamentally change our view of the structure of MS2. They represent an extension to all previous structural studies in this phage and presumably those of other RNA phages (Golmohammadi et al., 1996; Plevka et al., 2009; Tars et al., 1997, 2000). The asymmetric structure provides independent confirmation, with no prior assumptions, that the phage capsid is based upon a T = 3 icosahedral lattice. The additional insights arise from the ability to see, at any resolution, components of the virus that were previously invisible, namely the genome and the MP. Our interpretation of these structures is facilitated by the many previous X-ray and cryo-EM structures, and significantly by extensive biochemical and genetic studies.
Symmetry has been an essential feature of our understanding of virus structures since the realization that their limited coding capacity implied that capsids had to be built from repeating subunits (Hodgkin, 1950). This idea was reinforced by the proof that spherical viruses have icosahedral symmetry (Crick and Watson, 1956; Caspar, 1956) and was later refined by the theory of quasi-equivalence (Caspar and Klug, 1962) that accounts for capsid lattices made up of flexible proteins. These ideas were vindicated when Tomato Bushy Stunt virus was shown to have T = 3 geometry (Winkler et al., 1977). X-ray structures of other plant viruses (Abad-Zapatero et al., 1980; Liljas et al., 1982) and then animal viruses (Rossmann et al., 1985; see also Hogle et al., 1985) followed, starting an explosion in structural virology that has included the discovery of “nonallowed” triangulation numbers (Grimes et al., 1998), all pentamer capsids (Liddington et al., 1991; Rayment et al., 1982), and even the structures of capsids that include ordered membrane (Cockburn et al., 2004). In each case, symmetry averaging was applied, assuming that spherical viruses are icosahedral. More recently, advances in cryo-EM have permitted the determination of the structures of RNA viruses at resolutions that almost equal X-ray crystallography (Zhang et al., 2008, 2010), but again with icosahedral averaging. It is hard to overstate the biological insights that such work has provided. However, for the nonspecialist, it has had the effect of emphasizing symmetric features at the expensive of the asymmetric conformations of their genomes and minor protein components, which are crucial for function.
For MS2, our results show that this symmetry-dominated view is incorrect and fails to describe features essential for the viral life cycle. MS2 is asymmetric, both in the genome it protects, which is packaged in a single conformation, and in the protective protein layer itself, which contains a unique feature. Many other simple RNA viruses incorporate unique features. Virions of the Tombusviridae (such as TBSV) appear to contain a unique covalent dimer of their regular CP (Stockley et al., 1986), while picornaviruses often contain an uncleaved precursor protein (VP0) (Dunker and Rueckert, 1971). If these components are located within the CP shell, then they are equivalent to the packaged genome and are not routinely seen in averaged structures, and this includes extended arms of the CP subunits if present. Even if they replace one of the CP subunits in the protein shell, the statistics of averaging make it inevitable that such features will be lost in the final density map. For MS2, replacing a CP dimer with MP means that 1 dimer out of 90, or only ∼1% (∼1.8% by mass), of the capsid is changed.
The interpretation of the asymmetric structure that we describe here is independently validated by a wide range of biochemical observations. MS2 is a T = 3 capsid, and we have determined the mechanism that controls the formation of the dimeric quasi-conformers during its assembly (Basnak et al., 2010; Morton et al., 2010b; Stockley et al., 2007). In the absence of RNA, the CP dimer is symmetric (C/C-like; Figure 1) on the nuclear magnetic resonance time scale. Binding of an RNA stem loop triggers a conformational change to an A/B-like structure via dynamic allostery (Dykeman et al., 2010; Morton et al., 2010a). This effect is not sequence specific. This allows the multiple switching events (up to 60) needed to complete the T = 3 shell to occur with a genomic RNA that encodes a single high affinity binding site (Basnak et al., 2010; Rolfsson et al., 2010) but many other stem loop structures that could fulfill a similar function (Dykeman et al., 2013a). It appears that, as well as encoding viral proteins, the genome presents multiple packaging signals as stem loops. Recent single molecule fluorescence assays of assembly confirm this picture of assembly as a two-stage processes, the first of which is the rapid formation of a compacted, grand initiation complex in which multiple packaging signals have bound CPs (Borodavka et al., 2012, 2013). One consequence of the repeated CP-RNA interactions is that the genome follows a path around the inside surface of the capsid, where it is responsible for the switching of CP conformation. The consequence of this is that the genome must visit each A/B dimer position once and only once. Such a connected path, termed a Hamiltonian path in mathematics, is significantly facilitated by the action of the MP (Dykeman et al., 2011, 2013b). Binding to sites near its 5′ and 3′ ends, MP effectively circularizes the genomic RNA, dramatically reducing the combinatorial complexity of assembly. The asymmetric structure is fully consistent with these data that, in fact, predict a defined conformation of the encapsidated genome with respect to the MP (Dykeman et al., 2011, 2013a; Toropova et al., 2011). Note that assembly in the absence of MP is possible both in vitro (Rolfsson et al., 2010; see Figure 1C) and in vivo (Krahn et al., 1972). The preferred assembly pathway including the MP serves to make assembly highly efficient at in vivo concentrations (Borodavka et al., 2013). In addition, around 500 nucleotides (nt) at the 5′ end of the genome in in vivo assembled particles lacking MP are susceptible to nucleases (Argetsinger and Gussin, 1966), a result that is difficult to rationalize if the MP somehow spanned an otherwise intact shell of CPs.
The asymmetric virus-receptor complex described here is the means by which the virus captures its host; i.e., the first step in a viral infection. The mechanisms by which +ssRNA virus infections proceed beyond the primary receptor-binding event are poorly understood. The idea that the MP could be a structural component of the phage capsid suggests a plausible next step. E. coli that carry the F-plasmid grow and retract their pili seemingly at random (Clarke et al., 2008). The only part of the virion that is known to enter the bacterium is a complex between the proteolytically cleaved MP and the genomic RNA (Krahn et al., 1972). The natural retraction of the F-pilus could pull the MP from its location in the attached capsid, leaving a substantial pore through which the genome could exit into the cell, although the molecular mechanisms by which this entry occurs remain to be discovered. Again, these data are difficult to rationalize without the insights provided by the asymmetric structure.
The proposal that the MP replaces a CP dimer also suggests a tantalizing similarity between two classes of virus that have long been thought to be different. As described earlier, many large viruses including tailed dsDNA bacteriophages such as T4 and λ, and other large viruses such as PBCV and mimivirus, have a special vertex. Here, we show that a small, isometric virus, previously thought to be symmetric, also has the equivalent of a special vertex; in this case, a point in the capsid where a CP dimer has been replaced by the MP. Such ideas have been discussed before, based on the structure of an asymmetrically bound parvovirus-receptor complex (Hafenstein et al., 2007), but here, we show a structural feature that may explain this. Such observations conceptually bring the two classes of viruses together. Indeed, in this respect, the bacterial pilus may perform a similar function to that of the tails of the dsDNA phage.
These results underline the importance of obtaining additional asymmetric structures for other viral systems where infection initiates via receptor binding to, and/or genome extrusion from, one site within the capsid shell (Bakker et al., 2012; Brisco et al., 1985; Levy et al., 2010; Tuthill et al., 2009), including, very recently, across a membrane (Strauss et al., 2013). Such data are likely to throw new light on basic viral mechanisms, not to mention help identify novel drug targets in the future.
Experimental Procedures
Receptor Binding Studies
MS2 VLPs were assembled from recombinantly expressed MS2 CP as described previously (Mastico et al., 1993). Such particles lack the MP. To image the receptor binding properties of such particles, a 10-fold excess of either recombinant MS2 VLPs or wt, infectious MS2 virus (Lima et al., 2006) was mixed with purified F-pili (Brinton et al., 1964; Toropova et al., 2011; Valentine and Strand, 1965) and incubated at room temperature for ∼30 min. The final concentration of components was ∼0.2 mg · ml−1 F-pilus and ∼2 mg · ml−1 MS2 (or VLP) in 50 mM Tris-HCl (pH 7.8), 75 mM NaCl, and 2 mM EDTA. The sample was applied to lacey carbon grids (Agar Scientific), blotted, and plunged frozen into liquid ethane. Cryo-EM was carried out at liquid nitrogen temperatures using an Oxford CT3500 cryo-holder and a FEI Tecnai-F20 electron microscope. Images were recorded at 50,000× magnification on a Gatan US4000 CCD camera under low-dose conditions (∼20 e−/Å2).
Electron Tomography of the Virus-Receptor Complex
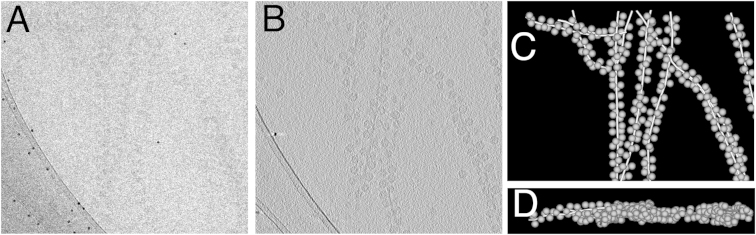
Purified MS2 and F-pili were mixed and allowed to adsorb over 30 min. Ten nanometer protein A-conjugated gold particles (AURION) were added, and the sample was applied to lacey carbon grids (Agar Scientific), blotted, and plunge frozen into liquid ethane. Cryo-electron tomography was carried out at liquid nitrogen temperatures using an Oxford CT3500 cryoholder and a FEI Tecnai G2-Spirit electron microscope. Single-axis tilt series were recorded using SerialEM (Kremer et al., 1996) at a magnification of 23,000×, resulting in an object sampling of 4.56 Å per pixel. Tilt series covered an angular range of −60° to +60° at either 2° or 3° intervals using a dose per frame of ∼1 e−/A2 (Figure 7A). Tomograms were recorded at a defocus of ∼3.0 μm.
Figure 7.
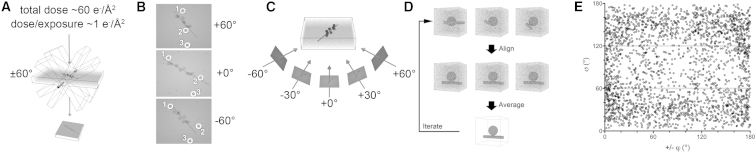
Electron Tomography Methodology
(A) Tilt series were collected between −60° and +60° at 2° intervals and with a dose of ∼1 e−/Å2 per image, i.e., a total accumulated dose of ∼61 e−/Å2.
(B) Alignment of frames in a tilt series was performed by tracking colloidal gold particles in the field of view (numbered white rings).
(C) 3D reconstruction was achieved by back projection.
(D) Individual virus particles were selected from the tomographic 3D reconstructions and iteratively aligned in 3D to a reference structure.
(E) Euler angles of particles going into the final 3D map. Angular coverage is not completely uniform, but the missing wedge is essentially filled by subtomographic averaging. This is apparent in the sphericalness of the resulting capsid structure.
Image Processing
Thirty-seven tilt series were aligned using fiducial markers, decimated by a factor of two, and reconstructed using IMOD (Kremer et al., 1996) at a final sampling of 9.12 Å per pixel (see Movie S1). Tomograms were low-pass filtered to exclude information at resolutions higher than the first zero of the electron microscope (∼31 Å). Tomogram quality was assessed, and those demonstrating unacceptable ice thickness (and poor image quality) were excluded. The locations of MS2 capsids and F-pili segments were modeled using 3DMOD (part of the IMOD package) after tomograms had been further decimated to a sampling of 18.42 Å per pixel and filtered to 72 Å per pixel (down-sampled and filtered). Particles were selected to avoid inclusion of edges of the carbon support or fiducial makers.
Initial Model Calculation
Coordinates from 22 tomograms were exported from IMOD and used to estimate the orientation of each virus-receptor complex by defining a vector (particle axis) between each capsid center and the nearest point along the associated F-pilus. This took into account the direction and polarity of the pilus and aligned each particle so that its axis was placed on the z axis, and the pilus long axis placed on the x axis. Because the features of the F-pilus are too fine to allow polarity to be observed directly in the tomograms, the polarity had to be determined by observing the direction in which the virions tilted on the F-pilus. Owing to the low S/N ratio of the tomograms as well as some out-of-plane tilting of the F-pili, the direction of the virion tilt was sometimes ambiguous when tomogram slices were observed directly. Consequently, subtomographic averages of the virus-receptor complex were generated for each of 85 pilus segments independently by averaging 10–40 subtomograms per pilus. Subtomogram 3D cross-correlation angular searches were carried out with missing wedge compensation using PEET (Nicastro et al., 2006). Orientation estimates were refined over four iterations, producing low-resolution subtomogram averages clearly showing the tilt of the virion on a well-defined F-pilus. The direction of tilt was used to ensure that the modeled coordinates for each pilus assigned the F-pilus “plus-end” as the direction of virion tilt, ensuring that all subtomograms were aligned with the F-pili pointing in the same direction. A reference model for 3D averaging of the whole data set was created by selecting a straight F-pilus that lay in the XY plane of the tomogram and averaging all 41 virus-receptor complexes that were bound to it.
Subtomographic Averaging
The aforementioned generated model was interpolated to a sampling of 9.12 Å per pixel and used as a reference template for alignment of 2,374 subtomograms (64 × 64 × 64 voxels) starting with an angular search range within −30° and +30° around each axis. This effectively prevents particles flipping around the point of attachment to the pilus, which would lead to averaging across a pseudo-2-fold symmetry axis at this point. The orientation estimates were refined to within 0.5° over five iterations that progressively increased the fineness of the angular search while limiting the angular range (Cope et al., 2010). Owing to the flexibility of the F-pili, a mask constructed from an appropriately sized sphere and cylinder was used to limit the length of pilus included in the alignment, as well as to exclude signal arising from closely neighboring capsids. Alignment parameters converged following three rounds of iteration and refinement in which the final subtomogram average was re-entered into the following alignment procedure. It was apparent that the F-pilus was preventing optimal alignment of the capsids owing to variable tilt of the capsids on the F-pilus. The alignment of the capsids was then further improved by refinement using a reference structure of the previous iteration but with the F-pilus masked. The final average consisted of the 1,500 highest correlating subtomograms (∼63% of the data set). Subtomogram averages were low-pass filtered to 30 Å and normalized. The handedness of the structure was confirmed by comparison with the X-ray crystal structure.
Acknowledgments
We thank Dr. Katerina Toropova and Alexander Borodavka for kind gifts of purified F-pili and MS2, respectively. We also thank Professors Simon Phillips (Research Complex at Harwell), Reidun Twarock (University of York), and Michael Rossmann (Purdue University) for their helpful comments on the manuscript and Professor James Hogle (Harvard Medical School) for discussion. K.C.D. was supported by a studentship from the Wellcome Trust (086774/Z/08/Z), and we thank the Biotechnology and Biological Sciences Research Council and Leverhulme Trust for support of various aspects of this research.
Published: June 27, 2013
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
Supplemental Information includes one figure and six movies and can be found with this article online at http://dx.doi.org/10.1016/j.str.2013.05.012.
Accession Numbers
The asymmetric structure of the MS2-pilus complex has been deposited in the EMDB as EMD-2365, and the atomic coordinates for the MS2 capsid (derived from 2ms2) (Golmohammadi et al., 1993) fitted into this electron density have been deposited in the Protein Data Bank as 4BP4, 4BP5, and 4BP6. A complete fitted capsid is available in mmCIF format as 4BP7.
Supplemental Information
XY slice through a reconstructed tomogram of a virus-decorated pilus.
The starting point is the side view of the asymmetric MS2-pilus complex at 1.3 σ, an identical view to that shown in Figure 3A. The electron density is radially colored with the same scheme used in all figures, and the X-ray crystal structure of the MS2 capsid (PDB ID: 2MS2) is shown in cartoon representation and colored according to quasi-equivalent conformation (A = blue, B = green, C = red), as in Figure 1.
Movie shows grayscale slices through the density for the asymmetric MS2-pilus structure.
Movie shows grayscale slices through the density for the asymmetric MS2-pilus structure.
Movie shows grayscale slices through the density for the asymmetric MS2-pilus structure.
The starting point is the side view of the asymmetric MS2-pilus complex at 1.3 σ, an identical view to that shown in Figure 3A. The electron density is radially colored with the same scheme used in all figures, and the X-ray crystal structure of the MS2 capsid (PDB ID: 2MS2) is shown in cartoon representation and colored according to quasi-equivalent conformation (A = blue, B = green, C = red), as in Figure 1.
References
- Abad-Zapatero C., Abdel-Meguid S.S., Johnson J.E., Leslie A.G.W., Rayment I., Rossmann M.G., Suck D., Tsukihara T. Structure of southern bean mosaic virus at 2.8 A resolution. Nature. 1980;286:33–39. doi: 10.1038/286033a0. [DOI] [PubMed] [Google Scholar]
- Abrescia N.G.A., Bamford D.H., Grimes J.M., Stuart D.I. Structure unifies the viral universe. Annu. Rev. Biochem. 2012;81:795–822. doi: 10.1146/annurev-biochem-060910-095130. [DOI] [PubMed] [Google Scholar]
- Argetsinger J.E., Gussin G.N. Intact ribonucleic acid from defective particles of bacteriophage R17. J. Mol. Biol. 1966;21:421–434. doi: 10.1016/0022-2836(66)90016-7. [DOI] [PubMed] [Google Scholar]
- Bakker S.E., Ford R.J., Barker A.M., Robottom J., Saunders K., Pearson A.R., Ranson N.A., Stockley P.G. Isolation of an asymmetric RNA uncoating intermediate for a single-stranded RNA plant virus. J. Mol. Biol. 2012;417:65–78. doi: 10.1016/j.jmb.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basnak G., Morton V.L., Rolfsson O., Stonehouse N.J., Ashcroft A.E., Stockley P.G. Viral genomic single-stranded RNA directs the pathway toward a T=3 capsid. J. Mol. Biol. 2010;395:924–936. doi: 10.1016/j.jmb.2009.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belyi V.A., Muthukumar M. Electrostatic origin of the genome packing in viruses. Proc. Natl. Acad. Sci. USA. 2006;103:17174–17178. doi: 10.1073/pnas.0608311103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borodavka A., Tuma R., Stockley P.G. Evidence that viral RNAs have evolved for efficient, two-stage packaging. Proc. Natl. Acad. Sci. USA. 2012;109:15769–15774. doi: 10.1073/pnas.1204357109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borodavka A., Tuma R., Stockley P.G. A two-stage mechanism of viral RNA compaction revealed by single molecule fluorescence. RNA Biol. 2013;10:481–489. doi: 10.4161/rna.23838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinton C.C., Jr., Gemski P., Jr., Carnahan J. A new type of bacterial pilus genetically controlled by the fertility factor of E. coli K 12 and its role in chromosome transfer. Proc. Natl. Acad. Sci. USA. 1964;52:776–783. doi: 10.1073/pnas.52.3.776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brisco M.J., Hull R., Wilson T.M. Southern bean mosaic virus-specific proteins are synthesized in an in vitro system supplemented with intact, treated virions. Virology. 1985;143:392–398. doi: 10.1016/0042-6822(85)90379-4. [DOI] [PubMed] [Google Scholar]
- Cardone G., Winkler D.C., Trus B.L., Cheng N., Heuser J.E., Newcomb W.W., Brown J.C., Steven A.C. Visualization of the herpes simplex virus portal in situ by cryo-electron tomography. Virology. 2007;361:426–434. doi: 10.1016/j.virol.2006.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspar D.L.D. Structure of bushy stunt virus. Nature. 1956;177:475–476. doi: 10.1038/177475a0. [DOI] [PubMed] [Google Scholar]
- Caspar D.L., Klug A. Physical principles in the construction of regular viruses. Cold Spring Harb. Symp. Quant. Biol. 1962;27:1–24. doi: 10.1101/sqb.1962.027.001.005. [DOI] [PubMed] [Google Scholar]
- Cherrier M.V., Kostyuchenko V.A., Xiao C., Bowman V.D., Battisti A.J., Yan X., Chipman P.R., Baker T.S., Van Etten J.L., Rossmann M.G. An icosahedral algal virus has a complex unique vertex decorated by a spike. Proc. Natl. Acad. Sci. USA. 2009;106:11085–11089. doi: 10.1073/pnas.0904716106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke M., Maddera L., Harris R.L., Silverman P.M. F-pili dynamics by live-cell imaging. Proc. Natl. Acad. Sci. USA. 2008;105:17978–17981. doi: 10.1073/pnas.0806786105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockburn J.J.B., Abrescia N.G.A., Grimes J.M., Sutton G.C., Diprose J.M., Benevides J.M., Thomas G.J., Jr., Bamford J.K.H., Bamford D.H., Stuart D.I. Membrane structure and interactions with protein and DNA in bacteriophage PRD1. Nature. 2004;432:122–125. doi: 10.1038/nature03053. [DOI] [PubMed] [Google Scholar]
- Cope J., Gilbert S., Rayment I., Mastronarde D., Hoenger A. Cryo-electron tomography of microtubule-kinesin motor complexes. J. Struct. Biol. 2010;170:257–265. doi: 10.1016/j.jsb.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick F.H., Watson J.D. Structure of small viruses. Nature. 1956;177:473–475. doi: 10.1038/177473a0. [DOI] [PubMed] [Google Scholar]
- Danziger R.E., Paranchych W. Stages in phage R17 infection. 3. Energy requirements for the F-pili mediated eclipse of viral infectivity. Virology. 1970;40:554–564. doi: 10.1016/0042-6822(70)90199-6. [DOI] [PubMed] [Google Scholar]
- Dunker A.K., Rueckert R.R. Fragments generated by pH dissociation of ME-virus and their relation to the structure of the virion. J. Mol. Biol. 1971;58:217–235. doi: 10.1016/0022-2836(71)90242-7. [DOI] [PubMed] [Google Scholar]
- Dykeman E.C., Stockley P.G., Twarock R. Dynamic allostery controls coat protein conformer switching during MS2 phage assembly. J. Mol. Biol. 2010;395:916–923. doi: 10.1016/j.jmb.2009.11.016. [DOI] [PubMed] [Google Scholar]
- Dykeman E.C., Grayson N.E., Toropova K., Ranson N.A., Stockley P.G., Twarock R. Simple rules for efficient assembly predict the layout of a packaged viral RNA. J. Mol. Biol. 2011;408:399–407. doi: 10.1016/j.jmb.2011.02.039. [DOI] [PubMed] [Google Scholar]
- Dykeman E.C., Stockley P.G., Twarock R. Packaging signals in two single-stranded RNA viruses imply a conserved assembly mechanism and geometry of the packaged genome. J. Mol. Biol. 2013 doi: 10.1016/j.jmb.2013.06.005. [DOI] [PubMed] [Google Scholar]
- Dykeman E.C., Stockley P.G., Twarock R. Building a viral capsid in the presence of genomic RNA. Phys Rev E Stat Nonlin Soft Matter Phys. 2013;87:022717. doi: 10.1103/PhysRevE.87.022717. [DOI] [PubMed] [Google Scholar]
- Fiers W., Contreras R., Duerinck F., Haegeman G., Iserentant D., Merregaert J., Min Jou W., Molemans F., Raeymaekers A., Van den Berghe A. Complete nucleotide sequence of bacteriophage MS2 RNA: primary and secondary structure of the replicase gene. Nature. 1976;260:500–507. doi: 10.1038/260500a0. [DOI] [PubMed] [Google Scholar]
- Frank J.A. Plenum Press; New York: 1992. Electron Tomography. [Google Scholar]
- Golmohammadi R., Valegård K., Fridborg K., Liljas L. The refined structure of bacteriophage MS2 at 2.8 A resolution. J. Mol. Biol. 1993;234:620–639. doi: 10.1006/jmbi.1993.1616. [DOI] [PubMed] [Google Scholar]
- Golmohammadi R., Fridborg K., Bundule M., Valegård K., Liljas L. The crystal structure of bacteriophage Q β at 3.5 A resolution. Structure. 1996;4:543–554. doi: 10.1016/s0969-2126(96)00060-3. [DOI] [PubMed] [Google Scholar]
- Grimes J.M., Burroughs J.N., Gouet P., Diprose J.M., Malby R., Ziéntara S., Mertens P.P.C., Stuart D.I. The atomic structure of the bluetongue virus core. Nature. 1998;395:470–478. doi: 10.1038/26694. [DOI] [PubMed] [Google Scholar]
- Hafenstein S., Palermo L.M., Kostyuchenko V.A., Xiao C., Morais M.C., Nelson C.D.S., Bowman V.D., Battisti A.J., Chipman P.R., Parrish C.R., Rossmann M.G. Asymmetric binding of transferrin receptor to parvovirus capsids. Proc. Natl. Acad. Sci. USA. 2007;104:6585–6589. doi: 10.1073/pnas.0701574104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison S.C., Olson A.J., Schutt C.E., Winkler F.K., Bricogne G. Tomato bushy stunt virus at 2.9 A resolution. Nature. 1978;276:368–373. doi: 10.1038/276368a0. [DOI] [PubMed] [Google Scholar]
- Hill H.R., Stonehouse N.J., Fonseca S.A., Stockley P.G. Analysis of phage MS2 coat protein mutants expressed from a reconstituted phagemid reveals that proline 78 is essential for viral infectivity. J. Mol. Biol. 1997;266:1–7. doi: 10.1006/jmbi.1996.0786. [DOI] [PubMed] [Google Scholar]
- Hodgkin D.C. X-ray analysis and protein structure. Cold Spring Harb. Symp. Quant. Biol. 1950;14:65–78. doi: 10.1101/sqb.1950.014.01.010. [DOI] [PubMed] [Google Scholar]
- Hogle J.M., Chow M., Filman D.J. Three-dimensional structure of poliovirus at 2.9 A resolution. Science. 1985;229:1358–1365. doi: 10.1126/science.2994218. [DOI] [PubMed] [Google Scholar]
- Jiang W., Chang J., Jakana J., Weigele P., King J., Chiu W. Structure of epsilon15 bacteriophage reveals genome organization and DNA packaging/injection apparatus. Nature. 2006;439:612–616. doi: 10.1038/nature04487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J.E., Chiu W. DNA packaging and delivery machines in tailed bacteriophages. Curr. Opin. Struct. Biol. 2007;17:237–243. doi: 10.1016/j.sbi.2007.03.011. [DOI] [PubMed] [Google Scholar]
- Koning R., van den Worm S., Plaisier J.R., van Duin J., Pieter Abrahams J., Koerten H. Visualization by cryo-electron microscopy of genomic RNA that binds to the protein capsid inside bacteriophage MS2. J. Mol. Biol. 2003;332:415–422. doi: 10.1016/s0022-2836(03)00846-5. [DOI] [PubMed] [Google Scholar]
- Kozak M., Nathans D. Fate of maturation protein during infection by coliphage MS2. Nat. New Biol. 1971;234:209–211. doi: 10.1038/newbio234209a0. [DOI] [PubMed] [Google Scholar]
- Krahn P.M., O’Callaghan R.J., Paranchych W. Stages in phage R17 infection. VI. Injection of A protein and RNA into the host cell. Virology. 1972;47:628–637. doi: 10.1016/0042-6822(72)90552-1. [DOI] [PubMed] [Google Scholar]
- Kremer J.R., Mastronarde D.N., McIntosh J.R. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- Lander G.C., Tang L., Casjens S.R., Gilcrease E.B., Prevelige P., Poliakov A., Potter C.S., Carragher B., Johnson J.E. The structure of an infectious P22 virion shows the signal for headful DNA packaging. Science. 2006;312:1791–1795. doi: 10.1126/science.1127981. [DOI] [PubMed] [Google Scholar]
- Levy H.C., Bostina M., Filman D.J., Hogle J.M. Catching a virus in the act of RNA release: a novel poliovirus uncoating intermediate characterized by cryo-electron microscopy. J. Virol. 2010;84:4426–4441. doi: 10.1128/JVI.02393-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liddington R.C., Yan Y., Moulai J., Sahli R., Benjamin T.L., Harrison S.C. Structure of simian virus 40 at 3.8-A resolution. Nature. 1991;354:278–284. doi: 10.1038/354278a0. [DOI] [PubMed] [Google Scholar]
- Liljas L., Unge T., Jones T.A., Fridborg K., Lövgren S., Skoglund U., Strandberg B. Structure of satellite tobacco necrosis virus at 3.0 A resolution. J. Mol. Biol. 1982;159:93–108. doi: 10.1016/0022-2836(82)90033-x. [DOI] [PubMed] [Google Scholar]
- Lima S.M.B., Vaz A.C.Q., Souza T.L.F., Peabody D.S., Silva J.L., Oliveira A.C. Dissecting the role of protein-protein and protein-nucleic acid interactions in MS2 bacteriophage stability. FEBS J. 2006;273:1463–1475. doi: 10.1111/j.1742-4658.2006.05167.x. [DOI] [PubMed] [Google Scholar]
- Lodish H.F., Horiuchi K., Zinder N.D. Mutants of the bacteriophage f2. V. On the production of noninfectious phage particles. Virology. 1965;27:139–155. doi: 10.1016/0042-6822(65)90154-6. [DOI] [PubMed] [Google Scholar]
- Mastico R.A., Talbot S.J., Stockley P.G. Multiple presentation of foreign peptides on the surface of an RNA-free spherical bacteriophage capsid. J. Gen. Virol. 1993;74:541–548. doi: 10.1099/0022-1317-74-4-541. [DOI] [PubMed] [Google Scholar]
- Morais M.C., Tao Y., Olson N.H., Grimes S., Jardine P.J., Anderson D.L., Baker T.S., Rossmann M.G. Cryoelectron-microscopy image reconstruction of symmetry mismatches in bacteriophage phi29. J. Struct. Biol. 2001;135:38–46. doi: 10.1006/jsbi.2001.4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton V.L., Burkitt W., O’Connor G., Stonehouse N.J., Stockley P.G., Ashcroft A.E. RNA-induced conformational changes in a viral coat protein studied by hydrogen/deuterium exchange mass spectrometry. Phys. Chem. Chem. Phys. 2010;12:13468–13475. doi: 10.1039/c0cp00817f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton V.L., Dykeman E.C., Stonehouse N.J., Ashcroft A.E., Twarock R., Stockley P.G. The impact of viral RNA on assembly pathway selection. J. Mol. Biol. 2010;401:298–308. doi: 10.1016/j.jmb.2010.05.059. [DOI] [PubMed] [Google Scholar]
- Nathans D., Oeschger M.P., Eggen K., Shimura Y. Bacteriophage-specific proteins in e. Coli infected with an RNA bacteriophage. Proc. Natl. Acad. Sci. USA. 1966;56:1844–1851. doi: 10.1073/pnas.56.6.1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicastro D., Schwartz C., Pierson J., Gaudette R., Porter M.E., McIntosh J.R. The molecular architecture of axonemes revealed by cryoelectron tomography. Science. 2006;313:944–948. doi: 10.1126/science.1128618. [DOI] [PubMed] [Google Scholar]
- Paranchych W. Stages in phage R17 infection: the role of divalent cations. Virology. 1966;28:90–99. doi: 10.1016/0042-6822(66)90309-6. [DOI] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Plevka P., Kazaks A., Voronkova T., Kotelovica S., Dishlers A., Liljas L., Tars K. The structure of bacteriophage phiCb5 reveals a role of the RNA genome and metal ions in particle stability and assembly. J. Mol. Biol. 2009;391:635–647. doi: 10.1016/j.jmb.2009.06.047. [DOI] [PubMed] [Google Scholar]
- Rayment I., Baker T.S., Caspar D.L., Murakami W.T. Polyoma virus capsid structure at 22.5 A resolution. Nature. 1982;295:110–115. doi: 10.1038/295110a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts J.W., Steitz J.E. The reconstitution of infective bacteriophage R17. Proc. Natl. Acad. Sci. USA. 1967;58:1416–1421. doi: 10.1073/pnas.58.4.1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolfsson O., Toropova K., Ranson N.A., Stockley P.G. Mutually-induced conformational switching of RNA and coat protein underpins efficient assembly of a viral capsid. J. Mol. Biol. 2010;401:309–322. doi: 10.1016/j.jmb.2010.05.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossmann M.G., Johnson J.E. Icosahedral RNA virus structure. Annu. Rev. Biochem. 1989;58:533–573. doi: 10.1146/annurev.bi.58.070189.002533. [DOI] [PubMed] [Google Scholar]
- Rossmann M.G., Arnold E., Erickson J.W., Frankenberger E.A., Griffith J.P., Hecht H.J., Johnson J.E., Kamer G., Luo M., Mosser A.G. Structure of a human common cold virus and functional relationship to other picornaviruses. Nature. 1985;317:145–153. doi: 10.1038/317145a0. [DOI] [PubMed] [Google Scholar]
- Shiba T., Suzuki Y. Localization of A protein in the RNA-A protein complex of RNA phage MS2. Biochim. Biophys. Acta. 1981;654:249–255. doi: 10.1016/0005-2787(81)90179-9. [DOI] [PubMed] [Google Scholar]
- Stockley P.G., Kirsh A.L., Chow E.P., Smart J.E., Harrison S.C. Structure of turnip crinkle virus. III. Identification of a unique coat protein dimer. J. Mol. Biol. 1986;191:721–725. doi: 10.1016/0022-2836(86)90456-0. [DOI] [PubMed] [Google Scholar]
- Stockley P.G., Rolfsson O., Thompson G.S., Basnak G., Francese S., Stonehouse N.J., Homans S.W., Ashcroft A.E. A simple, RNA-mediated allosteric switch controls the pathway to formation of a T=3 viral capsid. J. Mol. Biol. 2007;369:541–552. doi: 10.1016/j.jmb.2007.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss M., Levy H.C., Bostina M., Filman D.J., Hogle J.M. RNA transfer from poliovirus 135S particles across membranes is mediated by long umbilical connectors. J. Virol. 2013;87:3903–3914. doi: 10.1128/JVI.03209-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tars K., Bundule M., Fridborg K., Liljas L. The crystal structure of bacteriophage GA and a comparison of bacteriophages belonging to the major groups of Escherichia coli leviviruses. J. Mol. Biol. 1997;271:759–773. doi: 10.1006/jmbi.1997.1214. [DOI] [PubMed] [Google Scholar]
- Tars K., Fridborg K., Bundule M., Liljas L. The three-dimensional structure of bacteriophage PP7 from Pseudomonas aeruginosa at 3.7-A resolution. Virology. 2000;272:331–337. doi: 10.1006/viro.2000.0373. [DOI] [PubMed] [Google Scholar]
- Toropova K., Basnak G., Twarock R., Stockley P.G., Ranson N.A. The three-dimensional structure of genomic RNA in bacteriophage MS2: implications for assembly. J. Mol. Biol. 2008;375:824–836. doi: 10.1016/j.jmb.2007.08.067. [DOI] [PubMed] [Google Scholar]
- Toropova K., Stockley P.G., Ranson N.A. Visualising a viral RNA genome poised for release from its receptor complex. J. Mol. Biol. 2011;408:408–419. doi: 10.1016/j.jmb.2011.02.040. [DOI] [PubMed] [Google Scholar]
- Tsuruta H., Reddy V.S., Wikoff W.R., Johnson J.E. Imaging RNA and dynamic protein segments with low-resolution virus crystallography: experimental design, data processing and implications of electron density maps. J. Mol. Biol. 1998;284:1439–1452. doi: 10.1006/jmbi.1998.2231. [DOI] [PubMed] [Google Scholar]
- Tuthill T.J., Harlos K., Walter T.S., Knowles N.J., Groppelli E., Rowlands D.J., Stuart D.I., Fry E.E. Equine rhinitis A virus and its low pH empty particle: clues towards an aphthovirus entry mechanism? PLoS Pathog. 2009;5:e1000620. doi: 10.1371/journal.ppat.1000620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valegård K., Liljas L., Fridborg K., Unge T. The three-dimensional structure of the bacterial virus MS2. Nature. 1990;345:36–41. doi: 10.1038/345036a0. [DOI] [PubMed] [Google Scholar]
- Valegård K., Murray J.B., Stockley P.G., Stonehouse N.J., Liljas L. Crystal structure of an RNA bacteriophage coat protein-operator complex. Nature. 1994;371:623–626. doi: 10.1038/371623a0. [DOI] [PubMed] [Google Scholar]
- Valentine R.C., Strand M. Complexes of F-Pili and RNA Bacteriophage. Science. 1965;148:511–513. doi: 10.1126/science.148.3669.511. [DOI] [PubMed] [Google Scholar]
- Winkler F.K., Schutt C.E., Harrison S.C., Bricogne G. Tomato bushy stunt virus at 5.5-A resolution. Nature. 1977;265:509–513. doi: 10.1038/265509a0. [DOI] [PubMed] [Google Scholar]
- Xiao C., Rossmann M.G. Structures of giant icosahedral eukaryotic dsDNA viruses. Curr Opin Virol. 2011;1:101–109. doi: 10.1016/j.coviro.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao C., Kuznetsov Y.G., Sun S., Hafenstein S.L., Kostyuchenko V.A., Chipman P.R., Suzan-Monti M., Raoult D., McPherson A., Rossmann M.G. Structural studies of the giant mimivirus. PLoS Biol. 2009;7:e92. doi: 10.1371/journal.pbio.1000092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Settembre E., Xu C., Dormitzer P.R., Bellamy R., Harrison S.C., Grigorieff N. Near-atomic resolution using electron cryomicroscopy and single-particle reconstruction. Proc. Natl. Acad. Sci. USA. 2008;105:1867–1872. doi: 10.1073/pnas.0711623105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Jin L., Fang Q., Hui W.H., Zhou Z.H. 3.3 A cryo-EM structure of a nonenveloped virus reveals a priming mechanism for cell entry. Cell. 2010;141:472–482. doi: 10.1016/j.cell.2010.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
XY slice through a reconstructed tomogram of a virus-decorated pilus.
The starting point is the side view of the asymmetric MS2-pilus complex at 1.3 σ, an identical view to that shown in Figure 3A. The electron density is radially colored with the same scheme used in all figures, and the X-ray crystal structure of the MS2 capsid (PDB ID: 2MS2) is shown in cartoon representation and colored according to quasi-equivalent conformation (A = blue, B = green, C = red), as in Figure 1.
Movie shows grayscale slices through the density for the asymmetric MS2-pilus structure.
Movie shows grayscale slices through the density for the asymmetric MS2-pilus structure.
Movie shows grayscale slices through the density for the asymmetric MS2-pilus structure.
The starting point is the side view of the asymmetric MS2-pilus complex at 1.3 σ, an identical view to that shown in Figure 3A. The electron density is radially colored with the same scheme used in all figures, and the X-ray crystal structure of the MS2 capsid (PDB ID: 2MS2) is shown in cartoon representation and colored according to quasi-equivalent conformation (A = blue, B = green, C = red), as in Figure 1.