Abstract
Background
Huntington’s disease (HD) is caused by the abnormal expansion of the polyglutamine tract in the human Huntingtin protein (polyQ-hHtt). Although this mutation behaves dominantly, huntingtin loss of function also contributes to HD pathogenesis. Indeed, wild-type Huntingtin plays a protective role with respect to polyQ-hHtt induced defects.
Methodology/Principal Findings
The question that we addressed here is what part of the wild-type Huntingtin is responsible for these protective properties. We first screened peptides from the Huntingtin protein in HeLa cells and identified a 23 aa peptide (P42) that inhibits polyQ-hHtt aggregation. P42 is part of the endogenous Huntingtin protein and lies within a region rich in proteolytic sites that plays a critical role in the pathogenesis process. Using a Drosophila model of HD, we tested the protective properties of this peptide on aggregation, as well as on different polyQ-hHtt induced neuronal phenotypes: eye degeneration (an indicator of cell death), impairment of vesicular axonal trafficking, and physiological behaviors such as larval locomotion and adult survival. Together, our results demonstrate high protective properties for P42 in vivo, in whole animals. These data also demonstrate a specific role of P42 on Huntington’s disease model, since it has no effect on other models of polyQ-induced diseases, such as spinocerebellar ataxias.
Conclusions/Significance
Altogether our data show that P42, a 23 aa-long hHtt peptide, plays a protective role with respect to polyQ-hHtt aggregation as well as cellular and behavioral dysfunctions induced by polyQ-hHtt in vivo. Our study also confirms the correlation between polyQ-hHtt aggregation and neuronal defects. Finally, these results strongly suggest a therapeutic potential for P42, specific of Huntington’s disease.
Introduction
In Humans, Huntington’s disease (HD) is an inherited neurodegenerative disorder belonging to the polyglutamine pathologies. This disease is caused by an abnormal expansion of the CAG triplet in the huntingtin gene (htt). When the number of CAG repeats exceeds 35, the resulting mutant protein (polyQ-Htt) forms intracellular aggregates and causes neuronal degeneration, particularly affecting the neurons of the striatum. Consistent with a toxic role of aggregates in the disease, aggregation increases with the progression of the disease in HD mouse model [1]. In particular, when polyQ-Htt expression is turned off, aggregates disappear, leading to an improvement in the behavioral and cognitive deficits in the HD mouse model [2]. In Humans, HD affects 5 out of 100,000 people, with symptoms usually appearing in patients by their late 40s. HD is a fatal neurological disease with no effective treatment presently available.
Human Huntingtin (hHtt) is a large 3144 aa protein. An important advance in the study of this protein was the discovery of the importance of its proteolysis in apoptosis [3]. Proteolytic cleavages by caspases and calpains occur in both wild-type and mutant Huntingtin, but the frequency of these cleavages increase with the length of the polyQ [3] [4] [5] [6] [1]. This leads to an abnormal accumulation of short toxic N-terminal polyQ-hHtt fragments in an HD context, driving inappropriate apoptosis [3] [7] [8]. These fragments are able to aggregate with themselves and other proteins, and form large cytoplasmic and nuclear inclusions [9]. The production of these polyQ-containing fragments has several patho-physiological consequences for the affected cells, which can include proteasome impairment, mitochondrial dysfunction and oxidative stress, excitotoxicity, transcriptional dysregulation or cell death. The ultimate accumulation of N-truncated forms of polyQ-hHtt in insoluble protein aggregates constitutes an important pathological hallmark of HD [10] [11] [7] [8] [12]. In post-mortem brains of HD patients, aggregates are enriched with N-terminal truncated forms of polyQ-hHtt [9]. Therefore, blocking mutant Huntingtin proteolysis was found to be beneficial in treating HD mice [7] [10] [11]. In general, HD models with short truncations of the expanded Htt protein present more severe phenotypes that also appear earlier than hHtt full-length expressing models [8] [13]. Mouse [14] and Drosophila HD models [13] that express full-length mutant Huntingtin do not present all of the different hallmarks of HD. For instance, flies expressing full-length mutant polyQ-hHtt only show synaptic dysfunction [13]. Furthermore, a selective neurodegeneration of the striatum and the cortex is observed in full-length mutant Htt mouse models, which is correlated with a selective nuclear localization of mutant Htt in these neurons [8] [11]. However, models expressing full-length mutant Htt do not display those defects that appear later in pathogenesis, such as widespread nuclear inclusions in all the brain, and massive neuronal dysfunction and cell death. These late defects are associated with the release and accumulation of polyQ-hHtt N-terminal truncated forms, which amplifies the aggregation process. A reduced cleavage efficiency of the full-length Htt might explain why animal models with full-length Htt only partially reproduce HD pathogenesis [15]. Therefore, important advances in the study of HD came from the development of transgenic models in mice or Drosophila, where only N-terminal portions of human polyQ-hHtt were expressed [16] [17] [18].
In Drosophila, transgenic models have been constructed that express N-terminal portions of human hHtt carrying different lengths of the polyQ domain (Figure S1). These cover either exon 1, corresponding to the first 67 amino acids (hHtt67aa) [16] [19], or longer N-terminal regions covering 171 aa (hHtt171aa) [20], or 548 aa (hHtt548aa) [21]. In the presence of expanded polyQ (>35), all of these constructs behave similarly by forming aggregates and leading (for example) to eye degeneration, regardless to the length of the N-terminal hHtt sequence (67 aa, 171 aa or 548 aa).
These models have been particularly helpful in drug screening [22]. However, until now only a few drugs have been tested in HD patients, with limited impacts. Indeed, given the multiple pathogenic mechanisms involved in this disease, it is expected that a compound that targets one pathological mechanism may not be effective alone. An attractive option for treating HD is of course to specifically target the mutant polyQ-hHtt, which is a strategy now actively pursued [23] [24] [6].
Apart from the dominant function of polyQ-hHtt, loss of function of the wild-type protein may also contribute to the disease pathogenesis [25] [26] [27] [28] [20]. In support of this hypothesis, we previously showed that the ratio between the wild-type hHtt N-terminal fragment and the mutant polyQ-hHtt was critical for the onset of polyQ-hHtt associated-defects, in a Drosophila model of HD [20].
The question that we have addressed here is what part of the wild-type Huntingtin is important for this protective effect. We first screened for peptides, derived from human Htt, that were able to prevent polyQ-hHtt aggregation in HeLa cells. This allowed the identification of a 23 aa protective peptide (P42), whose protective role was tested in vivo in flies. In this report we illustrate how P42 was not only able to prevent aggregation, but also clearly improved all tested polyQ-hHtt induced-defects, such as axonal trafficking of vesicles, larval locomotion, eye degeneration and adult survival. Our data also reveal that P42 was protective for HD models only, and not for other polyQ disease models. Altogether, our data suggest that P42 is acting on polyQ-hHtt by preventing both its aggregation and the ensuing devastative effects to the organism.
Results
Identification of Intrinsic Huntingtin Peptides that Prevent Polyq-hHtt Aggregation
In a previous work, we determined that overexpression of a 548 aa N-terminal part of wild-type human Htt (0Q-hHtt548aa) was able to prevent 138Q-hHtt171aa aggregation in HeLa cells [20]. In order to identify the protective region of wild-type hHtt, we screened for peptides, within the human 0Q-hHtt548aa, that were able to prevent 138Q-hHtt171aa aggregation in HeLa cells. Within the 548 aa protein sequence, four peptides were designed according to known protein domains, such as the HEAT repeats that share homologies with Drosophila Htt [29] [20] (Figure 1A). These peptides (referred to as P1, P2, P3 and P4) were tested in HeLa cells co-transfected with a 138Q-hHtt171aa expressing construct (Figure S1). When transfected alone, 138Q-hHtt171aa formed cytoplasmic aggregates as visualized by immunocytochemical detection (Figures 2A, S2). When each of the four designed peptides was co-transfected with 138Q-hHtt171aa, we found that only P4 (aa 382–548 of hHtt) was able to rescue polyQ-hHtt aggregation (Figure 2B), whereas the other peptides had no effect (data not shown).
Figure 1. Schematic diagram of the 548 aa N-terminal part of human Huntingtin.
A- The different domains of human Huntingtin protein (hHtt) are indicated: Polyglutamine tract (PolyQ), N17 and Proline rich (PRR) domains covering exon 1, as well as the HEAT repeats that share homologies with Drosophila Huntingtin; the sites of cleavage by caspase (in red), calpain (in green) or metallomatrixprotease (MMP); and posttranslational modifications, such as sumoylation (S), palmitoylation (palm), acetylation (Ac) and some of the phosphorylation (P) sites [49]. The positions and/or sizes of P1, P2, P3, P4, P41, P42 and P43 are indicated. Note that P1 is 98aa long, and does not contain any polyQ (0Q). B- The amino acid sequence of P42 is shown (in blue), as well as the caspase (in red) and calpain (in green) sites that surround P42 and that are otherwise indicated in A. Note that the aa position is numbered according to a protein containing 23Q.
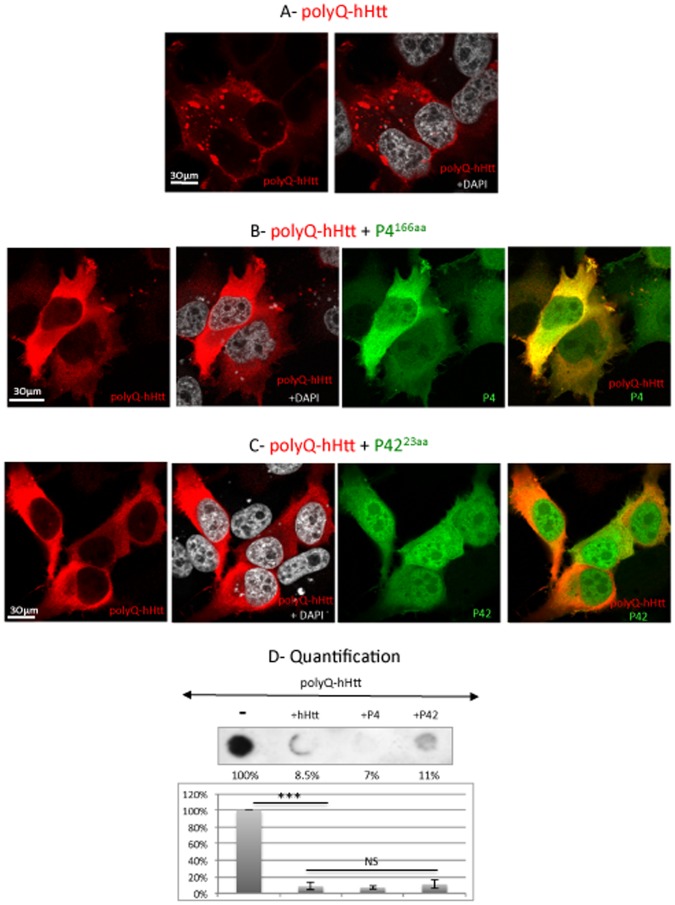
Figure 2. P42 reduces aggregate formation of polyQ-hHtt in HeLa cells.
HeLa cells were transfected with GFP-138Q-hHtt171aa (detected in red), in the presence of: A- empty vector; B- Cherry-P4166aa; or C- Cherry-P4223aa expressing vectors detected by the Cherry tag (in green). DAPI staining is shown in grey. D- Quantification of aggregation by dot-blot filtration retardation assay on cells transfected with GFP-138Q-hHtt171aa, in the presence of empty vector (-), 0Q-hHtt548aa, P4166aa or P4223aa as indicated. Aggregates were detected with an anti-GFP antibody. Quantification was performed on 3 independent experiments and is reported on a graph as percentages with respect to the control (-) set up at 100%. Data were analyzed by using the Student’s t-test: ***p<0.001. The presence of hHtt covering either 548 aa, 166 aa or 23 aa leads to equivalent rescues that are not significantly different (NS).
Since our previous work showed that the N-terminal part of Drosophila Htt covering 620 aa also prevents 138Q-hHtt171aa aggregation [20], we checked for conserved features present in both the 620 aa of Drosophila dHtt and the 166 aa of human P4 (aa 382-548 region of hHtt) (Figure S3). We thus identified a 23 aa region in the middle of P4 (aa 482-504; Figure 1B) that shares homologies between human and Drosophila Htt. Consequently, we further divided P4 into 3 fragments (P41, P42 and P43) with P42 corresponding to the 23 aa homology sequence. When these peptides were tested for their ability to rescue 138Q-hHtt171aa aggregation in HeLa cells, only P42 was able to completely rescue this defect (Figures 2C), whereas the other peptides had no such effect (Figure S5 and data not show). These results showed that, based on immunocytochemical detection of aggregates, P42 was able to inhibit 138Q-hHtt171aa aggregation in cultured human cells.
Inhibitory effects on aggregation were also quantified biochemically by filter retardation assays onto cellulose acetate membrane, in which polyQ-hHtt aggregates are specifically retained [30] (Figure 2D, and data not shown). Filter retardation assays indicated that the aggregation level falls from 100% (control) to 7% in the presence of P4, and to 11% in the presence of P42 (Figure 2D). This confirmed that the “AASSGVSTPGSAGHDIITEQPRS” 23 aa peptide (P42) was able to prevent 138Q-hHtt171aa aggregation as efficiently as hHtt548aa or P4166aa (Figure 2D).
Protective Effect of P42 Peptide in vivo on polyQ-hHtt Aggregation
We further tested the ability of P42 to rescue polyQ-hHtt aggregation in vivo in a whole organism, using a Drosophila model of HD. For this, we constructed transgenic flies that express P42 (either GFP or Myc tagged in its N-terminal end) under the control of UAS sequences. The HD model used for this experiment consisted of transgenic flies expressing a HA-tagged polyQ-hHtt (HA-138Q-hHtt171aa), also under the control of UAS sequences [20] (Figure S1). Using the UAS/Gal4 system [31] with the Gal4 driver MS1096, P42 was first examined for its ability to prevent aggregation in non-neuronal cells such as salivary gland cells; these cells are particularly large, which facilitated visualizing the aggregates. Expression of 138Q-hHtt171aa led to its aggregation (Figure 3A arrows, enlargement in C). It should be noted that this experiment was performed with transgenic flies also containing a neutral UAS transgene (UAS-mCD8-GFP) in order to homogenize the number of UAS constructs. This allowed for similar genetic conditions in all experiments, with the neutral UAS transgene being replaced by a UAS-P42 when needed. The 138Q-hHtt171aa aggregates were essentially localized in the cytoplasm, as assessed by immunocytochemistry (Figure 3A). When the UAS-mCD8-GFP neutral transgene was replaced by UAS-GFP-P42, 138Q-hHtt171aa aggregates were no longer detected (Figure 3B and quantification on Figure S4). In this case, 138Q-hHtt171aa was visible as a diffuse cytoplasmic staining (Figures 3B, 3C), similar to normal Htt staining which overlaps the microtubule staining (Figure S6), as we demonstrated in a previous work [20]. Notably, P42 is detectable in both the nucleus and the cytoplasm, and the cytoplasmic fraction also shows the same profile than Htt (Figures 3B, S6). Even though P42 is slightly concentrated in the nucleus of salivary glands, as found in HeLa cells (Figure 2C), its expression does not disturb the nucleo-cytoplasmic distribution of wild-type hHtt (Figure S6). This indicates that P42 was able to prevent polyQ-hHtt aggregation, and was able to restore a normal cytoplasmic repartition of Huntingtin, along the ß-tubulin network (Figure S6). Quantification of aggregates by filter retardation assays confirmed that 138Q-hHtt171aa aggregation in salivary glands was prevented by P42 (Figure 3D), which corroborates the results obtained in HeLa cells.
Figure 3. P42 reduces aggregate formation of polyQ-hHtt in salivary glands.
A–C: Anti-HA detection (in red) of HA-138Q-hHtt171aa expressed in salivary glands of MS1096-Gal4, UAS-HA-138Q-hHtt171aa/+ larva, in the presence of either A- one copy of a membrane-associated GFP neutral transgene (UAS-mGFP) (in green) or B- one copy of P42 (UAS-GFP-P42) (in green). Focal plan is at the level of the nuclei. Merged images are shown as indicated. A high magnification zoom from another focal plane is shown in C (HA staining only). D- Quantification of aggregation by dot-blot filtration assay performed on two independent sets of experiments; the percentage of aggregates formed in presence of P42 was determined with respect to the control (+mGFP) set up at 100%.
In conclusion, these data show that P42 is able to prevent polyQ-hHtt aggregation both in cultured human cells and in vivo in a whole organism.
Protective Effect of P42 Peptide on Impaired Axonal Transport Induced by polyQ-hHtt
We then studied in vivo, in Drosophila, the protective effect of P42 on polyQ-hHtt induced neuronal phenotypes that are distinct from protein aggregation alone. We first focused on axonal transport that is impaired by the presence of mutant polyQ-hHtt [32] [33]. Indeed, a defect in axonal transport could be one cause for the decrease in neurotrophic factor delivery (such as BDNF) and would participate in the selective striatal degeneration that occurs in HD patients [26].
Our axonal transport study focused on vesicular transport of GFP-tagged neuropeptide Y (NPY-GFP) within the Drosophila larval motoneurons, using an OK6-Gal4 driver [34] [35]. Neuropeptide-Y is a fast axonal transport cargo that is diffusely distributed in wild-type nerves, but accumulates when transport is compromised; forming vesicle clogs along the axons.
We first analyzed the global distribution of the NPY-GFP vesicles on fixed larvae with different genetic conditions (Figure 4). When expressed in motoneurons, the wild-type 0Q-hHtt548aa protein was found all along the axons, which corroborates previous studies that showed Htt particles along the microtubules [36] [37]. Interestingly, we observed that high expression of 0Q-hHtt548aa did not induce clogs of NPY-GFP vesicles (Figure 4A). On the contrary, mutant 128Q-hHtt548aa formed large aggregates along the axons (Figures 4B, S7). In addition, it induced the accumulation of NPY-GFP vesicles within clogs, especially at the location of the polyQ-hHtt aggregates [21] (Figure 4B arrows). This was performed in the presence of a control UAS-LacZ transgene, to ensure a similar number of UAS constructs under all tested conditions. When this control transgene was replaced by UAS-P42 to induce the expression of P42, we found that 128Q-hHtt548aa formed fewer aggregates (Figure 4C and quantification on Figure S7). In addition, trafficking of NPY-GFP vesicles along the axons was recovered, with fewer vesicular clogs (Figure 4C) and more individual, potentially moving, vesicles (Figure 4C, and quantifications on Figure 5B).
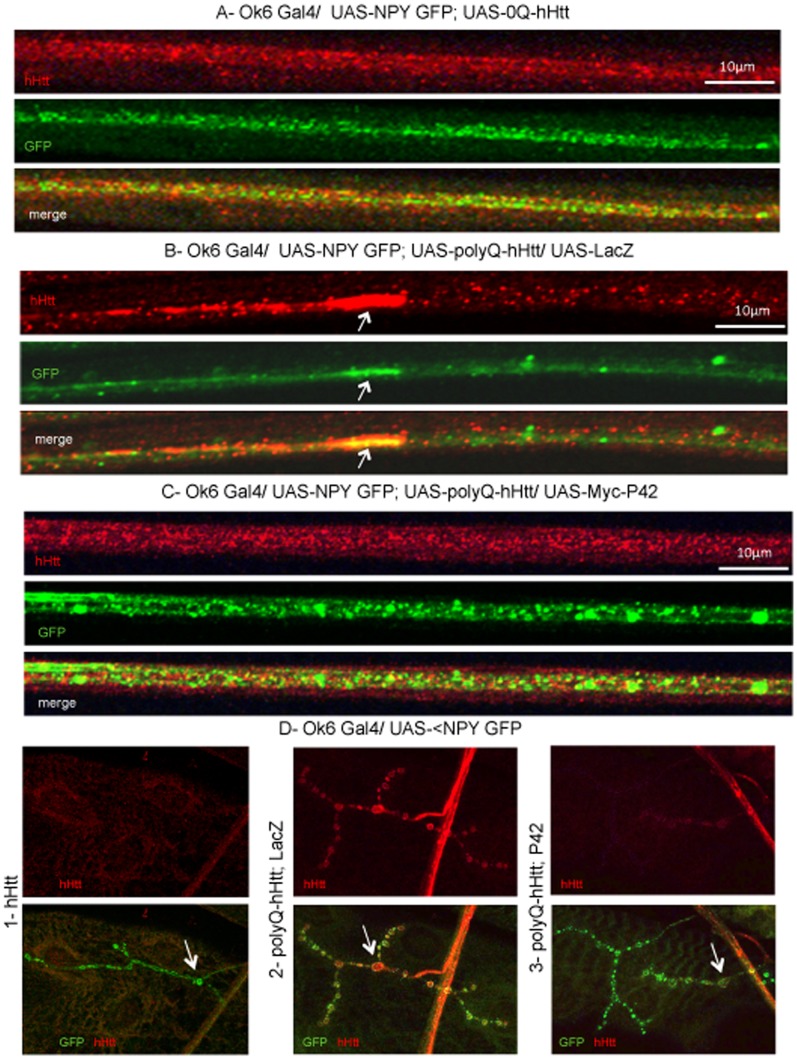
Figure 4. Distribution of neuropeptide Y-GFP vesicles along the axons and at the neuromuscular junctions (NMJ) of Drosophila larvae.
Vesicular neuropeptide-Y-GFP (NPY-GFP) expression was visualized by anti-GFP immunostaining (in green) in larval motoneurons expressing hHtt548aa detected by anti-hHtt (Hu-4C8) (in red), in different genotypes: A- OK6-Gal4/+; UAS-NPY-GFP/UAS-0Q-hHtt548aa (hHtt). B- OK6-Gal4/UAS-128Q-hHtt548aa; UAS-NPY-GFP;/UAS-LacZ (polyQ-hHtt; LacZ). C- OK6-Gal4/UAS-128Q-hHtt548aa; UAS-NPY-GFP/UAS-P42 (polyQ-hHtt; P42). Merged images are shown. D- Muscle 4 NMJs are shown in the different genetic backgrounds: 1- OK6-Gal4/+; UAS-NPY-GFP/UAS-0Q-hHtt548aa (hHtt). 2- OK6-Gal4/UAS-128Q-hHtt548aa; UAS-NPY-GFP;/UAS-LacZ (polyQ-hHtt; LacZ). 3- OK6-Gal4/UAS-128Q-hHtt548aa; UAS-NPY-GFP/UAS-P42 (polyQ-hHtt; P42). When expressing wild-type hHtt, NPY-GFP and 0Q-hHtt548aa are uniformly distributed along the axons and 0Q-hHtt548aa did not reach the NMJ boutons whereas NPY-GFP did (arrows in D1). In contrast to this control, 128Q-hHtt548aa is found in large aggregates (see arrows in B and Figure S7) and NPY-GFP vesicles form clogs along the axons, and 128Q-hHtt548aa accumulates at the NMJ boutons (arrows in D2). Note that the presence of P42 restores the normal distribution of NPY-GFP and 128Q-hHtt548aa both along the axons and at the NMJ boutons (arrows in D3).
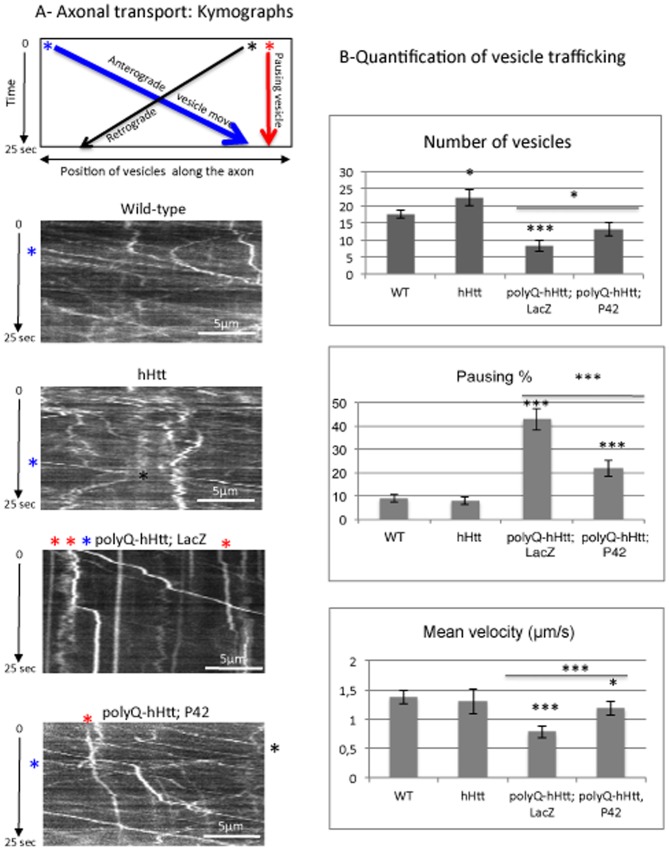
Figure 5. Axonal transport of neuropeptide Y-GFP vesicles.
A- A representative schematic kymograph of vesicular movement along the axons is shown. The data were obtained for vesicles going in anterograde (* in blue) or retrograde (* in black) directions, as well as for vesicles that do not move (* in red). The starting point of vesicles is shown as examples and follow a trajectory according to the color code. Kymographs of vesicular movement are shown for four genotypes: Wild-type, OK6-Gal4/+; UAS-NPY-GFP/UAS-0Q-hHtt548aa (hHtt), OK6-Gal4/UAS-128Q-hHtt548aa; UAS-NPY-GFP/UAS-LacZ (polyQ-hHtt; LacZ), and OK6-Gal4/UAS-128Q-hHtt548aa; UAS-NPY-GFP/UAS-P42 (polyQ-hHtt; P42). Note that only few vesicles are detectable in (polyQ-hHtt; LacZ) larvae and most of them are pausing (* in red), which is not the case in presence of P42. B- Quantification of different parameters (number of vesicles, % of pausing and mean velocity of vesicles) for NPY-GFP vesicular movement was performed in the genetic backgrounds described in A (n = 9-13 larvae). Mean values are compared to wild-type conditions (WT), or to polyQ-hHtt; LacZ. Data were analyzed with the Student’s t-test (*p<0.05, ***p<0.001). As compared to the control (WT), the presence of 128Q-hHtt548aa reduced vesicle number, increased the percentage of pausing, and reduced the mean velocity. Note that the presence of P42 rescued the defects induced by 128Q-hHtt548aa.
We also analyzed the Drosophila larval neuromuscular junction (NMJ), a glutamatergic synapse that is largely studied both developmentally and physiologically [38] (Figure 4D). When wild-type 0Q-hHtt548aa protein was expressed in motoneurons it was barely detectable at the synapse (Figure 4D1), although it was visible within the nerves (Figure 4A). On the contrary, expression of 128Q-hHtt548aa protein in motoneurons resulted in clear hHtt immunoreactivity in the nerves (Figure 4B) but also at the NMJ (Figure 4D2), indicating that the extended polyQ domain led to the accumulation of hHtt at the NMJ (Figure S8). The accumulation of 128Q-hHtt548aa did not have an obvious effect on the presence of NPY-GFP vesicles at the NMJ (Figure 4D2). As similarly observed for the aggregation phenotype, co-expression of P42 with the 128Q-hHtt548aa protein eliminated the accumulation of Huntingtin at the NMJ (Figure 4D3).
Altogether, these results show that P42, in addition to its rescuing effect on 128Q-hHtt548aa aggregates, is also able to rescue polyQ-hHtt mislocalization at the NMJ, as well as polyQ-hHtt-induced axonal vesicular clogs.
We next investigated in further detail the dynamic vesicle movement along the axons, using a non-invasive in vivo technique to follow the vesicle trajectories, through the cuticle of living larvae [34] [35]. Vesicle motion in motoneuron axons was captured in short movies and was recapitulated in representative kymographs for each genotype (Figure 5A). Vesicle counting and tracking allow the investigation of a range of characteristic parameters, including the number of vesicles, the proportion of vesicles that are pausing, and the mean velocity of mobile vesicles (Figure 5B). We first quantified the number of isolated, potentially moving vesicles within a fixed nerve length, for the first frame of each movie. In the control condition, a large number of NPY-GFP vesicles were traveling in motoneurons in both retrograde and anterograde directions (Figure 5A). The number of traveling vesicles was significantly enhanced in the presence of wild-type 0Q-hHtt548aa, probably because of its effect on microtubule-based transport, as previously described [37] [26] (Figures 5A, 5B). Expression of wild-type 0Q-hHtt548aa did not modify either the mean pausing time or the mean velocity of these traveling vesicles (Figure 5B). On the contrary, there were fewer vesicles in the presence of 128Q-hHtt548aa, and existing vesicles showed an increased pausing rate (Figures 5A, 5B), as well as a decreased mean velocity (Figure 5B).
When co-expressed with 128Q-hHtt548aa, P42 induced a rescue in the number of vesicles within the nerve to the level of wild-type larvae. In addition, P42 significantly rescued the dynamic vesicle movement affected by the presence of the mutant 128Q-hHtt548aa protein, by reducing the proportion of pausing vesicles and enhancing their mean velocity. Note, however, that this rescue was not complete and did not reach the wild-type level (Figures 5A, 5B).
In conclusion, we show that P42 is able to improve the vesicular trafficking defects induced by the mutant polyQ-hHtt within axons, by restoring the global number and motion of vesicles.
Protective Effect of P42 on Physiological Behaviors Affected by polyQ-hHtt
Next, we analyzed the protective effect of P42 on 128Q-hHtt548aa induced defects at a more integrated level, focusing on larval locomotion and adult lifespan.
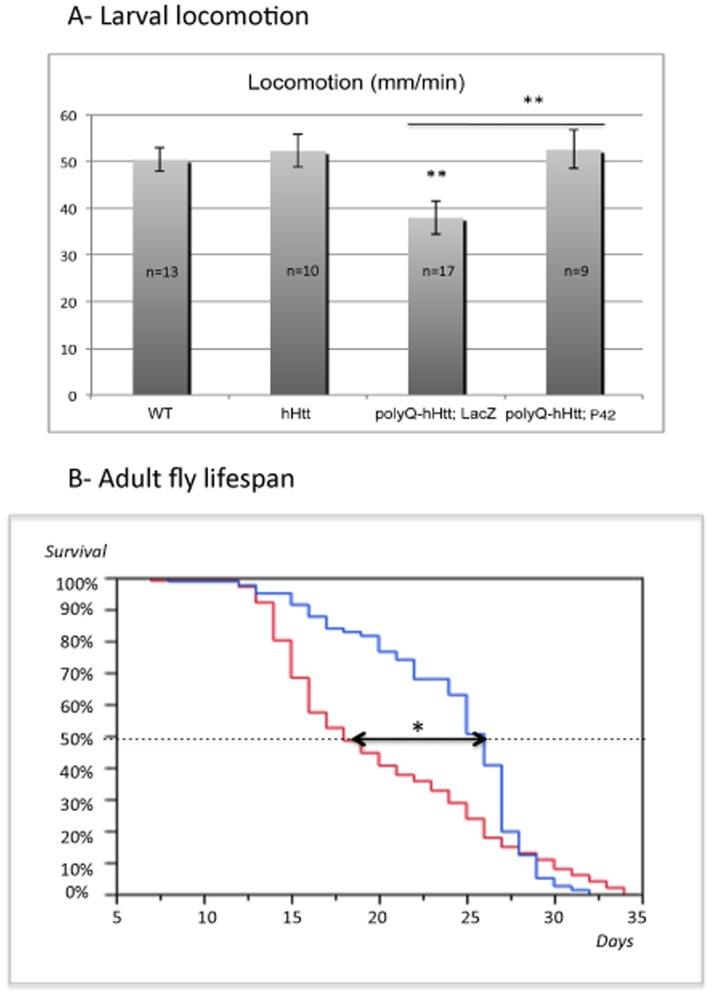
We analyzed the locomotion of larvae expressing wild-type or mutant 128Q-hHtt548aa in motoneurons, using the OK6-Gal4 driver (Figure 6A). The presence of wild-type 0Q-hHtt548aa protein did not affect larval locomotion (mean of 52 mm/min) (Figure 6A). On the contrary, expression of 128Q-hHtt548aa protein significantly impaired larval locomotion with a 25% speed reduction, in accordance with the presence of axonal transport defects. Remarkably, co-expression of P42 completely rescued this locomotor defect.
Figure 6. P42 rescues physiological behaviors induced by polyQ-hHtt.
A- Larval locomotion was examined during 2 min in four different genetic backgrounds: Wild-type (WT), OK6-Gal4/+; UAS-NPY-GFP/UAS-0Q-hHtt548aa (hHtt), OK6-Gal4/UAS-128Q-hHtt548aa; UAS-NPY-GFP/UAS-LacZ (polyQ-hHtt; LacZ), and OK6-Gal4/UAS-128Q-hHtt548aa; UAS-NPY-GFP/UAS-P42 (polyQ-hHtt; P42). Mean values are calculated on (n = 9 to 17), as indicated. Data were analyzed with the Student’s t-test (** p<0.01). Compared to the WT control, the presence of 128Q-hHtt548aa reduced larval locomotion, whereas P42 rescued the locomotion defect. B- Analysis of adult lifespan for UAS-128Q-hHtt548aa ; elav-Gal4/UAS-LacZ (polyQ-hHtt; LacZ in blue) and UAS-128Q-hHtt548aa ; elav-Gal4/UAS-P42 (polyQ-hHtt; P42 in red) flies are shown. The number of flies that survived from each cohort was evaluated once per day. Mean, median and maximum survival times (in days) were calculated from survival curves by using the Kaplan-Meier analysis; percentage of survival is provided. The arrow indicates the difference in age at which 50% of the flies have died; the median survival is increased in the presence of P42. P value was calculated by using log rank statistics (*p = 0.0148).
It has previously been reported that expression of polyQ-hHtt in all Drosophila neuronal cells reduces fly longevity [39] [40]. Therefore, we next examined the rescuing effect of P42 on adult fly viability (Figure 6B). To investigate this, we performed survival curves for different cohorts of flies expressing polyQ-hHtt (128Q-hHtt548aa) in all neurons (under control of the elav-Gal4 driver), either in the absence or presence of P42. The normal lifespan of wild-type flies ranges around 60 days [39] [40]. Expression of P42 alone did not alter the normal viability of the flies (data not shown), suggesting that its overexpression is not toxic. 128Q-hHtt548aa adults displayed a maximum lifespan of 33 days with a median lifespan (the age at which 50% of the flies have died) occurring at 18 days (Figure 6B). The maximal survival time did not change when P42 was co-expressed, although the median adult lifespan increased to 26 days, which corresponds to a 24% increase (Figure 6B). The mean viability of the flies was also significantly enhanced in the presence of P42. From these results, we can conclude that P42 significantly improves the survival of the HD flies, without enhancing their maximum lifespan. This indicates a partial rescue of P42 on longevity defects induced by mutant polyQ-hHtt.
To conclude, we found that P42 presents protective properties for a range of polyQ-hHtt induced phenotypes, from cellular defects to behavior.
Protective and Specific Effect of P42 on Cellular Death
Having confirmed the rescuing effect of P42 for a range of phenotypes, we finally focused on the most studied “degeneration” phenotype in the fly model, i.e. cell death in the eye. Because this phenotype is also common to many fly models of neurodegenerative diseases, this assay allowed us to examine whether P42 could additionally rescue neurodegeneration associated with other polyQ mutant proteins, such as Ataxins.
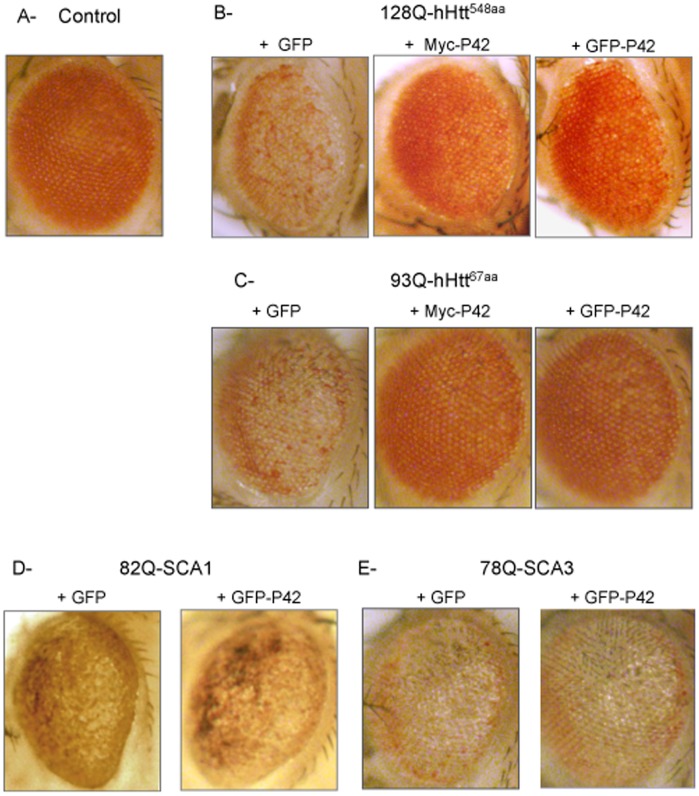
PolyQ-hHtt expression with the GMR-Gal4 driver in developing eyes leads to adults showing a loss of eye pigmentation, associated with abnormal ommatidial arrays (as detected by scanning electron microscopy) and a loss of photoreceptors [19] [21] [20] (compare Figure 7A to 7B or 7C and data not shown). Here, we used different polyQ-hHtt transgenes that encode different truncated forms of polyQ-hHtt (Figures S1, 7B, 7C). We first analyzed the rescuing activity of P42 on eye toxicity generated in flies expressing either 128Q-hHtt548aa [21], (containing the P42 sequence; Figures 7B, S1) or a short 93Q-hHtt67aa [19] that does not include the P42 sequence (Figures 7C, S1). Interestingly, the eye toxicity induced by both transgenes was rescued in the presence of P42 tagged with either Myc or GFP (Figures 7B, 7C). This indicates that P42 is able to rescue polyQ-hHtt toxicity induced by different forms of polyQ-hHtt N-terminal fragments. In particular, P42 can rescue eye toxicity induced by a polyQ-hHtt transgene containing only the first 67 aa (exon 1) of human Huntingtin (Figure 7C).
Figure 7. Influence of P42 on eye toxicity induced by different polyQ mutant proteins.
A- As a control to examine eye toxicity, we used GMR-Gal4/UAS-GFP flies that exhibit normal eyes. Adult eye phenotypes were analyzed in four different genetic backgrounds: B- GMR-Gal4/UAS-128Q-hHtt548aa, C- GMR-Gal4/+; UAS-93Q-hHtt67aa/+, D- GMR-Gal4/+; UAS-82Q-SCA1/+, and E- GMR-Gal4/+; UAS-78Q-SCA3/+, which was tested either in the presence of the UAS-GFP neutral transgene, or in the presence of P42 tagged with Myc or GFP, as indicated. PolyQ-hHtt flies carrying the UAS-GFP neutral transgene exhibited eye degeneration, with depigmented ommatidia. On the contrary, polyQ-hHtt flies exhibited normal eyes in presence of P42, regardless if this was tagged with Myc or GFP (in B and C). Note that other models of polyQ-induced ataxias exhibited eye degeneration in the presence of either a neutral UAS-GFP transgene or the UAS-P42 transgene (in D and E), indicating that the P42 rescue effect is specific to polyQ-hHtt.
We then wondered whether P42 could also rescue Drosophila models of other polyQ neurodegenerative diseases, such as spinocerebellar ataxia type 1 (82Q-SCA1) [41] (Figure 7D) or spinocerebellar ataxia type 3 (78Q-SCA3) [42] (Figure 7E). Interestingly, P42 was not able to rescue the eye degeneration induced by these other polyQ mutant proteins (Figures 7D, 7E).
This indicates that P42 has a protective effect that is specific to polyQ-hHtt induced toxicity.
Discussion
This study is a continuation of our previous observation that wild-type Htt (human Htt548aa and Drosophila Htt620aa) plays a protective role with respect to polyQ-hHtt induced toxicity [20]. We divided the human Htt548aa into four peptides and tested each one for their protective effect. We initially expected P1, P2 or P3 to play such a role because they contain important protein domains. Indeed, P1 contains the N17 and PRR domains known to influence the aggregation process [43] [44]. P2 and P3 contain HEAT domains that are known to participate in protein-protein interactions [45] and that are also found in Drosophila Htt. Surprisingly, we determined that only P4 was able to efficiently block 138Q-hHtt171aa aggregation in HeLa cells. Even though it does not contain any predicted domain, this 166 aa P4 peptide is particularly rich in proteolytic and phosphorylation sites that all impact the cleavage of the protein that plays important functions in the neurodegeneration process [5] [46] [47] [48] [49] [50]. We then searched for homologies between the 620 aa N-terminal part of Drosophila Htt and Human P4. This led to the identification of conserved features extending over a 23 aa peptide (P42) in the Human counterpart. A further partition of P4 revealed that indeed only P42 within P4 had efficiently retained the protective properties of the larger fragments (P4 or hHtt548aa). Whereas both P4 and P42 inhibit polyQ-hHtt aggregation, we could observe that their nucleo-cytoplasmic repartition is slightly different. They are both localized in the cytoplasm, which is in line with their role in prohibiting the formation of aggregates that are essentially detected in the cytoplasm of both culture cells and flies. However, P42 is also highly concentrated in the nucleus (Figures 2 and 3). As it has been demonstrated that nuclear inclusions of polyQ-hHtt appear in late stages of the disease [8] [11], the nuclear localization of P42 suggests that this peptide may also be protective for nuclear inclusions.
In flies, P42 was able to prevent aggregation induced by polyQ-hHtt, as it does in HeLa cells. Since the exact role of aggregates and their involvement in cell toxicity remain controversial [51], it was important to evaluate the impact of P42 on other cellular and functional phenotypes. When looking at polyQ-hHtt induced cellular defects, we observed that P42 was able to completely rescue some defects (including aggregation in salivary glands and polyQ-hHtt mislocalization at the NMJ), while others were only partially rescued (such as for instance the number of travelling vesicles along the axons). This incomplete recovery could result from the ability of soluble polyQ-hHtt to reduce kinesin-based axonal transport [33]. Thus, even when it is not involved in aggregates, the presence of soluble polyQ-hHtt might still affect axonal function. In accordance with our results at the cellular level, we could obtain either a full rescue (eye phenotypes, larval locomotion) or only a partial one (fly viability) at the physiological/behavioral level. Toxicity has been shown to be likely associated with both soluble and aggregated polyQ-hHtt levels [15]. Whereas aggregates are not formed in the presence of P42, the level of soluble polyQ-hHtt might be enhanced, which could explain the incomplete recovery. One way to improve these partial rescues obtained with P42, when considering therapeutic applications, could be to enhance the clearance of polyQ-hHtt through P42 targeting. To pursue this goal, fusion molecules comprising P42 and peptides required in macroautophagy [52] or in proteasomal degradation [53] could be engineered. Such processes have been shown to ameliorate the formation of aggregates in culture cells [53], as well as symptoms in mouse HD models [52] [54]. To ensure the safety of such a reagent, an important preliminary issue will be to verify that P42 preferentially binds polyQ-hHtt, and not the wild-type. Another way to better prevent the toxicity of polyQ-hHtt expression could be a combinatorial approach as already described [55], using the concomitant use of P42 and other potential therapeutic agents such as the QBP1 peptide that directly interferes with polyQ [40].
To explain P42 protective effect, we first excluded that P42 affects polyQ-hHtt level of expression both in cells (Figure S9), and in vivo in salivary glands (Figure S6). Hence, for its protective effect, P42 could act either indirectly by titrating proteins that are involved with the polyQ-hHtt in the disease, or more directly by interacting with the polyQ-hHtt. The fact that P42 was protective for polyQ-hHtt but not for other polyQ toxic proteins such as polyQ-SCA1 and polyQ-SCA3, and that it was able to rescue eye phenotypes induced by 93Q-hHtt67aa (Figure 7) strongly suggest that if direct, P42 interaction might not occur through the polyQ, but must occur with the first 67 aa of Htt (exon 1). It is interesting to notice that exon1 contains the N17, and the PRR (Proline rich region) domains (Figure 1), both of which play a role in polyQ-hHtt aggregation, although through different mechanisms [44] [56] [57]. In particular, aggregation process requires a nucleation step, involving the self-interaction of the N17 domains. During this nucleation event, the polyQ-hHtt N-terminus adopts a structure that is able to associate with itself, which leads to a local concentration and the oligomerization of the mutant hHtt. The presence of the N17 domain was shown to accelerate the aggregation process [56]. As a result, factors (such as intrabodies for instance) targeting the N17 domain are able to suppress aggregation and the associated toxicity by inhibiting this nucleation step [58] [39] [46] [59] [60]. Therefore, by targeting exon1, P42 might interfere on both the nucleation and aggregation processes, leading, as observed, to a complete disappearance of polyQ-hHtt aggregates.
We can also notice that P42, which is a part of the hHtt protein, is localized within a region rich in cleavage sites. One hypothesis is that an endogenous role of P42 could be to slow Htt cleavage. Therefore, blocking mutant Huntingtin proteolysis by P42 could be protective, which is known to be beneficial in treating HD mice [7] [10] [11].
In this report, we identified a short peptide that is protective in vivo of polyQ-hHtt induced defects. The identification of this 23 aa peptide endogenous to hHtt that saves neuronal defects induced by polyQ-hHtt in flies opens a new avenue for future therapeutic strategies. In accordance to a clear therapeutic potential for this peptide, the use of P42 combines on the one hand, a clear specificity of action towards hHtt and on the other hand, the advantages of peptide technologies such as low toxicity, mainly because it is a part of the Huntingtin protein.
Materials and Methods
Drosophila Stocks
Stocks MS1096-Gal4 (on X) [61]; OK6-Gal4 (on II) [62]; UAS-SCA1-82Q [41]; UAS-SCA3-78Q [42]; GMR-Gal4 (on II); elav-Gal4 (on III); UAS-GFP (on II); UAS-mCD8-GFP (on III); and UAS-LacZ (on III) were obtained from the Bloomington Drosophila stock center. UAS-NPY-GFP was obtained from I. Robinson (Cambridge, U.K.). For Drosophila models of Huntington’s disease, we used the following stocks: UAS-93Q-Htt67aa, obtained from Leslie Thompson [19]; UAS-HA-138Q-hHtt171aa [20], or UAS-128Q-Htt548aa, obtained from J. Troy Littleton [21]. UAS-0Q-Htt548aa [21] was used for normal human Huntingtin. Note that only hHtt171aa transgene is HA tagged at its N-terminal end. Since there is a direct link between polyQ-hHtt induced phenotypes and its level of expression, controls were always tested in the presence of a neutral UAS-transgene (UAS-LacZ or UAS-GFP or UAS-mCD8- GFP as indicated).
Antibodies
Primary antibodies used include: anti-HA antibody (SC805 rabbit polyclonal; Santa Cruz), anti-HA (3F10 rat monoclonal; Roche), anti-Huntingtin (Hu-4C8 mouse monoclonal; Chemicon), anti-GFP antibody (polyclonal rabbit; Invitrogen), and anti-DsRED antibody (polyclonal rabbit; Clontech). Secondary antibodies (from Jackson) include: anti-rabbit antibody coupled with HRP or with Alexa488, and anti-mouse Cy3-conjugated antibody.
Plasmid Constructions using Gateway
- pDon-P42 made by PCR on Human hHtt with two primers.
P42 up:GGGGACAAGTTTGTACAAAAAAGCAGGCTTCgctgcttcttcaggggtt, and P42 down: acagaacagccacggtcaGACCCAGCTTTCTTGTACAAAGTGGTCCCC.
PCR were recombined in Gateway destination vectors:
- The Cherry-TAG pCMV Gateway vector was provided by the MGC (Montpellier Genetic Collections).
- GFP-TAG is pDEST53 Gateway destination vector (Invitrogen).
- GFP-TAG; HA-TAG or MYC-TAG in pUASt gateway vectors were provided by the DGRC to construct Drosophila transgenic lines.
- Constructs were normally tagged at their 5′end.
Transfection of Cultured HeLa Cells
- Cells were transfected with 1.5 µg total DNA using JetPei reagent (Qbiogene) for 20 hrs. Cells were rinsed with 1x PBS and fixed with 4% PFA for immunocytochemistry.
Filter Retardation Assays
Protein extracts from L3 larval salivary glands or from HeLa cells were performed according to Mugat et al. (2008) [20]. Using a Bio-Rad dot-blot filtration unit, denatured protein samples were filtered through a cellulose acetate membrane (0.2 µm pore size, Schleicher & Schuell) that only retains aggregates. Membranes were then immunodetected with primary antibodies: rabbit polyclonal anti-HA antibody (1∶100), or rabbit polyclonal anti-GFP antibody (1∶5000;), as indicated. Subsequently, the membranes were incubated with the secondary antibody, anti-rabbit coupled HRP (1∶50,000). Immunoreactive spots were detected with chemiluminescent ECL substrate (Roche), and quantifications were performed using the ImageJ software.
Construction of Drosophila Transgenic Lines
UAS-Myc-P42 and UAS-GFP-P42 were cloned in the Gateway DGRC vector and injected into embryos (BestGene Inc.).
Immunostainings
Salivary glands of third instar male larvae were dissected and immunostained according to Mugat et al. (2008) [20]. Motoneurons and NMJs of third instar female larvae were dissected and immunostained according to Franco et al. (2004) [63]. Images were acquired by confocal microscopy.
Non-invasive Tracking of Vesicles in Segmental Nerves of Living Larvae
Following the protocol of Talmat-Amar et al. (2011) [35], we anaesthetized third instar female larvae expressing NPY-GFP in their motor neurons with ether during 2 min. Living larvae were mounted ventral side up on a slide in 1% agarose with a coverslip, and viewed with a fluorescent microscope to track vesicle motion in nerves between segments A2–A3. 100-frame movies were recorded every 280 ms, for one nerve per larva (n = 9–13). To quantify vesicle number, the total number of vesicles was counted along a distance of 21 µm in the first frame of each movie. For motion analysis, 20 vesicles were randomly selected in the first frame of each movie and tracked by ImageJ (using the plugin “manual vesicle tracking”). Vesicles with a velocity inferior to 0.01 µm/s were counted as pausing. Kymographs that permit visualizing the movement of all vesicles within a selected axonal segment versus time were created with imageJ (using the plugin “kymo”). The plugins were developed by F. Cordelières (Institut Curie, Orsay, France).
Larval Locomotion Analysis
Wandering third-instar female larvae were placed in the middle of a 13.5 cm wide Petri-dish filled with grape juice agar (classically used for Drosophila egg collection). A grid pattern consisting of 0.5 cm2 squares was placed underneath the plate, and the number of sides crossed by each larva during 2 minutes was recorded. We averaged the values obtained for all larvae of each genotype.
Eye Phenotype Analysis
We analyzed the eye degeneration and depigmentation in adult females where UAS transgene expression was induced by the GMR-Gal4 driver. Phenotypes were analyzed on 10-day old UAS-93Q-hHtt67aa flies that had been transferred to 29°C at L3; 15-day old UAS-128Q-hHtt548aa flies raised at 29°C; 2-day old UAS-SCA1-82Q flies that were raised at 29°C since L3; and 2day old UAS-SCA3-78Q flies raised at 25°C.
Longevity Assay
UAS-128Q-hHtt548aa/elav-Gal4 (III) flies were analyzed for mean and median survival in the presence of either LacZ (control) or GFP tagged P42. The cohorts were tested for each genotype (101 and 81 males, respectively), which were collected within 24 hrs after eclosion and maintained on food at 25°C. Survival of flies was evaluated every day.
Supporting Information
Diagram of N-truncated parts of human Huntingtin proteins expressed in the different Drosophila models used in this report.
(PDF)
Low magnification images of HeLa cells transfected with polyQ-hHtt.
(PDF)
Alignment between dHtt (1-620aa) and hHtt P4 (166aa).
(PDF)
Quantification of aggregates, using ImageJ software on confocal stacks.
(PDF)
PolyQ-hHtt expression in HeLa cells.
(PDF)
In salivary glands, hHtt intensity and pattern does not change in presence of P42.
(PDF)
Confocal images of third larval peripheral nerves expressing hHtt548aa.
(PDF)
Quantification of HHtt immunoreactivity at the NMJ.
(PDF)
Protein extracts from HeLa cells transfected by GFP-polyQ-hHtt.
(PDF)
Acknowledgments
We greatly acknowledge Laurent Soustelle, Yves Grau and Joel Bockaert for constructive comments on the manuscript. We thank Brandon Loveall from Improvence for English corrections. We warmly thank Henry Neel for his helpful advice throughout this work. We also thank the following individuals for providing transgenic Drosophila lines: Leslie Thompson for UAS-93Q-Htt67aa, and J. Troy Littleton for UAS-0Q-Htt548aa, and UAS-128Q-Htt548aa. We also acknowledge the MGC Montpellier Genetic collections for providing Gateway vectors, the Montpellier RIO imaging facilities and the Bloomington stock center. YA and YTA were supported by a fellowship from the French Ministry of Research and Technology.
Funding Statement
Source of funding: CNRS (French National center of scientific research), “soutien au transfert” and ANR Emergence (Agence Nationale pour la Recherche). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Cowin RM, Roscic A, Bui N, Graham D, Paganetti P, et al. (2012) Neuronal aggregates are associated with phenotypic onset in the R6/2 Huntington's disease transgenic mouse. Behav Brain Res 229: 308–319. [DOI] [PubMed] [Google Scholar]
- 2. Yamamoto A, Lucas JJ, Hen R (2000) Reversal of neuropathology and motor dysfunction in a conditional model of Huntington's disease. Cell 101: 57–66. [DOI] [PubMed] [Google Scholar]
- 3. Goldberg YP, Nicholson DW, Rasper DM, Kalchman MA, Koide HB, et al. (1996) Cleavage of huntingtin by apopain, a proapoptotic cysteine protease, is modulated by the polyglutamine tract. Nat Genet 13: 442–449. [DOI] [PubMed] [Google Scholar]
- 4. Martindale D, Hackam A, Wieczorek A, Ellerby L, Wellington C, et al. (1998) Length of huntingtin and its polyglutamine tract influences localization and frequency of intracellular aggregates. Nat Genet 18: 150–154. [DOI] [PubMed] [Google Scholar]
- 5. Wellington CL, Ellerby LM, Gutekunst CA, Rogers D, Warby S, et al. (2002) Caspase cleavage of mutant huntingtin precedes neurodegeneration in Huntington's disease. J Neurosci 22: 7862–7872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Southwell AL, Bugg CW, Kaltenbach LS, Dunn D, Butland S, et al. (2011) Perturbation with intrabodies reveals that calpain cleavage is required for degradation of huntingtin exon 1. PLoS One 6: e16676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gafni J, Ellerby LM (2002) Calpain activation in Huntington's disease. J Neurosci 22: 4842–4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Van Raamsdonk JM, Pearson J, Rogers DA, Bissada N, Vogl AW, et al. (2005) Loss of wild-type huntingtin influences motor dysfunction and survival in the YAC128 mouse model of Huntington disease. Hum Mol Genet 14: 1379–1392. [DOI] [PubMed] [Google Scholar]
- 9. DiFiglia M, Sapp E, Chase KO, Davies SW, Bates GP, et al. (1997) Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science 277: 1990–1993. [DOI] [PubMed] [Google Scholar]
- 10. Graham RK, Deng Y, Slow EJ, Haigh B, Bissada N, et al. (2006) Cleavage at the caspase-6 site is required for neuronal dysfunction and degeneration due to mutant huntingtin. Cell 125: 1179–1191. [DOI] [PubMed] [Google Scholar]
- 11. Tanaka Y, Igarashi S, Nakamura M, Gafni J, Torcassi C, et al. (2006) Progressive phenotype and nuclear accumulation of an amino-terminal cleavage fragment in a transgenic mouse model with inducible expression of full-length mutant huntingtin. Neurobiol Dis 21: 381–391. [DOI] [PubMed] [Google Scholar]
- 12. Miller JP, Holcomb J, Al-Ramahi I, de Haro M, Gafni J, et al. (2010) Matrix metalloproteinases are modifiers of huntingtin proteolysis and toxicity in Huntington's disease. Neuron 67: 199–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Romero E, Cha GH, Verstreken P, Ly CV, Hughes RE, et al. (2008) Suppression of neurodegeneration and increased neurotransmission caused by expanded full-length huntingtin accumulating in the cytoplasm. Neuron 57: 27–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Slow EJ, Graham RK, Osmand AP, Devon RS, Lu G, et al. (2005) Absence of behavioral abnormalities and neurodegeneration in vivo despite widespread neuronal huntingtin inclusions. Proc Natl Acad Sci U S A 102: 11402–11407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Weiss KR, Kimura Y, Lee WC, Littleton JT (2012) Huntingtin aggregation kinetics and their pathological role in a Drosophila Huntington's disease model. Genetics 190: 581–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mangiarini L, Sathasivam K, Seller M, Cozens B, Harper A, et al. (1996) Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell 87: 493–506. [DOI] [PubMed] [Google Scholar]
- 17. Schilling G, Becher MW, Sharp AH, Jinnah HA, Duan K, et al. (1999) Intranuclear inclusions and neuritic aggregates in transgenic mice expressing a mutant N-terminal fragment of huntingtin. Hum Mol Genet 8: 397–407. [DOI] [PubMed] [Google Scholar]
- 18. Jackson GR, Salecker I, Dong X, Yao X, Arnheim N, et al. (1998) Polyglutamine-expanded human huntingtin transgenes induce degeneration of Drosophila photoreceptor neurons. Neuron 21: 633–642. [DOI] [PubMed] [Google Scholar]
- 19. Steffan JS, Bodai L, Pallos J, Poelman M, McCampbell A, et al. (2001) Histone deacetylase inhibitors arrest polyglutamine-dependent neurodegeneration in Drosophila. Nature 413: 739–743. [DOI] [PubMed] [Google Scholar]
- 20. Mugat B, Parmentier ML, Bonneaud N, Chan HY, Maschat F (2008) Protective role of Engrailed in a Drosophila model of Huntington's disease. Hum Mol Genet 17: 3601–3616. [DOI] [PubMed] [Google Scholar]
- 21. Lee WC, Yoshihara M, Littleton JT (2004) Cytoplasmic aggregates trap polyglutamine-containing proteins and block axonal transport in a Drosophila model of Huntington's disease. Proc Natl Acad Sci U S A 101: 3224–3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fecke W, Gianfriddo M, Gaviraghi G, Terstappen GC, Heitz F (2009) Small molecule drug discovery for Huntington's Disease. Drug Discov Today 14: 453–464. [DOI] [PubMed] [Google Scholar]
- 23. Ona VO, Li M, Vonsattel JP, Andrews LJ, Khan SQ, et al. (1999) Inhibition of caspase-1 slows disease progression in a mouse model of Huntington's disease. Nature 399: 263–267. [DOI] [PubMed] [Google Scholar]
- 24. Gafni J, Hermel E, Young JE, Wellington CL, Hayden MR, et al. (2004) Inhibition of calpain cleavage of huntingtin reduces toxicity: accumulation of calpain/caspase fragments in the nucleus. J Biol Chem 279: 20211–20220. [DOI] [PubMed] [Google Scholar]
- 25. Dragatsis I, Levine MS, Zeitlin S (2000) Inactivation of Hdh in the brain and testis results in progressive neurodegeneration and sterility in mice. Nat Genet 26: 300–306. [DOI] [PubMed] [Google Scholar]
- 26. Gauthier LR, Charrin BC, Borrell-Pages M, Dompierre JP, Rangone H, et al. (2004) Huntingtin controls neurotrophic support and survival of neurons by enhancing BDNF vesicular transport along microtubules. Cell 118: 127–138. [DOI] [PubMed] [Google Scholar]
- 27. Van Raamsdonk JM, Pearson J, Murphy Z, Hayden MR, Leavitt BR (2006) Wild-type huntingtin ameliorates striatal neuronal atrophy but does not prevent other abnormalities in the YAC128 mouse model of Huntington disease. BMC Neurosci 7: 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cattaneo E, Zuccato C, Tartari M (2005) Normal huntingtin function: an alternative approach to Huntington's disease. Nat Rev Neurosci 6: 919–930. [DOI] [PubMed] [Google Scholar]
- 29. Li Z, Karlovich CA, Fish MP, Scott MP, Myers RM (1999) A putative Drosophila homolog of the Huntington's disease gene. Hum Mol Genet 8: 1807–1815. [DOI] [PubMed] [Google Scholar]
- 30. Ehrnhoefer DE, Duennwald M, Markovic P, Wacker JL, Engemann S, et al. (2006) Green tea (-)-epigallocatechin-gallate modulates early events in huntingtin misfolding and reduces toxicity in Huntington's disease models. Hum Mol Genet 15: 2743–2751. [DOI] [PubMed] [Google Scholar]
- 31. Brand AH, Perrimon N (1993) Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401–415. [DOI] [PubMed] [Google Scholar]
- 32. Feany MB, La Spada AR (2003) Polyglutamines stop traffic: axonal transport as a common target in neurodegenerative diseases. Neuron 40: 1–2. [DOI] [PubMed] [Google Scholar]
- 33. Morfini GA, You YM, Pollema SL, Kaminska A, Liu K, et al. (2009) Pathogenic huntingtin inhibits fast axonal transport by activating JNK3 and phosphorylating kinesin. Nat Neurosci 12: 864–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sinadinos C, Burbidge-King T, Soh D, Thompson LM, Marsh JL, et al. (2009) Live axonal transport disruption by mutant huntingtin fragments in Drosophila motor neuron axons. Neurobiol Dis 34: 389–395. [DOI] [PubMed] [Google Scholar]
- 35. Talmat-Amar Y, Arribat Y, Redt-Clouet C, Feuillette S, Bouge AL, et al. (2011) Important neuronal toxicity of microtubule-bound Tau in vivo in Drosophila. Hum Mol Genet 20: 3738–3745. [DOI] [PubMed] [Google Scholar]
- 36. Gutekunst CA, Levey AI, Heilman CJ, Whaley WL, Yi H, et al. (1995) Identification and localization of huntingtin in brain and human lymphoblastoid cell lines with anti-fusion protein antibodies. Proc Natl Acad Sci U S A 92: 8710–8714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gunawardena S, Her LS, Brusch RG, Laymon RA, Niesman IR, et al. (2003) Disruption of axonal transport by loss of huntingtin or expression of pathogenic polyQ proteins in Drosophila. Neuron 40: 25–40. [DOI] [PubMed] [Google Scholar]
- 38. Bogdanik L, Mohrmann R, Ramaekers A, Bockaert J, Grau Y, et al. (2004) The Drosophila metabotropic glutamate receptor DmGluRA regulates activity-dependent synaptic facilitation and fine synaptic morphology. J Neurosci 24: 9105–9116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wolfgang WJ, Miller TW, Webster JM, Huston JS, Thompson LM, et al. (2005) Suppression of Huntington's disease pathology in Drosophila by human single-chain Fv antibodies. Proc Natl Acad Sci U S A 102: 11563–11568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Popiel HA, Nagai Y, Fujikake N, Toda T (2007) Protein Transduction Domain-mediated Delivery of QBP1 Suppresses Polyglutamine-induced Neurodegeneration In Vivo. Mol Ther 15: 303–309. [DOI] [PubMed] [Google Scholar]
- 41. Fernandez-Funez P, Nino-Rosales ML, de Gouyon B, She WC, Luchak JM, et al. (2000) Identification of genes that modify ataxin-1-induced neurodegeneration. Nature 408: 101–106. [DOI] [PubMed] [Google Scholar]
- 42. Warrick JM, Paulson HL, Gray-Board GL, Bui QT, Fischbeck KH, et al. (1998) Expanded polyglutamine protein forms nuclear inclusions and causes neural degeneration in Drosophila. Cell 93: 939–949. [DOI] [PubMed] [Google Scholar]
- 43. Duennwald ML, Jagadish S, Muchowski PJ, Lindquist S (2006) Flanking sequences profoundly alter polyglutamine toxicity in yeast. Proc Natl Acad Sci U S A 103: 11045–11050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Southwell AL, Khoshnan A, Dunn DE, Bugg CW, Lo DC, et al. (2008) Intrabodies binding the proline-rich domains of mutant huntingtin increase its turnover and reduce neurotoxicity. J Neurosci 28: 9013–9020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Takano H, Gusella JF (2002) The predominantly HEAT-like motif structure of huntingtin and its association and coincident nuclear entry with dorsal, an NF-kB/Rel/dorsal family transcription factor. BMC Neurosci 3: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Southwell AL, Ko J, Patterson PH (2009) Intrabody gene therapy ameliorates motor, cognitive, and neuropathological symptoms in multiple mouse models of Huntington's disease. J Neurosci 29: 13589–13602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zala D, Colin E, Rangone H, Liot G, Humbert S, et al. (2008) Phosphorylation of mutant huntingtin at S421 restores anterograde and retrograde transport in neurons. Hum Mol Genet 17: 3837–3846. [DOI] [PubMed] [Google Scholar]
- 48. Warby SC, Doty CN, Graham RK, Shively J, Singaraja RR, et al. (2009) Phosphorylation of huntingtin reduces the accumulation of its nuclear fragments. Mol Cell Neurosci 40: 121–127. [DOI] [PubMed] [Google Scholar]
- 49. Zuccato C, Valenza M, Cattaneo E (2010) Molecular mechanisms and potential therapeutical targets in Huntington's disease. Physiol Rev 90: 905–981. [DOI] [PubMed] [Google Scholar]
- 50.Ehrnhoefer DE, Sutton L, Hayden MR (2011) Small Changes, Big Impact: Posttranslational Modifications and Function of Huntingtin in Huntington Disease. Neuroscientist. [DOI] [PMC free article] [PubMed]
- 51. Arrasate M, Mitra S, Schweitzer ES, Segal MR, Finkbeiner S (2004) Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature 431: 805–810. [DOI] [PubMed] [Google Scholar]
- 52. Bauer PO, Goswami A, Wong HK, Okuno M, Kurosawa M, et al. (2010) Harnessing chaperone-mediated autophagy for the selective degradation of mutant huntingtin protein. Nat Biotechnol 28: 256–263. [DOI] [PubMed] [Google Scholar]
- 53.Butler DC, McLear JA, Messer A (2011) Engineered antibody therapies to counteract mutant huntingtin and related toxic intracellular proteins. Prog Neurobiol. [DOI] [PMC free article] [PubMed]
- 54. Butler DC, McLear JA, Messer A (2012) Engineered antibody therapies to counteract mutant huntingtin and related toxic intracellular proteins. Prog Neurobiol 97: 190–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. McLear JA, Lebrecht D, Messer A, Wolfgang WJ (2008) Combinational approach of intrabody with enhanced Hsp70 expression addresses multiple pathologies in a fly model of Huntington's disease. FASEB J 22: 2003–2011. [DOI] [PubMed] [Google Scholar]
- 56. Thakur AK, Jayaraman M, Mishra R, Thakur M, Chellgren VM, et al. (2009) Polyglutamine disruption of the huntingtin exon 1 N terminus triggers a complex aggregation mechanism. Nat Struct Mol Biol 16: 380–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Dlugosz M, Trylska J (2011) Secondary structures of native and pathogenic huntingtin N-terminal fragments. J Phys Chem B 115: 11597–11608. [DOI] [PubMed] [Google Scholar]
- 58. Khoshnan A, Ko J, Patterson PH (2002) Effects of intracellular expression of anti-huntingtin antibodies of various specificities on mutant huntingtin aggregation and toxicity. Proc Natl Acad Sci U S A 99: 1002–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Tam S, Spiess C, Auyeung W, Joachimiak L, Chen B, et al. (2009) The chaperonin TRiC blocks a huntingtin sequence element that promotes the conformational switch to aggregation. Nat Struct Mol Biol 16: 1279–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Snyder-Keller A, McLear JA, Hathorn T, Messer A (2010) Early or late-stage anti-N-terminal Huntingtin intrabody gene therapy reduces pathological features in B6.HDR6/1 mice. J Neuropathol Exp Neurol 69: 1078–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Milan M, Cohen SM (2000) Temporal regulation of apterous activity during development of the Drosophila wing. Development 127: 3069–3078. [DOI] [PubMed] [Google Scholar]
- 62. Sanyal S (2009) Genomic mapping and expression patterns of C380, OK6 and D42 enhancer trap lines in the larval nervous system of Drosophila. Gene Expr Patterns 9: 371–380. [DOI] [PubMed] [Google Scholar]
- 63. Franco B, Bogdanik L, Bobinnec Y, Debec A, Bockaert J, et al. (2004) Shaggy, the homolog of glycogen synthase kinase 3, controls neuromuscular junction growth in Drosophila. J Neurosci 24: 6573–6577. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Diagram of N-truncated parts of human Huntingtin proteins expressed in the different Drosophila models used in this report.
(PDF)
Low magnification images of HeLa cells transfected with polyQ-hHtt.
(PDF)
Alignment between dHtt (1-620aa) and hHtt P4 (166aa).
(PDF)
Quantification of aggregates, using ImageJ software on confocal stacks.
(PDF)
PolyQ-hHtt expression in HeLa cells.
(PDF)
In salivary glands, hHtt intensity and pattern does not change in presence of P42.
(PDF)
Confocal images of third larval peripheral nerves expressing hHtt548aa.
(PDF)
Quantification of HHtt immunoreactivity at the NMJ.
(PDF)
Protein extracts from HeLa cells transfected by GFP-polyQ-hHtt.
(PDF)