Abstract
Slowly-cycling tumor cells that may be present in human tumors may evade cytotoxic therapies, which tend to be more efficient at destroying cells with faster growth rates. However, the proportion and growth rate of slowly-cycling tumor cells is often unknown in preclinical model systems used for drug discovery. Here we report a quantitative approach to quantitate slowly-cycling malignant cells in solid tumors, using a well-established mouse model of Kras-induced lung cancer (KrasG12D/+). Bromodeoxyuridine (BrdU) was administered to tumor-bearing mice and samples were collected at defined times during pulse and chase phases. Mathematical and statistical modeling of the label-retention data during the chase phase supported the existence of a slowly-cycling label-retaining population in this tumor model and permitted the estimation of its proportion and proliferation rate within a tumor. The doubling time of the slowly cycling population was estimated at ~5.7 weeks and this population represented ~31% of the total tumor cells in this model system. The mathematical modeling techniques implemented here may be useful in other tumor models where direct observation of cell cycle kinetics is difficult and may help evaluate tumor cell subpopulations with distinct cell-cycling rates.
Major Findings
Mathematical modeling of label-retention data from a mouse model of lung cancer provides quantitative evidence for the presence of two populations of tumor cells with distinct cell-cycling kinetics.
Keywords: mathematical modeling, label-retention in vivo, tumor cell proliferative kinetics, lung cancer, animal models of cancer
Quick Guide to Equations and Assumptions
Equations
The two mathematical models compared in the analysis of the chase data are the following:
-
exponential model (one-compartment model):
(1) Where W is the natural logarithm of the percentage of BrdU+ cells within a tumor, and ε ~ N (0, σ).
-
bi-exponential model (two-compartment model)
(2) Where W is the natural logarithm of the percentage of BrdU+ cells within a tumor and ε ~ N (0,σ).
The first equation uses an exponential decay function to model the percentage of tumor cells that are BrdU+. The parameters A and α are non-negative constants, and ε is an error term from a normal distribution with mean zero and standard deviation σ. The second equation models the same data as a bi-exponential decay function. The parameters A, α, B, β are non-negative constants, and ε is an error term from a normal distribution with mean zero and standard deviation σ.
Parameter descriptions and interpretation
The parameter t denotes the time elapsed from time zero (after all BrdU administration ended) during chase phase.
The parameters α and β model how fast the cycling cells lose BrdU labeling.
The parameters A and B give the initial percentage of BrdU+ cells for each population.
The parameter ratio A/B gives the relative ratio of fast- and slow-cycling population in tumors.
At given time t during the chase phase, gives the observed percentage of slow-cycling cells in BrdU+ cells.
Assumptions
The main assumptions include the following:
Proliferation rates are proportional to the number of cells present;
Each tumor cell has a cross-sectional area of approximately 121 μm2 (measurements of cell counts and areas of a random sample of 10 tumors gave a median value of 121 μm2, with a standard deviation of 25 μm2) (Supplemental Table S1).
Cell death rates are negligible for the time period of the experiment (we previously confirmed that cell death rates as determined by caspase-3 staining are indeed quite low (<1%) in this setting) ((1), also Supplemental Figure S1, Supplemental Table S2);
The variance of the residuals of the log-transformed data is approximately constant;
Proportional error structures best characterize the residual errors for both models (based on tests of proportional and additive error structures);
During BrdU pulse, every cell that undergoes cell division takes in enough BrdU to be detectable after only one cell division;
The incorporation of BrdU has negligible effect on the survival of cells and their rate of cycling;
All cells require the same number of cell divisions to reach undetectable label levels (which can actually undercount cells that slowly cycle);
The rate of loss of BrdU label is proportional to rate of proliferation of labeled cells;
All cells in the region labeled as tumor are actual tumor cells;
Tumors at the same time point but from different mice are comparable in age and size distributions;
Tumor measurements at the same time point are independent from one sample to another, whether from the same mouse or from different mice;
Selection of tumor slices to sample is random;
Tumor growth favors cells that cycle quickly over cells that cycle more slowly, in that any cells that cycle more slowly will become a smaller proportion of the total number of cells over time (for this analysis, the implication is that any population of slowly-cycling cells that is detected at later time points was likely a larger proportion of all cells at earlier time points);
Introduction and Background
Normal adult stem cells are thought to be relatively quiescent, a property which protects them from proliferative exhaustion (2). Because of this property, “label-retention” studies have been used to identify and characterize tissue-specific stem cells for decades, following the pioneering work by Potten and colleagues in the intestine (3). The existence of label-retaining cells has been proposed to be important for radiation response (3, 4). Label-retention approaches have also been used to identify stem cells in the interfollicular epidermis (5–9) and the hematopoietic system (10–12). In the hematopoietic stem cell (HSC) compartment, some studies have suggested the existence of a slowly-cycling stem cell population (9, 10, 12) whereas other investigators have not found label retention in this compartment (11). In cancer research, increasing attention has focused on the heterogeneity of tumor cells present within the tumor mass (distinct from the heterogeneity of non-tumor cells due to the presence of a tumor microenvironment) due to the hypothesis that certain subpopulations of tumor cells have increased capacity to propagate. These “cancer stem cells” (CSCs) or “tumor initiating cells (TICs) share with normal stem cells the ability to self-renew and “differentiate” into committed cell types with more limited proliferative capacity (13). Cancer stem cell populations have been identified and extensively characterized in several solid tumors (14–17). However, it is not clear whether these CSC populations share the property of “label-retention” that has been ascribed to some normal stem cells. Recent data in glioblastoma and in skin cancer suggest that indeed such slowly cycling CSCs may exist (18, 19). However, whether this is true for other tumor types remains to be determined. A limitation to carrying out a rigorous analysis of the cell cycle kinetics of solid tumor populations is the lack of robust and rigorous quantitative methods.
Cancer stem cells in solid tumors are traditionally identified by differential cell sorting with specific cell surface markers (20). The operational “stemness” property is defined as an increased ability to transplant tumors into an immunocompromised host. However, this definition of cancer stem cells remains controversial, as transplantation success rates can vary widely depending on a wide range of technical factors (21). In human lung cancer, several reports have suggested that a cancer stem cell population can be enriched using cell surface markers, yet the cell cycle kinetics of these subpopulations (or indeed, whether slowly-cycling cells are synonymous with cancer stem cells defined by transplantation assays) has not been explored (22, 23).
The KrasG12D/+ mouse model has been well-validated for the study of lung cancer initiation and progression in vivo (24). Previous work has demonstrated that this model closely recapitulates the molecular changes seen in human non-small cell lung cancer (NSCLC) (25). In this model, an oncogenic Kras allele is expressed only after delivery of Cre recombinase. Cre recombinase is delivered via nasal instillation using an Adenoviral vector (AdCre). Thus, timing of tumor initiation can be well-controlled. After Cre delivery, the lung epithelium begins to proliferate and over time the lung parenchyma is occupied by multiple adenomas, some of which then progress to adenocarcinoma. In the studies described here, BrdU incorporation was measured in tumors within the lungs in this KrasG12D/+ model. Previous work using this model identified a population of CD45−;PECAM−;Sca1+ cells enriched for double-positive SPC/CC10 cells (BASCs) that are activated after lung injury and thus thought to be CSCs (26). However, this population was not enriched for transplantation ability into immunocompromised mice in the KrasG12D/+ model(27). The fact that only a small percentage of cells in the KrasG12D/+ model can transplant tumors suggests that there is a subpopulation of cells with cancer stem-cell characteristics. “Label retention” of slowly-cycling cells provides an alternative method for FACs-sorting of surface markers to identify populations of cells within tumors with important biological properties that may bypass the limitations of the transplantation approach. An advantage of label-retention studies is that they allow for rigorous testing of the hypothesis that a slowly-cycling population exists without the need for complex crosses with lineage tracing reporters and other genetic tools (18, 19, 28). A priori, it cannot be assumed that CSCs are label retaining. However, if a label-retaining population can be identified by label-retention studies, this could justify further experimentation to allow for isolation of these cells. In addition, it may be important in some setting to determine whether the CSC population is synonymous with the slowly-cycling population or whether these are overlapping but distinct entities. Recent data isolating and characterizing a label-retaining population in breast tissue supports this approach (29).
A commonly-used method to identify label-retaining cells is to mark proliferating cells using the thymidine analogue 5-bromo-2-deoxyuridine (BrdU). After administration of BrdU ends, cycling cells will lose this DNA label as it will be diluted by half each complete cell cycle. Therefore, both BrdU uptake (“pulse”) and BrdU dilution (“chase”) data are useful in quantifying cell turnover rates. A key element of such an approach is to develop appropriate statistical and mathematical modeling techniques to demonstrate the presence of “label-retaining” cells. Mathematical modeling of BrdU label incorporation and retention can quantify the behavior of the labeled cells and enable identification of populations with different cell cycle kinetics. Rigorous mathematical approaches have been developed that use BrdU label retention to determine cell cycle kinetics in other settings. For example, Bonhoeffer et al. used a mathematical model of BrdU incorporation to describe the kinetics of CD4+ and CD8+ lymphocytes circulating in uninfected and SIV-infected macaques (30). The model was used to estimate proliferation and death rates of these specific lymphocyte subsets. Similarly, Kiel et al. modeled BrdU incorporation and retention data from mice to test the “immortal strand” hypothesis of asymmetric chromosome segregation during division of hematopoietic stem cells (HSC) and concluded that asymmetric segregation of chromosomes does not occur in the division of HSCs (3). Here we use mathematical and statistical analysis of in vivo BrdU labeling data to determine whether tumors in a mouse model of lung cancer contain a population of slowly cycling tumor cells. Our results strongly suggest that a population of slowly cycling tumor cells does exist in the KrasG12D/+ model. These studies establish the necessary justification for further analysis of this slowly cycling population in this model. More generally, we propose that the mathematical analysis of BrdU labeling in tumor models applied here may be broadly useful for other similar tumor models in order to elucidate the heterogeneity of tumor cell kinetics.
Materials and Methods
Mice
For all studies, the KrasG12D/+ mouse model was used. Mice were genotyped as previously described (24). Adenoviral Cre (AdCre) was delivered by intranasal instillation between 4 and 6 weeks of age. Mice were then allowed to age for 12 weeks prior to BrdU administration. All animal experiments were approved by the Stanford University School of Medicine Administrative Panel on Laboratory Animal Care (APLAC).
BrdU labeling
BrdU (Sigma-Aldrich) was administered to the mice for up to 15 days for the pulse experiment, and 7 days for the chase experiment. Administration was done simultaneously both through drinking water (1mg/mL) and daily i.p. injections (100 mg per kilogram body weight) in order to maximize delivery.
Immunohistochemistry and immunofluorescence
After sacrifice, mouse lungs were removed, fixed with Z-fix (Anatech Inc) and embedded in paraffin. Immunohistochemical or immunofluorescence staining was performed on 4 μm sections. For primary antibodies, anti-BrdU (BD Pharmigen clone 3d4, 1:200 dilution), anti-TTF1 (Abcam, 1:200 dilution), anti-E-Cadherin (BD Transduction Laboratories, clone 36, 1:200 dilution) and anti-cleaved caspase 3 (Cell signaling, 1:300 dilution) were used. Detection was performed with goat anti-rat Alexa Fluro® 594 (Invitrogen), donkey anti-mouse Alexa Fluro® 488 (Invitrogen), and donkey anti-rabbit Alexa Fluro® 488 (Invitrogen) for immunofluorescence, or HRP-conjugated goat anti-mouse and ABC goat-anti-rabbit kit for immunohistochemistry.
Quantification of immunohistochemistry
The area of each tumor cross-section was measured using Bioquant software and the number of BrdU-positive (BrdU+) cells within each tumor was assessed by manual counting. The number of BrdU+ cells per tumor cross-sectional area was recorded. A manual count of ten randomly-selected tumors gave an estimate of 121 μm2 area per tumor cell (standard deviation 24.9 μm2). This estimate was used to convert BrdU+ cells/area into units of BrdU+ cells/total tumor cells. Two mice at each time point were used to generate the pulse data. At each time point in the chase period, approximately 5 or 6 mice were selected at random (one time point had 7 mice, and another had 3). For each selected mouse, multiple slices of the tumor-bearing lung were obtained and placed at random positions on slides. Approximately five tumors were selected randomly from the available slices for each mouse for analysis (1 mouse with 1 tumor, 1 mouse with 2 tumors, 9 mice with 4 tumors and 39 mice with 5 tumors).
Mathematical modeling
At the end of the twelve weeks of the chase, labeled cells were still detected. This suggested the possibility of a label-retaining subpopulation with a slower cycling time than other cells. To test this hypothesis, we fit two mathematical models to the chase data and compared the goodness-of-fit of the models. The first was an exponential decay model, which assumes that there is a single cell-cycling rate. The second was a bi-exponential decay model, which assumes that there are two distinct cell-cycling rates. Each model included an error structure that was selected to give the best fit for that particular model, to account for residual variability. Different error structures were tested on each model, and a two-sided Kolmogorov-Smirnov test was performed to confirm that a proportional error structure resulted in residuals that were indistinguishable from a normal distribution for each of the models.
Due to the random selection of the slices and tumors for analysis (described above), we assume that the %BrdU values from a single mouse form a random sample. This is also supported by Supplemental Figure S2, which shows chase data plotted by mouse. Supplemental Figure S2 also provides graphical evidence that at each time point, the distributions of the %BrdU+ values are similar between mice. In addition, the Kruskal-Wallis test, an analog of the ANOVA test for non-normally distributed data, was performed to compare the median %BrdU+ values for all of the mice at a given time point. With a significance level of 0.05, five out of the nine time points showed no significant difference between those median values. For the remaining four time points, there was at least one mouse whose median differed from those of the other mice (although only one of those time points would have shown a difference at a significance level of 0.01). To further investigate possible differences between mice, we used the Wilcoxon rank sum test, a non-parametric analog of the t-test, to compare the median %BrdU+ values for each pair of mice at a time point. A Bonferroni correction was used to account for the multiple pairwise tests performed at each time point. For 118 out of the 119 possible comparisons, the median values for mice at a given time point were not pairwise different at a significance level of 0.05 (for the other comparison, the p-value was 0.045). Based on all of these results, we assume that all %BrdU+ values at a given time point form a random sample.
Goodness-of-fit testing
Both mathematical models were fit to the data using nonlinear regression. In both cases, a natural logarithm transformation of the data was used to account for the heteroscedasticity (nonuniform variance) of the data. Additive and proportional error structures were then tested on the models. For both models, proportional error structures gave the best fit, and were used throughout the analysis. Goodness-of-fit comparisons of the two models were made using mean square error (MSE) and Akaike information criterion (AIC) values.
Results
Determination of minimal sufficient length of labeling phase
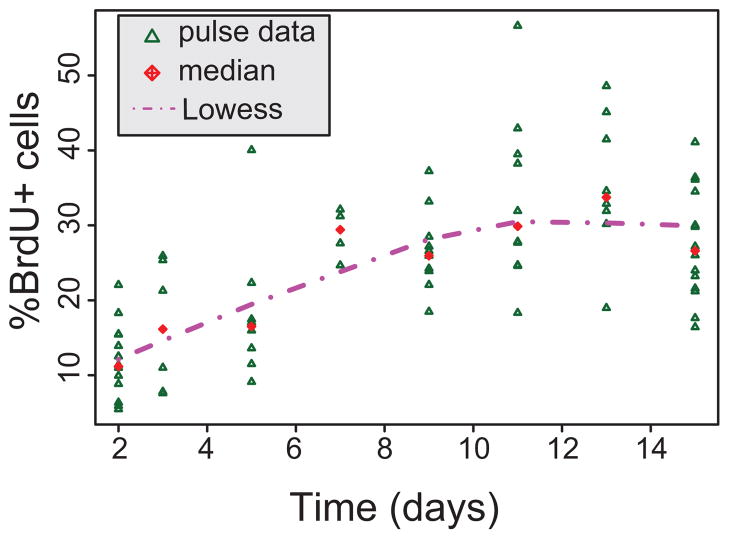
To determine how long BrdU label should be administered for the chase experiment, an extended pulse experiment was performed. KrasG12D/+ mice were treated with AdCre to initiate expression of oncogenic Kras. Twelve weeks after introduction of AdCre, when tumors were well-established, mice were given BrdU to label proliferating tumor cells. To determine the minimal sufficient length of labeling phase, an initial “pulse” of BrdU was delivered to mice for up to 15 days (see methods). Mice were sacrificed on days 2, 3, 5, 7, 9, 11, 13, and 15 during the labeling period and BrdU label incorporation was assessed by immunohistochemistry using an anti-BrdU antibody in paraffin-fixed sections of lung. As shown in Figure 1, the cell population was effectively fully labeled by Day 7 (chase time “0 week”), with a median Day 7 level of 29.4%. Therefore, for the chase data collection, a pulse phase of 7 days was used for the labeling of the mice.
Figure 1. Determination of minimal sufficient length of labeling phase.
All pulse data, expressed as the percentage of tumor cells that are BrdU-positive (%BrdU+). Times shown are the days since the first dose of BrdU administration began. We attempted to test a mono-exponential and a bi-exponential model for the pulse data, but the time period was insufficient to distinguish between the two models.
Detection of BrdU+ cells through a pulse-chase experiment
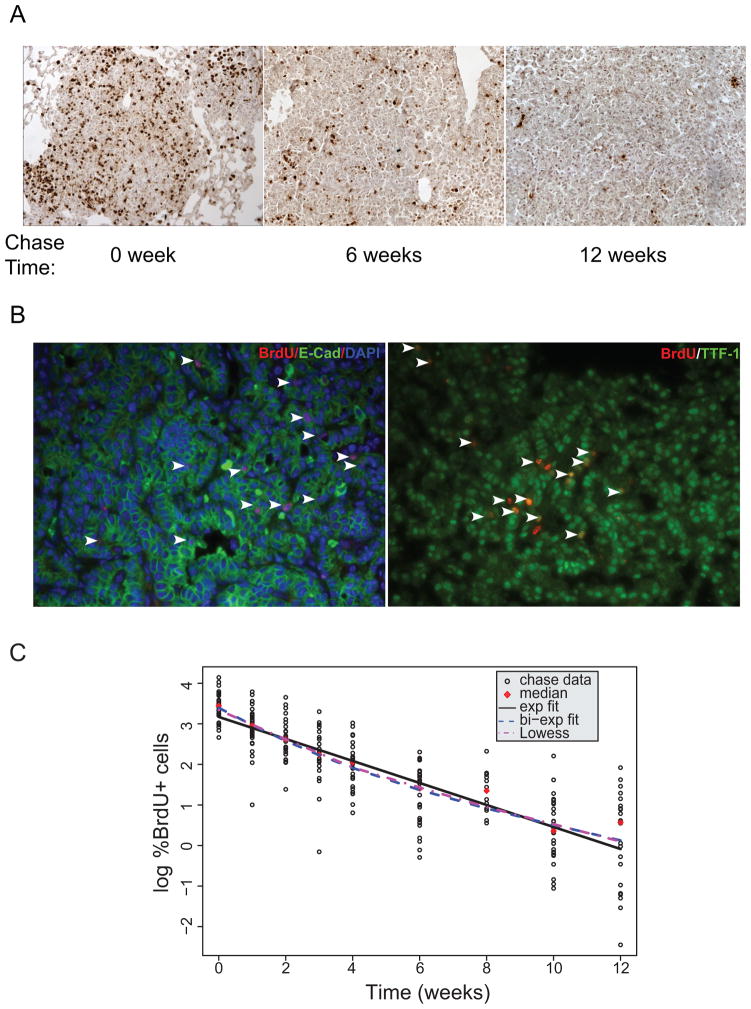
For the chase experiment, BrdU was administered over 7 days to a second cohort of KrasG12D/+ mice 12 weeks after tumor initiation (n=50). After seven days of BrdU “pulse”, mice were sacrificed at weeks 0, 1, 2, 3, 4, 6, 8, 10, and 12 (Figure 2). Four to six mice from this cohort were sacrificed at each specified time and the remaining number of BrdU+ cells was determined again by immunohistochemistry (Figure 2A). To confirm that BrdU label-retaining cells, and particularly those present at the end of the chase experiment, were indeed lung tumor cells and not cells from the surrounding stroma, we used double-labeling with immunofluorescence using an anti-BrdU antibody and an antibody against lung epithelial marker E-cadherin or TTF-1 (Figure 2B). The results demonstrate that the vast majority of BrdU+ cells are indeed of epithelial origin, and therefore are likely to be lung tumor cells.
Figure 2. Detection of BrdU+ cells through a pulse-chase experiment.
A. Representative examples of BrdU staining of mice at the indicated time points during the “chase” phase. B. Representative co-immunofluorescence staining of BrdU and epithelial markers E-Cadherin or TTF-1 at the end of “chase” phase. C. All chase data, expressed as the percentage of tumor cells that are BrdU-positive, in a log-linear plot. Time “0” represents when BrdU administration for chase experiment stopped.
Mathematical modeling supports the existence of a distinct, slow-cycling population
At the end of the twelve weeks of the chase, labeled tumor cells were still detected (Figure 2A, 12 week and Figure 2B). This suggested the possibility of a label-retaining tumor cell subpopulation with a slower cycling time than other tumor cells. To test this hypothesis, we fit two mathematical models to the chase data and compared the goodness-of-fit of the models (Figure 2C and Table 1). The first was an exponential decay model, which assumes that there is a single cell-cycling rate (Equation 1). The second was a bi-exponential decay model, which assumes that there are two distinct cell-cycling rates (Equation 2). Each model included an error structure (as described in Materials and Methods section) that was selected to give the best fit for that particular model, to account for residual variability. Different error structures were tested on each model, and a two-sided Kolmogorov-Smirnov test was performed to confirm that a proportional error structure resulted in residuals that were indistinguishable from a normal distribution for each of the models. We considered a few refinements for these two models, but finally used the simple models described above:
Table 1. Summary of parameter estimates and measures of “Goodness-of-fit” for exponential and bi-exponential models.
Percent relative standard error (%RSEs) for each parameter estimate is included in parentheses in left panel. Measures of “Goodness-of-fit” in the right panel are derived from either MSE or AIC method.
| A (%BrdU+ cells) | α (1/week) | B (%BrdU+ cells) | β (1/week) | MSE | AIC | |
|---|---|---|---|---|---|---|
| Exponential model | 23.4 (7.2%) | 0.269 (4.4%) | - | - | 0.476 | 496.46 |
| Bi-exponential model | 20.3 (43%) | 0.549 (49%) | 9.14 (110%) | 0.176 (50%) | 0.456 | 490.25 |
One refinement considered was allowing cells to take differing numbers of cell divisions to reach an undetectable level of label. Kiel et al. (11) estimated that approximately three rounds of cell division are needed to decrease the level of BrdU present in a cell below the threshold of detection after full labeling. Bonhoeffer et al. (30) estimated that the number of cell divisions needed to have the BrdU level drop below the lower limit of detection to be 5 to 6 divisions. We assumed that all cells require the same number of divisions during the chase phase to reach an undetectable level of label, which allowed us to use the simple models. This is a reasonable assumption if all cells begin the chase phase with equally high levels of label. However, if there are slower-cycling cells, they could begin the chase phase with lower levels of label than other cells, due to fewer cell divisions during the labeling period. Since BrdU is taken up into DNA during the S phase of cell division, the number of times a cell divides during the BrdU “pulse phase” affects the amount of BrdU incorporated at the end of the pulse phase. Cells that divide more rapidly can take up more label per time period than cells that divide more slowly (31). If the label period is not sufficiently long, slower-cycling cells may require fewer numbers of divisions for the label to become undetectable. This could lead to an undercounting of slower-cycling cells at later times during the chase phase, if such cells exist.
In contrast to the situation in normal tissues, the size of the compartment for solid tumors changes over time. To account for this, we normalized counts of BrdU+ cells by an area-based estimate of the total number of cells in each tumor sample. We did not include cell death in our mathematical models as tumors in the KrasG12D/+ model are highly proliferative and yet show very few apoptotic cells. Cell death rates as determined by caspase-3 staining are indeed quite low (<1%) in this setting (1). Therefore, cell death was not included in the mathematical models.
We also fit exponential and bi-exponential models to the pulse data and compared the goodness-of-fit of the models. However, the data were insufficient to be able to distinguish one model as significantly better than the other. Therefore we did not include a model for the pulse data in our results.
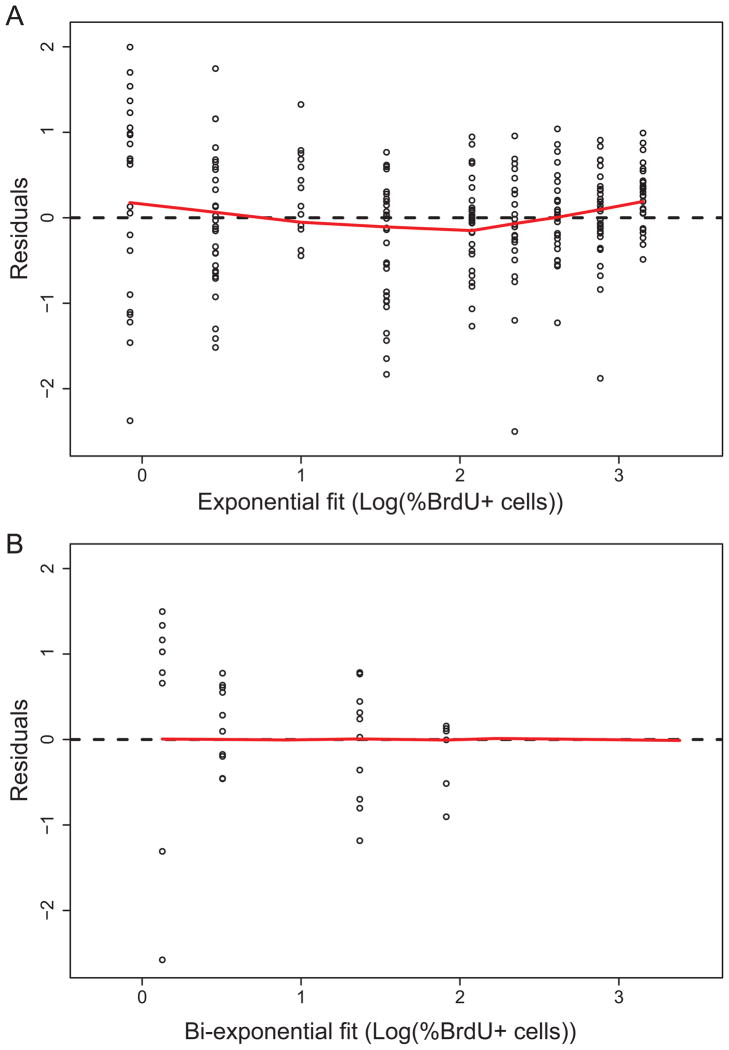
Mean squared error (MSE) and Akaike information criterion (AIC) were used to compare the models. Both MSE and AIC are measures of goodness of fit, with a smaller value indicating a better fit. However, AIC penalizes for extra parameters, taking into account that a model with more parameters has more degrees of freedom and will give a better fit to data because of this. As shown in Table 1, the MSE values were 0.476 and 0.456 for the exponential and bi-exponential models, respectively. The AIC values were 496.46 and 490.25 for the exponential and bi-exponential models, respectively. Therefore, the mathematical model that assumed two distinct cycling periods gave a measurably superior fit to the data than the model with a single cycling period using either criterion. Both models incorporated variability as described in the Methods section, so the difference was not simply due to random variability favoring a model with multiple cycling periods. In addition, a lowess fit was made for the residuals with a proportional error structure for each model, and visual comparison confirmed that the residuals for the bi-exponential model were more uniformly distributed than the residuals for the exponential model (Figures 3A and 3B).
Figure 3. Residual analysis for exponential and bi-exponential models.
A. Residuals for the exponential model, with a proportional error structure, fit to chase data. The red curve is a lowess fit. B. Residuals for the bi-exponential model, with a proportional error structure, fit to chase data. The red curve is a lowess fit.
Cross-validation supports that the two-compartment model is a superior model
To further verify that the bi-exponential model was indeed a better fit to the data, a number of cross-validation tests were performed. The cross-validations give a measure of the sensitivity of the models to the data, by testing whether the models over fit the data. For each cross-validation, a subset of data (the validation data) was removed from the full data set. The remaining data (the training data) were fit with the two models described in the Quick Guide to Equations and Assumptions. The structure of the training data sets for the various cross-validations is described below.
Cross validation 1 (omit one mouse): A training data set was formed by removing all data taken from a single mouse. This procedure was repeated so that each mouse was left out once, for a total of 50 cases. The fitting algorithm for the bi-exponential model did not converge in one of these cases, so there were only 49 cases in which the two models could be compared.
Cross validation 2 (omit one mouse per time point): A training set of data was formed by removing all data from one mouse at each time point. This was done 1000 times, with random selection of the mice.
Cross validation 3 (omit one tumor per mouse): Each training set of data was formed by randomly removing all of the data from one tumor from each mouse. This was done 1000 times.
Cross validation 4 (omit two tumors per mouse): Training sets were formed by removing two samples at random from the data from each mouse. This was done 1000 times.
Cross validation 5 (omit 20% of data per time point): Training sets were formed by removing approximately 20% of the samples at each time point at random. This was done 1000 times.
The mean square error (MSE) and the Akaike information criterion (AIC) were computed for each of the models when they were fit to a training data set. The percentages of times the bi-exponential model gave a superior fit (as measured by lower MSE or AIC) are recorded in Table 2. For all five types of cross-validations performed, the bi-exponential model gave a better fit than the exponential model as determined by both MSE and AIC. The MSE was lower for the bi-exponential model in 100% of cases. The AIC was lower for the bi-exponential model in more than 75% of cases for each of the five cross-validations.
Table 2. Summary of cross-validation results.
Upper panel shows the percentage of cases in which the bi-exponential model was superior to the exponential model, when both were fit to the training data sets. Lower panel is the percentage of cases in which the bi-exponential model was superior to the exponential model, as determined by lower value of MSE on the testing data set, for each model determined by fitting to the validation data sets.
| Percentage of training data sets for which the bi-exponential model fit was better | ||
|---|---|---|
| Cross-validation | According to MSE | According to AIC |
| 1 | 100 % | 100 % |
| 2 | 100 % | 98.2 % |
| 3 | 100 % | 97.9 % |
| 4 | 100 % | 75.3 % |
| 5 | 100 % | 100 % |
| Percentage of validation data sets for which the bi-exponential model fit was better | |
|---|---|
| Cross-validation | According to MSE |
| 1 | 53 % |
| 2 | 79.2 % |
| 3 | 80.9 % |
| 4 | 86.5 % |
| 5 | 100 % |
For each of the fitted models derived from the training data, the MSE was computed for the validation data. The percentages of cases in which the MSE value was better (lower) for the bi-exponential function than the exponential function are recorded in Table 2. In each of the cross-validation methods performed, the bi-exponential model had lower MSE values than the exponential model for the majority of cases (over 50%), and was therefore concluded to be the superior model.
Predictive checks supported selection of the bi-exponential model
As further model validation, two predictive checks were performed for the models. The best-fitting exponential and bi-exponential models to the full data set were used to produce simulated data.
Predictive check 1: The same number of data points at each time point as contained in the original data set were simulated for each time point, and this was done 1000 times, to give 1000 full data sets of simulated data for each of the two models.
Predictive check 2: A total of 500 data points were simulated at each time point. This was repeated 1000 times, to give 1000 simulated data sets (each with 500 data points at each time) for each of the two models.
For each predictive check, the distributions of the simulated and real data at each time point were compared. The two-sided Kolmogorov-Smirnov test was applied to determine statistical significance for differences between the distributions, with the null hypothesis being that any two distributions being compared cannot be distinguished (with a significance level of 0.05). Table 3 summarizes the results of the tests, with the entries giving the percentages of cases (out of 1000 simulations) in which the null hypothesis was rejected and the two distributions being compared were distinguishable according to the Kolmogorov-Smirnov test. In addition to the comparisons of simulated and real data, comparisons were made between simulated data sets for the two different models. Those results are included in the tables as well. In both predictive checks, for the majority of the points, the bi-exponential model was indistinguishable from the data more often than the exponential model was. This was true for six of nine time points in the first test (Table 3, upper panel) and eight of nine time points in the second test (Table 3, lower panel).
Table 3. The percentage of cases (out of 1000 simulations) in which the given distributions were found to be different at each time point with simulated data.
Predictive check 1 uses a sample size equal to the number of samples in the original data set at each time point. A sample size of 500 at each time point was used for predictive check 2.
| Predictive check 1 | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Time (weeks) | 0 | 1 | 2 | 3 | 4 | 6 | 8 | 10 | 12 |
| Data vs. log exp | 32.3 | 4.9 | 2.9 | 2.7 | 8.4 | 7.1 | 6.9 | 0.2 | 42 |
| Data vs. log bi-exp | 5.9 | 2.9 | 3.3 | 2.6 | 5.0 | 0.8 | 14.4 | 0.8 | 12.7 |
| Log exp vs. log bi-exp | 11.8 | 4.7 | 5.6 | 6.2 | 8.7 | 10.1 | 5.3 | 3.6 | 11.9 |
|
| |||||||||
| Predictive check 2 | |||||||||
| Time (weeks) | 0 | 1 | 2 | 3 | 4 | 6 | 8 | 10 | 12 |
|
| |||||||||
| Data vs. log exp | 98.8 | 0.1 | 0 | 0 | 0.6 | 0.8 | 0 | 0 | 100 |
| Data vs. log bi-exp | 0 | 0 | 0 | 0 | 0.4 | 0 | 2.3 | 0 | 15.4 |
| Log exp vs. log bi-exp | 99.5 | 36.3 | 10.8 | 61.9 | 89.2 | 92.2 | 40.3 | 14.2 | 97.5 |
Predicted values
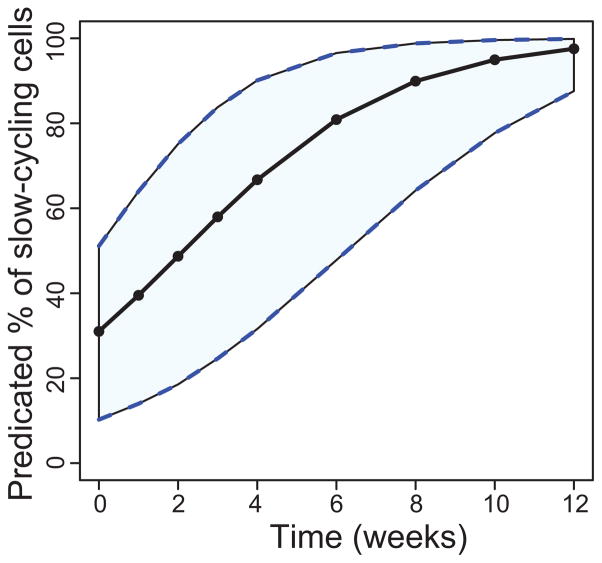
The parameter estimates for the two-compartment model in Table 1 indicate that approximately 31% of the cells were initially in the slow population, and that the mean turnover rates for the fast and slow populations were 1.8 and 5.7 weeks/cell, respectively. Figure 4 shows the bi-exponential function prediction of the slowly-cycling population over the course of the chase experiment. It was created by first bootstrapping the data sets to create 1000 new data sets, and fitting each data set to get a new set of parameter estimates. Those 1000 sets of parameter estimates were then used to calculate the fraction of slowly-cycling cells at each experimental time point. The 2.5, 50, and 97.5 percentiles of those values were plotted.
Figure 4. Percentage of labeled cells during the chase period in the slower-cycling population predicted by the bi-exponential model.
Time “0” represents the time at which BrdU administration was stopped. The solid curve is a median curve, and the shaded region shows the 95% bootstrapped confidence interval.
Discussion
We performed mathematical modeling and statistical analysis of BrdU label-retention data in tumors using a mouse model of human lung cancer. In every test performed, a model with two distinct cycling rates was found to fit the data better than a model with a single cycling rate. Multiple methods of cross-validation and predictive checks confirmed this conclusion. Residuals for both models were fit by lowess curves (Figures 3A and 3B), visually demonstrating a more uniform distribution of residuals for the bi-exponential model. The modeling and analysis provide quantitative evidence for the existence of a label-retaining population of cells, and thus indicate that there are likely two distinct proliferation rates in this solid tumor model. Further experimentation will be required to determine whether these label-retaining cells are indeed tumor-initiating “cancer stem cells”.
Conclusion
Mathematical modeling was applied to BrdU-labeled tumor cells in a mouse model of lung cancer. Measures of goodness-of-fit for the competing mathematical models demonstrated that two distinct proliferation rates are more likely than a single proliferation rate, providing quantitative evidence of a population of “label-retaining” tumor cells. The mathematical modeling also allowed us to estimate the proliferation rate of this label-retaining population as well as to estimate the proliferation rate of the faster-cycling, non-label-retaining population. Additionally, these estimated rates allow for the determination of the fraction of tumor cells that are label-retaining at various times during the chase phase. This provides an upper bound on the fraction of tumor cells in the samples that may have stem cell-like properties, and an estimate of their proliferation rate. The parameter estimates for the two-compartment model in Table 1 indicate that approximately 31% of the cells were initially in the slow population, and that the mean turnover rates for the fast and slow populations were 1.8 and 5.7 weeks/cell, respectively.
Supplementary Material
Acknowledgments
Grant Support
E.A.S.C. was funded by a Research Scholar Grant from the American Cancer Society and by NCI 5 R01 CA157510-03. Y.Z was supported by a Senior Research Fellowship from the American Lung Association (RT-82480-N)
Footnotes
Declaration of conflict of interest: the authors declare no conflict of interest.
References
- 1.Oliver TG, Mercer KL, Sayles LC, Burke JR, Mendus D, Lovejoy KS, et al. Chronic cisplatin treatment promotes enhanced damage repair and tumor progression in a mouse model of lung cancer. Genes Dev. 2010;24:837–52. doi: 10.1101/gad.1897010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Orford KW, Scadden DT. Deconstructing stem cell self-renewal: genetic insights into cell-cycle regulation. Nat Rev Genet. 2008;9:115–28. doi: 10.1038/nrg2269. [DOI] [PubMed] [Google Scholar]
- 3.Potten CS, Kovacs L, Hamilton E. Continuous labelling studies on mouse skin and intestine. Cell and tissue kinetics. 1974;7:271–83. doi: 10.1111/j.1365-2184.1974.tb00907.x. [DOI] [PubMed] [Google Scholar]
- 4.Potten CS. Extreme sensitivity of some intestinal crypt cells to X and gamma irradiation. Nature. 1977;269:518–21. doi: 10.1038/269518a0. [DOI] [PubMed] [Google Scholar]
- 5.Morris RJ, Fischer SM, Slaga TJ. Evidence that the centrally and peripherally located cells in the murine epidermal proliferative unit are two distinct cell populations. J Invest Dermatol. 1985;84:277–81. doi: 10.1111/1523-1747.ep12265358. [DOI] [PubMed] [Google Scholar]
- 6.Morris RJ, Fischer SM, Slaga TJ. Evidence that a slowly cycling subpopulation of adult murine epidermal cells retains carcinogen. Cancer Res. 1986;46:3061–6. [PubMed] [Google Scholar]
- 7.Cotsarelis G, Sun TT, Lavker RM. Label-retaining cells reside in the bulge area of pilosebaceous unit: implications for follicular stem cells, hair cycle, and skin carcinogenesis. Cell. 1990;61:1329–37. doi: 10.1016/0092-8674(90)90696-c. [DOI] [PubMed] [Google Scholar]
- 8.Morris RJ, Potten CS. Slowly cycling (label-retaining) epidermal cells behave like clonogenic stem cells in vitro. Cell Prolif. 1994;27:279–89. doi: 10.1111/j.1365-2184.1994.tb01425.x. [DOI] [PubMed] [Google Scholar]
- 9.Tumbar T, Guasch G, Greco V, Blanpain C, Lowry WE, Rendl M, et al. Defining the epithelial stem cell niche in skin. Science. 2004;303:359–63. doi: 10.1126/science.1092436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Foudi A, Hochedlinger K, Van Buren D, Schindler JW, Jaenisch R, Carey V, et al. Analysis of histone 2B-GFP retention reveals slowly cycling hematopoietic stem cells. Nat Biotechnol. 2009;27:84–90. doi: 10.1038/nbt.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kiel MJ, He S, Ashkenazi R, Gentry SN, Teta M, Kushner JA, et al. Haematopoietic stem cells do not asymmetrically segregate chromosomes or retain BrdU. Nature. 2007;449:238–42. doi: 10.1038/nature06115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilson A, Laurenti E, Oser G, van der Wath RC, Blanco-Bose W, Jaworski M, et al. Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell. 2008;135:1118–29. doi: 10.1016/j.cell.2008.10.048. [DOI] [PubMed] [Google Scholar]
- 13.Magee Jeffrey A, Piskounova E, Morrison Sean J. Cancer Stem Cells: Impact, Heterogeneity, and Uncertainty. Cancer Cell. 2012;21:283–96. doi: 10.1016/j.ccr.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100:3983–8. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hermann PC, Huber SL, Herrler T, Aicher A, Ellwart JW, Guba M, et al. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell. 2007;1:313–23. doi: 10.1016/j.stem.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 16.O’Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445:106–10. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- 17.Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, et al. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 18.Driessens G, Beck B, Caauwe A, Simons BD, Blanpain C. Defining the mode of tumour growth by clonal analysis. Nature. 2012 doi: 10.1038/nature11344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen J, Li Y, Yu TS, McKay RM, Burns DK, Kernie SG, et al. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012 doi: 10.1038/nature11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O’Brien CA, Kreso A, Jamieson CH. Cancer stem cells and self-renewal. Clin Cancer Res. 2010;16:3113–20. doi: 10.1158/1078-0432.CCR-09-2824. [DOI] [PubMed] [Google Scholar]
- 21.Quintana E, Shackleton M, Sabel MS, Fullen DR, Johnson TM, Morrison SJ. Efficient tumour formation by single human melanoma cells. Nature. 2008;456:593–8. doi: 10.1038/nature07567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang WC, Shyh-Chang N, Yang H, Rai A, Umashankar S, Ma S, et al. Glycine decarboxylase activity drives non-small cell lung cancer tumor-initiating cells and tumorigenesis. Cell. 2012;148:259–72. doi: 10.1016/j.cell.2011.11.050. [DOI] [PubMed] [Google Scholar]
- 23.Eramo A, Haas TL, De Maria R. Lung cancer stem cells: tools and targets to fight lung cancer. Oncogene. 2010;29:4625–35. doi: 10.1038/onc.2010.207. [DOI] [PubMed] [Google Scholar]
- 24.Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, Montoya R, et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 2001;15:3243–8. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sweet-Cordero A, Mukherjee S, Subramanian A, You H, Roix JJ, Ladd-Acosta C, et al. An oncogenic KRAS2 expression signature identified by cross-species gene-expression analysis. Nat Genet. 2005;37:48–55. doi: 10.1038/ng1490. [DOI] [PubMed] [Google Scholar]
- 26.Kim CF, Jackson EL, Woolfenden AE, Lawrence S, Babar I, Vogel S, et al. Identification of bronchioalveolar stem cells in normal lung and lung cancer. Cell. 2005;121:823–35. doi: 10.1016/j.cell.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 27.Curtis SJ, Sinkevicius KW, Li D, Lau AN, Roach RR, Zamponi R, et al. Primary tumor genotype is an important determinant in identification of lung cancer propagating cells. Cell Stem Cell. 2010;7:127–33. doi: 10.1016/j.stem.2010.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schepers AG, Snippert HJ, Stange DE, van den Born M, van Es JH, van de Wetering M, et al. Lineage tracing reveals Lgr5+ stem cell activity in mouse intestinal adenomas. Science. 2012;337:730–5. doi: 10.1126/science.1224676. [DOI] [PubMed] [Google Scholar]
- 29.Pece S, Tosoni D, Confalonieri S, Mazzarol G, Vecchi M, Ronzoni S, et al. Biological and molecular heterogeneity of breast cancers correlates with their cancer stem cell content. Cell. 2010;140:62–73. doi: 10.1016/j.cell.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 30.Bonhoeffer S, Mohri H, Ho D, Perelson AS. Quantification of cell turnover kinetics using 5-bromo-2′-deoxyuridine. J Immunol. 2000;164:5049–54. doi: 10.4049/jimmunol.164.10.5049. [DOI] [PubMed] [Google Scholar]
- 31.van der Wath RC, Wilson A, Laurenti E, Trumpp A, Lio P. Estimating dormant and active hematopoietic stem cell kinetics through extensive modeling of bromodeoxyuridine label-retaining cell dynamics. PLoS ONE. 2009;4:e6972. doi: 10.1371/journal.pone.0006972. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.