Abstract
Gene-expression profiling has had a considerable impact on our understanding of breast cancer biology, and more recently on clinical care. Two statistical approaches underlie these advancements. Supervised analyses have led to the development of gene-expression signatures designed to predict survival and/or treatment response, which has resulted in the development of new clinical assays. Unsupervised analyses have identified numerous biological signatures including signatures of cell type of origin, signaling pathways, and of cellular proliferation. Included within these biological signatures are the molecular subtypes known as the ‘intrinsic’ subtypes of breast cancer. This classification has expanded our appreciation of the heterogeneity of breast cancer and has provided a way to sub-classify the disease in a manner that might have clinical utility. In this Review, we discuss the clinical utility of gene-expression-based assays and their technical potential as clinical tools vis-a-vis the performance of breast cancer biomarkers that are the current standard of care.
Introduction
Breast cancer is a devastatingly common disease and there is a considerable need to improve approaches for prevention, diagnosis and treatment.1 One of the biggest barriers to progress is the extreme clinical and genetic heterogeneity of the disease. Rapid progress in diagnostic capabilities has occurred over the past decade, however, breast cancer management is caught between two worlds: the old world of familiar groupings defined by estrogen receptor (ER) and HER2 status and a new world of seemingly endless and complex ways to classify breast cancers for treatment individualization. Although enthusiasm for new genomic discovery tools should be tempered, many studies conducted using conventional gene-expression profiling should be vaunted as identifying successful biomarkers. Over the past decade, we have learnt many valuable lessons from gene-expression profiling studies. The clinical utility of gene-expression-based assays has been demonstrated with increasing confidence, particularly with respect to tailoring treatment for the current standard of care in chemotherapy and hormonal treatment.2–4 In this Review, we present the current status of clinically applicable gene-expression-based assays, and we discuss their reproducibility as well as the reproducibility of the biomarkers that are standard of care in breast cancer today.
Clinical gene expression-based assays
A plethora of prognostic and some predictive gene-expression signatures have been identified for breast cancer; however, most of these signatures identify breast cancer subtypes that have a similar biology, which encompasses ER status, HER2 status, and proliferation.5 Improvements offered by gene-expression-based assays must, therefore, go beyond these known variables, or at least encompass them in ways that are more quantitative, reproducible, and/or objective than the existing immunohistochemistry (IHC)-based assays. The main characteristics of five gene-expression-based predictors are shown in Table 1. Although there are considerable similarities between these predictors,6 there are enough statistically significant differences across them such that each should be considered to be unique.7
Table 1.
Main features of the commercially available multi-gene signatures in breast cancer
| Assay name | Genes (n) | Source | Platform | Training dataset | Output data |
|---|---|---|---|---|---|
| Biology driven | |||||
| PAM50 subtype-predictor | 55* | FFPE/FF | qRT-PCR/microarray/nCounter | Breast cancer-based cohort | Luminal A Luminal B HER2-E Basal-like Normal-like |
| Survival driven | |||||
| PAM50 risk of relapse (ROR) | 55* | FFPE/FF | qRT-PCR/microarray/nCounter | RFS; ER+/ER−/node− breast cancer patients receiving no adjuvant systemic treatment | Continuous Variable Low–medium–high |
| Oncotype DX® | 21 | FFPE | qRT-PCR | Survival: largely ER+/node− breast cancer patients receiving tamoxifen-only adjuvant treatment | Continuous Variable Low–intermediate–high |
| MammaPrint® | 70 | FF | Microarray | DRFS: largely ER+/node− breast cancer patients receiving no adjuvant systemic treatment | Continuous Variable Good–bad |
| Pathology driven | |||||
| MapQuant Dx™ | 97 | FF | Microarray | Grade 1 vs 3 in ER+ patients with breast cancer | Continuous Variable Low–high |
| Survival and pathology driven | |||||
| Breast Cancer Index | 7 | FFPE | qRT-PCR | MGI: grade 1 vs 3, HOXB13/IL17BR ratio DRFS: ER+ patients with breast cancer receiving tamoxifen-only treatment | Continuous Variable Low–intermediate–high |
5 genes included for expression normalization. Abbreviations: DRFS, distant relapse-free survival; FF, fresh-frozen; FFPE, formalin-fixed paraffin-embedded; MGI, molecular grade index; qRT-PCR, quantitative reverse transcription PCR; RFS, relapse-free survival.
Oncotype DX® recurrence score
Oncotype DX® (Genomic Health, Redwood City, CA) is a 21-gene quantitative reverse transcription (qRT)-PCR-based assay and is the most widely used clinical gene-expression assay in the USA. It is based on the levels of gene expression of 16 cancer-related genes and five genes for expression normalization using RNA from formalin fixed-paraffin embedded (FFPE) tumor tissues.8,9 The genes in the assay were selected from 250 candidates that were tested for association with survival in a cohort of 447 tumor samples, including 233 (52%) ER-positive samples from the tamoxifen-treated and node-negative cases of the National Surgical Adjuvant Breast and Bowel Project (NSABP) B-20 clinical trial.10 The final 16-gene list is weighted by biological processes including proliferation, HER2 and ER signaling. The resulting recurrence score (RS) is 0 to 100, which translates into three risk-group categories: low (RS <18), intermediate (RS from 18 to <31) and high (RS ≥31).
In a seminal study, Paik and colleagues11 validated Oncotype DX® in a large cohort of ER-positive, node-negative tamoxifen-treated patients with breast cancer enrolled in the NSABP B-14 trial; in this study, the rates of distant recurrence at 10 years were 6.8%, 14.3% and 30.5% for the low-risk, intermediate-risk and high-risk groups, respectively. In a subsequent validation study,12 Oncotype DX® RS was strongly associated with the risk of death from breast cancer in a similar cohort and also in patients who did not receive adjuvant systemic therapy. Importantly, the associations of Oncotype DX® with survival have been found to be independent from standard clinicopathological variables,11,12 thus showing that this assay adds information beyond ER and HER2 status, stage, and grade.
More recently, Oncotype DX® has been endorsed by ASCO3 and two expert panels: NCCN Breast Cancer Clinical Practice Guidelines2 and the 2011 St Gallen International Expert Consensus.4 These panels consider Oncotype DX® to be useful for patients with ER-positive, node-negative breast cancer as an aid to decision making for administering adjuvant chemotherapy. Since its commercialization, over 175,000 assays have been ordered by more than 7,500 physicians worldwide.
MammaPrint®
MammaPrint® is a microarray-based gene-expression profiling assay that was developed by Agendia (Amsterdam, the Netherlands) based on the Agilent (Agilent technologies, Santa Clara, CA) microarray-based platform that analyzed data from 78 patients with node-negative breast cancers.13 The vast majority of these 78 patients had ER-positive tumors and did not receive adjuvant systemic therapy. The 70 genes that comprise the MammaPrint® assay are proliferation genes and genes associated with invasion and angiogenesis. Since 2002, this signature has been validated on numerous cohorts of node-negative patients,14–16 and has been shown to provide independent prognostic information beyond standard clinico-pathological variables such as age, histological grade and pathological stage.14 MammaPrint® is also the first FDA-approved gene-expression-based assay to be used as a prognostic test for women with node-negative breast cancers. Unlike the Oncotype DX® assay, this test is based on microarrays results and hence requires high-quality RNA from freshly collected tissues.
MapQuant Dx™ genomic grade
MapQuant Dx™ (Ipsogen, Marseille, France) is a predictor test that was derived by identifying 97 differentially expressed genes from grade 1 and 3 breast tumors using a training set of 64 ER-positive tumors;17 most of the genes in this signature are involved in cell-cycle regulation and proliferation. In validation data sets, this ‘genomic grade index’ (GGI) was strongly associated with risk of recurrence among patients with grade 2 tumors.17,18 Similar to MammaPrint®, this assay is microarray based and requires freshly prepared tissues and high-quality RNA.
Breast Cancer IndexSM (BCI)
The Breast Cancer IndexSM (BCI; BioTheranostics, San Diego, CA) prognostic assay provides an assessment of the likelihood of distant recurrence in patients diagnosed with ER-positive, node-negative breast cancer. In development and validation studies, BCI stratified nearly 50% of tamoxifen-treated ER-positive, node-negative patients with breast cancer into a low-risk group for 10-year distant recurrence.19,20 Unlike the other commercialized tests, BCI has been developed from the combination of two indices: the HOXB13:IL17BR two-gene ratio, which predicts distant recurrence in patients with ER-positive disease treated with tamoxifen,21 and a proliferation-related five-gene molecular grade index (MGI),19 which discriminates grade 1 from grade 3 disease (similar to the MapQuant Dx™ assay). The BCI molecular assay is based on qRT-PCR using RNA from FFPE tissues.
Intrinsic subtypes and PAM50 assay
Over the past 12 years, global gene-expression analyses have identified at least four breast molecular subtypes known as the ‘intrinsic subtypes’ (luminal A, luminal B, HER2-enriched and basal-like),22–25 which show significant differences in epidemiological risk factors,26 baseline prognoses,6,27,28 and responses to systemic therapies.28–32 Information provided by the intrinsic subtypes, using an IHC-based approach, has also been adopted by the 2011 St Gallen Consensus Conference (Table 2).4 The biological and clinical features of the intrinsic subtypes of breast cancer have been extensively discussed in the literature and hence we will focus on the most recent version of the intrinsic subtyping assay, known as the PAM50 assay.28,33 The training set for the PAM50 subtype predictor was composed of tumor samples from 220 patients who had ER-positive or ER-negative tumors and HER2-positive or HER2-negative tumors and from whom the prototypic samples of each molecular subtype were identified using a list of 1,906 intrinsic genes; it also included 12 true normal breast samples that constituted the normal breast-like centroid.28 A final minimized set of 50 genes was derived from these prototypic samples, which comprises genes involved in proliferation, ER and ER-regulated genes, HER2, and basal and myoepithelial characteristics.
Table 2.
2011 St Gallen consensus recommendations of systemic treatment4
| IHC Subtype | Definition | Type of adjuvant therapy |
|---|---|---|
| Luminal A | HR+/HER2−/Ki67low | Endocrine therapy alone* |
| Luminal B | HR+/HER2−/Ki67high | Endocrine therapy ± cytotoxic therapy |
| Luminal B | HR+/HER2+ | Cytotoxics + anti-HER2 + hormonal therapy |
| HER2-positive | HR−/HER2+ | Cytotoxics + anti-HER2 therapy |
| Triple-negative | HR−/HER2− | Cytotoxic therapy |
A few patients require cytotoxics (such as high nodal status or other indicator of risk). Abbreviation: HR, hormone receptor.
The PAM50 assay provides a risk of relapse (ROR) score prognostic of relapse-free survival for patients with node-negative tumors who did not receive adjuvant systemic therapy.28 The ability of the ROR score to predict prognosis (as a continuous variable or as a three-class variable) has been confirmed in an independent test set of 786 patients with ER-positive disease treated only with adjuvant tamoxifen.33 In this cohort, the ROR together with tumor size outperformed standard clinicopathological variables that included expression of the proliferation marker Ki67, progesterone receptor (PR) and histological grade.33 In addition, a ROR model that contained a proliferation component (derived using 11 of 50 PAM50 genes associated with cell-cycle function) improved the original model, showing that the best model may contain variables for molecular subtype, tumor size and proliferation status. Interestingly, this 11-gene proliferation signature has been found to have more prognostic value than the expression of Ki67 assessed by IHC.33 The PAM50 assay is compatible with FFPE-derived RNA and is currently being developed for clinical use on the NanoString nCounter™ Analysis System (Nanostring technologies, Seattle, WA).34 It is also offered using a qRT-PCR format from ARUP Laboratories (Salt Lake City, UT) as a laboratory-developed test (that is, developed, evaluated, and validated within one particular laboratory) waived by the CLIA (Clinical Laboratory Improvement Amendments).
The intrinsic molecular subtypes are correlated with standard pathological markers based on gene-expression analysis.28,35–38 For example, most luminal tumors are ER-positive and/or PR-positive, and most HER2-enriched and basal-like tumors are typically HER2-amplified and triple-negative tumors, respectively.28,35–38 It should be noted that intrinsic subtyping by gene expression compared with standard pathological biomarkers are not interchangeable classifications,28,35,37 which demonstrates that intrinsic subtypes are not simple combinations of ER, PR, and HER2.
Predicting early-stage outcomes
Prognostic signatures in breast cancer consist mainly of changes in proliferation and/or ER-regulated genes.5,6,27 These two biological processes in breast cancer are critical for distinguishing the relatively good prognosis of the luminal A subtype (which is characterized by low proliferation and high expression of ER-regulated genes) from the worse prognosis of the luminal B, HER2-enriched, and basal-like subtypes (which are characterized by a higher proliferation status).22–25 Thus, the low-risk groups defined by Oncotype DX®, MammaPrint®, MapQuant Dx™ and PAM50-ROR are essentially composed of a subset of luminal A tumors.7,18 This observation explains why the prognostic ability of these different gene expression-based assays is similar, as most of them are differentiating luminal A from all other subtypes.6,27
Fan et al.27 evaluated 323 different gene signatures (also called modules), including research versions of Oncotype DX®, MammaPrint®, MapQuant Dx™ and PAM50-ROR, on a microarray-based cohort of 550 patients with node-negative breast cancer who did not receive adjuvant systemic therapy. This exploratory study showed that it is only within ER-positive, luminal disease that robust prognostic predictions can be made, but not within ER-negative, or within HER2-positive disease.27 Other investigators have identified immune-related signatures that might be able to predict survival within HER2-positive or triple-negative tumors; however, the ‘good’ prognostic group still showed a relatively high likelihood of relapse.39,40 Thus, from a clinical utility perspective, prognostic gene-expression-based assays are only indicated for patients with ER-positive, node-negative disease for whom it might be adequate to say that hormonal and local therapies are sufficient, and that chemotherapy may not be necessary.
The clinical utility of Oncotype DX® versus standard parameters in ER-positive, node-negative disease has been prospectively examined.41,42 In a relatively small multicenter study, treatment recommendations were changed for 31% of the patients based upon Oncotype DX® outcomes versus standard pathology-based risk assessments, with the largest change being the use of hormonal therapy only, with the omission of chemotherapy in 22.5% of cases. Similar data have been reported from an independent study of 135 patients.42 Thus, although the costs of these gene-expression-based tests might be high relative to the costs of ER, PR and HER2 testing, the additional reduction in adjuvant chemotherapy administration is likely to result in an overall cost saving,43,44 as well as reduced toxicity and quality-of-life improvements for patients.
The clinical utility of Oncotype DX® and the other gene-expression-based assays in ER-positive, node-positive breast cancer remains investigational. Although Oncotype DX®, MammaPrint® and PAM50-ROR are able to identify different predicted risk groups with statistically distinct outcomes, for those with ER-positive, node-positive disease, the low-risk groups do not show a sufficiently low rate of recurrence (that is fewer than 10% relapse rates at 8–10 years) to consider excluding adjuvant chemotherapy based on prognosis alone.7,15,33,45,46 However, when the subset of patients with one-to-three positive lymph nodes are considered, the low-risk groups of Oncotype DX® and MammaPrint® have shown outstanding outcomes with hormonal therapy alone (that is <10% relapse rate at 5–10 years), which is similar to the outcomes previously observed in similar groups with node-negative disease.45–47
Two large prospective clinical trials are underway to further establish the clinical utility of Oncotype DX® and MammaPrint®.9,16 In the fully accrued TAILORx trial, patients with ER-positive, HER2-negative, node-negative disease with a RS of 10 or less were given hormonal therapy alone, patients with a RS of 11–25 were randomly assigned to receive adjuvant hormonal therapy with or without chemotherapy, and those patients with a RS >25 received hormonal therapy and chemotherapy (standard of care). The primary end point is to determine if adjuvant chemotherapy plus hormonal therapy is better than hormonal therapy alone in those with a RS between 11 and 25. In addition, this trial will confirm whether patients with a low RS have an extremely low risk of recurrence that excludes the need for chemotherapy based on prognosis alone. In the MINDACT trial assessing the MammaPrint® assay in patients with ER-positive, node-negative disease in the adjuvant treatment setting will be assessed by standard clinicopathological prognostic factors using Adjuvant!Online and MammaPrint®. The primary objective of MINDACT is to confirm that patients with a ‘low risk’ prognosis according to MammaPrint® and ‘high risk’ prognosis according to clinical features can be spared chemotherapy without affecting survival outcomes. On the basis of the recent results of MammaPrint® in patients with positive lymph nodes,47 the MINDACT trial has also been amended to include patients with one-to-three positive lymph nodes.
Predicting treatment response
In the neoadjuvant setting, pathological complete response (pCR) in the breast has been used to determine the systemic response to chemotherapy agents, although pCR is only achieved in 20–30% of patients.48 Across the intrinsic subtypes, pCR rates to standard anthracycline-based and taxane-based chemotherapy have been found to be approximately 7% for luminal A, 17% for luminal B, 36% for HER2-enriched, and 43% for basal-like.48 Thus, the vast majority of patients that achieve a pCR have non-luminal A tumors. This explains, in part, why gene-expression signatures that largely discriminate luminal A from non-luminal A tumors can also predict pCR.49–51
The benefit of neoadjuvant hormonal therapies is also an area of interest, and a non-genomic prognostic index based on tumor size, nodal status, Ki67 levels and ER status after treatment with neoadjuvant hormonal therapy (known as the preoperative endocrine prognostic index [PEPI]52) has been developed. This index was developed in the P024 letrozole versus tamoxifen trial,53 and validated as predictive of relapse-free survival in an independent study of 203 postmenopausal women in the IMPACT trial.52 Interestingly, in the neoadjuvant aromatase inhibitor trial ACOSOG Z1031, luminal A tumors (defined using PAM50) achieved significantly higher rates of PEPI score zero (good prognosis) than the luminal B tumors,54 consistent with luminal A tumors being associated with superior outcome when treated with hormonal therapy alone.
In the adjuvant setting, prediction of chemotherapy and/or hormonal therapy efficacy relies on survival end points from clinical trials in which patients have been randomized to different treatments. The best data are from retrospective studies in which Oncotype DX® has been evaluated;45,55 in the NSABP-B20 trial,55 patients with ER-positive, node-negative breast cancer were randomly assigned to receive tamoxifen or tamoxifen plus chemotherapy (cyclophosphamide, methotrexate and 5-fluorouracil [CMF]). On the one hand, the low-RS Oncotype DX® group did not derive a survival benefit from the addition of chemotherapy and the relapse rates with or without chemotherapy were <5% at 12 years. On the other hand, the high-RS Oncotype DX® group had a large benefit from chemotherapy with an absolute decrease in 10-year distant recurrence of 27.6%. Less clear was the benefit of chemotherapy in the intermediate RS group in NASBP-B20, hence the design of the TAILORx trial, which randomized treatment to chemotherapy versus observation in this group.
In the SWOG 8814 trial,45 the Oncotype DX® assay was applied to 219 patients with ER-positive, node-positive disease randomly assigned to receive adjuvant fluorouracil, doxorubicin and cyclophosphamide (FAC) followed by tamoxifen versus tamoxifen alone. The benefit of FAC was only seen in the high-RS group. Importantly, further subanalysis within the group with one-to-three positive nodes indicated that the relapse rates at 5 years in the low-RS group was <10% regardless of the type of chemotherapy use. These results, together with the TransATAC findings in patients with one-to-three positive nodes,46 has led to the initiation of the RxPONDER clinical trial (NCT01272037) where around 4,000 patients with ER-positive breast cancer with one to three positive lymph nodes and a RS ≤25 will be randomized to hormonal therapy alone or chemotherapy followed by hormonal therapy.
Reproducibility of breast biomarkers
The reproducibility of genomic biomarkers has drawn considerable attention, which prompted us to investigate the issue of current biomarker reproducibility. Our review of the literature suggests that standardized gene-expression-based assays such as Oncotype DX® and MammaPrint® are more reproducible8,56,57 than current methodologies for ER, PR and HER2 testing.58,59 Currently offered gene-expression-based assays are single platform or single technology based (and sometimes even carried out within a single laboratory as is the case for Oncotype DX®) and follow a single protocol. This high level of standardization should be considered as a strength of these gene-expression-based assays as the existing breast cancer biomarker field is plagued by nonstandardized tests that do not require, or achieve, high levels of inter-laboratory concordance.
Since 2007, ASCO and the College of American Pathologists (CAP) have issued recommendation guidelines for ER, PR and HER2 testing.58,59 These recommendations summarized the various validated assays and provide a common scoring algorithm; however, many standardization issues remain. Indeed, ASCO and CAP note that 20% of ER, PR and HER2 measurements in the clinical setting are possibly inaccurate.58,59 This should be taken seriously as the status of these biomarkers dictates therapeutic decision making. In fact, if a new genomic or proteomic assay was to be put forth today with a 20% discordance rate, it likely would not be approved for clinical use.
In Table 3, we present a summary of the literature regarding reproducibility and concordance for the current breast cancer biomarker tests, and we distinguish the concordance between two identical protocols (inter-observer and inter-laboratory) and between two different methods (inter-method). A commonly used measure of reproducibility or concordance is the kappa (κ) statistic,60 which has an output range from −1 (total discordance) to +1 (total concordance). The following three categories based on κ values are used to define levels of concordance using concordance correlation coefficients: 0.40–0.60 (moderate concordance—defined here as level 3), 0.61–0.80 (substantial concordance—level 2) and ≥0.81 (almost perfect agreement—level 1).61–63
Table 3.
Reproducibility data of currently used biomarkers in breast cancer
| Level of agreement | Type of test | Identical protocols (inter-laboratory and/or inter-observer) | Different protocols or technologies (inter-method) |
|---|---|---|---|
| Level 1 (κ ≥0.81) Almost perfect agreement |
Pathology test | NA | |
| Gene-expression-based test | Basal-like by different gene list and microarray platform intrinsic subtype centroid predictors (microarray-based)63,107–109 | ||
| Level 2 (κ 0.60–0.80) Substantial agreement |
Pathology test | ||
| Gene-expression-based test | Overall intrinsic molecular subtypes by different gene list and microarray platform intrinsic subtype centroid predictors63,107–109 | ||
| Level 3 (κ 0.40–0.59) Moderate agreement |
Pathology test | Histological grade91–98 ER IHC with 4 or more groups64,67,90 PR IHC with 4 or more groups90 |
|
| Gene-expression-based test | NA |
|
All risk category groups have been calculated in the NKI295 dataset.15 For κ value calculation, low and intermediate groups of Oncotype DX® have been combined into a low-risk group when compared with two risk groups from MammaPrint®. For intrinsic molecular subtyping, a higher level of concordance is achieved when performing microarray platform to platform normalization, which was not performed by Weigelt et al.63
Abbreviations: ER, estrogen receptor; FISH, fluorescence in situ hybridization; HC, hierarchical clustering; HER2E, HER2-enriched; IHC, immunohistochemistry; LBA, ligand-binding assay; NA, not applicable; PR, progesterone receptor.
When two observers, or laboratories, use identical protocols (inter-observer and inter-laboratory studies), level 1 agreement is achieved for ER status defined by IHC and HER2 status defined by fluorescence in situ hybridization (FISH) when scored as a two-class variable (variable of two elements: positive versus negative).64–76 For HER2 status, which is usually scored as a three-class variable (0/1+, 2+ and 3+), level 1 agreement is observed in most inter-observer studies,65,77–79 whereas level 2 agreement is observed in inter-laboratory studies.71,73,76,79 The lower agreement achieved when the same HER2 IHC protocol is carried out across two different laboratories might be due to pre-analytical variables such as fixation times and/or in the critical area of subjectivity in IHC interpretation, especially for the HER2 2+ cases.80,81 In the NCCTG-N9831 trial, Perez et al.82 reported that 18.4% of 636 samples that were identified as HER2 3+ by the FDA-approved HercepTest™ (Dako, Glostrup, Denmark) were not confirmed centrally by the same assay; similar findings were noted in the NSABP-B-31 trial.83
Concordance between two different ER or PR testing methods—such as the concordance between the ligand-binding biochemical assay and IHC64,69,84–86—or between two different antibodies for ER,69,85,87 falls between level 2 and 3 (Table 3). It should be noted that different antibodies are allowed in the USA for ER and PR testing, and thus, level 2–3 is the current standard of reproducibility. For example, Rhodes et al.88 assessed histological sections from breast cancers with low, medium and high levels of ER expression and sent the samples to 200 laboratories for IHC staining where in-house techniques were used. The results were then centrally evaluated. The inter-method staining results showed that 80% of laboratories were able to demonstrate ER positivity in tumors with medium and high expressing ER levels, whereas only 37% of laboratories scored adequately for the low-expressing tumors. The low concordance within low ER-expressing tumors, together with data suggesting that these tumors might still derive benefit from hormonal therapy,86,89 has led the ASCO–CAP guidelines to recommend 1% positive cells or greater as the cutoff for identifying a sample as ER-positive.59 Multi-class scoring of ER and PR into four or more classes (using one antibody) by two pathologists shows level 3 concordance.64,67,90 Lastly, level 2 and 3 concordances are typical for histological grade as estimated by two or more pathologists;91–99 these levels of concordance are considered sufficient by some investigators to recommend using tumor grade to help make therapeutic decisions.100
Concordance of HER2 status by different IHC based-antibodies,69,85,87 or IHC versus FISH,77,101–104 also falls between level 2 and 3 (Table 3). In the NSABP B-31 trial, Paik et al.83 found that 45% of 24 samples that had a community-based HER2 3+ score using a non-HercepTest™ IHC test could not be confirmed by central HercepTest™. Interestingly, concordance of HER2 FISH and HER2 gene expression, as determined by central laboratories, falls within level 1 agreement,105,106 indicating that HER2 gene expression might be a more reliable surrogate of HER2 amplification than HER2 protein by IHC.
Overall, these data show that level 1 concordance can only be achieved in the clinical setting when identical protocols are used (for example, the same IHC antibody); however, this is not the current standard;58,59 instead, the current standards for ER, PR and HER2 testing allow for different antibodies to be used, or allow different technologies to be used (FISH versus IHC), and in these cases, level 2–3 concordance is observed and accepted for therapeutic decision making. These results highlight the need to improve the reproducibility of current diagnostic tests for patients with breast cancer, and we suggest that medical practice should strive for level 1 concordance.
Reproducibility of gene-expression biomarkers
The Oncotype DX® and MammaPrint® assays have shown level 1 κ concordance values and/or concordance correlation coefficients >0.90 when tested using the same starting materials and when repeated on different days, with different reagent batches, and by different technicians.,8,56,57 Clinical implementation of the 50-gene PAM50 subtype predictor is currently underway, and thus, no published data exist to judge its reproducibility; however, over the past decade, various research laboratory-based intrinsic subtype predictors have been developed using the most advanced DNA microarrays and statistical methods that are currently available. The identification of the intrinsic subtypes by multiple investigators across the world, using many different expression technologies, has been accomplished and has had an enormous impact, so that it is now commonplace to carry out research-based intrinsic subtyping as a means of anchoring new findings to the literature.
The inter-observer concordance of different intrinsic subtype classifications using DNA microarrays has been recently evaluated.62 Mackay et al.62 carried out 15 hierarchical clustering analyses using five independent breast cancer microarray datasets and three previously published intrinsic gene lists, and then estimated the agreement in identifying the intrinsic subtypes (luminal A, luminal B, HER2-enriched, basal-like and normal-like) across five observers. The results showed that the overall agreement was 0.64–0.70 (level 2), and was >0.81 (level 1) for the basal-like and HER2-enriched subtypes, and substantial (κ = 0.6–0.80, level 2) for the other subtypes (luminal A, luminal B and normal-like). Thus, the inter-observer concordance of the intrinsic subtypes in the research setting by hierarchical clustering analysis showed level 2 agreement (thus equivalent to current ER and HER2 testing), except for the basal-like and HER2-enriched subtypes that showed even better level 1 agreement.
The inter-method concordance of different molecular subtype classifications using DNA microarrays was evaluated by Weigelt et al.63 who compared three different intrinsic subtype prediction methods, each of which differed in three technological means (different gene lists, different tumor training data sets, and different microarray platforms); these analyses showed an overall κ value agreement of 0.24–0.74, and level 1 agreement for the basal-like subtype, whereas the other subtypes had level 2–3. This experiment would be similar to testing ER concordance using two different antibodies and the ligand-binding assay, determining cut points using three different patient training sets, and then comparing classification concordance results on yet other data sets. In addition, these substantial inter-method concordance values were obtained despite the fact that the Weigelt et al.63 analyses had several bioinformatic issues including a lack of microarray platform normalization.107–109 In the research setting, when using two different normalization procedures on the same data set before PAM50 intrinsic subtype classification, the κ concordance value for the five subtypes will drop from level 1 to level 2 (A. Prat and C. M. Perou, unpublished observations).
Similar to ER IHC and HER2 FISH, level 1 concordance for any gene-expression-based assay will only be achieved when using a single technology platform and a standardized protocol. This protocol is the current standard for Oncotype DX® and MammaPrint®, and the future protocol for intrinsic subtyping via the PAM50 assay. Overall, these results show that intrinsic subtyping, as carried out in the research setting, is reproducible across platforms and gene lists, and shows κ concordance values equivalent to current clinical testing for histological grade, ER, PR and HER2 testing. The next challenge for gene-expression- based assays will be to develop distributed assays that can maintain a similar level of analytical reproducibility and clinical utility, as has been demonstrated by single laboratories. Interestingly, a recent multisite reproducibility study of an FDA-approved microarray-based gene-expression test for identifying the tissue site of origin for tumors of unknown origins, known as Pathwork® (Pathwork Diagnostics, Redwood City, CA), obtained κ concordance values >0.81 when testing 149 paired samples.110
The next decade
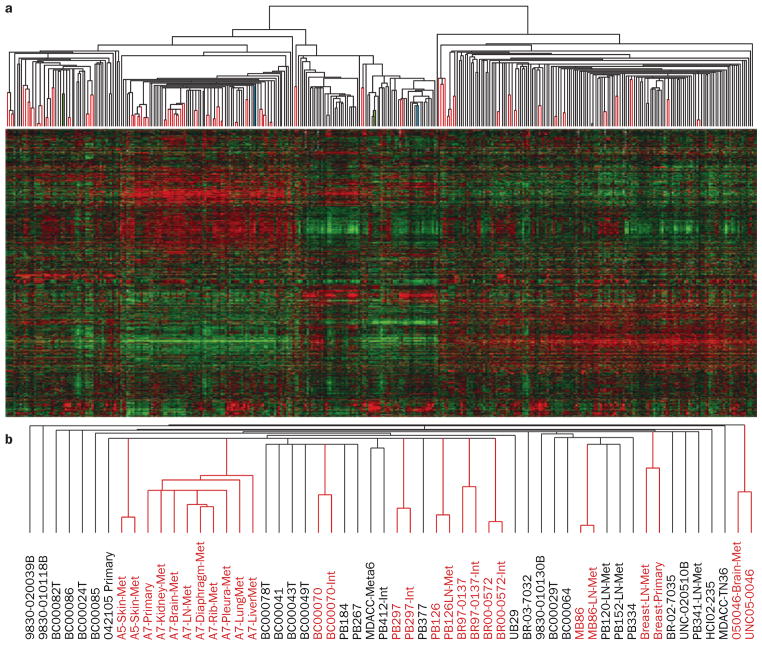
In the 12 years after the first publication of a gene-expression- based study on breast tumors,25 it is clear that the information provided by gene-expression-based assays is useful and deserves incorporation into clinical decision making. In fact, the term ‘molecular portraits’ was coined based on the fact that in the vast majority of tumors, different pieces of the same tumor, or primary tumor-metastasis pairs, are paired together when using gene-expression data and a hierarchical clustering analysis.22–24,111 To further illustrate this point, we re-analyzed 43 tumors with two or more distinct tumor aliquots or fragments (for example, one primary tumor cut into two fragments or a tumor-metastasis pair assessed with two RNA preps, and two microarrays) that were independently arrayed using data from Harrell et al.,112 and found that 41 of 43 were paired together (Figure 1). If we call pairing a successful classification (that is, inter-individual concordance), this 43 class classification analysis yields a κ value of 0.95 (near perfect agreement). Thus, gene-expression analysis not only accurately classified samples into common groups, but is also able to identify the true individuality of each tumor, thus showing that each tumor is in a unique and reproducible class by itself.
Figure 1.
Identification of tumor individuality using global gene-expression analyses. a | Supervised hierarchical clustering of breast cancer data from Harrell et al.112 of 367 breast samples using 1,900 intrinsic genes. Paired tumor samples are highlighted by the red lines in the array tree, with 41 out of 43 possible pairs being paired. The two paired samples that did not pair together are shown in green and light blue color. Note that one of each paired sample was a primary tumor sample that was present within the normal-like cluster, which suggests that both were highly contaminated with true normal breast tissue. b | A subset of the basal-like array tree showing the different array names. Note that some tumors had more than two samples, and in all cases, these were all grouped together. Abbreviations: INT, second partner of an intrinsic pair (that is, two distinct pieces of the same primary tumor); LN, lymph-node metastasis; Met, distant metastasis; Meta, metaplastic.
Conclusions
Current clinical recommendation for Oncotype DX® from ASCO–CAP and NCCN guidelines and the 2011 St Gallen consensus report support the use of gene- expression-based assays in the management of breast cancer. In the coming decade it is also likely that further improvements in gene-expression-based assays will occur and make a clinical impact, as will the discovery and implementation of new gene signatures. In addition, new and exciting technologies will also make a clinical diagnostic impact, with the most immediate possibilities being based on next-generation sequencing technologies, proteomics, or in the isolation and characterization of special cell populations such as circulating tumor cells. The first complete breast-cancer genomes have been published and revealed a myriad of somatic mutations,113,114 some of which may be therapeutically targetable. These are exciting times and we look forward to continued refinements of existing assays and new discoveries in the decade to come, so that outcomes for patients with breast cancer will continue to improve.
Key points.
Gene-expression-based assays provide independent prognostic information beyond standard clinical-pathological variables; however, tumor and nodal stage remain important and must be taken into account in the final prognostic assessment
Gene-expression-based assays identify patients with ER-positive node-negative disease at low risk of relapse after treatment with hormonal therapy and who might be spared from chemotherapy
Clinical use of gene-expression-based assays for the prediction of chemotherapy benefit in node-positive disease, and in ER-negative disease, is currently experimental
Current methodologies for ER, PR and HER2 testing might benefit from additional protocol standardizations, but may still be less reproducible than standardized gene-expression-based assays
Non-standardized research-based identification of the intrinsic subtypes shows concordance values equivalent to current clinical testing for histological grade, ER, PR and HER2
For daily clinical use, we recommend the highest level of reproducibility/concordance (Level 1), which will only be achieved for pathology and gene-expression-based tests by using a single platform and standardized protocol
Acknowledgments
We thank Cynthia Ma for reviewing this manuscript. This work was supported by funds from the NCI Breast SPORE program (P50-CA58223-09A1), by RO1-CA138255, by the Breast Cancer Research Foundation and the V Foundation for Cancer Research. A. Prat is affiliated to the Medicine PhD program of the Autonomous University of Barcelona (UAB), Spain.
Footnotes
Competing interests
M. J. Ellis and C. M. Perou declare associations with the following company: BioClassifier. See the article online for full details of the relationship. A. Prat declares no competing interests.
Contributor Information
Aleix Prat, Department of Genetics and Pathology, Lineberger Comprehensive Cancer Center, University of North Carolina, 450 West Drive, Chapel Hill, NC 27599, USA.
Matthew J. Ellis, Department of Medicine, Siteman Comprehensive Cancer Center, Washington University, 660 South Euclid Avenue, Saint Louis, MI 63110, USA
Charles M. Perou, Department of Genetics and Pathology, Lineberger Comprehensive Cancer Center, University of North Carolina, 450 West Drive, Chapel Hill, NC 27599, USA
References
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer Statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.NCCM Clinical Practicce Guidelines in Oncology. Breast Cancer. 2011 [online], http://www.nccn.org/professionals/physician_gls/pdf/breast.pdf.
- 3.Harris L, et al. American Society of Clinical Oncology 2007 update of recommendations for the use of tumor markers in breast cancer. J Clin Oncol. 2007;25:5287–5312. doi: 10.1200/JCO.2007.14.2364. [DOI] [PubMed] [Google Scholar]
- 4.Goldhirsch A, et al. Strategies for subtypes—dealing with the diversity of breast cancer: highlights of the St. Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2011. Ann Oncol. 2011;22:1736–1747. doi: 10.1093/annonc/mdr304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wirapati P, et al. Meta-analysis of gene expression profiles in breast cancer: toward a unified understanding of breast cancer subtyping and prognosis signatures. Breast Cancer Res. 2008;10:R65. doi: 10.1186/bcr2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fan C, et al. Concordance among gene-expression-based predictors for breast cancer. N Engl J Med. 2006;355:560–569. doi: 10.1056/NEJMoa052933. [DOI] [PubMed] [Google Scholar]
- 7.Prat A, et al. Concordance among gene- expression-based predictors for ER-positive breast cancer treated with adjuvant tamoxifen [abstract] J Clin Oncol. 2011;29 (Suppl):a502. doi: 10.1093/annonc/mds080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim C, Paik S. Gene-expression-based prognostic assays for breast cancer. Nat Rev Clin Oncol. 2010;7:340–347. doi: 10.1038/nrclinonc.2010.61. [DOI] [PubMed] [Google Scholar]
- 9.Sparano JA, Paik S. Development of the 21-gene assay and its application in clinical practice and clinical trials. J Clin Oncol. 2008;26:721–728. doi: 10.1200/JCO.2007.15.1068. [DOI] [PubMed] [Google Scholar]
- 10.Fisher B, et al. Tamoxifen and chemotherapy for lymph node-negative, estrogen receptor-positive breast cancer. J Natl Cancer Inst. 1997;89:1673–1682. doi: 10.1093/jnci/89.22.1673. [DOI] [PubMed] [Google Scholar]
- 11.Paik S, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 12.Habel LA, et al. A population-based study of tumor gene expression and risk of breast cancer death among lymph node-negative patients. Breast Cancer Res. 2006;8:R25. doi: 10.1186/bcr1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van’t Veer LJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 14.Buyse M, et al. Validation and clinical utility of a 70-gene prognostic signature for women with node-negative breast cancer. J Natl Cancer Inst. 2006;98:1183–1192. doi: 10.1093/jnci/djj329. [DOI] [PubMed] [Google Scholar]
- 15.van de Vijver MJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 16.Cardoso F, et al. Clinical application of the 70-gene profile: the MINDACT trial. J Clin Oncol. 2008;26:729–735. doi: 10.1200/JCO.2007.14.3222. [DOI] [PubMed] [Google Scholar]
- 17.Sotiriou C, et al. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J Natl Cancer Inst. 2006;98:262–272. doi: 10.1093/jnci/djj052. [DOI] [PubMed] [Google Scholar]
- 18.Loi S, et al. Definition of clinically distinct molecular subtypes in estrogen receptor-positive breast carcinomas through genomic grade. J Clin Oncol. 2007;25:1239–1246. doi: 10.1200/JCO.2006.07.1522. [DOI] [PubMed] [Google Scholar]
- 19.Ma XJ, et al. A five-gene molecular grade index and HOXB13:IL17BR are complementary prognostic factors in early stage breast cancer. Clin Cancer Res. 2008;14:2601–2608. doi: 10.1158/1078-0432.CCR-07-5026. [DOI] [PubMed] [Google Scholar]
- 20.Jerevall PL, et al. Prognostic utility of HOXB13:IL17BR and molecular grade index in early-stage breast cancer patients from the Stockholm trial. Br J Cancer. 2011;104:1762–1769. doi: 10.1038/bjc.2011.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma XJ, et al. A two-gene expression ratio predicts clinical outcome in breast cancer patients treated with tamoxifen. Cancer Cell. 2004;5:607–616. doi: 10.1016/j.ccr.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 22.Perou CM, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 23.Sorlie T, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci USA. 2003;100:8418–8423. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sørlie T, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perou CM, et al. Distinctive gene expression patterns in human mammary epithelial cells and breast cancers. Proc Natl Acad Sci USA. 1999;96:9212–9217. doi: 10.1073/pnas.96.16.9212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Millikan RC, et al. Epidemiology of basal-like breast cancer. Breast Cancer Res Treat. 2008;109:123–139. doi: 10.1007/s10549-007-9632-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fan C, et al. Building prognostic models for breast cancer patients using clinical variables and hundreds of gene expression signatures. BMC Med Genomics. 2011;4:3. doi: 10.1186/1755-8794-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parker JS, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27:1160–1167. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin M, et al. Genomic predictors of response to doxorubicin versus docetaxel in primary breast cancer. Breast Cancer Res Treat. 2011;128:127–136. doi: 10.1007/s10549-011-1461-y. [DOI] [PubMed] [Google Scholar]
- 30.Glück S, et al. TP53 genomics predict higher clinical and pathologic tumor response in operable early-stage breast cancer treated with docetaxel-capecitabine ± trastuzumab. Breast Cancer Res Treat. doi: 10.1007/s10549-011-1412-7. [DOI] [PubMed] [Google Scholar]
- 31.Carey LA, et al. The triple negative paradox: primary tumor chemosensitivity of breast cancer subtypes. Clin Cancer Res. 2007;13:2329–2334. doi: 10.1158/1078-0432.CCR-06-1109. [DOI] [PubMed] [Google Scholar]
- 32.Rouzier R, et al. Breast cancer molecular subtypes respond differently to preoperative chemotherapy. Clin Cancer Res. 2005;11:5678–5678. doi: 10.1158/1078-0432.CCR-04-2421. [DOI] [PubMed] [Google Scholar]
- 33.Nielsen TO, et al. A comparison of PAM50 intrinsic subtyping with immunohistochemistry and clinical prognostic factors in tamoxifen-treated estrogen receptor positive breast cancer. Clin Cancer Res. 2010;16:5222–5232. doi: 10.1158/1078-0432.CCR-10-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Geiss GK, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. 2008;26:317–325. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- 35.Prat A, Perou CM. Deconstructing the molecular portraits of breast cancer. Mol Oncol. 2011;5:5–23. doi: 10.1016/j.molonc.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cheang MC, et al. Ki67 index, HER2 status, and prognosis of patients with luminal B breast cancer. J Natl Cancer Inst. 2009;101:736–750. doi: 10.1093/jnci/djp082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nielsen TO, et al. Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin Cancer Res. 2004;10:5367–3574. doi: 10.1158/1078-0432.CCR-04-0220. [DOI] [PubMed] [Google Scholar]
- 38.Cheang MC, et al. Basal-like breast cancer defined by five biomarkers has superior prognostic value than triple-negative phenotype. Clin Cancer Res. 2008;14:1368–1368. doi: 10.1158/1078-0432.CCR-07-1658. [DOI] [PubMed] [Google Scholar]
- 39.Alexe G, et al. High expression of lymphocyte-associated genes in node-negative HER2+ breast cancers correlates with lower recurrence rates. Cancer Res. 2007;67:10669–10669. doi: 10.1158/0008-5472.CAN-07-0539. [DOI] [PubMed] [Google Scholar]
- 40.Teschendorff AE, et al. An immune response gene expression module identifies a good prognosis subtype in estrogen receptor negative breast cancer. Genome Biol. 2007;8:R157. doi: 10.1186/gb-2007-8-8-r157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lo SS, et al. Prospective multicenter study of the impact of the 21-gene recurrence score assay on medical oncologist and patient adjuvant breast cancer treatment selection. J Clin Oncol. 2010;28:1671–1676. doi: 10.1200/JCO.2008.20.2119. [DOI] [PubMed] [Google Scholar]
- 42.Geffen DB, et al. The impact of the 21-gene recurrence score assay on decision making about adjuvant chemotherapy in early-stage estrogen-receptor-positive breast cancer in an oncology practice with a unified treatment policy. Ann Oncol. 2011;22:2381–2386. doi: 10.1093/annonc/mdq769. [DOI] [PubMed] [Google Scholar]
- 43.Tsoi DT, Inoue M, Kelly CM, Verma S, Pritchard KI. Cost-effectiveness analysis of recurrence score-guided treatment using a 21-gene assay in early breast cancer. Oncologist. 2010;15:457–465. doi: 10.1634/theoncologist.2009-0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hornberger J, Cosler L, Lyman G. Economic analysis of targeting chemotherapy using a 21-gene RT-PCR assay in lymph-node-negative, estrogen-receptor-positive, early-stage breast cancer. Am J Manag Care. 2005;11:313–324. [PubMed] [Google Scholar]
- 45.Albain KS, et al. Prognostic and predictive value of the 21-gene recurrence score assay in postmenopausal women with node-positive, oestrogen-receptor-positive breast cancer on chemotherapy: a retrospective analysis of a randomised trial. Lancet Oncol. 2010;11:55–65. doi: 10.1016/S1470-2045(09)70314-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dowsett M, et al. Prediction of risk of distant recurrence using the 21-gene recurrence score in node-negative and node-positive postmenopausal patients with breast cancer treated with anastrozole or tamoxifen: a TransATAC study. J Clin Oncol. 2010;28:1829–1834. doi: 10.1200/JCO.2009.24.4798. [DOI] [PubMed] [Google Scholar]
- 47.Mook S, et al. The 70-gene prognosis-signature predicts disease outcome in breast cancer patients with 1–3 positive lymph nodes in an independent validation study. Breast Cancer Res Treat. 2009;116:295–302. doi: 10.1007/s10549-008-0130-2. [DOI] [PubMed] [Google Scholar]
- 48.Parker J, et al. Breast cancer molecular subtypes predict response to anthracycline/taxane-based chemotherapy [abstract] Cancer Res. 2009;69 (Suppl 3):a2019. [Google Scholar]
- 49.Liedtke C, et al. Genomic grade index is associated with response to chemotherapy in patients with breast cancer. J Clin Oncol. 2009;27:3185–3191. doi: 10.1200/JCO.2008.18.5934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Straver ME, et al. The 70-gene signature as a response predictor for neoadjuvant chemotherapy in breast cancer. Breast Cancer Res Treat. 2010;119:551–558. doi: 10.1007/s10549-009-0333-1. [DOI] [PubMed] [Google Scholar]
- 51.Gianni L, et al. Gene expression profiles in paraffin-embedded core biopsy tissue predict response to chemotherapy in women with locally advanced breast cancer. J Clin Oncol. 2005;23:7265–7277. doi: 10.1200/JCO.2005.02.0818. [DOI] [PubMed] [Google Scholar]
- 52.Ellis MJ, et al. Outcome prediction for estrogen receptor-positive breast cancer based on postneoadjuvant endocrine therapy tumor characteristics. J Natl Cancer Inst. 2008;100:1380–1388. doi: 10.1093/jnci/djn309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ellis MJ, et al. Letrozole is more effective neoadjuvant endocrine therapy than tamoxifen for ErbB-1- and/or ErbB-2-positive, estrogen receptor-positive primary breast cancer: evidence from a phase III randomized trial. J Clin Oncol. 2001;19:3808–3816. doi: 10.1200/JCO.2001.19.18.3808. [DOI] [PubMed] [Google Scholar]
- 54.Ellis MJ, et al. Randomized phase II neoadjuvant comparison between letrozole, anastrozole, and exemestane for postmenopausal women with estrogen receptor-rich stage 2 to 3 breast cancer: clinical and biomarker outcomes and predictive value of the baseline PAM50-based intrinsic subtype--ACOSOG Z1031. J Clin Oncol. 2011;29:2342–2349. doi: 10.1200/JCO.2010.31.6950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Paik S, et al. Gene expression and benefit of chemotherapy in women with node-negative, estrogen receptor-positive breast cancer. J Clin Oncol. 2006;24:3726–3734. doi: 10.1200/JCO.2005.04.7985. [DOI] [PubMed] [Google Scholar]
- 56.Cronin M, et al. Analytical validation of the Oncotype DX genomic diagnostic test for recurrence prognosis and therapeutic response prediction in node-negative, estrogen receptor-positive breast cancer. Clin Chem. 2007;53:1084–1091. doi: 10.1373/clinchem.2006.076497. [DOI] [PubMed] [Google Scholar]
- 57.Glas AM, et al. Converting a microarray breast cancer signature into a high throughput diagnostic test. BMC Genomics. 2006;7:278. doi: 10.1186/1471-2164-7-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wolff AC, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. J Clin Oncol. 2007;25:118–145. doi: 10.1200/JCO.2006.09.2775. [DOI] [PubMed] [Google Scholar]
- 59.Hammond ME, Hayes DF, Wolff AC, Mangu PB, Temin S. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J Clin Oncol. 2010;28:2784–2795. doi: 10.1200/JCO.2009.25.6529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cohen J. A coefficient of agreement for nominal scales. Educ Psychol Meas. 1960;20:37–46. [Google Scholar]
- 61.Randolph J. Free-marginal multirater kappa: an alternative to Fleiss’ fixed-marginal multirater kappa. Joensuu University Learning and Instruction Symp; October 14–15 2005. [Google Scholar]
- 62.Mackay A, et al. Microarray-based class discovery for molecular classification of breast cancer: analysis of interobserver agreement. J Natl Cancer Inst. 2011;103:662–673. doi: 10.1093/jnci/djr071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Weigelt B, et al. Breast cancer molecular profiling with single sample predictors: a retrospective analysis. Lancet Oncol. 2010;11:339–349. doi: 10.1016/S1470-2045(10)70008-5. [DOI] [PubMed] [Google Scholar]
- 64.Turbin DA, et al. Automated quantitative analysis of estrogen receptor expression in breast carcinoma does not differ from expert pathologist scoring: a tissue microarray study of 3,484 cases. Breast Cancer Res Treat. 2008;110:417–426. doi: 10.1007/s10549-007-9736-z. [DOI] [PubMed] [Google Scholar]
- 65.Bueno-de-Mesquita JM, et al. The impact of inter-observer variation in pathological assessment of node-negative breast cancer on clinical risk assessment and patient selection for adjuvant systemic treatment. Ann Oncol. 2010;21:40–47. doi: 10.1093/annonc/mdp273. [DOI] [PubMed] [Google Scholar]
- 66.Mudduwa L, Liyanage T. Immunohistochemical assessment of hormone receptor status of breast carcinoma: interobserver variation of the quick score. Indian J Med Sci. 2009;63:21–27. [PubMed] [Google Scholar]
- 67.Wells CA, et al. Consistency of staining and reporting of oestrogen receptor immunocytochemistry within the European Union—an inter-laboratory study. Virchows Arch. 2004;445:119–128. doi: 10.1007/s00428-004-1063-8. [DOI] [PubMed] [Google Scholar]
- 68.Badve SS, et al. Estrogen- and progesterone-receptor status in ECOG 2197: comparison of immunohistochemistry by local and central laboratories and quantitative reverse transcription polymerase chain reaction by central laboratory. J Clin Oncol. 2008;26:2473–2481. doi: 10.1200/JCO.2007.13.6424. [DOI] [PubMed] [Google Scholar]
- 69.Collins LC, Marotti JD, Baer HJ, Tamimi RM. Comparison of estrogen receptor results from pathology reports with results from central laboratory testing. J Natl Cancer Inst. 2008;100:218–221. doi: 10.1093/jnci/djm270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Parker RL, et al. Assessment of interlaboratory variation in the immunohistochemical determination of estrogen receptor status using a breast cancer tissue microarray. Am J Clin Pathol. 2002;117:723–728. doi: 10.1309/PEF8-GL6F-YWMC-AG56. [DOI] [PubMed] [Google Scholar]
- 71.Rydén L, et al. Reproducibility of human epidermal growth factor receptor 2 analysis in primary breast cancer: a national survey performed at pathology departments in Sweden. Acta Oncol. 2009;48:860–866. doi: 10.1080/02841860902862511. [DOI] [PubMed] [Google Scholar]
- 72.Press MF, et al. Diagnostic evaluation of HER-2 as a molecular target: an assessment of accuracy and reproducibility of laboratory testing in large, prospective, randomized clinical trials. Clin Cancer Res. 2005;11:6598–6607. doi: 10.1158/1078-0432.CCR-05-0636. [DOI] [PubMed] [Google Scholar]
- 73.Umemura S, et al. What causes discrepancies in HER2 testing for breast cancer? Am J Clin Pathol. 2008;130:883–891. doi: 10.1309/AJCP5UUMFMA5ZKII. [DOI] [PubMed] [Google Scholar]
- 74.Diaz LK, Gupta R, Kidwai N, Sneige N, Wiley EL. The use of TMA for interlaboratory validation of FISH testing for detection of HER2 gene amplification in breast cancer. J Histochem Cytochem. 2004;52:501–507. doi: 10.1177/002215540405200408. [DOI] [PubMed] [Google Scholar]
- 75.Di Palma S, et al. A quality assurance exercise to evaluate the accuracy and reproducibility of chromogenic in situ hybridisation for HER2 analysis in breast cancer. J Clin Pathol. 2008;61:757–760. doi: 10.1136/jcp.2007.053850. [DOI] [PubMed] [Google Scholar]
- 76.Bartlett JM, et al. Evaluating HER2 amplification and overexpression in breast cancer. J Pathol. 2001;195:422–428. doi: 10.1002/path.971. [DOI] [PubMed] [Google Scholar]
- 77.Turashvili G, et al. Inter-observer reproducibility of HER2 immunohistochemical assessment and concordance with fluorescent in situ hybridization (FISH): pathologist assessment compared to quantitative image analysis. BMC Cancer. 2009;9:165. doi: 10.1186/1471-2407-9-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.van der Vegt B, de Bock GH, Bart J, Zwartjes NG, Wesseling J. Validation of the 4B5 rabbit monoclonal antibody in determining Her2/neu status in breast cancer. Mod Pathol. 2009;22:879–886. doi: 10.1038/modpathol.2009.37. [DOI] [PubMed] [Google Scholar]
- 79.Tsuda H, et al. HER2 testing on core needle biopsy specimens from primary breast cancers: interobserver reproducibility and concordance with surgically resected specimens. BMC Cancer. 2010;10:534. doi: 10.1186/1471-2407-10-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Arena V, Pennacchia I, Monego G, Carbone A, Capelli A. Fluorescent in situ hybridization as a primary test for HER2 status in breast cancer: controversies. J Clin Oncol. 2009;28:e83–e84. doi: 10.1200/JCO.2009.25.1397. [DOI] [PubMed] [Google Scholar]
- 81.Sauter G, Lee J, Bartlett JM, Slamon DJ, Presse MJ. Guidelines for human epidermal growth factor receptor 2 testing: biologic and methodologic considerations. J Clin Oncol. 2009;27:1323–1333. doi: 10.1200/JCO.2007.14.8197. [DOI] [PubMed] [Google Scholar]
- 82.Perez EA, et al. HER2 testing by local, central, and reference laboratories in specimens from the North Central Cancer Treatment Group N9831 intergroup adjuvant trial. J Clin Oncol. 2006;24:3032–3038. doi: 10.1200/JCO.2005.03.4744. [DOI] [PubMed] [Google Scholar]
- 83.Paik S, et al. Real-world performance of HER2 testing--National Surgical Adjuvant Breast and Bowel Project experience. J Natl Cancer Inst. 2002;94:852–854. doi: 10.1093/jnci/94.11.852. [DOI] [PubMed] [Google Scholar]
- 84.Regan MM, et al. Re-evaluating adjuvant breast cancer trials: assessing hormone receptor status by immunohistochemical versus extraction assays. J Natl Cancer Inst. 2006;98:1571–1581. doi: 10.1093/jnci/djj415. [DOI] [PubMed] [Google Scholar]
- 85.Cheang MC, et al. Immunohistochemical detection using the new rabbit monoclonal antibody SP1 of estrogen receptor in breast cancer is superior to mouse monoclonal antibody 1D5 in predicting survival. J Clin Oncol. 2006;24:5637–5644. doi: 10.1200/JCO.2005.05.4155. [DOI] [PubMed] [Google Scholar]
- 86.Harvey JM, Clark GM, Osborne CK, Allred DC. Estrogen receptor status by immunohistochemistry is superior to the ligand-binding assay for predicting response to adjuvant endocrine therapy in breast cancer. J Clin Oncol. 1999;17:1474–1481. doi: 10.1200/JCO.1999.17.5.1474. [DOI] [PubMed] [Google Scholar]
- 87.Nassar A, Cohen C, Siddiqui M. Estimation of hormone receptor status and HER2 in cytologic cell blocks from breast cancer using the novel rabbit monoclonal antibodies (SP1, SP2, and SP3) Diagn Cytopathol. 2009;37:865–870. doi: 10.1002/dc.21121. [DOI] [PubMed] [Google Scholar]
- 88.Rhodes A, Jasani B, Barnes DM, Bobrow LG, Miller KD. Reliability of immunohistochemical demonstration of oestrogen receptors in routine practice: interlaboratory variance in the sensitivity of detection and evaluation of scoring systems. J Clin Pathol. 2000;53:125–130. doi: 10.1136/jcp.53.2.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Viale G, et al. Prognostic and predictive value of centrally reviewed expression of estrogen and progesterone receptors in a randomized trial comparing letrozole and tamoxifen adjuvant therapy for postmenopausal early breast cancer: BIG 1–98. J Clin Oncol. 2007;25:3846–3852. doi: 10.1200/JCO.2007.11.9453. [DOI] [PubMed] [Google Scholar]
- 90.Regitnig P, et al. Quality assurance for detection of estrogen and progesterone receptors by immunohistochemistry in Austrian pathology laboratories. Virchows Arch. 2002;441:328–334. doi: 10.1007/s00428-002-0646-5. [DOI] [PubMed] [Google Scholar]
- 91.Anderson TJ, Alexander FE, Lamb J, Smith A, Forrest AP. Pathology characteristics that optimize outcome prediction of a breast screening trial. Br J Cancer. 2000;83:487–492. doi: 10.1054/bjoc.2000.1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Frierson HF, Jr, et al. Interobserver reproducibility of the Nottingham modification of the Bloom and Richardson histologic grading scheme for infiltrating ductal carcinoma. Am J Clin Pathol. 1995;103:195–198. doi: 10.1093/ajcp/103.2.195. [DOI] [PubMed] [Google Scholar]
- 93.Ellis IO, et al. Impact of a national external quality assessment scheme for breast pathology in the UK. J Clin Pathol. 2006;59:138–145. doi: 10.1136/jcp.2004.025551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Longacre TA, et al. Interobserver agreement and reproducibility in classification of invasive breast carcinoma: an NCI breast cancer family registry study. Mod Pathol. 2005;19:195–207. doi: 10.1038/modpathol.3800496. [DOI] [PubMed] [Google Scholar]
- 95.Boiesen P, et al. Histologic grading in breast cancer--reproducibility between seven pathologic departments. South Sweden Breast Cancer Group. Acta Oncol. 2000;39:41–45. doi: 10.1080/028418600430950. [DOI] [PubMed] [Google Scholar]
- 96.Sloane JP, et al. Consistency achieved by 23 European pathologists from 12 countries in diagnosing breast disease and reporting prognostic features of carcinomas. Virchows Arch. 1999;434:3–10. doi: 10.1007/s004280050297. [DOI] [PubMed] [Google Scholar]
- 97.Adams AL, Chhieng DC, Bell WC, Winokur T, Hameed O. Histologic grading of invasive lobular carcinoma: does use of a 2-tiered nuclear grading system improve interobserver variability? Ann Diagn Pathol. 2009;13:223–225. doi: 10.1016/j.anndiagpath.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 98.Tsuda H, et al. Evaluation of the interobserver agreement in the number of mitotic figures of breast carcinoma as simulation of quality monitoring in the Japan National Surgical Adjuvant Study of Breast Cancer (NSAS-BC) protocol. Jpn J Cancer Res. 2000;91:451–457. doi: 10.1111/j.1349-7006.2000.tb00966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Reed W, et al. The prognostic value of p53 and c-erb B-2 immunostaining is overrated for patients with lymph node negative breast carcinoma. Cancer. 2000;88:804–813. doi: 10.1002/(sici)1097-0142(20000215)88:4<804::aid-cncr11>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 100.Rakha E, et al. Breast cancer prognostic classification in the molecular era: the role of histological grade. Breast Cancer Res. 2010;12:207. doi: 10.1186/bcr2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Dybdal N, et al. Determination of HER2 gene amplification by fluorescence in situ hybridization and concordance with the clinical trials immunohistochemical assay in women with metastatic breast cancer evaluated for treatment with trastuzumab. Breast Cancer Res Treat. 2005;93:3–11. doi: 10.1007/s10549-004-6275-8. [DOI] [PubMed] [Google Scholar]
- 102.Press MF, et al. HER-2 gene amplification, HER-2 and epidermal growth factor receptor mRNA and protein expression, and lapatinib efficacy in women with metastatic breast cancer. Clin Cancer Res. 2008;14:7861–7870. doi: 10.1158/1078-0432.CCR-08-1056. [DOI] [PubMed] [Google Scholar]
- 103.Dressler LG, et al. Comparison of HER2 status by fluorescence in situ hybridization and immunohistochemistry to predict benefit from dose escalation of adjuvant doxorubicin-based therapy in node-positive breast cancer patients. J Clin Oncol. 2005;23:4287–4297. doi: 10.1200/JCO.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 104.Powell WC, et al. A new rabbit monoclonal antibody (4B5) for the immunohistochemical (IHC) determination of the HER2 status in breast cancer: comparison with CB11, fluorescence in situ hybridization (FISH), and interlaboratory reproducibility. Appl Immunohistochem Mol Morphol. 2007;15:94–102. doi: 10.1097/pai.0b013e31802ced25. [DOI] [PubMed] [Google Scholar]
- 105.Noske A, et al. Comparison of different approaches for assessment of HER2 expression on protein and mRNA level: prediction of chemotherapy response in the neoadjuvant GeparTrio trial ( NCT00544765) Breast Cancer Res Treat. 2011;126:109–117. doi: 10.1007/s10549-010-1316-y. [DOI] [PubMed] [Google Scholar]
- 106.Baehner FL, et al. Human epidermal growth factor receptor 2 assessment in a case-control study: comparison of fluorescence in situ hybridization and quantitative reverse transcription polymerase chain reaction performed by central laboratories. J Clin Oncol. 2010;28:4300–4306. doi: 10.1200/JCO.2009.24.8211. [DOI] [PubMed] [Google Scholar]
- 107.Perou CM, Parker JS, Prat A, Ellis MJ, Bernard PS. Clinical implementation of the intrinsic subtypes of breast cancer. Lancet Oncol. 2010;11:718–719. doi: 10.1016/S1470-2045(10)70176-5. [DOI] [PubMed] [Google Scholar]
- 108.Sørlie T, et al. The importance of gene-centring microarray data. Lancet Oncol. 2010;11:719–720. doi: 10.1016/S1470-2045(10)70174-1. [DOI] [PubMed] [Google Scholar]
- 109.Dunning MJ, et al. The importance of platform annotation in interpreting microarray data. Lancet Oncol. 2010;11:717–717. doi: 10.1016/S1470-2045(10)70115-7. [DOI] [PubMed] [Google Scholar]
- 110.Pillai R, et al. Validation and reproducibility of a microarray-based gene expression test for tumor identification in formalin-fixed, paraffin-embedded specimens. J Mol Diagn. 2011;13:48–56. doi: 10.1016/j.jmoldx.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hu Z, et al. The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genomics. 2006;7:96. doi: 10.1186/1471-2164-7-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Harrell JC, et al. Genomic analysis identifies unique signatures predictive of brain, lung, and liver relapse. Breast Cancer Res Treat. doi: 10.1007/s10549-011-1619-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ding L, et al. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature. 2010;464:999–1005. doi: 10.1038/nature08989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Shah SP, et al. Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature. 2009;461:809–813. doi: 10.1038/nature08489. [DOI] [PubMed] [Google Scholar]
- 115.Nunes CB, et al. Comparative analysis of six different antibodies against Her2 including the novel rabbit monoclonal antibody (SP3) and chromogenic in situ hybridisation in breast carcinomas. J Clin Pathol. 2008;61:934–938. doi: 10.1136/jcp.2007.053892. [DOI] [PubMed] [Google Scholar]
- 116.Moelans CB, et al. Validation of a fully automated HER2 staining kit in breast cancer. Cell Oncol. 2010;32:149–155. doi: 10.3233/CLO-2010-0514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Tubbs RR, et al. Discrepancies in clinical laboratory testing of eligibility for trastuzumab therapy: apparent immunohistochemical false-positives do not get the message. J Clin Oncol. 2001;19:2714–2721. doi: 10.1200/JCO.2001.19.10.2714. [DOI] [PubMed] [Google Scholar]
- 118.Arihiro K, et al. Comparison of evaluations for hormone receptors in breast carcinoma using two manual and three automated immunohistochemical assays. Am J Clin Pathol. 2007;127:356–365. doi: 10.1309/4D1A04NCDK96WFY7. [DOI] [PubMed] [Google Scholar]