Abstract
We recently described a strategy to prepare DNA-tethered lipid membranes either to fixed DNA on a surface or to DNA displayed on a supported bilayer1, 2. With the latter system the DNA hybrids are laterally mobile; when orthogonal sense-antisense pairs of different lengths are used, the DNA hybrids segregate by height and the tethered membrane deforms to accommodate the height difference. This architecture is particularly useful for modelling interactions between membranes mediated by molecular recognition and resembles cell-to-cell junctions. The length, affinity and population of DNA hybrids between membranes are completely controllable. Interesting patterns of height segregation are observed by fluorescence interference contrast microscopy. Diverse behavior is observed in the segregation and pattern forming process and possible mechanisms are discussed. This model system captures some of the essential physics of synapse formation and is a step towards understanding lipid membrane behaviour in a cell-to-cell junction.
1 Introduction
Many biological processes involve tightly regulated interactions between two membrane surfaces. These are typically initiated by the recognition and binding of complementary molecules displayed on the two membrane surfaces. Examples include cell adhesion3, signalling cascades as in the case of the immunological T-cell response4, and neuronal fusion, where synaptic vesicles first dock to the presynaptic membrane, then undergo lipid mixing and content release5. The dynamic organization of ligand-receptors and membrane components and their reorganization during membrane engagement are essential for the assembly of inter- and intra-membrane protein complexes, and their emergent role in signal transduction6.
Several biologically relevant model or hybrid model-cellular systems have been described. In the case of cell adhesion and the T-cell response, supported lipid bilayers displaying some of the relevant components have been used to mimic at least one of the cell surfaces7-10, and this has given important insights into the clustering of membrane components for T-cell signalling. The reorganization of membrane-bound IgG in inter-membrane junctions that formed dense and sparse regions of IgG11 gives insight into the topological effect of spacers between two membranes. A theoretical model developed by Qi et al.12 to simulate these phenomena in membrane junctions can be particularly useful to develop mechanistic hypotheses that are experimentally testable.
We employed a DNA-tethered lipid membrane system developed in our group1 in which a planar free standing lipid bilayer is tethered to a fluid supported bilayer via DNA-hybridization (Figure 1) to simulate a cell-to-cell junction. DNA-lipid conjugates can be prepared with different lengths and sequences (see Table 1). In the cases discussed in the following, orthogonal sequences that are 24, 48 and 72-mers, corresponding approximately to 8, 16 and 24 nm hybrids, were employed. As illustrated schematically in Figure 2A, when DNA-lipid conjugates with different lengths are present, one expects that the largest number of hybrids can be formed by segregation into domains by length, involving a competition between binding kinetics, hybrid stability, lateral mobility and the curvature of the tethered membrane which must occur to accommodate the height difference. In the original report1, we presented evidence for this height segregation in a mixed 24-mer/48-mer system, and we refer to this in the following as topological domains. This was based on fluorescence interference contrast microscopy (FLIC13), and variable incidence angle FLIC (VIA-FLIC14), using fluorescent lipids in the tethered bilayer patch (see Figure 2A) to show that topological domains were formed and the distances separating the two membrane domains correspond to what is expected for 24- and 48-mer hybrids. Since the length, population and hybridization energy of the DNA hybrid tethers are controllable, and the DNA can be independently labeled with a fluorescent dye, we can now study the dependence of topological domain formation and evolution more systematically.
Fig. 1.
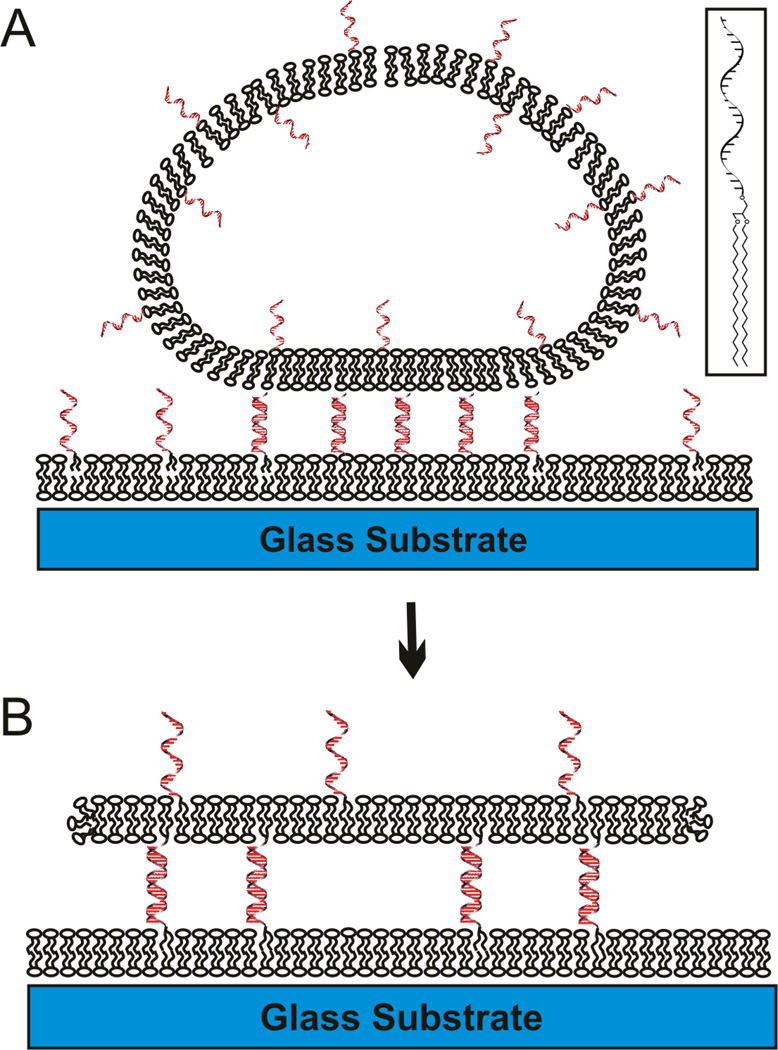
Schematic illustration of DNA-tethered lipid bilayer patches formation by GUV rupture on supported lipid bilayers, where DNA tethers are laterally mobile (not drawn to scale). When GUVs presenting the DNA are added onto a supported lipid bilayer presenting complementary DNA, GUVs are flattened (A) upon DNA binding, and some of them rupture to form a DNA-tethered membrane (B). The gap between membranes is precisely controllable by adjusting the DNA length. The inset shows the chemical structure of DNA-lipid conjugate.
Table 1.
Sequences of 24mer, 48mer, 72mer or Poly A/T (A/T) DNA oligonucleotides 5'-coupled to lipids as shown in the inset to Fig. 1A and used to tether lipid membranes.
| Name | Sequence (5' → 3') |
|---|---|
| 24-1 | TCG ACA CGG AAA TGT TGA ATA CTA |
| 24-2 | TAG TAT TCA ACA TTT CCG TGT CGA |
| 48-1 | TAA CTA CAG AAT TTA TAC TAT CCC GGG TCA CAG CAG AGA AAC AAG ATA - b |
| 48-2 | TAT CTT GTT TCT CTG CTG TGA CCC GGG ATA GTA TAA ATT CTG TAG TTA |
| 72-1 | TTG AGA TCT TCC ATA TGC TGA GAG GGA TTG TGT CGA GAT AAG GCT GTG GTG GTA TTG CGC TAT GAG TAC TAA - c |
| 72-2 | TTA GTA CTC ATA GCG CAA TAC CAC CAC AGC CTT ATC TCG ACA CAA TCC CTC TCA GCA TAT GGA AGA TCT CAA |
| poly24T | TTT TTT TTT TTT TTT TTT TTT TTT |
| poly24A | AAA AAA AAA AAA AAA AAA AAA AAA - a |
where Alexa 488 is conjugated
where Cy5 is conjugated
where Cy3.5 is conjugated
Fig. 2.
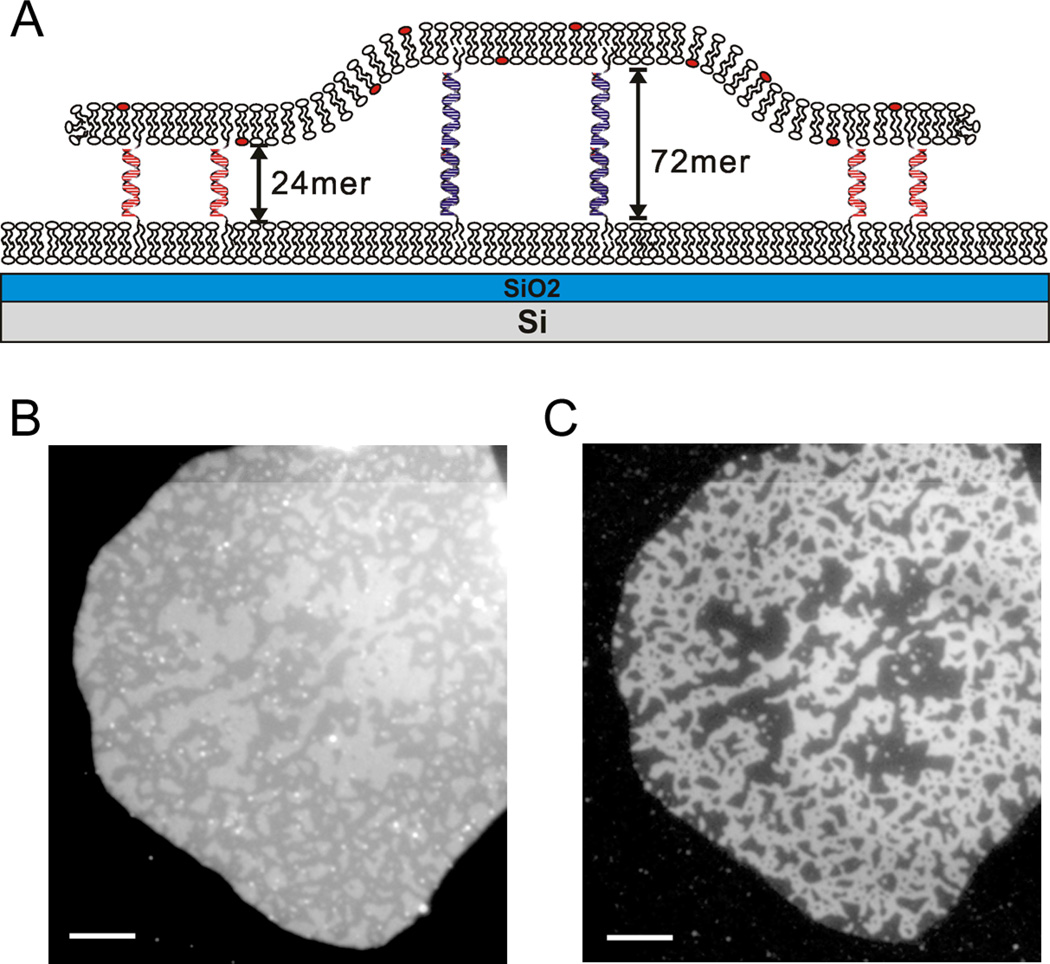
(A) Schematic illustration of a height segregated DNA-tethered bilayer patch with 24mer and 72mer DNAs. In some cases, as illustrated, the substrate is SiO2 grown on flat Si, a highly reflective mirror, to differentiate height differences by fluorescence intensity using FLIC (not drawn to scale). (B) The height variation in a tethered bilayer patch containing TR labeled lipid and containing a 1:1 mixture of Alexa 488 labeled 24mer and unlabeled 72mer DNA-lipid conjugate is visualized by FLIC (260 nm thickness of SiO2 on Si). The different height regions are distinguished by the intensity variation in TR emission in the tethered patch membrane: bright (higher) domains are the 72 mer and dim (lower) 24mer segregated domains are evident. (C) The same patch is visualized with Alexa 488 (right) attached on the membrane distal end of the 24mer. The dimmer 24mer regions in panel B completely overlap with Alexa 488 in panel C. Because fluorescence images offer more contrast than FLIC, Alexa 488 labeled DNA-lipid conjugates can be used as a surrogate marker for topological domains. Scale bar is 10 µm.
2 Experimental
2.1. Formation of supported lipid bilayers displaying DNA
Small unilamellar vesicles (SUVs) made of Egg yolk phosphatidylcholine (Avanti) were formed by the extrusion method as described extensively in the literature15, 16. The desired ratio of DNA-lipid conjugates (Fig. 1C), prepared as described17, with different DNA lengths and sequences (Table 1) were added to the SUVs to achieve overall 0.3 mol% portion of lipid molecules. Supported lipid bilayers (SLBs) displaying DNA were formed by adding the SUVs into a CoverWell chamber (Molecular Probes) on a cleaned glass cover slip or, for FLIC, a thermally grown flat SiO2 layer on highly reflective silicon (as illustrated in Fig. 2A). A 260 nm or 380 nm thick SiO2 layer was used in the latter so that the gradient in excitation intensity is large to assist distinguishing small height differences in the tens of nm range above the surface.1, 14
2.2. Giant unilamellar vesicle (GUV) preparation and tethered membrane patch formation
In order to form stable tethered bilayer patches2, a lipid mixture composed of cholesterol and saturated phospholipids – 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC), 1,2-dipentadecanoyl-sn-glycero-3-phosphocholine (D15PC) – was used with 0.5 mol% Texas Red 1,2-dihexadecanoyl-sn-glycero-3-phosphoethanolamine (TR-DHPE, Molecular Probes, Eugene, OR) for visualization. The molar ratios were 55:45 for DPPC:cholesterol and 65:35 for D15PC:cholesterol, respectively. Mixtures of DNA-lipid conjugates (see Table 1 for sequences and notation) were dried in a glass vial and re-dissolved in methanol, then added to the lipid mixture (also in methanol solution) to make overall 0.5 mol% DNA. When three labelled DNA-lipids were present – Alexa 488-poly24A, Cy5-48-1 and Cy3.5-72-1 – the TR lipid dye was not used to avoid overlap with Cy3.5. The fluorescence dye labelled DNA-lipid conjugates for tracking the location of the DNA tethers during the height segregation process were synthesized by adding the dye on the 3 ' end of the DNA, with the 5' end bound to the lipid17.
GUVs were grown by using the electroformation technique as previously described1, 18. The lipid and DNA mixture was coated on Pt electrodes and rehydrated in 0.5 M sucrose solution. Electroswelling with 2.5V and 10Hz alternating current was performed for 2hr at 60°C, above the chain melting temperature. Because all the components are mixed first, the DNA is displayed on both the inner and outer surface of the GUV (and the top and bottom of the resulting patch as illustrated in Figure 2). The GUVs were stored at 4°C and used within 2 days.
Patches are formed by rupture of the GUVs upon hybridization with the DNA presented on the SLB. When GUVs displaying the anti-sense DNA make contact with an SLB displaying the sense-strand, the GUVs bind, flatten and then rupture as DNA hybridization progresses. The lipid membrane of the ruptured GUV then rapidly spreads to form a planar tethered lipid bilayer patch (Fig. 1B). When a single length of DNA is present, flat tethered patches are formed, as seen by FLIC1 and illustrated in Fig. 1B. When different DNA lengths are present, the height segregation pattern is observed in the contact area between the SLB and the tethered patch, as illustrated in Fig. 2B. In some cases, the GUV does not rupture, and the evolution of the height segregation pattern can be observed in the flattened region between the GUV and the SLB.
Epifluorescence images were obtained on a Nikon Eclipse Ti-U inverted microscope with a 100x oil immersion objective (NA 1.49, Nikon) illuminating with a mercury arc lamp. Images were recorded using an Andor iXon897 CCD camera via the Metamorph imaging software. FLIC images were obtained using epifluorescence illumination by a 60x water dipping objective (NA 1.0, Nikon) on a Nikon 80i upright microscope equipped Andor Clara interline CCD.
2.3. Determination of domain area in segregated patches
The areas of domains in segregated patches were determined by analysis of the image TIFF files using a pixel count function of Matlab (The Mathworks, Inc.). Patches with two different lengths of DNA were allowed to form and become segregated, then washed by 10 mM phosphate and 100 mM NaCl, pH 7.4 buffer to remove bright GUV debris which can interfere with the analysis of patch area. Clean patches bigger than 15 µm diameter were selected because small patches can be biased by residual debris and edge effects. In the case of FLIC images, the intensity range for bright or dim regions was set and the number of pixels within each intensity range was counted. In the case of dye-labeled DNAs, the entire patch was imaged by the TR membrane dye, and the domain area was evaluated from the DNA dye fluorescence. In this case, the pixels of bright area were counted to obtain the area fraction by dividing the DNA dye area by the area of the entire patch.
3 Results
3.1. Characterization of height segregated DNA-tethered lipid bilayer patch
As described previously1, DNA-tethered lipid bilayer patches with mobile tethers are formed by GUV rupture on a supported lipid bilayer presenting complementary DNA (Figure 1). The lipid membrane of the GUV becomes the tethered patch, linked to the bottom fluid SLB membrane via DNA hybrids. The hydrophobic lipid tails of the DNA-lipid conjugates (Fig. 1A inset) are inserted into both the top and bottom fluid lipid membranes, thus the hybrid DNA tethers are laterally mobile and can reorganize and segregate when different lengths of DNAs are present. This behavior is not observed when the DNA is covalently to the glass support so that the hybrids cannot move once formed.1 We assume that the DNA-lipid conjugates with different DNA lengths are initially randomly distributed on both surfaces, but after they are constrained between two membranes by hybridization during binding of the GUVs and formation of patches, they segregate into regions with the same length, forming the interesting topological patterns with different heights, visualized either by FLIC (Fig. 2B) or directly by labeling the DNA (Fig. 2C). Though the intrinsic instability of tethered bilayer patches with mobile linkers limited systematic study of this phenomenon in the original paper1, more recent work led to a bilayer patch composition composed of saturated lipids and cholesterol which is stable,2 and this allows more time to observe the height segregation process and manipulate conditions.
Figure 2B and C shows a patch formed from a GUV labeled with a TR lipid dye and equal mol% of two different lengths of DNA: a 24mer (poly24A) labeled with Alexa 488 and a 72-mer (72-1; see Table 1). This was assembled on an SLB bilayer displaying poly24T and 72-2 on a FLIC surface (Fig. 2A). For the dimensions illustrated, and visualizing the TR lipid dye in the tethered patch, the low height region (24mer) is dimmer and the higher region (72mer) is brighter (Fig. 2B). In earlier work1, and for a mixture of 24mer and 48mer, the exact heights were measured by VIA-FLIC14 to be 10 ± 1 nm for the 24mer and 16 ± 1 nm for the 48mer, which correspond to the expected lengths of double-stranded DNA helices. The region of Alexa 488 (Fig. 2C) completely overlaps with the dimmer (by FLIC) 24mer region, proving that the height difference is truly caused by DNA tether segregation.
3.2. Domain area variations with ratio of DNA lengths or salt concentration
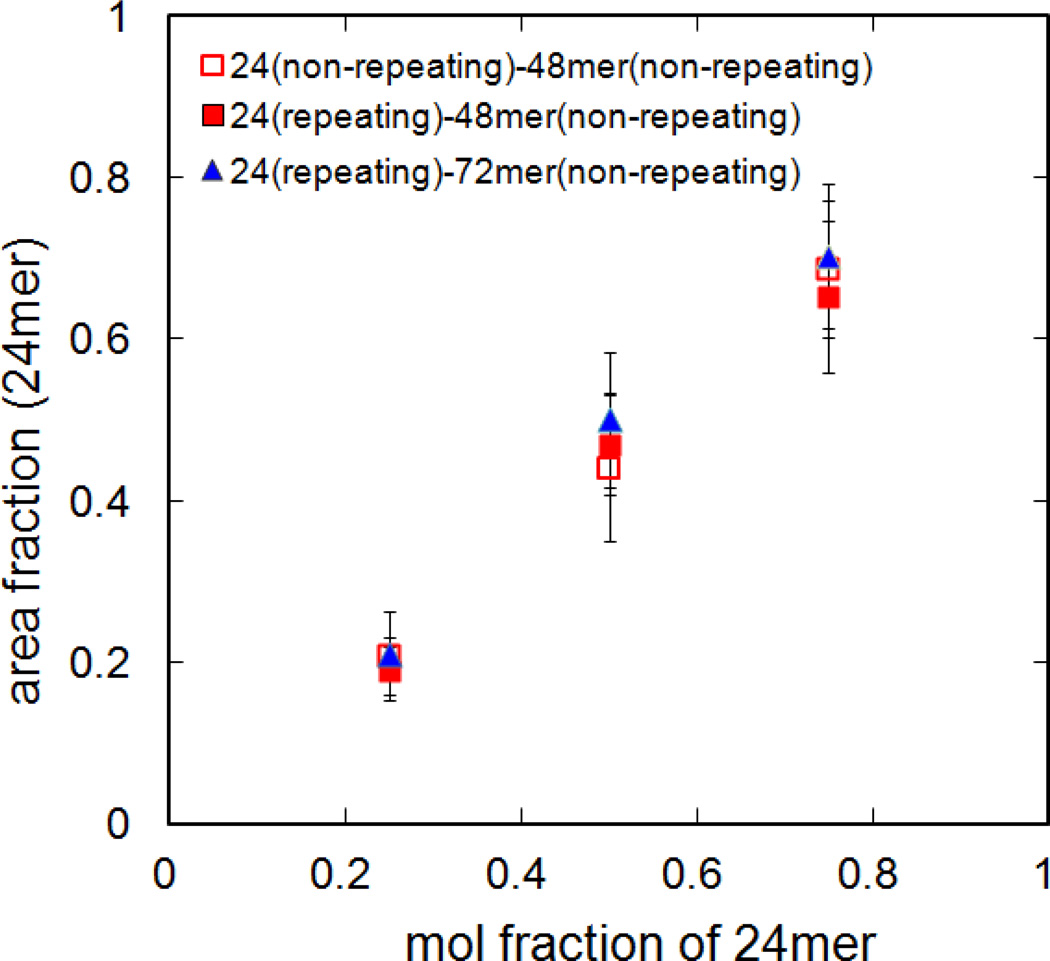
The dependence of domain area on the length ratio of each DNA and its sequence was measured. For the 24mer component, either a repeating sequence (poly24A/T, Table 1) or fully overlapping sequence (24-1/2, Table 1) was used to examine the dependence on the sequence of the complementary oligonucleotides, noting that the major difference is that the poly24A/T can be hybridized when any part of the two strands come into contact. The effect of height difference was tested by using a 48mer or 72mer DNA as the other component. Several ratios of short and long oligonucleotides (1:3, 1:1 and 3:1) were tested, and the area occupied by each kind of DNA was estimated by pixel count (see experimental) using either FLIC images or labeled DNA images. The area fraction of one length of DNA increased as its mole fraction increased, but longer DNA tends to occupy a larger area than its mol fraction. The 24mer sequence and height difference had no additional effect (Fig. 3). Although the lipid composition variation from GUV to GUV within a single GUV preparation is generally within 5mol%19, the standard deviation of the area fraction of 24mer domains was 10 ~ 15%, probably due to partial destruction of tethered patches. The patches composed of saturated lipids and cholesterol are relatively stable, but partial loss up to 20% of its original area was still observed over time2.
Fig. 3.
Area fraction of 24mer topological domains as a function of the mol fraction of the 24mer DNA. The domain height difference was varied by using either a 72mer (triangle) or 48mer (square), and the effect of DNA sequence was compared by using repeating sequence (poly24A/T, see table 1; solid symbols) or the fully overlapping sequence (24-1/2; open symbols). The 48mer (48-1/2) and 72mer (72-1/2) were fully overlapping sequences. The high and low regions were differentiated by either FLIC or labelled DNA. All data for patches in 10mM phosphate buffer with 100mM NaCl, pH 7.4.
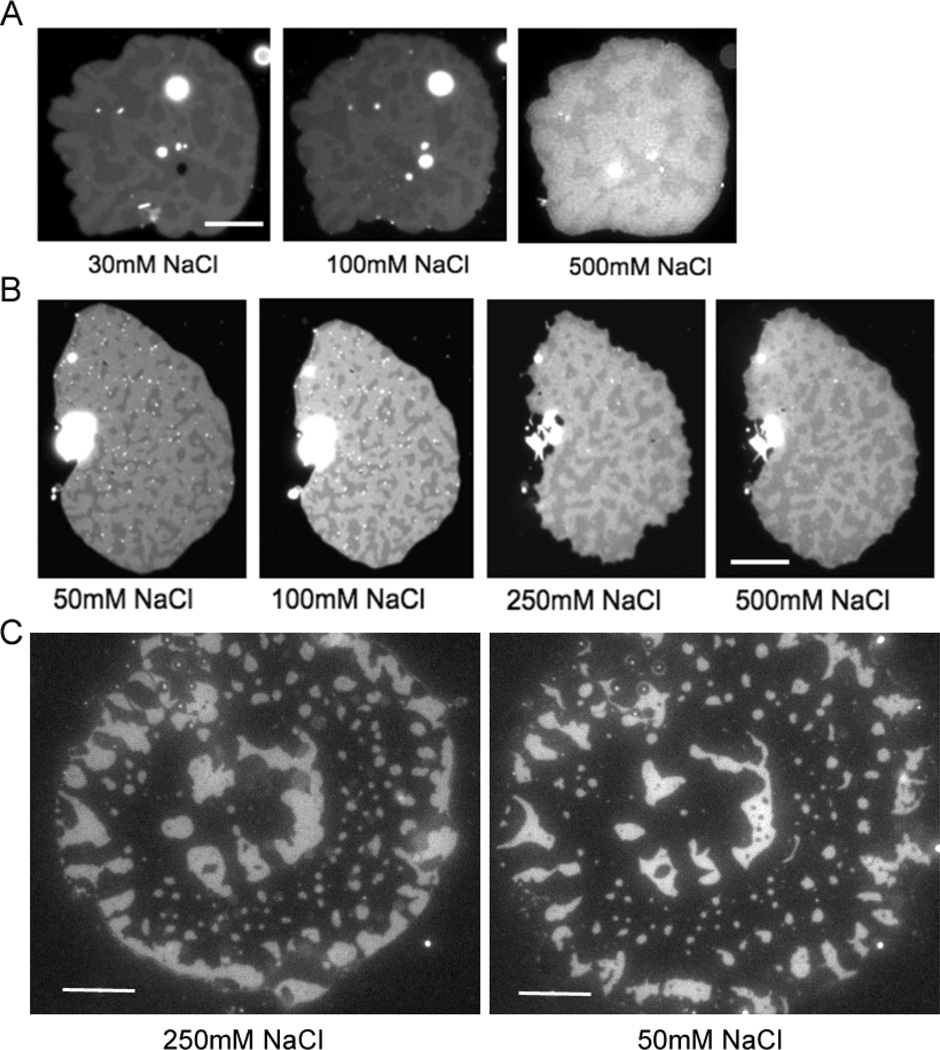
We next varied the ionic strength because this impacts the DNA duplex dissociation rate20 and the electric double layer between charged species. The tethered patches were typically formed in 10mM phosphate buffer and 240mM NaCl for osmotic balance with 0.5 mM sucrose solution inside the GUVs. After the patches were formed, the solution above the patches can be easily exchanged by washing the well on the cover glass with excess of a new solution. The solution in the thin gap under the patches equilibrates quickly (this has been observed directly when a dye was transferred into this space in membrane fusion experiments21). We initially focused on the low salt regime (< 100 mM) where a significant increase in the duplex dissociation rate and Debye length are expected. 30 mM or 50 mM NaCl concentration conditions were compared with 100 mM (Fig. 4A,B). Reduction in the salt concentration leads to expansion of the 48mer region accompanied by shrinkage of 24mer region. To our surprise, this effect continues upon returning to the high salt regime (> 100 mM). For the patch in Fig. 4A, the 24mer area fraction increased from 0.52 in 30 mM NaCl to 0.57 in 100 mM, and 0.66 in 500 mM. Another patch shown in Fig. 4B showed same trend: 0.32 in 50 mM, 0.36 in 100 mM, 0.43 in 250 mM, and 0.48 in 500 mM. This area change is reversible, that is, the area can expand and shrink many times by lowering and raising the salt concentration, while the domain shape and position remains similar. When the 24mer DNA is visualized by Alexa 488 one sees that the brightness increases as the domain shrinks with decreasing salt (Fig. 4C). Although complicated by photobleaching, it appears that the overall amount of 24mer DNA is conserved before and after ionic strength exchange. In this case, the increased brightness implies that the 24mer DNA tethers occupy a smaller area and are more densely packed in low salt concentration, while the 48mer DNA tethers can spread out.
Fig. 4.
Pattern change with salt concentration. (A) A patch containing 1:1 ratio of 24mer (repeating sequence, bright region) and 48mer (overlapping sequence, dark region) was imaged by FLIC on 260 nm SiO2 visualizing TR-labelled lipid. The pattern change was monitored with varying NaCl concentration. The contrast of the 500 mM picture was enhanced to compensate for photobleaching. (B) A patch contains 1:1 ratio of 24mer (overlapping sequence, dark region) and 48mer (overlapping sequence, bright region) and was imaged by FLIC on 380 nm SiO2. (C) The pattern change with decreasing salt from 250 mM to 50 mM NaCl was monitored by Alexa 488 labelled poly24A. The patch contains 1:1 ratio of 24mer and 48mer and was imaged by normal epi-fluorescence microscopy. Because the 250 mM image was captured when the patch is still spreading, the edge of patch became expanded in the 50 mM image. Scale bar is 10 µm.
3.3. Formation and evolution of height segregated patterns
As seen in Fig. 2B and Fig. 4, the size of height segregated domains was in the range of a few microns with a variety of shapes - some were connected with each other and some were entirely surrounded by regions with the other height. Once formed, the pattern barely changed over tens of minutes. We found that the segregated patterns can be broadly classified as two types – randomly distributed or concentric, depending on the patch forming process. Some GUVs rupture immediately after binding to the surface, but some stay as a flattened GUV for a while before they rupture and form patches1, 21. The former patches usually show randomly distributed domains, while the patches that stayed longer as a flattened GUV tend to show roughly concentric patterns (Fig. 4C).
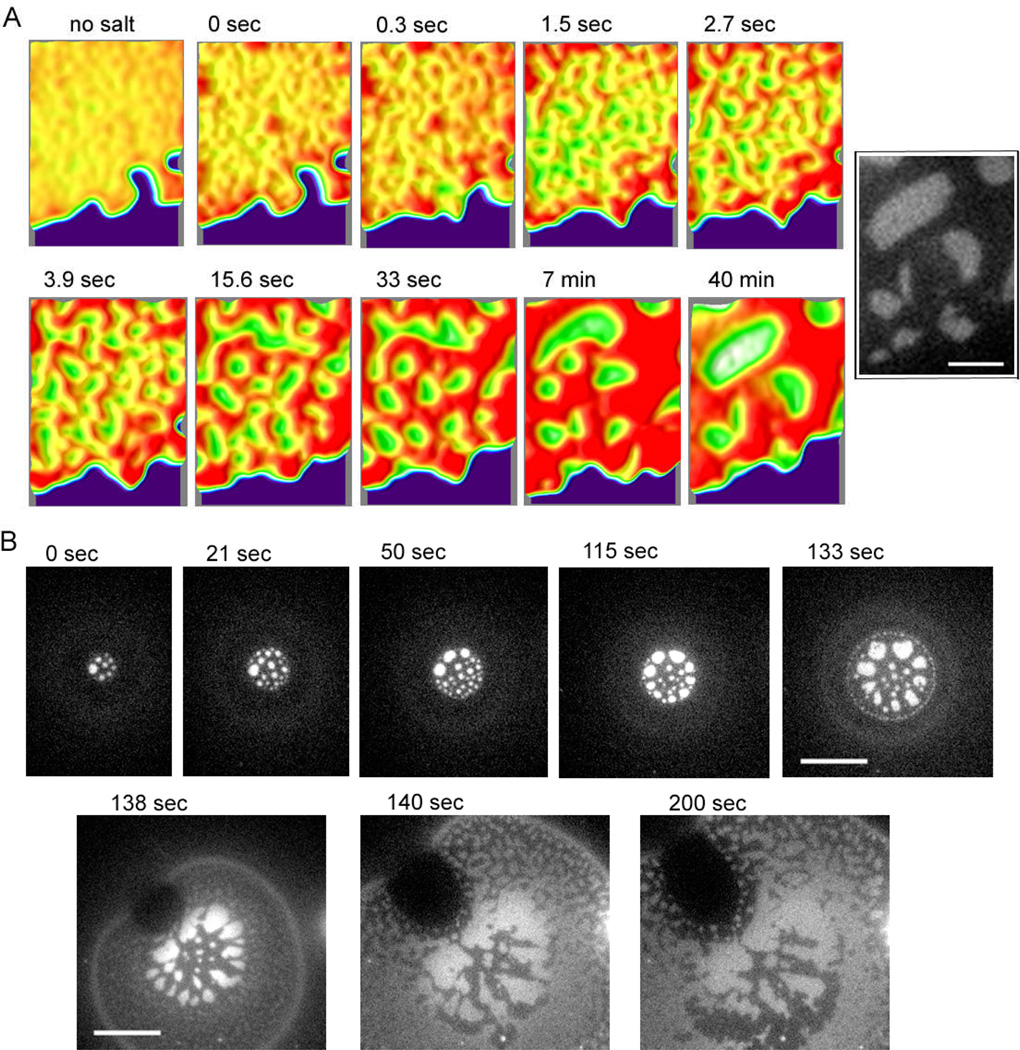
The evolution of the segregated patterns was monitored directly by using fluorescently labelled DNA as a surrogate for height, but with much better sensitivity than the contrast provided by FLIC. This was used to characterize the mechanism of pattern formation in both the tethered patches (Fig. 5A) and the flattened GUVs (Fig. 5B; pattern formation under flattened GUVs cannot be clearly observed by FLIC because the fluorescence from the upper membrane of the GUV overlaps with that from the bottom membrane). The flattened GUV and patch were formed by GUVs presenting an equal mol% of Alexa 488 labelled poly24A and 48-1 binding to unlabelled antisense partners on the target patch. The patches that form immediately are very hard to locate making it difficult to capture the initial state of the pattern forming process which occurs within tens of seconds. As an alternative we use the following approach to reset the pattern-forming process and gain some insight into the early stage. After a patch is formed (with an already segregated pattern), the salt of the solution is removed by gently adding distilled water to dissociate DNA hybrid tethers. If carefully handled to minimize flow stress, the patch remains in place due to weak electrostatic and van der Waals interactions11. At that point, the height segregation was immediately relaxed, but the DNAs were not fully mixed yet, showing fuzzy brighter regions where the bright 24mer domains were at higher salt. After the patch became uniform by diffusion (~ 10min), the higher salt buffer was added and DNA hybridization was initiated within a few seconds. The 24mer and 48mer immediately separate into small bright and dark domains (Fig. 5A, enhanced using color). The small domains were rapidly merged and coarsened within a few minutes, but after the 24mer and 48mer were separated and spaced, the pattern change became very slow, though the merging and rounding of domains continued.
Fig. 5.
Topological pattern evolution process during GUV adhesion and patch formation monitored with Alexa 488 labelled 24mer DNA by epi-fluorescence microscopy. GUVs displaying Alexa 488 labelled 24mer and unlabelled 48mer were bound on a supported bilayer displaying the complementary DNA where they flatten and rupture to form patches (see Fig. 1). (A) A tethered patch was formed, then the DNA tethers were un-hybridized by removing salt, and DNA hybridization was re-initiated by adding salt so that initial state of segregation in the tethered patches could be observed under controlled conditions. The DNAs become uniformly distributed after several minutes incubation in deionized water (no salt), then the salt was added, and after a few seconds, the patch became slightly rough, which is set as 0 sec. The segregation by tether length occurred rapidly, forming small 24mer (bright) and 48mer domains (~ 3 sec). The randomly distributed small domains gradually merged and became rounder in shape (~ 7 min); this segregation process continued, but became very slow after 10 min. The surface topology of the segregated patch is reconstituted for enhanced contrast by using ImageJ (v2.41) interactive 3D surface plot plug-in with thermal colour scale (red is high and blue is low). Inverted brightness was used for the patch area to depict realistic domain height – making the higher 48mer region brighter. The black and white inset is the original image at 40 min for reference. (B) Initial domain formation process as a GUV flattens and the binding area expands. Initially, circular bright 24mer domains were generated, arranged roughly concentrically (0~133sec). After the GUV ruptured and formed a patch (see Fig. 1), the circular 24mer domains became amorphous, and small segregated topological domains develop in new binding regions (138 sec). The new domains then started to merge with the already formed domains inside (~200 sec). Scale bar is 3 µm for (A) and 10 µm for (B).
The GUV binding and simultaneous DNA segregation was captured as soon as GUVs are added (Fig. 5B)†. Circular domains of 24mer were formed immediately and merged together to form a concentric pattern, which was completed in 2 minutes. The segregation seemed to occur as the binding area expands and the newly formed domains merged and arranged concentrically at the same time. Interestingly, the circular domain shape, reminiscent of lipid phase domains, and the concentric pattern was lost as soon as the GUV ruptured and became a patch. This suggests that membrane tension plays some role.
3.4. Tethered patches with three different tether lengths
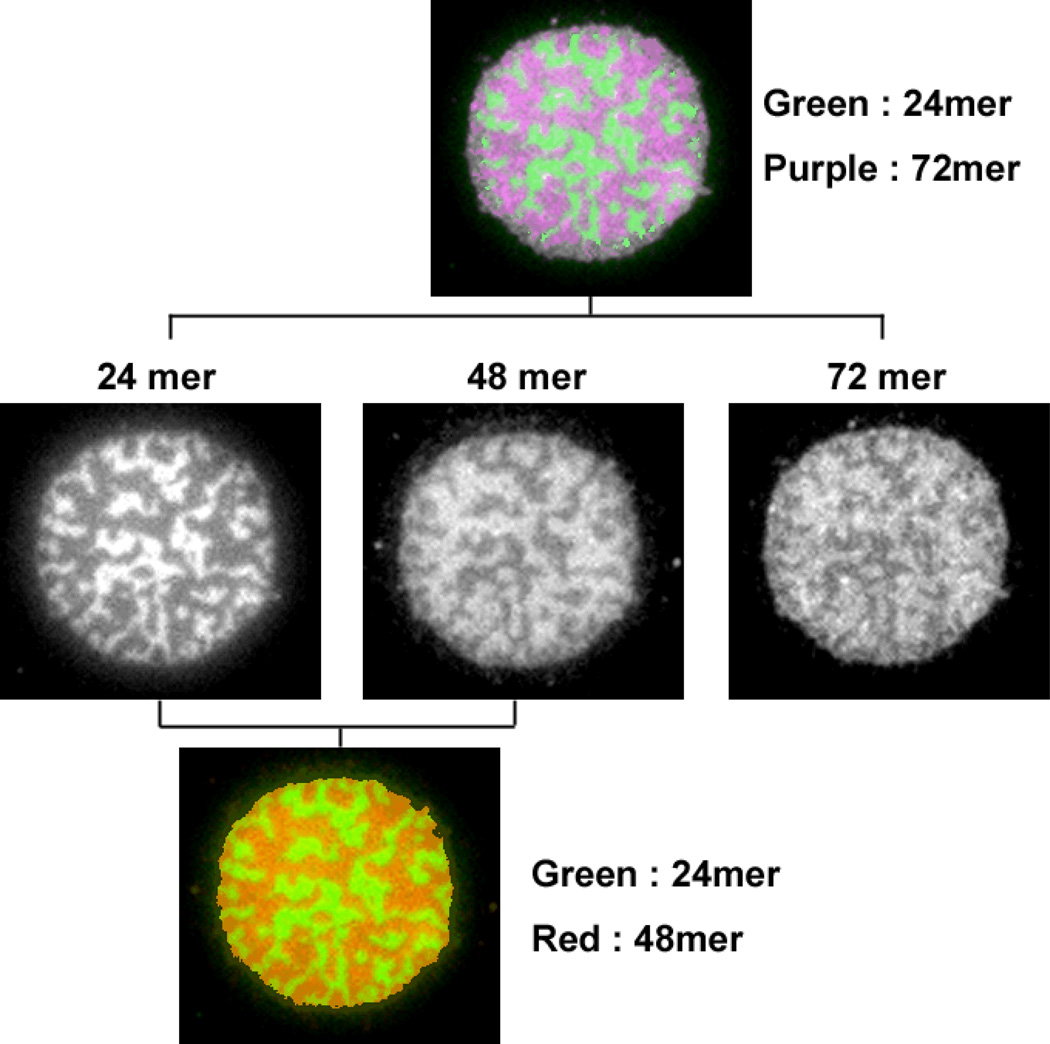
Figure 6 shows what happens when three different lengths of DNAs are present in the artificial membrane junction. In order to visualize each component, the 24mer was labeled with Alexa 488, the 48mer with Cy5 and the 72mer with Cy3.5 in 4:3:3 molar ratio, respectively. Surprisingly, only the 24mer (green) was segregated, while the 48mer (red) and 72mer (purple) were observed in almost identical regions. In a separate experiment, patches with 48mer and 72mer were observed not to segregate, but showed a uniform appearance (data not shown).
Fig. 6.
A patch with three different lengths of DNA that are labelled by fluorescence dye. Each DNA occupied regions imaged by corresponding fluorescence is shown for side-by-side comparison. Alexa 488 labelled 24mer is shown with green colour, and Cy5 labelled 48mer is red, and Cy3.5 labelled 72mer is purple in the overlay image.
4 Discussion
The images shown demonstrate topological pattern formation in the DNA-tethered lipid membrane with different lengths of mobile tethers and the effects of varying length, sequence and ratio of DNA tethers and ionic strength. Interestingly, the domain shape and segregation process resembles that of phase separated lipid domains of GUVs22 or planar lipid bilayers23, 24, for example, the initiation of phase separation induced by lowering temperature and domain ripening processes22 looks very similar to what is observed in Fig. 5. Nevertheless, the underlying mechanism is largely different. Lipid phase separation is compositional segregation determined by interactions between adjacent lipid molecules, while the segregation of DNA tethers is driven by the interplay of topographic size differences of the DNA tethers and membrane mechanics. Though the former has been extensively studied with various materials and conditions, the latter has received much less attention.
The mechanism of the segregated topological domain formation is likely to be governed by the same principles used in the theoretical model for the immunological synapse formation12. Accommodation of different length DNA tethers in the same region requires abrupt bending of the lipid membrane which is energetically unfavourable. This free energy cost drives rearrangement and segregation of DNA linkers with different heights. To minimize the energetic penalty of membrane bending, the lowest energy state may be achieved when the two lengths of linkers are completely separated and form two large domains at equilibrium. Slow coarsening is likely due to the fact that DNA hybrids, once formed, do not dissociate25, 26. This contrasts with the ligand-receptor binding in the immunological synapse where interactions are reversible. DNA hybrids could enter the surrounding region where the height is different height region, but this would cause unstable local membrane curvature for coarsening or movement of domains, thus the merging of small domains becomes very slow after they are spaced enough, as after 8 min in Fig. 5A.
Just as the composition of saturated or unsaturated lipids and temperature can be adjusted to investigate lipid phase separation behavior, the effects of length, sequence and ratio of DNA tethers and ionic strength were investigated. The domain area fraction of each length of DNA was roughly proportional to its mol fraction, but the area occupied by the longer DNA, whether measured by FLIC on the membrane dye or by using dye-labeled DNA, was always larger than its mol fraction (Fig. 3). The repeating sequences (polyA and polyT) could promote faster DNA binding, but the results indicate that these behave similarly to the fully overlapping sequence at equilibrium. The area of domains was also influenced by ionic strength. The 24mer domains reversibly expand with increasing salt concentration and shrink by returning to low salt concentration, while the 48mer area changes in opposite ways. All of the area measurements of Figure 3 were done at 100 mM NaCl concentration. Considering the 24mer area increase at high salt (> 100 mM) as shown in Fig. 4, the area occupied by the longer DNA would be smaller if it were measured at 250 mM NaCl or higher concentration.
The effect of changing the ionic strength is difficult to understand particularly in high salt regime since the average distance between DNA tethers (0.5 mol%) is about 10 nm which is longer than ~ 2 nm Debye length of the salt concentration used in this study27, in which case the ionic strength change should not have a noticeable effect. One plausible hypothesis is that the repulsion between 48mer hybrids would be greater than for 24mer hybrids due to the greater negative charge, thus more space is required than for short DNA. When the ionic strength increased, the 48mer undergoes less repulsion and allows relatively more space to the 24mer domains. We have not seen any evidence that the 24mers dissociated in low salt when the 24mer region shrinks. If this is the case, dissipation of fluorescence upon domain shrinkage or gathering upon expansion could be observed during the reversible salt change.
Another interesting, but puzzling observation was that the 48mer and 72mer DNA tethers were not segregated when in the same patch, while 24mer and 48mer, or 24mer and 72mer DNAs were segregated. This was clearly demonstrated for the patches with three different tether lengths examined that only the 24mer was separated from a region which has both the 48mer and 72mer, rather than three segregated domains with different height. Since the height difference between 48mer and 72mer is same as between the 24mer and 48mer, the membrane bending should be similar. Moreover, to occupy the same region adjacent to a different tether length, the free energy increase due to local curvature needs to be compensated in some way. One hypothesis to explain this observation would be that the 72mer DNA hybrid is more flexible and does not create enough local curvature to drive segregation. Double-stranded DNA is generally regarded as stiff with the persistence length being considerably longer28. However, unexpected length fluctuations in short ds DNA constructs have recently been reported29. Another possibility is that the 72mer DNA hybrid tether might tilt bringing the height into a range tolerable for the 48mer. In principle, the linker between the DNA and lipid, although short (Fig. 1 inset), couple provide sufficient flexibility,30 especially under tension due to membrane curvature. In this scenario, the 24mer DNAs are completely segregated because 24mer gap is too small for even tilted 48mer, while 48mer region can accommodate deformed 72mer.
We conclude with a comparison of our system to the immunological synapse which is formed by the sorting and clustering mechanism that leads to large-scale segregation of different types of receptors and ligands to different regions of the intercellular junction8. Based on theoretical/computational studies, with reasonable estimates for the length, binding constants, diffusion and membrane bending properties12, the segregation patterns can be explained only by considering the characteristics of the minima in the free energy functional, though other forces, such as coupling to the cytoskeleton may be important in vivo. Although the segregation process is basically governed by random nucleation and gathering of linkers, the physical properties of the membrane also affect the domain pattern. As seen in Figure 5, membrane tension appears to play some role for the concentric arrangement and circular shape of domains (Fig. 5). The domain size is expected to be larger and coarsening faster under conditions that promote migration of the linkers across domains. For example, a finite dissociation rate of the DNA hybrid (for example by introducing mismatches), high lipid mobility, low bending modulus of membrane and a smaller height difference between domains could all promote such reorganization. Systematic variations of this sort, using the model membrane system we have developed, should provide a starting point for thinking about the mechanisms underlying synapse assembly.
Acknowledgement
We thank Bettina van Lengerich for providing DNA-lipid conjugates, and Arup K. Chakraborty and Steve Abel for helpful discussions. This work was supported in part by grants from the NSF Biophysics Program, NIH GM069630, and by the MRSEC Program of the NSF under award DMR-0213618 (CPIMA).
Footnotes
Although the amount of DNA in GUVs (0.5 mol%) is more than on SLBs (0.3 mol%), the laterally mobile DNAs on SLBs gather and accumulate on patch area.
References
- 1.Chung M, Lowe RD, Chan YH, Ganesan PV, Boxer SG. J Struct Biol. 2009;168:190–199. doi: 10.1016/j.jsb.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chung M, Boxer SG. Langmuir. 2011;27:5492–5497. doi: 10.1021/la200234h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gumbiner BM. Cell. 1996;84:345–357. doi: 10.1016/s0092-8674(00)81279-9. [DOI] [PubMed] [Google Scholar]
- 4.Dustin ML, Bivona TG, Philips MR. Nat Immunol. 2004;5:363–372. doi: 10.1038/ni1057. [DOI] [PubMed] [Google Scholar]
- 5.Weber T, Zemelman BV, McNew JA, Westermann B, Gmachl M, Parlati F, Sollner TH, Rothman JE. Cell. 1998;92:759–772. doi: 10.1016/s0092-8674(00)81404-x. [DOI] [PubMed] [Google Scholar]
- 6.Hartman NC, Groves JT. Curr Opin Cell Biol. 2011;23:370–376. doi: 10.1016/j.ceb.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Groves JT, Dustin ML. J Immunol Methods. 2003;278:19–32. doi: 10.1016/s0022-1759(03)00193-5. [DOI] [PubMed] [Google Scholar]
- 8.Grakoui A, Bromley SK, Sumen C, Davis MM, Shaw AS, Allen PM, Dustin ML. Science. 1999;285:221–227. [PubMed] [Google Scholar]
- 9.Hartman NC, Nye JA, Groves JT. Proc Natl Acad Sci U S A. 2009;106:12729–12734. doi: 10.1073/pnas.0902621106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Manz BN, Jackson BL, Petit RS, Dustin ML, Groves J. Proc Natl Acad Sci U S A. 2011;108:9089–9094. doi: 10.1073/pnas.1018771108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parthasarathy R, Groves JT. Proc Natl Acad Sci U S A. 2004;101:12798–12803. doi: 10.1073/pnas.0402168101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qi SY, Groves JT, Chakraborty AK. Proc Natl Acad Sci U S A. 2001;98:6548–6553. doi: 10.1073/pnas.111536798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ajo-Franklin CM, Yoshina-Ishii C, Boxer SG. Langmuir. 2005;21:4976–4983. doi: 10.1021/la0468388. [DOI] [PubMed] [Google Scholar]
- 14.Ajo-Franklin CM, Ganesan PV, Boxer SG. Biophys J. 2005;89:2759–2769. doi: 10.1529/biophysj.105.066738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoshina-Ishii C, Boxer SG. J Am Chem Soc. 2003;125:3696–3697. doi: 10.1021/ja029783+. [DOI] [PubMed] [Google Scholar]
- 16.Yoshina-Ishii C, Miller GP, Kraft ML, Kool ET, Boxer SG. J Am Chem Soc. 2005;127:1356–1357. doi: 10.1021/ja043299k. [DOI] [PubMed] [Google Scholar]
- 17.Chan YH, van Lengerich B, Boxer SG. Biointerphases. 2008;3:FA17. doi: 10.1116/1.2889062. [DOI] [PubMed] [Google Scholar]
- 18.Longo ML, Ly HV. Methods Mol Biol. 2007;400:421–437. doi: 10.1007/978-1-59745-519-0_28. [DOI] [PubMed] [Google Scholar]
- 19.Veatch SL, Keller SL. Biochim Biophys Acta. 2005;1746:172–185. doi: 10.1016/j.bbamcr.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 20.Braunlin WH, Bloomfield VA. Biochemistry. 1991;30:754–758. doi: 10.1021/bi00217a026. [DOI] [PubMed] [Google Scholar]
- 21.Rawle RJ, van Lengerich B, Chung M, Bendix PM, Boxer SG. Biophys J. 2011;101:L37–L39. doi: 10.1016/j.bpj.2011.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Veatch SL, Keller SL. Biophys J. 2003;85:3074–3083. doi: 10.1016/S0006-3495(03)74726-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garg S, Ruhe J, Ludtke K, Jordan R, Naumann CA. Biophys J. 2007;92:1263–1270. doi: 10.1529/biophysj.106.091082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ratto TV, Longo ML. Biophys J. 2002;83:3380–3392. doi: 10.1016/S0006-3495(02)75338-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Starikov EB, Norden B. J Phys Chem B. 2009;113:11375–11377. doi: 10.1021/jp903924j. [DOI] [PubMed] [Google Scholar]
- 26.Huguet JM, Bizarro CV, Forns N, Smith SB, Bustamante C, Ritort F. Proc Natl Acad Sci U S A. 2010;107:15431–15436. doi: 10.1073/pnas.1001454107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Israelachvili JN. Intermolecular, Surface Forces. New York: Academic Press; 1985. [Google Scholar]
- 28.Becker NB, Everaers R. Phys Rev E Stat Nonlin Soft Matter Phys. 2007;76:021923. doi: 10.1103/PhysRevE.76.021923. [DOI] [PubMed] [Google Scholar]
- 29.Mathew-Fenn RS, Das R, Harbury PA. Science. 2008;322:446–449. doi: 10.1126/science.1158881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Becker NB, Everaers R. Science. 2009;325:538. doi: 10.1126/science.1168786. author reply 538. [DOI] [PubMed] [Google Scholar]