Abstract
The spontaneous generation of distinct phenotypes within a clonal population of cells allows for both bet-hedging at the population level and the division of labor among subpopulations. This is emerging as an important theme in bacterial pathogenesis, because bacterial pathogens exhibit phenotypic heterogeneity with respect to characteristics that impact virulence. The phenomenon of persister cells and models of Salmonella enterica serovar Typhimurium pathogenesis illustrate the importance of non-genetic diversity in the disease process. Such heterogeneity can arise from specific genetic architectures amplifying stochastic fluctuations in factors affecting gene expression, which drives variation in eukaryotic cells as well. Thus reproducible variation in both host and pathogen processes affects the outcome of infection.
Keywords: Regulation, Salmonella, infection, heterogeneity, bistability, bet-hedging
Diversification and disease
Recent advances in technologies that permit high-resolution analysis of events in single cells have revealed remarkable non-genetic diversity in populations of cells that were previously considered to be quite homogenous (Figure 1). Much of the observed diversity is currently attributed to stochastic variation, or noise, in the biochemical reactions of gene expression. Experiments using engineered Escherichia coli strains demonstrate that gene expression can vary substantially both between identical promoters within a single cell and among isogenic cells within a population [1], supporting the hypothesis that noise can give rise to substantial diversity.
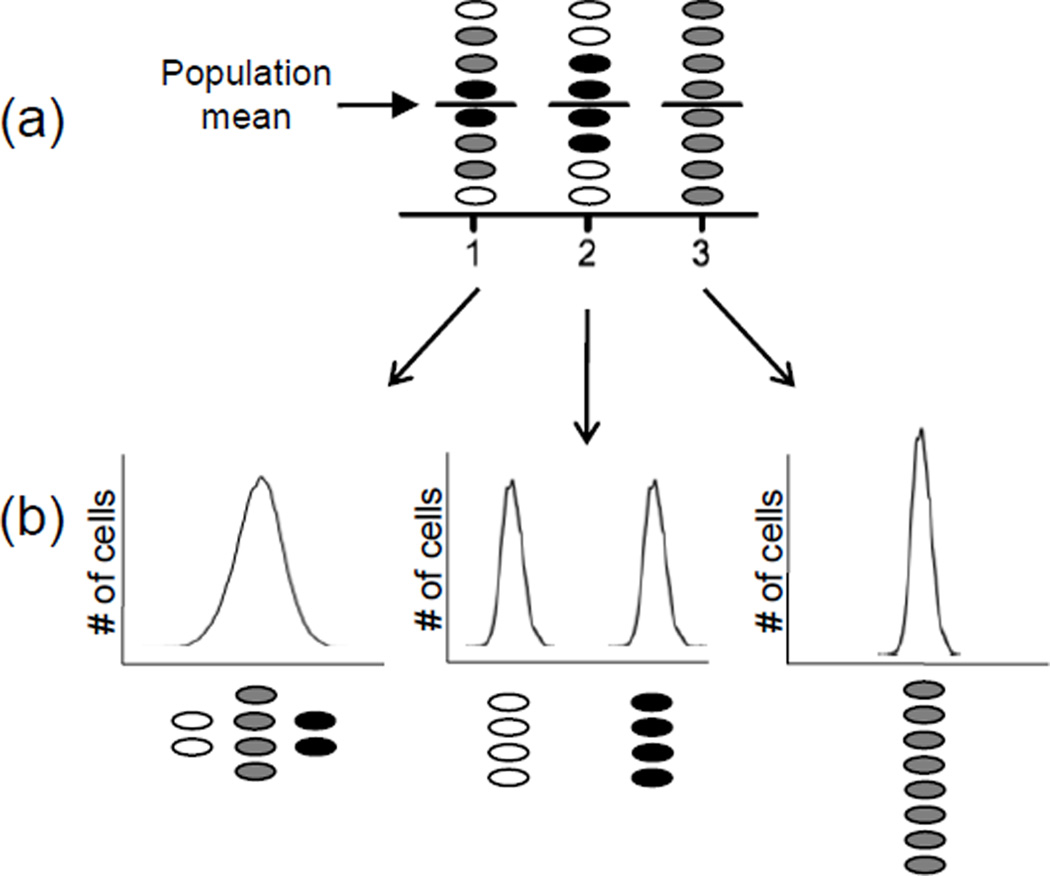
Figure 1. Surprises lurk within averaged data sets.
Colors represent cells with low (white), medium (gray) and high (black) levels of the measured parameter. (a) Bulk culture experimental techniques such as Western blots and β-galactosidase assays measure the average of a parameter across an entire population, but equal means may characterize very different populations as indicated by the different shading. (b) Single-cell analysis techniques such as flow cytometry or microscopy generate a more complete picture of the diversity that may be concealed by averaging.
The revelation that a given set of environmental conditions frequently gives rise to a range of cellular states is changing the way we view and study biology. Variation is more frequently recognized as an important biological phenomenon. In some cases, variation in the copy number of certain molecules results in strikingly different cell fates. This is best documented in prokaryotes, where noise plus specific genetic circuits producing net positive feedback can give rise to multiple stable and distinct phenotypes within a clonal population (Figure 2). When two stable phenotypes are present, the situation is referred to as bistability and the genetic architecture producing such a distribution of phenotypes is called a bistable switch [2, 3]. Many prokaryotic bistable switches have been described, and the mechanisms are diverse [4–9].
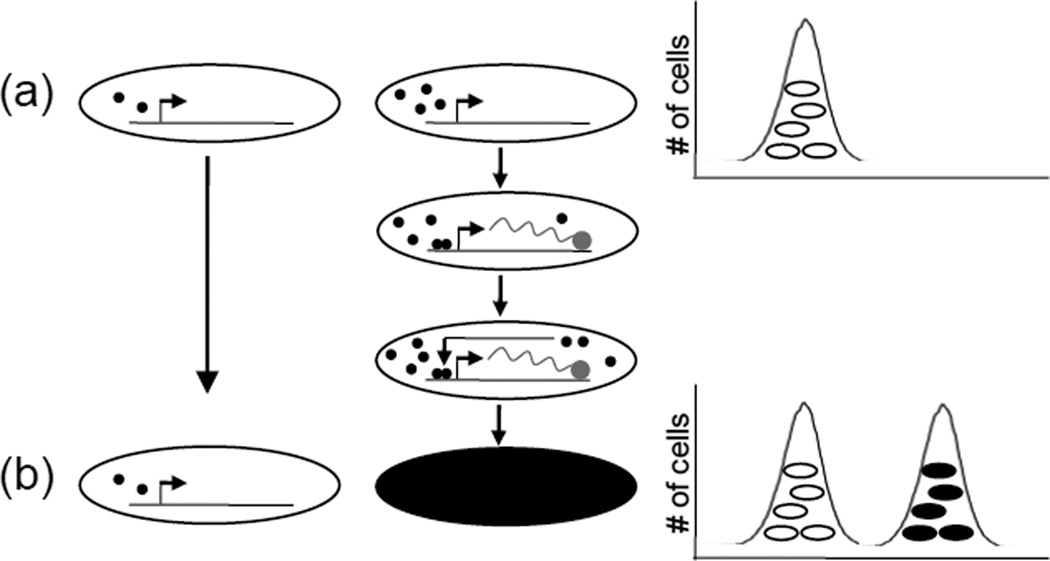
Figure 2. Noise and bistable gene expression circuits.
The biochemical reactions of gene expression are subject to a certain amount of randomness or stochasticity [47]. (a) A modest degree of population diversity in the concentration of factors that activate or repress gene expression (represented by black closed circles), especially when such factors are in low abundance, together with alternating permissive and non-permissive promoter DNA conformations, can lead to transcriptional bursting in a subset of cells. Above a critical threshold of activation, positive feedback and non-linear kinetics (such as dimerization of a regulator, shown) can accelerate gene expression, bifurcating a unimodal distribution into a bistable gene expression pattern (b) where intermediate levels of expression are rare events. This combination of noise, net positive feedback and non-linear kinetics has been documented in many bistable switches found in nature [2]; however, the mechanistic details of bistable switches are variable and may be exceedingly complex. Multiple positive feedback loops or even numbers of negative feedback loops (thus producing net positive feedback) may participate in the circuit, which can include many layers of regulation from transcriptional to post-translational.
To be successful, pathogenic microbes must either evade or exploit the immune system of their host. Disease progression can involve sequential colonization of different host compartments, and the immune response is a kinetic process that radically alters conditions within infected tissues, as does antimicrobial therapy. Additionally, many pathogens must live outside the host for varying lengths of time. Thus, pathogenic microbes must transition between diverse environments and endure a variety of stresses. Deterministic adaptation, whereby microbes sense environmental cues and respond accordingly, is a common solution to this problem. However, a pre-existing mixture of distinct physiologies that confer situation-specific selective advantages could be more adaptive than sensing and responding under some circumstances, and phenotypic heterogeneity provides another solution to the problem of survival in fluctuating environments. The ability to reproducibly generate multiple phenotypes from the same genotype allows for both bet-hedging and the division of labor within populations, while preserving the genotype of the clone. In bet-hedging scenarios, microbes differentiate into multiple phenotypes that improve the fitness of the clone across environments. Labor division allows multiple roles to be simultaneously fulfilled, each by a different subgroup of cells.
The study of phenotypic variation in the host is challenging. Subtle gradients in nutrient availability, moisture, or toxins, rather than stochastic variation of intracellular biochemical reactions, could trigger differential gene expression patterns in bacteria that are quite close together within tissues. What is clear is that medically important microbial characteristics such as the expression of virulence factors vary stochastically in vitro [e.g. for Pseudomonas aeruginosa [10], Vibrio cholerae [7], Salmonella enterica serovar Typhimurium (S. Typhimurium) [11]], suggesting the possibility that this occurs in vivo as well. Here we discuss the role of stochastically formed bacterial subpopulations in three disease scenarios. (i) Many bacterial species generate small subpopulations of transiently antibiotic-tolerant cells known as persisters, which could contribute to the problem of persistent bacterial infection. (ii) The presence of both cytotoxic and non-cytotoxic subpopulations of Salmonella in the gut tissue may contribute to systemic bacterial dissemination. (iii) Differential expression of Salmonella genes conferring motility and invasion generates subpopulations that play distinct roles during colitis. Mammalian cells in culture also display stochastic variation in characteristics that impact disease, such as activation of cell death pathways. Thus, evidence that phenotypic heterogeneity drives processes that impact disease is accumulating.
Bet-hedging: persister cells
Antibiotic treatment of sensitive bacterial cultures often uncovers a small subpopulation of cells with greater tolerance to antibiotics; these have been termed persisters [12]. Persisters are poorly understood and potentially very important, as subsequent outgrowth of this subpopulation may explain the failure to completely sterilize bacterial infections in some patients [13].
The progeny of persisters are as susceptible to antibiotics as the parent strain, indicating that persistence is not genetically determined. However, the propensity to form persisters is genetically regulated, since mutations that increase the size of the persister subpopulation have been reported in many bacterial species [14]. A clear, unified mechanism of persistence has not emerged from the data, indicating that many distinct mechanisms may exist by which antibiotic-sensitive cells can gain short-lived, reversible tolerance, often to multiple antibiotics [14].
Persisters are often functionally characterized as dormant or slow-growing subpopulations [15]. Antimicrobial strategies designed specifically to eliminate persisters focus upon minimizing heterogeneity by ‘waking up’ the dormant subpopulation to some extent, thus rendering the entire population uniformly susceptible to antimicrobial therapy. Re-establishing the proton motive force (PMF) in populations of E. coli and Staphylococcous aureus persistent to aminoglycosides resensitized the cells to the antibiotics [16]. PMF, which is required for aminoglycoside uptake, was restored by the addition of specific metabolites. Interestingly, addition of the metabolites did not restore normal growth; rather, the persisters entered an energized but non-dividing state. Kim et al. [17] used high-throughput chemical screening to identify a compound (C10) that did not alter the growth of E. coli when used alone, but potentiates killing of persisters by both ampicillin and norfloxacin. The lag time for persisters to resume growth after antibiotic treatment was considerably shortened in the presence of C10, suggesting that this compound sensitizes persisters to antibiotics by stimulating a return to normal growth physiology.
Thus, persisters are an example of a bet-hedging strategy hinging upon spontaneously generated phenotypic subpopulations. The presence of a few antibiotic-tolerant, metabolically-inactive cells spreads risk, positioning the population as a whole to both replicate vigorously in the short-term and survive threatening conditions should they arise.
Division of labor: Salmonella infection paradigms
Heterogeneous gene expression by S. Typhimurium also impacts pathogenesis. This organism lives freely within the environment and infects warm- and cold- blooded animals, causing gastroenteritis, systemic infection, or asymptomatic carriage, depending upon the host [18]. Salmonella are flagellated organisms, and can express the gene encoding the flagellar filament protein (fliC) in a heterogenous manner; this is controlled by ydiV, which prevents the transcription of the sigma factor required for fliC expression [9] (Figure 3). YdiV completely suppresses flagellin in some environments [19], deletion of ydiV results in a unimodal fliC-ON population, and intermediate levels of expression result in a heterogeneous mix of fliC-ON and fliC-OFF cells [9]. Thus, ydiV controls a mechanism by which the distribution of fliC expression in a population can be tuned.
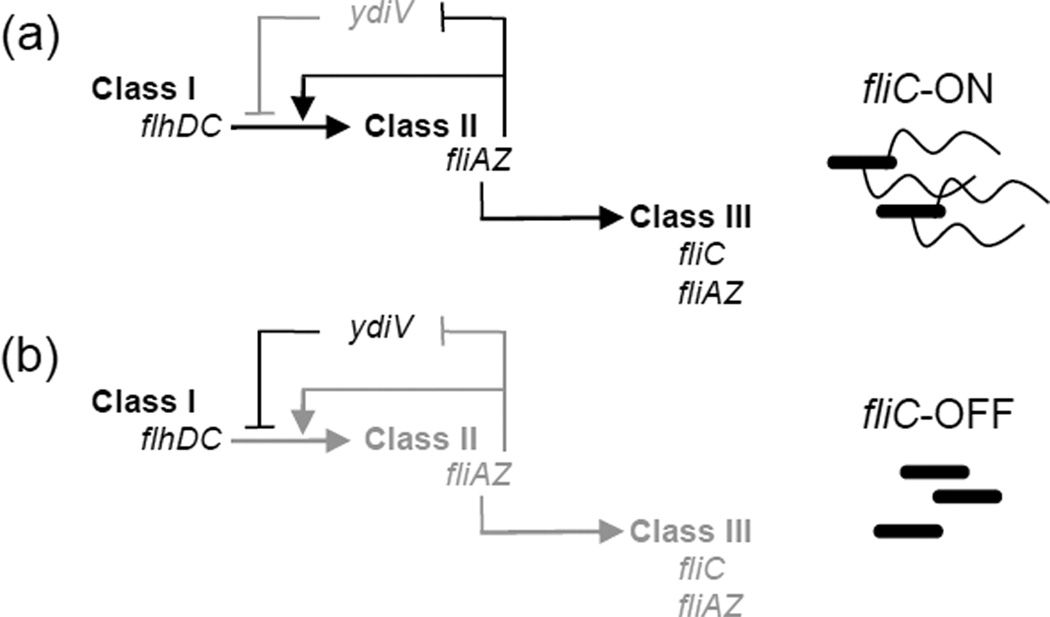
Figure 3. Bistable flagellar gene circuit in Salmonella.
In Salmonella, flagella are constructed via the sequential expression of three classes of genes [20]; ydiV blocks class II expression in a subset of cells, causing bistability [9]. Black arrows and text represent active regulatory relationships in the fliC-OFF or fliC-ON subpopulations, gray arrows and text represent inactive regulatory relationships in the subpopulations. (a) The proteins encoded by flhDC, the sole class I operon, form a complex that is required to activate class II gene expression. Class II genes encode structural proteins of the platform that anchors the flagellum within the bacterial cell envelope and of the flexible linker that connects the platform with the filament, as well as the important regulatory proteins FliZ and FliA. FliZ enhances class II gene expression [48], while FliA is the sigma factor required for expression of class III genes, including the flagellin filament protein FliC. Thus, the expression of the fliAZ operon constitutes a commitment step in the construction of flagella. fliZ-mediated class II transcriptional enhancement occurs via two mechanisms: increasing the level of the FlhC protein [49], and repressing ydiV [50]. Thus ydiV and fliZ form a system of mutually repressing repressors, a common motif in bistable gene expression circuits. (b) In fliC-OFF subpopulations, the balance is tipped toward ydiV expression and fliZ repression (whereas in the fliC-ON subpopulation, fliZ is expressed).
How could this be advantageous? The vast majority of Salmonella clinical isolates are flagellated organisms; this indicates that motility is important. However, constructing flagella requires considerable cellular resources: there are more than 60 structural and regulatory proteins involved, and up to 20,000 units of FliC in a single flagellum [20]. Thus flagellar gene expression could be a metabolic burden under some circumstances. Within the host, fliC expression poses other liabilities. FliC is the cognate ligand for multiple immune receptors (Nod-like [21, 22], Toll-like [23], B cell [24], and T cell [25] receptors). Binding of FliC to these receptors initiates inflammation and priming of adaptive immune responses. The impact of fliC expression upon fitness is therefore contextually determined; consistent with this, flagellar gene expression is tightly coupled to the environmental and metabolic state of the cell via the activity of many global regulators [26]. However, Salmonella has also evolved a mechanism that provides for heterogenous expression of flagellar genes. This suggests that responding to the environment is not always sufficient.
For a versatile bacterium such as S. Typhimurium, phenotypic heterogeneity could serve different functions in different environments. As a free-living organism, heterogeneous expression of fliC is a bet-hedging strategy. Motility is important, yet YdiV downregulates fliC in all members of a population under austere conditions [19], indicating that there are times when flagellar gene expression is less optimal. A mixed population with both flagellated and non- flagellated members can therefore maximize fitness in non-host environments. During infection, phenotypic heterogeneity of Salmonella populations allows for a division of labor that enhances virulence, as discussed below.
Roles of distinct populations in the host: systemic Salmonella infection
During infection, Salmonella disseminate from the gut to systemic tissues within host phagocytes; survival within macrophages is required for virulence [27, 28]. This poses an interesting potential conflict, because Salmonella also swiftly kills macrophages via the caspase-1-dependent pro-inflammatory cell death program pyroptosis [29]. Salmonella-triggered pyroptosis of macrophages requires the Salmonella pathogenicity island-1 (SPI-1) type III secretion system (T3SS), a molecular syringe that translocates molecules, including flagellin [30], from the bacteria to the host cell cytosol. Recognition of flagellin by host cytosolic receptors initiates caspase-1 activation and subsequent cell death [21].
In addition to this functional relationship between the flagellar genes and SPI-1, there is also a regulatory relationship: the flagellar class II gene fliZ activates SPI-1 gene expression [31] (Figure 4). Thus, these two important regulons are linked in such a way that FliC+ SPI-1+, FliC+ SPI-1−, and FliC− SPI-1− subpopulations could be formed. FliC+ SPI-1+ bacteria are both motile and pro- inflammatory, FliC+ SPI-1− bacteria would be motile but unable to cause pyroptosis, and FliC− SPI-1− bacteria are non-motile and non-inflammatory; these subpopulations may play specialized roles in the disease process.
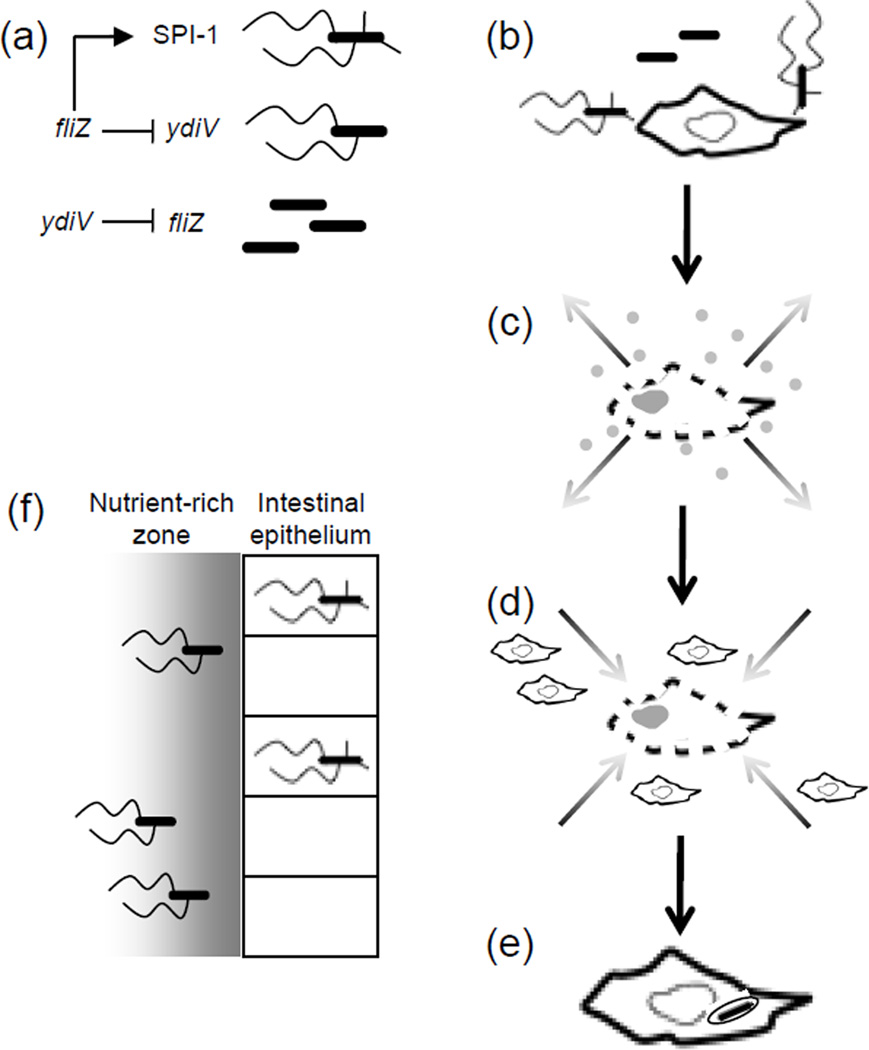
Figure 4. Divison of labor during Salmonella infection.
(a) The regulatory relationship between the flagellar genes and the Salmonella pathogenicity island-1 type III secretion system (SPI-1 T3SS) generates three subpopulations with distinct properties and roles during infection. In cells where ydiV represses flagellar class II transcription, neither the flagellin filament protein FliC nor SPI-1 is expressed. Cells expressing the flagellar class II gene fliZ could form two subsets: FliC+ SPI-1− and FliC+ SPI-1+. (b–e) Division of labor between specialized Salmonella subpopulations during systemic infection: (b) FliC+ SPI-1+ Salmonella encountering macrophages in the Peyer’s patches trigger pyroptosis. (c) The resulting cell lysis and release of pro-inflammatory cell contents into the surrounding tissue induces (d) migration of neighboring macrophages into the area. (e) Uptake of FliC− SPI-1− Salmonella by incoming macrophages then accelerates dissemination to systemic sites. (f) During Salmonella colitis, the FliC+ SPI-1+ cells invade the intestinal epithelium and cause inflammation, releasing nutrients that allow luminal Salmonella to outcompete the microbiota. While invading bacteria are most often killed by host defenses, the non-invasive subsets are protected from this fate; additionally, the FliC+ SPI-1− subset is able to chemotax to the nutrient-rich zone near the epithelium, providing these cells with an advantage over the non-motile and non- invasive FliC− SPI-1− cells.
Orally inoculated Salmonella breaches the gut mucosa, colonizes the Peyer’s patches (lymphoid tissue of the small intestine) [32], and then fashions a protected niche within phagocytes that transport the pathogen to systemic sites such as the spleen [33, 34]. Consistent with the organism’s ability to sense and adapt to its environment, Salmonella regulates the production of fliC in a host compartment-specific manner [35]. fliC is heterogeneously transcribed within the Peyer’s patches, but repressed in systemic tissues [35]. ydiV, which controls bistability of fliC expression in vitro, is required for full virulence of Salmonella [9, 36]. ΔydiV Salmonella are unable to completely repress fliC expression in the spleen; in fact, deletion of ydiV generates heterogeneity in splenic bacterial populations. ΔydiV bacteria are outcompeted at systemic sites by a ΔydiV ΔfliC strain, indicating that fliC expression by a subpopulation of cells reduces Salmonella fitness after dissemination [9]. Salmonella specifically engineered to activate fliC expression at systemic sites are attenuated [37], indicating that the pressure to repress this pro-inflammatory molecule after escape from the gut is strong.
The role of heterogeneity in fliC and SPI-1 expression by Salmonella populations colonizing the Peyer’s patches has not yet been elucidated. FliC+ strains reach the Peyer’s patches faster than non-flagellated strains after oral infection [9], which gives flagellated subpopulations a head start for systemic colonization. However, reaching systemic sites also depends upon the ability to live quiescently inside macrophages [27, 28]. FliC+ SPI-1+ subpopulations trigger pyroptosis upon internalization by macrophages, lysing host cells and activating host defenses; in contrast, SPI-1− cells transition smoothly to intracellular life. Heterogeneous fliC expression in Peyer’s patches therefore provides a division of labor: FliC+ SPI-1+ Salmonella reach the Peyer’s patches more quickly, stimulating inflammation that recruits phagocytes to the area. Those phagocytes then take up members of the SPI-1− subpopulation and transport them to other host compartments. While non-flagellated Salmonella can cause systemic infection on their own, this division of labor between inflammatory and non- inflammatory subpopulations could accelerate systemic spread while confining FliC-specific host responses to the gut.
Roles of distinct populations in the host: Salmonella colitis
A role for phenotypic heterogeneity has also been proposed in the murine model of Salmonella colitis. Inflammation in this model is highly dependent upon expression of the SPI-1 T3SS, which allows Salmonella to invade epithelial cells [38].
Ackermann et al. characterized the role of SPI-1+ cells during murine colitis as self-destructive cooperation [39]. SPI-1-dependent invasion of the intestinal epithelium triggers inflammation, which releases compounds from which Salmonella can preferentially benefit, allowing the pathogen to outcompete the gastrointestinal microbiota [40–42]. For example, Salmonella’s ability to use tetrathionate as an electron receptor potentiates growth on ethanolamine. This confers a growth advantage against many microbes in environments where both tetrathionate and ethanolamine are present, such as the inflamed intestine [40, 41]. However, bacteria which have invaded the epithelium can no longer access the enriched luminal environment. Therefore, the SPI-1+ subpopulation activates a process that benefits only other subpopulations, which could be construed as a cooperative act. Twelve hours after oral infection, 100% of bacteria found in the gut tissue were indeed SPI-1+, in contrast with only 15% of Salmonella found in the gut lumen. Intriguingly, Salmonella colonized gut tissue of mice lacking reactive oxygen- and nitrogen-based antibacterial defense systems (Cybb/Nos2 knockout mice) more easily than that of wild-type mice. There was no difference in colonization of the gut lumen between the two mouse strains, indicating that the defense systems specifically impinged upon bacteria within the intestinal mucosa [39]. This suggests that invasion exposes SPI-1+ cooperative bacteria to antibacterial defenses not encountered by microbes that remain in the lumen, and is therefore a self-destructive act.
The respective roles of the SPI-1+ subpopulation can be further subdivided into FliC+ and FliC− factions. Since flagellated Salmonella strains reach the Peyer’s patches before non-flagellated strains, the co-regulation of flagellar and SPI-1 genes generates a highly invasive and pro-inflammatory subpopulation. Intriguingly, Stecher et al. have demonstrated that motile Salmonella grow more quickly than non-motile strains in the inflamed intestine, because they are able to migrate to the nutrient-rich zone close to the epithelium [43]. Formation of a FliC+ SPI-1+ subpopulation that reaches the epithelium quickly, invades, and triggers inflammation, together with FliC+ SPI-1− cells able to reach the epithelium and benefit from the ensuing inflammation while remaining extracellular appears highly adaptive. Phenotypic heterogeneity may therefore provide a mechanism by which distinct, randomly generated subpopulations of Salmonella are primed to (i) trigger optimal conditions for colonization and (ii) benefit from the remodeling of the intestinal environment while staying out of danger.
Heterogeneity in host processes
Host cell death plays an important role in pathogenesis of many bacterial species, and mammalian cells in culture vary in their response to a lethal stimulus [44, 45]. Spencer et al. [45] quantified the time between exposure to a chemical inducer of apoptosis and mitochondrial outer membrane permeabilization (MOMP) in HeLa cell populations, and found that the time to MOMP correlated well between sister cells but poorly across randomly chosen pairs of cells. The correlation between sister cells decayed as the time from cell division increased, and improved when cycloheximide was added to prevent protein synthesis. Together, these data indicate that differences in pre-existing protein content of the cells across the population likely account for the differences in time to death.
The authors also note that when protein synthesis was allowed to proceed, a fraction of cells always survived, likely due to the induction of pro-survival pathways. This set of experiments, performed using a single cell type in culture, has potent implications for the study of biology in vivo. The variation observed in simple cell cultures is likely magnified in the host. Diffusion of cytokines and microbial products is a more complex process in tissues, and subtle differences in position could therefore result in huge differences in phenotype for cells engaging with invading pathogens. Such signals could tip the balance to favor host or pathogen during infection. For example, activation (by exposure to lipopolysaccharide) of Yersinia-infected macrophages redirects them from an apoptotic to a pyroptotic cell death program, thus mediating the difference between a non-inflammatory (advantageous to the pathogen) and a pro- inflammatory (advantageous to the host) response to infection [46].
Concluding remarks
Differential gene expression across many loci simultaneously would generate enormous diversity within bacterial populations, resulting in a broad array of unique physiologies and capabilities. Experimental techniques that generate a single cell census of physiological attributes across whole populations will illuminate the role of diversity in the disease process. Closer examination of individual cell processes on the part of the host could unveil previously unappreciated aspects of resistance mediated by minority cell populations. Creative methodologies may reveal that strategic heterogeneity lies behind many of the mysteries of biology.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Elowitz MB, et al. Stochastic gene expression in a single cell. Science. 2002;297:1183–1186. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- 2.Veening JW, et al. Bistability, epigenetics, and bet-hedging in bacteria. Annu Rev Microbiol. 2008;62:193–210. doi: 10.1146/annurev.micro.62.081307.163002. [DOI] [PubMed] [Google Scholar]
- 3.Dubnau D, Losick R. Bistability in bacteria. Mol Microbiol. 2006;61:564–572. doi: 10.1111/j.1365-2958.2006.05249.x. [DOI] [PubMed] [Google Scholar]
- 4.Chai Y, et al. Bistability and biofilm formation in Bacillus subtilis . Mol Microbiol. 2008;67:254–263. doi: 10.1111/j.1365-2958.2007.06040.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kearns DB, Losick R. Cell population heterogeneity during growth of Bacillus subtilis . Genes Dev. 2005;19:3083–3094. doi: 10.1101/gad.1373905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Keren I, et al. Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli . J Bacteriol. 2004;186:8172–8180. doi: 10.1128/JB.186.24.8172-8180.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nielsen AT, et al. A bistable switch and anatomical site control Vibrio cholera virulence gene expression in the intestine. PLoS Pathog. 2010;6:e1001102. doi: 10.1371/journal.ppat.1001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ozbudak EM, et al. Multistability in the lactose utilization network of Escherichia coli . Nature. 2004;427:737–740. doi: 10.1038/nature02298. [DOI] [PubMed] [Google Scholar]
- 9.Stewart MK, et al. Regulation of phenotypic heterogeneity permits Salmonella evasion of the host caspase-1 inflammatory response. Proc Natl Acad Sci U S A. 2011;108:20742–20747. doi: 10.1073/pnas.1108963108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rietsch A, Mekalanos JJ. Metabolic regulation of type III secretion gene expression in Pseudomonas aeruginosa . Mol Microbiol. 2006;59:807–820. doi: 10.1111/j.1365-2958.2005.04990.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Temme K, et al. Induction and relaxation dynamics of the regulatory network controlling the type III secretion system encoded within Salmonella pathogenicity island 1. J Mol Biol. 2008;377:47–61. doi: 10.1016/j.jmb.2007.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bigger JW. The bactericidal action of penicillin on staphylocccus pyogenes. Irish Journal of Medical Science. 1944;19:585–595. [Google Scholar]
- 13.Fauvart M, et al. Role of persister cells in chronic infections: clinical relevance and perspectives on anti-persister therapies. J Med Microbiol. 2011;60:699–709. doi: 10.1099/jmm.0.030932-0. [DOI] [PubMed] [Google Scholar]
- 14.Allison KR, et al. Heterogeneous bacterial persisters and engineering approaches to eliminate them. Curr Opin Microbiol. 2011;14:593–598. doi: 10.1016/j.mib.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Balaban NQ, et al. Bacterial persistence as a phenotypic switch. Science. 2004;305:1622–1625. doi: 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- 16.Allison KR, et al. Metabolite-enabled eradication of bacterial persisters by aminoglycosides. Nature. 2011;473:216–220. doi: 10.1038/nature10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim JS, et al. Selective killing of bacterial persisters by a single chemical compound without affecting normal antibiotic-sensitive cells. Antimicrob Agents Chemother. 2011;55:5380–5383. doi: 10.1128/AAC.00708-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andrews-Polymenis HL, et al. Taming the elephant: Salmonella biology, pathogenesis, and prevention. Infect Immun. 2010;78:2356–2369. doi: 10.1128/IAI.00096-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wada T, et al. EAL domain protein YdiV acts as an anti-FlhD4C2 factor responsible for nutritional control of the flagellar regulon in Salmonella enterica serovar Typhimurium. J Bacteriol. 2011;193:1600–1611. doi: 10.1128/JB.01494-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chevance FF, Hughes KT. Coordinating assembly of a bacterial macromolecular machine. Nat Rev Microbiol. 2008;6:455–465. doi: 10.1038/nrmicro1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miao EA, et al. Cytoplasmic flagellin activates caspase-1 and secretion of interleukin 1beta via Ipaf. Nat Immunol. 2006;7:569–575. doi: 10.1038/ni1344. [DOI] [PubMed] [Google Scholar]
- 22.Franchi L, et al. Cytosolic flagellin requires Ipaf for activation of caspase-1 and interleukin 1beta in salmonella-infected macrophages. Nat Immunol. 2006;7:576–582. doi: 10.1038/ni1346. [DOI] [PubMed] [Google Scholar]
- 23.Hayashi F, et al. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature. 2001;410:1099–1103. doi: 10.1038/35074106. [DOI] [PubMed] [Google Scholar]
- 24.Sbrogio-Almeida ME, et al. Host and bacterial factors affecting induction of immune responses to flagellin expressed by attenuated Salmonella vaccine strains. Infect Immun. 2004;72:2546–2555. doi: 10.1128/IAI.72.5.2546-2555.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bergman MA, et al. CD4+ T cells and toll-like receptors recognize Salmonella antigens expressed in bacterial surface organelles. Infect Immun. 2005;73:1350–1356. doi: 10.1128/IAI.73.3.1350-1356.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wozniak CE, et al. T-POP array identifies EcnR and PefI-SrgD as novel regulators of flagellar gene expression. J Bacteriol. 2009;191:1498–1508. doi: 10.1128/JB.01177-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller SI, et al. A two-component regulatory system (phoP phoQ) controls Salmonella typhimurium virulence. Proc Natl Acad Sci U S A. 1989;86:5054–5058. doi: 10.1073/pnas.86.13.5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller SI, Mekalanos JJ. Constitutive expression of the PhoP regulon attenuates Salmonella virulence and survival within macrophages. J Bacteriol. 1990;172:2485–2490. doi: 10.1128/jb.172.5.2485-2490.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fink SL, Cookson BT. Caspase-1-dependent pore formation during pyroptosis leads to osmotic lysis of infected host macrophages. Cell Microbiol. 2006;8:1812–1825. doi: 10.1111/j.1462-5822.2006.00751.x. [DOI] [PubMed] [Google Scholar]
- 30.Sun YH, et al. Injection of flagellin into the host cell cytosol by Salmonella enterica serotype Typhimurium. J Biol Chem. 2007;282:33897–33901. doi: 10.1074/jbc.C700181200. [DOI] [PubMed] [Google Scholar]
- 31.Lucas RL, et al. Multiple factors independently regulate hilA and invasion gene expression in Salmonella enterica serovar Typhimurium. J Bacteriol. 2000;182:1872–1882. doi: 10.1128/jb.182.7.1872-1882.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jepson MA, Clark MA. The role of M cells in Salmonella infection. Microbes Infect. 2001;3:1183–1190. doi: 10.1016/s1286-4579(01)01478-2. [DOI] [PubMed] [Google Scholar]
- 33.Salcedo SP, et al. Intracellular replication of Salmonella typhimurium strains in specific subsets of splenic macrophages in vivo. Cell Microbiol. 2001;3:587–597. doi: 10.1046/j.1462-5822.2001.00137.x. [DOI] [PubMed] [Google Scholar]
- 34.Vazquez-Torres A, et al. Extraintestinal dissemination of Salmonella by CD18-expressing phagocytes. Nature. 1999;401:804–808. doi: 10.1038/44593. [DOI] [PubMed] [Google Scholar]
- 35.Cummings LA, et al. In vivo, fliC expression by Salmonella enterica serovar Typhimurium is heterogeneous, regulated by ClpX, and anatomically restricted. Mol Microbiol. 2006;61:795–809. doi: 10.1111/j.1365-2958.2006.05271.x. [DOI] [PubMed] [Google Scholar]
- 36.Hisert KB, et al. A glutamate-alanine-leucine (EAL) domain protein of Salmonella controls bacterial survival in mice, antioxidant defence and killing of macrophages: role of cyclic diGMP. Mol Microbiol. 2005;56:1234–1245. doi: 10.1111/j.1365-2958.2005.04632.x. [DOI] [PubMed] [Google Scholar]
- 37.Miao EA, et al. Caspase-1-induced pyroptosis is an innate immune effector mechanism against intracellular bacteria. Nat Immunol. 2010;11:1136–1142. doi: 10.1038/ni.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Galan JE, Curtiss R., 3rd Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc Natl Acad Sci U S A. 1989;86:6383–6387. doi: 10.1073/pnas.86.16.6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ackermann M, et al. Self-destructive cooperation mediated by phenotypic noise. Nature. 2008;454:987–990. doi: 10.1038/nature07067. [DOI] [PubMed] [Google Scholar]
- 40.Thiennimitr P, et al. Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proc Natl Acad Sci U S A. 2011;108:17480–17485. doi: 10.1073/pnas.1107857108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Winter SE, et al. Gut inflammation provides a respiratory electron acceptor for Salmonella . Nature. 2010;467:426–429. doi: 10.1038/nature09415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stecher B, et al. Salmonella enterica serovar Typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol. 2007;5:2177–2189. doi: 10.1371/journal.pbio.0050244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stecher B, et al. Motility allows S. Typhimurium to benefit from the mucosal defence. Cell Microbiol. 2008;10:1166–1180. doi: 10.1111/j.1462-5822.2008.01118.x. [DOI] [PubMed] [Google Scholar]
- 44.Sharma SV, et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141:69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spencer SL, et al. Non-genetic origins of cell-to-cell variability in TRAIL-induced apoptosis. Nature. 2009;459:428–432. doi: 10.1038/nature08012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bergsbaken T, Cookson BT. Macrophage activation redirects yersinia-infected host cell death from apoptosis to caspase-1-dependent pyroptosis. PLoS Pathog. 2007;3:e161. doi: 10.1371/journal.ppat.0030161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Raser JM, O'Shea EK. Noise in gene expression: origins, consequences, and control. Science. 2005;309:2010–2013. doi: 10.1126/science.1105891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kutsukake K, et al. Two novel regulatory genes, flit and fliZ in the flagellar regulon of Salmonella . Genes Genet Syst. 1999;74:287–292. doi: 10.1266/ggs.74.287. [DOI] [PubMed] [Google Scholar]
- 49.Saini S, et al. FliZ Is a posttranslational activator of FlhD4C2-dependent flagellar gene expression. J Bacteriol. 2008;190:4979–4988. doi: 10.1128/JB.01996-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wada T, et al. FliZ acts as a repressor of the ydiV gene, which encodes an anti-FlhD4C2 factor of the flagellar regulon in Salmonella enterica serovar Typhimurium. J Bacteriol. 2011;193:5191–5198. doi: 10.1128/JB.05441-11. [DOI] [PMC free article] [PubMed] [Google Scholar]