Abstract
In this paper, we propose a novel method for segmentation of the left ventricle, right ventricle, and myocardium from cine cardiac magnetic resonance images of the STACOM database. Our method incorporates prior shape information in a graph cut framework to achieve segmentation. Poor edge information and large within-patient shape variation of the different parts necessitates the inclusion of prior shape information. But large interpatient shape variability makes it difficult to have a generalized shape model. Therefore, for every dataset the shape prior is chosen as a single image clearly showing the different parts. Prior shape information is obtained from a combination of distance functions and orientation angle histograms of each pixel relative to the prior shape. To account for shape changes, pixels near the boundary are allowed to change their labels by appropriate formulation of the penalty and smoothness costs. Our method consists of two stages. In the first stage, segmentation is performed using only intensity information which is the starting point for the second stage combining intensity and shape information to get the final segmentation. Experimental results on different subsets of 30 real patient datasets show higher segmentation accuracy in using shape information and our method's superior performance over other competing methods.
Keywords: Segmentation, Shape priors, Orientation histograms, Graph cuts, Cine cardiac, MRI
Introduction
Cardiovascular diseases are the leading cause of death in the Western world [1]. Diagnosis and treatment of these pathologies relies on numerous imaging modalities like echography, computed tomography (CT), coronary angiography, and magnetic resonance imaging (MRI). Noninvasive technique like MRI has emerged as a popular diagnostic tool for physicians to observe the behavior of the heart. It also gives reliable information on morphology, muscle perfusion, tissue viability, and blood flow. These parameters are obtained by segmenting the left ventricle (LV) and right ventricle (RV) from cardiac magnetic resonance (MR) images.
Manual segmentation is tedious and prone to intra- and interobserver variability. This has necessitated the development of automated/semiautomated segmentation algorithms. An exhaustive review of medical image segmentation algorithms can be found in Frangi et al. [2], while an excellent review of cardiac LV segmentation algorithms is given in Petitjean and Dacher [3]. While there are many methods for LV segmentation, the RV has not received so much attention [3] because of: (1) its complex crescent shape; (2) lower pressure to eject blood; (3) thinner structure than LV; and (4) less critical function than LV. However, Shors et al. [4] show that MR imaging also provides an accurate quantification of RV mass.
Most cardiac MRI images show poor contrast between LV blood pool and myocardium wall, thus giving minimal edge information. This, in addition to similar intensity distributions in different regions, makes segmentation of the LV a very challenging task when using only low level information (e.g., intensity, gradient, etc). RV segmentation poses challenges because of their reduced wall thickness in MR images and shape variations. In such a scenario, inclusion of prior shape information assumes immense significance in LV and RV segmentations. We propose a graph-cut-based method to segment the LV blood pool, RV, and myocardium from cine cardiac image sequences using distance functions and orientation histograms for prior shape information.
Over the years, many methods have addressed the problem of LV segmentation from cardiac sequences. Some of the approaches aim at coronary tree analysis [5–7], late enhancement detection [8, 9], and analysis of time intensity curves [10, 11]. These methods use low-level information from the data without incorporating prior knowledge. Statistical shape models have also been used extensively for LV segmentation [2]. Perperidis et al. proposed a four-dimensional (4D) model by including temporal information [12]. Besbes et al. [13] used a control point representation of the LV prior and other images were deformed to match the shape prior. Shape knowledge has also been combined with dynamic information to account for cardiac shape variability [14, 15]. Zhu et al. [14] use a subject specific dynamic model that handles intersubject variability and temporal dynamics (intrasubject) variability simultaneously. A recursive Bayesian framework is then used for segmenting each frame. In Sun et al. [15], the ardiac dynamics is learnt by using a second order dynamic model. Davies et al. [16] propose a method to automatically extract a set of optimal landmarks using the minimum description length (MDL). But it is not clear whether the landmarks thus extracted are optimal in the sense of anatomical correspondence.
Shape models are generally unable to capture variability outside the training set. Kaus et al. [17] combine a statistical model with coupled mesh surfaces for segmentation. However, their assumption that the heart is located in the center of the image is not always valid. In Jolly et al. [18], a LV blood pool localization approach is proposed which acts as an initialization for LV segmentation. A 4D probabilistic atlas of the heart and 3D intensity template was used in Lorenzo-Valdes et al. [19] to localize the LV. Many other methods have been proposed that segment the LV from short-axis images [20–22], multiple views [23, 24], and using registration information [25, 26]. Paragios [21] uses prior shape information in an active contour framework for segmenting the LV. Mitchell et al. [27] use a multistage active appearance model to segment LV and RV. Some of the works on LV segmentation also show results for RV segmentation. These include image-based approaches like thresholding [28–30], pixel classification approaches [31, 32], deformable models [33–36], active appearance models [27, 37], and atlas based methods [19, 23, 38].
Previous works on segmentation using shape priors include, among others, active contours [21, 39], Bayesian approaches [40], and graph cuts [41–43]. Graph cuts have the advantage of being fast, give globally optimal results, and are not sensitive to initialization, while active contours are sensitive to initialization and can get trapped in local minima. Shape information in graphs is based on interaction between graph nodes (or image pixels) and interpixel interaction is generally limited to the immediate neighborhood although graph cuts can also handle more complex neighborhoods [44]. Therefore prior shape models assume great significance. The first works to use prior shape information in graph cuts were [41, 42]. In Freedman and Zhang [41], the zero-level set function of a shape template of natural and medical images was used as the shape prior with the smoothness term. The smoothness term favors a segmentation close to the prior shape. Slabaugh et al. [42] used an elliptical shape prior, under the assumption that many objects can be modeled as ellipses. They then employ many iterations where a preinitialized binary mask is updated to get the final segmentation. Vu et al. [45] use a discrete version of shape distance functions to segment multiple objects, which can be cumbersome. A flux-maximization approach was used in Chittajallu et al. [46] to include prior shape information, while in Veksler [47], the smoothness cost was modified to include star shape priors. A graph cut method using parametric shape priors for segmenting the LV was given in Zhu-Jacquot and Zabih [48], while Ben Ayed et al. [49] employ a discrete distribution-matching energy. In other methods, Ali et al. [50] construct a shape prior with a certain degree of variability and use it to segment DCE-MR kidney images.
The above methods using graph cuts generally use a large amount of training data or assume that the prior shape is simplistic. We propose a method that has the capability to handle different shapes and uses a single image to get the shape prior, thus overcoming the constraints of the other methods. The novelties of our work are twofold. First, we determine the shape penalty based on a combination of distance functions and distribution of orientation angles between a pixel and edge points on the prior shape. Second, our method uses a single image from each dataset to get prior shape information and is flexible enough to be used on different datasets. Appropriate energy terms are formulated to assign correct labels to pixels near the prior boundary where deformation is observed. In “Materials and Methods,” we describe our method in greater detail. “Experiments and Results” presents results of our experiments on real datasets, and we list our conclusions in “Conclusions.”
Materials and Methods
Importance of Shape Information
Automatic segmentation of the LV in cine MR poses the following challenges: (1) the overlap between the intensity distributions of different regions of the heart; (2) the lack of edge information due to poor spatial contrast; (3) the shape variability of the endocardial and epicardial contours across slices; and (4) the intersubject variability of the above factors. RV segmentation poses challenges because of their reduced wall thickness in MR images and shape variations. Consequently, relying only on intensity information is bound to give inaccurate segmentation results. This requires use of some high-level information like prior shape information. But due to large intersubject variability, it is difficult to have a generalized shape model for all patients. To overcome this challenge, we propose to have a shape prior for each dataset which can easily be constructed. However, it is fairly common to observe large shape changes of the endocardium between images from the initial and later stages of cine MRI. Therefore, the cost function should be formulated in a manner that it can handle the large changes in shape. Our proposed method is based on this approach, i.e., to construct a simple prior shape model for each patient dataset and adapt it to handle the shape variations observed in a cine MR sequence.
We use a second-order Markov random field (MRF) energy function for our method. MRFs are suitable for discrete labeling problems (i.e., the solution is defined as a set of discrete labels) and enable us to include context-dependent information. Context-dependent information allows us to impose additional constraints such that neighboring pixels take similar labels and a smooth solution is favored. For two class image segmentation, the labels are object or background, and smoothness constraints minimize isolated outlier segmentations. Graph cuts can find the globally optimal solution for specific formulations of binary-labeled MRF binary functions [44]. The energy function of a second-order MRF is given as
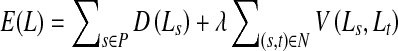 |
1 |
where P denotes the set of pixels, Ls denotes label of pixel s ∈ P, and N is the set of neighboring pixel pairs. The labels denote the segmentation class of a pixel (0 for background and 1 for object). The labels of the entire set of pixels are denoted by L. D(Ls), is a unary data penalty function derived from observed data and measures how well label Ls fits pixel s. V is a pairwise interaction potential that imposes smoothness and measures the cost of assigning labels Ls and Lt to neighboring pixels s and t. λ is a weight that determines the relative contribution of the two terms. Note that both D and V consist of two terms, each incorporating intensity and shape information.
Overview of Our Method
We describe our method in terms of LV segmentation but it can be easily extended for RV segmentation. Our method consists of the following steps to segment the LV in a new image: (1) manually identify a small region inside and outside the LV. These regions give the reference intensity histograms of object and background. (2) Segment the image using only intensity information. The obtained segmentation is used to update the intensity distributions of object and background. This initial segmentation need not be optimal; and 3) incorporate shape information in D and V to get the final segmentation. Before segmentation, the image sequence is corrected for any rotation or translation motion using the method in Mahapatra and Sun [51].
For every dataset, the shape prior is the first image of the sequence. Using this prior, we segment the second image of the sequence. The segmentation of the second image acts as the prior shape for segmenting the third image of the sequence and so on. This approach is necessary to account for the large degree of shape change of the LV due to its contraction (and subsequent expansion). If we use the shape prior of the first image to segment the third (or later) images, the magnitude of shape change is too large for an accurate segmentation. We shall first explain our method for two class segmentation and then extend it to multiclass segmentation.
Intensity Information
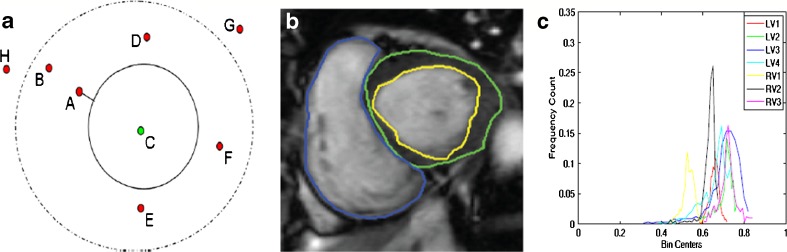
First, we manually identify image patches on the LV and background. The distribution of pixel intensities within the identified areas helps to determine a pixel's penalty for each label. Patches from the LV and RV belonging to training images show the intensity distributions follow a Gaussian. Figure 1c shows distributions of a few example patches, each obtained from a different patient. Since all of them are approximately unimodal Gaussians, we model the intensity distributions as Gaussians, and for each segmentation label, the mean and variance of intensities is estimated. The intensity penalty DI is the negative log likelihood and given as
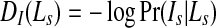 |
2 |
where Is is the intensity at pixel s, Pr is the likelihood, and Ls = object/background is the label. The smoothness term Vst assigns a low penalty at edge points based on the intensity of neighboring pixel pairs, and favors a piecewise constant segmentation result. It is defined as
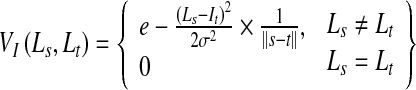 |
3 |
where σ determines the intensity difference up to which a region is considered as piecewise smooth. It is equal to the average intensity difference in a neighborhood and w.r.t pixel 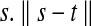 is the Euclidean distance between s and t.
is the Euclidean distance between s and t.
Fig. 1.
Illustration of shape prior segmentation using orientation information. (a) Orientation histograms for a synthetic image showing the different points outside and inside the shape; (b) reference cardiac image with LV endocardium highlighted in yellow, epicardium in green, and RV in blue; and (c) intensity distributions of the LV and RV. We refer the reader to the online version for a better interpretation of the figures
Shape Information
Figure 1a shows an illustration of the reference shape as a continuous circle and different points (A–F) inside and outside the shape. We shall explain the significance of the dotted line in LV segmentation later. Figure 1b shows the reference image from a typical dataset with the LV endocardium outlined in yellow and the epicardium outlined in green. First, we explain how the shape prior is used to segment the LV blood pool. Later, we extend our explanation to the segmentation of both blood pool and myocardium.
We define labels as background (label 0) and foreground (label 1). For point A (Fig. 1 (a)), we calculate the signed distance from the prior shape. We adopt the convention that pixels within the prior shape have a positive distance while pixels outside the prior have a negative distance. If the sign indicates the pixel to be outside the prior then it is likely to belong to the background and its penalty is defined as
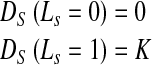 |
4 |
DS is the shape penalty and K is a variable which we shall describe later. If the sign of the distance indicates the pixel to be inside the prior, then it has greater probability of belonging to the LV and its penalty is defined as
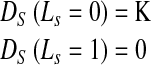 |
5 |
The above formulations make the segmented shape very similar to the reference shape. Cine cardiac images are acquired without gating over different phases of the cardiac cycle resulting in large degree of shape change of the LV. In order to segment the deformed LV, we relax the constraints on pixels near the prior’s boundary. Referring back to Fig. 1, we observe that pixel A is nearer to the reference shape than point B. We set a threshold normal distance of dth pixels from the shape within which the LV may deform and allow for a change in labels of pixels within this dth distance. Pixels which fall within this area are equally likely to take a particular label, and therefore have the same penalty. Therefore,
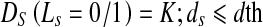 |
6 |
The above formulation indicates that both labels are equally likely. ds refers to the distance of pixel s from the shape. To determine dth, we adopt the following steps. After rigid alignment, we choose seven datasets having manual segmentations and calculate the Hausdorff distance (HD) between LV contours of successive frames. The HD gives a measure of shape variations, and the mean HD gives an idea of the average shape change over consecutive frames. It is observed that the maximum distance between LV contours of successive frames is 5 pixels, and the average distance is 3:5 pixels. Therefore, we set dth = 5 pixels. Note that for any point lying on the edge of the prior shape ds = 0. For most such edge points, the distribution of orientation angles may not be spread over four quadrants. But, since ds = 0, the penalty is assigned according to Eq. 5.
Another contribution of our work is the formulation of the smoothness penalty based on the prior shape. For every pixel, we calculate its distribution of orientation angles for all points on the prior shape. This is called shape context information and has been used for the purpose of shape matching and classification [52]. Let the histogram of orientation angles for point s with respect to the prior shape be denoted as hs. Similarly the corresponding histogram for neighboring point t is denoted as ht. The similarity between two histograms s; t is given by the χ2 metric as:
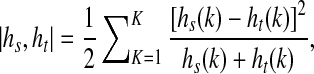 |
7 |
where k denotes the kth bin of the K-bin normalized histograms. K = 36 with 18 bins each positive and negative angle values (as the angle values are in the range (−180; 180)). The _2 metric gives values between 0 and 1. Note that other histogram similarity measures can be used (like Bhattacharyya measure) without any significant change in results. The smoothness cost due to prior shape information is given as
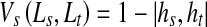 |
8 |
In Belongie et al. [52], it has been demonstrated that the above metric is robust to shape changes for the purpose of shape matching. Our objective for the above formulation is to check the similarity of a pair of neighboring points with respect to shape information. Similar pixels s and t will have low value of hs and ht and the corresponding VS is high. Dissimilar neighboring pixels s and t will have high value of hs and ht and the corresponding VS is low. Thus, the above formulation of VS serves our desired purpose.
Definition of K
The variable K is defined as
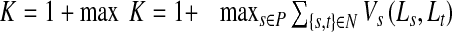 |
9 |
The value of K will always be between 1 and 2. Referring back to Eqs. 4 and 5, we observe that the value of K should be high compared with 0 (which is the penalty for the other label) so that the pixel s has less probability of being assigned the corresponding label. Moreover, the penalty should also be more than the weights of the links between neighboring pixels. The above formulation of K serves the purpose.
But pixels within ds distance of the prior shape are allowed to change their labels and their final labels will depend upon the values of VS(s; t). Considering this situation, the value of K should not be very high compared with VS such that there is very limited scope for change of labels. Considering all the above scenarios, we find that the current formulation of K also achieves this objective. Another advantage of such a formulation is that it avoids the need for assigning parameters on an ad hoc basis. The final cost function defined as a combination of intensity and shape information is
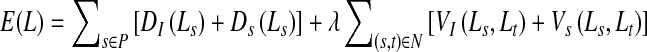 |
10 |
Extension to Multiple Classes
We describe our algorithm’s extension to segmentation of blood pool and myocardium. Consequently, its extension to RV segmentation is straightforward. Segmentation of the image into LV blood pool, myocardium, and background requires three labels (0 for background, 1 for myocardium (outside endoardium but within epicardium), and 2 for blood pool (within endocardium)). The intensity distribution is obtained by manually identifying a small patch in the different regions. For shape information, we define two prior shapes, one outlining the blood pool (endocardium) and the other outlining the myocardium (epicardium). The first stage for segmentation is accomplished using intensity distribution similar to Eq. 2. The difference is now we have three labels with each class defined by its mean and variance.
Figure 1b shows contours of manual segmentation for the epicardium, endocardium, and the RV. To include shape information we have two reference shapes. Let us denote the prior endocardium as shape A (continuous circle in Fig.1a) and the epicardium as shape B (dotted circle in Fig.1a). Any pixel within A is part of the endocardium, pixels outside A but within B are part of the myocardium, while pixels outside B belong to the background. Thus if pixel s ∈ A (s is within A), the penalty is defined as
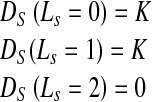 |
11 |
If pixel s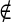 A and s ∈ B the penalty function is defined as
A and s ∈ B the penalty function is defined as
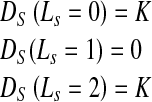 |
12 |
If pixel s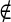 A and s
A and s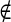 B the penalty function is defined as
B the penalty function is defined as
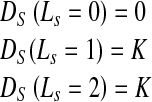 |
13 |
We also have to take into account the change in labels of pixels near the shape boundary. For pixels near the boundary of A, it can take either label 1 or 2. Thus, the penalty for such pixels is
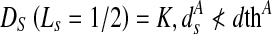 |
14 |
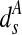 refers to the distance of pixel s from A and
refers to the distance of pixel s from A and 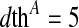 pixels is the threshold distance for shape A. Likewise, pixels near boundary of B will have labels either 0 or 1, and the penalty is defined as
pixels is the threshold distance for shape A. Likewise, pixels near boundary of B will have labels either 0 or 1, and the penalty is defined as
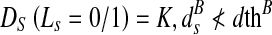 |
15 |
The shape smoothness cost VS is defined as in Eq. 8, where the distance is calculated from the nearest shape prior.
Optimization Using Graph Cuts
Pixels are represented as nodes on a graph which also consists of a set of directed edges that connect two nodes. The edge weight between two neighboring nodes is the smoothness term while the data penalty term is the edge weight for links between nodes and label nodes (terminal nodes). The optimum labeling set is obtained by severing the edge links in such a manner that the cost of the cut is minimum. The number of nodes is equal to the number of pixels and the number of labels is equal to the number of classes L. Details of graph construction and optimization can be found in Boykov and Veksler [44].
Validation of Segmentation Accuracy
To quantify the segmentation accuracy of our method, we use the dice metric (DM) [34] and HD. DM measures the overlap between the segmentation obtained by our algorithm and reference manual segmentation and is given by
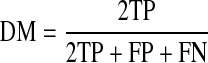 |
16 |
DM gives values between 0 and 1. The higher the DM, the better is the segmentation. Generally, DM values higher than 0.80 indicate good segmentation for cardiac images [34].
HD
The DM gives a measure of how much the actual manual segmentation was recovered by the automatic segmentation. But the boundaries of the segmented regions may be far apart. The HD aims to measure the distance between the contours corresponding to different segmentations.
We follow the definition of HD as given in Chalana and Kim [53]. If two curves are represented as sets of points A = {a1, a2,…} and B = {b1,b2,…}, where each ai and bi is an ordered pair of the x and y coordinates of a point on the curve, the distance to the closest point (DCP) for ai to the curve B is calculated. The HD is defined as the maximum of the DCPs between the two curves [54]
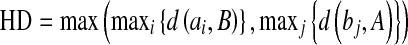 |
17 |
Experiments and Results
Description of Datasets
We have tested our algorithm on 30 training datasets from the STACOM 2011 4D LV Segmentation Challenge run by the Cardiac Atlas Project [55]. Although there are 100 training datasets, none of them have manual segmentations of the RV. Our radiologists manually segmented the RV in 30 of the datasets. All the data came from the Defibrillators to Reduce Risk by Magnetic Resonance Imaging Evaluation [56] cohort which consists of patients with coronary artery disease and prior myocardium infarction. The data were acquired using steady-state free precession MR imaging protocols with thickness of ≤10 mm, gap of ≤2 mm, TR of 30–50 ms, TE of 1:6 ms, flip angle of 60°, FOV of 360 mm, and 256 × 256 image matrix. MRI parameters varied between cases, giving a heterogeneous mix of scanner types and imaging parameters consistent with typical clinical cases. The images consist of two to six LA slices and on average 12 SA slices and 26 time frames. Possible breath-hold related misalignments between different LA and SA slices were corrected using the method of Elen et al. [57]. The ground truth images provided in this challenge were defined by an expert using an interactive guide point modeling segmentation algorithm [58].
The dataset was divided into training (15 patients) and test (15 patients) data. The training and test data were permuted to obtain 30 different groups such that each patient data were part of the training and test data. The reported results are the average of these 30 dataset groups. Automatic segmentations were obtained using three methods: graph cuts with intensity information alone (GC), our method using shape priors with graph cuts (GCSP); and the method in Lorenzo-Valdes et al. [19] (Met1). The automatic segmentations were compared with manual segmentation using DM and HD. For Met1, the same prior as our method was used. For implementing Met1, we calculate the appearance and location prior as described in their work, while for the shape prior, we use the flux maximization and template based star shape constraint only. We weight the different components of the energy function to get the best segmentation results for both Met1 and GC.
Segmentation Results
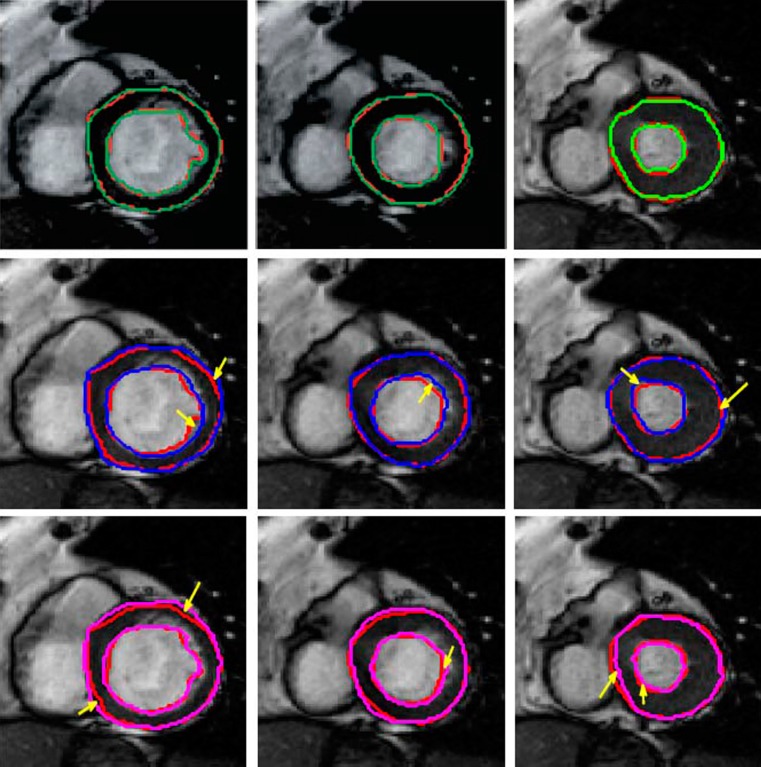
Figure 2 shows frames from different acquisition stages of a cine cardiac sequence along with the segmentation results of the LV from different methods. The first row shows results for GCSP followed by the results for GC (second row) and Met1 (third row). Each row shows three frames corresponding to different acquisition stages. The manual segmentations are shown as red contours. Inclusion of shape information leads to a definite improvement in segmentation results as the degree for mis-segmentation is highest for GC. GC is expected to give poor results because it relies only on intensity information. Among the two methods using shape information, GCSP shows a clearly superior performance. The areas of inaccurate segmentation for GC and Met1 are highlighted by yellow arrows.
Fig. 2.
Segmentation results of cine MRI using different methods. The manual segmentation is shown in red in all images, and the results for automatic segmentation are shown in different colors. First row shows result for GCSP, second row shows results for GC, and third row shows results for Met. We refer the reader to the online version for a better interpretation of the figures
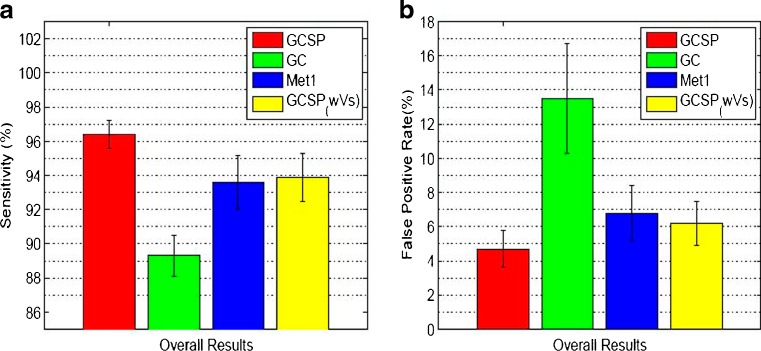
Comparative quantitative assessment of segmentations are made using different metrics like DM, HD, false-positive rates (Fpr), and sensitivity (Sen). Table 1 shows the DM and HD values for four methods, i.e., three methods mentioned before and GCSP without VS (GCSPwVS). Use of shape priors significantly improves the segmentation accuracy as intensity information alone is not sufficient for LV segmentation in MR images. Our method shows highest DM values and lowest HD, indicating that our approach of using shape prior information in both penalty and smoothness terms effectively accounts for shape changes due to deformations and improves segmentation accuracy. Furthermore, GCSPwVS shows lower segmentation accuracy than GCSP indicating that the importance of formulating a smoothness cost based on shape information. A paired t test on the DM values of GCSP and GCSPwVS gives p < 0.01 indicating statistically significant results. The mean and standard deviations for Sen and Fpr are shown in Fig. 3a, b. The choice of prior shape also determines the value of the different measures. If the other images of the dataset have a deformation magnitude more than dth then the segmentation accuracy decreases.
Table 1.
Comparative performance of segmentation accuracy of cine MRI using four methods and two metrics (DM and HD)
| Myocardium | LV blood pool | RV | |
|---|---|---|---|
| Dice metric (%) | |||
| GC | 84.2 ± 1.2 | 85.1 ± 0.9 | 84.7 ± 1.1 |
| GCSP | 91.6 ± 0.9 | 91.7 ± 1.1 | 92.2 ± 1.2 |
| GCSPwVs | 90.1 ± 1.0 | 89.3 ± 1.3 | 88.2 ± 1.1 |
| Met1 | 89.8 ± 1.3 | 88.5 ± 0.7 | 88.0 ± 1.1 |
| Hausdorff distance (pixel) | |||
| GC | 4.2 ± 0.9 | 3.3 ± 0.5 | 3.5 ± 0.6 (5.7) |
| GCSP | 1.9 ± 0.3 | 1.8 ± 0.4 | 2.0 ± 0.3 (1.2) |
| GCSPwVs | 2.4 ± 0.4 | 2.3 ± 0.3 | 2.8 ± 0.3 (1.6) |
| Met1 | 2.5 ± 0.3 | 2.2 ± 0.3 | 2.4 ± 0.3 (1.8) |
The values show the average measures over all datasets. Values indicate the mean and standard deviation. Entries enclosed in parentheses indicate maximum HD values
DM dice metric, HD Hausdorff distance, LV left ventricle, RV right ventricle, GC graph cut with intensity only, GCSP our method using graph cuts and shape priors, GCSPwVS GCSP without VS, Met1 method in Chittajallu et al. [46]
Fig. 3.
(a) Average sensitivity and (b) false-positive rates over 12 datasets. We refer the reader to the online version for a better interpretation of the figures
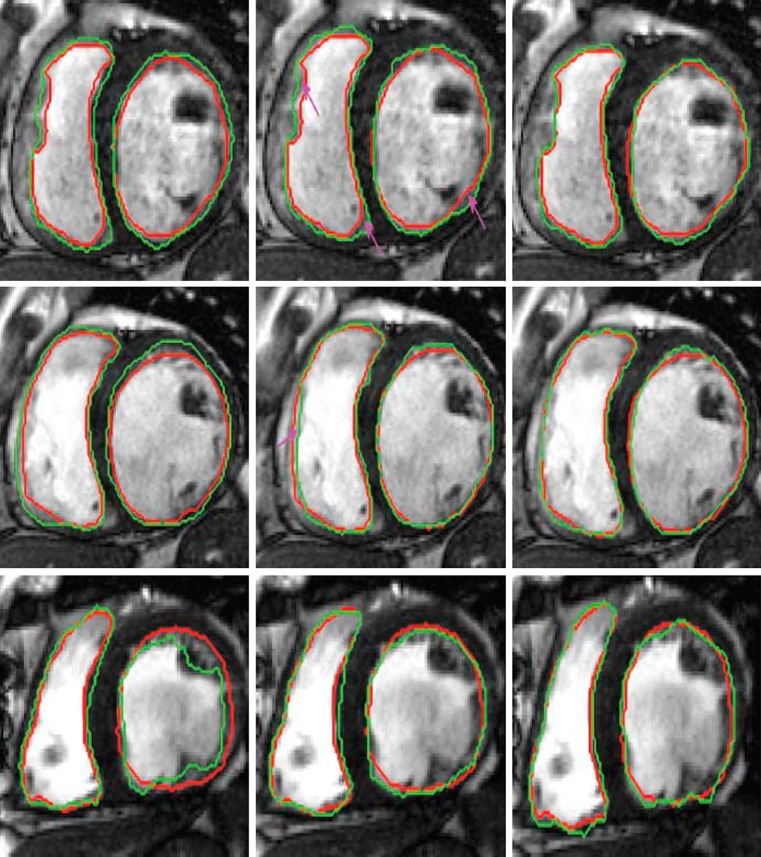
Except for Fpr and HD, higher values of other metrics indicate better agreement with manual segmentation. All metrics show that inclusion of shape information improves segmentation accuracy significantly. For all methods using prior shape information segmentation accuracy is similar for images which do not exhibit large shape changes with respect to the prior shape. However, for those datasets with large shape changes (as shown in Fig. 2), GCSP performs better than Met1. Paired t tests on the DM values of GCSP and Met1 give p < 0.01 indicating statistically significant results. Figure 4 shows segmentation results for RV and LV. Segmentations for only two parts are shown for clarity. In these examples, GC (column 1) gives better results than other patients because of fairly well defined intensity information. Still our method performs better than others, highlighting its advantages over GC and Met1 in various scenarios.
Fig. 4.
Segmentation results for different methods. Each row shows results of different patient datasets from the STACOM database. Red contour shows the manual segmentations, and green contours show results for automatic segmentations. Columns 1–3 show segmentation outputs for GC, Met1, and GCSP. We refer the reader to the online version for a better interpretation of the figures
The convergence time for each frame is shown in Table 2. GCSP takes the maximum time due to two stages while others have only one stage for segmentation. The time for GCSP includes the selection of patches, calculating the penalty values, and getting the final segmentation.
Table 2.
Convergence time for each frame of a cine cardiac MRI using different methods
| Convergence time (s) | |||
|---|---|---|---|
| GC | GCSP | GCSPwVs | Met1 |
| 4 | 11 | 9 | 6 |
GC graph cut with intensity only, GCSP our method using graph cuts and shape priors, GCSPwVS GCSP without VS, Met1 method in Chittajallu et al. [46]
Conclusions
In this paper, we have proposed a novel shape prior segmentation method using graph cuts for segmenting the LV from a sequence of cine cardiac MRI. We use a single frame to obtain prior shape information for each dataset. Manual intervention is limited to identifying the LV, myocardium, and RV in the first frame of the sequence. This serves as the prior shape based on which penalty values are calculated. The shape penalty is calculated using the distribution of orientation angles from every pixel to the edge points of the prior shape. Penalty and smoothness terms are formulated such that pixels near the boundary of the shape prior can change their labels to account for shape change due to deformations. Due to elastic deformations in cardiac images, pixels within a fixed distance from the prior boundary are equally likely to take any of the two labels. In this context, the formulation of the smoothness term acquires greater significance in determining the correct labels of the near-boundary pixels of the prior shape. The smoothness cost is inversely proportional to the pixel’s distance from the prior boundary. We also look at the relative importance of penalty values on segmentation results.
When shape information is combined with the intensity distributions of the object and background, our method results in accurate segmentation of the LV and RV. Experimental results on real patient datasets show the advantages of using shape priors and the superior performance of our method over a competing method.
References
- 1.S. Allender., European cardiovascular disease statistics. European Heart Network, 2008
- 2.Frangi AF, Niessen WJ, Viergever MA. Three dimensional modeling for functional analysis of cardiac images: a review. IEEE Trans Med. Imag. 2001;20(1):2–25. doi: 10.1109/42.906421. [DOI] [PubMed] [Google Scholar]
- 3.Petitjean C, Dacher J-N. A review of segmentation methods in short axis cardiac MR images. Med Imag Anal. 2011;15(2):169–184. doi: 10.1016/j.media.2010.12.004. [DOI] [PubMed] [Google Scholar]
- 4.Shors S, Fung C, Francois C, Finn P, Fieno D. Accurate quantification of right ventricular mass at MR imaging by using cine true fast imaging with steady state precession: study in dogs. Radiology. 2004;230(2):383–388. doi: 10.1148/radiol.2302021309. [DOI] [PubMed] [Google Scholar]
- 5.Frangi AF, Niessen WJ, Hoogeveen R, van Walsum T, Viergever MA. Model based quantization of 3-D magnetic resonance angiographic images. IEEE Trans Med Imag. 1999;18(10):946–956. doi: 10.1109/42.811279. [DOI] [PubMed] [Google Scholar]
- 6.Selle D, Preim B, Schnek A, Peitgen H. “Analysis of vasculature for liver surgical planning. IEEE Trans Med Imag. 2002;21(11):1344–1357. doi: 10.1109/TMI.2002.801166. [DOI] [PubMed] [Google Scholar]
- 7.Keegan J, Horkaew P, Buchanan T, Smart T, Yang G, Firmin D. Intra and interstudy reproducibility of coronary artery diameter measurements in magnetic resonance coronary angiography. J Magn Reson Imag. 2004;20(1):160–166. doi: 10.1002/jmri.20094. [DOI] [PubMed] [Google Scholar]
- 8.Kolipaka A, Chatzimavroudis G, White R, O’Donnell T, Setser R. Segmentation of non-viable myocardium in delayed magnetic resonance images. Int J Cadiovasc Imag. 2005;21(2):303–311. doi: 10.1007/s10554-004-5806-z. [DOI] [PubMed] [Google Scholar]
- 9.Noble N, Hill D, Breeuwer M et al: The automatic identification of hibernating myocardium. In: Proc. Intl. Conf. Med. Image Computing and Computer-Assisted Intervent (MICCAI), 2004, pp 890–898
- 10.Schwitter J. Myocardial perfusion. J Magn Reson Imag. 2006;24(5):953–963. doi: 10.1002/jmri.20753. [DOI] [PubMed] [Google Scholar]
- 11.Di Bella E, Sitek A et al: Time curve analysis techniques for dynamic contrast MRI studies. In: Intl. Conf Inf. Process. Med. Imag. (IPMI), 2001, pp 211–217
- 12.Perperidis D, Mohiaddin R, Rueckert D: Construction of a 4D statistical atlas of the cardiac anatomy and its use in classification. In: MICCAI, 2005, pp 402–410 [DOI] [PubMed]
- 13.Besbes A, Komodakis N, Paragios N: Graph-based knowledge-driven discrete segmentation of the left ventricle. In: ISBI, 2009, pp 49–52
- 14.Zhu Y, Papademetris X, Sinusas A et al: Segmentation of left ventricle from 3D cardiac mr image sequence using a subject specific dynamic model. In: Proc. CVPR, 2009, pp 1–8 [DOI] [PMC free article] [PubMed]
- 15.Sun W, Setin M, Chan R et al: Segmenting and tracking of the left ventricle by learning the dynamics in cardiac images. In: Proc. IPMI, , 2005, pp 553–565 [DOI] [PubMed]
- 16.Davies RH, Twining CJ, Cootes TF, Waterton JC, Taylor CJ. A minimum description length approach to statistical shape modelling. IEEE Trans Med Imag. 2002;21:525–537. doi: 10.1109/TMI.2002.1009388. [DOI] [PubMed] [Google Scholar]
- 17.Kaus MR, von Berg J, Weese J, Niessen W, Pekar V. Automated segmentation of the left ventricle in cardiac MRI. Med Image Anal. 2004;8(3):245–254. doi: 10.1016/j.media.2004.06.015. [DOI] [PubMed] [Google Scholar]
- 18.Jolly MP et al: Automatic recovery of the left ventricle blood pool in cardiac cine MR images. In: MICCAI, 2008, pp 110–118 [DOI] [PubMed]
- 19.Lorenzo-Valdes M, Sanchez-Ortiz GI, Elkington AG, Mohiaddin RH, Rueckert D. Segmentation of 4D cardiac MR images using a probabilistic atlas and the EM algorithm. Med Image Anal. 2004;8(3):255. doi: 10.1016/j.media.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 20.Niessen WJ, Romeny BMTH, Viergever MA. Geodesic deformable models for medical image analysis. IEEE Trans Med Imag. 1998;17(4):634–41. doi: 10.1109/42.730407. [DOI] [PubMed] [Google Scholar]
- 21.Paragios N. A variational approach for the segmentation of the left ventricle in cardiac image analysis. Intl J Comp Vis. 2002;50(3):345–362. doi: 10.1023/A:1020882509893. [DOI] [Google Scholar]
- 22.Jolly MP. Automatic segmentation of the left ventricle in cardiac MR and CT images. Int J Comp Vision. 2006;70(2):151–163. doi: 10.1007/s11263-006-7936-3. [DOI] [Google Scholar]
- 23.Ltjnen J, Kivist S, Koikkalainen J, Smutek D, Lauerma K. Statistical shape model of atria, ventricles and epicardium from short- and long-axis MR images. Med Image Anal. 2004;8(3):371–386. doi: 10.1016/j.media.2004.06.013. [DOI] [PubMed] [Google Scholar]
- 24.van Assen C, Danilouchkine MG, Frangi AF, Ords S, Westenberg JJ, Reiber JH, Lelieveldt BP. pasm: a 3D-asm for segmentation of sparse and arbitrarily oriented cardiac MRI data. Med Image Anal. 2006;10(2):286–303. doi: 10.1016/j.media.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 25.Mahapatra D, Sun Y: Joint registration and segmentation of dynamic cardiac perfusion images using MRFs. In: Proc. MICCAI, 2010, pp 493–501 [DOI] [PubMed]
- 26.Mahapatra D, Sun Y. Integrating segmentation information for improved MRF based elastic image registration. IEEE Trans Imag Proc. 2012;21(1):170–183. doi: 10.1109/TIP.2011.2162738. [DOI] [PubMed] [Google Scholar]
- 27.Mitchell SC, Lelieveldt BPF, et al. Multistage hybrid active appearance models: segmentation of cardiac MR and ultrasound images. IEEE Trans Med. Imag. 2001;20(5):415–423. doi: 10.1109/42.925294. [DOI] [PubMed] [Google Scholar]
- 28.Goshtasby A, Turner D. Segmentation of cardiac cine MR images for extraction of right and left ventricular chambers. IEEE Trans Med Imag. 1995;14(1):56–64. doi: 10.1109/42.370402. [DOI] [PubMed] [Google Scholar]
- 29.Weng J, Singh A, Chiu M. Learning based ventricle detection from cardiac MR and CT images. IEEE Trans Med Imag. 1997;16(4):378–391. doi: 10.1109/42.611346. [DOI] [PubMed] [Google Scholar]
- 30.Katouzian A, Konofagau E, Prakash A: A new automated technique for left and right ventricular segmentation in magnetic resonance imaging in IEEE. In: EMBS, 2006, pp 3074–3077 [DOI] [PubMed]
- 31.Gering D: Automatic segmentation of cardiac MRI. In: MICCAI, 2003, pp 524–532
- 32.Cocosco C, Niessen W, Netsch T, Vonken E-J, Lund G, Stork A, Viergever M. “Automatic image driven segmentation of the ventricles in cardiac cine MRI. J Magn Reson Imag. 2008;28(2):366–374. doi: 10.1002/jmri.21451. [DOI] [PubMed] [Google Scholar]
- 33.Battani R, Corsi C, Sarti A et al: Estimation of right ventricular volume without geometrical assumptions utilizing cardiac magnetic resonance data. In: Comput Cardiol, 2003, pp 81–84
- 34.Pluempitiwiriyawej C, Moura JMF, Wu YL, Ho C. STACS: new active contour scheme for cardiac MR image segmentation. IEEE Trans Med Imag. 2005;24(5):593–603. doi: 10.1109/TMI.2005.843740. [DOI] [PubMed] [Google Scholar]
- 35.Sermeanst M, Moireau P, Camara O, Sainte-Marie J, Adriantsimiavona R, Cimrman R, Hill DL, Chapelle D, Razavi R. Cardiac function estimation from MRI using a heart model and data assimilation: advances and difficulties. Med Image Anal. 2006;10(4):642–656. doi: 10.1016/j.media.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 36.Billet F, Sermeanst M,Delingette H et al: Cardiac motion recovery and boundary conditions estimation by coupling an electromechanical model and cine-MRI data. In: Functional imaging and modeling of the heart (FMIH), 2009, pp 376–385
- 37.Ordas S, Boisrobert L, Huguet M et al: Active shape models with invariant optimal features (IOFASM)—application to cardiac MRI segmentation. In: Comput Cardiol, 2003, pp 633–636
- 38.Lorenzo-Valdes M, Sanchez-Ortis G, Mohiaddin R et al: Atlas based segmentation and tracking of 3D cardiac MR images using non-rigid registration. In: MICCAI, 2002, pp 642–650
- 39.Cremers D, Tischhauser F, Weickert J, Schnorr C. Diffusion snakes: introducing statistical shape knowledge into the Mumford–Shah functional. Intl J Comp Vis. 2002;50(3):295–313. doi: 10.1023/A:1020826424915. [DOI] [Google Scholar]
- 40.Chang H, Yang Q, Parvin B: Bayesian approach for image segmentation with shape priors. In: CVPR, 2008, pp 1–8
- 41.Freedman D, Zhang T: Interactive graph cut based segmentation with shape priors. In CVPR, 2005, pp 755–762
- 42.Slabaugh G, Unal G: Graph cuts segmentation using an elliptical shape prior. In: ICIP, 2005, pp 1222–1225
- 43.Mahapatra D, Sun Y: Orientation histograms as shape priors for left ventricle segmentation using graph cuts. In: Proc MICCAI, 2011, pp 420–427 [DOI] [PubMed]
- 44.Boykov Y, Veksler O. Fast approximate energy minimization via graph cuts. IEEE Trans Pattern Anal Mach Intell. 2001;23:1222–1239. doi: 10.1109/34.969114. [DOI] [Google Scholar]
- 45.Vu N, Manjunath BS: Shape prior segmentation of multiple objects with graph cuts. in CVPR 2008
- 46.Chittajallu DR, Shah SK, Kakadiaris IA: A shape driven MRF model for the segmentation of organs in medical images. In: CVPR, 2010, pp 3233–3240
- 47.Veksler O: Star shape prior for graph cut segmentation. In: ECCV, 2008, pp 454–467
- 48.Zhu-Jacquot J, Zabih R: Segmentation of the left ventricle in cardiac mr images using graph cuts with parametric shape priors. In: ICASSP, 2008, pp 521–524
- 49.Ben Ayed I, Punithakumar K, Li S et al: Left ventricle segmentation via graph cut distribution matching. In: MICCAI 2009, pp 901–909 [DOI] [PubMed]
- 50.Ali AM, Farag AA, El-Baz AS: Graph cuts framework for kidney segmentation with prior shape constraints. In: MICCAI 2007 [DOI] [PubMed]
- 51.Mahapatra D, Sun Y: Registration of dynamic renal mr images using neurobiological model of saliency. In: Proc. ISBI, 2008, pp 1119–1122
- 52.Belongie S, Malik J, Puzicha J. Shape matching and object recognition using shape contexts. IEEE PAMI. 2002;24(24):509–522. doi: 10.1109/34.993558. [DOI] [Google Scholar]
- 53.Chalana V, Kim Y. A methodology for evaluation of boundary detection algorithms on medical images. IEEE Trans Med Imag. 1997;16(5):642–652. doi: 10.1109/42.640755. [DOI] [PubMed] [Google Scholar]
- 54.Huttenlocher DP, Klanderman GA, Rucklidge WJ. Comparing images using the Hausdorff distance. IEEE Trans Pattern Anal Machine Intell. 1993;15(9):850–863. doi: 10.1109/34.232073. [DOI] [Google Scholar]
- 55.Fonseca CG. The cardiac atlas project an imaging database for computational modeling and statistical atlases of the heart. Bioinformatics. 2011;27(16):2288–2295. doi: 10.1093/bioinformatics/btr360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kadish AH, Bello D, Finn JP, Bonow RO, Schaechte A, Subacius H, Albert C, Daubert JP, Fonseca CG, Goldberger JJ. Rationale and design for the defribrillators to reduce risk by magnetic resonance imaging evaluation (determine) trial. J Cardiovascular Electrophysiology. 2009;20(9):982–987. doi: 10.1111/j.1540-8167.2009.01503.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Elen A, Hermans J, Ganame J, Loeckx D, Bogaert J, Maes F, Suetens P. Automatic 3-D breath-hold related motion correction of dynamic multislice MRI. IEEE Trans Med Imag. 2010;29(3):868–878. doi: 10.1109/TMI.2009.2039145. [DOI] [PubMed] [Google Scholar]
- 58.Young AA, Cowan BR, Thrupp SF, Hedley WJ, DellItalia LJ. Left ventricular mass and volume: fast calculation with guide-point modeling on MR images. Radiology. 2000;202(2):597–602. doi: 10.1148/radiology.216.2.r00au14597. [DOI] [PubMed] [Google Scholar]