Abstract
The role of the growth hormone (GH)-insulin-like growth factor (IGF)-1 axis in the lifelong caloric restriction (CR)-associated remodeling of white adipose tissue (WAT), adipocyte size, and gene expression profiles was explored in this study. We analyzed the WAT morphology of 6–7-month-old wild-type Wistar rats fed ad libitum (WdAL) or subjected to CR (WdCR), and of heterozygous transgenic dwarf rats bearing an anti-sense GH transgene fed ad libitum (TgAL) or subjected to CR (TgCR). Although less effective in TgAL, the adipocyte size was significantly reduced in WdCR compared with WdAL. This CR effect was blunted in Tg rats. We also used high-density oligonucleotide microarrays to examine the gene expression profile of WAT of WdAL, WdCR, and TgAL rats. The gene expression profile of WdCR, but not TgAL, differed greatly from that of WdAL. The gene clusters with the largest changes induced by CR but not by Tg were genes involved in lipid biosynthesis and inflammation, particularly sterol regulatory element binding proteins (SREBPs)-regulated and macrophage-related genes, respectively. Real-time reverse-transcription polymerase chain reaction analysis confirmed that the expression of SREBP-1 and its downstream targets was upregulated, whereas the macrophage-related genes were downregulated in WdCR, but not in TgAL. In addition, CR affected the gene expression profile of Tg rats similarly to wild-type rats. Our findings suggest that CR-associated remodeling of WAT, which involves SREBP-1-mediated transcriptional activation and suppression of macrophage infiltration, is regulated in a GH–IGF-1-independent manner.
Electronic supplementary material
The online version of this article (doi:10.1007/s11357-012-9439-1) contains supplementary material, which is available to authorized users.
Keywords: Growth hormone, Insulin-like growth factor-1, Caloric restriction (CR), Lipid biosynthesis, Sterol regulatory element binding protein, DNA microarray
Introduction
Ames and Snell mutant dwarf mice, which lack growth hormone (GH), prolactin, and thyroid-stimulating hormone, live approximately 20–50 % longer than wild-type mice (Brown-Borg et al. 1996; Flurkey et al. 2001). Similarly, disrupted GH receptor/binding protein (GHR/BP) knockout (KO) mice live significantly longer than their wild-type controls (Coschigano et al. 2000). The longevity of heterozygous insulin-like growth factor (IGF)-1 receptor KO mice, and both heterozygous and homozygous insulin receptor substrate-1 KO mice (particularly females) show markedly extended lifespan compared with their counterparts (Holzenberger et al. 2003). We have also reported that heterozygous transgenic dwarf rats, bearing an anti-sense GH transgene, live longer than controls (Shimokawa et al. 2002). Based on these studies, the GH–IGF-1 axis and/or its related signaling pathways are important lifespan regulators (Bartke 2005).
Although new genetic interventions that extend the lifespan of mammals are emerging, caloric restriction (CR) remains the most robust, reproducible, and simple experimental manipulation known to extend both median and maximum lifespan, and to delay the onset of many age-associated pathophysiological changes in laboratory rodents (Weindruch and Walford 1988; Yu 1994). In general, attenuation of oxidative and other stresses, the modulation of glycemia and insulinemia, and the activation of sirtuin may be significant factors in the beneficial effects of CR (Weindruch and Walford 1988; Yu 1994; Sohal and Weindruch 1996; Masoro 2005; Sinclair 2005). Moreover, Nisoli et al. (2005) suggested that the enhanced mitochondrial biogenesis is also involved in the beneficial action of CR. Thereafter, several investigators reported that CR induces mitochondrial biogenesis (Anderson and Prolla 2009; López-Lluch et al. 2006; Shi et al., 2005). On the other hand, Hancock et al. (2011) and Gesing et al. (2011) demonstrated that CR does not increase it. Recently, we also reported that CR enhances mitochondrial biogenesis in white adipose tissue (WAT) but not in brown adipose tissue (Okita et al. 2012). Thus, the exact mechanisms underlying CR are still debated. CR animals share many characteristics with long-living dwarf mice, including smaller body size, and lower plasma insulin and IGF-1 levels (Sinclair 2005; Al-Regaiey et al. 2005). CR does not further extend the lifespan of the already long-lived GHR/BP KO mice (Bonkowski et al. 2006). In contrast, CR further extends the longevity of Ames dwarf mice and heterozygous transgenic dwarf rats bearing an anti-sense GH transgene, which live longer than their wild-type controls (Bartke et al. 2001; Shimokawa et al. 2003). Therefore, the beneficial effects of CR are not solely dependent on the GH–IGF-1 axis. In terms of the hepatic gene expression profile, CR mainly alters the expression of genes involved in the stress response, xenobiotic metabolism, and lipid metabolism. Most genes involved in stress response and xenobiotic metabolism are regulated in a GH–IGF-1-dependent manner, while those involved in lipid metabolism are regulated in a GH–IGF-1-independent manner. Moreover, CR enhances the expression of genes involved in fatty acid synthesis after feeding, and of genes encoding mitochondrial β-oxidation enzymes during food shortage, probably via transcriptional regulation by sterol regulatory element binding protein 1 (SREBP-1) and peroxisome proliferator-activated receptor (PPAR)-α, respectively (Higami et al. 2006b). Considering these findings together with serum biochemical parameters, we proposed that CR enhances lipid utilization via hepatic transcriptional changes and prevents hepatic steatosis in a GH–IGF-1-independent manner (Higami et al. 2006b).
Adipose tissue plays a central role in the regulation of both energy storage and expenditure (Saely et al. 2012). Numerous WAT-derived secretory molecules, including leptin, tumor necrosis factor (TNF)-α, and adiponectin, have been characterized, and some of these molecules play significant roles in obesity and insulin resistance (Torres-Leal et al. 2010; Ouchi et al. 2011). Therefore, WAT is now recognized as an endocrine organ rather than an inert tissue, and is implicated in the pathogenesis and complications of type 2 diabetes. It has been reported that fat-specific insulin receptor knockout mice live longer than their controls (Blüher et al. 2003). These mice show reduced adiposity and altered secretion of adipokines, including higher adiponectin and lower pro-inflammatory cytokine levels (Blüher et al. 2002). The transcription factors C/EBPα, C/EBPβ, and PPARγ are master regulators of adipocyte differentiation (Farmer 2006). Mice in which C/EBPα is replaced with C/EBPβ (β/β mice) live longer and have reduced adiposity compared with their wild-type controls (Chiu et al. 2004). In contrast, hetero-deficient PPARγ KO mice have a shortened lifespan (Argmann et al. 2009). Transgenic mice expressing adiponectin in the liver live longer than controls, and are resistant to high-calorie diet-induced obesity (Otabe et al. 2007). Thus, altered adipose tissue gene expression and modulation of adipokine secretion seem to influence the lifespan of rodents. CR reduces adiposity and reduces adipocyte size by altering the gene expression profile (Higami et al. 2004, 2006a). CR decreases plasma insulin and leptin levels, and increases plasma adiponectin levels (Higami et al. 2005; Yamaza et al. 2007). CR also reverses age-associated insulin resistance, possibly by decreasing adiposity (Barzilai et al. 1998). Moreover, Masternak et al. (2012) reported that visceral fat removal improved insulin sensitivity, suppressed fat accumulation in the skeletal muscle, and reduced body temperature and respiratory quotient in wild-type mice and had opposite effects on long-living GHR/BP KO mice. Therefore, we hypothesized that the beneficial actions of CR may be partially mediated by WAT remodeling as well as decreasing adiposity.
In the present study, to explore the role of the GH–IGF-1 axis in CR-associated remodeling of WAT, we compared adipocyte size and gene expression profiles of WAT between CR rats and transgenic dwarf rats, bearing an anti-sense GH transgene. We propose that CR-associated remodeling of WAT, which is transcriptionally regulated by SREBP-1 and modulated by macrophage infiltration, could be regulated in a GH–IGF-1-independent manner.
Materials and Methods
Animals
The present study was conducted in accordance with the provisions of the Ethics Review Committee for Animal Experimentation at Nagasaki University. The rat characteristics and animal care are described elsewhere (Higami et al. 2006a). Briefly, in this study we used ad libitum (AL)-fed male heterozygous transgenic dwarf rats, bearing the anti-sense growth hormone transgene (tg/−; Tg), and their genetic background, Jcl:Wistar (−/−; wild-type) rats. From 6 weeks of age, both wild-type and Tg rats were divided into two groups: AL and CR (70 % of the energy intake). CR was started without adjustment for food shortage. CR rats were fed every other day. Their 2-day food allotment was equal to 140 % of the mean daily intake of AL rats. Wild-type AL (WdAL) and CR (WdCR), and Tg AL (TgAL) and CR (TgCR) rats were killed at 6–7 months of age. The day before the rats were killed, they were all provided with their allocated food 30 min before the lights were turned off in the evening and were killed after the lights were turned on the following morning. Thus, all rats were not under fasting condition when killed. Immediately after killing the rats, epididymal white adipose tissue (WAT) was collected and its weight was measured. Some of the isolated WAT was fixed in a buffered formalin solution for histological examination and the rest was immediately diced, frozen in liquid nitrogen, and stored at −80°C. Total RNA was extracted from the stored WAT for DNA microarray analysis and quantitative real-time reverse-transcription polymerase chain reaction (RT-PCR).
Histological examination
Fixed tissues were processed routinely and embedded in paraffin. Tissue sections (5 μm thick) were stained with hematoxylin–eosin. The stained sections were scanned by microscopy with a charge-coupled device camera (Nikon, Tokyo, Japan). All images were recorded after precise focusing. ImageJ 1.43u/Java1.6.0_22 software was used for all tissue analyses. The size distribution of each white area in the black-and-white images, corresponding to lipid droplets, was counted and calculated. To avoid inter-rater variation, a single observer (YC) carried out tissue analyses.
Microarrays and data normalization
Total RNA was measured using an Affymetrix Rat Genome 230_2.0 GeneChip (Affymetrix, Santa Clara, CA, USA), with four biological repeats per group. The raw data were deposited in Gene Expression Omnibus (accession code: GSE30668). The perfect match data were normalized and the expression levels of each gene were estimated using the SuperNORM data service (Skylight Biotech Inc., Akita, Japan) according to a three-parameter log-normal distribution model (Konishi et al. 2008). To reduce noise effects, the analyses were focused on genes identified as positive by two-way analysis of variance (ANOVA), with a two-sided threshold of 0.001. Out of 31,099 genes on each chip, 6,641 were positive.
Principal component analysis
To compare the effects of CR (WdAL vs. WdCR) with those of Tg (WdAL vs. TgAL), we performed principal component (PC) analysis (Jackson 2005) on the ANOVA-positive genes. To reduce the effects of individual differences between samples, the axes of the PC analysis were estimated on a matrix of each group’s sample means and applied to all data, which were centered using the sample means of WdAL rats (the R scripts used are available in the Supplemental Materials and Methods). The methodology rotated the original data matrix around the center of the WdAL rats, to fit perpendicular axes toward which most of the variations in the data appeared. The distribution of each PC value was checked on a normal QQ plot and outlier genes that departed from the normality (PC1: >0.16 and < –016 in Supplemental Fig. 1; PC2: >0.1 and < –0.1 in Supplemental Fig. 2) were selected.
Evaluation of frequently occurring biological functions in gene annotations
To test the significance among categories of biological functions that appeared in the selected genes’ annotations, we applied binomial statistical tests based on the frequencies found in the Gene Ontology (GO) Biological Process annotations (release 31) provided by Affymetrix (Konishi et al. 2008).
Quantitative real-time RT-PCR
To obtain cDNA, 1 μg of RNA extracted from WAT of WdAL, WdCR, TgAL, and TgCR rats was reverse transcribed using PrimeScript Reverse Transcriptase (Takara, Shiga, Japan) with random hexamers (Takara). Quantitative real-time PCR was performed using an Applied Biosystems 7300 real-time PCR system (Applied Biosystems, Carlsbad, CA, USA) with SYBR Premix ExTaqII (Takara). The primer sequences for SREBP-1a, SREBP-1c, SREBP-2, fatty acid synthase (FASN), acetyl-CoA carboxylase 1 (ACC1), squalene epoxidase (Sqle), mevalonate kinase (Mvk), F4/80, monocyte chemotactic protein-1 (MCP-1), CD11c (also known as integrin alpha X), CD163, and TATA box binding protein (TBP) are shown in Table 1. TBP was used as a normalization control. The amount of target mRNA relative to TBP mRNA in the three groups was obtained. Data from three to six rats per group are expressed as means ± SEM and were compared using Tukey’s t test. Differences were considered statistically significant at P < 0.05.
Table 1.
List of primers for real-time RT-PCR
| Forward | Reverse | |
|---|---|---|
| SREBP-1aa | 5′-CCGAGGTGTGCGAAATGG-3′ | 5′-TTGATGAGCTGAAGCATGTCTTC-3′ |
| SREBP-1cb | 5′-GGAGCCATGGATTGCACATT-3′ | 5′-GGCCCGGGAAGTCACTGT-3′ |
| SREBP-2 | 5′-CGATCAAGTCAGCAGCCAAG-3′ | 5′-AATCCCACAGAGTCCACAAAAG-3′ |
| FASN | 5′-AGCAGGCACACACAATGGAC-3′ | 5′-GAAGAAGAAAGAGAGCCGGTTG-3′ |
| ACC1 | 5′-TGGATGAACCATCTCCGTTG-3′ | 5′-CATGTGAAAGGCCAAACC-3′ |
| Sqle | 5′-GTCTCCGGAAAGCAGCTATGG-3′ | 5′-CTCCTTGGTGTCCCCAGTCTC-3′ |
| Mvk | 5′-CAGAGCAATGGGAAAGTGAGC-3′ | 5′-TCTCCAGTTGCTCCAAGGTG-3′ |
| MCP-1 | 5′-CCAGCCAACTCTCACTGAAGC-3′ | 5′-CTTCTTTGGGACACCTGCTG-3′ |
| F4/80 | 5′-GGCCAAGATTCTCTTCCTCAC-3′ | 5′-TCACCACCTTCAGGTTTCTCAC-3′ |
| CD11c | 5′-AGCACACGGGGAAGGTTGTC-3′ | 5′-CAGGTCAGTGCTGCCATCTCTATC-3′ |
| CD163 | 5′-ACAAATACGTGGCTCTTTCCTG-3′ | 5′-ATGGGATTTCTCCTCCAACC-3′ |
| TBP | 5′-CAGTACAGCAATCAACATCTCAGC-3′ | 5′-CAAGTTTACAGCCAAGATTCACG-3′ |
Results
CR markedly reduced the body weight of both wild-type and Tg rats. Tg also significantly decreased the body weight of both AL and CR rats (WdAL, 486.9 ± 29.9 g; WdCR, 349.7 ± 15.6 g; TgAL, 310.3 ± 12.1 g; TgCR, 237.8 ± 25.7 g). Similarly, CR markedly reduced the epididymal WAT weight of both wild-type and Tg rats. Tg also significantly reduced it in both AL and CR rats (WdAL, 7.02 ± 1.04 g; WdCR, 4.96 ± 0.97 g; TgAL, 4.76 ± 0.74 g; TgCR, 4.32 ± 0.88 g). In contrast, WAT weight as a percentage of body weight, which represents adiposity, did not differ among WdAL, WdCR, and TgAL rats, but it was markedly increased in TgCR rats (WdAL, 1.44 ± 0.18 %; WdCR, 1.42 ± 0.27 %; TgAL, 1.53 ± 0.22 %; TgCR, 1.81 ± 0.26 %).
CR significantly reduced the size of white adipocytes, but this effect was predominantly found in wild-type rats compared with Tg rats (Fig. 1a–d). Tg also slightly reduced their size, but it was likely that the effect of Tg was less than that of CR (Fig. 1a and c). In fact, the median adipocyte size was significantly smaller in WdCR than in WdAL, but not in TgCR compared with TgAL. It was slightly but significantly smaller in TgAL than in WdAL (WdAL vs. WdCR: p = 0.001, TgAL vs. TgCR: p = 0.100, WdAL vs. TgAL: p = 0.026, Fig. 1f). The percentage of adipocytes showing >5,000 μm2 was also significantly lower in WdCR than WdAL, but not in TgCR compared with TgAL. It was slightly but significantly smaller in TgAL than in WdAL (WdAL vs. WdCR: p < 0.001, TgAL vs. TgCR: p = 0.090, WdAL vs. TgAL: p = 0.039, Fig. 1g). In WAT of WdCR and TgCR, the adipocytes were predominantly 1,000–3,000 μm2 in area, whereas those in WdAL and TgAL WAT showed a much greater size distribution. This pattern was more significant in WdAL than in TgAL rats (Fig. 1e).
Fig. 1.
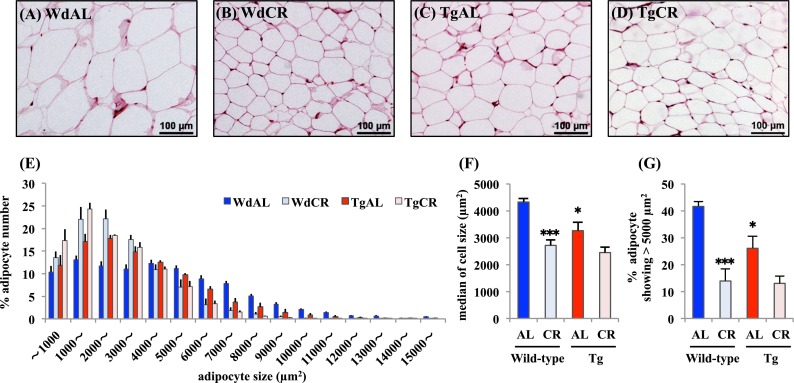
Effects of CR and Tg on morphologic features of white adipose tissue. Representative hematoxylin/eosin-stained histological sections of WAT from WdAL (a), WdCR (b), TgAL (c), and TgCR (d) rats (magnification ×40; scale bar 100 μm). e Distribution of adipocyte sizes in wild-type and tg rats. f Median of adipocyte sizes in wild-type and Tg rats. g The percentage of adipocytes showing >5,000 μm2 in wild-type and Tg rats. Adipocyte size was calculated based on a quantitative morphometric method using ImageJ 1.43u/Java1.6.0_22 software. Error bars are SEM for each group (n = 4). Approximately 1,000–2,100 adipocytes were counted per rat. *P < 0.05 and ***P < 0.001 vs. WdAL (Tukey’s t test)
Based on the DNA microarray data of WdAL, WdCR, and TgAL, using high-density oligonucleotide microarrays, 6,641 genes were positive based on two-way ANOVA (P < 0.001). These genes were applied to PC analysis. The PC scores for each group and their gene expressions are shown in a scatter plot (Fig. 2a). The PC1 and PC2 axes almost coincided with the effects of CR (WdAL vs. WdCR) and Tg (WdAL vs. TgAL), respectively. CR was more effective than Tg because the gene expression plots were more widely distributed along the PC1 axis than the PC2 axis. Indeed, 199 and 226 genes showing PC1 >0.16 and PC1 < –0.16 were upregulated and downregulated by CR, respectively. In contrast, only 65 and 118 genes showing PC2 >0.1 and PC2 < –0.1 were upregulated and downregulated by Tg, respectively (Table 2). Binomial tests were performed using the selected genes to compare the frequencies of genes matching specific categories or keywords in the GO Biological Process database; genes with P < 0.001 are presented in Table 2. Among the 199 genes with PC1 >0.16, several were involved in metabolic processes, particularly lipid biosynthesis (GO 0008610, 0006633, and 0019432). Among the 226 genes with PC1 < –0.16, several genes were related to inflammation (GO 0006955, 0006954, 0034097, and 0030593).
Fig. 2.
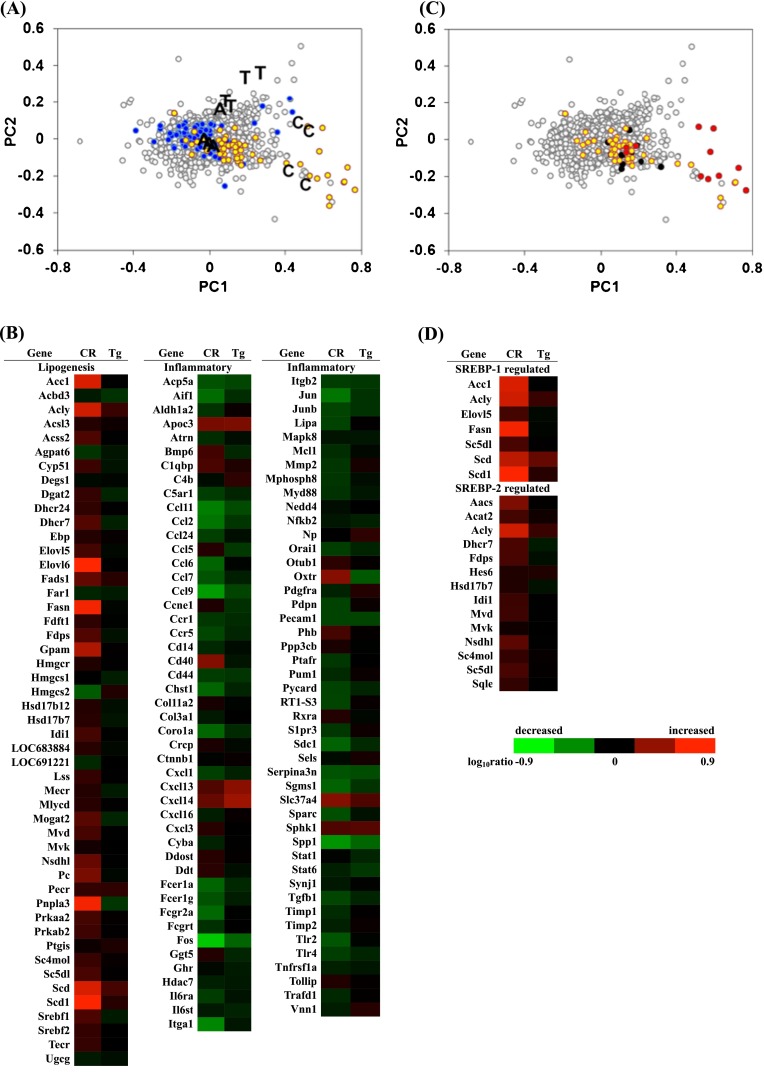
Principal component analysis of gene expression. a PC ordination of 6,641 ANOVA-positive genes based on the DNA microarray data of WdAL, WdCR, and TgAL (four biological repeats per group). A, C, and T represent WdAL, WdCR, and TgAL rats, respectively. The yellow and blue dots represent genes involved in lipid biosynthesis and inflammation, respectively. Gene function was defined based on annotations in the Gene Ontology (GO) Biological Process database (release 31) provided by the manufacturer. Genes involved in lipid biosynthesis were derived from the gene lists (GO: 0008610, 0006633, and 0019432). Genes involved in inflammation were derived from the gene lists (GO: 0006955, 0006954, 0034097, and 0030593). b Heat map of genes involved in lipid biosynthesis and inflammation. Genes involved in lipid biosynthesis were mostly upregulated by CR, while those involved in inflammation were predominantly downregulated by CR. The expression of these genes was not influenced by Tg. c Scatter plots of SREBP-1- (red) and SREBP-2- (black) regulated genes listed by Horton et al. (2003). All of the genes were included in the genes listed in GO 0008610, 0006633, and 0019432 (yellow). d Heat map of SREBP-1- and SREBP-2-regulated genes identified by Horton et al. (2003). Most of the SREBP-1-regulated genes were upregulated by CR, while most of the SREBP-2-regulated genes were not affected by CR
Table 2.
List of ratios and P values of the selected GO terms
| Gene ontology | Biological process | Ratio | P values |
|---|---|---|---|
| PC1 > 0 | 199 genes selected | ||
| 0008152 | Metabolic process | 37/756 | 7.9E-15 |
| 0055114 | Oxidation reduction | 23/591 | 8.2E-12 |
| 0008610 | Lipid biosynthetic process | 16/92 | 5.0E-14 |
| 0006629 | Lipid metabolic process | 13/232 | 5.2E-09 |
| 0006633 | Fatty acid biosynthetic process | 12/59 | 1.2E-14 |
| 0005975 | Carbohydrate metabolic process | 10/206 | 1.1E-06 |
| 0014070 | Response to organic cyclic substance | 9/274 | 8.1E-05 |
| 0008283 | Cell proliferation | 8/190 | 3.7E-05 |
| 0050873 | Brown fat cell differentiation | 7/34 | 3.5E-09 |
| 0006096 | Glycolysis | 7/45 | 2.3E-08 |
| 0016477 | Cell migration | 7/112 | 9.6E-06 |
| 0045471 | Response to ethanol | 7/153 | 6.8E-05 |
| 0010033 | Response to organic substance | 7/163 | 1.0E-04 |
| 0006090 | Pyruvate metabolic process | 6/21 | 6.9E-09 |
| 0009749 | Response to glucose stimulus | 6/118 | 1.3E-04 |
| 0006084 | Acetyl-CoA metabolic process | 5/11 | 1.3E-08 |
| 0019432 | Triglyceride biosynthetic process | 5/13 | 3.0E-08 |
| 0006641 | Triglyceride metabolic process | 5/40 | 7.1E-06 |
| 0005977 | Glycogen metabolic process | 5/42 | 9.0E-06 |
| 0006086 | Acetyl-CoA biosynthetic process from pyruvate | 4/7 | 1.6E-07 |
| 0009267 | Cellular response to starvation | 4/23 | 1.7E-05 |
| 0016126 | Sterol biosynthetic process | 4/28 | 3.6E-05 |
| 0006695 | Cholesterol biosynthetic process | 4/32 | 6.1E-05 |
| 0033574 | Response to testosterone stimulus | 4/41 | 1.6E-04 |
| 0009058 | Biosynthetic process | 4/58 | 5.8E-04 |
| 0042593 | Glucose homeostasis | 4/62 | 7.4E-04 |
| 0007595 | Lactation | 4/65 | 8.8E-04 |
| PC1 < 0 | 226 genes selected | ||
| 0006955 | Immune response | 10/243 | 1.5E-05 |
| 0043066 | Negative regulation of apoptosis | 9/284 | 2.7E-04 |
| 0006954 | Inflammatory response | 7/170 | 2.8E-04 |
| 0030509 | BMP signaling pathway | 6/65 | 9.8E-06 |
| 0034097 | Response to cytokine stimulus | 6/119 | 2.7E-04 |
| 0030900 | Forebrain development | 5/94 | 6.9E-04 |
| 0009612 | Response to mechanical stimulus | 5/102 | 9.9E-04 |
| 0016525 | Negative regulation of angiogenesis | 4/29 | 6.8E-05 |
| 0030593 | Neutrophil chemotaxis | 4/35 | 1.4E-04 |
| PC2 > 0 | 65 genes selected | ||
| No annotation was significantly marked | |||
| PC2 < 0 | 118 genes selected | ||
| 0051384 | Response to glucocorticoid stimulus | 5/163 | 4.3E-04 |
| 0048545 | Response to steroid hormone stimulus | 4/64 | 1.1E-04 |
| 0006006 | Glucose metabolic process | 4/79 | 2.6E-04 |
To better define the effects of CR and Tg, the selected genes are indicated with yellow dots (GO 0008610, 0006633, and 0019432) and blue dots (GO 0006955, 0006954, 0034097, and 0030593) in the scatter plot (Fig. 2a), and are shown in a heat map (Fig. 2b). CR enhanced the expression of several genes involved in lipid biosynthesis (GO 0008610, 0006633, and 0019432) and suppressed the expression of genes involved in inflammation (GO 0006955, 0006954, 0034097, and 0030593). In contrast, Tg did not affect the expression of most of these genes. The effects of Tg appeared in PC2 as the groups located next to the axis; however, very few genes had large scores (Fig. 2a). In fact, we found no biological processes that were only affected by Tg (Table 2).
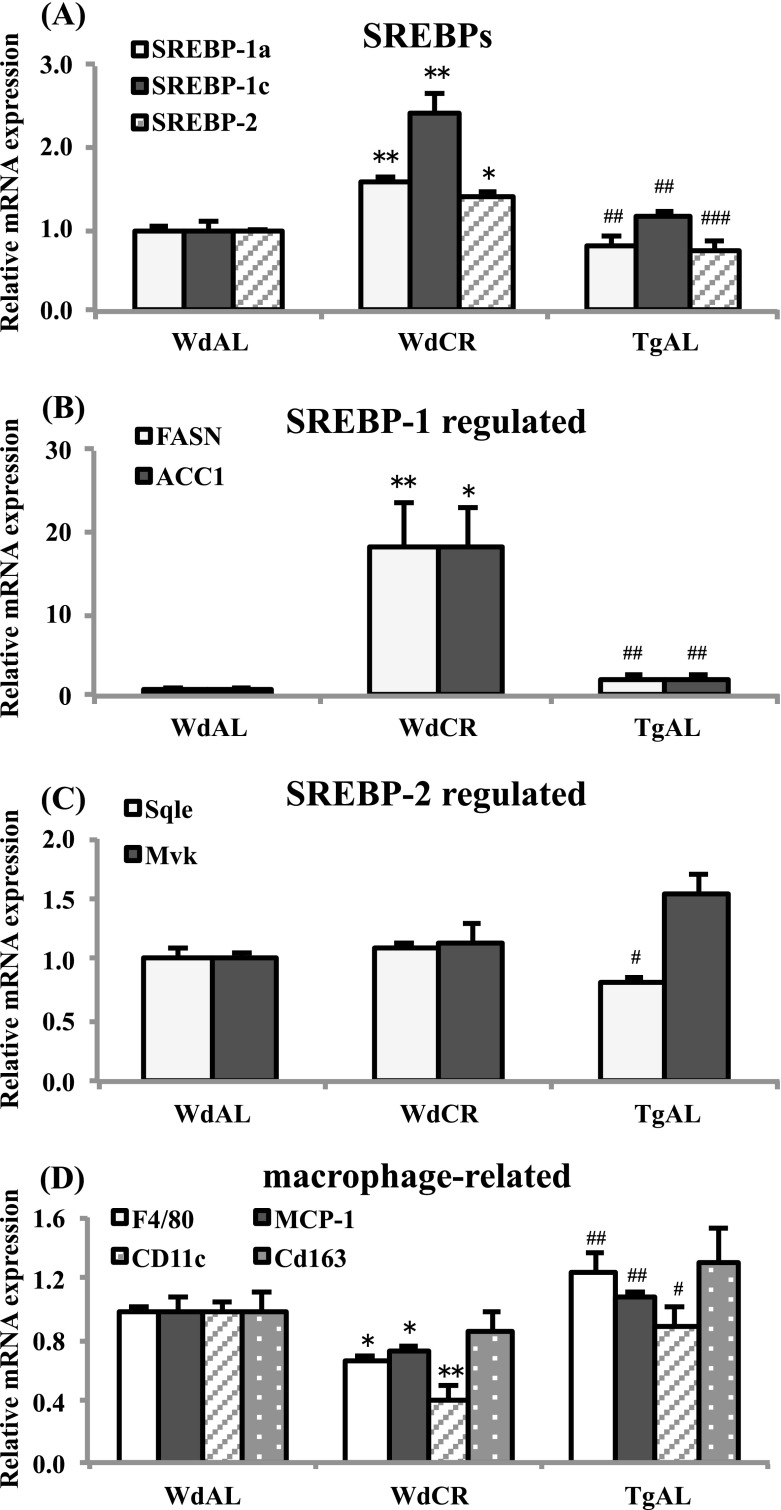
SREBPs are master transcriptional regulators of lipid metabolism and adipocyte differentiation. Three SREBP isoforms, 1a, 1c, and 2 have been identified (Osborne 2000; Osborne and Espenshade 2009). All three isoforms are synthesized as long inactive precursors, and SREBP cleavage-activating protein (SCAP) is required to convert these inactive precursors to transcriptionally active forms (Osborne 2000; Osborne and Espenshade 2009). Horton et al. identified and listed the genes regulated by SREBP-1 and SREBP-2 in vivo using transcriptome analysis of the liver of SREBP-1a transgenic, SREBP-2 transgenic, and SCAP knockout mice (Horton et al. 2003). We compared our data with theirs, and the SREBP-1- and SREBP-2-regulated genes are shown in a scatter plot with red and black dots, respectively (Fig. 2c). The SREBP-1- and SREBP-2-regulated genes identified by Horton et al. were observed in our heat map (Fig. 2d). SREBP-1-regulated genes were exclusively upregulated by CR, whereas SREBP-2-regulated genes were not, except for ATP-citrate lyase (Acly), which was also upregulated by SREBP-1 (Fig. 2c and d). Next, we examined the expression levels of SREBP-1a, 1c, and 2, and the genes regulated by SREBP-1 and SREBP-2 using real-time RT-PCR. We found that the expression of SREBPs was increased in WdCR, but not in TgAL, compared with WdAL. Among the SREBPs, it appears that CR had the strongest effect on SREBP-1c expression, followed by SREBP-1a, and the weakest effect on SREBP-2 expression (Fig. 3a). The SREBP-1-regulated genes, FASN and ACC1, were upregulated in WdCR, but not in TgAL (Fig. 3b). In contrast, the SREBP-2-regulated genes, Sqle and Mvk, were not significantly upregulated in either WdCR or TgAL (Fig. 3c).
Fig. 3.
Quantitative analysis of the mRNA expression of SREBPs, SREBP-1-regulated genes, SREBP-2-regulated genes, and macrophage-related genes in WdAL, WdCR, and TgAL rats. The mRNA expression levels in WAT of SREBPs (A: SREBP-1a, SREBP-1c, and SREBP-2), SREBP-1-regulated genes [B: fatty acid synthase (FASN) and acyl CoA carboxylase 1 (ACC1)], SREBP-2-regulated genes [C: squalene epoxidase (Sqle) and mevalonate kinase (Mvk)], and macrophage-related genes [D monocyte chemotactic protein-1 (MCP-1), F4/80, integrin alpha X (Itgax, CD11c), and CD163] were analyzed by real-time RT-PCR. Data were normalized against TBP expression. Values are means ± SEM (n = 3–6). *P < 0.05, **P < 0.01, and ***P < 0.001 vs. WdAL; #P < 0.05, ##P < 0.01, and ###P < 0.001 vs. WdCR (Tukey’s t test)
It is well-known that macrophages play a key role in inflammation in WAT of obese animals (Ouchi et al. 2011). An increased expression of MCP-1 [also known as chemokine (C-C motif) ligand 2] in WAT contributes to macrophage infiltration into WAT in obese mice (Kanda et al. 2006). Moreover, obesity leads to a shift from M2 (alternatively activated) macrophages to M1 (classically activated) macrophages in WAT (Lumeng et al. 2007). Therefore, we examined the expression of genes encoding the macrophage-specific transmembrane proteins, F4/80, MCP-1, the M1 macrophage-specific marker CD11c (also known as integrin alpha X), and the M2 macrophage-specific marker CD163 (Kawanishi et al. 2010). As expected, the expression of F4/80, MCP-1, and CD11c was downregulated in WdCR, but not in TgAL, compared with WdAL. In contrast, CR and Tg did not significantly affect the expression of CD163 (Fig. 3d).
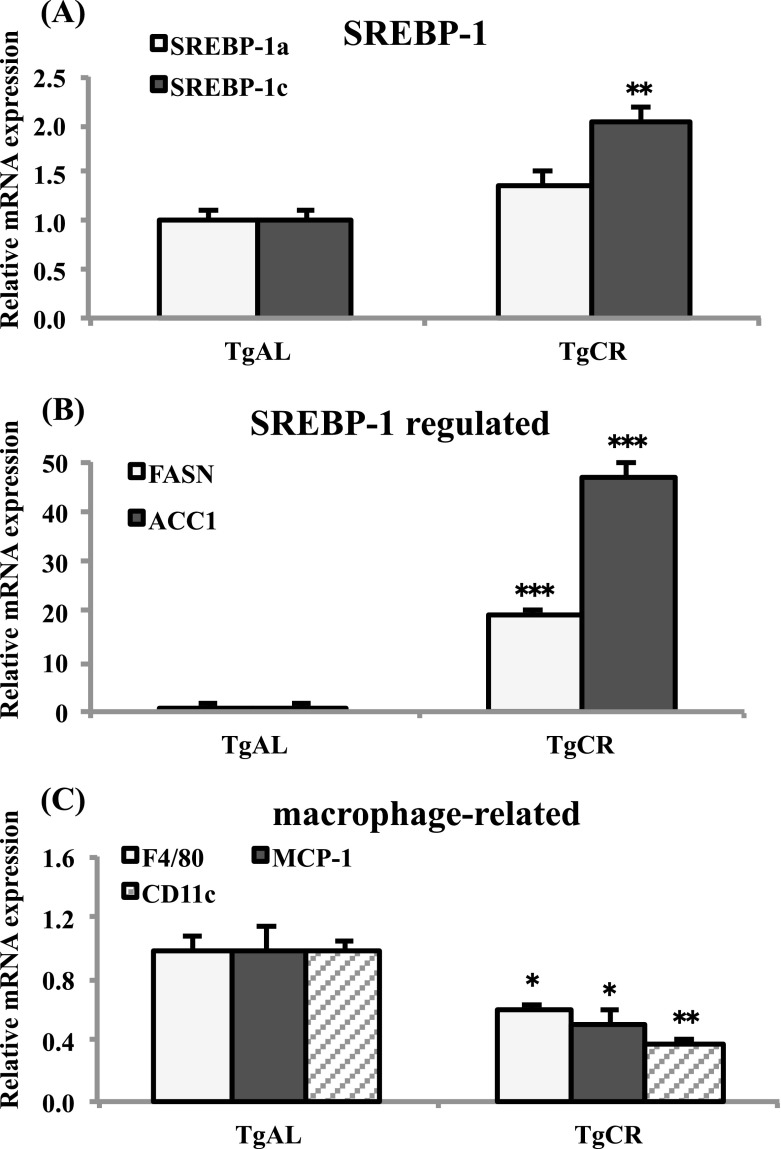
To confirm the CR-associated SREBP-1- and macrophage-related gene expression profiles observed in wild-type rats, we analyzed the effect of CR on Tg rats. Similarly to the wild-type rats, CR markedly upregulated the expression of SREBP-1c, and slightly increased the expression of SREBP-1a (P = 0.08; Fig. 4a). CR also markedly enhanced the expression of the SREBP-1-regulated genes, FASN and ACC1 (Fig. 4b), while it significantly downregulated the expression of the macrophage-involved genes, F4/80, MCP-1, and CD11c in Tg rats (Fig. 4c).
Fig. 4.
Quantitative analysis of the mRNA expression of SREBPs, SREBP-1-regulated genes, and macrophage-related genes in TgAL and TgCR rats. The mRNA expression levels in WAT of SREBPs (aSREBP-1a and SREBP-1c), SREBP-1-regulated genes [bfatty acid synthase (FASN) and acyl CoA carboxylase 1 (ACC1)], and macrophage-related genes [cF4/80, monocyte chemotactic protein-1 (MCP-1), integrin alpha X (Itgax, CD11c)] were analyzed by real-time RT-PCR. Data were normalized against TBP expression. Values are means ± SEM (n = 4–5). *P < 0.05, **P < 0.01, and ***P < 0.001 vs. TgAL (Tukey’s t test)
Discussion
In the current study, we found that CR markedly reduced the size of adipocytes in WAT of wild-type rats (Fig. 1a, b, f, and g), but this CR effect was blunted in Tg rats (Fig. 1c, d, f, and g). Tg also significantly reduced adipocyte size, but this effect of Tg was less than that of CR (Fig. 1a, b, c, f, and g). When looking at adiposity (WAT weight as a percentage of body weight), the effect of CR was predominantly found in wild-type rats rather than Tg rats. It is well-known that small adipocytes secrete more adiponectin and less pro-inflammatory cytokines, including TNF-α and leptin, and are generally more sensitive to insulin (Ouchi et al. 2011). Moreover, small adipocytes act as powerful buffers by absorbing lipids in the postprandial period. If this buffering action is impaired, lipids in the form of TG accumulate in non-adipose tissues, resulting in insulin resistance (Frayn 2002). Recently, it has been reported that inflammatory cells preferentially infiltrate into WAT containing large adipocytes (Ouchi et al. 2011). Therefore, the CR-associated adipokine profile, the inhibition of inflammatory cell infiltration, and the buffering activity in WAT may represent beneficial factors that contribute to the anti-aging and pro-longevity effects of CR. The plasma levels of IGF-1, insulin, adiponectin, and leptin in these rats have been reported elsewhere (Higami et al. 2006b; Yamaza et al. 2007). Briefly, the plasma IGF-1 and insulin concentrations were highest in WdAL rats, followed by WdCR rats, then TgAL, and lowest in TgCR rats, suggesting that plasma insulin concentrations correlate with plasma IGF-1 levels. In contrast, the levels of insulin and IGF-1 did not correlate with adipocyte size and adiposity. Therefore, adipocyte size and adiposity are not simply regulated in a GH–IGF-1- and/or insulin-dependent manner. Plasma adiponectin concentrations were higher in WdCR and TgAL rats than in WdAL rats. Plasma leptin concentrations were highest in WdAL rats, followed by TgAL rats, and lowest in WdCR rats. Because the plasma IGF-1 concentrations did not correlate with the plasma adiponectin or leptin concentrations among the three groups, it is likely that these parameters do not depend on IGF-1. However, continuous infusion of recombinant IGF-1 suppressed the CR-associated increase in plasma adiponectin levels (Yamaza et al. 2007). Moreover, CR and Tg equally enhanced glucose tolerance and insulin sensitivity (Yamaza et al. 2004). Therefore, the CR-associated adipokine profile, including adiponectin and leptin, and insulin sensitivity may be regulated, in part, in a GH–IGF-1-dependent manner.
Our transcriptome analysis combined with PC analysis revealed that CR upregulated several genes involved in lipid biosynthesis (GO 0008610, 0006633, and 0019432) and downregulated several genes associated with inflammation (GO 0006955, 0006954, 0034097, and 0030593; Fig. 2a and b and Table 2). These findings support our previous transcriptome analysis in mice (Higami et al. 2004, 2006a). We also found that CR was more effective than Tg in modulating these genes. Our data also demonstrated that the CR-associated changes in the expression of genes involved in lipid biosynthesis and inflammation occurred in a GH–IGF-1-independent manner (Fig. 2a and b).
Previously, we reported that in the liver of both wild-type and Tg rats, with the same background used in the present study, CR induced the expression of genes involved in fatty acid biosynthesis, probably via SREBP-1 (Higami et al. 2006b). SREBPs, transcription factors belonging to the basic helix-loop-helix-leucine zipper family, are master regulators of lipid metabolism and adipocyte differentiation. The three SREBP isoforms (1a, 1c, and 2) are expressed at varying levels in different tissues and act as homo- and hetero-dimers to activate gene expression (Osborne 2000; Osborne and Espenshade 2009). All three SREBPs are synthesized as long inactive precursors. They are bound to membranes of the endoplasmic reticulum (ER), where the C-terminal regulatory domain of SREBPs interacts with SCAP (Osborne and Espenshade 2009). SCAP escorts SREBPs from the ER to the Golgi apparatus, where they are cleaved sequentially by site-1 and site-2 proteases, to yield the active proteins. The active SREBPs consist of an NH2-terminal domain and can enter the nucleus to activate transcription (Osborne and Espenshade 2009). SREBP-1a and SREBP-1c are encoded by a single gene and are transcribed by alternate promoters. They stimulate the expression of genes that are preferentially involved in fatty acid and triglyceride biosynthesis. In contrast, SREBP-2 is encoded by a different gene and induces the expression of genes predominantly involved in cholesterol biosynthesis (Osborne 2000; Osborne and Espenshade 2009). When we compared our data with the SREBP-1- and SREBP-2-regulated genes listed by Horton et al. (2003), we found that in wild-type rats SREBP-1-regulated genes were exclusively upregulated by CR, whereas SREBP-2-regulated genes were not (Fig. 2c and d). Subsequent real-time RT-PCR analysis confirmed that CR, but not Tg, upregulated the expression of the SREBP-1-regulated genes, FASN and ACC1. In contrast, CR and Tg did not upregulate the SREBP-2-regulated genes, Sqle and Mvk (Fig. 3). The CR effect on the expression of SREBP-1 and the regulated genes was further confirmed in Tg rats (Fig. 4a and b). Therefore, we concluded that CR preferentially enhances the expression of SREBP-1-regulated genes, but not SREBP-2-regulated genes, in a GH–IGF-1-independent manner.
As described above, inflammatory cells, particularly macrophages, preferentially infiltrate into the WAT of obese animals (Ouchi et al. 2011). The majority of previous studies have examined F4/80 expression as a marker to identify adipose tissue-specific macrophages (Lumeng et al. 2007). MCP-1, which shows increased expression in WAT of obese animals, promotes adipose tissue inflammation and macrophage recruitment (Kanda et al. 2006). Therefore, we measured the expression of genes encoding F4/80 and MCP-1 in wild-type and Tg rats (Figs. 3d and 4c). We found that CR, but not Tg, reduced the expressions of F4/80 and MCP-1, suggesting that CR suppresses macrophage infiltration by decreasing the expression of MCP-1 in a GH–IGF-1-independent manner. Lumeng et al. (2007) reported that the F4/80+CD11c+ population of macrophages, a characteristic of M1 macrophages, was present in WAT of obese mice, but not in lean mice. In contrast, the macrophages in WAT of lean mice expressed many genes characteristic of M2 macrophages. Thus, obesity leads to a shift in the activation state of macrophages in WAT from an M2-polarized state in lean animals that may protect adipocytes from inflammation to an M1 pro-inflammatory state that contributes to insulin resistance (Lumeng et al. 2007). Unlike WAT in obese animals, our data suggest that the reduced infiltration of macrophages is mainly due to the decreased shift towards F4/80+CD11c+ M1 macrophages in WAT of both wild-type and Tg CR rats.
In this study, we demonstrated that CR-associated morphological changes preferentially found in WAT of wild-type rats rather than Tg rats are not solely regulated in a GH–IGF-1-dependent manner. Moreover, CR-enhanced lipid biosynthesis and CR-suppressed inflammation were only regulated in a GH–IGF-1-independent manner. In particular, CR-associated activation of lipid biosynthesis was predominantly regulated by SREBP-1. Recently, it has been hypothesized that CR-associated lipid utilization may reduce reactive oxygen species production (Guarente 2008). In fact, SREBP-1-regulated de novo lipid biosynthesis in WAT may play an important role in CR-associated lipid utilization (Okita et al. 2012). In addition, it has been reported that molecular inflammation, which is associated with nuclear factor-κB activation and enhanced expression of several pro-inflammatory cytokines, is involved in the aging process and is attenuated by CR (Chung et al. 2011). Therefore, we suggest that the activation of de novo lipid biosynthesis by SREBP-1 and the induction of anti-inflammatory conditions by reduced macrophage infiltration are pivotal regulators of CR-associated WAT remodeling, and may be important factors in the beneficial effects of CR.
Electronic supplementary material
(TXT 7 kb)
(JPEG 368 kb)
(JPEG 374 kb)
Acknowledgments
We thank Yutaka Araki and Yuko Moriyama (Department of Investigative Pathology, Nagasaki University Graduate School for Biomedical Sciences) for their technical assistance and cooperation. This work was supported by a Grant-in-Aid for Scientific Research (C) from the Japan Society for the Promotion of Science (no. 19590396).
Footnotes
Yoshikazu Chujo, Namiki Fujii, Naoyuki Okita, and Tomokazu Konishi contributed equally to this work.
References
- Al-Regaiey KA, Masternak MM, Bonkowski M, Sun L, Bartke A. Long-lived growth hormone receptor knockout mice: interaction of reduced insulin-like growth factor I/insulin signaling and caloric restriction. Endocrinology. 2005;146:851–860. doi: 10.1210/en.2004-1120. [DOI] [PubMed] [Google Scholar]
- Anderson R, Prolla T (2009) PGC-1alpha in aging and anti-aging interventions. Biochem Biophys Acta 1790:1059–1066 [DOI] [PMC free article] [PubMed]
- Argmann C, Dobrin R, Heikkinen S, Auburtin A, Pouilly L, Cock TA, Koutnikova H, Zhu J, Schadt EE, Auwerx J. Ppargamma2 is a key driver of longevity in the mouse. PLoS Genet. 2009;5:e1000752. doi: 10.1371/journal.pgen.1000752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartke A. Minireview: role of the growth hormone/insulin-like growth factor system in mammalian aging. Endocrinology. 2005;146:3718–3723. doi: 10.1210/en.2005-0411. [DOI] [PubMed] [Google Scholar]
- Bartke A, Wright JC, Mattison JA, Ingram DK, Miller RA, Roth GS. Extending the lifespan of long-lived mice. Nature. 2001;414:412. doi: 10.1038/35106646. [DOI] [PubMed] [Google Scholar]
- Barzilai N, Banerjee S, Hawkins M, Chen W, Rossetti L. Caloric restriction reverses hepatic insulin resistance in aging rats by decreasing visceral fat. J Clin Invest. 1998;101:1353–1361. doi: 10.1172/JCI485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blüher M, Michael MD, Peroni OD, Ueki K, Carter N, Kahn BB, Kahn CR. Adipose tissue selective insulin receptor knockout protects against obesity and obesity-related glucose intolerance. Dev Cell. 2002;3:25–38. doi: 10.1016/S1534-5807(02)00199-5. [DOI] [PubMed] [Google Scholar]
- Blüher M, Kahn BB, Kahn CR. Extended longevity in mice lacking the insulin receptor in adipose tissue. Science. 2003;299:572–574. doi: 10.1126/science.1078223. [DOI] [PubMed] [Google Scholar]
- Bonkowski MS, Rocha JS, Masternak MM, Al Regaiey KA, Bartke A. Targeted disruption of growth hormone receptor interferes with the beneficial actions of calorie restriction. Proc Natl Acad Sci U S A. 2006;103:7901–7905. doi: 10.1073/pnas.0600161103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown-Borg HM, Borg KE, Mellska CJ, Bartke A. Dwarf mice and the ageing process. Nature. 1996;384:33. doi: 10.1038/384033a0. [DOI] [PubMed] [Google Scholar]
- Chiu CH, Lin WD, Huang SY, Lee YH. Effect of a C/EBP gene replacement on mitochondrial biogenesis in fat cells. Genes Dev. 2004;18:1970–1975. doi: 10.1101/gad.1213104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung HY, Lee EK, Choi YJ, Kim JM, Kim DH, Zou Y, Kim CH, Lee J, Kim HS, Kim ND, Jung JH, Yu BP. Molecular inflammation as an underlying mechanism of the aging process and age-related diseases. J Dent Res. 2011;90:830–840. doi: 10.1177/0022034510387794. [DOI] [PubMed] [Google Scholar]
- Coschigano KT, Clemmons D, Bellush LL, Kopchick JJ. Assessment of growth parameters and life span of GHR/BP gene-disrupted mice. Endocrinology. 2000;141:2608–2613. doi: 10.1210/en.141.7.2608. [DOI] [PubMed] [Google Scholar]
- Farmer SR. Transcriptional control of adipocyte formation. Cell Metab. 2006;4:263–273. doi: 10.1016/j.cmet.2006.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flurkey K, Papaconstantinou J, Miller RA, Harrison DE. Lifespan extension and delayed immune and collagen aging in mutant mice with defects in growth hormone production. Proc Natl Acad Sci U S A. 2001;98:6736–6741. doi: 10.1073/pnas.111158898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frayn KN. Adipose tissue as a buffer for daily lipid flux. Diabetologia. 2002;45:1201–1210. doi: 10.1007/s00125-002-0873-y. [DOI] [PubMed] [Google Scholar]
- Gesing A, Masternak MM, Wang F, Joseph AM, Leeuwenburgh C, Westbrook R, Lewinski A, Karbownik-Lewinska M, Bartke A. Expression of key regulators of mitochondrial biogenesis in growth hormone receptor knockout (GHRKO) mice is enhanced but is not further improved by other potential life-extending interventions. J Gerontol A Biol Sci Med Sci. 2011;66:1062–1076. doi: 10.1093/gerona/glr080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L. Mitochondria—a nexus for aging, calorie restriction, and sirtuins? Cell. 2008;132:171–176. doi: 10.1016/j.cell.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock CR, Han DH, Higashida K, Kim SH, Holloszy JO. Does calorie restriction induce mitochondrial biogenesis? A reevaluation. FASEB J. 2011;25:785–791. doi: 10.1096/fj.10-170415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higami Y, Pugh TD, Page GP, Allison DB, Prolla TA, Weindruch R. Adipose tissue energy metabolism: altered gene expression profile of mice subjected to long-term caloric restriction. FASEB J. 2004;18:415–417. doi: 10.1096/fj.03-0678fje. [DOI] [PubMed] [Google Scholar]
- Higami Y, Yamaza H, Shimokawa I. Laboratory findings of caloric restriction in rodents and primates. Adv Clin Chem. 2005;39:211–237. doi: 10.1016/S0065-2423(04)39008-6. [DOI] [PubMed] [Google Scholar]
- Higami Y, Barger JL, Page GP, Allison DB, Smith SR, Prolla TA, Weindruch R. Energy restriction lowers the expression of genes linked to inflammation, the cytoskeleton, the extracellular matrix, and angiogenesis in mouse adipose tissue. J Nutr. 2006;136:343–352. doi: 10.1093/jn/136.2.343. [DOI] [PubMed] [Google Scholar]
- Higami Y, Tsuchiya T, Chiba T, Yamaza H, Muraoka I, Hirose M, Komatsu T, Shimokawa I. Hepatic gene expression profile of lipid metabolism in rats: impact of caloric restriction and growth hormone/insulin-like growth factor-1 suppression. J Gerontol A Biol Sci Med Sci. 2006;61:1099–1110. doi: 10.1093/gerona/61.11.1099. [DOI] [PubMed] [Google Scholar]
- Holzenberger M, Dupont J, Ducos B, Leneuve P, Géloën A, Even PC, Cervera P, Le Bouc Y. IGF-1 receptor regulates lifespan and resistance to oxidative stress in mice. Nature. 2003;421:182–187. doi: 10.1038/nature01298. [DOI] [PubMed] [Google Scholar]
- Horton JD, Shah NA, Warrington JA, Anderson NN, Park SW, Brown MS, Goldstein JL. Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc Natl Acad Sci U S A. 2003;100:12027–12032. doi: 10.1073/pnas.1534923100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson JE. A user’s guide to principal components. New York: Wiley; 2005. [Google Scholar]
- Kanda H, Tateya S, Tamori Y, Kotani K, Hiasa K, Kitazawa R, Kitazawa S, Miyachi H, Maeda S, Egashira K, Kasuga M. MCP-1 contributes to macrophage infiltration into adipose tissue, insulin resistance, and hepatic steatosis in obesity. J Clin Invest. 2006;116:1494–1505. doi: 10.1172/JCI26498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawanishi N, Yano H, Yokogawa Y, Suzuki K. Exercise training inhibits inflammation in adipose tissue via both suppression of macrophage infiltration and acceleration of phenotypic switching from M1 to M2 macrophages in high-fat-diet-induced obese mice. Exerc Immunol Rev. 2010;16:105–118. [PubMed] [Google Scholar]
- Konishi T, Konishi F, Takasaki S, Inoue K, Nakayama K, Konagaya A. Coincidence between transcriptome analyses on different microarray platforms using a parametric framework. PLoS One. 2008;3:e3555. doi: 10.1371/journal.pone.0003555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo H-Y, Miyashita M, Cho BH, Nakamura MT. Replacing dietary glucose with fructose increases ChREBP activity and SREBP-1 protein in rat liver nucleus. Biochem Biophys Res Commun. 2009;390:285–289. doi: 10.1016/j.bbrc.2009.09.109. [DOI] [PubMed] [Google Scholar]
- López-Lluch G, Hunt N, Jones B, Zhu M, Jamieson H, Hilmer S, Cascajo MV, Allard J, Ingram DK, Navas P, de Cabo R (2006) Calorie restriction induces mitochondrial biogenesis and bioenergetic efficiency. Proc Natl Acad Sci USA 103:1768–1773 [DOI] [PMC free article] [PubMed]
- Lumeng CN, Bodzin JL, Saltiel AR. Obesity induces a phenotypic switch in adipose tissue macrophage polarization. J Clin Invest. 2007;117:175–184. doi: 10.1172/JCI29881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masoro EJ. Overview of caloric restriction and ageing. Mech Ageing Dev. 2005;126:913–922. doi: 10.1016/j.mad.2005.03.012. [DOI] [PubMed] [Google Scholar]
- Masternak MM, Bartke A, Wang F, Spong A, Gesing A, Fang Y, Salmon AB, Hughes LF, Liberati T, Boparai R, Kopchick JJ, Westbrook R. Metabolic effects of intra-abdominal fat in GHRKO mice. Aging Cell. 2012;11:73–81. doi: 10.1111/j.1474-9726.2011.00763.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nisoli E, Tonello C, Cardile A, Cozzi V, Bracale R, Tedesco L, Falcone S, Valerio A, Cantoni O, Clementi E, Moncada S, Carruba MO. Calorie restriction promotes mitochondrial biogenesis by inducing the expression of eNOS. Science. 2005;310:314–317. doi: 10.1126/science.1117728. [DOI] [PubMed] [Google Scholar]
- Okita N, Hayashida Y, Kojima Y, Fukushima M, Yuguchi K, Mikami K, Yamauchi A, Watanabe K, Noguchi M, Nakamura M, Toda T, Higami Y. Differential responses of white adipose tissue and brown adipose tissue to caloric restriction in rats. Mech Ageing Dev. 2012;133:255–266. doi: 10.1016/j.mad.2012.02.003. [DOI] [PubMed] [Google Scholar]
- Osborne TF. Sterol regulatory element-binding proteins (SREBPs): key regulators of nutritional homeostasis and insulin action. J Biol Chem. 2000;275:32379–32382. doi: 10.1074/jbc.R000017200. [DOI] [PubMed] [Google Scholar]
- Osborne TF, Espenshade PJ. Evolutionary conservation and adaptation in the mechanism that regulates SREBP action: what a long, strange tRIP it’s been. Genes Dev. 2009;23:2578–2591. doi: 10.1101/gad.1854309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otabe S, Yuan X, Fukutani T, Wada N, Hashinaga T, Nakayama H, Hirota N, Kojima M, Yamada K. Overexpression of human adiponectin in transgenic mice results in suppression of fat accumulation and prevention of premature death by high-calorie diet. Am J Physiol Endocrinol Metab. 2007;293:E210–E218. doi: 10.1152/ajpendo.00645.2006. [DOI] [PubMed] [Google Scholar]
- Ouchi N, Parker JL, Lugus JJ, Walsh K. Adipokines in inflammation and metabolic disease. Nat Rev Immunol. 2011;11:85–97. doi: 10.1038/nri2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saely CH, Geiger K, Drexel H. Brown versus white adipose tissue: a mini-review. Gerontology. 2012;58:15–23. doi: 10.1159/000321319. [DOI] [PubMed] [Google Scholar]
- Shi T, Wang F, Stieren E, Tong Q (2005) SIRT3, a mitochondrial sirtuin deacetylase, regulates mitochondrial function and thermogenesis in brown adipocytes. J Biol Chem 280:13560–13567 [DOI] [PubMed]
- Shimokawa I, Higami Y, Utsuyama M, Tuchiya T, Komatsu T, Chiba T, Yamaza H. Life span extension by reduction in growth hormone-insulin-like growth factor-1 axis in a transgenic rat model. Am J Pathol. 2002;160:2259–2265. doi: 10.1016/S0002-9440(10)61173-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimokawa I, Higami Y, Tsuchiya T, Otani H, Komatsu T, Chiba T, Yamaza H. Life span extension by reduction of the growth hormone-insulin-like growth factor-1 axis: relation to caloric restriction. FASEB J. 2003;17:1108–1109. doi: 10.1096/fj.02-0819fje. [DOI] [PubMed] [Google Scholar]
- Sinclair DA. Toward a unified theory of caloric restriction and longevity regulation. Mech Ageing Dev. 2005;126:987–1002. doi: 10.1016/j.mad.2005.03.019. [DOI] [PubMed] [Google Scholar]
- Sohal RS, Weindruch R. Oxidative stress, caloric restriction, and aging. Science. 1996;273:59–63. doi: 10.1126/science.273.5271.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Leal FL, Fonseca-Alaniz MH, Rogero MM, Tirapegui J. The role of inflamed adipose tissue in the insulin resistance. Cell Biochem Funct. 2010;28:623–631. doi: 10.1002/cbf.1706. [DOI] [PubMed] [Google Scholar]
- Weindruch R, Walford RL. The retardation of aging and disease by dietary restriction. Springfield: Charles C Thomas; 1988. [Google Scholar]
- Yamaza H, Komatsu T, Chiba T, Toyama H, To K, Higami Y, Shimokawa I. A transgenic dwarf rat model as a tool for the study of calorie restriction and aging. Exp Gerontol. 2004;39:269–272. doi: 10.1016/j.exger.2003.11.001. [DOI] [PubMed] [Google Scholar]
- Yamaza H, Komatsu T, To K, Toyama H, Chiba T, Higami Y, Shimokawa I. Involvement of insulin-like growth factor-1 in the effect of caloric restriction: regulation of plasma adiponectin and leptin. J Gerontol A Biol Sci Med Sci. 2007;62:27–33. doi: 10.1093/gerona/62.1.27. [DOI] [PubMed] [Google Scholar]
- Yang J, Goldstein JL, Hammer RE, Moon YA, Brown MS, Horton JD. Decreased lipid synthesis in livers of mice with disrupted site-1 protease gene. Proc Natl Acad Sci. 2001;98:13607–13612. doi: 10.1073/pnas.201524598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu BP. Modulation of aging processes by dietary restriction. Boca Raton: CRC; 1994. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TXT 7 kb)
(JPEG 368 kb)
(JPEG 374 kb)