Abstract
The three core components of the ubiquitous bacterial chemosensory array – the transmembrane chemoreceptor, the histidine kinase CheA and the adaptor protein CheW – assemble to form a membrane-bound, hexagonal lattice in which receptor transmembrane signals regulate kinase activity. Both the regulatory domain of the kinase and the adaptor protein bind to overlapping sites on the cytoplasmic tip of the receptor (termed the protein interaction region). Notably, the kinase regulatory domain (P5) and the adaptor protein share the same fold constructed of two SH3-like domains. The present study focuses on the structural interface between the receptor and the kinase regulatory domain. Two models have been proposed for this interface: Model 1 is based on the crystal structure of a homologous Thermotoga complex between a receptor fragment and the CheW adaptor protein. This model has been used in current models of chemosensory array architecture to build the receptor-CheA kinase interface. Model 2 is based on a newly determined crystal structure of a homologous Thermotoga complex between a receptor fragment and the CheA kinase regulatory domain. Both models present unique strengths and weaknesses, and current evidence is unable to resolve which model best describes contacts in the native chemosensory arrays of Escherichia coli, Salmonella typhimurium and other bacteria. Here we employ disulfide mapping and TAM-IDS (Tryptophan and Alanine Mutation to Identify Docking Sites) to test Models 1 and 2 in well-characterized membrane-bound arrays formed from E. coli and S. typhimurium components. The results reveal that the native array interface between the receptor protein interaction region and the kinase regulatory domain is accurately described by Model 2, but not by Model 1. In addition, the results show that the interface possesses both a structural function that contributes to stable CheA kinase binding in the array, and a regulatory function central to transmission of the activation signal from receptor to CheA kinase. On-off switching alters the disulfide formation rates of specific Cys pairs at the interface, but not most Cys pairs, indicating that signaling perturbs localized regions of the interface. The findings suggest a simple model for the rearrangement of the interface triggered by the attractant signal, and for longer range transmission of the signal in the chemosensory array.
Keywords: two-component signaling pathway, chemotaxis, methyl-accepting chemotaxis protein, histidine kinase, Tsr, Tar, CheW, ultrastable
INTRODUCTION
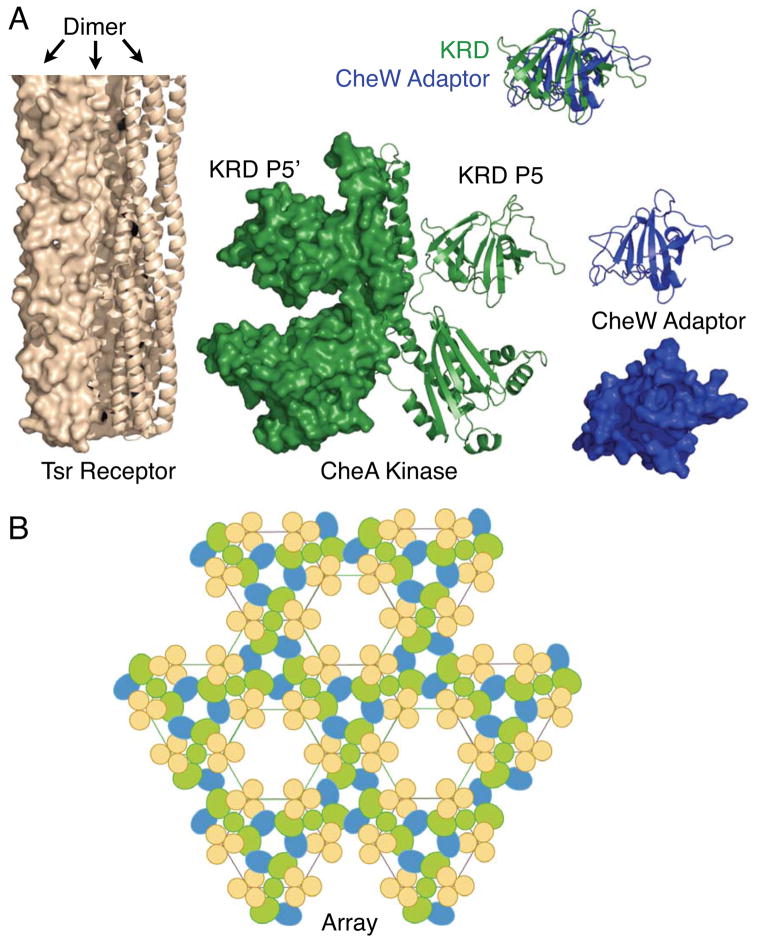
Two-component systems are the most prevalent signaling pathways in bacteria and typically consist of a receptor-linked histidine kinase that serves as a sensor and an aspartate kinase that acts as a response regulator (reviewed by 1–4). Bacterial chemotaxis utilizes a two-component pathway to sense chemical gradients and to control the swimming motor, enabling biased swimming up an attractant gradient or down a repellent gradient. In Escherichia coli and Salmonella typhimurium, and in other bacteria studied thus far 5, the three core components of the chemosensory pathway assemble into a hexagonal array composed of transmembrane chemoreceptors, the histidine kinase CheA, and the adaptor protein CheW, as illustrated in Figure 1. The receptor proteins form trimers-of-dimers located at the vertices of the hexagonal array 6–10, and the periplasmic domain of the receptor homodimers detect specific sugars or amino acids and trigger a transmembrane conformational signal 11–13,6,14,15. The CheA kinase and CheW adaptor proteins bind to the cytoplasmic tips of the receptor array where the receptor protein interaction region is located 6,16,8,17,5,18,9,10,19–23. The CheA kinase is a dimer of identical subunits, each possessing five structural and functional domains: the substrate domain (P1), the CheY/B binding domain (P2), the dimerization domain (P3), the catalytic domain (P4), and the regulatory domain (P5) 24–26,17,27. The CheW adaptor protein is a small monomeric protein that, notably, exhibits the same tandem SH3-like fold as the structurally homologous CheA kinase regulatory domain (backbone RSMD of ~2 Å) 25,28. In this shared fold, two distinct SH3-like domains define a central groove. The CheW adaptor protein and CheA kinase regulatory domain also bind to similar or identical sites on the receptor protein interaction region, sharing key contact points on the N-terminal helix of the receptor hairpin, and thus competitively inhibit each other’s binding 29,22,23.
Figure 1.
Core components and schematic model of the core bacterial chemosensory array 17,19,20. A) Receptor trimer-of-dimers (tan, one dimer shown as ribbon, PDB code 1QU7), homodimeric CheA kinase (green, one monomer shown as ribbon, PDB code 1B3Q), and CheW adaptor protein (blue, shown as both ribbon and spacefilling models, PDB code 2CH4) together serve as the three core components of the bacterial chemotaxis array. The CheA kinase regulatory domain (KRD or P5) and the CheW adaptor protein share the same fold constructed of two SH3-like domains, as illustrated by the superpositioning 25,28. All molecular graphics generated in MacPyMol 65. B) The architecture of the chemotactic array is defined by a hexagonal lattice in which one receptor trimer-of-dimers is located at each vertex (same protein colors as (A)). The homodimeric CheA kinase packs between the flatter faces of adjacent trimers and its two CheA kinase regulatory domains contact CheW adaptor proteins in a head-to-tail fashion forming two linked rings 19,20. Each CheA kinase regulatory domain and CheW adaptor proteins binds near the cytoplasmic tip of a receptor trimer-of-dimers 38,39,16,22,23. This basic hexagonal unit is propagated throughout the array, forming a network of linked rings.
The core chemosensory array can be assembled from the receptor, CheA kinase and CheW adaptor core components either in vivo or in vitro. When bacterial membranes containing assembled arrays are isolated, the resulting isolated arrays are functional and ultrastable, exhibiting attractant-triggered, receptor-mediated kinase regulation at 22°C for days or weeks 18,30,21.
The signaling state of the array is determined by both the ligand occupancy of the receptor periplasmic domain and the covalent modification state of the receptor cytoplasmic adaptation sites (reviewed by 14,31,1,3,4). In the apo state, receptors activate the CheA kinase triggering autophosphorylation of a specific histidine on its substrate domain; subsequently, the response regulator CheY catalyzes the transfer of this phosphate to its active site aspartate. Phospho-CheY then diffuses to the flagellar motor where it modulates the bacterial swimming state. Upon attractant binding to the receptor, the receptor transmembrane signal turns off the CheA histidine kinase, thereby leading to a reduced level of phospho-CheY and switching of the flagellar motor to the opposite swimming state. Within tens of seconds, covalent receptor adaptation by methyl esterification of its cytoplasmic adaptation glutamate residues generates an opposing signal that opposes the attractant signal, and leads to restoration of receptor-stimulated histidine kinase activity.
Extensive progress has been made in defining the structures of core array components. Crystal and/or NMR solution structures exist for specific domains of the receptor, CheA kinase, and CheW adaptor individually and in various complexes 32,11,24,13,33,25,6,26,28,34–37,19,22,23. Focusing on the interface between the receptor protein interaction region and the CheA kinase regulatory domain, extensive previous genetic, biochemical, PICM, NMR and crystallographic studies have highlighted the importance of this specific receptor-CheA kinase contact 38,39,6,40,16,8,17,41,9,10,19–23.
Two plausible models have been proposed for the interface between the receptor and the CheA kinase regulatory domain, but current experimental evidence is unable to differentiate between them 19,20,22,23. In both models, the N-terminal α-helix of the receptor hairpin sits in the long groove between the tandem SH3-like domains of the CheA kinase regulatory domain, but the translational and rotational registers of the helix differ significantly between models. Model 1 19 is based on the crystal structure of a Thermotoga complex between a receptor fragment and the tandem SH3-like domain of the CheW adaptor protein. It is plausible that this crystal structure could accurately represent the packing interface between receptor and tandem SH3-like motifs in general, including that of the CheA kinase regulatory domain. Indeed, NMR evidence suggests that the tandem SH3-like motifs of the CheA kinase regulatory domain and the CheW adaptor protein dock to a similar or identical site on the receptor 22,23. Model 1 was used to define the receptor-CheA kinase interface in the 3D model for array packing in E. coli, although the ~30 Å resolution of the cryo-EM data does not allow careful analysis of the protein-protein contacts 19,20. Model 2 is based on the crystal structure of a Thermotoga complex between a receptor fragment and the CheA kinase regulatory domain, presented in the companion manuscript by Li, Crane and coworkers 42. In this structure, the receptor fragment unfolds to form a non-native tertiary structure, although the local receptor-CheA kinase contacts are proposed to be native 42. Overall, both models display relative advantages (Model 1 - correct receptor tertiary structure; Model 2 - correct binding partner) and potential problems (Model 1 – different but homologous binding partner; Model 2 - incorrect receptor tertiary structure). Further studies are needed to determine whether one of these two models, or neither model, provides an accurate picture of the interface between receptor and the CheA kinase regulatory domain in functional chemoreceptor arrays.
The present study uses disulfide mapping and TAM-IDS to test Models 1 and 2 in a proven, bacterial chemosensory array assembled from well-characterized, interchangeable E. coli and S. typhimurium components. The goals are to (i) differentiate between Models 1 and 2 in functional, membrane-bound arrays formed in vitro, (ii) investigate whether a similar interface is present in arrays formed in vivo, and (iii) probe signal transduction through this interface. The results indicate that Model 2 best fits the receptor-CheA kinase interface in the functional, membrane-bound chemosensory array, and show that the interface plays important roles in both array structure and transmission of signals from receptor to CheA kinase.
MATERIALS AND METHODS
Materials
All chemicals were highly pure reagent-grade. Chemicals were obtained from Sigma Aldrich with the following exceptions: [γ-32P]ATP was from Perkin-Elmer, DTT was from Research Products International, Ni-NTA agarose resin was from Qiagen, Bicinchoninic Acid Assay (BCA) reagents were from Bio-rad, PVDF was from Millipore.
Protein Expression and Purification
S. typhimurium Cysless CheA kinase and Cysless CheW adaptor protein possessing 6xHis tags on their N-termini were expressed by plasmids pET6H-CheA and pET6H-CheW, respectively, in E. coli strain BL21(DE3) (Stratagene) 17. E. coli CheY with an N-terminal YFP and C-terminal 6xHis tag was expressed by plasmid pVSYFP-CheY-6H in E. coli strain-plasmid M15-pREP4 (Qiagen) 43. All soluble proteins were isolated as previously described by standard Ni-NTA agarose affinity chromatography 43,17. Protein concentrations were estimated by UV absorption using extinction coefficients calculated from protein sequences (all mM−1cm−1): 16.0 and 5.95 for CheA and CheW, both at 276 nm, and 18.0 for YFP-CheY at the YFP absorbance of 514 nm.
E. coli serine receptor (Tsr) was overexpressed in the gutted E. coli strain UU1581—which lacks all chemotaxis proteins including receptors and adaptation enzymes—using plasmid pJC3 40. Inside out, inner bacterial membrane vesicles containing Tsr were isolated as previously described 32,17. The total protein concentration in the membranes was determined by BCA assay, and the fraction of total protein represented by receptors was determined by ImageJ densitometry of SDS-PAGE gels, then receptor concentration was determined by combining these two values.
The indicated site-directed mutants of the receptor and CheA kinase were generated using the PCR-based QuickChange XLII mutagenesis kit (Agilent). All mutants were confirmed by DNA sequencing the entire coding region.
Reconstitution of Functional Signaling Arrays
Signaling arrays were reconstituted by combining 6.7 μM Tsr receptor, 5 μM CheA kinase, 10 μM CheW adaptor protein in either crosslinking buffer (160 mM NaCl, 6 mM MgCl2, 50 mM Tris, 3 mM EDTA, pH 7.5) or kinase assay buffer (160 mM NaCl, 5 mM MgCl2, 50 mM Tris, 0.5 mM EDTA, pH 7.5) for 1 hr at 22°C in the presence of 0.5 mg/mL BSA, 2 mM TCEP, and 2 mM PMSF. Samples were centrifuged at 21,000 × g for 7 min and pellets were washed twice by resuspending in a 10-fold excess of activity buffer (without BSA, TCEP, and PMSF) and re-pelleting. After the final wash, pellets were resuspended in the original volume of activity buffer resulting in functional, ultrastable arrays lacking unbound CheA kinase and CheW adaptor protein 18,21.
Quantitation of Attractant-Regulated CheA Kinase Activity
The relative kinase activities of arrays were measured as previously described with minor modifications 17. Five μL of washed and resuspended arrays were mixed with 5 μL of YFP-CheY yielding final concentrations of 3.3 μM receptor and 40 μM YFP-CheY, the latter sufficient to ensure that CheA kinase autophosphorylation was the rate determining step and not phosphotransfer from CheA~P to CheY 44. Sensitivity to attractant was measured by addition of 2 mM serine. Kinase reactions were carried out by addition of 1 mM [γ-32P]ATP (4000–8000 cpm/pmol) followed by incubation for 10 sec. Reactions were quenched in 30 μL of 2X Laemmli sample buffer containing 50 mM EDTA. Samples were resolved on denaturing SDS PAGE gels, extensively dried, and the γ-32P –labeled CheY band was imaged using phosphorimaging.
Oxidation Reactions
Washed ultrastable arrays (10 μL) prepared as described above were used for each reaction. Oxidation was initiated upon the addition of 2 μL of CuCl2 to a final concentration of 1, 2, or 5 mM. The reaction was quenched after 10 seconds by the addition of a 12 μL of 2X Laemmli sample buffer containing 50 mM EDTA and 80 mM NEM, incubated at 100°C for 1 min then snap frozen in liquid nitrogen.
Quantitation of Disulfide-Linked CheA Kinase-Receptor Heterodimer
The crosslinking products of CheA kinase were monitored by western blotting using a polyclonal antibody against CheA kinase (kindly provided by Dr. J. Stock). Briefly, dilute samples were resolved on 7.5% or 10% SDS PAGE gels and transferred to PVDF membranes. Membranes were blocked overnight in Tris-buffered saline Tween-20 (50 mM Tris, 160 mM NaCl, 0.1% Tween-20, TBST) with 3% dry milk powder before blotting with anti-CheA kinase primary and anti-rabbit, alkaline phosphatase conjugated secondary (Sigma). Extensive washing with TBST was performed after incubations. Washed membranes were incubated with ECF substrate and dried extensively before scanning with a Typhoon 9400 scanner. Blots were quantified by densitometry using ImageJ. The fraction CheA kinase-Receptor S-S heterodimer is defined as the density of the heterodimer divided by the sum of all CheA kinase densities (monomer, CheA kinase-CheA kinase S-S homodimer, and CheA kinase-Receptor S-S heterodimer).
TAM-IDS
To apply TAM-IDS in vivo, plasmid pDK28 expressing full length E. coli YFP-CheA kinase fusion (kindly provided by Dr. V. Sourjik) 45 was used to construct plasmids pSF2 and pSF5 that express the N-terminal YFP fusion with E. coli CheA CheY/B binding domain (P2) and CheA kinase regulatory domain (P5), respectively. Both plasmids were constructed by PCR subcloning of the P2 or P5 domain from pDK28, and ligating it into the purified empty pDK28 vector. Site-directed Ala and Trp mutants of P5 were generated using the PCR-based QuickChange XLII mutagenesis kit (Agilent). All mutants were confirmed by sequencing the entire coding region. The resulting YFP-P2 and YFP-P5 fusions were transformed in RP437 cells and single colonies were grown overnight at 30° C in VBC minimal growth medium supplemented with HMLTT containing 0.75% glycerol and 100 μg/mL ampicillin 46–49. The overnight culture was diluted 10-fold into fresh VBC media and grown until mid log phase (OD600 between 0.04 and 0.09). Induction was initiated by the addition of fresh 50 μM IPTG and allowed to proceed for 2 hours. The cells in 10 mL of culture were collected by centrifugation for 5 minutes at 3200 g and resuspended in 700 μL of imaging buffer (10 mM potassium phosphate, 0.1 mM EDTA, 1 mM L-methionine, 10 mM sodium lactate, pH 7.0) 45. The cells were then repelleted and resuspended in a final volume of 50 μL of imaging buffer. Finally, 20 μL of washed cells were pipetted onto a thin agarose pad (1% agarose in imaging buffer) and allowed to set for 3 minutes before a coverslip was added. Cells were immediately imaged on a Nikon TE-2000-E inverted microscope utilizing a 60X oil immersion objective, CFP/YFP/RFP dichroic mirror with corresponding single band excitation and emission filters, and a CoolSNAP ES camera with an exposure time of 1 second. Excitation was provided with a mercury lamp.
To apply TAM-IDS in vitro, Ala and Trp mutants were constructed in the Cysless CheA kinase background using site-directed mutagenesis as described above, and the effect of the mutations on CheA kinase incorporation and activity was measured in the reconstituted array. CheA kinase incorporation is defined as the density ratio of CheA kinase to receptor visualized on Coomassie gels as previously described 18. In vitro kinase assays were performed as described above.
Data and Error Analysis
All data points shown are averages of three to nine replicates. Error bars and ranges indicate the standard error unless otherwise specified. Asterisks indicate statistically significant changes (P < 0.05).
RESULTS
Two Working Models for the Interface Between Receptor and Kinase Regulatory Domain
The project began by building two working models of the interface based on the species components used in experiments. The present in vitro studies utilized a proven combination of interchangeable components from the Escherichia coli and Salmonella typhimurium chemosensory arrays: native E. coli serine receptor Tsr (which lacks Cys residues), Cysless S. typhimurium CheA kinase, and Cysless S. typhimurium CheW adaptor protein to investigate the two working models for the interface between the receptor and the CheA kinase regulatory domain. Together these three core proteins are known to generate a fully functional, ultrastable, Cysless array 18,21 into which pairs of Cys substitutions can be introduced for disulfide mapping, or Trp and Ala substitutions for TAM-IDS.
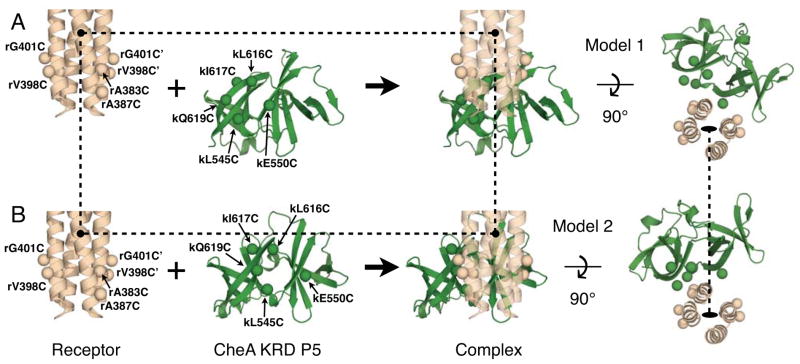
Figure 2 presents the two models. Working Model 1 is based on a structure from the Crane laboratory (PDB code 3UR1) for Thermotoga receptor bound to both SH3-like subdomains of the Thermotoga CheW adaptor protein 19. Model 1 was developed by (i) superimposing the crystallographic E. coli serine receptor dimer (1QU7) onto the crystallographic receptor dimer in the Thermotoga complex (3UR1); (ii) generating a homology model of S. typhimurium CheA kinase regulatory domain by threading its sequence onto the known structure (1B3Q) of the Thermotoga CheA kinase using Swiss Prot; and finally (iii) superpositioning the backbone structure of the regulatory domain homology model onto that of CheW adaptor protein in the Thermotoga receptor complex (3UR1), making use of the tandem SH3-like motif they both share.
Figure 2.
Working Models 1 and 2 for receptor-CheA kinase contacts in the chemosensory array, illustrating engineered Cys positions. Shown are the protein interaction region of a serine receptor homodimer (tan) and the kinase regulatory domain of CheA (green), with engineered Cys positions indicated as spheres. For simplicity, the receptor Cys residues located on the trimer interaction helix are shown only in the left-most homodimer – the other homodimers depict only the Cys residues located on the CheA kinase interaction helix. Circles and dashed lines represent fiduciary markers showing that receptor is in the same position in Model 1 and Model 2. (A) Working Model 1 for receptor-CheA kinase contacts between E. coli and S. typhimurium components 25,6, developed by homology modeling of the Thermotoga complex between the receptor protein interaction region and the adaptor protein CheW 19 (PDB Code 3UR1, homology modeling detailed in text). (B) Working Model 2 for receptor-CheA kinase contacts between E. coli and S. typhimurium components 25,6, developed by homology modeling of the newly determined Thermotoga complex between the receptor protein interaction region and the CheA kinase regulatory domain (KRD P5) (PDB Code 4JPB by Li et al, accompanying manuscript 42; homology modeling detailed in text). Of the five Cys residues on the CheA kinase regulatory domain (kL545, kE550, kL616, kI617, kQ619), all are oriented towards receptor except the negative control residue kI617, which points away from receptor. Looking through the fixed receptor (semi-transparent) towards the CheA kinase regulatory domain interface, one can see that translations and rotations are needed to move the CheA kinase regulatory domain between its contrasting locations in Model 1 and Model 2, thus explaining why the two models predict different distances between engineered Cys pairs (Table 1).
Working Model 2 is based on the new structure from the Crane laboratory (PDB code 4JPB) for the Thermotoga complex between receptor fragment and both SH3-like subdomains of the CheA kinase regulatory domain (Li et al, companion manuscript 42). This model was developed by (i) superimposing the crystallographic E. coli serine receptor dimer (1QU7) onto the crystallographic receptor dimer in the Thermotoga complex 42, then (ii) the same homology model of S. typhimurium CheA kinase regulatory domain utilized in Model 1 was superimposed onto the backbone structure of the CheA kinase regulatory domain in the Thermotoga complex 42.
Creating a Library of Functional Cys Pairs for Disulfide Mapping
The predicted contact surfaces of Working Models 1 and 2, together with relevant information from previous PICM, NMR, and crystallographic studies 38,39,16,17,19,22,23, were used to select positions for Cys substitution on both the receptor and CheA kinase regulatory domain. The goal was to generate ~20 Cys pairs that retained function in the membrane-bound array for disulfide mapping analysis of interface structure.
On the receptor, the four selected residues rA383, rA387, rV398, and rG401 (lower case “r” denotes receptor residues) are homologous to four surface-exposed positions on the aspartate receptor Tar where single Cys mutations retain 40–90% normal kinase activation and full attractant regulation in the functional array 38,39,16. All four were also implicated as contact residues in the functional array by PICM analysis in which modification of each Cys with a bulky fluorescein probe eliminated kinase activation 38,39,16. Moreover, NMR studies from Vu, Wang, Dahlquist and coworkers of the homologous fragment from the Thermotoga receptor TM001490-206 revealed significant chemical shift changes for these or nearby receptor residues triggered by the binding of the Thermotoga CheA kinase regulatory domain, or by binding of the Thermotoga CheW adaptor protein believed to compete for the same receptor surface 22,23. Both Working Models 1 and 2 predict all four residues lie within or nearby the contact surface for the CheA kinase regulator domain (Fig. 2). Thus, these four single Cys substitutions were introduced into the serine receptor by PCR site-directed mutagenesis (Methods).
On the CheA kinase regulatory domain, three of the five selected Cys substitutions kL545C, kE550C, and kI617C (lower case “k” denotes CheA kinase residues) at surface positions on free, full length CheA kinase are known to retain 60–90% normal kinase activity and full attractant regulation in the functional array, with kL545 implicated as a receptor contact residue by PICM chemical modification 17. NMR studies have detected chemical shift changes at these three or nearby homologous positions on a Thermotoga CheA kinase fragment upon binding of a Thermotoga receptor fragment, suggesting a direct role in the binding interface 22,23. For the present study, two new substitutions kL616C and kQ619C at surface positions were also introduced into the full length, Cysless CheA kinase protein. Of the five resulting Cys residues, four (kL545C, kE550C, kL616C, and kQ619C) span the predicted interfaces of Working Models 1 and 2 with their side chains oriented towards receptor. The fifth (kI617C) is a negative control residue predicted by both models to point away from receptor, decreasing the probability it will collide with receptor residues (Fig. 2).
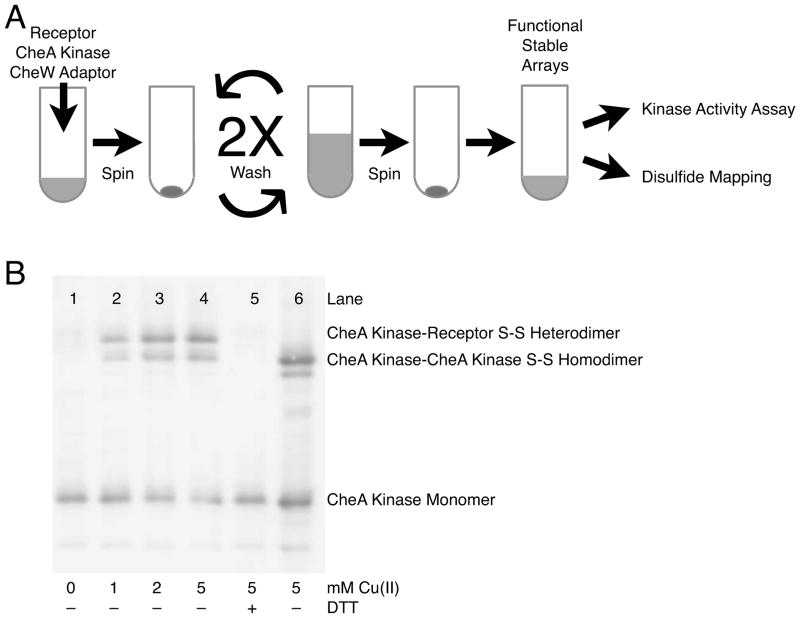
A library of reconstituted arrays was generated from all possible combinations of the four receptor and five CheA kinase regulatory domain Cys mutants, yielding 20 distinct arrays possessing different Cys pairs. As summarized in Figure 3, arrays were formed by incubating E. coli membranes containing a given single-Cys receptor with purified single-Cys CheA kinase and purified Cysless CheW adaptor protein to assemble the ultrastable, membrane-associated core array. Subsequently, unbound components were removed by pelleting and washing the membranes with buffer. Immediately following washing, the washed arrays were analyzed for receptor-regulated kinase activity and disulfide formation rates. The time required to carry out and quench the kinase and disulfide assays (approximately 10 min) is short compared to the remarkable kinetic stability of the arrays (dissociation of components from Cysless and Cys mutant arrays is undetectable over timescales of hours to weeks18,21), thereby ensuring that only array-bound components are analyzed. Figure 4 summarizes the receptor-stimulated kinase activity of each array. Only the rA387C, kI617C pair failed to retain at least 25% of the full CheA kinase activity of the Cysless array; this pair was deemed perturbing and not used further. The remaining 19 Cys pairs retained substantial CheA kinase activity as well as the native down-regulation of CheA kinase activity by addition of attractant serine (Fig. 4). Thus, each of these 19 Cys pairs retains the essential receptor-CheA kinase contacts needed for the formation of a functional, membrane-bound array capable of receptor-mediated CheA kinase on-off switching.
Figure 3.
Disulfide mapping protocol and representative western blot used to analyze disulfide-linked products. A) The functional, membrane bound array is reconstituted in vitro by combining E. coli membranes containing serine receptor with purified CheA kinase and CheW adaptor protein; where appropriate, the receptor and CheA kinase components possess engineered Cys residues. Self-assembly occurs during an incubation period, then unbound proteins are washed away to yield functional, ultrastable, membrane-bound arrays 18,21 employed in kinase activity assays and disulfide mapping experiments. B) Representative western blot using anti-CheA kinase polyclonal primary antibody. Increasing the Cu(II) concentration from 0 to 5 mM (lanes 1–4) shows an increase in formation of the disulfide-linked, receptor-CheA kinase heterodimer, which is reduced by addition of DTT as expected (lane 5). The heterodimer is easily resolved from the disulfide-linked CheA kinase homodimer, as confirmed by oxidation of pure CheA kinase in solution to generate the covalent homodimer (lane 6) that is reduced by DTT (lane 5).
Figure 4.
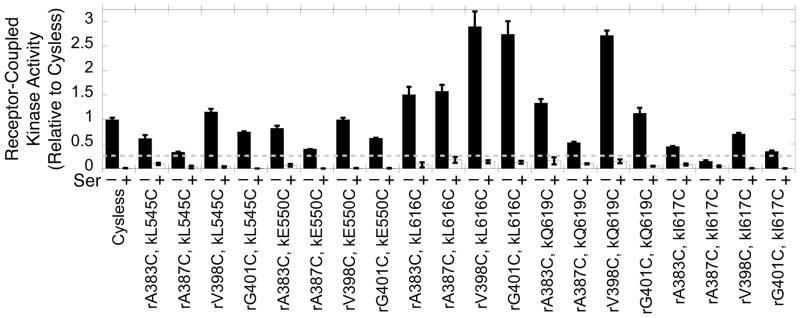
CheA kinase activities of the engineered, reduced Cys pairs in membrane-bound arrays. Reconstituted, membrane-bound arrays (Fig. 3) were mixed with CheY response regulator protein and ATP to initiate the kinase reaction (black bars) under reducing conditions. An identical sample contained serine to quantify attractant regulation of CheA kinase activity (white bars). The dashed line represents 25% normal CheA kinase activity relative to the fully functional, reconstituted Cysless array. Any Cys pair exhibiting activity below this threshold was dropped from further analysis. Error bars indicate the standard error of each triplicate mean.
Table 1 summarizes the Cβ-Cβ separations predicted by Working Models 1 and 2 for the 19 functional Cys pairs. For the three negative control pairs, in which kI617C points away from the interface, the separations are misleading because the unfavorable geometry does not allow simple backbone translations to generate productive collisions and disulfide formation. As a result, these three pairs are not classified as proximal or distal. For the remaining 16 pairs, Working Model 1 predicts there are 3 proximal and 13 distal pairs whereas Working Model 2 predicts 6 proximal and 10 distal pairs. Notably, when comparing the two models, four Cys pairs are distal in one model and proximal in the other, with a Cβ-Cβ distance difference of at least 2.5 Å between the two models: rA387C, kE550C is proximal in Model 1, while rA383C, kL545C; rA383C, kQ619C; and rG401C, kQ619C are each proximal in Model 2. These significant differences, which facilitate resolution of the two models by disulfide mapping, arise from the translational and rotational register shifts of the receptor helix relative to the CheA kinase regulatory domain that together distinguish Model 1 from 2 (Fig. 2).
Table 1.
Comparison of Model 1 and Model 2 Cβ-Cβ Distances
| Tsr, CheA Kinase Mutanta | Model 1 Cβ-Cβ (Å)b | Distance Designationc | Model 2 Cβ-Cβ (Å)b | Distance Designationc |
|---|---|---|---|---|
| rA383C, kL545C | 16.0 | Distal | 7.7 | Proximal |
| rA387C, kL545C | 9.9 | Proximal | 4.7 | Proximal |
| rV398C, kL545C | 22.1 | Distal | 18.3 | Distal |
| rG401C, kL545C | 22.5 | Distal | 22.6 | Distal |
| rA383C, kE550C | 13.4 | Distal | 11.6 | Distal |
| rA387C, kE550C | 9.6 | Proximal | 12.4 | Distal |
| rV398C, kE550C | 11.2 | Distal | 10.5 | Proximal |
| rG401C, kE550C | 16.5 | Distal | 15.2 | Distal |
| rA383C, kL616C | 12.4 | Distal | 11.2 | Distal |
| rA387C, kL616C | 11.7 | Distal | 15.1 | Distal |
| rV398C, kL616C | 17.3 | Distal | 19.8 | Distal |
| rG401C, kL616C | 19.5 | Distal | 22.0 | Distal |
| rA383C, kQ619C | 12.7 | Distal | 5.6 | Proximal |
| rA387C, kQ619C | 8.8 | Proximal | 8.3 | Proximal |
| rV398C, kQ619C | 20.5 | Distal | 12.2 | Distal |
| rG401C, kQ619C | 18.7 | Distal | 8.4 | Proximal |
| rA383C, kI617C | (15.7) | NA | (12.6) | NA |
| rV398C, kI617C | (25.3) | NA | (19.3) | NA |
| rG401C, kI617C | (22) | NA | (13.9) | NA |
Bold pairs switch from proximal to distal between the two models and exhibit a change of separation exceeding 2.5 Å.
In the case of Cys pairs possessing the receptor G401C substitution, the Gly was mutated to Ala prior to calculating Cβ-Cβ distances in MacPyMol 65.
Proximal and Distal pairs are defined as < 11 Å and > 11 Å Cβ-Cβ separation respectively.
Analyzing Interface Structure in Functional Arrays by Disulfide Mapping
The disulfide mapping approach is based on the observation that under the same oxidation conditions, proximal Cys pairs generally collide and form disulfide bonds more rapidly than distal Cys pairs 50–52. In order to form a disulfide bond, two Cys residues must collide with a Cβ-Cβ separation of ≤ 4.6 Å and the correct angular orientation. Thus, measurement of disulfide formation rates provides useful information about Cβ-Cβ proximity and local backbone dynamics.
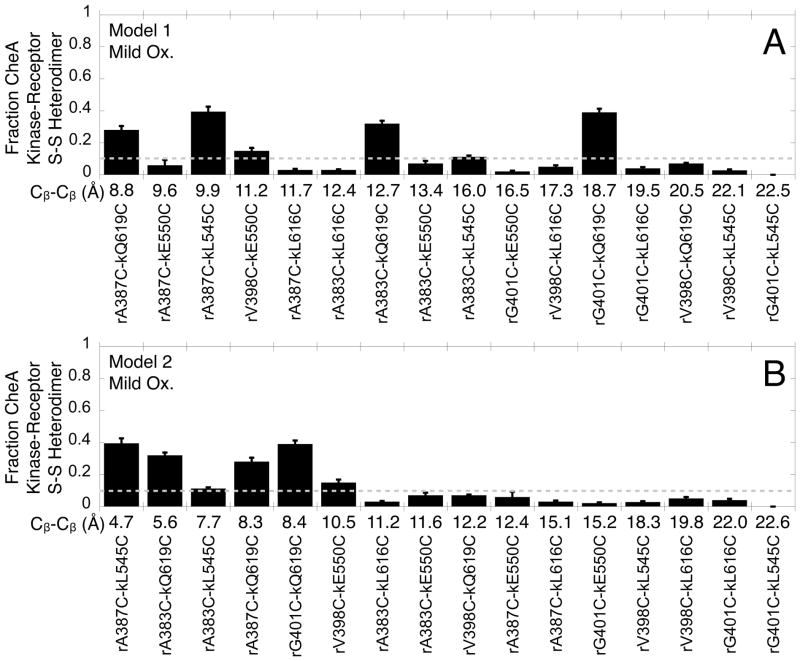
The method was applied to the 19 functional membrane-bound arrays containing distinct Cys pairs. Each washed, membrane-bound array was subjected to a highly controlled oxidation reaction, then the disulfide-linked products were quantified (Fig. 3B). Due to the unusually rapid disulfide formation observed for proximal Cys pairs, novel mild oxidation conditions were developed to enable analysis of disulfide formation reactions in the linear, initial rate limit. These ultra-mild conditions employed 1 mM Cu(II) as a redox catalyst buffered by 3 mM EDTA, yielding significantly slower oxidation than in previous studies. (Note the 1,10-phenanthroline ligand previously used to enhance Cu(II) catalytic efficiency and oxidation speed was omitted here). The excess EDTA ensured that a very small but consistent concentration of free Cu(II) (~ 1 picomolar for the mildest oxidation reaction 53) was present during the 10 sec reaction, which was followed by a rapid quench and quantification of products by SDS-PAGE and anti-CheA kinase western blot analysis. Densitometric analysis of the western blots yielded the fraction of total CheA kinase converted to disulfide linked, receptor-CheA kinase heterodimer. Figure 3B shows that the major products are the receptor-CheA kinase heterodimer and the CheA kinase-CheA kinase homodimer, and that both disulfide-containing products can be reduced. Since each reaction was in or near the initial rate limit, the final extent of heterodimer formation per 10 sec was equivalent to the disulfide formation rate. Figure 5 confirms that greater extents of disulfide formation are observed when the oxidation strength is increased to intermediate or strong conditions.
Figure 5.
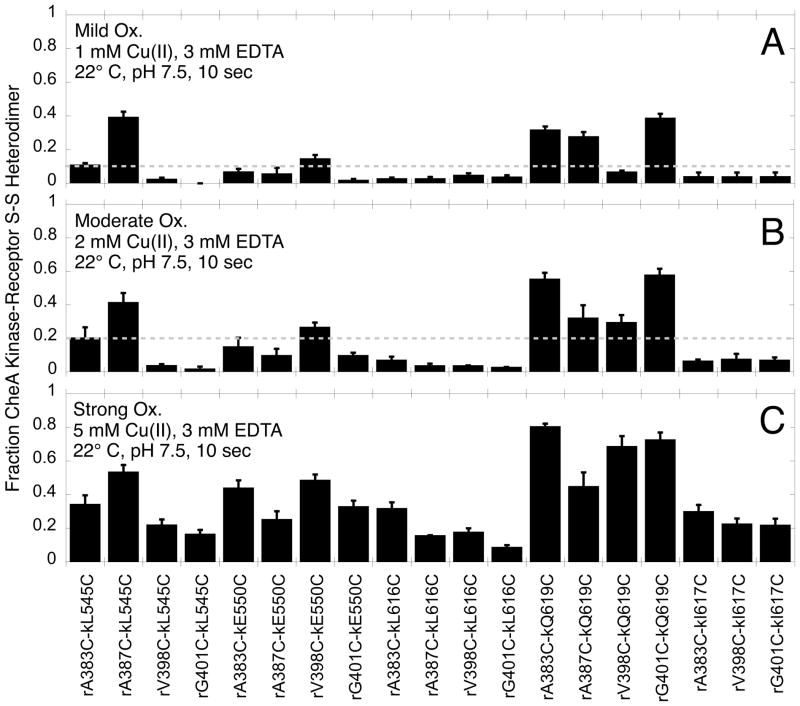
Relative disulfide formation rates measured for the engineered, functional Cys pairs in membrane-bound arrays. For each of the 19 active and regulated Cys pairs, the disulfide formation reaction was carried out for 10 sec under under mild, moderate, and strong oxidation conditions (1 mM, 2 mM, and 5 mM Cu(II), respectively, each buffered with 3 mM EDTA) prior to quenching and quantification of the heterodimer (Fig. 4). Under mild oxidizing conditions disulfide formation is far from completion and the extents of heterodimer production at 10 sec yield relative disulfide formation rates. Six Cys pairs, rA383C-kL545C, rA387C-kL545C, rV398C-kE550C, rA383C-kQ619C, rA387C-kQ619C, and rG401C-kQ619C, yield rapid disulfide formation rates, operationally defined as > 10% heterodimer under mild oxidation (dashed line, Fig. 4A). These six pairs also yield > 20% heterodimer formation under moderate oxidation (dashed line, Fig. 4B). Error bars indicate the standard error of each set of replicates, n = 3 to 9. Control experiments show that the 1 mM Cu(II) employed in mild oxidation does not significantly perturb the structure or function of the array, as evidenced by minimal effects on receptor-regulated CheA kinase activity in SI Figure S1.
Of the 19 functional Cys pairs, Figure 5A,B shows that six pairs yield the fastest formation of disulfide-linked, receptor-CheA kinase heterodimers: rA383C-kL545C, rA387C-kL545C, rV398C-kE550C, rA383C-kQ619C, rA387C-kQ619C, and rG401C-kQ619C. These pairs each yield at least 10% heterodimer under mild conditions, and at least 20% heterodimer under intermediate conditions. In the disulfide mapping approach, these Cys pairs are expected to possess the most proximal Cβ-Cβ distances.
Figure 6 orders the Cys pairs with increasing Cβ-Cβ distances from left to right for working Models 1 (Fig. 6A) and 2 (Fig. 6B), and presents the extents of disulfide-linked, receptor-CheA kinase heterodimer formation observed under mild oxidation conditions. In the case of Model 1, three of the six fastest Cys pairs are classified as proximal, but the other three are distal indicating poor agreement with the data. By contrast, in Model 2 all six of the fastest Cys pairs (rA383C-kL545C, rA387C-kL545C, rV398C-kE550C, rA383C-kQ619C, rA387C-kQ619C, and rG401C-kQ619C) are proximal, and in fact possess the six shortest Cβ-Cβ distances ranging from 4.7 to 10.5 Å. These disulfide mapping results provide strong evidence that Working Model 2 is an accurate depiction of the receptor-CheA kinase interface in functional, membrane-bound arrays.
Figure 6.
Comparing the disulfide formation rates measured in membrane-bound arrays with the Cys pair separations predicted by Working Models 1 and 2. (A,B) The disulfide formation rates measured in Figure 5 under mild oxidation conditions are re-sorted in order of increasing Cβ-Cβ separation as determined by Model 1 (A) or 2 (B). Error bars indicate the standard error of each set of replicates, n = 3 to 9.
Creating a Library of Trp and Ala Mutants for TAM-IDS Analysis
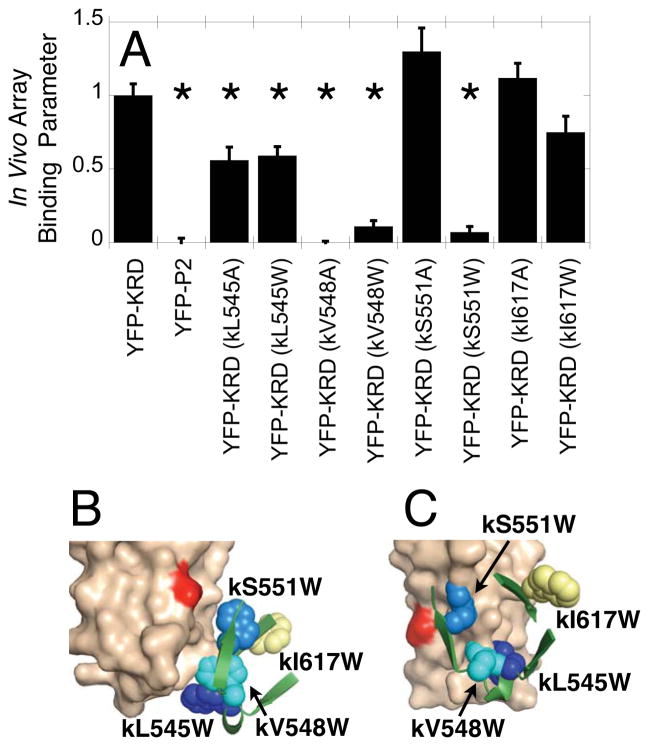
To further resolve Models 1 and 2, the method Tryptophan and Alanine Mutagenesis to Identify Docking Sites (TAM-IDS) was used. This method extends established approaches that employ tryptophan or alanine substitutions, or both, to probe interfaces in vivo or in vitro 54–56,40. TAM-IDS combines tryptophan and alanine replacements with quantitative measurements carried out both in vivo and in vitro to provide maximum information about binding and function. Figure 7 illustrates the four positions on the CheA kinase regulatory domain selected for TAM-IDS analysis (kL545, kV548, kS551, kI617). All four positions are located on the surface of the uncomplexed domain at exposed positions where Trp and Ala substitutions are unlikely to affect domain folding. In Model 1, none of the four selected native side chains directly contact the receptor, and at each of these positions most of the nine common conformers of a mutant Trp side chain do not contact receptor 65 (Fig. 7B). By contrast, in Model 2 three of the four native side chains (kL545, kV548, kS551) do contact the receptor, and at each of these three positions most common conformers of a mutant Trp side chain generate VDW clashes with receptor 65 (Fig. 7C). In this model the fourth position (kI617) lies well outside the interface where a Trp substitution yields no receptor clashes (Fig. 7C). No positions were selected on the receptor, since in the two-fold symmetric homodimer a given mutation often perturbs both the CheA kinase binding surface and the trimer interaction surface, complicating the interpretation.
Figure 7.
Testing the Model 2 receptor-CheA kinase interface using in vivo TAM-IDS: Tryptophan and Alanine Mutation to Identify Docking Sites. (A) The docking of the isolated CheA kinase regulatory domain to the native chemosensory array in live cells was quantified by measuring the array binding parameters (Eq. 1) for the indicated fluorescent constructs in which YFP is fused to the regulatory domain N-terminus. The array binding parameter scales the measured incorporation to the high level of binding observed for the wild type CheA kinase regulatory domain (YFP-KRD) and the low level of binding observed for a negative control domain (P2). Error bars indicate standard error for measurement of at least 20 cells in 2 separate experiments and asterisks indicate significant changes relative to WT YFP-KRD (P < 0.05). The kinase regulatory and P2 domains employed were isolated from E. coli CheA kinase 45. For consistency with other figures, S. typhimurium positions are presented here; the corresponding E. coli positions are summarized in SI Table S1. (B,C) Shown are the four mutant Trp residues in Models 1 and 2, respectively. In Model 1 most of the common Trp conformers at all four positions exhibit no VDW clashes with receptor 65 (B). In Model 2 most of the common Trp conformers at three positions (kL545W in dark blue, kV548W in cyan, kS551W in medium blue) do exhibit major VDW clashes with receptor, while the remaining Trp (kI617W in yellow) lacks clashes 65 (C).
To carry out TAM-IDS in vivo, Trp and Ala substitutions were incorporated at the four selected positions in the isolated CheA kinase regulatory domain fused to YFP. To carry out TAM-IDS in vitro, the same eight substitutions were introduced into Cysless, full length CheA kinase.
Analyzing Interface Structure in Live Cells by TAM-IDS
The TAM-IDS approach quantifies and compares the perturbations generated by Trp and Ala substitutions at a given position. When both substitutions yield large perturbations of the interface, the native side chain is deemed essential. When the Trp and Ala perturbations are large and small, respectively, the native side chain is deemed non-essential but is proposed to be located within the contact region, such that the bulky Trp substitution disrupts the interface. When both substitutions yield small perturbations, the native side chain is deemed outside the interface.
The TAM-IDS method was carried out in vivo using the E. coli CheA kinase regulatory domain fused to YFP. Kenter, Sourjik and coworkers previously demonstrated this minimal construct assembles into the chemosensory lattice of live cells to yield highly fluorescent, easily identified puncta on the bacterial membrane 45. Here we interrogate the ability of mutant fusion proteins to bind the native array via live cell fluorescence imaging. We define the array fluorescence (A) as:
| (Eq. 1) |
where FA is the integrated fluorescence intensity of a fixed area containing the array at the end of the cell, FC is the integrated fluorescence intensity of an equal fixed area of cytoplasm, and B is the background integrated intensity of an equal fixed area outside the cell. Use of the ratio corrects for deviations in the expression levels of the fusion protein in different cells. The array binding parameter (AB) of a given mutant is then defined as:
| (Eq. 2) |
where this AB parameter scales the array fluorescence of the mutant (AM) to the maximum array fluorescence of the WT CheA kinase regulatory domain (AWT) and the minimum array fluorescence measured for a negative control domain (A0, measured for P2 CheY/B binding domain of CheA kinase that does not bind to the array). Thus, when localization of the YFP-mutant to the array approaches the wild type level AB approaches unity, and when localization approaches the negative control level AB approaches zero.
Figure 7A shows the array binding parameters observed in live cells for the Trp and Ala mutations at the four selected positions (20 cells quantified for each; S. typhimurium numbering is used for consistency). (i) Both kL545W and kL545A retain some binding but show significant (P < 0.05) decreases relative to WT, indicating that Leu 545 is an important, though not essential, side chain at the docking interface. These findings support Model 2 wherein Leu 545 is a contact position, but not Model 1 in which the native side chain exhibits no receptor contacts (Fig. 7B,C). The fact that mutation to Trp does not completely disrupt binding of kL545W is consistent with the location of this position at the edge of the Model 2 interface, where some standard Trp rotamers can snorkel out of the interfacial region. (ii) Both the kV548A and kV548W mutations completely block binding of the CheA kinase regulatory domain to the array, indicating Val 548 is an essential binding residue that cannot tolerate a bulky substitution nor truncation. This finding strongly supports Model 2 wherein this position is a receptor contact site and Trp rotamers clash with receptor, but not Model 1 wherein Trp rotamers exhibit minimal receptor clashes (Fig. 7B,C). (iii) The kS551A mutant binds normally while kS551W exhibits no array binding, suggesting the native Ser 551 side chain is not essential for docking but does lie in the interface. This result strongly supports Model 2 which predicts receptor clashes for many Trp rotamers, but not Model 1 which predicts no such clashes (Fig. 7B,C). (iv) Finally, neither kI617A nor kI617W yield a significant effect on binding, as is expected since this negative control residue does not contact receptor in either model (Fig. 7B,C).
Overall, the in vivo TAM-IDS results (Fig. 7), like the in vitro disulfide mapping findings (Fig. 6), strongly support Model 2 for the structure of the interface between receptor and the CheA kinase regulatory domain. Since all three perturbing Trp substitutions are predicted to contact the N-terminal helix of the receptor hairpin, the findings highlight the importance of this helix interaction. Moreover, the E. coli receptor-E. coli CheA kinase interface (Fig. 7) is indistinguishable from the Escherichia coli receptor-Salmonella typhimurium CheA kinase interface (Fig. 6) within the resolution of the two methods.
Analyzing On-Off Switching in Functional Arrays by Disulfide Mapping
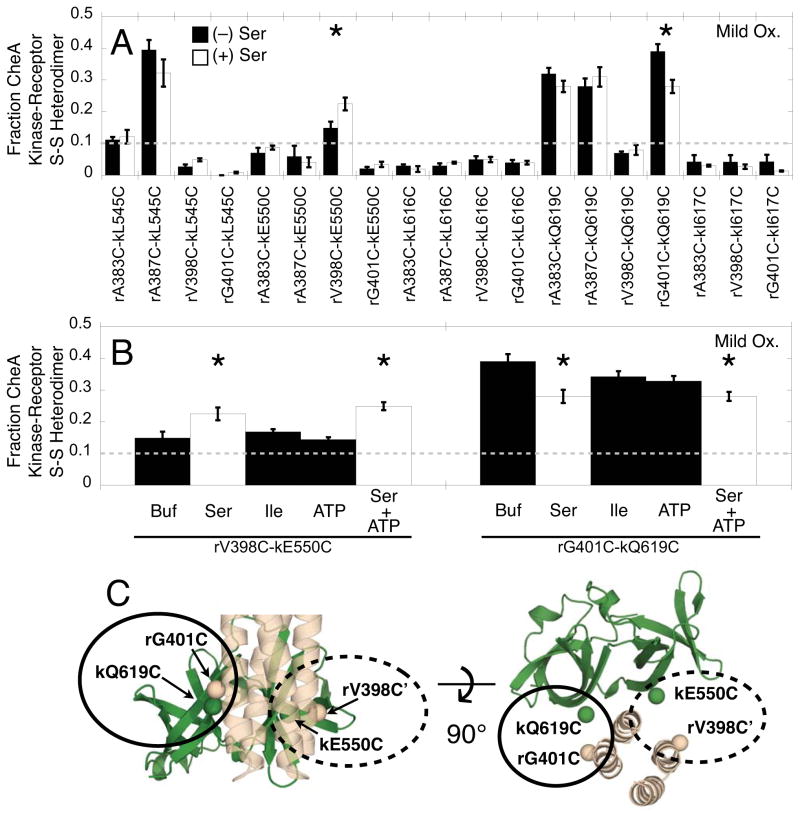
To identify regions of the interface perturbed by switching from the on-state to the off-state, disulfide mapping of the same 19 functional Cys pairs was carried out in the absence and presence of attractant serine. The apo array in the absence of serine is the on-state that exhibits maximal CheA kinase activity. Attractant serine binding to the array triggers switching to the off-state that exhibits minimal CheA kinase activity. The disulfide formation rate of a Cys pair is exquisitely sensitive to local changes in structure or dynamics that alter the Cys collision rate or accessibility to oxidizer, and is thus well-suited to detect even small attractant-triggered perturbations of the interface.
Figure 8 compares the resulting initial disulfide formation rates in the absence and presence of attractant. Only 2/19 of the Cys pairs exhibit a significant attractant effect (P < 0.05): upon addition of serine the rV398C, kE550C pair shows an increase in the rate of disulfide-linked heterodimer formation while the rG401C, kQ619C pair shows a decrease (Fig. 8A,B). In contrast, the negative control isoleucine, or the kinase substrate ATP, have no detectable effect on disulfide formation, thereby demonstrating that the attractant effect is highly specific for serine (Fig. 8B). The observation that attractant binding alters the disulfide formation rate for only 2/19 of the Cys pairs tested argues that attractant triggers a local, rather than global, structural or dynamical change in limited regions of the interface. Both of the rate changes involve contacts with the C-terminal helix of the receptor hairpin, while no rate changes are observed for contacts with the N-terminal helix, indicating that attractant binding does not alter the latter interaction.
Figure 8.
Effects of receptor-triggered on-off switching on the relative disulfide formation rates of engineered Cys pairs in membrane-bound arrays. A) As in Fig. 5A, relative disulfide formation rates were measured under mild oxidation conditions for the 19 active and regulated Cys pairs, both in the absence (black bars) and presence of 2 mM serine (white bars). Only two Cys pairs show significant attractant effects (asterisks, P < 0.05). Additional experiments (SI Fig. S2) measured disulfide formation rates under even milder oxidation conditions (0.5 mM Cu(II), 3 mM EDTA) for Cys pairs rA387C-kL545C, rA383C-kQ619C, and rA387C-kQ619C to ensure the reactions were not approaching saturation; again, no serine effects were detected. B) For the two attractant-sensitive disulfide formation reactions (rV398C-kE550C and rG401C-kQ619C), the attractant specificity was tested by comparison with buffer and a negative control ligand (isoleucine). These studies confirm the significant effect of Ser on disulfide formation rates (white bars, asterisks indicate P < 0.05), while no significant effects are observed for buffer, isoleucine, nor ATP (black bars). C) Mapping the attractant-sensitive Cys pairs (circled) on Model 2. Error bars indicate the standard error of each triplicate mean.
Analyzing Interface Function in Reconstituted Arrays by TAM-IDS
The above results suggest that the receptor-CheA kinase regulatory domain interface may play a role in both array structure and on-off switching. Unfortunately, the effects of the observed receptor-CheA kinase disulfide bonds on array stability and regulation cannot be directly studied, since the use of strong oxidation conditions to drive disulfide formation to completion yields disulfide bonds between receptor homodimers that damage the array, presumably by perturbing trimer contacts (refs 39,16,51).
Thus, in order to probe the structural and signaling roles of the interface the TAM-IDS method was carried out in vitro. To resolve structural and signaling functions, it is necessary to directly quantify the effects of mutations on both array assembly and on-off switching. Trp and Ala substitutions were incorporated at the same four CheA kinase regulatory domain positions employed in live cell studies (Fig. 7), this time in full length Cysless CheA kinase. Subsequently, each mutant CheA kinase was incorporated into reconstituted arrays with the receptor and adaptor components, and washed as usual to remove free or weakly bound components.
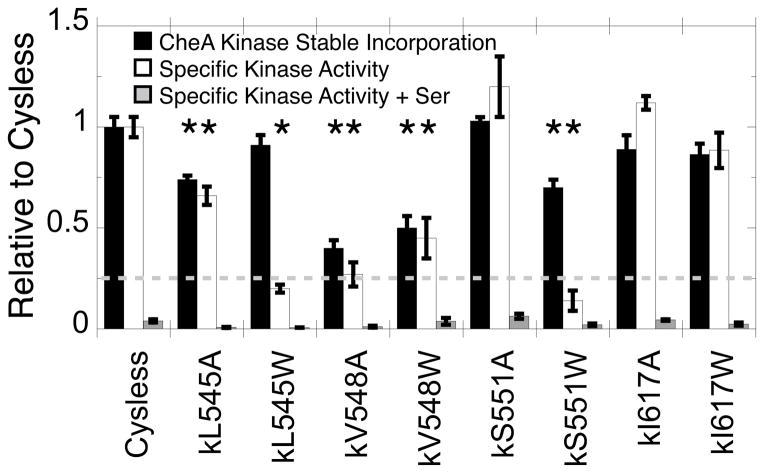
Figure 9 shows that all eight of the Trp and Ala CheA kinase mutants are stably incorporated into the array at significant levels, achieving at least 40% incorporation relative to the positive control Cysless CheA kinase. This result contrasts with the consistently lower incorporation observed for the CheA kinase regulatory domain mutants in live cell arrays (Fig. 7). The simplest explanation is that full length CheA kinase has additional array contacts and a dimeric structure that together provide binding stability not available to the isolated regulatory domain. Yet for four of the six interfacial mutants, the perturbation yields a statistically significant decrease (P < 0.5) in array incorporation relative to the positive control, arguing that the interface does play an important role in array stability and structure.
Figure 9.
Testing CheA kinase uptake and function in membrane-bound arrays using in vitro TAM-IDS. Reconstituted, washed membrane-bound arrays (Fig. 3) were prepared using the indicated CheA kinase proteins. Stable CheA kinase incorporation into the washed array was quantified as the ratio of the CheA kinase band to the receptor band on a Coomassie stained gel, normalized to the Cysless ratio (black bars). All kinase proteins, including the mutants, required CheW adaptor protein for stable incorporation as expected for normal array formation (SI Fig. S3). CheA kinase activity in the array was measured by adding with CheY response regulator protein and ATP to initiate the kinase reaction (white bars). An identical sample contained 2 mM serine to quantify attractant regulation of kinase activity (grey bars). The dashed line represents 25% normal CheA kinase uptake and activity relative to the fully functional, reconstituted Cysless array. Error bars indicate the standard error of each set of replicates, n = 6. Asterisks indicate significant changes relative to Cysless (P < 0.05).
Since all of the mutant CheA kinases are stably incorporated into the array at useful levels, their specific kinase activities can be quantified in the absence and presence of attractant. These specific activities are proportional to the turnover rate of the average bound CheA kinase molecule, and thus correct for differences in array incorporation between mutants. Figure 9 shows that at the three interfacial positions, five of the six mutants show significantly reduced (P < 0.05) specific activities in the apo/on-state, while the specific activities of mutations at the negative control position are unperturbed. The largest decreases are observed for the Trp substitutions at the three interfacial positions (kL545W, kV548W, kS551W), and for the Ala substitution at the essential interfacial position (kV548A). Since these positions are predicted to contact the N-terminal helix of the receptor hairpin, the findings highlight the importance of these contacts to receptor activation of the bound CheA kinase in the array apo/on-state. However, even the perturbed mutants retain attractant-triggered kinase inhibition, suggesting the native kinase-helix contacts are less important for CheA kinase inhibition. Overall, the results indicate that the native interface plays a significant role defining array structure and stability, and also is quite important to receptor-mediated CheA kinase activation.
DISCUSSION
The present results reveal that the new crystal structure of a complex between Thermotoga receptor and CheA kinase fragments (Li et al, accompanying manuscript 42) accurately represents the structure of the receptor-CheA kinase interface in functional, membrane-bound chemosensory arrays. Working Model 2 developed herein depicts this validated structure for the homologous E. coli/S. typhimurium arrays (Figs. 2B, 7C, 8C). In contrast, the present results show that a previous model used to describe this interface in current array models does not accurately depict the receptor-CheA kinase contacts, although it may provide an accurate view of receptor-CheW adaptor contacts 19. In the validated Model 2 the CheA kinase regulatory domain is translated ~2 helical turns along the receptor long axis towards the membrane, and translated ~50° around the cylinder defined by the receptor four-helix bundle relative to the previous model (compare Figs. 2A,B).
Validated Model 2 is supported by complementary evidence from both (i) disulfide mapping of the interface in functional, membrane-bound arrays in vitro, and (ii) TAM-IDS mapping of the interface in native arrays in vivo. The findings show that the main contact surface on the receptor protein interaction region is an exposed section of α-helix located just N-terminal to the receptor hairpin turn (Figs. 2B, 7C, 8C). The final two turns of this receptor helix dock into the helix-sized groove lying between the tandem SH3-like subdomains of the CheA kinase regulatory domain.
The validated model adequately explains previous evidence from chemical scanning studies of the interface between receptor and the CheA kinase regulatory domain 39,16,17,51. The chemical scan employed the Protein Interactions by Cysteine Modification (PICM) method to probe 20 positions on the surface of the CheA kinase regulatory domain. Model 2 predicts that only one of those 20 positions tested in the PICM scan, kL545, is located the receptor contact surface. Indeed this was the only one of the 20 positions where PICM chemical modification of the side chain with a bulky probe was observed to yield both (i) normal free CheA autokinase activity and (ii) normal CheW adaptor protein binding as well as (iii) loss of receptor-mediated CheA kinase activation, suggesting that the kL545 residue is located within or near the receptor docking site 17. Similarly, a PICM scan of the aspartate receptor tested five positions that Model 2 indicates lie on or near the docking site for the CheA kinase regulatory domain, such that chemical modification should perturb the receptor-CheA kinase interaction: rL380, rA383, rA387, rV398, rG401 (E. coli serine receptor residues). Indeed, PICM chemical modification of any one of the five residues was observed to block receptor-mediated CheA kinase activation, although some of those effects may stem from perturbation of receptor trimers-of-dimers contacts 38,39,16.
The validated model also explains genetic findings by Parkinson et al29 and recent NMR studies by Dahlquist and coworkers 22,23. The locations of suppressor mutations on receptor that counteract specific inhibitory mutations on the CheA kinase regulatory domain 29 are consistent with the model, as are the locations of inhibitory mutations lying on the receptor surface 41. The NMR studies analyzed complexes formed between a Thermotoga receptor fragment and (i) the Thermotoga CheA kinase regulatory domain or (ii) Thermotoga CheW adaptor protein. The results indicate both of these tandem SH3-like proteins bind to overlapping surfaces on the receptor protein interaction region, where receptor residues rA383, rA387 and rV398 (listed as the homologous E. coli serine receptor positions, each implicated in the present work) exhibit chemical shift changes upon docking of either protein. On the CheA kinase regulatory domain, residue kL545 – a position implicated in this work – exhibits a large chemical shift change upon docking to receptor. Thus, although previous PICM, genetic and NMR data lacked the resolution needed to resolve Models 1 and 2, the findings of those studies are fully consistent with the newly validated Model 2.
In principle, the observed contact between the receptor and the CheA kinase regulatory domain could possess one or more essential functions. (i) This interface could play an essential structural role in assembling and maintaining the hexagonal packing and ultrastability of the chemosensory array. (ii) The interface could play an essential signaling role in transmitting on-off signals from the receptor to the CheA kinase. (iii) The interface could play an essential role in longer range communication to other signaling units in the array, thereby generating the strong positive cooperativity of the highly networked chemosensory system. The present study directly tests (i) and (ii), providing direct evidence that the interface has both structural and signaling functions. A structural role is revealed by 5 of 6 interfacial mutations tested in vivo, and 4 of 6 interfacial mutations tested in vitro, that in each case significantly decrease array assembly (Figs. 7, 9). A signaling role, specifically in transmission of the activation signal from receptor to kinase, is demonstrated by 5 of 6 interfacial mutations tested in vitro that each significantly decrease the specific activity of CheA kinase molecules in the receptor-activated, on-state of the array.
Disulfide mapping analysis of interface regulation shows that 2 of 19 Cys pairs yield attractant-triggered changes in disulfide formation rates, indicating that the interface undergoes a local, attractant-triggered change in structure and/or dynamics during on-off switching. Careful examination of the spatial pattern exhibited by these attractant-triggered rate changes yields a testable working model for attractant-triggered rearrangement. Of the six Cys pairs that exhibit the fastest disulfide formation rates (Figs. 6, 8), four involve Cys residues on the N-terminal receptor helix that crosslink rapidly to the CheA kinase regulatory domain – these four pairs exhibit no rate changes upon addition of attractant even under the mildest oxidation conditions (Fig. 8, SI Fig. S2). By contrast, the remaining two involve Cys residues on the C-terminal receptor helix that crosslink rapidly to the CheA domain, and both exhibit significant rate changes upon addition of attractant (Fig. 8).
These observations provide strong evidence that the receptor N-terminal helix and CheA kinase regulatory domain form a tight, fixed complex that moves as a unit during attractant signaling, such that no changes in disulfide formation rates are observed within the unit (Figure 10). This local, high-affinity helix-domain interaction explains how the interface can form correctly even when the global receptor fold is non-native 42. On-off switching is proposed to alter the position or dynamics of this tight, helix-domain complex relative to the two adjacent C-terminal helices of the homodimeric, receptor four-helix bundle. The simplest trajectories consistent with the data are attractant-triggered rotational and/or translational movements of the helix-domain complex relative to the other helices of the receptor four-helix bundle. As Figure 10 illustrates, either rotational or translational movements of the helix-domain complex could maintain constant disulfide formation rates within the complex while (i) increasing the collision rate between subdomain 2 of the kinase regulatory domain and the C-terminal helix of the adjacent receptor subunit to generate the observed increase in the rV398C-kE550C disulfide formation rate, and (ii) decreasing the collision rate between subdomain 1 of the kinase regulatory domain and the C-terminal helix of the same receptor subunit to yield the observed decrease in the rG401C-kQ619C disulfide formation rate (Figs. 8, 10).
Figure 10.
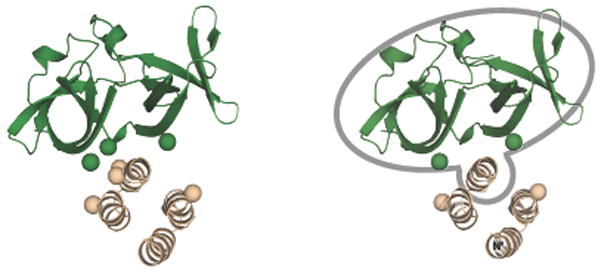
Model for interface on-off switching based on the effects of attractant on disulfide formation rates in the membrane-bound array. A) Shown is the cytoplasmic four-helix bundle of a homodimeric receptor – specifically, the hairpin region of the bundle at its cytoplasmic tip – as well as a single CheA kinase regulatory domain. The four pairs of Cys residues that exhibit rapid, attractant-insensitive disulfide formation rates are clustered in a well-defined region (black lines inside dashed circle). These attractant-insensitive crosslinks define a stable contact between the N-terminal helix of the receptor four-helix bundle and a localized interaction surface on kinase regulatory domain. The insensitivity of these disulfide reactions to attractant provides strong evidence that the stable helix-domain complex moves as a unit. The two pairs of Cys residues that exhibit rapid, attractant-sensitive disulfide formation rates are linked by blue lines. These attractant-sensitive crosslinks are formed between C-terminal helices of the receptor four-helix bundle and adjacent regions of the kinase regulatory domain and detect a change in local conformation or dynamics. B) The model proposes that the stable helix-domain complex (inside the grey border) undergoes an attractant-triggered change in equilibrium position or dynamics relative to the other three helices of the receptor four-helix bundle. Attractant-triggered switching of the on-state to the off-state yields rotation (R) or translation (T) of this helix-domain complex (blue arrows), speeding collisions with the C-terminal helix of the adjacent receptor subunit, and slowing collisions with the C-terminal helix of the same subunit. Thus the rV398C-kE550C and rG401C-kQ619C disulfide formation rates increase and decrease, respectively, in the presence of attractant as observed (Fig. 8).
The proposed attractant-triggered modulation of the tight, mobile complex between the kinase regulatory domain receptor helix and one helix of the receptor four-helix bundle (Fig. 10) suggest a possible mechanism for the long-range propagation of signals in the chemosensory array. The signal triggered by attractant binding to one receptor dimer is transmitted through the array to as many as 30 other receptors, yielding strong positive cooperativity in attractant regulation of kinase activity 57,58,7,59–62. Given that the kinase regulatory domain forms a 6-membered alternating ring with CheW adaptor protein, and that each ring is connected via dimeric CheA kinases to three other rings (Fig. 1B), the hypothesized change in the equilibrium position and/or mobility of the kinase regulatory domain could be transmitted through one or more rings to the multiple receptors contacting those rings. The transmission of signals from the receptor 4-helix bundle into the ring system is proposed to involve the same type of Yin-Yang, frozen-dynamic coupling mechanism observed to transmit signals through adjacent regions of the receptor cytoplasmic domain 63,57,52,64. In this picture, attractant binding to a receptor dimer switches the 4-helix bundle at its cytoplasmic tip between (i) a looser, relatively dynamic “on-state” bundle that imposes few constraints on the receptor helix tightly bound to a CheA kinase regulatory domain, allowing that kinase regulatory domain to adopt its most stable packing with the CheW adaptor protein to yield a relatively static 6-membered ring, and (ii) a tighter, more frozen “off-state” bundle in which the helix stably bound to the CheA kinase regulatory domain becomes more constrained, which in turn displaces the kinase regulatory domain from its preferred ring position, yielding a more dynamic ring.
Overall, the present disulfide mapping and TAM-IDS analysis of the interface between the receptor and the CheA kinase regulatory domain validates the crystallographic view of the interface in the accompanying study by Li, Crane and coworkers 42, and shows this view is an accurate depiction of the interface in the functional, intact signaling array. The validated structure will enable further refinement of recent models for array architecture, since both current array models propose receptor-CheA kinase interfaces that differ significantly from the validated structure 19,20. The resulting rearrangements are not expected to dramatically shift the overall array architecture (Fig. 1B), but will likely enable the CheA kinase-CheW adaptor protein ring to adopt the correct vertical register. Finally, the present study reveals for the first time the dual functional roles of the interface between receptor and CheA kinase regulatory domain: in its structural role the interface contributes to stable CheA kinase binding in the array, and in its signaling role it is central to receptor-stimulated CheA kinase activation.
Supplementary Material
Acknowledgments
The authors thank Dr. Annette Erbse and Andrew Natale for helpful discussions and technical advice, and HHMI/CU Boulder UROP for support of undergraduate research by Caleb Ulliman.
Support provided by NIH R01 GM-040731 (to JJF)
ABBREVIATIONS
- ATP
adenosine triphosphate
- PMSF
phenylmethylsulfonylfluoride
- YFP
yellow fluorescent protein
- DTT
dithiothreitol
- EDTA
ethylenediaminetetraacetic acid
- SDS
sodium dodecyl sulfate
- PAGE
polyacrylamide gel electrophoresis
- Tris
(2-Amino-2-hydroxymethyl-propane-1,3-diol
- TBS
Tris-buffered saline
- TBST
Tris buffered saline and Tween-20
- TCEP
tris(2-carboxyethyl)phosphine
- Ni-NTA
nickel-nitrilotriacetic acid, nickel-charged resin
- BCA
bicinchoninic acid
- PVDF
polyvinyldifluoride
- NEM
N-ethylmaleimide
- BSA
bovine serum albumin
- TAM-IDS
Tryptophan and Alanine Mutation to Identify Docking Sites
- PICM
Protein Interactions by Cysteine Modification
- SH3
SRC Homology 3
Footnotes
SUPPORTING INFORMATION AVAILABLE
Supporting Information is provided that describes (i) the minimal effect of Cu(II) on the function of chemosensory arrays (Fig. S1), (ii) the negligible effect of attractant on fast disulfide formation rates between the N-terminal helix of the receptor hairpin and CheA kinase regulatory domain (Fig. S2), (iii) the retention of the normal CheW adaptor protein requirement for stable binding of mutant CheA kinase to arrays (Fig. S3), and (iv) the correspondence between relevant sequence positions in CheA kinase regulatory domains, CheW adaptor proteins, and receptors from S. typhimurium, E. coli and T. maritime (Table S1). This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Hazelbauer GL, Falke JJ, Parkinson JS. Bacterial chemoreceptors: high-performance signaling in networked arrays. Trends Biochem Sci. 2008;33:9–19. doi: 10.1016/j.tibs.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gao R, Stock AM. Biological insights from structures of two-component proteins. Annu Rev Microbiol. 2009;63:133–154. doi: 10.1146/annurev.micro.091208.073214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bourret RB, Silversmith RE. Two-component signal transduction. Curr Opin Microbiol. 2010;13:113–115. doi: 10.1016/j.mib.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sourjik V, Wingreen NS. Responding to chemical gradients: bacterial chemotaxis. Curr Opin Cell Biol. 2012;24:262–268. doi: 10.1016/j.ceb.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Briegel A, Ortega DR, Tocheva EI, Wuichet K, Li Z, Chen S, Muller A, Iancu CV, Murphy GE, Dobro MJ, Zhulin IB, Jensen GJ. Universal architecture of bacterial chemoreceptor arrays. Proc Natl Acad Sci U S A. 2009;106:17181–17186. doi: 10.1073/pnas.0905181106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim KK, Yokota H, Kim SH. Four-helical-bundle structure of the cytoplasmic domain of a serine chemotaxis receptor. Nature. 1999;400:787–792. doi: 10.1038/23512. [DOI] [PubMed] [Google Scholar]
- 7.Li G, Weis RM. Covalent modification regulates ligand binding to receptor complexes in the chemosensory system of Escherichia coli. Cell. 2000;100:357–365. doi: 10.1016/s0092-8674(00)80671-6. [DOI] [PubMed] [Google Scholar]
- 8.Studdert CA, Parkinson JS. Crosslinking snapshots of bacterial chemoreceptor squads. Proc Natl Acad Sci U S A. 2004;101:2117–2122. doi: 10.1073/pnas.0308622100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li M, Hazelbauer GL. Core unit of chemotaxis signaling complexes. Proc Natl Acad Sci U S A. 2011;108:9390–9395. doi: 10.1073/pnas.1104824108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li M, Khursigara CM, Subramaniam S, Hazelbauer GL. Chemotaxis kinase CheA is activated by three neighbouring chemoreceptor dimers as effectively as by receptor clusters. Mol Microbiol. 2011;79:677–685. doi: 10.1111/j.1365-2958.2010.07478.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bowie JU, Pakula AA, Simon MI. The three-dimensional structure of the aspartate receptor from Escherichia coli. Acta crystallographica Section D, Biological crystallography. 1995;51:145–154. doi: 10.1107/S0907444994010498. [DOI] [PubMed] [Google Scholar]
- 12.Chervitz SA, Falke JJ. Molecular mechanism of transmembrane signaling by the aspartate receptor: a model. Proc Natl Acad Sci U S A. 1996;93:2545–2550. doi: 10.1073/pnas.93.6.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yeh JI, Biemann HP, Prive GG, Pandit J, Koshland DE, Jr, Kim SH. High-resolution structures of the ligand binding domain of the wild-type bacterial aspartate receptor. J Mol Biol. 1996;262:186–201. doi: 10.1006/jmbi.1996.0507. [DOI] [PubMed] [Google Scholar]
- 14.Falke JJ, Hazelbauer GL. Transmembrane signaling in bacterial chemoreceptors. Trends Biochem Sci. 2001;26:257–265. doi: 10.1016/s0968-0004(00)01770-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tajima H, Imada K, Sakuma M, Hattori F, Nara T, Kamo N, Homma M, Kawagishi I. Ligand specificity determined by differentially arranged common ligand-binding residues in bacterial amino acid chemoreceptors Tsr and Tar. J Biol Chem. 2011;286:42200–42210. doi: 10.1074/jbc.M111.221887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mehan RS, White NC, Falke JJ. Mapping out regions on the surface of the aspartate receptor that are essential for kinase activation. Biochemistry. 2003;42:2952–2959. doi: 10.1021/bi027127g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller AS, Kohout SC, Gilman KA, Falke JJ. CheA Kinase of bacterial chemotaxis: chemical mapping of four essential docking sites. Biochemistry. 2006;45:8699–8711. doi: 10.1021/bi060580y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Erbse AH, Falke JJ. The core signaling proteins of bacterial chemotaxis assemble to form an ultrastable complex. Biochemistry. 2009;48:6975–6987. doi: 10.1021/bi900641c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Briegel A, Li X, Bilwes AM, Hughes KT, Jensen GJ, Crane BR. Bacterial chemoreceptor arrays are hexagonally packed trimers of receptor dimers networked by rings of kinase and coupling proteins. Proc Natl Acad Sci U S A. 2012 doi: 10.1073/pnas.1115719109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu J, Hu B, Morado DR, Jani S, Manson MD, Margolin W. Molecular architecture of chemoreceptor arrays revealed by cryoelectron tomography of Escherichia coli minicells. Proc Natl Acad Sci U S A. 2012;109:E1481–1488. doi: 10.1073/pnas.1200781109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Slivka PF, Falke JJ. Isolated Bacterial Chemosensory Array Possesses Quasi- and Ultrastable Components: Functional Links between Array Stability, Cooperativity, and Order. Biochemistry. 2012;51:10218–10228. doi: 10.1021/bi301287h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vu A, Wang X, Zhou H, Dahlquist FW. The receptor-CheW binding interface in bacterial chemotaxis. J Mol Biol. 2012;415:759–767. doi: 10.1016/j.jmb.2011.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang X, Vu A, Lee K, Dahlquist FW. CheA-receptor interaction sites in bacterial chemotaxis. J Mol Biol. 2012;422:282–290. doi: 10.1016/j.jmb.2012.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McEvoy MM, Zhou H, Roth AF, Lowry DF, Morrison TB, Kay LE, Dahlquist FW. Nuclear magnetic resonance assignments and global fold of a CheY-binding domain in CheA, the chemotaxis-specific kinase of Escherichia coli. Biochemistry. 1995;34:13871–13880. doi: 10.1021/bi00042a019. [DOI] [PubMed] [Google Scholar]
- 25.Bilwes AM, Alex LA, Crane BR, Simon MI. Structure of CheA, a signal-transducing histidine kinase. Cell. 1999;96:131–141. doi: 10.1016/s0092-8674(00)80966-6. [DOI] [PubMed] [Google Scholar]
- 26.Mourey L, Da Re S, Pedelacq JD, Tolstykh T, Faurie C, Guillet V, Stock JB, Samama JP. Crystal structure of the CheA histidine phosphotransfer domain that mediates response regulator phosphorylation in bacterial chemotaxis. J Biol Chem. 2001;276:31074–31082. doi: 10.1074/jbc.M101943200. [DOI] [PubMed] [Google Scholar]
- 27.Gloor SL, Falke JJ. Thermal domain motions of CheA kinase in solution: Disulfide trapping reveals the motional constraints leading to trans-autophosphorylation. Biochemistry. 2009;48:3631–3644. doi: 10.1021/bi900033r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Griswold IJ, Zhou H, Matison M, Swanson RV, McIntosh LP, Simon MI, Dahlquist FW. The solution structure and interactions of CheW from Thermotoga maritima. Nat Struct Biol. 2002;9:121–125. doi: 10.1038/nsb753. [DOI] [PubMed] [Google Scholar]
- 29.Liu JD, Parkinson JS. Genetic evidence for interaction between the CheW and Tsr proteins during chemoreceptor signaling by Escherichia coli. J Bacteriol. 1991;173:4941–4951. doi: 10.1128/jb.173.16.4941-4951.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sferdean FC, Weis RM, Thompson LK. Ligand affinity and kinase activity are independent of bacterial chemotaxis receptor concentration: insight into signaling mechanisms. Biochemistry. 2012;51:6920–6931. doi: 10.1021/bi3007466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bourret RB, Stock AM. Molecular information processing: lessons from bacterial chemotaxis. J Biol Chem. 2002;277:9625–9628. doi: 10.1074/jbc.R100066200. [DOI] [PubMed] [Google Scholar]
- 32.Gegner JA, Graham DR, Roth AF, Dahlquist FW. Assembly of an MCP receptor, CheW, and kinase CheA complex in the bacterial chemotaxis signal transduction pathway. Cell. 1992;70:975–982. doi: 10.1016/0092-8674(92)90247-a. [DOI] [PubMed] [Google Scholar]
- 33.Welch M, Chinardet N, Mourey L, Birck C, Samama JP. Structure of the CheY-binding domain of histidine kinase CheA in complex with CheY. Nat Struct Biol. 1998;5:25–29. doi: 10.1038/nsb0198-25. [DOI] [PubMed] [Google Scholar]
- 34.Quezada CM, Gradinaru C, Simon MI, Bilwes AM, Crane BR. Helical shifts generate two distinct conformers in the atomic resolution structure of the CheA phosphotransferase domain from Thermotoga maritima. J Mol Biol. 2004;341:1283–1294. doi: 10.1016/j.jmb.2004.06.061. [DOI] [PubMed] [Google Scholar]
- 35.Park SY, Borbat PP, Gonzalez-Bonet G, Bhatnagar J, Pollard AM, Freed JH, Bilwes AM, Crane BR. Reconstruction of the chemotaxis receptor-kinase assembly. Nature structural & molecular biology. 2006;13:400–407. doi: 10.1038/nsmb1085. [DOI] [PubMed] [Google Scholar]
- 36.Pollard AM, Bilwes AM, Crane BR. The structure of a soluble chemoreceptor suggests a mechanism for propagating conformational signals. Biochemistry. 2009;48:1936–1944. doi: 10.1021/bi801727m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bhatnagar J, Borbat PP, Pollard AM, Bilwes AM, Freed JH, Crane BR. Structure of the ternary complex formed by a chemotaxis receptor signaling domain, the CheA histidine kinase, and the coupling protein CheW as determined by pulsed dipolar ESR spectroscopy. Biochemistry. 2010;49:3824–3841. doi: 10.1021/bi100055m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bass RB, Falke JJ. Detection of a conserved alpha-helix in the kinase-docking region of the aspartate receptor by cysteine and disulfide scanning. J Biol Chem. 1998;273:25006–25014. doi: 10.1074/jbc.273.39.25006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bass RB, Coleman MD, Falke JJ. Signaling domain of the aspartate receptor is a helical hairpin with a localized kinase docking surface: cysteine and disulfide scanning studies. Biochemistry. 1999;38:9317–9327. doi: 10.1021/bi9908179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ames P, Studdert CA, Reiser RH, Parkinson JS. Collaborative signaling by mixed chemoreceptor teams in Escherichia coli. Proc Natl Acad Sci U S A. 2002;99:7060–7065. doi: 10.1073/pnas.092071899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mowery P, Ostler JB, Parkinson JS. Different signaling roles of two conserved residues in the cytoplasmic hairpin tip of Tsr, the Escherichia coli serine chemoreceptor. J Bacteriol. 2008;190:8065–8074. doi: 10.1128/JB.01121-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li X, Fleetwood A, Bilwes AM, Ortega DR, Zhulin IB, Crane BR. The 3.2 Å resolution structure of a Receptor:CheA:CheW signaling complex defines overlapping binding sites and key residue interactions within bacterial chemosensory arrays. 2013 doi: 10.1021/bi400383e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miller AS, Falke JJ. Side chains at the membrane-water interface modulate the signaling state of a transmembrane receptor. Biochemistry. 2004;43:1763–1770. doi: 10.1021/bi0360206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chervitz SA, Lin CM, Falke JJ. Transmembrane signaling by the aspartate receptor: engineered disulfides reveal static regions of the subunit interface. Biochemistry. 1995;34:9722–9733. doi: 10.1021/bi00030a010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kentner D, Sourjik V. Spatial organization of the bacterial chemotaxis system. Curr Opin Microbiol. 2006;9:619–624. doi: 10.1016/j.mib.2006.10.012. [DOI] [PubMed] [Google Scholar]
- 46.Vogel HJ, Bonner DM. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956;218:97–106. [PubMed] [Google Scholar]
- 47.Parkinson JS, Houts SE. Isolation and behavior of Escherichia coli deletion mutants lacking chemotaxis functions. J Bacteriol. 1982;151:106–113. doi: 10.1128/jb.151.1.106-113.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weis RM, Koshland DE., Jr Reversible receptor methylation is essential for normal chemotaxis of Escherichia coli in gradients of aspartic acid. Proc Natl Acad Sci U S A. 1988;85:83–87. doi: 10.1073/pnas.85.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Winston SE, Mehan R, Falke JJ. Evidence that the adaptation region of the aspartate receptor is a dynamic four-helix bundle: cysteine and disulfide scanning studies. Biochemistry. 2005;44:12655–12666. doi: 10.1021/bi0507884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Careaga CL, Falke JJ. Structure and dynamics of Escherichia coli chemosensory receptors. Engineered sulfhydryl studies. Biophys J. 1992;62:209–216. doi: 10.1016/S0006-3495(92)81806-4. discussion 217–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bass RB, Butler SL, Chervitz SA, Gloor SL, Falke JJ. Use of site-directed cysteine and disulfide chemistry to probe protein structure and dynamics: applications to soluble and transmembrane receptors of bacterial chemotaxis. Methods Enzymol. 2007;423:25–51. doi: 10.1016/S0076-6879(07)23002-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Swain KE, Gonzalez MA, Falke JJ. Engineered socket study of signaling through a four-helix bundle: evidence for a yin-yang mechanism in the kinase control module of the aspartate receptor. Biochemistry. 2009;48:9266–9277. doi: 10.1021/bi901020d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schoenmakers TJ, Visser GJ, Flik G, Theuvenet AP. CHELATOR: an improved method for computing metal ion concentrations in physiological solutions. BioTechniques. 1992;12:870–874. 876–879. [PubMed] [Google Scholar]
- 54.Cunningham BC, Wells JA. High-resolution epitope mapping of hGH-receptor interactions by alanine-scanning mutagenesis. Science. 1989;244:1081–1085. doi: 10.1126/science.2471267. [DOI] [PubMed] [Google Scholar]
- 55.Sharp LL, Zhou J, Blair DF. Features of MotA proton channel structure revealed by tryptophan-scanning mutagenesis. Proc Natl Acad Sci U S A. 1995;92:7946–7950. doi: 10.1073/pnas.92.17.7946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sharp LL, Zhou J, Blair DF. Tryptophan-scanning mutagenesis of MotB, an integral membrane protein essential for flagellar rotation in Escherichia coli. Biochemistry. 1995;34:9166–9171. doi: 10.1021/bi00028a028. [DOI] [PubMed] [Google Scholar]
- 57.Bray D, Levin MD, Morton-Firth CJ. Receptor clustering as a cellular mechanism to control sensitivity. Nature. 1998;393:85–88. doi: 10.1038/30018. [DOI] [PubMed] [Google Scholar]
- 58.Bornhorst JA, Falke JJ. Attractant regulation of the aspartate receptor-kinase complex: limited cooperative interactions between receptors and effects of the receptor modification state. Biochemistry. 2000;39:9486–9493. doi: 10.1021/bi0002737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sourjik V, Berg HC. Functional interactions between receptors in bacterial chemotaxis. Nature. 2004;428:437–441. doi: 10.1038/nature02406. [DOI] [PubMed] [Google Scholar]
- 60.Endres RG, Oleksiuk O, Hansen CH, Meir Y, Sourjik V, Wingreen NS. Variable sizes of Escherichia coli chemoreceptor signaling teams. Molecular systems biology. 2008;4:211. doi: 10.1038/msb.2008.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hansen CH, Sourjik V, Wingreen NS. A dynamic-signaling-team model for chemotaxis receptors in Escherichia coli. Proc Natl Acad Sci U S A. 2010;107:17170–17175. doi: 10.1073/pnas.1005017107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Skoge M, Meir Y, Wingreen NS. Dynamics of cooperativity in chemical sensing among cell-surface receptors. Physical review letters. 2011;107:178101. doi: 10.1103/PhysRevLett.107.178101. [DOI] [PubMed] [Google Scholar]
- 63.Kim SH. “Frozen” dynamic dimer model for transmembrane signaling in bacterial chemotaxis receptors. Protein science: a publication of the Protein Society. 1994;3:159–165. doi: 10.1002/pro.5560030201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhou Q, Ames P, Parkinson JS. Biphasic control logic of HAMP domain signalling in the Escherichia coli serine chemoreceptor. Mol Microbiol. 2011;80:596–611. doi: 10.1111/j.1365-2958.2011.07577.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.The PyMOL Molecular Graphics System, Version 1.5.0.4. Schrödinger, LLC; [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.