Abstract
The evolution of antibiotic resistance can now be rapidly tracked with high-throughput technologies for bacterial genotyping and phenotyping. Combined with new approaches to evolve resistance in the laboratory and to characterize clinically evolved resistant pathogens, these methods are revealing the molecular basis and rate of evolution of antibiotic resistance under treatment regimens of single drugs or drug combinations. In this Progress article, we review these new tools to study the evolution of antibiotic resistance, and discuss how the genomic and evolutionary insights they provide could transform the diagnosis, treatment and predictability of antibiotic resistance in bacterial infections.
Introduction
The evolution and spread of antibiotic resistance in bacterial pathogens is a growing threat to public health. The frequency of antibiotic resistance in many bacterial pathogens is increasing around the world, and the resulting failures of antibiotic therapy cause hundreds of thousands of deaths annually1. The hope of addressing this crisis by developing new antibiotics is diminished both by the low rate of novel antibiotic discovery and by the likelihood that pathogens will evolve resistance to novel antibiotics just as they have to existing antibiotics. The long-term threat, therefore, is just as much the process of evolution as the microbial pathogens themselves. Although the use of antibiotics inevitably promotes resistance, the rate of evolution depends on the genomic background and treatment strategies. Thus, understanding the genomics and evolutionary biology of antibiotic resistance can inform therapeutic strategies that are both effective and mitigate the future potential to evolve resistance.
Antibiotic resistance can be acquired either by mutation or by the horizontal transfer of resistance-conferring genes, often in mobile genetic cassettes. The relative contribution of these factors depends on the class of antibiotic and on the genetic plasticity of the bacterial species. For example, Mycobacterium tuberculosis acquires antibiotic resistance primarily through nucleotide changes, while hospital-acquired Enterobacteriaceae infections often possess multi-drug resistance cassettes and may also acquire nucleotide changes that confer resistance to drugs that are not often resisted by mobile elements, such as quinolones2.
Progress in DNA sequencing and other genotyping technologies means that the genotypes of pathogens will soon be widely available in clinical as well as research settings. Genotype-based antibiotic resistance profiling is already faster and more economical than phenotypic profiling in select cases (for example, rifampicin resistance in M tuberculosis caused by nucleotide substitutions, and methicillin resistance in Staphylococcus aureus caused by a resistance cassette), and over time therapeutic and infection control strategies will more heavily rely on information derived from genome sequencing of the infecting agents3.
Importantly, genotypes can inform not only on the current drug susceptibility of a pathogen but also on its future potential to evolve resistance and spread. For example, sequencing could determine whether a drug susceptible strain carries precursors to resistance genes (which are termed proto-resistance genes4), such as drug degrading enzymes or efflux pumps that might be mutated to increase expression or strengthen activity. Sequencing could also determine whether resistance cassettes may be only one mutation away from increased potency or the capacity to resist other drugs related to the originally resisted drug4,5. Even a modest predictive power might improve therapeutic outcomes by informing the selection of drugs, the preference between monotherapy or combination therapy and the temporal dosing regimen to select genotype-based treatments that are most resilient to evolution of resistance. To realize such a potential will require new tools to explore how different treatment regimes affect the genotypic and phenotypic evolutionary paths to antibiotic resistance in the laboratory and in the clinic. Here we discuss new tools to select for drug resistance, strategies for identifying and characterizing adaptive mutations in the evolved genotypes, and approaches to study the genetic constraints on the evolution of resistance.
SELECTION FOR DRUG RESISTANCE
Drug resistance in laboratory experiments
Laboratory evolution6 can investigate how the rate and genotypic path to resistance varies across different controlled drug treatment regimens. In a traditional selection experiment, bacteria are exposed to fixed drug doses that permit only the growth of resistant mutants. Typically, this approach identifies only a single adaptive step and does not reveal how multiple mutations can accrue sequentially to confer strong resistance (Figure 1a). Technological innovations now facilitate rapid multi-step experimental evolution, revealing long-term evolutionary paths. Recurrent evolutionary patterns, such as the appearance of mutations in a preferred order, provide some level of predictability to a seemingly stochastic evolutionary process7. Devices for establishing spatial or temporal gradients of drug concentration allow evolving populations to be continuously challenged by effectively increasing the drug dosage to maintain selective pressure as stronger antibiotic resistance evolves. Continuous culture devices (for example, turbidostats) can be modified to increase drug dose steadily over time8, to implement automated feedback control of drug dosage in response to increasing levels of resistance7, or to mimic the antibiotic dosing regime experienced within a patient (Figure 1b). Multi-step experimental evolution can also be carried out in spatial drug gradients, as was demonstrated by a microfluidic device of connected chambers implementing a spatial drug gradient, allowing bacteria to expand throughout the device only as they evolve increasing levels of antibiotic resistance9 (Figure 1c). These experiments have revealed that although the evolution of resistance can follow similar phenotypic paths in replicate experiments, the underlying genotypic process can be variable for some drugs (for example, chloramphenicol, doxycycline) but reproducible for other drugs (for example, trimethoprim, ciprofloxacin)7,9. Substantial variability in rate is also observed: resistance to some drugs increases 1000-fold over 20 days, while resistance to other drugs may increase only 10-fold over the same period7. Therefore, for any specific genotype there could be vast differences between drugs in the propensity for resistance and the mechanisms by which resistance is acquired; these factors are crucial to the design of combination treatments that inhibit the evolution of resistance.
Figure 1. Selection of antibiotic resistant bacteria from experimental evolution.
Gradients of drug concentration over time or space facilitate multi-step experimental evolution. a, In a classical selection for antibiotic resistance, a uniform drug concentration selects for only a single mutation. b, A continuous culture device can select for multiple resistance-conferring mutations by dynamically increasing drug concentration in response to increasing drug resistance. c, If bacteria can migrate over a spatial gradient of drug concentration then they can explore larger regions of space only as they evolve increasing levels of drug resistance.
Combination therapy has the potential to slow the evolution of resistance, as a bacterial subpopulation with a mutation that renders it resistant to one drug may still be inhibited by a second drug, preventing the growth of a large drug-resistant population (that might subsequently evolve multi-drug resistance)10. However, the choice of an optimal combination to slow evolution can crucially depend on the details of the treatment regimen, drug interactions and cross-resistance10–13. Experimental evolution has facilitated the systematic analysis of evolution under different combination therapies and is revealing the principles behind their ability to slow down and possibly even reverse the evolution of resistance12–15 (reviewed in16). Several approaches have been used to select for drug resistance in multi-drug environments: mutants can be selected from a grid of drug concentrations across multiple agar dishes12 or in a microtitre plate13. Multiple mutations that confer strong multidrug resistance can be selected by serial passaging across such gradients13, or through the use of drug combinations in the continuous culture devices described above7.
Many questions about the evolution of multi-drug resistance remain, including: to what extent is resistance acquired by a series of drug specific mutations versus mutations that each confer resistance to multiple drugs (that is, positive cross-resistance); in which cases can resistance to one drug lead to sensitivity to another (that is, negative cross-resistance); and, even when resistance to one drug does not immediately confer positive or negative cross-resistance to a second drug, can it affect the future evolution of resistance to the second drug? These and other questions about the evolution of resistance to single or multiple drug treatments are being addressed by the systematic selection methodologies outlined above. Although these methods are often first applied to model organisms, they can and should be applied more widely to study pathogens isolated from human infections.
Drug resistance in clinical isolates
The increasing capacity to sequence whole bacterial genomes has enabled detailed analyses of large collections of clinical isolates. Various sampling approaches are available to view the evolution and spread of antibiotic resistant bacteria over different scales (Figure 2). Isolates collected from individual patients over the course of acute and chronic infections have revealed the within-patient evolution of antibiotic resistance, instances of cross-resistance between antibiotics, the evolution of compensatory mutations that alleviate the fitness costs of resistance, and the transmission of specific antibiotic resistant clones between organs17–19. Sampling during the spread of an epidemic has been used to identify the likely patient-to-patient transmission of antibiotic sensitive or antibiotic resistant bacteria and may reveal tradeoffs between infectivity and antibiotic resistance18. At the very largest scale, worldwide sampling of endemic infections over decades has been used to determine long-term trends in the evolution of antibiotic resistance and pathogenicity, and to determine transmission patterns across continents20,21
Figure 2. Selection of antibiotic resistant bacteria from clinical isolates.
The evolution and transmission of antibiotic resistant bacteria can be studied over scales ranging from continents to organs by different approaches from clinical sampling. Worldwide sampling of isolates reveals intercontinental transmission, sampling within a localized epidemic reveals patient to patient transmission networks, and sampling within a single patient can reveal transfer between sites of the body and possibly organ-specific evolution.
FINDING THE GENOTYPIC BASIS
Identifying adaptive mutations
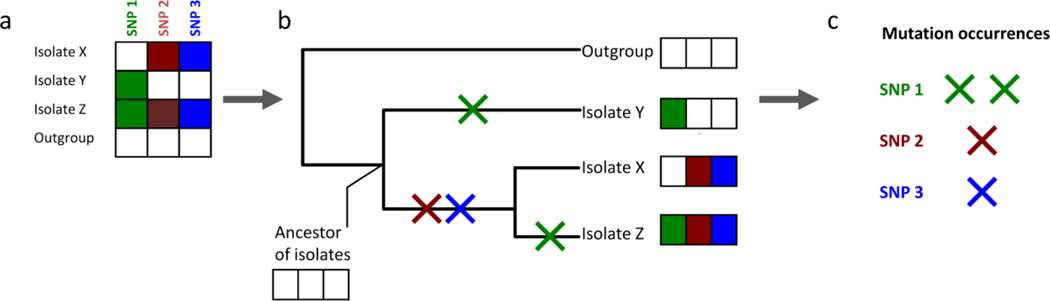
Comparing the genomes of ancestral and evolved strains identifies the precise genetic changes that underlie adaptive evolution. However, separating adaptive mutations from neutral or passenger mutations is challenging, particularly for clinical strains that may have been evolving antibiotic resistance over decades. In the context of contemporary bacterial evolution, tests for adaptive evolution based on rates of non-synonymous and synonymous substitutions (dN/dS) cannot be applied on a per gene basis as there are typically too few mutations for statistical power; also they should they be applied to a whole genome because different subsets of genes may have undergone adaptive, neutral, or purifying selection. Furthermore, such tests cannot determine whether an individual mutation is adaptive or neutral, and may neglect the possible role of adaptive non-coding or regulatory mutations. Fortunately, with increasing capacity to sequence many evolved strains, this challenge can be overcome by looking for parallel evolution. Parallel evolution provides a tool to distinguish adaptive mutations from neutral or deleterious mutations, as non-advantageous mutations should not independently arise and fix at the same loci as frequently as adaptive mutations. Additionally, the identification of adaptive mutations by parallel evolution is not biased against synonymous or regulatory mutations, even through the set of adaptive mutations will probably be enriched for non-synonymous substitutions. In a recent study of a bacterial epidemic in which parallel evolution occurred within multiple patients, the bacterial genome as a whole showed no statistical sign of adaptive evolution (the ratio of non-synonymous to synonymous substitutions, dN/dS, was as expected under drift), but examining dN/dS identified adaptive evolution against a background signal of purifying selection when genes were classified by whether they mutated only once, or whether repeatedly across the cohort. Genes that mutated only once showed signs of purifying selection (that is, unexpectedly few non-synonymous substitutions) and genes that mutated repeatedly showed a strong signature of adaptive evolution (that is, an unexpectedly high rate of non-synonymous substitutions)18. An important caveat applies to the identification of parallel evolution in clinical isolates: the repeated observation of a mutation could be a result of shared ancestry and does not necessarily imply that the same mutation arose repeatedly; a phylogenetic tree must be constructed to estimate the number of independent mutational events at each loci in the strains' histories (Figure 3).
Figure 3. Phylogenetic inference identifies parallel evolution.
a, A collection of related isolates will possess many shared mutations relative to a more distantly strain (an outgroup), but this does not necessarily imply that any of these mutations repeatedly occurred. b, Phylogenetic inference estimates the likely evolutionary history that connects the isolates and identifies when each mutation occurred. Note that many other mutations would need to have occurred for accurate phylogenetic inference; in this example only 3 mutations are shown to illustrate the principle. c, From the phylogenetic tree, the number of times that a gene mutated independently in separate lineages can be counted to distinguish mutations that are shared merely by common ancestry (red and blue) from mutations that are shared by parallel evolution (green), strongly indicating adaptive evolution. SNP, single-nucleotide polymorphism.
Phylogenetic trees describe the evolutionary history of related strains, providing crucial contributions to understanding mutation, selection, and transmission in the evolution of antibiotic resistance (see reference22 for a tutorial). A phylogenetic tree provides a view of transmission events across as large or as fine a scale as is represented by clinical isolates, from intercontinental transmission to transfer between the organs of a single patient. Specific genetic changes throughout the evolutionary history of bacterial strains can be correlated with the appearance of novel phenotypes including antibiotic resistance, changes in pathogenicity or fitness, or propensity for transmission. Whereas recombination amongst related bacterial strains can complicate the construction of a phylogenetic tree, maximum likelihood and Bayesian approaches can identify clusters of mutations that are more likely to be shared by recombination than point mutation23. Phylogenetic reconstruction can then be carried out only on vertically transmitted point mutations, as demonstrated in a recent study of worldwide isolates of the highly recombinogenic Streptococcus pneumoniae21.
Measuring the phenotypic effects of mutations
Even when adaptive mutations are identified, it is not necessarily straightforward to determine their specific phenotypic effects. That is, whether they increase antibiotic resistance, compensate for the fitness costs of antibiotic resistance, or confer adaptation to a host environment. Thus the lessons of high-throughput genotyping are limited unless combined with high-throughput phenotyping.
Fitness costs are a common feature of mutations that confer antibiotic resistance, which epidemiological models predict to substantially affect the spread of drug resistant pathogens24,25. The relative fitness of evolved versus ancestral strains can be measured by competition experiments in drug-free or antibiotic-containing environments. Throughput and precision were previously limited by the labor of counting colonies, but these experiments can now be automated using fluorescent labels for counting by flow cytometry or DNA barcodes for counting by next-generation sequencing14,15,26. Similar methods can also be applied to genetically intractable clinical isolates by deep-sequencing of mutated loci to measure allele frequencies27 Improvements in the precision of fitness measurements will probably be of benefit to epidemiological modeling25. Another high-throughput phenotyping tool, which is applicable to model organisms and clinical isolates alike, is automated imaging arrays built from flatbed scanners. These cheap custom systems acquire time-lapse videos of colony growth on large numbers of agar plates, that can be arranged to span ranges of antibiotic concentration or multiple antibiotics12,28. Studies using genetic complementation have also benefited from technological progress: the relative contributions to fitness of each mutated locus in an evolved strain can be simultaneously determined by a competition experiment between a mixture of strains, each transduced with a different fragment of the evolved genome29. Repeating such an experiment in the presence and absence of antibiotic could reveal both the degree of antibiotic resistance conferred by each mutation and the fitness cost during drug-free growth.
EVOLUTIONARY POTENTIAL AND CONSTRAINTS
The approaches described above are based on natural selection methodologies to identify adaptive mutations that spontaneously appear under drug treatments. Such evolutionary based approaches are powerful in determining the rate of adaptation and revealing its most likely genotypic paths, but it does not explicitly elucidate unlikely or ‘forbidden’ steps that can have the effect of directing evolution repeatedly along the few permitted paths. To systematically explore the effects of defined genetic changes or combinations thereof, whether advantageous or deleterious, a reverse genetics approach can be used. Here we review recent creative uses of reverse genetics to explore how systematic genetic perturbations, mutation combinations, and horizontal gene transfer can enhance or constrain evolutionary potential.
Systematic genetic perturbations
The genetic determinants of antibiotic resistance can be explored with pre-constructed libraries of mutant strains. For example, known resistance genes can be mutagenized to explore their adaptive potential and to measure the distribution of mutational effects. Applying this method to the most common beta-lactamase gene in gram-negative bacteria, TEM-1, has identified a long-tailed distribution with a few highly beneficial mutations30, potentially explaining the high degree of reproducibility often observed in the evolution of antibiotic resistance7,9,31. A genome-wide view can be taken with gene deletion libraries and Open Reading Frame expression libraries; although these were first constructed only for model organisms, advances in transposon mutagenesis are enabling the rapid construction of comparable libraries for clinically relevant pathogens32. These libraries can be screened in pools by employing next-generation sequencing to count the abundance of each strain in a mixture following drug selection32. Screening mutant strain libraries under antibiotic treatment identifies genes for which deletion or over-expression alters drug susceptibility, revealing the genetic basis of intrinsic antibiotic susceptibility or identifying paths to stronger antibiotic resistance33. Future studies could use these mutant strain libraries also as starting material for pooled evolution experiments, thereby identifying not only the immediate effects of the genetic perturbations, but also their effect on the potential to evolve yet higher levels of resistance.
Combinatorial genetic libraries
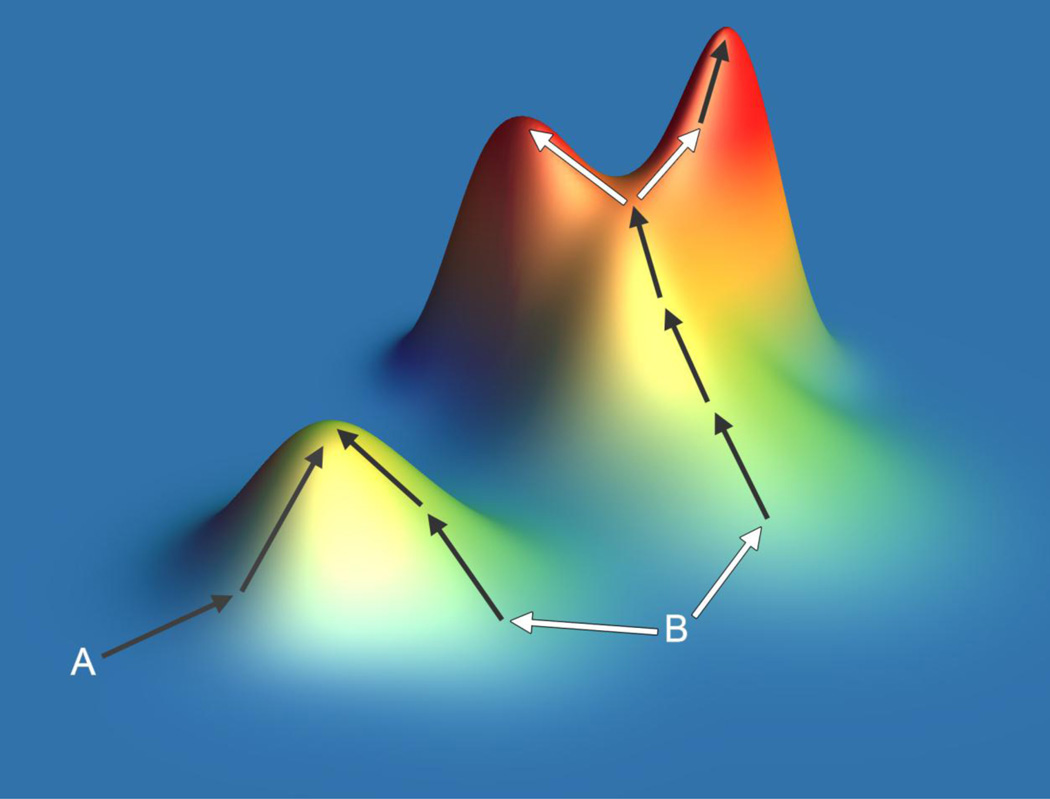
The effects of a mutation depend on the genetic background on which it arises. Genetic interactions between alleles impose constraints on the evolutionary pathways to antibiotic resistance, as a mutation may be beneficial only in the presence or absence of certain other mutations. The synthetic construction of different combinations of mutations that have previously been identified from the clinic or experimental evolution can reveal genetic constraints that would not be observed from studying only those mutation combinations favored in nature. This method has been applied to genes found in resistance cassettes as well as drug target genes, both being cases in which resistance can be increased by repeated mutation of the same gene. These studies have consistently observed strong constraints that can be responsible for the repeatability, and hence predictability, of evolutionary pathways34. Genetic interactions have been observed to limit the possible pathways to a few select sequences of mutations35,36 (Figure 4) and to limit the reversibility of evolution when switching between different drugs37. This approach has shown that in certain combinations, resistance mutations can also act as compensatory mutations that alleviate one another’s fitness costs, producing strongly drug-resistant or multi-drug resistant strains without substantial fitness costs38–40. Evolutionary experiments can also be carried out starting from different pre-built genotypes to investigate genetic influences on the reproducibility of evolution: one such study has revealed that different initial mutations in the TEM-1 beta-lactamase can define the subsequent evolutionary pathways31 (Figure 4). These approaches could identify those genotypes with a greater or lesser potential to evolve resistance to particular drugs, which could be valuable in selecting genotype-specific treatments that avoid the most harmful evolutionary outcomes.
Figure 4. Constrained evolutionary pathways to antibiotic resistance.
The properties of evolutionary processes can be illustrated by the concept of the ‘fitness landscape’. In this demonstration of several experimentally observed behaviors, height represents drug resistance. Different starting genotypes (A and B) may have a different propensity to evolve resistance owing to their proximities to drug-resistance peaks of varying height. The first genotypic step towards resistance can sometimes define the final genotype and level of resistance (arrows from B). The pathways to resistance can at times be constrained and predictable (dark arrows), but evolutionary pathways can diverge (light arrows) to distinct peaks separated by negative genetic interactions.
Horizontal transfer of environmental genes
The acquisition of resistance by horizontal gene transfer (HGT) provides evolutionary potential that cannot be predicted from an organism’s original (pre-transfer) genome. Instead, the potential for resistance by HGT can be investigated by sampling the extensive and ancient ability of genes in environmental or commensal microbes to resist a drug41–43. This approach has been implemented by extracting and cloning microbial DNA from soil samples or from human gut samples into a laboratory strain and plating on inhibitory concentrations of a range of antibiotics to identify novel microbial drug resistance genes that may in the future transfer into pathogens42,44. Although this approach is limited to identifying genes that can be successfully expressed in the laboratory strain, a more recent study demonstrated an expression-independent approach that directly assessed the capacity of environmental microbes to degrade the new and rarely clinically resisted antibiotic daptomycin45. A collection of environmental actinomycetes were screened for daptomycin resistance, and the supernatants of resistant cultures were analyzed by mass spectrometry to view the structures of daptomycin and its inactivation products. By precisely viewing the drug degradation products, the molecular mechanisms of resistance by degradation were inferred. This level of understanding has the potential to suggest structural variants of drugs that could resist environmental mechanisms of degradation.
TOWARDS THERAPIES INFORMED BY EVOLUTION
The short to medium-term challenge is to develop novel antibiotics to kill today’s antibiotic resistant bacteria, but the ‘arms race’ between antibiotic development and the evolution of resistance in pathogens is growing increasingly difficult. Addressing the long-term challenge posed by antibiotic resistant pathogens will require therapeutic strategies10 and compound development46,47 that considers ways to manipulate and slow the evolution of resistance.
The capacity for genome sequencing is approaching the point at which endemic pathogens can be extensively sampled and sequenced around the world, and evolution during bacterial outbreaks can be tracked in real time48–50. The ability to trace routes of bacterial transmission precisely is clearly of benefit to infection control efforts, which are especially critical for highly drug resistant infections for which few therapeutic options remain. Yet the value of genome sequencing extends beyond epidemic control: the genome of an organism defines its current antibiotic resistance and, to a large extent, its potential to evolve further resistance. The application of the methods reviewed here to genotype and phenotype drug resistant pathogens can identify resistance (or proto-resistance) genes and the ways that they might mutate to increase antibiotic resistance. Whole genome sequencing of pathogens thus has the potential to provide a catalog of the various pathways to resistance under different treatment regimens.
To treat a drug resistant infection, an understanding of interactions between drugs and resistance mutations can guide the selection of second-line therapies or combination therapies. Such therapies should have the weakest possible cross-resistance – negative cross-resistance if possible – and they should be chosen such that the genetic interactions between drug-specific resistance mutations lead to an accumulation of fitness costs rather than compensation. Finally, the specific genetic basis of resistance to a first-line drug may, through genetic interactions, alter the expected capacity to evolve resistance to second-line drugs. Such cases may identify sets of drugs that, when used in sequence or in combinations, minimize the risk that strong resistance will evolve. The widespread application of the new methods reviewed in this article might thus facilitate an evolutionary medicine paradigm in which pathogen genotyping coupled with evolutionary genetics guides the optimal choice of temporal and combinatorial drug treatment.
Acknowledgements
We thank T. Lieberman for discussions on phylogeny and comments on the manuscript. This work was supported in part by US National Institutes of Health grants R01GM081617 and US National Institute of General Medical Science Center grant P50GM068763, and the Novartis Institutes for BioMedical Research.
Glossary
- β-lactamase
An enzyme that can confer resistance to β-lactam antibiotics by catalysing their degradation.
- Commensal microbes
Microbes living on or in a host without causing disease, although they typically include opportunistic pathogens.
- Cross-resistance
The propensity of a genetic change that confers resistance to one drug also to affect resistance to a different drug (by either increasing or decreasing resistance).
- dN/dS
The ratio of mutation rates at nonsynonymous (N) and synonymous (S) sites. dN/dS is increased by selection for amino acid changes (a signature of adaptive selection) and decreased by selection against amino acid changes (purifying selection).
- Horizontal gene transfer
The acquisition of a gene by a means other than direct inheritance from a parent cell (vertical transfer). Common in many bacteria and archaea, mechanisms of horizontal gene transfer include transformation, conjugation and transduction.
- Maximum likelihood and Bayesian approaches
This definition applies to the context of phylogenetics. Phylogenetic trees can be constructed by maximum parsimony, maximum likelihood and Bayesian inference. Maximum parsimony methods select from all possible trees the one containing the fewest mutations. Trees chosen by maximum likelihood and other Bayesian methods may contain more mutations, as they weigh the relative probabilities of different mutations according to various models.
- Microfluidic device
Customized microscopic chambers in which fluid flows can be precisely controlled. Applied to microbiology these allow the study of bacterial behaviour in spatially and temporally controllable environments.
- Monotherapy
Chemical therapy by a single drug.
- Parallel evolution
When the same mutations (or a range of mutations in the same gene) repeatedly occur in independent lineages; this provides an indication that these mutations may have been fixed by positive selection rather than by chance.
- Proto-resistance genes
Evolutionary precursors to drug-resistance genes that do not yet contribute to drug resistance but may do so on mutation and selection by drug stress.
- Resistance cassettes
A genetic element containing one or more drug resistance genes, often carried in transposable elements or plasmids that facilitate horizontal gene transfer.
- Transposon mutagenesis
The insertion of transposons at random locations throughout a genome to generate a library of different gene disruptions. Transposons can be constructed with outward-facing promoters also to introduce gene overexpression into the library.
- Turbidostats
Devices that maintain constant cell density (turbidity) in a continuously growing microbial culture by routinely removing a small volume of culture and replacing it with fresh sterile media.
Footnotes
Competing interests statement
The authors declare no competing financial interests
References
- 1.The evolving threat of antimicrobial resistance: Options for action. Geneva: World Health Organization; 2012. [Google Scholar]
- 2.Paterson DL. Resistance in gram-negative bacteria: enterobacteriaceae. Am J Med. 2006;119:S20–S28. doi: 10.1016/j.amjmed.2006.03.013. discussion S62-70. [DOI] [PubMed] [Google Scholar]
- 3.Didelot X, Bowden R, Wilson DJ, Peto TE, Crook DW. Transforming clinical microbiology with bacterial genome sequencing. Nat Rev Genet. 2012;13:601–612. doi: 10.1038/nrg3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morar M, Wright GD. The genomic enzymology of antibiotic resistance. Annu Rev Genet. 2010;44:25–51. doi: 10.1146/annurev-genet-102209-163517. [DOI] [PubMed] [Google Scholar]
- 5.Novais A, et al. Evolutionary trajectories of beta-lactamase CTX-M-1 cluster enzymes: predicting antibiotic resistance. PLoS Pathog. 2010;6:e1000735. doi: 10.1371/journal.ppat.1000735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elena SF, Lenski RE. Evolution experiments with microorganisms: the dynamics and genetic bases of adaptation. Nat Rev Genet. 2003;4:457–469. doi: 10.1038/nrg1088. [DOI] [PubMed] [Google Scholar]
- 7.Toprak E, et al. Evolutionary paths to antibiotic resistance under dynamically sustained drug selection. Nat Genet. 2012;44:101–105. doi: 10.1038/ng.1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee HH, Molla MN, Cantor CR, Collins JJ. Bacterial charity work leads to population-wide resistance. Nature. 2010;467:82–85. doi: 10.1038/nature09354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Q, et al. Acceleration of emergence of bacterial antibiotic resistance in connected microenvironments. Science. 2011;333:1764–1767. doi: 10.1126/science.1208747. [DOI] [PubMed] [Google Scholar]
- 10.Torella JP, Chait R, Kishony R. Optimal drug synergy in antimicrobial treatments. PLoS Comput Biol. 2010;6:e1000796. doi: 10.1371/journal.pcbi.1000796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bonhoeffer S, Lipsitch M, Levin BR. Evaluating treatment protocols to prevent antibiotic resistance. Proc Natl Acad Sci U S A. 1997;94:12106–12111. doi: 10.1073/pnas.94.22.12106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Michel JB, Yeh PJ, Chait R, Moellering RC, Jr, Kishony R. Drug interactions modulate the potential for evolution of resistance. Proc Natl Acad Sci U S A. 2008;105:14918–14923. doi: 10.1073/pnas.0800944105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hegreness M, Shoresh N, Damian D, Hartl D, Kishony R. Accelerated evolution of resistance in multidrug environments. Proc Natl Acad Sci U S A. 2008;105:13977–13981. doi: 10.1073/pnas.0805965105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chait R, Craney A, Kishony R. Antibiotic interactions that select against resistance. Nature. 2007;446:668–671. doi: 10.1038/nature05685. [DOI] [PubMed] [Google Scholar]
- 15.Palmer AC, Angelino E, Kishony R. Chemical decay of an antibiotic inverts selection for resistance. Nat Chem Biol. 2010;6:105–107. doi: 10.1038/nchembio.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yeh PJ, Hegreness MJ, Aiden AP, Kishony R. Drug interactions and the evolution of antibiotic resistance. Nat Rev Microbiol. 2009;7:460–466. doi: 10.1038/nrmicro2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mwangi MM, et al. Tracking the in vivo evolution of multidrug resistance in Staphylococcus aureus by whole-genome sequencing. Proc Natl Acad Sci U S A. 2007;104:9451–9456. doi: 10.1073/pnas.0609839104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lieberman TD, et al. Parallel bacterial evolution within multiple patients identifies candidate pathogenicity genes. Nat Genet. 2011;43:1275–1280. doi: 10.1038/ng.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Comas I, et al. Whole-genome sequencing of rifampicin-resistant Mycobacterium tuberculosis strains identifies compensatory mutations in RNA polymerase genes. Nat Genet. 2012;44:106–110. doi: 10.1038/ng.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harris SR, et al. Evolution of MRSA during hospital transmission and intercontinental spread. Science. 2010;327:469–474. doi: 10.1126/science.1182395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Croucher NJ, et al. Rapid pneumococcal evolution in response to clinical interventions. Science. 2011;331:430–434. doi: 10.1126/science.1198545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baldauf SL. Phylogeny for the faint of heart: a tutorial. Trends Genet. 2003;19:345–351. doi: 10.1016/S0168-9525(03)00112-4. [DOI] [PubMed] [Google Scholar]
- 23.Didelot X, Falush D. Inference of bacterial microevolution using multilocus sequence data. Genetics. 2007;175:1251–1266. doi: 10.1534/genetics.106.063305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cohen T, Murray M. Modeling epidemics of multidrug-resistant M. tuberculosis of heterogeneous fitness. Nat Med. 2004;10:1117–1121. doi: 10.1038/nm1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Andersson DI, Hughes D. Antibiotic resistance and its cost: is it possible to reverse resistance? Nat Rev Microbiol. 2010;8:260–271. doi: 10.1038/nrmicro2319. [DOI] [PubMed] [Google Scholar]
- 26.Hegreness M, Shoresh N, Hartl D, Kishony R. An equivalence principle for the incorporation of favorable mutations in asexual populations. Science. 2006;311:1615–1617. doi: 10.1126/science.1122469. [DOI] [PubMed] [Google Scholar]
- 27.Chubiz LM, Lee MC, Delaney NF, Marx CJ. FREQ-Seq: A Rapid, Cost-Effective, Sequencing-Based Method to Determine Allele Frequencies Directly from Mixed Populations. PLoS One. 2012;7:e47959. doi: 10.1371/journal.pone.0047959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levin-Reisman I, et al. Automated imaging with ScanLag reveals previously undetectable bacterial growth phenotypes. Nat Methods. 2010;7:737–739. doi: 10.1038/nmeth.1485. [DOI] [PubMed] [Google Scholar]
- 29.Goodarzi H, Hottes AK, Tavazoie S. Global discovery of adaptive mutations. Nat Methods. 2009;6:581–583. doi: 10.1038/nmeth.1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schenk MF, Szendro IG, Krug J, de Visser JA. Quantifying the adaptive potential of an antibiotic resistance enzyme. PLoS Genet. 2012;8:e1002783. doi: 10.1371/journal.pgen.1002783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Salverda ML, et al. Initial mutations direct alternative pathways of protein evolution. PLoS Genet. 2011;7:e1001321. doi: 10.1371/journal.pgen.1001321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Opijnen T, Bodi KL, Camilli A. Tn-seq: high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat Methods. 2009;6:767–772. doi: 10.1038/nmeth.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Girgis HS, Hottes AK, Tavazoie S. Genetic architecture of intrinsic antibiotic susceptibility. PLoS One. 2009;4:e5629. doi: 10.1371/journal.pone.0005629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Poelwijk FJ, Kiviet DJ, Weinreich DM, Tans SJ. Empirical fitness landscapes reveal accessible evolutionary paths. Nature. 2007;445:383–386. doi: 10.1038/nature05451. [DOI] [PubMed] [Google Scholar]
- 35.Weinreich DM, Delaney NF, Depristo MA, Hartl DL. Darwinian evolution can follow only very few mutational paths to fitter proteins. Science. 2006;312:111–114. doi: 10.1126/science.1123539. [DOI] [PubMed] [Google Scholar]
- 36.Lozovsky ER, et al. Stepwise acquisition of pyrimethamine resistance in the malaria parasite. Proc Natl Acad Sci U S A. 2009;106:12025–12030. doi: 10.1073/pnas.0905922106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tan L, Serene S, Chao HX, Gore J. Hidden randomness between fitness landscapes limits reverse evolution. Phys Rev Lett. 2011;106:198102. doi: 10.1103/PhysRevLett.106.198102. [DOI] [PubMed] [Google Scholar]
- 38.Trindade S, et al. Positive epistasis drives the acquisition of multidrug resistance. PLoS Genet. 2009;5:e1000578. doi: 10.1371/journal.pgen.1000578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brown KM, et al. Compensatory mutations restore fitness during the evolution of dihydrofolate reductase. Mol Biol Evol. 2010;27:2682–2690. doi: 10.1093/molbev/msq160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hall AR, MacLean RC. Epistasis buffers the fitness effects of rifampicin-resistance mutations in Pseudomonas aeruginosa. Evolution. 2011;65:2370–2379. doi: 10.1111/j.1558-5646.2011.01302.x. [DOI] [PubMed] [Google Scholar]
- 41.D'Costa VM, McGrann KM, Hughes DW, Wright GD. Sampling the antibiotic resistome. Science. 2006;311:374–377. doi: 10.1126/science.1120800. [DOI] [PubMed] [Google Scholar]
- 42.Sommer MO, Church GM, Dantas G. The human microbiome harbors a diverse reservoir of antibiotic resistance genes. Virulence. 2010;1:299–303. doi: 10.4161/viru.1.4.12010. [DOI] [PubMed] [Google Scholar]
- 43.D'Costa VM, et al. Antibiotic resistance is ancient. Nature. 2011;477:457–461. doi: 10.1038/nature10388. [DOI] [PubMed] [Google Scholar]
- 44.Riesenfeld CS, Goodman RM, Handelsman J. Uncultured soil bacteria are a reservoir of new antibiotic resistance genes. Environ Microbiol. 2004;6:981–989. doi: 10.1111/j.1462-2920.2004.00664.x. [DOI] [PubMed] [Google Scholar]
- 45.D'Costa VM, et al. Inactivation of the lipopeptide antibiotic daptomycin by hydrolytic mechanisms. Antimicrob Agents Chemother. 2012;56:757–764. doi: 10.1128/AAC.05441-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chusri S, Villanueva I, Voravuthikunchai SP, Davies J. Enhancing antibiotic activity: a strategy to control Acinetobacter infections. J Antimicrob Chemother. 2009;64:1203–1211. doi: 10.1093/jac/dkp381. [DOI] [PubMed] [Google Scholar]
- 47.Lewis K. Antibiotics: Recover the lost art of drug discovery. Nature. 2012;485:439–440. doi: 10.1038/485439a. [DOI] [PubMed] [Google Scholar]
- 48.Koser CU, et al. Rapid whole-genome sequencing for investigation of a neonatal MRSA outbreak. N Engl J Med. 2012;366:2267–2275. doi: 10.1056/NEJMoa1109910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Snitkin ES, et al. Tracking a hospital outbreak of carbapenem-resistant Klebsiella pneumoniae with whole-genome sequencing. Sci Transl Med. 2012;4 doi: 10.1126/scitranslmed.3004129. 148ra116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Harris SR, et al. Whole-genome sequencing for analysis of an outbreak of meticillin-resistant Staphylococcus aureus: a descriptive study. Lancet Infect Dis. 2012 doi: 10.1016/S1473-3099(12)70268-2. [DOI] [PMC free article] [PubMed] [Google Scholar]