Abstract
Cholestatic liver diseases are caused by a range of hepatobiliary insults and involve complex interactions among environmental and genetic factors. Little is known about the pathogenic mechanisms of specific cholestatic diseases, which has limited our ability to manage patients with these disorders. However, recent genome-wide studies have provided insight into the pathogenesis of gallstones, primary biliary cirrhosis, and primary sclerosing cholangitis. A lithogenic variant in the gene that encodes the hepatobiliary transporter ABCG8 has been identified as a risk factor for gallstone disease; this variant has been associated with altered cholesterol excretion and metabolism. Other variants of genes encoding transporters that affect the composition of bile have been associated with cholestasis, namely ABCB11, which encodes the bile salt export pump, and ABCB4, which encodes hepatocanalicular phosphatidylcholine floppase. Many genes associated with gallstones have also been linked with vanishing bile duct syndromes and other cholestatic disorders. In contrast, studies have associated primary biliary cirrhosis and primary sclerosing cholangitis with genes encoding major histocompatibility complex proteins and identified loci associated with microbial sensing and immune regulatory pathways outside this region, such as genes encoding IL12, STAT4, IRF5, IL2 and its receptor (IL2R), CD28, and CD80. These discoveries have raised interest in the development of reagents that target these gene products. We review recent findings from genetic studies of patients with cholestatic liver disease. Future characterization of genetic variants in animal models, stratification of risk alleles by clinical course, and identification of interacting environmental factors will increase our understanding of these complex cholestatic diseases.
Complex cholestatic diseases include a range of disorders affecting small and large bile ducts and the gallbladder.1 To date, development of rational interventions for individuals with specific cholestatic disorders has been hampered by gaps in understanding disease pathogenesis. However, recent developments in identifying genetic influences (see Appendix for definitions) have begun to address an unmet need for rational treatment. The immune-mediated biliary disorders, primary biliary cirrhosis (PBC) and primary sclerosing cholangitis (PSC), represent the most important small and large bile duct diseases. The incidence and prevalence rates for PSC vary from 0 to 1.3 per 100,000 inhabitants/year and 0 to 16.2 per 100,000 inhabitants, respectively, whereas the incidence and prevalence of PBC range from 0.3 to 5.8 per 100,000 inhabitants/year and 1.9 to 40.2 per 100,000 inhabitants, respectively.2–4 PBC and PSC have been observed in all heritages, and geographic variations are evident with an increased prevalence in northern latitudes. Clustering of PBC has also been reported geographically, for example, in coastal First Nations of British Columbia, where disease has been recorded to be as high as 1 in 4 within generations of well-characterized multiplex families.5 In contrast, cholesterol gallstone disease is far more common and we have a much clearer understanding of the pathophysiology. Several factors combine to promote gallstone formation, such as supersaturation of bile with cholesterol or bilirubin, gallbladder hypomotility, and an imbalance of crystallization promoters (eg, mucin) and inhibitor proteins.6 Nevertheless, the incidence of gallstones differs markedly worldwide, reaching 50% in the American Indian population, 15% to 20% in the European population, approximately 10% in the Asian population, and less so in African populations.7 These differences are not fully explained by environmental factors such as physical inactivity or high-calorie, high-carbohydrate, and lowfiber diets or medications.8,9
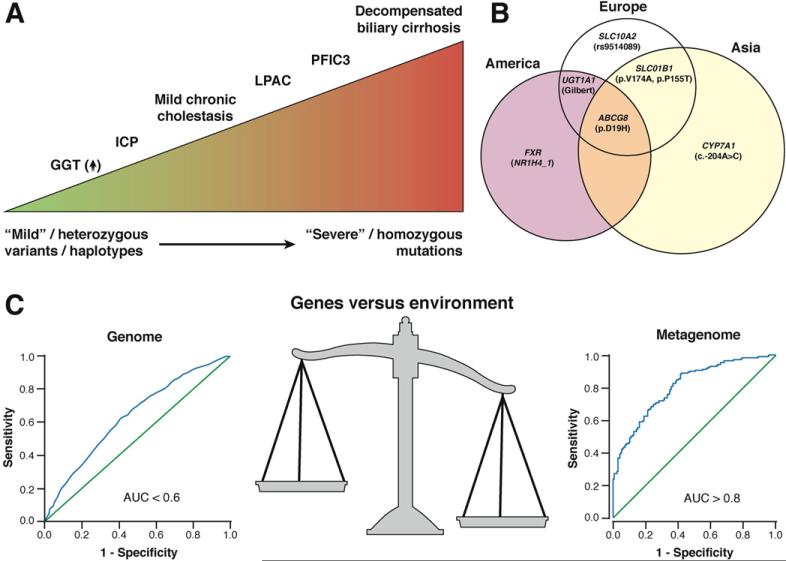
The dynamic genetic interactions that contribute to disease manifest at various levels. Some genes determining disease risk may only do so by imparting variability in how individuals respond to a particular environmental challenge. Others may express the consequence of genetic variation in a graded manner in as much as a single gene can be responsible for a wide phenotype spectrum depending on background genetic variability. A good example is provided by recognizing how ABCB4 variants can lead to disease ranging from mild elevations of γ-glutamyltransferase (GGT) levels to early development of biliary cirrhosis in childhood as well as cholestasis of pregnancy and intrahepatic gallstones with premature cholelithiasis (Figure 1A). The clinical spectrum of disease associated with this single gene aptly shows the mechanistic complexity of cholestatic disease. Genetic variation in structural proteins may also contribute to disease even if not directly related to the primary process mechanistically. For example, the association of keratin variants with PBC10 suggests how mutations in structural proteins can act as genetic modifiers and may be related to an accelerated liver disease phenotype alongside the overall genetic complexity of disease.
Figure 1.
Interaction of genes and environment in complex cholestatic disease. (A) Variations in severity of disease manifestation are observed with different genotypic variants in the ABCB4 encoding the biliary phosphatidylcholine transporter. Heterozygous ABCB4 variants encompass mild phenotypes, whereas homozygous deficiency leads to more severe diseases (ie, biliary cirrhosis and chronic liver failure). Specific genotypes might also contribute to chronic cholestasis and/or modify disease progression in patients with PBC and PSC. (B) Venn diagram illustrating variants that have been confirmed in replication studies. The size of each circle reflects the estimated number of adults with gallstones in each continent. Previous studies showed that Latin American populations have the highest (~30%) incidence of gallstone disease, with intermediate frequency of the disease in Europe (15%–20%) and lowest relative frequency in Asia (5%–6%). (C) The gut microbiome is more predictive of type 2 diabetes than current candidate loci derived by GWAS. A comparison of data derived from candidate type 2 diabetes loci108,109 and a selection of genes derived from metagenomic analyses of the gut microbiome from individuals with type 2 diabetes107 shows a superior correlation with the microbiome (based on the original cited data). ICP, intrahepatic cholestasis of pregnancy.
Heritability and the Role of the Major Histocompatibility Complex
Heritability is difficult to quantify and confounded by sharing of environmental triggers within close relatives. It is usually estimated by concordance rates in monozygotic versus dizygotic twins or by dividing the prevalence of disease among siblings with that of the general population (ie, sibling relative risk, λs). Collectively, such estimates suggest that the development of PBC and PSC is comparable to the 10-fold to 20-fold increased relative risk observed with other immune-mediated conditions such as inflammatory bowel disease (IBD). More precise estimates have been derived from countries with registries recording outcomes for disease for the population as a whole. For example, studies performed in ~30,000 Swedish twins with symptomatic gallstone disease have estimated the heritability to be approximately 25%.11
Before the advent of genome-wide association studies (GWAS), variants within the major histocompatibility complex (MHC) on chromosome 6p21 were the only robust candidates that were clearly associated with PBC and PSC. More than 30 years ago, PSC was linked with HLAB8, a known haplotypic marker for ancestral haplotype (AH) 8.1.12 For PBC, DRB1*0801 was shown to be over-represented in those of European descent and DRB1*0803 in Japanese subjects.13 A predominant role of the MHC region has subsequently been confirmed in genome-wide studies of PBC14 and PSC.15 Specific associations continue to be refined by ongoing studies.16,17 This is comparable to other autoimmune diseases such as type 1 diabetes, in which the genetic influence of the MHC has been estimated to contribute to more than 40% of the heritability.18
Some insight into risk-related alleles in the class II region of patients with PSC has been provided by fine mapping of the HLA-DRB1 genotypes with 3-dimensional modeling of the HLA-DRβ1 chain.15 These studies have identified key amino acids within the class II heterodimer that influence the range of peptides incorporated into the binding pocket of patients with PSC. Analogous studies in type 1 diabetes have provided a model whereby the disease-related alleles have an increased affinity for binding of insulin peptides into the HLA-DQ β1 chain with the capacity of promoting an autoimmune response.19 Another model of HLA-mediated immune injury has been suggested by abacavir hypersensitivity.20 Following the specific interaction of drug with HLA-B*57:01, the immune repertoire of endogenous peptides in the binding pocket is redirected, leading to activation of abacavirspecific T cells that drive polyclonal CD8 T-cell activation.21 In the context of cholestasis, the HLA-B*57:01 association identified in a genome-wide association study for flucloxacillin-induced liver injury provides another example.22 Intriguingly, while HLA is the strongest association for amoxicillin-clavulanate–induced liver injury,23 variants in a classic autoimmune-related gene, PTPN22, were also implicated.
A major distinction between PBC and PSC is the presence of highly conserved humoral and cellular immune responses to pyruvate dehydrogenase complex E2 (PDC-E2) in patients with PBC. The loss of tolerance to the mitochondrial proteins has been attributed to the aberrant expression of proteins resembling PDC-E2 on the cell surface of biliary epithelium.24 The immunodominant T-cell epitopes that bind with HLA-DRB4 have been mapped to the lipoyl domain of PDC-E225 and found to be similar to the immunodominant B-cell antigens, but the structural interaction of HLA-DRB8 and PDC-E2 has yet to be resolved. Also, our lack of knowledge of environmental factors that trigger the immune biliary diseases hinders our ability to study whether the disease-related HLA either lacks the ability to bind and help eradicate pathogens or specifically binds the environmental agent, triggering an autoimmune response.
Several GWAS studies have shown that the MHC is the major determinant in controlling viral infection with human immunodeficiency virus26 and chronic hepatitis B virus infection.27 This is not surprising given that more than 250 proteins encoded within the MHC region play a major role in response to pathogens. As a result, the complexity of the HLA and the complex pattern of linkage disequilibrium between disease-associated variants on the various extended HLA haplotypes make it difficult to discern the disease-related causal variants. For example, the PSC-associated AH8.1 has an extensive linkage throughout the MHC with HLA-A1, Cw7, B8, TNFAB*a2b3, TNFN*S, C2*C, Bf*s, C4A*Q0, C4B*1, DRB1*0301, DRB3*0101, DQA1*0501, and DQB1*0201.28 The AH8.1 has thus been linked with diminished complement levels, inability to clear immune complexes, increased susceptibility to infectious disease, and more rapid progression of bacterial and viral infections.28 However, it is not possible to determine the effects of single genes in the haplotype because of the strong linkage disequilibrium.
Of interest, AH8.1 is overrepresented in many other autoimmune diseases besides PSC, including autoimmune hepatitis. Patients with AH8.1 have differential cytokine responses to mitogens and lipopolysaccharide with increased tumor necrosis factor (TNF)-α levels, diminished interferon gamma levels, and predominant T-helper (Th)2 cytokine production. Although it is tempting to speculate that patients with AH8.1 have inefficient responses to pathogens and are more likely to have auto-immune responses, there are also data to suggest a potential benefit to survival. For example, the frequency of AH8.1 is approximately 10% in Europe and the haplotype is more frequently found in male nonagenarians.29 It could be argued that AH8.1 has been maintained within the population by the protective effects for men at the cost of increased risk of autoimmune disease in women. Thus, specific haplotypes may be beneficial by affecting the ability of the host to present antigens and clear infections but on the downside may contribute to autoimmune responses and “collateral” immune damage.
GWAS
The main goal of GWAS is to identify common genetic variation associated with well-phenotyped disease in a nonbiased fashion. Inherent to the properties of association-based statistics, the premise for conducting GWAS is based on the assumption that disease risk is associated with relatively common variants with a minor allele frequency of greater than 2% to 5%. The process is based on the principle of linkage disequilibrium whereby reasonable genomic coverage is accomplished by genotyping careful selections of 500,000 to 5,000,000 alleles at single nucleotide polymorphisms for any given individual. The readout is to identify frequency differences in polymorphisms between cases and controls. As a result, the studies mandate a large sample size to detect robust associations because false-positive findings will arise due to chance alone. The occurrence of linkage disequilibrium is used to advantage in that not all polymorphisms have to be genotyped to detect an association. On the downside, it is very difficult to ascertain which of a series of alleles in linkage disequilibrium is actually the causative variant on a haplotype. The larger the population studied for any given heterogeneous complex disorder, the more associated variants that can be confirmed. For example, GWAS in Crohn's disease and ulcerative colitis are now based on more than 75,000 cases and controls and have subsequently identified 163 inflammatory bowel disease loci.30 However, each locus contributes only a small risk to disease and the variants identified tend to be in noncoding regions of genes, implying subtle effects on gene expression rather than overt structural/functional changes.31
Not surprisingly, many of the risk alleles identified in immune-related conditions by GWAS are found in conjunction with genes related to immune function, both within and outside the MHC. Of note, disease-specific risk alleles related to the affected organ have seldom been identified with rare exceptions, such as insulin gene polymorphisms in type 1 diabetes.18 One obvious trend is that many risk loci are shared between multiple autoimmune diseases. For example, more than 160 risk loci have been found as common to both Crohn's disease and ulcerative colitis, suggesting a hypothesis that these variants represent a continuum in the risk of inflammatory bowel disease.30 Of interest, unrelated inflammatory diseases also share up to 50% of loci outside the MHC region. For instance, the detection of shared causal variants associated with the interleukin (IL)-12/IL-23 signaling pathway has been observed in patients with psoriasis, Crohn's disease, and multiple sclerosis (Figure 2).32 Although in most studies there is no obvious biological correlation with loci and disease, the studies provide a signpost of where to begin the search.
Figure 2.
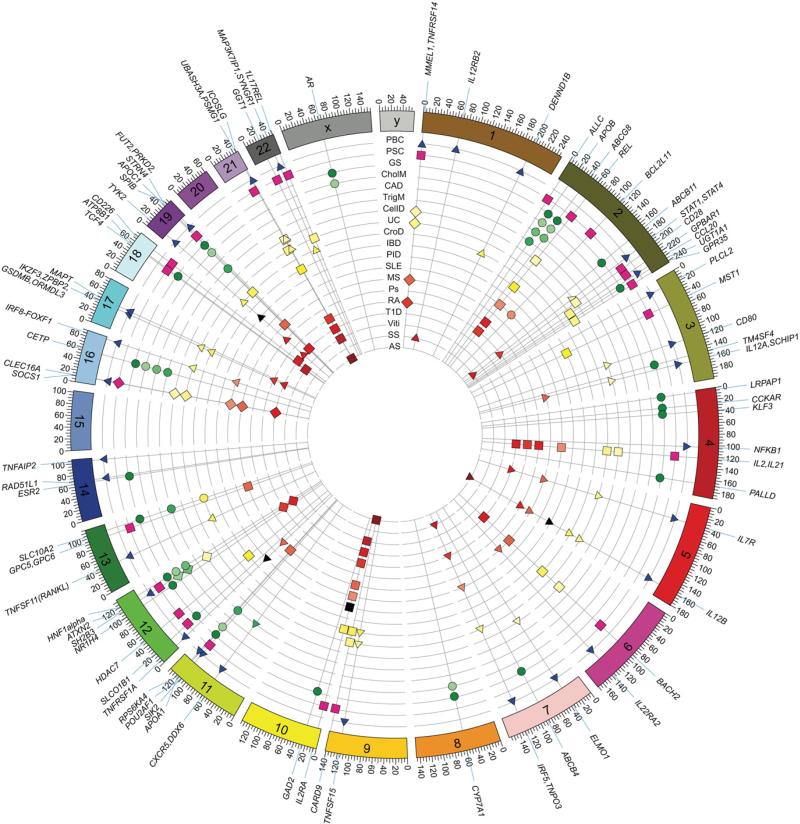
Circus plot of associations of cholestasis candidate genes with other diseases. Many of the candidate genes associated with PBC and PSC have previously been linked with other immune-mediated disorders and few, if any, specifically associated with biliary disease. Candidate loci found in patients with gallstone disease are predominantly associated with bile contents, lipid metabolism, and risk of coronary artery disease. Each radial line represents a PBC, PSC, or gallstone locus, ordered by genomic position and labeled around the rim, and each circular line represents a phenotype, with all points on a line colored according to the phenotype key given. Points sit at the intersection of radial and circular lines and represent sharing of a PBC, PSC, or gallstone locus with a given phenotype. The location of each locus is recorded in Table 1 and is shown as shapes in this figure, with triangles indicating PBC-specific disease, squares indicating PSC-specific disease, and circles indicating gallstone disease. AnkS, ankylosing spondylitis; CAD, coronary artery disease; CelD, celiac disease; CholM, cholesterol metabolism; CroD, Crohn's disease; GD, Graves’ disease; GS, gallstone disease; IBD, inflammatory bowel disease; MS, multiple sclerosis; PID, primary immunodeficiency syndromes; Ps, psoriasis; RA, rheumatoid arthritis; SLE, systemic lupus erythematosus; SS, systemic sclerosis; T1D, type 1 diabetes; TrigM, triglyceride metabolism; UC, ulcerative colitis; Viti, vitiligo.
GWAS in PBC
Several GWAS have been performed using subjects with PBC and controls from North America, Italy, the United Kingdom, and Japan.14,33–36 In keeping with other autoimmune diseases, it seems likely that there will be few, if any, PBC-specific genetic associations (Table 1). Many of the PBC-related variants have been identified in other GWAS of immune-related diseases, with a different mosaic of disease-specific risk contributing to the pathogenesis of PBC (Figure 2). Overall, the data suggest important contributions from a number of immune pathways to the development of PBC, particularly including a strong implied role for the interleukin (IL)-12 cytokine pathway and downstream JAK-STAT signaling (Figure 3).14,33–35,37 This needs to be seen in the context of a large preexisting body of evidence supporting immune models of disease. The present list of associated genes sustains a model heavily focused toward the effects of balancing immunoregulatory pathways, in particular Th cell lineages including Th1 and Th17 cells. Such unbiased, hypothesis-free generated data complement animal models (Table 2) and thereby focus research on teasing out the intricate balance of immunoregulatory triggers of disease.
Table 1.
Current Non-HLA Associations Recognized for PBC, PSC, and Gallstone Disease
| Chromosome (Position Mb) | Probable gene association | Primary cholestatic disease associations | Other disease associations from GWAS |
|---|---|---|---|
| 1 (2.5) | MMEL1, TNFRSF14 | PBC, PSC | Celiac disease, multiple sclerosis, RA, UC |
| 1 (67.7) | IL12RB2 | PBC | Systemic sclerosis |
| 1 (197.5) | DENND1B | PBC | Asthma, Crohn s disease |
| 2 (3.7) | ALLC | PSC | |
| 2 (21.2) | APOB | GS | Cholesterol metabolism, coronary artery disease, familial hypercholesterolemia, triglyceride metabolism |
| 2 (44.1) | ABCG8 | GS | GS, cholesterol metabolism, coronary artery disease |
| 2 (61.1) | REL | PSC | Celiac disease, IBD, lymphoma, psoriasis, SLE, RA |
| 2 (111.9) | BCL2L11 | PSC | Lymphoma |
| 2 (169.8) | ABCB11 | GS | Benign recurrent intrahepatic cholestasis 2, drug-induced liver injury, intrahepatic cholestasis of pregnancy, neonatal giant cell hepatitis, PFIC 2, hepatocellular carcinoma |
| 2 (191.8) | STAT1, STAT4 | PBC | Celiac disease, RA, systemic sclerosis, SLE |
| 2 (204.5) | CD28 | PSC | Celiac disease, chronic lymphocytic leukemia, Graves’ disease |
| 2 (219.1) | GPBAR1 | PSC | UC |
| 2 (228.7) | CCL20 | PBC | Hypertension |
| 2 (234.7) | UGT1A1 | GS | Bladder cancer, Gilbert's syndrome, irinotecan toxicity, neonatal jaundice |
| 2 (241.5) | GPR35 | PSC | UC |
| 3 (17.0) | PLCL2 | PBC | |
| 3 (49.7) | MST1 | PSC | IBD |
| 3 (119.2) | CD80 | PBC | Celiac disease |
| 3 (149.2) | TM4SF4 | GS | |
| 3 (159.7) | IL12A, SCHIP1 | PBC | Celiac disease, multiple sclerosis |
| 4 (3.5) | LRPAP1 | GS | |
| 4 (26.5) | CCKAR | GS | |
| 4 (38.7) | KLF3 | GS | |
| 4 (103.4) | NFKB1 | PBC | Schizophrenia |
| 4 (123.4) | IL2, IL21 | PSC | Celiac disease, Graves’ disease, psoriasis arthritis, primary immunodeficiency syndromes, RA, Sjögren's syndrome, SLE, type 1 diabetes, UC |
| 4 (169.4) | PALLD | GS | Nonalcoholic fatty liver disease, pancreatic cancer |
| 5 (35.9) | IL7R | PBC | UC, multiple sclerosis, type 1 diabetes |
| 5 (158.7) | IL12B | PBC | Ankylosing spondylitis, Crohn's disease, multiple sclerosis, psoriasis, primary immunodeficiency syndromes, UC |
| 6 (90.6) | BACH2 | PSC | Celiac disease, Crohn's disease, multiple sclerosis, type 1 diabetes |
| 6 (137.5) | IL22RA2 | PBC | Multiple sclerosis |
| 7 (36.9) | ELMO1 | PBC | Celiac disease |
| 7 (87.0) | ABCB4 | GS | Intrahepatic cholestasis of pregnancy, LPAC, PFIC 3, hepatocellular carcinoma |
| 7 (128.6) | IRF5, TNPO3 | PBC | Systemic sclerosis, SLE, RA, UC |
| 8 (59.4) | CYP7A1 | GS | Cholesterol metabolism, hypertension, neuromyelitis optica |
| 9 (117.5) | TNFSF15 | PBC | Crohn's disease, UC |
| 9 (139.2) | CARD9 | PSC | Ankylosing spondylitis, Crohn's disease, UC |
| 10 (6.1) | IL2RA | PSC | Alopecia areata, Crohn's disease, multiple sclerosis, RA, primary immunodeficiency syndromes, SLE, type 1 diabetes, vitiligo |
| 10 (26.5) | GAD2 | GS | Obesity |
| 11 (64.1) | RPS6KA4 | PBC | |
| 11 (111.2) | POU2AF1 | PBC | |
| 11 (111.5) | SIK2 | PSC | |
| 11 (116.7) | APOA1 | GS | Cholesterol metabolism, triglyceride metabolism |
| 11 (118.8) | CXCR5, DDX6 | PBC | Multiple sclerosis |
| 12 (6.4) | TNFRSF1A | PBC | Multiple sclerosis, primary immunodeficiency syndromes |
| 12 (21.2) | SLCO1B1 | PSC, GS | Hyperbilirubinemia, drug-induced liver injury, statin-induced myopathy |
| 12 (48.2) | HDAC7 | PSC | IBD |
| 12 (100.9) | NR1H4 | GS | |
| 12 (111.8) | SH2B3, ATXN2 | PBC, PSC | Celiac disease, cholesterol metabolism, coronary artery disease, RA, type 1 diabetes |
| 12 (121.4) | HNF1alpha | GS | Cholesterol metabolism, coronary artery disease, GGT levels, type 2 diabetes |
| 13 (43.1) | TNFSF11 (RANKL) | PBC | Crohn's disease |
| 13 (93.5) | GPC5, GPC6 | PSC | Multiple sclerosis |
| 13 (103.7) | SLC10A2 | GS | Bile salt malabsorption, Crohn's disease, triglyceride metabolism, prostate cancer |
| 14 (64.7) | ESR2 | GS | Gallbladder cancer |
| 14 (68.2) | RAD51L1 | PBC | |
| 14 (103.6) | TNFAIP2 | PBC | |
| 16 (11.3) | CLEC16A, SOCS1 | PBC, PSC | Celiac disease, immunoglobulin A deficiency, multiple sclerosis, SLE, type 1 diabetes, UC |
| 16 (57.0) | CETP | GS | Cholesterol metabolism, coronary artery disease, macular degeneration, triglyceride metabolism |
| 16 (85.9) | IRF8 - FOXF1 | PBC | SLE, UC |
| 17 (38.1) | IKZF3, ZPBP2, GSDMB, ORMDL3 | PBC | Asthma, Crohn's disease, RA, type 1 diabetes, UC |
| 17 (44.0) | MAPT | PBC | Parkinson's disease |
| 18 (52.9) | TCF4 | PSC | Corneal dystrophy, diabetic retinopathy, schizophrenia |
| 18 (55.3) | ATP8B1 | GS | Benign recurrent intrahepatic cholestasis 1, PFIC 1, vitiligo |
| 18 (67.5) | CD226 | PSC | Mean platelet volume, type 1 diabetes |
| 19 (10.5) | TYK2 | PBC | Crohn's disease, primary immunodeficiency syndromes, psoriasis, type 1 diabetes |
| 19 (45.4) | APOC1 | GS | Alzheimer's disease, cholesterol metabolism, triglyceride metabolism |
| 19 (47.2) | FUT2, PRKD2, STRN4 | PSC | Alkaline phosphatase levels, bipolar disorder, chronic lymphocytic leukemia, Crohn's disease, folate and vitamin B12 levels, multiple sclerosis, type 1 diabetes |
| 19 (50.9) | SPIB | PBC | |
| 21 (43.8) | UBASH3A, PSMG1 | PSC | Ankylosing spondylitis, celiac disease, IBD, RA, type 1 diabetes, vitiligo |
| 21 (45.6) | ICOSLG | PBC | Celiac disease, UC |
| 22 (25.0) | GGT1 | PSC | |
| 22 (39.8) | MAP3K7IP1, SYNGR1 | PBC | Crohn's disease |
| 22 (48.8) | 1L17REL | PSC | UC |
| X (66.8) | AR | GS | Cholesterol metabolism, male pattern baldness, prostate cancer |
NOTE. For gene name abbreviations, see http://www.ncbi.nlm.nih.gov/gene.
RA, rheumatoid arthritis; UC, ulcerative colitis; GS, gallstone disease; SLE, systemic lupus erythematosus.
Figure 3.
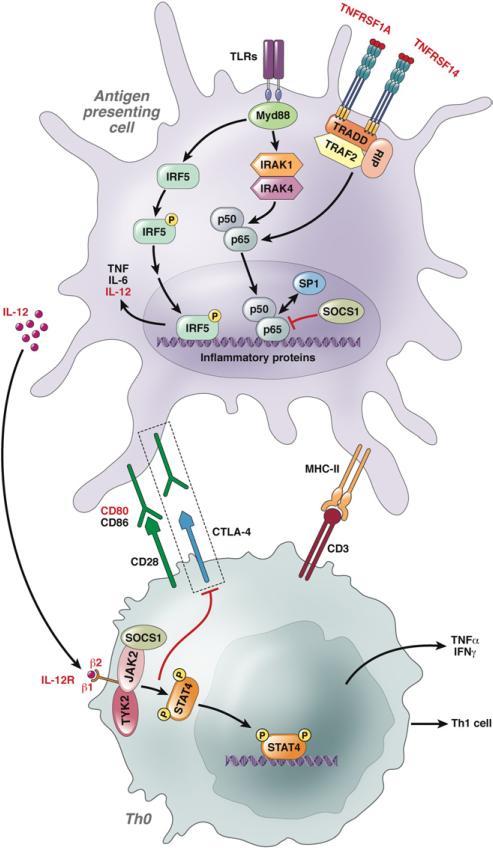
The putative role of IL-12 signaling in risk of PBC. IL-12 is a heterodimeric cytokine encoded by IL-12A and IL-12B, produced mainly by monocytes and macrophages, dendritic cells, and neutrophils. A strong role for IL-12 and related cytokines is implied in the pathogenesis of PBC by the identification of upstream and downstream mediators of IL-12 signaling as susceptibility loci in the PBC GWAS. CTLA-4, cytotoxic T-lymphocyte antigen 4; Foxp3, forkhead box P3; IFN-γ, interferon gamma; IκB, inhibitory κB; IRF-5, interferon regulatory factor 5; JAK, Janus kinase; NF-κB, nuclear factor κB; SOCS-1, suppressor of cytokine signaling; STAT, signal transducer and activator of transcription; TLR, toll-like receptor; TNFRSF, tumor necrosis factor receptor super-family; TYK2, tyrosine kinase 2.
Table 2.
Animal Models of Cholestasis
| Animal model |
Phenotype |
|||||||
|---|---|---|---|---|---|---|---|---|
| Human disease (reference) | Genotype background | Spontaneous versus inducible | Pathology | Portal inflammation | AMA production | Gallstone formation | Biliary strictures | Colitis |
| PBC92,93 | NOD.c3c4 mouse NOD strain with multiple innate and adaptive immune deficiencies |
Spontaneous Immunodeficient |
50% liver mortality from 9 to 11 mo | ++ | 50%–60% | – | Cystic dilatation of bile ducts | – |
| PBC94 | dnTGFβRII mouse C57Bl dominant negative TGFβ receptor II in lymphocytes |
Spontaneous Immunodeficient |
Inflammatory disease in lung, kidneys and intestinal tract Death ~25 wk |
+++ | 100% | – | ND | ++ |
| PBC95 | IL-2Rα–/– mouse C57Bl knockout of IL-2 receptor α |
Spontaneous Immunodeficient |
Inflammatory bowel disease within 12 wk Death ~25 wk |
+++ | 100% | – | ND | ++ |
| PBC96 | Scurfy mouse C57Bl knockout of FoxP3 |
Spontaneous Immunodeficient |
Multiorgan inflammation Death ~4 weeks |
+++ | 100% | – | NA | + |
| PBC104 | Ae2a,b–/– mouse FVB/N, 129/Sv knockout of AE2 Cl/HCO3 anion exchanger |
Spontaneous Altered bile composition |
Slow onset of AMA over 52 wk with biliary inflammation | + | 40%–80% | – | ND | – |
| PBC100 | 2-Octynoic acid–treated mouse C57Bll, NOD 1101 |
Inducible Xenobiotic with Freund's adjuvant |
Portal inflammation with granuloma formation | + | 100% | – | ND | – |
| PBC101 |
Novosphingobium aromaticivorans infection NOD 1101 mouse |
Inducible Bacterial infection |
Portal inflammation, cholangitis with granuloma formation | ++ | 80%–100% | – | ND | – |
| PSC78 | Retrograde biliary TNBS Sprague–Dawley rats |
Inducible Chemical cholangitis |
Pericholangitis and periductal onion skin fibrosis | + | ND | – | Ectatic extrahepatic ducts and beading intrahepatic ducts | – |
| PSC79 | Retrograde biliary TNBS alcohol and incomplete BD ligation Lewis rats |
Inducible Chemical cholangitis |
Pericholangitis and biliary fibrosis | + | ND | – | Strictures in intrahepatic and extrahepatic ducts | – |
| PSC77 | Lithocholic acid feeding Swiss albino mice |
Inducible Chemical cholangitis |
Bile infarcts, fibrosis, destructive cholangitis | + | ND | – | ND | – |
| PSC88 |
Helicobacter sp. intraperitoneal injection A/JCr, C3H/Hen, C57Bl/6 |
Inducible Bacterial infection |
Mild hepatitis, pericholangitis, and ductal proliferation | + | ND | – | ND | + |
| PSC89,90 |
Cryptosporidium parvum gavage BALB/c Nu/Nu, NHI-III Nu/Nu |
Inducible Bacterial infection |
Severe cholangitis, pericholangitis, biliary fibrosis, and hepatic necrosis | +++ | ND | – | ND | + |
| PSC87 | Small bowel bacterial overgrowth following blind loop construction Lewis and Wistar rats |
Inducible Bacterial products |
Cholangitis, pericholangitis, bile duct proliferation, and onion skin fibrosis | + | ND | – | Ectatic extrahepatic with irregular intrahepatic ducts | + |
| PSC80 | Rectal administration of fMLT+/– ascetic acid Lewis rats |
Inducible Bacterial products |
Cholangitis and pericholangitis | + | ND | – | Irregular, intrahepatic ducts | + |
| PSC, gallstones82,84,86 |
Abcb4–/– mouse Mdr2 knockout mice on 129, FVB/N, or BALB/c backgrounds |
Spontaneous Altered bile composition |
Cholangitis, periductal onion skin fibrosis, vanishing bile ducts, intrahepatic and extrahepatic gallstones | + | ND | + | + | – |
| PSC, gallstones81,83 | CFTR mouse models (KO, DF508) on 129/C57BL/6 background | Spontaneous Altered bile composition |
Cholangitis, periductal onion skin and fibrosis, black pigment stones | + | ND | + | ND | – |
| PSC85 | Ferrochelatase-deficient mouse BALB/c fch/fch model of erythropoietic protoporphyria |
Spontaneous Altered bile composition |
Cholangitis, periductal onion skin and fibrosis | + | ND | – | ND | – |
| Gallstones135,136 | Syrian hamster | Inducible EFA-deflcient diet (20% casein, 40%–74% sucrose/glucose) + 10% butter fat or 25% fiber |
Cholesterol or black pigment gallbladder stones; ductular proliferation and portal fibrosis | + | ND | + | ND | – |
| Gallstones137,138 | Prairie dog | Inducible Cholesterol diet (0.3%–1.2% cholesterol) |
Cholesterol gallbladder stones and mucin hypersecretion, polypoid hyperplasia of gallbladder mucosa and inflammation; microvesicular steatosis, ductular proliferation, and portal fibrosis | + | ND | + | ND | – |
| Gallstones139 | Inbred mouse strain C57L/J and others | Inducible Lithogenic diet (1% cholesterol, 0.5% cholic acid, 15% butter fat) |
Cholesterol gallbladder stones, gallbladder wall thickening with stromal granulocyte infiltration, fibrosis, and epithelial cell indentation | ND | + | ND | – | |
| Gallstones140 | Guinea pig | Inducible Chow supplemented with 0.5% cholesterol |
Calcium bilirubinate precipitates in enlarged gallbladders, fatty liver | + | ND | + | ND | – |
ND, not done; NA, not available; TNBS, trinitrobenzene sulfonic acid; fMLT, N-formyl l-methionine l-leucine l-tyrosine.
With regard to IL-12 signaling, the involvement is consistent with the major immunoregulatory roles of the gene products IL-12 p35 and IL-12 receptor β2, which associate with the IL-12 p40 and IL-12 receptor β1 chains, respectively, to generate the complex of IL-12 and its receptor. The IL-12 pathway has been implicated in other immune-mediated diseases,32 and it is central to generating Th1 immune responses directed toward clearance of intracellular pathogens.38 The engagement of IL-12 to its receptor is believed to modulate immune responses by evoking interferon gamma production, which in turn inhibits IL-23–driven induction of IL-17–producing helper T lymphocytes. A role for IL-35 is also worthy of investigation, given the subunit nature of the cytokine IL-35 and its receptor, that includes IL-12 p35 and IL-12R β2, respectively.39 It is also notable that the regulation of IL12RB2 expression in T regulatory cells appears to be important in determining Th cell lineage effects,40 because impaired expression of IL-12R β2 has been shown to help maintain T regulatory cell–suppressive function in the context of inflammatory Th1 cell responses.
A bidirectional dynamic interaction of T cells and biliary epithelial cells is likely relevant, with the “secretome” of the injured biliary cell having the potential to also contribute to the fate of the infiltrating lymphocytes. Many of the loci associated with PBC, such as IRF5, SOCS 1, and NF-κB, further suggest that Toll-like receptor signaling upstream to the IL-12 production may be relevant to disease (Figure 2). For example, IRF5 interacts with nuclear factor κB, which consequently causes expression of a number of potentially relevant Th1 cytokines, including IL-12. A putative role for IRF5 in macrophage polarization is also relevant and highlighted in the pathogenesis of systemic lupus erythematosus, a complex autoimmune disease also associated genetically with IRF5 variants.41 Additionally, IRF8 is a transcription factor that binds to the IL-12 promoter and directly modulates IL-12 and interferon gamma production. Downstream, engagement of IL-12 with the IL-12 receptor leads to production of another candidate gene in PBC, the STAT4 transcription factor, that leads to activation of Th1 cytokines (Figure 2).
It is relevant to reflect on the fact that genetic risk varies globally, such that a Japanese GWAS identifies a different panel of immunoregulatory pathways. This supports the concept of many pathways leading to disease. Genetic data ferment more precise biologic experimentation to refine understanding of the molecular immunology at play, whether through pathways associated with sensing of microbes or xenobiotics and antimicrobial responses. This is needed because we still lack sufficient clinical and experimental data concerning the specific role of IL-12 signaling, for instance, in PBC and animal models. In this regard, a case report documents the early onset of biliary cirrhosis in an IL-12–deficient child, although the immediate relevance to PBC is unclear.42 Collectively, more data will ensure we are better placed to guide future management strategies that seek to modulate IL-12 signaling and other relevant pathways. Pilot studies are under way to test the efficacy and safety of the human monoclonal anti–IL-12/IL-23 ustekinumab in patients with PBC (ClinicalTrials.gov identifier: NCT01389973).
GWAS in PSC
Several reports have documented results from GWAS including patients with PSC and controls mainly from northern Europe.15,43,44 The fibrotic strictures and intervening dilatations of the intrahepatic and extrahepatic bile ducts define the condition that results in the development of cholestatic cirrhosis in most cases.45 In 10% to 20% of patients, development of cholangiocarcinoma represents an important addition to the hepatobiliary phenotype. Up to 80% of patients with PSC from northern Europe have concurrent IBD, whereas the frequency of IBD in Southern Europe and Asia is approximately 30% to 50%.45 To date, most of the genetic studies in PSC have been performed in patients from northern Europe; therefore, the genetic data generated should be considered representative of the PSC and IBD phenotype. It is also important to note that 25% of patients with PSC of northern European origin with IBD have additional immune-mediated diseases.46 Therefore, the phenotypic expression of PSC can be divided into the hepatobiliary disorder and the associated extrahepatic disease involving other organ systems.47
The overlap between PSC and IBD as well as prototypical autoimmune diseases is reflected in the causal variants that have been detected to date (Figure 2).15,44,47 Similar to type 1 diabetes, celiac disease, and multiple sclerosis, the predominant attributable risk factors are found within the MHC on chromosome 6p21 and weaker associations are detected for multiple IBD-related and autoimmunity loci outside this region (Table 1). The overlap with IBD is far from complete, and even taking into account statistical power directly related to the number of cases of PSC available in the genetic studies, the majority of both ulcerative colitis and Crohn's disease risk loci do not confirm associations with PSC.45–49 This mirrors the clinical impression of IBD in PSC, which shows several distinguishing features when compared with regular UC, such as rectal sparing or backwash ileitis.50
Of note, shared risk loci for PSC and PBC have recently been confirmed with MMEL1/TNFRS14 on chromosome 1p36 and CLEC16A on chromosome 16p14. However, these risk loci are not specific to biliary disease (Table 1). Of interest, it appears that the MMEL1/TNFRS14 allele may confer differential risk, because it has opposite effects in PSC as compared with PBC.33,43MMEL1 encodes a membrane metalloendopeptidase-like protein of unknown function located within the biliary tree; these proteins have diverse roles in metabolizing peptide hormones and β-amyloid, among others. More is known about TNFRS14, which encodes a receptor for cytokines and membrane-bound ligands, including B and T lymphocyte attenuator.51 The receptor acts as a molecular switch to provide positive costimulation to lymphocytes with certain ligands and negative signals with others, such as B and T lymphocyte attenuator. Notably, the gene product of TNFRS14 also functions as an entry receptor for several herpes viruses, and in turn this engagement modulates immune responses to viral infection.51 Although the biology of this causal variant with PSC remains unexplored, it may be relevant that mice lacking B and T lymphocyte attenuator develop spontaneous autoimmune responses, cholangitis, irregular bile ducts, and histologic features comparable to PSC.52
Other causal variants have been identified in PSC GWAS that potentially interact directly with infectious agents. Specifically, the risk of PSC has recently been associated with the FUT2 gene encoding fucosyltransferase 2 (Table 1). The enzyme regulates expression of the ABO blood group antigens on the surface of epithelial cells and as a result determines the secretor status of the individual. Importantly, specific genotypes of FUT2 have been linked with both bacterial and viral infections. For example, subjects homozygous for the FUT2 alleles are nonsecretors who cannot express the H type 1 oligosaccharide ligand required for Norwalk virus binding53; these individuals are protected from viral infection. Similarly, expression of the secretor status has been shown to influence binding of bacteria as well, such as Helicobacter pylori.54 The direct implications for patients with PSC are unknown, but it is interesting to note that the nonsecretor genotype found in patients with PSC has been associated with differences in the biliary microbiome with increased Firmicutes and diminished Proteobacteria in the bile.43 An interesting parallel has also been observed in the gut microbiome in both healthy individuals and patients with Crohn's disease, where the nonsecretors also display a predominance of Firmicutes compared with Proteobacteria in the gut.55
Similar to PBC, mechanistic studies addressing PSC-specific aspects of the risk loci are lacking. A common theme from genetic association studies is that the genetic predisposition in PSC is likely to resemble that of other immune-mediated diseases. However, it remains unclear how this perception aligns with the general lack of efficiency of trials of immunosuppressive medication in PSC. Potentially, the diagnosis of PSC needs to be made at a preclinical, prefibrotic stage of bile duct injury for such a treatment to be efficient, but this has never been shown clinically. Moreover, there is the possibility that essential pathogenic mechanisms may also be relatively inert to drugs tested so far. The observation that causal variants influence functional receptors for microbes also requires further study. There is a need to explore the biliary and intestinal microbiome in the context of genetic findings for gene-microbial interactions.55,56 Because the genetics of PSC seem to put the disease somewhat at the intersection of prototypical autoimmunity and IBD (Table 1), queries into such aspects in PSC may even provide clues as to the role of gut microbial community composition in these disease groups. In turn, these studies may lead to a better understanding of the environmental factors that affect PSC.
GWAS in Gallstone Disease
The first GWAS for gallstones found a highly significant association with the p.D19H variant of ABCG8 (odds ratio, 2.2; population attributable fraction, 11%),57 which had been previously identified as a risk factor in animal models.58 This risk factor was confirmed globally.57,59–66 The ABCG5/G8 hepatocanalicular heterodimeric transporter for cholesterol is a member of the adenosine triphosphate–binding cassette (ABC) transporters. Three of these genes, ABCB4, ABCB11, and ABCG5/G8, are responsible for the biliary secretion of phosphatidylcholine, bile salts, and cholesterol, respectively (Figure 4). Accordingly, their secretory capacities determine the composition of mixed micelles and multilamellar vesicles, which form the “liquid crystals” where cholesterol precipitates in bile.
Figure 4.
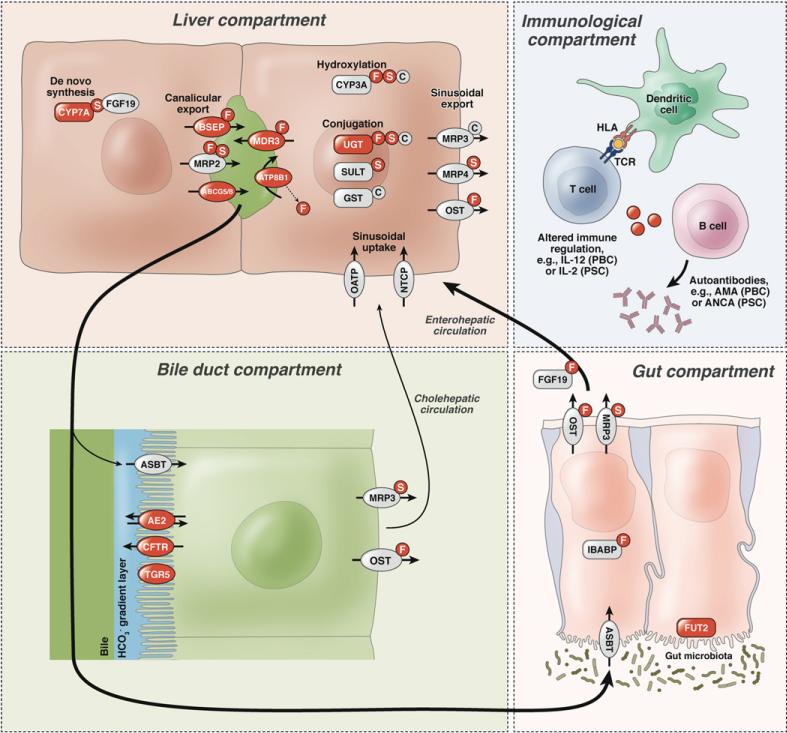
Modeling of genetic influence in cholestatic liver diseases. The liver compartment comprises genes involved in hereditary cholestatic syndromes and gallstone disease (highlighted in red). The same loci and loci in the biliary compartment may also serve as modifiers in immune-mediated injury, as exemplified by the immunologic compartment. Susceptibility to cholestatic liver disease has also recently been shown to be associated with genetic factors influencing microbial community composition, as illustrated by the gut compartment that also involves the enterohepatic circulation of bile acids. Cyp7A1, cytochrome P450, family 7, subfamily A, polypeptide 1; NTCP, Na/taurocholate cotransporting polypep-tide; OATP, organic anion-transporting polypeptide; BSEP, bile salt export pump; MRP2, multidrug resistance-associated protein 2; MDR3, P-glycoprotein-3/multiple drug resistance-3; ABCG5/8, adenosine triphosphate–binding cassette subfamily G member 5/8 heterodimer; ATP8B1, adenosine triphosphatase, class I, type 8B, member 1; CYP3A, cytochrome P450, family 3, subfamily A; UGT, uridine diphosphate glucoseglycoprotein glucosyltransferases; SULT, sulfotransferases; GST, glutathione S-transferase; MRP3/4, multidrug resistance-associated protein 3/4; OST, organic solute transporter; F, FXR, farnesoid X receptor; S, SXR/PXR, steroid and xenobiotic receptor/pregnane X receptor; C, CAR, constitutive androstane receptor; CFTR, cystic fibrosis transmembrane conductance regulator; TGR5, G protein–coupled bile acid receptor 1; ASBT, apical sodium-dependent bile acid transporter; AE2, anion exchange protein 2; TCR, T-cell receptor; ANCA, anti-neutrophil cytoplasmic antibodies; FUT2, fucosyltransferase 2; IBABP, ileal bile acid binding protein; FGF19, fibroblast growth factor 19.
Subsequent studies showed that obese women older than 60 years of age homozygous with the p.D19H risk allele had a 13% absolute 10-year risk of symptomatic gallstone disease compared with a range of 2% to 4% in noncarriers in the same risk stratum.66 It is likely that decreased intestinal cholesterol absorption and increased hepatic cholesterol synthesis represent the primary defect intensifying the susceptibility for the development of stones.67 This indicates that the lithogenic ABCG8 p.D19H variant confers decreased cholesterol absorption, higher cholesterol synthesis, and a trend toward lower serum cholesterol levels as well as a better response to statins. Accordingly, ABCG8 p.D19H might represent a “gain-of-function” mutation that increases clearance of sterol from the body.68–70 As a corollary, this genotype has also been shown to be protective against the development of atheromatous vascular disease by lowering cholesterol levels.71,72
Preliminary data from 2 other GWAS for gallstone disease have identified additional lithogenic candidate genes (Table 1).59,60 Data from a genome-wide association study of 4300 Sardinian subjects with increased serum bilirubin levels was used to identify the Gilbert syndrome variant in the promoter of the UGT1A1 gene as a candidate gene for gallstone disease73 that was verified in 2800 German and Chilean patients with gallstones.74 The link of diminished bilirubin conjugation with gallstone formation reinforces the concept of nucleation in supersaturated bile, because cholesterol gallstones usually contain small amounts of bilirubin pigment. The population-attributable fraction of the common ABCG8 and UGT1A1 variants in European subjects is 15% to 20%, approximating the total genetic gallstone risk stated in the preceding text (Figure 1B).
Other low-frequency variants with intermediate effects and rare Mendelian mutations have been identified in individual families with hereditary gallstones that show a geographic variance in prevalence (Figure 1B). These include ABCB11 associated with reduced bile salt secretion, APOB linked with increased biliary cholesterol secretion, farnesoid X receptor FXR regulating bile salt excretion, cholecystokinin-1 receptor CCKAR linked with gallbladder motility, and CYP7A1 linked with impaired cholesterol catabolism into bile salts (Table 1).22,75 Case reports and family linkage studies found that mutations of the ABCB4 gene might underlie a peculiar type of hereditary gallstone disease, referred to as low phospholipid-associated cholelithiasis (LPAC). Patients with LPAC usually present with symptoms before the age of 40 years and have cholesterol gallbladder stones and intrahepatic sludge or microlithiasis. As a result, biliary colic often recurs after cholecystectomy. These patients also frequently present with obstetric or mild chronic cholestasis (Figure 1). Hence, gallstones are part of the phenotypic spectrum of progressive familial intrahepatic cholestasis (PFIC) type 2 associated with low GGT levels and PFIC type 3, which are caused by ABCB11 and ABCB4 mutations, respectively. The genetic diagnosis is hampered by the genotype-tophenotype challenge in as much as a large number of gene variants await extensive functional studies.76
Animal Models of Cholestasis Parallel Genomic Efforts
After initial experimental studies in prairie dogs, hamsters, and guinea pigs, susceptible and resistant in-bred strains of mice played an important role in defining the heritability for gallstone disease before GWAS. Following challenge with a lithogenic diet of 15% fat, 1% cholesterol, and 0.5% cholic acid, lithogenic (Lith) loci were identified as quantitative trait regions within the mouse genome that were subsequently characterized as risk factors for gallstones (Table 2).75 Initially the Lith9 candidate was found to harbor the Abcg5/g8 genes, and subsequently other Lith loci in mice have been recognized that are relevant to human gallstone disease (Table 1).58
In contrast, animal models have played a minor role in identifying the heritability of PBC and PSC but have provided other insights into disease.21 For PSC, histologic evidence of cholangitis or large duct disease has been induced by chemical cholangitis77–80 or targeting of specific genes that alter the composition of bile.81–86 The resultant pathology of genetic manipulation usually exemplifies other biliary processes, however, such as vanishing bile syndrome (Table 2). PSC models have been generated by bacterial products87,88 and Cryptosporidium infection in immune-deficient strains.89,90 The latter provides close parallels with acquired immunodeficiency syndrome cholangiopathy91 and supports a model whereby biliary strictures occur at the intersection of infection and immunodeficiency. On a more global level, it is notable that some of the candidate genes for PSC and PBC are linked with primary immunodeficiency syndromes, as recently reported for patients with IBD30 (Figure 2).
In a similar vein, most of the mouse models of PBC that develop spontaneous antimitochondrial antibodies (AMA) and hepatic inflammation can be considered immune deficient (Table 2). These models include the NOD.c3c4 that develops granulomatous cholangitis92,93 as well as the T-cell transforming growth factor β receptor II dominant-negative (dnTGFBRII),94 IL-2Rα–/–,95 and the Scurfy mouse lacking T-regulatory cells96 that all die at a young age of diffuse extrahepatic inflammatory disease (Table 2). In the setting of immunodeficiency, these mice express mouse mammary tumor virus related to the betaretrovirus characterized in PBC; these viral proteins are located in biliary epithelium or lymphoid tissues that also display aberrant mitochondrial PDC-E2 expression, providing a mechanism for production of AMA.97–99 Other stimuli, including xenobiotics100 and bacteria,101 have also been shown to trigger AMA production in mouse models. With regard to the known genetic risk factors for PBC, conflicting results have been derived in a spontaneous mouse model of PBC, where mice lacking IL-12 p35 or IL-12 p40 developed either increased hepatic fibrosis or diminished autoimmune cholangitis, respectively.102,103 One exception of the spontaneous AMA-producing mice is the anion exchanger deficient AE2a,b–/– model, which provides an example of altered bile composition driving small duct disease (Figure 4).104 Collectively, these mouse models provide important information concerning immune, environmental, and genetic factors for disease.
Where Is the Missing Heritability?
Although GWAS have provided important genetic insights, concerns have been voiced that only a small proportion of the expected heritability has been uncovered by reported findings, referred to as the “missing heritability” of disease. Some have even gone as far as to state that the general hypothesis is incorrect and common variants do not explain the vast majority of genetic hereditability of any disease.105 This of course assumes that our previous estimates based on family studies of heritability for different diseases are correct; however, familial disease may be triggered by environmental and nongenetic elements, including the various microbial compartments. 106 In type 2 diabetes, for instance, a limited selection of bacterial genes within the gut microbiome107 has recently been found to be more predictive of developing disease than accumulated GWAS data108,109 (Figure 1C).
Others have argued that statistical approaches are incomplete and Bayesian inference analyses have the power to identify an additional 20% of heritability.110 It is also possible that a smaller number of undetected loci encompass rare causal variants exerting relatively high risk. Another reason for missing heritability may lie with nonadditive genetic effects such as epistasis, which is the effect of the interaction of 2 or more genes on the phenotype. Although a proportion of additive genetic variation can be shown when all polymorphisms are considered simultaneously,111 the extent of epistasis is difficult to determine using current methods. Suggestive evidence for epistasis arose from one study of PBC, for example, with demonstrated interaction between IL12RB2 and IRF5.112
The difficulty with detection of missing heritability may also be related to the structural intricacy of our genomes. GWAS are powered to examine common genomic variants in single nucleotide polymorphisms with minor allele frequency of 5% or greater. Therefore, rare genetic variants that may cause illness or contribute to disease escape detection because we simply lack sufficient numbers of patients.105 Also, copy number variants with insertions and deletions as well as inversions and translocations are difficult to detect using data from GWAS. Furthermore, GWAS are unable to discover de novo mutations occurring in the germline that have been uncovered by resequencing candidate genes associated with common disorders such as autism.113
Missing heritability may also result from epigenetic effects such as methylation of CpG islands that regulate gene expression. Studies of methylation status in twins with multiple sclerosis, for example, have been somewhat disappointing, largely because the differences between discordant twins with multiple sclerosis were found to be far less than in unrelated individuals.114 Another example of epigenetic modification involves the interesting phenomena of X chromosomal inactivation that ensures that all cells in male and female subjects have one active X chromosome. Enhanced X chromosome monosomy rates have been observed in patients with PBC,115 other autoimmune diseases such as systemic lupus erythematosus, and some hormone-responsive cancers.116 There is also the recent findings of increased Y chromosome loss in male subjects with PBC.117 The implications of chromosomal inactivation remain a mystery in chronic disease, and GWAS in PBC have not pinpointed sex chromosomal changes as relevant to date.
Overall, the accumulated data suggest that approximately 10% to 20% of the hereditability of complex immune-mediated diseases have been uncovered by GWAS, which is comparable to that seen with other complex diseases such as cancer.118 Therefore, it is likely that some of the missing risk will be accounted for by environmental agents, and some have gone as far to suggest that the major contributors to many complex diseases are nonhereditary.119
Beyond GWAS to the Microbiome
On a simplistic level, we cohabit a microecosystem contingent on an interaction with a staggeringly diverse microbiologic environment that has influenced our genetic variation over time. As a result, the application of genetics should reach beyond the host genome to so-called “meta-genetics” by investigating all the genes in the metagenome.120 Although we have already commented on the differences in the microbiome of patients with Crohn's disease and PSC associated with the FUT2 risk loci,43,55 these studies lack the design to directly implicate the changes in the microbiome with the disease process and animal models are often required to show the importance of the microbiome. One staggering example was recently described in a nonobese diabetic mice model, whereby the exchange of gut microflora from male to female mice transformed the disease to the male pheno-type and delayed the onset of diabetes.121
Other elegant animal models have shown the interaction of susceptibility genes with the microbiome to promote a disease-specific phenotype.122 For instance, mice missing key inflammasome genes fed a methionine-choline–deficient diet develop progressive nonalcoholic steatohepatitis.123 In this model, changes in the gut microbiome were associated with modulation of cytokine expression and development of the metabolic syndrome. Indeed, a direct role for the microbiota was inferred by cohousing the inflammasome-deficient mice with wildtype mice that subsequently developed increased obesity and hepatic steatosis as well.123 A similar study linking microbiome and host susceptibility genes has been conducted using a mouse model with a hypomorphic mutation in ATG16L1, a Crohn's disease susceptibility gene involved in autophagy. After infection with murine norovirus, the mice developed intestinal pathology comparable to IBD, and intriguingly the disease was preventable by administration of broad-spectrum antibiotics.124 This metagenetic process illustrates how the interaction of viral infection and Crohn's disease susceptibility genes disrupted the commensal flora to generate an IBD phenotype.
Another well-recognized role of the microbiome is prevention of pathogen invasion by organisms. One case in point is Clostridium difficile infection in patients receiving antibiotics who lose their ability to provide “colony resistance.” Similarly, the microbiome is altered in patients with global immunodeficiency to provide diminished protection against common pathogens.125 In these circumstances, complex infections with diverse organisms may arise that cause phenotypes resembling immune-mediated inflammatory diseases. One example is the various combinations of microbial infection with human immunodeficiency virus, cytomegalovirus, Cryptosporidium, and other pathogens that have been associated with the development of acquired immunodeficiency syndrome cholangiopathy.91 The study of bile microbiota is relatively unexplored as compared with other bodily compartments, and an unbiased metagenomic survey would seem to be an attractive approach to investigate the microbiome associated with cholestatic diseases.
Effect on Clinical Practice
With the falling costs of next-generation sequencing, whole exome and genome sequencing is starting to move into the clinic. In one remarkable report, the genome of a 4-year-old boy with severe inflammatory bowel disease was sequenced to reveal a loss-of-function polymorphism in the XIAP gene encoding the X-linked inhibitor of apoptosis protein.126 XIAP plays an important role in regulating immune responses to mucosal flora and directly interacts with the intracellular pattern recognition receptor encoded by NOD2, the first susceptibility gene found for Crohn's disease.111 The findings had translational implications and a bone marrow transplant corrected the pathology, presumably in part by restoring XIAP function.126 Indeed, the investigation of infants with very early-onset IBD has uncovered other novel candidate genes such as those encoding the IL-10 receptor or neutrophil cytosolic factor 2 in the reduced nicotinamide adenine dinucleotide phosphate oxidase complex that intersect along pathways previously uncovered by GWAS.127,128 Accordingly, the study of familial cases or highly characterized phenotypes of PBC, PSC, or gallstone disease might lead to the identification of rare, high-penetrant alleles at risk loci that shed light on pathogenesis. Phenotypes might include the biochemical response to ursodeoxycholic acid or the presence of pruritus.
With regard to genetic tests for gallstone disease, diagnostic tests in familial gallstone disease comprise resequencing of the ABCB4 and/or ABCB11 genes in patients who present with PFIC or LPAC phenotypes (Figure 1) and genotyping of the UGT1A1 promoter variant, which predisposes to unconjugated hyperbilirubinemia and gallstones129 but is also associated with decreased cardiovascular risk.130 In the future, it is possible that polygenic disease predisposition for gallstones and cholestasis might be assessed by gene tests. It is tempting to speculate that stratified polygenic risk scores131 based on genetic factors, including variants of ABCG8, UGT1A1, and ABCB4, and environmental factors, such as overeating, physical inactivity, and infections, might assist in defining normal and high-risk groups. Whereas lifestyle modifications with weight reduction, increased physical activity, and dietary alterations are appropriate for the normal-risk group, high-risk individuals might benefit from early prevention and diagnosis as well as targeted therapy with ursodeoxycholic acid, statins, ezetimibe, and/or modulators of bile salt signaling pathways.132 A similar process may be relevant in patients with PBC and PSC, with personal stratification of clinical events informed by genetic analysis of risk variants. Indeed, preliminary attempts are being made in regard to recurrence of PBC in liver allografts.133
Despite the optimism, there is a huge gap between the genetic information collected to date and the surprisingly small influence this knowledge has had on the clinical management of patients. Translation into real life is still pending, a fate that cholestatic liver diseases share with other complex diseases studied on a genome-wide level. Nevertheless, overall, the advances to date in understanding the genetics of cholestasis speak broadly to the ultimate goal of all such studies to guide personalized care that is rational and mechanism driven.
Acknowledgments
The authors thank the American Association for the Study of Liver Diseases for hosting the Special Interest Group Program on Genetics of Complex Cholestatic Disorders (2011), which generated many ideas and concepts driving the contents of our review, and Drs Evagelia Liaskou and Juan Jovel for assistance with figure preparation.
Funding
Supported in part by grants from the National Institutes of Health (DK 80670 and DK 84960 to K.N.L.), Canadian Institute for Health Research (to G.M.H. and A.L.M.), Canadian Liver Foundation and Alberta Innovates Health Solutions (to A.L.M.), and Deutsche Forschungsgemeinschaft (to F.L.).
Abbreviations used in this paper
- AH
ancestral haplotype
- AMA
antimitochondrial antibodies
- GGT
γ-glutamyltransferase
- GWAS
genome-wide association studies
- IBD
inflammatory bowel disease
- IL
interleukin
- LPAC
low phospholipid-associated cholelithiasis
- MHC
major histocompatibility complex
- PBC
primary biliary cirrhosis
- PDC-E2
pyruvate dehydrogenase complex E2
- PFIC
progressive familial intrahepatic cholestasis
- PSC
primary sclerosing cholangitis
- Th
T-helper
- TNF
tumor necrosis factor
Appendix
Concepts and Definitions Relevant to Genetic Studies in Cholestasis76,120,134
Allele: A particular nucleotide (or nucleotide sequence) at any type of polymorphism
Causal variant: A genetic variant that has a direct functional effect on disease risk or that causes the observed association signal (difficult to prove causality beyond reasonable doubt)
Epistasis: The phenomenon in which the effects of one genetic variant are modified by one or several other genetic variants
Exome: Protein coding regions (exons) of the genome
Genotype: The combination of alleles on the 2 chromosomes of an individual
Haplotype: A distinct combination of 2 or more alleles of polymorphisms that occur together on the same chromosome
Heritability (h2): The phenotypic variation in a population that is contributed by genetic variation
HLA: Antigen-presenting molecules encoded by genes within the major histocompatibility complex located on human chromosome 6p21
Linkage disequilibrium: Occurs when 2 alleles of polymorphisms on the same chromosome occur more frequently together than would be expected from their respective population frequencies
Meta-genetics: Genetic and genomic studies considering all of the genes in symbiotic organisms (the metagenome), for example, as opposed to considering host genes or microbial genes in isolation
Microbiome: The sum of all microbial genes in a given host comportment (eg, the gut microbiome)
Microbiota: The sum of all microbial species in a given host compartment (eg, the gut microbiota)
Polymorphisms: Genetic variants arising from mutational events in DNA; the variant should occur at a frequency >1% in the general population
Population attributable fraction: The proportional reduction in average disease risk over a specified time interval that would be achieved by eliminating the exposures of interest from the population while the distribution of other risk factors within the population remains unchanged
Single nucleotide polymorphisms: DNA polymorphisms that differ at only a single base
Footnotes
Conflicts of interest
The authors disclose the following: G.M.H. has served as a consultant for Intercept Pharma, Medigene, Centocor, and Lumena. A.L.M. has received research support from Abbott, Gilead, and Novartis. The remaining authors disclose no conflicts.
References
- 1.Hirschfield GM, Heathcote EJ, Gershwin ME. Pathogenesis of cholestatic liver disease and therapeutic approaches. Gastroenterology. 2010;139:1481–1496. doi: 10.1053/j.gastro.2010.09.004. [DOI] [PubMed] [Google Scholar]
- 2.Boonstra K, Beuers U, Ponsioen CY. Epidemiology of primary sclerosing cholangitis and primary biliary cirrhosis: a systematic review. J Hepatol. 2012;56:1181–1188. doi: 10.1016/j.jhep.2011.10.025. [DOI] [PubMed] [Google Scholar]
- 3.Hirschfield GM. Diagnosis of primary biliary cirrhosis. Best Pract Res Clin Gastroenterol. 2011;25:701–712. doi: 10.1016/j.bpg.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 4.Karlsen TH, Schrumpf E, Boberg KM. Genetic epidemiology of primary sclerosing cholangitis. World J Gastroenterol. 2007;13:5421–5431. doi: 10.3748/wjg.v13.i41.5421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arbour L, Rupps R, Field L, et al. Characteristics of primary biliary cirrhosis in British Columbia's First Nations population. Can J Gastroenterol. 2005;19:305–310. doi: 10.1155/2005/203028. [DOI] [PubMed] [Google Scholar]
- 6.Maurer KJ, Carey MC, Fox JG. Roles of infection, inflammation, and the immune system in cholesterol gallstone formation. Gastroenterology. 2009;136:425–440. doi: 10.1053/j.gastro.2008.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krawczyk M, Wang DQ, Portincasa P, et al. Dissecting the genetic heterogeneity of gallbladder stone formation. Semin Liver Dis. 2011;31:157–172. doi: 10.1055/s-0031-1276645. [DOI] [PubMed] [Google Scholar]
- 8.Lammert F, Sauerbruch T. Mechanisms of disease: the genetic epidemiology of gallbladder stones. Nat Clin Pract Gastroenterol Hepatol. 2005;2:423–433. doi: 10.1038/ncpgasthep0257. [DOI] [PubMed] [Google Scholar]
- 9.Koebnick C, Smith N, Black MH, et al. Pediatric obesity and gallstone disease. J Pediatr Gastroenterol Nutr. 2012;55:328–333. doi: 10.1097/MPG.0b013e31824d256f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhong B, Strnad P, Selmi C, et al. Keratin variants are overrep-resented in primary biliary cirrhosis and associate with disease severity. Hepatology. 2009;50:546–554. doi: 10.1002/hep.23041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Katsika D, Grjibovski A, Einarsson C, et al. Genetic and environmental influences on symptomatic gallstone disease: a Swedish study of 43,141 twin pairs. Hepatology. 2005;41:1138–1143. doi: 10.1002/hep.20654. [DOI] [PubMed] [Google Scholar]
- 12.Chapman RW, Varghese Z, Gaul R, et al. Association of primary sclerosing cholangitis with HLA-B8. Gut. 1983;24:38–41. doi: 10.1136/gut.24.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hirschfield GM, Invernizzi P. Progress in the genetics of primary biliary cirrhosis. Semin Liver Dis. 2011;31:147–156. doi: 10.1055/s-0031-1276644. [DOI] [PubMed] [Google Scholar]
- 14.Hirschfield GM, Liu X, Xu C, et al. Primary biliary cirrhosis associated with HLA, IL12A, and IL12RB2 variants. N Engl J Med. 2009;360:2544–2555. doi: 10.1056/NEJMoa0810440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Melum E, Franke A, Schramm C, et al. Genome-wide association analysis in primary sclerosing cholangitis identifies two non-HLA susceptibility loci. Nat Genet. 2011;43:17–19. doi: 10.1038/ng.728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Invernizzi P. Human leukocyte antigen in primary biliary cirrhosis: an old story now reviving. Hepatology. 2011;54:714–723. doi: 10.1002/hep.24414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Invernizzi P, Ransom M, Raychaudhuri S, et al. Classical HLADRB1 and DPB1 alleles account for HLA associations with primary biliary cirrhosis. Genes Immun. 2012;13:461–468. doi: 10.1038/gene.2012.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Todd JA, Walker NM, Cooper JD, et al. Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat Genet. 2007;39:857–864. doi: 10.1038/ng2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones EY, Fugger L, Strominger JL, et al. MHC class II proteins and disease: a structural perspective. Nat Rev Immunol. 2006;6:271–282. doi: 10.1038/nri1805. [DOI] [PubMed] [Google Scholar]
- 20.Illing PT, Vivian JP, Dudek NL, et al. Immune self-reactivity triggered by drug-modified HLA-peptide repertoire. Nature. 2012;486:554–558. doi: 10.1038/nature11147. [DOI] [PubMed] [Google Scholar]
- 21.Mason AL. An autoimmune biliary disease mouse model for primary biliary cirrhosis: something for everyone. Hepatology. 2006;44:1047–1050. doi: 10.1002/hep.21390. [DOI] [PubMed] [Google Scholar]
- 22.Daly AK, Donaldson PT, Bhatnagar P, et al. HLA-B*5701 genotype is a major determinant of drug-induced liver injury due to flucloxacillin. Nat Genet. 2009;41:816–819. doi: 10.1038/ng.379. [DOI] [PubMed] [Google Scholar]
- 23.Lucena MI, Molokhia M, Shen Y, et al. Susceptibility to amoxicillin-clavulanate-induced liver injury is influenced by multiple HLA class I and II alleles. Gastroenterology. 2011;141:338–347. doi: 10.1053/j.gastro.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Joplin R, Gershwin ME. Ductular expression of autoantigens in primary biliary cirrhosis. Semin Liver Dis. 1997;17:97–103. doi: 10.1055/s-2007-1007187. [DOI] [PubMed] [Google Scholar]
- 25.Shimoda S, Van de Water J, Ansari A, et al. Identification and precursor frequency analysis of a common T cell epitope motif in mitochondrial autoantigens in primary biliary cirrhosis. J Clin Invest. 1998;102:1831–1840. doi: 10.1172/JCI4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fellay J, Shianna KV, Ge D, et al. A whole-genome association study of major determinants for host control of HIV-1. Science. 2007;317:944–947. doi: 10.1126/science.1143767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kamatani Y, Wattanapokayakit S, Ochi H, et al. A genome-wide association study identifies variants in the HLA-DP locus associated with chronic hepatitis B in Asians. Nat Genet. 2009;41:591–595. doi: 10.1038/ng.348. [DOI] [PubMed] [Google Scholar]
- 28.Price P, Witt C, Allcock R, et al. The genetic basis for the association of the 8.1 ancestral haplotype (A1, B8, DR3) with multiple immunopathological diseases. Immunol Rev. 1999;167:257–274. doi: 10.1111/j.1600-065x.1999.tb01398.x. [DOI] [PubMed] [Google Scholar]
- 29.Listi F, Caruso C, Colonna-Romano G, et al. HLA and KIR frequencies in Sicilian Centenarians. Rejuvenation Res. 2010;13:314–318. doi: 10.1089/rej.2009.0984. [DOI] [PubMed] [Google Scholar]
- 30.Jostins L, Ripke S, Weersma RK, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dunham I, Kundaje A, Aldred SF, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cotsapas C, Voight BF, Rossin E, et al. Pervasive sharing of genetic effects in autoimmune disease. PLoS Genet. 2011;7:e1002254. doi: 10.1371/journal.pgen.1002254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Juran BD, Atkinson EJ, Larson JJ, et al. Carriage of a tumor necrosis factor polymorphism amplifies the cytotoxic T-lymphocyte antigen 4 attributed risk of primary biliary cirrhosis: evidence for a gene-gene interaction. Hepatology. 2010;52:223–229. doi: 10.1002/hep.23667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu X, Invernizzi P, Lu Y, et al. Genome-wide meta-analyses identify three loci associated with primary biliary cirrhosis. Nat Genet. 2010;42:658–660. doi: 10.1038/ng.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mells GF, Floyd JA, Morley KI, et al. Genome-wide association study identifies 12 new susceptibility loci for primary biliary cirrhosis. Nat Genet. 2011;43:329–332. doi: 10.1038/ng.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakamura M, Nishida N, Kawashima M, et al. Genome-wide association study identifies TNFSF15 and POU2AF1 as susceptibility loci for primary biliary cirrhosis in the Japanese population. Am J Hum Genet. 2012;91:721–728. doi: 10.1016/j.ajhg.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hirschfield GM, Liu X, Han Y, et al. Variants at IRF5-TNPO3, 17q12-21 and MMEL1 are associated with primary biliary cirrhosis. Nat Genet. 2010;42:655–657. doi: 10.1038/ng.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trinchieri G. Interleukin-12 and the regulation of innate resistance and adpative immunity. Nat Rev Immunol. 2003;3:133–146. doi: 10.1038/nri1001. [DOI] [PubMed] [Google Scholar]
- 39.Collison LW, Delgoffe GM, Guy CS, et al. The composition and signaling of the IL-35 receptor are unconventional. Nat Immunol. 2012;13:290–299. doi: 10.1038/ni.2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koch MA, Thomas KR, Perdue NR, et al. T-bet(+) Treg cells undergo abortive Th1 cell differentiation due to impaired expression of IL-12 receptor beta2. Immunity. 2012;37:501–510. doi: 10.1016/j.immuni.2012.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krausgruber T, Blazek K, Smallie T, et al. IRF5 promotes inflammatory macrophage polarization and TH1-TH17 responses. Nat Immunol. 2011;12:231–238. doi: 10.1038/ni.1990. [DOI] [PubMed] [Google Scholar]
- 42.Pulickal AS, Hambleton S, Callaghan MJ, et al. Biliary cirrhosis in a child with inherited interleukin-12 deficiency. J Trop Pediatr. 2008;54:269–271. doi: 10.1093/tropej/fmm119. [DOI] [PubMed] [Google Scholar]
- 43.Folseraas T, Melum E, Rausch P, et al. Extended analysis of a genome-wide association study in primary sclerosing cholangitis detects multiple novel risk loci. J Hepatol. 2012;57:366–375. doi: 10.1016/j.jhep.2012.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Karlsen TH, Schrumpf E, Boberg KM. Update on primary sclerosing cholangitis. Dig Liver Dis. 2010;42:390–400. doi: 10.1016/j.dld.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 45.Karlsen TH, Schrumpf E, Boberg KM. Primary sclerosing cholangitis. Best Pract Res Clin Gastroenterol. 2010;24:655–666. doi: 10.1016/j.bpg.2010.07.005. [DOI] [PubMed] [Google Scholar]
- 46.Saarinen S, Olerup O, Broome U. Increased frequency of autoimmune diseases in patients with primary sclerosing cholangitis. Am J Gastroenterol. 2000;95:3195–2199. doi: 10.1111/j.1572-0241.2000.03292.x. [DOI] [PubMed] [Google Scholar]
- 47.Janse M, Lamberts LE, Franke L, et al. Three ulcerative colitis susceptibility loci are associated with primary sclerosing cholangitis and indicate a role for IL2, REL, and CARD9. Hepatology. 2011;53:1977–1985. doi: 10.1002/hep.24307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Anderson CA, Boucher G, Lees CW, et al. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat Genet. 2011;43:246–252. doi: 10.1038/ng.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Karlsen TH, Franke A, Melum E, et al. Genome-wide association analysis in primary sclerosing cholangitis. Gastroenterology. 2010;138:1102–1111. doi: 10.1053/j.gastro.2009.11.046. [DOI] [PubMed] [Google Scholar]
- 50.Jorgensen KK, Grzyb K, Lundin KE, et al. Inflammatory bowel disease in patients with primary sclerosing cholangitis: clinical characterization in liver transplanted and nontransplanted patients. Inflamm Bowel Dis. 2012;18:536–545. doi: 10.1002/ibd.21699. [DOI] [PubMed] [Google Scholar]
- 51.Cheung TC, Humphreys IR, Potter KG, et al. Evolutionarily divergent herpesviruses modulate T cell activation by targeting the herpesvirus entry mediator cosignaling pathway. Proc Natl Acad Sci U S A. 2005;102:13218–13223. doi: 10.1073/pnas.0506172102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Oya Y, Watanabe N, Owada T, et al. Development of autoimmune hepatitis-like disease and production of autoantibodies to nuclear antigens in mice lacking B and T lymphocyte attenuator. Arthritis Rheum. 2008;58:2498–2510. doi: 10.1002/art.23674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lindesmith L, Moe C, Marionneau S, et al. Human susceptibility and resistance to Norwalk virus infection. Nat Med. 2003;9:548–553. doi: 10.1038/nm860. [DOI] [PubMed] [Google Scholar]
- 54.Falk PG, Bry L, Holgersson J, et al. Expression of a human alpha-1,3/4-fucosyltransferase in the pit cell lineage of FVB/N mouse stomach results in production of Leb-containing glycoconjugates: a potential transgenic mouse model for studying Helicobacter pylori infection. Proc Natl Acad Sci U S A. 1995;92:1515–1519. doi: 10.1073/pnas.92.5.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rausch P, Rehman A, Kunzel S, et al. Colonic mucosa-associated microbiota is influenced by an interaction of Crohn disease and FUT2 (Secretor) genotype. Proc Natl Acad Sci U S A. 2011;108:19030–19035. doi: 10.1073/pnas.1106408108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Spor A, Koren O, Ley R. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat Rev Microbiol. 2011;9:279–290. doi: 10.1038/nrmicro2540. [DOI] [PubMed] [Google Scholar]
- 57.Buch S, Schafmayer C, Volzke H, et al. A genome-wide association scan identifies the hepatic cholesterol transporter ABCG8 as a susceptibility factor for human gallstone disease. Nat Genet. 2007;39:995–999. doi: 10.1038/ng2101. [DOI] [PubMed] [Google Scholar]
- 58.Wittenburg H, Lyons MA, Li R, et al. FXR and ABCG5/ABCG8 as determinants of cholesterol gallstone formation from quantitative trait locus mapping in mice. Gastroenterology. 2003;125:868–881. doi: 10.1016/s0016-5085(03)01053-9. [DOI] [PubMed] [Google Scholar]
- 59.Schafmayer C, Teumer A, Buch S, et al. A genome-wide scan identifies TM4SF4 (intestine and liver tetraspan membrane protein 4) as a susceptibility locus for gallstone disease. Gastroenterology. 2010;138(Suppl 1):S–10. [Google Scholar]
- 60.Wittenburg H, Tönjes A, Mirzakhyl S, et al. Identifizierung neuer Gallensteingene (LITH-Gene) in einer genomweiten Assoziationsstudie basierend auf der “Gallensteinkarte” des Inzuchtmausmodells der Cholelithiasis. Z Gastroenterol. 2009:47. [Google Scholar]
- 61.Grunhage F, Acalovschi M, Tirziu S, et al. Increased gallstone risk in humans conferred by common variant of hepatic ATP-binding cassette transporter for cholesterol. Hepatology. 2007;46:793–801. doi: 10.1002/hep.21847. [DOI] [PubMed] [Google Scholar]
- 62.Katsika D, Magnusson P, Krawczyk M, et al. Gallstone disease in Swedish twins: risk is associated with ABCG8 D19H genotype. J Intern Med. 2010;268:279–285. doi: 10.1111/j.1365-2796.2010.02249.x. [DOI] [PubMed] [Google Scholar]
- 63.Kuo KK, Shin SJ, Chen ZC, et al. Significant association of ABCG5 604Q and ABCG8 D19H polymorphisms with gallstone disease. Br J Surg. 2008;95:1005–1011. doi: 10.1002/bjs.6178. [DOI] [PubMed] [Google Scholar]
- 64.Siddapuram SP, Mahurkar S, Duvvuru NR, et al. Hepatic cholesterol transporter ABCG8 polymorphisms in gallstone disease in an Indian population. J Gastroenterol Hepatol. 2010;25:1093–1098. doi: 10.1111/j.1440-1746.2010.06309.x. [DOI] [PubMed] [Google Scholar]
- 65.Srivastava A, Srivastava K, Choudhuri G, et al. Role of ABCG8 D19H (rs11887534) variant in gallstone susceptibility in northern India. J Gastroenterol Hepatol. 2010;25:1758–1762. doi: 10.1111/j.1440-1746.2010.06349.x. [DOI] [PubMed] [Google Scholar]
- 66.Stender S, Frikke-Schmidt R, Nordestgaard BG, et al. Sterol transporter adenosine triphosphate-binding cassette transporter G8, gallstones, and biliary cancer in 62,000 individuals from the general population. Hepatology. 2011;53:640–648. doi: 10.1002/hep.24046. [DOI] [PubMed] [Google Scholar]
- 67.Krawczyk M, Lutjohann D, Schirin-Sokhan R, et al. Phytosterol and cholesterol precursor levels indicate increased cholesterol excretion and biosynthesis in gallstone patients. Hepatology. 2012;55:1507–1517. doi: 10.1002/hep.25563. [DOI] [PubMed] [Google Scholar]
- 68.Teupser D, Baber R, Ceglarek U, et al. Genetic regulation of serum phytosterol levels and risk of coronary artery disease. Circ Cardiovasc Genet. 2010;3:331–339. doi: 10.1161/CIRCGENETICS.109.907873. [DOI] [PubMed] [Google Scholar]
- 69.Jakulj L, Vissers MN, Tanck MW, et al. ABCG5/G8 polymorphisms and markers of cholesterol metabolism: systematic review and meta-analysis. J Lipid Res. 2010;51:3016–3023. doi: 10.1194/jlr.M008128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Krawczyk M, Lutjohann D, Schirin-Sokhan R, et al. Phytosterol and cholesterol precursor levels indicate increased cholesterol excretion and biosynthesis in gallstone disease. Hepatology. 2012;55:1507–1517. doi: 10.1002/hep.25563. [DOI] [PubMed] [Google Scholar]
- 71.Szilvasi A, Andrikovics H, Pongracz E, et al. Frequencies of four ATP-binding cassette transporter G8 polymorphisms in patients with ischemic vascular diseases. Genet Test Mol Biomarkers. 2010;14:667–672. doi: 10.1089/gtmb.2010.0035. [DOI] [PubMed] [Google Scholar]
- 72.Koeijvoets KC, van der Net JB, Dallinga-Thie GM, et al. ABCG8 gene polymorphisms, plasma cholesterol concentrations, and risk of cardiovascular disease in familial hypercholesterolemia. Atherosclerosis. 2009;204:453–458. doi: 10.1016/j.atherosclerosis.2008.09.018. [DOI] [PubMed] [Google Scholar]
- 73.Sanna S, Busonero F, Maschio A, et al. Common variants in the SLCO1B3 locus are associated with bilirubin levels and unconjugated hyperbilirubinemia. Hum Mol Genet. 2009;18:2711–2718. doi: 10.1093/hmg/ddp203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Buch S, Schafmayer C, Volzke H, et al. Loci from a genome-wide analysis of bilirubin levels are associated with gallstone risk and composition. Gastroenterology. 2010;139:1942–1951. e2. doi: 10.1053/j.gastro.2010.09.003. [DOI] [PubMed] [Google Scholar]
- 75.Wittenburg H, Lammert F. Genetic predisposition to gallbladder stones. Semin Liver Dis. 2007;27:109–121. doi: 10.1055/s-2006-960174. [DOI] [PubMed] [Google Scholar]
- 76.Krawczyk M, Mullenbach R, Weber SN, et al. Genome-wide association studies and genetic risk assessment of liver diseases. Nat Rev Gastroenterol Hepatol. 2010;7:669–681. doi: 10.1038/nrgastro.2010.170. [DOI] [PubMed] [Google Scholar]
- 77.Fickert P, Fuchsbichler A, Marschall HU, et al. Lithocholic acid feeding induces segmental bile duct obstruction and destructive cholangitis in mice. Am J Pathol. 2006;168:410–422. doi: 10.2353/ajpath.2006.050404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mourelle M, Salas A, Vilaseca J, et al. Induction of chronic cholangitis in the rat by trinitrobenzenesulfonic acid. J Hepatol. 1995;22:219–225. doi: 10.1016/0168-8278(95)80432-3. [DOI] [PubMed] [Google Scholar]
- 79.Orth T, Neurath M, Schirmacher P, et al. A novel rat model of chronic fibrosing cholangitis induced by local administration of a hapten reagent into the dilated bile duct is associated with increased TNF-alpha production and autoantibodies. J Hepatol. 2000;33:862–872. doi: 10.1016/s0168-8278(00)80116-6. [DOI] [PubMed] [Google Scholar]
- 80.Yamada S, Ishii M, Liang LS, et al. Small duct cholangitis induced by N-formyl L-methionine L-leucine L-tyrosine in rats. J Gastroenterol. 1994;29:631–636. doi: 10.1007/BF02365447. [DOI] [PubMed] [Google Scholar]
- 81.Durie PR, Kent G, Phillips MJ, et al. Characteristic multiorgan pathology of cystic fibrosis in a long-living cystic fibrosis trans-membrane regulator knockout murine model. Am J Pathol. 2004;164:1481–1493. doi: 10.1016/S0002-9440(10)63234-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fickert P, Fuchsbichler A, Wagner M, et al. Regurgitation of bile acids from leaky bile ducts causes sclerosing cholangitis in Mdr2 (Abcb4) knockout mice. Gastroenterology. 2004;127:261–274. doi: 10.1053/j.gastro.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 83.Freudenberg F, Leonard MR, Liu SA, et al. Pathophysiological preconditions promoting mixed “black” pigment plus cholesterol gallstones in a DeltaF508 mouse model of cystic fibrosis. Am J Physiol Gastrointest Liver Physiol. 2010;299:G205–G214. doi: 10.1152/ajpgi.00341.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lammert F, Wang DQ, Hillebrandt S, et al. Spontaneous cholecysto- and hepatolithiasis in Mdr2-/- mice: a model for low phos pholipid-associated cholelithiasis. Hepatology. 2004;39:117–128. doi: 10.1002/hep.20022. [DOI] [PubMed] [Google Scholar]
- 85.Meerman L, Koopen NR, Bloks V, et al. Biliary fibrosis associated with altered bile composition in a mouse model of erythropoietic protoporphyria. Gastroenterology. 1999;117:696–705. doi: 10.1016/s0016-5085(99)70464-6. [DOI] [PubMed] [Google Scholar]
- 86.Smit JJ, Schinkel AH, Oude Elferink RP, et al. Homozygous disruption of the murine mdr2 P-glycoprotein gene leads to a complete absence of phospholipid from bile and to liver disease. Cell. 1993;75:451–462. doi: 10.1016/0092-8674(93)90380-9. [DOI] [PubMed] [Google Scholar]
- 87.Lichtman SN, Keku J, Clark RL, et al. Biliary tract disease in rats with experimental small bowel bacterial overgrowth. Hepatology. 1991;13:766–772. [PubMed] [Google Scholar]
- 88.Ward JM, Anver MR, Haines DC, et al. Chronic active hepatitis in mice caused by Helicobacter hepaticus. Am J Pathol. 1994;145:959–968. [PMC free article] [PubMed] [Google Scholar]
- 89.Stephens J, Cosyns M, Jones M, et al. Liver and bile duct pathology following Cryptosporidium parvum infection of immunodeficient mice. Hepatology. 1999;30:27–35. doi: 10.1002/hep.510300138. [DOI] [PubMed] [Google Scholar]
- 90.Ungar BL, Burris JA, Quinn CA, et al. New mouse models for chronic Cryptosporidium infection in immunodeficient hosts. Infect Immun. 1990;58:961–969. doi: 10.1128/iai.58.4.961-969.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cello JP. Acquired immunodeficiency syndrome cholangiopathy: spectrum of disease. Am J Med. 1989;86:539–546. doi: 10.1016/0002-9343(89)90381-1. [DOI] [PubMed] [Google Scholar]
- 92.Irie J, Wu Y, Wicker LS, et al. NOD.c3c4 congenic mice develop autoimmune biliary disease that serologically and pathogenetically models human primary biliary cirrhosis. J Exp Med. 2006;203:1209–1219. doi: 10.1084/jem.20051911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Koarada S, Wu Y, Fertig N, et al. Genetic control of autoimmunity: protection from diabetes, but spontaneous autoimmune biliary disease in a nonobese diabetic congenic strain. J Immunol. 2004;173:2315–2323. doi: 10.4049/jimmunol.173.4.2315. [DOI] [PubMed] [Google Scholar]
- 94.Oertelt S, Lian ZX, Cheng CM, et al. Anti-mitochondrial antibodies and primary biliary cirrhosis in TGF-beta receptor II dominant-negative mice. J Immunol. 2006;177:1655–1660. doi: 10.4049/jimmunol.177.3.1655. [DOI] [PubMed] [Google Scholar]
- 95.Wakabayashi K, Lian ZX, Moritoki Y, et al. IL-2 receptor alpha(-/-) mice and the development of primary biliary cirrhosis. Hepatology. 2006;44:1240–1249. doi: 10.1002/hep.21385. [DOI] [PubMed] [Google Scholar]
- 96.Zhang W, Sharma R, Ju ST, et al. Deficiency in regulatory T cells results in development of antimitochondrial antibodies and autoimmune cholangitis. Hepatology. 2009;49:545–552. doi: 10.1002/hep.22651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wasilenko S, Mason G, Mason A. Primary biliary cirrhosis, bacteria and molecular mimicry: what's the molecule and where's the mimic? Liver Int. 2009;29:779–782. doi: 10.1111/j.1478-3231.2009.02051.x. [DOI] [PubMed] [Google Scholar]
- 98.Xu L, Shen Z, Guo L, et al. Does a betaretrovirus infection trigger primary biliary cirrhosis? Proc Natl Acad Sci U S A. 2003;100:8454–8459. doi: 10.1073/pnas.1433063100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhang G, Chen M, Graham D, et al. Mouse mammary tumor virus in anti-mitochondrial antibody producing mouse models. J Hepatol. 2011;55:876–884. doi: 10.1016/j.jhep.2011.01.037. [DOI] [PubMed] [Google Scholar]
- 100.Wakabayashi K, Yoshida K, Leung PSC, et al. Induction of auto-immune cholangitis in non-obese diabetic (NOD).1101 mice following a chemical xenobiotic immunization. Clin Exp Immunol. 2009;155:577–586. doi: 10.1111/j.1365-2249.2008.03837.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mattner J, Savage PB, Leung P, et al. Liver autoimmunity triggered by microbial activation of natural killer T cells. Cell Host Microbe. 2008;3:304–315. doi: 10.1016/j.chom.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Tsuda M, Zhang W, Yang GX, et al. Deletion of IL-12p35 induces liver fibrosis in dominant negative transforming growth factor beta receptor type II mice. Hepatology. 2013;57:806–816. doi: 10.1002/hep.25829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yoshida K, Yang GX, Zhang W, et al. Deletion of interleukin-12p40 suppresses autoimmune cholangitis in dominant negative transforming growth factor beta receptor type II mice. Hepatology. 2009;50:1494–1500. doi: 10.1002/hep.23132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Salas JT, Banales JM, Sarvide S, et al. Ae2a,b-deficient mice develop antimitochondrial antibodies and other features resembling primary biliary cirrhosis. Gastroenterology. 2008;134:1482–1493. doi: 10.1053/j.gastro.2008.02.020. [DOI] [PubMed] [Google Scholar]
- 105.Manolio TA, Collins FS, Cox NJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Juran BD, Lazaridis KN. Genomics in the post-GWAS era. Semin Liver Dis. 2011;31:215–222. doi: 10.1055/s-0031-1276641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Qin J, Li Y, Cai Z, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490:55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- 108.Lyssenko V, Jonsson A, Almgren P, et al. Clinical risk factors, DNA variants, and the development of type 2 diabetes. N Engl J Med. 2008;359:2220–2232. doi: 10.1056/NEJMoa0801869. [DOI] [PubMed] [Google Scholar]
- 109.van Hoek M, Dehghan A, Witteman JC, et al. Predicting type 2 diabetes based on polymorphisms from genome-wide association studies: a population-based study. Diabetes. 2008;57:3122–3128. doi: 10.2337/db08-0425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Stahl EA, Wegmann D, Trynka G, et al. Bayesian inference analyses of the polygenic architecture of rheumatoid arthritis. Nat Genet. 2012;44:483–489. doi: 10.1038/ng.2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Visscher PM, Brown MA, McCarthy MI, et al. Five years of GWAS discovery. Am J Hum Genet. 2012;90:7–24. doi: 10.1016/j.ajhg.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Juran BD, Hirschfield GM, Invernizzi P, et al. Immunochip analyses identify a novel risk locus for primary biliary cirrhosis at 13q14, multiple independent associations at four established risk loci and epistasis between 1p31 and 7q32 risk variants. Hum Mol Genet. 2012;21:5209–5221. doi: 10.1093/hmg/dds359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.O'Roak BJ, Vives L, Fu W, et al. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science. 2012;338:1619–1622. doi: 10.1126/science.1227764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Baranzini SE, Mudge J, van Velkinburgh JC, et al. Genome, epigenome and RNA sequences of monozygotic twins discordant for multiple sclerosis. Nature. 2010;464:1351–1356. doi: 10.1038/nature08990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Invernizzi P, Miozzo M, Battezzati PM, et al. Frequency of monosomy X in women with primary biliary cirrhosis. Lancet. 2004;363:533–535. doi: 10.1016/S0140-6736(04)15541-4. [DOI] [PubMed] [Google Scholar]
- 116.Miozzo M, Selmi C, Gentilin B, et al. Preferential X chromosome loss but random inactivation characterize primary biliary cirrhosis. Hepatology. 2007;46:456–462. doi: 10.1002/hep.21696. [DOI] [PubMed] [Google Scholar]
- 117.Lleo A, Oertelt-Prigione S, Bianchi I, et al. Y chromosome loss in male patients with primary biliary cirrhosis. J Autoimmun. 2013;41:87–91. doi: 10.1016/j.jaut.2012.12.008. [DOI] [PubMed] [Google Scholar]
- 118.Lichtenstein P, Holm NV, Verkasalo PK, et al. Environmental and heritable factors in the causation of cancer—analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med. 2000;343:78–85. doi: 10.1056/NEJM200007133430201. [DOI] [PubMed] [Google Scholar]
- 119.Roberts NJ, Vogelstein JT, Parmigiani G, et al. The predictive capacity of personal genome sequencing. Sci Transl Med. 2012;4:133ra58. doi: 10.1126/scitranslmed.3003380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Virgin HW, Todd JA. Metagenomics and personalized medicine. Cell. 2011;147:44–56. doi: 10.1016/j.cell.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Markle JG, Frank DN, Mortin-Toth S, et al. Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science. 2013;339:1084–1088. doi: 10.1126/science.1233521. [DOI] [PubMed] [Google Scholar]
- 122.Maurer KJ, Ihrig MM, Rogers AB, et al. Identification of cholelithogenic enterohepatic helicobacter species and their role in murine cholesterol gallstone formation. Gastroenterology. 2005;128:1023–1033. doi: 10.1053/j.gastro.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 123.Henao-Mejia J, Elinav E, Jin C, et al. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature. 2012;482:179–185. doi: 10.1038/nature10809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Cadwell K, Patel KK, Maloney NS, et al. Virus-plus-susceptibility gene interaction determines Crohn's disease gene Atg16L1 phenotypes in intestine. Cell. 2010;141:1135–1145. doi: 10.1016/j.cell.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Saxena D, Li Y, Yang L, et al. Human microbiome and HIV/AIDS. Curr HIV/AIDS Rep. 2012;9:44–51. doi: 10.1007/s11904-011-0103-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Worthey EA, Mayer AN, Syverson GD, et al. Making a definitive diagnosis: successful clinical application of whole exome sequencing in a child with intractable inflammatory bowel disease. Genet Med. 2011;13:255–262. doi: 10.1097/GIM.0b013e3182088158. [DOI] [PubMed] [Google Scholar]
- 127.Muise AM, Xu W, Guo CH, et al. NADPH oxidase complex and IBD candidate gene studies: identification of a rare variant in NCF2 that results in reduced binding to RAC2. Gut. 2012;61:1028–1035. doi: 10.1136/gutjnl-2011-300078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Moran CJ, Walters TD, Guo CH, et al. IL-10R polymorphisms are associated with very-early-onset ulcerative colitis. Inflamm Bowel Dis. 2013;19:115–123. doi: 10.1002/ibd.22974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Milton JN, Sebastiani P, Solovieff N, et al. A genome-wide association study of total bilirubin and cholelithiasis risk in sickle cell anemia. PLoS One. 2012;7:e34741. doi: 10.1371/journal.pone.0034741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Schwertner HA, Vitek L. Gilbert syndrome, UGT1A1*28 allele, and cardiovascular disease risk: possible protective effects and therapeutic applications of bilirubin. Atherosclerosis. 2008;198:1–11. doi: 10.1016/j.atherosclerosis.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 131.Trusheim MR, Berndt ER, Douglas FL. Stratified medicine: strategic and economic implications of combining drugs and clinical biomarkers. Nat Rev Drug Discov. 2007;6:287–293. doi: 10.1038/nrd2251. [DOI] [PubMed] [Google Scholar]
- 132.Stokes CS, Krawczyk M, Lammert F. Gallstones: environment, lifestyle and genes. Dig Dis. 2011;29:191–201. doi: 10.1159/000323885. [DOI] [PubMed] [Google Scholar]
- 133.Carbone M, Mells G, Alexander G, et al. Calcineurin inhibitors and the IL12A locus influence risk of recurrent primary biliary cirrhosis after liver transplantation. Am J Transplant. 2013;13:1110–1111. doi: 10.1111/ajt.12132. [DOI] [PubMed] [Google Scholar]
- 134.Karlsen TH, Melum E, Franke A. The utility of genome-wide association studies in hepatology. Hepatology. 2010;51:1833–1842. doi: 10.1002/hep.23564. [DOI] [PubMed] [Google Scholar]
- 135.Rege RV, Ostrow JD. Animal models of pigment and cholesterol gallstone disease. In: Muraca M, editor. Methods of biliary research. CRC Press; Boca Raton, FL: 1995. pp. 203–242. [Google Scholar]
- 136.Dam H, Christensen F. Alimentary production of gallstones in hamsters. Acta Pathol Microbiol Scand. 1952;30:236–242. doi: 10.1111/j.1699-0463.1952.tb00179.x. [DOI] [PubMed] [Google Scholar]
- 137.Lee SP, Carey MC, LaMont JT. Aspirin prevention of cholesterol gallstone formation in prairie dogs. Science. 1981;211:1429–1431. doi: 10.1126/science.7466399. [DOI] [PubMed] [Google Scholar]
- 138.Brenneman DE, Connor WE, Forker EL, et al. The formation of abnormal bile and cholesterol gallstones from dietary cholesterol in the prairie dog. J Clin Invest. 1972;51:1495–1503. doi: 10.1172/JCI106946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Khanuja B, Cheah YC, Hunt M, et al. Lith1, a major gene affecting cholesterol gallstone formation among inbred strains of mice. Proc Natl Acad Sci U S A. 1995;92:7729–7733. doi: 10.1073/pnas.92.17.7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.LaMorte WW, Brotschi EA, Scott TE, et al. Pigment gallstone formation in the cholesterol-fed guinea pig. Hepatology. 1985;5:21–27. doi: 10.1002/hep.1840050106. [DOI] [PubMed] [Google Scholar]