Abstract
The neocortex represents the brain region that has undergone a major increase in its relative size during the course of mammalian evolution. The larger cortex results from a corresponding increase in progenitor cell number. The progenitors giving rise to neocortex are located in the ventricular zone of the dorsal telencephalon and highly express Lhx2, a LIM-homeodomain transcription factor. The neocortex fails to form in the Lhx2 constitutive knockout, indicating a role for Lhx2 in corticogenesis, but this outcome, and mid-embryonic lethality, requires use of conditional strategies for further study. Therefore, to explore Lhx2 function in neocortical progenitors, we generated mice with Lhx2 conditionally deleted from cortical progenitors at the onset of neurogenesis. We find that Lhx2 is critical for maintaining the proliferative state of neocortical progenitors during corticogenesis. In the conditional knockouts, the neocortex is formed but is significantly smaller than wild type. We find that deletion of Lhx2 leads to significantly decreased numbers of cortical progenitors and premature neuronal differentiation. A likely mechanism is indicated by our findings that Lhx2 is required for the expression of Hes1 in cortical progenitors, a key effector in the Notch signaling pathway that maintains the proliferative progenitor state. We conclude that Lhx2 regulates the balance between proliferation and differentiation in cortical progenitors and through this mechanism controls cortical size.
Keywords: cortical neurogenesis, Lhx2, Hes1
Introduction
The neocortex is the largest region of the mammalian cerebral cortex and is responsible for sensory perception, cognition and control of movements. The multilayered and interconnected neurons in the neocortex are formed through a tightly regulated series of cell divisions and migration (Aboitiz et al., 2001; Fish et al., 2008; Nadarajah and Parnavelas, 2002). All neocortical neurons are generated from progenitor cells located in the neuroepithelium lining the lateral ventricle. During development, these neural progenitor cells undergo both symmetric and asymmetric types of division (Noctor et al., 2004; Noctor et al., 2008). Cortical neural progenitors change their competency over time, giving rise to distinct types of progenitors or neurons dependent on developmental stage (Alvarez-Buylla et al., 2001; Fishell and Kriegstein, 2003; Gotz and Huttner, 2005; Miller and Gauthier, 2007). Before the onset of neurogenesis the early primary neural progenitor cells, neuroepithelial cells, expand the cortex via symmetric divisions to exponentially generate more progenitors. Around the onset of neurogenesis, the neuroepithelial cells differentiate into radial glia cells (RGCs), which are elongated progenitor cells. RGCs mainly divide asymmetrically to maintain the neural progenitor population while also producing daughter cells that will become either neurons or more restricted progenitors, the basal progenitors (or intermediate progenitors) (Gotz and Huttner, 2005; Kriegstein et al., 2006; Miyata et al., 2010; Noctor et al., 2008).
The precise regulation of cortical progenitor number is critical for determining the number of cortical neurons and cortical size. However, the molecular mechanisms underlying the determination of cortical size are largely unknown. The Notch signaling pathway was shown to play an important role in maintaining cortical neural progenitors in the proliferative state. The deletion of Hes1 and Hes5, two downstream effectors for Notch signaling pathway, leads to premature neurogenesis (Hatakeyama and Kageyama, 2006; Imayoshi et al., 2008; Ishibashi et al., 1995; Kageyama et al., 2008a; Kageyama et al., 2008b; Ross et al., 2003) and ectopic overexpression of Hes genes prevents neurogenesis (Nakamura et al., 2000; Ohtsuka et al., 1999; Ohtsuka et al., 2001). Thus, Hes proteins sustain progenitors in a proliferative state and inhibit differentiation.
We studied the role for LHX2, a LIM-homeodomain transcription factor, in the regulation of neocortical progenitor proliferation. Within the neocortex, Lhx2 is expressed throughout cortical neurogenesis by neocortical progenitors within the ventricular zone (VZ) of the dorsal telencephalon (dTel). It was previously shown that LHX2 is required for determination of cortical cell fate, as Lhx2 constitutive null mutants display an extremely diminished dTel with an expansion of the cortical hem (Bulchand et al., 2001; Porter et al., 1997). To study the function of Lhx2 in cortical progenitors after the cortex is determined, we generated an Lhx2 floxed line with LoxP sites flanking exons 1 to 3 of Lhx2. We reported previously that when Lhx2 is eliminated in dTel by Emx1-Cre at E10.5, the cortical hem does not expand and the neocortex forms (Chou et al., 2009; Mangale et al., 2008). With this conditional deletion at E10.5, the neocortex is disorganized and significantly smaller than WT neocortex, and the fate of progenitors of lateral neocortex is altered and they instead generate an ectopic olfactory cortex (Chou et al., 2009). Although in both the Lhx2 constitutive and Emx1-Cre conditional knockout models the size of the cortex is dramatically reduced, the altered fate of cortical progenitors in both models makes it difficult to determine the role of Lhx2 in regulation of cortical size.”
We found when Lhx2 is deleted by Nestin-Cre at E11.5, the fate of cortical progenitors is not changed (Chou et al., 2009). Using this conditional mutant, we demonstrate here that Lhx2 is required for maintaining cortical progenitors in the proliferative state. When Lhx2 is deleted by Nestin-Cre, the neocortex is reduced to less than 50% of the wild type (WT) size. The decreased cortical size is due to dTel neural progenitors exiting the cell cycle and generating neurons prematurely. We find that the deletion of Lhx2 results in a down-regulation of Hes1 expression, which likely contributes to the premature neurogenesis phenotype. Therefore, we have identified a novel molecular mechanism for maintaining cortical progenitors.
Results
Lhx2 controls cortical size
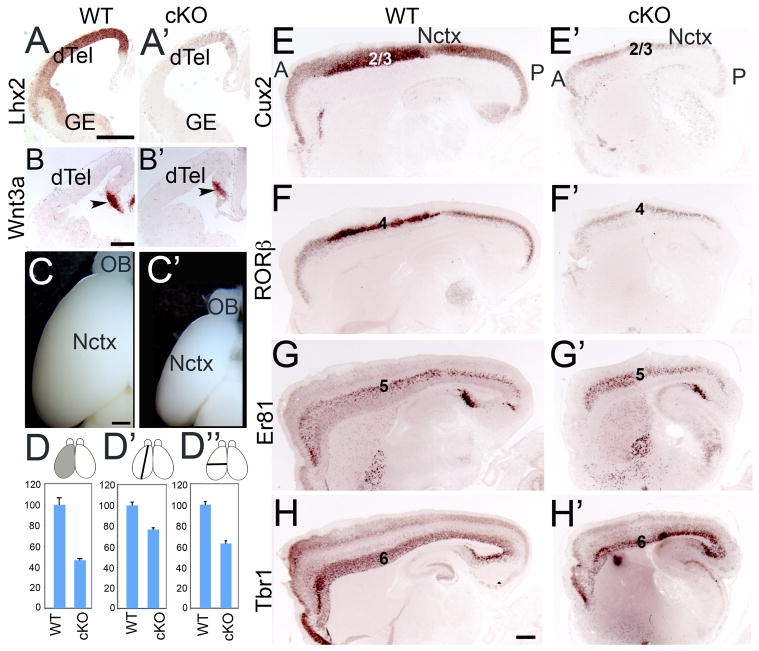
In the Lhx2 constitutive knockout animals, the cortical hem is expanded at the expense of cortical progenitors in the dTel VZ (Bulchand et al., 2001). The lack of neocortical progenitors in the Lhx2 null embryos makes it impossible to study the function of Lhx2 in these progenitors in dTel VZ. We therefore generated mice with an Lhx2 floxed allele (Lhx2 f/f) to enable deletion of Lhx2 after neocortical progenitors in dTel are specified (Chou et al., 2009). In this study, we crossed Lhx2 f/f mice with Nestin-Cre to delete Lhx2 in all neural progenitors (Tronche et al., 1999). In these Lhx2 cKO (Lhx2 f/f; Nestin-Cre), Lhx2 is deleted in dTel from E11.5 (Figure 1A, A′ and Chou et al., 2009). The cortical hem, marked by the expression of Wnt3a, is not expanded in the Lhx2 cKO (Figure 1B, B′). At P7, the size of the cortex in the Lhx2 cKO is significantly reduced to less than 50% of the WT size (Figure 1C, C′ and D, n=4; P<0.001), and both the length (Figure 1D′, n=4; P<0.005) and the width (Figure 1D″, n=4; P<0.001) of the cKO cortex are significantly decreased.
Figure 1. Conditional deletion of Lhx2 from progenitors at onset of neurogenesis with Nestin-Cre leads to a significantly smaller cortex.
(A) In situ hybridization with Lhx2 probe on coronal sections of E11.5 WT (A, Lhx2f/+) and Lhx2 conditional knockout (cKO) (A′, Lhx2f/f, Nestin-Cre) brains. Expression of Lhx2 is diminished in the ventricular zone (VZ) of telencephalon in the cKO (B) In situ hybridization with Wnt3a probe on coronal sections of E13.5 WT and cKO brains. Wnt3a labels cortical hem (arrowheads). The Wnt3a expression domain is similar between WT and cKO. (C) Dorsal view of P7 WT and cKO brains. (D) Histogram of relative surface area of the cerebral cortical hemisphere. The mean of the surface area of the WT cortex is set as 100. Compared with WT (mean ± SEM of 100±6.3, n=4), the surface area of the cKO cortex (46.4 ±1.4, n=4) is significantly decreased (P< 0.001 by unpaired Student’s t test). (D′) Histogram of relative length of the neocortex (Nctx; length from the rostral pole to the occipital pole). The mean of the length of WT neocortex is set as 100. Compared with WT (100±2.9, n=4), the length of the cKO neocortex (76.6±1.5, n=4) is significantly decreased (P<0.001). (D″) Histogram of relative width of the neocortex (length from the midline to the lateral side). The mean of the width of WT neocortex is set as 100. Compared with WT (100±2.6, n=4), the width of the cKO neocortex (62.6±2.2, n=4) is significantly decreased (P<0.001). (E–H) In situ hybridization with Cux2 (E, E′), RORβ (F. F′), Er81 (G, G′) and Tbr1 (H, H′) probes on saggital sections of P7 WT and Lhx2 cKO brains. The expression of RORb and Cux2 is greatly reduced in Lhx2 cKO brains. Scale bars: 0.2 mm. A, anterior; dTel, dorsal telencephalon; GE, ganglionic eminence; OB, olfactory bulb; P, posterior.
To determine if the six layers of neurons are formed properly in the small cortex of Lhx2 cKO, we performed a molecular analysis using layer-specific markers: Cux2, RORβ, Er81 and Tbr1 for layers 2/3, 4, 5 and 6 respectively (Hevner et al., 2003; Hevner et al., 2001; Nakagawa and O’Leary, 2003; Zimmer et al., 2004). The early born neurons, including theTbr1 expressing layer 6 and Er81 expressing layer 5 neurons, are slightly reduced in layer thickness in Lhx2 cKO (Figure 1G, G′, H, H′). However, the late born neurons, including the RORβ expressing layer 4 and Cux2 expressing layer 2/3 neurons are dramatically diminished in both their expression levels and the number of expressing neurons in Lhx2 cKO (Figure 1E, E′, F, F′).
These findings suggest that Lhx2 is a critical transcription factor regulating the size of cortex. Deleting Lhx2 using Nestin-Cre leads to a mutant cortex that is significantly smaller than the WT cortex. Yet, cortical neurons are generated in this Lhx2 cKO, unlike in Lhx2 null brains, though the number of superficial, late born neurons is considerably diminished.
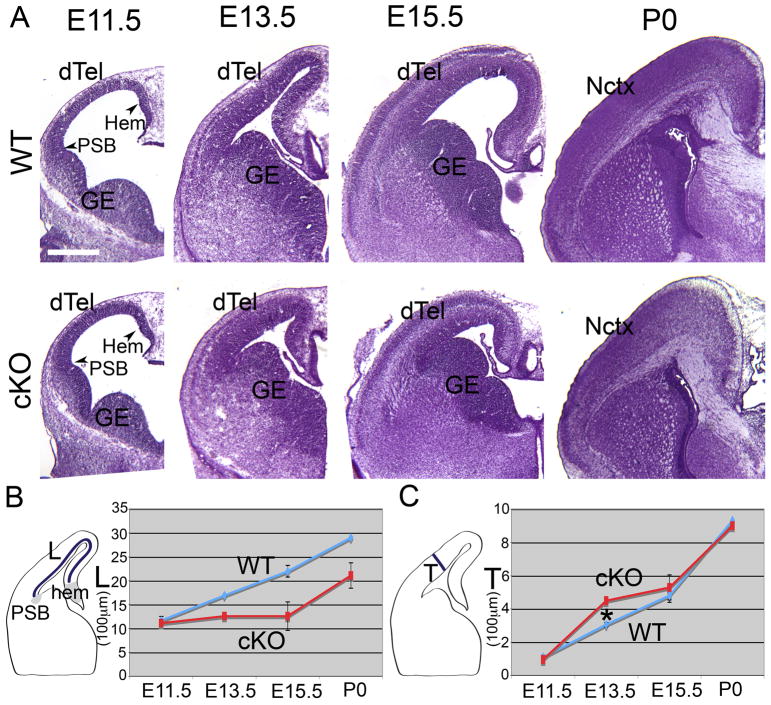
To further investigate the role of Lhx2 in controlling cortical size, we examined when during development the difference of cortical size is detectable. We find that the cKO cortex is notably smaller than the WT cortex from E13.5 (Figure 2). On Nissl stained coronal sections of WT and cKO brains from E11.5 to P0, we measured the size of the cortex by determining the length along the dTel VZ from hem to the pallium-subpallium boundary (PSB) (L, as indicated in Figure 2B) and the thickness of the cortex by measuring the length across dTel from the ventricle to pia (T, as indicated in Figure 2C). At E11.5, when Lhx2 is just deleted in the cKO dTel (Chou et al., 2009), both the size and the thickness of the cortex are indistinguishable between WT and cKO cortex (Figure 2). Starting from E13.5, the tangential extent of cKO cortex is significantly smaller than WT (Figure 2B, n=3, p<0.05). Remarkably, at E13.5 the cKO cortex is transiently thicker than WT (* in Figure 2C, n=3, p<0.001). By P7, as shown in Figure 1D–G, the WT cortex is thicker than the cKO cortex. The progression of these differences in cortical size and thickness suggest that the deletion of Lhx2 leads to premature neurogenesis and a concomitant depletion of neural progenitors.
Figure 2. Changes over embryonic corticogenesis in tangential extent and radial thickness of cortex following conditional deletion of Lhx2 from progenitors.
(A) Nissl stained coronal sections of E11.5, E13.5, E15.5 and P0 WT (Lhx2f/+) and Lhx2 cKO (A′, Lhx2f/f, Nestin-Cre) brains. (B) Histogram of size of the dTel. The size of the dTel is measured as L, the length between the cortical hem and the PSB. Compared with WT, the size of the cKO dTel is smaller from E13.5. (C) Histogram of thickness of dTel. The thickness of the dTel is measured as the length between the ventricle and the pia surface. Compared with WT, the thickness of the cKO dTel is transiently thicker at E13.5. Scale bars: 0.2 mm. A, anterior; dTel, dorsal telencephalon; GE, ganglionic eminence; OB, olfactory bulb; P, posterior; PSB, pallium-subpallium boundary.
Depletion of cortical progenitors in Lhx2 cKO
To investigate the reason for the decrease in cortex size in the Lhx2 cKO we analyzed cell cycle kinetics in the dTel VZ. We used BrdU to label dividing progenitors at E13.5 and analyzed the number of cells labeled in the cortical VZ one hour after BrdU exposure (Figure 3A and data not shown). Counts of BrdU labeled cells showed a significant 30% decrease in the number of dividing progenitors in cKO mice compared to their WT littermates at E13.5 (Figure 3A, A′, A″; n=9 for WT and n=6 for cKO, P<0.001). To confirm the loss of progenitors in the Lhx2 cKO cortex, we performed immunostaining to label mitotic cells with the phosphorylated histone 3 (pH3) antibody at E13.5 (Figure 3B, B′). We counted both the pH3 positive cells along the ventricle (the apical pH3 positive progenitors, A) as well as the ones located in the subventricular zone (the basal pH3 positive progenitors, B). We found that the apical pH3 positive progenitor cells were significantly diminished in the Lhx2 cKO at E13.5 (Figure 3B, B′, B″; n=9 for WT and n=6 for cKO, P<0.01) while the number of basal located pH3 positive progenitors was not significantly different between WT and cKO (Figure 3B″).
Figure 3. The numbers of actively proliferating progenitors are decreased in Lhx2 cKO.
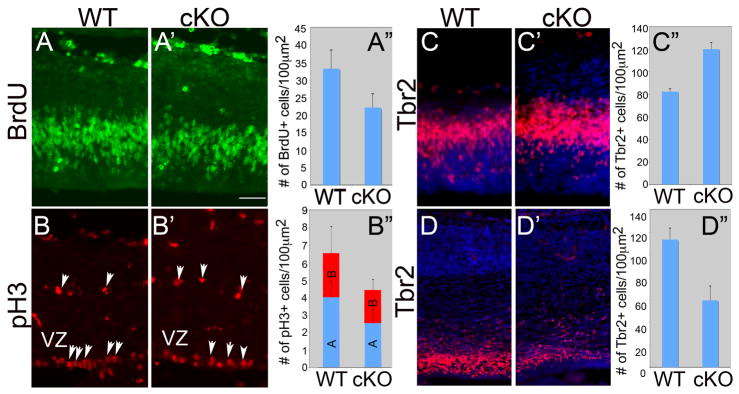
(A) Coronal sections of WT (A, Lhx2fl/+) and cKO (A′ Lhx2fl/f; Nestin-Cre) at E13.5 immunostained with anti-BrdU antibody. BrdU was administered at E13.5, 1hr before the brains were collected. (A″) Histogram of number of BrdU positive cells in 100-μm-wide sampling boxes of dTel. Compared with WT (33.3±5.2, n=9), the number of BrdU positive cells in the cKO (21.2±3.9, n=6) is significantly decreased (P<0.001). (B) Coronal sections of WT (B) and cKO (B′) at E13.5 immunostained with anti-phospho Histone3 (pH3) antibody. The two populations of pH3 positive cells, including that located along the ventricle (apical, A) and that in the subventricular zone (SVZ) (basal, B) are counted separately. (B″) Histogram of number of pH3 positive cells in 100-μm-wide sampling boxes of dTel. Compared with WT (4.2±0.9, n=9), the number of apical pH3 positive cells in the cKO (2.7±0.8, n=6) is significantly decreased (P<0.01). The basally located pH3 positive cells are not significantly different between WT and cKO (WT, 2.6±1.6; cKO, 2.0±0.6). (C) Coronal sections of WT (C) and cKO (C′) at E13.5 immunostained with anti-Tbr2 antibody (C″) Histogram of number of Tbr2 positive cells in 100-μm-wide sampling boxes of dTel. Compared with WT (82.3±2.5, n=3), the number of Tbr2 positive cells in the cKO (120.0±6.1, n=3) is significantly increased (P<0.001). (D) Coronal sections of WT (D) and cKO (D′) at E15.5 immunostained with anti-Tbr2 antibody (D″) Histogram of number of Tbr2 positive cells in 100 μm2 of dTel. Compared with WT (113.5±9.9, n=4), the number of BrdU positive cells in the cKO (59.6±12.4, n=5) is significantly decreased (P<0.001). Scale bar: 200 μm.
Basal progenitors reside in the SVZ, express Tbr2 exclusively, and are secondary cortical progenitors; they are exclusively neurogenic, and have limited self-renewal capacity (Englund et al., 2005; Haubensak et al., 2004; Miyata et al., 2004; Noctor et al., 2004). The number of Tbr2 expressing cells is similar between cKO and WT at E11.5 (data not shown), but is increased significantly in the cKO at E13.5 (Figure 3C, C′ and C″, n=3 for WT and cKO, P<0.001). However, the number of Tbr2 positive cells in the cKO is decreased significantly by E15.5 (Figure 3D, D′, D″, n=4 for WT; n=5 for cKO, P<0.001).
Furthermore, we used the TUNEL assay to observe apoptotic cell death at E11.5, E13.5 and E15.5 and find no evidence of increased cell death in the Lhx2 cKO dTel (data not shown). In summary, we observe a significantly decreased number of proliferating progenitors in the dTel of Lhx2 cKO. Moreover, we find that the deletion of Lhx2 leads to more neurogenic basal progenitors at E13.5. However, the depletion of neural progenitors leads to a decreased number of basal progenitors at E15.5. These results suggest that the deletion of Lhx2 leads to a premature differentiation of neural progenitors.
Premature neuronal differentiation in Lhx2 cKO
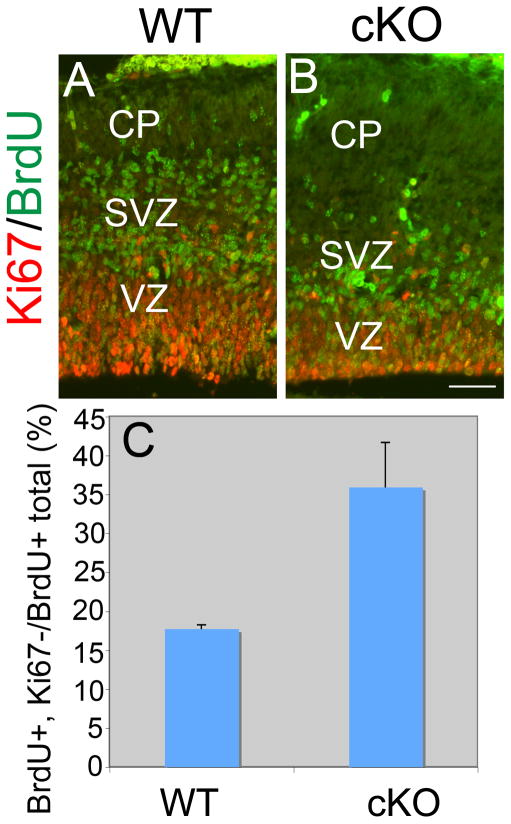
To address whether the premature thickening of the Lhx2 cKO dTel and loss of proliferating progenitors in Lhx2 cKO are due to changes in the mode of cell differentiation, we examined the ratio of cells that exit or reenter the cell cycle during neurogenesis. We labeled proliferating cells by a pulse of BrdU at E13.5 and collected the embryos 24 hr later at E14.5. We used antibodies against BrdU and Ki67 to mark cells proliferating at E13.5 and actively proliferating cells at E14.5, respectively. Cells that are labeled with BrdU but are not Ki67 positive have exited the cell cycle over the previous 24 hr period whereas those that are double labeled with BrdU and Ki67 have re-entered the cell cycle. In WT, fewer than 20% of the BrdU positive cells exit the cell cycle and are Ki67 negative. In the cKO, about 35% of the BrdU positive cells are Ki67 negative, a significant increase in the number of cortical progenitors exiting the cell cycle (Figure 4; n=3 for WT and n=5 for cKO, p<0.001).
Figure 4. Aberrant early exit of cells from cell cycle in dTel of Lhx2 cKO.
(A) Coronal sections of WT (A) and cKO (A′) at E14.5 immunostained with anti-BrdU and anti-Ki67 antibodies. BrdU was administered at E13.5, 24hr before the brains were collected. The cells re-entering cell cycle are positive for both BrdU and Ki67, while the cells exiting cell cycle are positive for BrdU but negative for Ki67. (A″) Histogram of the ratio of number of BrdU positive but Ki67 negative cells and total BrdU positive cells in 100-μm-wide sampling boxes of dTel. Compared with WT (0.18±0.01, n=3), the number of BrdU positive cells in the cKO (0.37±0.06, n=5) is significantly increased (P<0.001). Scare bar: 200um.
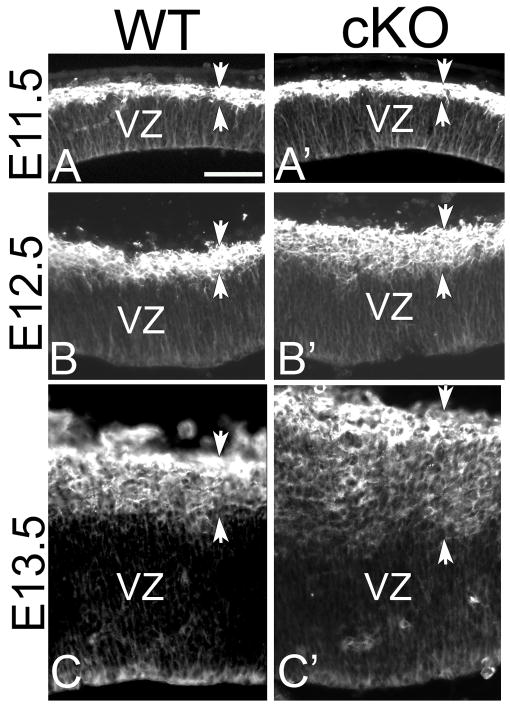
We further examined the expression of the neuronal marker TuJ1, which labels the neurons forming the cortical plate, in WT and cKO brains from E11.5 to E13.5. At E11.5, when early born neurons start to be formed outside the VZ, the TuJ1 positive region is similar in the WT and the cKO (Figure 5A, A′). The TuJ1 positive developing cortical plate is thicker in the cKO compared to WT by E12.5 (Figure 5B, B′), and this increase is more pronounced at E13.5 (Figure 5C, C′). These results confirm that the lack of Lhx2 results in a premature neurogenesis, leading to an overproduction of cortical neurons at the expense of progenitor expansion. Therefore, Lhx2 is required for the maintenance of neural progenitors in their proliferative state.
Figure 5. Precocious neurogenesis in cortex of Lhx2 cKO.
Coronal sections of WT (A, B, C) and cKO (A′, B′, C′) at E11.5 (A, A′), E12.5 (B, B′) and E13.5 (C, C′) immunostained with anti-TuJ1 antibody. TuJ1 labels differentiated neurons. At E11.5, the WT and cKO have similar level of TuJ1 positive neurons. At E12.5, and more dramatically at E13.5, more TuJ1 neurons are generated in the cKO dTel when compared with WT. VZ, ventricular zone. Scare bar: 200 um.
Lhx2 regulates the expression of Hes1
To understand the molecular mechanisms for Lhx2 to regulate cortical neurogenesis, we use a candidate approach to search for Lhx2 downstream factors. Apterous, the Lhx2 ortholog in Drosophila, was shown to interact with components in the Notch signaling pathway in wing disc (Milan and Cohen, 2000). Therefore, we examined the expression of Hes genes in Lhx2 cKO by in situ hybridization as they were shown to be Notch downstream factors critical for regulating neuron differentiation (Corbin et al., 2008; Kageyama et al., 2009).
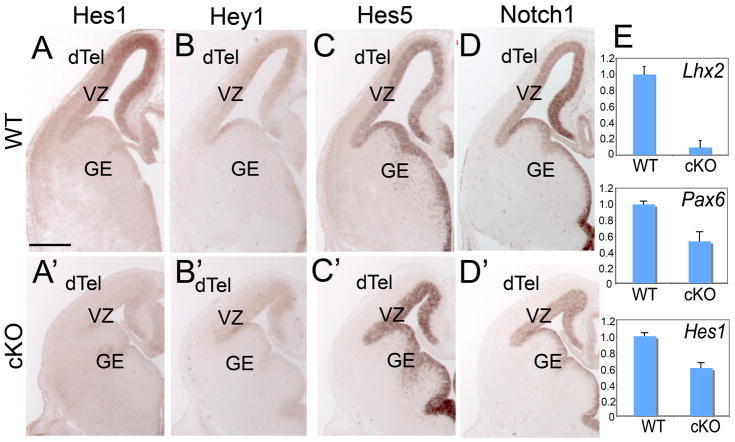
We found that the expression of both Hes1 and Hey1 was dramatically down-regulated in the forebrain of Lhx2 cKO at E13.5 (Figure 6A, A′ and 6B, B′), while the expression of Hes5 did not show a noticeable difference (Figure 6C, C′). Notch1 is expressed in WT dTel VZ in a high-medial to low-lateral gradient (Figure 6D). The expression level of Notch1 in the medial cortex in the cKO was also decreased, leading to a flattened Notch1 expression across the dTel VZ (Figue 6D′).
Figure 6. Expression of components of Notch signaling pathway is differentially affected in dTel ventricular zone by Lhx2 deletion from progenitors.
(A–D) In situ hybridization with Hes1, Hey1, Notch1 and Hes5 probes on coronal sections of E13.5 WT (A–D) and cKO (A′–D′) brains. In WT dTel, Hes1, Hey1, Notch1 and Hes5 are expressed in the VZ. The expression of Hes1 and Hey1 is decreased in cKO while the expression of Hes5 and Notch1 is maintained, although the medial-high to lateral-low expression of Notch1 appears flattened. (E) Quantitative PCR with Actin as internal control; the expression level of Lhx2, Pax6 and Hes1 is significantly decreased in cKO dTel relative to WT (n=3; p<0.001).
The relative Hes1 expression level in E13.5 WT and Lhx2 cKO dTel was further examined by quantitative PCR, using cyclophilin as well as mAct as internal controls. We confirmed that the expression of both Lhx2 and Pax6 is significantly down-regulated in Lhx2 cKO (Figure 6E; n=4; p<0.001) as we have shown previously (Chou et al., 2009). We found that the expression of Hes1 is also significantly down-regulated in the Lhx2 cKO (n=4; p<0.001).
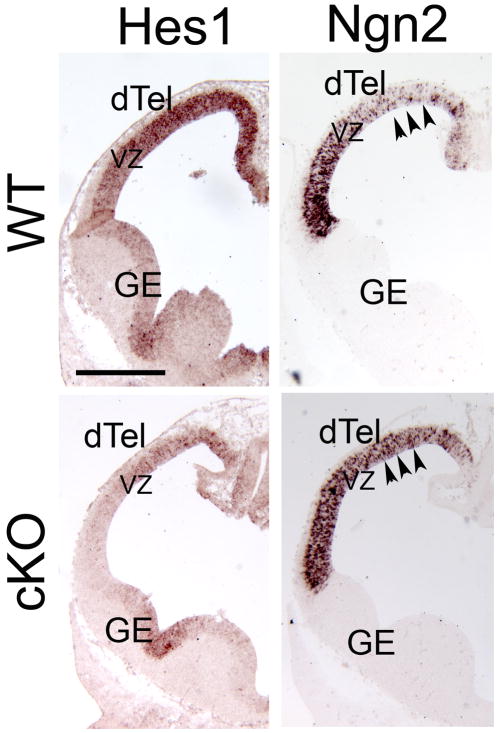
The down-regulation of Hes1 has been shown to accelerate neurogenesis by up-regulating proneural genes, such as Ngn2 (Hatakeyama and Kageyama, 2006; Ishibashi et al., 1995). We further investigated the expression of Ngn2 in Lhx2 cKO. Ngn2 is expressed in the VZ of dTel in a high-lateral to low-medial graded fashion, complementary to the expression gradient of Lhx2 and Hes1. We find at E11.5, when Lhx2 is deleted in the cKO dTel, Hes1 expression is down-regulated and the expression of Ngn2 in dTel of the cKO is up-regulated, especially in the medial dTel, resulting in a flattening of the Ngn2 expression gradient across the dTel VZ (Figure 7). This up-regulation of Ngn2 expression is consistent with the premature neurogenesis phenotype we observe in Lhx2 cKO.
Figure 7. Ngn2 expression is up-regulated in a pattern that parallels down-regulation of Hes1 expression in dTel of Lhx2 cKO.
In situ hybridization with Hes1 and Ngn2 probes on coronal sections of E12.5 WT and cKO brains. The expression of Hes1 is substantially down-regulated in the cKO brains compared to WT but maintains its high medial to low lateral graded expression, while the expression of Ngn2 is up-regulated in the cKO brains, being especially elevated more medially in mutant dTel compared to WT (arrowheads), leading to an overall more flattened expression relative to the high lateral to low medial expression observed in WT. dTel, dorsal telencephalon; GE, ganglionic eminence; VZ, ventricular zone. Scale bar: 200 um.
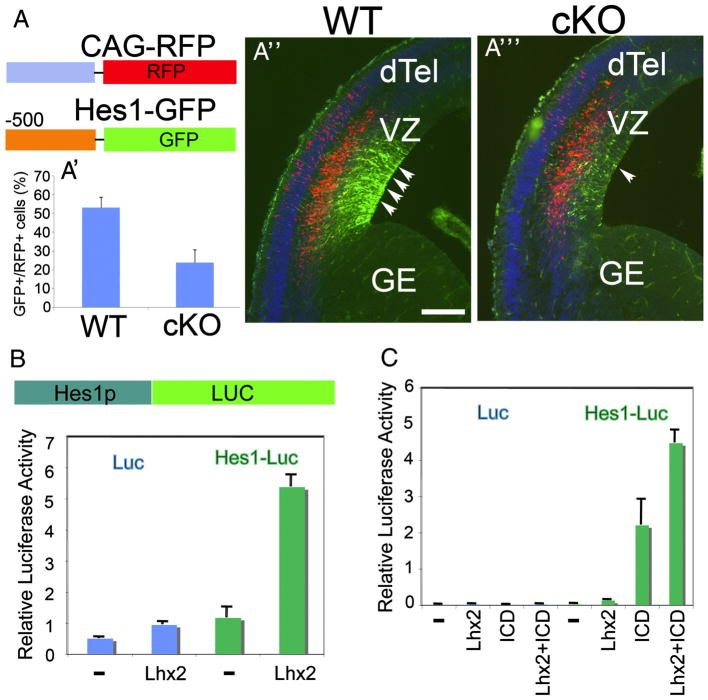
To further investigate if Lhx2 regulates transcription activity of the Hes1 gene, we first examined Hes1 promoter activity in Lhx2 cKO dTel. Using in utero electroporation, we transfected into E12.5 WT and cKO brains a GFP reporter construct driven by a 500-bp Hes1 promoter (Jarriault et al., 1995) and a RFP reporter construct driven by a constitutively active CAG promoter (Figure 8A). We visualized expression of GFP and RFP in the transfected brains analyzed 24 hrs after electroporation at E13.5. In both WT and cKO, strong RFP expressing cells are detected, while the GFP expression is prominent in the VZ in WT but sparse in the cKO (Figure 8A″, A‴, n=5 for WT and n=3 for cKO). We observed a significant decrease of the ratio of GFP positive cells (Hes1-expressing cells) and the RFP positive cells (all transfected cells) in cKO (Figure 8A′, p<0.001). This suggests that Lhx2 is an upstream regulator to promote Hes1 gene expression in the developing dTel.
Figure 8. Lhx2 directly regulates Hes1 promoter activity.
(A) Reporter constructs, CAG-RFP and Hes1-GFP, are cotransfected into developing dTel at E12.5 by in utero electroporation. The expression of RFP and GFP are visualized by E13.5. The ratio of GFP positive (Hes1 expressing) cells to RFP positive (transfected) cells is significantly decreased in cKO (A′; WT, 53.0±5.4%, cKO, 23.9±6.6%, P<0.001). When the numbers of cells expressing RFP are similar between WT and cKO, many GFP labeled cells are detected in WT dTel VZ (A″), while very few GFP positive cells are detected in cKO dTel (A‴). (B) Luciferase activity was measured 48hrs after reporter constructs transfected to N2A cells. The co-transfection of Lhx2 expression vector increases the Hes1 promoter activity. (C) The intracellular domain of Notch1 (ICD) activates Hes1 promoter activity and the co-transfection of Lhx2 and ICD can further activate Hes1 promoter activity.
In N2A cells, a neuroblastoma cell line, we confirmed the role of Lhx2 in regulating Hes1 promoter activity. We find Lhx2 can increase Hes1 promoter activity about 5 fold (Figure 8B). The intracellular domain (ICD) of Notch1 was shown to be able to activate the Hes1 promoter (Jarriault et al., 1995). We find that when Lhx2 is co-transfected with Notch1-ICD, the activity of the Hes1 promoter can be further activated (Figure 8C), suggesting that Lhx2 can cooperate with the Notch signaling pathway in regulating Hes1 expression. Therefore, we conclude that Lhx2 is an important regulator for Hes1 expression.
Discussion
In this study, we show that Lhx2 is critical for the determination of the size of cortical progenitor pools and number of cortical neurons, thereby ultimately regulating cortical size. Lhx2 belongs to the LIM homeodomain (LIM-HD) transcription factor (TF) family. It is known that LIM-HD TFs play important roles in many aspects of development, such as interneuron and motor neuron specification in spinal cord (reviewed by (Hunter and Rhodes, 2005)).
Lhx2 has been shown to play important roles in specifying neocortical progenitors. Lhx2 is strongly expressed in the neocortical progenitors in dTel VZ during corticogenesis. To determine the function of Lhx2 in these progenitors, we previously generated an Lhx2 floxed line with LoxP sites flanking exons 1 to 3 of Lhx2 and crossed it with several Cre-lines to conditionally delete Lhx2 from cortical cells at various developmental stages (Chou et al., 2009). We found that Lhx2 has distinct, stage-specific critical roles in corticogenesis throughout embryonic development. In contrast to the constitutive null, where the cortex is not formed due to the expansion of the cortical hem at the expense of cortical progenitors (Bulchand et al., 2001; Porter et al., 1997), when Lhx2 is eliminated in dTel by Emx1-Cre at E10.5, the cortical hem does not expand and the neocortex forms (Chou et al., 2009; Mangale et al., 2008). With this conditional deletion at E10.5, the neocortex is disorganized and significantly smaller than WT neocortex; moreover, the fate of lateral neocortex progenitors is altered, and they instead generate an ectopic olfactory cortex. However, Lhx2 deletion one day later, at E11.5 by Nestin-Cre, does not result in a re-fating of lateral neocortical progenitors, but results in a neocortex that is significantly smaller than WT (Chou et al., 2009). Therefore, we concluded that Lhx2 plays a crucial role in specifying neocortical progenitors in a narrow time window around E10.5.
In this study, we focused on the roles of Lhx2 in the neocortical progenitors after their neocortical fate is specified; we studied the defects in the mutant brains where Lhx2 is deleted by Nestin-Cre. In the dTel in the Nestin-Cre cKO, we find that the deletion of Lhx2 leads to decreased progenitor proliferation and increased neurogenesis and therefore, results in a dramatically smaller cortex. Due to the progressive diminishment of cortical progenitors, the later born cortical neurons in the layer 2/3 and 4 are more affected in Lhx2 cKO brains. We further demonstrate that Lhx2 regulates the proliferation of cortical progenitors, at least partly, through the regulation of Hes1, an important regulator for neural progenitors. Thus, we have identified a novel role for Lhx2 as a key regulator for the balance between proliferation and differentiation of neocortical progenitors.
Regulation of neurogenesis through Hes1
Hes1 is one of the Hes genes, mammalian homologues of Drosophila Hairy and Enhancer of Split. Hes genes are repressor-type basic helix-loop-helix (bHLH) genes and negatively regulate neurogenesis by antagonizing proneural genes such as the achaete-scute complex (Akazawa et al., 1992; Sasai et al., 1992). In mouse neural progenitor cells, the expression of the Hes1 oscillates and is required for maintenance of neural progenitors in the embryonic cortex (Kageyama et al., 2008a). Inactivation of Hes1 up-regulates expression of proneural genes, such as Ngn2, and accelerates neuronal differentiation (Hatakeyama and Kageyama, 2006; Imayoshi et al., 2008; Ishibashi et al., 1995; Kageyama et al., 2008a; Kageyama et al., 2008b; Ross et al., 2003), while misexpression of Hes1 inhibits neuronal differentiation (Ohtsuka et al., 2001). The down-regulation of Hes1 expression in the Lhx2 cKO explains the premature neurogenesis phenotype. However, in Hes1 mutants, the neurogenesis phenotype is relatively minor (Hatakeyama et al., 2004). Therefore, Lhx2 may regulate additional factors that are involved in cortical progenitor maintenance.
We find Hes1 expression is down-regulated in the Lhx2 cKO, but the expression of Hes5 is not changed. Previous studies have suggested that the expression of Hes1 and Hes5 might be differently regulated. Although Notch signaling regulates both Hes1 and Hes5 expression, the initial expression of Hes1 occurs in neuroepithelial cells in dTel before Notch signaling components are expressed, while Hes5 expression starts later and occurs together with the expression of Notch signaling components (Hatakeyama et al., 2004). This indicates that the initiation of Hes1 expression is independent of Notch signaling and the initiation of Hes5 expression may be dependent on Notch signaling. Lhx2 is expressed in the dTel neuroepithelial cells very early on (Nakagawa et al., 1999; Rincon-Limas et al., 1999). Our results suggest that Lhx2 is likely to regulate the pattern of Hes1 expression, which is different from Hes5.
Though it is well established that the expression of Hes1 is critical for neural progenitor proliferation, the mechanism by which the expression of Hes1 is regulated is still unclear. Recent studies have indicated that Gli2 and Nfia regulates Hes1 activity within neural progenitors (Piper et al., 2011; Wall et al., 2009), Hes1 also negatively regulates its own gene expression (Takebayashi et al., 1994) and the inhibitor of differentiation (Id) genes release Hes1 from autoregulatory inhibition (Bai et al., 2007). Moreover, it has been shown that the expression of Hes1 can be regulated by signaling pathways other than Notch. For example, the expression of Hes1 gene can be up-regulated by FGFs (Ogata et al., 2011; Shimizu et al., 2008; Tsai and Kim, 2005), Wnts (Shimizu et al., 2008) and Shh (Dave et al., 2011; Ingram et al., 2008) signaling pathways in neural progenitors.
Recently, it was shown that Lhx2 interacts with the Notch-Nfia signaling pathway to regulate astrogliogenesis in the developing hippocampus (Subramanian et al., 2011). Nfia is not expressed in the dTel VZ during early corticogenesis when we observe the phenotype in cKO cortex at E13.5 (Plachez et al., 2008). Therefore, in this study, we present a novel mechanism, which is independent of Nfia, for Lhx2 to regulate Hes1 expression and the Notch signaling pathway.
Multiple roles of Lhx2
We demonstrate an important role for Lhx2 in cortical neurogenesis. We find that Lhx2 is required for the maintenance of Hes1 expression. The diminished Hes1 expression in Lhx2 cKO dTel may contribute to the premature neurogenesis and the depletion of neural progenitors, leading to a dramatically smaller cortex.
Lhx2 is expressed in a graded fashion in dTel VZ, high medial to low lateral (Nakagawa et al., 1999). Hes1 and Notch1 expression show a similar high-medial to low-lateral gradient as Lhx2, while Ngn2 has a complimentary expression gradient as Lhx2, with high-lateral to low-medial. In the lateral dTel, with lower Lhx2 expression and higher Ngn2 expression among the progenitors in dTel VZ, neurogenesis starts early, while in the medial dTel VZ, where higher Lhx2 and lower Ngn2 are expressed, neurogenesis starts late. When Lhx2 is deleted, we find Hes1 is down-regulated, and the down-regulation of Hes1 in Lhx2 cKO is associated with flattened expression patterns of Ngn2 and Notch1. These results suggest that the graded expression pattern of Lhx2 may be an important factor for regulating the timing of neurogenesis among dTel progenitors.
Our and other studies have shown that Lhx2 regulates multiple events during cortical development. First, Lhx2 regulates the fate choice between neocortical and cortical hem fate (Bulchand et al., 2001); later, Lhx2 regulates the fate between lateral neocortex and piriform cortex (Chou et al., 2009). Here, we demonstrate that Lhx2 further regulates the proliferation and differentiation of neural progenitors. Much remains to be understood about the biochemical mechanisms that underlie these different Lhx2 actions. LIM-HD proteins have been demonstrated to recognize AT-rich DNA sequences, and promote (or repress) the expression of downstream genes. Interactions with partner proteins may modulate transcriptional activity of LIM-HD proteins (Matthews and Visvader, 2003). Furthermore, the post-translational modification of LIM-HD proteins or their interacting proteins may further influence their transcriptional activity or their subcellular locations (Ma et al., 2008; Petit et al., 2003). To achieve its diverse functions during cortical development, Lhx2 may regulate a variety of downstream genes, in part by interacting with different cofactors specific to each role for Lhx2 in corticogenesis.
Experimental methods
Animals
Lhx2 f/f, Nestin-Cre and Lhx2 f/+, Nestin-Cre mice are maintained in C56BL6 background. The day of insemination and the day of birth are designated as embryonic day 0.5 (E0.5) and postnatal day 0 (P0). All control animals used in our experiments were the littermates of the corresponding mutant mice. Mice were used in accordance with institution guidelines.
In Situ Hybridization
In situ hybridization was done using digoxigenin (DIG)-labeled riboprobes for Cux1, Er81, RORβ, and Tbr1, on whole brains or 20 μm cryostat sections as described previously. In situ hybridization was done as described previously.
Plasmids
The 500-bp Hes1 promoter was generated by PCR with C56BL6 genomic DNA. The Hes1 promoter was cloned into pGL3basic (Promega), a GFP reporter construct. Lhx2 cDNA and ICD of Notch1 were cloned into the CAG expression vector.
Quantitative PCR
Total RNA from dTel of E13.5 WT and cKO brains was extracted using Trizol (Invitrogen). Reverse transcription was performed using Superscript III (Invitrogen). 0.5 μg of total RNA was reverse transcribed with random hexamers. Quantitative PCRs (qPCRs) were performed using the SYBR Green PCR Master Mix (Invitrogen). When quantifying the mRNA expression levels, the housekeeping gene cyclophilin and actin was used as a relative standard. All the samples were tested in triplicate. For all qPCR analyses, RNA from three independent replicates for both wild-type and cKO mice were examined. Statistical analyses were performed using a two-tailed unpaired t test. Error bars represent the standard error of the mean. Primers used in the qPCR analyses:
Actin: 5′-GCCCTGAGGCTCTTTTTCC-3′ and 5′-TGCCACAGGATTCCATACCC-3′; Cyclophilin: 5′-TGTCTTTGGAACTTTGTCTGCAAAT-3′ and 5′-GGCCGATGACGAGCCC-3′; Lhx2: 5′-AAGCTCAACCTGGAGTCGGAA-3′ and 5′-TGAGGTGATAAACCAAGTCCCG-3′; Pax6: 5′-GCCCTTCCATCTTTGCTTGGGAAA-3′ and 5′-TAGCCAGGTTGCGAAGAACTCTGT-3′; Hes1: 5′-CAACACGACACCGGACAAAC-3′ and 5′-TCTTCTCCATGATAGGCTTTGATG-3′
In Utero Electroporation
Pregnant females were anesthetized with isoflurane, their uterine horns were exposed, and the lateral ventricles of embryos were injected through the uterus wall using pulled capillaries filled with DNA (0.75 μg/μl, expression constructs of Hes1-EGFP, and 0.25 μg/μl CAG-RFP) diluted in PBS and colored with 0.5% Fast Green (Sigma-Aldrich). DNA-injected embryos were electroporated using a 35–V/50 ms/950 ms/five pulse program (Nepa Gene). Embryos were collected 24 h after electroporation.
Histochemistry
For immunostaining on sections, brains were fixed in 4% PFA in PBS, cryoprotected in 20% sucrose/PBS, cut at 20 μm, and immunostained as described previously. The following antibodies were used: rat anti-BrdU (Accurate), rabbit anti-Ki67 (Vector), rabbit anti-Tbr2 (Chemicon), rabbit anti-phospho-Histone3 (Millipore) and mouse anti-TuJ1 (Covance). The size and the thickness of cortex were measured by NIH image J software.
Proliferation marker analyses
Pregnant females were sacrificed 60 minutes after injection with 10 μl/g body weight of 10 mg/ml bromodeoxyuridine (BrdU; Sigma). Coronal sections of embryonic brains were immunostained with anti-BrdU, anti-phosphoHistone3 and anti-Tbr2 antibodies. Cells were counted in 100-μm-wide sampling boxes along the ventricular zone of the dorsal telencephalon.
Cell culture and transfection
Neuro2A cells were cotransfected (Fugene; Roche) with the indicated plasmids at 1 μg/μl and incubated for 48 h.
Acknowledgments
We thank Berta Higgins and Haydee Gutierrez for technical assistance. This work was funded by NIH grants R01 NS31558 and R01 MH086147.
Abbreviations
- cKO
conditional knockout
- dTel
dorsal telencephalon
- subventricular zone
SVZ
- VZ
ventricular zone
- WT
wild type
Contributor Information
Shen-Ju Chou, Email: chou@salk.edu.
Dennis D.M. O’Leary, Email: doleary@salk.edu.
References
- Aboitiz F, Morales D, Montiel J. The inverted neurogenetic gradient of the mammalian isocortex: development and evolution. Brain research. 2001;38:129–139. doi: 10.1016/s0006-8993(01)02902-x. [DOI] [PubMed] [Google Scholar]
- Akazawa C, Sasai Y, Nakanishi S, Kageyama R. Molecular characterization of a rat negative regulator with a basic helix-loop-helix structure predominantly expressed in the developing nervous system. The Journal of biological chemistry. 1992;267:21879–21885. [PubMed] [Google Scholar]
- Alvarez-Buylla A, Garcia-Verdugo JM, Tramontin AD. A unified hypothesis on the lineage of neural stem cells. Nat Rev Neurosci. 2001;2:287–293. doi: 10.1038/35067582. [DOI] [PubMed] [Google Scholar]
- Bai G, Sheng N, Xie Z, Bian W, Yokota Y, Benezra R, Kageyama R, Guillemot F, Jing N. Id sustains Hes1 expression to inhibit precocious neurogenesis by releasing negative autoregulation of Hes1. Developmental cell. 2007;13:283–297. doi: 10.1016/j.devcel.2007.05.014. [DOI] [PubMed] [Google Scholar]
- Bulchand S, Grove EA, Porter FD, Tole S. LIM-homeodomain gene Lhx2 regulates the formation of the cortical hem. Mechanisms of development. 2001;100:165–175. doi: 10.1016/s0925-4773(00)00515-3. [DOI] [PubMed] [Google Scholar]
- Chou SJ, Perez-Garcia CG, Kroll TT, O’Leary DD. Lhx2 specifies regional fate in Emx1 lineage of telencephalic progenitors generating cerebral cortex. Nature neuroscience. 2009;12:1381–1389. doi: 10.1038/nn.2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbin JG, Gaiano N, Juliano SL, Poluch S, Stancik E, Haydar TF. Regulation of neural progenitor cell development in the nervous system. Journal of neurochemistry. 2008;106:2272–2287. doi: 10.1111/j.1471-4159.2008.05522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dave RK, Ellis T, Toumpas MC, Robson JP, Julian E, Adolphe C, Bartlett PF, Cooper HM, Reynolds BA, Wainwright BJ. Sonic hedgehog and notch signaling can cooperate to regulate neurogenic divisions of neocortical progenitors. PloS one. 2011;6:e14680. doi: 10.1371/journal.pone.0014680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englund C, Fink A, Lau C, Pham D, Daza RA, Bulfone A, Kowalczyk T, Hevner RF. Pax6, Tbr2, and Tbr1 are expressed sequentially by radial glia, intermediate progenitor cells, and postmitotic neurons in developing neocortex. J Neurosci. 2005;25:247–251. doi: 10.1523/JNEUROSCI.2899-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fish JL, Dehay C, Kennedy H, Huttner WB. Making bigger brains-the evolution of neural-progenitor-cell division. Journal of cell science. 2008;121:2783–2793. doi: 10.1242/jcs.023465. [DOI] [PubMed] [Google Scholar]
- Fishell G, Kriegstein AR. Neurons from radial glia: the consequences of asymmetric inheritance. Current opinion in neurobiology. 2003;13:34–41. doi: 10.1016/s0959-4388(03)00013-8. [DOI] [PubMed] [Google Scholar]
- Gotz M, Huttner WB. The cell biology of neurogenesis. Nature reviews. 2005;6:777–788. doi: 10.1038/nrm1739. [DOI] [PubMed] [Google Scholar]
- Hatakeyama J, Bessho Y, Katoh K, Ookawara S, Fujioka M, Guillemot F, Kageyama R. Hes genes regulate size, shape and histogenesis of the nervous system by control of the timing of neural stem cell differentiation. Development (Cambridge, England) 2004;131:5539–5550. doi: 10.1242/dev.01436. [DOI] [PubMed] [Google Scholar]
- Hatakeyama J, Kageyama R. Notch1 expression is spatiotemporally correlated with neurogenesis and negatively regulated by Notch1-independent Hes genes in the developing nervous system. Cereb Cortex. 2006;16(Suppl 1):i132–137. doi: 10.1093/cercor/bhj166. [DOI] [PubMed] [Google Scholar]
- Haubensak W, Attardo A, Denk W, Huttner WB. Neurons arise in the basal neuroepithelium of the early mammalian telencephalon: a major site of neurogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:3196–3201. doi: 10.1073/pnas.0308600100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hevner RF, Daza RA, Rubenstein JL, Stunnenberg H, Olavarria JF, Englund C. Beyond laminar fate: toward a molecular classification of cortical projection/pyramidal neurons. Developmental neuroscience. 2003;25:139–151. doi: 10.1159/000072263. [DOI] [PubMed] [Google Scholar]
- Hevner RF, Shi L, Justice N, Hsueh Y, Sheng M, Smiga S, Bulfone A, Goffinet AM, Campagnoni AT, Rubenstein JL. Tbr1 regulates differentiation of the preplate and layer 6. Neuron. 2001;29:353–366. doi: 10.1016/s0896-6273(01)00211-2. [DOI] [PubMed] [Google Scholar]
- Hunter CS, Rhodes SJ. LIM-homeodomain genes in mammalian development and human disease. Molecular biology reports. 2005;32:67–77. doi: 10.1007/s11033-004-7657-z. [DOI] [PubMed] [Google Scholar]
- Imayoshi I, Shimogori T, Ohtsuka T, Kageyama R. Hes genes and neurogenin regulate non-neural versus neural fate specification in the dorsal telencephalic midline. Development (Cambridge, England) 2008;135:2531–2541. doi: 10.1242/dev.021535. [DOI] [PubMed] [Google Scholar]
- Ingram WJ, McCue KI, Tran TH, Hallahan AR, Wainwright BJ. Sonic Hedgehog regulates Hes1 through a novel mechanism that is independent of canonical Notch pathway signalling. Oncogene. 2008;27:1489–1500. doi: 10.1038/sj.onc.1210767. [DOI] [PubMed] [Google Scholar]
- Ishibashi M, Ang SL, Shiota K, Nakanishi S, Kageyama R, Guillemot F. Targeted disruption of mammalian hairy and Enhancer of split homolog-1 (HES-1) leads to up-regulation of neural helix-loop-helix factors, premature neurogenesis, and severe neural tube defects. Genes & development. 1995;9:3136–3148. doi: 10.1101/gad.9.24.3136. [DOI] [PubMed] [Google Scholar]
- Jarriault S, Brou C, Logeat F, Schroeter EH, Kopan R, Israel A. Signalling downstream of activated mammalian Notch. Nature. 1995;377:355–358. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- Kageyama R, Ohtsuka T, Kobayashi T. Roles of Hes genes in neural development. Development, growth & differentiation. 2008a;50(Suppl 1):S97–103. doi: 10.1111/j.1440-169X.2008.00993.x. [DOI] [PubMed] [Google Scholar]
- Kageyama R, Ohtsuka T, Shimojo H, Imayoshi I. Dynamic Notch signaling in neural progenitor cells and a revised view of lateral inhibition. Nature neuroscience. 2008b;11:1247–1251. doi: 10.1038/nn.2208. [DOI] [PubMed] [Google Scholar]
- Kageyama R, Ohtsuka T, Shimojo H, Imayoshi I. Dynamic regulation of Notch signaling in neural progenitor cells. Current opinion in cell biology. 2009;21:733–740. doi: 10.1016/j.ceb.2009.08.009. [DOI] [PubMed] [Google Scholar]
- Kriegstein A, Noctor S, Martinez-Cerdeno V. Patterns of neural stem and progenitor cell division may underlie evolutionary cortical expansion. Nat Rev Neurosci. 2006;7:883–890. doi: 10.1038/nrn2008. [DOI] [PubMed] [Google Scholar]
- Ma YC, Song MR, Park JP, Henry Ho HY, Hu L, Kurtev MV, Zieg J, Ma Q, Pfaff SL, Greenberg ME. Regulation of motor neuron specification by phosphorylation of neurogenin 2. Neuron. 2008;58:65–77. doi: 10.1016/j.neuron.2008.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangale VS, Hirokawa KE, Satyaki PR, Gokulchandran N, Chikbire S, Subramanian L, Shetty AS, Martynoga B, Paul J, Mai MV, Li Y, Flanagan LA, Tole S, Monuki ES. Lhx2 selector activity specifies cortical identity and suppresses hippocampal organizer fate. Science (New York, NY. 2008;319:304–309. doi: 10.1126/science.1151695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews JM, Visvader JE. LIM-domain-binding protein 1: a multifunctional cofactor that interacts with diverse proteins. EMBO reports. 2003;4:1132–1137. doi: 10.1038/sj.embor.7400030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milan M, Cohen SM. Temporal regulation of apterous activity during development of the Drosophila wing. Development (Cambridge, England) 2000;127:3069–3078. doi: 10.1242/dev.127.14.3069. [DOI] [PubMed] [Google Scholar]
- Miller FD, Gauthier AS. Timing is everything: making neurons versus glia in the developing cortex. Neuron. 2007;54:357–369. doi: 10.1016/j.neuron.2007.04.019. [DOI] [PubMed] [Google Scholar]
- Miyata T, Kawaguchi A, Saito K, Kawano M, Muto T, Ogawa M. Asymmetric production of surface-dividing and non-surface-dividing cortical progenitor cells. Development (Cambridge, England) 2004;131:3133–3145. doi: 10.1242/dev.01173. [DOI] [PubMed] [Google Scholar]
- Miyata T, Kawaguchi D, Kawaguchi A, Gotoh Y. Mechanisms that regulate the number of neurons during mouse neocortical development. Current opinion in neurobiology. 2010;20:22–28. doi: 10.1016/j.conb.2010.01.001. [DOI] [PubMed] [Google Scholar]
- Nadarajah B, Parnavelas JG. Modes of neuronal migration in the developing cerebral cortex. Nat Rev Neurosci. 2002;3:423–432. doi: 10.1038/nrn845. [DOI] [PubMed] [Google Scholar]
- Nakagawa Y, Johnson JE, O’Leary DD. Graded and areal expression patterns of regulatory genes and cadherins in embryonic neocortex independent of thalamocortical input. The Journal of neuroscience: the official journal of the Society for Neuroscience. 1999;19:10877–10885. doi: 10.1523/JNEUROSCI.19-24-10877.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa Y, O’Leary DD. Dynamic patterned expression of orphan nuclear receptor genes RORalpha and RORbeta in developing mouse forebrain. Developmental neuroscience. 2003;25:234–244. doi: 10.1159/000072271. [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Sakakibara S, Miyata T, Ogawa M, Shimazaki T, Weiss S, Kageyama R, Okano H. The bHLH gene hes1 as a repressor of the neuronal commitment of CNS stem cells. J Neurosci. 2000;20:283–293. doi: 10.1523/JNEUROSCI.20-01-00283.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor SC, Martinez-Cerdeno V, Ivic L, Kriegstein AR. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nature neuroscience. 2004;7:136–144. doi: 10.1038/nn1172. [DOI] [PubMed] [Google Scholar]
- Noctor SC, Martinez-Cerdeno V, Kriegstein AR. Distinct behaviors of neural stem and progenitor cells underlie cortical neurogenesis. The Journal of comparative neurology. 2008;508:28–44. doi: 10.1002/cne.21669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata T, Ueno T, Hoshikawa S, Ito J, Okazaki R, Hayakawa K, Morioka K, Yamamoto S, Nakamura K, Tanaka S, Akai M. Hes1 functions downstream of growth factors to maintain oligodendrocyte lineage cells in the early progenitor stage. Neuroscience. 2011;176:132–141. doi: 10.1016/j.neuroscience.2010.12.015. [DOI] [PubMed] [Google Scholar]
- Ohtsuka T, Ishibashi M, Gradwohl G, Nakanishi S, Guillemot F, Kageyama R. Hes1 and Hes5 as notch effectors in mammalian neuronal differentiation. The EMBO journal. 1999;18:2196–2207. doi: 10.1093/emboj/18.8.2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuka T, Sakamoto M, Guillemot F, Kageyama R. Roles of the basic helix-loop-helix genes Hes1 and Hes5 in expansion of neural stem cells of the developing brain. The Journal of biological chemistry. 2001;276:30467–30474. doi: 10.1074/jbc.M102420200. [DOI] [PubMed] [Google Scholar]
- Petit MM, Meulemans SM, Van de Ven WJ. The focal adhesion and nuclear targeting capacity of the LIM-containing lipoma-preferred partner (LPP) protein. The Journal of biological chemistry. 2003;278:2157–2168. doi: 10.1074/jbc.M206106200. [DOI] [PubMed] [Google Scholar]
- Piper M, Barry G, Hawkins J, Mason S, Lindwall C, Little E, Sarkar A, Smith AG, Moldrich RX, Boyle GM, Tole S, Gronostajski RM, Bailey TL, Richards LJ. NFIA controls telencephalic progenitor cell differentiation through repression of the Notch effector Hes1. J Neurosci. 2011;30:9127–9139. doi: 10.1523/JNEUROSCI.6167-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plachez C, Lindwall C, Sunn N, Piper M, Moldrich RX, Campbell CE, Osinski JM, Gronostajski RM, Richards LJ. Nuclear factor I gene expression in the developing forebrain. The Journal of comparative neurology. 2008;508:385–401. doi: 10.1002/cne.21645. [DOI] [PubMed] [Google Scholar]
- Porter FD, Drago J, Xu Y, Cheema SS, Wassif C, Huang SP, Lee E, Grinberg A, Massalas JS, Bodine D, Alt F, Westphal H. Lhx2, a LIM homeobox gene, is required for eye, forebrain, and definitive erythrocyte development. Development (Cambridge, England) 1997;124:2935–2944. doi: 10.1242/dev.124.15.2935. [DOI] [PubMed] [Google Scholar]
- Rincon-Limas DE, Lu CH, Canal I, Calleja M, Rodriguez-Esteban C, Izpisua-Belmonte JC, Botas J. Conservation of the expression and function of apterous orthologs in Drosophila and mammals. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:2165–2170. doi: 10.1073/pnas.96.5.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross SE, Greenberg ME, Stiles CD. Basic helix-loop-helix factors in cortical development. Neuron. 2003;39:13–25. doi: 10.1016/s0896-6273(03)00365-9. [DOI] [PubMed] [Google Scholar]
- Sasai Y, Kageyama R, Tagawa Y, Shigemoto R, Nakanishi S. Two mammalian helix-loop-helix factors structurally related to Drosophila hairy and Enhancer of split. Genes & development. 1992;6:2620–2634. doi: 10.1101/gad.6.12b.2620. [DOI] [PubMed] [Google Scholar]
- Shimizu T, Kagawa T, Inoue T, Nonaka A, Takada S, Aburatani H, Taga T. Stabilized beta-catenin functions through TCF/LEF proteins and the Notch/RBP-Jkappa complex to promote proliferation and suppress differentiation of neural precursor cells. Molecular and cellular biology. 2008;28:7427–7441. doi: 10.1128/MCB.01962-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian L, Sarkar A, Shetty AS, Muralidharan B, Padmanabhan H, Piper M, Monuki ES, Bach I, Gronostajski RM, Richards LJ, Tole S. Transcription factor Lhx2 is necessary and sufficient to suppress astrogliogenesis and promote neurogenesis in the developing hippocampus. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:E265–274. doi: 10.1073/pnas.1101109108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takebayashi K, Sasai Y, Sakai Y, Watanabe T, Nakanishi S, Kageyama R. Structure, chromosomal locus, and promoter analysis of the gene encoding the mouse helix-loop-helix factor HES-1. Negative autoregulation through the multiple N box elements. The Journal of biological chemistry. 1994;269:5150–5156. [PubMed] [Google Scholar]
- Tronche F, Kellendonk C, Kretz O, Gass P, Anlag K, Orban PC, Bock R, Klein R, Schutz G. Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nature genetics. 1999;23:99–103. doi: 10.1038/12703. [DOI] [PubMed] [Google Scholar]
- Tsai RY, Kim S. Fibroblast growth factor 2 negatively regulates the induction of neuronal progenitors from neural stem cells. Journal of neuroscience research. 2005;82:149–159. doi: 10.1002/jnr.20627. [DOI] [PubMed] [Google Scholar]
- Wall DS, Mears AJ, McNeill B, Mazerolle C, Thurig S, Wang Y, Kageyama R, Wallace VA. Progenitor cell proliferation in the retina is dependent on Notch-independent Sonic hedgehog/Hes1 activity. The Journal of cell biology. 2009;184:101–112. doi: 10.1083/jcb.200805155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer C, Tiveron MC, Bodmer R, Cremer H. Dynamics of Cux2 expression suggests that an early pool of SVZ precursors is fated to become upper cortical layer neurons. Cereb Cortex. 2004;14:1408–1420. doi: 10.1093/cercor/bhh102. [DOI] [PubMed] [Google Scholar]