Abstract
Mycobacterium tuberculosis (Mtb) is the leading cause of death due to a bacterial infection. The success of the Mtb pathogen has largely been attributed to the nonreplicating, persistence phase of the life cycle, for which the glyoxylate shunt is required. In Escherichia coli flux through the shunt is controlled by regulation of isocitrate dehydrogenase (ICDH). In Mtb, the mechanism of regulation is unknown, and currently there is no mechanistic or structural information on ICDH. We optimized expression and purification to a yield high enough to perform the first detailed kinetic and structural studies for Mtb ICDH-1. A large solvent kinetic isotope effect (D2OV = 3.0 ± 0.2, D2O[V/Kisocitrate] = 1.5 ± 0.3) and a smaller primary kinetic isotope effect (DV = 1.3 ± 0.1, D[V/K[2R-2H]isocitrate] = 1.5 ± 0.2) allowed us to perform the first multiple kinetic isotope effect studies on any ICDH and suggest a chemical mechanism. In this mechanism, protonation of the enolate to form product α-ketoglutarate is the rate-limiting step. We report the first structure of Mtb ICDH-1 to 2.18 Å by X-ray crystallography with NADPH and Mn2+ bound. It is a homodimer in which each subunit has a Rossmann fold, and a common top domain of interlocking beta sheets. Mtb ICDH-1 is most structurally similar to the R132H mutant human ICDH found in glioblastomas. Similar to human R132H ICDH, Mtb ICDH-1 also catalyses the formation of α-hydroxyglutarate. Our data suggest that regulation of Mtb ICDH-1 is novel.
Keywords: isocitrate dehydrogenase, glyoxylate shunt, kinetics, isotope effects, tuberculosis, structure
Isocitrate dehydrogenase (ICDH), an enzyme in the tricarboxylic acid (TCA) cycle, catalyzes the oxidative decarboxylation of isocitrate using NAD(P)+ as a cosubstrate to form α-ketoglutarate (αKG), CO2, and NADPH. In organisms possessing the glyoxylate shunt, which is not present in humans(1), ICDH sits at a branch point of the TCA cycle and the glyoxylate shunt. Utilization of this shunt is necessary for the virulence and persistence of several organisms(2), notably in the difficult to treat, nonreplicating physiological phase of Mtb(3). This shunt bypasses the two decarboxylative steps of the TCA cycle, allowing organisms to survive under nutrient limiting conditions, for example in the persistence phase of the Mtb life cycle(3). Understanding ICDH, which presumably regulates flux into the glyoxylate shunt, will allow for inhibitor development against the shunt, and thus against persistent Mtb. Exploration of new drug targets in persistence is especially important because this phase of the life cycle is nonreplicating, and replication is currently the target of the majority of antibiotics(4). In Escherichia coli the fate of the isocitrate molecule, either through the glyoxylate shunt or through the TCA cycle, is regulated by the phosphorylation of ICDH by the kinase phosphatase AceK(5-7). Phosphorylation inactivates ICDH and isocitrate is funneled instead through isocitrate lyase, the first enzyme of the glyoxylate shunt. No analogous proteins to E. coli AceK, or any phosphorylation sites on Mtb ICDH, have been identified. This suggests an unknown mechanism is at play and creates a need and opportunity for research on Mtb ICDH.
The questions surrounding the Mtb TCA cycle do not stop there. Mtb has two ICDHs; the 45.5 kDa ICDH-1 and the 82.6 kDa ICDH-2, both of which are NADP+ dependent(8). Mtb ICDH-1 and ICDH-2 have differing kinetic parameters under several conditions including pH, temperature, and salt concentration, suggesting that each isoform may play a role in the diverse environmental conditions confronted by the bacilli(8). However, no in vivo studies have been performed on the necessity of the two ICDHs, and the functional relevance of the two isoforms is still unknown. Mtb was thought to lack αKG dehydrogenase(9), but recent studies have demonstrated αKG dehydrogenase activity(10). Even if an αKG dehydrogenase exits, it is unclear how or if intermediates are funneled through the usually well defined TCA cycle. Baughn et al.(11) proposed an α-ketoglutarate ferredoxin oxidoreductase that uses ferredoxin instead of NADP+ as the oxidizing agent in the conversion of αKG to succinyl-CoA . There is also the γ-aminobutyric acid (GABA) shunt in which αKG is aminated to glutamate, decarboxylated to GABA, transaminated to succinic semialdehyde, and oxidized to the TCA intermediate succinate(12, 13). However, metabolic labeling experiments by de Carvalho et al.(14) show a non-stoichiometric amount of flux in carbon sources between αKG and succinate, questioning the continuity of the generally defined TCA cycle in Mtb. If Mtb does not use the TCA cycle for metabolism as usual, the discovery of a unique pathway in central carbon metabolism will be fascinating to basic science, and provide a myriad of new drug targets.
In this paper we provide the first mechanistic study and structural data for Mtb ICDH-1. Currently, one third of the global population is latently infected with Mtb, 10% percent of whom will develop active TB in their lifetime(15). Without an increased understanding of the Mtb TCA cycle and the regulation of the glyoxylate shunt, humans will continue to play idle host for Mtb, harboring the disease for the next generation. Deducing the mechanism and crystal structure of Mtb ICDH-1 provides a molecular platform for the development of ICDH-1 as a potential drug target.
MATERIALS AND METHODS
Materials
Isocitrate for isotope effect studies was synthesized as described below, and concentrations were determined enzymatically. For the rest of the experiments isocitrate was purchased from Sigma Aldrich as the dl-isocitric acid trisodium salt hydrate and concentration was determined by weight. Cloning reagents were from New England Biolabs, and primers were from Invitrogen. DNase and complete EDTA-free protease inhibitor cocktail were from Roche. 99.9% Deuterium oxide and [2,3-2H2]fumarate were from Cambridge Isotope Laboratories. Crystallography reagents were from Hampton Research. The remainder of the chemicals were purchased from Sigma Aldrich.
Cloning, Expression, and Purification of ICDH-1
Cloning and expression were modified from a previously reported protocol(16). The Mtb icd1 gene (Rv3339c) was PCR amplified from H37Rv DNA using the forward primer 5’ GGAATTCCATATGTCCAACGCACCCAAGATA 3’ and the reverse primer 5’CCCAAGCTTCTAATTGGCCAGCTCCTTTTCC 3’. Restriction sites for NdeI and HindIII are underlined respectively. The PCR fragment was cloned into a pET-28a(+) vector containing a N-terminal His6-Tag. After sequence verification, the recombinant plasmid was transformed into T7 express competent E. coli cells. 6 L LB containing 30 Tg/mL kanamycin were inoculated with 10 mL overnight starter culture and incubated at 37°C while shaking at 200 RPM. At A600 ~ 0.35, cells were induced with 0.1 mM IPTG and incubated at 16°C overnight, shaking. Cells were harvested by centrifugation, and the pellets were resuspended in 25 mM triethanolamine (pH 7.5) containing 50 mM NaCl and 20 mM imidazole (buffer A), and 1 tablet EDTA-free protease inhibitor cocktail. The resuspended pellets were then emulsified (EmulsiFlex-C3, Avestin) and centrifuged at 17,000 RPM for 35 minutes. Batch purification was performed with a Ni-NTA agarose column. The column was washed with buffer A and the protein was eluted with a 0 - 300 mM imidazole gradient in buffer A. The His6-Tag was removed by overnight dialysis in 20 mM Tris pH 8.0 containing 150 mM NaCl, 2.5 mM CaCl2, 1 mM DTT, and 80 units of thrombin. After cleavage, the protein was concentrated and applied to a HiLoad 26/60 Superdex 200 prep grade size exclusion column run in 50 mM HEPES (pH 7.5), 150 mM NaCl, 5 mM MnCl2, and 5% glycerol. The protein was stored in this buffer at -80°C. The concentration of ICDH-1 was determined using A280 and an extinction coefficient of 67,965 M-1cm-1(17).
Measurement of Enzymatic Activity
Enzymatic activity for the primary reaction was determined by monitoring the conversion of NADP+ to NADPH at 340 nm. Experiments were conducted in a Shimadzu UV-2450 spectrophotometer in 100 mM HEPES (pH 7.5) containing 5 mM MnCl2, and varying amounts of isocitrate and NADP+, in a final reaction volume of 1 mL. Assays were performed at room temperature and 1 nM ICDH-1 was added to initiate the reaction. Rates were calculated using the molar extinction coefficient of NADPH, 6220 M-1cm-1(18) and the total enzyme concentration (E).
For the secondary reaction, preliminary kinetics were obtained by monitoring the decrease in NADPH and αKG at 340 nm. At high concentrations of αKG the absorbance of αKG at 340 nm overlaps with that of NADPH. Therefore we calculated a combined ε340 = 6260 M-1 cm-1 for NADPH and αKG, and we limited the “saturating” amount of αKG to keep A340 < 2. The αKG Michaelis-Menten experiment was done under steady-state conditions in an Applied Photophysics SX-20 stopped-flow in absorbance mode. Final concentrations were: 50 mM HEPES pH 7.5, 150 mM NaCl, 100 μM NADPH, varying (1-50 mM) αKG, 500 nM ICDH-1. Five replicates for each point were performed. Due to a non-reproducible signal observed during the first 10 s, data were collected from 15 - 30 s post mixing. The NADPH Michaelis-Menten experiment was performed in the Shimadzu UV-2450 spectrophotometer with 100 mM HEPES pH 7.5, 40 mM αKG, varying NADPH (1 – 50 μM), and initiated with 100 nM ICDH-1. Points were repeated in duplicate. Metal was not included in either of these assays due to the observance of a nonlinear rate upon ICDH-1 addition, which we hypothesize to be due to coordination of the metal ions by the large amounts of αKG. In both assays rates were calculated using ε340 = 6260 M-1 cm-1 and the total enzyme concentration (E).
Data Fitting
All data fitting was performed with GraphPad Prism version 5.0d. In all graphs the points are the mean of experimental duplicates, and the error bars are the standard deviation of the replicates. The solid lines are the result of fitting to the denoted equation. The error reported with the fitted values is the standard deviation of the fit and is always rounded up.
Initial Velocity of ICDH-1
Initial kinetic parameters were estimated by saturating with one substrate and varying the concentration of the other substrate. The resulting curve was fitted to eq 1 where V is the maximal velocity and S is the concentration of the varied substrate.
| (1) |
The initial velocity experiment was conducted at fixed concentrations of isocitrate and varying concentrations of NADP+. The resulting pattern was fitted globally to eq 2 for a sequential kinetic mechanism where V is the maximal velocity, A and B are the substrate concentrations, Ka and Kb are the respective Michaelis constants for each substrate, and Kia is the inhibition constant for substrate A.
| (2) |
Inhibition
Product inhibition by αKG and NADPH, and inhibition by malate were tested with variable amounts of inhibitor, and varying concentrations of both isocitrate and NADP+. Data were globally fitted to eq 3 for competitive inhibition and eq 4 for noncompetitive inhibition, where S is the varied substrate concentration, I is inhibitor concentration, Kis is the inhibition constant for the slope, Kii is the inhibition constant for the intercept, and Km is the Michaelis constant of substrate S.
| (3) |
| (4) |
pH Dependence of ICDH-1 Activity
The pH dependence of the kinetic parameters was determined in 100 mM buffer at the desired pH; MES pH 5.5 - 6.5 and HEPES pH 6.5 - 8.5. The resulting kcat and kcat/Km data were fitted to eq 5, which describes a single pKa, the negative log of the acid dissociation constant, where c is the pH independent plateau value, y is the rate, and x is the pH.
| (5) |
Primary Kinetic Isotope Effects
Primary kinetic isotope effects (KIEs) were measured using [2R-2H]isocitrate. Synthesis of [2R-2H]isocitrate was carried out first by chemical synthesis of [2-2H]glyoxylate via [2,3-2H2]tartaric acid, and then enzymatic synthesis of [2R-2H]isocitrate from [2-2H]glyoxylate. See Supporting Information Scheme 1 for a pictorial representation.
[2,3-2H2]Tartaric acid
N-methylmorpholine N-oxide (NMO, 1.00 g, 8.54 mmol) was dissolved in 45 mL of 1:1 THF:H2O under an argon atmosphere, and osmium tetroxide (4 wt % in H2O, 0.60 mL, 0.094 mmol) was added. [2,3-2H2]fumaric acid (1.00 g, 8.47 mmol) was added to the solution, and the mixture was stirred at 35°C overnight after which TLC analysis (5:4:2:1 n-BuOH:MeOH:NH4OH:H2O) showed the consumption of fumaric acid. Sodium sulfite (0.20 g, 1.59 mmol) was added to the mixture and THF and some H2O were removed in vacuo. The remaining solution was washed twice with EtOAc and the aqueous layer was subsequently concentrated in vacuo. The crude product was re-dissolved in H2O and applied to a 10 mL column of Dowex 1X-2 resin (formate form). The resin was washed with H2O (3 × 25 mL), then 0.7 N formic acid (3 × 25 mL) and finally 1.4 N formic acid (2 × 25 mL). Fractions containing tartaric acid were pooled and the solution volume was reduced in vacuo. Distilled water was added and the mixture was concentrated in vacuo; this procedure was repeated multiple times to remove any residual formic acid and gave [2,3-2H2]tartaric acid (0.70 g, 4.60 mmol, 54%) as a white solid. As expected, no proton signals were observed in the 1H NMR spectrum (D2O) and the 13C NMR spectrum signals matched literature values(19).
[2-2H]Glyoxylic acid
The synthesis of [2-2H]glyoxylic acid was carried out according to a modified form of an existing published procedure(19). [2,3-2H2]Tartaric acid (0.57 g, 3.75 mmol) was dissolved in 10 mL of H2O and the resultant solution was cooled to 0°C. Periodic acid (1.03 g, 4.50 mmol) was dissolved in 5 mL H2O and was added dropwise to the solution. After each 1 mL portion of periodic acid was added, a single ice chip was dropped into the reaction mixture. After the addition of periodic acid was complete, the cooling bath was removed and the reaction was stirred at room temperature and monitored for the disappearance of starting material by TLC analysis (5:4:2:1 n-BuOH:MeOH:NH4OH:H2O). After 20 min, the reaction was judged complete and the solution was concentrated to approximately 3 mL in vacuo and subsequently loaded onto a 5 mL column of Dowex 1X-2 resin (formate form). The resin was washed with H2O (60 mL) and then with 0.7 N formic acid (100 mL) to elute the glyoxylic acid. Fractions containing glyoxylic acid were pooled and the volume reduced in vacuo before additional distilled H2O was added to the solution; this procedure was repeated multiple times to remove any residual formic acid and give the fully protonated [2-2H]glyoxylic acid (225 mg, 3.00 mmol, 40%). No proton signals were observed in the 1H NMR spectrum (D2O) and the 13C NMR spectrum signals matched literature values(19). The product was dissolved in H2O and the solution was carefully titrated to pH 7.0 with dilute NaOH and then frozen and lyophilized to give the sodium salt of [2-2H]glyoxylic acid as a white powder.
[2R-2H]Isocitric acid
Isocitrate lyase (500 μL 37.5 μM) was added to a 20 mL solution containing 20 mM Tris (pH 7.5), 50 mM [2-2H]glyoxylate, 200 mM succinate, and 5 mM MgCl2. The mixture was left over the weekend at room temperature. Isocitrate lyase was removed by passing the solution through an Amicon centrifugal filtration device (10K MWCO), and the resulting filtrate was applied to a 85 mL column of Dowex 1X4 (formate form) and eluted with a gradient of 400 mL of 0 – 6 N formic acid. Fractions containing [2R-2H]isocitrate, as determined enzymatically using the Mtb ICDH-1 assay, were pooled and the volume of the solution reduced in vacuo. Distilled H2O was added and the solution concentrated in vacuo; this procedure was repeated multiple times to remove residual formic acid, giving [2R-2H]isocitrate (103 mg, 0.4 mmol, 80%). The mass spectrum was consistent with an increase in mass of one unit from the isotopically-unlabeled substrate (m/z = 192.1 [M - H]-). All mass spectrometry samples were collected on a 12 Tesla Fourier transform ion cyclotron resonance mass spectrometer (Agilent). Protiated isocitrate was enzymatically synthesized from Sigma Aldrich glyoxylate and subjected to identical purification procedures.
Isotope effect assays were performed with a saturating concentration of NADP+ and varying concentrations of isocitrate and [2R-2H]isocitrate. The results were globally fitted to eq 7 where V is the maximal velocity, S is the concentration of isocitrate or [2R-2H]isocitrate, Fi is the fraction of isotope (0 or 1), EV/K is the effect on (kcat/Km) 1 and EV is the effect on kcat - 1.
| (7) |
The pH dependence of the kinetic isotope effects (KIEs) was measured at pH 5.7 in MES, and 6.5, 7.5, and 8.5 in HEPES.
Solvent Kinetic Isotope Effects
Solvent KIEs were measured with saturating concentrations of NADP+ and varying concentrations of isocitrate, and globally fitted to eq 7 where Fi = 0 for H2O and Fi = 0.95 for D2O. A viscosity control of 9% glycerol(20) did not show any effect on either V or V/Kisocitrate (data not shown). A proton inventory was performed at saturating conditions of both isocitrate and NADP+ in 10% increments from 0 - 90% D2O.
Multiple Kinetic Isotope Effects
Multiple KIEs were measured with saturating concentrations of NADP+ and varying concentrations of [2R-2H]isocitrate in H2O and 95% D2O, and varying concentrations of isocitrate and [2R-2H]isocitrate in 95% D2O. Data were globally fitted to eq 7.
Transfer of 2H from [2R-2H]isocitrate to [4R-4-2H]NADPH
A 1 mL reaction was prepared containing 40 mM HEPES pH 7.5, 5 mM MnCl2, 5 mM isocitrate or [2R-2H]isocitrate, 5 mM NADP+, and a catalytic amount of ICDH-1. The enzyme was removed using an Amicon centrifugal filtration device (10K MWCO). 500 μL filtrate was loaded onto a MonoQ 4.6/100 PE column, washed with water, then eluted in 1 M ammonium bicarbonate (B) as follows: 0 - 50% B in 48 min, 50 - 100% B in min 48 – 49, 100% B in 59 - 64 min, 100% - 0% B in min 64 - 65, and then washed with water for 10 minutes. Fractions with absorbance at both 260 nm and 340 nm were pooled, lyophilized, and redissolved in D2O for 1H NMR spectroscopy.
Crystallization, X-ray Data Collection, and Refinement
Crystals of ICDH-1 in complex with NADPH and Mn2+ were grown at room temperature using the vapor diffusion method from hanging drops (1 μl of protein at 10 mg/ml, 5 mM NADPH and 5 mM MnCl2 mixed with 1 μl of reservoir solution) using a reservoir solution that contained 30% PEG2K, 0.1 M Tris HCl pH 8.0. Crystals were cryoprotected with mineral oil. Diffraction data were collected from a single frozen crystal using a RAXIS-IV++ detector mount on a Rigaku RH-200 rotating anode (copper anode) X-ray generator. Data were processed with HKL3000(21). The structure was solved by molecular replacement using the program Phenix(22) and the human ICDH structure with PDB ID 3MAS as a search model. The significantly built model (>90%) was completed by iterative cycles fitting in COOT(23) and refinement in Phenix. There is a dimer per asymmetric unit. Structure figures were synthesized using PyMOL(24). Atomic coordinates and experimental structure factors have been deposited in the Protein Data Bank (PDB ID code: 4HCX). Data collection and refinement statistics are presented in Table 2.
Table 2.
Summary of data collection and refinement statistics for the WT ICDH1-NADPH-Mn2+ Complex
| X-ray source | Rotating Anode |
| Wavelength (Å) | 1.5418 (Cu anode) |
| Temperature (K) | 100 |
| Resolution Range (Å) | 50.0-2.18 |
| Reflectiona | 45115 (2069) |
| Completeness (%) | 99.0 (92.0) |
| I/σ(I) | 24.04 (3.75) |
| Redundancy | 5.3(4.3) |
| Space group | P212121 |
| Unit cell (Å) | |
| a | 90.14 |
| b | 92.70 |
| c | 102.90 |
| α = β = γ | 90.0° |
| Molecules per a.u. | 2 |
| Refinement statistics | |
| Rwork (%) | 20.74 |
| Rfree (%) | 26.17 |
| Number of atoms | |
| Protein (Chain A) | 3130 |
| Protein (Chain B) | 3130 |
| NADPH (Chain A) | 48 |
| NADPH (Chain B) | 48 |
| Water | 310 |
| r.m.s. deviation | |
| Bond length (Å) | 0.008 |
| Bond angles (°) | 1.282 |
| Average B-factors (Å2) | |
| Overall (Chain A) | 32.45 |
| Protein main chain (Chain A) | 32.54 |
| Protein side chain (Chain A) | 32.35 |
| Overall (Chain B) | 35.96 |
| Protein main chain (Chain B) | 36.02 |
| Protein side chain (Chain B) | 35.89 |
| NADPH (Chain A) | 32.90 |
| NADPH (Chain B) | 39.42 |
| Water (Chain A) | 34.23 |
| Water (Chain B) | 34.68 |
| PDB accession code | 4HCX |
values for the highest resolution shell is in parenthesis
RESULTS AND DISCUSSION
Expression and Purification
Expression and purification of Mtb ICDH-1 has been notoriously difficult and low in yield (personal communication). With optimization we were able to modify the only other previously successful protocol(16) to obtain enough protein for X-ray crystallography. While our lab was not able to obtain as high a yield as Banerjee et al.(16), the most significant increase came after the substitution of emulsification instead of sonication for cell lysis. Expression yielded both soluble and insoluble proteins of the expected mass, with an increase in the soluble fraction after the post-induction temperature was decreased to 16°C. Initial purification attempts yielded unstable enzyme, which was remedied by the addition of 5 mM MnCl2 to the gel filtration and storage buffers. Enzymatic activity with several divalent cations was tested. The rate was significantly increased with the addition of 5 mM MgCl2 or MnCl2 compared to 5 mM CaCl2 or ZnCl2. When exogenous MnCl2 was added, the rate was increased 12% compared to the addition of exogenous MgCl2 (Table S1). Approximately 0.3 mg of soluble, pure ICDH-1 was obtained per liter of culture. SDS-PAGE was used to determine protein containing fractions of > 95% purity.
Kinetic Mechanism
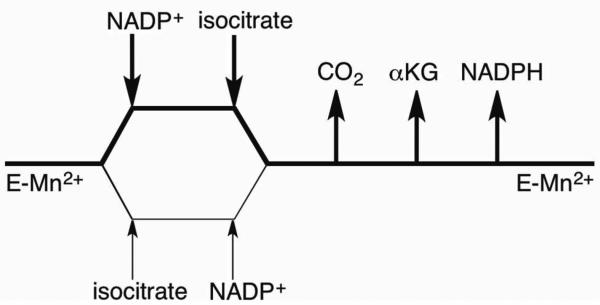
The first step to determine a chemical mechanism is to see whether the enzyme proceeds by a sequential mechanism, in which there is a central intermediate(s) containing all substrates, or a ping-pong mechanism in which one substrate is converted to one product before the next substrate is catalyzed. When varying NADP+ at fixed, variable isocitrate concentrations and fitted to a sequential mechanism (eq 2), the following parameters were determined: kcat = 33 ± 2 sec-1, KNADP+ = 15 ± 3 μM, Kisocitrate = 50 ± 7 μM, Kia = 144 ± 23 μM. Based on the intersecting lines in the double reciprocal plot, a sequential mechanism is proposed.
Inhibition studies were performed to determine the order of substrate binding and product release. All patterns and fitted values are listed in Table 1. Product inhibition with NADPH was competitive against NADP+ and noncompetitive against isocitrate suggesting NADP+ binds first and NADPH is released last. αKG was noncompetitive against NADP+. Attempts to determine CO2 inhibition with NaHCO3 and KHCO3 were unsuccessful due to precipitation at amounts lower than Ki. Our data support a kinetic mechanism in which NADP+ binds first followed by isocitrate to form a ternary complex before conversion to products. Since isocitrate could bind tightly to the divalent metal, we include the possibility that isocitrate can bind to the free enzyme (Scheme 1). Random sequential substrate binding has been proposed for porcine heart mitochondrial ICDH and E. coli ICDH(25-27). Malate was not a substrate and was used as a dead-end inhibitor. It inhibits both NADP+ and isocitrate noncompetitively. It appears that malate can bind to the metal ion in both the E-NADP+ and E-NADPH complexes, yielding effects on both the slope and the intercept of the reciprocal plots. Thus, malate inhibition was unable to distinguish between random or ordered substrate binding.
Table 1.
Product inhibition patterns for Mtb ICDH-1
| Experiment | Pattern | Fitted Parameters |
|---|---|---|
| NADPH vs. NADP+ | C | appKis = 6 ± 2 μM |
| NADPH vs. isocitrate | NC | appKis = 10 ± 2 μM appKii = 49 ± 8 μM |
| αKG vs. NADP+ | NC | appKis = 22 ± 6 mM appKii = 11 ± 1 mM |
| malate vs. NADP+ | NC | appKis = 80 ± 15 mM appKii = 198 ± 19 mM |
| malate vs. isocitrate | NC | appKis = 98 ± 15 mM appKii = 516 ± 114 mM |
C, competitive; NC, noncompetitive
SCHEME 1.
Proposed Kinetic Mechanism of Mtb ICDH-1
The noncompetitive pattern observed for αKG and NADP+ has two possible interpretations: random product release of CO2 and αKG, or that the ability of αKG to bind free enzyme with or without nucleotide introduces a slope effect to what would otherwise be uncompetitive inhibition. We prefer the later interpretation, which we are able to support by additional lines of reasoning. Our inability to demonstrate product inhibition by CO2, previous reports on the E. coli enzyme(27), and the demonstration of a large solvent KIE due to slow protonation of the metal-liganded αKG enolate anion all support CO2 as the first product released. Ordered release of αKG and NADPH complete the catalytic cycle.
pH dependence of ICDH-1
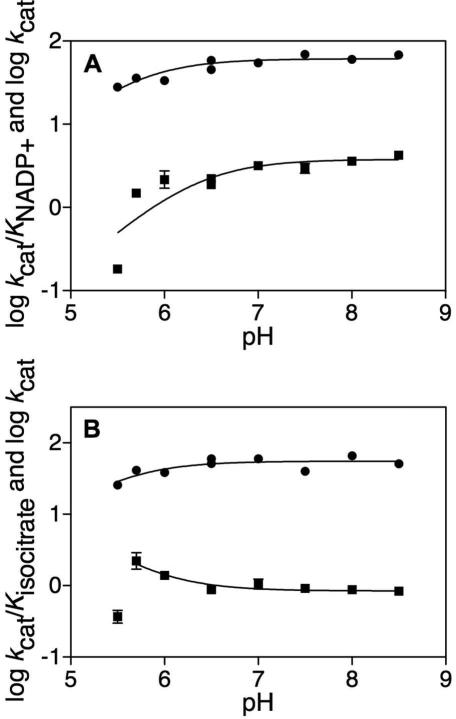
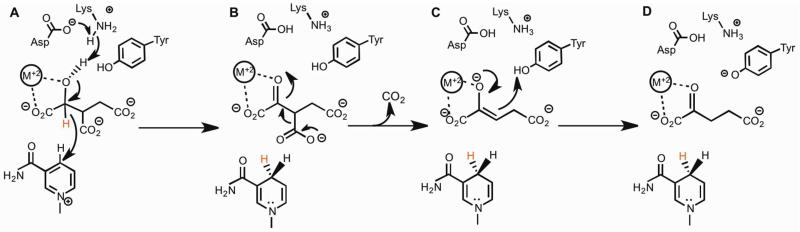
The pH dependence of kcat and kcat/Km for substrates were tested to explore the ionization states of groups involved in binding and catalysis. Testing was limited at both the upper and lower pH ranges. Above pH 8.5, a reddish brown precipitate formed. Below pH 5.5, enzyme stability and activity were significantly decreased. When varying NADP+ and fitting to eq (5) the pKa of the ionizable group observed for log kcat was 5.6 ± 0.1 and the pKa of the ionizable group observed for log kcat/KNADP+ was 6.3 ± 0.2 (Figure 1A). When varying isocitrate and fitting to eq (5) the pKa of the ionizable group observed for log kcat was 5.5 ± 0.2 (Figure 1B). When analyzing log kcat/Kisocitrate we fitted to the inverse of eq (5) and excluded data at pH 5.5 from the fitting. The pKa of the ionizable group observed for log kcat/Kisocitrate was 5.8 ± 0.1 (Figure 1B). The log kcat values report on the ionization behavior of groups involved in catalysis while the log kcat/Km values report on the ionization behavior of groups involved in catalysis and binding. We suggest that the kcat/KNADP+ pH profile is reporting on the binding of NADP+, whose 2’-phosphate is known to have a pKa = 6.1(18). When varying isocitrate, we suggest that the modest increase in V/Kisocitrate at pH 5.7 represents the preference of the enzyme for the dianion over the trianion of isocitrate. At the lowest pH values where reliable data could be obtained, kcat decreases modestly, suggesting possible general base catalysis of secondary alcohol oxidation. ICDHs are known to utilize tyrosine as a catalytic acid and lysine as a catalytic base(28). Aktas and Cook have suggested on the basis of the porcine heart mitochondrial ICDH studies that there is a catalytic triad in which Asp deprotonates Lys to deprotonate the isocitrate C2 hydroxyl, which is not rate-limiting(28). The Asp may be the source of the hint at the pKa in log kcat = 5.5 ± 0.2. Both the Lys 215 and Tyr 142 (Mtb numbering) components of the catalytic triad are conserved (Figure S4). For porcine ICDH-1, it has been suggested that Tyr protonates the intermediate enolate to form product αKG(29). This is plausible for Mtb ICDH-1 as well (Scheme 2C).
FIGURE 1.
“pH dependence of Mtb ICDH-1.” (A) kcat and kcat/KNADP+ determined at saturating isocitrate and varying NADP+. (A) (●) Log kcat fitting to eq 5 determined pKa = 5.6 ± 0.1. (■) Log kcat/KNADP+ fitting to eq 5 determined pKa = 6.3 ± 0.2. (B) kcat and kcat/Kisocitrate determined at saturating NADP+ and varying isocitrate. (●) Log kcat fitting to eq 5 determined pKa = 5.5 ± 0.2. (■) Log kcat/Kisocitrate fitting to the inverse of eq 5 determined pKa = 5.8 ± 0.1.
SCHEME 2.
Proposed Chemical Mechanism of Mtb ICDH-1
Primary Kinetic Isotope Effects
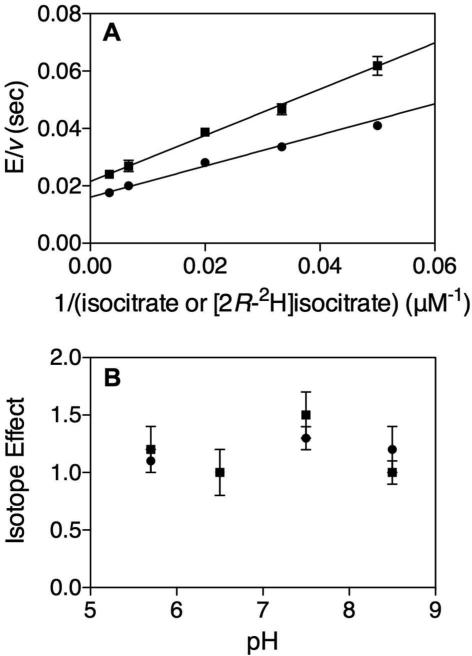
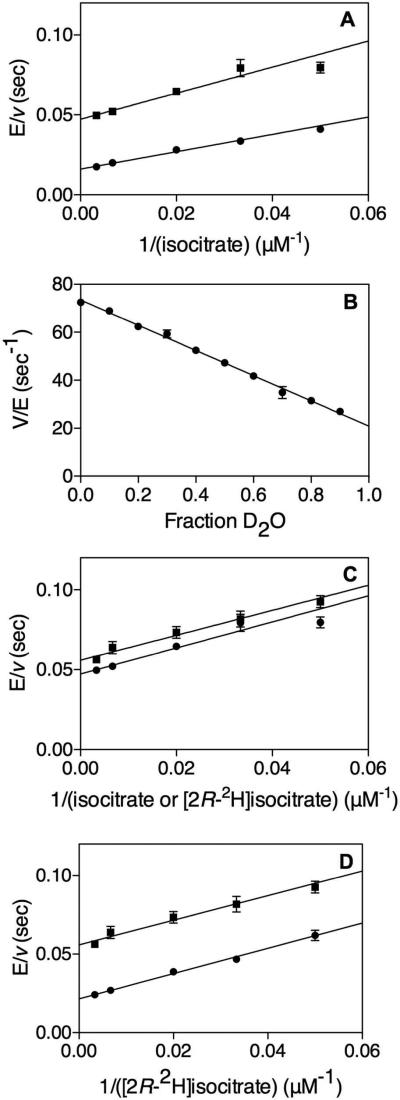
The primary KIEs with [2R-2H]isocitrate were DV = 1.3 ± 0.1 and D[V/K[2R-2H]isocitrate] = 1.5 ± 0.2 (Figure 2A). This indicates the transfer of the 2R hydrogen of isocitrate to NADP+ is involved in a partially rate-limiting step. In porcine heart ICDH, even smaller kinetic isotope effects with dl-isocitrate-2-(h,d) were observed where DV and D[V/KDL-Isocitratrate-2-(h,d)] were equivalent(30). However, a non-unity effect occurred at low (4.5) and high (9.5) pHs, with no effect observed at pH 7.45(30). Due to the pH limitations of the current Mtb assay, the pH dependence of the isotope effects was only tested from pH 5.7 - 8.5, and showed no significant pH dependence of the effect on V or V/K[2R-2H]isocitrate (Figure 2B).
FIGURE 2.
“Primary Kinetic Isotope Effects.” (A) Primary KIE at 500 μM NADP+ and varying (20-300 μM) (●) isocitrate or (■) [2R-2H]isocitrate. DV = 1.3 ± 0.1, D[V/K[2R-2H]isocitrate] = 1.5 ± 0.2. (B) pH dependence of (●) DV and (■) D[V/K[2R-2H]isocitrate] at 500 μM NADP+ and varying (5 – 150 μM) isocitrate or [2R-2H]isocitrate.
Solvent Kinetic Isotope Effects
Solvent KIEs were significantly larger, with D2OV = 3.0 ± 0.2 and D2O[V/Kisocitrate] = 1.5 ± 0.3, indicating that a water derived proton transfer is responsible for a rate-limiting step in catalysis (Figure 3A). A linear proton inventory (Figure 3B) shows that only a single solvent proton is giving rise to this effect. Additionally, when solving for 0% D2O in the line that was fitted to the data (y = -52.52x + 73.43) and dividing by the value at 95% D2O we get D2OV = 3.1, within the error of the experimentally determined D2OV.
FIGURE 3.
“Solvent and Multiple Kinetic Isotope Effects.” (A) Solvent KIE with 500 μM NADP+ varying (20-300 μM) isocitrate in (●) H2O or (■) 95 % D2O. D2OV = 3.0 ± 0.2, D2O[V/Kisocitrate] = 1.5 ± 0.3. (B) Proton inventory where kcat was measured with 500 μM isocitrate and 500 μM NADP+ at 10% increments of D2O and fitted to a straight line. (C) Multiple KIE with 500 μM NADP+ varying (20-300 μM) (●) isocitrate or (■) [2R-2H]isocitrate in 95% D2O. DV = 1.2 ± 0.1, D[V/K[2R-2H]isocitrate] = 1.0 ± 0.2. (D) Multiple KIE with 500 μM NADP+ varying (20-300 μM) [2R-2H]isocitrate in (●) H O or (■) 95% D2O. D2OV = 1.7 ± 0.2, D2O[V/K[2R-2H]isocitrate] = 1.0 ± 0.2.
There are two proton transfer reactions catalyzed by ICDH, the deprotonation of the secondary alcohol, concurrent with or prior to hydride transfer (Scheme 2A) and the protonation of the enolate of αKG after decarboxylation of the oxalosuccinate intermediate (Scheme 2C). For porcine heart ICDH, the authors’ proposed that the initial deprotonation of the secondary alcohol was the rate limiting step and source of the large solvent KIE(31). However, for there to be such a small primary KIE (DV = 1.3 ± 0.1 and D[V/K[2R-2H]isocitrate] = 1.5 ± 0.2) in Mtb ICDH-1, then the formation and collapse of the alkoxide must be completely uncoupled. Metal coordination of the formed alkoxide would be a stabilizing factor, but should also make its formation facile. This same argument applies to the stabilization of the enolate formed after decarboxylation of oxalosuccinate. However, if a strong metal interaction was present, the enolate would be protonated slowly, and this could be the source of the large solvent KIE that we observe. In order to distinguish between these two possibilities, we performed multiple KIE studies to determine if the deprotonation of the secondary alcohol, and the protonation of the enolate occurred in two discreet steps, or as a coupled, concerted step. Colman and Chu(31) did not have this luxury since their studies in 1969 predated the advent of multiple KIEs to deduce reaction mechanisms in 1982(32).
Multiple Kinetic Isotope Effects
Multiple KIEs were measured with isocitrate and [2R-2H]isocitrate in 95% D2O (Figure 3C). Values of DV = 1.2 ± 0.1, D[V/K[2R-2H]isocitrate] = 1.0 ± 0.2 were determined. Since these multiple KIEs are less than their value in H2O (DV = 1.3 ± 0.1 and D[V/K[2R-2H]isocitrate] = 1.5 ± 0.2) a stepwise mechanism is predicted where the primary KIE from [2R-2H]isocitrate and solvent KIE from D2O report on two separate steps. Multiple KIEs were also measured with [2R-2H]isocitrate in H2O and 95% D2O (Figure 3D). Values of D2OV = 1.7 ± 0.2, D2O[V/K[2R-2H]isocitrate] = 1.0 ± 0.2 were determined. These values are decreased compared to their value with isocitrate; D2OV = 3.0 ± 0.2 and D2O[V/Kisocitrate] = 1.5 ± 0.3, again indicating a stepwise mechanism. The clear data for a stepwise mechanism indicates either that the formation and collapse (Scheme 2A) of the isocitrate alkoxide are completely uncoupled, or that the origin of the Mtb ICDH-1 solvent KIE is the protonation of the αKG enolate formed after decarboxylation (Scheme 2C). Based on the likely strong stabilization of both anionic intermediates due to metal ion ligation, we believe that the results of our multiple KIEs are more compatible with protonation of the αKG enolate being the source of the large solvent KIE.
Transfer of 2H from [2R-2H]isocitrate to [4R-4-2H]NADPH
The transfer of the deuterium from [2R-2H]isocitrate to the A-side of the nicotinamide ring to form [4R-4-2H]NADPH was determined by 1H NMR spectroscopy (Figure S1A). This was the expected result, as other isocitrate dehydrogenases also catalyze an A-side specific transfer(33). A control reaction was performed with [2R-1H]isocitrate (Figure S2B). The assignment of stereochemistry of the transfer of deuterium to NADPH was based on previous work by Ottolina et al.(34).
Three-dimensional structure of Mtb ICDH-1
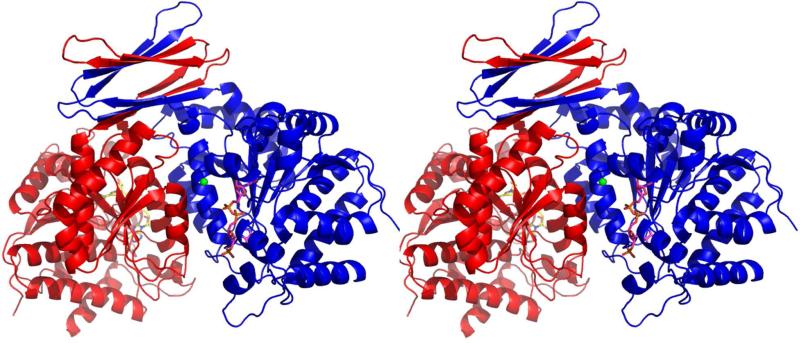
We crystallized Mtb ICDH-1 and solved the first three-dimensional structure of Mtb ICDH-1 by molecular replacement using human cytosolic (Hc) ICDH (PDB ID: 3MAS) as a template. It is interesting that Mtb ICDH-1 more closely resembles mammalian instead of bacterial ICDHs, and further suggests that Mtb ICDH-1 regulation may be drastically different from that in E. coli. After several rounds of minimization the structure was optimized with Rwork = 20.7 and Rfree = 26.2. The final structure is shown in Figure 4. The Ramachandran plot showed 89.4% of the residues in the most favored region and 9.1% of the residues in allowed regions. Each monomer of Mtb ICDH-1 consists of a bottom globular domain composed primarily of a Rossmann fold, a common dinucleotide binding domain. Above the Rossmann motif, each monomer has two β hairpins connected by a longer loop. Each of these hairpins interacts with the hairpin of the other monomer to form two anti parallel β sheets (Figure 4).
FIGURE 4.
“Global Structure of Mtb ICDH-1.” Stereo view of Mtb ICDH-1. Each monomer is bound to one Mn2+, shown as a green sphere, and one NADPH, shown in stick form.
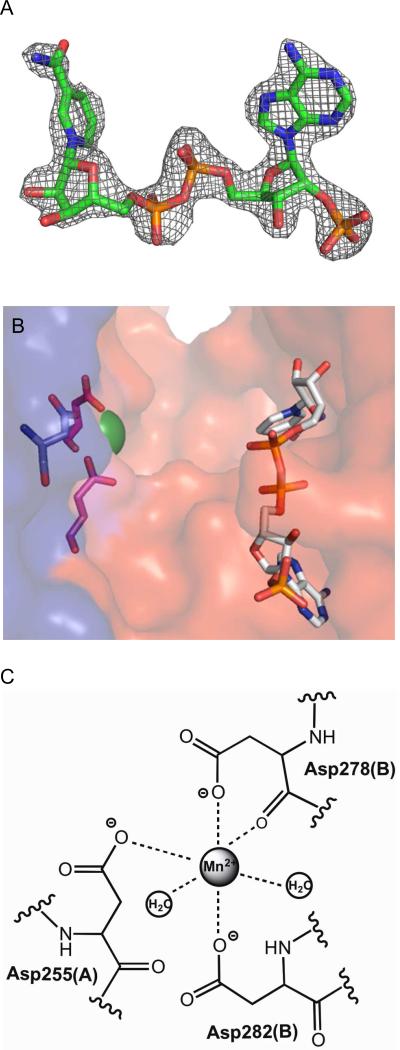
Mtb ICDH-1 exhibits planar symmetry with Mn2+ bound toward the center of the plane, represented as a green sphere in Figure 4. As shown for the B chain (Figure 5C), Mn2+ is hexagonally coordinated by Asp 282 and Asp 278 of chain B, Asp 255 of chain A, and two water molecules. We propose the coordination of the metal by both monomers enhances dimer formation and stability. In a sequence alignment (Figure S4) of the top three blast hits in the PDB, two of which are mammalian, plus Mtb ICDH-1 and E. coli ICDH, Asp 282 and Asp 278 are conserved in all aligned sequences and Asp 255 is conserved in all except E. coli.
FIGURE 5.
“Ligands in the Mtb ICDH-1 Structure.” (A) Electron density map for NADPH at 2σ. (B) A close up of one of the two the active sites showing Mn2+ as a green sphere and NADPH as sticks. Coordinating residues of the Mn2+ from the A chain are shown in pink, and the residue from the B chain is shown in blue.
(C) Metal binding site.
The NADPH molecules are exclusively bound by residues of the monomer to which they bind. All of the NADPH-Mtb ICDH-1 contacts are conserved in the top three blast hits (Figure S4). NADPH is primarily held in place by various electrostatic and hydrogen bonding interactions. A schematic showing all of the interactions of NADPH and Mtb ICDH-1 is shown in Figure S5. It is worth noting the side chain nitrogens from Arg 317 and His 318 coordinate the 2’-phosphate on the adenosine ribose. The importance of this interaction is highlighted by the inability of Mtb ICDH-1 to catalyze a detectable reaction using NAD+ as its cofactor, as shown by Banerjee et al.(8) and repeated in our own lab (data not shown). These interactions are generally on the order of 3 Å, and we predict a collapse of the enzyme active site upon the binding of isocitrate in the cleft (Figure 5B) to strengthen these interactions. The hypothesized structural rearrangement necessary for catalysis entertains the possibility of a structure based regulatory mechanism of Mtb ICDH-1.
Structural and Regulatory Comparison of Mtb and E. coli ICDH
The structural similarity of Mtb ICDH-1 to Hc ICDH, suggests that the E. coli method of regulation of the glyoxylate shunt may not be applicable to Mtb. E. coli ICDH is regulated by phosphorylation of Ser 113(35), which hydrogen bonds to isocitrate in the active site(36). In Mtb ICDH-1 Glu 88 corresponds to the phosphorylated Ser 113 in E. coli, and is located on the outer edge of the protein distant from the active site in the Mtb structure. Recently, the first structure of the “fully closed” E. coli ICDH has been solved, indicating E. coli ICDH undergoes conformational changes upon substrate binding, and providing a structural basis for the previously postulated mechanism(37).
Structural and Regulatory Comparison of Mtb and Human ICDH
Similar to E. coli, Hc ICDH is proposed to exist in three conformational states; open, semi open, and closed(38). However, Hc ICDH is self-regulated by the transition between these states and not by phosphorylation. It is also important to remember Hc ICDH regulation is for a different purpose, as the glyoxylate shunt is not present in humans. Initial work to search for the analogous phosphorylation site of E. coli ICDH in Hc ICDH proposed Ser 94 to be the corresponding regulation site to E. coli Ser 113. In Hc ICDH, Asp 278 interacts with Ser 94 to inhibit activity in the NADP+ bound, open form. In the active, closed form this interaction is disrupted by a conformational change, from a loop to a helix, of the segment bearing Asp 278, which then coordinates Ca2+ in the isocitrate-NADP+ Ca2+ bound form to allow enzymatic activity(38). In the Mtb enzyme, the equivalent of Hc Ser 94 is Ser 97 and this residue is located on a loop 7.6 Å from the bound NADPH and 12.8 Å directly across the cleft from Asp 278, which is coordinating the metal. The location of Mtb Ser 97 and Mtb Asp 278 across from one another, with no interference from any other residues, suggests that a similar regulatory mechanism, in which Asp 278 and Ser 97 could interact in a more compact conformation, is at least feasible. However, in our structure the regulatory segment exists as a helix without isocitrate bound. If this NADPH bound Mtb ICDH-1 structure represents an open conformation, an analogous Hc ICDH based regulatory mechanism would have to be tweaked so that this interaction occurred in a semi-open form, as Asp 278 and Ser 97 are currently too far away to interact in the Mtb structure. Three phosphorylation sites in Mtb ICDH-1 have been proposed using sequence-based computational tools(39), but to date there is no experimental evidence for phosphorylation. Neither the E. coli nor the Hc ICDH regulatory mechanisms are fully supported by this Mtb structure, suggesting Mtb ICDH-1 may be regulated by a novel mechanism.
The R132H mutation is responsible for the secondary activity of Hc ICDH converting αKG to α-hydroxyglutarate (αHG) in glioblastomas(40). R132 is conserved in all aligned ICDHs, and is equivalent to Mtb ICDH-1 R135. In our crystal structure the guanidinium nitrogens of R135 are situated 5.4 Å and 6.5 Å away from the Mn2+, in the cavity that we propose where isocitrate binds. This is not surprising, since human R132 is proposed to be involved in isocitrate binding and the transition to another conformation(41). Complexes of Mtb ICDH-1 with a catalytic or dead end complex, biochemical studies, and mutagenesis would be necessary to determine the exact role of R135 in Mtb ICDH-1.
Secondary Reaction
In 2009 Dang et al.(40) found that mutant forms of Hc ICDH identified in glioblastomas produced αHG. Subsequent studies by Pietrak et al.(42) found that αHG is also produced, albeit much slower, by the WT Hc ICDH and WT and mutant ICDH from Azotobacter vinelandii. This secondary reaction was also observed for WT Mtb ICDH-1 when αKG and NADPH were used as substrates. Subsequently, the formation of αHG was confirmed by mass spectrometry (Figure S2). Preliminary kinetic values were obtained: kcat ~ 0.1 - 0.2 sec-1, KαKG ~ 8 mM, KNADPH < 4 μM (see Figure S3 for measurements and associated errors). We are calling these kinetic parameters preliminary because we could not saturate with αKG and no MnCl2 was added. Addition of MnCl2 gave a nonlinear rate, presumably by coordinating αKG. Running the normal reaction without exogenous metal substantially decreased kcat (Table S1). These kinetic parameters support the mass spectrometry evidence for the enzymatic conversion of αKG and NADPH to αHG and NADP+. A detailed kinetic analysis of this reaction would be an intriguing follow-up. The significance of this reaction for WT Mtb ICDH-1 is unknown at this time.
Proposed Mechanism
As shown in Scheme 2A, isocitrate and NADP+ are both bound to the enzyme before the reaction can commence. Based on the pH-rate profile in which log kcat/Kisocitrate has an increase in rate at pH 5.7 (Figure 1B), and decreased activity without a divalent metal, isocitrate is proposed to coordinate the Mn2+ through its C1 carboxyl and C2 hydroxyl groups. There is a base assisted deprotonation from the C2 isocitrate hydroxyl, by the lysine group that is deprotonated by an aspartate residue whose pKa (Figure 1A, 1B) is observed in the log kcat pH-rate profile. Deprotonation of the C2 isocitrate hydroxyl is coupled to hydride transfer from C2 of isocitrate to the nicotinamide ring to generate the A-side (4R) product NADPH that remains bound on the enzyme (Scheme 2A, Figure S1). The intermediate oxalosuccinate (Scheme 2B) undergoes decarboxylation to generate CO2 and form the metal stabilized enolate depicted in Scheme 2C. The enolate is then protonated at the C3 position, by a catalytic acid, likely Tyr, bearing the solvent exchangeable proton giving rise to the large solvent KIE, to yield product αKG (Scheme 2D).
Conclusion
We have provided kinetic and structural evidence for the kinetic and chemical mechanism of Mtb ICDH-1. ICDH-1 catalyzes a sequential reaction in which NADP+ and isocitrate bind to the enzyme to form a ternary complex before conversion to product. Substrate binding is random, but NADP+ preferentially binds first. Product release is ordered with CO2 released first, followed by αKG, and finally NADPH. The first use of multiple KIE studies for any ICDH determined that protonation of the enolate of αKG, and not the initial deprotonation of the secondary alcohol, as reported for porcine heart ICDH, is the origin of the large solvent KIE. This suggests that catalysis is different in Mtb ICDH-1. The first X-ray crystallographic structure of Mtb ICDH-1 shows remarkable similarity to Hc ICDH, making it unlikely that regulation of this enzyme proceeds in the same manner as E. coli. Further similarity to Hc ICDH was observed by the presence of a secondary reaction to form αHG. The expression, purification, and crystallization conditions of Mtb ICDH-1 presented in this work should facilitate the structure determination of Mtb ICDH-1 in complex with Mn2+-isocitrate, Mn2+-isocitrate-NADPH, or an inhibitory or dead-end complex. All of these complexes are necessary in studying the regulatory mechanism of Mtb ICDH-1 through a structural approach. The biochemical and structural studies in this paper have allowed us to propose a chemical mechanism for Mtb ICDH-1. This is a critical building block for the full comprehension of the persistence phase of Mtb and for inhibitor development against the nonreplicating bacilli.
Supplementary Material
AWKNOWLEDGEMENTS
We would like to thank Dr. Hui Xiao for performing the mass spectrometric identification of αHG, and Dr. Michael Brenowitz for performing analytical ultracentrifugation studies not included in this paper. We would like to thank members of the laboratory of Dr. Steven Almo, particularly Dr. Matthew Vetting, Rafael Toro, and Rahul Bhosle for their helpful suggestions and expertise.
This work was supported by the National Institutes of Health Grant AI33696 to J.S.B. and T32 AI007501 to C.E.Q.
Abbreviations
- DTT
dithiothreitol
- EDTA
ethylenediaminetetraacetic acid
- HEPES
N-(2-hydroxyethyl)piperazine-N’-(2-ethanesulfonic acid)
- IPTG
isopropyl β-d-thiogalactopyranoside
- LB
Luria broth
- MES
2-(N-morpholino)ethanesulfonic acid
- MWCO
molecular weight cut off
- Ni-NTA
nickel nitriloacetic acid
- NMR
nuclear magnetic resonance
- PCR
polymerase chain reaction
- PDB
protein data bank
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- Tris
tris(hydroxymethyl)aminomethane
- WT
wildtype
Footnotes
SUPPORTING INFORMATION
A table of rates using different divalent cations (Table S1), the reaction scheme of [2R-2H]isocitrate synthesis (Scheme S1), product identification of [4R-4-2H]NADPH by NMR (Figure S1) and αHG by mass spectrometry (Figure S2), kinetics of the secondary reaction (Figure S3), a sequence alignment of other ICDHs (Figure S4), and NADPH-Mtb ICDH-1 contacts (Figure S5) are available in the supplemental material. Supplemental materials may be accessed free of charge online at http://pubs.acs.org.
Coordinates have been deposited under access code PDB ID: 4HCX
References
- 1.Kondrashov FA, Koonin EV, Morgunov IG, Finogenova TV, Kondrashova MN. Evolution of glyoxylate cycle enzymes in Metazoa: evidence of multiple horizontal transfer events and pseudogene formation. Biol Direct. 2006;1:31. doi: 10.1186/1745-6150-1-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dunn MF, Ramirez-Trujillo JA, Hernandez-Lucas I. Major roles of isocitrate lyase and malate synthase in bacterial and fungal pathogenesis. Microbiology. 2009;155:3166–3175. doi: 10.1099/mic.0.030858-0. [DOI] [PubMed] [Google Scholar]
- 3.McKinney JD, Honer zu Bentrup K, Munoz-Elias EJ, Miczak A, Chen B, Chan WT, Swenson D, Sacchettini JC, Jacobs WR, Jr., Russell DG. Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature. 2000;406:735–738. doi: 10.1038/35021074. [DOI] [PubMed] [Google Scholar]
- 4.Zhang Y. The magic bullets and tuberculosis drug targets. Annual review of pharmacology and toxicology. 2005;45:529–564. doi: 10.1146/annurev.pharmtox.45.120403.100120. [DOI] [PubMed] [Google Scholar]
- 5.LaPorte DC, Chung T. A single gene codes for the kinase and phosphatase which regulate isocitrate dehydrogenase. J Biol Chem. 1985;260:15291–15297. [PubMed] [Google Scholar]
- 6.Garnak M, Reeves HC. Phosphorylation of Isocitrate dehydrogenase of Escherichia coli. Science. 1979;203:1111–1112. doi: 10.1126/science.34215. [DOI] [PubMed] [Google Scholar]
- 7.LaPorte DC, Koshland DE., Jr. A protein with kinase and phosphatase activities involved in regulation of tricarboxylic acid cycle. Nature. 1982;300:458–460. doi: 10.1038/300458a0. [DOI] [PubMed] [Google Scholar]
- 8.Banerjee S, Nandyala A, Podili R, Katoch VM, Hasnain SE. Comparison of Mycobacterium tuberculosis isocitrate dehydrogenases (ICD-1 and ICD-2) reveals differences in coenzyme affinity, oligomeric state, pH tolerance and phylogenetic affiliation. BMC Biochem. 2005;6:20. doi: 10.1186/1471-2091-6-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tian J, Bryk R, Shi S, Erdjument-Bromage H, Tempst P, Nathan C. Mycobacterium tuberculosis appears to lack alpha-ketoglutarate dehydrogenase and encodes pyruvate dehydrogenase in widely separated genes. Mol Microbiol. 2005;57:859–868. doi: 10.1111/j.1365-2958.2005.04741.x. [DOI] [PubMed] [Google Scholar]
- 10.Wagner T, Bellinzoni M, Wehenkel A, O'Hare HM, Alzari PM. Functional plasticity and allosteric regulation of alpha-ketoglutarate decarboxylase in central mycobacterial metabolism. Chemistry & biology. 2011;18:1011–1020. doi: 10.1016/j.chembiol.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 11.Baughn AD, Garforth SJ, Vilcheze C, Jacobs WR., Jr. An anaerobic-type alpha-ketoglutarate ferredoxin oxidoreductase completes the oxidative tricarboxylic acid cycle of Mycobacterium tuberculosis. PLoS Pathog. 2009;5:e1000662. doi: 10.1371/journal.ppat.1000662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rhee KY, de Carvalho LP, Bryk R, Ehrt S, Marrero J, Park SW, Schnappinger D, Venugopal A, Nathan C. Central carbon metabolism in Mycobacterium tuberculosis: an unexpected frontier. Trends Microbiol. 2011;19:307–314. doi: 10.1016/j.tim.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Carvalho LP, Ling Y, Shen C, Warren JD, Rhee KY. On the chemical mechanism of succinic semialdehyde dehydrogenase (GabD1) from Mycobacterium tuberculosis. Archives of biochemistry and biophysics. 2011;509:90–99. doi: 10.1016/j.abb.2011.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Carvalho LP, Fischer SM, Marrero J, Nathan C, Ehrt S, Rhee KY. Metabolomics of Mycobacterium tuberculosis reveals compartmentalized co-catabolism of carbon substrates. Chemistry & biology. 2010;17:1122–1131. doi: 10.1016/j.chembiol.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 15.WHO Tuberculosis, Fact sheet 104. 2012 http://www.who.int/mediacentre/factsheets/fs104/en/index.html.
- 16.Banerjee S, Nandyala A, Podili R, Katoch VM, Murthy KJ, Hasnain SE. Mycobacterium tuberculosis (Mtb) isocitrate dehydrogenases show strong B cell response and distinguish vaccinated controls from TB patients. Proc Natl Acad Sci U S A. 2004;101:12652–12657. doi: 10.1073/pnas.0404347101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.ExPASy ProtParam Tool. 2011 Jun 15; http://ca.expasy.org/tools/protparam.html.
- 18.The Merck Index. Fourteenth Edition Merck & Co., Inc.; Whitehouse Station, NJ, USA: 2006. [Google Scholar]
- 19.Lobell M, Crout DHG. New insight into the pyruvate decarboxylase-catalysed formation of lactaldehyde from H-D exchange experiments: a ‘water proof’ site. J. Chem. Soc., Perkin Trans. 1996;1:1577–1581. [Google Scholar]
- 20.Karsten WE, Lai CJ, Cook PF. Inverse Solvent Isotope Effects in the NAD-Malic Enzyme Reaction Are the Result of the Viscosity Difference between D2O and H2O: Implications for Solvent Isotope Effect Studies. Journal of the American Chemical Society. 1995;117:5914–5918. [Google Scholar]
- 21.Otwinowski Z, Minor W. [20] Processing of X-ray diffraction data collected in oscillation mode. In: Carter Charles W., Jr., editor. Methods in Enzymology. Academic Press; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 22.Adams PD, Gopal K, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Pai RK, Read RJ, Romo TD, Sacchettini JC, Sauter NK, Storoni LC, Terwilliger TC. Recent developments in the PHENIX software for automated crystallographic structure determination. Journal of synchrotron radiation. 2004;11:53–55. doi: 10.1107/s0909049503024130. [DOI] [PubMed] [Google Scholar]
- 23.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta crystallographica. Section D, Biological crystallography. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 24.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; San Carlos, CA: 2002. [Google Scholar]
- 25.Uhr ML, Thompson VW, Cleland WW. The kinetics of pig heart triphosphopyridine nucleotide-isocitrate dehydrogenase. I. Initial velocity, substrate and product inhibition, and isotope exchange studies. J Biol Chem. 1974;249:2920–2927. [PubMed] [Google Scholar]
- 26.Northrop DB, Cleland WW. The kinetics of pig heart triphosphopyridine nucleotide-isocitrate dehydrogenase. II. Dead-end and multiple inhibition studies. J Biol Chem. 1974;249:2928–2931. [PubMed] [Google Scholar]
- 27.Dean AM, Koshland DE., Jr. Kinetic mechanism of Escherichia coli isocitrate dehydrogenase. Biochemistry. 1993;32:9302–9309. doi: 10.1021/bi00087a007. [DOI] [PubMed] [Google Scholar]
- 28.Aktas DF, Cook PF. A lysine-tyrosine pair carries out acid-base chemistry in the metal ion-dependent pyridine dinucleotide-linked beta-hydroxyacid oxidative decarboxylases. Biochemistry. 2009;48:3565–3577. doi: 10.1021/bi8022976. [DOI] [PubMed] [Google Scholar]
- 29.Kim TK, Lee P, Colman RF. Critical role of Lys212 and Tyr140 in porcine NADP-dependent isocitrate dehydrogenase. J Biol Chem. 2003;278:49323–49331. doi: 10.1074/jbc.M303781200. [DOI] [PubMed] [Google Scholar]
- 30.Cook PF, Cleland WW. pH variation of isotope effects in enzyme-catalyzed reactions. 1. Isotope- and pH-dependent steps the same. Biochemistry. 1981;20:1797–1805. doi: 10.1021/bi00510a014. [DOI] [PubMed] [Google Scholar]
- 31.Colman RF, Chu R. Deuterium solvent isotope effects in reactions catalyzed by isocitrate dehydrogenase. Biochemical and biophysical research communications. 1969;34:528–535. doi: 10.1016/0006-291x(69)90414-8. [DOI] [PubMed] [Google Scholar]
- 32.Hermes JD, Roeske CA, O'Leary MH, Cleland WW. Use of multiple isotope effects to determine enzyme mechanisms and intrinsic isotope effects. Malic enzyme and glucose-6-phosphate dehydrogenase. Biochemistry. 1982;21:5106–5114. doi: 10.1021/bi00263a040. [DOI] [PubMed] [Google Scholar]
- 33.Nelson DL, Cox MM. Lehninger Principles of Biochemistry. Fourth ed. W. H. Freeman and Company; New York: 2005. [Google Scholar]
- 34.Ottolina G, Riva S, Carrea G, Danieli B, Buckmann AF. Enzymatic synthesis of [4R-2H]NAD (P)H and [4S-2H]NAD(P)H and determination of the stereospecificity of 7 alpha- and 12 alpha hydroxysteroid dehydrogenase. Biochimica et biophysica acta. 1989;998:173–178. doi: 10.1016/0167-4838(89)90270-7. [DOI] [PubMed] [Google Scholar]
- 35.Thorsness PE, Koshland DE., Jr. Inactivation of isocitrate dehydrogenase by phosphorylation is mediated by the negative charge of the phosphate. J Biol Chem. 1987;262:10422–10425. [PubMed] [Google Scholar]
- 36.Hurley JH, Dean AM, Sohl JL, Koshland DE, Jr., Stroud RM. Regulation of an enzyme by phosphorylation at the active site. Science. 1990;249:1012–1016. doi: 10.1126/science.2204109. [DOI] [PubMed] [Google Scholar]
- 37.Goncalves S, Miller SP, Carrondo MA, Dean AM, Matias PM. Induced Fit and the Catalytic Mechanism of Isocitrate Dehydrogenase. Biochemistry. 2012 doi: 10.1021/bi300483w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu X, Zhao J, Xu Z, Peng B, Huang Q, Arnold E, Ding J. Structures of human cytosolic NADP-dependent isocitrate dehydrogenase reveal a novel self-regulatory mechanism of activity. J Biol Chem. 2004;279:33946–33957. doi: 10.1074/jbc.M404298200. [DOI] [PubMed] [Google Scholar]
- 39.Vinekar R, Ghosh I. Determination of phosphorylation sites for NADP-specific isocitrate dehydrogenase from mycobacterium tuberculosis. J Biomol Struct Dyn. 2009;26:741–754. doi: 10.1080/07391102.2009.10507286. [DOI] [PubMed] [Google Scholar]
- 40.Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, Marks KM, Prins RM, Ward PS, Yen KE, Liau LM, Rabinowitz JD, Cantley LC, Thompson CB, Vander Heiden MG, Su SM. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang B, Zhong C, Peng Y, Lai Z, Ding J. Molecular mechanisms of “off-on switch” of activities of human IDH1 by tumor-associated mutation R132H. Cell research. 2010;20:1188–1200. doi: 10.1038/cr.2010.145. [DOI] [PubMed] [Google Scholar]
- 42.Pietrak B, Zhao H, Qi H, Quinn C, Gao E, Boyer JG, Concha N, Brown K, Duraiswami C, Wooster R, Sweitzer S, Schwartz B. A tale of two subunits: how the neomorphic R132H IDH1 mutation enhances production of alphaHG. Biochemistry. 2011;50:4804–4812. doi: 10.1021/bi200499m. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.