Abstract
The Rb/E2F pathway is deregulated in virtually all human tumors. It is clear that, in addition to Rb itself, essential co-factors required for transcriptional repression and silencing of E2F target genes are mutated or lost in cancer. To identify novel co-factors required for Rb/E2F-mediated inhibition of cell proliferation, we performed a genome-wide shRNA screen. In addition to several known Rb co-factors, the screen identified components of the Mediator complex, a large multiprotein coactivator required for RNA polymerase II transcription. We show that the Mediator complex subunit MED13L is required for Rb/E2F control of cell growth, the complete repression of cell cycle target genes, and cell cycle inhibition.
Introduction
The retinoblastoma tumor suppressor (Rb) is a critical negative regulator of cellular proliferation that is frequently inactivated in human cancer (1, 2). In its active form, Rb binds to members of the E2F family of transcription factors and either suppresses their transactivation function or assembles an active repressor complex (3). E2F family members are essential regulators of genes required for DNA replication and mitosis (4-6). The transcriptional repression functions of Rb/E2F complexes have been shown to reflect the recruitment of transcriptional co-repressors (3, 7-12). In this manner, multi-protein transcriptional repressor complexes are guided to specific genes via the E2F binding sites in target gene promoters.
The ability of Rb to bind stably to E2F is regulated by phosphorylation. In response to mitogenic stimuli in G0, cyclin D/CDK4(6) complexes initiate the phosphorylation of Rb and disrupt Rb/E2F interactions (1, 5, 6). The alleviation of Rb-mediated repression of E2F enables activated transcription of requisite cell cycle genes and passage through the G1/S-transition. Deregulated Rb/E2F control of cell proliferation is a common feature of cancer and is underscored by the multitude of observed lesions in human tumors (1, 2, 6). Loss of Rb, loss of the CDK4(6) inhibitor, CDKN2A, and amplification/overexpression of CDK4 or cyclin D are examples of disruptions of cell growth control seen in human tumors. Importantly, the loss of key mediators of transcriptional repression and gene silencing has also been shown to confer resistance to Rb/E2F-mediated cell cycle arrest and been observed in human tumors (13, 14).
Previous studies from multiple independent laboratories have clearly implicated the Rb/E2F pathway in the maintenance of a stable cell cycle arrest state and the senescent phenotype in human cells (15). Cellular senescence was initially observed as a barrier limiting the proliferative capacity of cells in culture (16). In cultured human cells, the erosion of telomeres at chromosome ends leads to a p53-dependent response that arrests cell cycle progression (15, 17, 18). In addition to telomere shortening, senescence has been shown to be induced by oncogenic signaling, non-telomeric DNA damage, and other cellular stresses (15, 19). Typically, the levels of CDKN2A are greatly increased, leading to accumulation of active, hypophosphorylated Rb. Senescent cells characteristically exhibit flat-cell morphology and are positive for senescence-associated beta-galactosidase (SA-β-gal) activity (20). Additionally, Narita et al. (2003) described the formation of senescence-associated heterochromatic foci (SAHF) at E2F promoters during RasV12-induced senescence. Overexpression of the adenoviral E1A protein, which targets Rb and its related family members, bypassed the senescent phenotype, alleviated E2F target gene repression, and blocked formation of SAHF (21). The evidence for senescence as a relevant tumor suppressive mechanism in vivo is compelling (15, 22-24). Multiple independent investigators have observed senescence markers in premalignant murine lesions induced by activated oncogenes (25-29). Furthermore, human benign melanocytic nevi, premalignant lesions in the development of melanoma, possess multiple characteristics of a senescent phenotype (29). The loss of cooperating factors required for transcriptional repression and heterochromatin formation initiated by Rb/E2F complexes could represent a mechanism for the progression from benign neoplasia to more advanced malignancies. Here, we performed a genome-wide shRNA screen to identify factors critical in the induction and maintenance of the Rb/E2F-induced growth arrest state.
Results
A screen for Rb/E2F co-factors that maintain cell cycle arrest and senescence induction
In an effort to identify essential cofactors in the maintenance of Rb/E2F-mediated cellular senescence, an allele of the large pocket domain of Rb (amino acids 379-928) that harbors mutations in critical serine and threonine residues and is thereby refractory to CDK phosphorylation and inactivation (phosphorylation-site mutant-Rb, PSM-Rb) was expressed in the T98G glioblastoma cell line via retroviral transduction. The large pocket domain of Rb has been demonstrated to be the minimal domain required for E2F binding and growth suppression and for tumor suppression in mouse models (30, 31). Previous studies have demonstrated that this mutant form of Rb represses transcription of E2F target genes and strongly enforces cell cycle arrest (32, 33). Indeed, cells infected with PSM-Rb-producing retrovirus fail to undergo DNA replication, as measured by BrdU incorporation when compared to control cells (Fig. 1A). Consistent with prior observations, chronic PSM-Rb expression elicited stable growth arrest and characteristics of the senescent phenotype. After seven days of hygromycin selection, population of cells expressing PSM-Rb display a flat-cell morphology and are positive for senescence-associated β-galactosidase (SA- β-gal) activity (Fig. 1B).
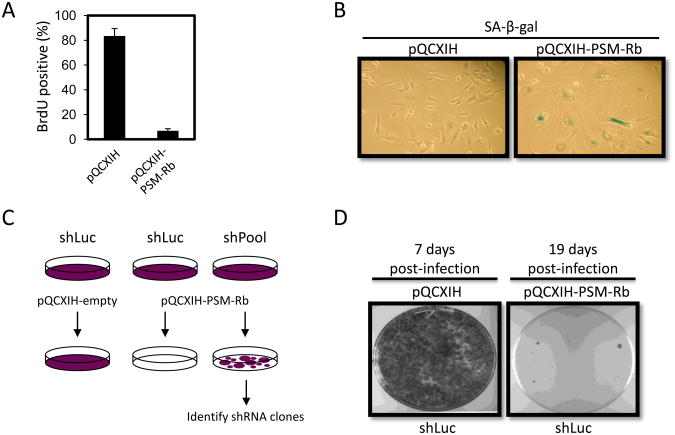
Figure 1. shRNA screen design: Active Rb expression induces stable cell cycle arrest and senescent phenotype.
(A) T98G glioblastoma cells expressing the murine ecotropic receptor (T98G-EcoR) were infected with empty retrovirus or retrovirus producing a PSM-Rb. Three days post-selection in hygromycin, cells were labeled with BrdU for 16 h, fixed and BrdU incorporation was detected by immunofluorescence. At least 200 cells were counted per condition. Bar graphs represent mean ± S.D. from two independent experiments for PSM-Rb. (B) T98G-EcoR cells treated as in (A) were stained to detect SA-beta-galatosidase activity. (C) The shRNA screen was designed to identify cells resistant to PSM-Rb expression in the shPool populations as compared to the control, shLuc (luciferase). (D) T98G-EcoR cells stably producing shRNA targeting firefly luciferase were infected with empty retrovirus or retrovirus producing PSM-Rb. Either seven days (empty retrovirus) or 19 days (PSM-Rb retrovirus) post-selection in hygromycin, dishes were fixed in methanol, stained with crystal violet, and images were captured.
Based on the stability of the arrest state induced by PSM-Rb expression, we designed a protocol for a genome-wide short hairpin RNA (shRNA) screen to identify factors required for the induction and maintenance of the arrest phenotype (Fig. 1C). The retroviral vector, pQCXIH, produces PSM-Rb and the hygromycin resistance gene on a bicistronic transcript, ensuring that hygromycin-resistant cells have not lost PSM-Rb expression. The use of the PSM-Rb allele was intended to reduce the complexity of hits from the screen by (1) eliminating factors that might affect Rb phosphorylation, thereby targeting components of the active Rb/E2F repressor complex, and (2) take advantage of the efficacy of PSM-Rb and the reduced background. Using the Open Biosystems human pSM2c shRNAmir library (version 1.15), thirty-two pools of shRNA constructs (∼2000 shRNA constructs per pool) were introduced via retroviral transduction into T98G cells expressing the murine ecotropic receptor (T98G-EcoR). As a control, a polyclonal population of T98G-EcoR cells was generated expressing an shRNA targeting firefly luciferase. Following the introduction of PSM-Rb into the thirty-two polyclonal T98G-EcoR shRNA populations, cells evading cell cycle arrest and senescence induction could be harvested and specific shRNA sequences identified by PCR and sequencing (Fig. 1C). As predicted, T98G-EcoR cells infected with an shRNA targeting luciferase (shLuc) formed very few colonies upon PSM-Rb introduction compared to control retrovirus (Fig. 1D).
Following the infection of T98G-EcoR populations harboring pools of shRNA clones, cells were selected with hygromycin for 19-21 days. As shown in Figure 2A, the proliferation of shLuc cells was inhibited, while colony outgrowth could be observed more readily in the pooled shRNA populations. Cells were harvested from each shRNA pool population and genomic DNA was prepared. PCR was performed to amplify candidate shRNAs and the resultant fragments were cloned and sequenced. Twenty to thirty sequencing reactions were performed from each shRNA pool (Fig. 2B) and two hundred and ninety-three specific candidate gene targets were identified by Human BLAT Search of the UCSC Genome Browser (www.genome.ucsc.edu) (Supplementary Table 1).
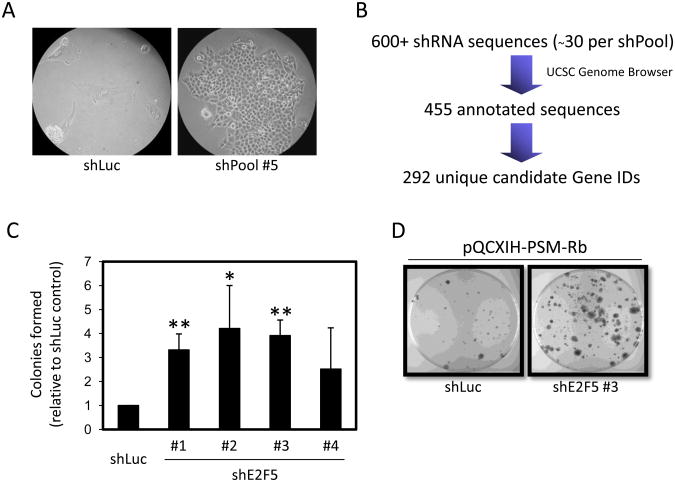
Figure 2. shRNA primary screen and validation identifies E2F5.
(A) T98G-EcoR cells expressing shLuc or expressing a pool of shRNAs from the Open Biosystems human shRNA library were infected with retrovirus producing PSM-Rb. Nineteen days post-selection in hygromycin, representative images of shLuc cells and shPool #5 cells were collected. (B) Cells that bypassed growth arrest were collected from the shPool populations and the shRNAs were identified by sequencing. UC Santa Cruz Genome browser was used to confirm the identity and validity of each shRNA sequence. In sum, 293 unique genes were identified from the primary screen. (C) T98G-EcoR cells expressing unique shRNAs targeting E2F5 or an shRNA targeting luciferase (shLuc) were generated by retroviral infection and selection. Polyclonal populations were subsequently infected with retrovirus producing PSM-Rb, selected in hygromycin for 19 days, methanol fixed, and stained with crystal violet. Colonies were counted and graphed relative to the control shLuc population. The mean ± S.D. is shown for at least two independent experiments. * indicates p<0.05. ** indicates p<0.01. (D) Representative images of dishes of shLuc cells and shE2F5 #3 as described in (C).
Validation of the screen: Identification of E2F5
With the expectation that the candidate genes included false positives, we initially sought to validate genes known to function in the Rb/E2F pathway. Confirming the success of the shRNA screen design, two sequences were recovered targeting the E2F family member E2F5. E2F5 is known to function with Rb family members to suppress transcription of cell cycle regulated genes (4). To validate these findings, several individual shRNA constructs targeting E2F5 were utilized in the generation of T98G-EcoR polyclonal populations and infected with retroviruses producing PSM-Rb (Fig. 2C). Consistent with the isolation of E2F5 shRNA clones from the screen, multiple shRNA sequences targeting E2F5 provided a growth advantage in the presence of PSM-Rb as compared to control cell populations. As shown in Figure 2C and D, cells expressing an shRNA targeting E2F5 formed more colonies in the presence of PSM-Rb expression when compared to cells expressing shLuc.
ZNF217 links Rb/E2F to known transcriptional repressor and histone-modifying complexes
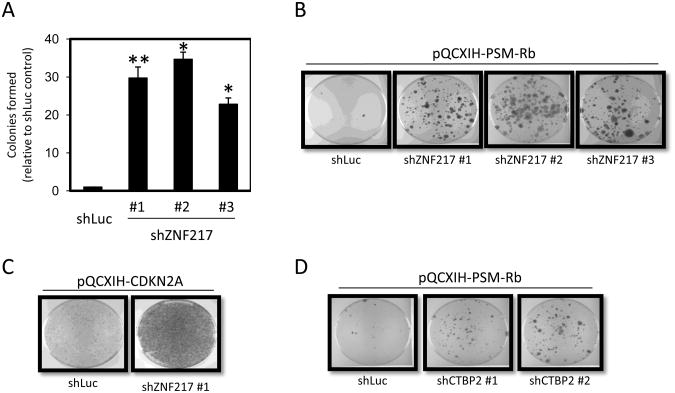
A further inspection of the candidate gene list provided by the screen identified two independent sequences targeting the zinc finger protein, ZNF217 (Supplementary Table 1). Biochemical studies have revealed that ZNF217 exists in large transcriptional repressor complexes with CoREST, histone deacetylases, SIN3B, and CtBP (34, 35). Previous studies have identified CtIP, a protein known to interact with CtBP, as a direct interaction partner with the pocket proteins, p130 and Rb (36, 37). The ability of CtBP to repress transcription suggested that Rb/E2F might recruit CtBP-containing complexes via an interaction with CtIP. To confirm that the loss of ZNF217 impairs the function of PSM-Rb in eliciting cell cycle arrest and senescence, we generated cells expressing shRNA sequences targeting ZNF217. As shown in Figure 3A and B, shZNF217 cell populations formed more colonies in the presence of PSM- Rb than shLuc control cells. As prior studies had identified Rb and p130 as interacting partners of CtIP, we utilized the CDK inhibitor, p16ink4a, to assess its potential for inducing growth arrest. The expression of p16ink4a blocks CDK4/6 activity and leads to the accumulation of endogenous pocket proteins (RB and p130) in their hypophosphorylated, active forms. As shown in Figure 3C, shZNF217 cells exhibited substantially greater resistance to p16ink4a expression relative to shLuc cells. To lend further credence to the involvement of the ZNF217 complex in the Rb/E2F-induced senescent phenotype, we established two polyclonal cell lines expressing shRNAs targeting CtBP2, as a core ZNF217 complex has been reported to co-purify with CtBP2 (38). As shown in Figure 3D, T98G-EcoR cells expressing shCtBP2 constructs formed colonies in the presence of PSM-Rb expression when compared to shLuc control cells. These data provide evidence that the ZNF217 repressor complex is required for Rb/E2F-mediated cell cycle arrest and senescence.
Figure 3. ZNF217 is required for Rb/E2F-mediated growth arrest.
(A) T98G-EcoR cells expressing unique shRNAs targeting ZNF217 or shRNA targeting luciferase (shLuc) were generated by retroviral infection and selection. Polyclonal populations were subsequently infected with retrovirus producing PSM-Rb, selected in hygromycin for 19 days, fixed, and stained with crystal violet. Colonies were counted and graphed relative to the control shLuc population. The mean ± S.D. is shown for at least two independent experiments. * indicates p<0.05. ** indicates p<0.01. (B) Representative dishes of shLuc and shZNF217 populations as described in (A). (B) T98G-EcoR cells expressing an shRNA targeting luciferase (shLuc) or ZNF217 were generated by retroviral infection and selection. Polyclonal populations were subsequently infected with retrovirus producing p16ink4a, selected in hygromycin for 7 days, fixed, and stained with crystal violet. (D) T98G-EcoR cells expressing shRNA targeting luciferase (Luc) or unique shRNAs targeting CtBP2 were generated by retroviral infection and selection. Polyclonal populations were subsequently infected with retrovirus producing PSM-Rb, selected in hygromycin for 19 days, fixed, and stained with crystal violet.
Discovery of novel RB/E2F co-factors – identification of Mediator complex subunits
The identification of E2F5 and ZNF217 demonstrated that the screen had the capacity to identify relevant functional Rb co-factors. However, the value of a genome-wide screen is the opportunity for discovery of novel co-factors with unexpected functional associations with Rb. As Rb/E2F recruits large, multi-subunit complexes such as SWI/SNF to repress transcription, we hypothesized that individual hits from the screen might represent subunits of a common module. To identify such complexes, we made use of the Ingenuity Pathways Analysis tools to scrutinize the candidate gene products (see Experimental Procedures). The analysis provided an unbiased means by which to interpret the data and potentially identify novel biological relationships that could be the basis for further study. Inspection of the individual networks revealed a highlyinterconnected hub containing six unique gene products that were each identified as independent hits in the shRNA screen (Fig. 4A). Interestingly, the interactions observed between the six gene products (MED13L, MED27, DDX52, ZCCHC10, TARDBP, and DNAJA1) were based upon a study designed to elucidate the subunit composition of the Mediator complex (39). The Mediator complex is comprised of 26-30 subunits and is collectively required for the activation of RNA polymerase II-dependent transcription (40, 41). While MED13L and MED27 are bona fide components of the Mediator complex, the possible role of DDX52, ZCCHC10, TARDBP, and DNAJA1 in Mediator function is unclear. Nevertheless, this analysis did suggest the potential role of Mediator complex components in Rb/E2F-mediated cell growth control.
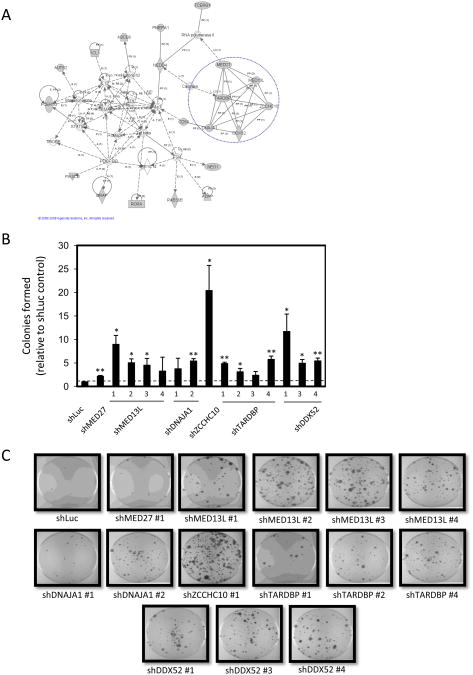
Figure 4. Involvement of Mediator complex subunits in Rb/E2F-mediated growth arrest.
(A) A highly interconnected node of candidate genes identified in the screen was revealed by Ingenuity Pathways Analysis. One identified network is shown. Shading indicates gene products identifed in the shRNA screen. MED13L, MED27, ZCCHC10, DDX52, TARDBP, and DNAJA1 were connected by reciprocal protein-protein interactions. (B) T98G-EcoR cells expressing shRNA sequences targeting luciferase (shLuc) or shRNAs targeting MED27, MED13L, DNAJA1, ZCCHC10, TARDBP, or DDX52 were generated by retroviral infection and selection. Polyclonal populations were subsequently infected with retrovirus producing PSM-Rb, selected in hygromycin for 19 days, fixed, and stained with crystal violet. The number of colonies formed was counted and graphed relative to the control (shLuc) plate. The mean ± S.D. is shown for two independent experiments. * indicates p<0.05. ** indicates p<0.01. (C) Representative images collected from dishes of cells as described in (B).
Initially, we sought to validate that knockdown of MED13L and MED27, as well as DDX52, ZCCHC10, TARDBP, and DNAJA1, conferred resistance to RB/E2F-induced growth arrest. Individual shRNAs targeting these gene products were obtained from the Open Biosystems library, introduced into T98G-EcoR cells, and the resultant populations were subsequently infected with PSM-Rb (Figure 4B and C). While additional shRNAs were not available for MED27 and ZCCHC10, multiple shRNA sequences targeting the Mediator complex associated proteins conferred resistance to the proliferation arrest induced by active Rb expression (Fig. 4B and C). Collectively, these findings confirm the role of multiple subunits of the Mediator complex (MED13L and MED27) in Rb/E2F-induced growth inhibition.
MED13L is a functional Rb/E2F co-factor
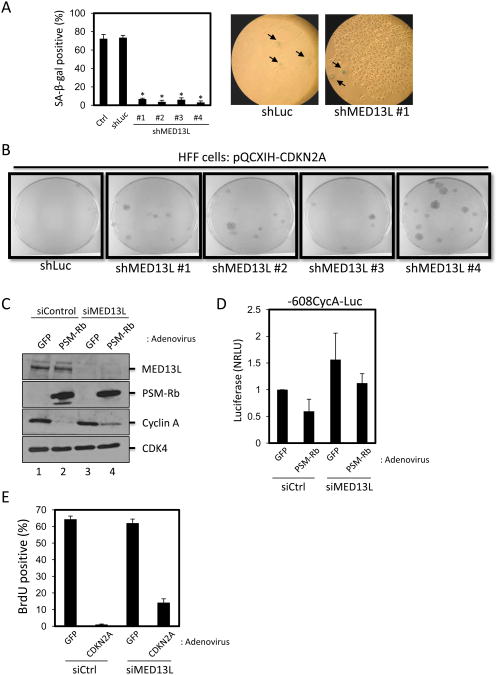
The various Mediator subunits have been grouped into structural modules: head, middle, tail, and kinase modules. The kinase, or CDK8, module contains the MED12, MED13, CDK8, and Cyclin C subunits (40, 41). The function of MED13L is less understood, but sequence homology and biochemical data support its classification as a Mediator subunit (39, 42). We first sought to determine whether the knockdown of MED13L might be required for the induction of the senescent phenotype by Rb/E2F. T98G cells (control) and polyclonal population of T98G cells expressing a control shRNA or shRNA sequences targeting MED13L were infected with retrovirus producing PSM-Rb. Approximately three weeks after infection, the cells were fixed and processed to detect SA-β-gal activity (Fig. 5A). Consistent with expectation, the colonies formed in shMED13L T98G populations were negative for SA-β-gal activity. To confirm the requirement for MED13L in Rb/E2F-mediated growth arrest in normal cells, we introduced the shLuc and shMED13L constructs into a diploid fibroblast cell line (human foreskin fibroblast, HFF) via retroviral transduction. Cells were then infected with retrovirus producing CDKN2A and selected with hygromycin for 19 days. Strikingly, each of the shMED13L HFF populations exhibited resistance to CDKN2A-mediated growth arrest when compared to shLuc cells (Fig. 5B).
Figure 5. MED13L is required for complete repression of Rb/E2F-regulated cell cycle targets and the inhibition of DNA synthesis.
(A) T98G-EcoR cells expressing shRNA sequences targeting luciferase (shLuc) or shRNAs targeting MED13L were infected with retrovirus producing PSM-Rb, selected in hygromycin for 19 days, fixed, and processed for SA-β-gal activity. Two hundred cells were scored for SA-β-gal staining. The mean ± S.D. is shown for two independent experiments. * indicates p<0.05. Representative images are shown at right. Arrows indicate SA-β-gal positive cells. (B) HFF cells expressing shRNA targeting luciferase (Luc) or MED13L were generated by retroviral infection and selection. Polyclonal populations were infected with retrovirus producing p16ink4a, selected in hygromycin for 19 days, fixed, and stained with crystal violet. (C) U2OS cells were transfected with siRNA pools, either a non-targeting siControl pool or a pool targeting MED13L. Forty-eight hours post-transfection, cells were infected with control adenovirus producing GFP or adenovirus producing GFP and PSM-Rb. Twenty-four hours post-infection, infected cells were harvested, whole cell lystates were prepared, and equal amounts of protein were separated by SDS-PAGE. The indicated proteins were detected by immunoblotting. CDK4 was used as a loading control. (D) U2OS cells were transfected with siRNA pools, either a non-targeting siControl pool or a pool targeting MED13L, Cyclin A-Luc reporter plasmid, and CMV-Renilla. Forty-eight hours post-transfection, cells were infected with control adenovirus producing GFP or adenovirus producing GFP and PSM-RB. Twenty-four hours post-infection, cells were harvested and luciferase activity was measured. Relative light units were normalized using Renilla luciferase activity (NRLU). (E) U2OS cells were transfected with siRNA pools, either a non-targeting siControl pool or a pool targeting MED13L. Forty-eight hours post-transfection, cells were infected with control adenovirus producing GFP or adenovirus producing GFP and CDKN2A. Twenty-four hours post-infection, cells were labeled with BrdU for 16 h. Cells were then fixed and BrdU incorporation was detected by immunofluorescence. At least 200 cells were counted per condition. Bar graphs represent mean ± S.D. from three independent experiments.
To determine whether MED13L played a functional role in PSM-Rb-mediated repression, we examined endogenous protein levels of cyclin A, a critical Rb/E2F target gene induced at G1/S of the cell cycle. Previous work has demonstrated that cyclin A overexpression could bypass the inhibition of DNA synthesis by PSM-Rb (43). U2OS cells were transfected with a control siRNA pool or an siRNA pool targeting MED13L. Forty-eight hours post-transfection, cells were infected with GFP or GFP and PSM-Rb adenovirus as previously described. Twenty-four hours post-infection, cells were harvested and protein levels were determined by SDS-PAGE and immunoblotting of whole cell lysates. Confirming the efficacy of the siRNA transfection, levels of MED13L protein were strongly reduced by the siMED13L pool relative to the siControl pool (Fig. 5C). Importantly, the knockdown of MED13L did not have an observable effect on the levels of CDK4 (loading control) or the ectopically-produced PSM-Rb protein. This suggested that the depletion of MED13L did not have a broad effect on cellular transcription. The expression of PSM-Rb in cells transfected with control siRNA resulted in the dramatic attenuation of cyclin A protein (Fig. 5C, compare lanes 1 and 2). In contrast, cells transfected with siRNA targeting MED13L retained detectable levels of cyclin A in the presence of PSM-Rb (Fig. 5C, compare lanes 2 and 4).
As further validation that MED13L functions as a requisite co-factor in the Rb/E2F-mediated repression of the cyclin A gene, we made use of a reporter construct under the control of the cyclin A promoter. Using the U2OS cell line, a control siRNA pool or an siRNA pool targeting MED13L, the reporter construct containing the cyclin A (-608CycA-Luc) promoter driving firefly luciferase expression, and a Renilla luciferase expression plasmid were introduced by transient transfection (Fig. 5D). Forty-eight hours later, cells were infected with adenovirus producing GFP as a control, or GFP and PSM-Rb. Twenty-four hours after adenoviral infection, luciferase activity was assessed. As shown in Figure 5D, while the cyclin A reporter activity could be repressed by PSM-Rb in the presence of control siRNA, the depletion of MED13L interfered with the attenuation of cyclin A activity in response to PSM-Rb. Taken together, these data demonstrate that MED13L is essential for the complete Rb/E2F-mediated attenuation of cyclin A promoter activity and endogenous protein level.
Given the data indicating a requirement of MED13L for Rb/E2F-mediated transcriptional repression of a G1/S gene encoding an important replication activity, we addressed the possibility that MED13L was similarly required by Rb to block cell cycle progression. As shown in Fig. 5E, U2OS cells transfected with siRNA pools targeting luciferase as a control or MED13L were subsequently infected with control adenovirus producing GFP or GFP and CDKN2A. Twenty four hours post-infection, cells were labeled with BrdU for 16 h to monitor DNA synthesis. Cells treated with control siRNA were strongly inhibited for BrdU incorporation by CDKN2A expression (Fig. 5E). In contrast, the siRNA-mediated knockdown of MED13L blocked the ability of CDKN2A to fully inhibit S-phase progression. These data indicate that the depletion of the Mediator subunit MED13L interferes with the function of the Rb/E2F pathway in the induction of senescence, transcriptional repression, and cell cycle arrest.
Discussion
The importance of cell proliferation control by the Rb/E2F pathway has been well established, as disruption of this control is common in human cancer. While Rb is frequently targeted in cancer, the loss of critical cofactors in Rb/E2F-mediated transcriptional repression has been observed in human tumors. For example, the SWI/SNF chromatin remodeling complex cooperates with Rb to repress transcription and loss or reduction of expression of multiple subunits has been reported in tumor samples (3, 14). In addition, co-factors that mediate the formation of heterochromatin at Rb/E2F target promoters, such as the histone methyltransferase Suv39H1, are important in tumor suppression. In Eμ-myc transgenic mice heterozygous for Suv39H1, T-cell lymphomas develop wherein SUV39H1 expression is lost (25). There is growing evidence that the senescent phenotype, initially observed in tissue culture cells, is induced in vivo in an Rb-dependent manner and acts as a protective barrier to tumor progression. A recent study has further illuminated the unique aspects of Rb function versus p107 and p130 in cellular senescence (44). Our intent here was to expand our understanding of Rb/E2F function so as to identify additional functional components that might be contributing to oncogenic phenotypes.
The design of our shRNA screen included the use of an ectopically produced phosphorylation-site mutant of Rb to elicit stable cell cycle arrest that exhibits a senescent phenotype. As demonstrated by the identification of E2F5, the screen was indeed effective in identifying critical Rb-binding partners. In addition, we identified multiple genes known to be important for the induction of cellular senescence, such as p27Kip1, CCAAT/enhancer binding protein beta, and RUNX1 (45-49) (Supplementary Table 1).
While we were able to validate the shRNA screen via confirmed hits with logical, or even well-established, roles in senescence induction and maintenance, further analysis uncovered novel links between Rb and repressor complexes. For example, ZNF217 has been purified in complexes with CtBP1, LSD1, CoREST, and other chromatin-modifying factors (34, 35, 50). Previous work from our laboratory had identified CtIP as an important binding partner of Rb and the related p130 (36, 37). The knockdown of ZNF217 and a known component of the ‘ZNF217 complex,’ CtBP2, resulted in an equivalent phenotype. Recently, Krig et al. (2007) performed ChIP analysis in multiple cell lines to identify ZNF217-binding sites in several tumor cell lines. In the embryonal carcinoma cell line Ntera2, ZNF217 and CtBP2 were detected at numerous promoters and the depletion of ZNF217 led to the upregulation of a specific subset of genes (51). Interestingly, ZNF217 has been proposed as a candidate oncogene in ovarian and breast cancer (38). Overexpression of ZNF217 can immortalize human mammary epithelial cells and its depletion in certain contexts can induce sensitivity to doxorubicin and reduce the growth and invasive capacity of an ovarian tumor cell line (52-54). The reconciliation of these studies with the apparent role of ZNF217 in transcriptional repressor complexes will be the subject of ongoing study.
Although the Mediator complex was initially thought to be a general required component for RNA polymerase II transcription, recent studies have elucidated the importance of several subunits in promoter-specific regulation. The presence of the CDK8 module can confer promoter-specific transcriptional repression or activation in a context-dependent manner (55-57). Specifically, Donner et al. (2007) showed that CDK8, MED12, and Cyclin C were recruited to the p21Cip1 promoter concurrent with its strong transcriptional activation. Furthermore, CDK8 association with multiple p53-responsive promoters correlated with strong transcriptional activity (55). In contrast, recent studies have demonstrated that MED12 is important in the repression of neuronal genes. Mutations in MED12 are associated with X-linked mental retardation disorders and these disease-associated mutations were shown to abrogate MED12-mediated repression (58). Interestingly, MED13L was identified as a gene harboring missense mutations and translocations in patients with transposition of the great arteries, a congenital heart defect (42). However, the effect of these mutations on MED13L transcriptional function has yet to be determined.
A connection between Mediator complex subunits, transcriptional control, and cancer was recently reported by Firestein et al. (2008) who showed that CDK8 is amplified in human colorectal cancer and exhibits oncogenic properties (59). CDK8 was required for activation of genes regulated by β-catenin, a pathway frequently activated in colorectal tumors. In a parallel study, Morris et al. (2008) shed light on the observation that colorectal tumors paradoxically tend to retain Rb during tumorigenesis. E2F1 was shown to strongly inhibit β-catenin activity, while both CDK8 and RB are able to oppose this function of E2F1 (60). These studies provided impetus to more thoroughly investigate possible connections between Mediator complex subunits and cancer. Here, we examined the contribution of the putative Mediator subunit MED13L to Rb/E2F function. We observed a requirement of MED13L for the full repression of the cyclin A levels and effective growth suppression by an activate Rb/E2F pathway. Consistent with this observation, Mediator has been proposed to confer promoter specificity and subunit composition may be a critical element of that control (61). Interestingly, the MED24 subunit has been recently identified as an essential gene product in the programmed cell death of the larval salivary gland during Drosophila development (62). This finding raises the obvious hypothesis that MED24 might play a role in apoptosis in human cells and its loss may promote tumorigenesis.
Collectively, our findings support the notion that disparate Mediator complex subunits have important contributions in a multitude of transcriptional signals. Moreover, further work is imperative to further elucidate their contribution to specific gene promoter activity, the subunit composition of specific Mediator complexes, and their potential significance in carcinogenesis. We present a functional screen for requisite cooperating factors in the cell cycle arrest program enforced by active Rb/E2F signaling. Based on this study, we submit that MED13L functions as a novel co-factor for Rb/E2F-mediated transcriptional repression and cell cycle arrest.
Materials and Methods
Cell culture and plasmids
T98G cells expressing the murine ecotropic receptor (T98G-EcoR) were provided by J. DeCaprio (Dana-Farber Cancer Institute, Boston, MA). T98G-EcoR cells were cultured in Dulbecco's Modified Eagle's Medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 200 μg/ml G418. Human foreskin fibroblast (HFF) and U2OS cells were maintained in DMEM supplemented with 10% FBS. Plat-E (ecotropic) and Plat-A (amphotropic) retrovirus packaging cells were provided by T. Kitamura (University of Tokyo) and cultured in DMEM supplemented with 10% FBS, 10 μg/ml blasticidin, and 1 μg/ml puromycin (63). The large pocket domain of Rb harboring mutations in the relevant CDK sites (PSM-Rb) was provided by E. Knudsen (Kimmel Cancer Center, Philadelphia, PA) and has been previously described (32, 64). PSM-Rb and CDKN2A were subcloned into the pQCXIH plasmid (Clontech). Individual Open Biosystems shRNA constructs were obtained either from the Duke RNAi Facility or directly from Open Biosystems.
Retroviral infections
To generate T98G-EcoR cells containing the Open Biosystems shRNA library (v. 1.16), individual shRNA constructs were divided into pools (∼2000 clones per pool) and 10 μg of each pooled plasmid DNA preparation was transfected into Plat-E ecotropic packaging cells using Lipofectamine 2000 according to manufacturer's instructions. Approximately 40 hours post-transfection, retroviral supernatant was collected, filtered using 0.45 μm HT Tuffryn syringe filters, and used to infect T98G-EcoR cells in the presence of 8 μg/ml polybrene. Forty-eight hours post-infection, cells were selected with 2.5 μg/ml puromycin. Individual Open Biosystems shRNA constructs (Supplementary Table 2) were used to infect T98G-EcoR cells in a similar manner. HFF cells were infected as above, using Plat-A amphotropic packaging cells, and selected in 1.25 μg/ml puromycin. Twenty-four hours post- infection with pQCXIH-CDKN2A or pQCXIH-PSM-Rb, cells were selected with 200 (HFF) or 500 μg/ml (T98G-EcoR) hygromycin.
Adenovirus infection
The cDNAs encoding PSM-Rb and CDKN2A were cloned into pAdTrackCMV plasmid. The pAdTrackCMV-PSM-Rb construct was recombined with pAdEasy-1 and used to generate recombinant adenovirus as previously described (65). U2OS cells were infected with GFP-producing, GFP and PSM-Rb-producing, or GFP and CDKN2A-producing adenovirus at a calculated MOI of ∼50 (infection efficiency was 95-100% as determined by GFP fluorescence) and harvested 24 h post-infection for western blotting or for 48 h for analysis of BrdU incorporation.
shRNA screen and candidate gene identification
T98G-EcoR cells stably expressing pooled shRNAs were infected with pQCXIH-PSM-Rb as described above. Approximately 19-21 days post-infection, surviving cells were collected and genomic DNA isolated using Qiagen DNeasy Blood and Tissue Kit according to manufacturer's protocol. Genomic DNA (200 ng) was amplified by PCR (30 cycles, forward primer: 5′ – GGGCCTATTTCCCATGATT – 3′ and reverse primer: 5′ – GCCTCAAAATGTTCTTTACGAT – 3′) and the resultant products purified using Qiaquick Gel Extraction kit (Qiagen). PCR products were cloned using TOPO TA cloning kit (Invitrogen) according to the manufacturer's protocol and at least thirty clones from each reaction were sequenced using M13rev primer and the target sequence was identified using UCSC Genome Browser.
Senescence-associated β-galactosidase assay
Cells were cultured in 6-cm dishes were fixed and stained using the Senescence β-galactosidase Staining Kit (Cell Signaling) according to the manufacturer's protocol.
Colony formation assay
Approximate 3×105 cells were seeded onto 60-mm dishes and infected with retroviruses, as indicated, 24 h later. Cells were washed with PBS, fixed with ice-cold methanol for 10 min, stained with 0.5% crystal violet (in 25% methanol) for 10 min, and rinsed with double-distilled water. Images were collected using a Fotodyne digital imaging system. The number of colonies per dish was counted and reported relative to control. Statistical significance was assessed using Student's t-test.
BrdU incorporation and immunofluorescence
Four days post-selection, cells grown on 25-mm glass cover slips were labeled with 10 μM BrdU for 16 hours, washed in PBS, fixed in 3.7% formaldehyde in PBS. DNA was denatured by incubating the cover slips in 1.5 N HCl for 15 min. Cover slips were washed with PBS and permeabilized in 0.5% Triton X-100 in PBS for 20 min. Anti-BrdU antibody (GE Healthcare) was used according to manufacturer's instructions and incorporated BrdU was detected using Texas-Red anti-mouse IgG (Vector). Nuclei were counterstained with Hoechst 33342 (Invitrogen). At least 200 nuclei were counted for BrdU staining per condition.
siRNA transfection
U2OS cells were plated on 6-cm dishes and transfected with 25 nM siGENOME Non-targeting siRNA pool #2 (Dharmacon cat. #D-001206-14-05) or siMED13L siGENOME SMARTpool (Dharmacon cat. #M-027126-01-0005) using Lipofectamine RNAiMAX (Invitrogen) according to manufacturer's protocol. Forty-eight hours post-transfection, cells were infected with adenovirus as described above.
Protein extraction and immunoblotting
U2OS cell pellets were lysed in ice-cold RIPA buffer (50 mM Tris, pH 8.0; 0.1% SDS; 150 mM NaCl; 1% Nonidet P-40; and 0.5% sodium deoxycholate) supplemented with protease inhibitor cocktail (Complete Mini, Roche). Samples were sonicated and insoluble material was pelleted by centrifugation. Protein concentration was determined by BCA Protein Assay (Thermo Scientific) and equal amounts of total protein were separated by SDS-PAGE, transferred to Immobilon-P PVDF membrane (Millipore), and blocked in 5% milk in TBS-T buffer. Primary antibodies used to detect cyclin A (sc-751), CDK4 (sc-601), and Rb (sc-50) were from Santa Cruz. MED13L antibody (A302-420A) was from Bethyl Laboratories. Secondary antibody used was goat anti-rabbit IgG coupled to horseradish peroxidase (Thermo Scientific) and detected using ECL Western Blotting Detection Reagents from Amersham.
Reporter assays
U2OS cells were transfected with 25 nM siRNA, -608-Cyclin A-Luc (provided by E. Knudsen), and 5 ng CMV-Renilla (Promega) using Gene Juice transfection reagent (Novagen) according to the manufacturer's protocol. Forty-eight hours post-transfection, cells were infected with adenovirus as described above. Twenty-four hours post-infection, cells were harvested and luciferase activity was measured using the Dual Luciferase Assay Kit (Promega), with firefly luciferase activity normalized against Renilla luciferase activity.
Ingenuity pathways analysis
As an initial query, the 292 Gene IDs corresponding to the genes identified in the screen were used to search the Ingenuity pathways database (www.ingenuity.com). Interactions were viewed as a single merged network or as individual networks.
Supplementary Material
Acknowledgments
All aspects of the research were supported under the National Cancer Institute Integrative Cancer Biology Program via grant National Institutes of Health 5-U54-CA112952. S.P.A. was supported by NIH F32CA113177. We thank Lazlo Jakoi for technical assistance and all members of the Nevins lab for critical feedback and suggestions. We are grateful to Bernard Mathey-Prevot and Jeffrey Chang for comments on the manuscript. We thank Kaye Culler for assistance in submitting the manuscript. We are grateful to T. Kitamura for providing Plat-E and Plat-A retroviral packaging cells, J. DeCaprio for providing the T98G-EcoR cells, and E. Knudsen for providing the -608Cyclin A-Luc and PSM-Rb plasmids.
References
- 1.Weinberg RA. The retinoblastoma protein and cell cycle control. Cell. 1995;81:323–30. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 2.Sherr CJ. Cancer cell cycles. Science. 1996;274:1672–7. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 3.Burkhart DL, Sage J. Cellular mechanisms of tumour suppression by the retinoblastoma gene. Nat Rev Cancer. 2008;8:671–82. doi: 10.1038/nrc2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Attwooll C, Denchi EL, Helin K. The E2F family: specific functions and overlapping interests. The EMBO J. 2004;23:4709–16. doi: 10.1038/sj.emboj.7600481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dyson N. The regulation of E2F by pRB-family proteins. Genes & Dev. 1998;12:2245–62. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- 6.Nevins JR. The Rb/E2F pathway and cancer. Hum Mol Genet. 2001;10:699–703. doi: 10.1093/hmg/10.7.699. [DOI] [PubMed] [Google Scholar]
- 7.Cobrinik D. Pocket proteins and cell cycle control. Oncogene. 2005;24:2796–809. doi: 10.1038/sj.onc.1208619. [DOI] [PubMed] [Google Scholar]
- 8.Nielsen SJ, Schneider R, Bauer UM, Bannister AJ, Morrison A, O'Carroll D, et al. Rb targets histone H3 methylation and HP1 to promoters. Nature. 2001;412:561–5. doi: 10.1038/35087620. [DOI] [PubMed] [Google Scholar]
- 9.Rayman JB, Takahashi Y, Indjeian VB, Dannenberg JH, Catchpole S, Watson RJ, et al. E2F mediates cell cycle-dependent transcriptional repression in vivo by recruitment of an HDAC1/mSin3B corepressor complex. Genes & Dev. 2002;16:933–47. doi: 10.1101/gad.969202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robertson KD, Ait-Si-Ali S, Yokochi T, Wade PA, Jones PL, Wolffe AP. DNMT1 forms a c omplex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nature Genetics. 2000;25:338–42. doi: 10.1038/77124. [DOI] [PubMed] [Google Scholar]
- 11.Strobeck MW, Knudsen KE, Fribourg AF, DeCristofaro MF, Weissman BE, Imbalzano AN, et al. BRG-1 is required for RB-mediated cell cycle arrest. Proc Natl Acad Sci U S A. 2000;97:7748–53. doi: 10.1073/pnas.97.14.7748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang HS, Gavin M, Dahiya A, Postigo AA, Ma D, Luo RX, et al. Exit from G1 and S phase of the cell cycle is regulated by repressor complexes containing HDAC-Rb-hSWI/SNF and Rb-hSWI/SNF. Cell. 2000;101:79–89. doi: 10.1016/S0092-8674(00)80625-X. [DOI] [PubMed] [Google Scholar]
- 13.Medina PP, Sanchez-Cespedes M. Involvement of the chromatin-remodeling factor BRG1/SMARCA4 in human cancer. Epigenetics. 2008;3:64–8. doi: 10.4161/epi.3.2.6153. [DOI] [PubMed] [Google Scholar]
- 14.Weissman B, Knudsen KE. Hijacking the chromatin remodeling machinery: impact of SWI/SNF perturbations in cancer. Cancer Res. 2009;69:8223–30. doi: 10.1158/0008-5472.CAN-09-2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Campisi J, d'Adda di Fagagna F. Cellular senescence: when bad things happen to good cells. Nat Rev Mol Cell Biol. 2007;8:729–40. doi: 10.1038/nrm2233. [DOI] [PubMed] [Google Scholar]
- 16.Hayflick L, Moorhead PS. The serial cultivation of human diploid cell strains. Exp Cell Res. 1961;25:585–621. doi: 10.1016/0014-4827(61)90192-6. [DOI] [PubMed] [Google Scholar]
- 17.Greider CW, Blackburn EH. Tracking telomerase. Cell. 2004;116:S83–6. doi: 10.1016/s0092-8674(04)00053-4. 1 p following S6. [DOI] [PubMed] [Google Scholar]
- 18.Serrano M, Blasco MA. Putting the stress on senescence. Curr Opin Cell Biol. 2001;13:748–53. doi: 10.1016/s0955-0674(00)00278-7. [DOI] [PubMed] [Google Scholar]
- 19.Serrano M, lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 20.Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci U S A. 1995;92:9363–7. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Narita M, Nunez S, Heard E, Narita M, Lin AW, Hearn SA, et al. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell. 2003;113:703–16. doi: 10.1016/s0092-8674(03)00401-x. [DOI] [PubMed] [Google Scholar]
- 22.Prieur A, Peeper DS. Cellular senescence in vivo: a barrier to tumorigenesis. Curr Opin Cell Biol. 2008;20:150–5. doi: 10.1016/j.ceb.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 23.Shay JW, Roninson IB. Hallmarks of senescence in carcinogenesis and cancer therapy. Oncogene. 2004;23:2919–33. doi: 10.1038/sj.onc.1207518. [DOI] [PubMed] [Google Scholar]
- 24.Collado M, Serrano M. Senescence in tumours: evidence from mice and humans. Nat Rev Cancer. 10:51–7. doi: 10.1038/nrc2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Braig M, Lee S, Loddenkemper C, Rudolph C, Peters AH, Schlegelberger B, et al. Oncogene-induced senescence as an initial barrier in lymphoma development. Nature. 2005;436:660–5. doi: 10.1038/nature03841. [DOI] [PubMed] [Google Scholar]
- 26.Chen Z, Trotman LC, Shaffer D, Lin HK, Dotan ZA, Niki M, et al. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature. 2005;436:725–30. doi: 10.1038/nature03918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Collado M, Gil J, Efeyan A, Guerra C, Schuhmacher AJ, Barradas M, et al. Tumour biology: senescence in premalignant tumours. Nature. 2005;436:642. doi: 10.1038/436642a. [DOI] [PubMed] [Google Scholar]
- 28.Ha L, Ichikawa T, Anver M, Dickins R, Lowe S, Sharpless NE, et al. ARF functions as a melanoma tumor suppressor by inducing p53-independent senescence. Proc Natl Acad Sci U S A. 2007;104:10968–73. doi: 10.1073/pnas.0611638104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Michaloglou C, Vredeveld LC, Soengas MS, Denoyelle C, Kuilman T, van der Horst CM, et al. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature. 2005;436:720–4. doi: 10.1038/nature03890. [DOI] [PubMed] [Google Scholar]
- 30.Qin XQ, Chittenden T, Livingston DM, Kaelin WG., Jr Identification of a growth suppression domain within the retinoblastoma gene product. Genes & Dev. 1992;6:953–64. doi: 10.1101/gad.6.6.953. [DOI] [PubMed] [Google Scholar]
- 31.Yang H, Williams BO, Hinds PW, Shih TS, Jacks T, Bronson RT, et al. Tumor suppression by a severely truncated species of retinoblastoma protein. Mol Cell Biol. 2002;22:3103–10. doi: 10.1128/MCB.22.9.3103-3110.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Knudsen ES, Buckmaster C, Chen TT, Feramisco JR, Wang JYJ. Inhibition of DNA synthesis by RB; effects on G1/S transition and S-phase progression. 1998;12:2278–92. doi: 10.1101/gad.12.15.2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Angus SP, Mayhew CN, Solomon DA, Braden WA, Markey MP, Okuno Y, et al. RB reversibly inhibits DNA replication via two temporally distinct mechanisms. Mol Cell Biol. 2004;24:5404–20. doi: 10.1128/MCB.24.12.5404-5420.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cowger JJ, Zhao Q, Isovic M, Torchia J. Biochemical characterization of the zinc-finger protein 217 transcriptional repressor complex: identification of a ZNF217 consensus recognition sequence. Oncogene. 2007;26:3378–86. doi: 10.1038/sj.onc.1210126. [DOI] [PubMed] [Google Scholar]
- 35.Quinlan KG, Nardini M, Verger A, Francescato P, Yaswen P, Corda D, et al. Specific recognition of ZNF217 and other zinc finger proteins at a surface groove of C-terminal binding proteins. Mol Cell Biol. 2006;26:8159–72. doi: 10.1128/MCB.00680-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meloni AR, Lai CH, Yao TP, Nevins JR. A mechanism of COOH-terminal binding protein-mediated repression. Mol Cancer Res. 2005;3:575–83. doi: 10.1158/1541-7786.MCR-05-0088. [DOI] [PubMed] [Google Scholar]
- 37.Meloni AR, Smith EJ, Nevins JR. A mechanism for Rb/p130-mediated transcription repression involving recruitment of the CtBP corepressor. Proc Natl Acad Sci USA. 1999;96:9574–9. doi: 10.1073/pnas.96.17.9574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Quinlan KG, Verger A, Yaswen P, Crossley M. Amplification of zinc finger gene 217 (ZNF217) and cancer: when good fingers go bad. Biochim Biophys Acta. 2007;1775:333–40. doi: 10.1016/j.bbcan.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 39.Sato S, Tomomori-Sato C, Parmely TJ, Florens L, Zybailov B, Swanson SK, et al. A set of consensus mammalian mediator subunits identified by multidimensional protein identification technology. Mol Cell. 2004;14:685–91. doi: 10.1016/j.molcel.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 40.Conaway RC, Sato S, Tomomori-Sato C, Yao T, Conaway JW. The mammalian Mediator complex and its role in transcriptional regulation. Trends Biochem Sci. 2005;30:250–5. doi: 10.1016/j.tibs.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 41.Kornberg RD. Mediator and the mechanism of transcriptional activation. Trends Biochem Sci. 2005;30:235–9. doi: 10.1016/j.tibs.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 42.Muncke N, Jung C, Rudiger H, Ulmer H, Roeth R, Hubert A, et al. Missense mutations and gene interruption in PROSIT240, a novel TRAP240-like gene, in patients with congenital heart defect (transposition of the great arteries) Circulation. 2003;108:2843–50. doi: 10.1161/01.CIR.0000103684.77636.CD. [DOI] [PubMed] [Google Scholar]
- 43.Sever-Chroneos Z, Angus SP, Fribourg AF, Wan H, Todorov I, Knudsen KE, et al. Retinoblastoma tumor suppressor protein signals through inhibition of cyclin-dependent kinase 2 activity to disrupt PCNA function in S phase. 2001;21:4032–45. doi: 10.1128/MCB.21.12.4032-4045.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chicas A, Wang X, Zhang C, McCurrach M, Zhao Z, Mert O, et al. Dissecting the unique role of the retinoblastoma tumor suppressor during cellular senescence. Cancer Cell. 17:376–87. doi: 10.1016/j.ccr.2010.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Acosta JC, O'Loghlen A, Banito A, Guijarro MV, Augert A, Raguz S, et al. Chemokine signaling via the CXCR2 receptor reinforces senescence. Cell. 2008;133:1006–18. doi: 10.1016/j.cell.2008.03.038. [DOI] [PubMed] [Google Scholar]
- 46.Alexander K, Hinds PW. Requirement for p27(KIP1) in retinoblastoma protein-mediated senescence. Mol Cell Biol. 2001;21:3616–31. doi: 10.1128/MCB.21.11.3616-3631.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kuilman T, Michaloglou C, Vredeveld LC, Douma S, van Doorn R, Desmet CJ, et al. Oncogene-induced senescence relayed by an interleukin-dependent inflammatory network. Cell. 2008;133:1019–31. doi: 10.1016/j.cell.2008.03.039. [DOI] [PubMed] [Google Scholar]
- 48.Sebastian T, Malik R, Thomas S, Sage J, Johnson PF. C/EBPbeta cooperates with RB:E2F to implement Ras(V12)-induced cellular senescence. Embo J. 2005;24:3301–12. doi: 10.1038/sj.emboj.7600789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wolyniec K, Wotton S, Kilbey A, Jenkins A, Terry A, Peters G, et al. RUNX1 and its fusion oncoprotein derivative, RUNX1-ETO, induce senescence-like growth arrest independently of replicative stress. Oncogene. 2009;28:2502–12. doi: 10.1038/onc.2009.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Banck MS, Li S, Nishio H, Wang C, Beutler AS, Walsh MJ. The ZNF217 oncogene is a candidate organizer of repressive histone modifiers. Epigenetics. 2009;4:100–6. doi: 10.4161/epi.4.2.7953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Krig SR, Jin VX, Bieda MC, O'Geen H, Yaswen P, Green R, et al. Identification of genes directly regulated by the oncogene ZNF217 using chromatin immunoprecipitation (ChIP)-chip assays. J Biol Chem. 2007;282:9703–12. doi: 10.1074/jbc.M611752200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang G, Krig S, Kowbel D, Xu H, Hyun B, Volik S, et al. ZNF217 suppresses cell death associated with chemotherapy and telomere dysfunction. Hum Mol Genet. 2005;14:3219–25. doi: 10.1093/hmg/ddi352. [DOI] [PubMed] [Google Scholar]
- 53.Nonet GH, Stampfer MR, Chin K, Gray JW, Collins CC, Yaswen P. The ZNF217 gene amplified in breast cancers promotes immortalization of human mammary epithelial cells. Cancer Res. 2001;61:1250–4. [PubMed] [Google Scholar]
- 54.Sun G, Zhou J, Yin A, Ding Y, Zhong M. Silencing of ZNF217 gene influences the biological behavior of a human ovarian cancer cell line. Int J Oncol. 2008;32:1065–71. [PubMed] [Google Scholar]
- 55.Donner AJ, Szostek S, Hoover JM, Espinosa JM. CDK8 is a stimulus-specific positive coregulator of p53 target genes. Mol Cell. 2007;27:121–33. doi: 10.1016/j.molcel.2007.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Knuesel MT, Meyer KD, Bernecky C, Taatjes DJ. The human CDK8 subcomplex is a molecular switch that controls Mediator coactivator function. Genes Dev. 2009;23:439–51. doi: 10.1101/gad.1767009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mo X, Kowenz-Leutz E, Xu H, Leutz A. Ras induces mediator complex exchange on C/EBP beta. Mol Cell. 2004;13:241–50. doi: 10.1016/s1097-2765(03)00521-5. [DOI] [PubMed] [Google Scholar]
- 58.Ding N, Zhou H, Esteve PO, Chin HG, Kim S, Xu X, et al. Mediator links epigenetic silencing of neuronal gene expression with x-linked mental retardation. Mol Cell. 2008;31:347–59. doi: 10.1016/j.molcel.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Firestein R, Bass AJ, Kim SY, Dunn IF, Silver SJ, Guney I, et al. CDK8 is a colorectal cancer oncogene that regulates beta-catenin activity. Nature. 2008;455:547–51. doi: 10.1038/nature07179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Morris EJ, Ji JY, Yang F, Di Stefano L, Herr A, Moon NS, et al. E2F1 represses beta-catenin transcription and is antagonized by both pRB and CDK8. Nature. 2008;455:552–6. doi: 10.1038/nature07310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Malik S, Roeder RG. The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat Rev Genet. 2010;11:761–72. doi: 10.1038/nrg2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang L, Evans J, Andrews HK, Beckstead RB, Thummel CS, Bashirullah A. A genetic screen identifies new regulators of steroid-triggered programmed cell death in Drosophila. Genetics. 2008;180:269–81. doi: 10.1534/genetics.108.092478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Morita S, Kojima T, Kitamura T. Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther. 2000;7:1063–6. doi: 10.1038/sj.gt.3301206. [DOI] [PubMed] [Google Scholar]
- 64.Knudsen KE, Fribourg AF, Strobeck MW, Blanchard JM, Knudsen ES. Cyclin A is a functional target of retinoblastoma tumor suppressor protein-mediated cell cycle arrest. J Biol Chem. 1999;274:27632–41. doi: 10.1074/jbc.274.39.27632. [DOI] [PubMed] [Google Scholar]
- 65.He TC, Zhou S, da Costa LT, Yu J, Kinzler KW, Vogelstein B. A simplified system for generating recombinant adenoviruses. Biochem Biophys Res Comm. 1998;95:2509–14. doi: 10.1073/pnas.95.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.