Short abstract
A hybrid approach combining fluctuating hydrodynamics with generalized Langevin dynamics is employed to study the motion of a neutrally buoyant nanocarrier in an incompressible Newtonian stationary fluid medium. Both hydrodynamic interactions and adhesive interactions are included, as are different receptor–ligand bond constants relevant to medical applications. A direct numerical simulation adopting an arbitrary Lagrangian–Eulerian based finite element method is employed for the simulation. The flow around the particle and its motion are fully resolved. The temperatures of the particle associated with the various degrees of freedom satisfy the equipartition theorem. The potential of mean force (or free energy density) along a specified reaction coordinate for the harmonic (spring) interactions between the antibody and antigen is evaluated for two different bond constants. The numerical evaluations show excellent comparison with analytical results. This temporal multiscale modeling of hydrodynamic and microscopic interactions mediating nanocarrier motion and adhesion has important implications for designing nanocarriers for vascular targeted drug delivery.
Keywords: nanocarrier motion, fluctuating hydrodynamics, generalized Langevin dynamics, hybrid numerical simulation, ligand–receptor interaction, targeted drug delivery
1. Introduction
The use of nanoparticles enables precise and successful delivery of drugs to target cells [1,2]. In general, nanoparticle drug-delivery systems have been shown to enhance drug efficacy and reduce the impact of drugs on nontarget tissues, thereby minimizing unwanted side effects. In order to more broadly integrate this technology into clinical medicine, a model-based design and optimization of nanoparticle transport in the vasculature and adhesion to target cells can prove effective. Toward achieving this goal, an important step is to determine the motion of a nanoparticle subject to hydrodynamic effects in the vasculature while simultaneously being subject to a constant temperature; this is crucial to accurately model the biological reactions (receptor–ligand interactions) mediating the adhesion of nanoparticle to the endothelial cell surface lining the vasculature [3–6]. The nanocarrier with a loaded cargo may be studied subsequently by extending this model.
A nanoparticle suspended in a fluid undergoes random motion due to the thermal fluctuations in the fluid. As a consequence, translational and rotational degrees of freedom become important. In determining the translational and rotational motions of the nanoparticle in an incompressible Newtonian medium, there exist two methods that couple the thermal fluctuations with the hydrodynamic interactions. These are the generalized Langevin method [7] and the fluctuating hydrodynamics method [8]. Either procedure would require numerical simulations for covering extensive parameter space.
In the fluctuating hydrodynamics approach, the nanoparticle motion incorporates both the Brownian motion and the effect of hydrodynamic force acting on its surface imparted from the surrounding fluid. This method essentially consists of adding stochastic stresses (random stress) to the stress tensor in the momentum equation of the fluid and stochastic fluxes to the heat flux where the energy equation is present [9]. The stochastic stress tensor depends on the temperature and the transport coefficients of the fluid medium [10,11]. Numerical simulations of the fluctuating hydrodynamics approach have been carried out employing the finite volume method [11–14], Lattice-Boltzmann method [15–21], finite element method [8,22,23] and stochastic immersed boundary method [24].
In the Langevin dynamics method, the effects of thermal fluctuations are incorporated as random forces and torques in the particle equation of motion [7,25–30]. The properties of these forces depend on the grand resistance tensor. The tensor in turn depends on the fluid properties, particle shape, and its instantaneous location such as its proximity to a wall or a boundary. This is a robust thermostat, which preserves equilibrium distributions at constant temperatures (i.e., adheres to the equipartition theorem). Clearly, coupling to a thermostat will alter the hydrodynamics of the nanoparticle system. The characterizations of the performance of the thermostat as well as how it alters the associated hydrodynamic correlations are important. Numerical schemes for studying the nanoparticle motion in a fluid must simultaneously consider the momentum (Langevin) equation for the particle and the Naiver–Stokes equation for the fluid. The random force/torque in the particle equation can then be related to the frictional force/torque via the generalized fluctuation–dissipation theorem [31,32]. The implementation can occur in two ways: (i) directly adjust the variance of the random force term in the classical Langevin equation to play the role of a thermostat. (ii) A second, more direct approach that preserves the structure of the generalized Langevin equation, is to consider the power spectrum for the variance of the random force term using a correlated or colored noise with a well defined characteristic memory time. Such a formalism simultaneously preserves the equipartition theorem and the nature of the long-time hydrodynamic correlations, and proves to be a versatile thermostat [7].
The fluctuating hydrodynamics approach in an incompressible fluid [8] captures the correct hydrodynamic correlations and conserves thermal equipartition only after adding the mass correction [10]. On the other hand, the generalized Langevin dynamics yields the correct thermal equipartition (without any mass correction), but modifies the nature of the hydrodynamics correlations (due to the coupling of the fluid equations with the thermostat degrees of freedom) [7].
Recently, we have formulated a novel hybrid approach combining Markovian fluctuating hydrodynamics of the fluid and the non-Markovian Langevin dynamics with the Ornstein–Uhlenbeck noise perturbing the translational and rotational equations of motion of the nanocarrier [33]. For this hybrid approach, we have verified the conservation of thermal equipartition and the nature of hydrodynamic correlations by comparisons with well-known analytical results [10]. This approach effectively produces a thermostat that also simultaneously preserves the true hydrodynamic correlations [33]. With this procedure, we have also evaluated adhesive interactions between a receptor and a ligand bond (tethered by a spring force [34,35]) very close to the cylindrical wall at a specified finite temperature [36].
In our earlier manuscripts, we have explored the sensitivity of parameters representing flow rates of the fluid [8,37] and density of the Brownian particle [37,38], validating our numerical schemes, the fluctuating hydrodynamics and the hybrid scheme. We have also validated our hybrid scheme for a nearly neutrally buoyant particle [38]. In this paper, we employ the hybrid approach to study the motion of a neutrally buoyant nanocarrier when subject to both hydrodynamic interactions and adhesive interactions at an endothelial cell wall for different receptor–ligand bond constants and at a specified temperature [6,39,40]. A direct numerical simulation procedure adopting an arbitrary Lagrangian–Eulerian based finite element method is employed to simulate the Brownian motion of a nanoparticle in an incompressible Newtonian fluid contained in a horizontal micron sized cylindrical vessel. The results for the attainment of thermal equilibrium between the particle, tethered spring, and the surrounding medium, and the potential of mean force (PMF) (or free energy density) along a specified reaction coordinate for two different bond constants are compared with analytical results. The comparisons are excellent and lend credibility to our novel numerical scheme.
2. Formulation of the Problem
Motion of a nanoparticle trapped in a harmonic potential residing in an incompressible Newtonian stationary fluid medium contained in a horizontal micron sized circular vessel (see Fig. 1) is considered. The fluid and particle equations are formulated in an inertial frame of reference. The radius, R, and the length, L, of the vessel (tube) are very large compared to the particle size, a, the radius of the particle. Antigen of length 19 nm is attached to the surface of a cell lying on the wall of the cylindrical tube () containing a quiescent Newtonian fluid and antibody of length 15 nm is attached to the surface of the neutrally buoyant solid spherical nanocarrier of radius a = 250 nm. The nanocarrier is placed close to the antigen such that the direction of antibody is initially pointing toward the antigen and the distance between them is . The tips of the antigen and the antibody are tethered by a simple harmonic (spring) potential with spring constant k [34,35]. Initially both the fluid and particle are at rest. No body force is considered either for the particle or for the fluid domain. At time t = 0, the nanocarrier experiences Brownian motion and harmonic motion. The motion of the nanocarrier is determined by the hydrodynamic and spring forces and torques acting on the particle. The numerical results are obtained from simulations of the fluid–particle system with physical parameters ; ; . The temperature of the fluid is initially set to T = 310 K.
Fig. 1.
Schematic representation of a nanoparticle in a cylindrical vessel (tube) (not to scale). Diameter of the tube: ; length of the tube: ; diameter of the nanoparticle: ; viscosity of the fluid: ; density of the fluid and the nanoparticle: ; and spring constant: .
2.1. Hybrid Scheme: Governing Equations and Boundary Conditions.
The motion of an incompressible Newtonian fluid satisfies the conservation of mass and momentum given by
| (1) |
| (2) |
where and are the velocity and density of the fluid, respectively, is the stress tensor, p is the pressure, is the identity tensor, and is the dynamic viscosity. The random stress tensor is assumed to be a Gaussian with
| (3) |
where is the ensemble average, is the Boltzmann constant, T is the absolute temperature, is the Kronecker delta, and the Dirac delta function denotes that the components of the random stress tensor are spatially uncorrelated (Markovian). The right hand side of Eq. (3) denotes the mean and variance of the thermal fluctuations chosen to be consistent with the fluctuation–dissipation theorem [9,10,41,42]. By including this stochastic stress tensor due to the thermal fluctuations in the governing equations, the macroscopic hydrodynamic theory is generalized to include the mesoscopic scales ranging from tens of nanometers to a few microns.
For a rigid particle suspended in an incompressible Newtonian fluid, the translational and rotational motions of the particle satisfy Newton's second law and the Euler equation, respectively. In the hybrid formulation, the time correlated noise is added into the particle equations of motion [33,36]
| (4) |
| (5) |
where m is the mass of the particle, is its moment of inertia, and are the translational and angular velocities of the particle, respectively, is the position of the centroid of the particle, is a vector from the center of the particle to a point on its surface, denotes the particle surface, is the unit normal vector on the surface of the particle pointing into the particle, and the random force and torque are given by
| (6) |
| (7) |
for the Ornstein–Uhlenbeck process. The time integral in Eqs. (4) and (5) excludes the frictional force and torque at the time instant t since it has already been accounted for in the hydrodynamic force and torque terms, respectively. The characteristic memory time for translational, , and rotational, , motions of the nanoparticle add certain amounts of memory from the previous history of fluctuations to the system. Here, is the time step for the numerical simulation, and correspond to the number of time steps required to adequately represent the memory effects. These are variable quantities and are determined on the basis of satisfying the equipartition theorem. The amount of memory required by translational and rotational motions of the nanoparticle in order to satisfy the equipartition theorem are different. Hence, is not a necessary condition for the temperature of the particle to attain the preset temperature of the fluid. Equations (6) and (7) are the random force and torque acting on the nanoparticle at time (a previous time instant). Since the random stress is Gaussian, and are also Gaussian with variance equivalent to the strength of the white noise in the Langevin equation. In the limit of the characteristic memory times (i.e., in the absence of memory), Eqs. (4) and (5) reduce to Newton's second law and Euler equations, respectively, which correspond to the Markovian fluctuating hydrodynamics. The spring force and torque acting on the particle are given by
| (8) |
where k is the spring constant, l is the length of the spring, is the unit vector pointing toward the tip of the antigen attached to the wall.
The initial and boundary conditions for the problem are
| (9) |
| (10) |
| (11) |
where is the domain occupied by the fluid and and are the inlet and outlet boundaries, respectively. The stochastic governing Eqs. (1)–(5) along with the initial and boundary conditions (9)–(11) are solved numerically. It is assumed that there is no body torque acting at any point in the fluid and the viscous stress tensor, , is symmetric.
A numerical simulation at a mesoscopic scale involving a particle in a fluid could be based on a discretization of the Eqs. (1)–(11). However, the discrete forms have to satisfy the fluctuation–dissipation theorem [11–13,41–44]. Español and Zúñiga [22] and Español et al. [23] have shown that a well behaved set of discrete equations obtained in terms of the finite element shape functions based on the Delaunay triangulation conserves mass, momentum and energy while ensuring thermodynamic consistency. In the present study, we obtain the discrete hydrodynamic equations using finite element shape functions based on the Delaunay–Voronoi tetrahedrizations. The computational domain is covered by a finite element mesh generated using Delaunay–Voronoi methods. The fluid domain is discretized by quadratic irregular tetrahedral elements. A typical element is shown in Fig. 2. Figure 3 shows a triangular mesh discretizing the surface of the fluid domain (cylinder) and the surface of the nanoparticle. The discretization of the fluid domain changes at each time step of the simulation due to the motion of the nanoparticle. The procedure for numerical simulation of the random stresses associated with the unstructured tetrahedron mesh while conserving the volume is described in detail in Uma et al. [8]. The details of combined fluid–solid weak formulation, spatial discretization, mesh movement techniques, and temporal discretization of time derivatives are discussed in Refs. [7,8]. These details will not be repeated here for brevity. Briefly, the fluid domain is approximated by quadratic tetrahedral finite-elements (ten nodes defined per tetrahedron with ten basis functions that are second-order polynomials). The discrete solution for the fluid velocity is approximated in terms of piecewise quadratic functions, and is assumed to be continuous over the domain (P2 elements). The discrete solution for the pressure is taken to be piecewise linear and continuous (P1 element). This P1/P2 element for the pressure and velocity is consistent with the Ladyzhenskaya–Babuska–Brezzi (LBB) or inf-sup condition and yields convergent solutions [45,46].
Fig. 2.
Representation of a ten-node tetrahedron
Fig. 3.
Finite element surface mesh of a cylindrical tube with one spherical nanoparticle
The time scales involved in this study are (i) , the Brownian relaxation time over which velocity correlations decay in the Langevin equation, (ii) , the Brownian diffusive time scale over which the nanoparticle diffuses over a distance equal to its own radius, (iii) , the hydrodynamic time scale for momentum to diffuse over a distance equal to the radius of the nanoparticle, and (iv) , the harmonic time for a single oscillation of spring, where is the Stokes dissipative friction force coefficient for a sphere, a is the radius of the nanoparticle, and is the kinematic viscosity of the fluid. The time step for the numerical simulation has been chosen to be smaller than all the relevant physical time scales described above. The simulations presented in this study have been carried out for long enough durations to allow for the temperature of the particle to equilibrate; i.e., if N is the number of simulated time steps then .
3. Numerical Results and Discussion
For a given nanoparticle of radius a, and tube radius R, a “realization” consists of N time steps (approximately 10 s wall clock time for each time step that is generally considered in this study). The number of time steps depends upon equilibration of particle temperature, or spring temperature. In order to ensure the uniqueness of the realizations, different initial seeds are chosen for a Gaussian random number generator. In this section, we numerically predict (i) the translational and rotational temperatures of the nanoparticle, where the temperature calculation is carried out until thermal equilibration is obtained for the particle; (ii) the translational and rotational velocity distributions of the nanoparticle motion; (iii) temperature of the spring; (vi) the translational and rotational velocity autocorrelations (VACFs); and (v) potential mean force along a specified reaction coordinate for different bond constants. We compare the various numerical predictions with known analytical results, where available.
3.1. Equipartition Theorem.
From the equipartition theorem, at thermal equilibrium, the translational and rotational temperatures of the nanoparticle are given by
| (12) |
Figure 4 shows that translational and rotational temperatures of neutrally buoyant Brownian particle trapped in a harmonic potential, thermally equilibrated, in a quiescent fluid medium are independent of the bond constant, k. The characteristic memory time for translational, , and rotational, , motions of the nanoparticle add certain amounts of memory from the previous history of fluctuations to the system. Here, and correspond to the number of time steps required to adequately represent the memory effects. These are variable quantities and are determined on the basis of satisfying the equipartition theorem. The amount of memory required by translational and rotational motions of the nanoparticle in order to satisfy the equipartition theorem are different. The nondimensionalized characteristic memory times are and (for more details, see Ref. [33]). The error bars have been plotted from standard deviations of the temperatures obtained with 15 different realizations, consisting of 100,000 time steps per realization.
Fig. 4.
Translational and rotational temperatures of the neutrally buoyant Brownian particle trapped in a harmonic potential in a stationary fluid medium as a function of the bond constant k. The nondimensionalized characteristic memory times are and . The error bars have been plotted from standard deviations of the temperatures obtained with 15 different realizations, consisting of 100,000 time steps per realization.
3.2. Maxwell–Boltzmann Distribution.
At thermal equilibrium, the probability density function of each velocity component (; ) of the fluctuating nanocarrier follows the Maxwell–Boltzmann distribution
| (13) |
| (14) |
The equilibrium statistics of the three components of U and along the three coordinate directions are independent of each other.
In Fig. 5, we plot the velocity distributions of the particle for each component of U (Figs. 5(a) and 5(c)) and (Figs. 5(b) and 5(d)) using hybrid scheme for bond constants k = 0.1 N/m (Figs. 5(a) and 5(b)) and k = 1.0 N/m (Figs. 5(c) and 5(d)). The nondimensionalized characteristic memory times are and [33]. For determining the velocity distribution of the nanocarrier, five realizations in each coordinate direction consisting of time steps have been computed. Thus, a total of 1,500,000 time steps have been computed. It is observed that each degree of freedom individually follows a Gaussian distribution. The mean and the variance obtained by using hybrid scheme agrees to within 6% error (see dotted line in Fig. 5) relative to that of the analytical Boltzmann distribution, implying an adherence to the equipartition theorem within statistical error (Fig. 5).
Fig. 5.
Equilibrium probability density of the (a) and (c) translational and (b) and (d) rotational velocities of the neutrally buoyant nanocarrier (a = 250 nm) trapped in a harmonic potential in a Newtonian fluid for bond constant k = 0.1 N/m (a) and (b) and k = 1.0 N/m (c) and (d). The nondimensionalized characteristic memory times are and . The distributions agree within 6% error (see dotted line) with that of the analytical Maxwell–Boltzmann distribution.
Figure 6 shows the probability distribution of spring length using hybrid scheme for bond constants k = 0.1 N/m and k = 1.0 N/m, respectively. The nondimensionalized characteristic memory times are and [33]. The equilibrium probability density of the displacement of spring in each Cartesian direction follows a Gaussian distribution. The agreement with the analytical Gaussian distribution with the mean zero and the variance is to within 6% error (see dotted line in Fig. 6) of statistical error using both the methods. These results demonstrate that our dynamic formalisms conserve the equilibrium distributions of the canonical (constant temperature) ensemble for different bond constants.
Fig. 6.
Equilibrium probability density of the displacement of spring length using hybrid scheme for bond constant (a) k = 0.1 N/m and (b) k = 1.0 N/m, where the standard deviation . The nondimensionalized characteristic memory times are and . The distributions agree within 6% error (see dotted line) with the analytical Maxwell–Boltzmann distribution.
3.3. Hydrodynamic Correlations.
A nanocarrier experiencing Brownian motion in a fluid is influenced by the hydrodynamic interactions. The fluid around the particle is dragged in the direction of motion of the particle. On the other hand, the motion of the particle is resisted by viscous forces arising due to its motion relative to the surrounding fluid. The momentum of the fluid surrounding the particle at any instant is related to its recent history. The friction coefficient is time dependent and is no longer given by the constant Stokes value. In this context, Zwanzig and Bixon [47] have shown that for constant friction coefficient , the VACF of the particle in a simple fluid obeys
| (15) |
| (16) |
which denote exponential decays, while for the time dependent friction coefficient, the decay of the VACF at long times obeys a power-law [10]
| (17) |
| (18) |
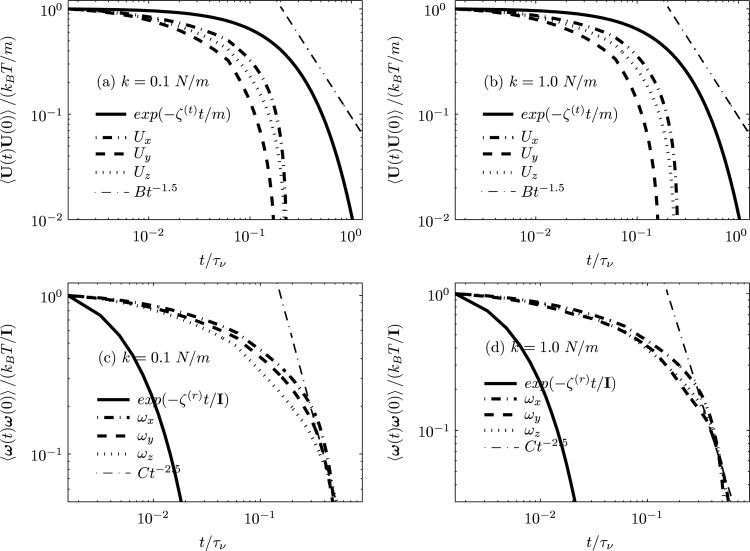
The nondimensionalized characteristic memory times are and [33]. For determining the VACF of the nanocarrier, five realizations in each coordinate directions have been employed with total computation of time steps. Figure 7 shows the VACF of the translational and rotational motions of a nanocarrier (a = 250 nm) in a quiescent fluid medium in a circular vessel as obtained from our numerical simulations. It may be observed that the translational and rotational VACFs of the Brownian particle have exponential and power-law decays () over long times, respectively. We note that the exponential decay of the translational velocity of the nanocarrier over long times is due to its proximity to the wall (due to the confining harmonic potential), again indicating that hydrodynamic correlations are correctly captured by our model. For a free nanocarrier, the long-time behavior of the translational and rotational VACFs both follow algebraic decays with time, as shown by us in previous studies [8,33].
Fig. 7.
Translational ((a) and (b)) and rotational ((c) and (d)) VACFs of the harmonically trapped Brownian particle of radius a = 250 nm through a circular vessel of radius obtained using hybrid scheme. The nondimensionalized characteristic memory times are and .
3.4. Potential of Mean Force.
In order to determine average properties corresponding to a given equilibrium distribution at a finite (fixed) temperature, we compute the PMF for the harmonic (spring) interactions between antibodies and antigens. We choose a reaction coordinate y, which is the vertical displacement between the tips of the antigen and the antibody; increase in y allows the nanocarrier to be displaced away from the wall while still being bound by the spring. Since, the maximum (and hence the average) displacements along y are limited by temperature and by the total time of the simulation, we perform umbrella sampling in multiple windows with harmonic biasing potentials to facilitate extensive sampling along y. The window size of the umbrella sampling is chosen as and the harmonic biasing potential in each window is chosen to be , where is the harmonic force constant and is the vertical coordinate of the center of window i. By updating , the tip of the antibody (on the average) is slowly varied relative to the antigen reaction tip. The weighted histogram analysis method (WHAM) algorithm [48] is used to unbias and combine the histograms in different windows to form a complete PMF (W(y)) profile using a tolerance factor of in the WHAM method. For determining the PMF profile using hybrid scheme, three realizations in each window have been computed with up to 100,000 time steps per realization (hence, yielding a total of time steps per window). All the relevant parameters including the window size , strength of the biasing potential and the sampling size in each window have been tested to ensure convergence.
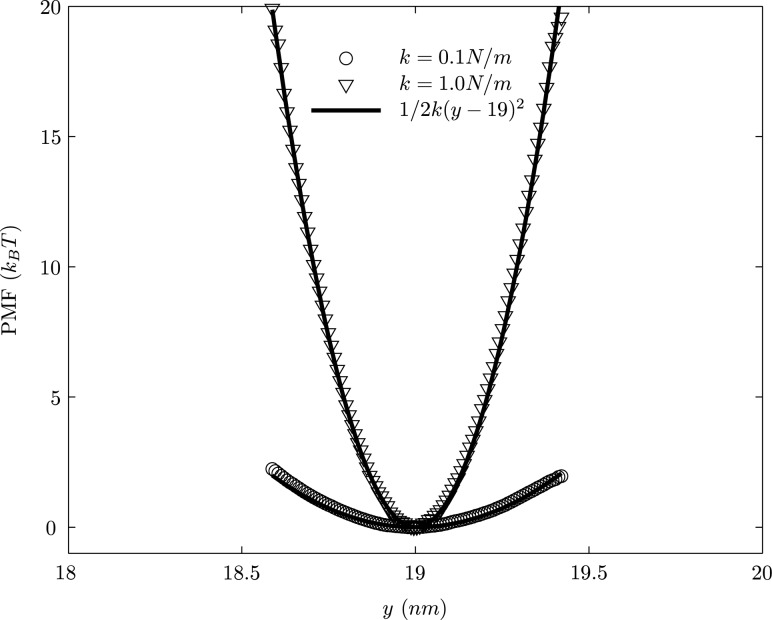
Figure 8 shows the calculated PMF profile for two different bond constants along reaction coordinate y using hybrid scheme at a temperature of 310 K. It is observed that our numerical results agree very well with the corresponding analytical solution. It is to be noted that the bond constant relevant to the biological applications are in the range 0.4 N/m to 2.5 N/m (Table 1, see, Table A3-1 of Refs. [35,39,49]). The procedure for fitting the data is outlined in detail in Ref. [39]. Briefly, the rupture force between a receptor and its ligand is measured in single molecule experiments (AFM or optical tweezers). Since the rupture force is inherently a stochastic variable, through repeated measurements (of 100 or more trials), the rupture force distribution is obtained experimentally (see, for instance, Ref. [6]). This distribution is reproducible within experimental error across multiple experiments. Then, using the Bell model (which includes the harmonic component introduced in our current study), the rupture force distribution is predicted using the theory of failure analysis: namely, the probability that a bond will rupture after time t in an interval delta t is given by the probability that the bond survived a rupture for time t and it subsequently ruptures in the interval delta t. Using a maximum likelihood estimate, we vary the parameters (namely the spring constant) and choose the value of k that best fits the experimental rupture force distribution.
Fig. 8.
The calculated PMF W(y) at a temperature of 310 K for different values of the bond constant, k
Table 1.
Fitting k to the experimental data of Zhang et al. [49] and Hanley et al. [35]. LFA-1: leukocyte function-associated antigen-1; iICAM-1: immobilized Intercellular Adhesion Molecule-1.
It is observed from Fig. 8 that if we choose the bond constant , the PMF becomes more linear and it is insensitive to the bond constant. On the other hand, for the values of , only 10% of bond constant values lay within the biological range. Therefore, in the present study, we choose to demonstrate PMF profile for the values k = 0.1 N/m and k = 1.0 N/m. The excellent agreement between the analytical and our numerical results reiterates the preservation of equilibrium distribution of the canonical ensemble by our dynamics method for different bond constants. The successful validation of the computed PMF using our hybrid scheme also highlights a concrete path for temporal multiscaling; namely, to reach a y-coordinate value of 0.4 nm corresponds to a PMF of , requires a time scale of , which is currently much outside the scope of conventional dynamics; however, the umbrella sampling strategy enables us to evaluate equilibrium probability distributions associated with rare-to-occur events.
4. Computational Time
The approximate number of CPU cycles required for the computation of 20,000 time steps for a particle of radius 250 nm in a cylindrical tube of radius 5 m (about 10,000 mesh nodes) is . All simulations were carried out on a 2.93 GHz processor in which the wall clock time for this typical run is h.
5. Conclusions
A hybrid approach based on Markovian fluctuating hydrodynamics of the fluid and a non-Markovian Langevin dynamics with the Ornstein–Uhlenbeck noise perturbing the translational and rotational equations of motion of a neutrally buoyant nanoparticle trapped in a harmonic potential is employed to simulate the Brownian motion in an incompressible Newtonian stationary fluid medium contained in a horizontal circular vessel. We demonstrate that the thermal equipartition of translation, rotation, and spring degrees of freedom are preserved by our hybrid scheme, while simultaneously resolving the nature of the hydrodynamic correlations. Our model shows that nanocarrier binding to the vessel wall is very sensitive to mechanical spring stiffness within the known biological range of spring constants for relevant adhesion molecules. This enables the determination of probability distributions and extensive conformational sampling of nanocarrier motion which is prohibitive by conventional dynamics. The framework we have presented in this article provides a comprehensive platform for temporal multiscale modeling of hydrodynamic and microscopic interactions mediating nanocarrier motion in vascular targeted drug delivery.
Acknowledgment
This work was sponsored by National Institute of Health (NIH) Grant No. R01 EB006818 (D.M.E.); National Science Foundation (NSF) Grant No. CBET-0853389. Computational resources were provided in part by the National Partnership for Advanced Computational Infrastructure under Grant No. MCB060006.
Glossary
Nomenclature
- R =
radius of the circular vessel
- L =
length of the circular vessel
- a =
radius of the nanoparticle
- =
velocity of the fluid
- =
density of the fluid
- =
density of the nanoparticle
- =
stress tensor
- p =
pressure
- =
identity tensor
- =
dynamic viscosity
- =
random stress tensor
- =
Boltzmann constant
- T =
absolute temperature
- =
Kronecker delta
- =
Dirac delta
- m =
mass of the particle
- =
moment of inertia of the particle
- =
translational and angular velocities of the particle
- =
position of the centroid of the particle
- =
vector from the center of the particle to a point on its surface
- =
the particle surface
- =
inlet boundary
- =
outlet boundary
- =
unit normal vector on the surface of the particle pointing into the particle
- =
random force
- =
random torque
- =
Brownian relaxation time
- =
Brownian diffusive time
- =
hydrodynamic time
- =
harmonic time for a single oscillation of spring
- =
Stokes dissipative friction force and torque coefficient
- =
characteristic memory times
- =
spring force
- =
spring torque
- k =
spring constant
- l =
length of the spring
- =
unit vector pointing toward the tip of the antigen attached to the wall
Contributor Information
B. Uma, Department of Anesthesiology and Critical Care, University of Pennsylvania, Philadelphia, PA 19104, e-mail: umab@seas.upenn.edu
R. Radhakrishnan, Department of Bioengineering, University of Pennsylvania, Philadelphia, PA 19104, e-mail: rradhak@seas.upenn.edu
D. M. Eckmann, Department of Anesthesiology and Critical Care, University of Pennsylvania, Philadelphia, PA 19104, e-mail: David.Eckmann@uphs.upenn.edu
P. S. Ayyaswamy, Department of Mechanical Engineering, and Applied Mechanics, University of Pennsylvania, Philadelphia, PA 19104, e-mail: ayya@seas.upenn.edu.
References
- [1]. Swaminathan, T. , Liu, J. , Uma, B. , Ayyaswamy, P. , Radhakrishnan, R. , and Eckmann, D. , 2011, “Dynamic Factors Controlling Carrier Anchoring on Vascular Cells,” IUBMB Life, 63(8), pp. 640–64710.1002/iub.475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2]. Muzykantov, V. , Radhakrishnan, R. , and Eckmann, D. , 2012, “Dynamic Factors Controlling Targeting Nanocarriers to Vascular Endothelium,” Curr. Drug Metab., 113, pp. 70–8110.2174/138920012798356916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3]. Calderon, A. J. , Muzykantov, V. , Muro, S. , and Eckmann, D. M. , 2009, “Flow Dynamics, Binding and Detachment of Spherical Carriers Targeted to ICAM-1 on Endothelial Cells,” Biorheology, 46, pp. 323–34110.3233/BIR-2009-0544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4]. Calderon, A. J. , Bhowmick, T. , Leferovich, J. , Burman, B. , Pichette, B. , Muzykantov, V. , Eckmann, D. M. , and Muro, S. , 2011, “Optimizing Endothelial Targeting by Modulating the Antibody Density and Particle Concentration of Anti-ICAM Coated Carriers,” J. Controlled Release, 150(1), pp. 37–4410.1016/j.jconrel.2010.10.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5]. Munn, L. L. , Melder, R. J. , and Jain, R. K. , 1996, “Role of Erythrocytes in Leukocyte-Endothelial Interactions: Mathematical Model and Experimental Validation,” Biophys. J., 71(1), pp. 466–47810.1016/S0006-3495(96)79248-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6]. Liu, J. , Weller, G. E. R. , Zern, B. , Ayyaswamy, P. S. , Eckmann, D. M. , Muzykantov, V. R. , and Radhakrishnan, R. , 2010, “A Computational Model for Nanocarrier Binding to Endothelium Validated Using In Vivo, In Vitro, and Atomic Force Microscopy Experiments,” Proc. Natl. Acad. Sci. U.S.A., 107, pp. 16530–1653510.1073/pnas.1006611107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7]. Uma, B. , Swaminathan, T. N. , Ayyaswamy, P. S. , Eckmann, D. M. , and Radhakrishnan, R. , 2011, “Generalized Langevin Dynamics of a Nanoparticle Using a Finite Element Approach: Thermostating With Correlated Noise,” J. Chem. Phys., 135, p. 114104.10.1063/1.3635776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8]. Uma, B. , Swaminathan, T. N. , Radhakrishnan, R. , Eckmann, D. M. , and Ayyaswamy, P. S. , 2011, “Nanoparticle Brownian Motion and Hydrodynamic Interactions in the Presence of Flow Fields,” Phys. Fluids, 23, p. 073602.10.1063/1.3611026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9]. Landau, L. D. , and Lifshitz, E. M. , 1959, Fluid Mechanics, Pergamon Press, London [Google Scholar]
- [10]. Hauge, E. H. , and Martin-Löf, A. , 1973, “Fluctuating Hydrodynamics and Brownian Motion,” J. Stat. Phys., 7(3), pp. 259–28110.1007/BF01030307 [Google Scholar]
- [11]. Serrano, M. , and Español, P. , 2001, “Thermodynamically Consistent Mesoscopic Fluid Particle Model,” Phys. Rev. E, 64(4), p. 046115.10.1103/PhysRevE.64.046115 [DOI] [PubMed] [Google Scholar]
- [12]. Sharma, N. , and Patankar, N. A. , 2004, “Direct Numerical Simulation of the Brownian Motion of Particles by Using Fluctuating Hydrodynamic Equations,” J. Comput. Phys., 201(2), pp. 466–48610.1016/j.jcp.2004.06.002 [Google Scholar]
- [13]. Serrano, M. , Gianni, D. , Español, P. , Flekkøy, E. , and Coveney, P. , 2002, “Mesoscopic Dynamics of Voronoi Fluid Particles,” J. Phys. A: Math. General, 35(7), pp. 1605–162510.1088/0305-4470/35/7/310 [Google Scholar]
- [14]. Donev, A. , Vanden-Eijnden, E. , Garcia, A. L. , and Bell, J. B. , 2010, “On the Accuracy of Explicit Finite-Volume Schemes for Fluctuating Hydrodynamics,” Commun. Appl. Math. Comput. Sci., 5(2), pp. 149–19710.2140/camcos.2010.5.149 [Google Scholar]
- [15]. Ladd, A. J. C. , 1993. “Short-Time Motion of Colloidal Particles: Numerical Simulation via a Fluctuating Lattice-Boltzmann Equation,” Phys. Rev. Lett., 70(9), pp. 1339–134210.1103/PhysRevLett.70.1339 [DOI] [PubMed] [Google Scholar]
- [16]. Ladd, A. J. C. , 1994, “Numerical Simulations of Particulate Suspensions via a Discretized Boltzmann Equation. Part 1. Theoretical Foundation,” J. Fluid Mech., 271, pp. 285–30910.1017/S0022112094001771 [Google Scholar]
- [17]. Ladd, A. J. C. , 1994, “Numerical Simulations of Particulate Suspensions via a Discretized Boltzmann Equation. Part 2. Numerical Results,” J. Fluid Mech., 271, pp. 311–33910.1017/S0022112094001783 [Google Scholar]
- [18]. Patankar, N. A. , 2002, “Direct Numerical Simulation of Moving Charged, Flexible Bodies With Thermal Fluctuations,” Technical Proceedings of the 2002 International Conference on Computational Nanoscience and Nanotechnology, Vol. 2, Nano Science and Technology Institute, pp. 93–96 [Google Scholar]
- [19]. Adhikari, R. , Stratford, K. , Cates, M. E. , and Wagner, A. J. , 2005, “Fluctuating Lattice–Boltzmann,” EPL, 71(3), pp. 473–47910.1209/epl/i2004-10542-5 [Google Scholar]
- [20]. Dünweg, B. , and Ladd, A. J. C. , 2008, “Lattice Boltzmann Simulations of Soft Matter Systems,” Adv. Polym. Sci., 221, pp. 89–16610.1007/978-3-540-87706-6_2 [Google Scholar]
- [21]. Nie, D. , and Lin, J. , 2009, “A Fluctuating Lattice-Boltzmann Model for Direct Numerical Simulation of Particle Brownian Motion,” Particuology, 7(6), pp. 501–50610.1016/j.partic.2009.06.012 [Google Scholar]
- [22]. Español, P. , and Zúñiga, I. , 2009, “On the Definition of Discrete Hydrodynamic Variables,” J. Chem. Phys., 131, p. 164106.10.1063/1.3247586 [DOI] [PubMed] [Google Scholar]
- [23]. Español, P. , Anero, J. , and Zúñiga, I. , 2009, “Microscopic Derivation of Discrete Hydrodynamics,” J. Chem. Phys., 131, p. 244117.10.1063/1.3274222 [DOI] [PubMed] [Google Scholar]
- [24]. Atzberger, P. J. , Kramer, P. R. , and Peskin, C. S. , 2007, “A Stochastic Immersed Boundary Method for Fluid-Structure Dynamics at Microscopic Length Scales,” J. Comput. Phys., 224(2), pp. 1255–129210.1016/j.jcp.2006.11.015 [Google Scholar]
- [25]. Ermak, D. L. , and McCammon, J. A. , 1978, “Brownian Dynamics With Hydrodynamic Interactions,” J. Chem. Phys., 69(4), pp. 1352–136010.1063/1.436761 [Google Scholar]
- [26]. Brady, J. F. , and Bossis, G. , 1988, “Stokesian Dynamics,” Ann. Rev. Fluid Mech., 20(1), pp. 111–15710.1146/annurev.fl.20.010188.000551 [Google Scholar]
- [27]. Foss, D. R. , and Brady, J. F. , 2000, “Structure, Diffusion and Rheology of Brownian Suspensions by Stokesian Dynamics Simulation,” J. Fluid Mech., 407, pp. 167–20010.1017/S0022112099007557 [Google Scholar]
- [28]. Banchio, A. J. , and Brady, J. F. , 2003, “Accelerated Stokesian Dynamics: Brownian Motion,” J. Chem. Phys., 118(22), pp. 10323–1033210.1063/1.1571819 [Google Scholar]
- [29]. Iwashita, T. , Nakayama, Y. , and Yamamoto, R. , 2008, “A Numerical Model for Brownian Particles Fluctuating in Incompressible Fluids,” J. Phys. Soc. Jpn, 77(7), p. 074007.10.1143/JPSJ.77.074007 [Google Scholar]
- [30]. Iwashita, T. , and Yamamoto, R. , 2009, “Short-Time Motion of Brownian Particles in a Shear Flow,” Phys. Rev. E, 79(3), p. 031401.10.1103/PhysRevE.79.031401 [DOI] [PubMed] [Google Scholar]
- [31]. Kubo, R. , 1966, “The Fluctuation-Dissipation Theorem,” Rep. Prog. Phys., 29(1), pp. 255–28410.1088/0034-4885/29/1/306 [Google Scholar]
- [32]. Kubo, R. , Toda, M. , and Hashitsume, N. , 1991, Nonequilibrium Statistical Mechanics, 2nd ed., Springer-Verlag, Berlin [Google Scholar]
- [33]. Uma, B. , Eckmann, D. M. , Ayyaswamy, P. S. , and Radhakrishnan, R. , 2012, “A Hybrid Formalism Combining Fluctuating Hydrodynamics and Generalized Langevin Dynamics for the Simulation of Nanoparticle Thermal Motion in an Incompressible Fluid Medium,” Mol. Phys., 110, pp. 1057–106710.1080/00268976.2012.663510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34]. Bell, G. , Dembo, M. , and Bongrand, P. , 1984, “Cell Adhesion. Competition Between Nonspecific Repulsion and Specific Bonding,” Biophys. J., 45(6), pp. 1051–106410.1016/S0006-3495(84)84252-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35]. Hanley, W. , McCarty, O. , Jadhav, S. , Tseng, Y. , Wirtz, D. , and Konstantopoulos, K. , 2003, “Single Molecule Characterization of P-Selectin/Ligand Binding,” J. Biol. Chem., 278(12), pp. 10556–1056110.1074/jbc.M213233200 [DOI] [PubMed] [Google Scholar]
- [36]. Radhakrishnan, R. , Uma, B. , Liu, J. , Ayyaswamy, P. , and Eckmann, D. , “Temporal Multiscale Approach for Nanocarrier Motion With Simultaneous Adhesion and Hydrodynamic Interactions in Targeted Drug Delivery,” J. Comput. Phys., Special Issue on Multiscale Modeling and Simulation of Biological Systems (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37]. Uma, B. , Radhakrishnan, R. , Eckmann, D. , and Ayyaswamy, P. , “Fluctuating Hydrodynamics Approach for the Simulation of Nanoparticle Brownian Motion in a Newtonian Fluid,” Proceedings of the 21st National and 10th ISHMT-ASME Heat and Mass Transfer Conference (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38]. Uma, B. , Radhakrishnan, R. , Eckmann, D. , and Ayyaswamy, P. , “A Hybrid Approach for the Simulation of the Thermal Motion of a Nearly Neutrally Buoyant Nanoparticle in an Incompressible Newtonian Fluid Medium,” ASME J. Heat Transfer (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39]. Agrawal, N. , and Radhakrishnan, R. , 2007, “The Role of Glycocalyx in Nanocarrier-Cell Adhesion Investigated Using a Thermodynamic Model and Monte Carlo Simulations,” J. Phys. Chem. C, 111(43), pp. 15848–1585610.1021/jp074514x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40]. Liu, J. , Agrawal, N. , Calderon, A. , Ayyaswamy, P. , Eckmann, D. , and Radhakrishnan, R. , 2011, “Multivalent Binding of Nanocarrier to Endothelial Cells Under Shear Flow,” Biophys. J., 101(2), pp. 319–32610.1016/j.bpj.2011.05.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41]. Grmela, M. , and Öttinger, H. , 1997, “Dynamics and Thermodynamics of Complex Fluids. I. Development of a General Formalism,” Phys. Rev. E, 56(6), pp. 6620–663210.1103/PhysRevE.56.6620 [Google Scholar]
- [42]. Öttinger, H. , and Grmela, M. , 1997, “Dynamics and Thermodynamics of Complex Fluids. II. Illustrations of a General Formalism,” Phys. Rev. E, 56(6), pp. 6633–665510.1103/PhysRevE.56.6633 [Google Scholar]
- [43]. Patankar, N. A. , Singh, P. , Joseph, D. D. , Glowinski, R. , and Pan, T. W. , 2000, “A New Formulation of the Distributed Lagrange Multiplier/Fictitious Domain Method for Particulate Flows,” Int. J. Multiphase Flow, 26, pp. 1509–152410.1016/S0301-9322(99)00100-7 [Google Scholar]
- [44]. Chen, Y. , Sharma, N. , and Patankar, N. , 2006, “Fluctuating Immersed Material (FIMAT) Dynamics for the Direct Simulation of the Brownian Motion of Particles,” Proceedings of the IUTAM Symposium on Computational Approaches to Multiphase Flow, S. Balachandar and Prosperetti A., eds., Springer, Dordrecht, The Netherlands, pp. 119–12910.1007/1-4020-4977-3_13 [Google Scholar]
- [45]. Hu, H. , 1996, “Direct Simulation of Flows of Solid-Liquid Mixtures,” Int. J. Multiphase Flow, 22(2), pp. 335–35210.1016/0301-9322(95)00068-2 [Google Scholar]
- [46]. Hu, H. H. , Patankar, N. A. , and Zhu, M. Y. , 2001, “Direct Numerical Simulations of Fluid-Solid Systems Using the Arbitrary Langrangian-Eulerian Technique,” J. Comput. Phys., 169(2), pp. 427–46210.1006/jcph.2000.6592 [Google Scholar]
- [47]. Zwanzig, R. , and Bixon, M. , 1970, “Hydrodynamic Theory of the Velocity Correlation Function,” Phys. Rev. A, 2(5), pp. 2005–201210.1103/PhysRevA.2.2005 [Google Scholar]
- [48]. Roux, B. , 1995, “The Calculation of the Potential of Mean Force Using Computer Simulations,” Comput. Phys. Commun., 91(1–3), pp. 275–28210.1016/0010-4655(95)00053-I [Google Scholar]
- [49]. Zhang, X. , Wojcikiewicz, E. , and Moy, V. , 2002, “Force Spectroscopy of the Leukocyte Function-Associated Antigen-1/Intercellular Adhesion Molecule-1 Interaction,” Biophys. J., 83(4), pp. 2270–227910.1016/S0006-3495(02)73987-8 [DOI] [PMC free article] [PubMed] [Google Scholar]