Embryo suspensor length is crucial for fast developmental progression in Arabidopsis seed.
Abstract
The first structure that differentiates during plant embryogenesis is the extra-embryonic suspensor that positions the embryo in the lumen of the seed. A central role in nutrient transport has been ascribed to the suspensor in species with prominent suspensor structures. Little is known, however, about what impact the size of the rather simple Arabidopsis (Arabidopsis thaliana) suspensor has on embryogenesis. Here, we describe mutations in the predicted exo-polygalacturonase gene NIMNA (NMA) that lead to cell elongation defects in the early embryo and markedly reduced suspensor length. Mutant nma embryos develop slower than wild-type embryos, and we could observe a similar developmental delay in another mutant with shorter suspensors. Interestingly, for both genes, the paternal allele has a stronger influence on the embryonic phenotype. We conclude that the length of the suspensor is crucial for fast developmental progression of the embryo in Arabidopsis.
Annual, self-pollinating weeds such as Arabidopsis (Arabidopsis thaliana) benefit from a short life cycle in their natural ephemeral habitats (Snell and Aarssen, 2005). A rapid progression through embryogenesis is a prerequisite for early seed maturation, and it is therefore not surprising that Arabidopsis sets up its body plan already after a very limited number of cell divisions (Aarssen, 2000; Lau et al., 2012).
Arabidopsis embryogenesis starts with the fertilized egg cell, the zygote, which elongates about 3-fold before it divides asymmetrically. The smaller apical cell is the founder of the embryo proper that contributes to most of the later seedling, while the larger basal cell develops into a support structure called the suspensor (Jeong et al., 2011a). The suspensor is formed by a series of transverse cell divisions followed by longitudinal cell expansion forming a stalk-like structure. Only the upper-most suspensor cell, the hypophysis, will contribute to parts of the root meristem, while the rest of the suspensor remains extraembryonic and will cease its growth at the heart stage of the embryo (Yeung and Meinke, 1993). The suspensor is thought to be important for pushing the embryo into the lumen of the seed, where the embryo is surrounded by the nourishing endosperm. In addition, a key function in nutrient and hormone transport to the embryo is assigned to the suspensor (Kawashima and Goldberg, 2010).
The Arabidopsis suspensor achieves its maximum length with a minimum number of cells by having a rod-shaped structure built by a single cell file. Although several mutants with distorted or shorter suspensors have been described in Arabidopsis, little is known about what impact suspensor length has on embryo development (Schwartz et al., 1994; Vernon and Meinke, 1994; Lukowitz et al., 2004; Breuninger et al., 2008; Bayer et al., 2009; Jeong et al., 2011b).
As in all plant cells, the size and shape of suspensor cells is primarily determined by the elasticity of the cell wall. While rigid cellulose microfibrils determine the direction of cell expansion, it is the pectin matrix that plays a major role in determining the stiffness of the cell wall (Peaucelle et al., 2012). Several pectin-modifying enzymes have been described that modulate the elasticity of the cell wall. Among these, pectin methyl esterases that determine the degree of homogalacturonan methylation seem to be major players (Palin and Geitmann, 2012). Pectin-degrading enzymes such as polygalacturonases (PGs) have been implicated with cell expansion, but their role in this process is much less clear (Hadfield and Bennett, 1998).
The pectin-rich middle lamella serves as cement to aid cell adhesion, and it is therefore not surprising that several PG genes are involved in dehiscence and abscission events (Rhee et al., 2003; González-Carranza et al., 2007, 2012; Ogawa et al., 2009). The expression profile of many PG genes correlates temporally and spatially with cell elongation and therefore implies a role in cell expansion (Sitrit et al., 1999). The exact function of PGs during cell elongation is still unclear, and the lack of elongation defects in loss-of-function mutants further complicates matters.
Here, we describe mutations in a PG gene, which we called NIMNA (NMA), that severely affect cell elongation in the embryo and suspensor.
We demonstrate that embryos with shorter suspensors show a significant delay in development during embryogenesis, possibly caused by reduced access to nutrients from the endosperm. Interestingly, reciprocal crosses reveal an unequal parental contribution to NMA function in cell elongation of the suspensor.
RESULTS
In a forward genetic screen for embryonic mutants, we isolated a candidate with abnormal suspensor development. In this mutant, suspensors were severely reduced in length and embryos were positioned in the micropylar niche, instead of being pushed into the lumen of the seed (Fig. 1). We therefore called the mutant nimna (nma) after the Sanskrit word for “sunken.”
Figure 1.
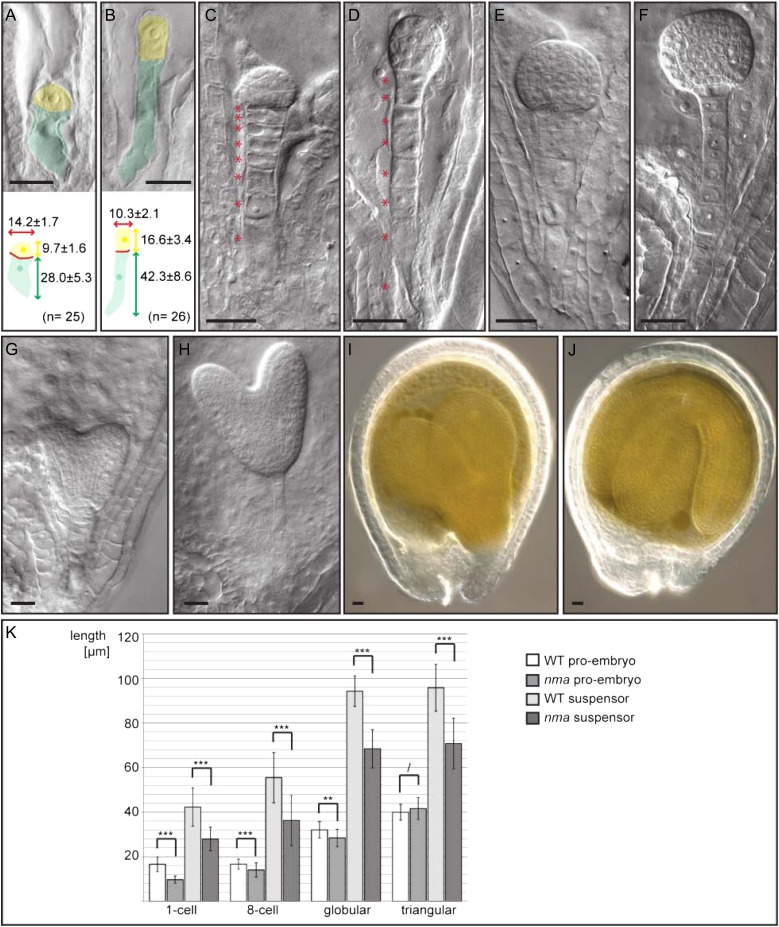
Embryonic phenotype of nma mutants compared with the wild type. A and B, Embryonic phenotype at one-cell stage of nma (A) and the wild type (B). Apical cells are false colored in yellow; basal cells are false colored in green. Measurements are given as average with sd in micrometers. C and D, Embryonic phenotype at 16-cell stage of nma (C) and the wild type (D). Suspensor cells are highlighted by accompanying asterisks. E and F, Late globular stage embryos of nma (E) and the wild type (F). G and H, Heart stage embryos of nma (G) and the wild type (H). I and J, Developing seed of nma (I) and the wild type (J) 7 dap. Bars = 20 µm. K, Measurements of embryo proper and suspensor length at different developmental stages. Number of analyzed embryos at one-cell stage: nma, n = 26 and the wild type, n = 26. Number at eight-cell stage: nma, n = 101 and the wild type, n = 98. Number at globular stage: nma, n = 36 and the wild type, n = 29. Number at triangular stage: nma, n = 54 and the wild type, n = 33. Mean values with sd are shown. Significant differences were determined in pairwise comparison by Mann-Whitney U test (* = P < 0.05, ** = P < 0.01, *** = P < 0.001, and / = P > 0.05). WT, Wild type.
Differences between mutant and wild-type embryos become apparent directly after fertilization. In nma mutants, the anisotropic cell elongation of the zygote is reduced, resulting in shorter but wider daughter cells (Fig. 1A). Initially, cells of the embryo proper and the suspensor are affected in cell elongation, but cells in the proembryo recover by early heart stage, while the cells of the suspensor remain short and wide (Fig. 1).
The short-suspensor phenotype is primarily caused by shorter cells and not by reduced cell number in the suspensor, pointing toward a function of NMA in cell elongation (Fig. 1, C and D).
We were able to recover homozygous seedlings, and adult plants show no apparent differences from the wild type (Supplemental Fig. S1).
Embryos with Shorter Suspensor Show a Lag in Developmental Progression
When studying the nma phenotype in detail, we noticed an apparent developmental delay in nma embryos compared with the wild type.
To determine the developmental stages of embryos at a defined time point, we emasculated and self pollinated nma-1–/– mutants and wild-type plants and analyzed the resulting embryos 4 d after pollination (dap). To ensure that fertilization of the analyzed embryos took place in a narrow time window, we only analyzed 10 to 20 neighboring ovules from the upper half of each silique. In our analysis, embryos were classified into six developmental stages: midglobular, late globular, triangular, early heart, late heart, and early torpedo (Fig. 2A).
Figure 2.
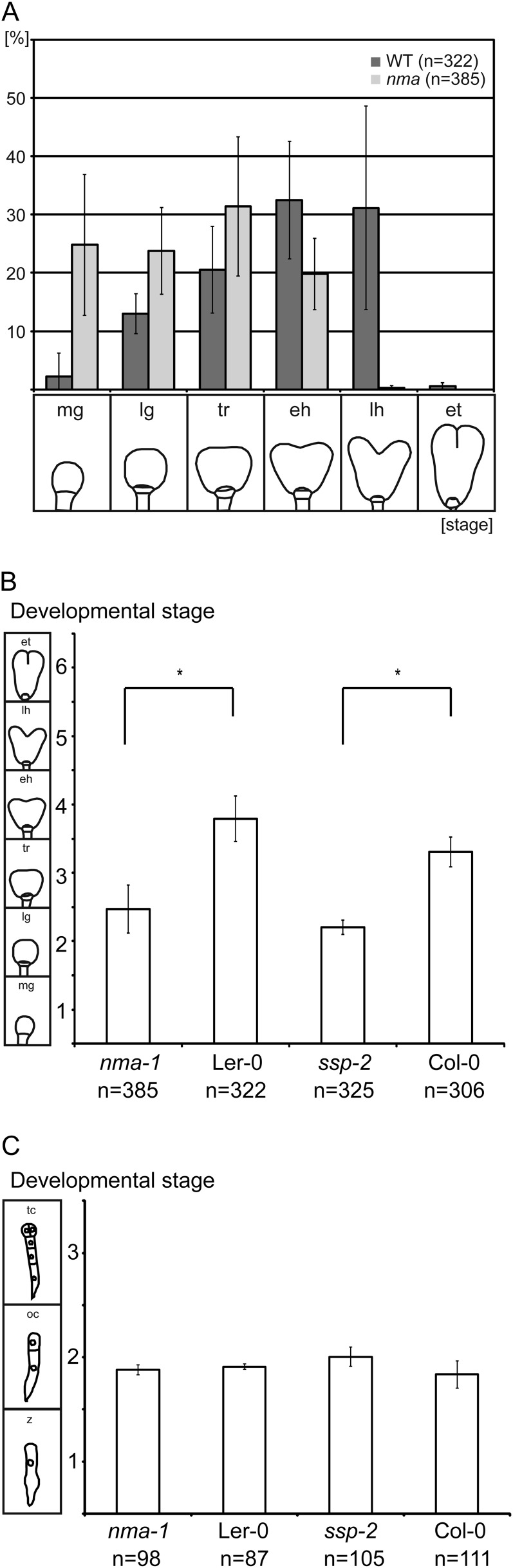
Developmental stages of embryos. A, Wild-type and nma embryos 4 dap were classified into six developmental stages: midglobular (mg), late globular (lg), triangular (tr), early heart (eh), late heart (lh), and early torpedo (et). Schematic depictions of the developmental stage are given below the graphs. Mean values of three independent biological replicates with sd are shown. B, Average developmental stage of embryos 4 dap in nma-1, Ler, ssp-2, and Col-0. Embryos were classified in six developmental stages as in Figure 2A. Numerical values were assigned to each stage. Averages and sds of three biological replicates are shown as bar graphs. Asterisks indicate sds in pairwise comparison (Mann-Whitney U test; P < 0.001). C, Average developmental stage of embryos 30 h after pollination in nma-1, Ler, ssp-2, and Col-0. Embryos were classified in three developmental stages: zygote (Z), one-cell (oc), and two- and four-cell (tc). Numerical values were assigned to each stage. Averages and sds of three biological replicates are shown as bar graphs.
Compared with the wild type, nma embryos consistently appeared to be at earlier developmental stages. At 4 dap, the majority of wild-type embryos reached early heart stage, while most nma embryos belonged to late globular or triangular stage. By using numerical values for the different developmental stages, the average for nma embryos (2.47 ± 0.35; n = 385) was significantly lower than the average for the wild type (3.79 ± 0.33; n = 322; Student’s t test, P < 0.01; Fig. 2B). This indicates that the nma mutation affects developmental progression of the embryo in addition to cell elongation. This might be a consequence of lower nutrient allocation to the embryo due to reduced suspensor or embryo surface in contact with the endosperm.
To test if this developmental delay is caused by the reduced suspensor size or depends on a NMA function in the embryo proper, we analyzed the developmental progression of embryos in another mutant with shorter suspensors.
The SHORT SUSPENSOR (SSP) gene functions in the YODA (YDA) mitogen-activated protein kinase pathway to determine aspects of suspensor identity (Lukowitz et al., 2004; Bayer et al., 2009). Although ssp embryos have shorter suspensors like nma embryos, the cause of the reduced size is different. The majority of ssp mutant suspensors are shorter than the wild type because they consist of fewer cells, while the average cell size is unaffected (Bayer et al., 2009). We therefore considered ssp mutants as good candidates to test if suspensor size affects the timing of embryonic development.
As with nma mutants, we self pollinated ssp-2–/– mutants and compared the developmental stages of embryos 4 dap to those of wild-type plants pollinated at the same time. As in nma mutants, we can observe a significant lag in developmental progression in mutant embryos (Fig. 2B).
Our classification of embryonic developmental stages was based on the morphology of the embryo. We therefore had to avoid possibly misinterpreting an altered morphology in the mutant embryo as a younger developmental stage. To make sure that ssp and nma embryos do show a delay in development and not just a change in overall shape, we counted, for each genotype, the cells in a central optical section of embryos 3 dap (Supplemental Fig. S2). Both nma and ssp mutant embryos show significantly fewer cells in the proembryo than the corresponding wild-type embryos, indicating that they develop slower than their wild-type counterparts (P < 0.001 in Mann-Whitney U test).
This could indicate that the reduced suspensor length in these mutants is responsible for the delay in development of the embryo. Alternatively, delayed fertilization in these mutants could cause the observed difference in development. Because NMA functions in cell elongation and is expressed in pollen, slower pollen tube growth in the mutant would be a likely scenario. We therefore measured the pollen tube length of in vitro-germinated nma-1 and wild-type pollen. After 6 h of inoculation in pollen tube growth medium, the average length of nma pollen tubes (134 ± 56 µm; n = 189) did not differ significantly from the average length of wild-type pollen tubes (129 ± 46 µm; n = 146; P > 0.79 in Mann-Whitney U test; Supplemental Fig. S3). In addition, we analyzed the segregation of the mutant allele in progeny of reciprocal crosses between wild-type plants and nma heterozygous plants. There was no significant difference to the expected one-to-one segregation ratio of nma+/– and wild-type plants in the offspring of nma+/– × wild-type crosses (90 nma+/– and 79 wild-type seedlings) and wild-type × nma+/– crosses (76 nma+/– and 97 wild-type seedlings; chi-square test showed no significant difference to expected segregation at 5% level), respectively. These results strongly suggest that the developmental delay we observe in nma embryos is not caused by slower pollen tube growth and delayed fertilization. For ssp, it was also shown that there is no significant divergence from Mendelian segregation in a F2 population, suggesting no effect of this mutation on pollen tube growth (Bayer et al., 2009). To test if the observed lag in development of ssp and nma mutants arises during embryogenesis and not during the fertilization process, we analyzed developmental stages of embryos 30 h after pollination in a similar manner as we have done before at 4 dap (Fig. 2C). Embryos were classified into three developmental stages with assigned numerical values: zygote stage (1), one-cell stage (2), and two-/four-cell stage (3). In differential interference contrast images of cleared ovules, it is not always easy to distinguish the two- and four-cell stage of the embryo proper. We therefore combined these in a single developmental stage to avoid possible mistakes.
At this early time point of embryogenesis, there is no significant difference in development between nma, ssp, and the corresponding wild-type embryos, respectively (nma-1/ecotype Landsberg erecta [Ler], P > 0.05; ssp-2/ecotype Columbia [Col-0], P > 0.05 in Mann-Whitney U test; Fig. 2C).
These results clearly indicate that the observed differences in developmental progression are not caused by delayed fertilization but arise during the course of embryogenesis.
NMA Function Has a Stronger Paternal Contribution
Mutations in ssp show a paternal effect on embryogenesis. This unusual parent-of-origin effect was discussed in the light of a parental conflict regarding nutrient allocation to the embryo (Bayer et al., 2009). Because nma embryos are also affected in developmental progression, we wondered if NMA function might also be under paternal control. To test for any parent-of-origin effects, we performed reciprocal crosses between nma-1 and wild-type plants. We could observe significantly shorter suspensors in crosses where nma was introduced from the paternal side (wild type × nma) compared with crosses from the maternal side (nma × wild type) or crosses of wild-type plants (wild type × wild type; P < 0.001 in Mann-Whitney U test; Supplemental Fig. S4). Homozygous nma embryos exhibit even shorter suspensors than the paternal cross. This argues for a dosage dependency of the suspensor phenotype with an unequal parental contribution (Supplemental Fig. S4).
NMA Codes for a Predicted Exo-PG
We identified the affected gene in nma-1 by map-based cloning (Lukowitz et al., 2000). Bulk segregant analysis as well as rough mapping positioned NMA on chromosome 2 in a 380-kb interval between marker T32F6 and F4P9. Screening 4,002 chromosomes for recombinations in this interval, we were able to recover 17 F2 plants that defined a 153-kb interval between markers 255-X (At2g33255) and 5B2-1 (At2g32870) with no further recombination (Supplemental Fig. S5).
Sequencing of coding regions in this interval revealed a G-to-A substitution at nucleotide position +836 in the protein coding sequence of At2g33160. This point mutation leads to an amino acid substitution of a conserved Gly to Asp at position 279 in the predicted PG protein sequence (Supplemental Fig. S6). We were able to confirm this candidate gene by analyzing additional transfer DNA (T-DNA) insertion alleles. SALK_126968 (nma-2) and SALK_015991 (nma-3) exhibit embryonic phenotypes that are basically indistinguishable from nma-1 (Supplemental Fig. S7). We could not detect transcripts downstream of the insertion site in nma-3 by reverse transcription (RT)-PCR, which indicated either low expression or that this line is possibly a null allele (data not shown). To confirm the predicted NMA gene model, we performed RT-PCR and 3′ RACE, which revealed an incorrectly annotated splice donor site in the TAIR10 release. The corrected gene model contains a single intron and codes for a protein with a predicted N-terminal signal peptide (SP) followed by a single PG domain (Supplemental Fig. S6). The corrected gene model will be submitted to The Arabidopsis Information Resource (TAIR).
We tested the mapping result by genetic complementation with a genomic fragment covering the coding region of At2g33160, including 2.5 kb upstream of the translational start codon and 1.6 kb downstream of the stop codon. In addition, we generated a variant of the above construct that carries a yellow fluorescent protein (YFP) at the N terminus of the mature protein after cleavage of the predicted SP. Both constructs complemented the embryonic phenotype of nma-1, further confirming that At2g33160 is the affected gene (all 17 transgenic lines carrying pNMA::NMA, and 27 out of 30 transgenic lines carrying pNMA::YFP-NMA complemented the phenotype; Supplemental Fig. S8).
NMA Is Predominantly Expressed in Reproductive Tissue
Mutant nma plants show obvious aberrant phenotypes only during embryogenesis. We therefore wondered if this is due to a very restricted expression pattern of NMA. Expression of redundant genes outside the embryo could also account for the temporal and spatial restriction of the phenotype. To monitor NMA expression with cellular resolution, we transcriptionally fused a 2.5-kb promoter fragment to nuclear-localized triple YFP (Fig. 3).
Figure 3.
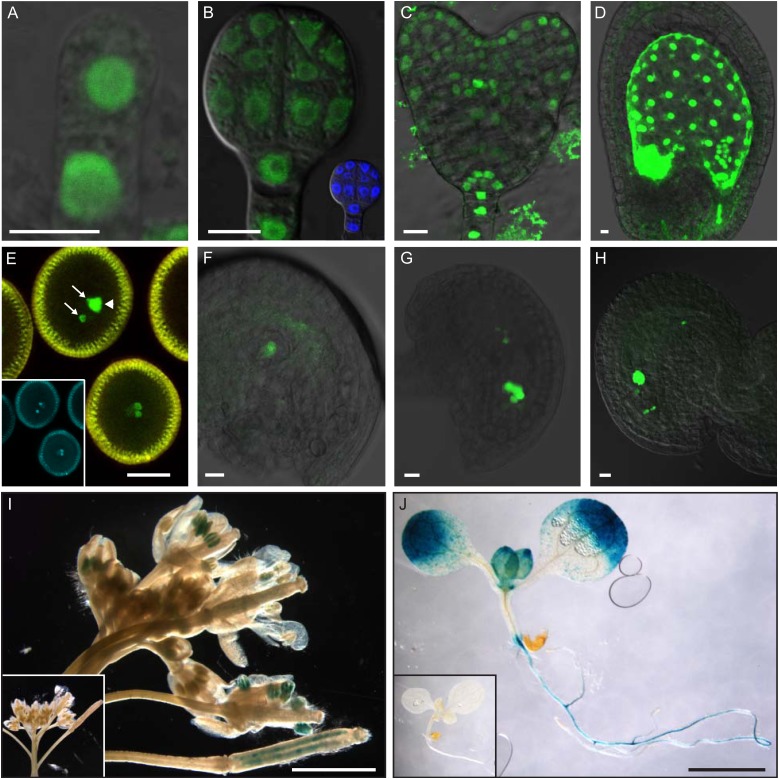
Expression analysis of NMA promoter::reporter gene constructs. A to H, Fluorescence micrographs of transgenic plants carrying a transcriptional fusion of a NMA promoter fragment to nuclear-localized triple Venus-YFP. A, Embryo after the first zygotic division. B, Globular-stage embryo, inset shows 4′,6-diamidino-2-phenylindole (DAPI) staining. C, Heart-stage embryo. D, Developing seed 2 dap. E, Pollen, arrows mark sperm cell nuclei and arrowhead indicates vegetative cell nucleus; inset shows DAPI staining. F, Female gametophyte after meiosis. G, Female gametophyte at eight-nuclei stage. H, Mature female gametophyte. Bars = 10 μm. I and J, GUS activity staining in transgenic plants carrying a transcriptional pNMA::GUS fusion construct. I, Inflorescence. J, Seedling. Insets in I and J show transgenic control plants without GUS gene. Bars = 2 mm.
After fertilization, NMA promoter activity can be observed in all cells of the embryo proper and the suspensor with similar fluorescence intensity (Fig. 3A). At late globular stage, the expression in the proembryo is reduced, while it stays strong in the suspensor (Fig. 3B). From heart stage onwards, NMA promoter activity is restricted to cells of the cotyledons, stele, quiescent center, and columella, as well as suspensor cells (Fig. 3C).
In the endosperm, pNMA::n3xYFP is initially expressed throughout at a very high level but becomes restricted to the chalazal endosperm from late heart stage onwards (Fig. 3D).
Because the expression of NMA can be detected already in the zygote, we also analyzed expression of the reporter gene in male and female gametophytes. YFP expression can be detected in all three cells of the mature pollen, with highest levels in the sperm cells (Fig. 3E). In the female gametophyte, YFP expression can be observed after meiosis in the functional megaspore and the nuclei of the developing gametophyte (Fig. 3, F and G). After cellularization, the central cell shows very strong signal, while egg cell, synergids, and antipodal remainder exhibit much weaker expression (Fig. 3H).
To observe NMA expression in seedlings and adult plants, we made a pNMA::GUS transcriptional fusion. Strong GUS expression can be detected as expected in pollen and young ovules (Fig. 3I). Additionally, GUS activity is also present in expanding leaf margins and root vasculature (Fig. 3J).
We corroborated this data by measuring NMA expression in various tissue types by quantitative RT-PCR (Supplemental Fig. S9). NMA transcripts accumulate to high levels in reproductive tissue, such as flowers, pollen, and siliques. Very low level of expression can be detected throughout the plant. This data are also in agreement with publicly available microarray data (Supplemental Fig. S10; Schmid et al., 2005; Winter et al., 2007).
NMA Acts Cell Autonomously in Suspensor Cell Elongation
The strong expression of NMA in the endosperm raises the question of whether the cell elongation defect in the embryo is caused in a cell-autonomous matter or if it is possibly an indirect effect of the central cell restricting the growth of the embryo.
We therefore expressed a translational YFP-fusion of NMA under the control of tissue-specific promoters in nma-1 background and tested for complementation of the mutant phenotype. The OBF BINDING PROTEIN1 (OBP1) promoter is active in the embryo proper and suspensor but not in the endosperm during early embryogenesis (Supplemental Fig. S11; Skirycz et al., 2008), and pOBP1::YFP-NMA expression rescued the cell elongation defects in the suspensor in nma-1 (Supplemental Fig. S8). On the other hand, YFP-NMA expression under the control of the SUCROSE PROTON SYMPORTER5 (SUC5) promoter, which is active in the endosperm but not the embryo during early stages of embryogenesis (Baud et al., 2005; Pommerrenig et al., 2013), could not complement the nma-1 suspensor phenotype. These results argue in favor of a cell-autonomous function of NMA in the embryo. We then expressed YFP-NMA under the control of the J10 promoter, which is exclusively active in the suspensor during early stages of development (Supplemental Fig. S11; J. Kong and G. Jürgens, personal communication). pJ10::YFP-NMA expression was sufficient to rescue the short-suspensor phenotype of nma-1 embryos (Supplemental Fig. S8). Again, this strongly suggests a cell-autonomous function of NMA in cell elongation of the suspensor.
Suspensor-Specific Expression of NMA Is Sufficient to Rescue the Developmental Delay of nma Embryos
Our results indicate that NMA acts cell autonomously in cell elongation. Expressing YFP-NMA suspensor specifically under the control of the J10 promoter in nma–/– mutants leads to wild-type-looking suspensors. The cells of the embryo proper, on the other hand, still display cell elongation defects because YFP-NMA is not expressed in this domain in the transgenic pJ10::YFP-NMA plants. We therefore considered the pJ10::YFP-NMA transgenic line as a good background to test our hypothesis that reduced suspensor length causes slower development of the embryo.
We analyzed the developmental stage of embryos in pJ10::YFP-NMA transgenic nma–/– plants compared with nma and wild-type plants 4 dap in a similar way as we had before. Expressing YFP-NMA only in the suspensors of nma–/– mutants rescues the developmental delay of the embryo proper (Supplemental Figure S12).
By using numerical values for the different developmental stages, the average for pJ10::YFP-NMA embryos (3.01 ± 0.09; n = 374) was significantly higher than for nma embryos (2.48 ± 0.29; n = 500; Student’s t test P < 0.05) but did not completely reach the wild-type level (3.61 ± 0.27; n = 1102).
This result indicates that in the context of developmental progression, NMA activity in the suspensor affects noncell autonomously the development of the embryo proper. We therefore conclude that the length of the suspensor critically influences the development of the embryo.
NMA Protein Localizes to the Cell Wall
The predicted pectin-modifying function of NMA would suggest an apoplastic localization of the protein. SignalP4.0 predicts a SP with cleavage site between amino acids 21 and 22 at the N terminus of the NMA protein sequence (discrimination score D = 0.798), which would target the protein to the secretory pathway (Petersen et al., 2011). To test the functional requirement of this predicted SP, we made two YFP-tagged versions of NMA under the control of the gene's own regulatory sequences. In one case, we fused the YFP directly at the N terminus (pNMA::YFP-NMA dSP), which displaces the SP from the N terminus and therefore disrupts its presumed function (Rapoport, 2007). In the other case, we introduced YFP after the predicted cleavage site (pNMA::YFP-NMA).
Only the latter version was able to complement the embryonic mutant phenotype when introduced in nma-1 plants (27 out of 30 transgenic lines). The former version did not show any complementation in 12 independent transgenic lines. To test whether this sequence is responsible for targeting the protein to the apoplast, we replaced the NMA promoter in these two constructs with the strong constitutive Cauliflower mosaic virus 35S promoter for transient expression in protoplasts (Fig. 4).
Figure 4.
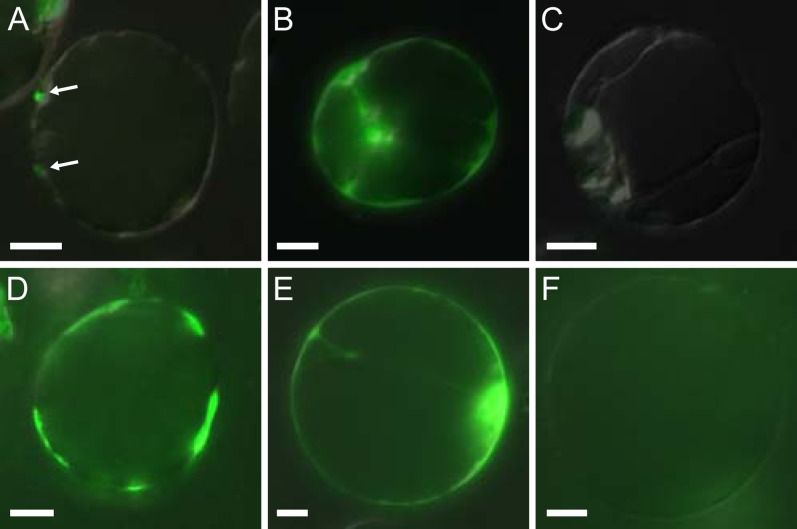
Transient expression of YFP-NMA fusion proteins in protoplasts. A to C, 24 h after transformation. D to F, 72 h after transformation. A and D, p35S::YFP-NMA fusion construct with intact SP sequence. Arrows in A point to dotted signals on protoplast surface. B and E, p35S::YFP-NMA dSP fusion construct with disrupted SP sequence. C and F, Untransformed control. Bars = 10 μm.
The version with intact SP was only detectable as tiny dots on the surface of the protoplasts, while the variant with disrupted SP gave strong cytoplasmic YFP fluorescence 24 h after transformation. Seventy-two hours after transformation, when the protoplasts were reforming cell walls, we could detect strong YFP fluorescence on the surface of the protoplasts in patches of newly formed call walls (Fig. 4) with the construct carrying an intact SP. This signal could not be detected if the SP was disrupted or in untransformed protoplasts. The variant with disrupted SP still showed cytoplasmic YFP localization at this time point (Fig. 4).
We were not able to detect the protein in vivo using the functional YFP-tagged version under the control of the native promoter, probably due to low steady-state protein levels. The cell wall localization of NMA is supported by a proteome study of cell wall proteins in etiolated hypocotyls. The authors of this study detected fragments of the NMA protein in cell wall extracts (Irshad et al., 2008).
DISCUSSION
Embryonic suspensors of flowering plants exist in a wide range of sizes and shapes. Historically, the suspensor was thought to just anchor the embryo within the seed, although some reports already speculated about a function as temporary embryonic root (for review, see Yeung and Meinke, 1993).
The suspensor is the first tissue that differentiates during embryogenesis, and biochemical, physiological, and anatomical studies suggest a role in nutrient supply to the embryo (for review, see Kawashima and Goldberg, 2010). Studies in Phaseolus vulgaris and Phaseolus coccineus clearly demonstrated active transport of nutrients and a growth-promoting function of the suspensor (Cionini et al., 1976; Yeung and Sussex, 1979; Yeung, 1980; Nagl, 1990). In cases where the endosperm supplies few nutrients to the embryo, the suspensor can be large and branched and form haustorium-like structures into the maternal tissue (Wardlaw, 1955); however, these are specialized cases with seeds that contain little endosperm. It is therefore questionable if the physiology and function of the relatively simple Arabidopsis suspensor can be compared to these large suspensors. Although molecular transporters for nutrients and hormones are also expressed in the Arabidopsis suspensor, it is still a matter of debate if, in this case, the suspensor also functions in allocating nutrients to the embryo (Friml et al., 2003; Meyer et al., 2004; Hruz et al., 2008).
We demonstrated that nma and ssp mutant embryos with shorter suspensors show an obvious developmental delay. The two genes have fundamentally different functions, and the mutant phenotypes do not resemble each other. The reduced size of the suspensor is the result of different developmental defects in the two mutants. While in nma, the reduced size is a consequence of failed cell elongation, ssp embryos possess suspensors with reduced cell number (Bayer et al., 2009). The observed developmental delay can therefore not be caused by a common function in the embryo proper. The one thing both mutations have in common is the diminished size of the suspensor. Furthermore, we could show that there is no obvious delay in fertilization in these mutants and that early embryos at 30 h after pollination show no difference in their development compared with wild-type embryos.
We therefore conclude that the developmental delay observed in both mutants is likely caused by the reduced suspensor length. This is supported by the fact that suspensor-specific expression of NMA in nma–/– background is sufficient to partially rescue the developmental delay of the nma embryo proper. This strongly argues in favor of the suspensor length influencing the developmental progression of the embryo. The J10 promoter becomes active only from the eight-cell stage of the embryo onwards (approximately 2 dap). This might explain why we observe only a partial rescue in this experiment.
An important role of the suspensor in nutrient and hormone transport to the embryo has been shown in Phaseolus spp. (Yeung, 1980). It is therefore possible that the reduced size of these mutant Arabidopsis suspensors impairs nutrient allocation to the embryo. In this scenario, a lack of nutrient availability could be the cause for slower growth of the embryo. It is unclear though if the lack of nutrients is a consequence of the embryo not being sufficiently pushed into the nourishing endosperm or because there is less suspensor surface to take up nutrient and transport them to the embryo.
Reciprocal crosses demonstrate that the paternal allele of NMA has a stronger contribution to suspensor length than the maternal allele. Although this effect is rather mild, this unequal parental contribution is unmasked by a dosage dependency. NMA is expressed at higher levels in sperm cells and much lower levels in the egg cell, based on the fluorescence intensity of promoter-reporter gene fusions. Unequal amounts of RNA and/or protein carried over from the gametes during fertilization might be an easy explanation for the different phenotypic strengths seen in reciprocal crosses. It is interesting to observe that the NMA gene that affects suspensor length also shows a paternal effect that seems to rely on a similar mechanism as the paternal effect of the ssp mutant (Bayer et al., 2009). Although this is far from conclusive evidence, it is tempting to speculate if there might be an evolutionary incentive to bring suspensor size and therefore efficient nutrient transport to the embryo under paternal control. It would be an elegant way to balance the maternal control over endosperm proliferation (Spillane et al., 2007). Further studies will be needed to address this question in the light of the parental conflict theory.
NMA displays highest expression levels in reproductive tissue and only low level in vegetative tissue. This expression pattern is common for many plant PGs that have been classified as clade C PGs (Torki et al., 2000). Based on the relative high exo-PG activity of pollen protein extracts, it was assumed that class C PGs are mainly exo-PGs (Pressey, 1991; Barakate et al., 1993). NMA carries all known conserved residues that characterize the protein as a putative PG. These are namely the NTD motif at position 201, GDD at position 223, GHG at position 246, and RIK at position 281, which are conserved residues found in all plant PGs and are diverged in other family 28 hydrolases, such as rhamnogalacturonases or xyloglucan hydrolases (Markovic and Janecek, 2001).
So far, all Arabidopsis PG genes for which a loss-of-function phenotype has been described have been classified as endo-PG and are involved in cell separation and abscission events (Rhee and Somerville, 1998; Rhee et al., 2003; González-Carranza et al., 2007; Ogawa et al., 2009). The lack-of-mutant phenotypes for exo-PG genes in Arabidopsis was thought to be the consequence of high genetic redundancy in this multigene family (Hadfield and Bennett, 1998; Kim et al., 2006).
Notably, nma loss-of-function mutations lead to a strong embryonic phenotype, although there seems to be a large number of PG genes that have generally overlapping expression patterns with NMA (Kim et al., 2006). This could indicate that in the embryo proper, for a short time period after fertilization and in the suspensor in general, there are no other active PGs present. Alternatively, this could also indicate different substrate preferences and/or functions for coexpressed PG family proteins. A more detailed study of expression patterns for these genes with cellular resolution would be necessary to address this question. The embryonic phenotype of nma makes it possible to study specificity of predicted exo-PG genes in the future in complementation assays by expressing various PG-coding regions under the control of NMA regulatory sequences in a nma mutant background.
CONCLUSION
NMA codes for a predicted exo- PG that is involved in cell elongation during early embryogenesis. Loss-of-function mutations in NMA lead to shorter suspensors and a developmental delay in the embryo. This lag in embryonic development can be seen in other embryonic mutants with shorter suspensors. We therefore conclude that the size of the suspensor is critical for efficient developmental progression during Arabidopsis embryogenesis. This study further suggests that there might be an incentive to bring development of the suspensor under paternal control.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana Col-0 and Ler) were used in this study. Plants were grown under long-day conditions (16 h at 3-kilolux illumination, 8-h-dark period) in walk-in chambers at 23°C and 65% relative humidity on commercial potting mix (Topferde CL T, Einheitserde) containing systemic insecticide added with the first watering (Confidor WG70, 200 µg L–1; Bayer CropScience).
T-DNA alleles of nma and ssp (nma-2, SALK_126968; nma-3, SALK_015991; ssp-2, SALK_051462; Alonso et al., 2003) were obtained from the Nottingham Arabidopsis Stock Center. T-DNA insertion lines were genotyped by PCR using primers LBb1.3 and 160-3 R for nma-3, LBb1.3 and SSP1-R for ssp-2, and SSP1-R and SSP1-I for the SSP allele. A derived cleaved amplified polymorphic sequences marker was designed to genotype nma-1 using primers NMA dC-F and NMA dC-R. The PCR product of the mutant allele carries a restriction site for SwaI. Sequences of primers can be found in the supplementary material (Supplemental Table S1).
Genetic Screen for Paternal Effect Mutations
nma-1 was found in a forward genetic screen for paternal effect mutations. Ethyl methanesulfonate-mutagenized M2 Ler plants (Lehle Seeds) were used individually as pollen donors to manually pollinate conditionally sterile oxophytodienoate-reductase3 plants (Stintzi and Browse, 2000). Seed set was monitored, and dissected immature seeds were cleared in Hoyer’s solution for microscopic analysis of embryonic phenotypes 3 dap as described previously (Bayer et al., 2009). A comprehensive summary of the screen will be published elsewhere.
Mapping of nma-1
Map-based cloning of nma-1 was performed as described by Lukowitz et al. (2000). Rough mapping positioned nma-1 on chromosome 2 in a 380-kb interval between marker T32F6 and F4P9. Screening 4,002 chromosomes for recombinations in this interval, we were able to recover 17 F2 plants that defined a 153,640-bp interval between markers 255-X (At2g33255) and 5B2-1 (At2g32870) with no further recombination. Coding regions in this interval were amplified by PCR and sequenced.
Microscopic Analysis of Embryos, Measurements, and Reporter Gene Analysis
Immature seeds were dissected and cleared in Hoyer’s solution as described previously (Bayer et al., 2009) and analyzed using a Zeiss Axio Imager.Z1 with an AxioCam HRc camera. Size measurements were performed using measurement tools of the Axiovision software. Fluorescence microscopy on Olympus FV1000 and Zeiss LSM510 and LSM780 NLO confocal microscopes and GUS activity staining was performed as described previously (Nawy et al., 2010). GUS staining patterns were analyzed using a Zeiss SteREO Discovery.V12 with the Axiovision software.
In Vitro Germination of Pollen and Measurements of Pollen Tubes
Pollen was germinated as described (Boavida and McCormick, 2007), except that the agarose concentration was reduced to 1%. Pollen germination was performed on microscope slides incubated in moist chambers at 23°C for 6 h. Measurements were performed as described above for immature seeds.
Transcript and Expression Analysis
RNA was extracted from various plant tissues using the RNeasy Plant Mini Kit (Qiagen). Synthesis of complementary DNA and quantitative real-time PCR was conducted as described previously (Lau et al., 2011) using primers qRT-F and qRT-R (Supplemental Table S1). For expression analysis in the wild type, nma-2, and nma-3, primers RT-5F and RT-5R, NRT-F3 and NRT-R4, and RT 160.2 F and RT 160.2 R were used (Supplemental Table S1). Three-prime RACE was conducted by using the ExactSTART Eukaryotic mRNA 5′- and 3′-RACE Kit (Epicentre).
Molecular Complementation and Plasmid Constructs
For molecular complementation, a 5.7-kb genomic fragment harboring At2g33160 was PCR-amplified using primers At2g33160P-FC and At2g33160-RC and subsequently cloned into pGreen II 0229 (Hellens et al., 2000). To be able to distinguish transgenes from the endogenous locus, in all other NMA constructs, the intron was removed by site-directed PCR mutagenesis using primers NMAi1-F and NMAi1-R. To obtain translational fusions of citrine YFP with NMA, AscI and PacI restriction sites were introduced either before or right after the predicted SP sequence by site-directed mutagenesis using primers NMAi1 AP-F and NMAi1 AP-R or NMAi1 ASP-F and NMAi1 ASP-R, respectively. Subsequently, an YFP fragment was cloned into these previously introduced restriction sites, generating pNMA::YFP-NMA dSP and pNMA::YFP-NMA. To express NMA in a tissue-specific manner, we replaced the endogenous promoter sequence in pNMA::YFP-NMA with PCR-amplified promoter fragments of OBP1 (Skirycz et al., 2008), SUC5 (Baud et al., 2005), and J10 (At5g42200; J. Kong and G. Jürgens, personal communication; Supplemental Table S1). For transient expression in rotoplasts, we replaced the endogenous promoter sequence in pNMA::YFP-NMA and pNMA::YFP-NMA dSP by a PCR-amplified Cauliflower mosaic virus 35S promoter (Table S1x).
The NMA promoter used in the complementation constructs was PCR-amplified with primers At2g33160P-FC and At2g33160P-RC and inserted in a pGreen variant carrying nuclear-localized 3xVenus-YFP or GUS, respectively.
Transient Expression in Protoplasts
Transient expression of p35S::YFP-NMA and p35S::YFP-NMA dSP was carried out in protoplasts derived from an Arabidopsis Col-0 dark-grown cell suspension culture, which were transfected as previously described (Schütze et al., 2009). YFP fluorescence was observed 24 and 72 h after transfection by confocal microscopy as described above.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Adult phenotype of nma-1 (Ler background) and nma-3 (Col-0 background).
Supplemental Figure S2. Number of cells in a central optical section of the embryo proper 3 dap.
Supplemental Figure S3. Pollen tube length of in vitro-germinated pollen.
Supplemental Figure S4. Measurements of suspensor length after reciprocal crosses of wild-type (Ler) and nma-1 mutant plants.
Supplemental Figure S5. Fine mapping of nma-1.
Supplemental Figure S6. Graphic representation of the NMA gene and protein.
Supplemental Figure S7. Embryonic phenotype of T-DNA insertion alleles of nma.
Supplemental Figure S8. Complementation assay based on suspensor length.
Supplemental Figure S9. Quantitative RT-PCR data of NMA expression in various tissue types.
Supplemental Figure S10. Graphic representation of NMA expression data in publicly available microarray data.
Supplemental Figure S11. Expression of promoter::reporter gene fusions.
Supplemental Figure S12. Average developmental stage of embryos 4 dap in pJ10::YFP-NMA, nma-1, and Ler-0.
Supplemental Table S1. Primer sequences.
Supplementary Material
Acknowledgments
We thank the Nottingham Arabidopsis Stock Centre for providing T-DNA insertion lines, Jixiang Kong, Daniel Slane, and Gerd Jürgens for sharing unpublished material, Caterina Brancato for protoplast transfections, Ancilla Neu and Michael Hothorn for support and helpful discussions, Wolfgang Lukowitz and Ueli Grossniklaus for support during the early phases of the project, and Gerd Juergens, Steffen Lau, Cameron Lee, and Rebecca Schwab for critical comments on the manuscript and helpful discussions.
Glossary
- PG
polygalacturonase
- dap
d after pollination
- Col-0
ecotype Columbia
- Ler
ecotype Landsberg erecta
- T-DNA
transfer DNA
- SP
signal peptide
References
- Aarssen LW. (2000) Why are most selfers annuals? A new hypothesis for the fitness benefit of selfing. Oikos 89: 606–612 [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Barakate A, Martin W, Quigley F, Mache R. (1993) Characterization of a multigene family encoding an exopolygalacturonase in maize. J Mol Biol 229: 797–801 [DOI] [PubMed] [Google Scholar]
- Baud S, Wuillème S, Lemoine R, Kronenberger J, Caboche M, Lepiniec L, Rochat C. (2005) The AtSUC5 sucrose transporter specifically expressed in the endosperm is involved in early seed development in Arabidopsis. Plant J 43: 824–836 [DOI] [PubMed] [Google Scholar]
- Bayer M, Nawy T, Giglione C, Galli M, Meinnel T, Lukowitz W. (2009) Paternal control of embryonic patterning in Arabidopsis thaliana. Science 323: 1485–1488 [DOI] [PubMed] [Google Scholar]
- Boavida LC, McCormick S. (2007) Temperature as a determinant factor for increased and reproducible in vitro pollen germination in Arabidopsis thaliana. Plant J 52: 570–582 [DOI] [PubMed] [Google Scholar]
- Breuninger H, Rikirsch E, Hermann M, Ueda M, Laux T. (2008) Differential expression of WOX genes mediates apical-basal axis formation in the Arabidopsis embryo. Dev Cell 14: 867–876 [DOI] [PubMed] [Google Scholar]
- Cionini PG, Bennici A, Alpi A, Damato F. (1976) Suspensor, gibberellin and invitro development of Phaseolus coccineus embryos. Planta 131: 115–117 [DOI] [PubMed] [Google Scholar]
- Friml J, Vieten A, Sauer M, Weijers D, Schwarz H, Hamann T, Offringa R, Jürgens G. (2003) Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426: 147–153 [DOI] [PubMed] [Google Scholar]
- González-Carranza ZH, Elliott KA, Roberts JA. (2007) Expression of polygalacturonases and evidence to support their role during cell separation processes in Arabidopsis thaliana. J Exp Bot 58: 3719–3730 [DOI] [PubMed] [Google Scholar]
- González-Carranza ZH, Shahid AA, Zhang L, Liu Y, Ninsuwan U, Roberts JA. (2012) A novel approach to dissect the abscission process in Arabidopsis. Plant Physiol 160: 1342–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadfield KA, Bennett AB. (1998) Polygalacturonases: many genes in search of a function. Plant Physiol 117: 337–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens RP, Edwards EA, Leyland NR, Bean S, Mullineaux PM. (2000) pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol Biol 42: 819–832 [DOI] [PubMed] [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P. (2008) Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinforma 2008: 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irshad M, Canut H, Borderies G, Pont-Lezica R, Jamet E. (2008) A new picture of cell wall protein dynamics in elongating cells of Arabidopsis thaliana: confirmed actors and newcomers. BMC Plant Biol 8: 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong S, Bayer M, Lukowitz W. (2011a) Taking the very first steps: from polarity to axial domains in the early Arabidopsis embryo. J Exp Bot 62: 1687–1697 [DOI] [PubMed] [Google Scholar]
- Jeong S, Palmer TM, Lukowitz W. (2011b) The RWP-RK factor GROUNDED promotes embryonic polarity by facilitating YODA MAP kinase signaling. Curr Biol 21: 1268–1276 [DOI] [PubMed] [Google Scholar]
- Kawashima T, Goldberg RB. (2010) The suspensor: not just suspending the embryo. Trends Plant Sci 15: 23–30 [DOI] [PubMed] [Google Scholar]
- Kim J, Shiu SH, Thoma S, Li WH, Patterson SE. (2006) Patterns of expansion and expression divergence in the plant polygalacturonase gene family. Genome Biol 7: R87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S, De Smet I, Kolb M, Meinhardt H, Jürgens G. (2011) Auxin triggers a genetic switch. Nat Cell Biol 13: 611–615 [DOI] [PubMed] [Google Scholar]
- Lau S, Slane D, Herud O, Kong J, Jürgens G. (2012) Early embryogenesis in flowering plants: setting up the basic body pattern. Annu Rev Plant Biol 63: 483–506 [DOI] [PubMed] [Google Scholar]
- Lukowitz W, Gillmor CS, Scheible WR. (2000) Positional cloning in Arabidopsis. Why it feels good to have a genome initiative working for you. Plant Physiol 123: 795–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukowitz W, Roeder A, Parmenter D, Somerville C. (2004) A MAPKK kinase gene regulates extra-embryonic cell fate in Arabidopsis. Cell 116: 109–119 [DOI] [PubMed] [Google Scholar]
- Markovic O, Janecek S. (2001) Pectin degrading glycoside hydrolases of family 28: sequence-structural features, specificities and evolution. Protein Eng 14: 615–631 [DOI] [PubMed] [Google Scholar]
- Meyer S, Lauterbach C, Niedermeier M, Barth I, Sjolund RD, Sauer N. (2004) Wounding enhances expression of AtSUC3, a sucrose transporter from Arabidopsis sieve elements and sink tissues. Plant Physiol 134: 684–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagl W. (1990) Translocation of putrescine in the ovule, suspensor and embryo of Phaseolus coccineus. J Plant Physiol 136: 587–591 [Google Scholar]
- Nawy T, Bayer M, Mravec J, Friml J, Birnbaum KD, Lukowitz W. (2010) The GATA factor HANABA TARANU is required to position the proembryo boundary in the early Arabidopsis embryo. Dev Cell 19: 103–113 [DOI] [PubMed] [Google Scholar]
- Ogawa M, Kay P, Wilson S, Swain SM. (2009) ARABIDOPSIS DEHISCENCE ZONE POLYGALACTURONASE1 (ADPG1), ADPG2, and QUARTET2 are polygalacturonases required for cell separation during reproductive development in Arabidopsis. Plant Cell 21: 216–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palin R, Geitmann A. (2012) The role of pectin in plant morphogenesis. Biosystems 109: 397–402 [DOI] [PubMed] [Google Scholar]
- Peaucelle A, Braybrook S, Höfte H. (2012) Cell wall mechanics and growth control in plants: the role of pectins revisited. Front Plant Sci 3: 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H. (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8: 785–786 [DOI] [PubMed] [Google Scholar]
- Pommerrenig B, Popko J, Heilmann M, Schulmeister S, Dietel K, Schmitt B, Stadler R, Feussner I, Sauer N. (2013) SUCROSE TRANSPORTER 5 supplies Arabidopsis embryos with biotin and affects triacylglycerol accumulation. Plant J 73: 392–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pressey R. (1991) Polygalacturonase in tree pollens. Phytochemistry 30: 1753–1755 [Google Scholar]
- Rapoport TA. (2007) Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature 450: 663–669 [DOI] [PubMed] [Google Scholar]
- Rhee SY, Osborne E, Poindexter PD, Somerville CR. (2003) Microspore separation in the quartet 3 mutants of Arabidopsis is impaired by a defect in a developmentally regulated polygalacturonase required for pollen mother cell wall degradation. Plant Physiol 133: 1170–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee SY, Somerville CR. (1998) Tetrad pollen formation in quartet mutants of Arabidopsis thaliana is associated with persistence of pectic polysaccharides of the pollen mother cell wall. Plant J 15: 79–88 [DOI] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Schölkopf B, Weigel D, Lohmann JU. (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37: 501–506 [DOI] [PubMed] [Google Scholar]
- Schütze K, Harter K, Chaban C. (2009) Bimolecular fluorescence complementation (BiFC) to study protein-protein interactions in living plant cells. Methods Mol Biol 479: 189–202 [DOI] [PubMed] [Google Scholar]
- Schwartz BW, Yeung EC, Meinke DW. (1994) Disruption of morphogenesis and transformation of the suspensor in abnormal suspensor mutants of Arabidopsis. Development 120: 3235–3245 [DOI] [PubMed] [Google Scholar]
- Sitrit Y, Hadfield KA, Bennett AB, Bradford KJ, Downie AB. (1999) Expression of a polygalacturonase associated with tomato seed germination. Plant Physiol 121: 419–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skirycz A, Radziejwoski A, Busch W, Hannah MA, Czeszejko J, Kwaśniewski M, Zanor MI, Lohmann JU, De Veylder L, Witt I, et al. (2008) The DOF transcription factor OBP1 is involved in cell cycle regulation in Arabidopsis thaliana. Plant J 56: 779–792 [DOI] [PubMed] [Google Scholar]
- Snell R, Aarssen LW. (2005) Life history traits in selfing versus outcrossing annuals: exploring the ‘time-limitation’ hypothesis for the fitness benefit of self-pollination. BMC Ecol 5: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spillane C, Schmid KJ, Laoueillé-Duprat S, Pien S, Escobar-Restrepo JM, Baroux C, Gagliardini V, Page DR, Wolfe KH, Grossniklaus U. (2007) Positive Darwinian selection at the imprinted MEDEA locus in plants. Nature 448: 349–352 [DOI] [PubMed] [Google Scholar]
- Stintzi A, Browse J. (2000) The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proc Natl Acad Sci USA 97: 10625–10630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torki M, Mandaron P, Mache R, Falconet D. (2000) Characterization of a ubiquitous expressed gene family encoding polygalacturonase in Arabidopsis thaliana. Gene 242: 427–436 [DOI] [PubMed] [Google Scholar]
- Vernon DM, Meinke DW. (1994) Embryogenic transformation of the suspensor in twin, a polyembryonic mutant of Arabidopsis. Dev Biol 165: 566–573 [DOI] [PubMed] [Google Scholar]
- Wardlaw CW (1955) Embryogenesis in Plants. Wiley, London. [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung EC. (1980) Embryogeny of Phaseolus: the role of the suspensor. Zeitschrift Fur Pflanzenphysiologie 96: 17–28 [Google Scholar]
- Yeung EC, Meinke DW. (1993) Embryogenesis in angiosperms: development of the suspensor. Plant Cell 5: 1371–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung EC, Sussex IM. (1979) Embryogeny of Phaseolus coccineus: the suspensor and the growth of the embryo-proper in vitro. Zeitschrift Fur Pflanzenphysiologie 91: 423–433 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.