A novel type of allosteric regulation in divergent cyclophilins engages disulfide bond formation and a loop-displacement mechanism.
Abstract
The citrus (Citrus sinensis) cyclophilin CsCyp is a target of the Xanthomonas citri transcription activator-like effector PthA, required to elicit cankers on citrus. CsCyp binds the citrus thioredoxin CsTdx and the carboxyl-terminal domain of RNA polymerase II and is a divergent cyclophilin that carries the additional loop KSGKPLH, invariable cysteine (Cys) residues Cys-40 and Cys-168, and the conserved glutamate (Glu) Glu-83. Despite the suggested roles in ATP and metal binding, the functions of these unique structural elements remain unknown. Here, we show that the conserved Cys residues form a disulfide bond that inactivates the enzyme, whereas Glu-83, which belongs to the catalytic loop and is also critical for enzyme activity, is anchored to the divergent loop to maintain the active site open. In addition, we demonstrate that Cys-40 and Cys-168 are required for the interaction with CsTdx and that CsCyp binds the citrus carboxyl-terminal domain of RNA polymerase II YSPSAP repeat. Our data support a model where formation of the Cys-40-Cys-168 disulfide bond induces a conformational change that disrupts the interaction of the divergent and catalytic loops, via Glu-83, causing the active site to close. This suggests a new type of allosteric regulation in divergent cyclophilins, involving disulfide bond formation and a loop-displacement mechanism.
Cyclophilins (Cyps) are ubiquitous proteins that belong to the immunophilin superfamily, exhibiting peptidyl prolyl cis-trans-isomerase (PPIase) activity, and are targets of the immunosuppressive drug cyclosporin A (CsA; Wang and Heitman, 2005). Cyp proteins act as chaperones or foldases and have been implicated in a wide range of biological processes, including cell division, transcription regulation, mRNA splicing, and stress tolerance (Fischer and Schmid, 1990; Wang and Heitman, 2005). Cyps are also known to play a key role in virus replication both in animal and plant cells (Luban et al., 1993; Nagy et al., 2011).
In plants, Cyps control transcription, gene silencing, and hormone signaling, affecting plant development and interaction with pathogens (Iki et al., 2012; Trupkin et al., 2012; Bannikova et al., 2013). Rotamase Cyp1 (ROC1), one of the best studied Cyps from Arabidopsis (Arabidopsis thaliana), affects plant architecture, hormone and phytochrome signaling, and is a target of the bacterial effector protein AvrRpt2 (Coaker et al., 2005; Trupkin et al., 2012; Ma et al., 2013). Similarly, citrus (Citrus sinensis) cyclophilin (CsCyp) was identified as a target of the bacterial effector protein PthA, one of the transcription activator-like effectors of Xanthomonas citri that is required to elicit cankers on citrus (Domingues et al., 2010). CsCyp is a nuclear protein that interacts with other PthA-interacting proteins, including the CsUev/CsUbc13 heterodimer and CsTdx, a TPR domain-containing thioredoxin (Domingues et al., 2010). Importantly, in addition to PthA and CsTdx, CsCyp also binds to the C-terminal domain of citrus RNA polymerase II (CTD). Like the yeast and mammalian CTDs, the citrus CTD consists of multiple tandem repeats of the consensus sequence YSPXSPX (Domingues et al., 2012), which play a key role in the transcriptional cycle (Buratowski, 2009).
In Saccharomyces cerevisiae, the heptad repeats of the CTD are the targets of Ess1 and Cpr1, two prolyl-isomerases that affect the progress of transcription and gene silencing (Arévalo-Rodríguez et al., 2000; Wu et al., 2000; Ma et al., 2012). We showed that CsCyp complements the function of both Ess1 and Cpr1 and that PthA inhibits its PPIase activity (Domingues et al., 2012). Since the depletion of CsCyp in citrus leaves enhances canker formation, we postulated that CsCyp acts as a negative regulator of cell growth through the isomerization of Pro residues of the CTD heptad repeats (Domingues et al., 2012).
CsCyp belongs to the subfamily of the so-called divergent Cyps. Members of this subgroup of Cyps are characterized by the presence of an additional loop with the consensus sequence XXGKXLH, two invariable Cys residues and a conserved Glu (Taylor et al., 1998; Dornan et al., 1999; Peterson et al., 2000). Previous studies have suggested that the divergent loop could mediate protein-protein interactions or be part of a P-loop or ATP-binding site, formed by residues 42-GEKGIGKS-49 and 163-VVIAD-167 (Dornan et al., 1999). In addition, due to their close proximity in the structure, it has also been postulated that the conserved Cys residues might form a disulfide bond or coordinate a metal ion via interaction with His-54 from the divergent loop (Dornan et al., 1999). Nevertheless, the function of these structural elements in protein-protein interactions or the regulation of enzyme activity has not yet been demonstrated.
Here, we show that the conserved Cys residues (Cys-40 and Cys-168) are targets of CsTdx and form a disulfide bond that regulates the CsCyp PPIase activity. We also demonstrate that the invariable Glu (Glu-83), which connects the divergent loop to the active site, is critical for protein activity, since it stabilizes the divergent loop in a conformation that favors the active site to remain open. Furthermore, we present evidence suggesting that, instead of binding ATP, the divergent loop functions as a trigger that transmits the redox status of the Cys residues to the active site of the enzyme via interaction with Glu-83. Finally, we show that CsCyp binds to the YSPSAP sequence of the citrus CTD, suggesting that, as in yeast cells, CsCyp modulates the progress of transcription.
RESULTS
CsCyp Belongs to the Subfamily of Divergent Cyps
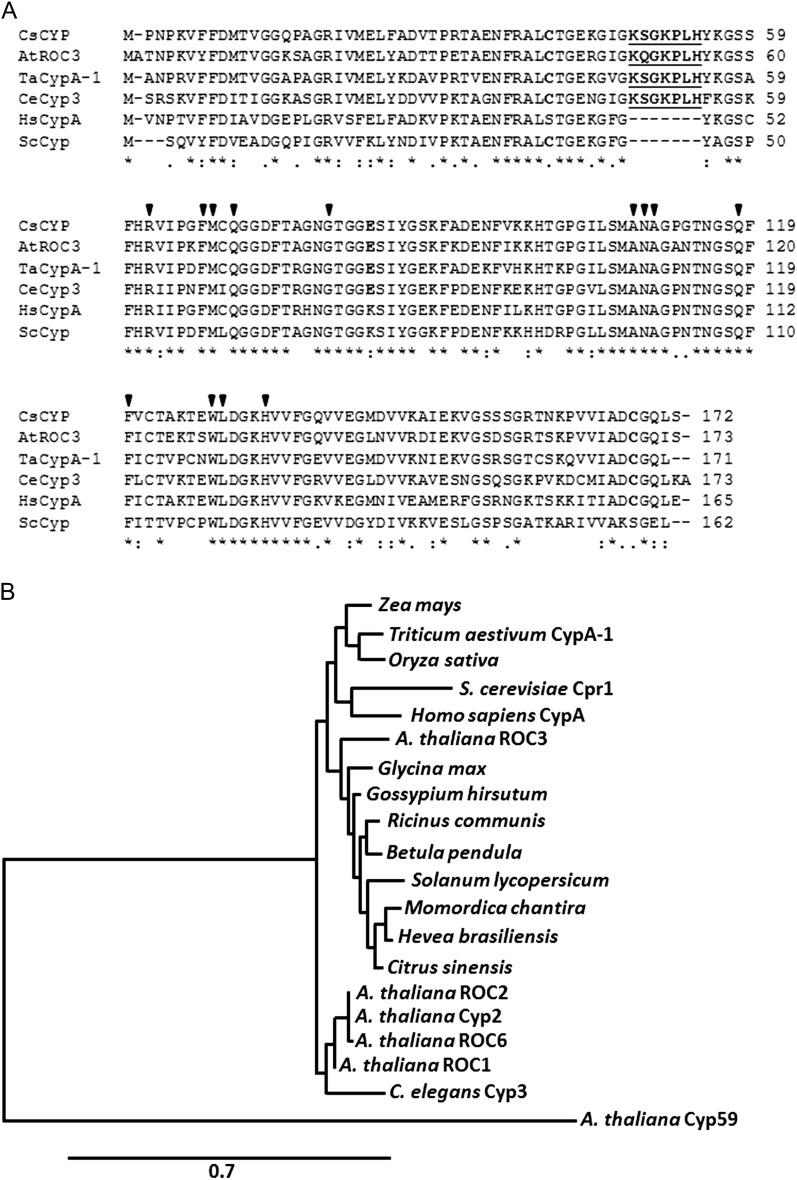
CsCyp is an 18-kD protein that has a single and typical PPIase domain; however, as a member of the divergent Cyp subfamily, CsCyp also carries the divergent loop (48-KSGKPLH-54), two invariable Cys residues (Cys-40 and Cys-168), and a conserved Glu (Glu-83) that are unique to this subfamily of Cyps (Fig. 1A). CsCyp is closely related to the Arabidopsis ROC3, wheat (Triticum aestivum) CypA-1, and Caenorhabditis elegans Cyp3 proteins, yet it also shares approximately 70% identity with human CypA and yeast Cpr1, despite both lacking the divergent loop, Glu-83, and one of the invariable Cys residues (Fig. 1A). A phylogenetic analysis shows that CsCyp forms a branch with several uncharacterized plant Cyps belonging to dicot species. Interestingly, CsCyp is not related to Arabidopsis Cyp59 (Fig. 1B), the only plant Cyp, besides CsCyp, known to interact with the CTD (Gullerova et al., 2006).
Figure 1.
CsCyp is a member of the divergent Cyp subgroup. A, Protein sequence alignment of CsCyp with Arabidopsis ROC3 (AtROC3), wheat CypA-1 (TaCypA-1), C. elegans Cyp3 (CeCyp3), human CypA (HsCypA), and yeast (Saccharomyces cerevisiae) Cpr1 (ScCpr1) performed with ClustalW. Arrows indicate the amino acid residues that are part of the active site of CsCyp and that bind CsA. Residues comprising the divergent loop are shown in boldface underlined, whereas the conserved Cys residues (Cys-40 and Cys-168) and Glu (Glu-83) are shown in boldface. B, Phylogenetic analysis of Cyp proteins from plants (monocots and dicots), humans, nematodes, and yeast, showing that CsCyp clusters with uncharacterized Cyp proteins from dicot plants. CsCyp is not related to Arabidopsis Cyp59. Only the PPIase domain of AtCyp59 was used in the alignment.
On the Structural (Dis)similarities between Classical and Divergent Cyps
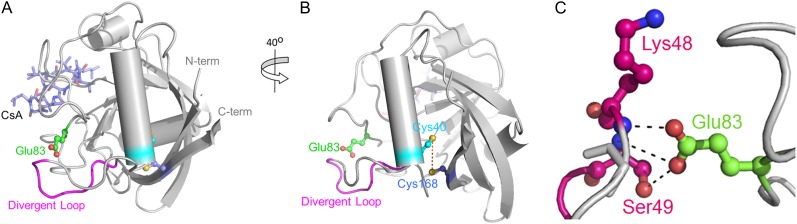
To gain insights into the regulation of CsCyp activity, the crystal structure of CsCyp in complex with CsA was solved. The overall architecture of CsCyp is highly similar to that of the C. elegans Cyp3 and human CypA structures in complex with CsA (Pflügl et al., 1993; Ke et al., 1994; Dornan et al., 1999) and comprises an eight-stranded antiparallel β-barrel capped at either end by two α-helices (Fig. 2, A and B). The CsCyp active site, composed of 13 residues that are also responsible for CsA binding (Fig. 1A), is identical to that of Cyp3, CypA, and TaCypA-1 (Pflügl et al., 1993; Ke et al., 1994; Dornan et al., 1999; Sekhon et al., 2013). Structure alignment of CsCyp with TaCypA-1, the only other plant divergent Cyp with known three-dimensional (3D) structure, showed no major structural differences (root mean square deviation = 0.43 Å). The CsCyp divergent loop (48-KSGKPLH-54), located above the active site, is tethered by the conserved Glu-83, through hydrogen bonds between the side-chain carboxyl group of Glu-83 and the main-chain amide groups of Lys-48 and Ser-49 (2.6 and 3.2 Å, respectively), and the hydroxyl group of Ser-49 (2.3 Å; Fig. 2C). In the Cyp3 structure (Dornan et al., 1999), the interactions between Glu-83 and the main-chain amide proton atoms of Lys-48 and Ser-49 are conserved (2.8 and 3.2 Å, respectively); however, the side chain of Ser-49 adopts a different orientation and is instead hydrogen bonded through its hydroxyl group to a neighboring water molecule. In addition, and different from CsCyp, Glu-83 in Cyp3 also exhibits a distinct rotameric conformation that disables the interaction with the hydroxyl group of Ser-49. These observations strongly support the hypothesis that Glu-83 is key in anchoring the divergent loop in a particular conformation in this subgroup of Cyps, as reported previously (Dornan et al., 1999; Taylor et al., 2001). Moreover, the two invariable Cys residues (Cys-40 and Cys-168), which under the conditions studied are found to be in their reduced forms in the crystal structure, are also in close spatial proximity to one another in CsCyp, as in Cyp3 (Dornan et al., 1999), with an interatomic sulfur-sulfur distance of 5.4 Å separating them (Fig. 2B).
Figure 2.
Overall structure of CsCyp in complex with CsA. A, Cartoon depicting the 3D structure of CsCyp (gray) showing the typical Cyp fold comprising eight β-strands that form a β-barrel closed by two α-helices. CsA is shown with light blue sticks. The divergent loop (pink) and conserved Glu-83 (green) are shown. B, A 40° rotated image showing the invariable Cys residues Cys-40 and Cys-168 (cyan and blue, respectively) and their thiol groups 5.4 Å apart (dashed yellow line). C, Detailed view of the hydrogen bond network between the side chain of Glu-83 and Lys-48 and Ser-49 of the divergent loop.
The Conserved Cys-40 and Cys-168 Form a Disulfide Bridge and Are Critical for CsCyp Activity
We and others have noticed that the two conserved Cys residues, which are in the reduced form and close to each other in the tertiary structure, could form a disulfide bond under oxidizing conditions (Dornan et al., 1999; Taylor et al., 2001). In addition, Dornan et al. (1999) postulated that the proximity of His-54 to the Cys residues Cys-40 and Cys-168 could alternatively form a metal coordination site involving these residues. It was thus suggested that formation of a disulfide bond or metal binding could represent a signaling mechanism in response to oxidative stress (Dornan et al., 1999; Taylor et al., 2001). However, since we found no electron density corresponding to metal ions in the CsCyp x-ray diffraction data, we anticipated that the disulfide bridge may in fact be part of a redox mechanism that controls the enzyme activity. To test this hypothesis, mass spectrometric analysis was conducted with purified CsCyp under nonreducing conditions. Even in the absence of a specific oxidizing agent, we detected a disulfide bond between Cys-40 and Cys-168 in our protein samples through identification of the linked peptides 38-ALCTGEK-44 and 159-TNKPVVIADCGQLS-172 (Supplemental Fig. S1). In addition, the same peptides were also identified in their reduced forms, suggesting that the recombinant CsCyp made in Escherichia coli under the conditions described in “Materials and Methods” is a mixture of reduced and oxidized forms of the protein. No alternative disulfide bridges involving either of the other two Cys residues in CsCyp were detected from our mass spectrometric analysis (Supplemental Fig. S1)
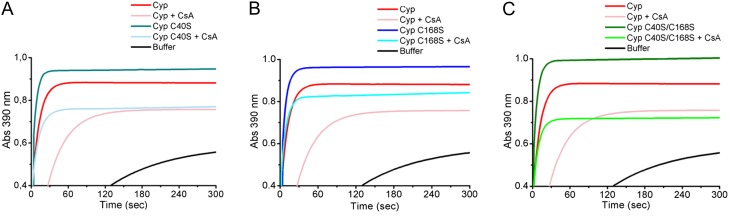
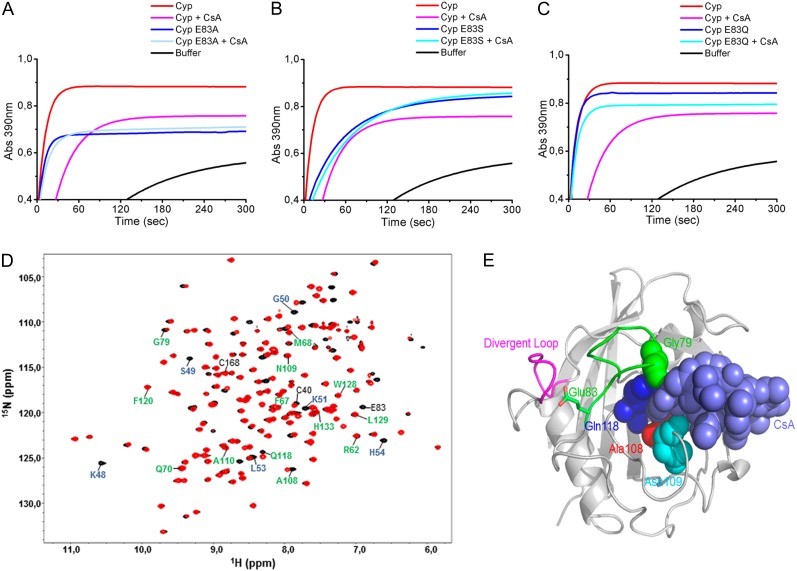
To establish the possible role of Cys-40 and Cys-168 in protein activity, the mutants C40S, C168S, and C40S/C168S were generated. We measured the PPIase activity of the mutated proteins relative to wild-type CsCyp and found that all the mutant proteins displayed a significantly higher PPIase activity than wild-type CsCyp (Fig. 3; Table I). The kcat/Km value for the wild-type CsCyp (5.6 s−1 µm−1) is consistent with values determined for other Cyp proteins (Dornan et al., 1999; Laxa et al., 2007; Sekhon et al., 2013). In addition, while the C40S and C168S mutants were similarly inhibited by CsA, the double mutant was more strongly inhibited by CsA relative to the wild-type protein (Fig. 3; Table I), indicating that CsCyp is active in its reduced form. This is consistent with the mass spectrometric analysis that shows that the wild-type protein preparation has a mixture of oxidized and reduced CsCyp.
Figure 3.
CsCyp is active in its reduced form. The PPIase activity of recombinant CsCyp, measured by the α-chymotrypsin-coupled assay, reaches a plateau within 30 to 60 s after the start of the reaction. The PPIase activity of wild-type CsCyp (red) is compared with those of the C40S (A), C168S (B), and C40S/C168S (C) mutants in the absence (dark cyan, blue, and green, respectively) or presence of 30 nm CsA (light blue, light cyan, and light green, respectively). The mutant proteins all show a higher PPIase activity than the wild-type CsCyp, and all are drastically inhibited by CsA. The PPIase activity of the buffer alone is shown in black.
Table I. kcat/Km values for PPIase activity of CsCyp and mutants.
| Cyp | kcat/Km | % |
|---|---|---|
| s−1 µm−1 | ||
| CsCyp | 5.6 | 100 |
| CsCyp + CsA | 1.3 | 24.1 |
| Cyp C40S | 5.9 | 105 |
| Cyp C40S + CsA | 3.5 | 62.7 |
| Cyp C168S | 6.7 | 118 |
| Cyp C168S + CsA | 3.1 | 55.7 |
| Cyp C40S/C168S | 8.1 | 142.9 |
| Cyp C40S/C168S + CsA | 3.9 | 69.7 |
| Cyp E83A | 2.9 | 51.8 |
| Cyp E83A + CsA | 2.6 | 45.9 |
| Cyp E83S | 3.3 | 59.5 |
| Cyp E83S + CsA | 3.0 | 53.7 |
| Cyp E83Q | 5.0 | 89 |
| Cyp E83Q + CsA | 4.5 | 79.9 |
| Cyp + CuSO4 | 4.3 | 76.3 |
| Cyp + MnSO4 | 5.4 | 96.6 |
| Cyp + ZnSO4 | 5.6 | 99 |
CsCyp Is Not a Zinc-Binding Protein
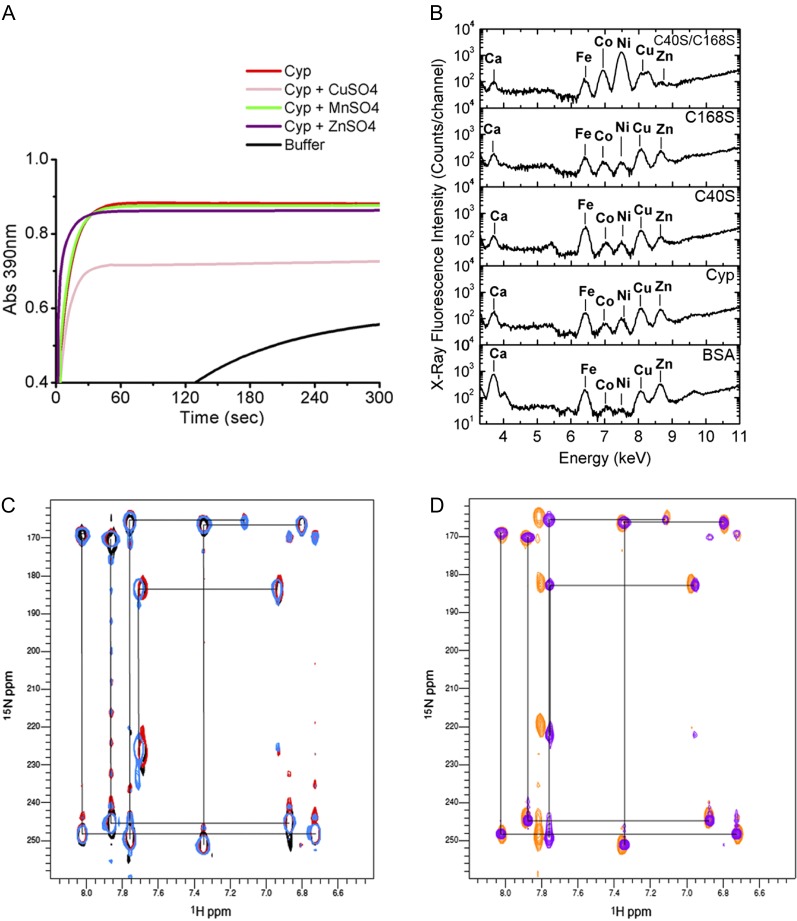
To investigate whether CsCyp activity could be regulated by a metal ion, the CsCyp PPIase activity was measured in the presence of various metal salts. With the exception of CuSO4, which significantly inhibited the CsCyp activity and which was shown to induce disulfide bond formation in nondivergent Cyps (Gourlay et al., 2007), the other metal ions tested, including Zn2+ and Mn2+, did not affect enzyme activity (Fig. 4A; Table I).
Figure 4.
CsCyp is not a zinc-binding protein. A, PPIase activity of CsCyp in the absence (red) or presence of CuSO4 (pink), ZnSO4 (purple), or MnSO4 (green), showing that CuSO4 inhibits CsCyp activity. The PPIase activity of the buffer alone is shown in black. B, X-ray fluorescence of purified CsCyp and mutants showing the presence of various elements in the samples. No major peak is observed for the wild-type protein compared with the mutant proteins, indicating that CsCyp does not coordinate a specific metal ion. C, Two-bond 2JNH region of 1H-15N SOFAST-HMQC spectra of wild-type CsCyp in the presence of DTT and EDTA (light blue), wild-type CsCyp with Zn2+ (black), and wild-type CsCyp with Zn2+ followed by the addition of EDTA (red). Black lines correlate resonances from atoms within a given imidazole ring. The correlation pattern indicates which of the tautomeric states the His side chain adopts (Pelton et al., 1993). All five His imidazole rings of CsCyp are observed. D, Two-bond 2JNH region of 1H-15N SOFAST-HMQC spectra of wild-type CsCyp in the absence of DTT and EDTA (blue) and of CsCyp C40S/C168S (orange). For clarity, black lines correlating resonances from atoms within a given imidazole ring of wild-type CsCyp only are shown. For details, see text.
We also investigated if CsCyp could bind a metal ion using x-ray fluorescence analysis. Among the elements detected, calcium, iron, cobalt, nickel, copper, and/or zinc were identified in the samples. However, with the exception of nickel and zinc in the C40S/C168S mutant, which were found at relatively higher and lower levels, respectively, no major peak was found in the wild-type protein that would indicate the presence of a specific metal ligand, relative to the C40S and C168S mutants, or to bovine serum albumin used as a negative control (Fig. 4B). Furthermore, we observed no significant differences in the His imidazole region of 1H-15N band-selective optimized-flip-angle-short-transient heteronuclear multiple quantum coherence (SOFAST-HMQC) spectra of CsCyp samples treated with ZnCl2 or EDTA, compared with the untreated CsCyp, or between wild-type CsCyp and the C40S/C168S mutant, indicating that Zn2+ does not bind to any His imidazole rings of CsCyp (Fig. 4, C and D, respectively). The small secondary chemical shifts observed between samples are most likely a consequence of small differences in pH between samples (change in pH approximately 0.2) or the effect of the Cys mutations. Although we observed that one His ring experiences a structural environment that differs from the others, as shown by its 15N chemical shifts resonating at 183 and 225 ppm (Fig. 4C), it is likely that in solution this His ring is protonated at the Nε2 nitrogen atom and coordinated to an as yet unidentified electrophile (though not Zn2+) at the Nδ1 nitrogen atom that is not observed in the x-ray diffraction electron density map. Even though none of the His imidazole rings have been unambiguously sequentially assigned, our results provide no evidence that a His imidazole ring is directly involved in zinc binding in CsCyp; consequently, His-54 is not likely to form a metal-binding site with Cys-40 and Cys-168, as suggested by Dornan et al. (1999). Taken together, our data suggest that coordination of a metal ion via interaction with His-54, Cys-40, and Cys-168 is not required and that protein activity is regulated solely by disulfide bond formation between the conserved Cys residues.
Cys-40 and Cys-168 Are Required for CsTdx Interaction
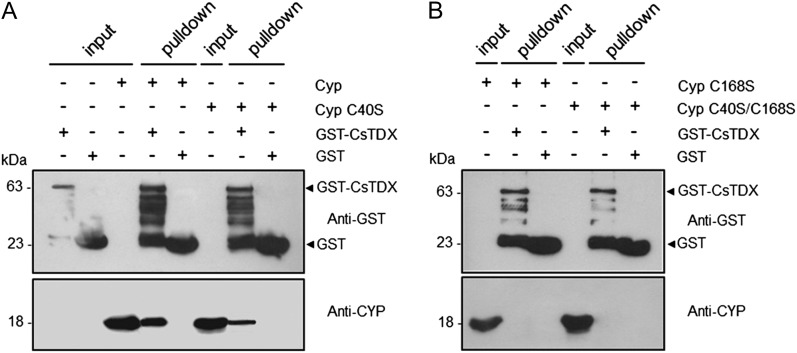
Considering that (1) CsCyp activity is affected by the disulfide bond formation between Cys-40 and Cys-168, (2) thioredoxins have been shown to reduce disulfide bonds in nondivergent Cyps (Motohashi et al., 2003), and (3) the citrus TPR domain-containing thioredoxin CsTdx is an interaction partner of not only CsCyp but also of PthA and the citrus CTD (Domingues et al., 2010, 2012), we decided to investigate whether Cys-40 and Cys-168 could be the targets of CsTdx. Glutathione S-transferase (GST) pull-down assays using GST-CsTdx as bait showed that the interaction between CsTdx and CsCyp is not only diminished by the C40S mutation but completely abolished by the C168S and C40S/C168S mutations (Fig. 5).
Figure 5.
The conserved Cys residues, Cys-40 and Cys-168, are required for the CsCyp-CsTdx interaction. A GST pull-down assay used GST-CsTdx as bait and the purified wild-type or CsCyp mutants without the 6xHis tag as prey. Protein samples were electrophoresed and probed with the anti-GST or anti-CsCyp serum. A, Wild-type CsCyp, and to a lesser extent the C40S mutant, bind to GST-CsTdx but not to GST alone. B, The interaction between CsCyp and CsTdx is abolished by the C168S and C40S/C168S mutations. Soluble cell extracts of GST-CsTdx, GST alone, or purified CsCyp proteins used as inputs are indicated. The molecular sizes of the corresponding proteins are shown on the left.
The Conserved Glu Is Also Critical for CsCyp Activity
We noticed from the CsCyp 3D structure that the conserved Glu-83 is connected to a loop that approaches the active site at one end of the substrate pocket. In fact, one of the residues of this loop, Gly-79, belongs to the active site of the protein (Fig. 1A). This suggested that the interaction of Glu-83 with the divergent loop could favor the active site to remain open and led us to investigate the possible role of Glu-83 as a key residue in controlling protein activity. To test this hypothesis, Glu-83 was mutated to Ala (E83A), Ser (E83S), or Gln (E83Q). It was expected from the 3D structure that a substitution to Ala or Ser would drastically disrupt all the hydrogen bonds with Lys-48 and Ser-49, allowing the divergent loop greater flexibility, whereas a substitution to Gln would probably maintain the divergent loop in its more rigid configuration (Fig. 2C). In accordance with this, the results show that the E83A and E83S mutants have a much lower PPIase activity than wild-type CsCyp or the E83Q mutant (Fig. 6; Table I). Interestingly, only the PPIase activity of the E83Q mutant could be still further inhibited by CsA (Fig. 6C; Table I), indicating that disruption of the hydrogen bonds between Glu-83 and the divergent loop dramatically affects the active site of CsCyp.
Figure 6.
Mutations at Glu-83 affect the PPIase activity of CsCyp. A to C, The PPIase activity of wild-type CsCyp in the absence (red) or presence of CsA (pink) is compared with those of the E83A (A), E83S (B), and E83Q (C) mutants in the absence (blue) or presence of CsA (light blue). Substitutions of Glu-83 to Ala or Ser, but not Gln, significantly reduce the PPIase activity of CsCyp. The PPIase activity of the buffer alone is shown in black. D, Superposition of 1H-15N HSQC spectra of CsCyp (black) and the Cyp E83A mutant (red). Residues of the divergent loop (48-KSGKPLH-54) and active site are labeled in blue and green, respectively. Cys-40, Cys-168, and Glu-83 are labeled in black. E, Cartoon of CsCyp in complex with CsA (light blue spheres) depicting the CsCyp residues (Gly-79, Ala-108, Asn-109, and Gln-118, represented by spheres) that showed differences in chemical shifts in the E83A mutant relative to the wild-type protein. The divergent (pink) and Glu-83 (green) loops are shown.
To investigate this further, 1H-15N HSQC experiments were performed with the E83A mutant as with the wild-type protein. Superposition of the two spectra revealed that most of the residues of the divergent loop, including Gly-50, Lys-51, Leu-53, and His-54, showed substantial chemical shift differences, indicating that the loop adopts a new conformation as a result of the disruption of the hydrogen bonds with Glu-83 (Fig. 6D). The loss of some of the loop signals (Lys-48 and Ser-49) and the weak intensity of others (G50), most likely due to line broadening resulting from chemical exchange, suggest a change in the dynamics of this region. Moreover, we observed changes in the chemical shifts of Gly-79, Ala-108, Asn-109, and Gln-118 of the active site and in Cys-40, indicating that loss of interaction between the divergent loop and Glu-83 affects both the active site and the Cys environment (Fig. 6D). As anticipated, some of the active-site residues whose environments are altered in the E83A mutant are the ones that make contacts with the ligand CsA at the end of the substrate pocket that is close to the Glu-83 loop (Fig. 6E), thus supporting the idea that disruption of the Glu-83 interaction with the divergent loop affects the active site of the protein.
CsCyp Binds Selectively to a CTD Heptad Repeat
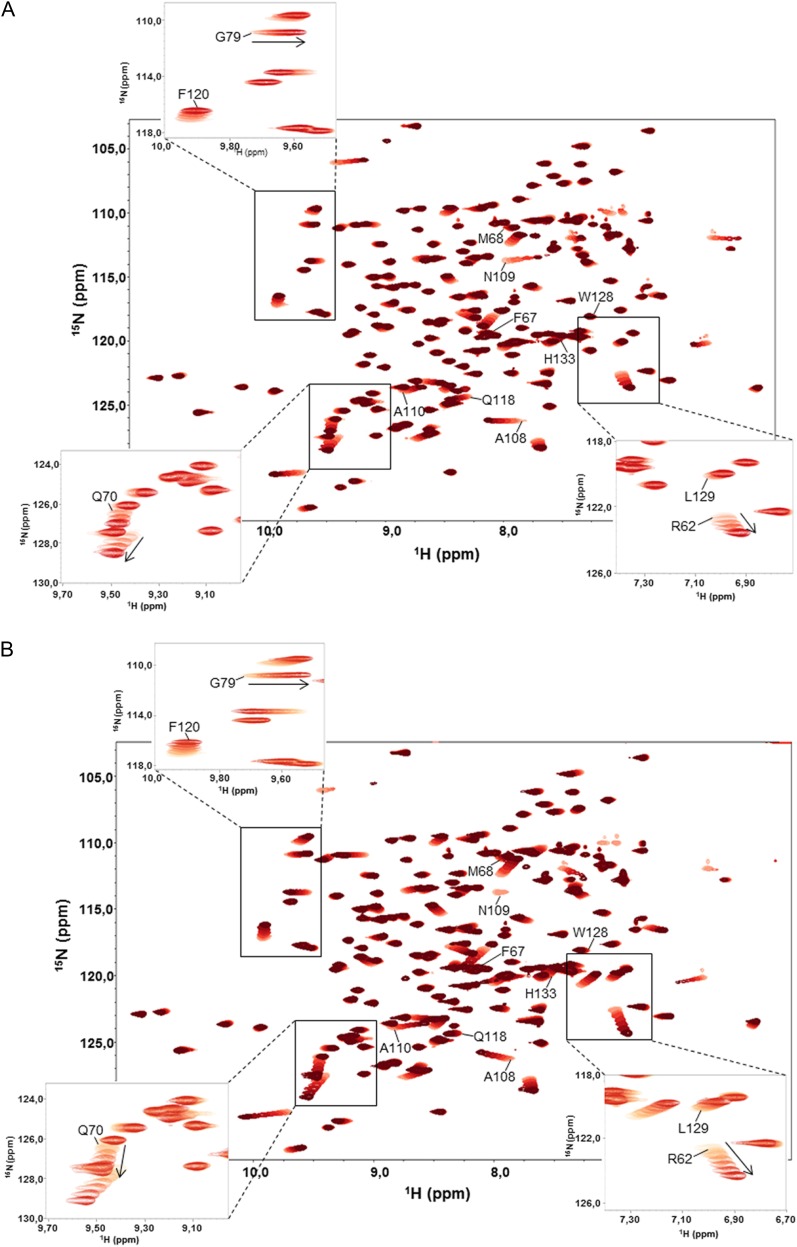
Specific peptide targets of divergent Cyps have not been described; nevertheless, CsCyp was shown to interact strongly with the citrus CTD, which is composed of numerous Pro-rich hepta repeats with the consensus sequence YSPXXPX (Domingues et al., 2012). To further understand this interaction and identify which repeat of the CTD CsCyp interacts preferably, we measured 1H-15N HSQC spectra using three peptides corresponding to the major heptad repeats of the citrus CTD. We observed that the interaction of CsCyp with peptides 1 (YSPTSP) and 2 (YSPSSP) was weak, and only Ala-108 and Asn-109, which belong to the active site, showed small chemical shift changes in the HSQC spectra (data not shown). However, strong interactions were observed with peptide 3, YSPSAP (Fig. 7A), as well as with the control peptide, FGPDLP (Fig. 7B), which is known to bind human CypA with high affinity (Piotukh et al., 2005). In both cases, major changes in the chemical shifts were observed for the active-site residues, including Arg-62, Glu-70, Gly-79, Phe-120, and Leu-129 (Fig. 7). Since the off rate of the ligand binding is fast with regard to the NMR time scale, the dissociation constant (Kd) of the interaction could be determined as Kd = 59 µm for peptide 3 and Kd = 104 µm for peptide 4. In comparison, a Kd of approximately 27 nm was calculated for the CsCyp-CsA interaction by isothermal titration calorimetry (Supplemental Fig. S2). Thus, the results indicate that CsCyp has a preference for the YSPSAP repeat of the citrus CTD.
Figure 7.
Binding dynamics of CsCyp to the YSPSAP and FGPDLP peptides. A, Superposition of 1H-15N HSQC spectra of free CsCyp (light red) and CsCyp titrated with peptide YSPSAP (darkening shades of red). B, Superposition of 1H-15N HSQC spectra of free CsCyp (light red) and CsCyp titrated with peptide FGPDLP (darkening shades of red). In both spectra, the arrows indicate the directions of the secondary chemical shifts of the backbone amide nuclei of the amino acids on addition of peptide. The labeled amino acids belong to the active site. The insets show signals for some of the amino acids that undergo a chemical shift perturbation and that were used to calculate the Kd of the interactions.
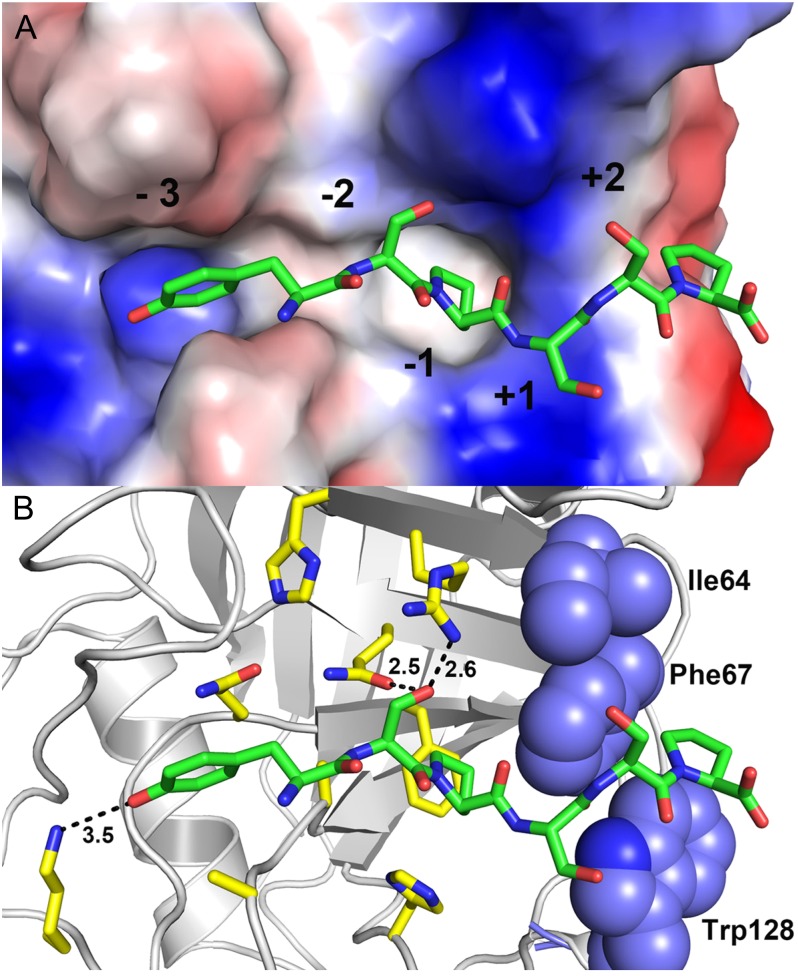
To further investigate the molecular basis of such peptide preference, docking studies were employed to support our experimental data. The results show that the substitution of Ala (YSPSAP) to Ser (YSPSSP) at the +2 position is detrimental for recognition and binding of the ligand peptide due to a hydrophobic microenvironment at this subsite, which is formed by the residues Ile-64, Phe-67, and Trp-128 of CsCyp (Fig. 8). Any polar residue at this subsite, such as Ser, is likely to disfavor or preclude binding of the ligand peptide. This idea is in accord with our NMR titration experiments and is also in agreement with literature data showing that polar residues at the +2 position negatively affect the binding of linear peptides to CypA (Piotukh et al., 2005).
Figure 8.
Molecular docking studies between the hexapeptide YSPSSP and CsCyp. A, Electrostatic surface representation of the active site with the docked peptide, illustrating the subsites. B, Interactions between the active-site residues and the peptide (carbon in green). The hydrophobic residues forming the +2 subsite are represented as blue spheres.
DISCUSSION
The 3D Structure of a Plant Cyp Is from a Revealing Divergent Cyp
In this work, we describe the 3D structure of a citrus Cyp that belongs to the divergent Cyp subfamily. Although 3D structures of divergent Cyps have been reported, the functional role of their unique structural elements (i.e. divergent loop, invariable Cys pair, and stabilizing Glu) remained unclear (Dornan et al., 1999; Taylor et al., 2001; Sekhon et al., 2013). Here, we show that, instead of coordinating a metal ion with His-54, as proposed previously (Dornan et al., 1999; Taylor et al., 2001), Cys-40 and Cys-168 form a disulfide bridge that blocks protein activity. In addition, the fact that the interaction between CsCyp and CsTdx requires the conserved Cys residues strongly indicates that the PPIase activity of CsCyp is regulated by a redox mechanism involving the thiol groups of Cys-40 and Cys-168. In fact, disulfide bond formation and interaction with thioredoxins has been reported to control the PPIase activity of nondivergent Cyps in animals and plants (Motohashi et al., 2003; Gourlay et al., 2007; Laxa et al., 2007). For instance, the activity of Arabidopsis CYP20-3 (ROC4), which links redox and light signals to Cys biosynthesis and stress responses in chloroplasts, is dependent on reduction by a thioredoxin (Motohashi et al., 2003; Dominguez-Solis et al., 2008).
It has also been suggested that the divergent loop could be part of an ATP-binding site formed by residues 42-GEKGIGKS-49 and 163-VVIAD-167 (Dornan et al., 1999). However, 1H-15N HSQC spectral analyses showed that ATP did not interact with the proposed P-loop but weakly interacted with a few residues of the active site with Kd > 1 mm, suggesting a nonspecific interaction (data not shown).
We also demonstrate that the conserved Glu-83, which connects the divergent loop to the active site of CsCyp, is critical for protein activity. Thus, since both Glu-83 and the disulfide bridge formation between Cys-40 and Cys-168 regulate the PPIase activity of CsCyp, and the Cys residues are structurally connected with the active site by the interaction of Glu-83 with the divergent loop, we propose a mechanistic model for the redox regulation of divergent Cyps.
A Mechanistic Model for the Redox Regulation of Divergent Cyps
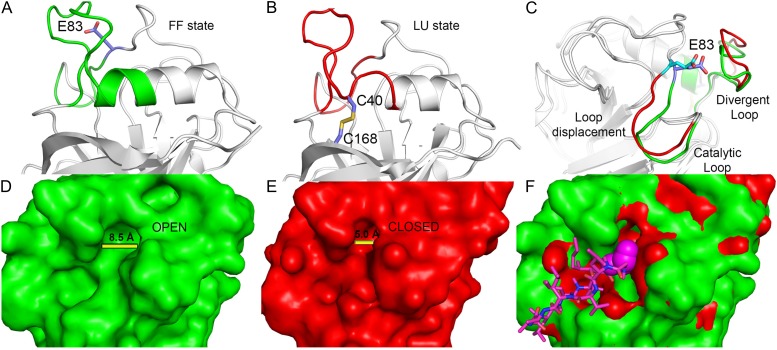
The divergent loop protrudes from the helix containing residue Cys-40, suggesting that disulfide bridge formation would promote structural rearrangements that would be transmitted to this loop. Interestingly, the process involved in such disulfide bridge formation in CsCyp seems to be a conserved redox 2-Cys mechanism observed in the catalytic cycle of peroxiredoxins (Hall et al., 2011). In this group of enzymes, a Cys referred to as peroxidatic Cys is located at the end of a helix (as Cys-40 is in divergent Cyps). When this Cys is in the reduced state, the helix adopts a fully folded (FF) conformation, whereas under oxidizing conditions (S-S bond), it adopts a locally unfolded (LU) conformation, which is required for disulfide bond formation between the peroxidatic Cys and the so-called resolving Cys located at the C terminus of the protein, just as Cys-168 is in CsCyp (Hall et al., 2011). Thus, analogously to peroxiredoxins, an FF→LU transition in CsCyp, mediated or stabilized by the disulfide bond formation between Cys-40 and Cys-168, would cause a significant change in the conformation of the divergent loop, as envisaged by our mechanistic model (Fig. 9). According to this model, transition from the FF to the LU state would lead to the disruption of the interaction between Glu-83 and the divergent loop, causing repositioning of the Glu-83 loop (segment 73-DFTAGNGTGGE-83) and a consequent narrowing of the active-site cleft (Fig. 9, A and B). The Glu-83 loop, which we suggest would function as a catalytic loop, is rich in Gly residues, and removal of its anchoring point with the divergent loop would provoke its displacement, thereby altering the substrate channel (Fig. 9, B and C). The fact that the E83A mutant does not bind CsA and shows chemical shift perturbations in Gly-79 and in three catalytic residues neighboring the catalytic loop (Fig. 6) supports our model. High crystallographic B factors for the divergent loop in wild-type CsCyp, together with conformational exchange-broadened residues Gly-77, Asn-78, Gly-79, and Thr-80 of the catalytic loop, evident in NMR spectra of CsCyp E83A, also corroborate our loop-displacement model. Thus, the model not only explains why the E83A and E83S mutants have reduced PPIase activity that cannot be further inhibited by CsA but also offers an elegant explanation for the redox control of protein activity. In Schistosoma mansoni Cyp, disulfide bond formation, which renders the enzyme inactive, was similarly suggested to cause changes in cavity areas. However, the molecular mechanism governing such allosteric regulation was not demonstrated (Gourlay et al., 2007). In any case, it does not involve a divergent loop or an FF→LU transition. Thus, to our knowledge, this is the first report of this type of enzyme regulation among Cyps.
Figure 9.
Schematic representation of the 2-Cys mechanism involved in the activity regulation of divergent Cyps. A, The reduced state of CsCyp with the helix containing Cys-40 in the FF state (green). B, The oxidized state of CsCyp with the same helix in the LU state (red), which is a requirement for disulfide bridge formation. C, Structural rearrangements of the divergent loop, induced by disulfide bridge formation, which disrupts the interaction with Glu-83, resulting in a perturbation of the catalytic loop, inactivating the enzyme. D and E, Molecular surface area indicating the cleft aperture in the reduced (D) and oxidized (E) states. F, Superposition of surface areas of both states with the CsA (pink) bound to the active site. The changes in the substrate-binding channel act as a physical barrier to the recognition and binding of CsA or substrate.
The Citrus CTD as a Target of CsCyp
Finally, we show that CsCyp binds with greater affinity to the YSPSAP sequence relative to the more abundant heptad repeats YSPTSP and YSPSSP found in the citrus CTD. The Kd value for the YSPSAP binding to CsCyp is comparable to that of the FGPDLP peptide binding to human CypA (Piotukh et al., 2005). Despite the facts that our molecular docking simulations offer a good explanation for such selective binding (Fig. 8) and that CypA preferentially binds Gly-Pro sequences (Howard et al., 2003; Piotukh et al., 2005), it is important to note that peptides with a Gly at position −1 bind Cyps with such great affinity that they actually function as inhibitors, as is the case for FGPDLP (Howard et al., 2003; Piotukh et al., 2005). Given the peculiar conformation Gly assumes at the −1 position, it has been suggested that suboptimal binding sequences may in fact be better substrates for isomerization (Howard et al., 2003). Thus, we cannot rule out the possibility that the YSPTSP and YSPSSP sequences are indeed also substrates of CsCyp.
MATERIALS AND METHODS
Protein Purification
The citrus (Citrus sinensis) CsCyp gene (ACX37092.1) was cloned into the NdeI/NotI sites of pET28a. The 6xHis-CsCyp was expressed in Escherichia coli BL21 (DE3) cells and purified by metal affinity chromatography. Cells were grown at 30°C in Luria-Bertani medium containing kanamycin (50 mg mL−1) to an optical density at 600 nm of 0.6, followed by induction with 0.4 mm isopropyl-thio-β-d-galactopyranoside (IPTG) for 3 h. After centrifugation, cells were suspended in 20 mm Tris-HCl, pH 8.0, 200 mm NaCl, 5 mm imidazole, and 20% glycerol, incubated on ice with lysozyme (1 mg mL−1) for 30 min, and sonicated. Clarified supernatants were incubated with cobalt resin for 2 h at room temperature. The beads were washed with 10 column volumes of 20 mm Tris-HCl, pH 8.0, 200 mm NaCl, 10 mm imidazole, and 20% glycerol, and the retained proteins were eluted with buffer containing 200 mm imidazole. The 6xHis tag was cleaved with thrombin at 25°C for 16 h. Protein was further purified on a Superdex 75 10/300 column equilibrated with 20 mm Tris-HCl, pH 8.0, 150 mm NaCl, and 2% glycerol. Purified CsCyp (11 mg mL−1) was stored at 4°C in 20 mm Tris-HCl, pH 8.0, 30 mm NaCl, and 0.4% glycerol. The CsTdx protein was expressed and purified as described previously (Domingues et al., 2010).
Crystallization
Despite numerous attempts to crystallize CsCyp in the absence of a ligand, or in complex with CTD peptides, only the CsCyp:CsA complex produced diffractable crystals. In addition, no crystals of oxidized CsCyp were obtained, since a protocol to fully oxidize the protein has not yet been established. Crystallization of the CsCyp:CsA complex (1:2 molar ratio) was performed using the sitting-drop vapor-diffusion method at 18°C. The drop contained 1 µL of the CsCyp:CsA complex plus 1 µL of the reservoir solution containing 100 mm Bis-Tris propane, pH 7.0, and 1.2 m sodium citrate. Crystals were refined using the seeding technique using the same crystallization conditions. Well-formed crystals grew within 1 d and were used for x-ray data collection.
X-Ray Data Collection, Structure Determination, and Refinement
Protein crystals were flash cooled in a stream of gaseous nitrogen at 100 K. The x-ray diffraction data were collected in the MX2 beamline (λ = 1.445 Å) of the Brazilian Synchrotron Light Laboratory using a MAR CCD detector (MARResearch GmbH), with a crystal-to-detector distance of 110 mm and 1° oscillations. Data images were processed with MOSFLM (Leslie, 1992), scaled, and merged with SCALA (Evans, 2006). The structure of the CsCyp:CsA complex was determined by molecular replacement with Phaser, using the structure of the Caenorhabditis elegans Cyp3 in complex with CsA (Protein Data Bank no. 1DYW) as a model. The structure was refined using Phenix and fit to generate electron density maps using Coot. All data were refined to give satisfactory final Rfactor and Rfree factors and geometric parameters. Information on data collection is summarized in Table II. The structure of CsCyp in complex with CsA has been deposited in the Protein Data Bank (no. 4JJM).
Table II. X-ray crystal structure data collection and refinement statistics for CsCyp: molecular replacement.
| Data Collection | Values |
|---|---|
| Space group | P3221 |
| Cell dimensions | |
| Unit-cell dimensions (Å) | a = b = 83.6, c = 85.0 |
| Resolution range (Å) | 37.52–2.09 (2.20–2.09) |
| Reflections | |
| Measured (n) | 108,111 |
| Unique (n) | 20,834 |
| I/σ (I) | 7.7 (2.4) |
| Completeness (%) | 100 (100) |
| Average redundancy | 5.2 (5.0) |
| Rmerge (%) | 19.6 (67.9) |
| Rpim (%) | 9.4 (33.5) |
| Rmeas (%) | 21.8 (75.9) |
| Refinement | |
| Resolution range (Å) | 37.5–2.09 |
| Reflections used (Rfree) | 20,811 (1,068) |
| Rfactor/Rfree (%) | 18.1/22.9 |
| r.m.s.d. bond lengths | 0.009 |
| r.m.s.d. bond angles | 1.029 |
| No molecules per asymmetric unit | 2 |
| Residues in (%) | |
| Preferred regions | 93.77 |
| Allowed regions | 5.67 |
| Outliers | 0.57 |
Mass Spectrometric Analysis
Purified CsCyp separated on polyacrylamide gels under nonreducing conditions was digested with trypsin, dried under vacuum, and reconstituted in 100 µL of 0.1% formic acid. The peptide mixture was analyzed on an LTQ Velos Orbitrap mass spectrometer (Thermo Fisher Scientific) coupled with liquid chromatography-tandem mass spectrometry by an EASY-nLC system (Proxeon Biosystem) through a Proxeon nanoelectrospray ion source, as described previously (Aragão et al., 2012). Peak lists (magellan storage file) were generated from the raw data files using Proteome Discoverer version 1.3 (Thermo Fisher Scientific) and the Sequest search engine and searched against the National Center for Biotechnology Information database using carbamidomethylation (+57.021 D) as a fixed modification, oxidation of Met (+15.995 D) as a variable modification, one trypsin missed cleavage, with a tolerance of 10 ppm for precursor and 0.02 D for fragment ions. Peak lists were filtered using Xcorr cutoffs (+1 > 1.8, +2 > 2.5, and +3 > 3.5).
For cross-link analysis, the raw data files generated by Xcalibur version 2.1 (Thermo Fisher Scientific) were converted to a peak list format (mgf) using Proteome Discoverer version 1.3. The Mascot generic file files were analyzed using the MassMatrix software (Xu and Freitas, 2007) to search automatically against the CsCyp sequence. The parameters for disulfide bond analysis were carbamidomethylation (+57.021 D) as a fixed modification, oxidation of Met (+15.995 D) as a variable modification, cross-linkage search with Cys-Cys, four trypsin missed cleavages, and a tolerance of 10 ppm for precursor and 0.02 D for fragment ions. Search results yielding high confidence (mass-matrix peptide score > 70) and potential cross-linked peptides were manually validated for b and y ion series containing α- and β-chains.
X-Ray Fluorescence Analysis
All measurements were performed at the XRF beamline of the Brazilian Synchrotron Light Laboratory, as described previously (Balan et al., 2006). A white beam 0.1 mm high × 5 mm wide was used to excite the CsCyp samples under total reflection conditions. Samples were prepared as follows: 5 µL of the protein solution at 8 mg mL−1 was dropped onto a Perspex support and dried using an infrared lamp for 15 min. Samples were measured for 300 s, and the collected x-ray fluorescence spectra were evaluated using the PyMCA program (Solé et al., 2007).
Site-Directed Mutagenesis
The Cys residues Cys-40 and Cys-168 were each replaced by Ser (C40S, C168S, and C40S/C168S), whereas Glu-83 was replaced by Ala, Ser, or Gln (E83A, E83S, and E83Q), by site-directed mutagenesis using the QuickChange Site-Directed Mutagenesis kit (Stratagene). The proteins, subcloned into pET28a, were expressed as 6xHis tag fusions and purified as described above.
PPIase Assay
The PPIase assay was performed as described previously (Domingues et al., 2012). Purified proteins were incubated in 50 mm HEPES, pH 8.0, and 100 mm NaCl and stabilized at 10°C for 5 min. CuSO4 and CsA were added to the reaction mix to final concentrations of 15 and 30 nm, respectively. The reaction was initiated by the addition of 1 mg of α-chymotrypsin followed by the peptide substrate N-succinyl-AAPF-p-nitroanilide (Sigma-Aldrich) to a final concentration of 100 μm. The PPIase reaction was monitored at 390 nm for a period of 5 min. Data were fitted to a first-order rate equation (A390 = A1 + A0 e−kt, with k as rate constant) and rate constants (kobs) derived as described by Motohashi et al. (2003). The kcat/Km values were calculated according to the equation kcat/Km = (kobs – k0)/[PPI], where k0 is the first-order rate constant for spontaneous cis-trans-isomerization (Motohashi et al., 2003).
GST Pull-Down Assays
GST-CsTdx, subcloned into the SalI/NotI sites of pGEX-4T, was expressed in BL21 (DE3) cells upon induction with 0.4 mm IPTG for 2 h at 30°C (Domingues et al., 2010). Cells were suspended in phosphate-buffered saline (PBS) containing 1 mm dithiothreitol (DTT) and lysozyme (1 mg mL−1). After sonication and centrifugation, soluble fractions of GST-CsTdx were immobilized on glutathione resin, and unbound proteins were removed with three PBS washes. Purified wild-type CsCyp, or CsCyp mutants (approximately 30 µg), were incubated with the resin containing GST or GST-CsTdx for 2 h at 4°C. The beads were washed four times with PBS, and bound proteins were resolved on 13% SDS-polyacrylamide gels. Proteins were transferred onto nylon membranes, probed with anti-CsCyp (1:3,000) or anti-GST (1:3,000) serum, and developed with the ImmunoCruz kit (Santa Cruz Biotechnology).
NMR Sample and Peptide Preparation
Uniformly 15N- and 13C,15N-labeled protein was prepared for NMR analysis by growing the CsCyp cell culture at 30°C in M9 minimal medium containing 15NH4Cl and/or [13C]Glc (Cambridge Isotopes) to an optical density at 600 nm of 0.6, followed by induction with 0.4 mm IPTG for 3 h. CsCyp was purified as described above. The protein (approximately 1 mm) was solubilized in 50 mm phosphate buffer, pH 6.8, containing 50 mm NaCl and 5% Deuterium oxide, and incubated with synthesized ligand hexapeptides derived from the heptad repeats of the citrus CTD (EY725107): peptide 1 (YSPTSP), peptide 2 (YSPSSP), and peptide 3 (YSPSAP). A fourth peptide (FGPDLP) was used as a control, since it binds human CypA (Piotukh et al., 2005).
NMR Spectroscopy
All NMR experiments were performed at 25°C using a Varian Inova 600-MHz spectrometer equipped with a triple-resonance HCN cryogenic probe, operating at a 1H frequency of 599.987 MHz, at the Brazilian Biosciences National Laboratory or with a four-channel Varian Unity Inova 900-MHz spectrometer operating at a 1H frequency of 899.794 MHz, equipped with a 5-mm triple-resonance HCN and Z-axis pulsed-field gradient cryogenic probe, at The Henry Wellcome Building for Biomolecular NMR Spectroscopy of the University of Birmingham. For sequential assignments, 3D HN(CA)CO, HNCO, HNCACB, and CBCACONH classic triple-resonance experiments (Sattler et al., 1999) were acquired using a 13C,15N-labeled sample. Titration experiments were performed by measuring a series of two-dimensional 1H-15N HSQC spectra, acquired with 2,048 complex data points in the acquisition domain and 256 increments in the time domain, on uniformly 15N-labeled samples at different equivalent ratios of ligand to protein. All spectra were processed using NMRPipe and NMRView software (Johnson and Blevins, 1994; Delaglio et al., 1995). Prior to Fourier transformation, the time domain data were zero filled in all dimensions. When necessary, a fifth-order polynomial baseline correction was applied after transformation and phasing. The Kd values for the CsCyp ligands were determined using the titration analysis module of the NMRView program.
To investigate the possibility of zinc binding to CsCyp, 1H-15N SOFAST-HMQC spectra were acquired at 900 MHz on 15N-labeled samples of (1) 1 mm CsCyp in the presence of (2) 14 mm DTT and 10 mm EDTA, (3) 10 mm ZnCl2, (4) 10 mm ZnCl2 followed by the addition of 10 mm EDTA (same sample as used in sample 3), and on (5) 1.4 mm 15N-labeled Cyp C40S/C168S. The standard pulse sequence, sofastNhmqc.c, from the Biopack sequence library was employed. A large spectral width in the 15N dimension (16,400 Hz) with the offset at 192 ppm, a JNH coupling constant of 22.7 Hz, and an Ernst excitation angle of 120° were used to enable optimal detection of the two-bond J-coupling constant between the amide proton and nitrogen nuclei in the His imidazole region of the spectrum (approximately 160–260 ppm). To compensate for the relatively large number of scans per free induction decay required to generate sufficient signal to noise (350, 864, and 1,200 scans for samples 2, 3, and 4, respectively), a recycle delay of 400 ms and an acquisition time of 40 ms were used to reduce the experimental time. Thirty-two complex data points were acquired in the 15N dimension. Spectra were processed using NMRPipe and analyzed using CcpNmr Analysis (Vranken et al., 2005). Prior to Fourier transformation, a cosine bell window function for apodization was applied in both dimensions as well as linear prediction in the indirect dimension.
Isothermal Titration Calorimetry
Isothermal titration calorimetry experiments were performed on a VP-ITC Microcal calorimeter at 25°C, according to Davis et al. (2010). Purified CsCyp was extensively dialyzed against 50 mm HEPES, pH 8.0, and 200 mm NaCl. Titrations consisted of 5-μL injections of dialyzed CsCyp into the sample cell containing the ligand CsA at 5 µm final concentration. The recorded data were analyzed using Origin 7 software (MicroCal). Binding constants were obtained from the average of three independent experiments.
Molecular Docking Simulations
Molecular docking studies between CsCyp and peptides YSPSAP and YSPSSP were carried out with the program Rosetta FlexPepDock (London et al., 2011). The peptides were roughly docked into the active site of CsCyp based on the binding mode of HIV-1 CA protein CypA (Howard et al., 2003) and then submitted to the high-resolution peptide-protein docking protocol, in which peptide backbone and rigid body orientation are optimized by the “Monte Carlo with minimization” approach. The complex was refined in 200 independent FlexPepDock simulations and ranked based on a Rosetta generic full-atom energy score.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Fragment ion spectra of reduced and cross-linked peptides.
Supplemental Figure S2. Interaction of CsCyp with CsA assessed by isothermal calorimetry, showing the raw injection heats and integrated data fit to a single-site binding model.
Supplementary Material
Acknowledgments
We thank the NMR facility at the HWB-NMR (University of Birmingham) for providing access to the Wellcome Trust-funded 900-MHz spectrometer. Special thanks also to Doug Ward (University of Birmingham) for help with initial mass spectrometry data collection and interpretation and to Romênia Ramos Domingues and Bianca Alves Pauletti for the mass spectrometry experiments performed at the Brazilian Biosciences National Laboratory.
Glossary
- Cyp
cyclophilin
- PPIase
peptidyl prolyl cis-trans-isomerase
- CsA
cyclosporin A
- CTD
C-terminal domain of citrus RNA polymerase II
- 3D
three-dimensional
- GST
glutathione S-transferase
- Kd
dissociation constant
- FF
fully folded
- LU
locally unfolded
- IPTG
isopropyl-thio-β-d-galactopyranoside
- PBS
phosphate-buffered saline
- DTT
dithiothreitol
References
- Aragão AZB, Nogueira MLC, Granato DC, Simabuco FM, Honorato RV, Hoffman Z, Yokoo S, Laurindo FR, Squina FM, Zeri AC, et al. (2012) Identification of novel interaction between ADAM17 (a disintegrin and metalloprotease 17) and thioredoxin-1. J Biol Chem 287: 43071–43082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arévalo-Rodríguez M, Cardenas ME, Wu X, Hanes SD, Heitman J. (2000) Cyclophilin A and Ess1 interact with and regulate silencing by the Sin3-Rpd3 histone deacetylase. EMBO J 19: 3739–3749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balan A, Santacruz CP, Moutran A, Ferreira RC, Medrano FJ, Pérez CA, Ramos CH, Ferreira LC. (2006) The molybdate-binding protein (ModA) of the plant pathogen Xanthomonas axonopodis pv. citri. Protein Expr Purif 50: 215–222 [DOI] [PubMed] [Google Scholar]
- Bannikova O, Zywicki M, Marquez Y, Skrahina T, Kalyna M, Barta A. (2013) Identification of RNA targets for the nuclear multidomain cyclophilin atCyp59 and their effect on PPIase activity. Nucleic Acids Res 41: 1783–1796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratowski S. (2009) Progression through the RNA polymerase II CTD cycle. Mol Cell 36: 541–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coaker G, Falick A, Staskawicz B. (2005) Activation of a phytopathogenic bacterial effector protein by a eukaryotic cyclophilin. Science 308: 548–550 [DOI] [PubMed] [Google Scholar]
- Davis TL, Walker JR, Campagna-Slater V, Finerty PJ, Jr, Paramanathan R, Bernstein G, MacKenzie F, Tempel W, Ouyang H, Lee WH, et al. (2010) Structural and biochemical characterization of the human cyclophilin family of peptidyl-prolyl isomerases. PLoS Biol 8: e1000439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6: 277–293 [DOI] [PubMed] [Google Scholar]
- Domingues MN, de Campos BM, de Oliveira ML, de Mello UQ, Benedetti CE. (2012) TAL effectors target the C-terminal domain of RNA polymerase II (CTD) by inhibiting the prolyl-isomerase activity of a CTD-associated cyclophilin. PLoS ONE 7: e41553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingues MN, De Souza TA, Cernadas RA, de Oliveira ML, Docena C, Farah CS, Benedetti CE. (2010) The Xanthomonas citri effector protein PthA interacts with citrus proteins involved in nuclear transport, protein folding and ubiquitination associated with DNA repair. Mol Plant Pathol 11: 663–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Solis JR, He Z, Lima A, Ting J, Buchanan BB, Luan S. (2008) A cyclophilin links redox and light signals to cysteine biosynthesis and stress responses in chloroplasts. Proc Natl Acad Sci USA 105: 16386–16391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dornan J, Page AP, Taylor P, Wu S, Winter AD, Husi H, Walkinshaw MD. (1999) Biochemical and structural characterization of a divergent loop cyclophilin from Caenorhabditis elegans. J Biol Chem 274: 34877–34883 [DOI] [PubMed] [Google Scholar]
- Evans PR. (2006) Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr 62: 72–82 [DOI] [PubMed] [Google Scholar]
- Fischer G, Schmid FX. (1990) The mechanism of protein folding: implications of in vitro refolding models for de novo protein folding and translocation in the cell. Biochemistry 29: 2205–2212 [DOI] [PubMed] [Google Scholar]
- Gourlay LJ, Angelucci F, Baiocco P, Boumis G, Brunori M, Bellelli A, Miele AE. (2007) The three-dimensional structure of two redox states of cyclophilin A from Schistosoma mansoni: evidence for redox regulation of peptidyl-prolyl cis-trans isomerase activity. J Biol Chem 282: 24851–24857 [DOI] [PubMed] [Google Scholar]
- Gullerova M, Barta A, Lorkovic ZJ. (2006) AtCyp59 is a multidomain cyclophilin from Arabidopsis thaliana that interacts with SR proteins and the C-terminal domain of the RNA polymerase II. RNA 12: 631–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A, Nelson K, Poole LB, Karplus PA. (2011) Structure-based insights into the catalytic power and conformational dexterity of peroxiredoxins. Antioxid Redox Signal 15: 795–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard BR, Vajdos FF, Li S, Sundquist WI, Hill CP. (2003) Structural insights into the catalytic mechanism of cyclophilin A. Nat Struct Biol 10: 475–481 [DOI] [PubMed] [Google Scholar]
- Iki T, Yoshikawa M, Meshi T, Ishikawa M. (2012) Cyclophilin 40 facilitates HSP90-mediated RISC assembly in plants. EMBO J 31: 267–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BA, Blevins RA. (1994) NMR View: a computer program for the visualization and analysis of NMR data. J Biomol NMR 4: 603–614 [DOI] [PubMed] [Google Scholar]
- Ke H, Mayrose D, Belshaw PJ, Alberg DG, Schreiber SL, Chang ZY, Etzkorn FA, Ho S, Walsh CT. (1994) Crystal structures of cyclophilin A complexed with cyclosporin A and N-methyl-4-[(E)-2-butenyl]-4,4-dimethylthreonine cyclosporin A. Structure 2: 33–44 [DOI] [PubMed] [Google Scholar]
- Laxa M, König J, Dietz KJ, Kandlbinder A. (2007) Role of the cysteine residues in Arabidopsis thaliana cyclophilin CYP20-3 in peptidyl-prolyl cis-trans isomerase and redox-related functions. Biochem J 401: 287–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie AG. (1992) Recent changes to the MOSFLM package for processing film and image plate data Joint CCP4 + ESF-EAMCB. Newsl Protein Crystallogr 26: 27–33 [Google Scholar]
- London N, Raveh B, Cohen E, Fathi G, Schueler-Furman O. (2011) Rosetta FlexPepDock Web server: high resolution modeling of peptide-protein interactions. Nucleic Acids Res 39: W249–W253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luban J, Bossolt KL, Franke EK, Kalpana GV, Goff SP. (1993) Human immunodeficiency virus type 1 Gag protein binds to cyclophilins A and B. Cell 73: 1067–1078 [DOI] [PubMed] [Google Scholar]
- Ma X, Song L, Yang Y, Liu D. (2013) A gain-of-function mutation in the ROC1 gene alters plant architecture in Arabidopsis. New Phytol 197: 751–762 [DOI] [PubMed] [Google Scholar]
- Ma Z, Atencio D, Barnes C, DeFiglio H, Hanes SD. (2012) Multiple roles for the Ess1 prolyl isomerase in the RNA polymerase II transcription cycle. Mol Cell Biol 32: 3594–3607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motohashi K, Koyama F, Nakanishi Y, Ueoka-Nakanishi H, Hisabori T. (2003) Chloroplast cyclophilin is a target protein of thioredoxin: thiol modulation of the peptidyl-prolyl cis-trans isomerase activity. J Biol Chem 278: 31848–31852 [DOI] [PubMed] [Google Scholar]
- Nagy PD, Wang RY, Pogany J, Hafren A, Makinen K. (2011) Emerging picture of host chaperone and cyclophilin roles in RNA virus replication. Virology 411: 374–382 [DOI] [PubMed] [Google Scholar]
- Pelton JG, Torchia DA, Meadow ND, Roseman S. (1993) Tautomeric states of the active-site histidines of phosphorylated and unphosphorylated IIIGlc, a signal-transducing protein from Escherichia coli, using two-dimensional heteronuclear NMR techniques. Protein Sci 2: 543–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson MR, Hall DR, Berriman M, Nunes JA, Leonard GA, Fairlamb AH, Hunter WN. (2000) The three-dimensional structure of a Plasmodium falciparum cyclophilin in complex with the potent anti-malarial cyclosporin A. J Mol Biol 298: 123–133 [DOI] [PubMed] [Google Scholar]
- Pflügl G, Kallen J, Schirmer T, Jansonius JN, Zurini MG, Walkinshaw MD. (1993) X-ray structure of a decameric cyclophilin-cyclosporin crystal complex. Nature 361: 91–94 [DOI] [PubMed] [Google Scholar]
- Piotukh K, Gu W, Kofler M, Labudde D, Helms V, Freund C. (2005) Cyclophilin A binds to linear peptide motifs containing a consensus that is present in many human proteins. J Biol Chem 280: 23668–23674 [DOI] [PubMed] [Google Scholar]
- Rupp B. (2009) Biomolecular Crystallography: Principles, Practice, and Application to Structural Biology, Ed 1. Garland Science, New York, NY. [Google Scholar]
- Sattler M, Schleucher J, Griesinger C. (1999) Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Prog Nucl Magn Reson Spectrosc 34: 93–158 [Google Scholar]
- Sekhon SS, Kaur H, Dutta T, Singh K, Kumari S, Kang S, Park SG, Park BC, Jeong DG, Pareek A, et al. (2013) Structural and biochemical characterization of the cytosolic wheat cyclophilin TaCypA-1. Acta Crystallogr D Biol Crystallogr 69: 555–563 [DOI] [PubMed] [Google Scholar]
- Solé VA, Papillon E, Cotte M, Walter P, Susini J. (2007) A multiplatform code for the analysis of energy-dispersive x-ray fluorescence spectra. Spectrochim Acta Part B At Spectrosc 62: 63–68 [Google Scholar]
- Taylor P, Dornan J, Carrello A, Minchin RF, Ratajczak T, Walkinshaw MD. (2001) Two structures of cyclophilin 40: folding and fidelity in the TPR domains. Structure 9: 431–438 [DOI] [PubMed] [Google Scholar]
- Taylor P, Page AP, Kontopidis G, Husi H, Walkinshaw MD. (1998) The x-ray structure of a divergent cyclophilin from the nematode parasite Brugia malayi. FEBS Lett 425: 361–366 [DOI] [PubMed] [Google Scholar]
- Trupkin SA, Mora-García S, Casal JJ. (2012) The cyclophilin ROC1 links phytochrome and cryptochrome to brassinosteroid sensitivity. Plant J 71: 712–723 [DOI] [PubMed] [Google Scholar]
- Vranken WF, Boucher W, Stevens TJ, Fogh RH, Pajon A, Llinas M, Ulrich EL, Markley JL, Ionides J, Laue ED. (2005) The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins 59: 687–696 [DOI] [PubMed] [Google Scholar]
- Wang P, Heitman J. (2005) The cyclophilins. Genome Biol 6: 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Wilcox CB, Devasahayam G, Hackett RL, Arévalo-Rodríguez M, Cardenas ME, Heitman J, Hanes SD. (2000) The Ess1 prolyl isomerase is linked to chromatin remodeling complexes and the general transcription machinery. EMBO J 19: 3727–3738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Freitas MA. (2007) A mass accuracy sensitive probability based scoring algorithm for database searching of tandem mass spectrometry data. BMC Bioinformatics 8: 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.