A ubiquitin ligase has dual functions in ABA-mediated drought stress and in amino acid export in Arabidopsis.
Abstract
Really Interesting New Gene (RING) E3 ubiquitin ligases have been implicated in cellular responses to the stress hormone abscisic acid (ABA) as well as to environmental stresses in higher plants. Here, an ABA-insensitive RING protein3 (atairp3) loss-of-function mutant line in Arabidopsis (Arabidopsis thaliana) was isolated due to its hyposensitivity to ABA during its germination stage as compared with wild-type plants. AtAIRP3 contains a single C3HC4-type RING motif, a putative myristoylation site, and a domain associated with RING2 (DAR2) domain. Unexpectedly, AtAIRP3 was identified as LOSS OF GDU2 (LOG2), which was recently shown to participate in an amino acid export system via interaction with GLUTAMINE DUMPER1. Thus, AtAIRP3 was renamed as AtAIRP3/LOG2. Transcript levels of AtAIRP3/LOG2 were up-regulated by drought, high salinity, and ABA, suggesting a role for this factor in abiotic stress responses. The atairp3/log2-2 knockout mutant and 35S:AtAIRP3-RNAi knockdown transgenic plants displayed impaired ABA-mediated seed germination and stomata closure. Cosuppression and complementation studies further supported a positive role for AtAIRP3/LOG2 in ABA responses. Suppression of AtAIRP3/LOG2 resulted in marked hypersensitive phenotypes toward high salinity and water deficit relative to wild-type plants. These results suggest that Arabidopsis RING E3 AtAIRP3/LOG2 is a positive regulator of the ABA-mediated drought and salt stress tolerance mechanism. Using yeast (Saccharomyces cerevisiae) two-hybrid, in vitro, and in vivo immunoprecipitation, cell-free protein degradation, and in vitro ubiquitination assays, RESPONSIVE TO DEHYDRATION21 was identified as a substrate protein of AtAIRP3/LOG2. Collectively, our data suggest that AtAIRP3/LOG2 plays dual functions in ABA-mediated drought stress responses and in an amino acid export pathway in Arabidopsis.
The ubiquitin (Ub)-26S proteasome system is an indispensable mechanism that regulates many crucial eukaryotic cellular processes, such as cell cycle progression, cell signaling, DNA repair, protein trafficking, and biotic and abiotic stress responses (Welchman et al., 2005; Dreher and Callis, 2007; Vierstra, 2009; Sadanandom et al., 2012). The ubiquitination pathway proceeds through a cascade of three enzymatic reactions catalyzed by E1 Ub-activating enzymes, E2 Ub-conjugating enzymes, and E3 Ub ligases (Guerra and Callis, 2012; Sadanandom et al., 2012). The presence of multiple isoforms of E3 Ub ligases in this system indicates that E3s determine the specificity of target proteins and facilitate the diverse functions of the ubiquitination system in many cellular processes (Vierstra, 2009; Sadanandom et al., 2012).
The E3 Ub ligases are divided into two groups. The first group is composed of Really Interesting New Gene (RING)/U-box and Homology to E6-AP Carboxyl Terminus E3s, which act as a single subunit (Dreher and Callis, 2007; Vierstra, 2009; Yee and Goring, 2009). The second group is composed of multisubunit Ub ligases, including the Skp1-Cullin-F-box complex and the anaphase-promoting complex (Vierstra, 2009; Park et al., 2011; Sadanandom et al., 2012; Seo et al., 2012b). In the Arabidopsis (Arabidopsis thaliana) genome, at least 477 genes encode putative RING E3 Ub ligases; thus, these ligases are the most abundant E3s among single subunit-type Ub ligases and the third largest gene family in Arabidopsis (Kraft et al., 2005; Stone et al., 2005; Vierstra, 2009). This abundance is characteristic of plants, suggesting a potential correlation between the diversity of RING E3s and the sessile life cycle of plants (Vierstra, 2009).
Recent reports indicate that RING E3s play roles in the cellular responses to the stress hormone abscisic acid (ABA) and to environmental stresses in higher plants (Lyzenga and Stone, 2012; Sadanandom et al., 2012). In particular, RING Ub ligases are involved in the drought stress responses. SDIR1 enhanced the drought stress response by positively regulating ABA signaling upstream of the ABF transcription factors in Arabidopsis (Zhang et al., 2007). CaRma1H1, which is an endoplasmic reticulum (ER)-localized RING E3, increased dehydration stress tolerance through down-regulation of the water channel protein aquaporin levels in transgenic Arabidopsis plants (Lee et al., 2009). Arabidopsis RING RHA2a/RHA2b and AtRDUF1/AtRDUF2 function as positive regulators in both ABA signaling and water stress responses (Bu et al., 2009; Li et al., 2011; Kim et al., 2012). On the other hand, DRIP-RING E3 serves as a negative regulator in the drought stress response in Arabidopsis (Qin et al., 2008). It ubiquitinates and induces proteasomal degradation of the drought-induced transcription factor DREB2A. Arabidopsis RGLG2 and rice (Oryza sativa) OsDSG1, OsRDCP1, and OsDIS1 were recently reported as either negative or positive regulators in response to dehydration stress (Park et al., 2010; Bae et al., 2011; Ning et al., 2011; Cheng et al., 2012). These studies are in agreement with the notion that different RING E3 Ub ligases participate in the defense mechanism against drought conditions in a positive or negative manner in both monocot and dicot model plants.
The subgroups of the above-mentioned RING E3 genes are either up-regulated or down-regulated by water deficit. In addition, the protein stability and subcellular localization of the RING E3s are also affected by environmental stresses (Lyzenga and Stone, 2012). Without ABA treatment, KEG1 is autoubiquitinated, and thereby, its cellular levels are regulated depending on the presence or absence of ABA (Liu and Stone, 2010). The RGLG2-GFP fusion protein translocates from the plasma membrane to the nucleus following salt stress treatment in Arabidopsis roots (Cheng et al., 2012). These observations suggest that the modes of action of plant RING E3s are modulated at various levels of cellular processes. Although the physiological roles of different RING E3s in response to environmental stresses have been elucidated, large numbers of RING Ub ligases still remain uncharacterized in higher plants.
In our previous studies, we collected 100 different Arabidopsis transfer DNA (T-DNA)-inserted loss-of-function mutants of RING E3 Ub ligases and investigated their phenotypes with respect to the ABA sensitivity of the germination stage. In those studies, the C3H2C3-type ABA-INSENSITIVE RING PROTEIN1 (AtAIRP1) and C3HC4-type AtAIRP2 RING E3s were found to play positive and combinatory roles in ABA-mediated drought stress responses (Ryu et al., 2010; Cho et al., 2011).
In this report, atairp3, a line with the At3g09770 locus knocked out, was isolated and investigated. Akin to atairp1 and atairp2, atairp3 progeny displayed significantly reduced ABA responses during germination and in postgermination stages. AtAIRP3 was a C3HC4-type RING E3 Ub ligase identified as LOSS OF GDU2 (LOG2), which was previously shown to be involved in amino acid export via interaction with GLUTAMINE DUMPER1 (GDU1; Pratelli et al., 2012). Transcript levels of AtAIRP3/LOG2 were up-regulated by drought, high salinity, and exogenous ABA treatments. The atairp3/log2-2 loss-of-function mutant lines exhibited impaired ABA-mediated stomata closure and hypersensitive phenotypes in response to high salinity and drought. Using yeast (Saccharomyces cerevisiae) two-hybrid, in vitro, and in vivo immunoprecipitation (IP), cell-free protein degradation, and in vitro ubiquitination assays, we identified RESPONSIVE TO DEHYDRATION21 (RD21) as a substrate protein of AtAIRP3. Collectively, our data indicate that the Arabidopsis C3HC4-type RING E3 Ub ligase AtAIRP3/LOG2 is a positive regulator of the ABA-mediated drought and salt stress tolerance mechanism via the ubiquitination of RD21. These results further suggest that AtAIRP3/LOG2 plays dual functions in drought stress response and amino acid transport in Arabidopsis.
RESULTS
The ABA-Induced RING E3 Ub Ligase AtAIRP3 Was Identified as the Recently Isolated LOG2
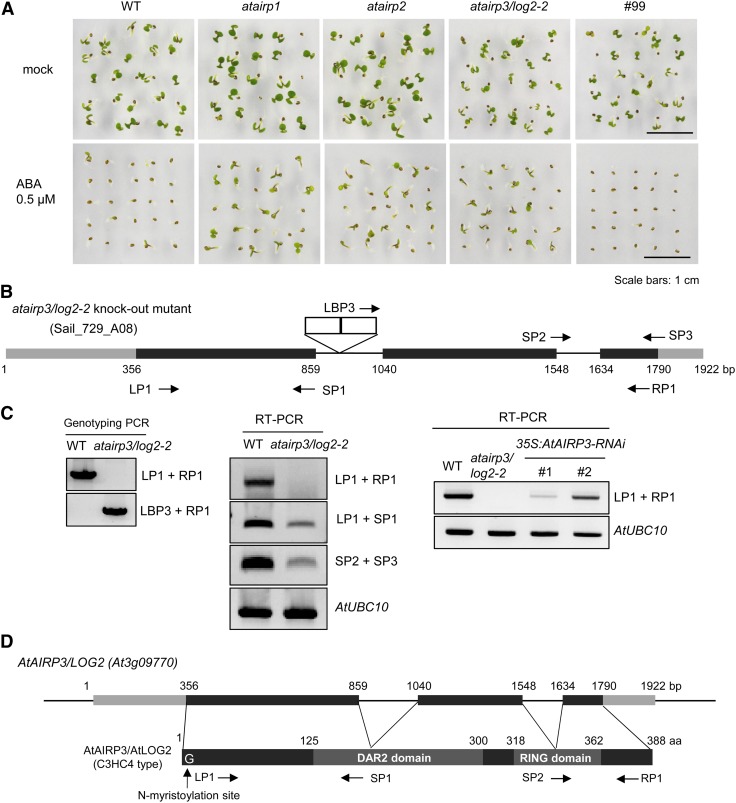
Previously, 100 different Arabidopsis T-DNA-inserted loss-of-function mutant lines of RING E3 Ub ligases were collected and examined in terms of ABA sensitivity. In those studies, the C3H2C3-type AtAIRP1 and C3HC4-type AtAIRP2 RING E3s were identified as positive regulators in ABA-mediated drought stress responses (Ryu et al., 2010; Cho et al., 2011). In this study, atairp3 was isolated and investigated. Similar to atairp1 and atairp2, the atairp3 progeny exhibited reduced sensitivity toward ABA. Figure 1A shows that the percentage of atairp3 seedlings (49.3%) that germinated was markedly higher than that of wild-type seedlings (6.7%) in the presence of 0.5 μm ABA for 5 d. The germination percentage of atairp3 with ABA was comparable to those of atairp1 (52.0%) and atairp2 (45.3%; Fig. 1A).
Figure 1.
Isolation of the atairp3/log2-2 T-DNA-inserted loss-of-function mutant line. A, Screening of an ABA-hyposensitive RING E3 Ub ligase mutant in the germination stage. Wild-type (WT) and knockout mutant seeds were germinated on MS growth medium in the absence (top panel) or presence (bottom panel) of 0.5 μm ABA. Germination rates with respect to cotyledon greening were determined after 5 d. The atairp1 and atairp2 mutants served as positive controls for ABA-insensitive phenotypes, while the #99 RING mutant served as a negative control to demonstrate the wild-type phenotype in response to ABA. Bars = 1 cm. B, Schematic structure of the atairp3/log2-2 mutant line (SAIL_729_A08). Gray bars represent the 5′ and 3′ untranslated regions, black bars show the coding regions, and solid lines indicate the introns of AtAIRP3/LOG2 (GenBank accession no. NC_003074). The atairp3/log2-2 mutant contains double T-DNA insertions in the first introns after nucleotides 873 and 887 in an antisense orientation. White boxes depict T-DNA insertions. Primers used for genotyping PCR and RT-PCR are shown with arrows. Nucleotide sequences of the primers are listed in Supplemental Table S1. C, Genotyping and RT-PCR analyses of the atairp3/log2-2 knockout mutant and 35S:AtAIRP3-RNAi knockdown transgenic plants. Left panel, genotyping PCR of wild-type and atairp3/log2-2 plants. Primers used for genomic PCR are shown on the right side of the agarose gel. Middle panel, RT-PCR of wild-type and atairp3/log2-2 plants. Primers used for RT-PCR are shown on the right side of the gel. The level of Arabidopsis AtUBC10 (E2 ubiquitin-conjugating enzyme) transcripts was used as a loading control. Right panel, RT-PCR of wild-type, atairp3/log2-2, and 35S:AtAIRP3-RNAi (independent transgenic T4 lines 1 and 2) plants. Primers used for RT-PCR are shown on the right. AtUBC10 was used as a loading control. D, Schematic structure of the AtAIRP3/LOG2 gene and its predicted protein. Gray bars represent 5′ and 3′ untranslated regions, black bars show coding regions, and solid lines indicate introns. A putative N-terminal myristoylation site, a DAR2 domain, and a single C3HC4-type RING motif are indicated. Primers used for RT-PCR are shown with arrows. [See online article for color version of this figure.]
The AtAIRP3 gene (GenBank accession no. NC_003074), which is located on chromosome 3, was composed of 1,922 bp with three exons and two introns. The atairp3 mutant line (SAIL_729_A08) contained double T-DNA insertions in the first intron after nucleotides 873 and 887 in the antisense orientation (Fig. 1B). Genotyping PCR verified homozygous atairp3 progeny, and reverse transcription (RT)-PCR further confirmed that, although partial mRNAs were still detectable, full-length AtAIRP3 transcripts were absent in the homozygous line (Fig. 1C). The predicted protein encoded by AtAIRP3 contains 388 amino acid residues with a molecular mass of 42.85 kD (Supplemental Fig. S1). AtAIRP3 harbors a single C3HC4-type RING motif in its C-terminal region, a putative myristoylation site in the N-terminal region, and a domain associated with RING2 (DAR2) domain in the central region (Fig. 1D). To examine whether AtAIRP3 contains an E3 Ub ligase activity, the MBP-AtAIRP3 fusion construct was expressed in Escherichia coli. The full-length MBP-AtAIRP3 was highly insoluble in bacterial cells under our experimental conditions. MBP-AtAIRP3101-388, an N-terminal partially deleted form, was expressed in the soluble fraction and used for an in vitro self-ubiquitination assay. As expected, MBP-AtAIRP3101-388 displayed in vitro Ub ligase activity in the presence of ATP, E1, and E2 (Supplemental Fig. S2). Search of The Arabidopsis Information Resource database (http://www.arabidopsis.org) revealed that AtAIRP3 is identical to LOG2, which was recently reported to be involved in amino acid export (Pratelli et al., 2012). LOG2 interacted with GDU1, and the LOG2-GDU1 complex was proposed to play a significant role in the regulation of amino acid export. The identity of AtAIRP3 as LOG2 was unexpected, as AtAIRP3 was highly inducible by the stress hormone ABA (Fig. 2), while LOG2 was identified as an interacting partner of a subunit of an amino acid exporter complex (Pratelli et al., 2012). AtAIRP3 was renamed as AtAIRP3/LOG2.
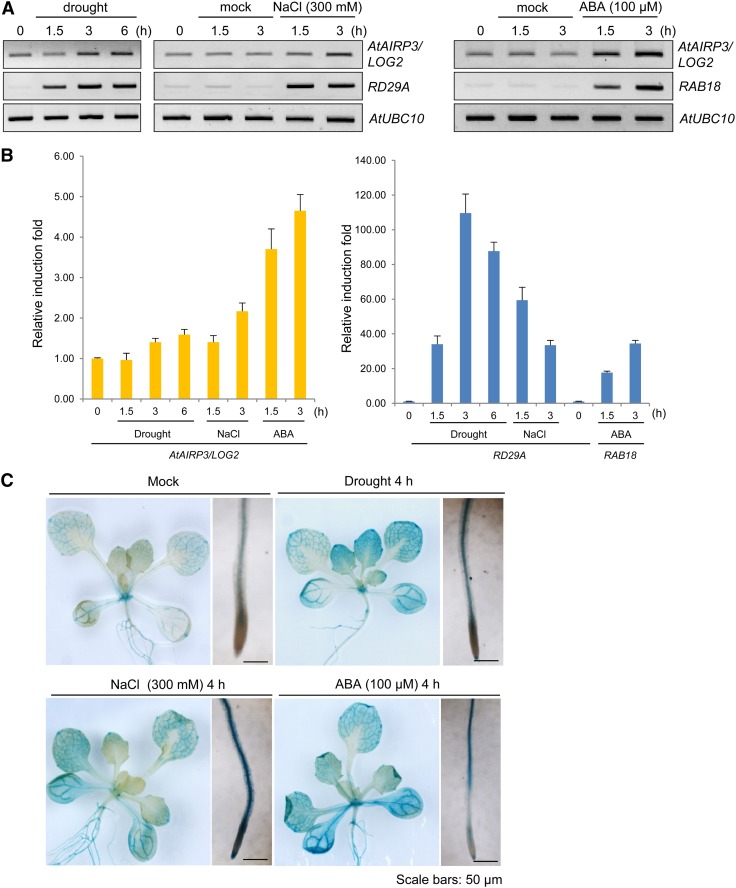
Figure 2.
Induction of AtAIRP3/LOG2 in response to drought, high salinity, and ABA. A, RT-PCR analysis of AtAIRP3/LOG2. Light-grown, 10-d-old Arabidopsis seedlings were subjected to drought (0, 1.5, 3, and 6 h), 300 mm NaCl (0, 1.5, and 3 h), or 100 μm ABA (1.5 and 3 h). Total RNA prepared from the treated tissues was analyzed by RT-PCR using gene-specific primer sets. RD29A served as a positive control for drought and salt treatments, whereas RAB18 served as a positive control for ABA induction. AtUBC10 was used as a loading control. B, Real-time qRT-PCR analysis of AtAIRP3/LOG2. Total RNA was isolated from the treated tissues and used for real-time qRT-PCR. Fold induction of AtAIRP3/LOG2 was normalized to levels of glyceraldehyde-3-phosphate dehydrogenase C subunit mRNA, which served as an internal control. Bars indicate means ± sd from three independent experiments. C, AtAIRP3/LOG2 promoter-GUS assay. Light-grown, 10-d-old AtAIRP3-promoter:GUS transgenic T3 plants were grown under drought conditions (4 h) with 300 mm NaCl (4 h) or with 100 μm ABA (4 h). Histochemical localization of GUS activities in leaf and root tissues was visualized by 5-bromo-4-chloro-3-indolyl β-D-glucoside staining for 1 h. Bars = 50 μm. [See online article for color version of this figure.]
AtAIRP3/LOG2 Was Induced by Drought, High Salinity, and ABA Treatment
The fact that the atairp3/log2-2 mutant seedlings were hyposensitive to the stress hormone ABA (Fig. 1A) raised the possibility that AtAIRP3/LOG2 is a stress-related gene. To follow up on this possibility, RT-PCR and real-time quantitative reverse transcription (qRT)-PCR experiments were conducted. The steady-state levels of AtAIRP3/LOG2 mRNAs were slightly enhanced by drought (1.3- to 1.6-fold) and high salinity (1.2- to 2.0-fold; Fig. 2, A and B). In addition, gene expression was elevated 3.5- and 4.5-fold after 1.5 and 3.0 h of treatment with ABA (100 μm), respectively. The magnitude of induction of AtAIRP3/LOG2, however, was less than those of the known marker genes RD29A for drought and salt and RAB18 for ABA (Fig. 2, A and B).
A promoter-GUS assay demonstrated that LOG2 was mainly expressed in vascular tissues in vegetative organs and in the style and pollen grains in reproductive organs (Pratelli et al., 2012). In addition to these basal levels of expression, AtAIRP3/LOG2 promoter activity increased in both leaves and roots in response to drought, salt, and ABA treatments (Fig. 2C). These results suggest that AtAIRP3/LOG2 is controlled at the transcriptional level in response to stress and ABA. Thus, we speculated that AtAIRP3/LOG2 is involved not only in the amino acid export system (Pratelli et al., 2012) but also in the ABA-mediated stress response (Figs. 1A and 2) in Arabidopsis.
Suppression of AtAIRP3/LOG2 Resulted in Hyposensitivity to ABA with Respect to Germination Rates and Stomatal Movement
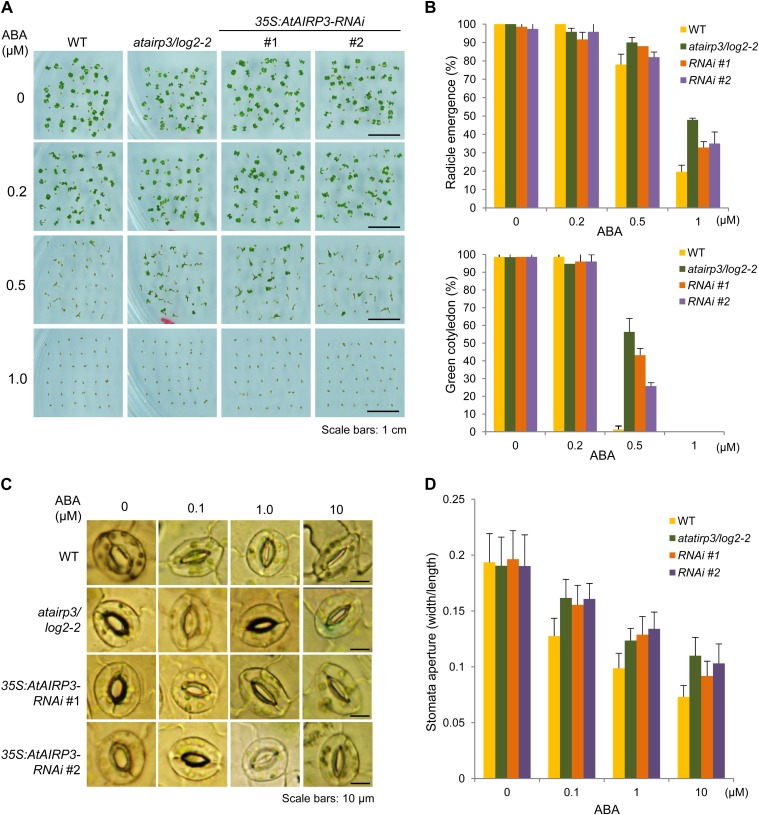
To examine the role of AtAIRP3/LOG2 in ABA and stress responses, the phenotype of the atairp3/log2-2 knockout line was tested. First, wild-type and mutant seeds were germinated in the absence or presence of ABA (0, 0.2, 0.5, or 1 μm), and germination percentages were counted in terms of radicle emergence and cotyledon greening after 3 and 5 d, respectively (Fig. 3A). In the presence of 0.5 μm ABA, 79.8% and 88.2% of the wild-type and atairp3/log2-2 seeds yielded radicles, respectively (Fig. 3B, top panel). In the presence of 1.0 μm ABA, only 19.7% of the wild-type seeds were able to germinate, while 47.9% of the mutant seeds germinated. Differences between the wild-type and mutant seeds were even more distinguishable in cotyledon greening. With 0.5 μm ABA, only approximately 2% of wild-type seeds developed normal green cotyledons (Fig. 3B, bottom panel). In contrast, 56.3% of atairp3/log2-2 seeds displayed functional green cotyledons. These results, along with those in Figure 1A, indicate that the atairp3/log2-2 knockout seeds were hyposensitive to ABA during germination as compared with wild-type seeds.
Figure 3.
ABA-insensitive phenotypes of atairp3/log2-2 knockout mutant and 35S:AtAIRP3/LOG2-RNAi knockdown transgenic plants with respect to germination rates and stomatal movements. A, ABA-mediated inhibition of germination. Wild-type (WT), atairp3/log2-2 mutant, and 35S:AtAIRP3-RNAi (T4 lines 1 and 2) seeds were germinated on MS growth medium supplemented with different concentrations (0, 0.2, 0.5, and 1.0 μm) of ABA. Germination rates were determined after 3 to 5 d. Photographs were taken at 5 d after germination. Bars = 1 cm. B, Germination percentages of wild-type, atairp3/log2-2 mutant, and 35S:AtAIRP3-RNAi (T4 lines 1 and 2) seeds in response to ABA. Top panel, germination percentages were determined with respect to radicle emergence at 3 d after ABA treatment. Bottom panel, germination percentages were measured in terms of cotyledon greening at 5 d after ABA treatment. Bars indicate means ± sd (n = 110) from three independent experiments. C, ABA-mediated stomatal closure. Light-grown, 4-week-old rosette leaves of wild-type, atairp3/log2-2 mutant, and 35S:AtAIRP3-RNAi (T4 lines 1 and 2) plants were immersed in stomatal opening solution for 4 h and in ABA solution (0, 0.1, 1.0, or 10 µm) for another 2 h. Bright-field microscopy was used to photograph the guard cells. Bars = 10 μm. D, Measurement of stomatal aperture after ABA treatment. Stomatal apertures of wild-type, atairp3/log2-2 mutant, and 35S:AtAIRP3-RNAi (T4 lines 1 and 2) leaves were measured as the ratio of width to length after ABA treatment. Bars indicate means ± sd (n ≥ 35). [See online article for color version of this figure.]
Second, ABA-mediated stomatal behavior of wild-type and atairp3/log2-2 plants was monitored. Light-grown, 4-week-old mature leaves were treated with ABA for 2 h, and stomatal movement profiles were measured as the ratio of width to length (Fig. 3C). Average stomatal apertures of wild-type leaves were concurrently reduced as concentrations of ABA increased (0, 0.1, 1.0, and 10 μm), from 0.19 ± 0.025 to 0.13 ± 0.015, 0.1 ± 0.013, and 0.07 ± 0.01, respectively (Fig. 3D). However, decreases in the stomatal apertures of atairp3/log2-2 mutant leaves in response to the same range of ABA concentrations appeared to be less evident. They were reduced from 0.19 ± 0.025 to 0.16 ± 0.016, 0.12 ± 0.01, and 0.11 ± 0.016, respectively.
To further determine the effects of AtAIRP3/LOG2-2 suppression, RNA interference (RNAi)-mediated knockdown transgenic lines (35S:AtAIRP3-RNAi) were constructed (Supplemental Fig. S3). The level of AtAIRP3/LOG2 mRNAs was significantly reduced in independent T4 35S:AtAIRP3-RNAi lines 1 and 2 as determined by RT-PCR (Fig. 1B). As shown in Figure 3, phenotypes of the 35S:AtAIRP3-RNAi knockdown plants in response to ABA were intermediate between wild-type and ataitp3/log2-2 plants with respect to both germination percentages and stomatal behaviors. These results indicate that ABA sensitivity during germination and postgermination growth is positively correlated with the expression level of AtAIRP3/LOG2.
Cosuppression and Complementation Experiments
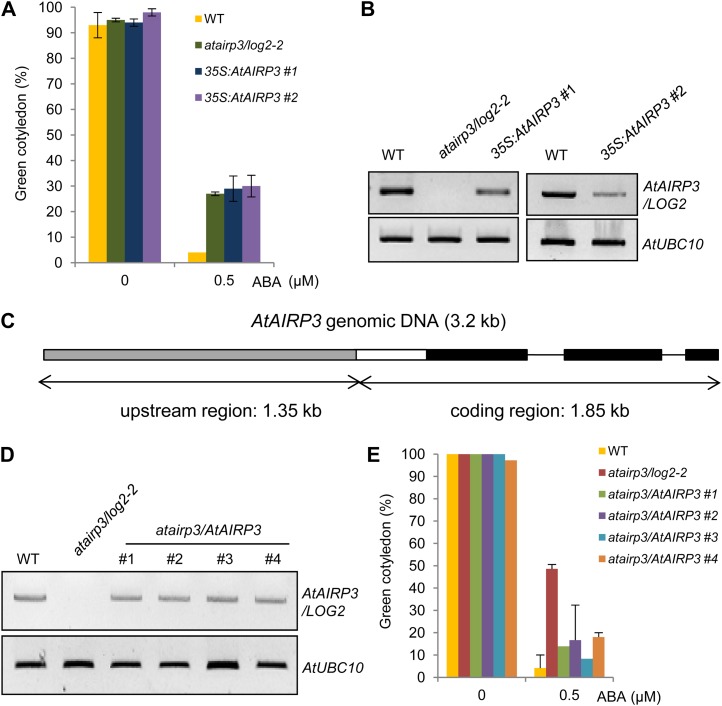
To obtain more detailed evidence for the roles of AtAIRP3/LOG2 in the response of Arabidopsis to ABA, we generated transgenic Arabidopsis plants that overexpressed AtAIRP3/LOG2 under the control of the cauliflower mosaic virus (CaMV) 35S promoter. Several independent T4 transgenic progeny (35S:AtAIRP3/LOG2) were obtained and used for phenotypic assays. In contrast to what we expected, the 35S:AtAIRP3/LOG2 seedlings (lines 1 and 2) displayed hyposensitivity toward ABA in the germination stage. Their germination percentages (cotyledon greening) were similar (28.7% for line 1 and 30.0% for line 2) to those of atairp3/log2-2 (27.3%) in the presence of 0.5 μm ABA (Fig. 4A). Under the same ABA concentration, less than 4.0% of the wild-type seeds successfully germinated. We repeated the overexpression experiments and obtained similar results. This led us to hypothesize that ectopic expression of AtAIRP3/LOG2 with the CaMV 35S promoter may cause cosuppression in Arabidopsis. RT-PCR analysis indeed demonstrated that the AtAIRP3/LOG2 transcripts were significantly reduced in the 35S:AtAIRP3/LOG2 lines relative to those in wild-type plants (Fig. 4B).
Figure 4.
Cosuppression and complementation experiments. A and B, Arabidopsis plants that expressed AtAIRP3/LOG2 under the control of the CaMV 35S promoter were constructed. Several independent T4 transgenic plants (35S:AtAIRP3/LOG2) were obtained and used for phenotypic assays. A, Inhibition of germination by ABA in wild-type (WT), atairp3/log2-2 mutant, and 35S:AtAIRP3/LOG2 transgenic T4 (independent lines 1 and 2) plants. Freshly harvested seeds of each plant were germinated on MS growth medium supplemented with 0.5 μm ABA. Percentages of green cotyledon development were counted after 5 d. B, AtAIRP3/LOG2 transcript levels of wild-type and 35S:AtAIRP3/LOG2 transgenic plants. AtUBC10 was used as an equal loading control. C to E, Generation and phenotypic analysis of atairp3/AtAIRP3 complementation transgenic plants. C, Cartoon of the AtAIRP3/LOG2 complementation construct. The plasmid construct (3.2 kb in length) is composed of a 1.35-kb upstream region and a 1.85-kb coding region. The gray bar indicates the upstream region. The black bars indicate coding regions, and the white bar indicates the 5′ untranslated region. Solid lines represent introns. D, Transcript levels of wild-type, atairp3/log2-2, and atairp3/AtAIRP3 T3 complementation transgenic plants. Total RNAs were isolated as indicated and subjected to RT-PCR analysis using gene-specific primer sets (Supplemental Table S1). E, Complementation of atairp3/log2-2 by the AtAIRP3/LOG2 transgene. Seeds of wild-type, atairp3/log2-2, and atairp3/AtAIRP3 complementation T3 (independent lines 1–4) plants were germinated on MS medium containing 0.5 μm ABA. Percentages of green cotyledon development of wild-type, atairp3/log2-2, and atairp3/AtAIRP3 complementation T3 plants were determined after 5 d. Bars are means ± sd (n = 72) from three independent experiments. [See online article for color version of this figure.]
We next carried out a complementation experiment. A genomic fragment that contained the 5′ upstream region (1.35 kb in length) and the entire coding region (1.85 kb in length) of AtAIRP3/LOG2 was transformed into an atairp3/log2-2 mutant plant (Fig. 4C). Independent T3 complementation transgenic plants (atairp3/AtAIRP3) were subsequently obtained. RT-PCR analysis showed that AtAIRP3/LOG2 mRNA levels were effectively restored in atairp3/AtAIRP3 transgenic plants (Fig. 4D). In addition, atairp3/AtAIRP3 transgenic seeds displayed increased sensitivity to ABA in germination as compared with the atairp3/log2-2 seeds (Fig. 4E). The ABA sensitivities of the complementation lines were intermediate between those exhibited by wild-type and atairp3/log2-2 seeds. Thus, introduction of the functional AtAIRP3/LOG2 gene into atairp3/log2-2 mutant plants partially rescued the loss-of-function ABA-insensitive phenotype. Collectively, the results of the cosuppression and complementation studies (Fig. 4) in combination with the results of the knockout experiments (Fig. 3) strongly suggest that AtAIRP3/LOG2 is positively involved in the response of Arabidopsis to ABA.
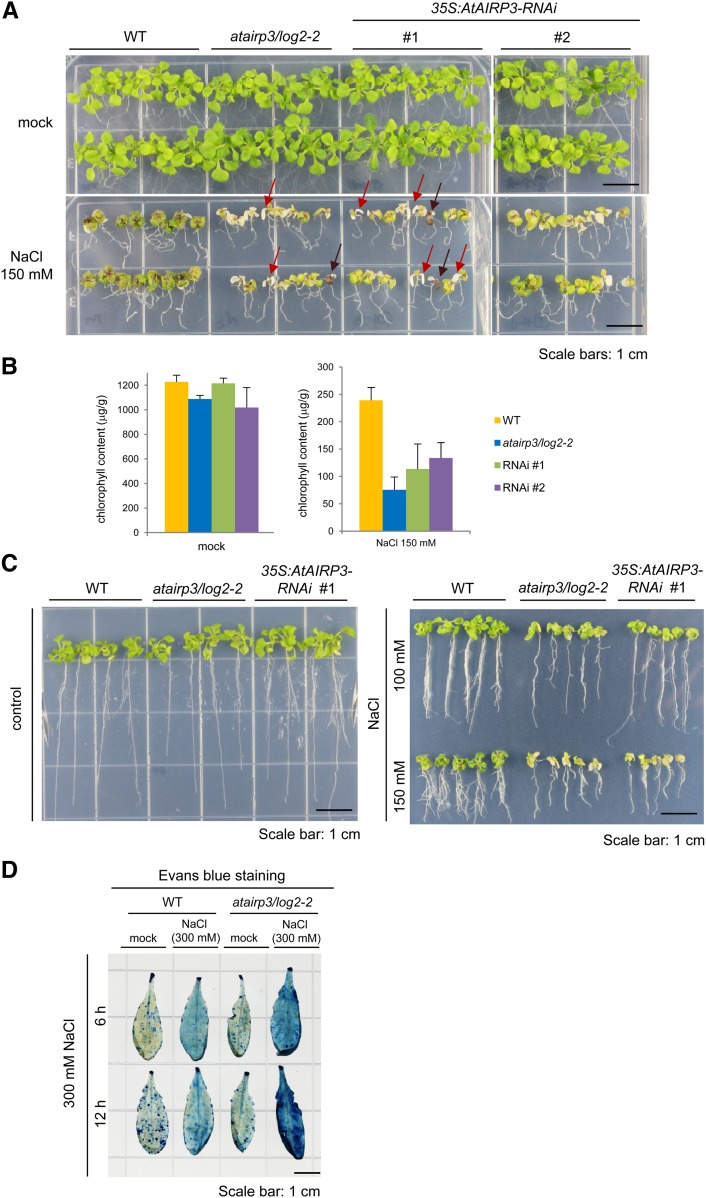
atairp3/log2-2 Mutant and 35S:AtAIRP3-RNAi Knockdown Transgenic Plants Were Hypersensitive to High Salinity
We went on to estimate the capacity of the atairp3/log2-2 mutant and the 35S:AtAIRP3-RNAi transgenic (lines 1 and 2) plants to respond to high salinity. Wild-type, mutant, and RNAi seedlings were grown for 5 d under normal growth conditions and then transported into medium supplemented with 150 mm NaCl. Their morphological changes were monitored after 14 d. In the presence of NaCl, the growth of wild-type plants was significantly retarded, and their leaves accumulated brown precipitates (Fig. 5A). The atairp3/log2-2 mutant plants showed even more evident developmental anomalies after salt treatment. Their leaves became pale green and yellowish with markedly reduced growth (Fig. 5A). The degree of aberrant growth of the RNAi knockdown leaves was intermediate between wild-type and knockout plants. Consistently, the average chlorophyll content in the mutant leaves was approximately 30% (75.4 ± 23.52 μg g−1 fresh weight) of that in wild-type leaves (239 ± 23.50 μg g−1 fresh weight) upon salt stress (Fig. 5B). Under the same salt conditions, the average chlorophyll levels in the RNAi leaves were 113.5 ± 45.8 to 133.7 ± 28.1 μg g−1 fresh weight.
Figure 5.
Hypersensitive phenotypes of atairp3/log2-2 knockout mutant and 35S:AtAIRP3/LOG2-RNAi knockdown transgenic plants in response to high salinity. A, NaCl-sensitive phenotypes of atairp3/log2-2 and 35S:AtAIRP3/LOG2-RNAi plants as compared with wild-type (WT) plants. Wild-type, mutant, and RNAi (independent T4 lines 1 and 2) seedlings were grown for 5 d under normal growth conditions and placed in medium supplemented without (top panel) or with (bottom panel) 150 mm NaCl. The morphological abnormalities were observed after 14 d. Some of the atairp3/log2-2 mutant leaves were severely discolored and became whitish or dark brown, as indicated by red and brown arrows, respectively. Bars = 1 cm. B, Chlorophyll content of leaves from wild-type, mutant, and RNAi transgenic plants after salt treatment. Wild-type, atairp3/log2-2, and 35S:AtAIRP3/LOG2-RNAi plants were identically treated as described above, and total leaf chlorophyll levels were measured. Bars indicate means ± sd from three independent experiments. C, Root growth analysis of wild-type, atairp3/log2-2, and 35S:AtAIRP3-RNAi plants in response to different concentrations (0, 100, and 150 mm) of NaCl. Wild-type, mutant, and RNAi (independent T4 lines 1 and 2) seedlings were grown for 5 d under normal growth conditions and placed in medium supplemented without (left panel) or with (right panel) 100 to 150 mm NaCl. The root growth profiles were observed after 14 d. Note that lateral root formation in atairp3/log2-2 and RNAi plants was greatly reduced by high salinity relative to wild-type plants. D, Evans blue staining. Mature, healthy leaves from wild-type and atairp3/log2-2 mutant plants were incubated with 300 mm NaCl for 6 to 12 h. Salt-treated leaves were subsequently treated with Evans blue staining solution. The degree of cell death was determined by the strength of dark blue color. Bar = 1 cm. [See online article for color version of this figure.]
In addition to leaf growth, the root growth of atairp3/log2-2 mutant and 35S:AtAIRP3-RNAi knockdown plants was also severely impaired by high salinity as compared with wild-type plants. Before salt treatment, the morphology of roots in wild-type, knockout, and knockdown plants was similar. Elongation of wild-type roots was significantly inhibited by 100 to 150 mm NaCl treatment for 14 d (Fig. 5C). While the elongation of mutant roots appeared to be somewhat similar to that of wild-type roots, lateral root formation in atairp3/log2-2 and RNAi plants was reduced more dramatically by high salinity than that in wild-type plants (Fig. 5C). Thus, both the leaf and root tissues of atairp3/log2-2 mutant and 35S:AtAIRP3-RNAi knockdown plants exhibited hypersensitive phenotypes in response to high salinity.
Some of the atairp3/log2-2 mutant leaves were severely discolored and became whitish or dark brown (Fig. 5A, red and brown arrows, respectively). To further evaluate the sensitivity of leaf tissue to high salinity, salt-treated leaves were subjected to Evans blue staining, which assesses cell death. Mature, healthy leaves of wild-type and atairp3/log2-2 plants were incubated with 300 mm NaCl for 6 to 12 h and subsequently treated with Evans blue staining solution. The degree of cell death was determined by the strength of the dark blue color (Watanabe and Lam, 2006). Upon salt treatment, cell death was significantly induced in wild-type leaves. In addition, more dramatic cell death was clearly detected in atairp3/log2-2 leaves (Fig. 5D), indicating that atairp3/log2-2 leaves were more susceptible to NaCl treatment than wild-type leaves. Thus, the suppression of AtAIRP3/LOG2 resulted in increased sensitivity to high salinity.
atairp3/log2-2 Knockout Mutant and 35S:AtAIRP3-RNAi Knockdown Transgenic Plants Were More Susceptible to a Water Deficit than Wild-Type Plants
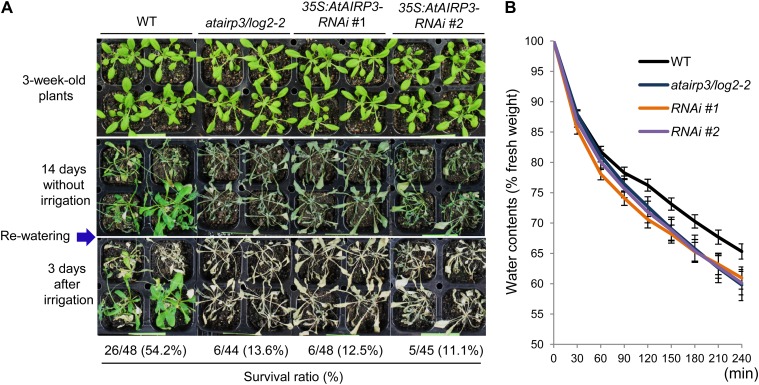
Because the atairp3/log2-2 and 35S:AtAIRP3-RNAi lines were hyposensitive to ABA (Fig. 3) and susceptible to high salinity (Fig. 5), we speculated that suppression of AtAIRP3/LOG2 alters the drought stress responses. Therefore, wild-type, atairp3/log2-2, and RNAi plants were subjected to dehydration stress. These plants were grown for 3 weeks in pots under normal growth conditions and grown for another 14 d without watering. Survival of these water-stressed plants was monitored 3 d after rewatering. Under these experimental conditions, 54.2% (26 of 48) of wild-type plants resumed their growth after rewatering (Fig. 6A); however, only 13.6% (six of 44) of the atairp3/log2-2 plants resumed growth after rewatering. In addition, the survival of 35S:AtAIRP3-RNAi plants was also very low (12.5% [six of 48] for line 1 and 11.1% [five of 45] for line 2). Thus, the suppression of AtAIRP3/LOG2 resulted in a greatly reduced tolerance to dehydration stress.
Figure 6.
Phenotypic analysis of atairp3/log2-2 knockout mutant and 35S:AtAIRP3/LOG2-RNAi knockdown transgenic plants in response to drought stress. A, Hypersensitive phenotypes of atairp3/log2-2 and 35S:AtAIRP3/LOG2-RNAi plants in response to water deficit. Wild-type (WT), mutant, and RNAi (independent T4 transgenic lines 1 and 2) plants were grown for 3 weeks in pots under normal growth conditions. The plants were grown for another 14 d without watering. Survival of these water-stressed plants was monitored 3 d after rewatering. B, Measurement of leaf water loss rates. The aerial parts of 4-week-old wild-type, atairp3/log2-2, and 35S:AtAIRP3/LOG2-RNAi plants were incubated at room temperature for different time periods. Changes in the fresh weight of leaves were measured at the given time points. Bars indicate means ± sd of 10 plants. Similar results were obtained from four biologically independent replicates. [See online article for color version of this figure.]
Furthermore, mutant and RNAi transgenic leaves lost their water content more rapidly than wild-type leaves. After a 2-h incubation at room temperature, detached wild-type and mutant leaves lost approximately 23.8% and 27.3% to 30.4% of their fresh weight, respectively (Fig. 6B). After 4 h of incubation, the fresh weight of wild-type leaves decreased approximately 34.7%, while that of mutant and knockdown leaves was reduced by 39.1% to 40.9% (Fig. 6B). These results indicate that atairp3/log2-2 knockout and 35S:AtAIRP3-RNAi knockdown plants were more susceptible to a water deficit than wild-type plants. Overall, our phenotypic analyses suggest that suppression of AtAIRP3/LOG2 results in hyposensitivity to ABA (Fig. 3) and markedly reduces tolerance not only to high salinity (Fig. 5) but also to drought (Fig. 6) in Arabidopsis.
AtAIRP3/LOG2 Interacts with RD21
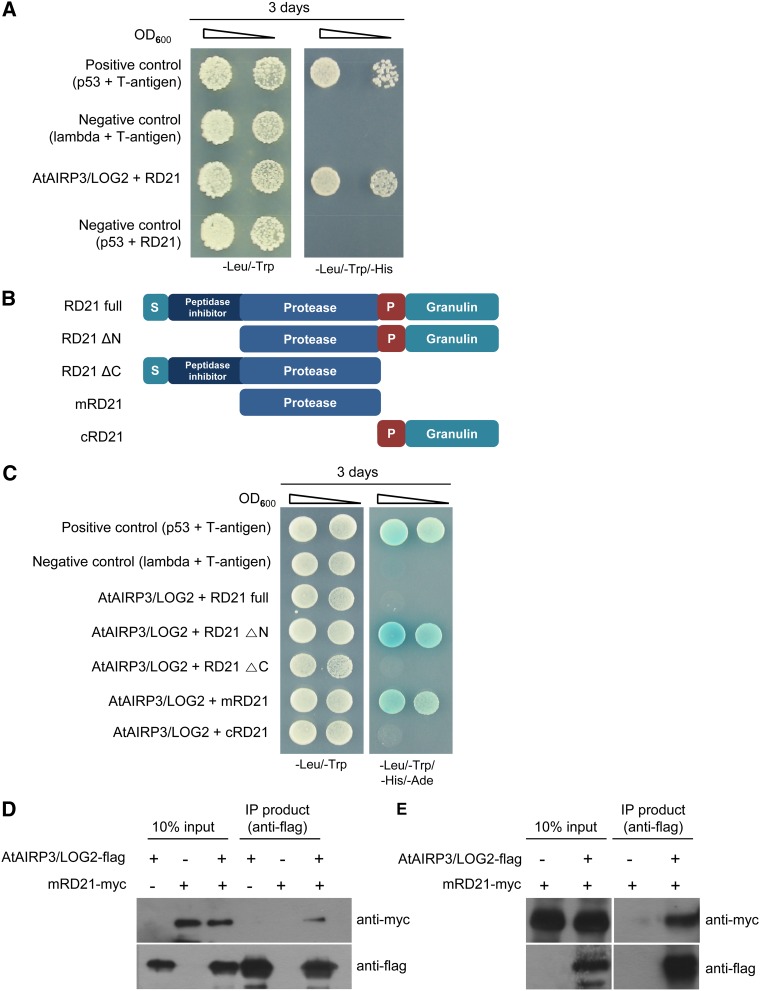
To uncover the cellular mechanism by which AtAIRP3/LOG2 participates in drought stress responses, we performed yeast two-hybrid screening using a complementary DNA (cDNA) library prepared from 3-d-old etiolated seedlings and a full-length AtAIRP3/LOG3 cDNA as bait. Several positive colonies that grew in the presence of AtAIRP3/LOG2 in the three-minus selection medium (SD/−Trp/−Leu/−His) with 10 mm 3-amino-1,2,4,-triazole (3-AT) were identified. One of the positive yeast cells contained a partial cDNA encoding RD21 (At1g47128; Fig. 7A). Arabidopsis RD21 was initially identified as a drought-inducible Cys proteinase of the papain family (Koizumi et al., 1993). It is produced as a 57-kD preproprotein and posttranslationally cleaved to a 30- to 33-kD mature protein (Yamada et al., 2001; Gu et al., 2012). The intermediate and mature forms of RD21 localize to the ER body and lytic vacuole in senescing Arabidopsis leaves (Hayashi et al., 2001; Yamada et al., 2001; Lampl et al., 2013).
Figure 7.
AtAIRP3/LOG2 interacts with RD21. A, Yeast two-hybrid screening using a cDNA library prepared from 3-d-old etiolated seedlings and a full-length AtAIRP3/LOG3 cDNA. Arabidopsis cDNAs were cloned into the pACT2 vector (prey), and AtAIRP3/LOG2 was cloned into the pGBKT7 vector (bait). The cDNA-pACT2 and AtAIRP3/LOG2-pGBKT7 plasmids were cotransformed into yeast AH109 cells. Yeast cells were plated on SD/−Trp/−Leu/−His medium that contained 10 mm 3-AT and allowed to grow for 5 d at 30°C. The yeast cells containing RD21-pACT2 + AtAIRP3/LOG2-pGBKT7 grew efficiently in SD/−Trp/−Leu/−His medium in the presence of 3-AT. p53 + T-antigen were used as the positive control. Lambda + T-antigen and p53 + RD21 were used as the negative control. OD600, Optical density at 600 nm. B, Schematic representation of full-length (RD21 full), intermediate (RD21 ΔN and RD21 ΔC), mature (mRD21), and deletion (cRD21) forms of RD21. P, Pro-rich domain; S, signal peptide. C, Yeast two-hybrid assay. Different forms of RD21 were cloned into pGADT7. The RD21-pGADT7 and AtAIRP3/LOG2-pGBKT7 constructs were cotransformed into yeast AH109 cells. Yeast cells were grown in SD/−Leu/−Trp/−His/−Ade growth medium at 30°C for 3 d. p53 + T-antigen were used as the positive control. Lambda + T-antigen were used as the negative control. D, In vitro IP assay. Bacterially expressed AtAIRP3/LOG2-flag and mRD21-myc fusion proteins were coincubated in the presence of an anti-flag affinity gel matrix. After extensive washing, the bound proteins were eluted with Gly (100 mm) buffer and subjected to immunoblot analysis using anti-flag and anti-myc antibodies. E, In vivo IP assay. The AtAIRP3/LOG2-flag and mRD21-myc fusion genes were transiently expressed in tobacco leaves. Leaf crude extracts (1 mg of protein) were immunoprecipitated with anti-flag affinity gel matrix. The bound proteins were eluted, separated by SDS-PAGE, and detected with anti-flag or anti-myc antibody. [See online article for color version of this figure.]
To confirm the interaction between AtAIRP3/LOG3 and RD21 in yeast cells, we constructed four different truncated forms of RD21 (Fig. 7B) and repeated the yeast two-hybrid assay using RD21 as prey and AtAIRP3/LOG3 as bait under four-minus selection medium (SD/−Leu/−Trp/−His/−Ade) growth conditions. Full-length RD21 was unable to interact with AtAIRP3/LOG2. In contrast, RD21 ΔN (the intermediate form), which contains the protease, Pro-rich, and granulin domains, and mRD21 (the mature form), which consists of only the protease domain, interacted with AtAIRP3/LOG2 (Fig. 7C). These results indicate that the intermediate and mature forms of RD21 interact with AtAIRP3/LOG2 in yeast cells.
To corroborate the association of AtAIRP3/LOG2 and RD21, an in vitro IP experiment was carried out. AtAIRP3/LOG2-flag and mRD21-myc fusion proteins were expressed in E. coli. Purified recombinant proteins were coincubated in the presence of an anti-flag affinity gel matrix. After extensive washing, the bound proteins were eluted with Gly (100 mm) buffer and subjected to immunoblot analysis using anti-flag and anti-myc antibodies. The results in Figure 7D revealed that mRD21A-myc protein was coimmunoprecipitated with AtAIRP3/LOG2-flag by anti-flag antibody. In contrast, mRD21-myc alone failed to bind the anti-flag affinity matrix. These results indicate that AtAIRP3/LOG2 and mRD21 interact in vitro.
Interaction between AtAIRP3/LOG2 and RD21 was further substantiated by an in vivo IP assay. The AtAIRP3/LOG2-flag and mRD21-myc gene constructs were expressed in tobacco (Nicotiana benthamiana) leaves using an Agrobacterium tumefaciens-meditated transient expression method. Leaf crude extracts (1 mg of protein) were immunoprecipitated with anti-flag affinity gel matrix. The bound proteins were eluted, separated by SDS-PAGE, and detected using anti-flag and anti-myc antibodies. AtAIRP3/LOG2-flag and mRD21-myc were coimmunoprecipitated by anti-flag antibody in leaf crude extracts expressing AtAIRP3/LOG2-flag and mRD21-myc fusion genes (Fig. 7E); however, mRD21-myc was not immunoprecipitated with the anti-flag antibody in tobacco leaf extracts expressing the mRD21-myc fusion gene alone. These yeast two-hybrid and IP assays suggest that RD21 is one of the target proteins of AtAIRP3/LOG2.
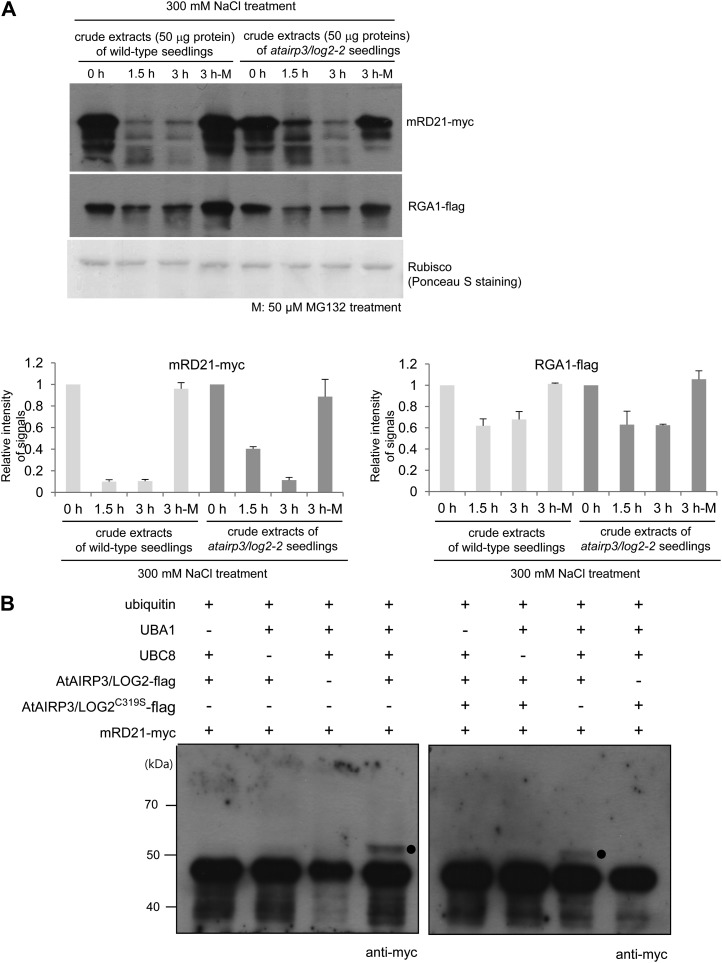
Ubiquitination of RD21 by AtAIRP3/LOG2
The interaction between AtAIRP3/LOG2 and RD21 raised the possibility that the cellular level of RD21 is subject to control by the Ub-26S proteasome pathway. To test this possibility, an in vitro cell-free degradation assay was performed. Bacterially expressed mRD21-myc protein was incubated for different time periods (0, 1.5, and 3 h) with crude extracts prepared from salt-treated (300 mm NaCl) 10-d-old wild-type seedlings. The protein levels were monitored over time by immunoblotting with anti-myc antibody. The mRD21-myc level was rapidly reduced with wild-type cell-free extracts. After 1.5 h of incubation, approximately 90.1% of the protein was already degraded (Fig. 8A). However, mRD21-myc was stable during incubation with the wild-type crude extracts in the presence of MG132, an inhibitor of the proteasome complex. After 3 h of incubation with 50 μm MG132, only 5% of mRD21-myc was degraded (Fig. 8A, lane 3 h-M). These results revealed that MG132 hinders reduction of the mRD21-myc level, indicating proteasome-dependent degradation of RD21 with cell-free extracts. An identical experiment was conducted with crude extracts prepared from salt-treated atairp3/log2-2 mutant seedlings. After a 1.5-h incubation, more than 40% of mRD21-myc was still detected in the reaction mixture (Fig. 8A). Thus, the degradation of mRD21-myc was significantly slowed down with atairp3/log2-2 cell-free extracts as compared with wild-type extracts. This finding suggests that the stability of RD21 is dependent, at least in part, on AtAIRP3/LOG2. Again, the degradation of mRD21-myc was strongly hampered by MG132 with atairp3/log2-2 extracts. RGA1 and Rubisco proteins were used for control experiments. RGA1 is regulated by the Ub-26S proteasome pathway (Dill et al., 2004; Lee et al., 2010), whereas Rubisco served as a loading control. The results showed that the proteasome-dependent fluctuation patterns of the RGA1-flag protein were very similar in the wild-type and atairp3/log2-2 cell-free extracts, indicating that its degradation was independent of AtAIRP3/LOG2. As expected, the level of Rubisco remained constant in all reaction mixtures examined (Fig. 8A). Overall, these results suggest that the stability of RD21 is subject to control by the 26S proteasome complex in an AtAIRP3/LOG2-dependent manner.
Figure 8.
AtAIRP3/LOG2-dependent degradation of RD21. A, Cell-free degradation assay for mRD21. The mRD21-myc protein was incubated for 0 to 3 h with crude extracts of salt-treated (300 mm NaCl) 10-d-old wild-type or atairp3/log2-2 mutant seedlings in the absence (lanes labeled 0 h, 1.5 h, and 3 h) or presence (lanes labeled 3 h-M) of 50 μm MG132. The time-dependent protein levels were examined by immunoblotting with anti-myc antibody. RGA1, which is known to be regulated by the Ub-26S proteasome pathway, was used as a positive control for proteasome-dependent degradation. The RGA1-flag protein was detected with an anti-flag antibody. Rubisco served as a loading control and was detected by Ponceau S staining. The protein levels were quantified using Multi Gauge version 3.1 software (Fuji Film). B, In vitro ubiquitination of mRD21 by AtAIRP3/LOG2. Recombinant AtAIRP3/LOG2-flag or the AtAIRP3/LOG2-flagC319S derivative was coincubated with mRD21-myc in the presence or absence of Ub, ATP, E1 (Arabidopsis UBA1), and E2 (Arabidopsis UBC8) at 30°C for 1 h. The reaction mixture was analyzed by immunoblotting with an anti-myc antibody. Black circles indicate the shifted high-molecular-mass ubiquitinated protein band.
We next performed an in vitro ubiquitination assay. Recombinant mRD21-myc and AtAIRP3/LOG2-flag were coincubated in the presence or absence of Ub, ATP, E1 (Arabidopsis UBA1), and E2 (Arabidopsis UBC8) at 30°C for 1 h. The reaction mixture was analyzed by immunoblotting using anti-myc antibody. Coincubation of mRD21-myc and AtAIRP3/LOG2-flag gave rise to a high-molecular-mass band (Fig. 8B, left panel). Exclusion of E1 or E2 from the reaction mixture abrogated the ubiquitinated band. In addition, the AtAIRP3/LOG2-flagC319S derivative, in which the conserved Cys residue in the RING motif was replaced by Ser, was unable to ubiquitinate mRD21-myc (Fig. 8B, right panel). Taken together, the data presented in Figure 8, together with those in Figure 7, further support the view that RD21 is one of the substrate proteins of AtAIRP3/LOG2 in Arabidopsis.
DISCUSSION
As sessile organisms, land plants encounter diverse environmental stresses, including drought, high salinity, heavy metals, and extreme temperatures, during their life cycle (Boyer, 1982; Cushman and Bohnert, 2000). The prompt and appropriate responses of plants to these adverse growth conditions are inevitable factors that determine whether the plants survive and flourish. With respect to agriculture, the defense mechanisms by which plants retaliate to detrimental environmental stresses are closely tied with crop yield (Ahuja et al., 2010; Hirayama and Shinozaki, 2010). Thus, examination of the cellular stress responses and the development of stress-tolerant transgenic crops have steadily increased (Verslues and Juenger, 2011; Deikman et al., 2012).
Considerable evidence indicates that plant RING E3 Ub ligases mediate cellular responses to abiotic stresses (Lyzenga and Stone, 2012; Sadanandom et al., 2012). The modes of action of RING E3s appear to be diverse. These ligases work as either positive or negative factors and/or in either an ABA-dependent or -independent manner. RING E3s also function individually or in combination with other homologous RING E3s. Furthermore, these proteins localize to different cellular sites within plant cells.
In this report, 100 different Arabidopsis T-DNA-inserted loss-of-function mutants of RING E3 Ub ligases were screened with regard to ABA sensitivity during germination. The atairp3 mutant was isolated due to its hyposensitivity to ABA (Fig. 1). AtAIRP3 gene activity was up-regulated by high salinity, drought, and ABA treatment, suggesting a role for this factor in abiotic stress responses (Fig. 2). AtAIRP3 is a RING E3 Ub ligase with a single C3HC4-type RING motif, a putative myristoylation site, and a DAR2 domain. Unexpectedly, AtAIRP3 turned out to be identical to LOG2, which is involved in amino acid export at the plasma membrane (Pratelli et al., 2012). LOG2 interacts with and ubiquitinates GDU1, which is considered a subunit of the amino acid export complex. The association of LOG2 with GDU1 was proposed to stabilize GDU1 localization at the plasma membrane (Pratelli et al., 2012). Therefore, AtAIRP3 was renamed AtAIRP3/LOG2.
Detailed phenotypic analyses revealed that atairp3/log2-2 and 35S:AtAIRP3-RNAi plants exhibited markedly reduced sensitivity to ABA for both germination rates and stomatal movements (Fig. 3). Cosuppression and complementation data were also in good agreement with those of the knockout and RNAi experiments (Fig. 4). In addition, suppression of AtAIRP3/LOG2 resulted in decreased tolerance to high salinity (Fig. 5) and drought conditions (Fig. 6). Therefore, AtAIRP3/LOG2 most likely regulates water stress responses and serves as a positive regulator in an ABA-dependent fashion. Based on these results, we speculate that AtAIRP3/LOG2 has dual functions in Arabidopsis: this protein participates in the ABA-mediated drought stress responses and in an amino acid export pathway.
Pratelli et al. (2012) showed that AtAIRP3/LOG2 was localized to the membrane fractions, particularly to the plasma membrane. Their results indicated that myristoylation was important for the localization of AtAIRP3/LOG2 to the plasma membrane and facilitated the association of AtAIRP3/LOG2 with GDU1. The AtAIRP3/LOG2-GDU1 complex may play a role in the regulation of amino acid export (Pratelli et al., 2012). With these in mind, we hypothesized that the mechanism by which AtAIRP3/LOG3 regulates the drought stress response may be also linked to the plasma membrane. Membrane-associated RING E3s have recently been implicated in stress responses in plants (Guerra and Callis, 2012). For example, SDIR1 is an endomembrane-localized RING E3 that functions positively in ABA-associated drought stress tolerance (Zhang et al., 2007). The ER-localized RING E3 CaRma1H1 enhances dehydration stress tolerance by ubiquitinating the water channel protein aquaporin PIP2;1 isoform in transgenic Arabidopsis plants (Lee et al., 2009). The mode of action of CaRma1H1 appeared to be ABA independent. KEG RING E3 exhibits subcellular localization at both the endosomes and the nucleus. At the endosomes, it is involved in programmed cell death (PCD) in response to pathogen invasion (Gu and Innes, 2011), whereas, at the nucleus, this protein down-regulates the ABA-responsive transcription factor ABI5 (Stone at al., 2006; Liu and Stone, 2010). More recently, RGLG2 RING E3 was shown to localize to the plasma membrane under normal growth conditions, but this protein moved into the nucleus in response to high salinity, where it negatively modulates salt stress responses (Cheng et al., 2012).
Yeast two-hybrid screening identified RD21 as a protein that interacts with AtAIRP3/LOG2. Subsequently, mRD21, the 30-kD mature form of RD21, was shown to physically bind AtAIRP3/LOG2 on the basis of the yeast two-hybrid and in vitro and in planta IP assays (Fig. 7). The mRD21 protein was degraded by a 26S proteasome complex in an AtAIRP3/LOG2-dependent manner in the cell-free degradation assay (Fig. 8A). Moreover, mRD21 was ubiquitinated in vitro by AtAIRP3/LOG2 (Fig. 8B). Notably, RD21 was previous identified as a ubiquitinated protein via matrix-assisted laser-desorption ionization time of flight + liquid chromatography-mass spectrometry proteomic analysis of Arabidopsis proteins (Manzano et al., 2008). Taking these results into account, it seems highly likely that RD21 is one of the target proteins of AtAIRP3/LOG2.
RD21 was initially isolated as a drought-induced Cys proteinase (Koizumi et al., 1993). It localizes to the ER body and lytic vacuoles in senescing leaves and may act to aid in the cell death process under stress conditions and during leaf senescence by facilitating nitrogen recycling (Hayashi et al., 2001; Yamada et al., 2001). PROTEIN DISULFIDE ISOMERASE5 (PDI) is concomitantly targeted with RD21 from the ER through the Golgi to vacuoles and inhibits the proteinase activity of RD21 in the endothelial cells of developing Arabidopsis seeds (Ondzighi et al., 2008). Loss of PDI, which in turn resulted in increased RD21 proteinase activity, initiated premature PCD of the endothelium in developing seeds, suggesting a role for RD21 in PCD. C14 is the tomato (Solanum lycopersicum) homolog of RD21. Ectopic expression of C14 hindered Phytophthora infestans infection at the apoplast in tobacco (Bozkurt et al., 2011). All these results are consistent with the view that RD21 may be functional under stress conditions (e.g. drought, senescence, PCD, or pathogen attack) to remobilize the building blocks of macromolecules.
Currently, however, the functional relationships between AtAIRP3/LOG2 and RD21 in drought stress tolerance remain to be solved. Because RD21 contains Cys proteinase activity, excessive release of RD21 from the vacuole may be harmful to cells. In this aspect, we are tempted to propose that AtAIRP3/LOG2 ubiquitinates and thus leads to the proteasomal degradation of RD21 in response to drought to ward off the undesirable degradation of cellular proteins facilitated by RD21. AtAIRP3/LOG2 is a plasma membrane-bound protein through its myristoylation without membrane-spanning domains (Pratelli et al., 2012). Hence, AtAIRP3/LOG2 may meet RD21 released from the vacuole in response to water stress at the cytosolic surfaces of the plasma membrane. In fact, a recent report suggested that RD21 that was sequestered in the ER body and vacuole is released to the cytoplasmic fractions during plant-pathogen (necrotropic fungi) interactions (Lampl et al., 2013). RD21 is then associated with cytoplasmic AtSerpin1, a protease inhibitor. The AtSerpin-RD21 complex may limit the cellular damage caused during cell death. These results are in concordance with our current scenario of functional correlations of AtAIRP3/LOG2 and RD21 in drought stress responses. Alternatively, AtAIRP3/LOG2 and RD21 may interact with each other during the trafficking processes to the plasma membrane and vacuole, respectively, under drought stress conditions. Yamada et al. (2001) showed that the intermediate (the protease + Pro-rich + granulin domains) and mature (the protease domain) forms of RD21 were present in the vacuoles. AtAIRP3/LO2 is able to interact with the intermediate and mature forms but not with the proprotein form of RD21 (Fig. 7, B and C). Thus, interaction between AtAIRP3/LOG2 and RD21 during their trafficking pathway appears to be unlikely. Therefore, additional studies are required to uncover how AtAIRP3/LOG2 and RD21 are functionally correlated to expedite defense mechanism against drought conditions.
The 35S:AtAIRP3-RNAi knockdown plants exhibited phenotypes that were intermediate between those of wild-type and atairp3/log2-2 knockout mutant plants in terms of ABA-mediated inhibition of seed germination (Fig. 3, A and B) and salt stress responses (Fig. 5). However, it should be noted that the RNAi and knockout plants displayed comparable insensitivities in ABA-mediated stomatal closure (Fig. 3, C and D). In addition, the RNAi plants showed greatly reduced tolerance to drought stress, and their survival rates (Fig. 6A) and leaf water loss rates (Fig. 6B) were very similar to those of knockout mutant plants. These may indicate that AtAIRP3/LOG2 is more intimately associated with drought stress responses than with either ABA or salt stress responses.
The roles of AtAIRP3/LOG2 in exporting amino acids and in degrading RD21 do not seem to be unrelated with regard to the regulation of intracellular free amino acid levels. AtAIRP3/LOG2, along with GDU1, promotes amino acid export at the plasma membrane, and AtAIRP3/LOG2 ubiquitinates RD21, which increases amino acid levels through its proteinase activity. Thus, two different functions of AtAIRP3/LOG2 may be interconnected. In conclusion, the data presented in this report suggest that AtAIRP3/LOG2 plays dual functions in the ABA-mediated drought stress response and in the amino acid export system in Arabidopsis.
MATERIALS AND METHODS
Plant Materials
Arabidopsis (Arabidopsis thaliana ecotype Columbia-0) wild-type and T-DNA-inserted atairp3/log2-2 loss-of-function mutant seeds were collected from the Arabidopsis Biological Resource Center. The T-DNA-inserted mutation was confirmed by genotyping PCR and RT-PCR. Seeds were treated with 30% bleach solution and washed 10 times with sterilized water to remove residual bleach solution. Seedlings were grown in medium containing Murashige and Skoog (MS) salt (Duchefa Biochmie) and Suc (1%–3%) in phytoagar (0.8%) or in soil (Sunshine mix5; Sun Gro Horticulture) at 22°C under long-day conditions (16 h of light/8 h of dark).
RT-PCR and Real-Time qRT-PCR Analysis
Light-grown 10-d-old seedlings were subjected to dehydration by opening the lid of a plant culture plate at the growth chamber (22°C) for 1.5, 3, and 6 h. For NaCl and exogenous ABA treatments, seedlings were transferred to one-half-strength MS liquid medium (Duchefa Biochmie) containing 300 mm NaCl or 100 μm ABA (Sigma-Aldrich) for 1.5 and 3 h.
Total RNA was isolated from stress- or ABA-treated 10-d-old wild-type, knockout mutant, and RNAi knockdown seedlings with an Easy Spin plant total RNA extraction kit (Intron Biotechnology) according to the manufacturer’s protocol. The cDNA was synthesized using a Moloney murine leukemia virus cDNA synthesis kit (Enzynomics) with 1.5 μg of total RNA. RT-PCR was performed according to the established protocol described in the previous study (Seo et al., 2012a). The DNA sequences of the primers used in this study are listed in Supplemental Table S1. PCR products were separated with agarose gel electrophoresis and visualized with Gel-doc (Bio-Rad Laboratories). Real-time qRT-PCR was performed using an IQ5 light cycler (Bio-Rad Laboratories) as described by Ryu et al. (2010). The relative levels of the transcripts were normalized to the glyceraldehyde-3-phosphate dehydrogenase C subunit mRNA level, which was included as an internal control.
Amino Acid Sequence Alignment
The predicted amino acid sequences for AtAIRP3/LOG2 and its homologs were obtained from The Arabidopsis Information Resource (http://www.arabidopsis.org). Amino acid alignment was performed using ClustalX2.0 software (http://www.clustal.org), and the results were edited using Genedoc software (http://www.nrbsc.org/gfx/genedoc). Functional domains in the predicted AtAIRP3/LOG2 were analyzed as described by Stone et al. (2005) and Marchler-Bauer et al. (2011).
Construction of AtAIRP3-Promoter:GUS, 35S:AtAIRP3-RNAi Knockdown, 35S:AtAIRP3 Cosuppression, and atairp3/AtAIRP3 Complementation Transgenic Plants
For the promoter-GUS assay, the upstream region (1.5 kb) of AtAIRP3/LOG2 was amplified from genomic DNA by PCR. The PCR product was ligated into the pJET blunt vector (Fermentas) and subcloned into the pCAMBIA1381 vector using the BamHI and NcoI sites.
For the RNAi suppression study, the C-terminal region (1,126–1,557 bp) of the AtAIRP3/LOG2 cDNA was amplified by PCR using RNAi primer sets (Supplemental Table S1), and the product was ligated into the pENTR vector (Invitrogen), which harbored a GUS intron. The RNAi construct was inserted into the pEarleyGate100 vector (Earley et al., 2006), which contains the CaMV 35S promoter, using an LR Clonase II enzyme (Invitrogen) reaction. For the 35S:AtAIRP3 cosuppression study, a full-length AtAIRP3/LOG2 cDNA was obtained by RT-PCR using gene-specific primers (Supplemental Table S1). The PCR product was cloned into the pJET blunt cloning vector (Fermentas). After confirming the correct reading frame of the gene construct by DNA sequencing, the plasmid containing a full-length AtAIRP3/LOG2 cDNA was ligated into the pENTR vector using the BamHI restriction enzyme. The cDNA clone was again subcloned into the pEarleyGate100 vector using the LR Clonase II enzyme.
For the complementation study, genomic DNA that contained the 5′ upstream region (1.35 kb) and the entire coding region (1.85 kb) of AtAIRP3/LOG2 was amplified by gene-specific primers (Supplemental Table S1) and ligated into the pTOP blunt vector (Enzynomics). The construct was ligated into the pENTR vector and reinserted into the pMDC163 vector (Curtis and Grossniklaus, 2003) via an LR reaction.
The constructed plasmids were transformed into Arabidopsis plants by the floral dip method (Clough and Bent, 1998) using Agrobacterium tumefaciens. Glufosinate-ammonium (BASTA; 25 μg mL−1; Duchefa Biochmie) and hygromycin (35 μg mL−1; Duchefa Biochmie) were used for the selection of transgenic lines. RT-PCR and genomic Southern-blot analyses were performed to confirm the expression and copy number of the transgene in transgenic plants.
Germination Assay and Stomatal Aperture Measurement
Seeds of wild-type, atairp3/log2-2 mutant, and 35S:AtAIRP3-RNAi (independent transgenic lines 1 and 2) plants were sterilized with a 30% bleach solution and sown on MS growth medium with various concentrations (0, 0.1, 0.5, or 1.0 μm) of ABA (Sigma-Aldrich). Percentages of radicle emergence and cotyledon greening were counted at 3 and 5 d after sowing, respectively.
For stomatal aperture analysis, rosette leaves of 4-week-old wild-type, atairp3/log2-2, and 35S:AtAIRP3-RNAi (transgenic lines 1 and 2) plants were detached and incubated for 4 h in stomata-opening solution (10 mm MES [pH 6.1], 100 μm CaCl2, and 10 mm KCl) and transferred into the ABA-containing solution (0, 0.1, 1.0, or 10 μm) for 2 h. The adaxial side of the leaf epidermis was peeled off using 3M tape, mounted on glass slides, and observed with an Olympus CX41 microscope. Stomata images were photographed with a JUJAK560 CCD camera (Dixi Optics) and analyzed using Photoshop CS4 software (Adobe Systems). At least 35 stomata were analyzed for each experimental group.
Phenotype Analysis in Response to High Salinity and Drought
Wild-type, mutant, and RNAi (independent transgenic T4 lines 1 and 2) seedlings were grown for 5 d under normal growth conditions and transferred into medium supplemented with 100 or 150 mm NaCl. Morphological abnormalities were observed after 14 d.
Light-grown, 3-week-old wild-type, atairp3/log2-2, and 35S:AtAIRP3-RNAi (transgenic lines 1 and 2) plants were grown for 14 d without irrigation as described by Kim et al. (2010). After 3 d of irrigation, survival rates of drought-stressed plants were counted. To analyze the leaf water loss rates, the aerial parts of the plants were detached from the roots and incubated for 4 h on the bench at room temperature. Fresh weights of detached plants were measured by electric microbalance at various time points. In each assay, at least 10 plants were used. The experiments were performed four times with four different replicates.
Quantification of Chlorophyll Content, Evans Blue Staining, and GUS Staining
Quantification of chlorophyll content was performed based on the method described by Page et al. (2001). Salt-treated leaves of wild-type, atairp3/log2-2 mutant, and 35S:AtAIRP3/LOG2-RNAi plants were pulverized in liquid nitrogen using a Retsch MM301 mixer mill homogenizer. Total chlorophyll was extracted with 80% (v/v) acetone and analyzed with a spectrophotometer (model DU800; Beckman Coulter). Chlorophyll a was measured at 663.6 nm, and chlorophyll b was measured at 646.6 nm. Total chlorophyll content was calculated by the following formula: chlorophyll (μg mL−1) = 17.34 × A663.6 + 17.76 × A646.6. The absorbance was normalized by measurement at A750.
Evans blue staining was performed by the method of Watanabe and Lam (2006). Light-grown, 4-week-old rosette leaves were incubated with 300 mm NaCl for 6 or 12 h. The salt-treated leaves were incubated with 0.1% Evans blue solution overnight. Leaf chlorophyll was eliminated with a destaining solution (95% ethanol:lactophenol, 2:1). The degree of cell death was determined by the strength of the dark blue color.
Histochemical GUS staining was performed as described previously by Cho et al. (2011). AtAIRP3-promoter:GUS T3 transgenic plants were incubated with a GUS staining solution {2 mm 5-bromo-4-chloro-3-indolyl β-D-glucoside [cyclohexylammonium salt; Duchefa Biochemie], 0.5 mm K3Fe(CN)6, and 0.5 mm K4Fe(CN)6 in 50 mm Tris buffer [pH 7.5]} for 1 h at 37°C. To remove the chlorophyll, tissues were treated with 70% ethanol for 5 h.
Yeast Two-Hybrid Screening
The Arabidopsis Lambda-ACT two-hybrid cDNA library was obtained from the Arabidopsis Biological Resource Center (stock no. CD4-22). AtAIRP3/LOG2 was cloned into the pGBKT7 vector. The cDNA clones in the pACT vector and the AtAIRP3/LOG2-pGBKT7 construct were cotransformed into yeast strain AH109 (Clontech) as recently described (Bae and Kim, 2013) and allowed to grow for 5 d at 30°C. Approximately 5.0 × 105 transformants were screened on the SD/−Trp/−Leu/−His growth medium supplemented with 10 mm 3-AT. To check the interaction between AtAIRP3/LOG2 and various forms of RD21, RD21-pGADT7 and AtAIRP3/LOG2-pGBKT7 constructs were cotransformed into yeast AH109 cells. Yeast cells were grown on SD/−Leu/−Trp/−His/−Ade growth medium supplemented with p-nitrophenyl-α-d-galactoside at 30°C for 3 d. In addition, p53 (murine p5372-390) + T-antigen (SV40 large T-antigen87–708) were used as a positive control. Lambda (human lamin C66-230) + T-antigen were used as a negative control.
In Vitro and In Vivo Coimmunoprecipitation Experiments
The in vitro coimmunoprecipitation experiment was conducted as described by Cho et al. (2008) with modifications. Bacterially expressed AtAIRP3/LOG2-2xflag and mRD21-6xmyc recombinant proteins were coincubated in 50 μL of anti-flag M2 affinity gel matrix (Sigma-Aldrich) for 2 h at 4°C in IP buffer (phosphate-buffered saline, 0.5 m EDTA, 0.2 m phenylmethylsulfonyl fluoride, protease inhibitor cocktail VI [AG Scientific], and 0.5% Triton X-100). The affinity gel was washed three times with IP buffer, and bound proteins were eluted with 0.1 m Gly buffer, pH 2.3. The eluted proteins were resolved by 8% SDS-PAGE, transferred to polyvinylidene difluoride membranes, and probed with anti-flag (1:10,000 dilution; Sigma-Aldrich) and anti-myc (1:4,000 dilution; Applied Biological Materials) antibodies.
The in vivo coimmunoprecipitation experiment was performed as described by Son et al. (2010) with modifications. The 35S:p19, 35S:AtAIRP3/LOG2-flag, and 35S:mRD21-myc constructs were transformed into A. tumefaciens (GV3101) and cultured overnight at 28°C in yeast extract peptone medium containing 50 μg mL−1 kanamycin (Sigma-Aldrich) and 50 μg mL−1 rifampicin (Sigma-Aldrich). The cultured bacterial cells were harvested and resuspended in tobacco infiltration medium (10 mm MES, pH 5.7, 10 mm MgCl2, and 0.5 mm acetosyringone [MB Cell]) at a final optical density of 1.0. Light-grown, 3-week-old tobacco (Nicotiana benthamiana) leaves were infiltrated with A. tumefaciens suspension mixtures (35S:p19 + 35S:mRD21-myc and 35S:p19 + 35S:AtAIRP3/LOG2-flag + 35S:mRD21-myc) and incubated for 4 d at 25°C. Infiltrated leaves were ground in a mixer mill, and total protein was extracted in extraction buffer (50 mm Tris [pH 7.5], 100 mm NaCl, 0.5% Triton X-100, 2 mm EDTA, 1 mm MgCl2, protease inhibitor cocktail VI [AG Scientific], 1 mm phenylmethylsulfonyl fluoride, 20 mm sodium fluoride, and 10% glycerol) in the presence of 10 μm MG132 (AG Scientific). Leaf extract (1 mg of total proteins) was incubated with anti-flag gel matrix for 2 h, and precipitated proteins were detected with immunoblot analysis using anti-flag (1:10,000 dilution; Sigma-Aldrich) and anti-myc (1:4,000 dilution; Applied Biological Materials) antibodies.
Cell-Free Degradation and In Vitro Ubiquitination Assays
Cell-free crude extracts were prepared from salt (300 mm NaCl)-treated 10-d-old wild-type and atairp3/log2-2 seedlings using extraction buffer (25 mm Tris-HCl [pH 7.5], 10 mm MgCl2, 5 mm dithiothreitol, 0.1% Triton X-100, 10 mm ATP, and 10 mm NaCl). The mRD21-6xmyc and RGA1-2xflag recombinant proteins were incubated with cell-free crude extracts (50 μg of total protein) for 1.5 and 3 h in the presence or absence of 50 μm MG132 (AG Scientific). SDS sample buffer was added to the reaction mixture. The protein degradation profiles were analyzed by immunoblotting using anti-flag and anti-myc antibodies. RGA1 was used as a positive control for proteasome-dependent degradation (Lee et al., 2010). Rubisco served as the loading control.
An in vitro self-ubiquitination assay was conducted as described by Cho et al. (2006). MBP-AtAIRP3/LOG2101-388 recombinant protein was incubated for 1 h in the presence or absence of E1 (Arabidopsis UBA1), E2 (Arabidopsis UBC2), ATP, and Ub. The reaction products were subjected to immunoblotting using anti-MBP and anti-Ub antibodies. For the in vitro mRD21 ubiquitination assay, AtAIRP3/LOG2-flag and mRD21-myc proteins were coincubated as described above. The reaction products were analyzed using an anti-myc antibody.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers: AtAIRP3/LOG2 (At3g09770), AtLUL1 (At5g03200), AtLUL2 (At3g53410), AtLUL3 (At5g19080), AtLUL4 (At5g06140), and RD21 (At1g47128).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Sequence analysis of AtAIRP3/LOG2.
Supplemental Figure S2. In vitro self-ubiquitination assay.
Supplemental Figure S3. Construction of 35S:AtAIRP3-RNAi transgenic plants.
Supplemental Table S1. PCR primer sequences used for this article.
Supplementary Material
Glossary
- ABA
abscisic acid
- ER
endoplasmic reticulum
- T-DNA
transfer DNA
- IP
immunoprecipitation
- RT
reverse transcription
- qRT
quantitative reverse transcription
- RNAi
RNA interference
- CaMV
cauliflower mosaic virus
- cDNA
complementary DNA
- SD/−Trp/−Leu/−His
three-minus selection medium
- 3-AT
3-amino-1,2,4,-triazole
- SD/−Leu/−Trp/−His/−Ade
four-minus selection medium
- PCD
programmed cell death
- MS
Murashige and Skoog
References
- Ahuja I, de Vos RCH, Bones AM, Hall RD. (2010) Plant molecular stress responses face climate change. Trends Plant Sci 15: 664–674 [DOI] [PubMed] [Google Scholar]
- Bae H, Kim SK, Cho SK, Kang BG, Kim WT. (2011) Overexpression of OsRDCP1, a rice RING domain-containing E3 ubiquitin ligase, increased tolerance to drought stress in rice (Oryza sativa L.). Plant Sci 180: 775–782 [DOI] [PubMed] [Google Scholar]
- Bae H, Kim WT. (2013) The N-terminal tetra-peptide (IPDE) short extension of the U-box motif in rice SPL11 E3 is essential for the interaction with E2 and ubiquitin-ligase activity. Biochem Biophys Res Commun 433: 266–271 [DOI] [PubMed] [Google Scholar]
- Boyer JS. (1982) Plant productivity and environment. Science 218: 443–448 [DOI] [PubMed] [Google Scholar]
- Bozkurt TO, Schornack S, Win J, Shindo T, Ilyas M, Oliva R, Cano LM, Jones AME, Huitema E, van der Hoorn RAL, et al. (2011) Phytophthora infestans effector AVRblb2 prevents secretion of a plant immune protease at the haustorial interface. Proc Natl Acad Sci USA 108: 20832–20837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu Q, Li H, Zhao Q, Jiang H, Zhai Q, Zhang J, Wu X, Sun J, Xie Q, Wang D, et al. (2009) The Arabidopsis RING finger E3 ligase RHA2a is a novel positive regulator of abscisic acid signaling during seed germination and early seedling development. Plant Physiol 150: 463–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng MC, Hsieh EJ, Chen JH, Chen HY, Lin TP. (2012) Arabidopsis RGLG2, functioning as a RING E3 ligase, interacts with AtERF53 and negatively regulates the plant drought stress response. Plant Physiol 158: 363–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SK, Chung HS, Ryu MY, Park MJ, Lee MM, Bahk Y-Y, Kim J, Pai HS, Kim WT. (2006) Heterologous expression and molecular and cellular characterization of CaPUB1 encoding a hot pepper U-box E3 ubiquitin ligase homolog. Plant Physiol 142: 1664–1682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SK, Ryu MY, Seo DH, Kang BG, Kim WT. (2011) The Arabidopsis RING E3 ubiquitin ligase AtAIRP2 plays combinatory roles with AtAIRP1 in abscisic acid-mediated drought stress responses. Plant Physiol 157: 2240–2257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SK, Ryu MY, Song C, Kwak JM, Kim WT. (2008) Arabidopsis PUB22 and PUB23 are homologous U-box E3 ubiquitin ligases that play combinatory roles in response to drought stress. Plant Cell 20: 1899–1914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U. (2003) A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cushman JC, Bohnert HJ. (2000) Genomic approaches to plant stress tolerance. Curr Opin Plant Biol 3: 117–124 [DOI] [PubMed] [Google Scholar]
- Deikman J, Petracek M, Heard JE. (2012) Drought tolerance through biotechnology: improving translation from the laboratory to farmers’ fields. Curr Opin Biotechnol 23: 243–250 [DOI] [PubMed] [Google Scholar]
- Dill A, Thomas SG, Hu J, Steber CM, Sun TP. (2004) The Arabidopsis F-box protein SLEEPY1 targets gibberellin signaling repressors for gibberellin-induced degradation. Plant Cell 16: 1392–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreher K, Callis J. (2007) Ubiquitin, hormones and biotic stress in plants. Ann Bot (Lond) 99: 787–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS. (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45: 616–629 [DOI] [PubMed] [Google Scholar]
- Gu C, Shabab M, Strasser R, Wolters PJ, Shindo T, Niemer M, Kaschani F, Mach L, van der Hoorn RAL. (2012) Post-translational regulation and trafficking of the granulin-containing protease RD21 of Arabidopsis thaliana. PLoS ONE 7: e32422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y, Innes RW. (2011) The KEEP ON GOING protein of Arabidopsis recruits the ENHANCED DISEASE RESISTANCE1 protein to trans-Golgi network/early endosome vesicles. Plant Physiol 155: 1827–1838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra DD, Callis J. (2012) Ubiquitin on the move: the ubiquitin modification system plays diverse roles in the regulation of endoplasmic reticulum- and plasma membrane-localized proteins. Plant Physiol 160: 56–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi Y, Yamada K, Shimada T, Matsushima R, Nishizawa NK, Nishimura M, Hara-Nishimura I. (2001) A proteinase-storing body that prepares for cell death or stresses in the epidermal cells of Arabidopsis. Plant Cell Physiol 42: 894–899 [DOI] [PubMed] [Google Scholar]
- Hirayama T, Shinozaki K. (2010) Research on plant abiotic stress responses in the post-genome era: past, present and future. Plant J 61: 1041–1052 [DOI] [PubMed] [Google Scholar]
- Kim EY, Seo YS, Lee H, Kim WT. (2010) Constitutive expression of CaSRP1, a hot pepper small rubber particle protein homolog, resulted in fast growth and improved drought tolerance in transgenic Arabidopsis plants. Planta 232: 71–83 [DOI] [PubMed] [Google Scholar]
- Kim SJ, Ryu MY, Kim WT. (2012) Suppression of Arabidopsis RING-DUF1117 E3 ubiquitin ligases, AtRDUF1 and AtRDUF2, reduces tolerance to ABA-mediated drought stress. Biochem Biophys Res Commun 420: 141–147 [DOI] [PubMed] [Google Scholar]
- Koizumi M, Yamaguchi-Shinozaki K, Tsuji H, Shinozaki K. (1993) Structure and expression of two genes that encode distinct drought-inducible cysteine proteinases in Arabidopsis thaliana. Gene 129: 175–182 [DOI] [PubMed] [Google Scholar]
- Kraft E, Stone SL, Ma L, Su N, Gao Y, Lau OS, Deng XW, Callis J. (2005) Genome analysis and functional characterization of the E2 and RING-type E3 ligase ubiquitination enzymes of Arabidopsis. Plant Physiol 139: 1597–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampl N, Alkan N, Davydov O, Fluhr R. (2013) Set-point control of RD21 protease activity by AtSerpin1 controls cell death in Arabidopsis. Plant J 74: 498–510 [DOI] [PubMed] [Google Scholar]
- Lee HK, Cho SK, Son O, Xu Z, Hwang IH, Kim WT. (2009) Drought stress-induced Rma1H1, a RING membrane-anchor E3 ubiquitin ligase homolog, regulates aquaporin levels via ubiquitination in transgenic Arabidopsis plants. Plant Cell 21: 622–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J-H, Yoon HJ, Terzaghi W, Martinez C, Dai M, Li J, Byun MO, Deng XW. (2010) DWA1 and DWA2, two Arabidopsis DWD protein components of CUL4-based E3 ligases, act together as negative regulators in ABA signal transduction. Plant Cell 22: 1716–1732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Jiang H, Bu Q, Zhao Q, Sun J, Xie Q, Li C. (2011) The Arabidopsis RING finger E3 ligase RHA2b acts additively with RHA2a in regulating abscisic acid signaling and drought response. Plant Physiol 156: 550–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Stone SL. (2010) Abscisic acid increases Arabidopsis ABI5 transcription factor levels by promoting KEG E3 ligase self-ubiquitination and proteasomal degradation. Plant Cell 22: 2630–2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyzenga WJ, Stone SL. (2012) Abiotic stress tolerance mediated by protein ubiquitination. J Exp Bot 63: 599–616 [DOI] [PubMed] [Google Scholar]
- Manzano C, Abraham Z, López-Torrejón G, Del Pozo JC. (2008) Identification of ubiquitinated proteins in Arabidopsis. Plant Mol Biol 68: 145–158 [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, et al. (2011) CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res 39: D225–D229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning Y, Jantasuriyarat C, Zhao Q, Zhang H, Chen S, Liu J, Liu L, Tang S, Park CH, Wang X, et al. (2011) The SINA E3 ligase OsDIS1 negatively regulates drought response in rice. Plant Physiol 157: 242–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ondzighi CA, Christopher DA, Cho EJ, Chang SC, Staehelin LA. (2008) Arabidopsis protein disulfide isomerase-5 inhibits cysteine proteases during trafficking to vacuoles before programmed cell death of the endothelium in developing seeds. Plant Cell 20: 2205–2220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page T, Griffiths G, Buchanan-Wollaston V. (2001) Molecular and biochemical characterization of postharvest senescence in broccoli. Plant Physiol 125: 718–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park GG, Park JJ, Yoon J, Yu SN, An G. (2010) A RING finger E3 ligase gene, Oryza sativa Delayed Seed Germination 1 (OsDSG1), controls seed germination and stress responses in rice. Plant Mol Biol 74: 467–478 [DOI] [PubMed] [Google Scholar]
- Park HJ, Park HC, Lee SY, Bohnert HJ, Yun D-J. (2011) Ubiquitin and ubiquitin-like modifiers in plants. J Plant Biol 54: 275–285 [Google Scholar]
- Pratelli R, Guerra DD, Yu S, Wogulis M, Kraft E, Frommer WB, Callis J, Pilot G. (2012) The ubiquitin E3 ligase LOSS OF GDU2 is required for GLUTAMINE DUMPER1-induced amino acid secretion in Arabidopsis. Plant Physiol 158: 1628–1642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin F, Sakuma Y, Tran LS, Maruyama K, Kidokoro S, Fujita Y, Fujita M, Umezawa T, Sawano Y, Miyazono K, et al. (2008) Arabidopsis DREB2A-interacting proteins function as RING E3 ligases and negatively regulate plant drought stress-responsive gene expression. Plant Cell 20: 1693–1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu MY, Cho SK, Kim WT. (2010) The Arabidopsis C3H2C3-type RING E3 ubiquitin ligase AtAIRP1 is a positive regulator of an abscisic acid-dependent response to drought stress. Plant Physiol 154: 1983–1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadanandom A, Bailey M, Ewan R, Lee J, Nelis S. (2012) The ubiquitin-proteasome system: central modifier of plant signalling. New Phytol 196: 13–28 [DOI] [PubMed] [Google Scholar]
- Seo DH, Ryu MY, Jammes F, Hwang JH, Turek M, Kang BG, Kwak JM, Kim WT. (2012a) Roles of four Arabidopsis U-box E3 ubiquitin ligases in negative regulation of abscisic acid-mediated drought stress responses. Plant Physiol 160: 556–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo K-I, Song E, Chung S, Lee J-H. (2012b) Roles of various Cullin-RING E3 ligases in hormonal and stress responses in plants. J Plant Biol 55: 421–428 [Google Scholar]
- Son OR, Cho SK, Kim SJ, Kim WT. (2010) In vitro and in vivo interaction of AtRma2 E3 ubiquitin ligase and auxin binding protein 1. Biochem Biophys Res Commun 393: 492–497 [DOI] [PubMed] [Google Scholar]
- Stone SL, Hauksdóttir H, Troy A, Herschleb J, Kraft E, Callis J. (2005) Functional analysis of the RING-type ubiquitin ligase family of Arabidopsis. Plant Physiol 137: 13–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone SL, Williams LA, Farmer LM, Vierstra RD, Callis J. (2006) KEEP ON GOING, a RING E3 ligase essential for Arabidopsis growth and development, is involved in abscisic acid signaling. Plant Cell 18: 3415–3428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verslues PE, Juenger TE. (2011) Drought, metabolites, and Arabidopsis natural variation: a promising combination for understanding adaptation to water-limited environments. Curr Opin Plant Biol 14: 240–245 [DOI] [PubMed] [Google Scholar]
- Vierstra RD. (2009) The ubiquitin-26S proteasome system at the nexus of plant biology. Nat Rev Mol Cell Biol 10: 385–397 [DOI] [PubMed] [Google Scholar]
- Watanabe N, Lam E. (2006) Arabidopsis Bax inhibitor-1 functions as an attenuator of biotic and abiotic types of cell death. Plant J 45: 884–894 [DOI] [PubMed] [Google Scholar]
- Welchman RL, Gordon C, Mayer RJ. (2005) Ubiquitin and ubiquitin-like proteins as multifunctional signals. Nat Rev Mol Cell Biol 6: 599–609 [DOI] [PubMed] [Google Scholar]
- Yamada K, Matsushima R, Nishimura M, Hara-Nishimura I. (2001) A slow maturation of a cysteine protease with a granulin domain in the vacuoles of senescing Arabidopsis leaves. Plant Physiol 127: 1626–1634 [PMC free article] [PubMed] [Google Scholar]
- Yee D, Goring DR. (2009) The diversity of plant U-box E3 ubiquitin ligases: from upstream activators to downstream target substrates. J Exp Bot 60: 1109–1121 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Yang C, Li Y, Zheng N, Chen H, Zhao Q, Gao T, Guo H, Xie Q. (2007) SDIR1 is a RING finger E3 ligase that positively regulates stress-responsive abscisic acid signaling in Arabidopsis. Plant Cell 19: 1912–1929 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.