Background: PIKKs regulate cellular processes such as growth, DNA repair, and stress responses.
Results: The ATM, ATR, DNA-PKcs, SMG-1, and TRRAP FATC domains interact with membrane mimetics, resulting in an increase of α-helical secondary structure.
Conclusion: All PIKK FATC domains may function as membrane anchors.
Significance: The regulation of PIKK localization by a network of interactions may allow a specific and localized cellular signaling output.
Keywords: Lipids, Membrane Bilayer, NMR, Phospholipid Vesicle, Protein Conformation, PIKK, Bicelle, Micelle, Protein-Lipid Interactions, Protein-Membrane Interactions
Abstract
The FATC domain is shared by all members of the family of phosphatidylinositol-3 kinase-related kinases (PIKKs). It has been shown that the FATC domain plays an important role for the regulation of each PIKK. However, other than an involvement in protein-protein interactions, a common principle for the action of the FATC domain has not been detected. A detailed characterization of the structure and lipid binding properties of the FATC domain of the Ser/Thr kinase target of rapamycin (TOR) revealed that it contains a redox-sensitive membrane anchor in its C terminus. Because the C-terminal regions of the FATC domains of all known PIKKs are rather hydrophobic and especially rich in aromatic residues, we examined whether the ability to interact with lipids and membranes might be a general property. Here, we present the characterization of the interactions with lipids and different membrane mimetics for the FATC domains of human DNA-PKcs, human ATM, human ATR, human SMG-1, and human TRRAP by NMR and CD spectroscopy. The data indicate that all of these can interact with different membrane mimetics and may have different preferences only for membrane properties such as surface charge, curvature, and lipid packing. The oxidized form of the TOR FATC domain is well structured overall and forms an α-helix that is followed by a disulfide-bonded loop. In contrast, the FATC domains of the other PIKKs are rather unstructured in the isolated form and only significantly populate α-helical secondary structure upon interaction with membrane mimetics.
Introduction
Members of the family of phosphatidylinositol 3-kinase-related kinases (PIKK)3 regulate various cellular signaling pathways in response to different stresses and nutrient availability (1–3). Ataxia-telangiectasia mutated (ATM), ataxia- and Rad3-related (ATR), and the DNA-dependent protein kinase catalytic subunit (DNA-PKcs) are important for DNA damage response and signaling in the presence of DNA damages (3). These PIKK family members phosphorylate proteins that regulate processes such as DNA repair, cell cycle progression, cellular senescence, and apoptosis (3–6). DNA repair by ATM and DNA-PKcs is activated by the presence of DNA double strand breaks, whereas ATR can respond to single-stranded DNA gaps or more generally to DNA replication blocks (2, 3). The importance of ATM in the oxidative stress response and its role in regulating mitochondrial functions and metabolism have been reviewed recently (4, 7). In addition to DNA repair, DNA-PKcs is involved in processes that concern the inflammatory response via NF-κB (nuclear factor κB light-chain enhancer) (8) and plays an important role in metabolic gene regulation and the regulation of the homeostasis of cell proliferation (6). Furthermore, DNA-PKcs has been suggested to play a role in the signaling response to ionizing radiation (IR), which involves phosphorylation of lipid raft proteins (9, 10). Very recently, it has further been shown that treatment of HT-29 cells with cis-9,trans-11-conjugated linoleic acid (c9,t11-CLA), substances that can alter the properties of membranes, delays double strand break repair following X-radiation, which corresponds with insufficient DNA-PKcs activation (11). Interestingly, ATM has also been associated with the response to IR and has been found to localize in the nucleus and at cytoplasmic vesicles (12). In addition, ATM has been detected at the plasma membrane (13).
Suppressor of morphogenesis in genitalia-1 (SMG-1) is involved in nonsense-mediated mRNA decay as well as in oxidative stress and cell survival (2, 3, 14–16). The target of rapamycin (TOR), the best studied family member, regulates cellular growth in response to nutrient and energy supply and to stress conditions such as hypoxia (17, 18). Processes controlled by TOR include translation, transcription, ribosome biogenesis, lipid metabolism, and autophagy (17, 18). The function of TOR as well as ATM has further been related to redox signaling (4, 7). Transformation/transcription domain-associated protein (TRRAP) scaffolds several histone acetyltransferase complexes, thereby regulating gene transcription of target genes such as, for example, mitotic checkpoint genes or liver receptors that play a role in lipid metabolism (2, 19, 20). In addition, TRRAP is also suggested to be involved in double strand break repair processes (21).
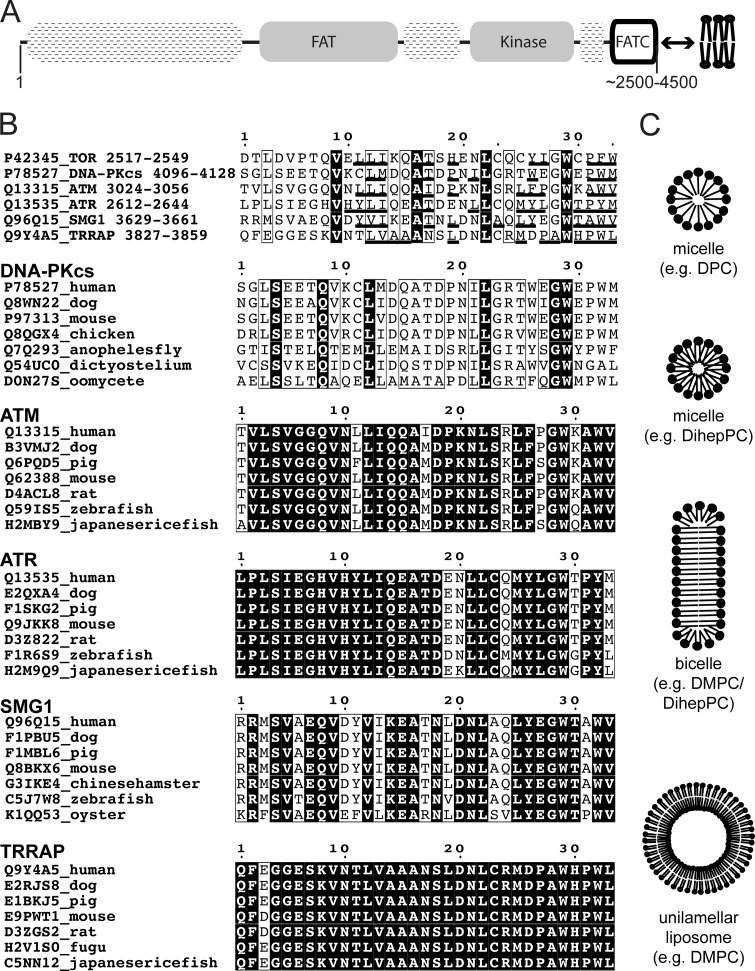
All PIKK family members have a similar domain organization (Fig. 1A). The total length of the amino acid sequences ranges from about 2500 to 4500 residues. The defining element is the kinase domain close to the C terminus that, despite a high homology to lipid kinases, phosphorylates serine and threonine residues in target proteins (2). Only TRRAP is not catalytically active (2, 3). N-terminally the kinase domain is flanked by the FRAP-ATM-TRRAP (FAT) domain (22). The FAT domain and the preceding N-terminal region with only low sequence homology between different PIKKs are suggested to be composed mostly of α-helical repeat motifs that typically form platforms for protein-protein interactions (22, 23). A more detailed analysis of the TOR sequence indicates that its N-terminal region contains mainly HEAT (huntingtin, elongation factor 3, regulatory subunit A of PP2A, yeast TOR1) repeats, whereas the FAT domain is composed of tetratricopeptide repeats (24). The linker region between the kinase and the FAT C-terminal (FATC) domain has been referred to as the PIKK regulatory domain (2, 3, 25). However, this region varies significantly in length and sequence composition among different PIKKs (2, 25). The well conserved region of the FATC domain corresponds to the C-terminal ∼35 amino acids (PFAM domain database entry PF02660) (1, 22). This domain has been shown to serve an important function in the regulation of the kinase domain (2, 16, 25–28). The FATC domain is highly evolutionarily conserved among each family member, as illustrated for DNA-PKcs, ATM, ATR, SMG-1, and TRRAP in Fig. 1B and supplemental Fig. 1. The FATC domains of ATM, DNA-PKcs, and ATR are proposed to mediate protein-protein interactions (2, 28). The FATC domain of TOR contains two conserved cysteines that form a disulfide bond (29). The structure of the free oxidized FATC domain (Protein Data Bank ID 1w1n) consists of an α-helix and a C-terminal hydrophobic disulfide-bonded loop (29). Based on NMR-monitored binding studies with different lipids and membrane mimetics, the FATC domain interacts with lipids above the critical micelle concentration (CMC) as well as with bicelles (30) and small unilamellar vesicles (SUVs).4 The structures of the oxidized and reduced dodecylphosphocholine (DPC) micelle-associated forms (Protein Data Bank ID 2kio and 2kit) are rather similar to that of the free protein (30). However, the α-helix extends further toward the C terminus, which is more pronounced for the reduced form (30). Although not restricted by a disulfide bond, the C terminus of the reduced micelle-associated state also folds back toward the α-helix. Overall the data suggest that the FATC domain of TOR may function as a redox-sensitive membrane anchor that could participate in regulating the localization of the known TOR complexes to different cellular membranes (30). Mammalian TOR has been localized at the plasma membrane and the membranes of the endoplasmic reticulum, Golgi, mitochondria, and lysosomes as well as in the nucleus and associated with ribosomes (31–35). An analysis of the sequences of the FATC domains of the best known PIKKs shows that they are all rather hydrophobic and, especially in the C-terminal region, rich in aromatic residues (Fig. 1B). Thus, not only the FATC domain of TOR but also those of the other PIKKs may interact with lipids and membranes. Localization in the nucleus as well as at cellular membranes has already been suggested for ATM and DNA-PKcs (9, 36). Here, we present a characterization of the interactions of the FATC domains of human DNA-PKcs, ATM, ATR, SMG-1, and TRRAP with lipids and different membrane mimetics by NMR and CD spectroscopy. Overall the data show that the FATC domain of all tested PIKKs could interact with membrane mimetics, albeit with different preferences for specific membrane features such as surface charge, curvature, and packing of the fatty acid acyl chains.
FIGURE 1.
Domain organization of PIKKs, sequence conservation of their FATC domain, and membrane mimetics used. A, the general domain organization of PIKKs. All share the kinase, FAT, and FATC domain. Furthermore, all contain additional functional regions of variable sizes, indicated by the round-cornered patterned rectangles. B, sequence alignment of the highly conserved part of the FATC domain (PFAM entry PF02260). The sequences of the FATC domains of different human PIKKs are aligned in the topmost panel. Below that, the sequence conservation of the FATC domains of DNA-PKcs, ATM, ATR, SMG-1, and TRRAP is illustrated by alignments of the respective sequences from different organisms. For ATM, ATR, SMG-1, and TRRAP the alignments shown include only the sequences from higher eukaryotes. Supplemental Fig. S1 shows the same type of alignments for some PIKKs including additional sequences for lower eukaryotes and plants. The sequence alignments were generated using the program ESPript (82). C, schematic representations of the membrane mimetics used. Chemical structures for some of the lipids used are given in Fig. 2C.
EXPERIMENTAL PROCEDURES
Plasmid Cloning and Protein Expression and Purification
The constructs of all tested FATC domains included the well conserved C-terminal 33 residues (Fig. 1, A and B).
Residues 4096–4128 of human DNA-PKcs (UniProt ID P78527) corresponding to its FATC domain (hDNAPKfatc) were cloned into GEV2 (37) using the XhoI and BamHI sites. An additional factor Xa cleavage site was introduced by the PCR primers used. In addition, a variant with an enterokinase (hDNAPKfatc-gb1ent) instead of a factor Xa (hDNAPKfatc-gb1xa) cleavage site was prepared by site-directed mutagenesis. The success of the mutagenesis was verified by DNA sequencing. Either variant was overexpressed in Escherichia coli BL21(DE3) in LB, 15N, or 15N-13C M9 minimal medium at 37 °C. When the A600 of the culture was ∼0.7–0.9, protein expression was induced with 1 mm IPTG for 3 h. Both the fusion protein, consisting of the B1 domain of protein G (GB1, 56 residues) and a thrombin and a factor Xa site, and hDNAPKfatc (= hDNAPKfatc-gb1xa, 99 residues) or the one with enterokinase instead of the factor Xa site (= hDNAPKfatc-gb1ent, 100 residues) were obtained mainly from the soluble fraction after sonication of the cells in 50 mm Tris, 2 mm benzamidine, 2 mm EDTA, pH 7.5. The GB1 fusion proteins were purified using an IgG-Sepharose column (GE Healthcare) as described in the manufacturer's manual. The purified protein was lyophilized, resuspended in 50 mm Tris, 100 mm NaCl, pH 6.5, and washed several times with the same buffer using centrifugal filter devices (Amicon Ultra, Merck Millipore, MWCO 3000). To prevent intermolecular disulfide bond formation by the single cysteine, 10 mm TCEP was added to the final protein stock solution.
To obtain some pure 15N hDNAPKfatc, the cell pellet was resuspended in PBS and the cell suspension sonicated. The resulting cell lysate was centrifuged for 30 min at 35,000 rpm. The supernatant was transferred to a new tube and incubated for 5 min in an 80 °C water bath (38) followed by incubation for 10 min on ice and centrifugation for 30 min at 17,500 rpm. The supernatant was purified using IgG-Sepharose as described in the manufacturer's manual (GE Healthcare). Fractions containing hDNAPKfatc-gb1ent were pooled and lyophilized, resuspended in 50 mm Tris, 100 mm NaCl, 8 m urea, pH 8.0, dialyzed to 50 mm Tris, 100 mm NaCl, pH 8.0, and digested with enterokinase. The GB1 tag and undigested fusion protein were removed by reverse phase HPLC in an acetonitrile/trifluoroacetic acid buffer system. Before the reverse phase HPLC run, the sample was incubated at room temperature with 10 mm TCEP to remove cellular glutathione from the single cysteine. Fractions containing pure hDNAPKfatc were lyophilized, resuspended in 20 mm NaPi, 50 mm NaCl, 10 mm TCEP, pH 6.5 (initial structural characterization), or 50 mm Tris, 100 mm NaCl, pH 6.5 (NMR titrations), and concentrated. Mass spectrometry confirmed the correct molecular weight. Because the protease digest with factor Xa was not successful and the one with enterokinase not very efficient, most NMR studies were done using the GB1 fusion protein directly, as suggested recently (39).
Residues 3024–3056 of human ATM FATC (UniProt ID 13315) were cloned into GEV2 (37) using the BamHI and XhoI sites and overexpressed in E. coli BL21(DE3) (Novagen) in LB or M9 minimal medium. An additional factor Xa or enterokinase protease site was introduced by the PCR primers used or by site-directed mutagenesis. Expression was done at 37 °C. Cells were grown to an A600 of ∼0.9 and ∼0.7, respectively, and induced with 1 mm IPTG for 3 h. Cells were harvested by centrifugation at 4 °C. The cell pellet was resuspended in 25 ml of 50 mm Tris, 2 mm EDTA, 2 mm benzamidine, pH 7.5, thoroughly vortexed, and stored at −20 °C. The fusion protein consisting of the B1 domain of protein G (GB1), a thrombin, and an enterokinase or factor Xa site as well as the human ATM FATC domain (= hATMfc-gb1ent, 100 residues in total, and hATMfc-gb1xa, 99 residues in total) was mainly expressed as soluble protein. The fusion protein was extracted by incubating the cell suspension for 5 min at 80 °C. This heating step also resulted in an initial purification, as many E. coli proteins precipitate under these conditions (38). Afterward the cell suspension was cooled on ice for 10 min. Following centrifugation at 4 °C for 30 min at 20,000 × g, the supernatant containing the fusion protein was further purified by IgG affinity chromatography as described in the manufacturer's manual for IgG-Sepharose (GE Healthcare). The purified fusion protein was lyophilized, resuspended in ∼10 ml of 50 mm Tris, 100 mm NaCl, pH 6.5, washed two times, and concentrated using a centrifugal filter device (Amicon Ultra, Millipore Merck, MWCO 3000). The digestion of hATMfatc-gb1xa, with either factor Xa or thrombin, or of hATMfatc-gb1ent, with enterokinase or thrombin, was inefficient. Therefore, we followed a recently published procedure and used the purified GB1 fusion protein directly for the interaction studies with lipids and membrane mimetics (39). Uniformly 15N-labeled protein was prepared in M9 minimal medium containing 15NH4Cl as the sole nitrogen source. The purity and identity of the purified protein was analyzed by SDS-PAGE and mass spectrometry. Protein concentrations were determined by UV absorption measurements.
Chemically synthesized peptides (HPLC purified, purity > 90%) corresponding to the FATC domains of human SMG-1 (residues 3629–3661 = hSMG1fatc, 33 residues, molecular mass 3870.3 Da, ϵ280 = 13,980 m−1cm−1; UniProt ID Q96Q15), human ATR (residues 2612–2644 = hATRfatc, 33 residues, molecular mass 3879.4 Da, ϵ280 = 9970 m−1cm−1; UniProt ID Q13535), and human TRRAP (residues 3827–3859 = hTRRAPfatc, 33 residues, molecular mass 3671.1, ϵ280 = 11000 m−1cm−1; UniProt ID Q9Y4A5) were obtained from Thermo Scientific. 7.3 mg of hSGM1fatc peptide were initially dissolved in ∼9 ml of 50 mm Tris, 100 mm NaCl, pH 6.5. This peptide solution was concentrated using a centrifugal filter device (Millipore, Amicon Ultra, MWCO 3000). Because the solution got slightly turbid when the volume was reduced to ∼2.5 ml, the salt concentration was increased to 300 mm and the pH to 8.0 to improve the solubility of the peptide. This peptide solution was washed twice with 50 mm Tris, 300 mm NaCl, pH 8.0, and finally concentrated to a 1.44 mm stock solution. 5 mg of hATRfatc peptide were initially dissolved in ∼2.6 ml of 50 mm Tris, 100 mm NaCl, 10 mm TCEP, pH 6.5. The high turbidity of the solution indicated that this peptide was not very soluble in aqueous buffer. Increasing the pH to 8.0 and heating for 10 min at 42 °C as well as lowering the NaCl concentration to 50 mm by dilution improved the solubility. The resulting peptide solution was concentrated with a centrifugal filter device (Millipore, Amicon Ultra, MWCO 3000) to obtain a 1.42 mm stock solution. However, because of aggregation problems, it had to be diluted again for the NMR measurements. The final pH for the NMR and CD measurement was 7.7. Five mg of hTRRAPfatc peptide were dissolved in 50 mm Tris, 100 mm NaCl, pH 6.5, to obtain a 1.0 mm stock solution.
Preparation of Membrane Mimetics
DPC, 1,2-dioleoyl-sn-glycero-3-phosphate (DOPA), and 1,2-dioctanoyl-sn-glycero-3-phosphate (DioctPA) were purchased from Avanti Polar Lipids. 1,2-Diheptanoyl-sn-glycero-3-phosphocholine (DihepPC) was bought from Avanti Polar Lipids and Affymetrix. 1,2-Dimyristoyl-sn-glycero-3-phosphocholine (DMPC) was obtained from Genzyme Pharmaceuticals and Affymetrix and deuterated DPC (d38-DPC) from Cambridge Isotopes.
Generally, lipid stock solutions for the titrations were prepared as follows. A defined amount of lipid from a concentrated stock in chloroform was placed in a glass vial and dried under a stream of nitrogen gas. The dried lipid was then dissolved in buffer or a protein sample. Only DihepPC was weighted and then dissolved directly in buffer or in the peptide solution (only hTRRAPfatc). Micelles formed above the CMC, which is 1.1 mm for DPC (40) and 1.4–1.8 mm for DihepPC (41, 42).
For the preparation of bicelles, the appropriate amount of a DMPC stock solution in chloroform was placed in a glass vial and dried under a stream of nitrogen gas. Bicelles were formed by the stepwise addition of the appropriate amount of a DihepPC stock solution in buffer and vigorous vortexing after each step. Finally, the protein solution was added, and the pH was adjusted to 6.5.
For the preparation of liposomes, an appropriate amount of DMPC in chloroform to obtain a 50 or 100 mm solution, respectively, was placed in a glass vial and dried under a stream of nitrogen and resuspended in buffer. The lipid was dissolved by seven cycles of freezing in liquid nitrogen, incubation in a water bath at 40 °C, and vigorous vortexing. The formation of SUVs was induced by incubation in an ultrasonic bath for about one-half hour. Centrifugation for 5 min at maximum speed in a tabletop centrifuge allowed the separation of the remaining large uni- and multilamellar vesicles, which formed a fluffy white precipitate. Only the clear supernatant containing the SUVs was used for the NMR and CD sample preparation. Using the 50 mm stock solution, the sample contained <30 mm DMPC and using the 100 mm stock solution, <60 mm. Note that the exact amount of DMPC left after centrifugation of the ultrasonicated liposome mixture is not known.
NMR Sample Preparation
For the NMR resonance assignment and structural characterization in the absence and presence of micelles, samples containing ∼0.4 mm 15N or 15N-13C hDNAPKfatc-gb1ent in 50 mm Tris, 100 mm NaCl, 10 mm TCEP, 0.02% NaN3 (95% H2O/5% D2O), pH 6.5, with or without 150 mm d38-DPC, were used. The sample that was used for the resonance assignment and initial structural characterization of pure hDNAPKfatc contained ∼0.4 mm 15N-labeled protein in 20 mm NaPi, 50 mm NaCl, 10 mm TCEP, 0.02% NaN3 (95% H2O/5% D2O), pH 6.5. For the titrations and interaction studies of pure hDNAPKfatc or hDNAPKfatc-gb1 (ent or xa) with different lipids or lipid mixtures, the protein concentration was in the range of ∼65 to 100 μm in 50 mm Tris, 100 mm NaCl, pH 6.5 (95% H2O/5% D2O). The concentrations of the DPC stocks in buffer used for the titrations were 50 mm, 250 mm, and 1 m. For titration with the negatively charged lipid phosphatidic acid, a 4:1 mixture of DioctPA and DOPA was used (in total 50 mm lipid, 40 mm DioctPA, and 10 mm DOPA). The bicelles for the sample of 15N hDNAPKfatc-gb1xa were composed of DMPC and DihepPC (q = 0.3, [DMPC] = 0.0625 m, and [DihepPC] = 0.21 m, cL ∼14.4% w/w).
All hATMfatc-gb1ent NMR samples contained 50 mm Tris, 100 mm NaCl, 0.02% NaN3 (95% H2O/5% D2O), pH 6.5. The samples in the absence or presence of 150 mm d38-DPC had a concentration of ∼0.46 mm 15N-labeled protein, and those in the absence or presence of <30 mm DMPC liposomes had a concentration of ∼0.12 mm. The sample used for the titration with DPC and those in the presence and absence of DMPC/DihepPC bicelles (q = 0.2, [DMPC] = 0.04 m, and [DihepPC] = 0.20 m, cL = 12.3% w/w) had a concentration of ∼0.2 mm 15N hATMfatc-gb1ent. The concentrations of the DPC stocks used for the NMR-monitored titration of hATMfatc-gb1ent were 2.5 and 500 mm. For the final titration step (10 to 50 mm DPC), an appropriate amount of DPC in chloroform was dried under a stream of nitrogen gas and dissolved by adding the NMR sample from the second to last titration step.
All hSMG1fatc peptide NMR samples contained 50 mm Tris, 300 mm NaCl, 0.02% NaN3 (95% H2O/5% D2O), pH 8. The peptide concentration in the samples of the free peptide or in the presence of 50 mm d38-DPC or 50 mm DihepPC was ∼1 mm.
All hATRfatc peptide NMR samples contained 50 mm Tris, 50 mm NaCl, 0.02% NaN3 (95% H2O/5% D2O), pH 7.7. The peptide concentration in the samples of the free peptide or in the presence of 50 mm d38-DPC was ∼0.23 mm.
All hTRRAPfatc peptide NMR samples contained 50 mm Tris, 100 mm NaCl, 0.02% NaN3 (95% H2O/5% D2O), pH 6.5. The peptide concentration in the samples of the free peptide or in the presence of 50 mm d38-DPC or 48 mm DihepPC was ∼0.92 mm.
NMR Spectroscopy
NMR spectra were acquired at 298 K on Bruker Avance 750 and 500 spectrometers, the latter equipped with a cryogenic probe. NMR data for the pure hDNAPKfatc domain were recorded on a Bruker DRX600. Data were processed with NMRPipe (43) and analyzed using NMRView (44).
Assignments for the 13C, 15N, and 1H nuclei of hDNAPKfatc-gb1ent were based on three-dimensional HNCA, CCONH-TOCSY, and HCCH-TOCSY, and 15N- and 13C-edited NOESY spectra. Pure free hDNAPKfatc was assigned based on three-dimensional 15N-edited NOESY and TOCSY spectra, as well as three-dimensional HNHA (45) and HNHB data sets. Information about backbone dynamics was derived from the measurement of 15N relaxation experiments, including T1, T2, and {1H}-15N NOE at 500 MHz.
Assignments for the 15N, and 1H nuclei of micelle-immersed hATMfatc-gb1ent were based on three-dimensional 15N-edited NOESY and TOCSY as well as HNHA (45) data sets. Information about backbone dynamics was derived from {1H}-15N NOE data at 500 MHz. For the calculation of 1H, 15N, and 13C secondary shifts, random coil values from the literature were used (46).
The interaction with membrane mimetics was monitored based on 1H-15N HSQC and one-dimensional 1H NMR spectra, and for hATMfatc-gb1ent it was also based on {1H}-15N NOE data. For hSMG1fatc and hTRRAPfatc the 1H-15N HSQC had to be recorded at natural abundance. For hSMG1fatc in the absence and presence of d38-DPC, this was done using the SOFAST-HMQC pulse program (47). The average chemical shift change for the backbone amide nitrogen and proton Δδ(N,H)av for hDNAPKfatc due to the presence of DPC micelles or DihepPC/DMPC bicelles was calculated as [(ΔδHN)2 + (ΔδN/5)2]1/2. For hATRfatc, natural abundance 1H-15N HSQC spectra could not be recorded because the peptide concentration could not be increased sufficiently without suffering aggregation problems. NMR diffusion measurements for hSMG1fatc and hTRRAPfatc were measured using the DOSY tool in Bruker Topspin 3.1 as described in the manufacturer's manual.
CD Spectroscopy
All CD spectra were recorded at 298 K on a Jasco J715 or J720 spectropolarimeter using a cuvette with a path length of 0.1 cm. Spectra were generally recorded with an acquisition time of 50 nm/min (8 s response time) and five scans. The sample for the CD measurement of pure hDNAPKfatc was 25 μm in 20 mm NaPi, 50 mm NaCl, 10 mm TCEP, pH 6.5. The peptide concentration of the hSMG1fatc samples was 56.8 μm, that of the hATRfatc samples 16.4 μm, and that of the hTRRAPfatc samples 25.2 μm. The buffer for the peptide samples was the same as for the NMR measurements.
RESULTS
The Amino Acid Sequences of All PIKK FATC Domains Share Characteristic Features
An alignment of the FATC domains of different human PIKKs is given at the top of Fig. 1B. Shown are the well conserved C-terminal 33 residues based on the PFAM domain database entry PF02260. Similar to the FATC domain of TOR, which has been shown to interact with membrane mimetics (30), the C-terminal halves of the FATC domains of DNA-PKcs, ATM, ATR, SMG-1, and TRRAP are also rich in hydrophobic aromatic and aliphatic residues. For many of these hydrophobic residues, a positive free energy contribution for the transfer of a model peptide from a lipid bilayer to water has been shown (48). All share a conserved tryptophan at position 29 that is preceded by a small residue (mostly Gly, only in TRRAP Ala). In addition, all contain at least another tryptophan and/or one or more tyrosine residues. Tryptophan-rich peptides have been shown to have a high affinity for membrane mimetics and, based on an analysis of the membrane regions of several proteins, are often found at the interface between the apolar interior and the polar aqueous environment (49). Tyrosine, with its side chain hydroxyl group, may favor a similar location if present in a membrane binding region. The bottom four panels of Fig. 1B show the amino acid sequence alignments of the FATC domains of DNA-PKcs from different organisms and for ATM, ATR, SMG-1, and TRRAP from different higher eukaryotes. As can be seen, the FATC domain of each PIKK has been highly evolutionary conserved, especially among higher eukaryotes. A bit more variation can be seen if lower eukaryotes and/or plants are included (Fig. 1B, DNA-PKcs, and supplemental Fig. S1, ATM, ATR, SMG-1, and TRRAP). However, there is again a very high degree of sequence conservation if all plant, insect, or other subgroups of species are considered, suggesting that the exact composition may be important for the regulation of the function and possibly for a specific localization pattern.
The FATC Domain of DNA-PKcs Interacts with Membrane Mimetics
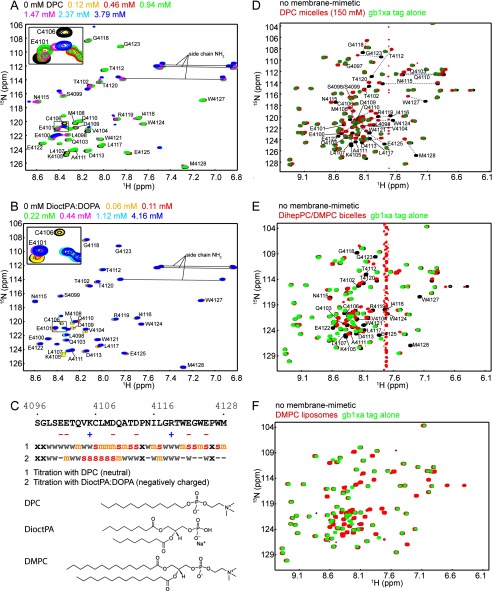
The six C-terminal residues of human DNA-PKcs (human: GWEPWM (Fig. 1B)) exhibit a similarity to the predicted lipid binding motif of the FATC domain of TOR (human: GWCPFW) (30). To estimate the ability of the FATC domain of DNA-PKcs to interact with lipids or membrane-mimetic particles, 1H-15N HSQC spectra of the pure FATC in the presence of increasing concentrations of DPC were acquired (Fig. 2A). Up to a concentration of 0.94 mm DPC, no significant spectral changes could be observed. However, at the next titration point, corresponding to 1.47 mm DPC, which is just above the CMC of DPC (1.1 mm), several resonances disappeared or shifted significantly. At 3.79 mm only a few resonances in the N-terminal region and around residues 4115–4120 were still well visible. Fig. 2C illustrates the spectral changes as a function of the hDNAPKfatc sequence. As expected based on previous lipid binding studies with the FATC domain of TOR (30), several resonances in the tryptophan-rich C terminus (Trp-4121–Met-4128) showed strong chemical shifts or disappeared in the presence of 3.79 mm DPC. Consistent with this, the side chain amide protons of the three tryptophans also showed strong chemical shift changes (see the full spectrum in supplemental Fig. S3A). In addition several residues in the central region from Cys-4106 to Asp-4113, which contains several charged as well as hydrophobic but no aromatic residues, also showed medium to strong spectral changes. The side chain amides in this region (Gln-4103, Gln-4111, and Asn-4115) showed weak chemical shift changes.
FIGURE 2.
Interaction of the human DNA-PKcs FATC domain with different membrane mimetics. A and B, 1H-15N HSQC spectra of hDNAPKfatc in the presence of increasing amounts of DPC or a 4:1 mixture of DioctPA/DOPA, respectively. The lipid concentrations with their respective color coding are indicated above each plot. The inserts in the upper left corners shows an enlarged view of the region highlighted by a black square. C, summary of the chemical shift differences observed in the NMR titrations shown in A and B. In all cases spectral changes were observed around the estimated CMC. Residues that disappeared just above the CMC are marked with a red letter s. Residues that disappeared or shifted significantly above the CMC are colored with an orange m, and those that disappeared or shifted at higher lipid concentrations are labeled with a gray w. Residues that were not significantly affected by the addition of lipid are marked with a minus sign, and an x represents an amino acid that shows no 1H-15N HSQC peak. In addition the chemical structures of DPC, DioctPA, and DMPC are shown. D–F, superposition of the 1H-15N HSQC spectra of hDNAPKfatc-gb1ent in the absence (black) or presence of a high concentration of DPC micelles, DihepPC/DMPC bicelles (∼270 mm total lipid concentration), or DMPC liposomes (<30 mm DMPC), respectively (red). To better discriminate the peaks corresponding to the FATC part, the spectrum of the GB1 tag (including a thrombin and a factor Xa site = GB1-xa) is shown in green at the top. Because the GB1 tag does not interact with membrane mimetics, its signals do not shift (39). The assignment is indicated by the single-letter amino acid code and the sequence position. See also supplemental Figs. S2–S6 for more information about the chemical shift assignments, a quantification of the observed shifts in E and F, and additional NMR-monitored lipid binding data.
As cellular membranes contain several lipids with negatively charged head groups, we further analyzed the interaction of hDNAPKfatc with a 4:1 mixture of the short-chain lipid DioctPA (C8) and the long-chain lipid DOPA (C18) (Fig. 2, B and C). Significant spectral changes were already visible at the second titration point (0.11 mm total lipid). No values could be found in the literature for the CMCs of DioctPA and DOPA. The CMC for DioctPA was expected to be on the same order as that for dioctanoylphosphocholine (CMC 0.27 mm) and for dioctanoylphosphoglycerol (CMC 1.21 mm) as well as for DOPA in the range of the CMCs for dipalmitoylphosphocholine (CMC 0.46 nm) and dimyristoylphosphoglycerol (CMC 0.011 mm). All CMC values were found at the Web site of Avanti Polar Lipids. A mixture of DioctPA and DOPA is expected to form micelles at lower lipid concentrations than DioctPA alone, because the CMC of DOPA is much smaller compared with that for DioctPA due to the longer fatty acid chain (C18 versus C8). Thus, the CMC of the mixture is expected to be below ∼0.1 mm, and hDNAPKfatc may as observed for DPC interact only with membrane-mimetic particles but not with single lipids. In line with the presence of several negatively charged amino acids in the sequence (Fig. 2C), the observed spectral changes were smaller overall than with the neutral DPC. This can be explained by electrostatic repulsion between the negative charges in the phosphatidic acid head group and the side chains of the present aspartates and glutamates (Fig. 2C). In contrast to DPC, the strongest chemical shift changes with DioctPA/DOPA occurred around the positively charged residue Lys-4105. The backbone amide resonances of Val-4104 to Gln-4110 disappeared or showed significant chemical shift changes (Fig. 2, B and C), whereas the C-terminal tryptophan-rich region, which contains two negatively charged glutamates, showed only very weak to no changes for the backbone resonances as well as for the side chain amides of the three tryptophans (Fig. 2, B and C, and supplemental Fig. 3B).
Because the yield of hDNAPKfatc following the protease digest of the GB1 fusion protein was very low, we used the GB1 fusion protein directly with an additional enterokinase recognition site (hDNAPKfatc-gb1ent) for further NMR-monitored lipid binding experiments and for the structural characterization of the micelle-immersed state. We recently showed that the GB1 tag itself does not interact with membrane-mimetic micelles, bicelles, or liposomes (39), and thus the observed spectral changes should only confer to the attached FATC domain of DNA-PKcs. To ensure that the presence of the GB1 tag did not influence the affinity of the interaction with DPC micelles, we titrated 15N hDNAPKfatc-gb1ent with DPC (supplemental Fig. S4A) similar to what was done for the pure FATC domain (Fig. 2A). To shift the equilibrium more to the micelle-immersed state, the final DPC concentration was significantly higher (∼60 mm). As for the pure FATC domain (Fig. 2A), several peaks of hDNAPKfatc-gb1ent start to shift at ∼1.5 mm (supplemental Fig. 4A), which is just above the CMC of DPC (1.1 mm). Until ∼10 mm there were still significant shifts. Between ∼10 and 60 mm only small further shifts occurred. Because the enterokinase cleavage site contains four negatively charged aspartates and only one positively charged lysine (DDDDK), we further did a titration of the fusion protein variant containing an overall neutral factor Xa site (IEGR). The titration of 15N hDNAPKfatc-gb1xa with DPC is shown in supplemental Fig. S4B. Overall the same resonances show about the same strength and direction of chemical shift changes as observed for 15N hDNAPKfatc-gb1ent (supplemental Fig. S4A) and for untagged hDNAPKfatc (Fig. 2A). This indicates that neither the GB1 tag nor the enterokinase or factor Xa site had a significant influence on the affinity for DPC micelles.
As below described for the structural characterization of the micelle-immersed state that was done using a higher protein concentration (∼0.4 mm), we increased the DPC concentration to 150 mm to ensure that the equilibrium was shifted to the micelle-associated state. A superposition of the respective 1H-15N HSQC spectrum with that of the free protein at about the same concentration is given in Fig. 2D. The change of the chemical shifts between the free and the micelle-immersed state as a function of the sequence is displayed in supplemental Fig. S6; to better discriminate the FATC peaks from that of the GB1 tag, a spectrum of the latter is additionally shown at the top. The assignments for the FATC part in the presence and absence of DPC micelles are indicated. Assignments for the full fusion protein including the GB1 tag are given in supplemental Fig. S2. Compared with the spectrum at ∼60 mm DPC, the spectral appearance of the micelle-immersed form is about the same.
hDNAPKfatc Also Interacts with Bicelles but Shows No Significant Affinity for Liposomes
DPC forms rather small spherical micelles (∼54 × 351.5 Da = ∼19 kDa; Fig. 1C, top schematic representation) with a high curvature (50, 51). Moreover, DPC has only one fatty acid tail (Fig. 2C) and thus resembles more a lysolipid. This should influence the packing mode of the fatty acid chains in the micelle. To evaluate the influence of the curvature and the packing density of the membrane mimetic on the interaction with hDNAPKfatc, additional NMR-monitored interaction studies with neutral bicelles and liposomes were performed (Fig. 2, E and F, respectively). In bicelles, the rather planar bilayer is formed by a long chain lipid, whereas the rim is formed by a short chain lipid or a bile acid (52). Moreover, bicelles are significantly larger (>250 kDa) (52) than for example DPC micelles (∼19 kDa) (51). In our studies we used bicelles composed of DMPC (C14) (Figs. 1C and 2C) and DihepPC (C7), which both share the neutral phosphocholine head group with DPC (Fig. 2C). Fig. 2E shows a superposition of the 1H-15N HSQC spectra of hDNAPKfatc-gb1ent in the absence and presence of DMPC/DihepPC bicelles. Based on a plot of the chemical shift changes as a function of the sequence, overall the same residues shift as in the presence of DPC micelles (supplemental Fig. S6). However, the magnitude of the changes with DMPC/DihepPC bicelles is lower than with DPC micelles, either because of a lower affinity for bicelles and/or because the number of membrane mimetic particles at the lipid concentrations used is smaller in the bicelle compared with the micelle sample. As observed with DPC micelles (Fig. 2D), the GB1 tag showed also no significant changes in the presence of bicelles (Fig. 2E). Liposomes are large to very large spherical particles that are composed of one or more bilayers. To analyze the interaction of hDNAPKfatc with liposomes, we used SUVs composed of DMPC. In contrast to DPC micelles or DMPC/DihepPC bicelles, the presence of liposomes induced no significant spectral changes (Fig. 2F). Using SUVs that were prepared starting from a more highly concentrated DMPC suspension (final concentration in sample <60 instead of <30 mm) did not change the result (supplemental Fig. S5). It should be noted that the final lipid concentration in the liposome samples was remarkably lower (<30 or 60 mm, respectively) than in the bicelle sample (∼270 mm). However, the fact that hDNAPKfatc shows significant chemical shift changes in the presence of small concentrations of DPC or even negatively charged lipids as well as DMPC/DihepPC bicelles but not with DMPC SUVs may rather be explained based on the different curvature and fatty acid chain packing properties (see schematic representations in Fig. 1C). Although bicelles have a rather planar bilayer area, they also have a rather curved rim region, which may show a curvature that is comparable to that of micelles.
Further NMR Interaction Studies Suggest That All PIKK FATC Domains Can Interact with Membrane Mimetics
To find out whether membrane binding is a general property of the FATC domain of all PIKKs, the interaction of the FATC domains of ATM, ATR, SMG-1, and TRRAP with different membrane mimetics was also probed by specific NMR interaction studies.
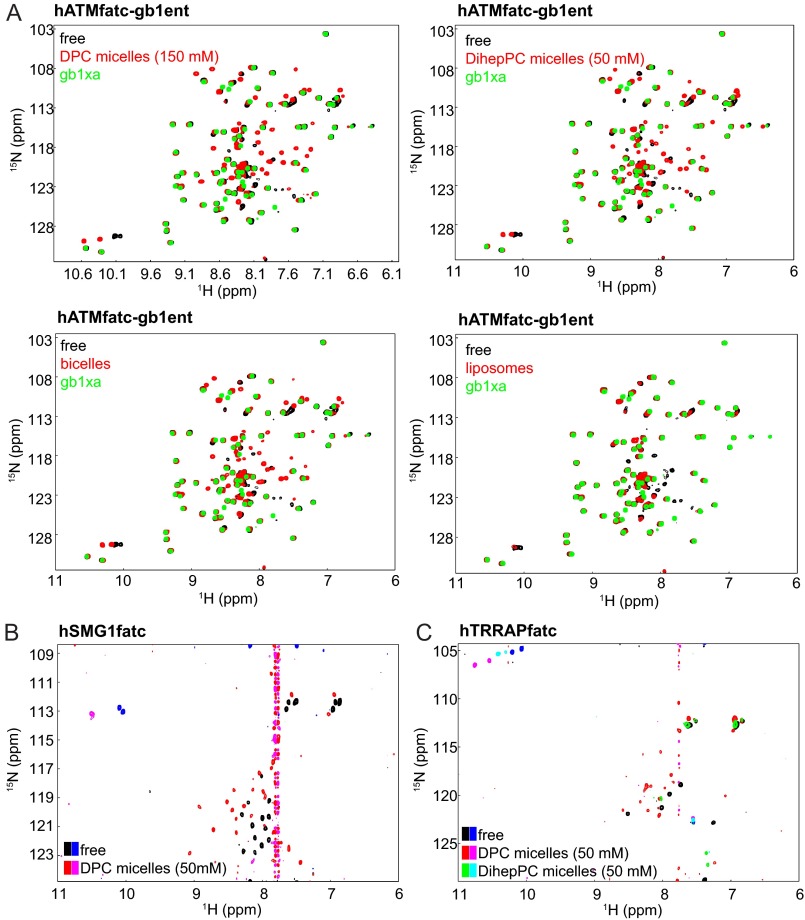
The interaction of the FATC domain of human ATM (hATMfatc) with different membrane mimetics was analyzed in more detail by using, as for the FATC domain of DNA-PKcs, a recently published procedure that is based on the use of 15N-labeled GB1 fusion proteins for the NMR binding assays (39). Fig. 3A shows the superpositions of the 1H-15N HSQC spectra of hATMfatc-gb1ent in the absence and presence of either DPC micelles or DihepPC micelles or of DMPC/DihepPC bicelles or DMPC liposomes (see schematic representations in Fig. 1C). To better differentiate the peaks arising from the FATC part from those arising from the GB1 tag, the spectrum of the GB1 tag containing a thrombin and a factor Xa site (= gb1xa) is shown on top in each plot of Fig. 1C. With all four tested membrane mimetics, a major change of the spectral appearance of the FATC peaks can be observed. The addition of 150 mm DPC, 50 mm DihepPC, or DMPC/DihepPC bicelles (total lipid concentration ∼270 mm) results in the disappearance of resonances characteristic for the free form and the appearance of new peaks for the membrane mimetic-associated state. As for the other analyzed FATC domains, spectral shifts can be seen for many backbone amide groups as well as for the side amides of the two tryptophans and those of some glutamines and asparagines, respectively. The titration of hATMfatc-gb1ent with increasing amounts of DPC is shown in supplemental Fig. S7A. In the presence of 0.1 mm DPC, no significant spectral changes can be seen. At 1.1 mm DPC, which corresponds to the CMC of DPC, some peaks of the FATC part get weaker and/or show small shifts. Increasing the DPC concentration to 5 mm resulted in further shifts. The spectra at ∼10 and ∼49 mm DPC are about the same as at 5 mm. This indicates that hATMfatc interacts only with the membrane-mimetic micelles and not with single DPC molecules. If DMPC liposomes are added (<30 mm DMPC), only the disappearance of peaks can be seen. The peaks of the bound form are presumably not visible because of the significantly larger size of SUVs compared with bicelles and micelles, which is expected to result in a significant line broadening for liposome-associated hATMfatc-gb1ent. In addition, the total lipid concentration in the sample with SUVs is much lower (<30 mm DMPC). Therefore the equilibrium is not shifted as much to the bound form. Finally, the affinity for liposomes may be lower than for micelles or bicelles because of differences in the surface curvature and packing density.
FIGURE 3.
NMR analysis of the interaction of the FATC domains of human ATM, SMG-1, and TRRAP with different membrane mimetics. A, superpositions of the 1H-15N HSQC spectra of hATMfatc-gb1ent in the absence and presence of either DPC or DihepPC micelles, DMPC/DihepPC bicelles, or DMPC liposomes. The spectrum of the free form is always shown in black and the one with the respective membrane mimetic in red. To better identify the signals of the ATM FATC part, the spectrum of the GB1 tag including an additional factor Xa site (= gb1xa) is additionally shown in green on top in each plot. Accordingly, all peaks that are green on top belong to the GB1 tag. As the GB1 tag does not interact with membrane mimetics, its signals do not shift (39). The superposition of the 1H-15N HSQC spectra corresponding to the titration of 15N hATMfatc-gb1ent with increasing amounts of DPC (0–50 mm) is displayed in supplemental Fig. S7A. B, superposition of the natural abundance 1H-15N SOFAST-HMQC spectra of hSMG1fatc in the absence and presence of DPC micelles. A superposition of the natural abundance 1H-15N HSQC spectra of the same protein in the absence and presence of DihepPC micelles is shown in supplemental Fig. S7B. C, superposition of the natural abundance 1H-15N HSQC spectra of hTRRAPfatc in the absence and presence of DPC micelles. Positive signals in B and C are shown in black, red, and green and negative ones in blue, magenta, and cyan. Because of the low solubility of hATRfatc, its interaction with membrane-mimetic DPC micelles could only be monitored based on one-dimensional NMR spectra (supplemental Fig. S8).
Fig. 3B shows a superposition of the natural abundance 1H-15N SOFAST-HMQC spectra of a 33-residue peptide corresponding to the FATC domain of human SMG-1 (hSMG1fatc) in the absence and presence of 50 mm d38-DPC. As the CMC of DPC is 1.1 mm (40), DPC should be present mostly in membrane-mimetic micelles (Fig. 1C top schematic representation). The peak pattern in the two spectra is clearly different, indicating an interaction of hSMG1fatc with the membrane-mimetic micelles, which results in a change of the chemical environment for all detectable peaks. In addition, the interaction of hSMG1fatc with also neutral DihepPC micelles (Fig. 1C, second from top schematic representation) was analyzed by recording natural abundance 1H-15N HSQC spectra (supplemental Fig. S7B). Also in this case, the spectral appearance changes significantly if 50 mm DihepPC are present. The CMC of DihepPC is about 1.4–1.8 mm (41, 42). Thus, hSMG1fatc appears also to interact with DihepPC micelles. With both types of micelles, all detectable backbone amides can be found at new positions, accompanied by an overall increase in the dispersion of the signals. The latter finding indicates that the peptide may become more structured upon interaction with micelles. Strong shifts can be observed further for the side amide Hϵ1 protons of both tryptophans, which are characterized by proton chemical shifts around 10–11 ppm (Fig. 3B and supplemental Fig. S7B). The intensity of these peaks is negative because of spectral folding in the 15N dimension. Finally, smaller shifts are visible for the side amide protons of glutamines and asparagines (1H ∼6.5–7.5 ppm, 15N ∼111–114 ppm). The fact that the number of backbone amide resonances is smaller than expected based on the number of residues in the sequences can be explained by the following reasons. First, the sensitivity of a 1H-15N HSQC/HMQC spectrum recorded at natural abundance is significantly lower than one recorded of a 15N-enriched sample. Thus, weaker resonances may not be detected. Second, some resonances may be very broad and thus weak because of motional averaging and/or exchange between the free and the micelle-immersed state.
The interaction of the FATC domain of human TRRAP (hTRRAPfatc) with membrane mimetics has, as for SMG-1, been analyzed by recording 1H-15N HSQC spectra in the absence and presence of either DPC or DihepPC micelles at natural abundance (Fig. 3C). Also in this case, the spectral appearance changes significantly for almost all detectable resonances. In the case of DPC, the number of detectable peaks as well as their dispersion increases, indicating that hTRRAPfatc may become more structured upon interaction with DPC micelles. Both DPC and DihepPC micelles induce significant shifts of the tryptophan side chain Hϵ1 protons, albeit the effect is stronger with DPC than with DihepPC. Shifts for the side chain amide protons of glutamine and asparagines can be seen only in the presence of DPC micelles. This and the overall smaller shifts for the two tryptophan side chain protons as well as the smaller number of peaks in the presence of DihepPC micelles suggest that a higher concentration of DihepPC may be needed to induce spectral changes of similar strength as observed with DPC micelles. The reason for this could be either a lower affinity for DihepPC compared with DPC micelles or because the number of DihepPC micelles at a concentration of 50 mm (53) is lower than that of DPC micelles (51) at the same concentration.
The interaction of a peptide corresponding to the FATC domain of human ATR (hATRfatc) with membrane-mimetic DPC micelles could only be monitored based on one-dimensional 1H-NMR spectra (supplemental Fig. S8). Overall the spectral appearance of the one-dimensional 1H-NMR spectrum changes dramatically in the presence of 50 mm d38-DPC (Fig. supplemental S8, top panel). Looking only at the amide region (∼6–11 ppm) (supplemental Fig. S8, middle panel), the number of resonances as well as their intensity and dispersion increases in the presence of micelles. This includes the appearance of a small signal for the side chain amide proton of the single tryptophan (∼10.4 ppm). An increase in the signal intensity is also evident for the aliphatic region (∼5–0 ppm, supplemental Fig. S8, bottom panel). However, in this chemical shift range, signals from the buffer (Tris and TCEP) and residual signals from d38-DPC disturb the interpretation. Overall the data suggest that hATRfatc interacts with DPC micelles and thereby may become more structured. In summary, the NMR-monitored binding studies indicate that the FATC domains of all of the PIKKs tested can interact with membrane mimetics.
The Presence of Membrane Mimetics Results in a Significant Population of α-Helical Secondary Structure in the Tested FATC Domains
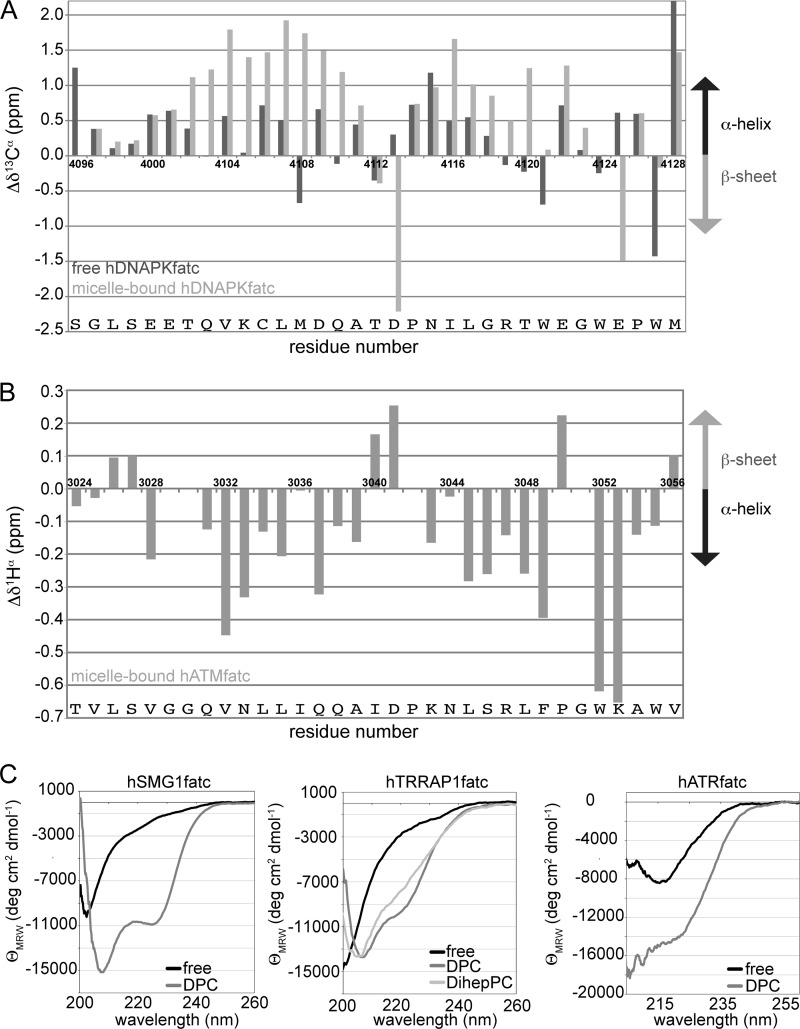
The structure of hDNAPKfatc was initially characterized using untagged 15N-labeled protein. Based on the CD spectrum (supplemental Fig. S9, top) and the observed 3JHNHα coupling constants (supplemental Fig. S9, bottom), the free, isolated FATC domain is rather unstructured. A comparison of the assigned 1Hα-chemical shift values with the respective random coil values indicated for the majority of the residues only a small tendency to populate α-helical secondary structure. This was overall confirmed by the analysis of 13Cα- secondary shifts of the FATC part of free hDNAPKfatc-gb1ent (Fig. 4A, black bars). The interaction with DPC micelles significantly increases the population of α-helical secondary structure (Fig. 4A, gray bars). Based on the shown 13Cα- and 1Hα- secondary shifts (Fig. 4A and supplemental Fig. S10), the FATC part of micelle-immersed hDNAPKfatc-gb1ent contains roughly two helical stretches that are disturbed around residues Thr-4112–Asp-4113 preceding Pro-4114 and finish around Thr-4120. The presence of helical secondary structure in the micelle-immersed state is further confirmed by the observation of helix-typical NOE correlations, which are listed in tabular form in supplemental Fig. S10D. The C-terminal tryptophan-rich region contains residues that have 13Cα secondary shifts that are typical for either α-helix or β-sheet. This has been observed similarly for the 13Cα secondary shifts of the corresponding region of the TOR FATC domain (30). Thus, this region may, as observed for the free oxidized as well as the oxidized and reduced micelle-immersed states of the TOR FATC domain (29, 30), fold back toward the preceding helical region. This folding-back or loop formation is facilitated by the glycine at position 4123. This glycine is conserved not only in the FATC domain of DNA-PKcs (Fig. 1B) and TOR (30) but also in those of other PIKKs (Fig. 1B and supplemental Fig. S1).
FIGURE 4.
Analysis of changes in the secondary structure upon the interaction of selected PIKK FATC domains with membrane mimetics by NMR and CD spectroscopy. A, estimate of the secondary structure content of free and micelle-immersed hDNAPKfatc based on its 13Cα secondary shifts (83). The difference between the measured 13Cα chemical shift and the random coil value for the respective amino acid was plotted as a function of the amino acid sequence (dark gray, free state; lighter gray, micelle-immersed state induced by the presence of 150 mm DPC). Values significantly higher than the random coil value indicate the presence of α-helical and those significantly lower of β-sheet secondary structure. The data were measured using 15N hDNAPKfatc-gb1ent. Additional secondary shifts and a table listing the presence of 1H-1H NOE correlations typical for helical regions can be found in supplemental Fig. S10. Additional information about the structure of the free pure hDNAPKfatc can be found in supplemental Fig. S9 (CD spectrum and 3JHNHα values). B, estimate of the secondary structure content of micelle-immersed hATMfatc based on its 1Hα secondary shifts (83). The difference between the measured 1Hα chemical shift and the random coil value for the respective amino acids was plotted as a function of the amino acid sequence (lighter gray, micelle-immersed state induced by the presence of 150 mm DPC). Values significantly lower than the random coil value indicate the presence of α-helical structure, and those significantly higher indicate the presence of β-sheet secondary structure. Additional 1H secondary shifts and 3JHNHα data can be found in supplemental Fig. S12. C, superposition of the CD spectra of hSMG1fatc (left), hTRRAPfatc (middle), and hATRfatc (right) in the absence (black) and presence of the DPC micelles (dark gray) and additionally for hTRRAPfatc with DihepPC micelles (light gray). The DPC or DihepPC concentration was ∼50 mm.
Also for the FATC domains of the other four PIKKs analyzed, the signal dispersion in the NMR spectra increased upon the addition of a membrane mimetic (Fig. 3 and supplemental Figs. S7 and S8), which may indicate that the respective proteins also have become more structured, resulting in a more diverse chemical environment for the different micelle-immersed residues. The presence of helical secondary structure in hATMfatc was derived based on an analysis of the 1Hα secondary shifts and the 3JHNHα values (Fig. 4B and supplemental Fig. S12). The data indicate the presence of two helical stretches in about the same regions as for DNA-PKcs (Fig. 4, A and B). However, residues ∼3051–3054 at the C-terminal hATMfatc may form another short helix or helical turn (Fig. 4B). In ATM this may be possible because ATM does not have a proline in the third to last position (residue 3054) as DNA-PKcs does (Pro-4126) but at position 3050. The change in the secondary structure content of hSMG1fatc, hTRRAPfatc, and hATRfatc was monitored by circular dichroism spectroscopy (Fig. 4C). The hSMG1fatc peptide shows a spectrum that is characteristic of an unfolded protein in the free form (Fig. 4C, left). In the presence of 50 mm DPC the absolute signal intensity increases, and the minimum shifts from about 205 to 208 nm. Additionally a second minimum becomes visible at about 222 nm. Minima at 208 and 222 nm are typically observed for proteins containing a significant amount of α-helical secondary structure. The higher absolute mean residue ellipticity at 208 compared with 222 nm may be explained by the presence of a remaining short unstructured stretch. The spectrum of free hTRRAPfatc is also typical for an unfolded protein with a minimum around 200 nm, whereas the spectra in the presence of membrane-mimetic DPC micelles show minima at about 208 and 222 nm, which indicates the presence of α-helical secondary structure (Fig. 4C, middle). The presence of DihepPC micelles also results in significant spectral change and a minimum at about 208 nm; however the minimum at 222 nm is not well resolved. Thus the amount of helical secondary structure in the DihepPC micelle-immersed state is expected to be lower than in the DPC micelle-immersed state. This is consistent with the fact that fewer NMR signals are visible for the DihepPC micelle-associated form than for the DPC micelle-associated form (Fig. 3C). The spectrum of the free form of hATRfatc shows a minimum at 215 nm, which is typically observed for proteins containing β-sheet structure (Fig. 4C, right). This β-sheet-typical appearance may be related to the observation that the free hATRfatc peptide has a tendency to aggregate. The addition of 50 mm DPC results in a change of the spectral shape, which is characterized by the appearance of minima at about 208 and 222 nm that are typical for α-helical secondary structure. Because of the bad spectral quality, the two minima can however not be as clearly detected as for SMG1 and TRRAP. The quality of the CD spectra of hATRfatc is lower because smaller protein concentrations had to be used, as the protein had to be diluted to lower the TCEP concentration to reduce too strong distortions from its presence at higher concentrations. For the FATC domains of SMG-1, TRRAP, and ATR, a future detailed structural characterization needs to be done to confirm that they also significantly populate the helical secondary structure in the presence of membrane mimetics.
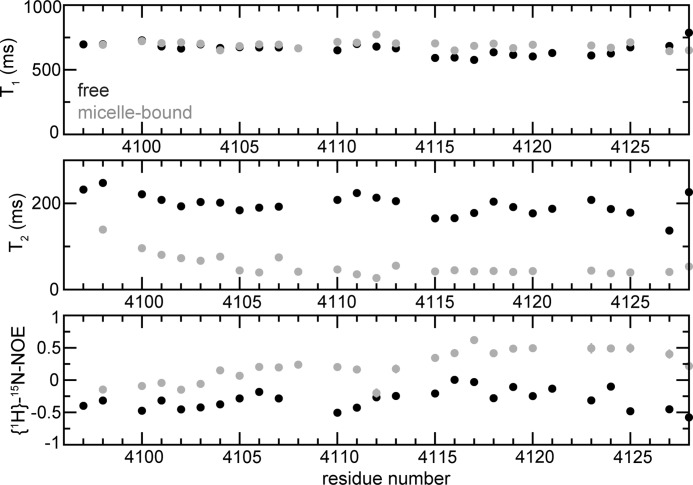
A Comparison of the 15N Relaxation Data of the Free and Micelle-immersed Forms Confirms That hDNAPKfatc Associates with Micelles, Which Results in Reduced Backbone Dynamics
The backbone dynamics of free and micelle-immersed hDNAPKfatc-gb1ent were studied by 15N relaxation experiments. The presence of the GB1 tag makes the analysis of the relaxation data for the FATC part a bit more difficult, because exchange effects arising from motions of the two proteins with respect to each other may have to be considered. Based on the relaxation data presented for the whole fusion protein in the absence and presence of DPC micelles (supplemental Fig. S11), the FATC and the GB1 part tumble rather independently in both states, and the thrombin and enterokinase sites act as a flexible linker. The average 15N T1 and T2 and {1H}-15N NOE values for the GB1 tag (residues 1–56) are 479 ± 35 ms, 73 ± 11 ms, and 0.66 ± 0.07 for the free protein and 496 ± 40 ms, 635 ± 11 ms, and 0.65 ± 0.08 for the micelle-associated fusion protein, respectively. The 15N T1 and the {1H}-15N NOE values are in the range reported for the isolated GB1 domain (54). The 15N T2 values for the GB1 tag, especially in the absence of micelles, are significantly lower than expected for a completely unhindered isotropic reorientation of a 6-kDa protein. Using a model system based on GB1 domains connected by different linker regions, it has been shown that each domain exhibits different rotational diffusion and alignment properties even if the linker is 18 residues long (55). In the absence of micelles, the presence of the FATC domain in a mostly unstructured, flexible form may further result in an additional viscous drag. A similar effect has been observed for the N-terminal domain of formin C, which contains a large unstructured loop (56).
The 15N T1 and T2 and {1H}-15N NOE data for only the FATC part of the free and micelle-immersed fusion protein are shown in Fig. 5 (in black and gray, respectively). The average 15N T1 and T2 and {1H}-15N NOE values for residues 4108–4128 of the FATC part of the fusion protein are 645 ± 51 ms, 191 ± 23 ms, and −0.27 ± 0.17 for the free form and 689 ± 31 ms, 42 ± 6 ms, and 0.34 ± 0.18 for the micelle-associated form, respectively. The association of hDNAPKfatc-gb1ent with DPC micelles results in only a small increase of the average T1 value. Overall, a stronger increase of the T1 values would be expected upon interaction with the ∼19-kDa DPC micelles (51). Unstructured proteins or protein regions show overall higher T1 values than the respective folded state, which has for example been shown for an SH3 domain (57). Thus, a smaller increase in T1 than expected can at least in part be accounted for by the fact that the FATC domain in the free state is rather unstructured and flexible and becomes significantly structured only upon interaction with micelles. Therefore, the increase in T1 due to an increase in the molecular weight upon complex formation is partially compensated for by the FATC domain becoming more structured. Another factor influencing the 15N relaxation parameters may be the time scale of the exchange between the free and the micelle-immersed state. The T2 times of most residues decrease strongly from about ∼190 ms in the free form to ∼40 ms in the micelle-immersed state. This confirms that the FATC domain interacts with the DPC micelles. Overall the T2 values of the bound state are in the range expected for a ∼23–30-kDa complex (∼19-kDa DPC micelle and ∼4–11-kDa protein) (30, 51). The increase in the {1H}-15N NOE values for most of the FATC domain confirms that it binds to the larger DPC micelles (∼19 kDa) (51) and is consistent with a more structured state, indicated by the observed secondary chemical shifts and 1H-1H NOE correlations (Fig. 4A and supplemental Fig. S10).
FIGURE 5.
Backbone dynamics of free and micelle-immersed FATC domain of DNA-PKcs. 15N T1 (first panel) and T2 (second panel) relaxation times and {1H}-15N NOE values (third panel) were plotted as a function of the sequence. The data for the free form are shown as filled black circles and that of the micelles-immersed form as filled gray circles. The data were measured using 15N hDNAPKfatc-gb1ent. The data for the whole fusion protein including the GB1 tag are displayed in supplemental Fig. S11. For comparison, see also the {1H}-15N NOE data for free and micelle-immersed hATMfatc-gb1ent shown in supplemental Fig. S13.
Association with DPC Micelles Also Reduces the Backbone Dynamics or Diffusion Constants of Other FATC Domains
For hATMfatc-gb1ent the association with membrane mimetics was also further confirmed by {1H}-15N NOE data (supplemental Fig. S13). Already the fact that several of the backbone resonances of the free FATC domain of human ATM appear not to be visible in the 1H-15N HSQC spectra (Fig. 3A), indicates increased backbone dynamics, presumably due to motional averaging, which may broaden some of the signals beyond detection. Because the sequential assignment of the free state is hampered by the lack of several 1H-15N correlations, the {1H}-15N NOE data for free hATMfatc-gb1ent was only qualitatively interpreted making use of the fact that the signals of the GB1 tag can be differentiated from those of the FATC domain based on spectral superpositions (Fig. 3A). As indicated by the presence of zero to negative intensity peaks in the spectrum with the NOE effect of the free form, most of the FATC part must be rather flexible (supplemental Fig. S11, top). For the well resolved peaks visible for the linker and the FATC part, the NOE values range between −0.6 and 0.2. In contrast, in the presence of DPC micelles most of the peaks for the FATC part are visible and show a positive intensity, thus also indicating positive NOE values (supplemental Fig. S13, bottom). The NOE values for residues Val-3028–Val-3056 range between ∼0.3 and 0.75. This confirms that the ATM FATC domain interacts with DPC micelles. Considering the range of NOE values typically observed for structured and flexible regions of proteins or protein complexes of a similar size (∼5–25 kDa) (30, 56, 57), most of the micelle-immersed ATM FATC domain appears rather well structured, with NOE values of >0.5.
For the FATC domains of ATR, SMG-1, and TRRAP, we had no 15N-labeled peptides available and thus could not record 15N relaxation data. However, as the NMR samples of hSMG1fatc and hTRRAPfatc in the absence and presence of 50 mm d38-DPC had rather high protein concentrations (∼1 mm), reliable diffusion constants could be obtained for both proteins (Table 1). The diffusion constants of the free forms are ∼1.7–1.8 × 10−10 m2/s. The addition of deuterated DPC micelles lowers the diffusion constant significantly to ∼1–1.1 × 10−10 m2/s for hSMG1fatc and ∼0.9–1 × 10−10 m2/s for hTRRAPfatc (Table 1). This is in the range expected for a complex of ∼23 kDa arising from the association of a ∼4-kDa peptide with a ∼19-kDa DPC micelle and using a DPC concentration of only 50 mm (30, 51). The diffusion constant for the reduced and oxidized TOR FATC domain in the presence of 30 mm DPC, which corresponds to a partially micelle-bound situation, is 1.32 and 1.13 × 10−10 m2/s, and in the presence of 170 mm DPC, which corresponds to the bound state, it is 0.59 and 0.72 × 10−10 m2/s (30). The diffusion constant of the DPC micelle alone at 170 mm is 1.10 × 10−10 m2/s (30). Thus the diffusion data confirm the association of the SMG-1 and TRRAP FATC domains with membrane-mimetic DPC micelles. Diffusion measurements in the presence of DihepPC were hampered by the huge signal from the undeuterated lipid, which impairs the reliable detection and analysis of the peptide signals.
TABLE 1.
NMR-derived diffusion coefficients (D) of free and micelle-associated FATC domains
| Protein | Free |
50 mm d38-DPC |
||
|---|---|---|---|---|
| c.s.a | D | c.s.a | D | |
| ppm | ×10−10 m2/s | ppm | ×10−10 m2/s | |
| hSMG1fatc | 1.124 | 1.76 | 1.861 | 1.12 |
| 0.832 | 1.76 | 1.490 | 1.03 | |
| 0.784 | 1.78 | 0.901 | 1.09 | |
| hTRRAPfatc | 1.940 | 1.87 | 1.489 | 0.95 |
| 1.311 | 1.69 | 0.873 | 0.95 | |
| 0.843 | 1.72 | 0.740 | 0.92 | |
| 0.778 | 1.77 | |||
a Chemical shift.
DISCUSSION
It was recognized early that PIKKs share a unique conserved C-terminal domain and that this FATC domain plays an important role in the regulation of their function (1, 22). This was confirmed for several family members based on mutagenesis studies (16, 26–28, 58). In addition, the FATC domains of some PIKKs appear to be functionally equivalent, at least with respect to some interactions. For example if the FATC domain of ATM is replaced by that of DNA-PKcs, ATR, or TRRAP, it can still interact with the protein Tip60. However, the FATC domain of ATM cannot replace that of ATR (25) or TOR (26). Although it has been shown that some FATC domains mediate protein-protein interactions (28, 58, 59), the question remained as to whether there is a further more general common property of the FATC domain that influences PIKK function. Based on the NMR- and CD-monitored lipid binding data presented for the FATC domains of human DNA-PKcs, ATM, ATR, SMG-1, and TRRAP, as well as on the earlier published data for TOR, the ability to interact with membrane mimetics and thus a potential role for the mediation or regulation of interactions at cellular membranes may be one function that is common to all PIKK FATC domains. TOR, which plays a central role in the regulation of cell growth, has been localized at different cellular membranes and in the nucleus (31–34). This suggests that the outcome of TOR signaling may be influenced by its localization, which may depend on specific cellular signals (30, 32). Besides the FATC domain, the FRB domain and the N-terminal HEAT repeat region have been suggested to mediate membrane interactions based on interactions with membrane lipids or membrane-associated proteins, respectively (33, 60–63). Thus, the membrane localization of TOR appears to be regulated by a network of interactions. For PIKKs involved in DNA repair, such as ATM, ATR, and DNA-PKcs, a predominant nuclear localization is expected. However, for ATM and DNA-PKcs additional functions as well as membrane localization have been suggested, which are outlined in the following text. DNA-PKcs shows a high abundance in mammalian cells, and thus it is not astonishing that it also plays a role in the signaling response to IR, metabolic gene regulation, and the regulation of the homeostasis of cell proliferation (6). Moreover, DNA-PKcs and the two Ku proteins have been suggested as separately localizing to lipid rafts of mammalian cells (9). Because DNA-PKcs has no evident transmembrane domain and is also not known to have a glycosylphosphatidylinositol or fatty acid anchor, it was further proposed that recruitment to the membrane might be mediated by protein-protein interactions (9). Based on the NMR-monitored interaction studies presented (Fig. 2 and supplemental Figs. 3–5), DNA-PKcs may directly interact with membrane regions via its FATC domain.
For ATM, a role in the oxidative stress response and thus as a linker of genome stability and carbon metabolism has been proposed (4, 7). Earlier, it has been shown that ATM is localized not only in the nucleus but also at cytoplasmic vesicles (12). In addition, it has been shown that the protein CKIP-1 (casein kinase-2 interaction protein-1), which is involved in muscle differentiation and the regulation of the actin cytoskeleton and cell morphology, recruits ATM to the plasma membrane and that this interaction is mediated by the C-terminal region including the catalytic and the FATC domains (36). Besides protein-protein interactions with CKIP-1, the FATC domain of ATM may therefore mediate direct membrane interactions. Similarly, ATR may be involved in a broad spectrum of cellular processes that go well beyond DNA replication and repair (5). SMG-1 has also been suggested to be involved in additional processes other than RNA surveillance, such as cell survival during TNFα-induced stress (15), life span regulation and oxidative stress resistance in Caenorhabditis elegans (14), the regulation of hypoxia-inducible factor-1α (HIF-1α) (64), and in finally the response to injury and in growth control in planarian flatworms, possibly by suppressing mTOR signaling (65). Thus its cellular localization may, as observed for TOR, ATM, and DNA-PKcs, vary and may possibly involve interactions with membrane regions.
Immunoprecipitated human SMG-1 with the mutation L3646A in the FATC domain shows strongly reduced kinase activity, whereas the mutant W3653F reduces activity to about 50% (16). Trp-3653 and Leu-3646 may possibly participate in interactions with membranes and/or regulator proteins, which may influence the catalytic activity. Because the lysis buffer for the isolation of FLAG-tagged SMG-1 contains detergents, potential membrane-localized SMG-1 including membrane-localized regulators could also have been isolated by this procedure (16). Although the immunoprecipitated SMG-1 was washed with detergent-free kinase assay buffer (16), potential complex partners including detergents/lipids may not have been removed. Future biochemical and biophysical studies will be needed to analyze the influence of specific mutations on the affinity for membrane mimetics and regulator proteins of SMG-1 as well as intramolecular interactions in SMG-1.
TRRAP appears to fulfill most of its functions as a component of histone acetyltransferase complexes (20). For the yeast homolog Tra1 it has been shown that the mutations L3733A and F3744A result in transcriptional changes and phenotypes similar to those caused by mutations in the kinase-related domain (27). The L3733A mutations further result in significantly reduced steady-state levels of Tra1 (27). Reduced cellular stability, however, due to a cysteine to serine mutation that disables the formation of an intramolecular disulfide bond in the FATC domain, has also been observed for TOR (29). Finally, an additional glycine at the C terminus of yeast Tra1 results in the loss of cellular viability (27). In principle, all of these mutations may also influence the interactions with membranes, the protein stability, and/or the interactions with specific membrane-localized proteins important for additional functions that are not related to transcriptional regulation. Future localization studies in cells will be needed to clarify the cellular localization patterns of TRRAP and other PIKKs and how they vary in response to specific signals. In addition, the influence of mutations on the ability to interact with membrane mimetics has to be analyzed as well as the effect of mutations that abrogate these interactions on cellular localization, cellular stability, or response to specific signals.
Because membrane-mimetic interactions of the FATC domain of DNA-PKcs and the other PIKKs were studied using the isolated FATC domain or a fusion to a GB1 tag, the question of the accessibility in the context of the full-length protein arises. Recent EM studies of DNA-PKcs alone suggest that the region assigned to the FATC domain resembles a “protruding finger” (66, 67), which therefore is amenable to interactions with membranes or regulatory proteins. Analogous observations were made for the accessibility of the FATC domain of TOR in the full-length protein (68).
Because all tested FATC domains interact with different membrane mimetics, it may be considered that any small protein or peptide may interact nonspecifically with membrane mimetics or may just be solubilized by the detergent-like properties of some of them. We recently showed that the well structured GB1 tag, including the linker region containing protease sites, does not show any significant NMR spectral changes if DPC or DihepPC micelles, DMPC/DihepPC bicelles, or DMPC liposomes are added (39). This finding was further confirmed here by the NMR data of GB1 fusion proteins (Figs. 2, D–F, and 3A and supplemental Figs. S4, S5, S7A, S11, and S13). In another former study, we analyzed the interaction of a largely unstructured 26-mer peptide with DPC micelles (56). This peptide corresponds to the large unstructured loop of the N-terminal domain of the protein formin C. To mimic the restriction of mobility in the full-length protein, this peptide could further be circularized by oxidation of the two terminal cysteines. However, neither the reduced nor the oxidized peptide showed any spectral changes in CD or NMR spectra upon the addition of 50 mm DPC (56).
The titrations of pure hDNAPKfatc with DPC and a 4:1 mixture of DioctPA and DOPA (Fig. 2B and supplemental Fig. S3B) and of hDNAPKfatc-gb1ent/xa (Fig. 2A and supplemental Figs. S3A and S4) and hATMfatc-gb1ent (supplemental Fig. 7A) with DPC indicate that the respective FATC domains interact only with micelles, as no significant shifts could be observed below the known or estimated CMC. This is consistent with the more detailed NMR-monitored binding studies of the yeast TOR1 FATC domain with DPC and different lipids (30). For the FATC domain of yeast TOR1, the interaction with different lipids has also been analyzed using DPC and a 4:1 mixture of DioctPA and DOPA but also DioctPA alone as well as the highly negatively charged lipid DihexPIP345. In all cases spectral changes occurred only above the respective CMC or, in the case of DihexPIP345, if DPC was added, to induce micelle formation (30). Thus, the FATC domains of PIKKs appear not to recognize specific lipid head groups and/or interact with single lipids but only with membrane-mimetic structures such as micelles, bicelles, or liposomes. Titrations with DPC for the peptides used here corresponding to the FATC domains of TRRAP, SMG-1, and ATR could not be monitored by 1H-15N HSQC experiments because that would have been too time consuming without 15N labeling.
The interaction of hDNAPKfatc with DPC micelles results in strong spectral changes for the C-terminal tryptophan-rich region (Trp-4121–Met-4128) as well as for residues Cys-4106–Asp-4113 (Fig. 2A, C), whereas negatively charged mixed phosphatidic acid membrane-mimetic particles induced strong changes in a region harboring a positively charged lysine (Val-4104–Gln-4111) (Fig. 2, B and C). This is in contrast to the results for the FATC domain of TOR, which show similar spectral changes with neutral and negatively charged membrane mimetics (30). Because the N terminus of the TOR FATC is rather acidic, the affinity for negatively charged membrane-mimetic particles is nevertheless also expected to be slightly lower. Compared with the FATC domain of TOR, that of DNA-PKcs in higher eukaryotes contains two negatively charged residues (Figs. 1B and 2C and supplemental Fig. 1) in the C-terminal tryptophan-rich region, which can weaken the interaction with negatively charged membrane-mimetic particles due to electrostatic repulsion. Whereas the FATC domain of DNA-PKcs interacts with neutral micelles and bicelles (Fig. 2, D and E), it has no apparent affinity for neutral DMPC SUVs (Fig. 2F). This is in further contrast to the results for the FATC domains of TOR4 as well as ATM (Fig. 3A), which both interact with DMPC liposomes. The observation that the FATC domain DNA-PKcs has no significant affinity for liposomes indicates that its interactions with the membrane regions may be more sensitive to the surface curvature and lipid packing than those of TOR and ATM.
Based on the analysis of the chemical shifts, the CD data, the determined 3JHNHα coupling constants, and the 15N relaxation data (Figs. 4 and 5 and supplemental Figs. S9–S13), the isolated free FATC domains of DNA-PKcs, ATM, and TRRAP were found to be rather unstructured and flexible. This may be either because they were analyzed without the preceding domains, which may stabilize their fold, or because they are generally natively unfolded without an interaction partner. In contrast, the oxidized form of the TOR FATC domain is rather well structured and consists of an α-helix and a disulfide-bonded loop (29). However, reduction of the disulfide bond increases the backbone dynamics of the region containing the cysteines (29). The addition of DPC micelles significantly increases the population of the α-helical secondary structures in the FATC domains of DNA-PKcs, SMG-1, TRRAP, and ATR. Accretion of or a higher population of α-helical secondary structures in the presence of a membrane mimetic or, in other words, folding upon binding to membrane mimetics has also been observed for other small proteins, e.g. very recently for the 30-residue-long glucagon-like peptide 1 (GLP-1), which targets a G protein-coupled receptor (69). According to the membrane catalysis hypothesis, signaling peptides targeting membrane-resident receptors must first interact with the bilayer before interacting with the protein receptor, which increases their local concentration and allows them to adopt a structure that is recognized by the target protein (69). Similarly, the observed formation and/or stabilization of α-helical structure could enable PIKKs to interact with specific membrane-localized proteins. As an alternative to a mechanism whereby interactions with specific membrane regions induce the formation of an α-helical structure, a selection of conformers that populate the α-helical secondary structure in the free form may be considered (70).
In line with TOR already being more structured in the free form, the interaction with micelles results only in a stabilization of the α-helical secondary structure and a slight extension toward the C terminus (30). The respective structures of the oxidized and reduced micelle-immersed states of the yeast TOR1 FATC domain (Protein Data Bank ID 1kio and 1kit) show a distortion of the α-helix around Ala-2453, which was not present in the free form and which has been suggested to arise because this alanine is at the interface between the solvent and the micelle (30). The α-helical structure of hDNAPKfatc in the presence of DPC micelles is also distorted at the equivalent alanine (Fig. 4A, Ala-4111). Based on the 13Cα secondary shifts presented, the C-terminal region may further, as observed for the micelle-immersed oxidized and reduced forms of TOR, fold back onto the preceding region. This is suggested to be facilitated by a conserved glycine (30), which is shared by the FATC domains of all PIKKs but TRRAP (Fig. 1) and which also has a rather small alanine at the equivalent position. Based on the presented secondary structure for the FATC domain of ATM (Fig. 4B and supplemental Fig. S12), the helical regions are interrupted after Ala-3039 and Pro-3050/Gly-3051. Thus the two residues (Ala-4111 and Gly-4123 in DNA-PKcs and Ala-3039 and Gly-3051 in ATM (Fig. 1)) may play a role in the formation of similar membrane-bound structures and/or in positioning in the membrane, as in the proposed model for the TOR FATC membrane association (30). The positioning is however expected to be controlled mostly by the distribution of charged and hydrophobic, especially aromatic, residues along the sequence. For tryptophans in membrane-binding proteins, it has been shown that they favor a position at the interface between the lipid head groups and the membrane interior (71). Polar or charged residues are located rather at the rim of a membrane anchor and/or mediate ionic interactions with charged head groups (30, 72).
Based on the current data, the ability to interact with membrane mimetics appears to be a general property of the FATC domain of all PIKKs. Differences in the distribution of hydrophobic and charged side chains in the FATC domains of the various PIKKs (Fig. 1 and supplemental Fig. S1) may thus result in different preferences for specific membrane properties such as surface charge and curvature or the packing density of the lipid acyl chains as well as the presence of cholesterol or certain proteins. This is expected to result in different binding specificities for different cellular membranes or membrane regions, which would be consistent with the different localization patterns and localized specific activities of the various PIKKs in cells. Future detailed interaction studies with differently composed membrane mimetics and different mutant proteins as well as structural studies of the membrane mimetic-bound forms will need to be targeted for each individual PIKK FATC domain to clarify their specific membrane preferences as well as structural differences in the membrane-associated states.
A role for the FATC domain as a temporal membrane anchor does not exclude additional contacts with membrane components or membrane-localized proteins mediated by other domains of DNA-PKcs. For TOR, it has been suggested that the HEAT repeats in the N-terminal region mediate protein-protein interactions important for the localization of TOR to the plasma membrane (33). Moreover, the FRB domain of TOR may mediate direct interactions with membrane regions (60, 63). Finally, TOR supposedly interacts with the small GTPase Rheb, which is farnesylated and thereby localizes to endosomal membranes, with the protein FKBP38, which has a transmembrane domain that targets it to the mitochondrial membrane, and with the so called Ragulator-Rag complex at the lysosomal surface (73–75). It has been proposed that ATM interacts with the protein CKIP-1 (casein kinase-2-interacting protein-1) at the plasma membrane (36). For DNA-PKcs it is speculated that it may interact with the protein C1B, a calcium-binding protein that is N-terminally myristoylated and interacts with two different plasma membrane proteins (9). As it becomes more and more apparent that all PIKKs intercept various signaling pathways and/or regulate different processes, a tight control of their cellular localization by a network of interactions may be one possibility to ensure a specific signaling output in response to the signaling state of the cell.
Another aspect that may be considered for future membrane interaction studies is the effect of reactive oxygen species or oxidized lipids on cysteines in the highly conserved region of the FATC domain, as shown in Fig. 1B (TOR, DNA-PKcs, ATR, and TRRAP), or about 60–80 residues N-terminal to that region (ATM, SMG-1). As mentioned above, ATM has been shown to function in cellular redox signaling (4). Oxidation of Cys-2991 in the less conserved part of the FATC region results in the formation of a dimer (76) in which the FATC domain may show different accessibility for interactions with regulators and membrane patches. Also, DNA-PKcs, TOR, and SMG-1 have been related to redox-influenced cellular processes or states such as the mitochondrial metabolisms and/or hypoxia (64, 77–80). Finally, posttranslational modifications may influence the protein and/or membrane interactions of the FATC domain, such as for example acetylation of lysine 3016 in the FATC domain of ATM (58, 81).
Supplementary Material
Acknowledgments
We thank several Technische Universität München biochemistry bachelor students for their contributions. We thank Melanie Meier as well as Sarah Hammes and Verena Kanoldt for help with the purification and/or with some of the NMR and CD measurements and Tanja Kraus and Ann-Kathrin Flörsheimer for the preparation of the bicelle samples of DNAPKfatc-gb1ent. We acknowledge Prof. Dr. Stephan Grzesiek at the Biozentrum of the University of Basel for allowing measurement time on his Bruker DRX600 NMR spectrometer.
This work was supported by Grant DA 1195/3-1 from the German Research Foundation (to S. A. D.).

This article contains supplemental Figs. S1–S13.
L. A. M. Sommer and S. A. Dames, unpublished results.
- PIKK
- phosphatidylinositol-3 kinase-related kinase
- ATM
- ataxia-telangiectasia mutated
- ATR
- ataxia- and Rad3-related
- CMC
- critical micelle concentration
- DihepPC
- 1,2-diheptanoyl-sn-glycero-3-phosphocholine
- DioctPA
- 1,2-dioctanoyl-sn-glycero-3-phosphate
- DMPC
- 1,2-dimyristoyl-sn-glycero-3-phosphocholine
- DNA-PK
- DNA-dependent protein kinase
- DNA-PKcs
- DNA-PK catalytic subunit
- DOPA
- 1,2-dioleoyl-sn-glycero-3-phosphate
- DPC
- dodecylphosphocholine
- FATC
- FRAP-ATM-TRRAP C-terminal domain
- GB1
- B1 immunoglobulin-binding domain of streptococcal protein G (56 residues)
- MWCO
- molecular weight cut-off
- SUV
- small unilamellar vesicle
- TCEP
- Tris(2-carboxylethyl)phospin-HCl
- TOCSY
- total correlation spectroscopy
- TOR
- target or rapamycin
- TRRAP
- transformation/transcription domain-associated protein.
REFERENCES
- 1. Keith C. T., Schreiber S. L. (1995) PIK-related kinases: DNA repair, recombination, and cell cycle checkpoints. Science 270, 50–51 [DOI] [PubMed] [Google Scholar]
- 2. Lempiäinen H., Halazonetis T. D. (2009) Emerging common themes in regulation of PIKKs and PI3Ks. EMBO J. 28, 3067–3073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lovejoy C. A., Cortez D. (2009) Common mechanisms of PIKK regulation. DNA Repair (Amst.) 8, 1004–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ditch S., Paull T. T. (2012) The ATM protein kinase and cellular redox signaling: beyond the DNA damage response. Trends Biochem. Sci. 37, 15–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Flynn R. L., Zou L. (2011) ATR: a master conductor of cellular responses to DNA replication stress. Trends Biochem. Sci. 36, 133–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kong X., Shen Y., Jiang N., Fei X., Mi J. (2011) Emerging roles of DNA-PK besides DNA repair. Cell. Signal. 23, 1273–1280 [DOI] [PubMed] [Google Scholar]
- 7. Krüger A., Ralser M. (2011) ATM is a redox sensor linking genome stability and carbon metabolism. Sci. Signal. 4, pe17. [DOI] [PubMed] [Google Scholar]
- 8. Ju J., Naura A. S., Errami Y., Zerfaoui M., Kim H., Kim J. G., Abd Elmageed Z. Y., Abdel-Mageed A. B., Giardina C., Beg A. A., Smulson M. E., Boulares A. H. (2010) Phosphorylation of p50 NF-κB at a single serine residue by DNA-dependent protein kinase is critical for VCAM-1 expression upon TNF treatment. J. Biol. Chem. 285, 41152–41160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lucero H., Gae D., Taccioli G. E. (2003) Novel localization of the DNA-PK complex in lipid rafts: a putative role in the signal transduction pathway of the ionizing radiation response. J. Biol. Chem. 278, 22136–22143 [DOI] [PubMed] [Google Scholar]
- 10. Toulany M., Lee K. J., Fattah K. R., Lin Y. F., Fehrenbacher B., Schaller M., Chen B. P., Chen D. J., Rodemann H. P. (2012) Akt promotes post-irradiation survival of human tumor cells through initiation, progression, and termination of DNA-PKcs-dependent DNA double-strand break repair. Mol. Cancer Res. 10, 945–957 [DOI] [PubMed] [Google Scholar]
- 11. Grdzka I., Sochanowicz B., Brzóska K., Wójciuk G., Sommer S., Wojewódzka M., Gasińska A., Degen C., Jahreis G., Szumiel I. (2013) Cis-9,trans-11-conjugated linoleic acid affects lipid raft composition and sensitizes human colorectal adenocarcinoma HT-29 cells to X-radiation. Biochim. Biophys. Acta 1830, 2233–2242 [DOI] [PubMed] [Google Scholar]
- 12. Watters D., Khanna K. K., Beamish H., Birrell G., Spring K., Kedar P., Gatei M., Stenzel D., Hobson K., Kozlov S., Zhang N., Farrell A., Ramsay J., Gatti R., Lavin M. (1997) Cellular localisation of the ataxia-telangiectasia (ATM) gene product and discrimination between mutated and normal forms. Oncogene 14, 1911–1921 [DOI] [PubMed] [Google Scholar]
- 13. Yan J., Khanna K. K., Lavin M. F. (2000) Defective radiation signal transduction in ataxia-telangiectasia cells. Int. J. Radiat. Biol. 76, 1025–1035 [DOI] [PubMed] [Google Scholar]
- 14. Masse I., Molin L., Mouchiroud L., Vanhems P., Palladino F., Billaud M., Solari F. (2008) A novel role for the SMG-1 kinase in lifespan and oxidative stress resistance in Caenorhabditis elegans. PloS One 3, e3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Oliveira V., Romanow W. J., Geisen C., Otterness D. M., Mercurio F., Wang H. G., Dalton W. S., Abraham R. T. (2008) A protective role for the human SMG-1 kinase against tumor necrosis factor-α-induced apoptosis. J. Biol. Chem. 283, 13174–13184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Morita T., Yamashita A., Kashima I., Ogata K., Ishiura S., Ohno S. (2007) Distant N- and C-terminal domains are required for intrinsic kinase activity of SMG-1, a critical component of nonsense-mediated mRNA decay. J. Biol. Chem. 282, 7799–7808 [DOI] [PubMed] [Google Scholar]
- 17. Wullschleger S., Loewith R., Hall M. N. (2006) TOR signaling in growth and metabolism. Cell 124, 471–484 [DOI] [PubMed] [Google Scholar]
- 18. Laplante M., Sabatini D. M. (2012) mTOR signaling in growth control and disease. Cell 149, 274–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Unno A., Takada I., Takezawa S., Oishi H., Baba A., Shimizu T., Tokita A., Yanagisawa J., Kato S. (2005) TRRAP as a hepatic coactivator of LXR and FXR function. Biochem. Biophys. Res. Commun. 327, 933–938 [DOI] [PubMed] [Google Scholar]
- 20. Herceg Z., Wang Z. Q. (2005) Rendez-vous at mitosis: TRRAPed in the chromatin. Cell Cycle 4, 383–387 [DOI] [PubMed] [Google Scholar]
- 21. Robert F., Hardy S., Nagy Z., Baldeyron C., Murr R., Déry U., Masson J. Y., Papadopoulo D., Herceg Z., Tora L. (2006) The transcriptional histone acetyltransferase cofactor TRRAP associates with the MRN repair complex and plays a role in DNA double-strand break repair. Mol. Cell. Biol. 26, 402–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bosotti R., Isacchi A., Sonnhammer E. L. (2000) FAT: a novel domain in PIK-related kinases. Trends Biochem. Sci. 25, 225–227 [DOI] [PubMed] [Google Scholar]
- 23. Perry J., Kleckner N. (2003) The ATRs, ATMs, and TORs are giant HEAT repeat proteins. Cell 112, 151–155 [DOI] [PubMed] [Google Scholar]
- 24. Knutson B. A. (2010) Insights into the domain and repeat architecture of target of rapamycin. J. Struct. Biol. 170, 354–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mordes D. A., Glick G. G., Zhao R., Cortez D. (2008) TopBP1 activates ATR through ATRIP and a PIKK regulatory domain. Genes Dev. 22, 1478–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Takahashi T., Hara K., Inoue H., Kawa Y., Tokunaga C., Hidayat S., Yoshino K., Kuroda Y., Yonezawa K. (2000) Carboxyl-terminal region conserved among phosphoinositide-kinase-related kinases is indispensable for mTOR function in vivo and in vitro. Genes Cells 5, 765–775 [DOI] [PubMed] [Google Scholar]
- 27. Hoke S. M., Irina Mutiu A., Genereaux J., Kvas S., Buck M., Yu M., Gloor G. B., Brandl C. J. (2010) Mutational analysis of the C-terminal FATC domain of Saccharomyces cerevisiae Tra1. Curr. Genet. 56, 447–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jiang X., Sun Y., Chen S., Roy K., Price B. D. (2006) The FATC domains of PIKK proteins are functionally equivalent and participate in the Tip60-dependent activation of DNA-PKcs and ATM. J. Biol. Chem. 281, 15741–15746 [DOI] [PubMed] [Google Scholar]
- 29. Dames S. A., Mulet J. M., Rathgeb-Szabo K., Hall M. N., Grzesiek S. (2005) The solution structure of the FATC domain of the protein kinase target of rapamycin suggests a role for redox-dependent structural and cellular stability. J. Biol. Chem. 280, 20558–20564 [DOI] [PubMed] [Google Scholar]
- 30. Dames S. A. (2010) Structural basis for the association of the redox-sensitive target of rapamycin FATC domain with membrane-mimetic micelles. J. Biol. Chem. 285, 7766–7775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Berchtold D., Walther T. C. (2009) TORC2 plasma membrane localization is essential for cell viability and restricted to a distinct domain. Mol. Biol. Cell 20, 1565–1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Drenan R. M., Liu X., Bertram P. G., Zheng X. F. (2004) FKBP12-rapamycin-associated protein or mammalian target of rapamycin (FRAP/mTOR) localization in the endoplasmic reticulum and the Golgi apparatus. J. Biol. Chem. 279, 772–778 [DOI] [PubMed] [Google Scholar]
- 33. Kunz J., Schneider U., Howald I., Schmidt A., Hall M. N. (2000) HEAT repeats mediate plasma membrane localization of Tor2p in yeast. J. Biol. Chem. 275, 37011–37020 [DOI] [PubMed] [Google Scholar]
- 34. Zhang X., Shu L., Hosoi H., Murti K. G., Houghton P. J. (2002) Predominant nuclear localization of mammalian target of rapamycin in normal and malignant cells in culture. J. Biol. Chem. 277, 28127–28134 [DOI] [PubMed] [Google Scholar]
- 35. Zinzalla V., Stracka D., Oppliger W., Hall M. N. (2011) Activation of mTORC2 by association with the ribosome. Cell 144, 757–768 [DOI] [PubMed] [Google Scholar]
- 36. Zhang L., Tie Y., Tian C., Xing G., Song Y., Zhu Y., Sun Z., He F. (2006) CKIP-1 recruits nuclear ATM partially to the plasma membrane through interaction with ATM. Cell. Signal. 18, 1386–1395 [DOI] [PubMed] [Google Scholar]
- 37. Huth J. R., Bewley C. A., Jackson B. M., Hinnebusch A. G., Clore G. M., Gronenborn A. M. (1997) Design of an expression system for detecting folded protein domains and mapping macromolecular interactions by NMR. Protein Sci. 6, 2359–2364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Koenig B. W., Rogowski M., Louis J. M. (2003) A rapid method to attain isotope labeled small soluble peptides for NMR studies. J. Biomol. NMR 26, 193–202 [DOI] [PubMed] [Google Scholar]
- 39. Sommer L. A., Meier M. A., Dames S. A. (2012) A fast and simple method for probing the interaction of peptides and proteins with lipids and membrane-mimetics using GB1 fusion proteins and NMR spectroscopy. Protein Sci. 21, 1566–1570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Stafford R. E., Fanni T., Dennis E. A. (1989) Interfacial properties and critical micelle concentration of lysophospholipids. Biochemistry 28, 5113–5120 [DOI] [PubMed] [Google Scholar]
- 41. Weschayanwiwat P., Scamehorn J. F., Reilly P. J. (2005) Surfactant properties of low molecular weight phospholipids. J. Surfactants Deterg. 8, 65–72 [Google Scholar]
- 42. Tausk R. J., Karmiggelt J., Oudshoorn C., Overbeek J. T. (1974) Physical chemical studies of short-chain lecithin homologues. I. Influence of the chain length of the fatty acid ester and of electrolytes on the critical micelle concentration. Biophys. Chem. 1, 175–183 [DOI] [PubMed] [Google Scholar]
- 43. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 44. Johnson B. A. (2004) Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol. Biol. 278, 313–352 [DOI] [PubMed] [Google Scholar]
- 45. Vuister G. W., Bax A. (1993) Quantitative J correlation: a new approach for measuring homonuclear three-bond J (HNH.α.) coupling constants in 15N-enriched proteins. J. Am. Chem. Soc. 115, 7772–7777 [Google Scholar]
- 46. Wishart D. S., Bigam C. G., Holm A., Hodges R. S., Sykes B. D. (1995) 1H, 13C, and 15N random coil NMR chemical shifts of the common amino acids. I. Investigations of nearest-neighbor effects. J. Biomol. NMR 5, 67–81 [DOI] [PubMed] [Google Scholar]
- 47. Schanda P., Kupce E., Brutscher B. (2005) SOFAST-HMQC experiments for recording two-dimensional heteronuclear correlation spectra of proteins within a few seconds. J. Biomol. NMR 33, 199–211 [DOI] [PubMed] [Google Scholar]
- 48. Wimley W. C., White S. H. (1996) Experimentally determined hydrophobicity scale for proteins at membrane interfaces. Nat. Struct. Biol. 3, 842–848 [DOI] [PubMed] [Google Scholar]
- 49. Liu W., Caffrey M. (2006) Interactions of tryptophan, tryptophan peptides, and tryptophan alkyl esters at curved membrane interfaces. Biochemistry 45, 11713–11726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lazaridis T., Mallik B., Chen Y. (2005) Implicit solvent simulations of DPC micelle formation. J. Phys. Chem. B 109, 15098–15106 [DOI] [PubMed] [Google Scholar]
- 51. Tieleman D. P., van der Spoel D., Berendsen H. J. C. (2000) Molecular dynamics simulations of dodecylphosphocholine micelles at three different aggregate sizes: Micellar structure and chain relaxation. J. Phys. Chem. B 104, 6380–6388 [Google Scholar]
- 52. Whiles J. A., Deems R., Vold R. R., Dennis E. A. (2002) Bicelles in structure-function studies of membrane-associated proteins. Bioorg. Chem. 30, 431–442 [DOI] [PubMed] [Google Scholar]
- 53. Tausk R. J., van Esch J., Karmiggelt J., Voordouw G., Overbeek J. T. (1974) Physical chemical studies of short-chain lecithin homologues. II. Micellar weights of dihexanoyl- and diheptanoyllecithin. Biophys. Chem. 1, 184–203 [DOI] [PubMed] [Google Scholar]
- 54. Seewald M. J., Pichumani K., Stowell C., Tibbals B. V., Regan L., Stone M. J. (2000) The role of backbone conformational heat capacity in protein stability: temperature dependent dynamics of the B1 domain of streptococcal protein G. Protein Sci. 9, 1177–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Walsh J. D., Meier K., Ishima R., Gronenborn A. M. (2010) NMR studies on domain diffusion and alignment in modular GB1 repeats. Biophys. J. 99, 2636–2646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Dames S. A., Junemann A., Sass H. J., Schönichen A., Stopschinski B. E., Grzesiek S., Faix J., Geyer M. (2011) Structure, dynamics, lipid binding, and physiological relevance of the putative GTPase-binding domain of Dictyostelium formin C. J. Biol. Chem. 286, 36907–36920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Farrow N. A., Zhang O., Forman-Kay J. D., Kay L. E. (1997) Characterization of the backbone dynamics of folded and denatured states of an SH3 domain. Biochemistry 36, 2390–2402 [DOI] [PubMed] [Google Scholar]
- 58. Sun Y., Jiang X., Chen S., Fernandes N., Price B. D. (2005) A role for the Tip60 histone acetyltransferase in the acetylation and activation of ATM. Proc. Natl. Acad. Sci. U.S.A. 102, 13182–13187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Nakada D., Hirano Y., Tanaka Y., Sugimoto K. (2005) Role of the C terminus of Mec1 checkpoint kinase in its localization to sites of DNA damage. Mol. Biol. Cell 16, 5227–5235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Fang Y., Vilella-Bach M., Bachmann R., Flanigan A., Chen J. (2001) Phosphatidic acid-mediated mitogenic activation of mTOR signaling. Science 294, 1942–1945 [DOI] [PubMed] [Google Scholar]
- 61. Foster D. A. (2009) Phosphatidic acid signaling to mTOR: signals for the survival of human cancer cells. Biochim. Biophys. Acta 1791, 949–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Veverka V., Crabbe T., Bird I., Lennie G., Muskett F. W., Taylor R. J., Carr M. D. (2008) Structural characterization of the interaction of mTOR with phosphatidic acid and a novel class of inhibitor: compelling evidence for a central role of the FRB domain in small molecule-mediated regulation of mTOR. Oncogene 27, 585–595 [DOI] [PubMed] [Google Scholar]
- 63. Rodriguez Camargo D. C., Link N. M., Dames S. A. (2012) The FKBP-rapamycin binding domain of human TOR undergoes strong conformational changes in the presence of membrane mimetics with and without the regulator phosphatidic acid. Biochemistry 51, 4909–4921 [DOI] [PubMed] [Google Scholar]
- 64. Chen R. Q., Yang Q. K., Chen Y. L., Oliveira V. A., Dalton W. S., Fearns C., Lee J. D. (2009) Kinome siRNA screen identifies SMG-1 as a negative regulator of hypoxia-inducible factor-1α in hypoxia. J. Biol. Chem. 284, 16752–16758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. González-Estévez C., Felix D. A., Smith M. D., Paps J., Morley S. J., James V., Sharp T. V., Aboobaker A. A. (2012) SMG-1 and mTORC1 act antagonistically to regulate response to injury and growth in planarians. PLoS Genet. 8, e1002619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Spagnolo L., Rivera-Calzada A., Pearl L. H., Llorca O. (2006) Three-dimensional structure of the human DNA-PKcs/Ku70/Ku80 complex assembled on DNA and its implications for DNA DSB repair. Mol. Cell 22, 511–519 [DOI] [PubMed] [Google Scholar]
- 67. Rivera-Calzada A., Maman J. D., Spagnolo L., Pearl L. H., Llorca O. (2005) Three-dimensional structure and regulation of the DNA-dependent protein kinase catalytic subunit (DNA-PKcs). Structure 13, 243–255 [DOI] [PubMed] [Google Scholar]
- 68. Adami A., García-Alvarez B., Arias-Palomo E., Barford D., Llorca O. (2007) Structure of TOR and its complex with KOG1. Mol. Cell 27, 509–516 [DOI] [PubMed] [Google Scholar]
- 69. Myers G. A., Gacek D. A., Peterson E. M., Fox C. B., Harris J. M. (2012) Microscopic rates of peptide-phospholipid bilayer interactions from single-molecule residence times. J. Am. Chem. Soc. 134, 19652–19660 [DOI] [PubMed] [Google Scholar]
- 70. Lange O. F., Lakomek N. A., Farès C., Schröder G. F., Walter K. F., Becker S., Meiler J., Grubmüller H., Griesinger C., de Groot B. L. (2008) Recognition dynamics up to microseconds revealed from an RDC-derived ubiquitin ensemble in solution. Science 320, 1471–1475 [DOI] [PubMed] [Google Scholar]
- 71. Sun H., Greathouse D. V., Andersen O. S., Koeppe R. E., 2nd. (2008) The preference of tryptophan for membrane interfaces: insights from N-methylation of tryptophans in gramicidin channels. J. Biol. Chem. 283, 22233–22243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kutateladze T. G., Capelluto D. G., Ferguson C. G., Cheever M. L., Kutateladze A. G., Prestwich G. D., Overduin M. (2004) Multivalent mechanism of membrane insertion by the FYVE domain. J. Biol. Chem. 279, 3050–3057 [DOI] [PubMed] [Google Scholar]
- 73. Buerger C., DeVries B., Stambolic V. (2006) Localization of Rheb to the endomembrane is critical for its signaling function. Biochem. Biophys. Res. Commun. 344, 869–880 [DOI] [PubMed] [Google Scholar]
- 74. Bai X., Ma D., Liu A., Shen X., Wang Q. J., Liu Y., Jiang Y. (2007) Rheb activates mTOR by antagonizing its endogenous inhibitor, FKBP38. Science 318, 977–980 [DOI] [PubMed] [Google Scholar]
- 75. Sancak Y., Bar-Peled L., Zoncu R., Markhard A. L., Nada S., Sabatini D. M. (2010) Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 141, 290–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Guo Z., Kozlov S., Lavin M. F., Person M. D., Paull T. T. (2010) ATM activation by oxidative stress. Science 330, 517–521 [DOI] [PubMed] [Google Scholar]
- 77. Wouters B. G., Koritzinsky M. (2008) Hypoxia signalling through mTOR and the unfolded protein response in cancer. Nat. Rev. Cancer 8, 851–864 [DOI] [PubMed] [Google Scholar]
- 78. Chen B. P., Li M., Asaithamby A. (2012) New insights into the roles of ATM and DNA-PKcs in the cellular response to oxidative stress. Cancer Lett. 327, 103–110 [DOI] [PubMed] [Google Scholar]
- 79. Pan Y., Nishida Y., Wang M., Verdin E. (2012) Metabolic regulation, mitochondria and the life-prolonging effect of rapamycin: a mini-review. Gerontology 58, 524–530 [DOI] [PubMed] [Google Scholar]
- 80. Dazert E., Hall M. N. (2011) mTOR signaling in disease. Curr. Opin. Cell Biol. 23, 744–755 [DOI] [PubMed] [Google Scholar]
- 81. Sun Y., Xu Y., Roy K., Price B. D. (2007) DNA damage-induced acetylation of lysine 3016 of ATM activates ATM kinase activity. Mol. Cell. Biol. 27, 8502–8509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Gouet P., Courcelle E., Stuart D. I., Métoz F. (1999) ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics 15, 305–308 [DOI] [PubMed] [Google Scholar]
- 83. Wishart D. S., Sykes B. D., Richards F. M. (1992) The chemical shift index: a fast and simple method for the assignment of protein secondary structure through NMR spectroscopy. Biochemistry 31, 1647–1651 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.