Abstract
Proteomics-based clinical studies have been shown to be promising strategies for the discovery of novel biomarkers of a particular disease. Here, we present a study of hepatocellular carcinoma (HCC) that combines complementary two-dimensional difference in gel electrophoresis (2D-DIGE) and liquid chromatography (LC-MS)-based approaches of quantitative proteomics. In our proteomic experiments, we analyzed a set of 14 samples (7 × HCC versus 7 × nontumorous liver tissue) with both techniques. Thereby we identified 573 proteins that were differentially expressed between the experimental groups. Among these, only 51 differentially expressed proteins were identified irrespective of the applied approach. Using Western blotting and immunohistochemical analysis the regulation patterns of six selected proteins from the study overlap (inorganic pyrophosphatase 1 (PPA1), tumor necrosis factor type 1 receptor-associated protein 1 (TRAP1), betaine-homocysteine S-methyltransferase 1 (BHMT)) were successfully verified within the same sample set. In addition, the up-regulations of selected proteins from the complements of both approaches (major vault protein (MVP), gelsolin (GSN), chloride intracellular channel protein 1 (CLIC1)) were also reproducible. Within a second independent verification set (n = 33) the altered protein expression levels of major vault protein and betaine-homocysteine S-methyltransferase were further confirmed by Western blots quantitatively analyzed via densitometry. For the other candidates slight but nonsignificant trends were detectable in this independent cohort. Based on these results we assume that major vault protein and betaine-homocysteine S-methyltransferase have the potential to act as diagnostic HCC biomarker candidates that are worth to be followed in further validation studies.
Hepatocellular carcinoma (HCC)1 currently is the fifth most common malignancy worldwide with an annual incidence up to 500 per 100,000 individuals depending on the geographic region investigated. Whereas 80% of new cases occur in developing countries, the incidence increases in industrialized nations including Western Europe, Japan, and the United States (1). To manage patients with HCC, tumor markers are very important tools for diagnosis, indicators of disease progression, outcome prediction, and evaluation of treatment efficacy. Several tumor markers have been reported for HCC, including α-fetoprotein (AFP) (2), Lens culinaris agglutinin-reactive fraction of AFP (AFP-L3) (3), and des-γ-carboxyl prothrombin (DCP) (4). However, none of these tumor markers show 100% sensitivity or specificity, which calls for new and better biomarkers.
To identify novel biomarkers of HCC, many clinical studies using “omics”-based methods have been reported over the past decade (5–6). In particular, the proteomics-based approach has turned out to be a promising one, offering several quantification techniques to reveal differences in protein expression that are caused by a particular disease. In most studies, the well-established 2D-DIGE technique has been applied for protein quantification followed by identification via mass spectrometry (7–15). Even if the quantification is very accurate and sensitive in this gel-based approach, the relatively high amount of protein sample necessary for protein identification is the major disadvantage of this technique. Several mass-spectrometry-based quantitative studies using labeling-techniques like SILAC (stable isotope labeling by amino acids in cell culture) or iTRAQ (isobaric tags for relative and absolute quantification) have also been carried out for biomarker discovery of HCC (16–18). Here, the concomitant protein quantification and identification in a mass spectrometer allows high-throughput analyses. However, such experiments imply additional labeling reactions (in case of iTRAQ) or are limited to tissue culture systems (in case of SILAC). In the latter case, one can overcome the limitation by using the isotope-labeled proteins obtained from tissue culture as an internal standard added to a corresponding tissue sample. This approach is known as CDIT (culture-derived isotope tags) and was applied in a HCC study, very recently (19). Label-free proteomics approaches based on quantification by ion-intensities or spectral counting offer another possibility for biomarker discovery. These approaches are relatively cheap compared with the labeling approaches, because they do not require any labeling reagents and furthermore they allow for high-throughput and sensitive analyses in a mass spectrometer. A quantitative study of HCC using spectral counting has been reported (20), whereas to our knowledge an ion-intensity-based study has not been performed yet. Apart from these quantification strategies, protein alterations in HCC have been studied by MALDI imaging, as well. Here, the authors could show that based on its proteomic signature, hepatocellular carcinoma can be discriminated with high accuracy from liver metastasis samples or other cancer types (21) as well as liver cirrhosis (22). Based on these results, it could be assumed that MALDI imaging might be a promising alternative to standard histological methods in the future.
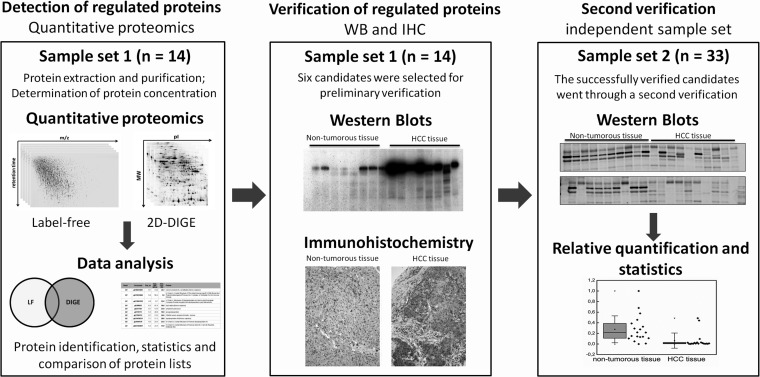
Here, we report a quantitative proteomic study that combines two different techniques, namely the well-established 2D-DIGE approach and a label-free ion-intensity-based quantification via mass spectrometry and liquid chromatography. To our knowledge this is the first time such a combined study was performed with regard to hepatocellular carcinoma. By comparing the results of both studies, we aim to identify high-confident biomarker candidates of HCC, as gel- and LC-MS-based techniques are complementary. To verify the differential protein expressions detected in our proteomic studies we performed additional immunological verifications for selected proteins within two different sample sets (Fig. 1).
Fig. 1.
Schematic representation of the applied workflow.
METHODS
Clinical Data
Tissue samples from hepatocellular carcinoma and nontumorous liver were collected from 26 patients suffering from primary liver cancer (19 males and 7 females). Normal liver and HCC tumor samples were acquired in the course of hepatic resections or liver transplantations. The age of the patients ranged from 21 years to 76 years (mean 62.1). The tumors were classified according to the pathologic TNM (pTNM) system (seventh edition) (23). The tumor grading ranged from G1 to G3, and all tumors showed clear surgical margins. None of the patients in the first sample set that was used for the quantitative proteomics study had liver cirrhosis or hepatitis B or C infection. Further details about patients and tumor characteristics for all samples used during discovery and verification phases are shown in Table I. Informed consent was obtained from every patient and the study protocol conforms to the ethical guidelines of the 1975 Declaration of Helsinki.
Table I. Patient and tumor characteristics. NASH, non alcoholic steatohepatitis; HepB, hepatitis B infection; HepC, hepatitis C infection; n.k., not known; NX, Regional lymph nodes cannot be assessed.
| ID | Gender | Age | T | N | G | V | R | Underlying liver disease |
|---|---|---|---|---|---|---|---|---|
| Discovery and verification set | ||||||||
| 1 | Female | 57 | T1 | N0 | G3 | V0 | R0 | n.k. |
| 2 | Female | 42 | T1 | NX | G3 | V0 | R0 | n.k. |
| 3 | Male | 51 | T1 | N0 | G1 | V0 | R0 | n.k. |
| 4 | Male | 21 | T2 | N1 | G2 | V1 | R0 | n.k. |
| 5 | Male | 71 | T1 | NX | G2–3 | V0 | R0 | NASH |
| 6 | Male | 69 | T1 | NX | G1 | V0 | R0 | n.k. |
| 7a | Male | 65 | T1 | NX | G2–3 | V0 | R0 | NASH |
| 8b | Male | 74 | T1 | NX | G1 | V0 | R0 | n.k. |
| Independent verification set | ||||||||
| 9c | Female | 59 | T3 | NX | G2 | V0 | R0 | n.k. |
| 10 | Female | 76 | T2 | NX | G2–3 | V1 | R0 | n.k. |
| 11 | Male | 53 | T3 | NX | G2 | V1 | R0 | HepC |
| 12 | Male | 62 | T2 | NX | G2 | V1 | R0 | HepC |
| 13 | Male | 72 | T1 | n.k. | G1 | n.k. | R1 | HepB |
| 14 | Female | 67 | T1 | N0 | G1 | V0 | R0 | n.k. |
| 15 | Male | 75 | T1 | N0 | G1 | V0 | R0 | HepB, liver cirrhosis |
| 16d | Male | 65 | T2 | NX | G3 | V1 | R1 | n.k. |
| 17c | Male | 67 | T3 | NX | G3 | V0 | R0 | n.k. |
| 18 | Male | 57 | T2 | NX | G2 | V0 | R0 | Liver cirrhosis |
| 19 | Male | 66 | T1 | NX | G2 | V0 | R0 | Liver cirrhosis |
| 20 | Female | 56 | T3 | NX | G3 | V0 | R0 | n.k. |
| 21 | Male | 76 | T1 | NX | G2 | V0 | R0 | n.k. |
| 22 | Female | 68 | T3 | N0 | G2 | V1 | RX | n.k. |
| 23 | Male | 37 | T2 | N0 | G2 | V1 | R0 | HepB |
| 24 | Male | 71 | T2 | NX | G2 | V0 | R0 | n.k. |
| 25 | Male | 65 | T1 | NX | G2 | V0 | R0 | HepC |
| 26 | Male | 73 | T1 | NX | G2 | V0 | R0 | HepC |
a From this patient only tumor tissue was used in the proteomic study.
b From this patient only nontumor tissue was used in the proteomic study.
c Tumor tissue sample of this patient was not included in the validation because of sample degradation as shown by 1D-PAGE.
d Nontumor tissue sample of this patient was not included in the validation due to sample degradation as shown by 1D-PAGE.
Tissue Preparation
Liver tumor and nontumor tissue were collected and fixed in 4% buffered formalin, paraffin embedded and prepared for pathological examination and immunohistochemical evaluation. For the proteomics study and the verification via Western blotting, the samples were immediately placed on ice, snap-frozen and stored at −80 °C. The tissue samples were lysed by sonication (6 × 10s pulses on ice) in sample buffer (30 mm Tris HCl; 2 m thiourea; 7 m urea; 4% 3-[(3-cholamidopropyl)dimethylammonio]propanesulfonate, pH 8.5). After centrifugation at 15,000 × g for 5 min the supernatant was collected and protein concentration was determined using the Bio-Rad Protein Assay (Bio-Rad, Hercules, CA).
2D-DIGE Analysis
Protein Labeling
Proteins were labeled using cyanine dyes in the ratio 50 μg protein to 400 pmol dyes (minimal labeling dyes (GE Healthcare, Munich, Germany)). The labeling reaction was performed according to the manufacturer's instructions. Samples of HCC-tissue and nontumor tissue were randomized by labeling with Cy3 dye or Cy5 dye to avoid any dye biases. The internal standard, which is a mixture of same amounts of all analyzed samples, was labeled with Cy2 dye.
2D Electrophoresis
Before 2D electrophoresis the seven sample mixtures including appropriate Cy3- and Cy5-labeled pairs and a Cy2-labeled internal standard were generated. The corresponding sample pairs were mixed (each labeled 50 μg) together and 50 μg of labeled internal standard were added to each of the seven mixtures. Per 100 μl cell lysate, 10 μl dithiotreitol (DTT) (1.08 g/ml) and 10 μl Ampholine 2–4 (GE Healthcare, Munich, Germany) were added. For the isoelectric focusing (IEF) the whole 150 μg per mixture were loaded. The focusing was performed using tube gels (20 cm × 0.9 mm) containing carrier ampholytes (CA-IEF). A voltage gradient was applied in an IEF-chamber produced in house (24). After IEF, the ejected tube gels were incubated in equilibration buffer (125 mm Tris, 40% (w/v) glycerol, 3% (w/v) SDS, 65 mm DTT, pH 6.8) for 10 min. The second dimension (SDS-PAGE) was performed on polyacrylamide gels (15.2% total acrylamide, 1.3% bisacrylamide) using a Desaphor VA 300 system. IEF tube gels were placed onto the polyacrylamide gels (20 cm × 30 cm × 0.7 mm) and fixed using 1.0% (w/v) agarose containing 0.01% (w/v) bromphenol blue dye (Riedel de-Haen, Seelze, Germany). For identification of proteins by MS, an amount of 250 μg total protein of the internal standard was applied to IEF tube gels (20 cm × 1.5 mm) and subsequently to preparative SDS-PAGE gels (20 cm × 30 cm × 1.5 mm). Silver post-staining was performed after gel scanning using a MS-compatible protocol as described elsewhere (25). Gel images of the Cy3- and Cy5-labeled samples as well as an example of a Cy2-labeled internal standard are shown in supplemental Data S6.
Scanning, Image Analysis, and Statistics
SDS-PAGE gels were scanned using a Typhoon 9400 scanner (GE Healthcare, Munich, Germany). Excitation and emission wavelengths were chosen specifically for each of the dyes according to recommendations of the manufacturer. Images were preprocessed using the ImageQuantTM software (GE Healthcare, Munich, Germany). Intragel spot detection, inter-gel matching, and normalization of spot intensities were performed using the Differential In-gel Analysis (DIA) mode and Biological Variation Analysis (BVA) mode of DeCyder 2DTM software (GE Healthcare, Munich, Germany), respectively. Spot intensities were normalized to the internal standard. The Extended Data Analysis tool (EDA), implemented in the DeCyder 2DTM software package was used for the statistical analysis of the 2D-DIGE experiments. Here, only spots appearing in at least 70% of all analyzed and matched spot maps were chosen for further analysis. Significantly regulated proteins were identified by Student's t test including a false-discovery-rate correction. Protein spots differentially expressed (p ≤ 0.05, Average Ratio ≥ 1.5) between HCC and nontumor samples were identified using MALDI-TOF-MS or nano-HPLC-ESI-MS/MS.
Digestion and Protein Identification
In-gel digestion of proteins was performed with trypsin following standard protocols and the obtained peptides were extracted from the gel matrix (26). MALDI-TOF-MS analyses were performed on an UltraFlexTM II instrument (Bruker Daltonics, Bremen, Germany). For nano-HPLC-ESI-MS/MS experiments an Ultimate 3000 RSLCnano system (Dionex, Idstein, Germany) online coupled to a Bruker Daltonics HCT plus ion trap instrument equipped with a nanoelectrospray ion source (Bruker Daltonics, Bremen, Germany) was used. For protein identification, database searches against IPI human database (ver. 3.87, #sequences with shuffled decoy sequences: 91,464) were performed using Mascot (ver. 2.3.0.2) (Matrix Science Ltd., London, UK) (27). Further details regarding the experimental setup, search parameters or identification threshold were described earlier (26). PMF spectra of identified proteins and peak lists containing peptide annotations are listed in supplemental Data S1.
Label-free Analysis
In-gel Digestion and Sample Preparation
Before LC-MS analysis, 5 μg of each protein sample were loaded on a 4–20% SDS-PAGE gel (Anamed, Groβ-Bieberau, Germany) and allowed to run into the gel for about 1 cm (15 min at 50 V). After Coomassie-staining, in-gel trypsin digestion was performed following a previously described protocol (26). The generated peptides were extracted by sonication (15 min, ice cooling) of the gel pieces in ∼20 μl of 50% acetonitrile in 0.1% trifluoroacetic acid, twice. Afterward, acetonitrile was removed by vacuum centrifugation. Peptide concentration of the resulting solution was determined by amino acid analysis performed on an ACQUITY-UPLC with an AccQ Tag Ultra-UPLC column (Waters, Eschborn, Germany) calibrated with Pierce Amino Acid Standard (Thermo Scientific, Bremen, Germany). Prior to LC-MS analysis, samples were diluted with 0.1% TFA to adjust a peptide concentration of 23.3 ng/μl.
LC-MS/MS Analysis
Quantitative label-free analyses were performed on an Ultimate 3000 RSLCnano system (Dionex, Idstein, Germany) online coupled to a LTQ Orbitrap Velos instrument (Thermo Scientific, Bremen, Germany). For each analysis 15 μl of sample were injected, corresponding to an amount of 350 ng tryptic digested proteins. The peptides were preconcentrated with 0.1% TFA on a trap column (Acclaim® PepMap 100, 75 μm × 2 cm, nano Viper, C18, 2 μm, 100 Å) at a flow rate of 7 μl/min for 10 min. Subsequently, the peptides were transferred to the analytical column (Acclaim® PepMap RSLC, 75 μm × 50 cm, nano Viper, C18, 2 μm, 100 Å) and separated using a 90 min gradient from 5–40% solvent B at a flow rate of 300 nl/min (solvent A: 0.1% formic acid, solvent B: 0.1% formic acid 84% acetonitrile). The column oven temperature was set to 60 °C. The mass spectrometer was operated in a data-dependent mode. Full scan MS spectra were acquired at a mass resolution of 30,000 in the Orbitrap analyzer. Tandem mass spectra of the twenty most abundant peaks were acquired in the linear ion trap by peptide fragmentation using collision-induced dissociation.
Peptide Quantification and Filtering
Progenesis LC-MSTM software (version 4.0.4265.42984, Nonlinear Dynamics Ltd., Newcastle upon Tyne, UK) was used for the ion-intensity-based label-free quantification. After importing the .raw files, one sample was selected as a reference run to which the retention times of the precursor masses in all other samples were aligned to. In the following, a list of features was generated including the m/z values of all eluted peptides at given retention times. For further analysis, only features comprising charges of 2+ and 3+ were selected. Subsequently, the raw abundances of each feature were automatically normalized for correcting experimental variations. In this step, for all features in a LC-MS run quantitative abundance ratios between the run to be aligned and a reference run are calculated and logarithmized. Logarithmic transformations are necessary at this point to ensure that up- and down-regulated features are taken into account to the same extent. The anti-log of the average of these log(ratio) values gives a global scaling factor for the particular LC-MS run that is then used to align the run and to correct experimental and technical variations. Such as, differences in protein quantity loaded into the instrument or differences in ionization. In a following step, the samples were grouped corresponding to the selected experimental design, in this case a two-group comparison between “nontumor” and “HCC.” Differences of peptide abundances between both groups were assigned to be significant if the following filter criteria were satisfied in the following statistical analysis: ANOVA p value ≤ 0.05 and q-value ≤ 0.05. Because of the fact, that multiple MS/MS spectra were acquired for the same features, only fragment-ion spectra of the ten most intense precursors of a feature were selected for the generation of a peak list exported to a Mascot generic file.
Protein Identification
The generated .mgf file was searched against IPI human database (ver. 3.87, #sequences with shuffled decoy sequences: 91,464) using Mascot (ver. 2.3.0.2) (Matrix Science Ltd., London, UK) (27). The following search parameters were applied: variable modifications propionamide (C) and oxidation (M), tryptic digestion with up to one missed cleavage, 13C = 1, precursor ion mass tolerance of 5 ppm and fragment ion mass tolerance of 0.4 Da. For further analysis, only peptides with mascot ion scores >36 (p ≤ 0.01 identity threshold) were chosen. Above this threshold a false discovery rate of <1% (67 decoy hits within 6909 PSMs) was estimated. By importing the list of identified peptides in Progenesis LC-MSTM, the previously quantified features were matched to the corresponding peptides. A summary of the search results and detailed information concerning fragment assignments in the case of single-peptide-based protein identifications are listed in supplemental Data S2 and S3, respectively. Additionally, results of protein searches are shared via the PRoteomics IDEntifications database (PRIDE) (28).
Protein Quantification and Filtering
For the protein quantification, only nonconflicting peptides (i.e., peptides occurring in only one protein) were chosen, and the protein-grouping function implemented in Progenesis LC-MSTM was disabled. At the protein level, the significance of expression changes was again tested by calculating an ANOVA p value and a q-value. Proteins not satisfying the significance criteria (ANOVA p value ≤ 0.05 and q-value ≤ 0.05) were filtered out. Finally, proteins showing less than 1.5-fold change of expression were discarded as well.
Analysis of Regulated Proteins
Ingenuity Pathway Analysis software (Version 12402621, Ingenuity Systems, www.ingenuity.com) was used to assign the localizations as well as the molecular and cellular functions of the regulated proteins.
Western Blotting
Western Blots (ECL)
Prior to Western blotting, protein concentration of the samples was determined by amino acid analysis. Equal amounts of 15 μg protein per sample were separated by SDS-PAGE on a 4–20% polyacrylamide gel (Criterion TGX, Bio-Rad, Hercules, CA). Proteins were subsequently transferred onto nitrocellulose membrane (Trans-Blot Turbo, Bio-Rad) and membranes were blocked with StartingBlock blocking buffer (Thermo Scientific, Bremen, Germany) for one hour at room temperature. Primary antibodies anti-CLIC1 (Clone 2D4, Abnova, Heidelberg, Germany, dilution 1:1000), anti-MVP (Clone 1032, Acris, Herford, Germany, dilution 1:1000), anti-PPA1 (ab96099, Abcam, Cambridge, UK, dilution 1:5000), anti-TRAP1 (clone EPR5381, Abcam, Cambridge, UK, dilution 1:15000), anti-GSN (clone GS-2C4, Sigma-Aldrich, Munich, Germany, dilution 1:1000), and anti-BHMT (clone EPR6782, Epitomics, Burlingame, CA, dilution 1:20000) were diluted in StartingBlock and incubated with membranes over night at 4 °C. Horseradish peroxidase-labeled secondary antibodies (Jackson ImmunoResearch, Newmarket, UK) were used for detection for one hour at room temperature. Bound antibodies were visualized by enhanced chemiluminescence and exposure to hyperfilm (GE Healthcare, Munich, Germany).
Densitometric Analysis
For densitometric analysis of Western blots, infrared fluorescence detection was performed using IRDye®-labeled secondary antibodies (Rockland, Gilbertsville, USA) with the same incubation conditions as described above. Odyssey imaging system was used for detection, and densitometric analysis was done with Odyssey application software version 3.0.21 (both from Li-Cor Biosciences, Lincoln, NE). For densitometric analysis, total lane background was subtracted from measured band average intensity. Equal loading amounts were verified by Coomassie staining (supplemental Data S7). No normalization to a housekeeping protein was performed, because the commonly used housekeeping proteins (i.e., β-actin, β-tubulin, GAPDH) were identified in the proteomics studies with expression changes higher than 1.5-fold between the experimental groups.
Immunohistochemistry
Paraffin embedded 4 μm slides were dewaxed and pretreated in EDTA buffer (pH 9) at 95 °C for 30 min. All Immunohistochemical stains were performed with an automated staining device (Dako Autostainer, Glostrup, Denmark). Both, the source of the primary antibodies and the technical staining details of the automatically performed stainings are listed in Table II. All stains were developed using Polymer Kit (ZytoChemPlus (HRP), POLHRS-100, Zytomed Systems). Replacement of the various primary antibodies by mouse or rabbit immunoglobulin served as negative controls.
Table II. Antibodies used for immunohistochemistry. AB, antibody; RT, room temperature.
| Antibody | Distributor | Code | Source | AB concentration |
|---|---|---|---|---|
| TRAP 1 | abcam | ab109323 | Rabbit monoclonal | 1:200, 30 min. RT |
| LRP/MVP | Kamiya | MC-603 | Mouse monoclonal | 1:100, 30 min. RT |
| Pyrophosphatase-1 | abcam | ab96099 | Rabbit polyclonal | 1:500, 30 min. RT |
| CLIC1 | Abnova | H00001192-M01 | Mouse monoclonal | 1:9000, 30 min. RT |
| Gelsolin | Sigma | G4896 | Mouse monoclonal | 1:3000; 30 min. RT |
| BHMT | abcam | Ab124992 | Rabbit monoclonal | 1:100; 30 min. RT |
RESULTS
Quantitative Proteomic Analysis
To identify novel biomarker candidates of hepatocellular carcinoma we performed a study that combined two complementary techniques of quantitative proteomics, namely the gel-based 2D-DIGE and the label-free LC-MS-based approaches. Following a straightforward workflow (Fig. 1), we analyzed the differential protein expression in primary liver cancer tissue (n = 7) in comparison to adjacent nontumorous liver tissue (n = 7).
In the gel-based approach, a total of 1,366 protein spots represented in at least 70% of all investigated spot maps were detected. Of these, only protein spots showing significant expression changes between nontumor and malignant tissue specimens (p ≤ 0.05 and 1.5-fold change of expression) have been isolated and analyzed. By the means of MALDI-MS and nano-LC-ESI-MS/MS analyses 240 proteins (148 nonredundant proteins) were successfully identified. Among these, 55 proteins were found to be up- and 83 proteins down-regulated in HCC tumor tissue. Ten proteins showed variable regulation directions within several detected isoforms (supplemental Data S4).
In the label-free approach 31,673 features comprising charges of 2+ or 3+ were detected. Significant differences in abundance between the two experimental groups were observed for 3507 of the features. Of these, 1038 regulated features have been assigned to peptide matches by the acquired tandem mass spectra. These identifications resulted in 476 significantly regulated proteins of which 284 were found to be up-regulated in tumor tissue and 194 down-regulated, respectively (supplemental Data S5).
In summary, a total of 573 differentially expressed proteins were found, whereas 97 proteins were exclusively identified in the 2D-DIGE study and 425 proteins in the LC-MS study. Hence, only 51 differential proteins were identified irrespective of the applied quantification technique, which clearly shows that both approaches are complementary (Table III). Except of eight proteins, the regulation directions of the proteins identified in both studies were the same. In four of the eight cases of inconsistent regulations, the protein expressions already vary between several isoforms detected in the gel-based approach.
Table III. Regulated proteins identified in both proteomic studies.
| No. | IPI accession | Gene | Protein | Fold changes |
Reported association to HCC |
||
|---|---|---|---|---|---|---|---|
| DIGE | LC-MS | Regulation | References | ||||
| Up-regulated in HCC tissue | |||||||
| 1 | IPI00015018 | PPA1 | Inorganic pyrophosphatase | 2.0 | 5.9 | ↓ | (29) |
| 2 | IPI00448925 | IGHG1;IGHV4–31 | 44 kDa protein | 2.4 | 3.9 | - | - |
| 3 | IPI00553177 | SERPINA1 | Alpha-1-antitrypsin (isoform 1) | 2.7–3.7 | 3.6 | ↑ | (30–35) |
| 4 | IPI00418471 | VIM | Vimentin | 3.1 | 2.9 | ↑ | (29, 36) |
| 5 | IPI00021405 | LMNA | Prelamin-A/C (isoform A) | 2.8–3.7 | 2.7 | - | - |
| 6 | IPI00554788 | KRT18 | Keratin, type I cytoskeletal 18 | 1.7 | 2.4 | ↑ | (37–38) |
| 7 | IPI00219018 | GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | 2.0–3.1 | 2.4 | ↑ | (39–40) |
| 8 | IPI00479186 | PKM2 | Pyruvate kinase isozymes M1/M2 (isoform M2) | 3.2 | 2.3 | ↑ | (41) |
| 9 | IPI00007765 | HSPA9 | HSPA9 Stress-70 protein, mitochondrial | 2.7–2.8 | 2.3 | ↑ | (9, 42–43) |
| 10 | IPI00003362 | HSPA5 | 78 kDa glucose-regulated protein | 3.8 | 2.2 | ↑ | (9, 43–46) |
| 11 | IPI00030275 | TRAP1 | Heat shock protein 75 kDa, mitochondrial | 3.0 | 2.2 | - | - |
| 12 | IPI00017855 | ACO2 | Aconitate hydratase, mitochondrial | 2.3–2.1 | 1.7 | - | - |
| 13 | IPI00003865 | HSPA8 | Heat shock cognate 71 kDa protein (isoform 1) | 1.7–2.7 | 1.6 | ↑ | (9, 43) |
| 14 | IPI00010720 | CCT5 | T-complex protein 1 subunit epsilon | 1.8 | 1.5 | - | - |
| Down-regulated in HCC tissue | |||||||
| 15 | IPI00011416 | ECH1 | Delta(3,5)-Delta(2,4)-dienoyl-CoA isomerase, mitochondrial | −2.0 | −1.5 | - | - |
| 16 | IPI00218733 | SOD1 | Superoxide dismutase [Cu-Zn] | −1.9 | −1.8 | ↓ | (47–49) |
| 17 | IPI00218414 | CA2 | Carbonic anhydrase 2 | −2.3 | −1.8 | ↓ | (50) |
| 18 | IPI00014439 | QDPR | Dihydropteridine reductase | −1.8 | −2.0 | - | - |
| 19 | IPI00009367 | AGXT | Serine–pyruvate aminotransferase | −2.3 | −2.2 | - | - |
| 20 | IPI00216057 | SORD | Sorbitol dehydrogenase | −2.4 | −2.3 | - | - |
| 21 | IPI00016801 | GLUD1 | Glutamate dehydrogenase 1, mitochondrial | −3.4–−1.7 | −2.4 | - | - |
| 22 | IPI00889534 | CPS1 | Carbamoyl-phosphate synthase [ammonia], mitochondrial (isoform a precursor) | −5.1–−4.4 | −2.4 | ↓ | (51) |
| 23 | IPI00024990 | ALDH6A1 | Methylmalonate-semialdehyde dehydrogenase (acylating), mitochondrial | −3.5–−2.1 | −2.4 | ↓ | (52) |
| 24 | IPI00037448 | GRHPR | Glyoxylate reductase/hydroxypyruvate reductase | −1.9 | −2.5 | - | - |
| 25 | IPI00329331 | UGP2 | UTP-glucose-1-phosphate uridylyltransferase (isoform 1) | −2.0 | −2.5 | - | - |
| 26 | IPI00006663 | ALDH2 | Aldehyde dehydrogenase, mitochondrial | −2.5–−2.4 | −2.6 | - | (53) |
| 27 | IPI00024993 | ECHS1 | Enoyl-CoA hydratase, mitochondrial | −3.5–−2.2 | −2.7 | ↓ | (54–55) |
| 28 | IPI00289524 | AKR1C4 | Aldo-keto reductase family 1 member C4 | −2.0 | −2.7 | - | - |
| 29 | IPI00218914 | ALDH1A1 | Retinal dehydrogenase 1 | −1.7 | −2.7 | - | - |
| 30 | IPI00165360 | MPST | 3-Mercaptopyruvate sulfurtransferase | −2.5 | −2.8 | - | - |
| 31 | IPI00020632 | ASS1 | Argininosuccinate synthase | −2.0–−1.8 | −2.8 | ↓ | (56) |
| 32 | IPI00027701 | ACADS | Short-chain specific acyl-CoA dehydrogenase, mitochondrial | −2.1 | −2.8 | - | - |
| 33 | IPI00218407 | ALDOB | Fructose-bisphosphate aldolase B | −3.5–−2.4 | −3.0 | ↓ | (6, 8, 51–52, 57) |
| 34 | IPI00024623 | ACADSB | Short/branched chain specific acyl-CoA dehydrogenase, mitochondrial | −2.1–−1.6 | −3.0 | - | - |
| 35 | IPI00216136 | KHK | Ketohexokinase (isoform C) | −1.7 | −3.2 | ↓ | (8) |
| 36 | IPI00034308 | SARDH | Sarcosine dehydrogenase, mitochondrial | −3.3–−2.8 | −3.3 | ↓ | (58) |
| 37 | IPI00001441 | FTCD | Formimidoyltransferase-cyclodeaminase (isoform A) | −3.4 | −3.3 | ↓ | (13, 59) |
| 38 | IPI00010180 | CES1 | Liver carboxylesterase 1 (isoform 1) | −1.9 | −3.3 | ↓ | (52, 60) |
| 39 | IPI00025341 | BDH1 | d-beta-hydroxybutyrate dehydrogenase, mitochondrial | −2.4 | −3.5 | - | - |
| 40 | IPI00024896 | PBLD | Phenazine biosynthesis-like domain-containing protein | −2.8 | −3.6 | ↓ | (61) |
| 41 | IPI00073772 | FBP1 | Fructose-1,6-bisphosphatase 1 | −2.3–−1.8 | −4.0 | ↓ | (62) |
| 42 | IPI00004101 | BHMT | Betaine-homocysteine S-methyltransferase 1 | −3.7–−3.0 | −5.6 | ↓ | (63–64) |
| 43 | IPI00215925 | GNMT | Glycine N-methyltransferase | −3.1 | −6.3 | ↓ | (64–65) |
| Inconsistent regulation | |||||||
| 44 | IPI00745872 | ALB | Serum albumin (isoform 1) | −3.9–3.8 | 2.4 | ↑↓ | (6) |
| 45 | IPI00419585 | PPIA | Peptidyl-prolyl cis-trans isomerase A | −2.9–1.7 | 1.7 | ↑ | (58, 66) |
| 46 | IPI00218342 | MTHFD1 | C-1-tetrahydrofolate synthase, cytoplasmic | 1.8 | −2.1 | - | - |
| 47 | IPI00030363 | ACAT1 | Acetyl-CoA acetyltransferase, mitochondrial | 1.8 | −2.3 | - | - |
| 48 | IPI00797038 | PCK2 | Phosphoenolpyruvate carboxykinase [GTP], mitochondrial (isoform 1) | 2.1–3.3 | −2.9 | ↓ | (67) |
| 49 | IPI00032103 | GATM | Glycine amidinotransferase (isoform 1), mitochondrial | 1.8 | −3.2 | - | - |
| 50 | IPI00473031 | ADH1B | Alcohol dehydrogenase 1B | −6.3–3.1 | −3.4 | - | - |
| 51 | IPI00218899 | ADH4 | Alcohol dehydrogenase 4 (isoform 2) | −2.9–2.3 | −3.6 | ↑ | (68) |
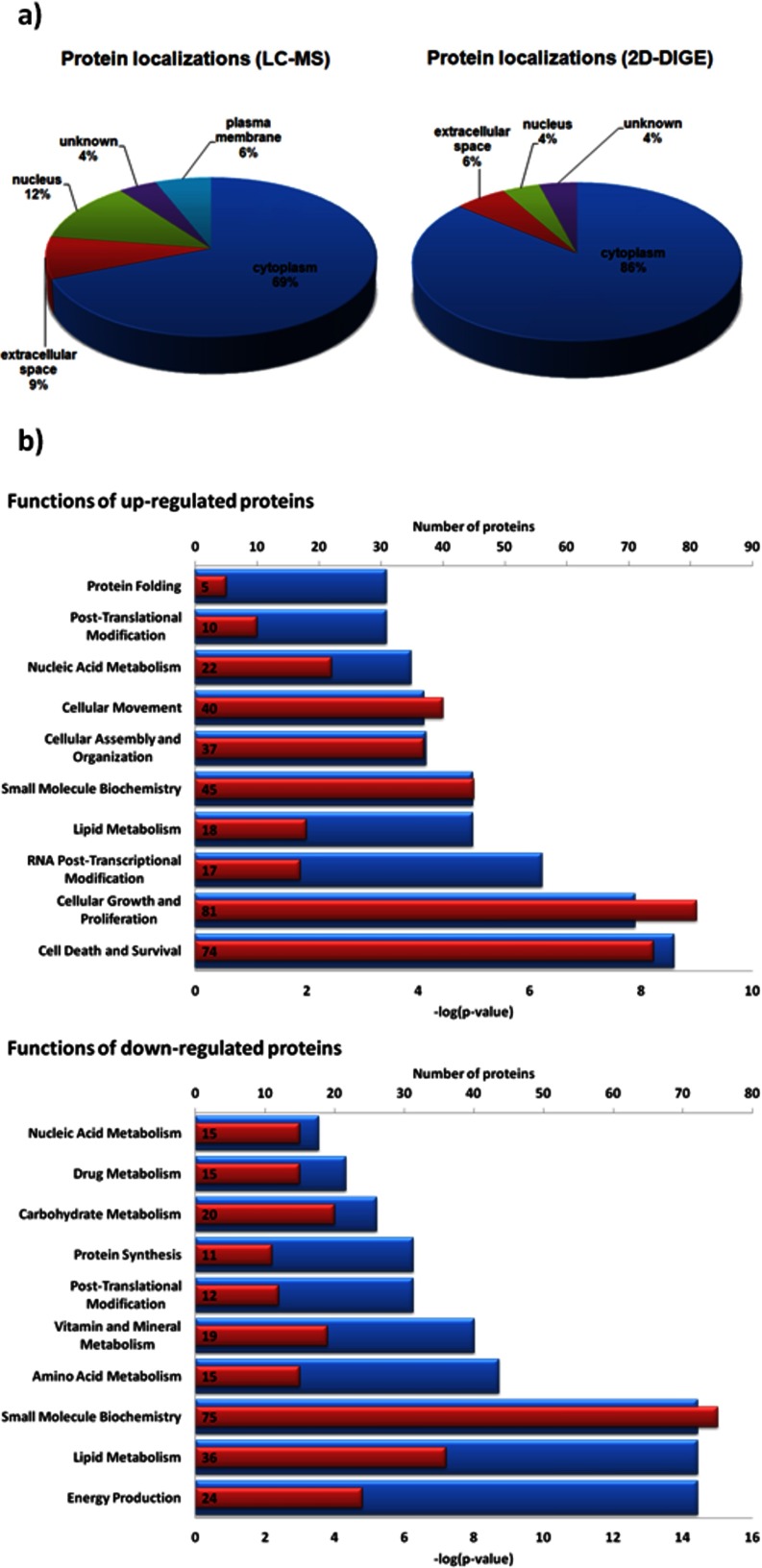
An analysis of the protein localizations revealed, that by using a gel-based approach mainly cytoplasmic proteins were detected. Whereas the proteins detected in label-free approach widespread over a broader range of cellular localizations, in particular the plasma membrane (Fig. 2A). The up-regulated proteins identified in the whole study were found to be mainly involved in molecular and cellular processes like cell death and survival, cellular growth and proliferation as well as RNA post-transcriptional modification. Contrary, the down-regulated proteins are involved in energy production, lipid metabolism, and small molecule biochemistry (Fig. 2B).
Fig. 2.
A, Localizations of the differentially expressed proteins detected in the 2D-DIGE or LC-MS-based approach. B, Molecular and cellular functions of up- and down-regulated proteins identified in the whole study. Numbers of proteins per functional class are shown as red bars including the exact number of annotated proteins. Corresponding p values are shown as blue bars.
Selection of Biomarker Candidates for Further Verification
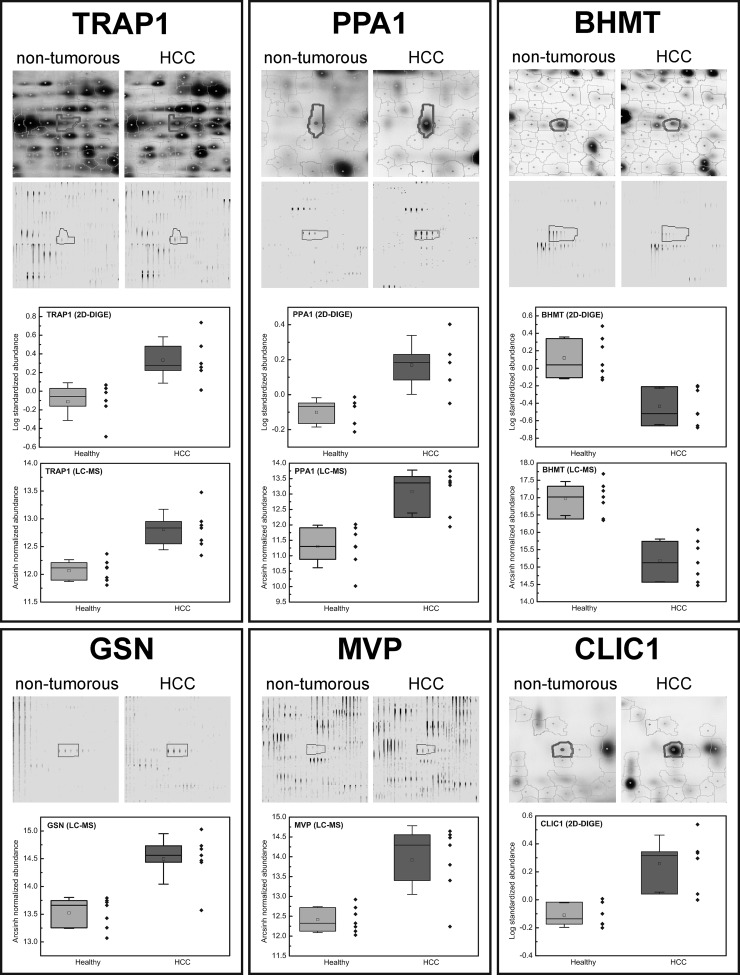
To verify the observed complementarity of the applied techniques and to identify proteins that might act as biomarker candidates for HCC, we have chosen several regulated proteins for further validations that were identified either in the 2D-DIGE study, the label-free study or in both studies. From the proteins exclusively identified in the gel-based 2D-DIGE approach we have chosen chloride intracellular channel protein 1 (CLIC1), comprising a 2.5-fold over-expression in tumor tissue. From the complement of the label-free LC-MS based approach, major vault protein (MVP), which showed a 5.4-fold over-expression based on quantification with six peptides, as well as gelsolin (GSN) with a 2.8-fold higher expression (quantified with three peptides) were selected. The first regulated protein chosen from the overlap of both studies was tumor necrosis factor type 1 receptor-associated protein 1 (TRAP1), also known as heat shock protein 75 (HSP75). For this protein, fold changes of 3.0 and 2.2 were observed in the gel- and LC-MS-based approaches, respectively. As a second candidate from this group we selected inorganic pyrophosphatase 1 (PPA1), which was detected with fold changes of 2.0 in the 2D-DIGE experiment and 5.9 in the label-free approach. As an example for a candidate protein down-regulated in tumor tissue in comparison to nontumor tissue we have chosen betaine-homocysteine S-methyltransferase 1 (BHMT) for further verification. BHMT was found to be down-regulated in both studies with fold changes ranging from -3.0 to -3.7 in the gel-based approach and -5.6 in the label-free study (Fig. 3).
Fig. 3.
Regulation patterns of selected proteins. Depending on the study in which the protein was detected, spot volume of the protein (2D-DIGE) and/or feature intensity of a representative peptide (LC-MS) in HCC and nontumorous tissue samples are shown. Additionally, protein regulations within the investigated patient cohort are shown in the box plots (boxes represent 25th and 75th percentile, whiskers indicate the standard deviation, the median is shown as a black bar and the mean value as an empty square within box).
Methodological Verification via Western Blotting and Immunohistochemistry
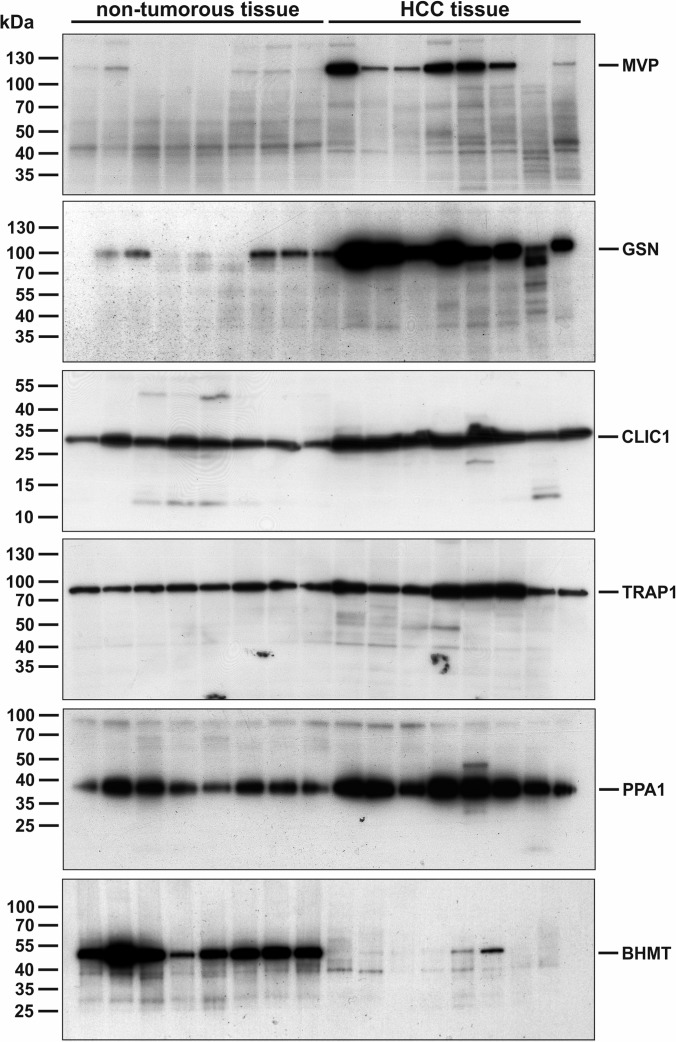
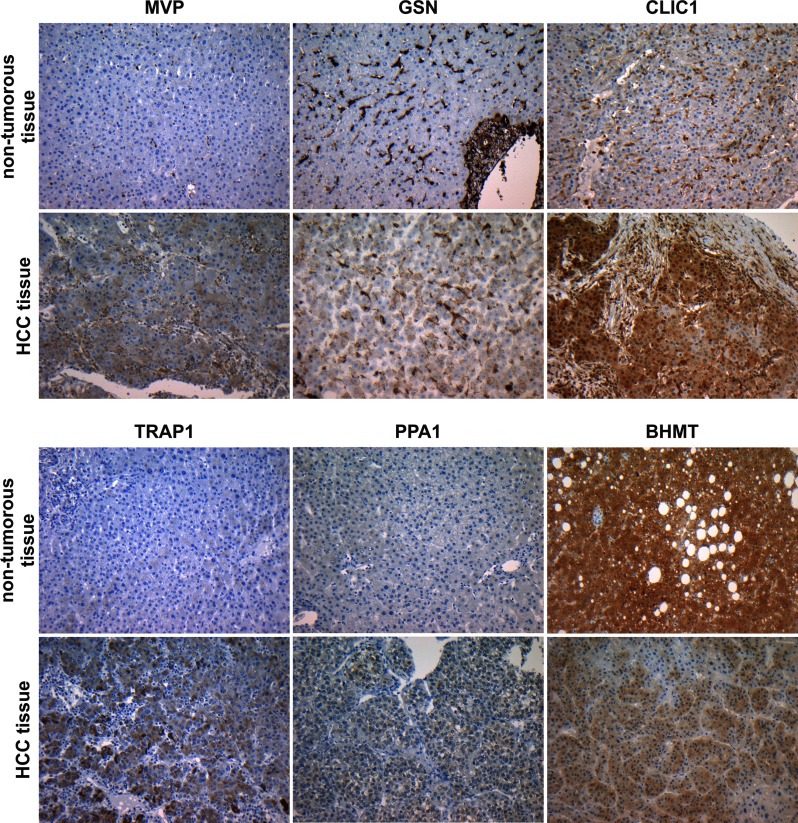
The selected candidate proteins were first investigated by Western blot analysis of eight sample pairs of HCC and nontumorous liver tissues that were also used in label-free and 2D-DIGE differential proteomics studies (patient ID 1–8). Here, the analysis showed differential expression of all candidates in tumorous tissue compared with nontumor tissue. MVP showed strong expression in six of eight tumor samples whereas weak or no expression was observed in nontumor tissue. Gelsolin was found with general high expression levels in HCC tissue and only weak expression in the control group. For CLIC1, enhanced expression levels were observed in all tumor samples. TRAP1 and PPA1 also showed higher expression levels in four of eight and five of eight HCC tissue samples, respectively. For BHMT only little expression was detected in HCC-tissue compared with strong expression in all samples of nontumor tissue (Fig. 4). In addition to the Western blot analysis, immunohistochemical stainings of the six candidate proteins were performed as an additional methodological verification using an alternative immunological method. Again, the previously observed alterations in protein expression were reproducible. In normal liver tissue, CLIC1 showed positive reactions in nonhepatocytes, whereas hepatocytes were completely negative. In HCC the tumor cells displayed a strong positive signal in the cytoplasm and in the nuclei. In addition, stroma cells were also positive for CLIC1. The antibody against MVP showed an immunoreactive signal in the cytoplasm of HCC cells but was negative in normal hepatocytes. TRAP1 was located in the cytoplasm of HCC cells but was negative in the nontumor liver tissue. Using an antibody against pyrophosphatase 1, the tumor cells were slightly positive in the cytoplasm while the nontumor liver cells were negative. Using an antibody against gelsolin in the nontumor liver sinusoidal cells were immunopositive as well as fibrous tissue of portal tracts and cholangiocytes, but the hepatocytes were negative. In contrast, in HCC the tumor cells were positively stained. In accordance to the Western blot results, the immunohistochemical staining against BHMT showed a strong signal in nontumor hepatocytes while tumor cells stained weaker (Fig. 5).
Fig. 4.
Western blot analysis of nontumorous tissue and HCC tumor tissue, respectively. Up-regulation of biomarker candidates MVP, GSN, CLIC1, TRAP1, and PPA1 in HCC tissue could been validated using immunoblots. All up-regulated candidates show stronger signals in HCC samples in comparison to nontumorous tissue samples. BHMT as an example for a biomarker candidate down-regulated in HCC shows only weak or no signal in tumor tissue but strong signal in nontumorous tissue samples.
Fig. 5.
Immunohistochemical staining of HCC and corresponding nontumorous liver from the same patient. MVP: In the normal liver MVP is located in some nucleated blood cells but hepatocytes are negative in contrast to HCC with positive signals in the cytoplasm of tumor cells. GSN: For GSN a positive reactivity in HCC tumor cells is observed in contrast to a negative antibody reaction in normal hepatocytes. CLIC1: Immunohistochemistry against CLIC1 shows reactivity in sinusoidal lining cells but shows no signal in hepatocytes. In HCC strong reactivity is present in the cytoplasm and nuclei of tumor cells and also in nontumor stroma cells. TRAP1: Immunohistochemistry against TRAP1 shows strong reactivity in HCC cells, but is negative in the normal liver. PPA1: The antibody against PPA1 shows a faint reactivity in HCC cells, but is completely negative in the normal liver. BHMT: The antibody against BHMT shows a strong reactivity in normal hepatocytes and a faint reaction in HCC tumor cells. Original magnification: ×200.
Verification of Differentially Expressed Proteins as Biomarker Candidates
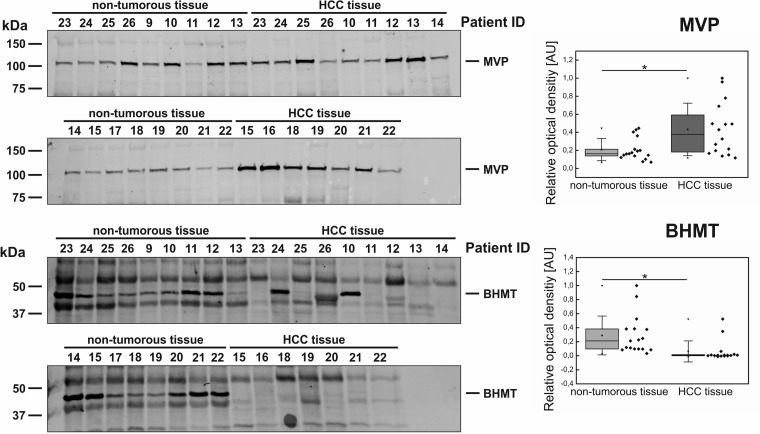
To determine whether a regulated protein might act as a biomarker candidate of a particular disease or not, further verifications using and independent and larger patient cohorts are required. Hence, the expression changes of all six candidate proteins were further investigated within an independent verification set containing 33 samples (16 HCC and 17 nontumorous tissues from patient ID 9–26). To assess the significance of protein-expression changes between HCC and nontumorous tissue Western blots were analyzed via densitometry. In this larger sample set only two of the six candidates showed apparent regulations between both experimental groups. MVP was found to be significantly up-regulated in HCC tumor tissue compared with nontumorous liver tissue (Student's t test, unpaired, two-sided, unequal variances, p = 0.0102). Furthermore, BHMT was confirmed to be down-regulated in HCC tissue (p = 0.0067). Only in two samples of tumor tissue BHMT expression was observed (patient ID 10 and 24) (Fig. 6). For PPA1 and CLIC1, the up-regulation in HCC tissue could be reproduced as a slight but nonsignificant trend. However, for TRAP1 and GSN the previously observed regulation patterns were not detected at all (supplemental Data S7).
Fig. 6.
Western blot analysis of the independent verification cohort and corresponding densitometric evaluations. Biomarker candidates were tested with 17 independent samples of HCC-tumor tissue and 16 samples of nontumorous liver tissue, respectively. MVP and BHMT show differential expression between both experimental groups. Box plots show the results of densitometric analyses (rhombs depict individual data points, boxes represent 25th and 75th percentile, whiskers indicate standard deviation, median is shown as a black bar and the mean value as a square within boxes,). MVP was found to be significantly higher abundant in tumor tisssue (Student's t test, unpaired, two-sided, unequal variances, p = 0.0102) whereas BHMT was found to be significantly down-regulated in HCC (p = 0.0067).
DISCUSSION
To identify novel biomarker candidates of HCC, a study combining complementary gel-based and LC-MS based quantification methods was performed. By comparing the lists of dysregulated proteins obtained from both approaches the assumed complementarity could be clearly shown. In particular, 51 proteins were identified in both studies corresponding to only 9% of the total number of proteins. This result clearly shows the benefit of using different techniques in combination, which leads to greater proteome coverage. Furthermore, the fact that a regulated protein is identified in both studies necessarily increases the probability that this particular biomarker candidate may be more clinically relevant. This assumption is corroborated by the fact that the overlap includes many proteins that have already been associated to hepatocellular carcinoma and whose disease-related dysregulations have been reported in numerous independent studies (see references in Table I). Especially, among the up-regulated proteins which are most interesting with respect to a use as a biomarker. However, the overlap also includes several proteins that to our knowledge were not associated to HCC earlier (e.g. TRAP1) and therefore they might be new biomarker candidates for this particular disease.
In some cases, the comparison of protein regulations showed different results in the label-free and gel-based approach. However, in at least four of eight cases, this result is caused by the detection of several up- or down-regulated isoforms of the same protein in the 2D-DIGE experiment. In such cases, it can be assumed that the regulations determined by the label-free bottom-up approach are more reliable regarding the overall expression change of a protein as they are not influenced by any isoform effects. For example, the over-expressions of alcohol dehydrogenase 4 (ADH4) or peptidylprolyl isomerase A (PPIA) in HCC tissue specimens, as observed in the label-free approach, are in line with previously published data (58, 68). However, inconclusive results were obtained in the current 2D-DIGE study.
For a first methodological verification of the applied proteomics approaches six proteins were selected for further analyses, namely TRAP1, MVP, CLIC1, GSN, PPA1, and BHMT. In addition, these proteins were further investigated in an independent patient cohort to assess their potential applications as HCC biomarker candidates. The proteins were chosen with respect to their currently known or unknown association with HCC as well as the part of our proteomic study they were identified in (e.g. complements of the label-free and 2D-DIGE approaches or the overlap of both).
TRAP1 and GSN: TRAP1 is a member of the HSP90 family of molecular chaperones which consists of three other major homologues, namely HSP90α, HSP90β, and 94kDa glucose-regulated protein (GRP94). In the present study, each of the four HSP90 homologues was found to be significantly over-expressed in HCC tissue, whereas only TRAP1 was identified irrespective of the applied quantification technique. For the homologues HSP90α, HSP90β, and GRP94 the observed up-regulation has already been reported regarding several carcinoma types including HCC. However, the mitochondrial TRAP1 has not yet been investigated to such an extent. TRAP1 is involved in processes like drug resistance, cell survival, stress response, mitochondrial homeostasis and protein folding. Earlier, it was found to be over-expressed in colorectal (69) and nasopharyngeal carcinoma (70) as well as cisplatin-resistant ovarian cancer cells (71–72). In the prior case, the involvement of TRAP1 in drug-resistance was additionally studied by inhibiting TRAP1 activity with shepherdin resulting in higher drug sensitivity (69). Hence, TRAP1 would be not only a promising tumor marker candidate, but moreover a potential drug target for improved cancer therapies. The ubiquitous Ca2+-regulated actin-binding protein gelsolin (GSN) was also found to be over-expressed in tumorous tissue compared with adjacent nontumor tissue. The protein exists in two major isoforms, namely the intracellular cytoplasmic one (cGSN) and a secreted form, also known as plasma gelsolin (pGSN). The three regulated peptides detected in the label-free approach are shared between those forms (for sequences see: supplemental Data S5) which makes a clear decision between both forms impossible at this point. Dysregulation of gelsolin in cause of several malignancies has been reported in numerous studies. In a high number of cancer types, including human breast, colorectal, gastric, bladder, lung, prostate, kidney, ovarian, pancreatic, or oral cancers, gelsolin was down-regulated leading to the assumption that gelsolin might act as a tumor suppressor. However, in a subset of nonsmall cell lung cancers gelsolin was found to be over-expressed. Furthermore, increased gelsolin levels have been associated to tumor recurrence and progression in urothelial tumors (73). In our current proteomic study, TRAP1 and GSN were found to be up-regulated in HCC tissue and their regulation patterns were successfully verified by immunological methods within the same sample set. However, the experiments performed with a second independent sample set revealed no significant up-regulations in HCC tissue. Instead, only a small subpopulation of four samples shows a higher protein expression in HCC tissue. Hence, the summarized results are not necessarily supportive for the assumption that TRAP1 and GSN could act as biomarker candidates of hepatocellular carcinoma.
CLIC1: In the gel-based approach, chloride intracellular channel protein 1 (CLIC1) was found to be up-regulated in HCC tumor tissue. Members of the CLIC protein family are widely expressed and involved in several cellular processes like apoptosis, cell division or secretion. An HCC-related up-regulation of CLIC1 has already been reported in a proteomic study of hepatocellular carcinoma developed in patients with chronic hepatitis C infection as an underlying disease (74). Earlier, transcriptomics data were published that also revealed an over-expression of CLIC1 related to HCC, which is in agreement with our data (75). Within the patient cohort investigated in our proteomics study, none of the patients had hepatitis B or C infections. Hence, the observed over-expression of CLIC1 in HCC seems to be irrespective of the underlying disease. During a second verification within an independent sample set, the over-expression of CLIC1 was observable as a slight but nonsignificant trend. Nonetheless, the following fact supports that CLIC1 can be denoted as a diagnostic HCC biomarker candidate for histopathological purposes worth to be followed in ongoing validation studies. Samples used in the verification with Western blots were full lysates of nonmicrodissected tissues samples and therefore heterogeneous. As shown by the immunohistochemical analysis CLIC1 clearly can distinguish HCC tumor cells and normal hepatocytes. However, CLIC1 is also positive for nonhepatocytes in normal liver tissue, which could produce misleading or nonsignificant results if full tissue lysates are used.
PPA1: Inorganic pyrophosphatase (PPA1) was identified as a regulated protein in the label-free and 2D-DIGE approaches. It catalyzes the hydrolysis of pyrophosphate to orthophosphate and is ubiquitously expressed. It has been shown to be differentially expressed in various types of cancer including enhanced expression in primary colorectal cancer (76), lung adenocarcinoma (77), and prostate cancer (78) and has also been shown to be expressed in a hepatocellular carcinoma cell line (79). However, in a proteomic pilot study of HCC in which tissue samples of only three patients have been analyzed using 2D gel electrophoresis, PPA1 has been found to be down-regulated (29). In our current proteomic study, we can show with a larger sample set and two different quantification methods that PPA1 is significantly up-regulated in HCC. Furthermore, we verified this result using immunological methods within the same sample set. In the independent set of samples the up-regulation was solely reproducible as a nonsignificant trend. However, in comparison to CLIC1, PPA1 remains an interesting candidate protein for diagnostic purposes that should be investigated in further validation studies.
MVP: In previous studies, MVP has been found to be over-expressed in several human cancers such as pancreatic, breast, ovarian, urinary bladder carcinomas, melanomas, sarcomas, and leukemias (80). However, in case of liver carcinomas a variable expression has been reported (81). Contrary, our recent results clearly verify an over-expression of MVP in tumorous HCC tissue. MVP is the main constituent of the so called vaults, which are ribonucleoprotein particles with masses of ∼13 MDa (82). It has been associated with several signaling pathways, including PI3K/Akt, MAPK, and STAT suggesting regulatory roles in these signaling processes (83–85). More recently, MVP has been found to be involved in resistance to epidermal growth factor inhibition of several HCC-derived cell lines (86). In our current study, we observed a significant up-regulation of MVP in HCC tissue that was verified in two independent patient cohorts. The relatively large variance of expression levels that was detected within the HCC experimental groups is in line with previous observations and is caused by an interindividual heterogeneity of MVP expression in liver tissue that has already been described (81). In summary, we conclude that MVP is a robust HCC diagnostic biomarker candidate that can be used to distinguish normal liver and HCC tumor tissues irrespective of the underlying liver disease or tumor characteristics. Hence, follow-up studies for the validation of MVP as a diagnostic HCC marker in larger patients cohorts should be envisaged.
BHMT: Earlier, a strong decrease of BHMT expression in HCC tumor tissue has already been shown in gel-based proteomic studies (13, 64) as well as on the transcript level (63). Very recently, the transcription of an aberrant splicing variant has been described as mechanism leading to decreased BHMT levels in HCC (87). BHMT is involved in homocysteine metabolism where it catalyzes the synthesis of methionine from betaine and homocysteine. Loss of BHMT function therefore leads to impaired hemostasis of 1-carbon metabolism and is directly associated with various diseases including hepatocellular carcinogenesis (88). In our study, the decreased expression of BHMT in HCC was confirmed for the first time using a label-free quantification method that was successfully verified by immunological methods within two independent patient cohorts. Even if the down-regulation of BHMT has been described in several studies, its use as diagnostic HCC marker for histopathological purposes has not been tested by immunohistochemical methods in large patient cohorts yet. Such a comprehensive validation study should definitely be envisaged in future projects to assess the use of BHMT as a diagnostic HCC biomarker.
CONCLUSION
In our study we compared the proteomic profile of HCC tumor tissue (n = 7) with nontumorous liver tissue (n = 7) by following a 2D-DIGE approach and a label-free LC-MS-based quantification strategy. As expected, our results clearly show that both techniques gave complementary results, as only 51 of the 573 regulated proteins have been identified irrespective of the applied technique, which in turn shows the benefit of using both techniques in combination. For a methodological verification, HCC-related regulations of six proteins (MVP, BHMT, CLIC1, GSN, TRAP1, PPA1) were investigated by Western blots and immunohistochemistry using the same sample set. Here, for each protein the previously observed regulation pattern was successfully reproduced. To assess the applicability of these proteins as biomarker candidates of HCC, a second verification was performed with samples derived from a larger and independent patient cohort (16 HCC samples and 17 nontumorous tissue samples). Here, a significant regulation was only detectable for two of the six candidate proteins. For MVP a significant up-regulation and for BHMT a down-regulation in HCC tissues was detected by Western blots analyzed via densitometry. In summary, the results of the current study suggest a potential applicability of these proteins as diagnostic HCC biomarkers. Hence, a validation of these marker candidates in larger patient cohorts will be envisaged in future projects. In particular, the potential use of MVP and BHMT as parts of biomarker panel will be investigated. Such multiple marker panels are commonly used for the diagnosis of the very heterogeneous HCC, and show a higher sensitivity and selective than single markers (89).
Supplementary Material
Acknowledgments
We would like to thank Kristin Rosowski, Birgit Korte, and Stephanie Tautges for their excellent technical assistance as well as Marius Loscha and Xi Liu for the help during the preparation of the manuscript.
Footnotes
* The project was supported by the operational program cofinanced by the European Regional Development Fund (ERDF) Objective 2 “Regional Competitiveness and Employment” 2007–2013), North Rhine-Westphalia (Germany). A part of this study was funded from P.U.R.E. (Protein Unit for Research in Europe), a project of North Rhine-Westphalia, a federal state of Germany as well.
 This article contains supplemental Data Files S1 to S7.
This article contains supplemental Data Files S1 to S7.
1 The abbreviations used are:
- HCC
- hepatocellular carcinoma
- SILAC
- stable isotope labeling by amino acids in cell culture
- iTRAQ
- isobaric tags for relative and absolute quantification
- CDIT
- culture-derived isotope tags
- NASH
- nonalcoholic steatohepatitis
- IEF
- isoelectric focusing
- ECL
- Enhanced chemiluminescence
- AB
- antibody
- RT
- room temperature.
REFERENCES
- 1. El-Serag H. B., Mason A. C. (1999) Rising incidence of hepatocellular carcinoma in the United States. N. Engl. J. Med. 340, 745–750 [DOI] [PubMed] [Google Scholar]
- 2. Di Bisceglie A. M., Sterling R. K., Chung R. T., Everhart J. E., Dienstag J. L., Bonkovsky H. L., Wright E. C., Everson G. T., Lindsay K. L., Lok A. S., Lee W. M., Morgan T. R., Ghany M. G., Gretch D. R. (2005) Serum alpha-fetoprotein levels in patients with advanced hepatitis C: results from the HALT-C Trial. J. Hepatol. 43, 434–441 [DOI] [PubMed] [Google Scholar]
- 3. Oka H., Saito A., Ito K., Kumada T., Satomura S., Kasugai H., Osaki Y., Seki T., Kudo M., Tanaka M. (2001) Multicenter prospective analysis of newly diagnosed hepatocellular carcinoma with respect to the percentage of Lens culinaris agglutinin-reactive alpha-fetoprotein. J. Gastroenterol. Hepatol. 16, 1378–1383 [DOI] [PubMed] [Google Scholar]
- 4. Liebman H. A., Furie B. C., Tong M. J., Blanchard R. A., Lo K. J., Lee S. D., Coleman M. S., Furie B. (1984) Des-gamma-carboxy (abnormal) prothrombin as a serum marker of primary hepatocellular carcinoma. N. Engl. J. Med. 310, 1427–1431 [DOI] [PubMed] [Google Scholar]
- 5. Pei Y., Zhang T., Renault V., Zhang X. (2009) An overview of hepatocellular carcinoma study by omics-based methods. Acta Biochim. Biophys. Sin. 41, 1–15 [DOI] [PubMed] [Google Scholar]
- 6. Liu Z., Ma Y., Yang J., Qin H. (2011) Upregulated and downregulated proteins in hepatocellular carcinoma: a systematic review of proteomic profiling studies. OMICS 15, 61–71 [DOI] [PubMed] [Google Scholar]
- 7. Orimo T., Ojima H., Hiraoka N., Saito S., Kosuge T., Kakisaka T., Yokoo H., Nakanishi K., Kamiyama T., Todo S., Hirohashi S., Kondo T. (2008) Proteomic profiling reveals the prognostic value of adenomatous polyposis coli-end-binding protein 1 in hepatocellular carcinoma. Hepatology 48, 1851–1863 [DOI] [PubMed] [Google Scholar]
- 8. Yokoyama Y., Kuramitsu Y., Takashima M., Iizuka N., Toda T., Terai S., Sakaida I., Oka M., Nakamura K., Okita K. (2004) Proteomic profiling of proteins decreased in hepatocellular carcinoma from patients infected with hepatitis C virus. Proteomics 4, 2111–2116 [DOI] [PubMed] [Google Scholar]
- 9. Takashima M., Kuramitsu Y., Yokoyama Y., Iizuka N., Toda T., Sakaida I., Okita K., Oka M., Nakamura K. (2003) Proteomic profiling of heat shock protein 70 family members as biomarkers for hepatitis C virus-related hepatocellular carcinoma. Proteomics 3, 2487–2493 [DOI] [PubMed] [Google Scholar]
- 10. Zhang D., Lim S. G., Koay E. S. (2007) Proteomic identification of down-regulation of oncoprotein DJ-1 and proteasome activator subunit 1 in hepatitis B virus-infected well-differentiated hepatocellular carcinoma. Int. J. Oncol. 31, 577–584 [PubMed] [Google Scholar]
- 11. Corona G., De Lorenzo E., Elia C., Simula M. P., Avellini C., Baccarani U., Lupo F., Tiribelli C., Colombatti A., Toffoli G. (2010) Differential proteomic analysis of hepatocellular carcinoma. Int. J. Oncol. 36, 93–99 [PubMed] [Google Scholar]
- 12. Zubaidah R. M., Tan G. S., Tan S. B., Lim S. G., Lin Q., Chung M. C. (2008) 2-D DIGE profiling of hepatocellular carcinoma tissues identified isoforms of far upstream binding protein (FUBP) as novel candidates in liver carcinogenesis. Proteomics 8, 5086–96 [DOI] [PubMed] [Google Scholar]
- 13. Sun W., Xing B., Sun Y., Du X., Lu M., Hao C., Lu Z., Mi W., Wu S., Wei H., Gao X., Zhu Y., Jiang Y., Qian X., He F. (2007) Proteome analysis of hepatocellular carcinoma by two-dimensional difference gel electrophoresis: novel protein markers in hepatocellular carcinoma tissues. Mol. Cell. Proteomics 6, 1798–1808 [DOI] [PubMed] [Google Scholar]
- 14. Lee I. N., Chen C. H., Sheu J. C., Lee H. S., Huang G. T., Yu C. Y., Lu F. J., Chow L. P. (2005) Identification of human hepatocellular carcinoma-related biomarkers by two-dimensional difference gel electrophoresis and mass spectrometry. J. Proteome Res. 4, 2062–2069 [DOI] [PubMed] [Google Scholar]
- 15. Minagawa H., Yamashita T., Honda M., Tabuse Y., Kamijo K., Tsugita A., Kaneko S. (2008) Comparative analysis of proteome and transcriptome in human hepatocellular carcinoma using 2D-DIGE and SAGE. Protein J. 27, 409–419 [DOI] [PubMed] [Google Scholar]
- 16. Sun Y., Mi W., Cai J., Ying W., Liu F., Lu H., Qiao Y., Jia W., Bi X., Lu N., Liu S., Qian X., Zhao X. (2008) Quantitative proteomic signature of liver cancer cells: tissue transglutaminase 2 could be a novel protein candidate of human hepatocellular carcinoma. J. Proteome Res. 7, 3847–3859 [DOI] [PubMed] [Google Scholar]
- 17. Chen N., Sun W., Deng X., Hao Y., Chen X., Xing B., Jia W., Ma J., Wei H., Zhu Y., Qian X., Jiang Y., He F. (2008) Quantitative proteome analysis of HCC cell lines with different metastatic potentials by SILAC. Proteomics 8, 5108–5118 [DOI] [PubMed] [Google Scholar]
- 18. Chaerkady R., Harsha H. C., Nalli A., Gucek M., Vivekanandan P., Akhtar J., Cole R. N., Simmers J., Schulick R. D., Singh S., Torbenson M., Pandey A., Thuluvath P. J. (2008) A quantitative proteomic approach for identification of potential biomarkers in hepatocellular carcinoma. J. Proteome Res. 7, 4289–4298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li C., Ruan H. Q., Liu Y. S., Xu M. J., Dai J., Sheng Q. H., Tan Y. X., Yao Z. Z., Wang H. Y., Wu J. R., Zeng R. (2012) Quantitative proteomics reveal up-regulated protein expression of the SET complex associated with hepatocellular carcinoma. J. Proteome Res. 11, 871–885 [DOI] [PubMed] [Google Scholar]
- 20. Lee S., Kwon M. S., Lee H. J., Paik Y. K., Tang H., Lee J. K., Park T. (2011) Enhanced peptide quantification using spectral count clustering and cluster abundance. BMC Bioinformatics 12, 423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Meding S., Nitsche U., Balluff B., Elsner M., Rauser S., Schöne C., Nipp M., Maak M., Feith M., Ebert M. P., Friess H., Langer R., Höfler H., Zitzelsberger H., Rosenberg R., Walch A. (2012) Tumor classification of six common cancer types based on proteomic profiling by MALDI imaging. J. Proteome Res. 11, 1996–2003 [DOI] [PubMed] [Google Scholar]
- 22. Le Faouder J., Laouirem S., Chapelle M., Albuquerque M., Belghiti J., Degos F., Paradis V., Camadro J. M., Bedossa P. (2011) Imaging mass spectrometry provides fingerprints for distinguishing hepatocellular carcinoma from cirrhosis. J. Proteome Res. 10, 3755–3765 [DOI] [PubMed] [Google Scholar]
- 23. Sobin L. H., Gospodarowicz M. K., Wittekind C. (2009) TNM Classification of Malignant Tumours, 7th ed., Wiley-Blackwell, New-York [Google Scholar]
- 24. Klose J., Kobalz U. (1995) Two-dimensional electrophoresis of proteins: an updated protocol and implications for a functional analysis of the genome. Electrophoresis 16, 1034–1059 [DOI] [PubMed] [Google Scholar]
- 25. Sitek B., Lüttges J., Marcus K., Klöppel G., Schmiegel W., Meyer H. E., Hahn S. A., Stühler K. (2005) Application of fluorescence difference gel electrophoresis saturation labelling for the analysis of microdissected precursor lesions of pancreatic ductal adenocarcinoma. Proteomics 5, 2665–2679 [DOI] [PubMed] [Google Scholar]
- 26. Poschmann G., Sitek B., Sipos B., Ulrich A., Wiese S., Stephan C., Warscheid B., Klöppel G., Vander Borght A., Ramaekers F. C., Meyer H. E., Stuhler K. (2009) Identification of proteomic differences between squamous cell carcinoma of the lung and bronchial epithelium. Mol. Cell. Proteomics 8, 1105–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Perkins D. N., Pappin D. J., Creasy D. M., Cottrell J. S. (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567 [DOI] [PubMed] [Google Scholar]
- 28. Vizcaino J. A., Cote R. G., Csordas A., Dianes J. A., Fabregat A., Foster J. M., Griss J., Alpi E., Birim M., Contell J., O'Kelly G., Schoenegger A., Ovelleiro D., Perez-Riverol Y., Reisinger F., Rios D., Wang R., Hermjakob H. (2013) The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 41, D1063–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Matos J. M., Witzmann F. A., Cummings O. W., Schmidt C. M. (2009) A pilot study of proteomic profiles of human hepatocellular carcinoma in the United States. J. Surg. Res. 155, 237–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tzonou A., Sparos L., Kalapothaki V., Zavitsanos X., Rebelakos A., Trichopoulos D. (1990) Alpha 1-antitrypsin and survival in hepatocellular carcinoma. Br. J. Cancer 61, 72–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fargion S., Klasen E. C., Lalatta F., Sangalli G., Tommasini M., Fiorelli G. (1981) Alpha 1-antitrypsin in patients with hepatocellular carcinoma and chronic active hepatitis. Clin. Genet. 19, 134–139 [DOI] [PubMed] [Google Scholar]
- 32. Tartaglino B., Macri G., Molino G., Furlani D., Sansalvadore F. (1980) Behavior of serum alpha 1-antitrypsin in chronic hepatopathies and its diagnostic significance. Quad. Sclavo. Diagn. 16, 451–458 [PubMed] [Google Scholar]
- 33. Kawakami T., Hoshida Y., Kanai F., Tanaka Y., Tateishi K., Ikenoue T., Obi S., Sato S., Teratani T., Shiina S., Kawabe T., Suzuki T., Hatano N., Taniguchi H., Omata M. (2005) Proteomic analysis of sera from hepatocellular carcinoma patients after radiofrequency ablation treatment. Proteomics 5, 4287–4295 [DOI] [PubMed] [Google Scholar]
- 34. Kang X., Sun L., Guo K., Shu H., Yao J., Qin X., Liu Y. (2010) Serum protein biomarkers screening in HCC patients with liver cirrhosis by ICAT-LC-MS/MS. J Cancer Res. Clin Oncol 136, 1151–9 [DOI] [PubMed] [Google Scholar]
- 35. Lee H. B., Yoo O. J., Ham J. S., Lee M. H. (1992) Serum alpha 1-antitrypsin in patients with hepatocellular carcinoma. Clin. Chim. Acta 206, 225–2230 [DOI] [PubMed] [Google Scholar]
- 36. Sun S., Poon R. T., Lee N. P., Yeung C., Chan K. L., Ng I. O., Day P. J., Luk J. M. (2010) Proteomics of hepatocellular carcinoma: serum vimentin as a surrogate marker for small tumors (<or=2 cm). J. Proteome Res. 9, 1923–1930 [DOI] [PubMed] [Google Scholar]
- 37. Song H. Y., Liu Y. K., Feng J. T., Cui J. F., Dai Z., Zhang L. J., Feng J. X., Shen H. L., Tang Z. Y. (2006) Proteomic analysis on metastasis-associated proteins of human hepatocellular carcinoma tissues. J. Cancer Res. Clin. Oncol. 132, 92–98 [DOI] [PubMed] [Google Scholar]
- 38. Srisomsap C., Sawangareetrakul P., Subhasitanont P., Panichakul T., Keeratichamroen S., Lirdprapamongkol K., Chokchaichamnankit D., Sirisinha S., Svasti J. (2004) Proteomic analysis of cholangiocarcinoma cell line. Proteomics 4, 1135–1144 [DOI] [PubMed] [Google Scholar]
- 39. Gao Q., Wang X. Y., Fan J., Qiu S. J., Zhou J., Shi Y. H., Xiao Y. S., Xu Y., Huang X. W., Sun J. (2008) Selection of reference genes for real-time PCR in human hepatocellular carcinoma tissues. J. Cancer Res. Clin. Oncol. 134, 979–986 [DOI] [PubMed] [Google Scholar]
- 40. Lau W. Y., Lai P. B., Leung M. F., Leung B. C., Wong N., Chen G., Leung T. W., Liew C. T. (2000) Differential gene expression of hepatocellular carcinoma using cDNA microarray analysis. Oncol. Res. 12, 59–69 [DOI] [PubMed] [Google Scholar]
- 41. Kitamura K., Hatano E., Higashi T., Narita M., Seo S., Nakamoto Y., Yamanaka K., Nagata H., Taura K., Yasuchika K., Nitta T., Uemoto S. (2011) Proliferative activity in hepatocellular carcinoma is closely correlated with glucose metabolism but not angiogenesis. J. Hepatol. 55, 846–857 [DOI] [PubMed] [Google Scholar]
- 42. Yi X., Luk J. M., Lee N. P., Peng J., Leng X., Guan X. Y., Lau G. K., Beretta L., Fan S. T. (2008) Association of mortalin (HSPA9) with liver cancer metastasis and prediction for early tumor recurrence. Mol. Cell. Proteomics 7, 315–325 [DOI] [PubMed] [Google Scholar]
- 43. Kuramitsu Y., Nakamura K. (2005) Current progress in proteomic study of hepatitis C virus-related human hepatocellular carcinoma. Expert Rev. Proteomics 2, 589–601 [DOI] [PubMed] [Google Scholar]
- 44. Luk J. M., Lam C. T., Siu A. F., Lam B. Y., Ng I. O., Hu M. Y., Che C. M., Fan S. T. (2006) Proteomic profiling of hepatocellular carcinoma in Chinese cohort reveals heat-shock proteins (Hsp27, Hsp70, GRP78) up-regulation and their associated prognostic values. Proteomics 6, 1049–1057 [DOI] [PubMed] [Google Scholar]
- 45. Lim S. O., Park S. G., Yoo J. H., Park Y. M., Kim H. J., Jang K. T., Cho J. W., Yoo B. C., Jung G. H., Park C. K. (2005) Expression of heat shock proteins (HSP27, HSP60, HSP70, HSP90, GRP78, GRP94) in hepatitis B virus-related hepatocellular carcinomas and dysplastic nodules. World J. Gastroenterol. 11, 2072–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Su R., Li Z., Li H., Song H., Bao C., Wei J., Cheng L. (2010) Grp78 promotes the invasion of hepatocellular carcinoma. BMC Cancer 10, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Liaw K. Y., Lee P. H., Wu F. C., Tsai J. S., Lin-Shiau S. Y. (1997) Zinc, copper, and superoxide dismutase in hepatocellular carcinoma. Am. J. Gastroenterol. 92, 2260–2263 [PubMed] [Google Scholar]
- 48. Goldenberg D., Ayesh S., Schneider T., Pappo O., Jurim O., Eid A., Fellig Y., Dadon T., Ariel I., de Groot N., Hochberg A., Galun E. (2002) Analysis of differentially expressed genes in hepatocellular carcinoma using cDNA arrays. Mol. Carcinog. 33, 113–124 [PubMed] [Google Scholar]
- 49. Li Y., Wan D., Wei W., Su J., Cao J., Qiu X., Ou C., Ban K., Yang C., Yue H. (2008) Candidate genes responsible for human hepatocellular carcinoma identified from differentially expressed genes in hepatocarcinogenesis of the tree shrew (Tupaia belangeri chinesis). Hepatol. Res. 38, 85–95 [DOI] [PubMed] [Google Scholar]
- 50. Kuo W. H., Chiang W. L., Yang S. F., Yeh K. T., Yeh C. M., Hsieh Y. S., Chu S. C., The differential expression of cytosolic carbonic anhydrase in human hepatocellular carcinoma. In Life Sci., 2003/08/21 ed.; 2003; Vol. 73, 2211–2223 [DOI] [PubMed] [Google Scholar]
- 51. Kinoshita M., Miyata M. (2002) Underexpression of mRNA in human hepatocellular carcinoma focusing on eight loci. Hepatology 36, 433–438 [DOI] [PubMed] [Google Scholar]
- 52. Liu Y., Zhu X., Zhu J., Liao S., Tang Q., Liu K., Guan X., Zhang J., Feng Z. (2007) Identification of differential expression of genes in hepatocellular carcinoma by suppression subtractive hybridization combined cDNA microarray. Oncol. Rep. 18, 943–951 [PubMed] [Google Scholar]
- 53. Park K. S., Cho S. Y., Kim H., Paik Y. K. (2002) Proteomic alterations of the variants of human aldehyde dehydrogenase isozymes correlate with hepatocellular carcinoma. Int. J. Cancer 97, 261–265 [DOI] [PubMed] [Google Scholar]
- 54. Hu Y., Pang E., Lai P. B., Squire J. A., MacGregor P. F., Beheshti B., Albert M., Leung T. W., Wong N. (2004) Genetic alterations in doxorubicin-resistant hepatocellular carcinoma cells: a combined study of spectral karyotyping, positional expression profiling and candidate genes. Int. J. Oncol. 25, 1357–1364 [PubMed] [Google Scholar]
- 55. Kurokawa Y., Matoba R., Takemasa I., Nakamori S., Tsujie M., Nagano H., Dono K., Umeshita K., Sakon M., Ueno N., Kita H., Oba S., Ishii S., Kato K., Monden M. (2003) Molecular features of non-B, non-C hepatocellular carcinoma: a PCR-array gene expression profiling study. J. Hepatol. 39, 1004–1012 [DOI] [PubMed] [Google Scholar]
- 56. Dillon B. J., Prieto V. G., Curley S. A., Ensor C. M., Holtsberg F. W., Bomalaski J. S., Clark M. A. (2004) Incidence and distribution of argininosuccinate synthetase deficiency in human cancers: a method for identifying cancers sensitive to arginine deprivation. Cancer 100, 826–833 [DOI] [PubMed] [Google Scholar]
- 57. Peng S. Y., Lai P. L., Pan H. W., Hsiao L. P., Hsu H. C. (2008) Aberrant expression of the glycolytic enzymes aldolase B and type II hexokinase in hepatocellular carcinoma are predictive markers for advanced stage, early recurrence and poor prognosis. Oncol. Rep. 19, 1045–1053 [PubMed] [Google Scholar]
- 58. Lim S. O., Park S. J., Kim W., Park S. G., Kim H. J., Kim Y. I., Sohn T. S., Noh J. H., Jung G. (2002) Proteome analysis of hepatocellular carcinoma. Biochem. Biophys. Res. Commun. 291, 1031–1037 [DOI] [PubMed] [Google Scholar]
- 59. Seimiya M., Tomonaga T., Matsushita K., Sunaga M., Oh-Ishi M., Kodera Y., Maeda T., Takano S., Togawa A., Yoshitomi H., Otsuka M., Yamamoto M., Nakano M., Miyazaki M., Nomura F. (2008) Identification of novel immunohistochemical tumor markers for primary hepatocellular carcinoma; clathrin heavy chain and formiminotransferase cyclodeaminase. Hepatology 48, 519–530 [DOI] [PubMed] [Google Scholar]
- 60. Na K., Lee E. Y., Lee H. J., Kim K. Y., Lee H., Jeong S. K., Jeong A. S., Cho S. Y., Kim S. A., Song S. Y., Kim K. S., Cho S. W., Kim H., Paik Y. K. (2009) Human plasma carboxylesterase 1, a novel serologic biomarker candidate for hepatocellular carcinoma. Proteomics 9, 3989–3999 [DOI] [PubMed] [Google Scholar]
- 61. Long J., Lang Z. W., Wang H. G., Wang T. L., Wang B. E., Liu S. Q. (2010) Glutamine synthetase as an early marker for hepatocellular carcinoma based on proteomic analysis of resected small hepatocellular carcinomas. Hepatobiliary Pancreat. Dis. Int. 9, 296–305 [PubMed] [Google Scholar]
- 62. Chen M., Zhang J., Li N., Qian Z., Zhu M., Li Q., Zheng J., Wang X., Shi G. (2011) Promoter hypermethylation mediated downregulation of FBP1 in human hepatocellular carcinoma and colon cancer. PLoS One 6, e25564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Avila M. A., Berasain C., Torres L., Martin-Duce A., Corrales F. J., Yang H., Prieto J., Lu S. C., Caballeria J., Rodés J., Mato J. M. (2000) Reduced mRNA abundance of the main enzymes involved in methionine metabolism in human liver cirrhosis and hepatocellular carcinoma. J. Hepatol. 33, 907–914 [DOI] [PubMed] [Google Scholar]
- 64. Liang C. R., Leow C. K., Neo J. C., Tan G. S., Lo S. L., Lim J. W., Seow T. K., Lai P. B., Chung M. C. (2005) Proteome analysis of human hepatocellular carcinoma tissues by two-dimensional difference gel electrophoresis and mass spectrometry. Proteomics 5, 2258–2271 [DOI] [PubMed] [Google Scholar]
- 65. Chen Y. M., Shiu J. Y., Tzeng S. J., Shih L. S., Chen Y. J., Lui W. Y., Chen P. H. (1998) Characterization of glycine-N-methyltransferase-gene expression in human hepatocellular carcinoma. Int. J. Cancer 75, 787–793 [DOI] [PubMed] [Google Scholar]
- 66. Iizuka N., Tsunedomi R., Tamesa T., Okada T., Sakamoto K., Hamaguchi T., Yamada-Okabe H., Miyamoto T., Uchimura S., Hamamoto Y., Oka M. (2006) Involvement of c-myc-regulated genes in hepatocellular carcinoma related to genotype-C hepatitis B virus. J. Cancer Res. Clin. Oncol. 132, 473–481 [DOI] [PubMed] [Google Scholar]
- 67. Krapivner S., Chernogubova E., Ericsson M., Ahlbeck-Glader C., Hamsten A., van 't Hooft F. M. (2007) Human evidence for the involvement of insulin-induced gene 1 in the regulation of plasma glucose concentration. Diabetologia 50, 94–102 [DOI] [PubMed] [Google Scholar]
- 68. Wei R. R., Zhang M. Y., Rao H. L., Pu H. Y., Zhang H. Z., Wang H. Y. (2012) Identification of ADH4 as a novel and potential prognostic marker in hepatocellular carcinoma. Med. Oncol. 29, 2737–2743 [DOI] [PubMed] [Google Scholar]
- 69. Costantino E., Maddalena F., Calise S., Piscazzi A., Tirino V., Fersini A., Ambrosi A., Neri V., Esposito F., Landriscina M. (2009) TRAP1, a novel mitochondrial chaperone responsible for multi-drug resistance and protection from apoptotis in human colorectal carcinoma cells. Cancer Lett. 279, 39–46 [DOI] [PubMed] [Google Scholar]
- 70. Fang W., Li X., Jiang Q., Liu Z., Yang H., Wang S., Xie S., Liu Q., Liu T., Huang J., Xie W., Li Z., Zhao Y., Wang E., Marincola F. M., Yao K. (2008) Transcriptional patterns, biomarkers and pathways characterizing nasopharyngeal carcinoma of Southern China. J. Transl. Med. 6, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Alvero A. B., Chen R., Fu H. H., Montagna M., Schwartz P. E., Rutherford T., Silasi D. A., Steffensen K. D., Waldstrom M., Visintin I., Mor G. (2009) Molecular phenotyping of human ovarian cancer stem cells unravels the mechanisms for repair and chemoresistance. Cell Cycle 8, 158–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Landriscina M., Amoroso M. R., Piscazzi A., Esposito F. (2010) Heat shock proteins, cell survival and drug resistance: the mitochondrial chaperone TRAP1, a potential novel target for ovarian cancer therapy. Gynecol. Oncol. 117, 177–182 [DOI] [PubMed] [Google Scholar]
- 73. Li G. H., Arora P. D., Chen Y., McCulloch C. A., Liu P. (2012) Multifunctional roles of gelsolin in health and diseases. Med. Res. Rev. 32, 999–1025 [DOI] [PubMed] [Google Scholar]
- 74. Blanc J. F., Lalanne C., Plomion C., Schmitter J. M., Bathany K., Gion J. M., Bioulac-Sage P., Balabaud C., Bonneu M., Rosenbaum J. (2005) Proteomic analysis of differentially expressed proteins in hepatocellular carcinoma developed in patients with chronic viral hepatitis C. Proteomics 5, 3778–3789 [DOI] [PubMed] [Google Scholar]
- 75. Huang J. S., Chao C. C., Su T. L., Yeh S. H., Chen D. S., Chen C. T., Chen P. J., Jou Y. S. (2004) Diverse cellular transformation capability of overexpressed genes in human hepatocellular carcinoma. Biochem. Biophys. Res. Commun. 315, 950–958 [DOI] [PubMed] [Google Scholar]
- 76. Tomonaga T., Matsushita K., Yamaguchi S., Oh-Ishi M., Kodera Y., Maeda T., Shimada H., Ochiai T., Nomura F. (2004) Identification of altered protein expression and post-translational modifications in primary colorectal cancer by using agarose two-dimensional gel electrophoresis. Clin. Cancer Res. 10, 2007–2014 [DOI] [PubMed] [Google Scholar]
- 77. Chen G., Gharib T. G., Huang C. C., Thomas D. G., Shedden K. A., Taylor J. M., Kardia S. L., Misek D. E., Giordano T. J., Iannettoni M. D., Orringer M. B., Hanash S. M., Beer D. G. (2002) Proteomic analysis of lung adenocarcinoma: identification of a highly expressed set of proteins in tumors. Clin. Cancer Res. 8, 2298–2305 [PubMed] [Google Scholar]
- 78. Lexander H., Palmberg C., Auer G., Hellstrom M., Franzen B., Jornvall H., Egevad L. (2005) Proteomic analysis of protein expression in prostate cancer. Anal. Quant. Cytol. Histol. 27, 263–272 [PubMed] [Google Scholar]
- 79. Liang R. C., Neo J. C., Lo S. L., Tan G. S., Seow T. K., Chung M. C. (2002) Proteome database of hepatocellular carcinoma. J. Chromatogr. B 771, 303–328 [DOI] [PubMed] [Google Scholar]
- 80. Sitek B., Sipos B., Alkatout I., Poschmann G., Stephan C., Schulenborg T., Marcus K., Luttges J., Dittert D. D., Baretton G., Schmiegel W., Hahn S. A., Klöppel G., Meyer H. E., Stühler K. (2009) Analysis of the pancreatic tumor progression by a quantitative proteomic approach and immunhistochemical validation. J. Proteome Res. 8, 1647–1656 [DOI] [PubMed] [Google Scholar]
- 81. Raidl M., Berger W., Schulte-Hermann R., Kandioler-Eckersberger D., Kappel S., Wrba F., Micksche M., Grasl-Kraupp B. (2002) Expression of the lung resistance-related protein in human and rat hepatocarcinogenesis. Am. J. Physiol. Gastrointest. Liver Physiol. 283, G1117–G1124 [DOI] [PubMed] [Google Scholar]
- 82. Lara P. C., Pruschy M., Zimmermann M., Henriquez-Hernández L. A. (2011) MVP and vaults: a role in the radiation response. Radiat. Oncol. 6, 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Steiner E., Holzmann K., Pirker C., Elbling L., Micksche M., Sutterlüty H., Berger W. (2006) The major vault protein is responsive to and interferes with interferon-gamma-mediated STAT1 signals. J. Cell Sci. 119, 459–469 [DOI] [PubMed] [Google Scholar]
- 84. Kolli S., Zito C. I., Mossink M. H., Wiemer E. A., Bennett A. M. (2004) The major vault protein is a novel substrate for the tyrosine phosphatase SHP-2 and scaffold protein in epidermal growth factor signaling. J. Biol. Chem. 279, 29374–29385 [DOI] [PubMed] [Google Scholar]
- 85. Berger W., Steiner E., Grusch M., Elbling L., Micksche M. (2009) Vaults and the major vault protein: novel roles in signal pathway regulation and immunity. Cell. Mol. Life Sci. 66, 43–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Losert A., Lötsch D., Lackner A., Koppensteiner H., Peter-Vörösmarty B., Steiner E., Holzmann K., Grunt T., Schmid K., Marian B., Grasl-Kraupp B., Schulte-Hermann R., Krupitza G., Berger W., Grusch M. (2012) The major vault protein mediates resistance to epidermal growth factor receptor inhibition in human hepatoma cells. Cancer Lett. 319, 164–172 [DOI] [PubMed] [Google Scholar]
- 87. Pellanda H., Namour F., Fofou-Caillierez M., Bressenot A., Alberto J. M., Chery C., Ayav A., Bronowicki J. P., Gueant J. L., Forges T. (2012) A splicing variant leads to complete loss of function of betaine-homocysteine methyltransferase (BHMT) gene in hepatocellular carcinoma. Int. J. Biochem. Cell Biol. 44, 385–392 [DOI] [PubMed] [Google Scholar]
- 88. Teng Y. W., Mehedint M. G., Garrow T. A., Zeisel S. H. (2011) Deletion of betaine-homocysteine S-methyltransferase in mice perturbs choline and 1-carbon metabolism, resulting in fatty liver and hepatocellular carcinomas. J. Biol. Chem. 286, 36258–36267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Di Tommaso L., Franchi G., Park Y. N., Fiamengo B., Destro A., Morenghi E., Montorsi M., Torzilli G., Tommasini M., Terracciano L., Tornillo L., Vecchione R., Roncalli M. (2007) Diagnostic value of HSP70, glypican 3, and glutamine synthetase in hepatocellular nodules in cirrhosis. Hepatology 45, 725–734 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.