Abstract
Although adipogenesis is associated with induction of endoplasmic reticulum (ER) stress, the role of selenoprotein S (SEPS1), an ER resident selenoprotein known to regulate ER stress and ER-associated protein degradation, is unknown. We found an inverse relationship between SEPS1 level in adipose tissue and adiposity in mice. While SEPS1 expression was increased during adipogenesis, a markedly reduced SEPS1 protein level was found in the early phase of adipogenesis due to dexamethasone (DEX)-induced proteosomal degradation of SEPS1. Overexpression of SEPS1 in the early phase of cell differentiation resulted in impairment of adipogenesis with reduced levels of CCAAT/enhancer binding protein α and other adipocyte marker genes during the course of adipogenesis. Conversely, knockdown of SEPS1 resulted in the promotion of adipogenesis. Additionally, altered SEPS1 expression was associated with changes in expression of ER stress marker genes in the early phase of adipogenesis, and ubiquitin-proteasome system (UPS)-related ubiquitination and proteasome function. Our study reveals that SEPS1 is a novel anti-adipogenic selenoprotein that modulates ER stress- and UPS-dependent adipogenesis. Our results also identifies a novel function of DEX in the regulation of adipogenesis through induction of SEPS1 degradation. Taken together, DEX-dependent degradation of SEPS1 in the early phase of adipogenesis is necessary for initiating ER stress- and UPS-dependent maturation of adipocytes.
Keywords: 3T3-L1, adipocytes, glucocorticoid, endoplasmic reticulum stress, ubiquitin-proteasome system, endoplasmic reticulum-associated protein degradation, selenoprotein S
An excessive gain of adipose mass contributes to the development of obesity and its related insulin resistance. While an increase in both adipocyte size and number results in adipose tissue mass gain, adipocyte hyperplasia appears to control the whole body's lipid storing potential throughout life in humans (1). In vitro adipogenesis is possibly accelerated by treating with dexamethasone (DEX), a synthetic glucocorticoid; methylisobutylxanthine (MIX), a phosphodiesterase inhibitor; and insulin. In particular, DEX is necessary to induce adipogenesis. This is largely through glucocorticoid receptor-dependent modulation of adipogenesis-associated genes such as transcriptional activation of CCAAT/enhancer binding protein (C/EBP)δ (2), acetylation of C/EBPβ (3), and transcriptional suppression of preadipocyte factor-1 (pref-1) (a preadipocyte-specific inhibitor of adipogenesis) (4). In addition, DEX appears to modulate protein stability, as shown by its role in glucocorticoid-induced protein degradation in muscle atrophy via the ubiquitin-proteasome system (UPS) (5–9). However, DEX-degraded proteins required for adipogenesis are poorly characterized.
Selenoprotein S (SEPS1), also referred to as valosin-containing protein-interacting membrane protein (VIMP) (10), SelS (11), and Tanis (12), was identified as a gene involved in modulating inflammation and endoplasmic reticulum (ER) stress (11, 13, 14). As an ER resident selenoprotein, SEPS1 participates in nuclear factor-κB (NF-κB)-dependent inflammatory response (14). Moreover, suppression of SEPS1 is known to promote pro-inflammatory activation in macrophages (13). Additionally, other metabolic stresses, such as hypoglycemia and ER stress, are shown to induce SEPS1 expression through ER stress response elements (ERSEs) found in the SEPS1 promoter (11). We previously demonstrated that SEPS1 overexpression protects macrophages from ER stress-induced apoptosis, thereby promoting cell survival (15). This finding indicates that SEPS1 is likely to be an ER stress sensing protein that potentially acts as a negative feedback regulator of ER stress-induced cell death. Supporting the role of SEPS1 in ER stress, SEPS1 has been identified as a binding protein to p97 ATPase, also called valosin-containing protein (VCP) and cdc48 in yeast. The SEPS1 and VCP complex contributes to the formation of retrotranslocation/ER-associated protein degradation (ERAD) machinery for maintaining protein quality control and waste disposal process by proteasome-dependent degradation of accumulated misfolded proteins in ER (10).
ER stress is positively correlated with body mass index and obesity (16). Increased ER stress is found in both high-fat diet-induced obese and genetically obese animals. High levels of ER stress in obese animals are associated with the development of insulin resistance (17). Moreover, elevated ER stress and activation of the unfolded protein response (UPR) appear to be necessary for initiation of adipogenesis. Indeed, X-box binding protein 1 (XBP1), an ER stress-inducible transcription factor, was shown to mediate C/EBPβ-induced transcriptional activation of C/EBPα in the early phase of adipogenesis (18). Conversely, reducing ER stress with chemical chaperones or weight loss is likely to contribute to systemic improvement of energy homeostasis in obese animals (19) and humans (20), respectively.
Despite the protective role of SEPS1 in ER stress and inflammation, the role of SEPS1 in adipose development and obesity has not yet been elucidated. Here, we examine the role of SEPS1 in adipogenesis and its associated ER stress. We report an inverse relationship between SEPS1 level and adiposity in mice. We also observe a dramatic decrease in SEPS1 protein level in the early phase of adipogenesis. We further demonstrate an anti-adipogenic function of SEPS1 and this is through modulation of ER stress and its related UPS. We also found that SEPS1 is a proteasome-dependent DEX-degraded protein in the early phase of adipogenesis, which is required for an adequate level of adipogenesis.
MATERIALS AND METHODS
Materials and reagents
Fetal calf serum (FCS) and fetal bovine serum (FBS) were purchased from PAA Cell Culture Company. Dulbecco's Modified Eagle's medium (DMEM), 0.25% trypsin-EDTA, sodium pyruvate, and penicillin/streptomycin were obtained from Thermo Scientific. DEX, MIX, insulin, cycloheximide (CHX), MG132, and Oil Red O (ORO) were purchased from Sigma-Aldrich.
Animals
Five week old male C57BL/6 mice weighing 19.54 ± 0.52 g were purchased from Jackson Laboratory and maintained in a controlled environment with 40–60% humidity, 22–24°C, and a 12 h light/dark cycle. Animal experiments were approved by the Institutional Animal Care and Use Committee at Purdue University. After 1 week of acclimation to the facility, mice were fed a regular chow diet or a high-fat diet (60% calories from fat) for 6 weeks with free access to water and food.
Cell culture and differentiation of 3T3-L1 preadipocytes
3T3-L1 preadipocytes purchased from American Type Culture Collection were maintained in DMEM containing 10% (v/v) FCS at 37°C in a humidified 5% CO2 atmosphere. At day 0, when 3T3-L1 preadipocytes were 2 days postconfluent, adipogenesis was initiated with DMEM containing 10% FBS and an adipogenic cocktail (167 nM insulin, 0.5 mM MIX, and 5 μM DEX) for 2 days. Medium was then changed to 10% FBS-DMEM supplemented with insulin for another 2 days. Cells were then treated with 10% FBS-DMEM for an additional 2 days until day 6. All media contained final concentrations of 100 U/ml penicillin, 100 μg/ml streptomycin, and 0.11 g/l sodium pyruvate. Human embryonic kidney 293 (HEK293) cells were also cultured in 10% FBS-DMEM.
ORO staining
In order to visualize the intracellular lipid accumulation in differentiated 3T3-L1 cells, as described elsewhere (21), adipocytes were fixed with 3% formaldehyde for 30 min at room temperature and then incubated with fresh ORO solution for 1 h. The ORO stained cells were photographed. Spectrophotometric quantification of the ORO stained intracellular lipid was estimated by measuring optical density of isopropanol-extracted ORO dye from ORO stained cells at 490 nm.
Isolation of total RNA, RT-PCR, and quantitative RT-PCR
Total RNAs were extracted from 3T3-L1 cells and epididymal fat pads from lean and obese mice using Trizol® (Invitrogen) according to the manufacturer's instructions. Reverse transcription was performed to generate cDNA using 1 μg of isolated RNA and a SuperScriptII kit (Invitrogen). Generated cDNA was subjected to quantitative RT-PCR with SYBR premixed Taq reaction mixture and RT-PCR using thermocyclers purchased from Applied Biosystems and Bio-Rad Laboratories, respectively. Quantitative RT-PCR data were presented as relative expression level (mean ± SEM) using the ΔΔCT method. RT-PCR products were separated by agarose gel electrophoresis and visualized under UV light using BenchtopTransilluminator (UVP). The sequence of primers used in quantitative RT-PCR are as follows: SEPS1 (forward, 5′-GCT GGA CCA AGC CGA GAC T-3′; reverse, 5′-CTA GAA GCA TTT GCG GTG GAC GAT GGA GGG-3′); fatty acid synthase (FAS) (forward, 5′-ACC ACT GCA TTG ACG GCC GG-3′; reverse, 5′-GGG TCA GGC GGG AGA CCG AT-3′); peroxisome proliferator-activated receptor γ (PPARγ) (forward, 5′-CCC AAT GGT TGC TGA TTA CAA AT-3′; reverse, 5′-CTA CTT TGA TCG CAC TTT GGT ATT CT-3′); adiponectin (forward, 5′-GAT GCA GGT CTC TTG GTC CTA A-3′; reverse, 5′-GGC CCT TCA GCT CCT GTC A-3′); fatty acid binding protein 4 (FABP4) (forward, 5′-GCG TGG AAT TCG ATG AAA TCA-3′; reverse, 5′-CCC GCC ATC TAG GGT TAT GA-3′); adipsin (forward, 5′-GCT ATC CCA GAA TGC CTC GTT-3′; reverse, 5′-TTC CAC TTC TTT GTC CTC GTA TTG-3′); C/EBP homologous protein (CHOP) (forward, 5′-GTC CTG TCC TCA GAT GAA ATT GG-3′; reverse, 5′-AAG GTT TTT GAT TCT TCC TCT TCG T-3′); XBP1 (forward, 5′-GGA TTT GGA AGA AGA GAA CCA CAA-3′; reverse, 5′-CCG TGA GTT TTC TCC CGT AAA A-3′); spliced form of XBP1 (XBP1s) (forward, 5′-CAG CAC TCA GAC TAT GTG CA-3′; reverse, 5′-GTC CAT GGG AAG ATG TTC TGG-3′); glucose-regulated protein78 (GRP78) (forward, 5′-CGG ACG CAC TTG GAA TGA C-3′; reverse, 5′-AAC CAC CTT GAA TGG CAA GAA-3′); and β-actin (forward, 5′-TGA CGG GGT CAC CCA CAC TGT GCC CAT CTA-3′; reverse, 5′-CTA GAA GCA TTT GCG GTG GAC GAT GGA GGG-3′). Primers used in RT-PCR analysis are as follows: SEPS1 (forward, 5′-ATG GAT CGC GAT GAG GAA CCT-3′; reverse, 5′-TCA GCC GCC AGA TGA T-3′) and β-actin (forward, 5′-TGA CGG GGT CAC CCA CAC TGT GCC CAT CTA-3′; reverse, 5′-CTA GAA GCA TTT GCG GTG GAC GAT GGA GGG-3′).
Immunoblot analysis
After collecting cell pellets by scraping, cell lysates were prepared in cell lysis buffer containing 100 mM Tris-Cl (pH 7.5), 100 mM NaCl, 0.5% TritonX-100, protease inhibitor cocktails (Pierce), 1 mM Na3VO4, and 10 mM NaF. After 10 min of incubation on ice, cell lysates were centrifuged at 13,000 rpm for 15 min, and supernatants were harvested. The protein concentrations of cell lysates were assessed by Bradford reagent (Bio-Rad Laboratories). Proteins in the cell lysates were separated by 10% SDS-PAGE and then transferred to nitrocellulose membrane for performing immunoblot assay using anti-β-actin (Santa Cruz Biotechnology), anti-SEPS1/VIMP (Sigma-Aldrich), anti-ATF6 (IMGENEX), anti-Flag (Covance), anti-GRP78 (Genscript), anti-ubiquitin (Ub), and anti-GAPDH (Santa Cruz Biotechnology) antibodies, followed by incubation with horseradish peroxidase-conjugated secondary antibodies (Santa Cruz Biotechnology). Protein levels were visualized using an enhanced chemiluminescence reagent (22) by autoradiography. Films were scanned and processed using the National Institutes of Health ImageJ software (version 1.45S) for quantification of SEPS1 protein band intensity by normalization to the band intensity of β-actin.
Generation and transfection of Flag-SEPS1 plasmid
Expression vector for Flag-SEPS1 was generated by PCR reaction using a pair of primers: 5′-GCC ACC ATG GAT TAC AAG GAT GAC GAC GAT AAG GAA CGC CAA GA-3′, and 3′ primer, 5′′-GAA GTC CAT AAATCT CCT TG-3′, as previously described (15). The amplified PCR product was ligated into pTARGET vector (Promega), and then subcloned into pcDNA3.1 vector (Invitrogen). Approximately 50–70% confluent 3T3-L1 preadipocytes and HEK293 cells were transiently transfected with pcDNA3.1 vector or Flag-SEPS1/pcDNA3.1 expression vector using Lipofectamine 2000 (Invitrogen) by following the manufacturer's protocol. Transfected cells with the vectors were selected by culturing in 10% FCS-DMEM.
Construction of lentiviral plasmid, production of viral vector, and transfection
The lentivirus vector (pLKO.1) containing mouse SEPS1 (GenBank accession number NM_024439.3) short hairpin RNA (shRNA) sequence of AAT CAG GAA GGC CTC AGG AAG AAC T was generated as previously described (23). pLKO.1-SEPS1 or pLKO.1-empty plasmid was transfected to HEK293 cells with virus package plasmids of pHR′-CMV-ΔR8.20vpr and pHR′-CMV-VSV-G using Lipofectamine 2000 (Invitrogen). Viruses were harvested every 12 h from 24 to 72 h of posttransfection, cell culture medium. Collected medium was filtered through a 0.45 μm pore size filter and then centrifuged at 167,000 rpm for 90 min. The viral pellets were resuspended in 500 μl of TNE buffer (50 mM Tris-HCl, pH 7.8, 130 mM NaCl, and 1 mM EDTA) and then incubated overnight at 4°C. Approximately 50–70% confluent 3T3-L1 preadipocytes were transfected with the virus in serum-free DMEM medium including 10 μg/ml of polybrene and 10 mM HEPES. After 1 day of transduction, cells were selected with 0.8 μg/ml of puromycin for 16 days and the live cells were selected for the study.
Proteasome activity assay
For preparing a crude proteasome fraction, HEK293 cells or 3T3-L1 preadipocytes transfected with pcDNA3.1 or Flag-SEPS1/pcDNA3.1 vector were incubated with a lysis buffer containing 50 mM Tris (pH 7.5), 1 mM DTT, 0.25 mM sucrose, 5 mM MgCl2, 2 mM ATP, and 0.5 mM EDTA for 10 min at 4°C. Cells were sonicated at maximum power for 2–3 s followed by centrifugation at 3,000 rpm for 5 min at 4°C. To measure trypsin-, chymotrypsin-, and caspase-like proteasome activities, 5 μg of crude proteasome fraction was incubated with assay buffer [20 mM Tris (pH 7.5), 0.1% BSA, 2 mM MgCl2, and 1 mM ATP] at 37°C for 1 h in a black-bottomed 96-well plate in the presence of their corresponding fluorogenic substrates; 60 μM Boc-LRR-AMC (EMD Millipore), 60 μM Suc-LLVY-AMC (Fisher Scientific), and 15 μM AC-YVAD-AMC (Calbiochem), respectively. The fluorescence signal was then detected by a fluorescence spectrometer (Molecular Devices) using an excitation wavelength of 355 nm and an emission wavelength of 460 nm.
Statistical analysis
Data are shown as mean ± SEM. Statistical analysis was performed using SAS9.2 software. One-way ANOVA was used to determine the significance of treatment effect and interactions. Significant differences between group means were assessed by Dunnette's multiple comparison which is accepted at P < 0.05.
RESULTS
SEPS1 expression in diet-induced obese mice and during adipogenesis of 3T3-L1 preadipocytes
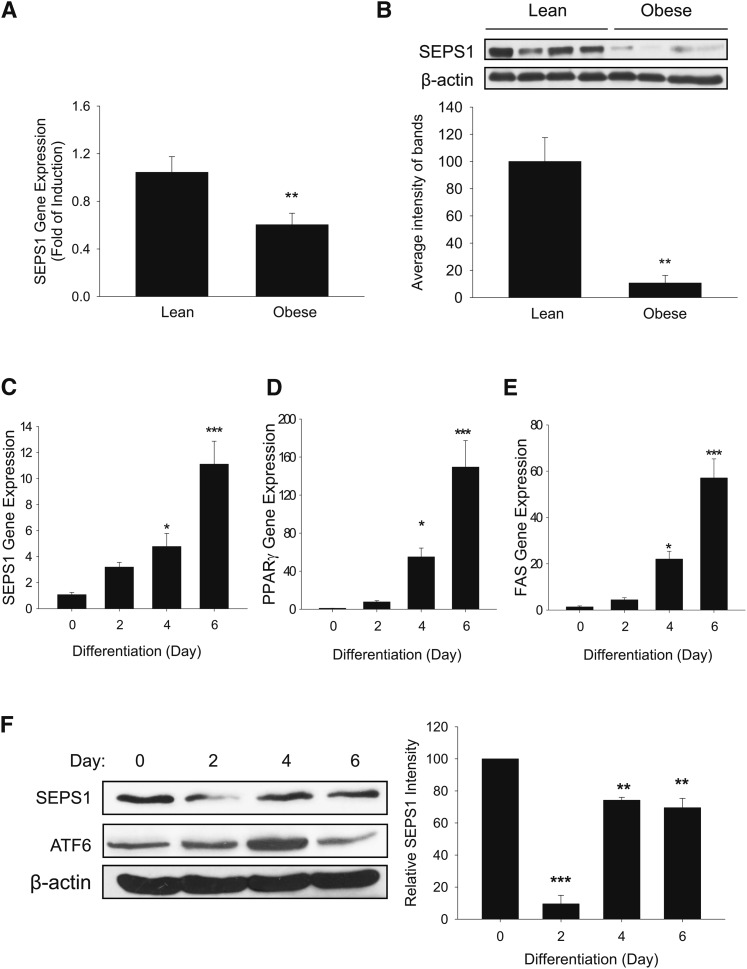
To investigate the role of SEPS1 in adipose development, we first examined the levels of SEPS1 mRNA and protein in epididymal white adipose tissue from lean and high-fat diet-induced obese mice. Feeding of a chow diet and a high-fat diet over 6 weeks resulted in an increase in body weight of lean mice (25.16 ± 0.59 g) and obese mice (33.84 ± 0.59 g) (P = 0.000024), respectively. The level of SEPS1 mRNA in the epididymal fat pad of obese mice was approximately 40% lower than that of lean mice as judged by quantitative PCR analysis (Fig. 1A). Consistently, the epididymal fat pad in obese mice exhibited an approximately 90% lower protein level of SEPS1 than that in lean mice as judged by immunoblot analysis (Fig. 1B). These data indicate that obesity status is inversely correlated with SEPS1 expression in adipose tissue.
Fig. 1.
SEPS1 gene and protein expression in adipose tissue from lean and obese mice, and during adipogenesis of 3T3-L1 preadipocytes. Epididymal white adipose tissue was isolated from lean and high-fat diet-induced obese mice for 6 weeks (n = 4/group). RNA and tissue homogenates prepared from white adipose tissue were subjected to quantitative RT-PCR and immunoblot analysis, respectively, for analyses of mRNA (A) and protein (B) levels of SEPS1. Adipogenesis was initiated in 2 day postconfluent 3T3-L1 preadipocytes for 6 days by treating with an adipogenic cocktail from day 0 to day 2, insulin-containing medium from day 2 to day 4, and growth medium from day 4 to day 6. Differentiating cells were harvested at days 0, 2, 4, and 6 for analyses of SEPS1 (C), PPARγ (D), and FAS (E) mRNA levels by quantitative RT-PCR. F: Protein levels of SEPS1 were estimated by immunoblot analysis. β-actin was used as a control. Band intensities of SEPS1 normalized by β-actin were quantified using ImageJ 1.45S software. Data represent mean ± SEM (n = 3). *P < 0.05; **P < 0.01; ***P < 0.001.
To further examine the role of SEPS1 in adipogenesis, the expression of SEPS1 was monitored at different stages of adipogenesis in 3T3-L1 preadipocytes. As shown in Fig. 1C, an increase in SEPS1 mRNA level was observed during adipogenesis with its maximum level at day 6. Adipogenesis of these cells was confirmed by monitoring expression patterns of PPARγ and FAS (Fig. 1D, E). In contrast, SEPS1 protein level was dramatically decreased within 2 days after initiation of adipogenesis. This was followed by a rapid increase in SEPS1 protein at days 4 and 6 (Fig. 1F). Consistent with a previous report (24), the level of ATF6, a marker of ER stress, was increased during the first 4 days of adipogenesis followed by a subsequently decreased level at day 6 (Fig. 1F). Collectively, these results suggest that SEPS1 protein level in the early phase of adipogenesis is likely regulated at the posttranslational level.
DEX-induced SEPS1 degradation in the early phase of adipogenesis is mediated by proteosomal activity
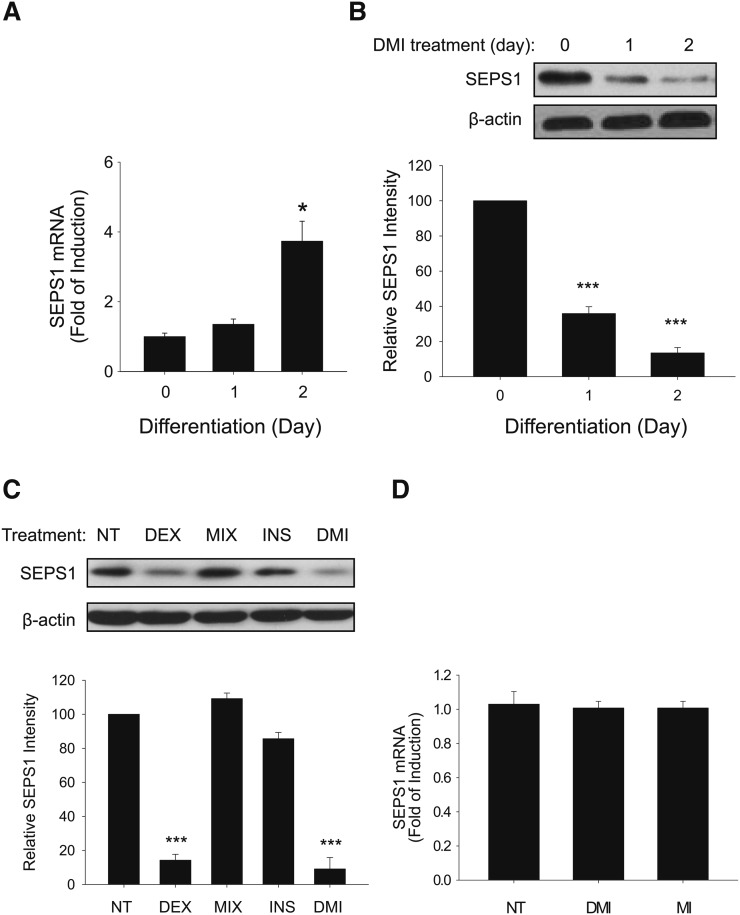
We further examined the degradation profile of SEPS1 protein in the early phase of adipogenesis at day 0 to day 2. We first found that the SEPS1 mRNA level was significantly increased from day 1 to day 2 to approximately 4-fold in 3T3-L1 preadipocytes treated with adipogenic cocktail (Fig. 2A). Contrary to this, the SEPS1 protein level was markedly decreased at days 1 and 2 (Fig. 2B). Because standard protocol of in vitro adipogenesis requires a 2 day incubation of postconfluent preadipocytes with an adipogenic cocktail (a mixture of DEX, MIX, and insulin), we hypothesized that components in the adipogenic cocktail promote SEPS1 degradation in the early phase of adipogenesis. To test this, we examined the SEPS1 level in 3T3-L1 preadipocytes after 2 days of individual or combined treatments of the components in adipogenic cocktail. As shown in Fig. 2C, a 2 day treatment of preadipocytes with 5 μM DEX alone markedly lowered the SEPS1 level to a similar extent to that in DMI treatment. However, MIX or insulin alone showed no effect on SEPS1 level (Fig. 2C). While SEPS1 protein in 3T3-L1 preadipocytes was reduced by the treatments with DEX alone and DEX containing adipogenic cocktail, this was not a consequence of lower SEPS1 mRNA expression by DEX (Fig. 2D), supporting the possibility of the existence of a DEX-dependent regulatory mechanism of SEPS1 protein stability.
Fig. 2.
DEX treatment lowers SEPS1 protein level during the early stage of adipogenesis of 3T3-L1 preadipocytes. Two day postconfluent 3T3-L1 preadipocytes were incubated with the adipogenic cocktail containing DEX, MIX, and insulin (INS) for 1 and 2 days. A: SEPS1 mRNA levels were examined in the early stage of adipogenesis from day 0 to day 2 by quantitative RT-PCR. The signal of β-actin was used for normalization. B: Levels of SEPS1 protein at days 0, 1 and 2 were detected by immunoblot analysis. β-actin was used as a loading control. C: Two day postconfluent 3T3-L1 preadipocytes were stimulated by DEX, MIX, INS, or adipogenic cocktail (DMI) for 48 h. NT, nontreated. Protein samples were subjected to immunoblot analysis for detection of SEPS1 and β-actin levels. Intensity of the protein bands was quantified using ImageJ 1.45S software. Relative SEPS1 intensity was presented after normalization by β-actin band intensity. D: SEPS1 mRNA levels in 3T3-L1 cells stimulated by adipogenic cocktail (DMI) or adipogenic cocktail without DEX (MI) for 2 days were determined by quantitative RT-PCR. Data represent mean ± SEM (n = 3). *P < 0.05; ***P < 0.001.
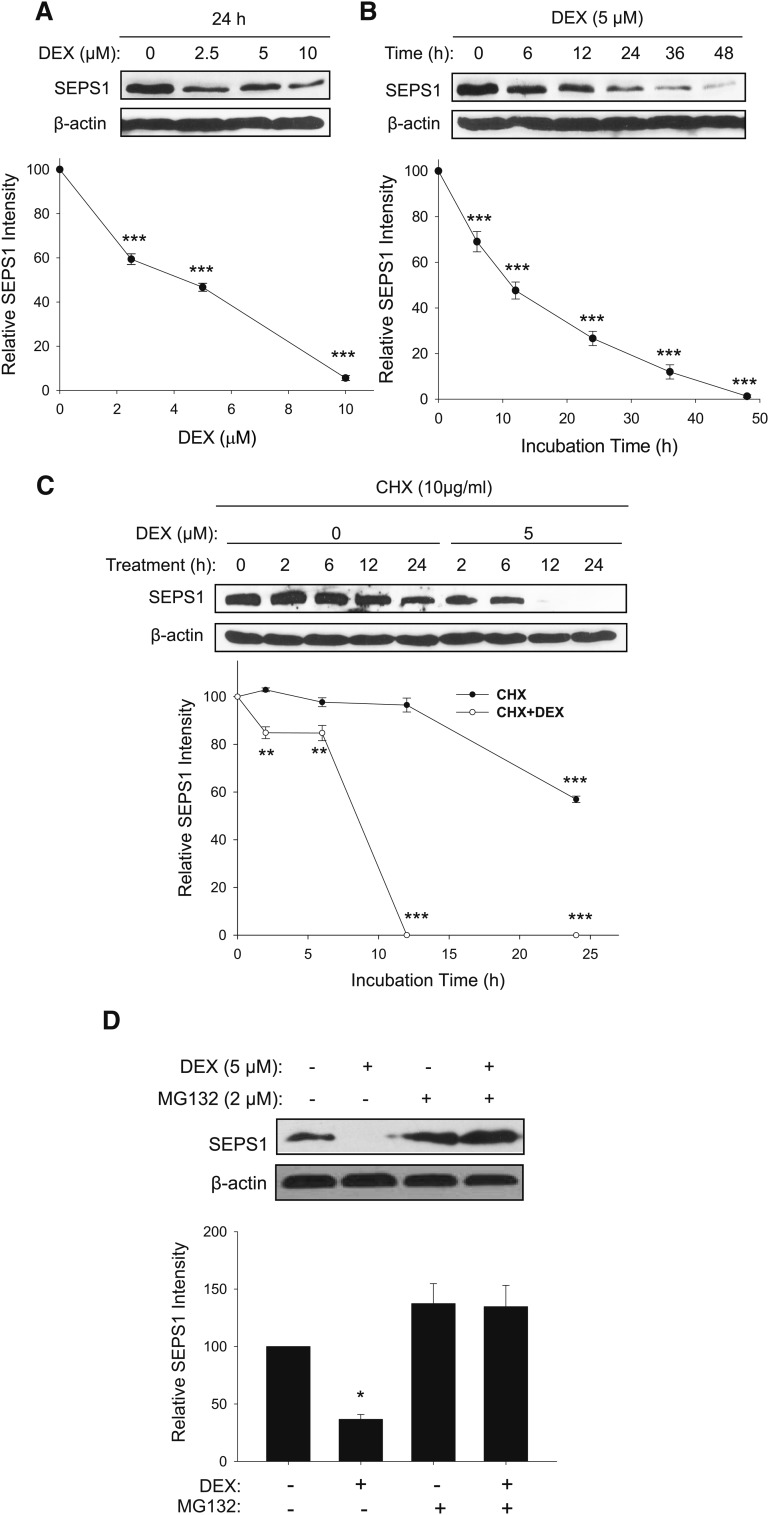
We further examined the DEX-dependent degradation kinetics of SEPS1 in preadipocytes. DEX induced SEPS1 degradation in a dose-dependent manner with an approximate half-maximal inhibitory concentration (IC50) of 4.5 μM after 24 h of treatment (Fig. 3A). In addition, preadipocytes exposed to 5 μM DEX showed degradation of SEPS1 in a time-dependent manner with an approximate half-life of 13 h (Fig. 3B). To ascertain protein stability, SEPS1 levels in preadipocytes treated with DEX (0–5 μM) for 24 h were then examined in the presence or absence of CHX, an inhibitor of protein synthesis. The SEPS1 level in CHX-treated preadipocytes remained unchanged until 12 h, followed by a marked decrease after 24 h with an approximate half-life of 30 h (Fig. 3C). However, addition of 5 μM DEX to CHX-treated preadipocytes resulted in a rapid degradation of SEPS1 with a complete degradation after 12 h and an approximate half-life of 7.5 h (Fig. 3C). Because glucocorticoids have been shown to induce protein degradation partly through UPS-dependent proteolytic pathway in muscle cells (25, 26), we tested whether DEX-induced SEPS1 degradation in the early phase of adipogenesis could be through activation of proteasome activity in preadipocytes. While 24 h of DEX treatment lowered the SEPS1 level in preadipocytes, this DEX-induced SEPS1 degradation was blunted by treatment with MG132, a proteasome inhibitor (Fig. 3D). However, MG132 alone showed a slight, but not significant, increase in SEPS1 level in preadipocytes. Collectively, our results indicate that DEX is responsible for SEPS1 degradation in the early phase of adipogenesis, and that this appears to be through DEX-induced proteasome activity in preadipocytes.
Fig. 3.
DEX in the adipogenic cocktail causes SEPS1 protein degradation in dose- and time-dependent manners with reduced half-life. 3T3-L1 preadipocytes were exposed to various concentrations of DEX (0–10 μM) for 24 h (A), or 5 μM of DEX for various time points (0–48 h) (B). These cells were then subjected to immunoblot analysis to examine SEPS1 levels. C: 3T3-L1 preadipocytes were treated with 10 μg/ml of CHX in the presence (+) or absence (−) of 5 μM of DEX. After 2, 6, 12, and 24 h of incubation, protein samples were harvested for immunoblot analysis of SEPS1. D: 3T3-L1 preadipocytes treated with 5 μM of DEX in the presence or absence of 2 μM of MG132 for 24 h were subjected to immunoblot analysis to determine SEPS1 levels. Intensity of the protein bands was quantified using ImageJ 1.45S software. Relative SEPS1 intensity was presented after normalization by β-actin band intensity. Data represent mean ± SEM (n = 3). *P < 0.05; **P < 0.01; ***P < 0.001.
SEPS1 protein modulates adipogenesis and its associated ER stress
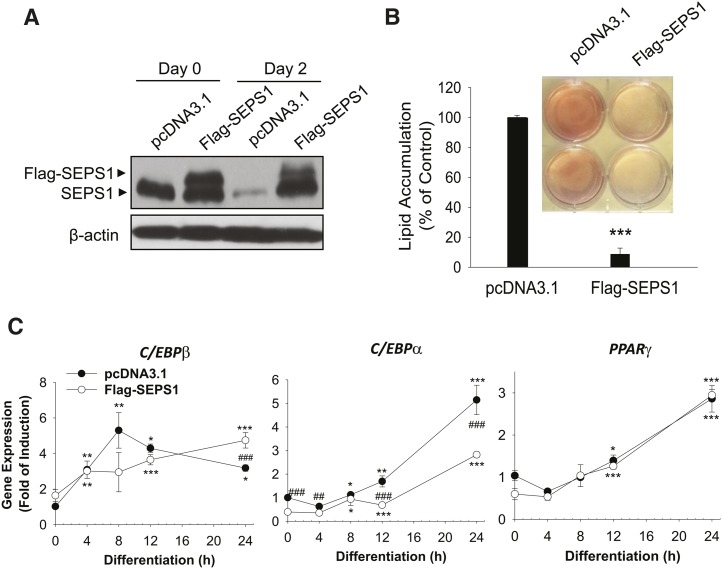
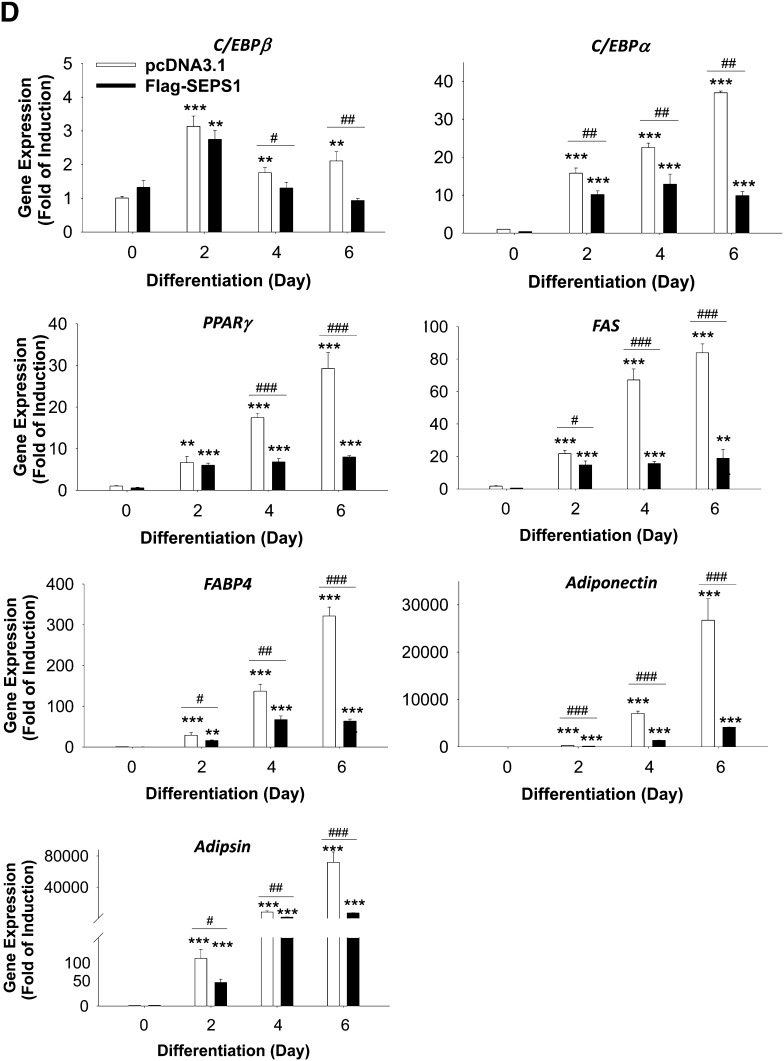
To investigate the impact of SEPS1 on adipogenesis, we studied the effects of altered SEPS1 expression on adipogenesis. We first examined the effect of SEPS1 overexpression in the early phase of adipogenesis on overall adipogenesis. While control vector transfected 3T3-L1 preadipocytes exhibited a reduced level of endogenous SEPS1 at day 2, cells transfected with Flag-SEPS1 expression vector exhibited an elevated level of Flag-tagged SEPS1 fusion protein to the similar extent of the endogenous SEPS1 level at day 0, and both endogenous SEPS1 and Flag-SEPS1 proteins at day 2 of adipogenesis as judged by immunoblot analysis (Fig. 4A). Interestingly, Flag-SEPS1 overexpression resulted in an elevated level of endogenous SEPS1 at day 2 of adipogenesis in 3T3-L1 cells (Fig. 4A). This overexpression of Flag-SEPS1 and elevated endogenous SEPS1 in differentiating preadipocytes in the early phase of adipogenesis resulted in an inhibition of adipogenesis by 90% as judged by ORO staining of accumulated lipid in adipocytes at day 6 (Fig. 4B). To further characterize the effect of SEPS1 overexpression on the transcription program of adipogenesis, we examined the mRNA expression levels of adipocyte marker genes in various phases of adipogenesis in differentiating 3T3-L1 cells. Quantitative PCR revealed that SEPS1 overexpression resulted in an altered expression of C/EBPβ with a significant suppression of C/EBPα within the first 24 h of adipogenesis; however, SEPS1 overexpression did not affect PPARγ expression in the early phase of adipogenesis (Fig. 4C). Consistently, C/EBPα expression was markedly decreased in SEPS1 overexpressing cells during the course of adipogenesis with an approximately 75% decrease in gene expression at day 6 (Fig. 4D). We also found that mRNA levels of C/EBPβ and PPARγ were significantly lower in SEPS1 overexpressing cells at intermediate (i.e., day 4) and late (i.e., day 6) phases of adipogenesis (Fig. 4D). Consequently, expression of other adipocyte marker genes such as FAS, FABP4, adiponectin, and adipsin was also dramatically decreased in SEPS1 overexpressing cells at various phases of adipogenesis (Fig. 4D). These data further demonstrate that SEPS1 plays an inhibitory role in adipogenesis by primarily targeting expression of the early adipogenic transcription factors such as C/EBPβ and C/EBPα, and subsequently their downstream adipocyte marker genes during the course of cell differentiation.
Fig. 4.
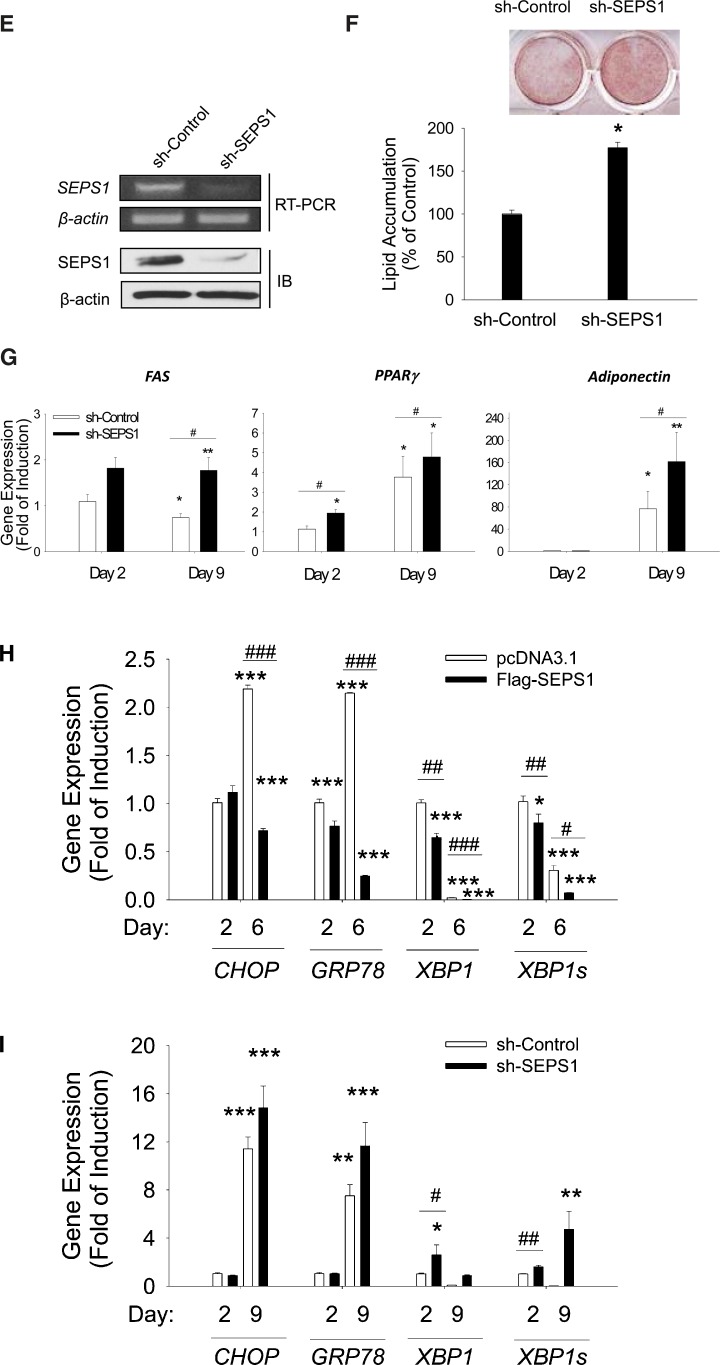
Altered SEPS1 level in preadipocytes modulates adipogenesis and its associated ER stress. A: Levels of endogenous SEPS1 and Flag-SEPS1 fusion protein in 3T3-L1 preadipocytes transfected with pcDNA3.1 or Flag-SEPS1/pcDNA3.1 vector were confirmed by immunoblot analysis at day 0 and day 2 of adipogenesis using anti-SEPS1 antibody. B: After 6 days of adipogenesis, intracellular lipid droplets in differentiated adipocytes were stained with ORO for an image analysis (top panel). ORO stained lipid droplets were extracted for a quantification of lipid accumulation as described in Materials and Methods (bottom panel). The mRNA levels of adipogenic transcription factors (C/EBPβ, C/EBPα, and PPARγ) and adipocyte marker genes (FAS, adiponectin, FABP4, and adipsin) in pcDNA 3.1 or Flag-SEPS1/pcDNA3.1 vector transfected 3T3-L1 preadipocytes were examined by quantitative RT-PCR at 0, 4, 8, 12, and 24 h (C), or at days 0, 2, 4, and 6 of adipogenesis (D). E: 3T3-L1 preadipocytes infected with lentivirus-containing control shRNA (sh-Control) or SEPS1 shRNA (sh-SEPS1) were subjected to RT-PCR and immunoblot analysis (IB) to verify SEPS1 deficiency. F: These cells were incubated with adipogenic cocktail to initiate adipogenesis for 9 days, followed by ORO staining to visualize (top panel) and quantify (bottom panel) accumulated intracellular lipid droplets. G: The mRNA levels of FAS, PPARγ, and adiponectin in 3T3-L1 preadipocytes infected with lentivirus-containing control shRNA (sh-Control) or SEPS1 shRNA (sh-SEPS1) were measured by quantitative RT-PCR at day 2 and day 9 of adipogenesis. H, I: mRNA levels of ER stress marker genes, including CHOP, GRP78, XBP1, and XBP1s in cells in panels (C) and (F) were examined by quantitative RT-PCR analysis. Data represent mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001; #P < 0.05; ##P < 0.01; ###P < 0.001 versus pcDNA3.1 transfected or sh-Control cells. Data represent mean ± SEM (n = 3). The experiment was conducted at least three times.
Next, we investigated the requirement of SEPS1 gene for adipogenesis by examining the effect of SEPS1 knockdown on differentiating 3T3-L1 cells infected with lentivirus expressing SEPS1 shRNA. RT-PCR and immunoblot analysis confirmed the downregulation of SEPS1 mRNA and its gene product in 3T3-L1 preadipocytes, respectively, after RNA interference (Fig. 4E). Lentivirus-mediated SEPS1 knockdown in preadipocytes markedly promoted adipogenesis as evidenced by elevation of intracellular lipid accumulation at day 9 by ∼177% relative to control adipocytes (Fig. 4F). This was associated with significant induction of FAS and adiponectin mRNA at day 9, and PPARγ mRNA both at day 2 and day 9 in differentiating cells transduced with lentivirus expressing SEPS1 shRNA compared with control cells (Fig. 4G).
Given that activation of ER stress is required for adipogenesis (18), and that SEPS1 has been shown to modulate ER stress in nonadipocytes (14, 15, 27), we questioned whether SEPS1-regulated adipogenesis is associated with altered ER stress. To test this, we examined the effects of altered SEPS1 expression on expression of ER stress marker genes in the early and late phases of adipogenesis in 3T3-L1 preadipocytes. As shown in Fig. 4H, Flag-SEPS1 overexpression resulted in suppression of both full-length XBP1 and XBP1s genes by approximately 30% and 40% respectively at day 2, whereas no significant changes in CHOP and GRP78 levels were observed. Moreover, Flag-SEPS1 overexpression led to marked reduction of CHOP, GRP78, XBP1, and XBP1s gene expression at day 6 of adipogenesis (Fig. 4H). Conversely, lentiviral SEPS1 knockdown resulted in an increase in XBP1 and XBP1s mRNA levels by ∼2.5-fold and 1.7-fold, respectively, at day 2 of adipogenesis with no effect on CHOP and GRP78 levels (Fig. 4I). Lentiviral SEPS1 knockdown further resulted in a significant induction of CHOP, GRP78, XBP1, and XBP1s gene expression at day 9 of adipogenesis (Fig. 4I).
Taken together, these results unequivocally demonstrate that SEPS1 is a novel regulator of adipogenesis in that SEPS1 exhibits an anti-adipogenic property and DEX-induced degradation of SEPS1 protein in the early phase of adipogenesis is required for adipogenesis and its related ER stress in vitro.
SEPS1 modulates ER stress-associated UPS
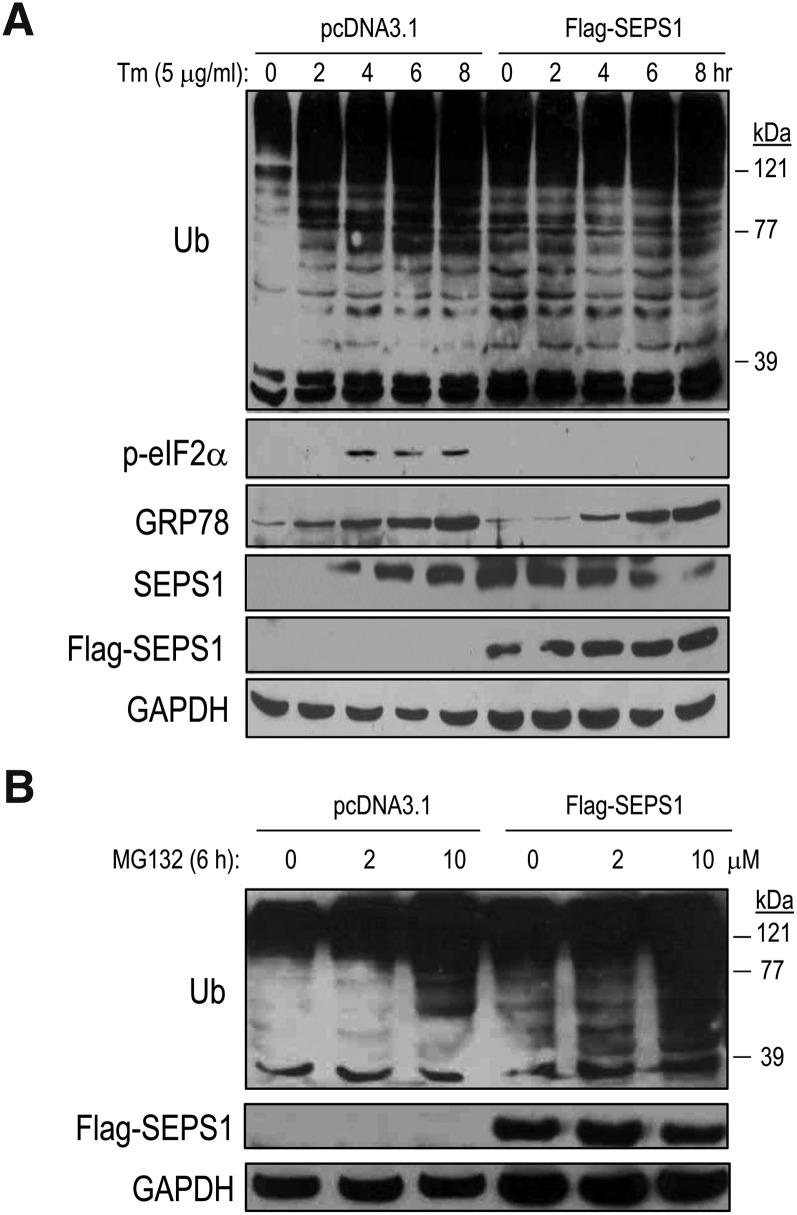
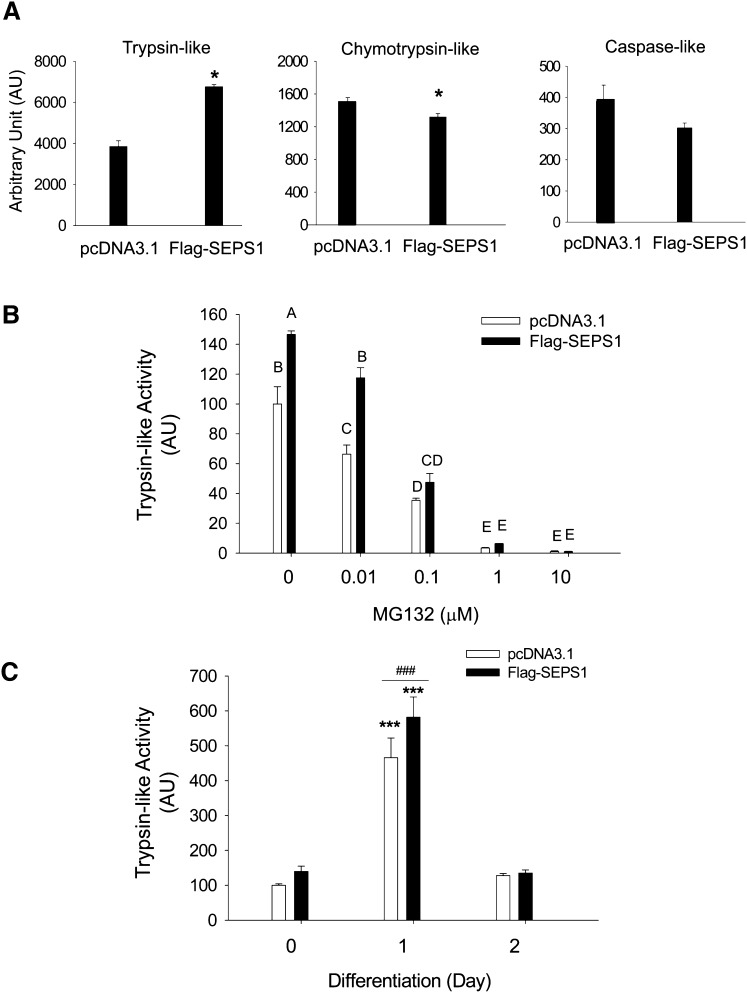
Because SEPS1 is known to participate in VCP-mediated ERAD (10), we further sought to determine the effect of SEPS1 on ER stress-related UPS. Using HEK293 cells transiently transfected with Flag-SEPS1/pcDNA3.1 vector, we investigated the effect of SEPS1 overexpression on ER stress-induced protein ubiquitination by immunoblot analysis. Control vector-transfected cells treated with tunicamycin (Tm), an N-glycosylation inhibitor and a pharmacological ER stress inducer (28, 29), displayed a time-dependent induction of ER stress marker proteins. This is evidenced by increased levels of phosphorylated eukaryotic translation initiation factor 2 (eIF2)α, GRP78, and endogenous SEPS1, and generation of ubiquitinated proteins in HEK293 cells (Fig. 5A). In contrast, Flag-SEPS1 overexpression resulted in elevated levels of ubiquitinated proteins even in the absence of Tm. Consistent with our previous study in macrophages (15), we also observed that Flag-SEPS1 overexpression resulted in inhibition of Tm-induced p-eIF2α and delayed induction of GRP78 expression in HEK293 cells (Fig. 5A). Intriguingly, Flag-SEPS1 overexpression impacted the endogenous SEPS1 level in HEK293 cells. Moreover, Flag-SEPS1 overexpressing cells treated with MG132 displayed a greater level of ubiquitinated proteins than control cells treated with MG132 (Fig. 5B). These results suggest that elevated levels of both Flag-SEPS1 and endogenous SEPS1 facilitate protein ubiquitination. This is likely to be through SEPS1-induced ubiquitination-dependent protein degradation. To test the latter possibility, we examined the effect of SEPS1 on proteasome activity by performing fluorogenic proteasome function assays using SEPS1 overexpressing HEK293 cells and 3T3-L1 preadipocytes. Because the 26S proteasome has at least three distinct proteosomal activities, including trypsin-, chymotrypsin-, and caspase-like activities, we examined these enzyme activities using their corresponding fluorogenic substrates: Boc-LRR-AMC, Suc-LLVY-AMC, and AC-YVAD-AMC, respectively. Trypsin-like activity was significantly increased by ∼1.75-fold in SEPS1 overexpressing HEK293 cells (Fig. 6A). In contrast, SEPS1 overexpression resulted in a slight decrease in chymotrypsin-like activity and no significant effect on caspase-like activity (Fig. 6A). SEPS1 overexpressing HEK293 cells were resistant to dose-dependent inhibition of trypsin-like activity by MG132 (Fig. 6B). Consistent with these results, SEPS1 overexpression in differentiating 3T3-L1 preadipocytes caused an increase in trypsin-like proteasome activity of approximately 20% at day 1 of adipogenesis (Fig. 6C). Taken together, our studies identify SEPS1 as an ER stress sensing protein with the capacity to potentiate ER stress-associated UPS.
Fig. 5.
SEPS1 overexpression modulates ER stress-induced protein ubiquitination and ER marker protein expression. HEK293 cells transfected with pcDNA3.1 or Flag-SEPS1/pcDNA3.1 vector were incubated with 5 μg/ml of Tm for 0, 2, 4, 6, and 8 h (A) or 0–10 μM of MG132 for 6 h (B). Cells were then subjected to immunoblot analysis to detect ubiquitinated proteins, phosphorylated eIF2α (p-eIF2α), GRP78, endogenous SEPS1, and/or Flag-SEPS1 proteins using anti-ubiquitin (Ub), anti-GRP78, anti-SEPS1, and/or anti-Flag antibodies. GAPDH was used as a loading control.
Fig. 6.
SEPS1 overexpression increases trypsin-like proteasome activity. A: HEK293 cells transfected with pcDNA3.1 or Flag-SEPS1/pcDNA3.1 vector were subjected to trypsin-, chymotrypsin-, and caspase-like proteosomal activity assays using their corresponding fluorogenic substrates as described in Materials and Methods. *P < 0.05. B: Trypsin-like activity was determined in pcDNA3.1 or Flag-SEPS1/pcDNA3.1 vector transfected HEK293 cells exposed to 0–10 μM of MG132 for 6 h. Values without a common letter differ, P < 0.05. C: 3T3-L1 preadipocytes transfected with pcDNA3.1 or Flag-SEPS1/pcDNA3.1 vector were incubated with adipogenic cocktail for 1 or 2 days. These cells were then subjected to trypsin-like proteasome activity assay. ***P < 0.001; ###P < 0.001 versus pcDNA3.1 transfected cells. Data represent mean ± SEM (n = 3). The experiment was conducted at least three times. AU, arbitrary unit.
DISCUSSION
Selenocysteine-containing selenoproteins (25 found in human and 24 in mouse) are largely known to participate in maintaining cellular redox homeostasis. Moreover, many selenoproteins such as selenoprotein W, selenoprotein P, glutathione peroxidase 4, thioredoxin reductase, and selenoprotein T are known to modulate various cellular processes such as muscle development (30), bone remodeling (31), neurodegeneration (32), epithelial growth (33), and neuroendocrine secretion (34), respectively. While a recent study indicated a role of selenoprotein P in promoting adipocyte differentiation and its related inflammation (35), the role of SEPS1 in adipogenesis remained elusive. SEPS1 is an ER resident protein and it is known to act as a negative regulator in ER stress signaling pathway and inflammation (13, 15). Furthermore, SEPS1 is shown to participate in modulating ERAD, a ubiquitin-proteasome-dependent cellular survival mechanism against ER stress-induced cellular dysfunctions (10, 36, 37). Because elevated ER stress and inflammation were reported to promote the development of obesity and its related insulin resistance, we hypothesized that SEPS1 plays a critical role in adipose development through its negative function in ER stress and its dependent ERAD.
In this study, we showed an inverse correlation i) between SEPS1 expression and obesity in vivo and ii) between SEPS1 mRNA and protein levels in the early phase of adipogenesis in vitro. We also demonstrated that the inconsistency between SEPS1 mRNA and protein levels in the early phase of adipogenesis is due to DEX-induced degradation of SEPS1 protein. The level of functional SEPS1 in the early phase of adipogenesis appears to be tightly regulated for adequate adipogenesis because our study of overexpression of SEPS1 and in preadipocytes resulted in impaired adipogenesis. This is likely through a SEPS1-induced marked suppression of C/EBPα in the early phase of adipogenesis, followed by a subsequent reduction of mRNA levels of adipocyte marker genes during the course of adipogenesis. Supporting our finding of the anti-adipogenic function of SEPS1, knockdown of SEPS1 resulted in an enhancement of adipogenesis. These results implicate SEPS1 as a novel selenoprotein that could play an essential role in the development of adipose tissue. It should be noted that our results also revealed a previously unknown function of DEX in adipogenesis. We found that DEX helped maintain a low level of SEPS1 protein by promoting SEPS1 degradation in the early phase of differentiation, thereby contributing to an adequate level of adipogenesis. While more studies are needed to understand the molecular basis underlying DEX-dependent proteolytic regulation of adipose development, DEX has been reported to play a key role in muscle atrophy and muscle wasting-associated cancer cachexia (9). These studies indicated that DEX participates in induction of ubiquitin-dependent protein degradation (6–8). It should also be noted that SEPS1 overexpression appears to impact the endogenous SEPS1 level through an unknown mechanism because Flag-SEPS1 expression resulted in an elevated level of endogenous SEPS1 in adipocytes (Fig. 4A) and HEK293 cells (Fig. 5A). Supporting this notion, Flag-SEPS1 expression has been previously shown to increase the endogenous SEPS1 level in hepatoma cells (38) and macrophages (15).
Based on the previously suggested role of SEPS1 in ER stress and ERAD, it would be plausible to assume that the anti-adipogenic function of SEPS1 is likely through modulation of ER stress and ERAD-associated UPS during the differentiation of preadipocytes. Indeed, our study demonstrates that ER stress-inducible SEPS1 plays a critical role in the expression of XBP1, an ER stress-induced transcription factor required for initiation of the early adipogenic transcription program (18), in the early phase of adipogenesis. We also provided evidence of a potential role of SEPS1 in UPS: SEPS1 overexpression resulted in accumulation of ubiquitinated proteins and activation of trypsin-like proteasome activity. To our knowledge, this is the first report to place SEPS1 in a key position for UPR-regulated adipogenesis.
While understanding of the mechanism underlying SEPS1-regulated ER stress and UPS remains elusive, it is possible that this is through a binding of SEPS1 to other protein components in ERAD machinery, such as VCP, a cytosolic AAA-ATPase protein, and Derlin-1, an ER membrane bound protein important for ERAD substrate recognition. This VCP/SEPS1/Derlin-1 protein complex might then participate in eliminating ER accumulated misfolded proteins by facilitating polyubiquitination and retrotranslocation of these proteins into the cytosol for their proteasomal degradation (10, 36, 39, 40). Although the role of SEPS1 in ERAD is still unclear, the importance of VCP in ERAD has been demonstrated as inactivation of VCP (41) and RNA interference of VCP (42) caused accumulation of misfolded proteins with apparently little effect on proteasome activity, respectively. Moreover, inactivation of Derlin-1 impaired elimination of misfolded proteins in the ER and caused ER stress (10), whereas overexpression of Derlin-2 (a Derlin-1 homologous protein) or Derlin-3 accelerated degradation of misfolded glycoproteins, and their knockdown blocked ERAD (43). Thus, it cannot be ruled out that SEPS1-regulated ER stress and ubiquitin-proteasome function during adipogenesis is a mixed consequence of SEPS1-dependent alteration of VCP and/or Derlin-1 function in the ERAD machinery.
In humans, increased SEPS1 levels were found in adipose tissue from insulin-infused type 2 diabetic subjects (44). Genetic variations of human SEPS1 located on chromosome 15q26.3, which was suggested to contain quantitative trait loci influencing inflammation-related disorders such as type 2 diabetes, Alzheimer's disease, and celiac disease (45–47), were reported to be strongly associated with inflammation in Mexican American individuals (13). Recently, variation in the SEPS1 gene locus has been reported to be associated with coronary heart disease (48), preeclampsia (49), gastric cancer (50), non-small cell lung cancer (51), and aggressive periodontitis (52). Some of the single nuclear polymorphisms in the SEPS1 gene were found in the conserved ERSE in the promoter region: this appears to alter inducibility of SEPS1 by ER stress and its potential anti-inflammatory function (13). Because development of adipose tissue and obesity is accompanied by elevated levels of ER stress and accumulation of misfolded/unfolded proteins in the ER with presumably overloaded ERAD machinery, it would be of great interest to explore the function of SEPS1 and its interacting ERAD machinery proteins in obesity and its related metabolic and inflammatory disorders.
Increased levels of glucocorticoids are implicated to impact energy balance, adipose dysfunction, and obesity (53–57). This appears to be largely due to 11β-hydroxysteroid dehydrogenase 1 (11β-HSD1)- and glucocorticoid receptor-dependent action of glucocorticoids in adipose tissue. Central glucocorticoid infusion (55) and overexpression of 11β-HSD1 in adipose tissue (58) resulted in visceral obesity and metabolic syndrome in animals. Conversely, mice with adipose tissue-specific inhibition of glucocorticoids (54) and 11β-HSD1 knockout mice (59) displayed resistance to diet-induced obesity with improved energy balance. In line with this important role of glucocorticoids in adipose dysfunction, our findings of a reduced level of SEPS1 protein in adipose tissue from obese mice (Fig. 1B) and DEX-induced degradation of anti-adipogenic SEPS1 protein in preadipocytes (Fig. 2) reveal SEPS1 as a novel target of glucocorticoid-induced proteolysis in mediating adiposity and the development of obesity. Future studies will be directed to uncover the physiological function of SEPS1 in glucocorticoid-dependent adipose dysfunction in vivo.
Overall, we have identified SEPS1 as a novel anti-adipogenic selenoprotein and that DEX-induced degradation of SEPS1 in the early phase of adipogenesis is required for ER stress- and UPS-dependent adipocyte differentiation. Our results provide new insights into ER stress- and UPS-regulated development of adipose tissue.
Acknowledgments
The authors thank Chih-Yu Chen, Yuyan Zhu, and Jonathan Kershaw for critical review of the manuscript.
Footnotes
Abbreviations:
- C/EBP
- CCAAT/enhancer binding protein
- CHOP
- CCAAT/enhancer binding protein homologous protein
- CHX
- cycloheximide
- DEX
- dexamethasone
- eIF2
- eukaryotic translation initiation factor 2
- ER
- endoplasmic reticulum
- ERAD
- endoplasmic reticulum-associated protein degradation
- ERSE
- endoplasmic reticulum stress response element
- FCS
- fetal calf serum
- GR
- glucocorticoid receptor
- GRP78
- glucose-regulated protein 78
- HEK293
- human embryonic kidney 293
- 11β-HSD1
- 11β-hydroxysteroid dehydrogenase 1
- MIX
- methylisobutylxanthine
- ORO
- Oil Red O
- PPARγ
- peroxisome proliferator-activated receptor γ
- SEPS1
- selenoprotein S
- shRNA
- short hairpin RNA
- Tm
- tunicamycin
- Ub
- ubiquitin
- UPR
- unfolded protein response
- UPS
- ubiquitin-proteasome system
- VCP
- valosin-containing protein
- VIMP
- valosin-containing protein-interacting membrane protein
- XBP1
- X-box binding protein 1
- XBP1s
- spliced form of X-box binding protein 1
This work was supported by a start-up fund from Purdue University (K-H.K.).
REFERENCES
- 1.Spalding K. L., Arner E., Westermark P. O., Bernard S., Buchholz B. A., Bergmann O., Blomqvist L., Hoffstedt J., Naslund E., Britton T., et al. 2008. Dynamics of fat cell turnover in humans. Nature. 453: 783–787 [DOI] [PubMed] [Google Scholar]
- 2.Wu Z., Bucher N. L., Farmer S. R. 1996. Induction of peroxisome proliferator-activated receptor gamma during the conversion of 3T3 fibroblasts into adipocytes is mediated by C/EBPbeta, C/EBPdelta, and glucocorticoids. Mol. Cell. Biol. 16: 4128–4136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wiper-Bergeron N., Salem H. A., Tomlinson J. J., Wu D., Hache R. J. 2007. Glucocorticoid-stimulated preadipocyte differentiation is mediated through acetylation of C/EBPbeta by GCN5. Proc. Natl. Acad. Sci. USA. 104: 2703–2708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smas C. M., Chen L., Zhao L., Latasa M. J., Sul H. S. 1999. Transcriptional repression of pref-1 by glucocorticoids promotes 3T3–L1 adipocyte differentiation. J. Biol. Chem. 274: 12632–12641 [DOI] [PubMed] [Google Scholar]
- 5.Yang H., Wei W., Menconi M., Hasselgren P. O. 2007. Dexamethasone-induced protein degradation in cultured myotubes is p300/HAT dependent. Am. J. Physiol. Regul. Integr. Comp. Physiol. 292: R337–R344 [DOI] [PubMed] [Google Scholar]
- 6.Isozaki U., Mitch W. E., England B. K., Price S. R. 1996. Protein degradation and increased mRNAs encoding proteins of the ubiquitin-proteasome proteolytic pathway in BC3H1 myocytes require an interaction between glucocorticoids and acidification. Proc. Natl. Acad. Sci. USA. 93: 1967–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shah O. J., Kimball S. R., Jefferson L. S. 2000. Glucocorticoids abate p70(S6k) and eIF4E function in L6 skeletal myoblasts. Am. J. Physiol. Endocrinol. Metab. 279: E74–E82 [DOI] [PubMed] [Google Scholar]
- 8.Sun L., Trausch-Azar J. S., Muglia L. J., Schwartz A. L. 2008. Glucocorticoids differentially regulate degradation of MyoD and Id1 by N-terminal ubiquitination to promote muscle protein catabolism. Proc. Natl. Acad. Sci. USA. 105: 3339–3344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Polge C., Heng A. E., Jarzaguet M., Ventadour S., Claustre A., Combaret L., Bechet D., Matondo M., Uttenweiler-Joseph S., Monsarrat B., et al. 2011. Muscle actin is polyubiquitinylated in vitro and in vivo and targeted for breakdown by the E3 ligase MuRF1. FASEB J. 25: 3790–3802 [DOI] [PubMed] [Google Scholar]
- 10.Ye Y., Shibata Y., Yun C., Ron D., Rapoport T. A. 2004. A membrane protein complex mediates retro-translocation from the ER lumen into the cytosol. Nature. 429: 841–847 [DOI] [PubMed] [Google Scholar]
- 11.Gao Y., Feng H. C., Walder K., Bolton K., Sunderland T., Bishara N., Quick M., Kantham L., Collier G. R. 2004. Regulation of the selenoprotein SelS by glucose deprivation and endoplasmic reticulum stress - SelS is a novel glucose-regulated protein. FEBS Lett. 563: 185–190 [DOI] [PubMed] [Google Scholar]
- 12.Walder K., Kantham L., McMillan J. S., Trevaskis J., Kerr L., De Silva A., Sunderland T., Godde N., Gao Y., Bishara N., et al. 2002. Tanis: a link between type 2 diabetes and inflammation? Diabetes. 51: 1859–1866 [DOI] [PubMed] [Google Scholar]
- 13.Curran J. E., Jowett J. B., Elliott K. S., Gao Y., Gluschenko K., Wang J., Abel Azim D. M., Cai G., Mahaney M. C., Comuzzie A. G., et al. 2005. Genetic variation in selenoprotein S influences inflammatory response. Nat. Genet. 37: 1234–1241 [DOI] [PubMed] [Google Scholar]
- 14.Gao Y., Hannan N. R., Wanyonyi S., Konstantopolous N., Pagnon J., Feng H. C., Jowett J. B., Kim K. H., Walder K., Collier G. R. 2006. Activation of the selenoprotein SEPS1 gene expression by pro-inflammatory cytokines in HepG2 cells. Cytokine. 33: 246–251 [DOI] [PubMed] [Google Scholar]
- 15.Kim K. H., Gao Y., Walder K., Collier G. R., Skelton J., Kissebah A. H. 2007. SEPS1 protects RAW264.7 cells from pharmacological ER stress agent-induced apoptosis. Biochem. Biophys. Res. Commun. 354: 127–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berghöfer A., Pischon T., Reinhold T., Apovian C. M., Sharma A. M., Willich S. N. 2008. Obesity prevalence from a European perspective: a systematic review. BMC Public Health. 8: 200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ozcan U., Cao Q., Yilmaz E., Lee A. H., Iwakoshi N. N., Ozdelen E., Tuncman G., Gorgun C., Glimcher L. H., Hotamisligil G. S. 2004. Endoplasmic reticulum stress links obesity, insulin action, and type 2 diabetes. Science. 306: 457–461 [DOI] [PubMed] [Google Scholar]
- 18.Sha H., He Y., Chen H., Wang C., Zenno A., Shi H., Yang X., Zhang X., Qi L. 2009. The IRE1alpha-XBP1 pathway of the unfolded protein response is required for adipogenesis. Cell Metab. 9: 556–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ozcan U., Yilmaz E., Ozcan L., Furuhashi M., Vaillancourt E., Smith R. O., Gorgun C. Z., Hotamisligil G. S. 2006. Chemical chaperones reduce ER stress and restore glucose homeostasis in a mouse model of type 2 diabetes. Science. 313: 1137–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gregor M. F., Yang L., Fabbrini E., Mohammed B. S., Eagon J. C., Hotamisligil G. S., Klein S. 2009. Endoplasmic reticulum stress is reduced in tissues of obese subjects after weight loss. Diabetes. 58: 693–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim K. A., Kim J. H., Wang Y., Sul H. S. 2007. Pref-1 (preadipocyte factor 1) activates the MEK/extracellular signal-regulated kinase pathway to inhibit adipocyte differentiation. Mol. Cell. Biol. 27: 2294–2308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cornelius L. A., Nehring L. C., Harding E., Bolanowski M., Welgus H. G., Kobayashi D. K., Pierce R. A., Shapiro S. D. 1998. Matrix metalloproteinases generate angiostatin: effects on neovascularization. J. Immunol. 161: 6845–6852 [PubMed] [Google Scholar]
- 23.Liu X., Lei M., Erikson R. L. 2006. Normal cells, but not cancer cells, survive severe Plk1 depletion. Mol. Cell. Biol. 26: 2093–2108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lowe C. E., Dennis R. J., Obi U., O'Rahilly S., Rochford J. J. 2012. Investigating the involvement of the ATF6α pathway of the unfolded protein response in adipogenesis. Int. J. Obes. (Lond.). 36: 1–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dardevet D., Sornet C., Taillandier D., Savary I., Attaix D., Grizard J. 1995. Sensitivity and protein turnover response to glucocorticoids are different in skeletal muscle from adult and old rats. Lack of regulation of the ubiquitin-proteasome proteolytic pathway in aging. J. Clin. Invest. 96: 2113–2119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang L., Luo G. J., Wang J. J., Hasselgren P. O. 1998. Dexamethasone stimulates proteasome- and calcium-dependent proteolysis in cultured L6 myotubes. Shock. 10: 298–306 [DOI] [PubMed] [Google Scholar]
- 27.Kelly E., Greene C. M., Carroll T. P., McElvaney N. G., O'Neill S. J. 2009. Selenoprotein S/SEPS1 modifies endoplasmic reticulum stress in Z variant alpha1-antitrypsin deficiency. J. Biol. Chem. 284: 16891–16897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liao W., Chan L. 2001. Tunicamycin induces ubiquitination and degradation of apolipoprotein B in HepG2 cells. Biochem. J. 353: 493–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Menéndez-Benito V., Verhoef L. G., Masucci M. G., Dantuma N. P. 2005. Endoplasmic reticulum stress compromises the ubiquitin-proteasome system. Hum. Mol. Genet. 14: 2787–2799 [DOI] [PubMed] [Google Scholar]
- 30.Yeh J. Y., Ou B. R., Forsberg N. E., Whanger P. D. 1997. Effects of selenium and serum on selenoprotein W in cultured L8 muscle cells. Biometals. 10: 11–22 [DOI] [PubMed] [Google Scholar]
- 31.Dreher I., Schutze N., Baur A., Hesse K., Schneider D., Kohrle J., Jakob F. 1998. Selenoproteins are expressed in fetal human osteoblast-like cells. Biochem. Biophys. Res. Commun. 245: 101–107 [DOI] [PubMed] [Google Scholar]
- 32.Wirth E. K., Conrad M., Winterer J., Wozny C., Carlson B. A., Roth S., Schmitz D., Bornkamm G. W., Coppola V., Tessarollo L., et al. 2010. Neuronal selenoprotein expression is required for interneuron development and prevents seizures and neurodegeneration. FASEB J. 24: 844–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nalvarte I., Damdimopoulos A. E., Nystom C., Nordman T., Miranda-Vizuete A., Olsson J. M., Eriksson L., Bjornstedt M., Arner E. S., Spyrou G. 2004. Overexpression of enzymatically active human cytosolic and mitochondrial thioredoxin reductase in HEK-293 cells. Effect on cell growth and differentiation. J. Biol. Chem. 279: 54510–54517 [DOI] [PubMed] [Google Scholar]
- 34.Grumolato L., Ghzili H., Montero-Hadjadje M., Gasman S., Lesage J., Tanguy Y., Galas L., Ait-Ali D., Leprince J., Guerineau N. C., et al. 2008. Selenoprotein T is a PACAP-regulated gene involved in intracellular Ca2+ mobilization and neuroendocrine secretion. FASEB J. 22: 1756–1768 [DOI] [PubMed] [Google Scholar]
- 35.Zhang Y., Chen X. 2011. Reducing selenoprotein P expression suppresses adipocyte differentiation as a result of increased preadipocyte inflammation. Am. J. Physiol. Endocrinol. Metab. 300: E77–E85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ye Y., Shibata Y., Kikkert M., van Voorden S., Wiertz E., Rapoport T. A. 2005. Recruitment of the p97 ATPase and ubiquitin ligases to the site of retrotranslocation at the endoplasmic reticulum membrane. Proc. Natl. Acad. Sci. USA. 102: 14132–14138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ye Y. 2005. The role of the ubiquitin-proteasome system in ER quality control. Essays Biochem. 41: 99–112 [DOI] [PubMed] [Google Scholar]
- 38.Gao Y., Pagnon J., Feng H. C., Konstantopolous N., Jowett J. B., Walder K., Collier G. R. 2007. Secretion of the glucose-regulated selenoprotein SEPS1 from hepatoma cells. Biochem. Biophys. Res. Commun. 356: 636–641 [DOI] [PubMed] [Google Scholar]
- 39.Ye Y., Meyer H. H., Rapoport T. A. 2001. The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature. 414: 652–656 [DOI] [PubMed] [Google Scholar]
- 40.Stolz A., Hilt W., Buchberger A., Wolf D. H. 2011. Cdc48: a power machine in protein degradation. Trends Biochem. Sci. 36: 515–523 [DOI] [PubMed] [Google Scholar]
- 41.Griciuc A., Aron L., Roux M. J., Klein R., Giangrande A., Ueffing M. 2010. Inactivation of VCP/ter94 suppresses retinal pathology caused by misfolded rhodopsin in Drosophila. PLoS Genet. 6: e1001075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wójcik C., Yano M., DeMartino G. N. 2004. RNA interference of valosin-containing protein (VCP/p97) reveals multiple cellular roles linked to ubiquitin/proteasome-dependent proteolysis. J. Cell Sci. 117: 281–292 [DOI] [PubMed] [Google Scholar]
- 43.Oda Y., Okada T., Yoshida H., Kaufman R. J., Nagata K., Mori K. 2006. Derlin-2 and Derlin-3 are regulated by the mammalian unfolded protein response and are required for ER-associated degradation. J. Cell Biol. 172: 383–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Karlsson H. K., Tsuchida H., Lake S., Koistinen H. A., Krook A. 2004. Relationship between serum amyloid A level and Tanis/SelS mRNA expression in skeletal muscle and adipose tissue from healthy and type 2 diabetic subjects. Diabetes. 53: 1424–1428 [DOI] [PubMed] [Google Scholar]
- 45.Field L. L., Tobias R., Magnus T. 1994. A locus on chromosome 15q26 (IDDM3) produces susceptibility to insulin-dependent diabetes mellitus. Nat. Genet. 8: 189–194 [DOI] [PubMed] [Google Scholar]
- 46.Blacker D., Bertram L., Saunders A. J., Moscarillo T. J., Albert M. S., Wiener H., Perry R. T., Collins J. S., Harrell L. E., Go R. C., et al. 2003. Results of a high-resolution genome screen of 437 Alzheimer's disease families. Hum. Mol. Genet. 12: 23–32 [DOI] [PubMed] [Google Scholar]
- 47.Susi M., Holopainen P., Mustalahti K., Maki M., Partanen J. 2001. Candidate gene region 15q26 and genetic susceptibility to coeliac disease in Finnish families. Scand. J. Gastroenterol. 36: 372–374 [DOI] [PubMed] [Google Scholar]
- 48.Alanne M., Kristiansson K., Auro K., Silander K., Kuulasmaa K., Peltonen L., Salomaa V., Perola M. 2007. Variation in the selenoprotein S gene locus is associated with coronary heart disease and ischemic stroke in two independent Finnish cohorts. Hum. Genet. 122: 355–365 [DOI] [PubMed] [Google Scholar]
- 49.Moses E. K., Johnson M. P., Tommerdal L., Forsmo S., Curran J. E., Abraham L. J., Charlesworth J. C., Brennecke S. P., Blangero J., Austgulen R. 2008. Genetic association of preeclampsia to the inflammatory response gene SEPS1. Am. J. Obstet. Gynecol. 198: 336.e1–336.e5 [DOI] [PubMed] [Google Scholar]
- 50.Shibata T., Arisawa T., Tahara T., Ohkubo M., Yoshioka D., Maruyama N., Fujita H., Kamiya Y., Nakamura M., Nagasaka M., et al. 2009. Selenoprotein S (SEPS1) gene -105G>A promoter polymorphism influences the susceptibility to gastric cancer in the Japanese population. BMC Gastroenterol. 9: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hart K., Landvik N. E., Lind H., Skaug V., Haugen A., Zienolddiny S. 2011. A combination of functional polymorphisms in the CASP8, MMP1, IL10 and SEPS1 genes affects risk of non-small cell lung cancer. Lung Cancer. 71: 123–129 [DOI] [PubMed] [Google Scholar]
- 52.Scapoli C., Mamolini E., Carrieri A., Guarnelli M. E., Annunziata M., Guida L., Romano F., Aimetti M., Trombelli L. 2011. Gene–gene interaction among cytokine polymorphisms influence susceptibility to aggressive periodontitis. Genes Immun. 12: 473–480 [DOI] [PubMed] [Google Scholar]
- 53.Veyrat-Durebex C., Deblon N., Caillon A., Andrew R., Altirriba J., Odermatt A., Rohner-Jeanrenaud F. 2012. Central glucocorticoid administration promotes weight gain and increased 11beta-hydroxysteroid dehydrogenase type 1 expression in white adipose tissue. PLoS ONE. 7: e34002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kershaw E. E., Morton N. M., Dhillon H., Ramage L., Seckl J. R., Flier J. S. 2005. Adipocyte-specific glucocorticoid inactivation protects against diet-induced obesity. Diabetes. 54: 1023–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zakrzewska K. E., Cusin I., Stricker-Krongrad A., Boss O., Ricquier D., Jeanrenaud B., Rohner-Jeanrenaud F. 1999. Induction of obesity and hyperleptinemia by central glucocorticoid infusion in the rat. Diabetes. 48: 365–370 [DOI] [PubMed] [Google Scholar]
- 56.Dallman M. F., Warne J. P., Foster M. T., Pecoraro N. C. 2007. Glucocorticoids and insulin both modulate caloric intake through actions on the brain. J. Physiol. 583: 431–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chapman K. E., Seckl J. R. 2008. 11beta-HSD1, inflammation, metabolic disease and age-related cognitive (dys)function. Neurochem. Res. 33: 624–636 [DOI] [PubMed] [Google Scholar]
- 58.Masuzaki H., Paterson J., Shinyama H., Morton N. M., Mullins J. J., Seckl J. R., Flier J. S. 2001. A transgenic model of visceral obesity and the metabolic syndrome. Science. 294: 2166–2170 [DOI] [PubMed] [Google Scholar]
- 59.Kotelevtsev Y., Holmes M. C., Burchell A., Houston P. M., Schmoll D., Jamieson P., Best R., Brown R., Edwards C. R., Seckl J. R., et al. 1997. 11beta-hydroxysteroid dehydrogenase type 1 knockout mice show attenuated glucocorticoid-inducible responses and resist hyperglycemia on obesity or stress. Proc. Natl. Acad. Sci. USA. 94: 14924–14929 [DOI] [PMC free article] [PubMed] [Google Scholar]