Abstract
Plant-associated bacteria provide important services to host plants. Environmental factors such as cultivar type and pedoclimatic conditions contribute to shape their diversity. However, whether these environmental factors may influence the plant growth promoting (PGP) potential of the root-associated bacteria is not widely understood. To address this issue, the diversity and PGP potential of the bacterial assemblage associated with the grapevine root system of different cultivars in three Mediterranean environments along a macrotransect identifying an aridity gradient were assessed by culture-dependent and independent approaches. According to 16S rRNA gene PCR-DGGE, the structure of endosphere and rhizosphere bacterial communities was highly diverse (P = 0.03) and was associated with a cultivar/latitudinal/climatic effect. Despite being diverse, the bacterial communities associated with Egyptian grapevines shared a higher similarity with the Tunisian grapevines than those cultivated in North Italy. A similar distribution, according to the cultivar/latitude/aridity gradients, was observed for the cultivable bacteria. Many isolates (23%) presented in vitro multiple stress resistance capabilities and PGP activities, the most frequent being auxin synthesis (82%), insoluble phosphate solubilisation (61%), and ammonia production (70%). The comparable numbers and types of potential PGP traits among the three different environmental settings indicate a strong functional homeostasis of beneficial bacteria associated with grape root.
1. Introduction
Grapevine is among the most ancient crops grown in the Mediterranean basin. Historically, grape and derived products held an important economic role in the area, and this importance persists to the present day. Vineyards are extremely sensitive to phytopathogen attacks, and a huge interest has been devoted to understanding the mechanisms of virulence [1] and environmental-friendly biocontrol approaches [2]. Certain biocontrol strategies rely on the exploitation of beneficial traits of plant growth-promoting (PGP) microorganisms that are naturally associated with plants [3].
Little attention has been dedicated to bacteria associated with grapevine, and, subsequently, relatively few studies are available. It was shown that grapevine tissues, including flowers, berries pulp, and seeds, host an endophytic community that encompasses, among others, bacteria affiliated to the Bacillus, Burkholderia, and Pseudomonas genera [4–9]. Despite these studies, little information is currently available on the taxonomic and functional diversity of the bacterial communities associated with the endosphere and root system of grapevine. Recently, we observed that Enterobacteriaceae were dominant in the root system of five different Barbera cv. rootstocks cultivated in Oltrepò Pavese, with a differential genera distribution and colonization of the rhizosphere and endosphere [10].
Even fewer studies have investigated the potential of grapevine-associated bacteria in promoting plant resistance to abiotic stresses [11, 12]. Water deprivation reduces fruit yield remarkably and causes quality losses, and most of the grapevine-growing regions worldwide suffer seasonal periods of drought [13]. Although generally referred to as a drought-resistant plant, grapevine is severely affected in terms of fruit yield and quality by the cooccurrence of elevated temperatures and high evaporation rates that reduce carbohydrate content in berries and cause wilting of leaves. Even at temperate latitudes and cool climates, extensive periods of drought coupled with certain soil types and topography in the vineyard may determine anticipated harvests and influence phenology [14]. In both managed and natural ecosystems, the interaction of plants with the associated native-drought resistant microbiome is an important factor in supporting plant health and physiology under water stress [15, 16]. As sessile organisms, plants adopt different strategies to persist in the face of unfavourable environmental settings, including phenotypic plasticity or “escape and migrate” mechanisms. Rapid adaptation of plants to low soil moisture is achieved through changes in the structure and functionality of the belowground microbiome [17]. The selective force exerted by water scarcity combined with unfavourable harsh environmental conditions in arid lands enriches the rhizosphere of beneficial bacteria that exhibit antagonistic activity against phytopathogens and promote plant resistance to drought [16, 18]. Recently, we observed that the treatment of grape plantlets with selected bacteria determined an increase in epigeal biomass and the formation of a larger root system during drought stress [10].
It is well known that bacteria play key roles in promoting plant growth in conventional and extreme ecosystems [16, 19], and that a plethora of environmental factors such as the cultivar type or pedoclimatic conditions can affect and modulate the structure of bacterial microbiomes [20]. However, the influence of such environmental factors on the PGP potential of root-associated bacteria is poorly understood.
The present study aims to assess the range of bacterial diversity and functional PGP potential of culturable bacteria associated with grapevine roots growing in three different agrosystems in the Mediterranean basin, in order to evaluate whether such PGP potential is independent from the specific environmental conditions. The sampling sites were located in North Italy, North Tunisia, and North Egypt, drawing a latitudinal/aridity macro-transect in the Mediterranean basin. The structure of the bacterial communities associated with the root endosphere and soil of grapevines in the three sites was dissected by 16S rRNA gene-based PCR-DGGE (denaturing gradient gel electrophoresis) analysis. The results were compared with the diversity of the culturable bacteria and their PGP potential.
2. Materials and Methods
2.1. Study Area and Sample Collection
Three different agroclimatic regions of the Mediterranean basin were chosen for sampling. During July 2008, grapevine root samples were collected from vineyards located in Mornag, North Tunisia, (TN) and in an arid farm 30 km north-west of Cairo, Egypt (ET). In July 2009, grapevine roots were collected from a vineyard of the “Le Frecce” farm, North Italy (IT). The sites were located along a latitude gradient, from 44° to 30°N (IT: 44°57′N, TN: 36°84′N, ET: 30°2′N). The roots (root tissues and rhizosphere) of healthy grapevine plants were collected at 50–60 cm depth. After removing the roots, the root-surrounding soil was collected and the bulk soil was sampled at a distance of 4 m from the grapevine plants. All soil and root samples were collected under sterile conditions using sterile tools. Recovered samples were stored at −20°C for molecular analysis or at 4°C for isolation and processed the following day in the laboratory.
2.2. Total DNA Extraction and PCR from All Root System Fractions
Grapevine roots with attached soil particles were placed in a 50 mL screw-cap tube containing 9 mL of physiological solution (9 g/L NaCl) to separate the rhizosphere soil by vortexing. The root tissues removed from the 50 mL screw-cap tube were sterilized as described by Sun et al. [21] by immersion in 70% ethanol for 3 min, sodium hypochlorite solution (2.5% available Cl−) for 5 min, and 70% ethanol for 30 seconds. After these treatments, the tissues were washed five times with sterile distilled water. The efficacy of the sterilization method was verified by plating the water of the last washing step on PAF medium (10 g/L proteose peptone, 10 g/L hydrolyzed casein, 3 g/L MgSO4, 1.5 g/L K2HPO4, 10 mL/L glycerol, and 15 g/L agar for solid medium). Total DNA was extracted from the soil fractions (rhizosphere, root-surrounding soil, and bulk soil) and root tissues using a Power Soil kit (MoBio) and DNeasy Plant kit (Qiagen), respectively, according to the manufacturer's procedure. The DNA was quantified and stored at −20°C until use. PCR amplification of the 16S rRNA gene was performed using the 907R and 357F primers, adding a GC-clamp to the forward primer [22]. PCR reaction was performed in 0.2 mL tubes in a final volume of 50 μL containing the 1x diluted buffer, 1.5 mM MgCl2, 5% DMSO, 0.12 mM of a mixture of dNTPs, 0.3 µM of each primer, 1 U Taq polymerase, and 10 ng of template. When necessary, DNA was properly diluted. The amplification program consisted of an initial denaturing step at 94°C for 4 min, followed by 10 cycles of 94°C for 0.5 min, 61°C for 1 min, and 72°C for 1 min, followed by further 20 cycles at 94°C for 0.5 min, 56°C for 1 min, 72°C for 1 min, and a final extension at 72°C for 7 min. Two μL of the PCR products was analyzed by electrophoresis in 1% agarose gels.
2.3. PCR-DGGE and Profile Analysis
DGGE was performed using polyacrylamide gel (8% of a 37 : 1 acrylamide-bisacrylamide mixture in a Tris acetate EDTA (TAE) 1x buffer, 0.75 mm thick, 16 × 10 cm) with a 45–60% denaturant gradient. Gels were run overnight at 90 V in TAE 1x buffer at 60°C in DCode apparatus (Bio-Rad, Italy). The gels were stained with 1x Sybr Green (Life Technologies) and scanned with gel photo GS-800 system. The DGGE bands were excised from the gels using a sterile scalpel and eluted in 50 μL water at 37°C for 3 hours. The DNA eluted from DGGE bands was amplified using 907R and 357F primers (without the GC-clamp) [22]. The PCR was performed in a final volume of 50 μL with the same conditions as above and using the following protocol: 95°C for 5 min, 30 cycles of 95°C for 1 min, 61°C for 1 min, 72°C for 1 min, and a final extension at 72°C for 7 min. The PCR products obtained were sequenced by Macrogen Inc. (Korea). The DGGE band patterns were converted to a binary dataset by using ImageJ software [23]. Principal component analysis (PCA) was carried out using XLSTAT (version 7.5.2 Addinsoft, France). Analysis of variance along the PCA axis was evaluated using the statistical test ANOVA and Student's t test with significance at P ≤ 0.05.
The nonparametric statistical test PERMANOVA [24] was used to test the null hypothesis, in which there were no differences between microbial assemblages of sites; PERMANOVA was conducted with the factors of site (fixed, orthogonal, and three levels IT, TN, and ET) and microbial community (fixed, orthogonal, two levels, endophyte, and rhizosphere).
To test for significant relationships among the microbiological assemblage and climate traits [25], a distance-based multivariate analysis for a linear model (DistLM) and distance-based redundancy analysis (dbRDA) [26] were used. PERMANOVA and DistLM were performed with software PERMANOVA + for PRIMER 6 [27].
2.4. Isolation of Cultivable Bacteria
One gram of each soil fraction (rhizosphere, root-surrounding soil, and bulk soil) and one gram of triturated root were used as inoculum for ACC-deaminase enrichment culture as described by Penrose and Glick [28]. The medium was supplemented with 100 μg/mL of the fungicide cycloheximide. CFUs per gram of sample were calculated, and 50 colonies from each fraction were randomly selected and propagated on PAF medium plates. One gram of each fraction sample, suspended in 9 mL of sterile physiological solution (9 g/L NaCl), was diluted in 10-fold series and plated in triplicate onto KB medium (20 g/L peptone, 1.5 g/L dipotassium sulphate, 3.2 g/L magnesium dichloride, 10 mL/L glycerol, and pH = 7.2) [29] and on R2A medium (Oxoid). After three days of incubation at 30°C, a total count was performed, and twelve colonies per medium per fraction were randomly selected. For colony purification, all the isolated colony types were spread three times on the original medium. A total of 769 purified isolates were frozen in 25% glycerol at −80°C until use.
2.5. DNA Extraction of Isolates, Dereplication and PCR of 16S rRNA Gene
Purified bacterial colonies were resuspended in 50 μL of sterile TE (10 mM Tris/HCl, pH 8, and 1 mM EDTA) in 1.5 mL tubes. Tubes were incubated at 95°C for 8 min and centrifuged at 13000 rpm for 10 min. The supernatant containing the DNA was stored at −20°C and used for PCR. Isolates were dereplicated using the ITS-PCR fingerprinting protocol [30–32]. Two μL of the PCR products was checked by electrophoresis in 1.5% agarose gel and stained with ethidium bromide. Gel images were captured using Gel Doc 2000 system (Bio-Rad, Milan, Italy), and bacteria redundancy was reduced by evaluating the different ITS profiles. One strain per each ITS haplotype was used in the phylogenetic analysis and for further experiments. A total of 331 strains isolated on ACCd [28], R2A (Oxoid), and KB [29] media were characterized by 16S rRNA gene sequencing. The reaction mixture contained the diluted buffer, 1.5 mM of MgCl2, 0.12 mM of a mixture of dNTPs, 0.3 μM of each primer, 1 U of Taq polymerase, and 10 ng of template. The universal primers were 27F (3′-AGAGTTTGATCMTGGCTCAG-5′) and 1492R (3′-CTACGGCTACCTTGTTACGA-5′). Conditions for amplification consisted of an initial denaturation at 94°C for 4 min, followed by 35 cycles of 94°C for 0.5 min, 55°C for 1 min, 72°C for 2 min, and a final extension at 72°C for 10 min. The PCR products were checked by electrophoresis in 1% agarose gel, and sequencing service was performed by Macrogen Inc., South Korea.
2.6. Characterization of Plant Growth Promoting Activity and Abiotic Stress Resistance
The 331 bacterial strains identified were screened for production of indole acetic acid (IAA), siderophores, exopolysaccharide (EPS) and ammonia productions, mineral phosphate solubilization, protease activity and tolerance to drought, salt, and osmotic stress. The ability of isolated strains to produce IAA was evaluated in the original liquid medium supplemented with L-tryptophan (100 mg/L) as described by Bric et al. [33]. Strains were considered as IAA-producers for concentrations higher than 2 μg/mL. Pure IAA (Sigma-Aldrich Co., Italy) was used to prepare the standard curve and to quantify the amount of IAA produced. Siderophore release was detected in a modified PAF medium (without Fe) using the Chrome Azurol S (CAS) method described by Schwyn and Neilands [34]. Bacterial culture was streaked on a half plate containing growth media. Plates were incubated at 30°C for 7 days, and the formation of orange or pink halos indicated the presence of siderophore. The mineral P-solubilizing ability of the strains was determined on Pikovskaya's liquid medium amended with 0.5% tricalcium phosphate [Ca3(PO4)2] as inorganic P [35]. Exopolysaccharides (EPSs) production was estimated as described by Santarella et al. [36], using the modified Weaver mineral media enriched with 20 g/L sucrose. Bacterial isolates were tested for the production of ammonia in peptone water (peptone 5 g/L). Freshly grown cultures were inoculated in 5 mL peptone water in each tube and incubated for 72 h at 30°C. Nessler's reagent (0.5 mL) was added in each tube. Development of yellow-brownish color indicated NH3 production [37]. Proteolytic activity (casein degradation) was determined from clearing zones in skim milk agar after 4 days of incubation at 30°C as described by Nielsen and Sørensen [38]. Resistance to salt was assessed by adding 5, 8, and 10% sodium chloride to culture media and incubating the plate at 30°C for 7 days. The ability to grow at 4, 42, and 50°C was verified in solid media placed in incubators set at the indicated temperatures, and the growth was qualitatively scored after 7 days. Tolerance to osmotic stress was evaluated by adding 10–20% polyethylene glycol (PEG) to the original liquid media.
2.7. Nucleotide Sequence Identification and Accession Numbers
Analysis of sequences was performed with the basic sequencing alignment BLAST program run against the database (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The sequences of the partial 16S rRNA genes for isolates were deposited in the GeneBank database under the accession numbers from HF584760 to HF585082, from HE610893 to HE610899, and from HF562892 to HF562897. The DGGE sequences were submitted under the accession numbers from HF678228 to HF678357.
3. Results and Discussion
3.1. Cultivation-Independent Analysis of Grapevine Root-Associated Bacterial Communities
The diversity of bacterial communities associated with the grapevine root system from each of the three studied regions was investigated through the analysis of the diversity of the 16S rRNA gene in the root tissues (E), rhizosphere (R), and root-surrounding soil (S) fractions of three replicate plants including bulk soil (B) as a comparison (Figure 1(a)). Multiple-band PCR-DGGE profiles were observed in all fractions (E, R, and S) of the three grapevine root systems and in the respective bulk soils (Figure 1(a)). The endophyte fractions from the soil samples were different, showing the simplest profile composed of a limited number of bands compared to the other root system samples (Figure 1(a)). This observation was confirmed by PCA of the PCR-DGGE band patterns that showed a sharp separation between E-associated bacterial communities and those of soil fractions (Figures 1(b) and 1(c)). The reduced microbial diversity in root tissues may reflect specific physiological requirements to enter the interior of the roots and establish as endophytic populations [39]. The soil fractions showed a multiple-band profile in the DGGE gels, indicating the hosting of a high number of different bacterial taxa. Several shared bands were observed among the R, S, and B fractions, suggesting that similar bacteria may have colonized different soil portions (Figure 1(a)). This was observed in the case of grapevines cultivated in North Italy and Tunisia where R, S, and B fractions tended to cluster together according to the PCA (Figure 1(b)). On the contrary, a clear rhizosphere effect was observed in the root system of grapevines growing in Egypt; the structure of the rhizosphere bacterial communities diverged from the S and B samples (Figure 1(c)), according to the scores of axis 1 of PCA, whereas a statistically significant difference was detected among the bacterial communities associated with the endosphere and soil fractions in Egypt, according to the scores of axis 2 of PCA (Figure 1(d)). Generally, the rhizosphere is defined as a transition zone between the root surface and soil, where the released exudates favour microbial proliferation, inducing changes in the structure and chemical-physical features of the soil [40]. The rhizosphere effect is especially pronounced in nutrient-poor soils and under severe abiotic stresses, as previously observed for herbaceous and arboreal plants grown in arid lands [16, 41].
Figure 1.
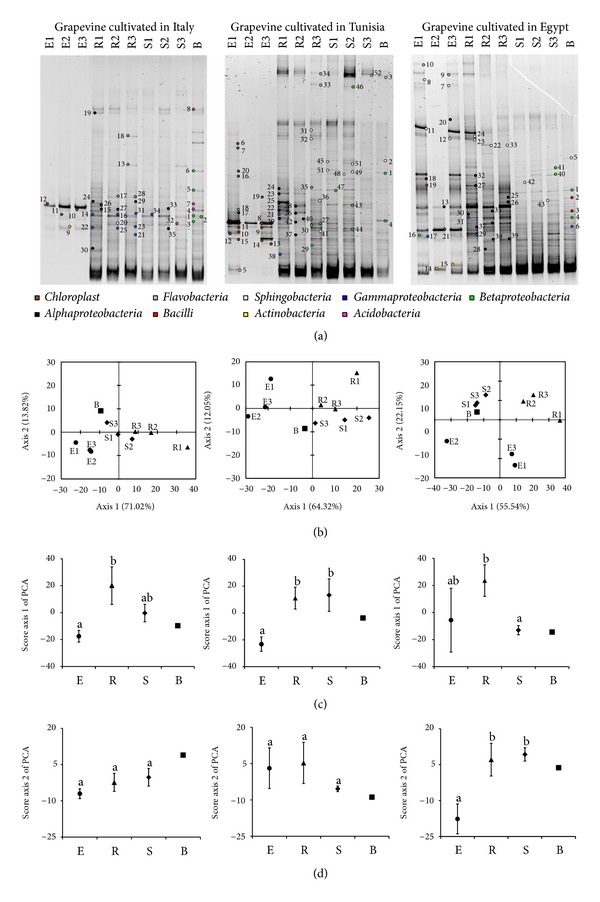
DGGE analysis of grapevine root-associated microbial communities. (a) Representative DGGE gels of the separation of 16S rRNA gene fragments along a denaturing gradient. The analyzed fractions were root tissues (E), rhizosphere (R), root-surrounding soil (S), and bulk soil (B) of three replicate plants of grapevine from a vineyard in Italy (left panel), Tunisia (central panel), and Egypt (right panel). (b) Principal component analysis (PCA) of the plot line profiles that were obtained from DGGE fingerprinting of the bacterial community. E replicates are represented by a circle, R samples by a triangle, S samples by a rhombus, and B by a square. (c)-(d) ANOVA analysis was performed on the average values of the line plot score along axis 1 and 2, respectively, of PCA analysis in order to assess the degree of similarities among plant and soil-associated bacterial communities in the three study sites. Different letters (a and b), shown at the top of the scatter plots in the graph, indicate a statistical significance at P ≤ 0.05 according to ANOVA analysis.
We focused on the comparison of the structure of endosphere and rhizosphere-associated microbial communities of grapevines along the investigated transect (Figure 2(a)). A differentiation of the bacterial diversity among the two different fractions was observed by PCA of the PCR-DGGE profiles along axis 1 (P = 0.0032) and axis 2 (P = 0.0006) (Figures 2(b) and 2(c)). The separation between the rhizospheric and endophytic communities along the transect was confirmed by statistical analysis (PERMANOVA, F = 30.36; df = 5; P = 0.0001). A latitudinal gradient effect was observed in the distribution of the bacterial community in E and R (Figure 2(a)). Bacterial communities associated with grapevines cultivated in Egypt clustered with those of plants cultivated in Tunisia, while the rhizosphere community of grapevine cultivated in Italy was separated in a different cluster, showing a significant difference from the Egyptian and Tunisian communities along axis 2 (P < 0.05). Furthermore, the PERMANOVA analysis performed considering the geographical origin showed a significant statistical difference in the distribution of the bacterial communities, indicating an influence of the site of origin in shaping the microbial community of both fractions (PERMANOVA, F = 30.45; df = 5; P = 0.0078). As previously reported, aridity seems to be the driving force influencing the structure of both Archea and bacterial communities in bare soils [42].
Figure 2.
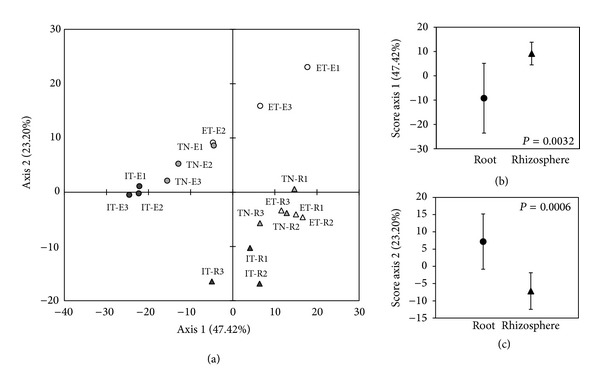
Comparison of the structure of endosphere and rhizosphere bacterial communities associated with grapevines along an aridity transect in the Mediterranean basin. (a) Principal component analysis (PCA) of the PCR-DGGE profiles of endophyte (E) and rhizosphere (R) fractions associated with grapevine cultivated in Italy (IT), Tunisia (TN), and Egypt (ET). Samples were run and analyzed in triplicate. (b)-(c) Statistical analysis was applied to the average values of endophyte and rhizosphere samples along axis 1 and axis 2, respectively, of the PCA analysis. Statistical significance (P) was evaluated according to the Student's t-test and is indicated in every graph.
DISTLM multivariate analysis was performed in order to correlate the differences in the structure of microbial communities in the different agroclimatic sites with abiotic environmental parameters (Table 1). The selection of soil microorganisms by plants is a complex process controlled by several factors, often not easily correlated to geochemical settings [43]. Nevertheless, we observed that among abiotic factors, annual rainfall exerted a statistical significant influence in determining the structure of the root system-associated bacterial community (Table 1). It is possible that a cooccurrence of harsh environmental factors, including a hot and dry climate and reduced rainfall, may reflect the differences among the rhizosphere fractions in the three analysed sites (Table 1).
Table 1.
Relationships between endophytic/rhizosphere bacterial assemblages and climate features of different regions using nonparametric multivariate multiple regression analysis (DISTLM). Partial (conditional) test where the amount explained by each variable added to the model is conditional on variables already in the model.
| Variable | IT | TN | ET | AIC | F | P | % Var | % Cumul | Res. df |
|---|---|---|---|---|---|---|---|---|---|
| Annual rainfall (mm) | 809.1 | 561.1 | 39.7 | 110.09 | 27.749 | 0.0048 | 28.388 | 28.388 | 7 |
| Summer rainfall (mm) | 101.4 | 7.4 | 0.1 | 109.66 | 18.639 | >0.05 | 16.973 | 45.362 | 6 |
| Average rainfall (mm) | 9 | 1 | 0 | 109.66 | 0 | >0.05 | <0.0001 | 45.362 | 6 |
| Max temperature (°C) | 31 | 30 | 35 | 109.66 | 0 | >0.05 | <0.0001 | 45.362 | 6 |
| Min temperature (°C) | 19 | 21 | 23 | 109.66 | 0 | >0.05 | <0.0001 | 45.362 | 6 |
| Average temperature (°C) | 25 | 25.5 | 29 | 109.66 | 0 | >0.05 | <0.0001 | 45.362 | 6 |
| Latitude | 44.6 | 36.8 | 30.2 | 109.66 | 0 | >0.05 | <0.0001 | 45.362 | 6 |
| Longitude | 9.1 | 10.1 | 31.2 | 109.66 | 0 | >0.05 | <0.0001 | 45.362 | 6 |
| Irradiation (w/m2) | 720 | 870 | 880 | 109.66 | 0 | >0.05 | <0.0001 | 45.362 | 6 |
AIC: coefficient of regression; F: value of pseudo F; P: significance of F; % Var: percentage of variance explained by each single variable; % Cumul: cumulative percentage of variance explained; Res. df: residual degrees of freedom.
3.2. Phylogenetic Affiliation of DGGE Bands Representative of Grapevine Root-Associated Microbial Diversity
One hundred and twenty-nine bands, representative of all fractions, were excised from the DGGE gel and affiliated to five bacterial phyla by sequencing: Acidobacteria, Actinobacteria, Firmicutes, Proteobacteria, and Bacteroidetes (Figures 1(a) and 3, and Table 2). Fifty-one percent of the sequences from E fractions were affiliated to plant chloroplasts, indicating a contamination in DNA extracts with the plastid DNA, as observed in other studies characterizing the endophytic population of grapevine [8, 9] and other plants [21, 44]. Despite plant material contamination, the root tissues from the grapevines grown in Egypt presented a higher bacterial diversity with the Gammaproteobacteria, Sphingobacteria, and Flavobacteria being specific classes of this site. The remaining bands from the endophytic fraction were attributed to Actinobacteria, Alphaproteobacteria, and Betaproteobacteria (Figures 1(a) and 3), which were also dominant in the rhizosphere fractions, confirming that endophytes could represent a subgroup of rhizobacteria which have the ability to enter and establish in the root interior [4, 5, 39]. Although other portals for endophyte entrance into plant tissues cannot be excluded, considering both stomata on the phylloplane of grape flowers and xylem sap [4, 7] or endophyte transmission through seeds [4, 45], root cracks are considered the “hot spot” for bacteria endophytic colonization [37, 46, 47]. Several studies have suggested that the diversity of endophytic bacteria depends on the cultivar and age of the host plant [48], on geochemical and physiological conditions of soil [49] and on climatic variables, such as soil moisture and chemical features [50]. Alphaproteobacteria were the best represented class in all rhizospheric soils collected, while Betaproteobacteria, which were not detected in Egypt, were equally represented in the other soil fractions in Italy and Tunisia (Figures 1(a) and 3). Despite their diffusion in both reproductive and vegetative plant organs [51], no sequences affiliated to Burkholderia spp. were retrieved in our study. Among Betaproteobacteria, sequences affiliated to Massilia spp. were detected (Table 2). These bacteria were already detected in grape endora during flowering [4] and display a copiotrophic lifestyle at the root niche [52]. Some members affiliated to this genus have been shown to efficiently colonize cucumber seed coats and radicles and are found to be associated with the hyphae of mycorrhizae infecting germinating seeds [52]. Sequences related to Chryseobacterium spp. were to be found associated with grape plants in Tunisia and Egypt (Table 2). A C. balustinum strain was characterized by its stimulation of the release of flavonoids in Phaseolus vulgaris, specifically produced during plant-bacteria interaction, that can presumably be metabolized by the bacterium [53]. Gammaproteobacteria sequences were retrieved in rhizosphere samples from the vineyard in Italy, while its relative abundance was lower in grapevine rhizosphere samples from Tunisia (Figure 3). Interestingly, Rhizobiales spp. was observed only in R fractions, in agreement with previous findings regarding the isolation of Rhizobium spp. from grape rhizosphere during flowering [4]. Their role in supporting plant growth or nutrient assimilation remains to be elucidated, considering that under low soil moisture plants suffer from a disturbed bioavailability of nutrients [17]. Indeed, as a consequence of its ability to release EPS, a Rhizobium sp. strain contributed to improve root-adhering soil (RAS) aggregation in sunflower plantlets grown both in drained and dry soils [54]. Acidobacteria were revealed only in soil fractions loosely or nonassociated with roots (S and B) in grapevines cultivated in Italy. No bacteria affiliated to the Firmicutes phylum were observed in the root system (Figure 3). Members of the Flavobacteriaceae class, that were retrieved in Egypt (endosphere) and Tunisia (rhizosphere), were classified among the most abundant bacteria in root and phyllosphere of Arabidopsis thaliana plants grown in the wild [55]. A larger genome size has been advocated as a possible reason for the success of Flavobacterium spp. in plant organ colonization, supporting a higher metabolic flexibility for the use of complex sugar compounds secreted by plants [56].
Figure 3.
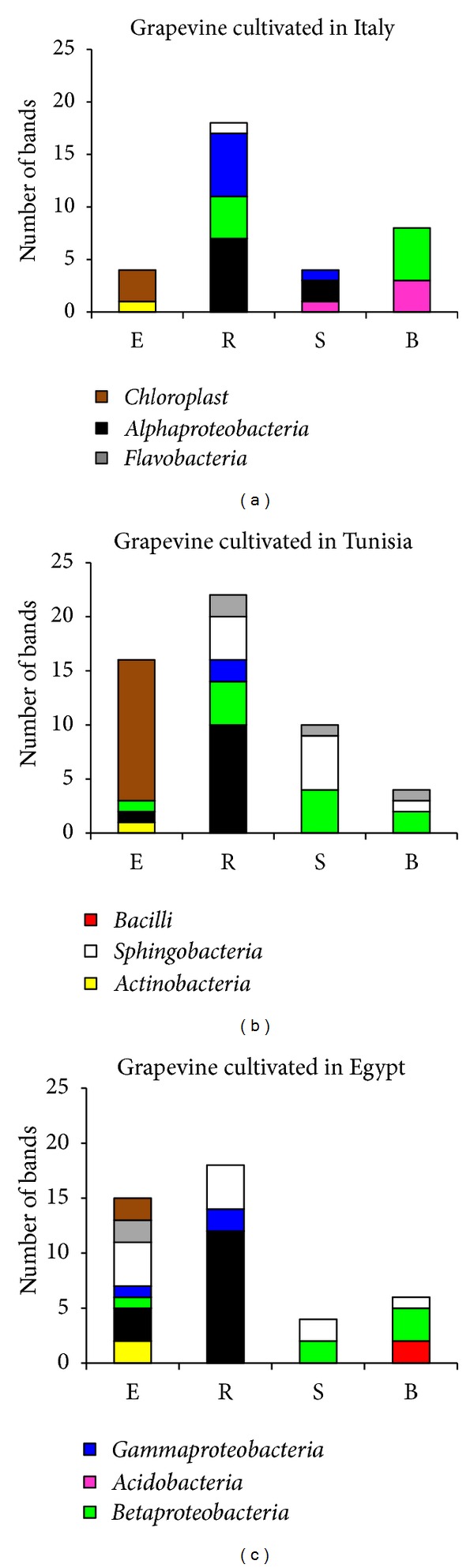
Phylogenetic identification of DNA fragments that were excised from the DGGE gel and successfully amplified and sequenced.
Table 2.
Identification of the dominant bands in the PCR-DGGE fingerprinting profiles (marked in Figure 1). The codes of the different fractions of the grapevine root systems are as follows: E, Endosphere; R, rhizosphere; S, root-surrounding soil; B, bulk soil.
| Frac. | DGGE band | Closest relative (NCBI database) |
Acc. N° | % | Closest describe relative (Ez Taxon database) |
Acc. N° | % | Class |
|---|---|---|---|---|---|---|---|---|
| B | IT-01 | Ramlibacter sp. | AY429716 | 88 | Curvibacter fontanus | AB120963 | 87 | Betaproteobacteria |
| B | IT-02 | Unc. Comamonadaceae | AY360707 | 86 | Curvibacter fontanus | AB120963 | 85 | Betaproteobacteria |
| B | IT-03 | Unc. Variovorax sp. | JN590646 | 90 | Xylophilus ampelinus | AF078758 | 89 | Betaproteobacteria |
| B | IT-04 | Unc. Acidobacteria | EF651005 | 95 | Desulfomonile limimaris | AF230531 | 81 | Acidobacteria |
| B | IT-05 | Unc. bacterium | HQ393158 | 98 | Massilia plicata | AY966000 | 96 | Betaproteobacteria |
| B | IT-06 | Massilia sp. | JF279920 | 99 | Massilia niabensis | EU808006 | 99 | Betaproteobacteria |
| B | IT-07 | Unc. bacterium | AY274152 | 97 | Solibacter usitatus | CP000473 | 92 | Acidobacteria |
| B | IT-08 | Unc. Acidobacteria | GU257870 | 94 | Chloracidobacterium thermophilum | CP002514 | 80 | Acidobacteria |
| E | IT-09 | Micromonospora sp. | FR692086 | 92 | Micromonospora peucetia | X92603 | 90 | Actinobacteria |
| E | IT-10 | Chloroplast | U189132 | 99 | Chloroplast | AM711640 | 99 | Eukarya |
| E | IT-11 | Chloroplast | HQ325745 | 100 | Chloroplast | DQ386163 | 99 | Eukarya |
| E | IT-12 | Chloroplast | HQ325745 | 100 | Chloroplast | DQ386163 | 100 | Eukarya |
| R | IT-13 | Unc. Rhodocyclaceae | EF643420 | 97 | Thiobacter subterraneus | AB180657 | 91 | Betaproteobacteria |
| R | IT-14 | Unc. bacterium | FR853277 | 91 | Steroidobacter denitrificans | EF605262 | 89 | Gammaproteobacteria |
| R | IT-15 | Unc. bacterium | JN855310 | 98 | Bradyrhizobium pachyrhizi | AY624135 | 97 | Alphaproteobacteria |
| R | IT-16 | Unc. Pseudomonadales | FJ889292 | 97 | Steroidobacter denitrificans | EF605262 | 96 | Gammaproteobacteria |
| R | IT-17 | Unc. bacterium | HM445266 | 92 | Rubrivivax gelatinosus | D16213 | 90 | Betaproteobacteria |
| R | IT-18 | Unc. bacterium | EU881322 | 99 | Thiobacter subterraneus | AB180657 | 90 | Betaproteobacteria |
| R | IT-19 | Unc. Rhizobium sp. | EF074979 | 92 | Rhizobium giardinii | U86344 | 91 | Alphaproteobacteria |
| R | IT-20 | Sphingobacterium sp. | EU580525 | 97 | Dyadobacter hamtensis | AJ619978 | 93 | Sphingobacteria |
| R | IT-21 | Unc. bacterium | EF019453 | 86 | Methylogaea oryzae | EU672873 | 79 | Gammaproteobacteria |
| R | IT-22 | Unc. bacterium | EF392989 | 96 | Steroidobacter denitrificans | EF605262 | 87 | Gammaproteobacteria |
| R | IT-23 | Unc. bacterium | FR853277 | 95 | Steroidobacter denitrificans | EF605262 | 93 | Gammaproteobacteria |
| R | IT-24 | Unc. bacterium | GU568879 | 87 | Novosphingobium resinovorum | EF029110 | 82 | Alphaproteobacteria |
| R | IT-25 | Unc. bacterium | FJ479326 | 93 | Steroidobacter denitrificans | EF605262 | 91 | Gammaproteobacteria |
| R | IT-26 | Unc. bacterium | JN855310 | 99 | Bradyrhizobium pachyrhizi | AY624135 | 98 | Alphaproteobacteria |
| R | IT-27 | Unc. bacterium | JN855310 | 95 | Nitrobacter hamburgensis | CP000319 | 94 | Alphaproteobacteria |
| R | IT-28 | Unc. bacterium | DQ643675 | 93 | Rubrivivax gelatinosus | D16213 | 91 | Betaproteobacteria |
| R | IT-29 | Unc. bacterium | GU291531 | 94 | Novosphingobium resinovorum | EF029110 | 94 | Alphaproteobacteria |
| R | IT-30 | Unc. Alphaproteobacteria | JN371328 | 81 | Blastochloris sulfoviridis | D86514 | 78 | Alphaproteobacteria |
| R | IT-31 | Unc. Steroidobacter | FN297970 | 100 | Steroidobacter denitrificans | EF605262 | 98 | Gammaproteobacteria |
| S | IT-32 | Unc. Acidobacteria | HQ597613 | 97 | Chloracidobacterium thermophilum | CP002514 | 81 | Acidobacteria |
| S | IT-33 | Unc. bacterium | JN855310 | 91 | Nitrobacter winogradskyi | CP000115 | 90 | Alphaproteobacteria |
| S | IT-34 | Unc. bacterium | FJ479326 | 99 | Steroidobacter denitrificans | EF605262 | 97 | Gammaproteobacteria |
| S | IT-35 | Unc. Alphaproteobacteria | FJ568851 | 95 | Donghia mobilis | FJ455532 | 91 | Alphaproteobacteria |
|
| ||||||||
| B | TN-01 | Unc. bacterium | KC541101 | 100 | Massilia aurea | AM231588 | 99 | Betaproteobacteria |
| B | TN-02 | Unc. bacterium | HM186197 | 99 | Ohtaekwangia koreensis | GU117702 | 93 | Sphingobacteria |
| B | TN-03 | Chryseobacterium indoltheticum | AY468448 | 99 | Chryseobacterium indoltheticum | AY468448 | 98 | Flavobacteria |
| B | TN-04 | Unc. bacterium | HF546519 | 83 | Xenophilus azovorans | AF285414 | 77 | Betaproteobacteria |
| E | TN-05 | Unc. bacterium | FN667504 | 97 | Streptomyces sodiiphilus | AY236339 | 96 | Actinobacteria |
| E | TN-06 | Chloroplast | HQ336404 | 99 | Chloroplast | DQ386163 | 99 | Cyanobacteria |
| E | TN-07 | Chloroplast | HQ336404 | 99 | Chloroplast | DQ386163 | 99 | Cyanobacteria |
| E | TN-08 | Chloroplast | HQ336404 | 100 | Chloroplast | DQ386163 | 99 | Cyanobacteria |
| E | TN-09 | Chloroplast | EU189132 | 99 | Chloroplast | AM711640 | 99 | Cyanobacteria |
| E | TN-10 | Chloroplast | HQ336404 | 99 | Chloroplast | DQ386163 | 99 | Cyanobacteria |
| E | TN-11 | Chloroplast | HQ336404 | 100 | Chloroplast | DQ386163 | 92 | Cyanobacteria |
| E | TN-12 | Chloroplast | HQ336404 | 99 | Chloroplast | DQ386163 | 99 | Cyanobacteria |
| E | TN-13 | Chloroplast | EU118126 | 100 | Chloroplast | EU118126 | 99 | Cyanobacteria |
| E | TN-14 | Chloroplast | EU118126 | 100 | Chloroplast | EU118126 | 99 | Cyanobacteria |
| E | TN-15 | Chloroplast | EU118126 | 100 | Chloroplast | EU118126 | 100 | Cyanobacteria |
| E | TN-16 | Chloroplast | EU118126 | 99 | Chloroplast | EU118126 | 98 | Eukarya |
| E | TN-17 | Rhizobium radiobacter | AJ389904 | 100 | Rhizobium radiobacter | AJ389904 | 100 | Alphaproteobacteria |
| E | TN-18 | Hydrogenophilus thermoluteolus | AB680730 | 98 | Hydrogenophilus hirschii | FR749905 | 98 | Betaproteobacteria |
| E | TN-19 | Chloroplast | HQ325745 | 99 | Chloroplast | L37580 | 96 | Eukarya |
| E | TN-20 | Chloroplast | HQ325745 | 99 | Chloroplast | DQ386163 | 99 | Eukarya |
| R | TN-21 | Unc. bacterium | JF198689 | 89 | Steroidobacter denitrificans | EF605262 | 84 | Gammaproteobacteria |
| R | TN-22 | Rhizobium sullae | NR_029330 | 89 | Rhizobium sullae | Y10170 | 82 | Alphaproteobacteria |
| R | TN-23 | Rhizobium giardinii | JX869993 | 91 | Rhizobium selenitireducens | EF440185 | 74 | Alphaproteobacteria |
| R | TN-24 | Rhizobium huautlense | HQ538618 | 99 | Rhizobium huautlense | AF025852 | 99 | Alphaproteobacteria |
| R | TN-25 | Unc. bacterium | HM328693 | 99 | Novosphingobium resinovorum | EF029110 | 95 | Alphaproteobacteria |
| R | TN-26 | Rhizobium etli | NR_029184 | 100 | Rhizobium vallis | FJ839677 | 99 | Alphaproteobacteria |
| R | TN-27 | Unc. Betaproteobacteria | EU266802 | 92 | Hydrogenophaga palleronii | AF019073 | 78 | Betaproteobacteria |
| R | TN-28 | Rhizobium giardinii | EU410948 | 99 | Rhizobium endophyticum | EU867317 | 98 | Alphaproteobacteria |
| R | TN-29 | Unc. bacterium | HF546519 | 85 | Piscinibacter aquaticus | DQ664244 | 81 | Betaproteobacteria |
| R | TN-30 | Unc. bacterium | HE586741 | 85 | Hydrogenophaga palleronii | AF019073 | 78 | Betaproteobacteria |
| R | TN-31 | Unc. bacterium | JF175892 | 93 | Niastella populi | EU877262 | 81 | Sphingobacteria |
| R | TN-32 | Unc. bacterium | JF175892 | 93 | Chitinophaga arvensicola | D12657 | 84 | Sphingobacteria |
| R | TN-33 | Unc. Flavobacterium sp. | EU017400 | 99 | Flavobacterium reichenbachii | AM177616 | 84 | Flavobacteria |
| R | TN-34 | Flavobacterium sp. | EF601822 | 99 | Flavobacterium pectinovorum | AM230490 | 98 | Flavobacteria |
| R | TN-35 | Unc. Betaproteobacteria | EF662768 | 97 | Azoarcus buckelii | AJ315676 | 88 | Betaproteobacteria |
| R | TN-36 | Sphingobacterium sp. | EU580525 | 96 | Dyadobacter hamtensis | AJ619978 | 94 | Sphingobacteria |
| R | TN-37 | Reichenowi ornate | AY316684 | 82 | Aurantimonas altamirensis | DQ372921 | 80 | Alphaproteobacteria |
| R | TN-38 | Unc. Gammaproteobacteria | JN648252 | 97 | Steroidobacter denitrificans | EF605262 | 88 | Gammaproteobacteria |
| R | TN-39 | Rhizobium sp. | FN546874 | 98 | Rhizobium endophyticum | EU867317 | 97 | Alphaproteobacteria |
| R | TN-40 | Rhizobium sp. | AM922181 | 99 | Rhizobium endophyticum | EU867317 | 98 | Alphaproteobacteria |
| R | TN-41 | Unc. bacterium | FJ719038 | 97 | Dyadobacter hamtensis | AJ619978 | 95 | Sphingobacteria |
| R | TN-42 | Rhizobium leguminosarum | HQ853453 | 99 | Rhizobium vallis | FJ839677 | 98 | Alphaproteobacteria |
| S | TN-43 | Unc. bacterium | AF423222 | 99 | Thiobacter subterraneus | AB180657 | 89 | Betaproteobacteria |
| S | TN-44 | Unc. Comamonadaceae | AY360707 | 99 | Acidovorax konjaci | AF078760 | 97 | Betaproteobacteria |
| S | TN-45 | Unc. Sphingobacteriales | AM934931 | 99 | Ohtaekwangia koreensis | GU117702 | 93 | Sphingobacteria |
| S | TN-46 | Unc. bacterium | GQ023702 | 91 | Hydrogenophilus hirschii | FR749905 | 91 | Betaproteobacteria |
| S | TN-47 | Unc. Burkholderiaceae | AM935484 | 99 | Thiobacter subterraneus | AB180657 | 90 | Betaproteobacteria |
| S | TN-48 | Pedobacter sp. | AY599662 | 85 | Pedobacter metabolipauper | AM491370 | 84 | Sphingobacteria |
| S | TN-49 | Pedobacter africanus | NR_028977 | 99 | Pedobacter africanus | AJ438171 | 98 | Sphingobacteria |
| S | TN-50 | Unc. bacterium | HM049699 | 88 | Pedobacter africanus | AJ438171 | 87 | Sphingobacteria |
| S | TN-51 | Unc. bacterium | HM049699 | 99 | Pedobacter steynii | AM491372 | 98 | Sphingobacteria |
| S | TN-52 | Flavobacterium sp. | HM149210 | 99 | Flavobacterium pectinovorum | AM230490 | 98 | Flavobacteria |
|
| ||||||||
| B | ET-01 | Unc. Betaproteobacteria | JF806989 | 99 | Thiobacter subterraneus | AB180657 | 90 | Betaproteobacteria |
| B | ET-02 | Bacillus sp. | FM992819 | 99 | Bacillus pocheonensis | AB245377 | 98 | Bacilli |
| B | ET-03 | Bacillus sp. | FM992819 | 87 | Bacillus alcalophilus | X60603 | 85 | Bacilli |
| B | ET-04 | Unc. bacterium | DQ398884 | 86 | Hydrogenophaga flava | AF078771 | 84 | Betaproteobacteria |
| B | ET-05 | Unc. Sphingobacteriales | AM934931 | 99 | Ohtaekwangia koreensis | GU117702 | 92 | Sphingobacteria |
| B | ET-06 | Unc. bacteria | JF681924 | 90 | Sterolibacterium denitrificans | AJ306683 | 84 | Gammaproteobacteria |
| E | ET-07 | Unc. bacterium | JQ357881 | 99 | Chitinophaga pinensis | CP001699 | 98 | Sphingobacteria |
| E | ET-08 | Unc. bacterium | JQ358300 | 99 | Chitinophaga sancti | AB078066 | 98 | Sphingobacteria |
| E | ET-09 | Chryseobacterium wanjuense | AB682410 | 100 | Chryseobacterium wanjuense | DQ256729 | 100 | Flavobacteria |
| E | ET-10 | Flavobacterium sp. | JF915324 | 99 | Flavobacterium subsaxonicum | AM934666 | 96 | Flavobacteria |
| E | ET-11 | Chitinophaga sp. | JQ659659 | 98 | Chitinophaga ginsengisoli | AB245374 | 96 | Sphingobacteria |
| E | ET-12 | Chitinophaga sp. | JQ659659 | 98 | Chitinophaga ginsengisoli | AB245374 | 96 | Sphingobacteria |
| E | ET-13 | Novosphingobium sp. | EU984513 | 99 | Novosphingobium resinovorum | EF029110 | 99 | Alphaproteobacteria |
| E | ET-14 | Unc. bacterium | FN667504 | 97 | Streptomyces sodiiphilus | AY236339 | 96 | Actinobacteria |
| E | ET-15 | Glycomyces scopariae | JQ342894 | 99 | Glycomyces scopariae | EU200682 | 99 | Actinobacteria |
| E | ET-16 | Variovorax paradoxus | AB680784 | 99 | Variovorax paradoxus | D88006 | 99 | Betaproteobacteria |
| E | ET-17 | Pseudoxanthomonas sp. | JF703645 | 99 | Pseudoxanthomonas | AB008507 | 98 | Gammaproteobacteria |
| E | ET-18 | Rhizobium sp. | AM922181 | 99 | Rhizobium radiobacter | AJ389904 | 99 | Alphaproteobacteria |
| E | ET-19 | Chloroplast | HQ325745 | 99 | Chloroplast | DQ386163 | 99 | Eukarya |
| E | ET-20 | Rhizobium sp. | AM922181 | 85 | Rhizobium radiobacter | AJ389904 | 83 | Alphaproteobacteria |
| E | ET-21 | Chloroplast | HQ325745 | 100 | Chloroplast | DQ386163 | 99 | Eukarya |
| R | ET-22 | Chitinophaga terrae | AB267724 | 98 | Chitinophaga niabensis | EU714259 | 95 | Sphingobacteria |
| R | ET-23 | Chitinophaga sp. | GQ281772 | 99 | Chitinophaga niabensis | EU714259 | 97 | Sphingobacteria |
| R | ET-24 | Chitinophaga sp. | JQ659659 | 97 | Chitinophaga sancti | AB078066 | 96 | Sphingobacteria |
| R | ET-25 | Novosphingobium sp. | AB453877 | 90 | Novosphingobium resinovorum | EF029110 | 88 | Alphaproteobacteria |
| R | ET-26 | Unc. bacterium | GQ074926 | 99 | Novosphingobium resinovorum | EF029110 | 97 | Alphaproteobacteria |
| R | ET-27 | Unc. bacterium | GQ169020 | 90 | Blastochloris sulfoviridis | D86514 | 88 | Alphaproteobacteria |
| R | ET-28 | Unc. bacterium | DQ814032 | 99 | Mesorhizobium thiogangeticum | AJ864462 | 99 | Alphaproteobacteria |
| R | ET-29 | Pseudoxanthomonas mexicana | JQ660737 | 100 | Pseudoxanthomonas japonensis | AB008507 | 99 | Gammaproteobacteria |
| R | ET-30 | Mesorhizobium sp. | JN688938 | 94 | Sinorhizobium americanum | AF506513 | 93 | Alphaproteobacteria |
| R | ET-31 | Unc. bacterium | AB540382 | 82 | Steroidobacter denitrificans | EF605262 | 79 | Gammaproteobacteria |
| R | ET-32 | Sphingopyxis chilensis | JF459975 | 98 | Sphingopyxis panaciterrae | AB245353 | 98 | Alphaproteobacteria |
| R | ET-33 | Chitinophaga sp. | JN680879 | 89 | Chitinophaga niabensis | EU714259 | 79 | Sphingobacteria |
| R | ET-34 | Unc. bacterium | GU291531 | 100 | Novosphingobium resinovorum | EF029110 | 99 | Alphaproteobacteria |
| R | ET-35 | Devosia chinhatensis | EF433462 | 99 | Devosia chinhatensis | EF433462 | 99 | Alphaproteobacteria |
| R | ET-36 | Ensifer sp. | JF450188 | 99 | Ensifer adhaerens | AM181733 | 98 | Alphaproteobacteria |
| R | ET-37 | Sinorhizobium fredii | HQ836172 | 98 | Sinorhizobium americanum | AF506513 | 97 | Alphaproteobacteria |
| R | ET-38 | Mesorhizobium loti | HQ424911 | 93 | Mesorhizobium plurifarium | Y14158 | 92 | Alphaproteobacteria |
| R | ET-39 | Mesorhizobium loti | HQ424911 | 88 | Mesorhizobium plurifarium | Y14158 | 87 | Alphaproteobacteria |
| S | ET-40 | Cupriavidus sp. | AB266608 | 99 | Cupriavidus oxalaticus | AF155567 | 98 | Betaproteobacteria |
| S | ET-41 | Unc. Burkholderiaceae | JF681924 | 95 | Thiobacter subterraneus | AB180657 | 79 | Betaproteobacteria |
| S | ET-42 | Unc. Bacteroidetes | FM877553 | 87 | Ohtaekwangia koreensis | GU117702 | 85 | Sphingobacteria |
| S | ET-43 | Unc. Bacteroidetes | FM877553 | 99 | Ohtaekwangia kribbensis | GU117703 | 92 | Sphingobacteria |
Frac.: Fraction analysed; Unc.: Uncultured.
Our findings on the grapevine-associated bacterial community structure are in agreement with other studies carried out on grape roots. Few bacterial groups were found in grapevine tissues using a diverse array of methods including 16S rRNA gene libraries, length heterogeneity PCR, and FISH hybridization [4, 9, 57]. Among other bacteria, FISH analysis detected cells affiliated to Firmicutes and Gammaproteobacteria adhering on epidermal cells, associated with xylem elements and colonizing different organs of flowers, including ovaries [4]. ARDRA profiles demonstrated that flowers, fruits, and seeds host a rather low diversity of endophytes compared to rhizosphere and root tissues, suggesting that specific metabolic skills are required to translocate from the root interior to other plant tissues [4]. Among the genetic determinants affecting the endophyte lifestyle, a gene codifying for β-galactosidase and two genes for the expression of acyl homoserine-lactones (AHL) were recently found in the genome of Methylobacterium sp. strain GFX4 [58]. This strain, associated with the xylem of Riesling grapevines, could communicate with other xylem-associated endophytes, potentially influencing their behaviours; by partially digesting galactan, this strain could also improve its colonization in the xylem through modification of the plant cell wall [58]. Endophyte communication through the release of AHL has been documented for strains isolated from both grapevine and sugarcane and could play a, still unknown, role for bacteria establishment in plant tissues [59].
3.3. Diversity of Culturable Bacteria Associated with Grapevine Root Systems
The isolation of native bacterial species associated with grapevine cultivated in North Italy, Tunisia, and Egypt was performed using different media in order to select for oligotrophic bacteria, Pseudomonadaceae, and ACC deaminating bacteria, already well documented as plant growth promoters [60–62]. The highest number of cultivable bacteria expressed as colony-forming units (CFUs) per gram of sample was in the rhizosphere (108-109) and progressively decreased passing from the S (107–109) to the B fractions (105–107). The root tissues presented the lowest values (105–107), supporting previous data obtained on grapevine-associated bacterial communities [4, 7, 63]. It is noteworthy that bulk soil and endosphere host similar bacterial communities, in terms of size, suggesting that the ability to thrive in plant tissues requires specific genetic requirements [39]. A total of 769 isolates were obtained from the selected agar media as representative of the different populations/morphologies. To reduce genotypic redundancy, the ACCd bacterial collection was dereplicated using ITS-PCR fingerprinting, and a representative bacterium from each haplotype was selected for further identification and characterization. In total, 331 strains were identified by partial sequencing of the 16S rRNA gene. The phylogenetic identification of culturable bacteria highlighted the diversity in terms of composition of the different fractions, revealing a predominance of Gram-negative bacteria (66%), belonging to the Gammaproteobacteria (63%), Alphaproteobacteria (2%), and Betaproteobacteria (1%) subclasses. The remaining Gram-positive isolates were affiliated to the Firmicutes (31%), Actinobacteria (2%), and Bacteroidetes (1%) classes (Figure 4(a)). Members of these taxa have been found to be associated with other grape plants cultivated in Italy [9, 57], France [4, 63], Turkey [64], Nova Scotia [7], and Australia [8]. A high Shannon-Weaver diversity index, calculated from the number of individuals per genus, was found within the rhizosphere of grapes cultivated in Italy (H′ = 1.52) and Tunisia (H′ = 1.36) and in the endosphere of ungrafted Barbera in Italy (H′ = 1.37). On the contrary, the bacterial communities associated with the root system of grapes cultivated in Egypt presented lower diversity values (H′ = 1.037 in E and H′ = 1.07 in R). The high genetic diversity of grape root systems presumably resulted from the combined effects of root exudates and agricultural management practices, particularly at the lower latitude site where the arid pedoclimatic condition may have influenced the bacterial community composition [43]. The microbial community in the bulk soil, not directly influenced by the root system or agricultural practices, showed the lowest diversity indexes, particularly in the samples collected from the Southern Mediterranean sites (H′ = 0.69 in Tunisia and H′ = 0.88 in Egypt). On the contrary, the bulk soil collected in vineyards in Italy recorded a higher Shannon index (H′ = 1.158), probably because of a more structured soil texture that is able to host a richer microbial community [43]. Significant differences were observed in the structure of the bacterial communities in the analyzed vineyards, in particular for the differential distribution pattern of the major bacterial genera (Figure 4(b)). According to the cluster analysis at the genus level performed on the entire strain collection, the composition of the cultivable communities associated with grapes cultivated in Egypt and Tunisia shared a higher similarity (82%) than those in Italy (68%). A similar profile of bacteria distribution was observed in the root systems of all grapevines studied, with the dominance of Gammaproteobacteria followed by Firmicutes. Even at the genus level, differences are highlighted in the bacteria distribution in plant and soil fractions and within the studied sites, particularly among Tunisia and Egypt. The rhizosphere of grapes cultivated in these countries differed for the different percentages of bacteria affiliated to Pseudomonas (40% in Tunisia and 8% in Egypt) and Enterobacter (10% in Tunisia and 65% in Egypt). Endophytes from the Italian plants were dominated by Pantoea (40%), a genus that was not detected in grapes grown at lower latitudes, followed by Pseudomonas (34%), Bacillus (14%), Enterobacter (8%), Arthrobacter (3%), and Rhodococcus (1%). The genus Pantoea has been frequently associated with grape tissues and may contribute to prime plants for accelerated phytoalexin production after B. cinerea challenge [65]; its plant growth potential has already been documented for several model plants [66, 67]. Egypt and Tunisia grape root tissues hosted a higher percentage of isolates affiliated to Pseudomonas and Bacillus genera, confirming previous findings on bacteria community composition in grape tissues as assessed through isolation and culture-independent methods [4, 8]. Pseudomonas spp. in particular was abundant in the soil fractions from Tunisia (55%), although it was also observed in the other two sites (25% in Egypt and 14% in Italy), confirming the widespread diffusion of this genus in root-influenced soils [68]. Despite the presence of sequences affiliated to Rhizobiales in all three vineyards, as observed by DGGE analysis, isolates belonging to the Rhizobium genus were retrieved only in the root tissues of grapes cultivated in Egypt (13%). The role of these bacteria remains to be elucidated, although their association with other crops has been proposed for field applications [69]. The most abundant genera that were associated with the endosphere were also retrieved from the rhizosphere, although at lower percentages. This observation strongly supports the theory regarding endophytic bacteria entry from the root system to spread in plant tissues through xylem translocation, as previously documented for both beneficial and pathogenic strains such as Agrobacterium tumefaciens [70], Burkholderia sp. strain PsJN [5], Yersinia enterocolitica strain [71], and Xylella fastidiosa [72].
Figure 4.
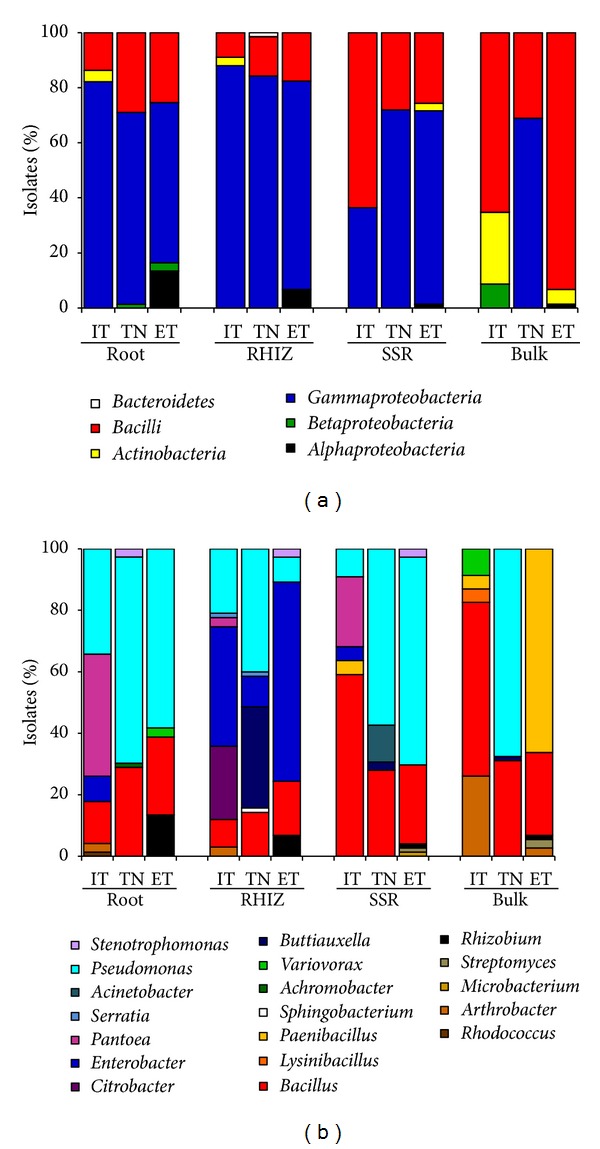
Phylogenetic identification of culturable bacteria associated with grapevine plant and soil fractions. (a)-(b) Bacteria repartition in endosphere (ROOT), rhizosphere (RHIZ), root-surrounding soil (SSR), and bulk soil (BULK) of grapevine grown in Italy (IT), Tunisia (TN), and Egypt (ET) according to the class and genus level, respectively.
Isolates from the R fraction were mainly affiliated to the Enterobacteriaceae family. Enterobacter that was shared among the rhizosphere soils of the three sites was found at a higher percentage in the soils in Egypt (65%) and Italy (39%). In the rhizosphere of grapes from Tunisia, Buttiauxiella (33%) was the main genus of the Enterobacteriaceae, while in Italy Citrobacter accounted for 24% of the local collection (Figure 3(b)). The predominance of bacteria affiliated to the Enterobacteriaceae family could be attributed to the application of crop management techniques based on the use of natural fertilizers such as manure and plant residues [16, 18]. Nevertheless, representative species of Enterobacteriaceae were widespread in several plant systems, suggesting a role in plant colonization and plant promotion also during stressful conditions [73]. In the S fractions, less influenced by root exudates, the isolation frequency of Bacillus increased. This shift in the bacterial community composition was evident in the bulk soil, not subjected to amendment and irrigation processes. In both S and B fractions, the Enterobacteriaceae disappeared, while the spore-forming bacteria (Firmicutes and Actinobacteria), typical of poorly structured soil, increased in incidence reaching 98% in the bulk soil in Egypt, 91% in Italy, and 31% in Tunisia (Figure 3(b)). The prevailing genera Arthrobacter, Bacillus, and Paenibacillus can survive as resting cells or spores under adverse environmental conditions, hence, making them typical taxa of uncultivated and arid soils [43].
The observed diversity of the culturable fraction is in agreement with the findings provided by PCR-DGGE analysis. Indeed, the same microbial taxa were retrieved with the exception of Acidobacteria that were detected only through molecular analysis. Despite the fact that the cultivation approach generally favours some taxa [74] and that the PCR-based techniques of metagenomes are biased by the preferential amplification of certain bacterial groups [75], the combination of cultivation independent and dependent techniques revealed sharp differences among the structure of microbial communities associated with root systems of grapes cultivated in three Mediterranean regions.
3.4. Determining the PGP Potential of Grapevine-Associated Bacteria
One hundred and seventy-five isolates of the de-replicated collection, 93 from the rhizosphere and 82 endophytes, were further screened in vitro for the presence of PGP traits. To characterize their PGP potential, auxin (IAA) production, phosphate solubilization, ammonia and siderophores productions, protease activity, and exopolysaccharide (EPS) release were evaluated. The majority (95%) of isolates showed multiple PGP activities, which may promote plant growth directly, indirectly, or synergistically. In particular, none of the rhizobacteria and only 4% of the endophytes showed only one or no activity, while about 80% of isolates from both fractions displayed more than three PGP activities. Interestingly, the distribution of the number of PGP activities displayed by the strains in the three study sites revealed a similar distribution profile (96% in Italy, 97% in Tunisia, and 94% in Egypt), supporting the hypothesis that a huge functional PGP potential is maintained in grapevine root systems (Figures 5(a), 5(b), and 5(c)). Among the most common PGP abilities was the production of auxin-like compounds (82%), followed by the synthesis of ammonia (70%) and the solubilization of insoluble phosphates (61%). In terms of auxins, IAA role in the stimulation and elongation of the root apparatus is well documented, extending the root surface involved in nutrient and water uptake [76]. In our bacterial collection, the IAA production was equally distributed among endophytic (84%) and rhizospheric (80%) bacteria, and a similar trend was observed in all three sites along the latitude transect (79% in Italy, 82% in Tunisia, and 85% in Egypt) (Figures 5(d), 5(e), and 5(f)), in agreement with previous observations that IAA synthesis is a widespread PGP trait [77]. The IAA production ranged from 2.11 to 36.2 μgmL−1, with the highest amount produced by the isolates from the rhizosphere and endosphere of grape cultivated in Italy (14.4 and 18.6 μgmL−1 in E and R fractions, resp.). The production of ammonia can indirectly influence plant growth through the supply of nitrogen [78]. In the present investigation, 70% of isolates displayed ammonia production. A similar distribution among the endophytic and rhizospheric bacteria was observed (Figures 5(d), 5(e), and 5(f)), with the exception of strains isolated from Italian root tissues (48%). Similarly, phosphate solubilisation ability was exhibited by 61% of the isolates collected. Phosphorous is a key nutrient for plant growth, representing one of the main factors limiting plant development and productivity [79]. The ability of rhizobacteria to solubilize phosphate (79% in Egypt, 78% in Tunisia, and 56% in Italy) through the production of organic acids or phytases can support plant growth in nutrient-poor soils in drought-prone ecosystems, such as those studied in this work [80–82]. Moreover, the isolates showed protease (46%), siderophore production (47%), and EPS release (41%). The synthesis of protease presented a similar pattern of distribution (about 50%) along all the fractions of grape root system analyzed, except for the endophytes associated with grapevine from Italy (12%). Several siderophore-producing bacteria were observed mainly in the rhizosphere (78% in Italy, 65% in Tunisia, and 35% in Egypt), probably because this PGP trait confers competitive colonization ability in iron-limiting soil and exerts a biocontrol role, reducing iron-dependent spore germination of fungi. A high percentage of siderophore-releasing bacteria was recorded only among the endophytic bacteria isolated from grapes cultivated in Italy (64%). Finally, EPS production was qualitatively evaluated. Only 49% of endophyte and 32% of rhizobacteria were able to produce EPS, with the highest percentages observed for the isolates associated with grape roots from Egypt (65% in R and 52% in E). Bacteria adapted to arid environments are well known to protect themselves from extreme climate conditions, producing EPS-rich biofilms that entrap water molecules and thus retaining moisture [83]. Bacterial EPS production in clay-rich soils, such as those in Italy, may presumably play an additional role for favoring root penetration in hard soils such as dry clay soils [84].
Figure 5.
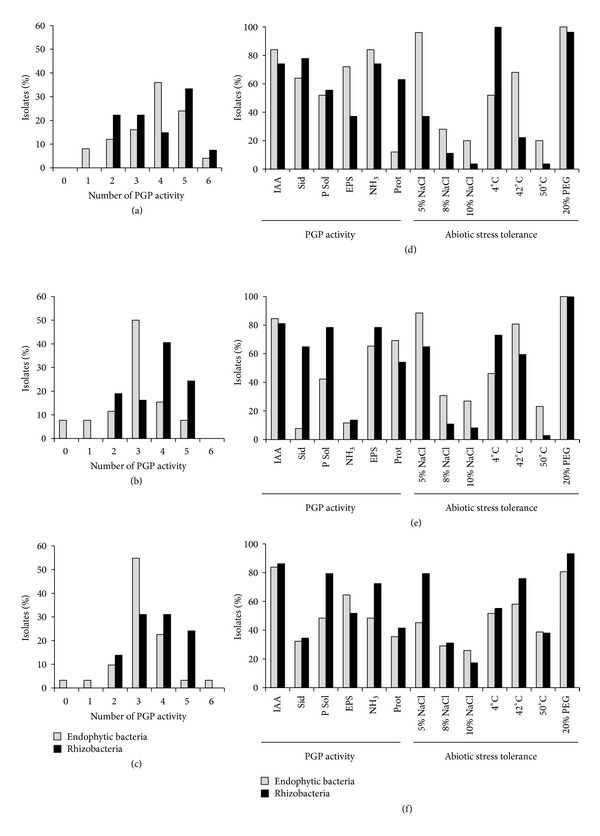
PGP potential of grapevine-associated bacteria. (a)–(c) Percentage of isolates showing an increasing number of PGP abilities in the strain collection isolated from grapevine grown in vineyards located in Italy, Tunisia, and Egypt, respectively. (d)–(f) Percentage of isolates displaying the assayed PGP traits and abiotic stress tolerance in the bacterial collection of strains associated with grapevine cultivated in Italy, Tunisia, and Egypt, respectively.
Further analyses were performed to evaluate bacteria resistance to abiotic stresses that are often associated with drought, such as increased salinization of soils and air temperatures that rise up to 50°C in daytime and drop down during the night. Thus, we analyzed the ability of bacteria to survive in the presence of increasing concentrations of salt, to grow despite temperature fluctuations (4, 42 and 50°C), and to thrive in conditions of low water availability (Figures 5(d), 5(e), and 5(f)). Salinization of dry soils, together with drought and temperature variations, deeply hamper plant physiology and development [85]. As expected, the number of strains resistant to salt decreased with increasing NaCl concentration (5%, 8%, and 10%). While 67% of the isolates were able to grow on media containing 5% NaCl, this percentage decreased to 22% and 16% at 8% and 10% NaCl, respectively. As shown in Figure 4, even at lower NaCl concentrations, bacteria from the endophytic fraction showed sensitivity to salt, particularly among bacteria isolated from Egyptian grape root, where 55% of isolates could not grow in the presence of salt. On the contrary, rhizobacteria isolated from Egypt included the highest proportion of isolates resistant to salinity (79%), followed by those isolated from Tunisia (64%) and Italy (37%). At increasing concentrations of salt, the percentage of resistant isolates decreased, with only 17% of isolates able to grow at 10% NaCl. In particular, the capacity to tolerate high salt concentration followed the latitudinal/aridity transect from the south to the north (Figures 5(a), 5(b), and 5(c)), with percentages ranging from 31% in the rhizosphere of grapes from Egypt to 4% in the rhizosphere from Italy. Although halotolerance has been studied in bacteria affiliated to Halomonas spp. [86], this study highlights that even under soil dryness bacteria with moderate halotolerance can be observed. Interestingly, almost all of the isolates were able to grow under low water availability induced by PEG [87]. All the strains isolated in Tunisia and 98% of those associated with Italian grapevine root systems (100% in E and 96% in R) were able to grow at 20% of PEG, while slightly lower percentages of tolerance were observed for bacteria isolated from grapes cultivated in Egypt, that is, 80% in the endosphere and 93% in the rhizosphere (Figures 5(d), 5(e), and 5(f)). During drought, belowground microbiomes survive by using water reserves in soils that, in turn, are rapidly depleted by earlier development of plants [88]. Thus, osmotic tolerance is a key feature for microbial survival.
Finally, we observed that 63% and 61% of the isolates could grow at 4°C and 42°C, respectively. The majority (52% in E and 100% in R) of the strains isolated from Italian vineyards were able to grow at 4°C (Figures 5(d), 5(e), and 5(f)), presumably adapted to the cold temperatures in autumn and winter when the average air temperatures can be as low as −1°C [25]. On the contrary, only bacteria associated with grapevines grown in Egypt presented resistance to high temperatures, with 39% and 38% of strains isolated from root and rhizosphere, respectively, capable of growing at 50°C, probably being adapted to hot summer temperatures [89]. Global warming is predicted to affect microbial communities, hampering their physiology and growth [90]. The 29% of the collected strains were able to grow both at low and high temperatures, confirming that these isolates were adapted to the peculiar temperature fluctuations of the studied environments. The 23% of isolates presented the potential to express their PGP ability in unfavourable environmental conditions influenced by drought, simultaneously showing halotolerance, resistance to a variable temperature range, and low water availability.
4. Conclusions
The Mediterranean is a closed basin, encompassing subtropical, arid, and continental climates that, to date, are rapidly changing through the increase in length and extent of dry periods [91]. Among the most cultivated and economically relevant crops, Vitis vinifera is widespread at all the basin latitudes, and grape quality and yields are affected by prolonged drought events. The findings reported in the present study contribute to expand the knowledge on the diversity and PGP potential of grapevine-associated bacteria under three different agroclimatic conditions. Culture-dependent and independent techniques highlighted that, according to a specific and yet undefined selection mediated by the different cultivars and rainfall and temperature regimes, the rhizosphere and endosphere microbial communities are different among the three different sites. Indeed, many environmental factors may explain such diversity, such as soil moisture and temperature [79]. While summer temperatures in Italy, Tunisia, and Egypt are quite high and of a similar magnitude, in winter, grapevines in the different study sites experience different levels of low and freezing temperatures that presumably contribute to the modelling of the grape-associated microbial communities. Despite the different biopedoclimatic conditions of the three studied sites (in terms of different cultivars, soil types, and climate conditions), a large set of PGP abilities is still displayed by the respective collections of isolates, independently from the site of origin, and with a similar profile in terms of the number of PGP traits and activities. A redundant functional capability of the isolates from the root systems has been recorded in all the three sites, indicating that in these three environments, the root bacterial communities are adapted to the respective conditions. For instance, insoluble phosphate solubilisation, auxin synthesis, and ammonia production were exhibited by multiple strains in all three sites, highlighting that despite site-specific chemical settings, grapevines in the three investigated agroclimatic regions share similar physiological requirements that are, at least partially, provided by PGP-associated bacteria. Such results suggest that the great diversity of the bacterial world has enough resources to provide functional redundancy in rather different environments, such as those examined, and that a functional homeostasis of the root system bacterial communities may sustain grapevine life in rather different environmental settings.
Authors' Contribution
Ramona Marasco and Eleonora Rolli contributed equally to the work.
Acknowledgments
This work was supported by the project BIODESERT GA-245746 “Biotechnology from Desert Microbial Extremophiles for Supporting Agriculture Research Potential in Tunisia and Southern Europe” (European Union), Fondazione Bussolera Branca, and the project BIOGESTECA n° 15083/RCC “Fondo per la Promozione di Accordi Istituzionali” (Regione Lombardia, Italy) through a fellowship to R. Marasco. E. Rolli was supported by Università degli Studi di Milano, DeFENS, European Social Fund (FSE), and Regione Lombardia (contract “Dote Ricerca”). The authors thank Erika Corretto for the excellent technical assistance.
References
- 1.Shi XY, Bi JL, Morse JG, Toscano NC, Cooksey DA. Effect of xylem fluid from susceptible and resistant grapevines on developmental biology of Xylella fastidiosa . European Journal of Plant Pathology. 2013;135:127–135. [Google Scholar]
- 2.Aballay E, Mårtensson A, Persson P. Screening of rhizosphere bacteria from grapevine for their suppressive effect on Xiphinema index Thorne & Allen on in vitro grape plants. Plant and Soil. 2011;347(1):313–325. [Google Scholar]
- 3.Balloi A, Rolli E, Marasco R, et al. The role of microorganisms in bioremediation and phytoremediation of polluted and stressed soils. Agrochimica. 2010;54(6):353–369. [Google Scholar]
- 4.Compant S, Mitter B, Colli-Mull JG, Gangl H, Sessitsch A. Endophytes of grapevine flowers, berries, and eeds: identification of cultivable bacteria, comparison with other plant parts, and visualization of niches of colonization. Microbial Ecology. 2011;62(1):188–197. doi: 10.1007/s00248-011-9883-y. [DOI] [PubMed] [Google Scholar]
- 5.Compant S, Reiter B, Sessitsch A, Nowak J, Clément C, Barka EA. Endophytic colonization of Vitis vinifera L. by plant growth-promoting bacterium Burkholderia sp. strain PsJN. Applied and Environmental Microbiology. 2005;71(4):1685–1693. doi: 10.1128/AEM.71.4.1685-1693.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Compant S, Kaplan H, Sessitsch A, Nowak J, Ait Barka E, Clément C. Endophytic colonization of Vitis vinifera L. by Burkholderia phytofirmans strain PsJN: from the rhizosphere to inflorescence tissues. FEMS Microbiology Ecology. 2008;63(1):84–93. doi: 10.1111/j.1574-6941.2007.00410.x. [DOI] [PubMed] [Google Scholar]
- 7.Bell CR, Dickie GA, Harvey WLG, Chan JWYF. Endophytic bacteria in grapevine. Canadian Journal of Microbiology. 1995;41(1):46–53. [Google Scholar]
- 8.West ER, Cother EJ, Steel CC, Ash GJ. The characterization and diversity of bacterial endophytes of grapevine. Canadian Journal of Microbiology. 2010;56(3):209–216. doi: 10.1139/w10-004. [DOI] [PubMed] [Google Scholar]
- 9.Bulgari D, Casati P, Brusetti L, et al. Endophytic bacterial diversity in grapevine (Vitis vinifera L.) leaves described by 16S rRNA gene sequence analysis and length heterogeneity-PCR. Journal of Microbiology. 2009;47(4):393–401. doi: 10.1007/s12275-009-0082-1. [DOI] [PubMed] [Google Scholar]
- 10.Rolli E, Marasco R, Ettoumi B, et al. The plant growth promoting microbiome increases grapevine resistance to drought stress: a collaborative study between Fondazione Bussolera Branca, Le Fracce farm and Milan universities. BOIV. 2012;85:189–199. [Google Scholar]
- 11.Fernandez O, Theocharis A, Bordiec S, et al. Burkholderia phytofirmans PsJN acclimates grapevine to cold by modulating carbohydrate metabolism. Molecular Plant-Microbe Interactions. 2012;25(4):496–504. doi: 10.1094/MPMI-09-11-0245. [DOI] [PubMed] [Google Scholar]
- 12.Theocharis A, Bordiec S, Fernandez O, et al. Burkholderia phytofirmans PsJN primes Vitis vinifera L. and confers a better tolerance to low nonfreezing temperatures. Molecular Plant-Microbe Interactions. 2012;25(2):241–249. doi: 10.1094/MPMI-05-11-0124. [DOI] [PubMed] [Google Scholar]
- 13.Chaves MM, Zarrouk O, Francisco R, et al. Grapevine under deficit irrigation: hints from physiological and molecular data. Annals of Botany. 2010;105(5):661–676. doi: 10.1093/aob/mcq030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seguin B, de Cortazar IG. Climate warming: consequences for viticulture and the notion of ‘terroirs’ in Europe. Acta Horticulturae. 2005;689:61–69. [Google Scholar]
- 15.Hayat R, Ali S, Amara U, Khalid R, Ahmed I. Soil beneficial bacteria and their role in plant growth promotion: a review. Annals of Microbiology. 2010;60(4):579–598. [Google Scholar]
- 16.Marasco R, Rolli E, Ettoumi B, et al. A drought resistance-promoting microbiome is selected by root system under desert farming. PLoS One. 2012;7 doi: 10.1371/journal.pone.0048479.e48479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lau JA, Lennon JT. Rapid responses of soil microorganisms improve plant fitness in novel environments. Proceedings of the National Academy of Sciences. 2012;109:14058–14062. doi: 10.1073/pnas.1202319109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Köberl M, Müller H, Ramadan EM, Berg G. Desert farming benefits from microbial potential in arid soils and promotes diversity and plant health. PLoS ONE. 2011;6(9) doi: 10.1371/journal.pone.0024452.e24452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ambrosini A, Beneduzi A, Stefanski T, Pinheiro FG, Vargas LK, Passaglia LMP. Screening of plant growth promoting rhizobacteria isolated from sunflower (Helianthus annuus L.) Plant Soil. 2012;356:245–264. [Google Scholar]
- 20.Bachar A, Soares MIM, Gillor O. The effect of resource islands on abundance and diversity of bacteria in arid soils. Microbial Ecology. 2012;63(3):694–700. doi: 10.1007/s00248-011-9957-x. [DOI] [PubMed] [Google Scholar]
- 21.Sun L, Qiu F, Zhang X, Dai X, Dong X, Song W. Endophytic bacterial diversity in rice (Oryza sativa L.) roots estimated by 16S rDNA sequence analysis. Microbial Ecology. 2008;55(3):415–424. doi: 10.1007/s00248-007-9287-1. [DOI] [PubMed] [Google Scholar]
- 22.Muyzer G, De Waal EC, Uitterlinden AG. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Applied and Environmental Microbiology. 1993;59(3):695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rasband WS. US National Institutes of Health. Bethesda, Md, USA: ImageJ; 1997–2012. [Google Scholar]
- 24.Anderson MJ. A new method for non-parametric multivariate analysis of variance. Austral Ecology. 2001;26(1):32–46. [Google Scholar]
- 25. http://www.worldweatheronline.com.
- 26.Anderson MJ, Gorley RN, Clarke KR. PERMANOVA for PRIMER, Guide to Software and Statistical Methods. Plymouth, UK: PRIMER E; 2008. [Google Scholar]
- 27.Anderson MJ. DISTLM V.5: A FORTRAN Computer ProgrAm to Calculate a Distance-Based Multivariate Analysis for a Linear Model. Auckland , New Zealand: Department of Statistics, University of Auckland; 2005. [Google Scholar]
- 28.Penrose DM, Glick BR. Methods for isolating and characterizing ACC deaminase-containing plant growth-promoting rhizobacteria. Physiologia Plantarum. 2003;118(1):10–15. doi: 10.1034/j.1399-3054.2003.00086.x. [DOI] [PubMed] [Google Scholar]
- 29.King EO, Ward MK, Raney DE. Two simple media for the demonstration of pyocyanin and fluorescin. The Journal of Laboratory and Clinical Medicine. 1954;44(2):301–307. [PubMed] [Google Scholar]
- 30.Daffonchio D, De Biase A, Rizzi A, Sorlini C. Interspecific, intraspecific and interoperonic variability in the 16S rRNA gene of methanogens revealed by length and single-strand conformation polymorphism analysis. FEMS Microbiology Letters. 1998;164(2):403–410. doi: 10.1111/j.1574-6968.1998.tb13116.x. [DOI] [PubMed] [Google Scholar]
- 31.Daffonchio D, Borin S, Frova G, Manachini PL, Sorlini C. PCR fingerprinting of whole genomes: the spacers between the 16s and 23S rRNA genes and of intergenic tRNA gene regions reveal a different intraspecific genomic variability of Bacillus cereus and Bacillus licheniformis . International Journal of Systematic Bacteriology. 1998;48(1):107–116. doi: 10.1099/00207713-48-1-107. [DOI] [PubMed] [Google Scholar]
- 32.Daffonchio D, Cherif A, Borin S. Homoduplex and heteroduplex polymorphisms of the amplified ribosomal 16S-23S internal transcribed spacers describe genetic relationships in the “Bacillus cereus group”. Applied and Environmental Microbiology. 2000;66(12):5460–5468. doi: 10.1128/aem.66.12.5460-5468.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bric JM, Bostock RM, Silverstone SE. Rapid in situ assay for indoleacetic acid production by bacteria immobilized on a nitrocellulose membrane. Applied and Environmental Microbiology. 1991;57(2):535–538. doi: 10.1128/aem.57.2.535-538.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwyn B, Neilands JB. Universal chemical assay for the detection and determination of siderophores. Analytical Biochemistry. 1987;160(1):47–56. doi: 10.1016/0003-2697(87)90612-9. [DOI] [PubMed] [Google Scholar]
- 35.Mehta S, Nautiyal CS. An efficient method for qualitative screening of phosphate-solubilizing bacteria. Current Microbiology. 2001;43(1):51–56. doi: 10.1007/s002840010259. [DOI] [PubMed] [Google Scholar]
- 36.Santaella C, Schue M, Berge O, Heulin T, Achouak W. The exopolysaccharide of Rhizobium sp. YAS34 is not necessary for biofilm formation on Arabidopsis thaliana and Brassica napus roots but contributes to root colonization. Environmental Microbiology. 2008;10(8):2150–2163. doi: 10.1111/j.1462-2920.2008.01650.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cappuccino JC, Sherman N. Microbiology: A Laboratory Manual. 3rd edition. New York, NY, USA: Benjamin/Cummings Publishing Company; 1992. [Google Scholar]
- 38.Nielsen P, Sørensen J. Multi-target and medium-independent fungal antagonism by hydrolytic enzymes in Paenibacillus polymyxa and Bacillus pumilus strains from barley rhizosphere. FEMS Microbiology Ecology. 1997;22(3):183–192. [Google Scholar]
- 39.Hardoim PR, van Overbeek LS, Elsas JDV. Properties of bacterial endophytes and their proposed role in plant growth. Trends in Microbiology. 2008;16(10):463–471. doi: 10.1016/j.tim.2008.07.008. [DOI] [PubMed] [Google Scholar]
- 40.Walker TS, Bais HP, Grotewold E, Vivanco JM. Root exudation and rhizosphere biology. Plant Physiology. 2003;132(1):44–51. doi: 10.1104/pp.102.019661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bachar A, Soares MIM, Gillor O. The effect of resource islands on abundance and diversity of bacteria in arid soils. Microbial Ecology. 2012;63(3):694–700. doi: 10.1007/s00248-011-9957-x. [DOI] [PubMed] [Google Scholar]
- 42.Angel R, Soares MIM, Ungar ED, Gillor O. Biogeography of soil archaea and bacteria along a steep precipitation gradient. ISME Journal. 2010;4(4):553–563. doi: 10.1038/ismej.2009.136. [DOI] [PubMed] [Google Scholar]
- 43.Bachar A, Al-Ashhab A, Soares MIM, et al. Soil microbial abundance and diversity along a low precipitation gradient. Microbial Ecology. 2010;60(2):453–461. doi: 10.1007/s00248-010-9727-1. [DOI] [PubMed] [Google Scholar]
- 44.Garcias-Bonet N, Arrieta JM, de Santana CN, Duarte CM, Marbà N. Endophytic bacterial community of a Mediterranean marine angiosperm (Posidonia oceanica) Front Microbiol. 2012;3(article 342) doi: 10.3389/fmicb.2012.00342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hardoim PR, Hardoim CCP, van Overbeek LS, van Elsas JD. Dynamics of seed-borne rice endophytes on early plant growth stages. PLoS ONE. 2012;7(2) doi: 10.1371/journal.pone.0030438.e30438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sørensen J, Sessitsch A. Plant-associated bacteria lifestyle and molecular interactions. In: van Elsas JD, Jansson JK, Trevors JT, editors. Modern Soil Microbiology. 2nd edition. CRC Press; 2006. pp. 211–236. [Google Scholar]
- 47.Ryan RP, Germaine K, Franks A, Ryan DJ, Dowling DN. Bacterial endophytes: recent developments and applications. FEMS Microbiology Letters. 2008;278(1):1–9. doi: 10.1111/j.1574-6968.2007.00918.x. [DOI] [PubMed] [Google Scholar]
- 48.Siciliano SD, Germida JJ. Taxonomic diversity of bacteria associated with the roots of field-grown transgenic Brassica napus cv. Quest, compared to the non-transgenic B. napus cv. Excel and B. rapa cv. Parkland. FEMS Microbiology Ecology. 1999;29(3):263–272. [Google Scholar]
- 49.Izumi H, Anderson IC, Killham K, Moore ERB. Diversity of predominant endophytic bacteria in European deciduous and coniferous trees. Canadian Journal of Microbiology. 2008;54(3):173–179. doi: 10.1139/w07-134. [DOI] [PubMed] [Google Scholar]
- 50.Long HH, Sonntag DG, Schmidt DD, Baldwin IT. The structure of the culturable root bacterial endophyte community of Nicotiana attenuata is organized by soil composition and host plant ethylene production and perception. New Phytologist. 2010;185(2):554–567. doi: 10.1111/j.1469-8137.2009.03079.x. [DOI] [PubMed] [Google Scholar]
- 51.Compant S, Nowak J, Coenye T, Clément C, Ait Barka E. Diversity and occurrence of Burkholderia spp. in the natural environment. FEMS Microbiology Reviews. 2008;32(4):607–626. doi: 10.1111/j.1574-6976.2008.00113.x. [DOI] [PubMed] [Google Scholar]
- 52.Ofek M, Hadar Y, Minz D. Ecology of root colonizing Massilia (Oxalobacteraceae) PLoS One. 2012;7 doi: 10.1371/journal.pone.0040117.e40117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dardanellia MS, Fernández de Córdobab FJ, Estéveza J, et al. Changes in flavonoids secreted by Phaseolus vulgaris roots in the presence of salt and the plant growth-promoting rhizobacterium Chryseobacterium balustinum . Applied Soil Ecology. 2012;57:31–38. [Google Scholar]
- 54.Alami Y, Achouak W, Marol C, Heulin T. Rhizosphere soil aggregation and plant growth promotion of sunflowers by an exopolysaccharide-producing Rhizobium sp. strain isolated from sunflower roots. Applied and Environmental Microbiology. 2000;66(8):3393–3398. doi: 10.1128/aem.66.8.3393-3398.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bodenhausen N, Horton MW, Bergelson J. Bacterial communities associated with the leaves and the roots of Arabidopsis thaliana . PLoS One. 2012;8 doi: 10.1371/journal.pone.0056329.e56329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kolton M, Green SJ, Harel YM, Sela N, Elad Y, Cytryna E. Draft genome sequence of Flavobacterium sp. strain F52, isolated from the rhizosphere of bell pepper (Capsicum annuum L. cv. Maccabi) Journal of Bacteriology. 2012;194:5462–5463. doi: 10.1128/JB.01249-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bulgari D, Casati P, Crepaldi P, et al. Restructuring of endophytic bacterial communities in grapevine yellows-diseased and recovered Vitis vinifera L. plants. Applied and Environmental Microbiology. 2011;77(14):5018–5022. doi: 10.1128/AEM.00051-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gan HM, Chew TH, Hudson AO, Savkac MA. Genome sequence of Methylobacterium sp. strain GXF4, a xylem-associated bacterium isolated from Vitis vinifera L. grapevine. Journal of Bacteriology. 2012;194:5157–5158. doi: 10.1128/JB.01201-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hudson AO, Ahmad NH, Van Buren R, Savka MA. Sugarcane and grapevine endophytic bacteria: isolation, detection of quorum sensing signals and identification by 16S v3 rDNA sequence analysis. In: Vilas AM, editor. Current Research, Technology and Education Topics in Applied Microbiology and Microbial bioTechnology. 2010. pp. 801–806. [Google Scholar]
- 60.Bertrand H, Nalin R, Bally R, Cleyet-Marel J-C. Isolation and identification of the most efficient plant growth-promoting bacteria associated with canola (Brassica napus) Biology and Fertility of Soils. 2001;33(2):152–156. [Google Scholar]
- 61.Siddikee MA, Chauhan PS, Anandham R, Han G-H, Sa T. Isolation, characterization, and use for plant growth promotion under salt stress, of ACC deaminase-producing halotolerant bacteria derived from coastal soil. Journal of Microbiology and Biotechnology. 2010;20(11):1577–1584. doi: 10.4014/jmb.1007.07011. [DOI] [PubMed] [Google Scholar]
- 62.Glick BR, Penrose DM, Li J. A model for the lowering of plant ethylene concentrations by plant growth-promoting bacteria. Journal of Theoretical Biology. 1998;190(1):63–68. doi: 10.1006/jtbi.1997.0532. [DOI] [PubMed] [Google Scholar]
- 63.Trotel-Aziz P, Couderchet M, Biagianti S, Aziz A. Characterization of new bacterial biocontrol agents Acinetobacter, Bacillus, Pantoea and Pseudomonas spp. mediating grapevine resistance against Botrytis cinerea . Environmental and Experimental Botany. 2008;64(1):21–32. [Google Scholar]
- 64.Karagöz K, Ateş F, Karagöz H, Kotan R, Çakmakçı R. Characterization of plant growth-promoting traits of bacteria isolated from the rhizosphere of grapevine grown in alkaline and acidic soils. European Journal of Soil Biology. 2012;50:144–150. [Google Scholar]
- 65.Verhagen B, Trotel-Aziz P, Jeandet P, Baillieul F, Aziz A. Improved resistance against Botrytis cinerea by grapevine-associated bacteria that induce a prime oxidative burst and phytoalexin production. Phytopathology. 2011;101(7):768–777. doi: 10.1094/PHYTO-09-10-0242. [DOI] [PubMed] [Google Scholar]
- 66.Quecine MC, Araujo WL, Rossetto PB, et al. Sugarcane growth promotion by the endophytic bacterium Pantoea agglomerans 33.1. Applied and Environmental Microbiology. 2012;78(21):7511–7518. doi: 10.1128/AEM.00836-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Taurian T, Anzuay MS, Angelini JG, et al. Phosphate-solubilizing peanut associated bacteria: screening for plant growth-promoting activities. Plant and Soil. 2010;329(1):421–431. [Google Scholar]
- 68.Chaparro JM, Badri DV, Bakker MG, Sugiyama A, Manter DK, Vivanco JM. Root exudation of phytochemicals in Arabidopsis follows specific patterns that are developmentally programmed and correlate with soil microbial functions. PLoS One. 2013;2 doi: 10.1371/journal.pone.0055731.e55731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bhattacharjee RB, Singh A, Mukhopadhyay SN. Use of nitrogen-fixing bacteria as biofertiliser for non-legumes: prospects and challenges. Applied Microbiology and Biotechnology. 2008;80(2):199–209. doi: 10.1007/s00253-008-1567-2. [DOI] [PubMed] [Google Scholar]
- 70.Tarbah FA, Goodman RN. Rapid detection of Agrobacterium tumefaciens in grapevine propagation material and basis for an efficient indexing system. Plant Disease. 1986;70:566–568. [Google Scholar]
- 71.Thorne ET, Young BM, Young GM, et al. The structure of xylem vessels in grapevine (Vitaceae) and a possible passive mechanism for the systemic spread of bacterial disease. American Journal of Botany. 2006;93(4):497–504. doi: 10.3732/ajb.93.4.497. [DOI] [PubMed] [Google Scholar]
- 72.Chatelet DS, Matthews MA, Rost TL. Xylem structure and connectivity in grapevine (Vitis vinifera) shoots provides a passive mechanism for the spread of bacteria in grape plants. Annals of Botany. 2006;98(3):483–494. doi: 10.1093/aob/mcl124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nadeem SM, Zahir ZA, Naveed M, Nawaz S. Mitigation of salinity-induced negative impact on the growth and yield of wheat by plant growth-promoting rhizobacteria in naturally saline conditions. Annals of Microbiology. 2013;63:225–232. [Google Scholar]
- 74.Hamaki T, Suzuki M, Fudou R, et al. Isolation of novel bacteria and actinomycetes using soil-extract agar medium. Journal of Bioscience and Bioengineering. 2005;99(5):485–492. doi: 10.1263/jbb.99.485. [DOI] [PubMed] [Google Scholar]
- 75.Suzuki MT, Giovannoni SJ. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Applied and Environmental Microbiology. 1996;62(2):625–630. doi: 10.1128/aem.62.2.625-630.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kochar M, Srivastava S. Surface colonization by Azospirillum brasilense SM in the indole-3-acetic acid dependent growth improvement of sorghum. Journal of Basic Microbiology. 2012;52(2):123–131. doi: 10.1002/jobm.201100038. [DOI] [PubMed] [Google Scholar]
- 77.Barazani O, Friedman J. Is IAA the major root growth factor secreted from plant-growth-mediating bacteria? Journal of Chemical Ecology. 1999;25(10):2397–2406. [Google Scholar]
- 78.Wani PA, Khan MS, Zaidi A. Synergistic effects of the inoculation with nitrogen-fixing and phosphate-solubilizing rhizobacteria on the performance of field-grown chickpea. Journal of Plant Nutrition and Soil Science. 2007;170(2):283–287. [Google Scholar]
- 79.Schachtman DP, Reid RJ, Ayling SM. Phosphorus uptake by plants: from soil to cell. Plant Physiology. 1998;116(2):447–453. doi: 10.1104/pp.116.2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vyas P, Rahi P, Gulati A. Stress tolerance and genetic variability of phosphate-solubilizing fluorescent Pseudomonas from the cold deserts of the trans-himalayas. Microbial Ecology. 2009;58(2):425–434. doi: 10.1007/s00248-009-9511-2. [DOI] [PubMed] [Google Scholar]
- 81.Ndung’u-Magiroi KW, Herrmann L, Okalebo JR, Othieno CO, Pypers P, Lesueur D. Occurrence and genetic diversity of phosphate-solubilizing bacteria in soils of differing chemical characteristics in Kenya. Annals of Microbiology. 2011;62(3):897–904. [Google Scholar]
- 82.Mapelli F, Marasco R, Balloi A, et al. Mineral-microbe interactions: biotechnological potential of bioweathering. Journal of Biotechnology. 2012;157(4):473–481. doi: 10.1016/j.jbiotec.2011.11.013. [DOI] [PubMed] [Google Scholar]
- 83.Branda SS, Vik Å, Friedman L, Kolter R. Biofilms: the matrix revisited. Trends in Microbiology. 2005;13(1):20–26. doi: 10.1016/j.tim.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 84.Timmusk S, Grantcharova N, Wagner EGH. Paenibacillus polymyxa invades plant roots and forms biofilms. Applied and Environmental Microbiology. 2005;71(11):7292–7300. doi: 10.1128/AEM.71.11.7292-7300.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bartels D, Sunkar R. Drought and salt tolerance in plants. Critical Reviews in Plant Sciences. 2005;24(1):23–58. [Google Scholar]
- 86.Jha B, Gontia I, Hartmann A. The roots of the halophyte Salicornia brachiata are a source of new halotolerant diazotrophic bacteria with plant growth-promoting potential. Plant and Soil. 2011;356(1-2):265–277. [Google Scholar]
- 87.Sandhya V, Z. AS, Grover M, Reddy G, Venkateswarlu B. Alleviation of drought stress effects in sunflower seedlings by the exopolysaccharides producing Pseudomonas putida strain GAP-p45. Biology and Fertility of Soils. 2009;46(1):17–26. [Google Scholar]
- 88.Parmesan C. Ecological and evolutionary responses to recent climate change. Annual Review of Ecology, Evolution, and Systematics. 2006;37:637–669. [Google Scholar]
- 89.Thiel V. Extreme environments. Encyclopedia of Geobiology, 2012, http://www.springerreference.com/docs/html/chapterdbid/187271.html.
- 90.Sheik CS, Beasley WH, Elshahed MS, Zhou X, Luo Y, Krumholz LR. Effect of warming and drought on grassland microbial communities. ISME Journal. 2011;5(10):1692–1700. doi: 10.1038/ismej.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Vicente-Serrano SM, Zouber A, Lasanta T, Pueyo Y. Dryness is accelerating degradation of vulnerable shrublands in semiarid Mediterranean environments. Ecological Monographs. 2012;4:407–428. [Google Scholar]


