Abstract
The Cdc5L (cell division cycle 5-like) complex is a spliceosomal subcomplex that also plays a role in DNA repair. The complex contains the splicing factor hPrp19, also known as SNEV or hPso4, which is involved in cellular life-span regulation and proteasomal breakdown. In a recent large-scale proteomics analysis for proteins associated with this complex, proteins involved in transcription, cell-cycle regulation, DNA repair, the ubiquitin–proteasome system, chromatin remodelling, cellular aging, the cytoskeleton and trafficking, including four members of the exocyst complex, were identified. In the present paper we report that Exo70 interacts directly with SNEVhPrp19/hPso4 and shuttles to the nucleus, where it associates with the spliceosome. We mapped the interaction site to the N-terminal 100 amino acids of Exo70, which interfere with pre-mRNA splicing in vitro. Furthermore, Exo70 influences the splicing of a model substrate as well as of its own pre-mRNA in vivo. In addition, we found that Exo70 is alternatively spliced in a cell-type- and cell-age-dependent way. These results suggest a novel and unexpected role of Exo70 in nuclear mRNA splicing, where it might signal membrane events to the splicing apparatus.
Keywords: alternative splicing, exocyst, Exo70, protein interaction, Prp19, SNEV
INTRODUCTION
In higher eukaryotes, the vast majority of protein-coding genes contain introns, which must be removed from pre-mRNA in a process called splicing. Splicing is carried out by the spliceosome, a highly dynamic multiple megadalton molecular machine. The spliceosome consists of five snRNPs (small nuclear ribonucleoproteins) called U1, U2, U4, U5 and U6, and a large number of non-snRNP splicing factors. For efficient splicing to take place, snRNPs and other splicing factors must associate on the pre-mRNA in a highly ordered manner, and several structural rearrangements are necessary before the spliceosome adopts its catalytically active composition (reviewed in [1]). Among the >35 non-snRNP proteins recruited during formation of the active spliceosome is the Prp19–Cdc5L (cell division cycle 5-like) complex. The mammalian Prp19–Cdc5L core complex consists of Prp19, also called SNEVhPrp19/hPso4 in humans, Cdc5L, Plrg1 (pleiotropic regulator 1) and Spf27. Recently, the molecular architecture of this complex has been elucidated [2]. Additional so-called Prp19/Cdc5L-related proteins physically interact with either SNEVhPrp19/hPso4 or Cdc5L, including the protein SKIP (skeletal muscle- and kidney-enriched inositol phosphatase) [3,4].
We were able to confirm that SNEVhPrp19/hPso4 plays an essential role in splicing by showing that SNEVhPrp19/hPso4 interacts with itself, and that inhibition of this self interaction blocks spliceosome assembly [5]. Its necessity for pre-mRNA splicing might also explain why SNEVhPrp19/hPso4 deletion is early embryonic lethal in mice [6]. Furthermore, by analysing directly interacting proteins we recently identified a novel pre-mRNA splicing factor termed Blom7α [7].
In addition to its presence in the spliceosome, SNEVhPrp19/hPso4 has been described as a DNA repair factor that is induced in response to DNA-damaging agents and that, together with Cdc5L and WRN (Werner), is necessary for repair of DNA interstrand cross-links [8]. It recruits the DNA repair factor Metnase to DNA double-strand breaks [9]. Recently it was found that Cdc5L interacts with the major damage response mediator ATR (ataxia telangiectasia mutated- and Rad3-related) and is necessary for activation of ATR downstream effectors [10]. Probably it is this activity that increases stress resistance and replicative life span upon SNEVhPrp19/hPso4 overexpression in human endothelial cells [11].
SNEVhPrp19/hPso4 also has catalytic activity as an E3 ubiquitin ligase in vitro [12] and directly interacts with the proteasome [13,14]. In addition, SNEVhPrp19/hPso4 ubiquitinates Prp3, a component of the U4 snRNP. The modification of Prp3 with Lys63-linked ubiquitin chains increases its affinity for the U5 component Prp8 and thus stabilizes the U4/U6/U5 snRNP [15].
In an attempt to further elaborate on the multiple cellular roles of the Cdc5L–Prp19 complex, a large-scale pull-down from HeLa nuclear extracts using antibodies against Cdc5L and against SKIP was recently performed [16]. Consistent with the known functions of the proteins, we found, besides pre-mRNA splicing factors, proteins known to be involved in DNA repair, protein turnover by the proteasome, cell-cycle regulation and cellular aging. Moreover, we identified proteins with functions not directly related to splicing such as trafficking, chromatin remodelling, the cytoskeleton, or protein folding and assembly. However, one novel interaction partner was identified that also came up in a recently performed yeast two-hybrid screening using SNEVhPrp19/hPso4 as the bait [7]: the exocyst subunit Exo70, also termed EXOC7. For Cdc5L and SKIP, additional exocyst components were found as interactors: Sec5 (or EXOC2), Sec8 (or EXOC4) and Sec10 (or EXOC5), as well as Sec13-like protein, a constituent of the endoplasmic reticulum and the nuclear pore complex that is required for vesicle biogenesis. These novel interactions were unexpected because the main function of the exocyst complex is vesicular transport, where it mediates the tethering and spatial targeting of post-Golgi vesicles to the plasma membrane. Up to now, only cytoplasmic localization has been observed for exocyst components, although the Sec13-like protein was already found to shuttle to the nucleus [17].
In the present study, we show a novel facet of Exo70. It is not only a direct and physical interactor of SNEV that shuttles between the cytoplasm and the nucleus, we also present data that suggests a role for Exo70 in pre-mRNA splicing. Furthermore, we found previously unknown splicing isoforms of Exo70, which are differentially regulated in various tissues and during cellular aging.
MATERIALS AND METHODS
Construction of plasmids
The coding sequence of SNEVhPrp19/hPso4 was amplified by RT (reverse transcriptase)–PCR, therefore total RNA was prepared using TRIzol® reagent (Invitrogen). Total RNA (1 μg) from HUVECs (human umbilical vein endothelial cells) was reverse-transcribed using oligo(dT)25 primers, and standard PCR with the primers SNEV NdeI sense 5′- GTACACATATGTCCCTAATCTGCTCC-3′ and SNEV SmaI antisense 5′-CATTGACCCGGGCAGGCTGTAGAACTTGAGG-3′ was performed. Exo70 and its truncated forms CC (coiled-coil) and ΔCC were amplified by PCR from the clone isolated by yeast two-hybrid screening [7]. SNEV cDNA was ligated into pGADT7 and pGBKT7 for yeast two-hybrid analysis, and SNEVhPrp19/hPso4 and Exo70 cDNA was ligated into pEYFP-C1, -N1 [EYFP is enhanced YFP (yellow fluorescent protein)] and pECFP-C1, -N1 [ECFP is enhanced CFP (cyan fluorescent protein)] for FRET (fluorescence resonance energy transfer) analysis (Clontech Laboratories). Exo70 in pEGFP-N1 and -C1 were kindly provided by Dr R. H. Scheller (Department of Biological Sciences, Stanford University, Stanford, CA, U.S.A.). For GST (glutathione transferase) pull-down assays, SNEVhPrp19/hPso4 was inserted into pGEX-6P-1 (Amersham Biosciences), whereas Exo70, CC and ΔCC were cloned into pET28a (Novagen) The His6-tagged cDNAs of Exo70, CC and ΔCC were cut and religated into pCIneo (Promega) for transient mammalian expression, as well as into pET28a for recombinant protein expression in Escherichia coli. For in vivo splicing assays, Exo70 was amplified from HeLa cDNA using the primers Exo70_start_EcoRI_long 5′-CCCTTTGAATTCATGATTCCCCCACAGGAGGC-3′ and Exo70_stop_ BamHI_long 5′-CCCTTTGGATCCTCAGGCAGAGGTGTCG-3′ and cloned into pEGFP-C2 by EcoRI and BamHI restriction sites. pMT-E1A was kindly provided by Dr J. F. Caceres (MRC Human Genetics Unit, Western General Hospital, Edinburgh, Scotland, U.K.). All cDNAs contained in the genetic constructs were confirmed to contain no mutations by sequence analysis [IBL (Innovationen Biochemie und Labor Manegement), Vienna, Austria]. The His6–SNEVhPrp19/hPso4-containing plasmid for baculoviral expression was kindly provided by Dr S. Hatakeyama (Hokkaido University, Sapporo, Japan).
Directed yeast two-hybrid assay
Screening for SNEV-interacting proteins was performed using the MATCHMAKER GAL4 Two-Hybrid System3 (Clontech Laboratories) according to the manufacturer’s protocol. As bait, pGBKT7-SNEVhPrp19/hPso4 was co-transformed into the yeast reporter strain AH109 together with human Exo70, and truncation mutants containing CC or ΔCC were inserted into pGADT7. Yeast co-transformation was performed using the lithium acetate method [18]. Interactions were confirmed on plates containing high stringency 4 × drop-out medium [SD (synthetic drop-out)/ – Leu/ – Trp/ – His/ – Ade]. Autoactivation of the constructs was excluded by transformation of plasmids alone and co-transformation with the respective empty vector plasmid.
Recombinant protein expression and purification of His–Exo70 and generation of monoclonal antibodies
The plasmids pET28a-Exo70, -CC and -ΔCC were introduced into E. coli BL21LysDE. Bacteria were grown in 1 litre of LB (Luria–Bertani) broth containing 25 μg/ml kanamycin at 37 °C on a shaker until a D600 of 0.6 was reached. Thereupon recombinant protein expression was induced by the addition of IPTG (isopropyl β-d-thiogalactopyranoside) to a final concentration of 1 mM, and incubation at 37 °C with shaking was continued for 4 h. Bacteria were sonicated in lysis buffer [50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole and 0.05% Tween 20 (pH 8.0)] using six pulses of 30 s with a VibraCell VC300 ultrasound device (Sonics and Materials; 300 W, 200 V, 3 A and 20 kHz). His6-tagged proteins were purified under native conditions using Ni-NTA (Ni2+ -nitrilotriacetate) Sepharose beads (Amersham Biosciences). Beads were washed three times with wash buffer [50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole and 0.05% Tween 20 (pH 8.0)]. Bound proteins were eluted with elution buffer [50 mM NaH2PO4, 300 mM NaCl, 250 mM imidazole and 0.05% Tween 20 (pH 8.0)]. Polyclonal sera and monoclonal antibodies were generated following our previously published protocol for SNEV [19]. The pGEX-6P-1-SNEV plasmid was transformed into E. coli BL21 cells, and the fusion protein was expressed using Overnight Express Autoinduction System 1 (Novagen) and purified using the Micro Spin GST Purification Module (Amersham Biosciences). Recombinant proteins were dialysed against 20% glycerol in PBS. Aliquots (20 μl) were snap-frozen in liquid nitrogen and stored at −80 °C. Purified proteins were used for immunization of mice and-antibodies were generated as described in [19]. All animal experimentation was carried out according to the appropriate institutional guidelines.
CoIP (co-immunoprecipitation)
The purified His6-tagged Exo70 was immobilized on Ni-NTA agarose beads. In vitro translation of SNEVhPrp19/hPso4 was performed using the TnT® Quick-Coupled Transcription/Translation System (Promega) and the pGBKT7-derived plasmid as a template, resulting in a c-Myc-tagged SNEVhPrp19/hPso4. After mixing the in vitro translate with His6-tagged Exo70 and incubation at 37 °C for 1 h, precipitation was performed by the addition of the Ni-NTA agarose beads and incubation at room temperature (approximately 23 °C) for 3 h in binding buffer [20 mM Tris/HCl (pH 7.4), 140 mM NaCl, 10% (v/v) glycerol, 1 mM CaCl2, 0.1% Triton X-100 and Complete protease inhibitor cocktail (Roche)]. After washing four times with binding buffer, the proteins were eluted by heating for 10 min in SDS sample buffer. After SDS/PAGE, proteins were detected by Western blotting using an anti-c-Myc antibody (Clontech Laboratories) and an anti-mouse peroxidase conjugate as the secondary antibody (Sigma–Aldrich).
For the GST pull-down assays, equimolar amounts of the purified proteins were mixed and incubated for 2 h at 4 °C in CoIP buffer [200 mM NaCl, 25 mM Tris/HCl (pH 7.4) and 0.5% Triton X-100], followed by the addition of glutathione–Sepharose 4B beads (Amersham Biosciences) equilibrated with CoIP buffer and a 2 h incubation at 4 °C. The beads were washed three times with CoIP buffer, and proteins were eluted by heating for 10 min in SDS sample buffer. After SDS/PAGE, precipitated SNEVhPrp19/hPso4 was detected using an anti-His4 antibody (Qiagen). GST was detected using an anti-GST antibody (Amersham) and an anti-goat peroxidase conjugate as the secondary antibody (Sigma–Aldrich).
SDS/PAGE and Western blot analysis
Protein samples were separated on a NuPAGE 4–12% Bis/Tris polyacrylamide gel (Invitrogen). Electrophoresis and blotting to PVDF membrane (Roth) were performed in accordance with the manufacturer’s protocol (Invitrogen). After blocking with 3% (w/v) dried skimmed milk powder in PBS, the membranes were incubated with the primary antibodies followed by incubation with the appropriate secondary antibody.
Endogenous SNEV was detected using anti-SNEV_867 and goat anti-rabbit IgG peroxidase-conjugated antibody as the secondary antibody (Sigma). Endogenous Exo70 was detected using the monoclonal anti-Exo70 antibody generated as described for SNEV [19] and an anti-mouse IgG peroxidase-conjugated antibody (Sigma–Aldrich).
To determine the purity of subcellular fractionation, an anti-(lamin A/C) rabbit polyclonal antibody and an anti-GAPDH (glyceraldehyde-3-phosphate dehydrogenase) rabbit antibody (both Santa Cruz Biotechnology) were used in combination with anti-rabbit-IRDye 800 (from goat, H + L, Licor).
Detection was performed with ECL (enhanced chemiluminescence) Plus (Amersham Biosciences) and the LumiImager (Roche Applied Science), or with the Odyssey Infrared Scanner (Licor), depending on the type of secondary antibody used.
Cell culture
HeLa cells were grown in RPMI 1640 (Biochrom) supplemented with 4 mM l-glutamine and 10% FBS (fetal bovine serum). HEK (human embryonic kidney)-293, HDF (human dermal fibroblast) and MDCK (Madin–Darby canine kidney) cells were grown in DMEM (Dulbecco’s modified Eagle’s medium)/Ham’s F12 (1:1; Biochrom) supplemented with 4 mM l-glutamine and 10% FBS. HUVECs were grown in M199 medium (Biochrom) supplemented with 4 mM l-glutamine, 15% FBS, 90 μg/ml heparin and 200 μg/ml ECGS (endothelial cell growth supplement). RPTECs (renal proximal tubule epithelial cells) were grown as described previously [20]. Cells were fixed for immunostaining after 6 h or overnight exposure to 10 pM LMB (leptomycin B) (Calbiochem). Cells are considered senescent after undergoing an irreversible growth arrest due to telomere shortening [21,22]. HDFs enter senescence at approximately PD (population doubling) 60, HUVECs at approximately PD 45–50 [23] and RPTECs at approximately PD 20–25 [20]. Cells are considered ‘young’ throughout the first half of their replicative life span.
Preparation of nuclear extracts
Splicing-competent nuclear extracts were prepared as described previously [24]. In brief, HeLa cells were harvested at approximately 80% confluence and washed twice with PBS. The cells were resuspended in NE1 buffer [10 mM Hepes (pH 8.0), 1.5 mM MgCl2, 10 mM KCl and 1 mM DTT (dithiothreitol)] and incubated for 15 min on ice. Cells were passed through a 23-gauge syringe five times. After centrifugation (14000 g for 20 s at 4 °C) the pellet was resuspended in NE2 buffer [20 mM Hepes (pH 8.0), 1.5 mM MgCl2, 25% glycerol, 420 mM NaCl, 0.2 mM EDTA, 1 mM DTT and 0.5 mM PMSF] and incubated on ice for 30 min. Nuclear debris was pelleted by centrifugation (14000 g for 5 min at 4 °C) and supernatant was dialysed against NE2 buffer.
FRET
SNEVhPrp19/hPso4 and Exo70 inserted into pECFP-N1, pECFP-C1 and pEYFP-N1, pEYFP-C1 respectively were transiently co-transfected into COS-1 cells by Lipofectamine™ 2000 (Invitrogen) according to the manufacturer’s protocol. After 24 h, FRET images from living cells were generated by the MicroFRET method as described by Youvan et al. [25] and as previously published [5,7,13]. Photos were captured on a Nikon Diaphot TMD microscope with a cooled charge-coupled device camera (Kappa), with the YFP, CFP and FRET filter sets (Omega Optical), under identical conditions and processed with Scion Image software version beta 4.0.2 (Scion Corporation). The images were aligned by pixel shifting, inverted and background was subtracted. Images from the YFP and CFP filter sets were multiplied with their previously assessed correction factors (0.19 for YFP and 0.59 for CFP) and subtracted from the FRET filter set picture. The remaining signal was multiplied by three for better visualization and represents the corrected FRET. Representative pictures are shown. In total, ten out of ten cells analysed showing similar expression of CFP and YFP fusion proteins also gave a FRET signal.
Cell staining and immunofluorescence analyses
Cells were washed in PBS and fixed for 5 min in 3.7% (w/v) paraformaldehyde in CSK buffer [10 mM Pipes (pH 6.8), 10 mM NaCl, 300 mM sucrose, 3 mM MgCl2 and 2 mM EDTA] at room temperature. Permeabilization was performed with 1% Triton X-100 in PBS for 15 min at room temperature. Cells were incubated with primary antibodies diluted in PBS with 1% goat serum for 1 h, washed three times for 10 min with PBS, incubated for 1 h with the appropriate secondary antibodies diluted in PBS with 1% goat serum, and washed three times for 10 min with PBS. Antibodies used were rabbit anti-SNEVhPrp19/hPso4 antibody (Prp19-867) and anti-Sec6 (Calbiochem). As secondary antibodies, TRITC (tetramethylrhodamine β-isothiocyanate)-labelled anti-rabbit or anti-mouse antibodies (Jackson Immunoresearch Laboratories) were used. Microscopy and image analysis was carried out using a Zeiss DeltaVision Restoration microscope as described previously [5,7,26].
Preparation of spliceosomal complexes
Human spliceosomal complexes were prepared as described previously [7,27]. Briefly, a mixture of spliceosomal complexes was assembled on biotinylated radioactively labelled RNA. As the splicing substrate the AD1 (adenovirus) transcript was used. The substrate was biotin-labelled and incubated under splicing conditions with HeLa nuclear extracts in 1 ml reaction mixtures at 30 °C for 1 h, forming both active spliceosomes and assembly intermediates. After incubation the samples were immediately loaded on to a 2.5 cm × 75 cm S-500 gel-filtration column, and pooled fractions from the spliceosome peak were affinity-selected on streptavidin beads [28]. Proteins bound to the beads were washed three times in wash buffer [100 mM NaCl and 20 mM Tris/HCl (pH 7.5)], and then eluted in 0.3 ml of elution buffer [2% SDS, 20 mM Tris/HCl (pH 7.5) and 20 mM DTT]. Eluted proteins were precipitated with 1 ml of methanol together with 12 μg of slipper limpet glycogen carrier and finally resuspended in 50 μl of elution buffer. This procedure was repeated 12 times, and the resulting samples were pooled separately for each of the pre-mRNA substrates. Based on the staining with Coomassie Blue, we estimate that each fraction contained ~6–10 μg of protein in total. For the background control, nuclear extract was incubated without labelled RNA, followed by gel filtration as described above. Beads were mixed with the fractions that corresponded to the ones that contained labelled RNA in the above-described experiment. Beads were washed, and the bound material was eluted as above.
In vitro splicing assay
Nuclear extracts used in the splicing assays were obtained commercially from Dundee Cell Products. Splicing assays were performed using uniformly labelled, capped pre-mRNAs incubated with nuclear extract as described previously [24]. Purified recombinant proteins (80, 160 or 320 pmol) were added to the splicing reactions. The adeno-pre-mRNA was transcribed from Sau3AI-digested plasmid pBSAd1 [29]. The splicing reactions were loaded on to a 10% polyacrylamide and 8 M urea denaturing gel and run in 1 × TBE [Tris/borate/EDTA (1 × TBE = 45 mM Tris/borate and 1 mM EDTA)] to separate the splicing products.
Alternative splicing assays
HeLa cells were transiently co-transfected in triplicate with pMT-E1A and 0.5, 1 or 2 μg of pEGFFP-C2 containing either full-length Exo70, ΔCC or CC or empty pEGFP-C1 using Metafectene Pro transfection reagent (Biontex). Total RNA was isolated 48 h post-transfection and reverse transcribed using SuperScript™ III reverse transcriptase. To exclude the possibility that the RNA isolation was contaminated with pMT-E1A vector DNA, RNA was treated with DNase I (Ambion) prior to cDNA synthesis. E1A alternatively spliced isoforms were amplified from 3 μl of cDNA by PCR in 50 μl reaction mixtures with the KAPA 2G Robust Polymerase (Peqlab) using primers E1A_ Exon1_forward 5′-GTTTTCTCCTCCGAGCCGCTCCGA-3′ and E1A_Exon2_reverse 5′-CTCAGGCTCAGGTTCAGACACAGG-3′ at final concentrations of 500 nM. After 5 min of denaturation, 30 PCR cycles (30 s at 95 °C, 30 s at 71 °C and 30 s at 72 °C) were performed. For the quantification of E1A isoforms generated by in vivo splicing, 1 μl of each PCR product was subjected to chip-based capillary electrophoresis with the Agilent 2100 Bioanalyzer, and the proportional amount of each fragment to the whole yield of splicing products was calculated.
For the detection of Exo70 alternatively spliced isoforms, cDNA was prepared as described above. In this case, primers Exo70_SpliceVar_for 5′-CCCCAACAAGAGGAAAGACA-3′ and Exo70_SpliceVar_rev 5′-TTGACGAAGGCACTGACG-3′ were used for amplification of Exo70 in different cell lines, and primer Exo70_SpliceVar_for was used together with the reverse primer 3 5′-TGCTTGTCGTTCAAGGCC-3′ when using cDNA from Exo70-overexpressing cells as the template. All other steps were carried out as described above.
One-way ANOVA F-tests were performed using R version 2.9.1 for comparing levels of splice isoforms.
RESULTS
Identification of proteins interacting with the SNEVPrp19/Pso4–Cdc5L associated complex
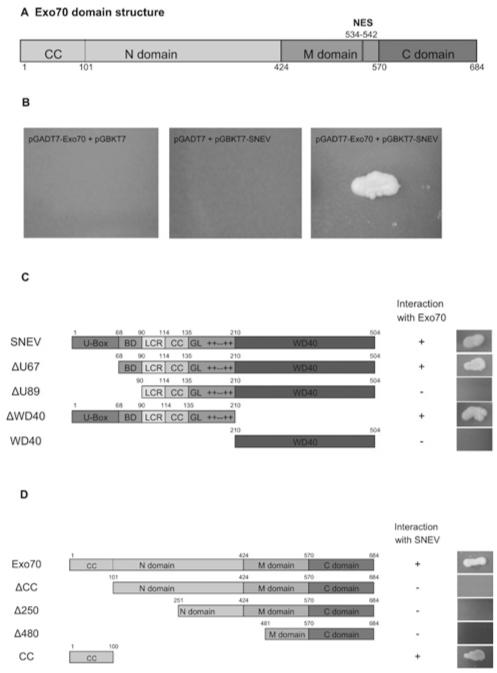
In a recent yeast two-hybrid screening using SNEVPrp19/Pso4 as bait, we found Exo70 as prey [7]. In addition, co-precipitations of the core complex, using antibodies against Cdc5L, a member of the core complex, and SKIP, which is described as associated with the core complex [4], have been performed [16]. Thereby, a large number of proteins involved in a variety of cellular functions were identified, including factors involved in cell-cycle regulation, DNA repair, trafficking, the ubiquitin–proteasome system, chaperones, chromatin remodelling, aging and the cytsoskeleton, besides factors involved in pre-mRNA splicing and transcription [16]. Interestingly, four out of eight members of the exocyst complex were identified. Since the nuclear localization of the exocyst has not yet been reported, we decided to analyse in more detail the interaction of Exo70 and SNEVhPrp19/hPso4. First, we looked at the domain structure and putative nuclear localization signals of Exo70 (Figure 1A). The protein consists of 19 α-helices connected by loops of various lengths and can be subdivided into an N-terminal domain, a middle (M) domain and a C-terminal domain [30]. Several regions with polar compositional bias (especially residues 160–180, 240–320 and 500–550) might represent a flexible hinge. Directed yeast two-hybrid tests were performed to exclude autoactivation and to confirm the library screening result (Figure 1B). In order to narrow down the interaction domain, several truncated forms of SNEV, as well as of Exo70, were cloned into the yeast two-hybrid vectors. First we tested several truncation mutants of SNEV against full-length Exo70 (Figure 1C). Colony formation of yeast transformants was observed only with SNEV constructs containing amino acids 68–90. From our previous findings, the amino acids between 68 and 90 are also involved in SNEVhPrp19/hPso4 homo-oligomerization [5], as well as for binding the alternative splicing factor Blom7α, a novel splicing factor interacting with SNEVhPrp19/hPso4 [7]. Taken together, these results suggest that either this domain is indeed an interaction ‘hot-spot’, or that Exo70 does not interact with monomeric SNEVhPrp19/hPso4, but associates with a structure that forms by SNEVhPrp19/hPso4 oligomerization only. For the mapping of the interaction domain on Exo70 we used N-terminal deletion mutants together with full-length SNEV in a directed yeast two-hybrid screen (Figure 1D). The CC domain was found to be necessary and sufficient for SNEV binding. In addition, we found that the exocyst subunit Sec6 and the chaperone TCP1 (T-protein complex 1) were also present in the immunoprecipitates of SNEVhPrp19/hPso4, SKIP and Blom7α, further confirming these protein associations (Supplementary Figures S1 and S2 at http://www.BiochemJ.org/bj/438/bj4380081add.htm).
Figure 1. Yeast two-hybrid screening identifies Exo70 as a putative interaction partner of SNEVhPrp19/hPso4.
(A) Scheme representing the domain structure of Exo70. Each domain consists of a bundle of α-helices. Exo70 contains a putative nuclear export signal. A CC structure in the N-terminal domain provides a protein-binding site. (B) SNEVhPrp19/hPso4 was used as bait protein in a yeast two-hybrid cDNA library screening and identified Exo70 as a putative interaction partner. Although neither SNEVhPrp19/hPso4 nor Exo70 co-transformed together with the respective second empty vector allowed colony formation on high-stringency drop-out medium, co-transformation of GAL4–BD-SNEV and GAL4–AD-Exo70 into yeast two-hybrid reporter strain AH109 resulted in colony formation. (C) Co-transformation of different truncated forms of SNEVhPrp19/hPso4 together with Exo70 showed that yeast colony growth depends on the presence of the amino acids 68–90. U-box, U-box domain; BD, binding domain; LCR, low complexity region; GL, globular domain; + + − + +, charged region; WD40, 7 × WD40 repeats. (D) Several truncated forms of Exo70 were tested against SNEVhPrp19/hPso4 in order to map the interaction site. The 100 N-terminal amino acids of Exo70 are necessary and sufficient for the interaction. ‘+’ and ‘−’ indicate colony upon co-transformation with SNEV.
The interaction between SNEVhPrp19/hPso4 and Exo70 is direct
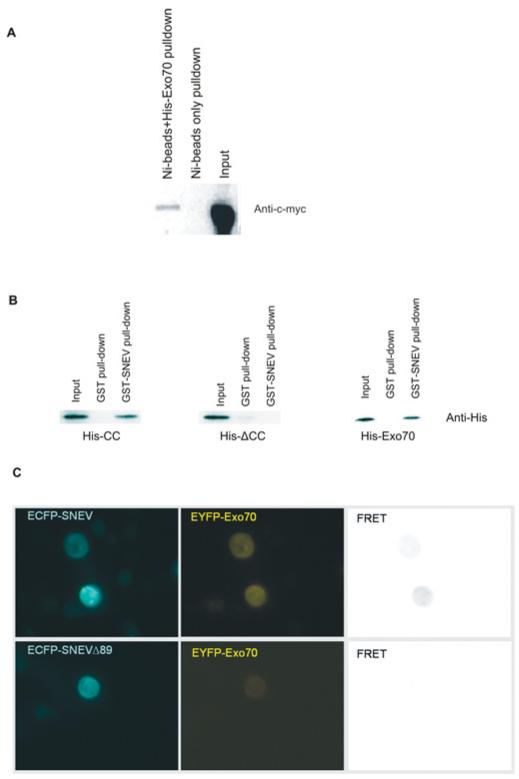
In order to confirm the interaction between SNEVhPrp19/hPso4 and Exo70, we performed pull-down experiments. Myc–SNEVhPrp19/hPso4 was co-precipitated by His6–Exo70 bound to Ni-NTA agarose beads, whereas in the absence of His6–Exo70, no SNEV was detectable by Western blot analysis (Figure 2A). Furthermore, we pulled down full-length His6–Exo70 as well as His6–CC, but not the His6–ΔCC mutant, using GST–SNEVhPrp19/hPso4 immobilized on glutathione–agarose beads (Figure 2B). Thus we conclude that the interaction between SNEVhPrp19/hPso4 and Exo70 is direct and mediated by the CC domain of Exo70. Finally, we inserted the cDNAs of SNEVhPrp19/hPso4 and Exo70 into pECFP and pEYFP vectors and performed MicroFRET analysis [25] in COS-1 cells. A clear FRET signal was detected throughout the cells, whereas SNEV lacking the N-terminal 89 amino acids that comprise the putative Exo70-interacting domain did not yield a detectable FRET signal (Figure 2C). Negative controls using co-transfection with either SNEVhPrp19/hPso4 or Exo70 fusion constructs with the corresponding empty vector did not result in detectable FRET signals (results not shown). Although these results further confirm the interaction between SNEVhPrp19/hPso4 and Exo70, their significance in terms of localizing the interaction within the cell is limited. The cellular localization of endogenous SNEVhPrp19/hPso4 is usually nuclear and was probably altered either by overexpression or by the interaction with free EYFP–Exo70 that might interfere with the import of SNEV into the nucleus.
Figure 2. Interaction of SNEVhPrp19/hPso4 and Exo70 is direct.
(A) The yeast two-hybrid screening results were confirmed by coIP. Although Ni2+ beads alone did not pull down Myc–SNEVhPrp19/hPso4, CoIP using His6–Exo70-loaded beads resulted in detectable amounts of Myc–SNEVhPrp19/hPso4 on Western blots detected using the anti-c-Myc antibody. A 50% input was loaded. (B) GST–SNEVhPrp19/hPso4, in contrast with GST alone, pulled down His6-tagged Exo70 as well as the His6-CC domain of Exo70, whereas ΔCC was not pulled down by GST or GST–SNEVhPrp19/hPso4. A 25% input was loaded. (C) FRET analysis confirms the SNEV–Exo70 interaction. COS-1 cells were co-transfected with ECFP–SNEV and EYFP–Exo70, and FRET analysis was performed as described in the Materials and methods section, resulting in a detectable signal. Control FRET analysis using a SNEV deletion mutant lacking the 89 N-terminal amino acids necessary for the interaction with Exo70 yielded no detectable signal.
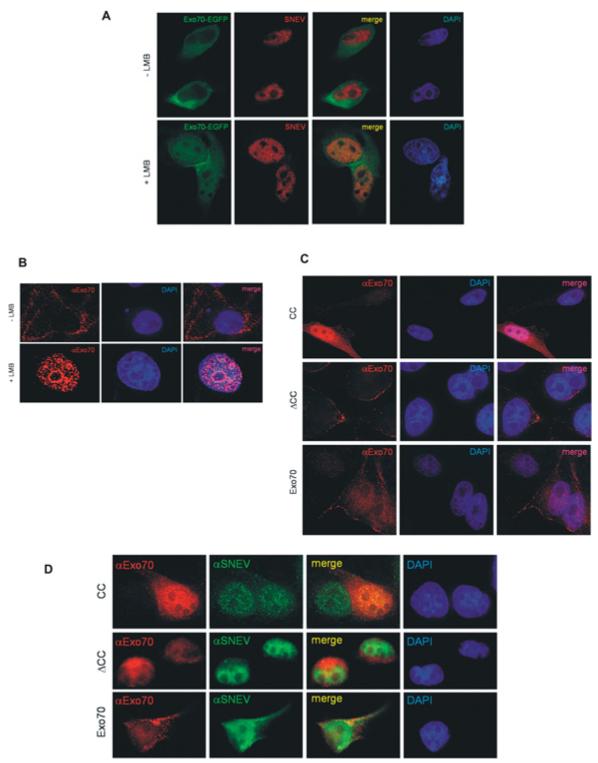
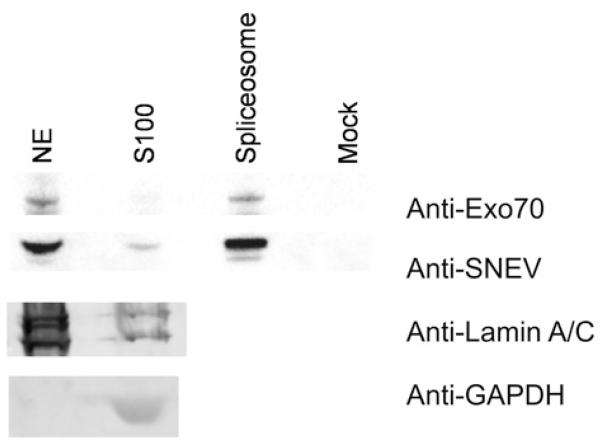
Exo70 shuttles to the nucleus in HeLa cells and co-localizes with SNEVhPrp19/hPso4. Although the predicted localization for Exo70 by PSORT2 [31] is nuclear, it has not yet been observed as a nuclear protein. A reason for this might be the presence of a putative Crm1/exportin 1-dependent leucine-rich nuclear export signal predicted by NetNES 1.1 [32]. Therefore we tested whether the localization of Exo70–EGFP [enhanced GFP (green fluorescent protein)] changes within HeLa cells after blocking nuclear export by incubation with LMB, a reagent that alkylates and inhibits Crm1, a protein required for nuclear export of proteins containing a nuclear export sequence [33]. At 2 h after transfection with pExo70–EGFP, cells were incubated with LMB for 8 h and then fixed for immunostaining. Indeed, Exo70–EGFP accumulates in the nucleus upon LMB treatment and to some extent co-localizes with SNEVhPrp19/hPso4, whereas in untreated control cells it was predominantly located in the cytoplasm and at the membranes (Figure 3A, top panels). However, there are specific regions of the nucleus where there is only SNEVhPrp19/hPso4 or EGFP–Exo70, which might indicate additional functions of both proteins that are not dependent on their interaction or might be caused by overexpression of EGFP–Exo70 and LMB treatment. Using our monoclonal antibody against endogenous Exo70, the nuclear accumulation of Exo70 upon LMB treatment was confirmed (Figure 3B). Finally, we overexpressed Exo70, ΔCC and CC and detected them using monoclonal anti-Exo70 antibodies. Without LMB treatment, CC and Exo70 were detectable in nuclei (Figure 3C), whereas, upon LMB treatment, all co-localized with SNEV in the nucleus (Figure 3D). Surprisingly, another subunit of the exocyst, Sec6 (EXOC3), also localizes to the nucleus upon blocking nuclear export (see Supplementary Figure S3 at http://www.BiochemJ.org/bj/438/bj4380081add.htm). These results suggest that at least two members of the exocyst indeed shuttle between cytoplasm and the nucleus. Together with the results of the large-scale immunoprecipitation, the possibility is raised that the whole exocyst shuttles, although the function of this movement remains unclear. In order to confirm that endogenous Exo70 is present in the nucleus, we probed nuclear extract, cytoplasmic extract S100 from HeLa cells and purified mature spliceosomes for the presence of Exo70. Indeed we were able to detect Exo70 in all cellular fractions tested (Figure 4). In particular, the presence of Exo70 together with SNEV in the mature spliceosome suggested that this interaction might play a role in pre-mRNA splicing.
Figure 3. Exo70 co-localizes with SNEV in the nucleus of HeLa cells upon inhibition of nuclear export.
(A) Exo70–EGFP (green) is located in the cytoplasm and at the plasma membrane of HeLa cells, whereas SNEV (red) is located in the nucleus. Upon inhibition of nuclear export by LMB, Exo70 accumulates within the nucleoplasm in a similar pattern as SNEV, sparing the nucleoli. (B) Immunofluorescence analysis with an antibody against endogenous Exo70 confirms accumulation of Exo70 in the nucleus upon inhibition of nuclear export. (C) Overexpression of His6-tagged Exo70 and two truncated forms (ΔCC and CC, see Figure 1C) and immunofluorescence staining using an anti-Exo70 antibody. (D) Immunofluorescence analysis with an antibody against endogenous Exo70 confirms co-localization with SNEVhPrp19/hPso4 in the nucleus upon LMB treatment.
Figure 4. Exo 70 and SNEV are contained in the same complex in nuclear extracts.
Endogenous SNEVhPrp19/hPso4 and Exo70 are present in nuclear extract (NE) and cytoplasmic S100 extract (S100), as well as in the mature spliceosome, whereas no signal is detected in the mock spliceosome purification. Our protocol for subcellular fractionation was established and tested for purity by Western blotting using antibodies against nuclear [anti-(lamin A/C)] and cytoplasmic (anti-GAPDH) marker proteins (bottom panels). In a further experiment, fractions were prepared as described and tested for the presence of SNEVhPrp19/hPso4 and Exo70 (top panels).
The CC domain of Exo70 inhibits mRNA splicing in vitro
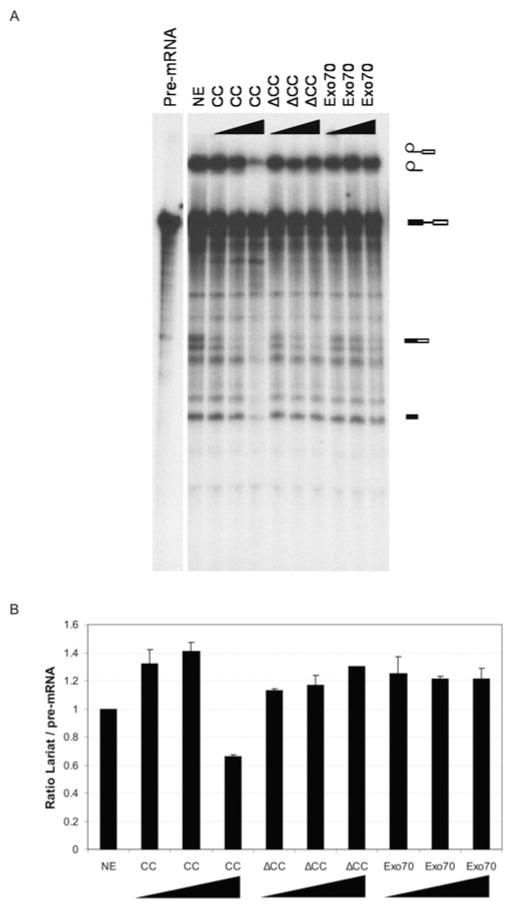
In order to test whether the interaction between Exo70 and SNEVhPrp19/hPso4 is important for pre-mRNA splicing, we added the recombinantly expressed proteins Exo70, CC and ΔCC to in vitro splicing assays. Indeed, the splicing reaction was inhibited by 44% compared with the control splicing reaction with the addition of increasing amounts of CC (Figure 5). These results indicate that the interaction of Exo70 with SNEVhPrp19/hPso4 might be involved in pre-mRNA splicing in vitro. However, these results could also be explained by a non-specific titration effect. To further test whether Exo70 is indeed involved in pre-mRNA splicing, we examined a possible function of Exo70 and its deletion mutants on splicing in vivo.
Figure 5. The CC domain of Exo70 inhibits splicing in vitro.
(A) Recombinant Exo70, ΔCC and CC were added in equimolar amounts to in vitro splicing reactions. Neither full-length Exo70 nor the truncated form lacking the 100 N-terminal amino acids influenced the efficiency of the splicing assay, whereas the CC deletion mutant inhibited the splicing reaction, resulting in a decrease of the lariat form. Symbols on the right-hand side of the gel represent (from the top to the bottom): lariat fused to exon 2, free lariat, pre-mRNA, spliced mRNA and Exon 1. (B) Quantification of the ratio between lariat and pre-mRNA band intensities. S.D.s are calculated from three independent experiments.
Exo70 and its deletion mutants influence mRNA splicing in vivo
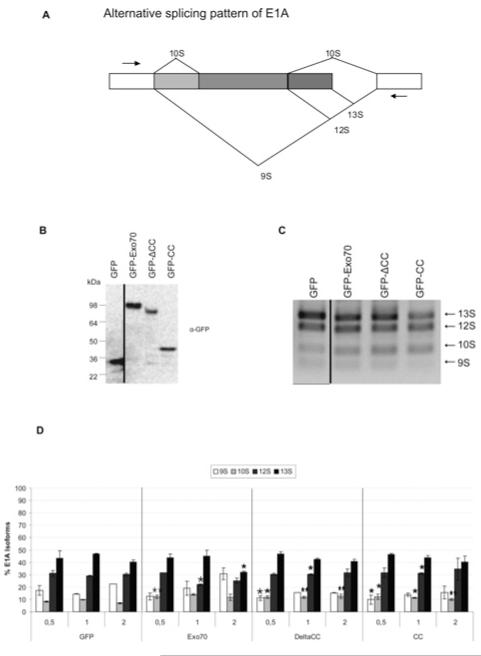
Since the CC domain of Exo70 turned out to influence mRNA splicing in vitro, we investigated the effect of Exo70 on splicing of a model substrate in vivo. Therefore we used an in vivo splicing assay based on the alternative splicing of the adenovirus E1A gene. Alternative splicing of E1A gives rise to five isoforms (9S, 10S, 11S, 12S and 13S) (Figure 6A) [34]. We co-transfected HeLa cells with the E1A minigene together with increasing amounts of EGFP–Exo70 or its deletion mutants ΔCC and CC, or empty vector. As a negative control we used empty pEGFP. E1A isoforms were detected by semi-quantitative PCR. Exo70, as well as the two deletion mutants, significantly altered the E1A splicing pattern when compared with the EGFP control (Figure 6C). PCR products were quantified by microfluidics-based capillary gel electrophoresis using the Bioanalyzer (Agilent), where molar ratios were calculated from three independent experiments and in turn converted into percentage fractions of total E1A isoforms (Figure 6D). Although we see a significant dose-dependent change using full-length Exo70, a small change was also seen using the truncated isoforms which does not seem to be dose-dependent. For example, the 13S isoforms decreases to 32% of all isoforms in the highest dose of Exo70 overexpression, compared with 40.5% in the corresponding negative control. In order to control for correct size and comparable expression levels of Exo70 and its mutants, cell lysates were analysed by Western blotting with an anti-GFP antibody (Figure 6B).
Figure 6. Exo70 influences splicing in vivo.
(A) Schematic representation of the alternative splicing pattern of the E1A minigene. Only major isoforms are depicted. Arrows denote binding sites for primers E1A exon 1 forward and E1A exon 2 reverse. (B) HeLa cells were co-transfected with pEGFP-C2-Exo70, -ΔCC or -CC. Empty pEGFP-C2 vector was used as a negative control. Correct expression of transfected constructs was confirmed by resolving 50 μg of total protein extract on SDS/PAGE and probing with an anti-GFP antibody. GFP, 29 kDa; GFP-Exo70, 103 kDa, GFP–ΔCC, 90 kDa; and GFP-CC, 42 kDa. (C) Overexpression of GFP-tagged full-length and truncated forms of Exo70 influences E1A splicing in vivo. Total RNA was isolated 48 h post-transfection, reverse-transcribed and used as template in PCRs with E1A-specific primers. PCR products (10 μl) were resolved on a 1% agarose gel. (D) Exo70 has a dose-dependent effect on splicing of E1A. Molar amounts of PCR products were calculated using the Agilent 100 Bioanalyzer and converted into percentage fractions. Error bars represent the S.D. of the mean calculated across three individual experiments. Hela cells were transfected with different amounts of Exo70, its deletion mutants or empty vector. Asterisks indicate significant differences to the amount of the same product in the control transformed with an equivalent amount of GFP (*P < 0.05; **P <0.01 using a one-way ANOVA F-test).
Alternative splicing of Exo70 is cell-type-dependent and changes during cellular aging
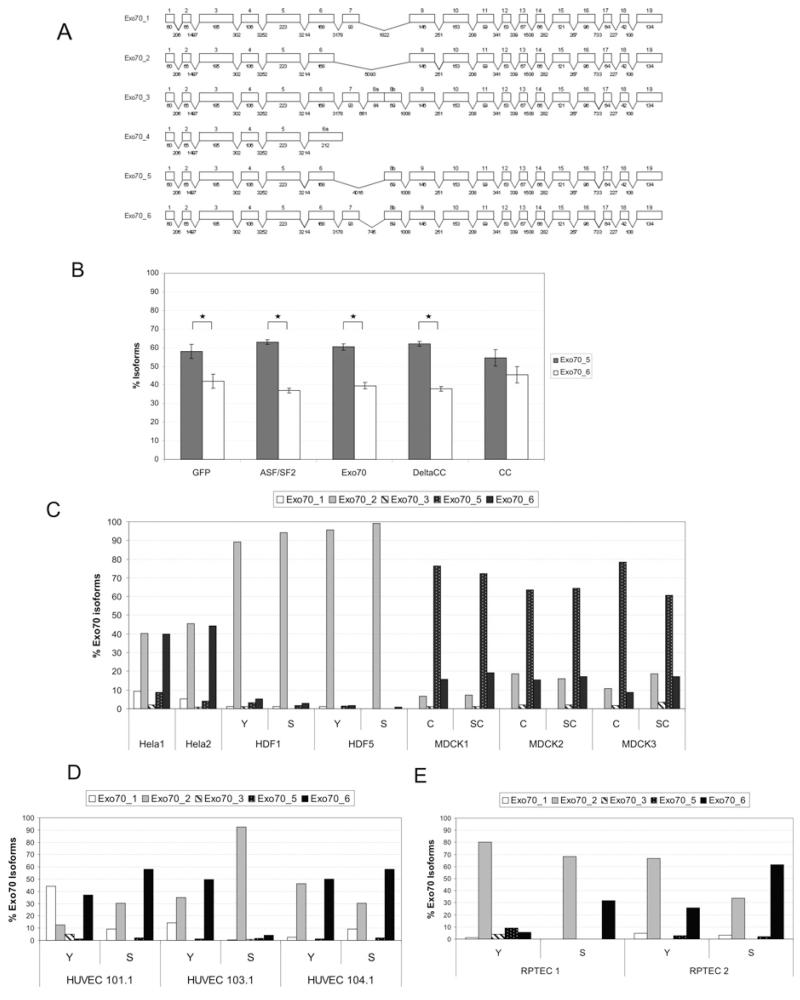
During our attempts to amplify the cDNA coding for Exo70, we discovered at least four alternatively spliced Exo70 isoforms in HeLa cells. Sequence analysis and databank mining showed that only two of them were known isoforms, whereas two were novel. These sequence data have been termed Exo70_5 and Exo70_6 and submitted to the GenBank® database under accession numbers FJ457119 and FJ457120. Figure 7(A) gives an overview of the exon–intron structure of all known Exo70 isoforms, including the two novel isoforms Exo70 isoform 5 and Exo70 isoform 6. Interestingly, although the secondary structure of the Exo70 protein consist of a series of α-helices, the region varying between isoforms is predicted to be largely unstructured [30] and thus the isoforms might differ by the flexibility of the domains. Since several alternative splicing factors change their own splicing pattern, we tested whether Exo70 influences alternative splicing of its own mRNA. Therefore we overexpressed Exo70 and its deletion mutants ΔCC and CC in HeLa cells and quantified the differentially spliced Exo70 isoforms (Figure 7B). To avoid the predominant amplification of mRNA transcribed from the vector, which corresponds to isoform 1, we designed an alternative reverse primer that binds to a non-constitutive exon that is spliced out in isoform 1. Here, the proportion between isoform 5 and isoform 6 is not changed by ASF/SF2 (alternative splicing factor/splicing factor 2), indicating that Exo70 mRNA might not be a target of ASF/SF2. The same holds true for Exo70 and ΔCC, whereas the CC domain alone causes a small, but significant, shift in such a way that levels of isoforms 5 and 6 are no longer significantly different. In order to find out whether our newly identified isoforms are specific for HeLa cells, we designed primers that bind upstream and downstream of the variable region.
Figure 7. Exo70 is itself present as multiple alternatively spliced isoforms.
(A) Exon–intron organization of the Exo70 gene. Boxes denote exons. Exons are drawn to scale. Numbers below boxes indicate exon lengths (in bp). Numbers in between exons indicate intron lengths (in bp, not drawn to scale). In Exo70_4, the omission of the 5′ splice site of the intron following exon 6 leads to a longer alternative 6a exon with a premature stop codon. Exo70_5 and Exo70_6 were not previously known in databases. (B) The CC deletion mutant of Exo70 influences splicing of Exo70 mRNA. To avoid detection of the predominant mRNA transcribed from the vector and allow only detection of endogenous Exo70 isoforms, we used a reverse primer binding to exon 9, which is not present in the vector sequence that corresponds to Exo70 isoform 1. Overexpression of the CC deletion mutant significantly altered the relative abundances of Exo70 isoform 5 and isoform 6 as they are no longer significantly different in levels as they are in the other transfections. Asterisks (*) indicate significant differences (P < 0.05 using a one-way ANOVA F-test). (C) Distribution of Exo70 isoforms is cell-type-dependent. In HeLa and MDCK cells, the distribution of Exo70 isoforms is constant over several months in culture. In fibroblasts, the pattern is very similar across different donors. (D) In endothelial cells, distribution of Exo70 isoforms is donor-specific and probably dependent on aging. (E) The same is true for RPTECs. Y, young; S, senescent; C, confluent; SC, subconfluent.
In a variety of different cell strains and conditions, we did not detect isoform 4, whereas isoform 3 showed very low to low abundance in HeLa cells, young and senescent fibroblasts from two different donors, and three independent RNA isolations from confluent and subconfluent MDCK cells (Figure 7C). The high consistency among the results derived from HeLa cells, fibroblasts or MDCK cells indicates that our assay is reliable and reproducible. Since Exo70 was first characterized in MDCK cells where the exocyst complex, targeted by Exo70, is recruited from the cytosol to cell–cell contacts [35,36], we wondered whether the confluence state of these cells might be associated with a shift in the distribution of Exo70 isoforms, but this does not seem to be the case (see Figure 7C). However, it is remarkable that the splicing pattern does not significantly change during 5 months of continuous culture between the time points of RNA isolation. Figure 7(D) shows the distribution of Exo70 isoforms in HUVECs derived from three different donors. Here, the situation is different. There is high variability between the different donors, and within the donors, the splicing pattern clearly changes when the cells have reached senescence. This is also true for RPTECs (Figure 7E), indicating a so far unknown donor-dependent variation in alternative splicing.
DISCUSSION
The exocyst is an evolutionarily conserved octameric protein complex that directs post-Golgi vesicles to specific localizations at the plasma membrane prior to vesicle fusion. It consists of subunits Sec3, Sec5, Sec6, Sec8, Sec10, Sec15, Exo70 and Exo84, also termed EXOC1, EXOC2, EXOC3, EXOC4, EXOC5, EXOC6, EXOC7 and EXOC8. The function of the exocyst complex is essential for many cellular processes (reviewed in [37]). Also of major importance for exocyst function is the subunit Exo70, which is critical for assembly of the complex and its recruitment to the plasma membrane upon insulin signalling [38]. Exo70 plays a key role in cytoskeleton co-ordination by interacting with the Arp2/3 complex, a master regulator of actin polymerization [39]. The tight link between the cytoskeleton and Exo70 might also be reflected by our co-precipitation of a large number of cytoskeletal proteins [16].
Although so far no nuclear localization has been reported for human Exo70, at least some of the 23 Exo70 proteins known in Arabidopsis thaliana are present in the nucleus [40]. Several additional indications support our finding that it shuttles to the nucleus. First, putative nuclear localization and Crm1-dependent export signals are present in Exo70 [41], and indeed Exo70 accumulated in the nuclei upon blocking Crm1-mediated export by LMB. Furthermore, an interaction with Nup62, which localizes to the central channel of the nuclear pore complex and occasionally binds cargo directly to support nuclear import, has been observed, although the authors report no nuclear localization of Exo70. Finally, one other subunit of the exocyst, Exo84 (EXOC8), has been found as an interaction partner of the splicing factor Snp1, the yeast homologue of the 70K protein [42]. While Exo84 directly binds to Exo70 in yeast, Snp1 is considered to be close to the SNEVhPrp19/hPso4 orthologue Prp19, since both are interaction partners of Prp8 and Smx3 [43,44]. The interaction of Exo84 and Snp1 has been shown to be important in splicing, since a temperature-sensitive mutant of Exo84 shows decreased splicing activity [42]. An independent yeast two-hybrid screening identified Exo84 as a Prp8-binding partner [45]. The authors assume that Exo84 might mediate interactions of Prp8 and the U1 snRNP and hypothesize that even though Exo84 is primarily located to the cytoplasm, a small fraction might localize to the nucleus and function in splicing.
In addition to the nuclear localization, endogenous Exo70 seems to accumulate in the granular compartment of the nucleoli upon inhibition of nuclear export (see Figure 3C). This agrees with its involvement in splicing, since the nucleolar proteome is known to contain many splicing factors [46], and hypophosphorylated serine/argininie-rich proteins localize around active NORs (nucleolar organizing regions) during telophase [47]. Together with its presence in affinity-purified spliceosomes, this encouraged us to test whether Exo70 might be involved in human pre-mRNA splicing. Indeed, the purified CC domain of Exo70 resulted in a decrease of splicing intermediates and spliced products, whereas ΔCC or full-length Exo70 did not. The CC domain may thus represent a dominant-negative mutant. Although clearly confirming the SNEVhPrp19/hPso4–Exo70 interaction and demonstrating its importance, we cannot exclude the possibility that CC inhibits splicing by merely titrating away SNEVhPrp19/hPso4 from its other interaction partners, and thus inhibits splicing in vitro without any in vivo relevance.
Therefore we used a minigene as a splicing reporter construct and indeed found that overexpression of Exo70 significantly alters the splicing pattern in a dose-dependent manner and favours exon skipping of the E1A minigene, suggesting that Exo70 might be involved in alternative splice site selection.
Alternative splicing is a potent regulator of protein function: it specifies their binding properties, intracellular localization, enzymatic activity, protein stability and post-translational modifications, whereupon effects range from very subtle attenuations to a complete abolition of function or introduction of a new function [48]. Several splicing factors regulate splicing of their own mRNA [49,50], Indeed, overexpression of Exo70 also influences the relative amounts of its own isoforms 5 and 6.
In addition, we observed different splice patterns depending on the cell lines used and their replicative age. Age-related alternative splicing of some mRNAs is already known (reviewed in [51]), specifically also in senescent endothelial cells [52], although only very few results on the mechanistic basis is available.
Furthermore, we also identified two previously unknown isoforms of Exo70. These isoforms differ in a putatively unstructured region between α-helices 6 and 7 [30], probably resulting in different flexibility and binding affinities of Exo70, or providing sites for post-translational modifications.
Still the function of this interaction remains unclear. One intriguing possibility is that it provides a direct signalling link between membrane events and nuclear splicing, allowing for rapid changes in the pattern of alternatively spliced proteins. Insulin signalling especially might be such a membrane event, since Exo70 mediates extracellular exposure of GLUT4 (glucose transporter 4) protein upon insulin signalling, possibly by directing vesicles to the site of membrane fusion [53]. This is remarkable, since in plants another member of the Cdc5L-associated complex, PRL1, the A. thaliana homologue of Plrg1, is reported to affect glucose response and uptake [54].
A further link between membrane and splicing factors is the spreading initiation centre, a structure at the cell membrane that is formed during spreading and adherence of human fibroblasts and that contains numerous RNA-binding proteins [55].
Besides this signalling function, a transport function is as plausible, as the cargo of exocyst might also be mRNA. In this regard it is of note that the exocyst has been found at the translocon, co-localizing with the translation machinery of the endoplasmic reticulum in yeast [56], as well as in mammalian cells, where it might help to transport mRNAs whose proteins are destined for export to the endoplasmic reticulum membrane [57]. This scenario seems possible since mRNA transport is coupled to splicing [58], and the results of the present study show the presence of Exo70 at the spliceosome.
Another question of interest with regard to Exo70 shuttling is to determine which physiological signal induces this movement. One possible signal is cell confluence. Although Exo70 is suggested to be cytoplasmic (although we think that there is nuclear localization in Figure 4I of [36]) in contact-naïve MDCK cells, it translocates to the plasma membrane upon cell–cell contact [36]. Therefore it might act as a direct communicator of cell contact events to the splicing apparatus.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr J. F. Caceres (MRC Human Genetics Unit, Western General Hospital, Edinburgh, Scotland, U.K.), Dr S. Hatakeyama (Hokkaido University, Sapporo, Japan) and Dr R. H. Scheller (Department of Biological Sciences, Stanford University, Stanford, CA, U.S.A.) for expression plasmids, Dr S. C. Hsu (Department of Cell Biology and Neuroscience, Rutgers University, Piscataway, NJ, U.S.A.) for antibodies, Hannes Schmid for helpful advice with FRET analysis, Boris Ferko for immunization of rabbits and mice, Matthias Hackl for help with statistical analysis, and Gustav Ritter and Martina Wostry for excellent technical support.
FUNDING
This work was supported by the Austrian Science Fund [NRN grant number S93]; Genome Research Austria GEN-AU [Project 820982 “Non-coding RNAs”]; the Herzfelder’sche Familienstiftung and Centre de Recherches e d’Investigations Epidermiques et Sensorielle de Chanel. H. D. is a recipient of a DOC-fFORTE Fellowship of the Austrian Academy of Sciences.
Abbreviations used
- ASF/SF2
alternative splicing factor/splicing factor 2
- ATR
ataxia telangiectasia mutated- and Rad3-related
- CC
coiled-coil
- Cdc5L
cell division cycle 5-like
- CFP
cyan fluorescent protein
- CoIP
co-immunoprecipitation
- DTT
dithiothreitol
- ECFP
enhanced CFP
- EGFP
enhanced green fluorescent protein
- EYFP
enhanced yellow fluorescent protein
- FBS
fetal bovine serum
- FRET
fluorescence resonance energy transfer
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- GFP
green fluorescent protein
- GST
glutathione transferase
- HDF
human dermal fibroblast
- HUVEC
human umbilical vein endothelial cell
- LMB
leptomycin B
- MDCK
Madin–Darby canine kidney
- Ni-NTA
Ni2+ -nitrilotriacetate
- PD
population doubling
- Plrg1
pleiotropic regulator 1
- RPTEC
renal proximal tubule epithelial cell
- SKIP
skeletal muscle- and kidney-enriched inositol phosphatase
- snRNP
small nuclear ribonucleoprotein
- YFP
yellow fluorescent protein
Footnotes
REFERENCES
- 1.Wahl M, Will C, Lührmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 2.Grote M, Wolf E, Will C, Lemm I, Agafonov D, Schomburg A, Fischle W, Urlaub H, Lührmann R. Molecular architecture of the human Prp19/CDC5L complex. Mol. Cell. Biol. 2010;30:2105–2119. doi: 10.1128/MCB.01505-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ajuh P, Kuster B, Panov K, Zomerdijk JC, Mann M, Lamond AI. Functional analysis of the human CDC5L complex and identification of its components by mass spectrometry. EMBO J. 2000;19:6569–6581. doi: 10.1093/emboj/19.23.6569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Makarova OV, Makarov EM, Urlaub H, Will CL, Gentzel M, Wilm M, Luhrmann R. A subset of human 35S U5 proteins, including Prp19, function prior to catalytic step 1 of splicing. EMBO J. 2004;23:2381–2391. doi: 10.1038/sj.emboj.7600241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grillari J, Ajuh P, Stadler G, Löscher M, Voglauer R, Ernst W, Chusainow J, Eisenhaber F, Pokar M, Fortschegger K, et al. SNEV is an evolutionarily conserved splicing factor whose oligomerization is necessary for spliceosome assembly. Nucleic Acids Res. 2005;33:6868–6883. doi: 10.1093/nar/gki986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fortschegger K, Wagner B, Voglauer R, Katinger H, Sibilia M, Grillari J. Early embryonic lethality of mice lacking the essential protein SNEV. Mol. Cell. Biol. 2007;27:3123–3130. doi: 10.1128/MCB.01188-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grillari J, Löscher M, Denegri M, Lee K, Fortschegger K, Eisenhaber F, Ajuh P, Lamond A, Katinger H, Grillari-Voglauer R. Blom7α is a novel heterogeneous nuclear ribonucleoprotein K homology domain protein involved in pre-mRNA splicing that interacts with SNEVPrp19-Pso4. J. Biol. Chem. 2009;284:29193–29204. doi: 10.1074/jbc.M109.036632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang N, Kaur R, Lu X, Shen X, Li L, Legerski R. The Pso4 mRNA splicing and DNA repair complex interacts with WRN for processing of DNA interstrand cross-links. J. Biol. Chem. 2005;280:40559–40567. doi: 10.1074/jbc.M508453200. [DOI] [PubMed] [Google Scholar]
- 9.Beck B, Park S, Lee Y, Roman Y, Hromas R, Lee S. Human Pso4 is a metnase (SETMAR)-binding partner that regulates metnase function in DNA repair. J. Biol. Chem. 2008;283:9023–9030. doi: 10.1074/jbc.M800150200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang N, Kaur R, Akhter S, Legerski R. Cdc5L interacts with ATR and is required for the S-phase cell-cycle checkpoint. EMBO Rep. 2009;10:1029–1035. doi: 10.1038/embor.2009.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Voglauer R, Chang M, Dampier B, Wieser M, Baumann K, Sterovsky T, Schreiber M, Katinger H, Grillari J. SNEV overexpression extends the life span of human endothelial cells. Exp. Cell Res. 2006;312:746–759. doi: 10.1016/j.yexcr.2005.11.025. [DOI] [PubMed] [Google Scholar]
- 12.Hatakeyama S, Yada M, Matsumoto M, Ishida N, Nakayama KI. U-Box proteins as a new family of ubiquitin-protein ligases. J. Biol. Chem. 2001;276:33111–33120. doi: 10.1074/jbc.M102755200. [DOI] [PubMed] [Google Scholar]
- 13.Löscher M, Fortschegger K, Ritter G, Wostry M, Voglauer R, Schmid JA, Watters S, Rivett AJ, Ajuh P, Lamond AI, et al. The U-box E3 ligase SNEV interacts with the 7 subunit of the 20S proteasome. Biochem. J. 2005;388:593–603. doi: 10.1042/BJ20041517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sihn C, Cho S, Lee J, Lee T, Kim S. Mouse homologue of yeast Prp19 interacts with mouse SUG1, the regulatory subunit of 26S proteasome. Biochem. Biophys. Res. Commun. 2007;356:175–180. doi: 10.1016/j.bbrc.2007.02.134. [DOI] [PubMed] [Google Scholar]
- 15.Song E, Werner S, Neubauer J, Stegmeier F, Aspden J, Rio D, Harper J, Elledge S, Kirschner M, Rape M. The Prp19 complex and the Usp4Sart3 deubiquitinating enzyme control reversible ubiquitination at the spliceosome. Genes Dev. 2010;24:1434–1447. doi: 10.1101/gad.1925010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Llères D, Denegri M, Biggiogera M, Ajuh P, Lamond A. Direct interaction between hnRNP-M and CDC5L/PLRG1 proteins affects alternative splice site choice. EMBO Rep. 2010;11:445–451. doi: 10.1038/embor.2010.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Enninga J, Levay A, Fontoura B. Sec13 shuttles between the nucleus and the cytoplasm and stably interacts with Nup96 at the nuclear pore complex. Mol. Cell. Biol. 2003;23:7271–7284. doi: 10.1128/MCB.23.20.7271-7284.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gietz D, St. Jean A, Woods RA, Schiestl RH. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Böhm E, Grillari J, Voglauer R, Gross S, Ernst W, Ferko B, Kunert R, Katinger H, Borth N. Establishment of a strategy for the rapid generation of a monoclonal antibody against the human protein SNEV (hNMP200) by flow-cytometric cell sorting. J. Immunol. Methods. 2005;307:13–23. doi: 10.1016/j.jim.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 20.Wieser M, Stadler G, Jennings P, Streubel B, Pfaller W, Ambros P, Riedl C, Katinger H, Grillari J, Grillari-Voglauer R. hTERT alone immortalizes epithelial cells of renal proximal tubules without changing their functional characteristics. Am. J. Physiol. Renal Physiol. 2008;295:F1365–F1375. doi: 10.1152/ajprenal.90405.2008. [DOI] [PubMed] [Google Scholar]
- 21.Hayflick L, Moorhead P. The serial cultivation of human diploid cell strains. Exp. Cell Res. 1961;25:585–621. doi: 10.1016/0014-4827(61)90192-6. [DOI] [PubMed] [Google Scholar]
- 22.Hayflick L. The limited in vitro lifetime of human diploid cell strains. Exp. Cell Res. 1965;37:614–636. doi: 10.1016/0014-4827(65)90211-9. [DOI] [PubMed] [Google Scholar]
- 23.Chang M, Grillari J, Mayrhofer C, Fortschegger K, Allmaier G, Marzban G, Katinger H, Voglauer R. Comparison of early passage, senescent and hTERT immortalized endothelial cells. Exp. Cell Res. 2005;309:121–136. doi: 10.1016/j.yexcr.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 24.Lamond AI, Konarska MM, Sharp PA. A mutational analysis of spliceosome assembly: evidence for splice site collaboration during spliceosome formation. Genes Dev. 1987;1:532–543. doi: 10.1101/gad.1.6.532. [DOI] [PubMed] [Google Scholar]
- 25.Youvan DC, Silva CM, Bylina EJ, Coleman WJ, Dilworth MR, Yang MM. Calibration of fluorescence resonance energy transfer in microscopy using genetically engineered GFP derivatives on nickel chelating beads. Biotech. et alia. 1997;3:1–18. [Google Scholar]
- 26.Platani M, Goldberg I, Swedlow JR, Lamond AI. In vivo analysis of Cajal body movement, separation, and joining in live human cells. J. Cell Biol. 2000;151:1561–1574. doi: 10.1083/jcb.151.7.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rappsilber J, Ryder U, Lamond A, Mann M. Large-scale proteomic analysis of the human spliceosome. Genome Res. 2002;12:1231–1245. doi: 10.1101/gr.473902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Calvio C, Neubauer G, Mann M, Lamond A. Identification of hnRNP P2 as TLS/FUS using electrospray mass spectrometry. RNA. 1995;1:724–733. [PMC free article] [PubMed] [Google Scholar]
- 29.Konarska MM, Sharp PA. Interactions between small nuclear ribonucleoprotein particles in formation of spliceosomes. Cell. 1987;49:763–774. doi: 10.1016/0092-8674(87)90614-3. [DOI] [PubMed] [Google Scholar]
- 30.Moore BA, Robinson HH, Xu Z. The crystal structure of mouse Exo70 reveals unique features of the mammalian exocyst. J. Mol. Biol. 2007;371:410–421. doi: 10.1016/j.jmb.2007.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakai K, Horton P. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem. Sci. 1999;24:34–36. doi: 10.1016/s0968-0004(98)01336-x. [DOI] [PubMed] [Google Scholar]
- 32.la Cour T, Kiemer L, Mølgaard A, Gupta R, Skriver K, Brunak S. Analysis and prediction of leucine-rich nuclear export signals. Protein Eng. Des. Sel. 2004;17:527–536. doi: 10.1093/protein/gzh062. [DOI] [PubMed] [Google Scholar]
- 33.Hamamoto T, Seto H, Beppu T. Leptomycins A and B, new antifungal antibiotics. II. Structure elucidation. J. Antibiot. (Tokyo) 1983;36:646–650. doi: 10.7164/antibiotics.36.646. [DOI] [PubMed] [Google Scholar]
- 34.Cáceres J, Stamm S, Helfman D, Krainer A. Regulation of alternative splicing in vivo by overexpression of antagonistic splicing factors. Science. 1994;265:1706–1709. doi: 10.1126/science.8085156. [DOI] [PubMed] [Google Scholar]
- 35.Grindstaff K, Yeaman C, Anandasabapathy N, Hsu S, Rodriguez-Boulan E, Scheller R, Nelson W. Sec6/8 complex is recruited to cell-cell contacts and specifies transport vesicle delivery to the basal-lateral membrane in epithelial cells. Cell. 1998;93:731–740. doi: 10.1016/s0092-8674(00)81435-x. [DOI] [PubMed] [Google Scholar]
- 36.Matern HT, Yeaman C, Nelson WJ, Scheller RH. The Sec6/8 complex in mammalian cells: characterization of mammalian Sec3, subunit interactions, and expression of subunits in polarized cells. Proc. Natl. Acad. Sci. U.S.A. 2001;98:9648–9653. doi: 10.1073/pnas.171317898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He B, Guo W. The exocyst complex in polarized exocytosis. Curr. Opin. Cell Biol. 2009;21:537–542. doi: 10.1016/j.ceb.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bao Y, Lopez J, James D, Hunziker W. Snapin interacts with the Exo70 subunit of the exocyst and modulates GLUT4 trafficking. J. Biol. Chem. 2008;283:324–331. doi: 10.1074/jbc.M706873200. [DOI] [PubMed] [Google Scholar]
- 39.Zuo X, Zhang J, Zhang Y, Hsu S, Zhou D, Guo W. Exo70 interacts with the Arp2/3 complex and regulates cell migration. Nat. Cell Biol. 2006;8:1383–1388. doi: 10.1038/ncb1505. [DOI] [PubMed] [Google Scholar]
- 40.Chong Y, Gidda S, Sanford C, Parkinson J, Mullen R, Goring D. Characterization of the Arabidopsis thaliana exocyst complex gene families by phylogenetic, expression profiling, and subcellular localization studies. New Phytol. 2010;185:401–419. doi: 10.1111/j.1469-8137.2009.03070.x. [DOI] [PubMed] [Google Scholar]
- 41.Fornerod M, Ohno M, Yoshida M, Mattaj IW. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 42.Awasthi S, Palmer R, Castro M, Mobarak CD, Ruby SW. New roles for the Snp1 and Exo84 proteins in yeast pre-mRNA splicing. J. Biol. Chem. 2001;276:31004–31015. doi: 10.1074/jbc.M100022200. [DOI] [PubMed] [Google Scholar]
- 43.Gavin A, Bösche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick J, Michon A, Cruciat C, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 44.Ho Y, Gruhler A, Heilbut A, Bader G, Moore L, Adams S, Millar A, Taylor P, Bennett K, Boutilier K, et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 45.Kuhn A, Brow D. Suppressors of a cold-sensitive mutation in yeast U4 RNA define five domains in the splicing factor Prp8 that influence spliceosome activation. Genetics. 2000;155:1667–1682. doi: 10.1093/genetics/155.4.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boisvert F, van Koningsbruggen S, Navascués J, Lamond A. The multifunctional nucleolus. Nat. Rev. Mol. Cell Biol. 2007;8:574–585. doi: 10.1038/nrm2184. [DOI] [PubMed] [Google Scholar]
- 47.Bubulya PA, Prasanth KV, Deerinck TJ, Gerlich D, Beaudouin J, Ellisman MH, Ellenberg J, Spector DL. Hypophosphorylated SR splicing factors transiently localize around active nucleolar organizing regions in telophase daughter nuclei. J. Cell Biol. 2004;167:51–63. doi: 10.1083/jcb.200404120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stamm S, Ben-Ari S, Rafalska I, Tang Y, Zhang Z, Toiber D, Thanaraj T, Soreq H. Function of alternative splicing. Gene. 2005;344:1–20. doi: 10.1016/j.gene.2004.10.022. [DOI] [PubMed] [Google Scholar]
- 49.Jumaa H, Nielsen P. The splicing factor SRp20 modifies splicing of its own mRNA and ASF/SF2 antagonizes this regulation. EMBO J. 1997;16:5077–5085. doi: 10.1093/emboj/16.16.5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Malygin A, Parakhnevitch N, Ivanov A, Eperon I, Karpova G. Human ribosomal protein S13 regulates expression of its own gene at the splicing step by a feedback mechanism. Nucleic Acids Res. 2007;35:6414–6423. doi: 10.1093/nar/gkm701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meshorer E, Soreq H. Pre-mRNA splicing modulations in senescence. Aging Cell. 2002;1:10–16. doi: 10.1046/j.1474-9728.2002.00005.x. [DOI] [PubMed] [Google Scholar]
- 52.Blanco FJ, Grande MT, Langa C, Oujo B, Velasco S, Rodriguez-Barbero A, Perez-Gomez E, Quintanilla M, López-Novoa JM, Bernabeu C. S-endoglin expression is induced in senescent endothelial cells and contributes to vascular pathology. Circ. Res. 2008;103:1383–1392. doi: 10.1161/CIRCRESAHA.108.176552. [DOI] [PubMed] [Google Scholar]
- 53.Inoue M, Chang L, Hwang J, Chiang SH, Saltiel AR. The exocyst complex is required for targeting of Glut4 to the plasma membrane by insulin. Nature. 2003;422:629–633. doi: 10.1038/nature01533. [DOI] [PubMed] [Google Scholar]
- 54.Nemeth K, Salchert K, Putnoky P, Bhalerao R, Koncz-Kalman Z, Stankovic-Stangeland B, Bako L, Mathur J, Okresz L, Stabel S, et al. Pleiotropic control of glucose and hormone responses by PRL1, a nuclear WD protein, in Arabidopsis. Genes Dev. 1998;12:3059–3073. doi: 10.1101/gad.12.19.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.de Hoog C, Foster L, Mann M. RNA and RNA binding proteins participate in early stages of cell spreading through spreading initiation centers. Cell. 2004;117:649–662. doi: 10.1016/s0092-8674(04)00456-8. [DOI] [PubMed] [Google Scholar]
- 56.Toikkanen JH, Miller KJ, Soderlund H, Jantti J, Keranen S. The β subunit of the Sec61p endoplasmic reticulum translocon interacts with the exocyst complex in Saccharomyces cerevisiae. J. Biol. Chem. 2003;278:20946–20953. doi: 10.1074/jbc.M213111200. [DOI] [PubMed] [Google Scholar]
- 57.Lipschutz JH, Lingappa VR, Mostov KE. The exocyst affects protein synthesis by acting on the translocation machinery of the endoplasmic reticulum. J. Biol. Chem. 2003;278:20954–20960. doi: 10.1074/jbc.M213210200. [DOI] [PubMed] [Google Scholar]
- 58.Reed R. Coupling transcription, splicing and mRNA export. Curr. Opin. Cell Biol. 2003;15:326–331. doi: 10.1016/s0955-0674(03)00048-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.