The authors report the crystal structure of a negative strand virus nucleocapsid protein in complex with RNA. This virus appears to sequester RNA via a novel mechanism.
Keywords: Schmallenberg virus, orthobunyavirus, bunyavirus, nucleoprotein structure, RNA replication and transcription
Abstract
Schmallenberg virus (SBV) is a newly emerged orthobunyavirus (family Bunyaviridae) that has caused severe disease in the offspring of farm animals across Europe. Like all orthobunyaviruses, SBV contains a tripartite negative-sense RNA genome that is encapsidated by the viral nucleocapsid (N) protein in the form of a ribonucleoprotein complex (RNP). We recently reported the three-dimensional structure of SBV N that revealed a novel fold. Here we report the crystal structure of the SBV N protein in complex with a 42-nt-long RNA to 2.16 Å resolution. The complex comprises a tetramer of N that encapsidates the RNA as a cross-shape inside the protein ring structure, with each protomer bound to 11 ribonucleotides. Eight bases are bound in the positively charged cleft between the N- and C-terminal domains of N, and three bases are shielded by the extended N-terminal arm. SBV N appears to sequester RNA using a different mechanism compared with the nucleoproteins of other negative-sense RNA viruses. Furthermore, the structure suggests that RNA binding results in conformational changes of some residues in the RNA-binding cleft and the N- and C-terminal arms. Our results provide new insights into the novel mechanism of RNA encapsidation by orthobunyaviruses.
INTRODUCTION
Schmallenberg virus (SBV) was first identified in November 2011 in northern Germany, and has since spread across Europe (Hoffmann et al. 2012; Beer et al. 2013), probably vectored by culicoid midges (De Regge et al. 2012; Rasmussen et al. 2012). SBV infects cattle, sheep, and goats, causing transient fever, diarrhea, reduction in milk yield, and abortion or malformations in offspring, leading to considerable economic losses (Garigliany et al. 2012; Tarlinton et al. 2012). No vaccines are yet available to control the disease.
SBV is most closely related to Sathuperi virus (Goller et al. 2012) in the Simbu serogroup of the genus Orthobunyavirus in the family Bunyaviridae. There are more than 350 recognized bunyaviruses that are divided into five genera, Orthobunyavirus, Hantavirus, Nairovirus, Phlebovirus, and Tospovirus (Plyusnin et al. 2012). The orthobunyaviruses make up the largest genus in Bunyaviridae, comprising about 170 members. Some orthobunyaviruses, such as La Crosse virus (LACV) and Oropouche virus, are significant human pathogens, whereas others like Akabane and Cache Valley viruses cause economically important disease in ruminants. Like all bunyaviruses, SBV is an enveloped negative-stranded RNA virus with a tripartite genome. The large (L) segment encodes a RNA-dependent RNA polymerase (L protein), and the middle segment (M) encodes two surface glycoproteins (Gc and Gn) and a nonstructural protein (NSm), while the small segment encodes a nucleoprotein (N) and, for most orthobunyaviruses, a second nonstructural protein called NSs (Elliott et al. 2011). The nucleoprotein interacts with viral genomic and antigenomic (replicative intermediate) RNA species to form ribonucleoprotein complexes (RNP) that are the functional templates for RNA replication and transcription (Elliott et al. 2011). We recently determined the structure of SBV N protein, which showed a novel fold, and proposed a new mechanism for genome RNA encapsidation and replication (Dong et al. 2013). Although the potential RNA-binding cleft was identified, details of the interaction between N and the viral RNA to form the ribonucleoprotein complex, essential for understanding viral RNA encapsidation, replication, and transcription, were not elucidated. We now report the structure of the SBV nucleoprotein in complex with a 42-base-long RNA.
RESULTS
SBV N forms a tetramer when complexed with 42-nt RNA
The SBV N protein was expressed in bacteria, and then either purified in the native form or the protein was denatured and refolded as previously described (Dong et al. 2013). As natively purified N binds host RNA nonspecifically and heterogeneously, we attempted to reconstitute a homogenous N–RNA complex by incubating RNA-free N with chemically synthesized RNAs (polyU) of 21, 28, or 42 bases in length. Only crystals of SBV N in complex with the 42-base-long RNA diffracted at high resolution (see below). The oligomeric state of the N–42-nt RNA complex was examined by chemical cross-linking and SDS-PAGE and compared with that of the natively purified N or RNA-free refolded N, suggesting that the RNA-free refolded N protein and the N–42-nt RNA complex exist as trimer and tetramer, respectively. This correlates with the tetrameric state of natively purified SBV N that contains host RNA (Dong et al. 2013).
Structure of tetrameric N in complex with 42-nt RNA
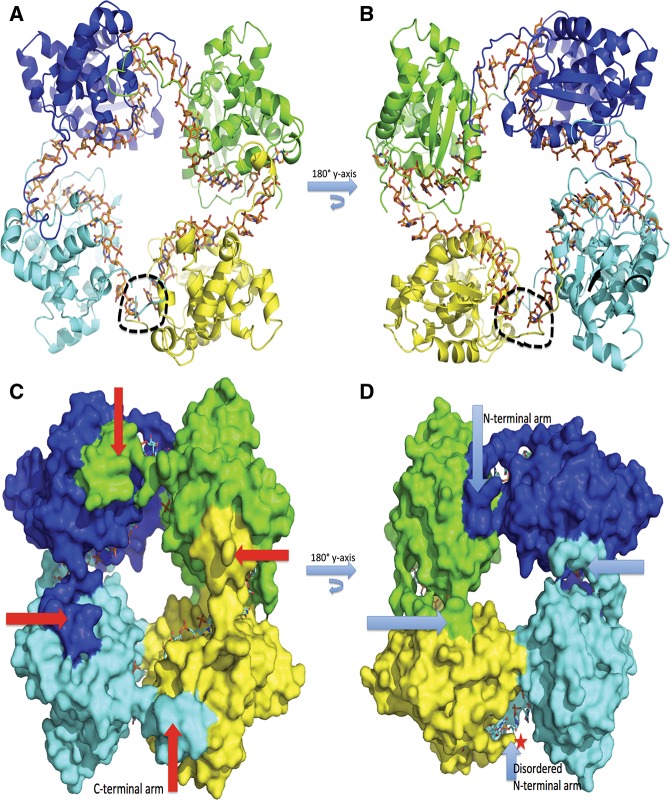
Crystals of SBV N complexed with 42-nt RNA were obtained in space group P21 with unit cell dimensions a = 76.50 Å, b = 86.05 Å, c = 77.46 Å, and β = 101.98°. The complex structure was determined by molecular replacement using Phaser (McCoy et al. 2007) and the native SBV N structure (PDB access code 4IDU) as the searching model. The N–RNA complex formed a tetrameric ring structure (Fig. 1A,B), in which the C-terminal arm of each protomer stretches out to bind to a hydrophobic region in the C-terminal domain of the neighboring N molecule (Fig. 1C), as previously described (Dong et al. 2013). Meanwhile, the N-terminal arms of three N protomers move away from their own or neighboring RNA-binding clefts and interact with their neighboring N molecules to further stabilize the oligomeric structure, while the fourth N-terminal arm is disordered (Fig. 1D). The RNA molecule is bent extensively between the protomers to form a cross-shape inside the tetrameric ring (Fig. 1A,B).
FIGURE 1.
Tetrameric structure of SBV N–RNA complex. (A) Cartoon representation of the tetrameric structure of the SBV N–RNA complex. Four protomers are shown in blue, green, yellow, and cyan. The 42-nt-long RNA is shown in stick form in orange bound inside the tetrameric ring, forming a cross shape. The black dotted circle shows the gap in the RNA. (B) Same as in A but rotated along the y-axis at 180°. (C) Surface representation of the tetrameric structure. The red arrows point to the C-terminal arms, which bind to the neigboring protomer’s C-terminal binding sites, thus playing an essential role for oligomerization. (D) Same as in C but rotated along the y-axis at 180°. The light blue arrows point to the N-terminal arms, which bind RNAs and to neighboring protomers. One of the N-terminal arms, indicated by a star, is disordered.
RNA is mainly bound at the central RNA-binding cleft
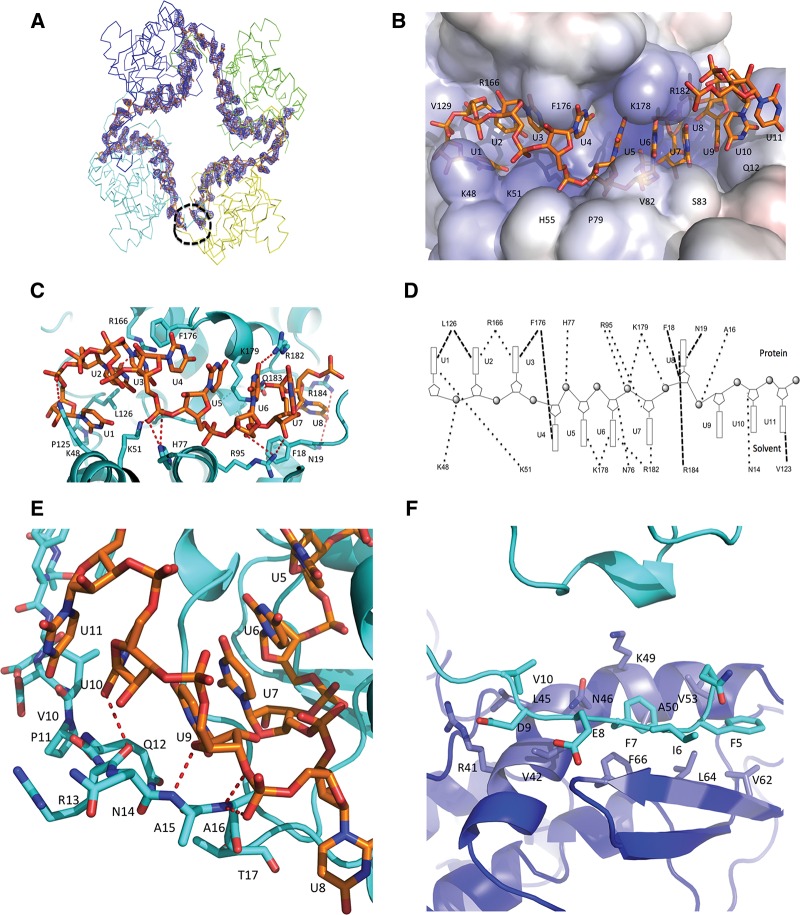
The electron density of the 42-nt RNA is clearly visible alongside the inner face of the tetramer (Fig. 2A). We previously suggested that the central positively charged cleft between the N- and C-domains of N is used for RNA binding (Dong et al. 2013). Indeed, our structure of the N–RNA complex clearly shows that the cleft can host eight RNA bases and that residues K48, K51, R166, H77, R95, K178, K179, R182, and R184 play a crucial role in binding the RNA (Fig. 2B–D). This is consistent with our previous observations that mutations of residues K48 and K51 impaired RNA-binding activity (Dong et al. 2013). It is also noticeable that the protrusion of the hydrophobic residues F176 and F18 between the bases of the RNA changes the orientation of the RNA bases and results in an “S” shape for the RNA chain in the cleft (Fig. 2B,C). In detail, bases U1, U2, and U3 face toward the inside of the RNA-binding cleft, interacting with positively charged residues K48, K51, and R166, and form hydrophobic interactions with residues L126 and F176 (Fig. 2B,D); F176 lies between bases U3 and U4. Bases U4, U5, U6, and U7, which are facing outward to solvent, stack together at the surface of the cleft, where these bases interact with positively charged residues R95, K178, K179, and R182 (Fig. 2B–D). Meanwhile, base U8 sits deeply in the cleft and is sandwiched by residues R184 and N19 (Fig. 2B–D), whereas residue F18 sits between bases U7 and U8.
FIGURE 2.
SBV N–RNA interactions. (A) Unbiased Fo–Fc electron density map of the 42-nt polyU contoured at 3 σ. The electron density is shown in blue and the RNA is shown in stick form in orange. The four protomers are shown in ribbon form. The black dotted circle shows the RNA gap. (B) Electrostatic potential surface map showing RNA binding in a SBV N protomer. The RNA is shown as stick form in orange. One protomer binds 11 nt of RNA with 8 nt in the RNA-binding cleft and 3 nt to the N-terminal arm. Bases U1 to U3 face into protein inside the RNA-binding cleft, bases U4 to U7 point outside the RNA-binding cleft and are exposed to solvent, while base U8 is bent into the bottom of the binding cleft and faces the protein. (C) Interaction between the RNA and residues of the RNA-binding cleft. The RNA forms an “S” shape in the RNA-binding cleft. The RNA is shown as stick form in orange, while amino acids of the RNA-binding cleft are shown in cyan. (D) Schematic diagram showing interactions between RNA and amino acids in SBV N. Bases oriented to the top indicate those that face into protein in the RNA-binding cleft, while the bases oriented toward the bottom indicate the bases exposed to solvent. The dark dotted lines show the interactions. (E) Interaction of the N-terminal arm and the RNA. Bases U9–U11 interact with atoms of the main chain of the N-terminal arm. Base U9 is stacked above base U7. The RNA is shown in orange and the protein in cyan. (F) The N-terminal arm binds to another hydrophobic area of a neighboring protomer. In particular, residues F7 and F66 form π–π interaction. The N-terminal arm is in cyan and the hydrophobic area of the neighboring protomer is in blue.
The N-terminal arm is also involved in RNA binding
Beside the RNA-binding cleft, the N-terminal arm of N also contributes to RNA binding. The arm, which is located between two neighboring N protomers, can host the remaining three bases (U9–11) (Fig. 2B,D,E). In particular, residues A15, A16, and Q12 on the N-terminal arm interact with bases U9, U10, and U11 to shield the RNA from exposure to the outside of the tetramer. This is consistent with our previous observation that truncation of the N-terminal arm completely abrogated RNA-binding activity and the N-terminal arm was disordered in the refolded RNA-free N structure (Dong et al. 2013). The N-terminal arm binds to a hydrophobic area consisting of residues V42, V53, V62, L64, and F66 in a neighboring protomer (Fig. 2F) and, in particular, residues F7 and F66 form π–π hydrophobic interactions. As each N protomer can bind 11 bases, the tetramer is able to bind 44-nt RNA. However, because we used a 42-nt RNA to generate the complex for crystallization, this results in a gap that is visible in the tetrameric complex structure (Fig. 1A,B), and more clearly seen in the electron density map of the RNA (Fig. 2A).
EM structure of SBV N
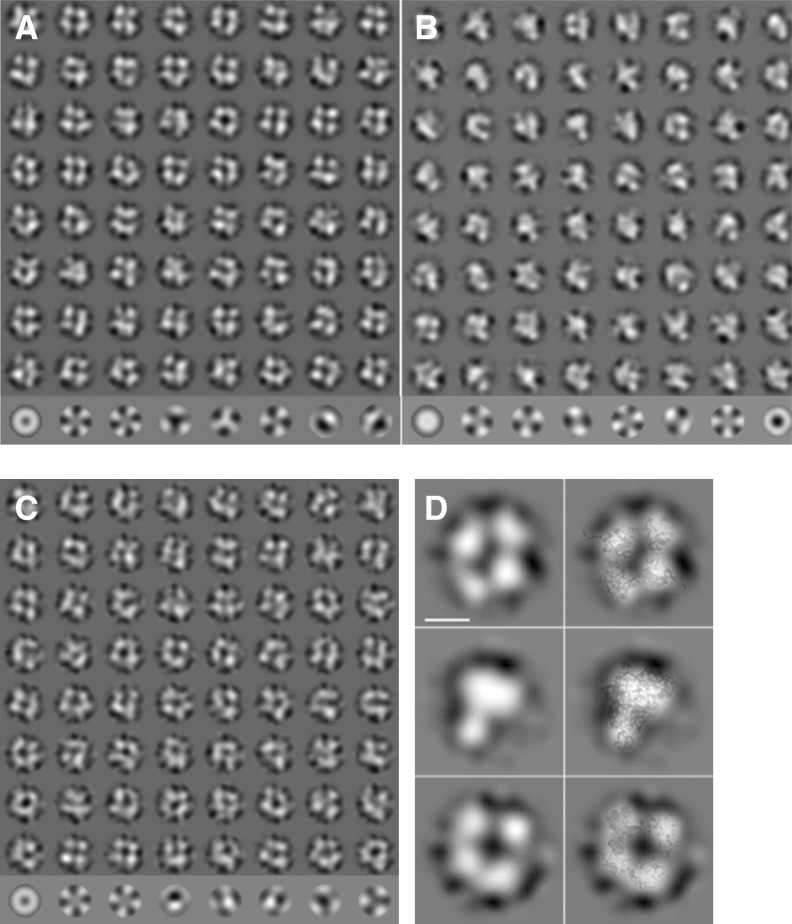
The oligomeric states of the natively purified N, RNA-free refolded N, and the refolded N complexed with 42-nt RNA were also examined by electron microscopy. The natively purified N and the N–RNA complex were mainly seen as tetramers but minor amounts of other oligomeric states, such as trimers or pentamers, were observable (Fig. 3A,C). The oligomeric states of the RNA-free refolded N were heterogeneous and irregular compared with the natively purified N (Fig. 3B). However, the predominant population of the refolded protein was identified as trimeric, although tetramers and pentamers were also present (Fig. 3B). The EM projection maps also match well with the crystal structures, as shown in overlays of the crystal structures to selected class averages (Fig. 3D). Thus, the EM observations were largely in agreement with our chemical cross-linking analysis and crystal structure data.
FIGURE 3.
Electron microscopy of negatively stained SBV N protein preparations. (A) Class averages (rows 1–8) and eigenimages (row 9) of SBV N native particles. Most of the class averages are consistent with a tetramer. However, there are also a few other oligomeric states such as trimer (row 8, column 3) or pentamer (row 8, column 6). (B) Class averages of refolded SBV N particles (rows 1–8) and eigenimages (row 9). The class averages show a heterogeneous particle population with trimers (e.g., row 1, column 1; row 2, column 8; row 3, column 6; row 7, column 1) as major component and some tetramers (e.g., row 1, column 4; row 1, column 6). (C) Class averages of the refolded SBV N-RNA complex (rows 1–8) and eigenimages (row 9). Most class averages show a tetramer. However, some class averages are consistent with other stoichiometries. (D) Selected class averages (left) and class average with the respective crystal structures overlaid (right). (Top) native SBV N; (middle) refolded SBV N; (bottom) refolded SBV N RNA complex. The overlay shows a good match between the crystal structures and the electron microscopic class averages. The length of the scale bar equals 5 nm.
RNA-free SBV N undergoes conformational changes upon RNA binding
We noticed that the RNA-free refolded N was unstable and liable to aggregation during the denaturation and refolding process, suggesting that N requires RNA binding to stabilize its structure. In solution the refolded SBV N is heterogeneous with a trimer as the predominant species (Figs. 3B, 4A), and the hexameric form may be required to form crystals based on the observed crystal packing. The hexameric structure was generated according to symmetry-related molecules (Dong et al. 2013).
FIGURE 4.
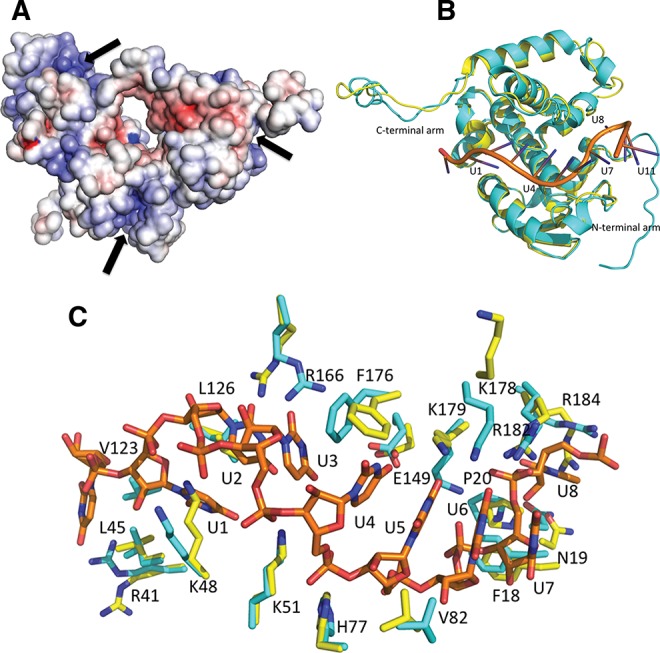
RNA binding is a prerequisite for the oligomeric transformation of refolded RNA-free SBV N. (A) Electrostatic potential surface map of the trimeric RNA-free SBV N. The potential RNA-binding clefts are on the outside of the trimer and available for RNA interaction. The dark arrows indicate the RNA-binding clefts. (B) Overall conformational changes of the SBV N before and after RNA binding. The refolded RNA-free SBV N is shown in yellow, the refolded SBV N–RNA complex is in cyan, and the RNA is shown in orange. (C) RNA-binding residues at the cleft undergo conformational changes for RNA binding. The residues prior to RNA binding are shown in yellow, and after RNA binding are shown in cyan.
In the structure of trimeric refolded N without bound RNA (Fig. 4A) (PDB code 4IDX) all of the positively charged residues in the RNA-binding clefts face toward the solution phase and, therefore, would be accessible to RNA in solution. When incubated with the 42-nt polyU RNA, the refolded SBV N converted from a trimer to a tetramer, where the RNA-binding clefts now face toward the inside of the ring, thus shielding the RNA from the outside (Fig. 1A,B). The overall structure of the SBV N in the N–RNA complex is similar to the refolded RNA-free SBV N with an RMSD of 1.05 over 201 residues (Cα backbones) (Fig. 4B). The most notable changes are that three of the N-terminal arms in the tetramer become ordered in the SBV N–RNA complex structure (Fig. 1D) and that there are conformational changes in the C-terminal arms. Furthermore, the side chains of the RNA-binding residues R41, K48, L45, K51, H77, V82, F18, N19, P20, R184, R182, K178, K179, F176, and R166 undergo conformational changes in order to bind the RNA (Fig. 4C).
DISCUSSION
The genomic RNA of negative-stranded viruses is encapsidated by nucleoprotein in the form of RNP. The RNP not only serves as a template for viral RNA replication and transcription by the cognate viral RNA-dependent RNA polymerase (RdRp), but also protects the viral genome from ribonucleases and helps to avoid alerting the host innate immune response (Qi et al. 2010; Jiang et al. 2013). Although the nucleoproteins have similar functions in different families of negative-sense RNA viruses, their structures seem to be very variable, as exemplified by our previous publication reporting the crystal structure of the SBV N protein (Dong et al. 2013). Here, we have extended this work by determining the structure of the SBV N protein in complex with RNA at high resolution, which sheds new light on details of the N–RNA interaction for orthobunyaviruses in general.
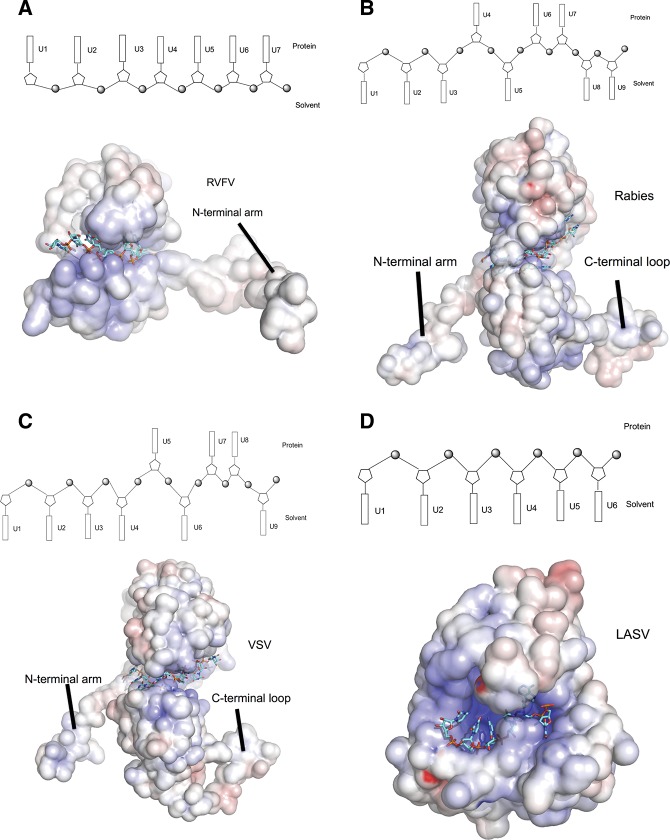
Rift Valley fever virus (RVFV) and SBV are classified in different genera in the family Bunyaviridae and have similarly sized N proteins, but their N protein folds are different (Raymond et al. 2010; Ferron et al. 2011). The nucleoprotein of RVFV uses its flexible N-terminal arm for N multimerization (Fig. 5A), and the RNPs form irregular structures due to this flexibility. Both the N- and C-terminal arms of SBV N are highly flexible and our data show that the SBV N–RNA complex forms a ring-like tetrameric structure mediated by these arms. Intriguingly, this is reminiscent of the nucleoproteins of rabies and vesicular stomatitis viruses (VSV), two nonsegmented genome viruses that form ring structures by multimerization mediated by the N-terminal arm and a loop in the C-terminal domain (Fig. 5B,C). This arrangement allows the RNP to form a super helix. Whether the same pertains for SBV RNPs is currently unknown. Although the majority of the native SBV N forms tetramers as observed by EM and chemical cross-linking, further study is needed to determine whether the building block for RNP assembly is the tetramer or some other form, such as the monomer.
FIGURE 5.
Comparison of RNA-binding modules of negative-stranded RNA virus nucleoproteins. Electrostatic potential maps are shown for all proteins, with RNA depicted in stick form in cyan. The RNA-binding modules are shown in schematic diagrams above each eletrostatic potential map. Bases oriented to the top indicate that the bases face into protein, while bases oriented to the bottom suggest that the bases are exposed to solvent. (A) RVFV N, showing that only the N-terminal arm is available for N multimerization. The RNA-binding cleft hosts four ribonucleotides with all bases facing into the protein and three bases between two N subunits. (B) Rabies virus N. The N-terminal arm and the loop of the C-terminal domain are involved in mutimerization. The RNA-binding cleft of one protomer binds six ribonucleotides with bases 1, 2, 3, and 5 exposed to solvent, bases 4 and 6 facing into the protein, and three bases between protomers (bases 8 and 9 exposed to solvent and base 7 facing to the protein). (C) Vescular stomatitis virus N. The features of the multimerization and the RNA binding of the vescular stomatitis virus N is similar to that of the rabies virus with the bases 1, 2, 3, 4, 6, and 9 facing to solvent, and bases 5, 7, and 8 turning to protein. (D) N-terminal domain of Lassa fever virus nucleoprotein. The RNA-binding cleft binds six ribonucleotides, with all bases exposed to solvent.
A common feature of negative-sense RNA virus nucleoproteins is the presence of an RNA-binding cleft or groove that is formed by mainly positively charged residues, but to date few structures of RNA bound to nucleoprotein have been determined. The available structures indicate that the mechanisms by which the nucleoproteins sequester RNAs are different. For both rabies virus and VSV, each N subunit binds nine ribonucleotides (six bases in RNA-binding cleft and three bases between N subunits) with six bases exposed to solvent and three bases facing into the protein (Fig. 5B,C) (Albertini et al. 2006; Green et al. 2006). The Lassa fever virus nucleoprotein binds six RNA bases that face the outside of the RNA-binding cleft and are exposed to solvent (Fig. 5D; Hastie et al. 2011), while in RVFV nucleoprotein, four RNA bases locate to the bottom of the RNA-binding cleft and face into the protein (Fig. 5A); an additional two or three bases are bound between N–N subunits (Raymond et al. 2012). In a further variation, we show that each SBV N subunit sequesters eight bases in the RNA-binding cleft with bases U1, U2, U3, and U8 located to the bottom of the RNA-binding cleft to face into the protein, bases U4–U7 exposed to solvent, and the N-terminal arm binding another three bases in the N–N interface (Fig. 2D).
For genomic RNA replication and transcription, the bases of the RNA have to be “read” by the viral RNA-dependent RNA polymerase. As the genomic RNA is closely encapsidated in the N oligomeric ring, the question arises as to how the RNA becomes accessible to the polymerase. In the SBV N–RNA complex there are four bases facing into the protein in the RNA-binding cleft, which suggests that the encapsidated RNA has to be partially released from the RNA-binding cleft or that the N undergoes a conformational change (or rotation) to permit polymerase access. Comparing the RNA-free N and RNA–N complex structures, we observe conformational change in some of the RNA-binding residues in the RNA-binding cleft and N-terminal and C-terminal arms. To achieve this, the flexible N- and C-terminal arms may play an essential role. Indeed, truncation of the N-terminal arm of SBV N completely abrogates its RNA-binding activity, truncation of the C-terminal arm decreases RNA-binding ability, while truncations of either or both arms results in loss of RNA replication activity (Dong et al. 2013).
In summary, we have determined the SBV N–RNA complex structure to high resolution that suggests a new RNA sequestration mechanism in orthobunyavirus RNP formation. These data will help to further our understanding of viral RNA encapsidation, replication, and transcription, and may aid in the development of antiviral compounds against diseases caused by medically important orthobunyaviruses.
Note added in Revision: While responding to the reviewers’ comments on this manuscript, lower resolution structures of N protein–ssRNA complexes from other viruses in the Orthobunyavirus genus, including La Crosse (3.4 Å resolution), Leanyer (2.78 Å resolution), and Bunyamwera viruses (3.2 Å resolution) were reported (Ariza et al. 2013; Li et al. 2013; Niu et al. 2013; Reguera et al. 2013). They all share similar structural features and proposed mode of RNA binding. For example, all complexes form tetrameric structures that encapsidate the RNA inside the ring. Each N protomer binds 10 or 11 ribonucleotides, and the C-terminal and N-terminal arms are involved in N multimerization.
MATERIALS AND METHODS
Protein expression, purification, and N–RNA complex (RNP) reconstitution
Construction of bacterial expression plasmid p14SBVN and purification of natively purified SBV N (with host RNA bound) and refolded RNA-free N have been described previously (Dong et al. 2013). The natively purified and refolded RNA-free SBV N was concentrated to 9 mg/mL in 20 mM Tris (pH 7.5), 300 mM NaCl, and 10% glycerol. To reconstitute RNP, the refolded N was mixed in a 1:1 molar ratio with RNA (poly U of 21, 28, or 42 bases in length; Eurogentec) and incubated on ice for 90 min. The preparation was screened for crystallization conditions.
Chemical cross-linking of SBV N protein
Cross-linking of natively purified SBV N, refolded RNA-free N, and the refolded N–RNA complex was performed using dithiobis (succinimidyl propionate; DSP), as previously reported (Dong et al. 2013). Briefly, samples containing 10–20 μg of protein were treated with 1 mM DSP for 30 min at room temperature in PBS buffer and then analyzed by SDS-PAGE under reducing or nonreducing conditions, followed by Coomassie blue staining.
Negatively stained EM structures of SBV N
For electron microscopy the refolded SBV N, native SBV N, or refolded SBV N–42-nt RNA complex were diluted to concentrations between 2 and 20 µg/mL. The protein was incubated for 1–2 min on carbon-coated grids, and then excess sample was removed. The grids were washed twice with water and twice with 2% uranyl acetate before staining for 5 min with 2% uranyl acetate. Excess staining solution was blotted off and the grids were air dried.
For imaging, grids were transferred to a FEI F20 electron microscope equipped with a 4k × 4k CMOS camera (TVIPS F416). Micrographs were recorded with EM-Tools (TVIPS GmbH) under low-dose conditions at 200 kV, a primary magnification of 66,000 (calibrated pixel size of 1.73Å), and an approximate dose of 20 e/Å2.
For image processing, particles were semiautomatically selected from the micrographs using e2boxer (Tang et al. 2007). Further image processing was done with IMAGIC (Van Heel et al. 2011). Particle images were binned 3 × 3, giving a calibrated pixel size of 5.2 Å. The binned images were band-pass filtered and normalized in their gray value distribution. Filtered particle images were classified using multivariate statistical analysis. Class averages were centered, rotationally aligned with respect to each, and averaged. The filtered particle images were aligned to this average, followed by classification of the 70% of particle images with the highest cross correlation.
Structure determination
Crystals of SBV N complexed with the 42-nt RNA were obtained in 0.1 M Bis-Tris (pH5.7), 0.3 M NaCl, and 25% PEG3350, and were cryo-protected by adding glycerol to 20% in the crystallization conditions. The structure of the N–RNA complex was determined by molecular replacement using Phaser (McCoy et al. 2007) with the native SBV N as a search model (4IDU). The model was built using Coot (Emsley et al. 2010) and the structure was refined using REFMAC5 (Murshudov et al. 2011) with the noncrystallographic symmetry restraints. The structure was validated by MolProbity (Chen et al. 2010). The statistics of data collection and structure refinement are listed in Table 1.
TABLE 1.
Statistics of data collection and structure refinement

DATA DEPOSITION
The atomic coordinates and the structure factors have been deposited in the Protein Data Bank with PDB access code 4JNG for Schmallenberg virus N–42-nt RNA complex.
ACKNOWLEDGMENTS
We thank the staff of IO4 and I24 beam stations at Diamond Light Resources UK and Dr. Xiaohong Shi for critical reading of the manuscript. This work was supported by the Medical Research Council (grant No G1100110/1 to C.D.) and Wellcome Trust (Career Development Fellowship WT083501MA to C.D., and Programme Grant 079810 and Project Grant 091783 to R.M.E.). R.M.E. is a Wellcome Trust Senior Investigator. B.B. was supported by the Darwin Trust of Edinburgh. The use of the electron microscopy facility was supported by the Wellcome Trust Equipment grant WT087658 and by the Scottish University Life Sciences Alliance (SULSA).
REFERENCES
- Albertini AA, Wernimont AK, Muziol T, Ravelli RB, Clapier CR, Schoehn G, Weissenhorn W, Ruigrok RW 2006. Crystal structure of the rabies virus nucleoprotein-RNA complex. Science 313: 360–363 [DOI] [PubMed] [Google Scholar]
- Ariza A, Tanner SJ, Walter CT, Dent KC, Shepherd DA, Wu W, Matthews SV, Hiscox JA, Green TJ, Luo M, et al. 2013. Nucleocapsid protein structures from orthobunyaviruses reveal insight into ribonucleoprotein architecture and RNA polymerization. Nucleic Acids Res 10.1093/nar/gkt268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beer M, Conraths FJ, van der Poel WH 2013. ‘Schmallenberg virus’—a novel orthobunyavirus emerging in Europe. Epidemiol Infect 141: 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen VB, Arendall WB III, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC 2010. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66: 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Regge N, Deblauwe I, De Deken R, Vantieghem P, Madder M, Geysen D, Smeets F, Losson B, van den Berg T, Cay AB 2012. Detection of Schmallenberg virus in different Culicoides spp. by real-time RT-PCR. Transbound Emerg Dis 59: 471–475 [DOI] [PubMed] [Google Scholar]
- Dong H, Li P, Elliott RM, Dong C 2013. Structure of Schmallenberg orthobunyavirus nucleoprotein suggests a novel mechanism for genome encapsidation. J Virol 87: 5593–5601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott RM, Blakqori G 2011. Molecular biology of orthobunyaviruses. Caister Academic Press, Norfolk, UK [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66: 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferron F, Li Z, Danek EI, Luo D, Wong Y, Coutard B, Lantez V, Charrel R, Canard B, Walz T, et al. 2011. The hexamer structure of Rift Valley fever virus nucleoprotein suggests a mechanism for its assembly into ribonucleoprotein complexes. PLoS Pathog 7: e1002030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garigliany MM, Bayrou C, Kleijnen D, Cassart D, Jolly S, Linden A, Desmecht D 2012. Schmallenberg virus: A new Shamonda/Sathuperi-like virus on the rise in Europe. Antiviral Res 95: 82–87 [DOI] [PubMed] [Google Scholar]
- Goller KV, Hoper D, Schirrmeier H, Mettenleiter TC, Beer M 2012. Schmallenberg virus as possible ancestor of Shamonda virus. Emerg Infect Dis 18: 1644–1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green TJ, Zhang X, Wertz GW, Luo M 2006. Structure of the vesicular stomatitis virus nucleoprotein-RNA complex. Science 313: 357–360 [DOI] [PubMed] [Google Scholar]
- Hastie KM, Liu T, Li S, King LB, Ngo N, Zandonatti MA, Woods VL Jr, de la Torre JC, Saphire EO 2011. Crystal structure of the Lassa virus nucleoprotein–RNA complex reveals a gating mechanism for RNA binding. Proc Natl Acad Sci 108: 19365–19370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann B, Scheuch M, Hoper D, Jungblut R, Holsteg M, Schirrmeier H, Eschbaumer M, Goller KV, Wernike K, Fischer M, et al. 2012. Novel orthobunyavirus in Cattle, Europe, 2011. Emerg Infect Dis 18: 469–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Huang Q, Wang W, Dong H, Ly H, Liang Y, Dong C 2013. Structures of arenaviral nucleoproteins with triphosphate dsRNA reveal a unique mechanism of immune suppression. J Biol Chem 10.1074/jbc.M112.420521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Wang Q, Pan X, Fernandez de Castro I, Sun Y, Guo Y, Tao X, Risco C, Sui SF, Lou Z 2013. Bunyamwera virus possesses a distinct nucleocapsid protein to facilitate genome encapsidation. Proc Natl Acad Sci 110: 9048–9053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ 2007. Phaser crystallographic software. J Appl Crystallogr 40: 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA 2011. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr 67: 355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu F, Shaw N, Wang YE, Jiao L, Ding W, Li X, Zhu P, Upur H, Ouyang S, Cheng G, et al. 2013. Structure of the Leanyer orthobunyavirus nucleoprotein–RNA complex reveals unique architecture for RNA encapsidation. Proc Natl Acad Sci 110: 9054–9059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plyusnin A, Beaty B, Elliott RM, Goldbach R, Kormelink R, Lundkvist Å, Schmaljohn CS, Tesh RB 2012. Bunyaviridae. Elsevier Academic Press, London, UK [Google Scholar]
- Qi X, Lan S, Wang W, Schelde LM, Dong H, Wallat GD, Ly H, Liang Y, Dong C 2010. Cap binding and immune evasion revealed by Lassa nucleoprotein structure. Nature 468: 779–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen LD, Kristensen B, Kirkeby C, Rasmussen TB, Belsham GJ, Bodker R, Botner A 2012. Culicoids as vectors of Schmallenberg virus. Emerg Infect Dis 18: 1204–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond DD, Piper ME, Gerrard SR, Smith JL 2010. Structure of the Rift Valley fever virus nucleocapsid protein reveals another architecture for RNA encapsidation. Proc Natl Acad Sci 107: 11769–11774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond DD, Piper ME, Gerrard SR, Skiniotis G, Smith JL 2012. Phleboviruses encapsidate their genomes by sequestering RNA bases. Proc Natl Acad Sci 109: 19208–19213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reguera J, Malet H, Weber F, Cusack S 2013. Structural basis for encapsidation of genomic RNA by La Crosse Orthobunyavirus nucleoprotein. Proc Natl Acad Sci 110: 7246–7251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ 2007. EMAN2: An extensible image processing suite for electron microscopy. J Struct Biol 157: 38–46 [DOI] [PubMed] [Google Scholar]
- Tarlinton R, Daly J, Dunham S, Kydd J 2012. The challenge of Schmallenberg virus emergence in Europe. Vet J 194: 10–18 [DOI] [PubMed] [Google Scholar]
- Van Heel M, Portugal R, Rohou A, Linnemayr C, Bebeacua C, Schmidt R, Grant T, Schatz M 2011. Four-dimensional cryo electron microscopy at quasi atomic resolution: “IMAGIC 4D”. Wiley, New York [Google Scholar]