Cells that survive fractional killing by TRAIL or FasR agonists enter a state of resistance accompanied by inflammatory phenotypes. This state is transient, decaying over the course of several days, but can be sustained by periodic TRAIL treatments. This finding has implications for optimal dosing strategies of extrinsic cell death agents.
Abstract
When clonal populations of human cells are exposed to apoptosis-inducing agents, some cells die and others survive. This fractional killing arises not from mutation but from preexisting, stochastic differences in the levels and activities of proteins regulating apoptosis. Here we examine the properties of cells that survive treatment with agonists of two distinct death receptors, tumor necrosis factor–related apoptosis-inducing ligand (TRAIL) and anti-FasR antibodies. We find that “survivor” cells are highly resistant to a second ligand dose applied 1 d later. Resistance is reversible, resetting after several days of culture in the absence of death ligand. “Reset” cells appear identical to drug-naive cells with respect to death ligand sensitivity and gene expression profiles. TRAIL survivors are cross-resistant to activators of FasR and vice versa and exhibit an NF-κB–dependent inflammatory phenotype. Remarkably, reversible resistance is induced in the absence of cell death when caspase inhibitors are present and can be sustained for 1 wk or more, also without cell death, by periodic ligand exposure. Thus stochastic differences in cell state can have sustained consequences for sensitivity to prodeath ligands and acquisition of proinflammatory phenotypes. The important role played by periodicity in TRAIL exposure for induction of opposing apoptosis and survival mechanisms has implications for the design of optimal therapeutic agents and protocols.
INTRODUCTION
Tumor necrosis factor (TNF)–related apoptosis-inducing ligand (TRAIL) is a member of the TNF family of death ligands that binds to transmembrane DR4/5 receptors and induces apoptosis via the extrinsic cell death pathway; TRAIL and DR4/5 agonist antibodies are in phase II trials as anticancer drugs (Ashkenazi and Dixit, 1999). TRAIL is believed to play a role in tumor immune surveillance but might have other, less-well-understood, physiological activities (Takeda et al., 2002; Wilson et al., 2009). The promise of recombinant TRAIL ligand and agonist anti-DR4/5 antibodies as cancer therapies reflects their selectivity in killing tumor cells (Ashkenazi, 2008; Ashkenazi and Herbst, 2008; Johnstone et al., 2008). The molecular basis of sensitivity and resistance to TRAIL, however, remains relatively poorly understood (Falschlehner et al., 2007; Gonzalvez and Ashkenazi, 2010), and clinical development has proven challenging in part because many cancers exhibit a fractional response in which only a subset of cells dies even at saturating ligand doses.
During TRAIL-induced apoptosis, binding of ligand to DR4/5 leads to formation of death-inducing signaling complexes (DISCs) and activation of initiator procaspases-8 and -10 (C8/10; Kischkel et al., 1995; Martin et al., 1998), a process that is inhibited by the DISC protein c-FLIP (Scaffidi et al., 1999). In some cell types (type I cells) cell death requires only C8/10 cleavage and activation of effector caspases-3 and -7 (C3/7), but in most cells (type II cells), C3/7 activity is held in check by the inhibitor of apoptosis protein XIAP (Deveraux et al., 1997). In these cells, death requires mitochondrial outer membrane permeabilization (MOMP; Deng et al., 2002; Sun et al., 2002; Barnhart et al., 2003). The induction of MOMP involves C8/10-mediated cleavage of Bid to tBid, which translocates to the mitochondria and activates Bcl-2 family members Bax/Bak, thereby promoting self-assembly into transmembrane pores (Eskes et al., 2000). Pore formation is possible only when levels of active Bax/Bak exceed those of antiapoptotic Bcl-2 proteins. MOMP allows the translocation of Smac and cytochrome c through pores into the cytosol (Luo et al., 1998; Li et al., 2002). Cytosolic cytochrome c serves as a platform for assembly of the apoptosome, whereas Smac binds XIAP and relieves XIAP-mediated C3/7 inhibition (Riedl and Salvesen, 2007). After MOMP, rapid activation of C3/7 leads to processing of ICAD nucleases, digestion of the cellular proteome and genome, and cell death (Rehm et al., 2006; Albeck et al., 2008).
In addition to apoptosis, ligands of the TNF family such as TRAIL induce prosurvival and inflammatory pathways, many of which involve NF-κB (Chaudhary et al., 1997; Jeremias and Debatin, 1998). NF-κB is normally sequestered in the cytoplasm by IκBα, but after activation of IKK at the DISC, IκBα is phosphorylated and degraded, releasing NF-κB into the nucleus (Luo et al., 2005). TRAIL also induces phosphorylation of kinases involved in cell survival and inflammation such as extracellular signal–regulated protein kinase (ERK), Akt, p38, and Jun-N-terminal kinase (JNK; Varfolomeev et al., 2005; Kim et al., 2008; Son et al., 2010; Sun et al., 2010a), and these kinases, acting in combination with NF-κB, contribute to TRAIL resistance under at least some circumstances (Falschlehner et al., 2007; Guicciardi and Gores, 2009).
Even in clonal populations of sensitive cells, substantial heterogeneity exists in TRAIL responsiveness: only a subset of cells dies, and the interval between TRAIL exposure and death varies from ∼40 min to >12 h (Rehm et al., 2002; Albeck et al., 2008; Hellwig et al., 2008). Lineage studies show that recently born sister cells are much more similar in the timing and probability of death than are random pairs of cells but that the similarity between sisters decays rapidly. Such “transiently heritable” variability is believed to arise from natural fluctuations in the levels of proapoptotic and antiapoptotic proteins (Bhola and Simon, 2009; Rehm et al., 2009; Spencer et al., 2009) and in the rates or efficiency with which prodeath and prosurvival responses are activated (Zauli et al., 2005; Fricker et al., 2010). NF-κB is postulated to play a particularly important role in the heterogeneity of receptor-mediated apoptosis (Neumann et al., 2010), even though inhibiting NF-κB does not always sensitize cells to TRAIL (Ganten et al., 2005; Diessenbacher et al., 2008).
Insensitivity to TRAIL is a natural feature of some cell types, but resistance can also be acquired, either by selection for resistance mutations or through adaptive changes that follow prolonged or repeated exposure to ligand (Zhang and Fang, 2005; Lane et al., 2006; Li et al., 2011). These changes can involve down-regulation of proapoptotic proteins such as caspase-8 or Bax or up-regulation of antiapoptotic proteins such as FLIP, Bcl-2, or XIAP (Falschlehner et al., 2007). Short-term adaptive changes have also been observed, for example, in conjunction with receptor down-regulation (Song et al., 2010) or death ligand–mediated activation of survival signals (Jang et al., 2008), and these may occur in both resistant and fractionally sensitive cell populations (Song et al., 2007b). In resistant cell lines, TRAIL-induced signaling can further result in inflammatory phenotypes and cancer progression (Malhi and Gores, 2006), but how cell lines that exhibit fractional killing are affected by these pathways has been less explored.
In this article we examine the origins and consequences of fractional responsiveness to death ligands, in particular as they relate to “nondeath” signaling pathways induced by TRAIL. We ask whether cells that survive an initial exposure to TRAIL are sensitive or resistant to a second dose of death ligand applied at various later times. We find that TRAIL survivors are highly resistant to a second dose of ligand at 24 h but that resistance disappears after several days in culture, regenerating the same degree of fractional killing observed in naive cell populations. NF-κB–mediated inflammatory genes are activated in resistant survivor cells, leading to an inflammatory phenotype, but ligand resistance itself is NF-κB independent. Resistance does not involve down-regulation of TRAIL receptors or insensitivity to inducers of intrinsic apoptosis but results from impaired DISC assembly and thus extends to other death ligands: TRAIL survivors are cross-resistant to FasR agonists and vice versa. Resistance and inflammation can be sustained with periodic TRAIL treatments, but both are reversible when cells are grown for several days in the absence of TRAIL. The idea that a transiently resistant state can be maintained in a subset of cells by repeated exposure without further cell killing shows that cell-to-cell differences within clonal cell populations can be amplified and sustained for extended periods. Transiently heritable nongenetic differences may therefore have long-term influence on the fates of cells and tissues.
RESULTS
Cells that survive treatment with death ligands are transiently resistant to a second death-ligand treatment
What distinguishes cells that survive exposure to TRAIL from those that do not? To address this question, we exposed a population of TRAIL-sensitive MCF10A cells (immortalized but nontransformed mammary epithelial cells) to sufficient TRAIL to kill 70–90% of the cell population (50 ng/ml for 6 h). Cells that survived TRAIL exposure (“survivor” cells) remained attached to the dish, whereas dead cells detached, allowing survivors to be recovered by trypsinization. Survivors were replated into fresh medium in the absence of TRAIL and were observed to divide normally. Survivor cells were then challenged with a second dose of TRAIL 1, 3, 6, or 9 d later and monitored for extent of cell death. A population of naive cells that had not been previously exposed to TRAIL served as a control (Figure 1A). All cultures were split regularly to avoid confluency, and cells were replated at equivalent densities. Apoptosis was assessed via multiple methods, including 1) using flow cytometry with antibodies selective for the cleaved form of the caspase-3 substrate PARP (cPARP), 2) measuring the fraction of surviving cells using a vital dye (allowing the fraction of dead cells to be calculated relative to a control population), and 3) measuring the fraction of cells positive for annexin V staining, a specific marker for loss of membrane asymmetry during apoptosis. The three assays yielded similar data.
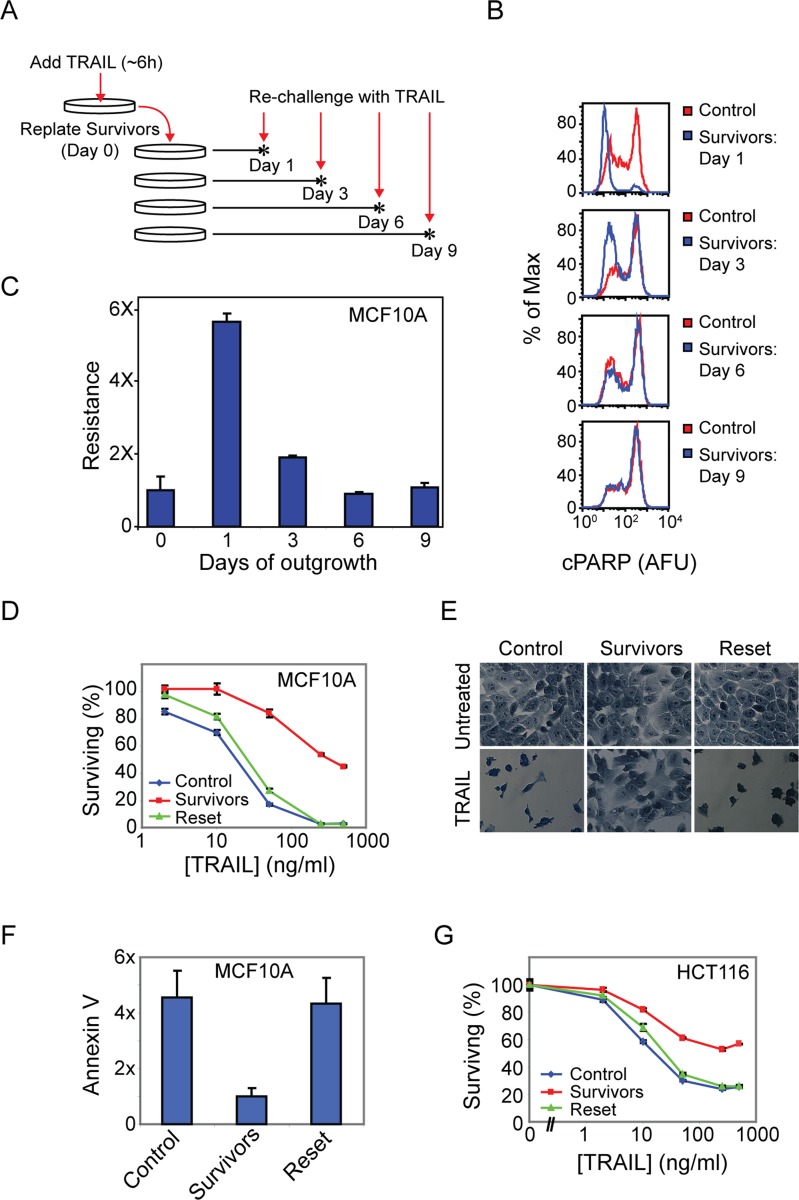
FIGURE 1:
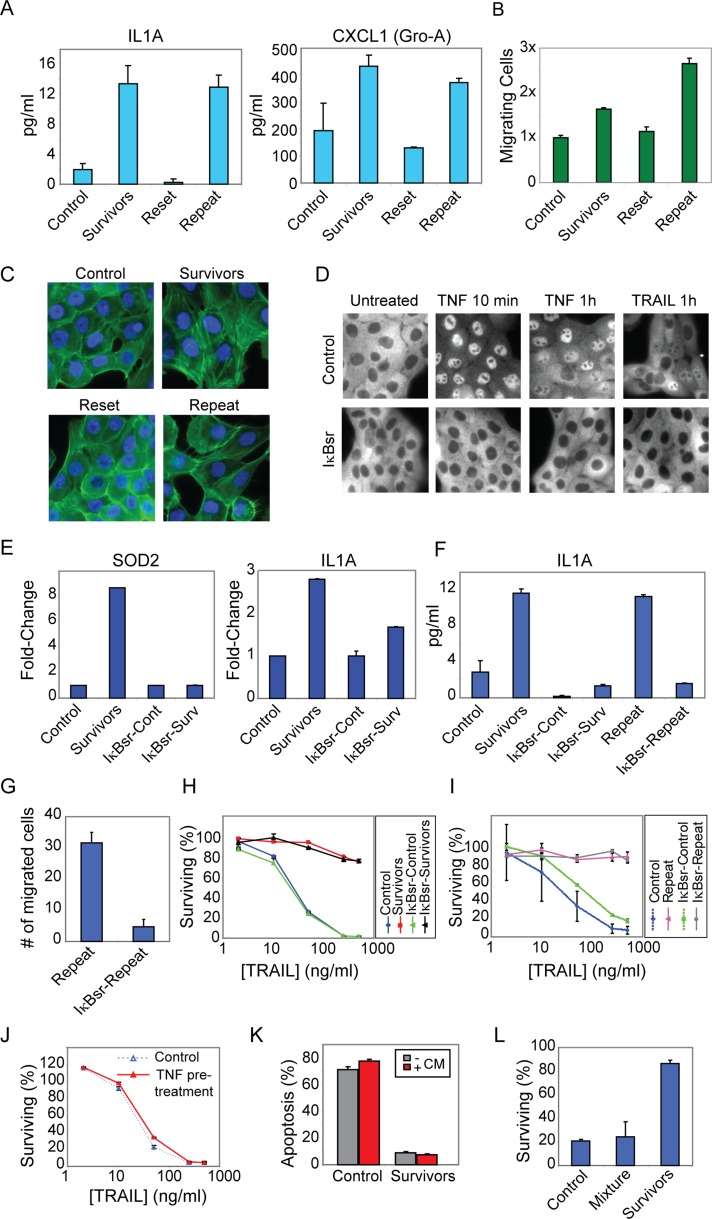
Cells that survive TRAIL-mediated apoptosis exhibit transient resistance to a second TRAIL challenge. (A) Experimental setup showing rechallenge of survivor cells on subsequent days after an initial TRAIL treatment (50 ng/ml for 6 h). (B) Flow cytometry histograms of PARP cleavage in TRAIL-treated control (naive) and survivor MCF10A cells. (C) Quantitation of B showing the fraction of cleaved PARP-negative cells (Resistance) in TRAIL-treated survivor cells following the indicated days of outgrowth, normalized to that of TRAIL-treated control cells. (D) Cell viability assay showing the percentage of surviving control, survivors (day 1), and reset (day 7) MCF10A cells after a 6-h TRAIL treatment. (E) Methylene blue staining of untreated or treated (50 ng/ml TRAIL for 6 h) control, survivor, and reset MCF10A cells; dead cells were washed off before staining. (F) Annexin V labeling of TRAIL-treated (50 ng/ml TRAIL for 6 h) control, survivor, and reset MCF10A cells; levels were normalized to TRAIL-treated survivor cells. (G) Cell viability assay of TRAIL-treated (50 ng/ml TRAIL for 7 h) control, survivor, and reset HCT116 cells (clonal population). Error bars in all plots represent the SE of triplicate samples.
When survivors of a first round of TRAIL treatment were challenged with TRAIL 1 d later (“day 1 survivors”), very few cells (∼10%) were positive for cPARP or annexin V staining, as compared with 85% of naive control cells (Figure 1B and Supplemental Figure S1A), representing a sixfold increase in TRAIL resistance (i.e., 90 vs. 15% survival; Figure 1C). TRAIL-mediated apoptosis is dose-dependent, and resistance was observed in day 1 survivors across a wide range of TRAIL doses, with a maximum of ninefold relative resistance at the highest ligand dose tested, 500 ng/ml (Figure 1D). When survivor cells were passaged over a period of 3–9 d, however, sensitivity to TRAIL gradually returned, giving rise to “reset” cells that were as sensitive to TRAIL as naive cells (Figure 1, B–F). The phenomenon of initial sensitivity to TRAIL followed by a period of resistance and subsequent regaining of sensitivity was observed with multiple clonal populations of MCF10A cells (established by single-cell cloning; Supplemental Figure S1B), in multiple oncogenic derivatives of the MCF10A cell line (Bargmann et al., 1986; Santner et al., 2001; Debnath et al., 2003; Supplemental Figure S1C), in a clonal population of the colon carcinoma HCT116 cell line (Figure 1G), and to a greater or lesser extent in a panel of unrelated transformed and nontransformed cell lines (Supplemental Figure S1D). Because cells that survive treatment with TRAIL regenerate a population of cells with the same TRAIL sensitivity as the starting population, fractional killing is a stable property of clonal cell lines, and genetic mutation is eliminated as a significant factor.
Reversible ligand resistance was also observed after treatment of cells with an antibody agonist of the FasR/CD95 receptor (Apo-1-3; Trauth et al., 1989). Fas is an essential inducer of cell death in immune cells that engages a receptor-mediated apoptotic signaling pathway similar to that engaged by TRAIL (Siegel and Fleisher, 1999; Wilson et al., 2009). Control populations of MCF10A cells exhibited a dose-dependent response to Apo-1-3, with ∼60% of cells dying at the highest antibody concentration tested. Cells that survived exposure to Apo-1-3 (5 μg/ml for 6 h) were almost completely resistant to a second dose of Apo-1-3 delivered 1 d later, regardless of dose (Figure 2A, blue and green lines). In contrast, when MCF10A cells were exposed for 6 h to 1 μM staurosporine, sufficient drug to induce intrinsic apoptosis in ∼40% of cells, survivors were no more resistant than control cells to a second dose of staurosporine, TRAIL, or Apo-1-3 applied 24 h later (Figure 2, A–C, pink and blue lines). Thus staurosporine survivors exhibited regeneration of a mixed population of sensitive and resistant cells within 24 h. In contrast, resistance on day 1 was specific to survivors of extrinsic cell death induced by molecules such as TRAIL and Apo-1-3.
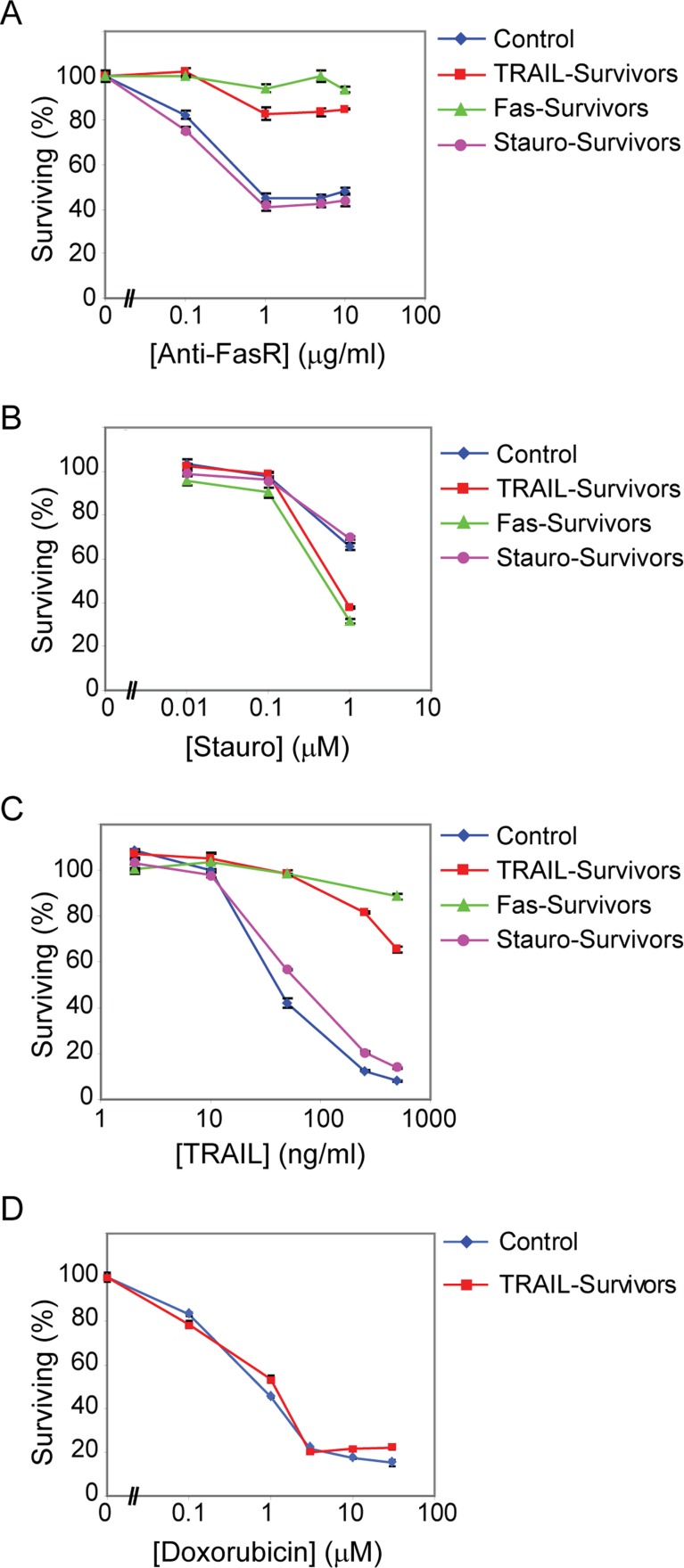
FIGURE 2:
TRAIL survivors are cross-resistant to Fas agonists and vice versa, but are sensitive to inducers of intrinsic cell death. (A–C) Cell viability assay showing the sensitivity of control cells vs. TRAIL, Anti-FasR, or staurosporine survivors (day 1) to a subsequent 6-h treatment with the indicated doses of Anti-FasR (A), staurosporine (B), or TRAIL (C). (D) Sensitivity of control vs. TRAIL-survivor cells (day 1) to an 18-h treatment with the indicated doses of doxorubicin.
To determine whether cells transiently resistant to TRAIL were cross-resistant to agonists of FasR and vice versa, we treated TRAIL survivors with Apo-1-3 and Apo-1-3 survivors with TRAIL and measured the extent of cell death. TRAIL survivors were threefold less sensitive to Apo-1-3 than naive control cells (Figure 2A, red line), and Apo-1-3 survivors were >10-fold more resistant to TRAIL than naive cells at the highest TRAIL dose tested (Figure 2C, green line). In contrast, cells transiently resistant to TRAIL or Apo-1-3 were not cross-resistant to inducers of intrinsic cell death such as staurosporine or doxorubicin. We observed more, not less killing by staurosporine in TRAIL and Apo-1-3 survivors than in control cells across a range of drug doses (Figure 2B) and equivalent cell killing by doxorubicin in control cells and TRAIL survivors (Figure 2D). We conclude that TRAIL survivors are Fas-resistant and Fas survivors are TRAIL-resistant. Survivors of both TRAIL and Fas are apoptosis competent, however, since they can be killed to the same extent as control cells by exposure to staurosporine or doxorubicin.
TRAIL induces transient resistance even in the absence of initial cell death
The observation that survivors of staurosporine treatment regenerate a mixed population of sensitive and resistant cells within 24 h but survivors of TRAIL and Fas require several days to reset caused us to speculate that TRAIL and Fas ligand might actively prolong the resistant state. To test this idea, we treated cells for 18 h with TRAIL plus sufficient caspase inhibitor to prevent cell death, changed the medium, and then measured sensitivity to a second treatment with TRAIL. Cells subjected to this procedure were approximately five times more TRAIL-resistant than control cells, implying that TRAIL exposure can induce resistance without actually killing cells. As a control we showed that cells exposed to caspase inhibitor alone for 18 h were as sensitive as untreated cells to a subsequent treatment with TRAIL, demonstrating that the caspase inhibitor could be effectively washed out (Figure 3A). We conclude that reversible ligand resistance is induced directly by TRAIL in the absence of cell death and therefore does not involve conventional selection. Moreover, reversible resistance is not mediated by factors released from dying cells.
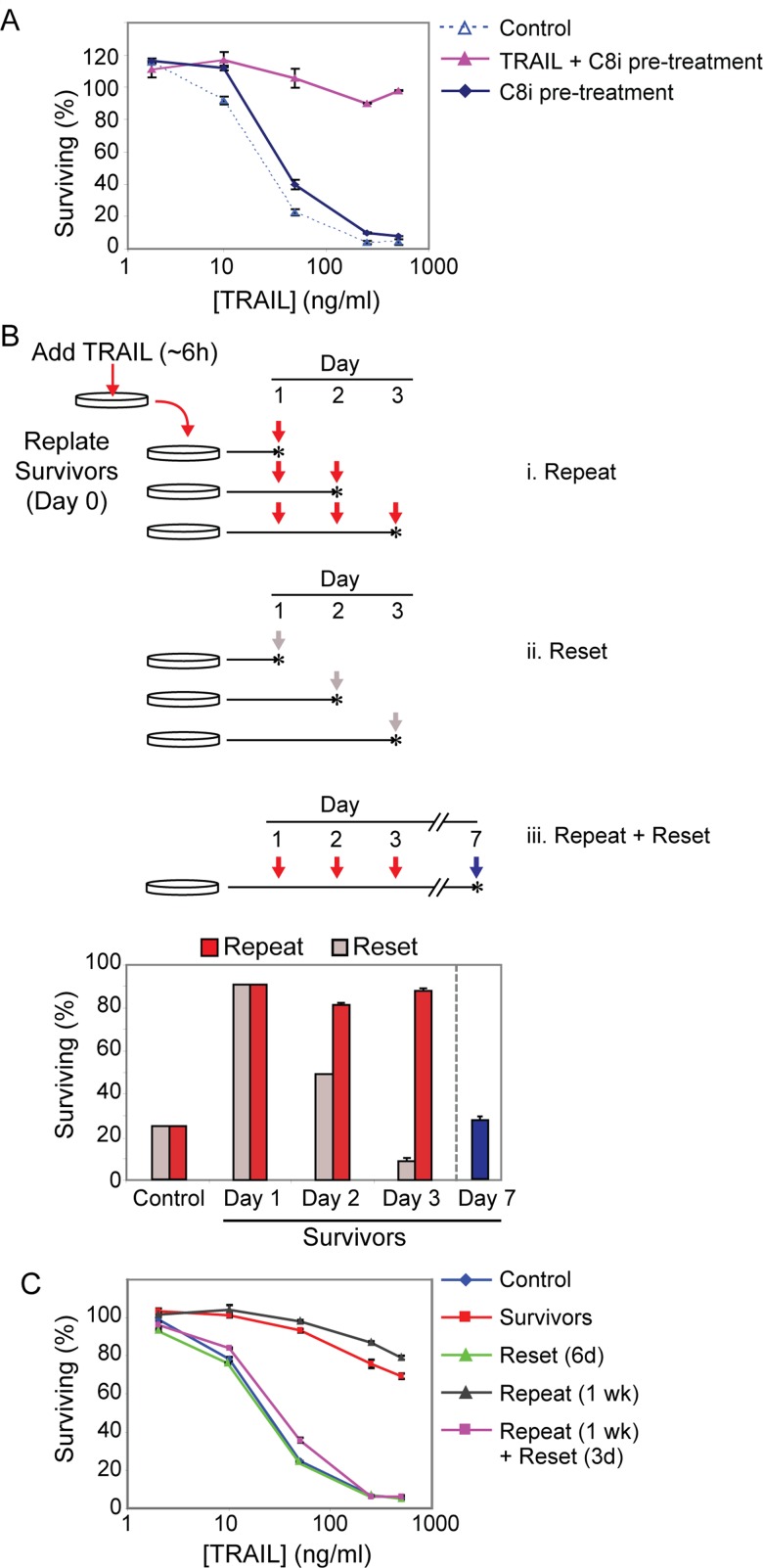
FIGURE 3:
Reversible resistance is induced even in the absence of cell death and is sustained by periodic TRAIL treatments. (A) Cell viability assay showing the sensitivity of control cells, cells pretreated for 18 h with TRAIL (50 ng/ml) + caspase-8 inhibitor (25 μM), or cells pretreated for 18 h with caspase inhibitor alone to a subsequent 6-h treatment with the indicated doses of TRAIL (after wash-off of the initial treatment). (B) Plot showing percentage of surviving (cleaved PARP-negative) cells after TRAIL (50 ng/ml for 6 h) treatment in a “repeat” experiment (i, red arrows and red bars), or in a parallel “reset” experiment (ii, gray arrows and gray bars). The plot shows the percentage of surviving control and day 1 survivor cells after TRAIL treatment for both experiments, followed by percentage of surviving cells after “retreatment” (50 ng/ml TRAIL for 6 h) on consecutive days as shown in the schematic (red scheme) or after allowing cells to “reset” for the number of days indicated before the final treatment (gray scheme). iii, Percentage of surviving (cleaved PARP-negative) cells after TRAIL (50 ng/ml for 6 h) treatment of cells allowed to recover for 4 additional days after three successive “repeat” treatments (blue arrow and blue bar). Vertical arrows in the schematic represent treatments (50 ng/ml TRAIL for 6 h), and stars represent collection times after treatment. Data are mean ± SE of triplicate samples. (C) Cell viability plot of the percentage of surviving control, survivors (day 1), reset (day 6), repeat (treated daily for 1 wk with 50 ng/ml for 6 h), and repeat + reset (treated daily for 1 wk with 50 ng/ml for 6 h and then allowed to recover for 3 d in the absence of TRAIL) MCF10A cells after a 6-h TRAIL (50 ng/ml) treatment.
Repeated TRAIL exposure sustains transient ligand resistance
To determine whether resistance to TRAIL can be sustained, we exposed “survivor” cells to TRAIL on 3 successive days and remeasured the fraction of dying cells (Figure 3B). Whereas 80% of cells were killed by an initial exposure to TRAIL, subsequent treatment of survivor cells on days 1, 2, and 3 resulted in very low levels of killing. Such “repeat” treatment cells regained sensitivity only after a subsequent period of outgrowth in the absence of TRAIL (Figure 3B, day 7, blue arrow and blue bar). TRAIL resistance could be sustained for at least 1 wk (the longest time tested) by periodic exposure to TRAIL but was still reversible after 3 d of outgrowth in the absence of TRAIL (Figure 3C). Thus, periodic TRAIL exposure sustains a transiently resistant state in virtually all cells, arguing in favor of an induced survival response rather than selection for a population of inherently resistant cells. Consistent with this, when day 1 survivor cells were exposed to TRAIL plus cycloheximide, all cells were sensitized, arguing that protein synthesis is necessary to maintain resistance (Supplemental Figure S2A). We conclude that TRAIL and Fas induce a resistant state in surviving cells and that this state can be sustained by further TRAIL treatments even in the absence of cell death.
Survivor and repeat cells exhibit distinct expression states compared with naive and reset cells
To uncover relationships among naive (control), survivor, repeat, and reset cell states, we performed microarray-based expression analysis. RNA was collected from similar numbers of each cell population (Figure 4A). This revealed ∼200 genes up-regulated in day 1 TRAIL survivors as compared with control cells (assuming a threshold of 1.5-fold change and 5% false discovery rate; we confirmed a portion of the expression changes detected on arrays by quantitative PCR [qPCR]; Supplemental Figure S3A). A subset (∼42) of the genes up-regulated in survivors was also up-regulated in repeat cells (Figure 4B and Supplemental Table S1). Survivor and repeat cells also each contained ∼50 down-regulated genes, but these had minimal overlap with each other (Figure 4C and Supplemental Table S1). As a simple means to compare patterns of expression we performed principal components analysis (PCA). The first three PCA components captured 83% of the variance in the data, validating the approach. In PCA space, control cells clustered with reset cells and away from survivors (Figure 4D). Repeat cells clustered into a separate group, more closely related to controls along principal component 1 (PC1) and in between control and survivor cells along PC2 (Supplemental Table S2); supervised hierarchical clustering confirmed this grouping (Supplemental Figure S3B). We conclude that reset cells are highly similar if not identical to naive cells despite having been exposed to high doses of TRAIL. In contrast, day 1 survivor cells and repeat cells exhibit distinct expression states that separate them from naive cells and from reset cells that regained TRAIL sensitivity.
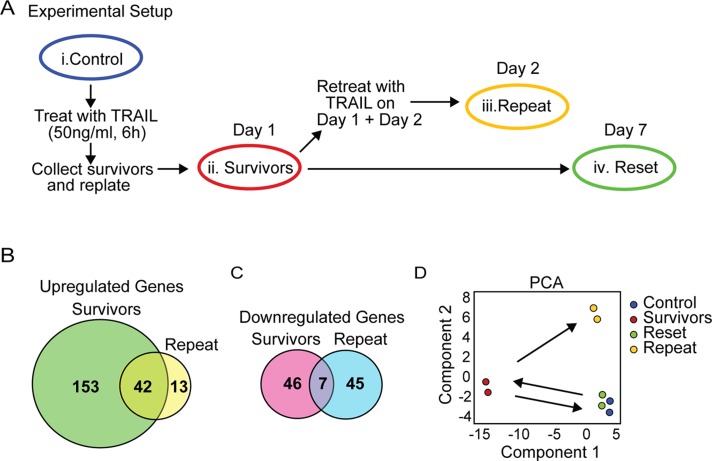
FIGURE 4:
Microarray analysis reveals distinct gene expression profiles for survivors and repeat cells, but control and reset cells cluster together. (A) Experimental setup for RNA collection from control (i), survivors (ii), repeat (iii), and reset cells (iv). (B) Venn diagram of genes significantly up-regulated in survivor and repeat cells compared with control cells. (C) Venn Diagram of genes down-regulated in survivor and repeat cells compared with control cells. (D) Principal components analysis of the gene expression data. Groups (representing duplicate samples) are plotted along the two components that account for the most variation (74.7%) in the data. The first component represents genes that differ most between control and survivor cells; the second component represents genes that differ most between control and repeat cells.
Survivor and repeat cells exhibit changes in morphology, increased cell migration, and secretion of inflammatory cytokines
In addition to cell death, Fas and TRAIL can induce cell migration, proliferation, and secretion of inflammatory cytokines (Ehrhardt et al., 2003; Wajant et al., 2003; Barnhart et al., 2004; Ishimura et al., 2006; Kleber et al., 2008; Li et al., 2011). Consistent with this, genes upregulated in survivor and repeat cells were highly enriched in Gene Ontology (GO) terms suggestive of an inflammatory response (Supplemental Tables S3 and S4). Analysis of conditioned media using enzyme-linked immunosorbent assay (ELISA) also revealed elevated levels of inflammatory cytokines such as IL-1A and CXCL1/GroA in the media of survivors as compared with control cells (Figure 5A). To determine whether survivor and repeat cells exhibited phenotypic changes consistent with increased cell migration, we performed a Transwell chemotaxis assay. Survivor cells were observed to have approximately twofold increase in migration toward epidermal growth factor (EGF) and repeat cells approximately threefold increase relative to control cells (Figure 5B). Survivor and repeat cells also exhibited a change in morphology consistent with a migratory phenotype, including increased formation of actin stress fibers and lamellipodia (Figure 5C). Cell proliferation was slightly reduced in day 1 survivor cells, but the proliferation rates of survivor cells were identical to those of naive cells within 3 d of outgrowth in TRAIL-free medium or after 3 d of “repeat” TRAIL exposure (Supplemental Figure S4, A and B). We conclude that cell state changes observed in survivor and repeat cells involve not only resistance to death ligands, but also inflammatory and migratory phenotypes such as elevated cytokine secretion, acquisition of an active morphology, and increased chemotactic activity.
FIGURE 5:
NF-κB mediates activation of inflammatory genes and phenotypes in survivor cells but does not mediate survival or reversible resistance. (A) ELISA of cytokine secretion (IL1A and CXCL1) in 8-h-conditioned media from MCF10A control, survivor, reset, and repeat cells. Cytokine secretion was normalized to the number of cells in each well, determined using the methylene blue cell viability assay. Error bars represent SE of replicate samples. (B) Migration assay of MCF10A cells toward EGF (100 ng/ml). Error bars indicate SE of triplicate wells; shown is a representative of three independent experiments. (C) Actin stress fiber staining of MCF10A cells (phalloidin, green; Hoechst 33342, blue). (D) NF-κB (p65) immunostaining of MCF10A ± IκBsr either untreated or treated with 50 ng/ml TNF or TRAIL for the indicated times. (E) qPCR analysis of SOD2 and IL1A gene expression in survivor cells compared with control cells and in survivor cells expressing IκBsr (IκBsr-Surv) compared with control cells expressing IκBsr (IκBsr-Cont), normalized to glyceraldehyde-3-phosphate dehydrogenase levels (mean ± SE of replicate samples). (F) ELISA of secreted IL1A in 8-h-conditioned medium from control, survivor, and repeat cells ± IκBsr expression. (G) Migration assay of repeat cells ± IκBsr expression. Number of migrated cells was counted and averaged for three independent fields. (H) Cell viability assay of control, survivors, IκBsr-Control, and IκBsr-Survivors treated with TRAIL for 6 h. (I) Cell viability assay of control, repeat, IκBsr-Control, and IκBsr-Repeat cells treated with TRAIL for 6 h. (J) Sensitivity of control MCF10A or MCF10A cells pretreated with TNF-α (100 ng/ml) for 18 h to a subsequent 6-h treatment with the indicated doses of TRAIL. Cell survival was measured using the methylene blue viability assay. (K) Percentage apoptosis in control and day 1 survivor cells either untreated (gray bars) or pretreated for 12 h with 6-h-conditioned media from another set of day 1 survivor cells (red bars) and then treated with 50 ng/ml TRAIL for 6 h. Apoptosis was assessed by staining with an antibody to cleaved PARP. (L) Fluorescently labeled control cells (control), fluorescently labeled survivor cells (survivors), or a 1:1 mixture of fluorescently labeled control cells and unlabeled survivor cells (mixture) either untreated or treated with 50 ng/ml TRAIL for 6 h. Dead cells were washed off, and fluorescence intensity was measured for each well using a fluorescence plate reader. Fraction of cells surviving was measured as a ratio of fluorescence intensity for each treated condition relative to its untreated control. The total cell density plated for each condition was held constant.
NF-κB mediates expression of inflammatory genes and phenotypes but not induced ligand resistance
The NF-κB transcription factor plays a central role in inflammatory signaling in general and survival signaling by TNF family members in particular. Many genes upregulated in survivor cells (Supplemental Tables S1 and S4) are known to be NF-κB targets (Pahl, 1999), and we observed NF-κB (p65) to translocate into the nucleus of TRAIL-treated MCF10A cells in a dose-dependent manner (TRAIL was less potent as an NF-κB inducer than TNF, in agreement with previous reports; Supplemental Figure S4, C and D; Lin et al., 2000; Varfolomeev et al., 2005). To determine whether NF-κB–mediated transcription was necessary for induction of reversible TRAIL resistance and/or inflammatory phenotypes, we stably transfected MCF10A cells with a nondegradable, dominant-negative IκBα (the IκB “superrepressor” [IκBsr]; Brown et al., 1995; Boehm et al., 2007). Clones in which TRAIL and TNF exposure failed to cause NF-κB nuclear translocation (Figure 5D) were analyzed for sensitivity to TRAIL and changes in gene expression. In IκBsr-expressing survivor cells nearly two-thirds of the genes up-regulated in parental survivor cells were no longer induced (Supplemental Tables S1, S5, and S6), including known NF-κB targets involved in inflammation and wounding, such as IL1A and SOD2 (array data were confirmed by qPCR; Figure 5E and Supplemental Figure S4E). This confirms that IκBsr expression effectively inhibits NF-κB–mediated transcription and that NF-κB is responsible for many of the gene expression changes in survivor cells. Cell migration and secretion of inflammatory cytokines were also suppressed in IκBsr-expressing survivor and repeat cells (Figure 5, F and G, and Supplemental Figure S4F), demonstrating that both processes are mediated in large part by NF-κB. Reversible resistance of survivor and repeat cells was not inhibited by IκBsr expression, however, nor were IκBsr-expressing naive cells rendered more sensitive to TRAIL than parental MCF10A cells (Figure 5, H and I). Furthermore, TNF pretreatment, which strongly activates NF-κB (Supplemental Figure S4, C and D), failed to protect MCF10A cells against subsequent TRAIL-induced death (Figure 5J and Supplemental Figure S4G). Thus, whereas the NF-κB pathway mediates TRAIL-induced inflammatory signaling in MCF10A survivor and repeat cells, resistance is NF-κB independent.
To test whether resistance is mediated by secreted factors, we incubated control cells for 12 h with conditioned medium from day 1 survivor cells and tested their sensitivity to TRAIL. We found that conditioned medium did not protect cells against TRAIL-induced death (Figure 5K). Moreover, coculture of control and survivor cells failed to protect the control cells after addition of TRAIL (Figure 5L). Finally, blocking the IL1A/B receptor (IL1R) with a neutralizing antibody failed to protect cells against induction of resistance or to sensitize survivor cells (Supplemental Figure S4H). Thus resistance is cell autonomous and not mediated by secreted factors. Because up-regulation of most secreted factors in survivor cells was NF-κB dependent (including IL1A/B; Supplemental Table S6), this further supports our conclusion that resistance and inflammatory phenotypes are mediated by two separate pathways.
Changes in the expression of individual genes do not explain reversible resistance
To narrow our search for genes whose change in expression might contribute to reversible resistance of survivor cells, we examined the list of genes up-regulated in both parental and IκBsr-expressing survivor cells (i.e., NF-κB–independent genes up-regulated in resistant cells; Supplemental Table S7). Of note, levels of most genes associated with apoptosis (e.g., Bcl-2 family members) did not significantly change in parental survivor or IκBsr survivor cells, and this was confirmed by Western blotting for various apoptosis regulators in parental control and survivor cells (Supplemental Figure S5A). Only three genes up-regulated in both survivors and IκBsr survivors were associated with the GO category “antiapoptosis.” Of these, the gene for FLIP (CFLAR) was the most interesting because of its known role in preventing apoptosis in response to death ligands (Sharp et al., 2005; Supplemental Tables S6 and S7). FLIP was also up-regulated approximately twofold at the protein level in survivor cells (Supplemental Figure S5B). FLIP mRNA expression was only slightly upregulated in repeat cells (below our cut-off threshold of 1.5-fold change), however, and we did not detect any up-regulation of FLIP protein in repeat cells (Supplemental Figure S5C). Consistent with this observation, when FLIP expression was knocked down in MCF10A cells using specific short hairpin RNA (shRNA) constructs, the cells were not sensitized to TRAIL, nor was reversible resistance inhibited in survivor cells (Supplemental Figure S5, D and E). However, induced cytokine secretion was inhibited in FLIP shRNA–expressing survivor cells (Supplemental Figure S5F), confirming a role for FLIP in NFκB-mediated inflammation (Kataoka and Tschopp, 2004).
The second “antiapoptosis” gene up-regulated in both survivor cells and IκBsr-expressing survivor cells is the cross-linking enzyme TGM-2, which was recently shown to mediate resistance to TRAIL-induced cell death (Frese-Schaper et al., 2010; Li et al., 2011). TGM-2 protein was up-regulated in both survivor and repeat MCF10A cells (Supplemental Figure S5C); however, cotreatment with the TGM-2 inhibitor cystamine only slightly sensitized naive and survivor cells to TRAIL-induced apoptosis (Supplemental Figure S6A). The third gene, HSPB1 (hsp27), was not upregulated in repeat cells (Supplemental Table S6). Thus we could not specifically link changes in the expression of known “resistance” genes with reversible resistance, and it is likely that this phenotype is mediated instead by changes at the protein level and/or by a multifactorial change in cell state.
Codrugging partially sensitizes resistant survivor cells
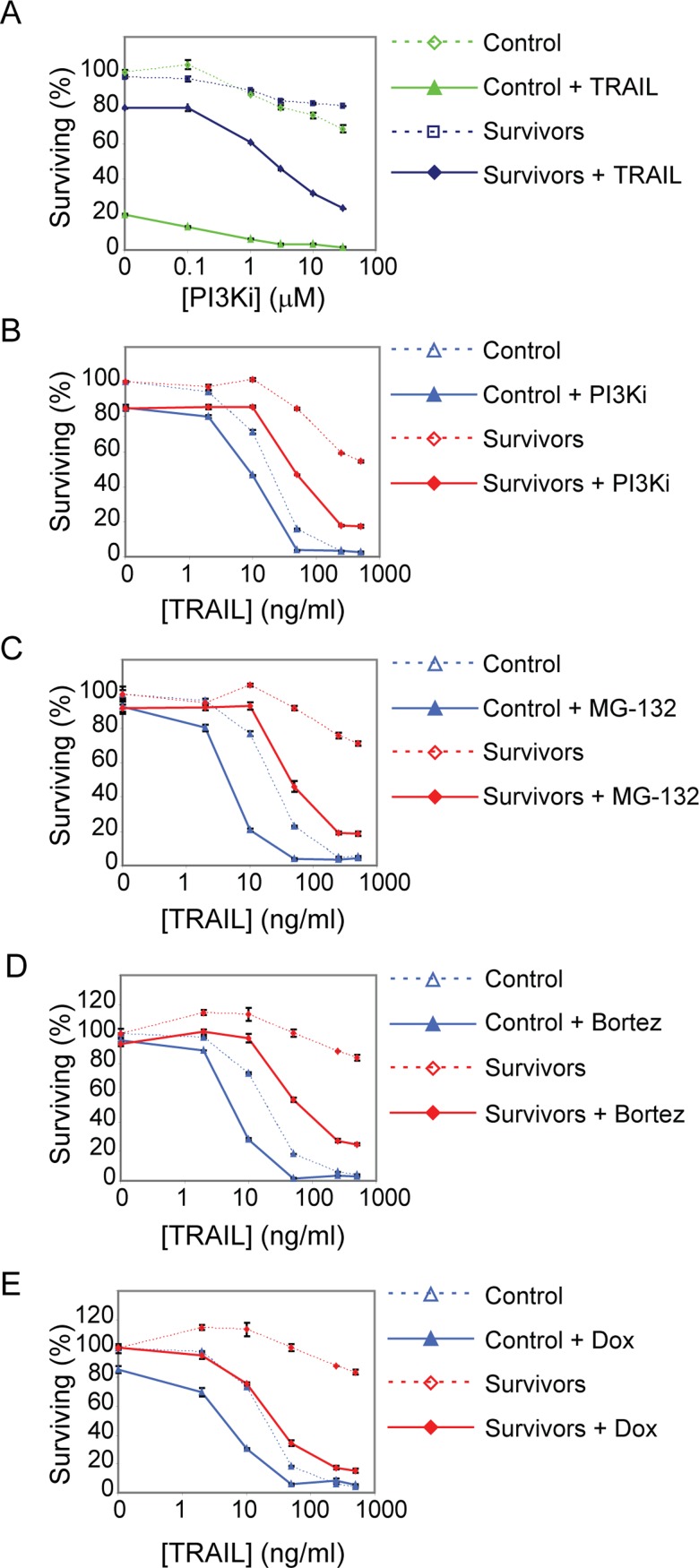
In addition to NF-κB, kinase signaling has been shown to protect cells against TRAIL-induced apoptosis, and codrugging with kinase inhibitors can sensitize resistant cells to TRAIL-mediated death (Lee et al., 2005; Zauli et al., 2005; Song et al., 2009; Son et al., 2010; Opel et al., 2011; Hellwig and Rehm, 2012). To determine whether kinase signaling was involved in reversible resistance, we treated MCF10A cells with inhibitors of several kinases previously shown to play a role in TRAIL signaling (Song et al., 2009; Sun et al., 2010b). Whereas inhibitors of mitogen-activated protein/extracellular signal-related kinase (MEK), p38, and JNK had little effect on MCF10A cells in combination with TRAIL at a range of doses, an inhibitor of phosphatidylinositol-3 kinase (PI3K; PI-103) was potent at sensitizing both naive and survivor MCF10A cells to TRAIL-induced death: <5% of naive cells survived TRAIL plus PI-103 treatment, as compared with 20% with TRAIL alone (Figure 6A and Supplemental Figure S6B); survivor cells were sensitized approximately threefold to fourfold to TRAIL (using a combination of 50 ng/ml TRAIL and 1–10 μM PI-103; Figure 6, A and B, and Supplemental Figure S6B). However, this level of killing was still lower than that of naive or reset cells treated with TRAIL alone (Figure 6B, red solid line and blue dotted line). Thus, even the most potent kinase inhibitor we assayed (PI-103) was less effective at killing survivor cells than simply waiting for a few days. This demonstrates that optimizing the interval between exposures to TRAIL can be at least as, if not more, effective a means to kill cells as “codrugging” with inhibitors of known survival pathways.
FIGURE 6:
Codrugging sensitizes survivor cells to TRAIL but is not as effective as waiting for cells to reset. (A) Cell viability plot of control and survivor cells treated with a range of doses of the PI3K inhibitor PI-103 with or without TRAIL (50 ng/ml) for 6 h. (B) Cell viability plot of control and survivor cells treated with a range of doses of TRAIL with or without PI-103 (1 μM) for 6 h. (C) Cell viability plot of control and survivor cells treated with a range of doses of TRAIL with or without MG-132 (1 μM) for 6 h. (D) Cell viability plot of control and survivor cells treated with a range of doses of TRAIL with or without bortezomib (0.1 μM) for 6 h. (E) Cell viability plot of control and survivor cells treated with a range of doses of TRAIL with or without doxorubicin (10 μM) for 6 h.
To see whether survivor cells could be sensitized to TRAIL using other drugs, we examined agents previously shown to sensitize resistant cell lines to TRAIL-mediated cell death (Johnstone et al., 2008). We observed that cotreating survivor cells with TRAIL plus gefitinib or TRAIL plus ABT-737 resulted in only a low level of killing (Supplemental Figure S6C). Cotreating the same cells with TRAIL plus the proteasome inhibitor MG-132 or bortezomib, however, had a much larger sensitizing effect (Figure 6, C and D, and Supplemental Figure 6, D and E), consistent with the known effect of these agents on many cellular targets (Menke et al., 2011). Doxorubicin also strongly sensitized survivor cells to TRAIL-mediated apoptosis, but etoposide had a much smaller effect (Figure 6E and Supplemental Figure S6F). Of note, the drugs we tested had a stronger effect on survivor cells than on control cells, demonstrating that it might be valuable to test codrugging approaches for their ability to reverse transiently resistant states rather than simply look for enhanced killing in the basal state.
Resistance is mediated by impaired DISC assembly and reduced cleavage of caspase-8 substrates
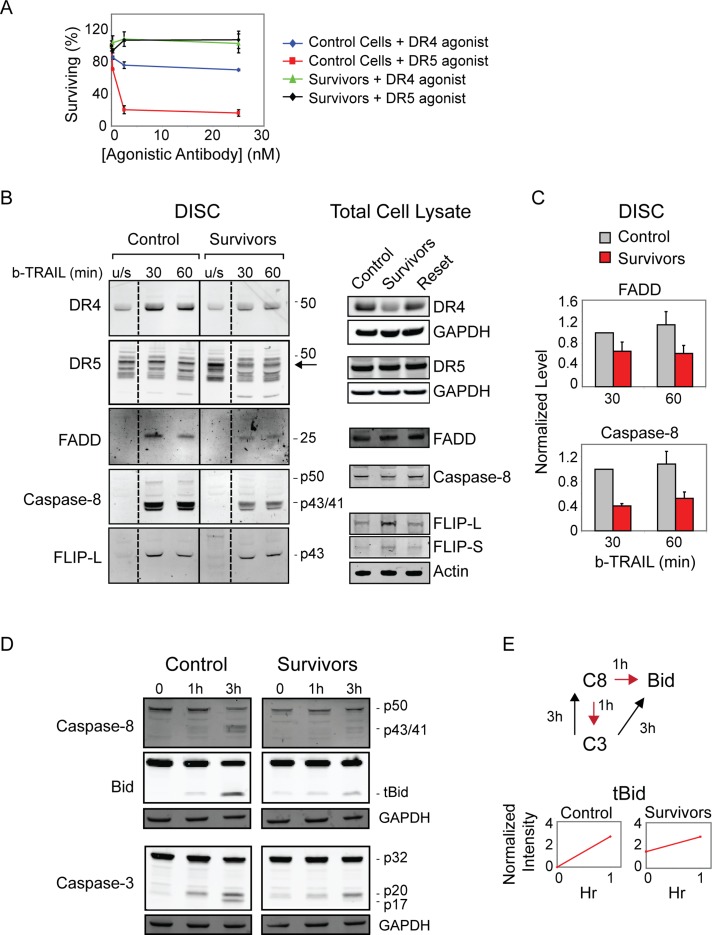
To explain resistance of survivor cells at the level of signaling, we examined different components of the apoptosis pathway. First we tested the role of receptor down-regulation or degradation, phenomena responsible for ligand-mediated adaptation by many transmembrane receptors (Le Roy and Wrana, 2005), including death ligands (Yoshida et al., 2009; Song et al., 2010). When cell-surface expression of DR4/5 receptors was measured in control and survivor cells by flow cytometry, however (using antibodies previously shown to be highly specific; Wagner et al., 2007), we detected only modest changes: cell surface levels of DR5 were unchanged in survivor cells, and levels of DR4 were reduced less than twofold (Supplemental Figure S7A). Levels of the two decoy receptors, DcR1 and DcR2, were also unchanged (Supplemental Figure S7B). DR5 is believed to be the primary TRAIL receptor in MCF10A cells (Herrero-Martin et al., 2009), and we found MCF10A cells to be threefold more sensitive to saturating concentrations of anti-DR5 agonist antibodies than anti-DR4 antibodies (Pukac et al., 2005; Adams et al., 2008; Figure 7A, red and blue lines). Moreover, TRAIL survivors were highly resistant to both agonists (Figure 7A, green and black lines), demonstrating that the observed twofold reduction in DR4 levels in survivor cells is not sufficient to explain resistance to TRAIL. TRAIL survivors were also cross-resistant to FasR agonists (as described earlier), without any detectable change in the levels of FasR (Supplemental Figure S7C), further arguing that changes in receptor expression do not explain reversible resistance to death ligands.
FIGURE 7:
Impaired DISC signaling mediates resistance of survivor cells. (A) Cell viability assay of MCF10A control and survivor cells treated for 6 h with agonist DR4 or DR5 antibodies. (B) Immunoblot of precipitated DISC proteins in control and survivor cells stimulated with biotinylated-TRAIL (500 ng/ml). For unstimulated (u/s) controls, biotinylated-TRAIL was added directly to cell lysates before pull down. The arrow marks the DR5 band corresponding to the single band observed by immunoblot analysis of total proteins (right). (C) Quantitation of DISC protein levels in B normalized to control cells at 30 min. (D) Immunoblot detection of caspase-8, Bid, and caspase-3 in control and survivor cells treated with TRAIL (50 ng/ml) for the indicated times. Dead cells were collected and lysed together with live cells. (E) Normalized intensity of tBid bands in D representing the accumulation of tBid cleaved by caspase-8 during the pre-MOMP interval (0–1 h, indicated by red arrows in schematic) in control and survivor cells. At the 3-h time point, feedback from caspase-3 amplifies the amount of cleaved substrates in dying cells (black arrows in schematic) and is therefore excluded from the quantitation.
To measure the ability of ligand-bound DR4/5 receptors to assemble functional DISC complexes, we used a pull-down assay involving biotinylated TRAIL (b-TRAIL; Harper and MacFarlane, 2008). As described earlier, DR4 was less abundant in survivors than naive cells, but overall levels of DR5 receptor were nearly identical (Figure 7B and Supplemental Figure S7D). After stimulation with b-TRAIL, levels of DISC-bound FADD adaptor protein and cleaved caspase-8 were approximately twofold to threefold lower in survivors than in control cells, demonstrating less efficient DISC formation in survivors (Figure 7, B and C). DISC-associated FLIP levels were constant in control and survivor cells, perhaps because the protein was up-regulated approximately twofold in the latter (Figure 7B and Supplemental Figure S5B). Consistent with these changes, cleavage of caspase-8 and its substrate Bid was reduced in survivor cells. Whereas naive MCF10A cells processed Bid to tBid within 1 h of TRAIL treatment, survivors contained residual tBid but little further tBid production (1 h is the most relevant time point for measuring receptor-mediated activation of initiator C8/10 because caspase feedback loops are not yet active; Albeck et al., 2008; Figure 7, D and E). Cleavage of caspase-3 was also reduced in survivors compared with control cells, with survivor cells exhibiting some cleavage of caspase-3 to the p20 fragment but not the p17 fragment, indicating pre-MOMP rather than post-MOMP cleavage (Albeck et al., 2008). These changes are consistent with the hypothesis that DISC formation and signaling from the DISC to the mitochondria are impaired in survivor cells in a manner that is common to both classes of death receptors.
DISCUSSION
Across a range of cell lines with different intrinsic sensitivities to TRAIL or FasL, a fraction of cells survives exposure to death ligands. The number of dying cells varies with dose and cell line, but fractional response is a fundamental feature of apoptosis. We previously ascribed fractional response in naive cell populations to stochastic fluctuations in protein abundance and activity (Spencer et al., 2009). Moreover, we showed that cells in a clonal population that survive fractional killing regenerate both the sensitivity and the death-time distribution of the starting population, in agreement with a role for stochastic fluctuations in cell state and protein “resetting” (Flusberg et al., 2013). In this article we investigate the properties of cells that survive initial exposure to TRAIL or anti-FasR antibodies and ask how these survivors respond to subsequent treatment with death ligand at various times. We find that survivor cells are highly resistant to TRAIL applied 24 h later and that their transcriptional profiles differ significantly from those of pretreatment (naive) cells. When survivor cells are cultured in the absence of ligand for ∼3–7 d, however, their transcriptional profiles return to the pretreatment state and cells exhibit the same fractional sensitivity to death ligands as naive cells. Although TRAIL survivors (on day 1) are highly and reversibly cross-resistant to killing by activators of the Fas pathway and vice versa, this property does not extend to inducers of intrinsic apoptosis such as staurosporine. Moreover, resistance can be sustained for 1 wk or more by periodically exposing cells to TRAIL without any cell killing (and is induced in naive cells even when death is blocked by caspase inhibitors). Like survivor cells, such “repeat” treatment cells regain TRAIL sensitivity when allowed to recover for several days in the absence of death ligand. Thus, cells in a TRAIL-sensitive population can enter a highly resistant state that is maintained by periodic exposure to TRAIL but resets to the drug-naive state over several days in the absence of ligand. Among the cell lines we studied, reversible resistance is most prominent for transformed and nontransformed breast epithelia, but it is also observed to a greater or lesser extent with other cell types. These observations have significant implications for maximizing cell killing by therapeutic agents targeting TRAIL receptors.
In addition to TRAIL insensitivity, survivor cells exhibit altered cell morphology, increased motility, induced secretion of cytokines, and elevated expression of inflammatory genes. These responses have been shown to be features of TRAIL or FasL responses in cell lines resistant to apoptosis (Barnhart et al., 2004; Ishimura et al., 2006; Trauzold et al., 2006; Chen et al., 2010; Li et al., 2011). We show that similar morphological and inflammatory changes are also exhibited by the relatively resistant subpopulation of cells in a TRAIL-sensitive cell line. Moreover, many of the genes induced by TRAIL in survivor cells are similar to those previously shown to be activated by TNF in epithelial cells, representing a response to injury that both protects against future insult (through inhibition of apoptosis) and initiates healing of the injury through activation of cytokines, cellular migration, and differentiation (Banno et al., 2004, 2005). It is possible that transient TRAIL and FasL resistance is adaptive in the same way. Motile and invasive phenotypes and inflammatory gene expression are undesirable properties for an anticancer drug, however, particularly if these properties are sustained by successive treatments.
Origins of reversible resistance
How do cells become transiently resistant to death ligand? A simple possibility is internalization and degradation of cell-surface receptors, phenomena common to hormone and growth factor receptors (Le Roy and Wrana, 2005) that are reported to dampen responsiveness to TRAIL (Austin et al., 2006; Kohlhaas et al., 2007; Zhang et al., 2009; Song et al., 2010). We detect small changes in the levels of cell-surface DR4 in TRAIL survivors, but DR5 levels do not change. Because survivors are resistant to anti-DR5 agonist antibodies, DR4 degradation or internalization is unlikely to explain transient TRAIL resistance. Moreover, resistance to TRAIL can be induced by FasR agonists (and vice versa), and there is no evidence for changes in DR4/5 abundance under these conditions. A second straightforward explanation for reversible resistance, namely inhibition of the core apoptosis machinery, is also ruled out: survivor cells can be effectively killed with TRAIL in the presence of cycloheximide and are sensitive to inducers of intrinsic cell death such as staurosporine and doxorubicin.
Some studies suggest that NF-κB signaling plays an important role in inhibiting TRAIL- or Fas-mediated apoptosis (Jeremias and Debatin, 1998; Neumann et al., 2010), but others suggest that it is relatively unimportant (Ganten et al., 2005; Diessenbacher et al., 2008). TRAIL does induce a strong NF-κB–dependent transcriptional response in MCF10A cells, and blocking NF-κB activation with IκBsr prevents induction of three-fourths of the genes up-regulated in survivors (many of which are known NF-κB targets; Pahl, 1999). This block largely inhibits acquisition of motile and inflammatory phenotypes, consistent with reports in other cell types (Ehrhardt et al., 2003; Li et al., 2003; Ishimura et al., 2006; Malhi and Gores, 2006; Trauzold et al., 2006; Tang et al., 2009). IκBsr expression does not alter the sensitivity of naive cells to TRAIL, however, nor does it impair the acquisition of reversible TRAIL resistance. Thus, in MCF10A cells NF-κB plays an important role in TRAIL-mediated inflammation but not in the regulation of cell death: NF-κB–dependent signaling is unmasked in survivor cells but does not itself cause resistance.
Examination of NF-κB–independent changes in gene expression in survivor and repeat cells reveals a very short list of genes that are up-regulated in both situations. The majority of these genes are involved in epidermal tissue repair and have been reported to be regulated by TNF in epithelial cells (Banno et al., 2004). Of interest, epidermal differentiation has also been associated with TRAIL resistance (Jansen et al., 2003). Cell cycle genes comprise the largest class of genes down-regulated in survivor cells (and in IκBsr survivor cells; Supplemental Table S1), and slow growth rates were associated with drug resistance in previous studies (Balaban et al., 2004; Roesch et al., 2010; Sharma et al., 2010). Repeat cells exhibit proliferation rates and expression of cell cycle genes equivalent to control cells, however, demonstrating that cell cycle does not mediate the reversibly resistant phenotype, although it might predispose cells initially to survival. Knockout of at least some antiapoptosis genes on the list of coordinately regulated genes (e.g., CFLAR) does not inhibit the acquisition of transient resistance, and we speculate that multifactorial changes associated with differentiation and with a reduced capacity for functional DISC formation are a likely explanation for the resistance phenotype.
At the level of signaling, we found that a ligand-induced change in DISC activity is associated with reversible resistance to TRAIL and FasR agonists. The composition of the DISC clearly differs between naive and survivor cells, with twofold to threefold less DISC-bound FADD and caspase-8 in survivor than control cells. Because DISCs assembled on DR4/5 and FasR are very similar (Wilson et al., 2009), changes in the abundance or activity of DISC components would account for cross-resistance of TRAIL survivors to FasR agonists and vice versa. In contrast, control and survivor cells are equally competent to undergo MOMP when exposed to inducers of intrinsic cell death (Figure 2, B and D, and Supplemental Figure S7F). Several studies demonstrated that subtle changes in DISC composition can affect the balance between death and survival signaling (Muppidi et al., 2004; Varfolomeev et al., 2005; Lavrik et al., 2007; Kleber et al., 2008; Hughes et al., 2009; Fricker et al., 2010; Neumann et al., 2010). FLIP is one such protein that has been shown to shift life–death outcomes in cells exposed to death ligands (Chang et al., 2002; Song et al., 2007a; Fricker et al., 2010; Neumann et al., 2010), and we observed that FLIP was up-regulated in survivor cells; however, knockdown of FLIP using shRNA was not sufficient to prevent reversible resistance in MCF10A cells. Other changes in survivor cells leading to reduced DISC activity could include those affecting receptor modification or aggregation state (Tang et al., 2006; Feig et al., 2007; Song et al., 2007a; Wagner et al., 2007; Rossin et al., 2009; Mazurek et al., 2011), reduced affinity of DISC components for intracellular tails of receptors (Harper et al., 2003), or increased activity of inhibitory kinase pathways (Varfolomeev et al., 2005; Kleber et al., 2008; Yan et al., 2013). Although the precise molecular mechanism awaits further testing, we postulate that these changes are likely to be multifactorial: for example, total protein expression of both caspase-8 and FADD were lower in survivor cells under certain lysis conditions (Supplemental Figure S7E). In further support of this idea, proteasome inhibition sensitizes survivor cells to TRAIL and was previously shown to reverse multifactorial acquired TRAIL resistance (Menke et al., 2011). In principle, proteasome inhibition could sensitize cells via stabilization of proapoptotic proteins such as caspase-3 and tBid (Breitschopf et al., 2000; Albeck et al., 2008), thereby increasing the likelihood of death even when these proteins are activated more slowly due to reduced DISC function, or through direct alteration of the DISC (Sayers and Murphy, 2006; Brooks et al., 2010). The PI3K/mTOR inhibitor PI-103 was similarly shown to sensitize resistant cell types to TRAIL via alteration in the levels and phosphorylation states of multiple downstream proteins, effectively “priming” cells and reducing the threshold for cell death (Opel et al., 2011). Thus transient resistance to TRAIL has some of the hallmarks of conventional acquired resistance and can in principle be reversed using some of the same treatments. No cotreatment, however, appears as effective as simply waiting several days for cells to reset. We previously showed that relatively small but coordinated changes in protein levels are sufficient to account for variability in cell fate, time to death, and whether a cell undergoes type I or type II apoptosis (Aldridge et al., 2011; Gaudet et al., 2012), and we hypothesize that similar subtle but coordinated changes in DISC composition are sufficient to explain reversible TRAIL resistance. We are developing a quantitative computational model of DISC biochemistry to test this hypothesis.
The role of preexisting versus induced variability in cellular responses to TRAIL
We previously demonstrated that preexisting differences in the levels or activities of apoptotic regulators among cells in a clonal population determine the time and probability of TRAIL-induced death (Spencer et al., 2009). These differences are believed to arise from the stochastic fluctuations that are a fundamental feature of the reactions involved in synthesis and degradation of proteins (Sigal et al., 2006; Raj and van Oudenaarden, 2008; Eldar and Elowitz, 2010). Stochastic fluctuations impinging on the transcriptional states of cells can also give rise to changes in fate and cell-to-cell variation in drug sensitivity (Sharma et al., 2010; Singh et al., 2010; Gupta et al., 2011). These findings raised the question of whether reversible resistance is induced de novo by death ligand treatment or preexists in a subset of naive cells, as might be predicted from time scales of protein “remixing” (Sigal et al., 2006). Clearly, preexisting differences affect whether cells live or die when first exposed to death ligands. However, two lines of evidence support the idea that the reversibly resistant state we observe is induced and is not merely a result of preexisting differences among cells: first, all cells enter the resistant state when cell death is blocked using a caspase inhibitor, and second, the resistant state is maintained in the absence of cell killing by exposing cells to periodic doses of TRAIL. Transcriptional profiling reveals that such “repeat” cells enter a state that is in between that of survivor and naive cells in gene expression space.
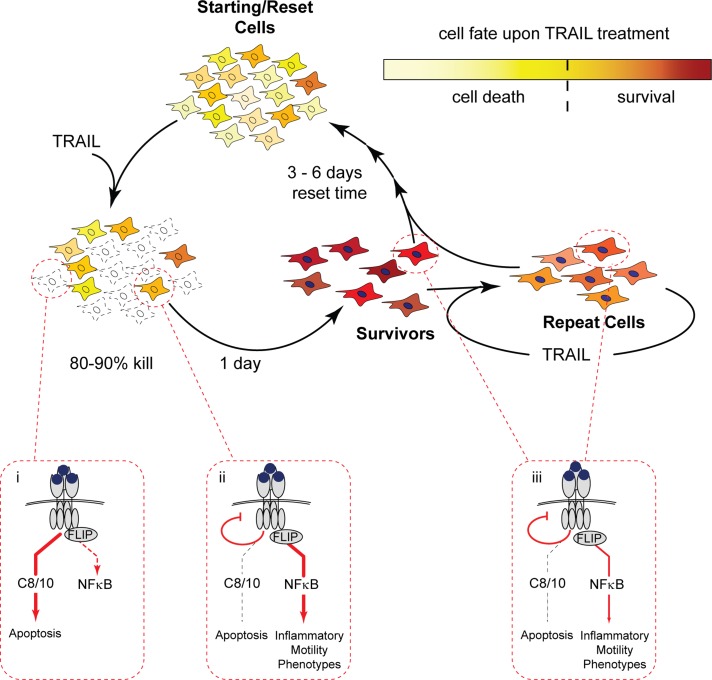
The distinction between preexisting difference and induced difference may not be so simple, however: the extent to which survival pathways are activated will itself be subject to cell-to-cell variability. Cohen et al. (2008) showed, for example, that fractional killing of cancer cells by a topoisomerase inhibitor involves differential induction of survival pathways in some cells and not others, and this may also be true of death ligands (Zauli et al., 2005; Neumann et al., 2010). The reversible resistance induced by a 6-h treatment with TRAIL plus caspase inhibitor was less than that induced by an 18-h treatment under the same conditions, whereas 6 h was sufficient for full resistance in the absence of caspase inhibition (Supplemental Figure S2C). We ascribe this difference to the fact that both induced survival and selection are at work in the latter case. Thus both preexisting and induced differences are likely to play a role in reversible resistance to TRAIL, and the two are almost certainly interwoven: stochastic variation in the levels of proapoptotic and antiapoptotic factors is expected to determine the rates at which competing death and survival mechanisms act on effector caspases in individual cells that are more or less prone to die. Cells that are relatively resistant due to natural fluctuation will be pushed into a highly resistant state by TRAIL-induced changes that affect DISC signaling; activation of an NF-κB–mediated inflammatory response is then unmasked in these cells. In the case of repeated TRAIL treatments, feedback loops are presumed to sustain resistance and inflammatory phenotypes as long as the stimulus is administered more frequently than the natural decay time of the resistant state (Figure 8).
FIGURE 8:
TRAIL induces reversible resistance and inflammatory pathways in cells that survive an initial treatment. In this schematic, yellow shadings depict nongenetic heterogeneity in protein levels or other factors in a naive cell population. After treatment, the sensitive fraction of cells dies by apoptosis via a caspase-8/10 (C8/10) pathway, and the less sensitive fraction survives (dark yellow cells). TRAIL-induced NF-κB signaling is activated in both sensitive and resistant cells but is cut short in cells that die (see blow-ups i and ii). Within hours, survivor cells activate a transcriptional program and enter a state of induced reversible resistance whose peak lasts for ∼24 h (survivors; red cells, filled nuclei); resistance involves attenuated DISC assembly that prevents activation of sufficient C8/10 to initiate apoptosis when cells are retreated with TRAIL (blow-up ii). Resistant cells exhibit activation of a FLIP-dependent, NF-κB–mediated inflammatory response, although resistance is independent of both NF-κB and FLIP. When TRAIL is removed, survival and inflammation signals decay as cells divide, and within several days protein levels redistribute such that the new cell population is equivalent to the starting control population (reset cells). In contrast, if survivors are reexposed to TRAIL treatment during the resistance stage, resistance and inflammatory phenotypes are sustained (repeat cells). NF-κB activation is submaximal upon repeated treatment of survivor/repeat cells due to attenuated DISC assembly but is sufficient to sustain inflammatory phenotypes (blow-up iii). Repeat cells (orange shading) have an intermediate gene expression profile that has characteristics of both control and survivor cells.
The existence of reversible adaptations to therapeutic drugs is an emerging theme in cancer biology (Sabnis et al., 2008; Gupta et al., 2010; Sharma et al., 2010; Muranen et al., 2012), and it seems probable that such responses are a major factor limiting the effectiveness of chemotherapy. “Preconditioning” or “tolerance” effects ascribed to TNF-family proteins may also play a role in protecting surviving cells from exposure to a future insult (Jang et al., 2008; Rakoff-Nahoum and Medzhitov, 2009). We have observed that the most effective method for overcoming reversible resistance in cultured cells is simply to wait for the resistant state to decay. If this holds true in animals and patients, it has implications for the optimal pharmacokinetics of TRAIL-like drugs and the interval between successive drug doses. Alternatively, it may suggest the need to identify drugs such as bortezomib that, when used in combination with TRAIL, can minimize confounding prosurvival responses.
MATERIALS AND METHODS
Cell lines and materials
MCF10A cells were obtained from J. Brugge (Harvard Medical School, Boston, MA) and were cultured as described (Debnath et al., 2003). HCT116 cells were obtained from the American Type Culture Collection (Manassas, VA); single-cell clones were generated by serial dilution and were cultured as described (Aldridge et al., 2011). MCF10A cells stably expressing the IκB superrepressor (Boehm et al., 2007) were generated using a retrovirus produced by cotransfection (FuGENE 6 Transfection Reagent; Roche, Indianapolis, IN) of 293T cells (American Type Culture Collection) with pCL-ampho and pBabe-Puro-IκBalpha-mut (plasmid 15291; Addgene, Cambridge, MA) and selection with puromycin. Single-cell clones generated by serial dilution were tested for complete (<0.1%) lack of nuclear translocation of NF-κB after stimulation with TNFα (PeproTech, Rocky Hill, NJ). SuperKiller TRAIL and Apo-1-3 were obtained from Alexis Biochemicals (San Diego, CA). Agonistic DR4 and DR5 antibodies were a gift from Merrimack Pharmaceuticals (Cambridge, MA). Staurosporine and doxorubicin were obtained from Sigma-Aldrich (St. Louis, MO), PI-103 from Cayman Chemical Company (Ann Arbor, MI), caspase-8 inhibitor IETD-fmk and MG-132 from EMD Biosciences (San Diego, CA), and bortezomib from Santa Cruz Biotechnology (Santa Cruz, CA). Biotinylated TRAIL was a gift from M. MacFarlane (University of Leicester, Leicester, United Kingdom). Additional materials can be found in the Supplemental Materials and Methods.
Challenge–recover (survivors) experiment
MCF10A cells were treated with 50 ng/ml SuperKiller TRAIL for 6 h. Dead cells were washed off, and surviving cells were collected by trypsinization and replated without TRAIL. On subsequent days cells were rechallenged with TRAIL and analyzed for extent of cell death; see Supplemental Materials and Methods for additional details.
Cell death measurements
For detection of cleaved PARP by flow cytometry, live and dead cells were combined and analyzed as previously described (Albeck et al., 2008). For cell viability (cell count) measurements, dead cells were washed off and live cells were labeled with 5 mg/ml methylene blue (MB; Sigma-Aldrich) in 50% EtOH, rinsed with H2O, and air dried. MB was dissolved in 10 mg/ml sarcosine (Sigma-Aldrich) and 620 – 405 nm absorbance was read on a Victor3V Platereader (PerkinElmer, Waltham, MA); see Supplemental Materials and Methods for details. For annexin V measurements, cells were labeled with annexin V and Sytox Green according to the manufacturer's protocol (Molecular Probes, Eugene, OR) and analyzed by flow cytometry on a FACSCalibur (BD Biosciences, San Diego, CA).
Cell migration assay
Fluorescently labeled MCF10A cells were seeded in the top chambers of a 96-well, 8-μm-pore Fluoroblok Transwell cell migration device (BD Biosciences) in MCF10A media lacking EGF. Cell migration toward media containing 100 ng/ml EGF in the bottom chamber was detected after 24 h by measuring fluorescence using a Victor3V Platereader or by counting the number of migrated cells. Samples were run in triplicate, and the experiment was repeated at least twice.
Cytokine analysis
Control, survivor, reset, and repeat were replenished with fresh media 1 d after seeding. The 8-h-conditioned medium was collected, and cytokine secretion was analyzed by ELISA (Quantikine human CXCL1 immunoassay, R&D Systems, Minneapolis, MN; and Human IL1A ELISA, Biolegend, San Diego, CA). Concentration of secreted cytokines was normalized to cell density determined by staining cells with methylene blue.
Gene expression microarray
RNA was isolated using a NucleoSpin RNA II kit (Macherey-Nagel, Bethlehem, PA). Biotin-labeled cRNA was generated from reverse-transcribed cDNA, hybridized onto Illumina Human Ref-8 Beadchip arrays (Illumina, San Diego, CA), stained with Cy3-streptavidin, and scanned on an Illumina BeadArray Reader (Microarray Core Facility, Children's Hospital, Boston, MA). Duplicate samples of control, survivors, reset, and repeat cells from three individual experiments (at least six replicates of each condition) and duplicate samples of IκB superrepressor–expressing control and survivor cells were submitted for analysis. See Supplemental Materials and Methods for further details. Tables of raw and normalized data can be found in the National Center for Biotechnology Information's Gene Expression Omnibus (GEO; Edgar et al., 2002) and are accessible through GEO Series Accession Number GSE33340 (www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE33340). Full lists of differentially expressed genes from several different analyses are given in Supplemental Table S1.
Western blotting
Cells were lysed in Triton lysis buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 10% glycerol, 1% Triton X-100) or in RIPA buffer containing Mini Complete protease inhibitors (EDTA-free; Roche Molecular Biochemicals, Indianapolis, IN). Equal protein amounts determined using a BCA assay (Pierce, Rockford, IL) were boiled in sample buffer (50 mM Tris, pH 6.8, 10% glycerol, 2% SDS, 0.004% bromphenol blue, 2% β-mercaptoethanol), separated by SDS–PAGE, and transferred onto polyvinylidene fluoride or nitrocellulose membranes. Membranes were blocked with Odyssey Blocking Buffer (LI-COR Biosciences, Lincoln, NE) and probed with primary antibodies at 4°C overnight. Information on antibodies and detection is provided in the Supplemental Materials and Methods.
DISC pull down
DISC precipitations were performed using biotin-labeled human recombinant TRAIL (b-TRAIL) as previously described (Harper and MacFarlane, 2008). MCF10A cells were treated with b-TRAIL (500 ng/ml) for the indicated times (30 or 60 min); unbound b-TRAIL was removed by washing the cells three times with ice-cold PBS. Cells were resuspended in Triton lysis buffer and lysed for 60 min on ice, followed by centrifugation at 15,000 × g for 30 min at 4°C. For unstimulated control cells, b-TRAIL was added directly to the lysates of untreated cells. Receptor complexes were precipitated from samples containing equal amounts of protein (bicinchoninic acid assay; Pierce) by incubation with 40 μl of streptavidin-coated magnetic beads (Dynabeads, Invitrogen, Carlsbad, CA) at 4°C overnight. Precipitates were washed with lysis buffer, and receptor complexes were eluted with sample buffer and analyzed by Western blot.
Supplementary Material
Acknowledgments
We thank A. Ashkenazi (Genentech, South San Francisco, CA), M. MacFarlane, W. Hahn, T. Bagci-Onder, K. Shah, J. Brugge, I. Lavrik, and Merrimack Pharmaceuticals (Cambridge, MA) for reagents; and T. Vo, A. Letai, H. Nguyen, B. Millard, J. Sims, and V. Becker for technical assistance and helpful discussions. Microarray studies were performed by the Molecular Genetics Core Facility at Children's Hospital Boston, which is supported by NIH-P50-NS40828 and NIH-P30-HD18655. For assistance with microarray data analysis, we thank Charlie Whittaker from the Koch Institute Bioinformatics and Computing Core Facility (Cambridge, MA), who is supported in part by Cancer Center Support (Core) Grant P30-CA14051 from the National Cancer Institute; and Oliver Hofmann, whose contribution was supported by National Institutes of Health Award UL1 RR 025758. This work was supported by National Institutes of Health Grant P01-CA139980 to P.K.S. and National Institutes of Health Pre-doctoral Training Grant GM07226.
Abbreviations used:
- b-TRAIL
biotinylated TRAIL
- C3/7
caspase-3/7
- C8/10
caspase-8/10
- cPARP
cleaved PARP
- DISC
death-inducing signaling complex
- EGF
epidermal growth factor
- ELISA
enzyme-linked immunosorbent assay
- ERK
extracellular signal-regulated protein kinase
- GO
Gene Ontology
- IkBa
inhibitor of kappa B alpha
- IkBsr
IkBa superrepressor
- IKK
IkB kinase
- IL1R
IL1A/B receptor
- JNK
Jun-N-terminal kinase
- MEK
mitogen-activated protein/extracellular signal-related kinase
- MOMP
mitochondrial outer membrane permeabilization
- PCA
principal components analysis
- PI3K
phosphatidylinositol-3 kinase
- qPCR
quantitative PCR
- shRNA
short hairpin RNA
- TNF
tumor necrosis factor
- TRAIL
TNF-related apoptosis-inducing ligand
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E12-10-0737) on May 22, 2013.
REFERENCES
- Adams C, et al. Structural and functional analysis of the interaction between the agonistic monoclonal antibody Apomab and the proapoptotic receptor DR5. Cell Death Differ. 2008;15:751–761. doi: 10.1038/sj.cdd.4402306. [DOI] [PubMed] [Google Scholar]
- Albeck JG, Burke JM, Aldridge BB, Zhang M, Lauffenburger DA, Sorger PK. Quantitative analysis of pathways controlling extrinsic apoptosis in single cells. Mol Cell. 2008;30:11–25. doi: 10.1016/j.molcel.2008.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldridge BB, Gaudet S, Lauffenburger DA, Sorger PK. Lyapunov exponents and phase diagrams reveal multi-factorial control over TRAIL-induced apoptosis. Mol Syst Biol. 2011;7:553. doi: 10.1038/msb.2011.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashkenazi A. Targeting the extrinsic apoptosis pathway in cancer. Cytokine Growth Factor Rev. 2008;19:325–331. doi: 10.1016/j.cytogfr.2008.04.001. [DOI] [PubMed] [Google Scholar]
- Ashkenazi A, Dixit VM. Apoptosis control by death and decoy receptors. Curr Opin Cell Biol. 1999;11:255–260. doi: 10.1016/s0955-0674(99)80034-9. [DOI] [PubMed] [Google Scholar]
- Ashkenazi A, Herbst RS. To kill a tumor cell: the potential of proapoptotic receptor agonists. J Clin Invest. 2008;118:1979–1990. doi: 10.1172/JCI34359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin CD, et al. Death-receptor activation halts clathrin-dependent endocytosis. Proc Natl Acad Sci USA. 2006;103:10283–10288. doi: 10.1073/pnas.0604044103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. Bacterial persistence as a phenotypic switch. Science. 2004;305:1622–1625. doi: 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- Banno T, Gazel A, Blumenberg M. Effects of tumor necrosis factor-alpha (TNF alpha) in epidermal keratinocytes revealed using global transcriptional profiling. J Biol Chem. 2004;279:32633–32642. doi: 10.1074/jbc.M400642200. [DOI] [PubMed] [Google Scholar]
- Banno T, Gazel A, Blumenberg M. Pathway-specific profiling identifies the NF-kappa B-dependent tumor necrosis factor alpha-regulated genes in epidermal keratinocytes. J Biol Chem. 2005;280:18973–18980. doi: 10.1074/jbc.M411758200. [DOI] [PubMed] [Google Scholar]
- Bargmann CI, Hung MC, Weinberg RA. Multiple independent activations of the neu oncogene by a point mutation altering the transmembrane domain of p185. Cell. 1986;45:649–657. doi: 10.1016/0092-8674(86)90779-8. [DOI] [PubMed] [Google Scholar]
- Barnhart BC, Alappat EC, Peter ME. The CD95 type I/type II model. Semin Immunol. 2003;15:185–193. doi: 10.1016/s1044-5323(03)00031-9. [DOI] [PubMed] [Google Scholar]
- Barnhart BC, Legembre P, Pietras E, Bubici C, Franzoso G, Peter ME. CD95 ligand induces motility and invasiveness of apoptosis-resistant tumor cells. EMBO J. 2004;23:3175–3185. doi: 10.1038/sj.emboj.7600325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhola PD, Simon SM. Determinism and divergence of apoptosis susceptibility in mammalian cells. J Cell Sci. 2009;122:4296–4302. doi: 10.1242/jcs.055590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm JS, et al. Integrative genomic approaches identify IKBKE as a breast cancer oncogene. Cell. 2007;129:1065–1079. doi: 10.1016/j.cell.2007.03.052. [DOI] [PubMed] [Google Scholar]
- Breitschopf K, Zeiher AM, Dimmeler S. Ubiquitin-mediated degradation of the proapoptotic active form of bid. A functional consequence on apoptosis induction. J Biol Chem. 2000;275:21648–21652. doi: 10.1074/jbc.M001083200. [DOI] [PubMed] [Google Scholar]
- Brooks AD, Jacobsen KM, Li W, Shanker A, Sayers TJ. Bortezomib sensitizes human renal cell carcinomas to TRAIL apoptosis through increased activation of caspase-8 in the death-inducing signaling complex. Mol Cancer Res. 2010;8:729–738. doi: 10.1158/1541-7786.MCR-10-0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown K, Gerstberger S, Carlson L, Franzoso G, Siebenlist U. Control of I kappa B-alpha proteolysis by site-specific, signal-induced phosphorylation. Science. 1995;267:1485–1488. doi: 10.1126/science.7878466. [DOI] [PubMed] [Google Scholar]
- Chang DW, Xing Z, Pan Y, Algeciras-Schimnich A, Barnhart BC, Yaish-Ohad S, Peter ME, Yang X. c-FLIP(L) is a dual function regulator for caspase-8 activation and CD95-mediated apoptosis. EMBO J. 2002;21:3704–3714. doi: 10.1093/emboj/cdf356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhary PM, Eby M, Jasmin A, Bookwalter A, Murray J, Hood L. Death receptor 5, a new member of the TNFR family, and DR4 induce FADD-dependent apoptosis and activate the NF-kappaB pathway. Immunity. 1997;7:821–830. doi: 10.1016/s1074-7613(00)80400-8. [DOI] [PubMed] [Google Scholar]
- Chen L, et al. CD95 promotes tumour growth. Nature. 2010;465:492–496. doi: 10.1038/nature09075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen AA, et al. Dynamic proteomics of individual cancer cells in response to a drug. Science. 2008;322:1511–1516. doi: 10.1126/science.1160165. [DOI] [PubMed] [Google Scholar]
- Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30:256–268. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- Deng Y, Lin Y, Wu X. TRAIL-induced apoptosis requires Bax-dependent mitochondrial release of Smac/DIABLO. Genes Dev. 2002;16:33–45. doi: 10.1101/gad.949602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveraux QL, Takahashi R, Salvesen GS, Reed JC. X-linked IAP is a direct inhibitor of cell-death proteases. Nature. 1997;388:300–304. doi: 10.1038/40901. [DOI] [PubMed] [Google Scholar]
- Diessenbacher P, et al. NF-kappaB inhibition reveals differential mechanisms of TNF versus TRAIL-induced apoptosis upstream or at the level of caspase-8 activation independent of cIAP2. J Invest Dermatol. 2008;128:1134–1147. doi: 10.1038/sj.jid.5701141. [DOI] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrhardt H, Fulda S, Schmid I, Hiscott J, Debatin KM, Jeremias I. TRAIL induced survival and proliferation in cancer cells resistant towards TRAIL-induced apoptosis mediated by NF-kappaB. Oncogene. 2003;22:3842–3852. doi: 10.1038/sj.onc.1206520. [DOI] [PubMed] [Google Scholar]
- Eldar A, Elowitz MB. Functional roles for noise in genetic circuits. Nature. 2010;467:167–173. doi: 10.1038/nature09326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskes R, Desagher S, Antonsson B, Martinou JC. Bid induces the oligomerization and insertion of Bax into the outer mitochondrial membrane. Mol Cell Biol. 2000;20:929–935. doi: 10.1128/mcb.20.3.929-935.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falschlehner C, Emmerich CH, Gerlach B, Walczak H. TRAIL signalling: decisions between life and death. Int J Biochem Cell Biol. 2007;39:1462–1475. doi: 10.1016/j.biocel.2007.02.007. [DOI] [PubMed] [Google Scholar]
- Feig C, Tchikov V, Schutze S, Peter ME. Palmitoylation of CD95 facilitates formation of SDS-stable receptor aggregates that initiate apoptosis signaling. EMBO J. 2007;26:221–231. doi: 10.1038/sj.emboj.7601460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flusberg DA, Sorger PK. Modulating cell-to-cell variability and sensitivity to death ligands by co-drugging. Phys Biol. 2013;10:035002. doi: 10.1088/1478-3975/10/3/035002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frese-Schaper M, Schardt JA, Sakai T, Carboni GL, Schmid RA, Frese S. Inhibition of tissue transglutaminase sensitizes TRAIL-resistant lung cancer cells through upregulation of death receptor 5. FEBS Lett. 2010;584:2867–2871. doi: 10.1016/j.febslet.2010.04.072. [DOI] [PubMed] [Google Scholar]
- Fricker N, Beaudouin J, Richter P, Eils R, Krammer PH, Lavrik IN. Model-based dissection of CD95 signaling dynamics reveals both a pro- and antiapoptotic role of c-FLIPL. J Cell Biol. 2010;190:377–389. doi: 10.1083/jcb.201002060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganten TM, Koschny R, Haas TL, Sykora J, Li-Weber M, Herzer K, Walczak H. Proteasome inhibition sensitizes hepatocellular carcinoma cells, but not human hepatocytes, to TRAIL. Hepatology. 2005;42:588–597. doi: 10.1002/hep.20807. [DOI] [PubMed] [Google Scholar]
- Gaudet S, Spencer SL, Chen WW, Sorger PK. Exploring the contextual sensitivity of factors that determine cell-to-cell variability in receptor-mediated apoptosis. PLoS Comput Biol. 2012;8:e1002482. doi: 10.1371/journal.pcbi.1002482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalvez F, Ashkenazi A. New insights into apoptosis signaling by Apo2L/TRAIL. Oncogene. 2010;29:4752–4765. doi: 10.1038/onc.2010.221. [DOI] [PubMed] [Google Scholar]
- Guicciardi ME, Gores GJ. Life and death by death receptors. FASEB J. 2009;23:1625–1637. doi: 10.1096/fj.08-111005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A, et al. Autophagy inhibition and antimalarials promote cell death in gastrointestinal stromal tumor (GIST) Proc Natl Acad Sci USA. 2010;107:14333–14338. doi: 10.1073/pnas.1000248107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta PB, Fillmore CM, Jiang G, Shapira SD, Tao K, Kuperwasser C, Lander ES. Stochastic state transitions give rise to phenotypic equilibrium in populations of cancer cells. Cell. 2011;146:633–644. doi: 10.1016/j.cell.2011.07.026. [DOI] [PubMed] [Google Scholar]
- Harper N, Hughes MA, Farrow SN, Cohen GM, MacFarlane M. Protein kinase C modulates tumor necrosis factor-related apoptosis-inducing ligand-induced apoptosis by targeting the apical events of death receptor signaling. J Biol Chem. 2003;278:44338–44347. doi: 10.1074/jbc.M307376200. [DOI] [PubMed] [Google Scholar]
- Harper N, MacFarlane M. Recombinant TRAIL and TRAIL receptor analysis. Methods Enzymol. 2008;446:293–313. doi: 10.1016/S0076-6879(08)01618-2. [DOI] [PubMed] [Google Scholar]
- Hellwig CT, Kohler BF, Lehtivarjo AK, Dussmann H, Courtney MJ, Prehn JH, Rehm M. Real time analysis of tumor necrosis factor-related apoptosis-inducing ligand/cycloheximide-induced caspase activities during apoptosis initiation. J Biol Chem. 2008;283:21676–21685. doi: 10.1074/jbc.M802889200. [DOI] [PubMed] [Google Scholar]
- Hellwig CT, Rehm M. TRAIL signaling and synergy mechanisms used in TRAIL-based combination therapies. Mol Cancer Therapeut. 2012;11:3–13. doi: 10.1158/1535-7163.MCT-11-0434. [DOI] [PubMed] [Google Scholar]
- Herrero-Martin G, Hoyer-Hansen M, Garcia-Garcia C, Fumarola C, Farkas T, Lopez-Rivas A, Jaattela M. TAK1 activates AMPK-dependent cytoprotective autophagy in TRAIL-treated epithelial cells. EMBO J. 2009;28:677–685. doi: 10.1038/emboj.2009.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes MA, Harper N, Butterworth M, Cain K, Cohen GM, MacFarlane M. Reconstitution of the death-inducing signaling complex reveals a substrate switch that determines CD95-mediated death or survival. Mol Cell. 2009;35:265–279. doi: 10.1016/j.molcel.2009.06.012. [DOI] [PubMed] [Google Scholar]
- Ishimura N, Isomoto H, Bronk SF, Gores GJ. Trail induces cell migration and invasion in apoptosis-resistant cholangiocarcinoma cells. Am J Physiol Gastrointest Liver Physiol. 2006;290::G129–G136. doi: 10.1152/ajpgi.00242.2005. [DOI] [PubMed] [Google Scholar]
- Jang JH, Moritz W, Graf R, Clavien PA. Preconditioning with death ligands FasL and TNF-alpha protects the cirrhotic mouse liver against ischaemic injury. Gut. 2008;57:492–499. doi: 10.1136/gut.2007.137703. [DOI] [PubMed] [Google Scholar]
- Jansen BJ, van Ruissen F, Cerneus S, Cloin W, Bergers M, van Erp PE, Schalkwijk J. Tumor necrosis factor related apoptosis inducing ligand triggers apoptosis in dividing but not in differentiating human epidermal keratinocytes. J Invest Dermatol. 2003;121:1433–1439. doi: 10.1046/j.1523-1747.2003.12636.x. [DOI] [PubMed] [Google Scholar]
- Jeremias I, Debatin KM. TRAIL induces apoptosis and activation of NFkappaB. Eur Cytokine Netw. 1998;9:687–688. [PubMed] [Google Scholar]
- Johnstone RW, Frew AJ, Smyth MJ. The TRAIL apoptotic pathway in cancer onset, progression and therapy. Nat Rev Cancer. 2008;8:782–798. doi: 10.1038/nrc2465. [DOI] [PubMed] [Google Scholar]
- Kataoka T, Tschopp J. N-terminal fragment of c-FLIP(L) processed by caspase 8 specifically interacts with TRAF2 and induces activation of the NF-kappaB signaling pathway. Mol Cell Biol. 2004;24:2627–2636. doi: 10.1128/MCB.24.7.2627-2636.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Choi C, Benveniste EN, Kwon D. TRAIL induces MMP-9 expression via ERK activation in human astrocytoma cells. Biochem Biophys Res Commun. 2008;377:195–199. doi: 10.1016/j.bbrc.2008.09.095. [DOI] [PubMed] [Google Scholar]
- Kischkel FC, Hellbardt S, Behrmann I, Germer M, Pawlita M, Krammer PH, Peter ME. Cytotoxicity-dependent APO-1 (Fas/CD95)-associated proteins form a death-inducing signaling complex (DISC) with the receptor. EMBO J. 1995;14:5579–5588. doi: 10.1002/j.1460-2075.1995.tb00245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleber S, et al. Yes and PI3K bind CD95 to signal invasion of glioblastoma. Cancer Cell. 2008;13:235–248. doi: 10.1016/j.ccr.2008.02.003. [DOI] [PubMed] [Google Scholar]
- Kohlhaas SL, Craxton A, Sun XM, Pinkoski MJ, Cohen GM. Receptor-mediated endocytosis is not required for tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)-induced apoptosis. J Biol Chem. 2007;282:12831–12841. doi: 10.1074/jbc.M700438200. [DOI] [PubMed] [Google Scholar]
- Lane D, Cote M, Grondin R, Couture MC, Piche A. Acquired resistance to TRAIL-induced apoptosis in human ovarian cancer cells is conferred by increased turnover of mature caspase-3. Mol Cancer Therapeut. 2006;5:509–521. doi: 10.1158/1535-7163.MCT-05-0362. [DOI] [PubMed] [Google Scholar]
- Lavrik IN, Golks A, Riess D, Bentele M, Eils R, Krammer PH. Analysis of CD95 threshold signaling: triggering of CD95 (FAS/APO-1) at low concentrations primarily results in survival signaling. J Biol Chem. 2007;282:13664–13671. doi: 10.1074/jbc.M700434200. [DOI] [PubMed] [Google Scholar]
- Le Roy C, Wrana JL. Clathrin- and non-clathrin-mediated endocytic regulation of cell signalling. Nat Rev Mol Cell Biol. 2005;6:112–126. doi: 10.1038/nrm1571. [DOI] [PubMed] [Google Scholar]
- Lee MW, Bach JH, Lee HJ, Lee DY, Joo WS, Kim YS, Park SC, Kim KY, Lee WB, Kim SS. The activation of ERK1/2 via a tyrosine kinase pathway attenuates trail-induced apoptosis in HeLa cells. Cancer Invest. 2005;23:586–592. doi: 10.1080/07357900500283036. [DOI] [PubMed] [Google Scholar]
- Li JH, Kirkiles-Smith NC, McNiff JM, Pober JS. TRAIL induces apoptosis and inflammatory gene expression in human endothelial cells. J Immunol. 2003;171:1526–1533. doi: 10.4049/jimmunol.171.3.1526. [DOI] [PubMed] [Google Scholar]
- Li S, Zhao Y, He X, Kim TH, Kuharsky DK, Rabinowich H, Chen J, Du C, Yin XM. Relief of extrinsic pathway inhibition by the Bid-dependent mitochondrial release of Smac in Fas-mediated hepatocyte apoptosis. J Biol Chem. 2002;277:26912–26920. doi: 10.1074/jbc.M200726200. [DOI] [PubMed] [Google Scholar]
- Li Z, Xu X, Bai L, Chen W, Lin Y. EGFR-mediated tissue transglutaminase overexpression couples acquired TRAIL-resistance and migration through c-FLIP and MMP-9 in lung cancer cells. J Biol Chem. 2011;286:21164–21172. doi: 10.1074/jbc.M110.207571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Devin A, Cook A, Keane MM, Kelliher M, Lipkowitz S, Liu ZG. The death domain kinase RIP is essential for TRAIL (Apo2L)-induced activation of IkappaB kinase and c-Jun N-terminal kinase. Mol Cell Biol. 2000;20:6638–6645. doi: 10.1128/mcb.20.18.6638-6645.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo JL, Kamata H, Karin M. IKK/NF-kappaB signaling: balancing life and death–a new approach to cancer therapy. J Clin Invest. 2005;115:2625–2632. doi: 10.1172/JCI26322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X, Budihardjo I, Zou H, Slaughter C, Wang X. Bid, a Bcl2 interacting protein, mediates cytochrome c release from mitochondria in response to activation of cell surface death receptors. Cell. 1998;94:481–490. doi: 10.1016/s0092-8674(00)81589-5. [DOI] [PubMed] [Google Scholar]
- Malhi H, Gores GJ. TRAIL resistance results in cancer progression: a TRAIL to perdition. Oncogene. 2006;25:7333–7335. doi: 10.1038/sj.onc.1209765. [DOI] [PubMed] [Google Scholar]
- Martin DA, Siegel RM, Zheng L, Lenardo MJ. Membrane oligomerization and cleavage activates the caspase-8 (FLICE/MACHalpha1) death signal. J Biol Chem. 1998;273:4345–4349. doi: 10.1074/jbc.273.8.4345. [DOI] [PubMed] [Google Scholar]
- Mazurek N, Byrd JC, Sun Y, Hafley M, Ramirez K, Burks J, Bresalier RS. Cell-surface galectin-3 confers resistance to TRAIL by impeding trafficking of death receptors in metastatic colon adenocarcinoma cells. Cell Death Differ. 2011;19:523–533. doi: 10.1038/cdd.2011.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menke C, Bin L, Thorburn J, Behbakht K, Ford HL, Thorburn A. Distinct TRAIL resistance mechanisms can be overcome by proteasome inhibition but not generally by synergizing agents. Cancer Res. 2011;71:1883–1892. doi: 10.1158/0008-5472.CAN-10-2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muppidi JR, Tschopp J, Siegel RM. Life and death decisions: secondary complexes and lipid rafts in TNF receptor family signal transduction. Immunity. 2004;21:461–465. doi: 10.1016/j.immuni.2004.10.001. [DOI] [PubMed] [Google Scholar]
- Muranen T, Selfors LM, Worster DT, Iwanicki MP, Song L, Morales FC, Gao S, Mills GB, Brugge JS. Inhibition of PI3K/mTOR leads to adaptive resistance in matrix-attached cancer cells. Cancer Cell. 2012;21:227–239. doi: 10.1016/j.ccr.2011.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann L, Pforr C, Beaudouin J, Pappa A, Fricker N, Krammer PH, Lavrik IN, Eils R. Dynamics within the CD95 death-inducing signaling complex decide life and death of cells. Mol Syst Biol. 2010;6:352. doi: 10.1038/msb.2010.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opel D, Naumann I, Schneider M, Bertele D, Debatin KM, Fulda S. Targeting aberrant PI3K/Akt activation by PI103 restores sensitivity to TRAIL-induced apoptosis in neuroblastoma. Clin Cancer Res. 2011;17:3233–3247. doi: 10.1158/1078-0432.CCR-10-2530. [DOI] [PubMed] [Google Scholar]
- Pahl HL. Activators and target genes of Rel/NF-kappaB transcription factors. Oncogene. 1999;18:6853–6866. doi: 10.1038/sj.onc.1203239. [DOI] [PubMed] [Google Scholar]
- Pukac L, et al. HGS-ETR1, a fully human TRAIL-receptor 1 monoclonal antibody, induces cell death in multiple tumour types in vitro and in vivo. Br J Cancer. 2005;92:1430–1441. doi: 10.1038/sj.bjc.6602487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, van Oudenaarden A. Nature, nurture, or chance: stochastic gene expression and its consequences. Cell. 2008;135:216–226. doi: 10.1016/j.cell.2008.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakoff-Nahoum S, Medzhitov R. Toll-like receptors and cancer. Nat Rev Cancer. 2009;9:57–63. doi: 10.1038/nrc2541. [DOI] [PubMed] [Google Scholar]
- Rehm M, Dussmann H, Janicke RU, Tavare JM, Kogel D, Prehn JH. Single-cell fluorescence resonance energy transfer analysis demonstrates that caspase activation during apoptosis is a rapid process. Role of caspase-3. J Biol Chem. 2002;277:24506–24514. doi: 10.1074/jbc.M110789200. [DOI] [PubMed] [Google Scholar]
- Rehm M, Huber HJ, Dussmann H, Prehn JH. Systems analysis of effector caspase activation and its control by X-linked inhibitor of apoptosis protein. EMBO J. 2006;25:4338–4349. doi: 10.1038/sj.emboj.7601295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehm M, Huber HJ, Hellwig CT, Anguissola S, Dussmann H, Prehn JH. Dynamics of outer mitochondrial membrane permeabilization during apoptosis. Cell Death Differ. 2009;16:613–623. doi: 10.1038/cdd.2008.187. [DOI] [PubMed] [Google Scholar]
- Riedl SJ, Salvesen GS. The apoptosome: signalling platform of cell death. Nat Rev Mol Cell Biol. 2007;8:405–413. doi: 10.1038/nrm2153. [DOI] [PubMed] [Google Scholar]
- Roesch A, Fukunaga-Kalabis M, Schmidt EC, Zabierowski SE, Brafford PA, Vultur A, Basu D, Gimotty P, Vogt T, Herlyn M. A temporarily distinct subpopulation of slow-cycling melanoma cells is required for continuous tumor growth. Cell. 2010;141:583–594. doi: 10.1016/j.cell.2010.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossin A, Derouet M, Abdel-Sater F, Hueber AO. Palmitoylation of the TRAIL receptor DR4 confers an efficient TRAIL-induced cell death signalling. Biochem J. 2009;419:185–192. doi: 10.1042/BJ20081212. [DOI] [PubMed] [Google Scholar]
- Sabnis GJ, Macedo LF, Goloubeva O, Schayowitz A, Brodie AM. Stopping treatment can reverse acquired resistance to letrozole. Cancer Res. 2008;68:4518–4524. doi: 10.1158/0008-5472.CAN-07-5999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santner SJ, Dawson PJ, Tait L, Soule HD, Eliason J, Mohamed AN, Wolman SR, Heppner GH, Miller FR. Malignant MCF10CA1 cell lines derived from premalignant human breast epithelial MCF10AT cells. Breast Cancer Res Treat. 2001;65:101–110. doi: 10.1023/a:1006461422273. [DOI] [PubMed] [Google Scholar]
- Sayers TJ, Murphy WJ. Combining proteasome inhibition with TNF-related apoptosis-inducing ligand (Apo2L/TRAIL) for cancer therapy. Cancer Immunol Immunother. 2006;55:76–84. doi: 10.1007/s00262-005-0676-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaffidi C, Schmitz I, Krammer PH, Peter ME. The role of c-FLIP in modulation of CD95-induced apoptosis. J Biol Chem. 1999;274:1541–1548. doi: 10.1074/jbc.274.3.1541. [DOI] [PubMed] [Google Scholar]
- Sharma SV, et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141:69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp DA, Lawrence DA, Ashkenazi A. Selective knockdown of the long variant of cellular FLICE inhibitory protein augments death receptor-mediated caspase-8 activation and apoptosis. J Biol Chem. 2005;280:19401–19409. doi: 10.1074/jbc.M413962200. [DOI] [PubMed] [Google Scholar]
- Siegel RM, Fleisher TA. The role of Fas and related death receptors in autoimmune and other disease states. J Allergy Clin Immunol. 1999;103:729–738. doi: 10.1016/s0091-6749(99)70412-4. [DOI] [PubMed] [Google Scholar]
- Sigal A, Milo R, Cohen A, Geva-Zatorsky N, Klein Y, Liron Y, Rosenfeld N, Danon T, Perzov N, Alon U. Variability and memory of protein levels in human cells. Nature. 2006;444:643–646. doi: 10.1038/nature05316. [DOI] [PubMed] [Google Scholar]
- Singh DK, Ku CJ, Wichaidit C, Steininger RJ 3rd, Wu LF, Altschuler SJ. Patterns of basal signaling heterogeneity can distinguish cellular populations with different drug sensitivities. Mol Syst Biol. 2010;6:369. doi: 10.1038/msb.2010.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son JK, Varadarajan S, Bratton SB. TRAIL-activated stress kinases suppress apoptosis through transcriptional upregulation of MCL-1. Cell Death Differ. 2010;17:1288–1301. doi: 10.1038/cdd.2010.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JH, Tse MC, Bellail A, Phuphanich S, Khuri F, Kneteman NM, Hao C. Lipid rafts and nonrafts mediate tumor necrosis factor related apoptosis-inducing ligand induced apoptotic and nonapoptotic signals in non small cell lung carcinoma cells. Cancer Res. 2007a;67:6946–6955. doi: 10.1158/0008-5472.CAN-06-3896. [DOI] [PubMed] [Google Scholar]
- Song JJ, An JY, Kwon YT, Lee YJ. Evidence for two modes of development of acquired tumor necrosis factor-related apoptosis-inducing ligand resistance. Involvement of Bcl-xL. J Biol Chem. 2007b;282:319–328. doi: 10.1074/jbc.M608065200. [DOI] [PubMed] [Google Scholar]
- Song JJ, Kim JH, Sun BK, Alcala MA, Jr, Bartlett DL, Lee YJ. c-Cbl acts as a mediator of Src-induced activation of the PI3K-Akt signal transduction pathway during TRAIL treatment. Cell Signal. 2009;22:377–385. doi: 10.1016/j.cellsig.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JJ, Szczepanski MJ, Kim SY, Kim JH, An JY, Kwon YT, Alcala MA, Jr, Bartlett DL, Lee YJ. c-Cbl-mediated degradation of TRAIL receptors is responsible for the development of the early phase of TRAIL resistance. Cell Signal. 2010;22:553–563. doi: 10.1016/j.cellsig.2009.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer SL, Gaudet S, Albeck JG, Burke JM, Sorger PK. Non-genetic origins of cell-to-cell variability in TRAIL-induced apoptosis. Nature. 2009;459:428–432. doi: 10.1038/nature08012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun BK, Kim JH, Nguyen HN, Kim SY, Oh S, Lee YJ, Song JJ. TRAIL-induced caspase/p38 activation is responsible for the increased catalytic and invasive activities of Akt. Int J Oncol. 2010a;38:249–256. [PMC free article] [PubMed] [Google Scholar]
- Sun BK, Kim JH, Nguyen HN, Oh S, Kim SY, Choi S, Choi HJ, Lee YJ, Song JJ. MEKK1/MEKK4 are responsible for TRAIL-induced JNK/p38 phosphorylation. Oncol Rep. 2010b;25:537–544. doi: 10.3892/or.2010.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun XM, Bratton SB, Butterworth M, MacFarlane M, Cohen GM. Bcl-2 and Bcl-xL inhibit CD95-mediated apoptosis by preventing mitochondrial release of Smac/DIABLO and subsequent inactivation of X-linked inhibitor-of-apoptosis protein. J Biol Chem. 2002;277:11345–11351. doi: 10.1074/jbc.M109893200. [DOI] [PubMed] [Google Scholar]
- Takeda K, Smyth MJ, Cretney E, Hayakawa Y, Kayagaki N, Yagita H, Okumura K. Critical role for tumor necrosis factor-related apoptosis-inducing ligand in immune surveillance against tumor development. J Exp Med. 2002;195:161–169. doi: 10.1084/jem.20011171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Wang W, Zhang Y, Liu S, Liu Y, Zheng D. Tumour necrosis factor-related apoptosis-inducing ligand (TRAIL)-induced chemokine release in both TRAIL-resistant and TRAIL-sensitive cells via nuclear factor kappa B. FEBS J. 2009;276:581–593. doi: 10.1111/j.1742-4658.2008.06809.x. [DOI] [PubMed] [Google Scholar]
- Tang Z, Bauer JA, Morrison B, Lindner DJ. Nitrosylcobalamin promotes cell death via S nitrosylation of Apo2L/TRAIL receptor DR4. Mol Cell Biol. 2006;26:5588–5594. doi: 10.1128/MCB.00199-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trauth BC, Klas C, Peters AM, Matzku S, Moller P, Falk W, Debatin KM, Krammer PH. Monoclonal antibody-mediated tumor regression by induction of apoptosis. Science. 1989;245:301–305. doi: 10.1126/science.2787530. [DOI] [PubMed] [Google Scholar]
- Trauzold A, Siegmund D, Schniewind B, Sipos B, Egberts J, Zorenkov D, Emme D, Roder C, Kalthoff H, Wajant H. TRAIL promotes metastasis of human pancreatic ductal adenocarcinoma. Oncogene. 2006;25:7434–7439. doi: 10.1038/sj.onc.1209719. [DOI] [PubMed] [Google Scholar]
- Varfolomeev E, Maecker H, Sharp D, Lawrence D, Renz M, Vucic D, Ashkenazi A. Molecular determinants of kinase pathway activation by Apo2 ligand/tumor necrosis factor-related apoptosis-inducing ligand. J Biol Chem. 2005;280:40599–40608. doi: 10.1074/jbc.M509560200. [DOI] [PubMed] [Google Scholar]
- Wagner KW, et al. Death-receptor O-glycosylation controls tumor-cell sensitivity to the proapoptotic ligand Apo2L/TRAIL. Nat Med. 2007;13:1070–1077. doi: 10.1038/nm1627. [DOI] [PubMed] [Google Scholar]
- Wajant H, Pfizenmaier K, Scheurich P. Non-apoptotic Fas signaling. Cytokine Growth Factor Rev. 2003;14:53–66. doi: 10.1016/s1359-6101(02)00072-2. [DOI] [PubMed] [Google Scholar]
- Wilson NS, Dixit V, Ashkenazi A. Death receptor signal transducers: nodes of coordination in immune signaling networks. Nat Immunol. 2009;10:348–355. doi: 10.1038/ni.1714. [DOI] [PubMed] [Google Scholar]
- Yan J, Xiang J, Lin Y, Ma J, Zhang J, Zhang H, Sun J, Danial NN, Liu J, Lin A. Inactivation of BAD by IKK inhibits TNFalpha-induced apoptosis independently of NF-kappaB activation. Cell. 2013;152:304–315. doi: 10.1016/j.cell.2012.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Zhang Y, Rivera Rosado LA, Zhang B. Repeated treatment with subtoxic doses of TRAIL induces resistance to apoptosis through its death receptors in MDA-MB-231 breast cancer cells. Mol Cancer Res. 2009;7:1835–1844. doi: 10.1158/1541-7786.MCR-09-0244. [DOI] [PubMed] [Google Scholar]
- Zauli G, Sancilio S, Cataldi A, Sabatini N, Bosco D, Di Pietro R. PI-3K/Akt and NF-kappaB/IkappaBalpha pathways are activated in Jurkat T cells in response to TRAIL treatment. J Cell Physiol. 2005;202:900–911. doi: 10.1002/jcp.20202. [DOI] [PubMed] [Google Scholar]
- Zhang L, Fang B. Mechanisms of resistance to TRAIL-induced apoptosis in cancer. Cancer Gene Ther. 2005;12:228–237. doi: 10.1038/sj.cgt.7700792. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Yoshida T, Zhang B. TRAIL induces endocytosis of its death receptors in MDA-MB-231 breast cancer cells. Cancer Biol Ther. 2009;8:917–922. doi: 10.4161/cbt.8.10.8141. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.