The WASH complex, an endosomal activator of the Arp2/3 complex involved in branched actin polymerization, is identified as a new factor in vesicle traffic mediated by the Hermansky–Pudlak syndrome complex BLOC-1.
Abstract
Vesicle biogenesis machinery components such as coat proteins can interact with the actin cytoskeleton for cargo sorting into multiple pathways. It is unknown, however, whether these interactions are a general requirement for the diverse endosome traffic routes. In this study, we identify actin cytoskeleton regulators as previously unrecognized interactors of complexes associated with the Hermansky–Pudlak syndrome. Two complexes mutated in the Hermansky–Pudlak syndrome, adaptor protein complex-3 and biogenesis of lysosome-related organelles complex-1 (BLOC-1), interact with and are regulated by the lipid kinase phosphatidylinositol-4-kinase type IIα (PI4KIIα). We therefore hypothesized that PI4KIIα interacts with novel regulators of these complexes. To test this hypothesis, we immunoaffinity purified PI4KIIα from isotope-labeled cell lysates to quantitatively identify interactors. Strikingly, PI4KIIα isolation preferentially coenriched proteins that regulate the actin cytoskeleton, including guanine exchange factors for Rho family GTPases such as RhoGEF1 and several subunits of the WASH complex. We biochemically confirmed several of these PI4KIIα interactions. Of importance, BLOC-1 complex, WASH complex, RhoGEF1, or PI4KIIα depletions altered the content and/or subcellular distribution of the BLOC-1–sensitive cargoes PI4KIIα, ATP7A, and VAMP7. We conclude that the Hermansky–Pudlak syndrome complex BLOC-1 and its cargo PI4KIIα interact with regulators of the actin cytoskeleton.
INTRODUCTION
Vesicular trafficking is a general cellular mechanism by which secretory and endocytic pathway organelles selectively exchange components. This exchange mechanism requires coordinated steps, which include sorting and concentration of membrane protein cargo into nascent vesicles, membrane deformation and scission, directional movement through the cytosol, and fusion at the target organelle (Bonifacino and Glick, 2004). Many of these steps require mechanical force, which is generated within vesicular trafficking pathways by association of specialized molecular machines. These molecular machines include coat proteins that sort membrane protein cargo, BAR-domain proteins to sense or induce membrane deformation, the dynamin GTPase for membrane scission, and tethers and soluble N-ethylmaleimide–sensitive factor attachment protein receptor (SNARE) complexes for membrane fusion (Kaksonen et al., 2005; Schmid and McMahon, 2007; McMahon and Boucrot, 2011; Wickner and Schekman, 2008; Brocker et al., 2010; Taylor et al., 2011). Whereas some of these machines are intrinsically capable of force generation, such as dynamin and SNARE fusion complexes (Schmid and Frolov, 2011; Ferguson and De Camilli, 2012; Gao et al., 2012), others require association with the actin or microtubule cytoskeleton. For example, coat proteins sort membrane protein cargo into nascent vesicles. In addition, however, some coat proteins also bind cytoskeletal components for both vesicle biogenesis and directional movement (Nakagawa et al., 2000; Styers et al., 2004; Kaksonen et al., 2006; Delevoye et al., 2009; Anitei et al., 2010; Anitei and Hoflack, 2012; Mooren et al., 2012).
A fundamental unanswered question concerns the extent to which this cytoskeletal interaction is a general principle for coat protein function. One of the best-characterized interactions between vesicle trafficking machinery and the cytoskeleton is the formation of vesicles at the plasma membrane by the clathrin and AP-2 adaptor coats. In this vesicle biogenesis pathway, the coat and coat-associated factors, such as BAR-domain proteins, locally regulate the polymerization and organization of actin filaments. Actin polymerization is nucleated in large part by the Arp2/3 complex and its activator, the nucleation-promoting factor neural Wiskott–Aldrich syndrome protein (N-WASP; Conner and Schmid, 2003; Kaksonen et al., 2005, 2006; Schmid and McMahon, 2007; McMahon and Boucrot, 2011; Taylor et al., 2011; Mooren et al., 2012). The recent identification of a nucleation-promoting factor that specifically localizes to endosomes, the WASP and SCAR homologue (WASH; Derivery et al., 2009; Gomez and Billadeau, 2009), suggests that coat association with the actin cytoskeleton may be a general principle operating in the generation of vesicles. Supporting this possibility, depletion of WASH leads to functional defects in recycling, endosome-to–Golgi, and endosome-to-lysosome trafficking pathways (Derivery et al., 2009; Gomez and Billadeau, 2009; Gomez et al., 2012; Duleh and Welch, 2010; Carnell et al., 2011; Zech et al., 2011; Harbour et al., 2012; Hao et al., 2013; Piotrowski et al., 2013). To date, however, WASH has been only been described to interact with one endosomal coat complex, the retromer complex, which primarily sorts cargo into the endosome-to-Golgi pathway (Gomez and Billadeau, 2009; Gomez et al., 2012; Harbour et al., 2012; Jia et al., 2012; Seaman, 2012; Hao et al., 2013; Helfer et al., 2013). Therefore whether endosomal coats coordinate with the actin cytoskeleton as a general principle has been untested.
We provide evidence supporting this hypothesis through the unbiased identification of an interaction between the WASH complex and sorting complexes associated with the Hermansky–Pudlak syndrome. This syndrome is genetically defined by defects in four molecular complexes: the adaptor protein complex-3 (AP-3) coat and the biogenesis of lysosome-related organelles complexes-1 to -3 (BLOC-1 to BLOC-3). In addition, genetic defects in specific Rabs and regulatory enzymes trigger Hermansky–Pudlak syndrome–like phenotypes (Bultema and Di Pietro, 2013). Defects in these complexes in mammals and invertebrates lead to the hypopigmentation, platelet dysfunction, pulmonary fibrosis, and, in some cases, immune dysfunction and neurologic phenotypes (Newell-Litwa et al., 2007; Raposo and Marks, 2007; Raposo et al., 2007; Huizing et al., 2008; Dell'Angelica, 2009; Wei and Li, 2013). These phenotypes are a consequence of impaired cargo trafficking between early endosomes and target organelles such as lysosome-related organelles and synaptic vesicles (Newell-Litwa et al., 2007; Raposo and Marks, 2007; Raposo et al., 2007; Huizing et al., 2008; Dell'Angelica, 2009; Wei and Li, 2013). Therefore exploring the molecules and functional mechanisms of these complexes both increases our understanding of the molecular basis of the Hermansky–Pudlak syndrome and allows us to test fundamental questions concerning principles of vesicular trafficking processes.
To identify functional mechanisms of complexes associated with Hermansky–Pudlak syndrome, we focus on a membrane-anchored and endosome-localized lipid kinase, phosphatidylinositol-4-kinase type IIα (PI4KIIα; Balla et al., 2002; Guo et al., 2003; Minogue et al., 2006; Craige et al., 2008). Biochemical and genetic evidence indicates that PI4KIIα regulates and binds two complexes mutated in Hermansky–Pudlak syndrome, AP-3 and BLOC-1 (Craige et al., 2008; Salazar et al., 2009; Larimore et al., 2011; Gokhale et al., 2012a). We therefore hypothesized that PI4KIIα would interact with novel proteins modulating the function of BLOC-1 and AP-3. To test this hypothesis, we isolated PI4KIIα and its interactors by quantitative immunoaffinity purification from in vivo cross-linked cell lysates. PI4KIIα preferentially coisolated with proteins that regulate actin cytoskeleton polymerization, including the WASH complex and RhoGEF1. Depletion of PI4KIIα, BLOC-1 subunits, RhoGEF1, or the WASH complex modified the content of other components in the PI4KIIα interactome, thereby establishing genetic interactions in addition to physical interactions. Further, depletion of the WASH complex resulted in changes in PI4KIIα endosomal morphology and mislocalization of two BLOC-1 cargoes, the Menkes disease copper transporter ATP7A and a lysosomal SNARE, VAMP7. We propose that diverse coats such as clathrin-AP-2, retromer, and AP-3–BLOC-1 use common principles to garner force or create discrete membrane domains by localized actin polymer assembly.
RESULTS
The PI4KIIα interactome enriches actin-regulatory proteins
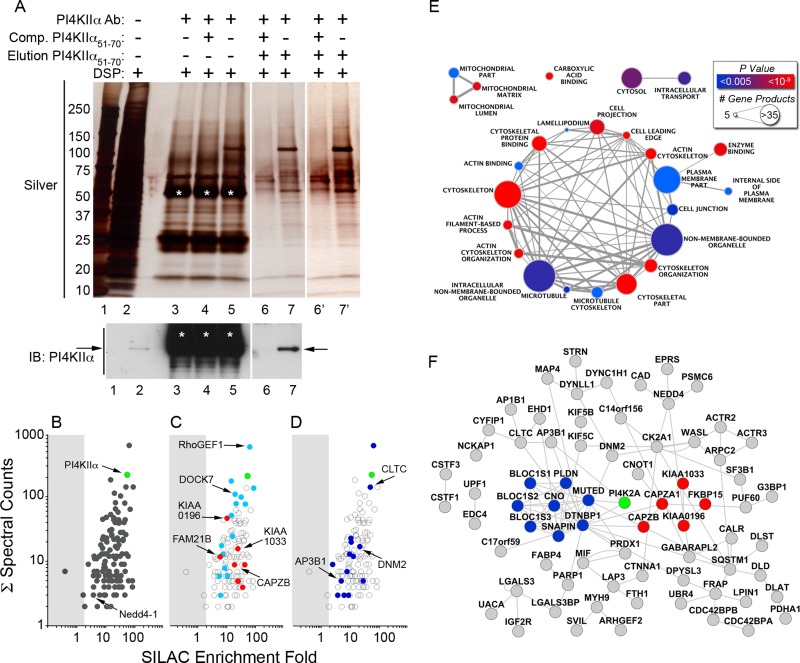
PI4KIIα binds the AP-3 and BLOC-1 complexes, and these interactions regulate its subcellular distribution (Craige et al., 2008; Salazar et al., 2009; Larimore et al., 2011; Gokhale et al., 2012a). In addition, the lipid kinase activity of PI4KIIα regulates the recruitment of the AP-3 and BLOC-1 coat complex to endosomal membranes (Craige et al., 2008; Salazar et al., 2009). Therefore we hypothesized that interactors of PI4KIIα are previously unrecognized regulators of the Hermansky–Pudlak syndrome–associated AP-3 and BLOC-1 complexes. We tested this hypothesis by isolation of endogenous PI4KIIα and interacting proteins. To isolate PI4KIIα, we raised an antibody to a peptide containing the dileucine sorting motif that is required for interaction of PI4KIIα with the AP-3 and BLOC-1 complexes (Supplemental Figure S1A; Craige et al., 2008). Mutation of the dileucine sorting motif abrogates the recognition of PI4KIIα by this antibody by immunoblotting (Supplemental Figure S1A). Therefore we premised that this antibody would compete with AP-3 for binding to PI4KIIα and thereby preferentially enrich for PI4KIIα that is not bound to AP-3. This design strategy allowed for the enrichment of PI4KIIα-interacting proteins that are upstream or downstream of AP-3 binding. After establishing that this antibody specifically recognizes PI4KIIα, we immunoprecipitated PI4KIIα from minimally cross-linked SHSY-5Y neuroblastoma cell lysates (Supplemental Figure S1A and Figure 1A). As we extensively characterized, this cross-linking strategy 1) stabilizes protein interactions to allow rigorous biochemical isolation strategies, as described later; 2) uses a cell-permeable and chemically reducible cross-linker, dithiobis succinimidyl propionate (DSP; Lomant and Fairbanks, 1976), that allows for in vivo cross-linking but is amenable to protein identification by mass spectrometry and immunoblot; and 3) is nonsaturating to minimize stabilization of unrelated protein complexes (Salazar et al., 2009; Zlatic et al., 2010; Gokhale et al., 2012a, b; Perez-Cornejo et al., 2012). Silver stain of immunoprecipitated protein complexes revealed several polypeptides that coisolate with PI4KIIα (Figure 1A, lane 5). As expected, several of these coisolated polypeptides were not detected in control reactions where an excess of the PI4KIIα antigenic peptide outcompeted the endogenous protein for binding to the immunoprecipitation antibody (Figure 1A, lane 4). However, eluting protein complexes from antibody-coated beads with Laemmli sample buffer released background polypeptides that precluded the identification of PI4KIIα and interacting proteins by silver stain (Figure 1A, lanes 3–5). For this reason, we eluted protein complexes after immunoprecipitation by incubating the washed beads with an excess of the PI4KIIα antigenic peptide (Figure 1A, lane 7). This immunoaffinity purification strategy greatly reduced background contaminants and immunoglobulins, as revealed by silver stain and immunoblot (Figure 1A, compare lanes 4 and 5 to lanes 6 and 7). Finally, immunoblot demonstrated that our immunoaffinity purification approach highly enriched for PI4KIIα (Figure 1A, compare lanes 2 and 7).
FIGURE 1:
The PI4KIIα interactome enriches actin-regulatory proteins. (A) PI4KIIα was immunoaffinity purified from DSP–cross-linked SHSY-5Y neuroblastoma cell homogenates. PI4KIIα and coisolating proteins were resolved by SDS–PAGE and silver stained (lane 5). Some polypeptides are absent from controls, which include antibody-coated beads without lysate (lane 3) or incubations in which an excess of antigenic PI4KIIα peptide was included (lane 4). Bead elution was performed with Laemmli sample buffer (lanes 3–5). Lanes 6 and 7 depict antigenic peptide eluates from beads similar to those in lanes 4 and 5. Immunoaffinity purification allowed for the selective elution of PI4KIIα-coisolating proteins against low background (compare lanes 6 and 7; lanes 6′ and 7′ depict contrast-enhanced lanes 6 and 7). An arrow marks the band containing PI4KIIα. Silver stain is a representative image of two independent experiments. Bottom, immunoblot of PI4KIIα in a parallel set of assays as those in the silver-stained SDS–PAGE. Immunoglobulin G chains are marked by asterisks. Input represents 1 and 0.33% for immunoprecipitation and immunoaffinity purifications, respectively. (B) Plots represent PI4KIIα and copurifying proteins (see Supplemental Table S1 for details). The x- and y-axes show SILAC fold of enrichment and the total number of spectral counts used for protein identification, respectively. Reference dots highlight PI4KIIα (green) and Nedd4-1. (C) Teal and red dots highlight actin-related proteins coisolated with PI4KIIα. These include GDP exchange factors RhoGEF1, DOCK7, and GEF-H1. Red dots indicate the actin-related proteins that are subunits or interactors of the WASH complex: strumpellin (KIAA0196), SWIP (KIAA1033), Fam21B, FKBP15, capping protein α, and capping protein β. (D) Blue dots highlight membrane-trafficking related proteins coisolated with PI4KIIα, including clathrin heavy chain (CLTC), dynamin-2 (DNM2), and β3A subunit of the AP-3 complex (AP3B1). (E) Functional annotation analysis of the identified proteins using Gene Ontology annotations revealed a preponderance of actin-related proteins. Enrichment of categories such as cytoskeleton (30/124 interactors) and actin cytoskeleton (13/124) were significant at p = 1.62 × 10−7 and 7.67 × 10−7, respectively. See Supplemental Table S2 for details. (F) Network analysis of putative PI4KIIα interactome components and BLOC-1 subunits. PI4KIIα (green), the BLOC-1 subunits (blue), and WASH complex subunits (red) are highlighted.
Having analytically established a selective approach to isolate PI4KIIα and interacting proteins, we performed a preparative immunoaffinity purification of cross-linked PI4KIIα complexes. We identified interactors by quantitative mass spectrometry using stable isotope labeling with amino acids in cell culture (SILAC; Ong et al., 2002; Mann, 2006). SHSY-5Y neuroblastoma cells were grown to equilibrium in either a light-label medium in which the amino acids arginine and lysine contain 12C and 14N isotopes (R0K0 medium) or a heavy-label medium in which these amino acids contain 13C and 15N (R10K8 medium; Supplemental Figure S1B). After incorporation of these amino acid isotope labels, PI4KIIα was immunoprecipitated from light-labeled cell lysates. As a control, immunoprecipitations from heavy-labeled cell lysates were performed in the presence of an excess of PI4KIIα antigenic peptide (Supplemental Figure S1B). As described earlier, both samples were then competitively eluted from the beads with an excess of PI4KIIα antigenic peptide. Light- and heavy-labeled eluted polypeptides were mixed 1:1 for tandem mass spectrometry analysis. This purification enriched PI4KIIα by ∼55-fold, and its relative abundance was represented by 220 spectral counts (Figure 1, B–D, green dot). In addition, we identified 701 coisolating proteins with two or more unique peptides. Of the total 702 proteins, 268 were enriched more than twofold by PI4KIIα immunoaffinity purification over control, a SILAC cutoff that we previously demonstrated enriches for interactions that can be confirmed genetically (Gokhale et al., 2012a, b; Perez-Cornejo et al., 2012). However, one known limitation of bead-based purifications is the tendency of proteins to bind nonspecifically to the antibody-coated beads (Trinkle-Mulcahy et al., 2008). To overcome this limitation, we filtered out any polypeptides that bind nonspecifically to either beads alone or beads coated with unrelated antibody. Applying these filters identified 123 coisolating proteins in addition to PI4KIIα for further study (Figure 1, B–D and F, and Supplemental Table S1).
We first characterized the PI4KIIα interactome by performing functional annotation analysis of the 124 identified proteins using Gene Ontology annotations (Figure 1E). This analysis revealed that cytoskeleton proteins (30/124) and actin cytoskeleton proteins (13/124) were enriched in the PI4KIIα interactome as compared with a random sampling of the human genome (p = 1.62 × 10−7 and 7.67 × 10−7, respectively). For example, actin-related proteins within the PI4KIIα interactome include regulatory proteins for small GTPases, such as RhoGEF1 (ARHGEF1), dedicator of cytokinesis 7 (DOCK7), and GEF-H1 (ARHGEF2), which are guanine exchange factors for RhoA, cdc42, and Rho-Rac GTPases, respectively (Figure 1C, teal dots; Rossman et al., 2005; Birkenfeld et al., 2008; Blasius et al., 2009; Aittaleb et al., 2010). In addition, functional annotation analysis indicated a highly significant enrichment of proteins annotated as “vesicle-mediated transport proteins” (13/124), as predicted, given the role of PI4KIIα in regulating AP-3–dependent vesicle biogenesis (Figure 1E; p = 1.19 × 10−3; Supplemental Table S2). These vesicle-mediated transport proteins include clathrin heavy chain, the β subunit of the AP-3 complex (AP3B1), and dynamin-2 (DYN-2; Figure 1D, blue dots). In addition to functional annotation, we performed network analysis to identify previously published direct interactions among these copurifying proteins. Our goal was to identify functional subcomplexes and hubs of high network connectivity that copurify with PI4KIIα and might play a role in AP-3– and BLOC-1–mediated vesicle biogenesis (Figure 1F; Gokhale et al., 2012b). For this reason, we included the eight subunits of the BLOC-1 complex in this analysis. Network analysis revealed the presence of three of the five subunits of the WASH complex (FAM21B, KIAA0196, and KIAA1033) and three known interactors of the WASH complex (FKBP15, CAPZA1, and CAPZB; Figure 1C, red dots; Campellone and Welch, 2010; Jia et al., 2010; Rottner et al., 2010; Rotty et al., 2012). The WASH complex localizes to early endosomes, where, it is suggested, it polymerizes actin for cargo sorting or vesicle scission, suggesting a role of the WASH complex in PI4KIIα-regulated, BLOC-1–mediated vesicle biogenesis.
Biochemical and genetic confirmation of PI4KIIα -interacting proteins
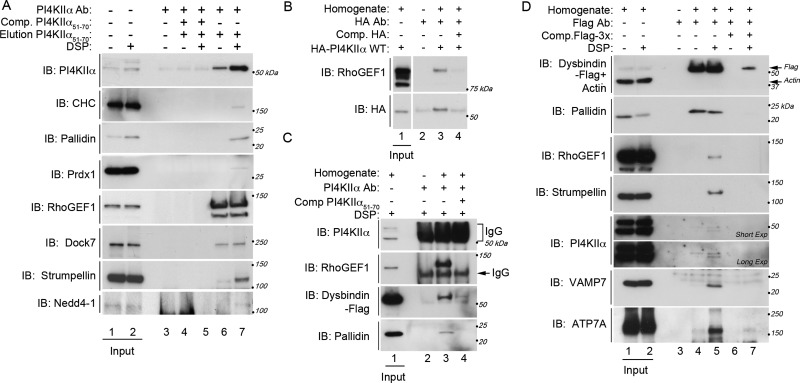
We confirmed PI4KIIα interactions identified by SILAC mass spectrometry by independent immunoaffinity purification of PI4KIIα followed by immunoblotting to test for the presence of proteins of interest. We found that PI4KIIα interacts with vesicle-mediated transport proteins such as clathrin heavy chain and the pallidin subunit of BLOC-1 (Figure 2A, lanes 6 and 7). Interacting proteins were found only in PI4KIIα isolations and not in controls, where an excess of antigenic peptide was used to outcompete immunoprecipitation of PI4KIIα complexes (Figure 2A, lanes 4 and 5). Consistent with our previous reports, detection of interactions between PI4KIIα and vesicle-mediated transport proteins required treatment with the cross-linker DSP, presumably because these associations are labile and/or transient. Further, we found that PI4KIIα interacts with two known interactors of the BLOC-1 and AP-3 complexes, respectively—the antioxidant enzyme peroxiredoxin-1 (Prdx-1) and a ubiquitin ligase Nedd4-1 (Mossinger et al., 2012; Figure 2A, lanes 6 and 7; also see Salazar et al., 2009; Gokhale et al., 2012a). Nedd4-1 represents the lower boundaries of our significance thresholds in terms of spectral count and enrichment cut-offs for our SILAC quantification, thereby validating that these thresholds minimize inclusion of false negatives in the PI4KIIα interactome (three spectral counts, 3.69-fold enrichment; Figure 1B). Finally, we tested the interaction between PI4KIIα and proteins that regulate the actin cytoskeleton, since functional annotation and network analysis suggested that these interactions were highly significant. We confirmed the interaction between PI4KIIα and the strumpellin subunit of the WASH complex and two guanine exchange factors for small GTPases RhoGEF1 and Dock7. Of note, these copurifications did not require the DSP cross-linker, suggesting that PI4KIIα interacts with these proteins with either higher affinity or a longer half-life than with the membrane-trafficking related proteins (Figure 2A, lanes 6 and 7). To further test the specificity of these interactions, we isolated hemagglutinin (HA)-tagged PI4KIIα from HEK293T cells by HA immunoprecipitation. We confirmed the presence of RhoGEF1 in these protein complexes, demonstrating that these interactions can be detected regardless of purification strategy (Figure 2B, lane 3). Taken together, these results indicate that the PI4KIIα interactome enriches proteins whose gene ontology annotation significantly implicates them in actin function.
FIGURE 2:
Biochemical confirmation of PI4KIIα-interacting proteins. (A) PI4KIIα was immunoaffinity purified from un-cross-linked (odd lanes) and DSP–cross-linked (even lanes) detergent-soluble SHSY-5Y neuroblastoma cell extracts. Immune complexes were resolved by SDS–PAGE and analyzed by immunoblot with antibodies against PI4KIIα, clathrin heavy chain (CHC), pallidin, peroxiredoxin-1 (Prdx-1), RhoGEF1, Dock7, and the WASH complex subunit strumpellin and Nedd4-1. Coisolating proteins were not detected in control reactions where an excess of PI4KIIα antigenic peptide was included to outcompete the endogenous PI4KIIα (lanes 4 and 5). Lane 3 shows antibody-coated beads incubated in the absence of cell lysate. Inputs represent 0.25%. (B) HEK293T cells transiently expressing HA-tagged PI4KIIα were DSP–cross-linked (lanes 3 and 4), and detergent-soluble extracts were immunoprecipitated with HA antibodies. Immune complexes were resolved by SDS–PAGE and analyzed by immunoblot with the indicated antibodies. RhoGEF1 is present in HA immunoprecipitations (lane 3) but not in peptide outcompetition controls (lane 4). Lane 2 shows antibody-coated beads incubated in the absence of cell lysate. Inputs represent 0.5%, and n = 3. (C, D) SHSY-5Y cells stably expressing a FLAG-tagged subunit of the BLOC-1 complex (dysbindin) were incubated in the absence (D, lanes 1, 4, and 6) or presence (C, all lanes, and D, lanes 2, 5, and 7) of DSP cross-linker, and detergent-soluble extracts were immunoprecipitated with antibodies against PI4KIIα (C) or the FLAG epitope (D). Immune complexes were resolved by SDS–PAGE and analyzed by immunoblot with antibodies against the indicated proteins identified in the PI4KIIα interactome. Also included are two BLOC-1 membrane protein cargoes, the v-SNARE VAMP7 and the Menkes disease copper transporter ATP7A. As in A and B, controls are antigenic peptide outcompetition (C, lane 4, and D, lanes 6 and 7). Lanes C2 and D3 show antibody-coated beads incubated in the absence of cell lysate. Inputs represent 0.5%.
Our network analysis of the PI4KIIα interactome and BLOC-1 subunits suggested connections between PI4KIIα-interacting proteins and BLOC-1 subunits (Figure 1F). Moreover, BLOC-1 regulates the subcellular localization of PI4KIIα (Larimore et al., 2011). For these reasons, we predicted that these previously unrecognized actin-related PI4KIIα interactors would coisolate with BLOC-1. To test this prediction, we exploited an SHSY-5Y cell line that stably expresses a FLAG-tagged dysbindin subunit of BLOC-1. This tagged subunit incorporates into functional BLOC-1 complexes (Larimore et al., 2011; Gokhale et al., 2012a). First, we confirmed that this BLOC-1 subunit could be detected in PI4KIIα immunoaffinity purifications (Figure 2C, lane 3). We next tested whether PI4KIIα interactors would be present in BLOC-1 protein complexes isolated by FLAG immunoaffinity purification (Figure 2D, lanes 4 and 5). As previously reported, PI4KIIα can be detected by this method (Figure 2D, lane 5; Larimore et al., 2011). In addition, two PI4KIIα interactors, RhoGEF1 and the WASH complex subunit strumpellin, coisolate with BLOC-1 complexes (Figure 2D, lane 5). Further, we also detected the presence of two known BLOC-1 cargoes, the Menkes disease copper transporter ATP7A and the lysosomal SNARE VAMP7 (Salazar et al., 2006; Setty et al., 2008; Newell-Litwa et al., 2009, 2010). These interactions were not detected in controls in which an excess of FLAG peptide was included in the reaction to outcompete FLAG-dysbindin for binding to the immunoprecipitation antibody (Figure 2D, compare lanes 5 and 7). Our results indicate that isolating PI4KIIα or BLOC-1 protein complexes robustly copurifies an actin-regulatory machinery. The copurification of PI4KIIα, strumpellin, RhoGEF1, and BLOC-1 subunits suggests that these proteins might act in concert to regulate BLOC-1–dependent traffic.
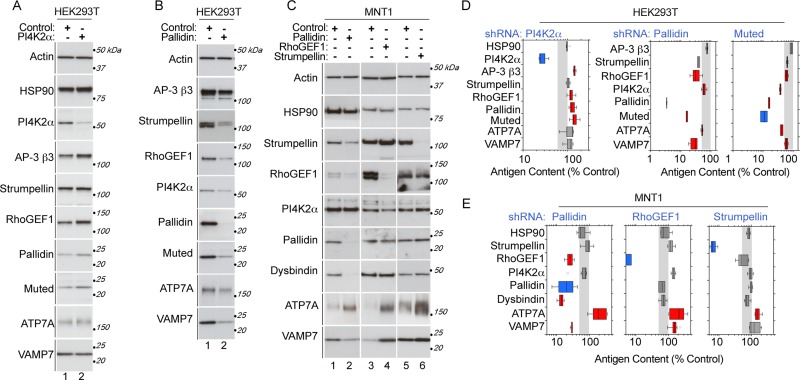
Mutation or depletion of a membrane trafficking complex subunit usually correlates with down-regulation of the other complex subunits and with changes in the total cellular content of pathway cargoes and associated factors (Peden et al., 2002; Kantheti et al., 2003; Li et al., 2003; Starcevic and Dell'Angelica, 2004; Jia et al., 2010). We used this principle to independently test components of the PI4KIIα interactome. We predicted that perturbation of either PI4KIIα or BLOC-1 would affect the content of PI4KIIα interactome components if these proteins were regulators of a BLOC-1–dependent vesicle biogenesis pathway. PI4KIIα and two BLOC-1 subunits, muted and pallidin, were independently depleted by short hairpin RNA (shRNA) in HEK293T cells (Figure 3, A, B, and D). Semiquantitative immunoblotting analysis demonstrated that PI4KIIα depletion up-regulated the content of RhoGEF1, as well as that of subunits of AP-3 and BLOC-1. The latter observation is consistent with the existence of a tripartite complex between PI4KIIα, AP-3, and BLOC-1 (Salazar et al., 2009; Figure 3, A and D). These relationships among components of the PI4KIIα interactome were also observed after depletion of the BLOC-1 complex (Figure 3, B and D). The total cellular content of PI4KIIα, RhoGEF1, and BLOC-1 subunits was significantly reduced in response to the depletion of either muted or pallidin (Figure 3, B and D). We then analyzed the content of two well-characterized BLOC-1 cargoes, a lysosomal SNARE, VAMP7, and the copper transporter mutated in Menkes disease, ATP7A (OMIM 309400; Salazar et al., 2006; Setty et al., 2008; Newell-Litwa et al., 2009, 2010). The total cellular content of both of these cargoes was significantly decreased in BLOC-1–depleted cells but not in PI4KIIα-depleted cells (Mann–Whitney U test p < 0.05; Figure 3, A, B, and D, red boxes). The normal content of VAMP7 and ATP7A in PI4KIIα-depleted cells suggests that the up-regulation of BLOC-1 complex subunits may be compensatory.
FIGURE 3:
Genetic confirmation of PI4KIIα-interacting proteins. (A, D) HEK293T cells were treated with scramble siRNA control or siRNA targeting PI4KIIα. (B, D) HEK293T cells were treated with lentiviruses carrying scramble shRNA control or shRNA targeting the pallidin or muted subunits of the BLOC-1 complex. (C, E) MNT-1 human melanoma cells were treated with lentiviruses carrying scramble shRNA control or shRNA targeting the BLOC-1 subunit pallidin, RhoGEF1, or the WASH complex subunit strumpellin. HEK293T and MNT-1 cell lysates were resolved by SDS–PAGE, and the total cellular content of proteins of interest was measured by semiquantitative immunoblot. Box plots in D and E depict quantifications where the targeted protein is highlighted in blue and antigens significantly changed are labeled in red. Gray shading represents 50–100% of control. All experiments were done in biological triplicates with at least one determination per replicate. Significant p values are all <0.025 and were determined by a nonparametric analysis using a group Kruskal–Wallis rank sum test followed by Wilcoxon–Mann–Whitney rank sum test. The AP-3 subunit β3A (D) and Hsp90 (D, E) were used as controls for comparisons.
Next we tested whether shRNA-mediated depletion of BLOC-1, strumpellin, or RhoGEF1 would affect the total cellular content of other PI4KIIα interactome components. First, we explored the consequences of BLOC-1 depletion in the melanoma cell line MNT-1, a cell type in which localization of BLOC-1 to tubular endosomes has been defined at the subcellular level by electron microscopy (Di Pietro et al., 2006). Consistent with our finding in HEK293T cells, the total cellular content of the PI4KIIα interactor RhoGEF1 decreased in MNT1 cells depleted of BLOC-1 (Figure 3, C and E). Depletion of the PI4KIIα interactome components RhoGEF1 and strumpellin did not modify the total cellular content of BLOC-1 subunits (Figure 3, C and E). However, the BLOC-1 cargoes VAMP7 and ATP7A were significantly changed (Figure 3, C and E). The total cellular content of ATP7A increased in response to BLOC-1, RhoGEF1, or strumpellin depletion in MNT1 cells (Figure 3E; 2.3 ± 0.12–, 2.2 ± 0.46–, and 1.6 ± 0.13–fold increase compared with controls, respectively). In addition, VAMP7 levels were significantly decreased in BLOC-1 depletions, significantly increased in RhoGEF1 depletions, and unchanged in strumpellin depletions. These data suggest a disruption to the regulation of steady-state levels of these BLOC-1 cargoes upon depletion of PI4KIIα interactome components and thereby establish genetic interactions between PI4KIIα interactome components.
The WASH complex and filamentous actin reside on PI4KIIα-positive early endosomes
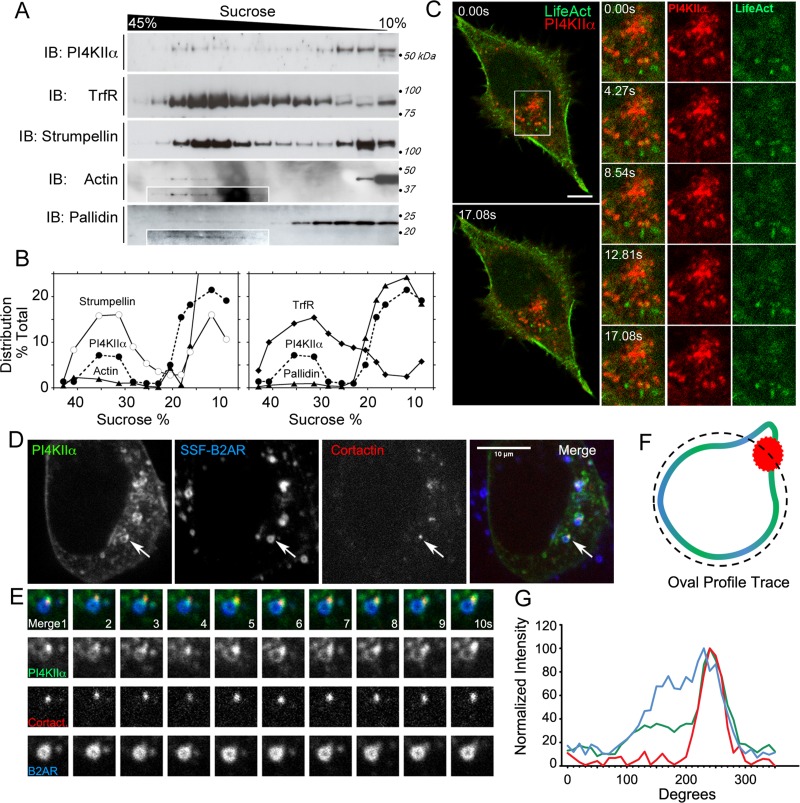
The WASH complex is an actin-regulatory complex that localizes to early endosomes (Derivery et al., 2009; Gomez and Billadeau, 2009; Gomez et al., 2012; Duleh and Welch, 2010; Carnell et al., 2011; Zech et al., 2011; Harbour et al., 2012). Similarly, PI4KIIα localizes to early endosomes, where it binds to BLOC-1 (Balla et al., 2002; Guo et al., 2003; Minogue et al., 2006; Craige et al., 2008; Salazar et al., 2009). Therefore we predicted that PI4KIIα-positive endosomes contain BLOC-1, the WASH complex, and actin. To test this prediction, we first separated endosomal compartments from HEK293T cell homogenates by sucrose velocity sedimentation (Clift-O'Grady et al., 1998; Lichtenstein et al., 1998). As expected, PI4KIIα cosedimented to 35% sucrose with transferrin receptor, an endosomal marker (Figure 4, A and B). In addition, the WASH complex subunit strumpellin, the BLOC-1 complex subunit pallidin, and actin were present in these fractions (Figure 4, A and B). Pallidin and actin were predominantly found at the top of gradients where free cytosolic proteins sediment (Clift-O'Grady et al., 1998; Lichtenstein et al., 1998). The presence of filamentous actin on PI4KIIα-containing endosomes was confirmed by live-cell imaging of fluorescently tagged EGFP-PI4KIIα with the LifeAct fluorescent probe (Figure 4C; Riedl et al., 2008). We tracked fluorescently labeled FLAG antibodies bound to an exofacial tag engineered in the β2-adrenergic receptor (SSF-B2AR; Figure 4D). We stimulated internalization of receptor-FLAG antibody complexes by addition of the adrenergic receptor agonist isoproterenol to functionally define early endosomes (Puthenveedu et al., 2010). In these early endosomes, branched actin networks were detected by cortactin-dsRed (Figure 4, D–G), which was concentrated at the base of small, digit-like projections that were enriched for PI4KIIα. The small amounts of pallidin and actin cosedimenting and/or colocalizing with PI4KIIα and endosomal markers reflects the transient nature of these associations when analyzed in the absence of the cross-linker DSP. Thus we unequivocally identified that PI4KIIα and branched actin colocalize in functionally defined early endosomes.
FIGURE 4:
PI4KIIα, the WASH complex subunit strumpellin, and actin cytoskeleton components coreside at early endosomes. (A, B) HEK293T cells were homogenized, and low-speed supernatants were resolved by sucrose velocity sedimentation on a 10–45% sucrose gradient. PI4KIIα and the WASH complex subunit strumpellin cosedimented to 35% sucrose with endosomal organelles, as identified by the transferrin receptor (TrfR). Cytosolic fractions sediment at the top of the gradient (10% sucrose). (B) Relative distribution of the indicated proteins for the fractionation shown in A. Images and distribution profile are representative of two independent experiments. (C) Live-cell imaging of fluorescently tagged PI4KIIα and LifeAct to visualize filamentous actin in HEK293T cells confirmed the presence of actin at PI4KIIα-positive organelles (n = 12 cells). (D) HEK293T cells stably expressing PI4KIIα-GFP and Signal-Sequence FLAG-tagged β2-adrenergic receptors (SSF-B2AR) were transiently transfected with mCortactin-dsRed and imaged 10 min after addition of 10 μM isoproterenol. A single confocal stack was captured in three channels at 1-s intervals. (E) Expanded image of endosome indicated by an arrow in A over a 10-s period. (F) Diagram of the oval profile used to generate traces of fluorescence intensity around endosomes. (G) Representative trace of normalized fluorescence intensity for each protein in the endosome above.
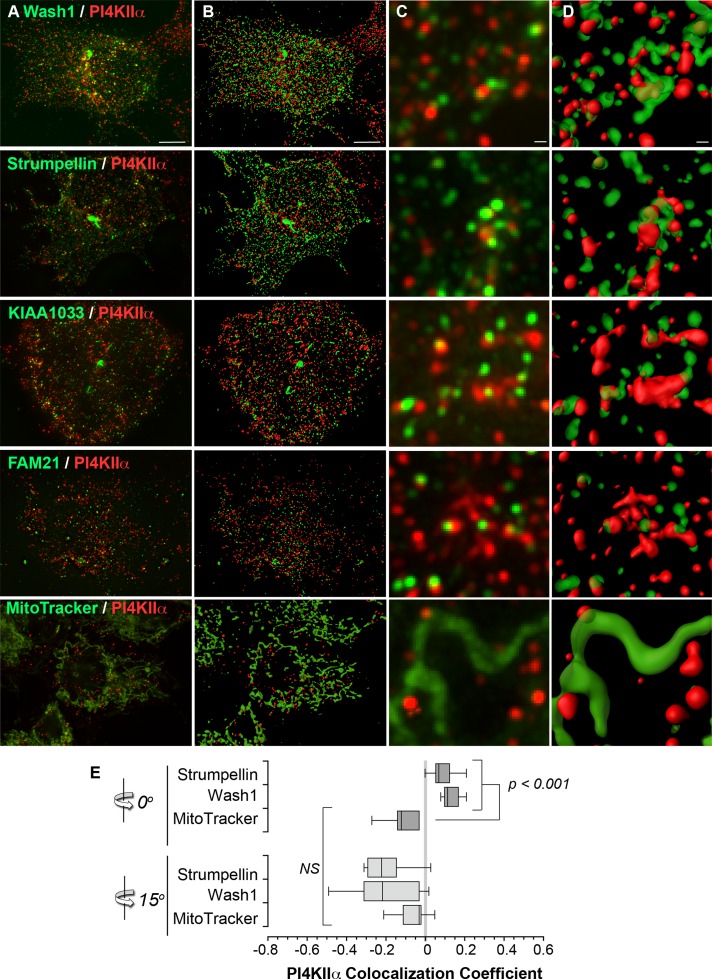
We tested the presence of the WASH complex in PI4KIIα-positive endosomes by visualizing EGFP-tagged subunits of the WASH complex and endogenous PI4KIIα using high-resolution deconvolution microscopy. Wash1-, strumpellin-, SWIP/KIAA1033-, and Fam21-EGFP were all closely apposed to the PI4KIIα signal (Figure 5, A and C). This physical proximity was especially apparent in three-dimensional projections of the z-stack data, where the WASH complex subunits appear to wrap about PI4KIIα-containing organelles (Figure 5, B and D). We observed a low degree of overlap between the highest-intensity signals and thus measured the colocalization of strumpellin-EGFP and Wash1-EGFP with endogenous PI4KIIα using Pearson's colocalization coefficient. Pearson's coefficient was 9.1 ± 2.5 and 13.1± 1.9 for strumpellin-EGFP and Wash1-EGFP, respectively, confirming that there was a low correlation of intensity between overlapping signals (Figure 5E). However, we verified the specificity of this calculated coefficient by rotating one channel 15° and analyzing the colocalization of PI4KIIα with the mitochondrial probe MitoTracker. Pearson's colocalization coefficient between PI4KIIα and MitoTracker was negative, reflecting a complete lack of overlap of these signals (Figure 5E). This observation was confirmed because channel rotation did not modify the negative Pearson's coefficient between PI4KIIα and MitoTracker. In contrast, rotating either strumpellin-EGFP or Wash1-EGFP by 15° dropped Pearson's coefficient to values indistinguishable from those of MitoTracker colocalization (p ≥ 0.05; Figure 5E). These results indicate that the WASH complex and actin filaments reside in subdomains of PI4KIIα-containing endosomes. These findings suggest that the Wash1-actin machinery is poised to regulate the trafficking of PI4KIIα and other BLOC-1 cargoes from early endosomes.
FIGURE 5:
PI4KIIα colocalizes with the WASH complex. (A, C) HEK293T cells were either transiently transfected with EGFP-tagged subunits of the WASH complex or stained with MitoTracker green, fixed, and processed for indirect immunofluorescence microscopy. Images were acquired by high-resolution deconvolution microscopy. The z-stack projections are shown. (B, D) Isosurface renderings of the z-stack data created using Imaris software. (E) Calculation of Pearson's correlation coefficient between signal intensities of strumpellin-EGFP, WASH-EGFP, or MitoTracker and endogenous PI4KIIα. Correlation was lost when one channel was rotated by 15°. Here n = 6 for WASH-EGFP, n = 7 for strumpellin-EGFP, and n = 9 for MitoTracker-A488. Statistical comparisons were performed by nonparametric analysis using a group Kruskal–Wallis rank sum test followed by individual Wilcoxon–Mann–Whitney rank sum tests.
The WASH complex modulates the targeting of BLOC-1 cargoes
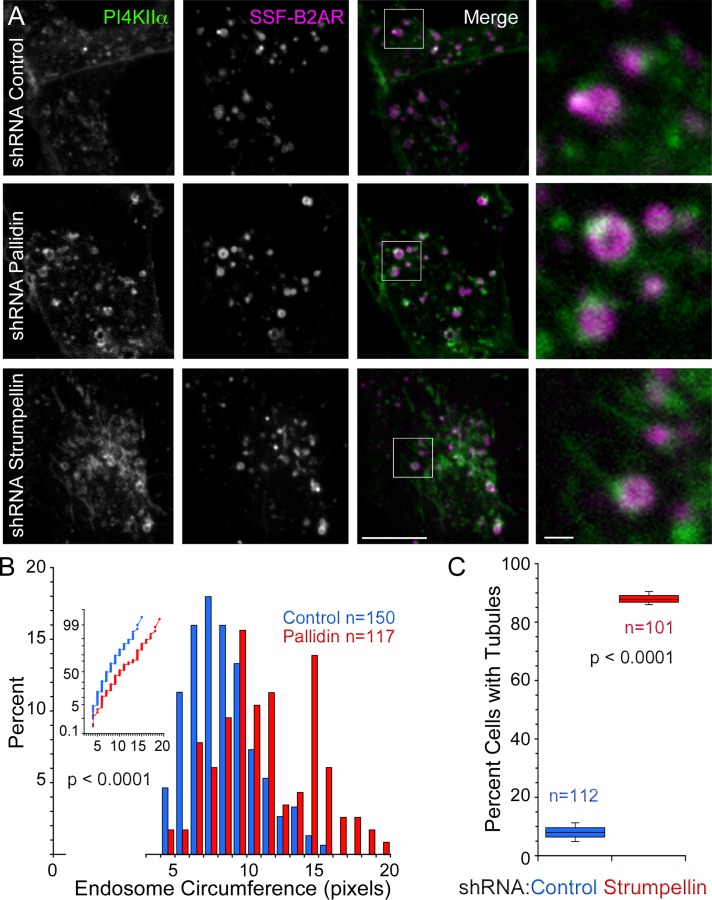
WASH complex depletion is characterized by the appearance of enlarged cargo-laden tubules from endosomes and cargo missorting (Gomez and Billadeau, 2009; Gomez et al., 2012; Harbour et al., 2012; Jia et al., 2012; Seaman, 2012; Helfer et al., 2013). Therefore we hypothesized that PI4KIIα is present in tubular structures whose length is regulated by the WASH complex. To test this hypothesis, we expressed PI4KIIα-EGFP in HEK293T cells stably expressing FLAG-tagged β2-adrenergic receptor and imaged cells by live-cell microscopy. Endosomes were unequivocally identified by ligand-induced internalization of fluorescent FLAG antibodies bound to the exofacial domain of the β2-adrenergic receptor. Tubules containing PI4KIIα and/or β2-adrenergic receptor emerged from early endosomes, and their size increased in WASH complex–depleted cells (Figure 6, A and C). Overall the size of early endosomes containing PI4KIIα and β2-adrenergic receptor was unaffected by WASH complex deficiency (Figure 6A). In contrast, BLOC-1 depletion by shRNA targeting of pallidin was characterized by an enlargement of these early endosomes without enlargement of tubules (Figure 6, A and B). The changes in the architecture of early endosomes containing PI4KIIα and β2-adrenergic receptor by depletion of either BLOC-1 or WASH complexes support a model in which WASH complexes in concert with BLOC-1 regulate the targeting of BLOC-1 cargoes from early endosomes.
FIGURE 6:
Depletion of pallidin and strumpellin alter endosomal morphology. (A) HEK293T cells stably expressing Signal-Sequence FLAG-tagged β2-adrenergic receptors (SSF-B2AR) and PI4KIIα-EGFP were transduced with scrambled, pallidin, or strumpellin shRNA lentiviruses. Pallidin-depleted cells were imaged 8 d from initiation of transduction, and strumpellin-depleted cells were imaged 5 d after transduction. Cells were grown under selection after day 2 of transduction. All knockdowns were compared with parallel transduction with scrambled shRNA. Images are maximum projections of three confocal slices taken with 0.3-μm intervals. (B) Frequency histogram of endosomal circumference from cells treated with either scrambled (blue) or pallidin (red) shRNA. Insert to the left depicts a probability plot of the same data. The p value determined by nonparametric Kolmogorov–Smirnov test. Here n = 150 for control cells and n = 117 for pallidin-depleted cells. (C) Percentage of cells expressing scrambled or strumpellin shRNA displaying endosomal tubulation. Cell selection was performed in the SSF-B2AR channel and blinded for PI4KIIα before FLAG antibody internalization, then imaged 15 min after addition of 10 μM isoproterenol. The p value was determined by nonparametric Fisher exact test. Here n = 112 for control cells and n = 101 for pallidin-depleted cells.
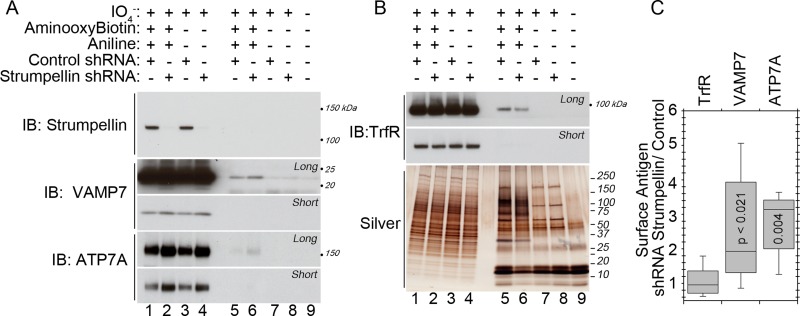
We predicted that WASH complex deficiencies would alter the subcellular distribution of BLOC-1 cargoes if these complexes act on the same pathway. To test this prediction, we treated MNT1 melanoma cells with shRNA targeting the strumpellin subunit of the WASH complex or scramble control. We assessed the cell surface level of two BLOC-1 cargoes: the Menkes copper transporter ATP7A and the lysosomal SNARE protein VAMP7 (Salazar et al., 2006; Setty et al., 2008; Newell-Litwa et al., 2009, 2010). As previously described, we observed an ∼1.5-fold increase in the total cellular content of ATP7A in WASH complex depletion (Figure 7A, lanes 1–4). Cell surface levels of ATP7A, however, increased 3.3 ± 0.8–fold in WASH complex depletion (Figure 7A, lanes 5 and 6, and 7C; average ± SEM). Similarly, cell surface levels of VAMP7 increased 2.5 ± 0.6–fold in WASH complex depletions as compared with control, whereas the total cellular levels of VAMP7 were moderately but not significantly affected (Figure 7A, lanes 5 and 6, and Figure 3, C and D). These cell surface increases were specific to BLOC-1 cargoes rather than global perturbations, as the total cellular content and cell surface levels of transferrin receptor were unaffected in WASH complex–depleted cells (Figure 7, B and C). Moreover, silver stain of total biotinylated cell surface protein showed no global increases in cell surface proteins from WASH complex depletions (Figure 7B). Thus BLOC-1 cargoes are missorted to the cell surface in WASH complex depletion. These data support a model in which WASH complexes modify the traffic of BLOC-1 cargoes.
FIGURE 7:
WASH complex depletion alters the subcellular distribution of BLOC-1 cargoes. (A) MNT1 melanoma cells were treated with shRNA directed against the strumpellin subunit of the WASH complex or scramble control. Cell surface proteins were labeled by surface biotinylation and isolated by streptavidin bead pull-down. Representative immunoblots show an increase in the surface content of ATP7A and Vamp7, two BLOC-1 cargoes, in WASH complex depletions. (B) Transferrin receptor (TrfR) was measured as control. Silver stain of isolated proteins revealed no global decreases in the cell surface content of proteins (compare lanes 5 and 6). (C) Quantifications of the surface expression of proteins of interest in WASH complex depletion relative to scramble control. Data are averages ± SEM. We found significant increases in the BLOC-1 cargoes ATP7A and Vamp7 of 3.3 ± 0.8–fold and 2.5 ± 0.6–fold, respectively. Transferrin receptor was not significantly changed. The p value was determined by nonparametric Wilcoxon–Mann–Whitney rank sum test. There were n = 8 determinations from four independent experiments.
DISCUSSION
In this study we identified previously unrecognized components of a membrane protein sorting pathway affected in the Hermansky–Pudlak syndrome by isolating the membrane-anchored lipid kinase PI4KIIα. PI4KIIα binds and regulates two complexes mutated in the Hermansky–Pudlak syndrome, the AP-3 and BLOC-1 complexes, by means of a dileucine sorting motif and its kinase activity (Craige et al., 2008; Salazar et al., 2009; Larimore et al., 2011; Gokhale et al., 2012a). We therefore hypothesized that PI4KIIα interacts with unrecognized components of a vesicle transport pathway requiring the BLOC-1–AP-3–PI4KIIα complex. PI4KIIα could interact with these components either concurrently with binding to AP-3 and BLOC-1 and/or at steps either preceding or following the binding to these complexes. We therefore designed a strategy to enrich PI4KIIα interactors upstream and downstream of the PI4KIIα dileucine sorting motif recognition event. To achieve this goal, we raised an antibody against a 20-residue peptide of PI4KIIα that contains the ERQPLL sorting motif. Mutation of this motif to ERQPAA is sufficient to prevent recognition of PI4KIIα by this antibody by immunoblot. Although the ERQPLL motif is present in five other proteins encoded in the human genome, none of these proteins is enriched in the PI4KIIα interactome, ensuring that this antibody specifically recognizes PI4KIIα (www.mrc-lmb.cam.ac.uk/genomes/madanm/harvey/). We used this antibody to immunoaffinity purify PI4KIIα from isotope-labeled cell lysates (SILAC), allowing for the quantitative identification of interacting proteins over nonspecific contaminants and immunoglobulins (Ong et al., 2002; Mann, 2006; Zlatic et al., 2010; Gokhale et al., 2012a). Strikingly, PI4KIIα isolation preferentially coenriched proteins that regulate the actin cytoskeleton, including guanine exchange factors for Rho family GTPases, RhoGEF1, DOCK7, and GEF-H1, and several subunits of the WASH complex. In addition, PI4KIIα coisolated with membrane trafficking protein complexes, such as clathrin heavy chain and the β3A subunit of the AP-3 complex. These membrane-trafficking complexes coisolated with PI4KIIα at a lower stoichiometry as compared with actin regulators, as intended with our isolation strategy. We biochemically confirmed several of these PI4KIIα interactions. Further, we predicted that if PI4KIIα interactome components interact, then perturbing these proteins would change the total cellular content of other interactome components. With this guiding rationale, we established that genetic depletion of PI4KIIα itself, the BLOC-1 complex, RhoGEF1, or the WASH complex altered the total cellular content of either PI4KIIα interactome components or BLOC-1 cargoes, the Menkes disease copper transporter ATP7A and the lysosomal SNARE VAMP7 (Salazar et al., 2006; Setty et al., 2008; Newell-Litwa et al., 2009, 2010). We conclude that these novel PI4KIIα interactors biochemically and genetically interact with components of the actin cytoskeleton. We favor a model in which membrane protein cargoes are sorted by a BLOC-1–dependent mechanism into tubular carriers whose size is controlled by the WASH complex, possibly by generating force for membrane scission.
We focused our efforts to characterize the consequences of these interactions on the WASH complex. Wash1 is a type I nucleation-promoting factor that binds the Arp2/3 complex to locally nucleate and organize the polymerization of branched actin filaments (Derivery et al., 2009; Jia et al., 2010; Veltman and Insall, 2010; Rotty et al., 2012). The WASH complex localizes to tubular domains of early endosomes, where actin polymerization is proposed to create localized membrane domains for cargo sorting and/or regulate tubule morphology by membrane scission (Gomez and Billadeau, 2009; Gomez et al., 2012; Duleh and Welch, 2010, 2012; Harbour et al., 2012; Helfer et al., 2013). Our work identifies the WASH complex as a previously unrecognized interactor of BLOC-1 and PI4KIIα. This assertion is founded on the following findings and rationale. We found that WASH complex subunits localize to PI4KIIα-containing organelles and coprecipitate with BLOC-1 complex subunits, which is consistent with a previous report that Wash1 interacts with the BLOS2 subunit of the BLOC-1 complex by yeast two-hybrid analysis and immunoprecipitation of tagged subunits (Monfregola et al., 2010). Further, we found that PI4KIIα is contained in small tubular structures that emanate from early endosomes, which were functionally labeled by internalized β-adrenergic receptors. If BLOC-1 and the WASH complex coordinate to regulate early endosomal tubule morphology and/or cargo sorting, then we predicted morphological and missorting phenotypes would arise upon perturbation of protein complexes. First, we predicted early endosomal morphological changes in BLOC-1 and WASH complex depletions, leading to either enlarged endosomes or endosomal tubules. Indeed, we found that BLOC-1 depletion increases the size of functionally identified early endosomes without increasing tubule size. This phenotype is consistent with our previous findings that endosomes increase in size after AP-3, BLOC-1, or PI4KIIα depletion (Salazar et al., 2009). Further, in WASH complex depletion we found elongation of early endosomal PI4KIIα-containing tubules, which is consistent with previous reports demonstrating increases in endosomal tubule length upon Wash1 depletion (Derivery et al., 2009; Gomez and Billadeau, 2009). Second, we predicted missorting of BLOC-1 cargo in WASH complex depletions, which would suggest an impaired flow of cargoes or membrane out of endosomes. We concentrated on the missorting of specific endolysosomal cargoes to the cell surface, which is a characteristic phenotype in AP-3, BLOC-1, and PI4KIIα deficiencies (Dell'Angelica et al., 1999; Peden et al., 2002; Di Pietro et al., 2006; Salazar et al., 2006; Setty et al., 2007; Baust et al., 2008). Two BLOC-1 cargoes, ATP7A and VAMP7, mislocalize to the cell surface in WASH complex depletions. This phenotype was specific to endolysosomal cargo, as cell surface levels of a receptor that undergoes constitutive recycling, the transferrin receptor, were unchanged.
How do BLOC-1 and the WASH complex mechanistically coordinate to regulate endosomal morphology and cargo sorting? Actin polymerization fulfills many roles in membrane protein sorting and trafficking, including creating domains to spatially organize sorting machinery, providing mechanical support for membrane deformation during vesicle budding, generating force to aid in membrane scission of vesicles, and propelling carriers after budding is completed (Qualmann et al., 2000). The BLOC-1 cargo missorting phenotype observed after WASH complex depletion suggests a role for Wash1 in steps occurring at or upstream of the membrane scission of vesicles. Actin polymerization by the WASH complex is proposed to generate force required for membrane scission, a hypothesis that awaits testing by in vitro biochemical reconstitution (Bear, 2009; Derivery et al., 2009; Gomez and Billadeau, 2009; Rotty et al., 2012). However, this model is consistent with the binary biochemical association of WASH complex subunits, PI4KIIα, BLOC-1, and the BLOC-1 cargoes ATP7A and VAMP7 documented here and in our previous work (Salazar et al., 2009; Larimore et al., 2011). In addition, this model predicts that these components interact concurrently, which remains to be documented. An alternative hypothesis is that PI4KIIα and the BLOC-1 complex independently associate with the WASH complex to regulate its activity. Irrespective of whether BLOC-1 and PI4KIIα act in concert or as independent factors, we speculate that the WASH complex acts as a generic mechanism to nucleate branched actin polymerization in endosomes, with spatial or cargo specificity dictated by different upstream accessory factors such as coats and/or lipid modifications.
PI4KIIα is an interesting candidate upstream activator of the WASH complex because of its capacity to produce PI4P (Balla et al., 2002; Barylko et al., 2002). The WASH complex Fam21 subunit, a component of the PI4KIIα interactome, is proposed to localize WASH to endosomal membranes by binding phosphoinositides and membrane protein sorting complex subunits, such as the Vps35 subunit of the retromer complex (Gomez and Billadeau, 2009; Harbour et al., 2012; Helfer et al., 2013). Fam21 binds PI4P in vitro, the lipid product of PI4KIIα (Jia et al., 2010). PI4KIIα and PI4P may thereby either recruit the WASH complex to early endosomal membranes or stabilize the complex at these membranes for function within the AP-3–BLOC-1 or multiple sorting pathways. As discussed later, genetic disruption of a WASH complex subunit and PI4K2A cause common neurodegenerative phenotypes in human patients and mouse models, supporting a regulatory role for PI4KIIα in WASH localization (Valdmanis et al., 2007; Simons et al., 2009; Clemen et al., 2010). In addition, PI4KIIα interactome components may regulate WASH complex activation. A common feature of nucleation-promoting factors such as Wash1 is their regulation and activation by small GTPases. The regulatory GTPase for the WASH complex has remained elusive in mammalian cells. In Drosophila melanogaster genetic interactions between Rho and Wash1 have been reported (Liu et al., 2009). The high-stoichiometry association of a RhoA guanine exchange factor, RhoGEF1, with PI4KIIα and the similarities between strumpellin depletion and RhoGEF1 depletion in MNT1 cells suggest a regulatory mechanism between these two PI4KIIα interactors. Future experiments are needed to test precise molecular and functional relationships between PI4KIIα, RhoGEF1, the WASH complex, and the BLOC-1 complex.
In addition to expanding our conception of the vesicular trafficking pathway controlled by the Hermansky–Pudlak syndrome complex BLOC-1, our findings have novel implications for the understanding of neurodegenerative diseases that affect central or peripheral motor neuron axons, such as hereditary spastic paraplegia (HSP). Point mutations in the WASH complex subunit strumpellin cause autosomal-dominant HSP (OMIM 610657), which is characterized by the degeneration of axons projecting to the spinal cord from the brain motor cortex (Valdmanis et al., 2007; Clemen et al., 2010). More than 50 genes have been linked to HSP in humans (Finsterer et al., 2012). The common theme of this disease is degeneration of central neurons with long axons, presumably due to the high burden of maintaining a synapse distant from the corresponding cell body. Consistent with this theme, many of the 50 identified genes are endosomal membrane trafficking complexes (Blackstone, 2012). No interactions between strumpellin and other HSP-causative genes have been described, making it difficult to place strumpellin and the WASH complex within a cellular framework to understand how mutations in the strumpellin protein lead to disease. A recent study found that strumpellin subunits containing HSP disease–causing mutations assemble into WASH complexes that localize to early endosomes in neurons and have no apparent consequence for retromer complex–mediated recycling of the β-adrenergic receptor (Freeman et al., 2013). Our data suggest that PI4KIIα interactome components could be an alternative pathway disrupted by these mutations. This hypothesis is supported by the HSP-like phenotype that results after targeted disruption of PI4KIIα in mice (Simons et al., 2009). In addition to a role of the PI4KIIα interactome in degeneration of central motor neurons, our data suggest connections with disease mechanisms in peripheral motor neuron axonal degeneration. Point mutations to the BLOC-1 cargo ATP7A in human patients cause a degeneration of peripheral motor axons (Kennerson et al., 2010; Yi et al., 2012). In this article, we found that ATP7A is mislocalized in strumpellin-depleted cells. In addition, PI4KIIα itself is a BLOC-1 cargo, suggesting that it too might mislocalize in strumpellin perturbations that cause HSP in humans. Therefore we propose that disruption to PI4KIIα interactome components might contribute to the pathophysiology of central and peripheral motor neuron axon degeneration.
MATERIALS AND METHODS
Antibodies and cell culture
Antibodies used in this study are listed in Supplemental Table S3. The PI4KIIα antibody used here for immunoprecipitation, Western blot, and immunofluorescence microscopy experiments was raised against the sequence 51-PGHDRERQPLLDRARGAAAQ-70 and was described in Larimore et al. (2011) We obtained this antigenic peptide by custom peptide synthesis (Bio-Synthesis, Lewisville, TX). The antigenic peptide was diluted to a 20 mM stock in 0.5 M MOPS, pH 7.4, and stored at −80°C.
HEK293T and SHSY-5Y cells (American Type Culture Collection, Manassas, VA) were maintained in DMEM with 4.5 g/l glucose, l-glutamine, and sodium pyruvate (Cellgro, Manassas, VA) supplemented with 10% fetal bovine serum (FBS; HyClone, Logan, UT) and 100 μg/ml penicillin/streptomycin (HyClone) at 37°C and 10% CO2. MNT1 cells were maintained in DMEM supplemented with 20% AIM-V medium (Life Technologies, Carlsbad, CA), 10% FBS (heat inactivated at 65°C for 60 min), and 100 μg/ml penicillin/streptomycin at 37°C and 5% CO2.
For SILAC labeling, cells were grown in either a light medium containing only 12C and 14N in amino acids arginine and lysine (R0K0) or a heavy medium containing 13C and 15N in these amino acids (R10K8). All reagents for SILAC labeling were obtained from Dundee Cell Products (Dundee, United Kingdom). Cells were grown for seven passages to ensure maximal incorporation of these amino acids. Our previous work found that these conditions lead to >97.5% incorporation of these amino acids into the total cellular pool (Ong and Mann, 2006; Gokhale et al., 2012a; Perez-Cornejo et al., 2012).
DNA expression constructs
The following PI4KIIα expression constructs were used: PI4KIIα-EGFP WT, PI4KIIα-HA WT, PI4KIIα-HA D308A, and PI4KIIα-HA LL60,61AA (Craige et al., 2008). The FLAG-tagged dysbindin construct is described in previous work (Gokhale et al., 2012a). Wash1-EGFP, Strumpellin-EGFP, KIAA1033-EGFP, and FAM21-EGFP were described previously (Harbour et al., 2010). Cortactin-dsRed and Signal-Sequence FLAG tagged β2-adrenergic receptors (SSF-B2AR) were previously described (Kaksonen et al., 2000; Puthenveedu et al., 2010).
Immunoprecipitation and immunoaffinity chromatography
SHSY-5Y neuroblastoma cells were grown to confluence and minimally cross-linked as previously described (Zlatic et al., 2010; Gokhale et al., 2012a, b; Perez-Cornejo et al., 2012). Briefly, cells were rapidly moved from the incubator to an ice bath and washed two times with ice-cold phosphate-buffered saline (PBS) supplemented with CaCl2 (0.1 mM) and MgCl2 (1 mM) (PBS-Ca-Mg). After this wash, cells were incubated for 2 h in 1 mM DSP diluted in PBS-Ca-Mg or a dimethyl sulfoxide vehicle–only control. After cross-linking, the DSP reaction was quenched by addition of a 25 mM Tris, pH 7.4, solution for 15 min. Cells were then washed two times in ice-cold PBS-Ca-Mg and lysed for 30 min in buffer A (150 mM NaCl, 10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid [HEPES], 1 mM ethylene glycol tetraacetic acid [EGTA], 0.1 mM MgCl2) with 0.5% Triton X-100 supplemented with Complete antiprotease (Roche, Indianapolis, IN). After lysis, cell debris were scraped from the plates, and lysates were spun at 16,100 × g for 15 min. The supernatant was recovered and diluted to 1 mg/ml. For immunoprecipitation, 500 μg of this clarified cell lysate was applied to 30 μl of Dynal magnetic beads (Invitrogen, Carlsbad, CA) coated with the immunoprecipitation antibody of interest. These immunoprecipitation reactions were incubated for 2 h with end-over-end rotation at 4°C. For PI4KIIα peptide competition controls, the PI4KIIα antigenic peptide was included in the immunoprecipitation reaction at a concentration of 40 μM. For FLAG competition controls, the 3X FLAG peptide was included at 340 μM, as previously described (Gokhale et al., 2012a; Perez-Cornejo et al., 2012). For HA competition controls, the HA peptide was included at 10 μM. After the 2-h incubation, beads were washed five times in buffer A with 0.1% Triton X-100. Proteins were then eluted from the beads either by boiling in Laemmli sample buffer at 75°C for 5 min or incubating with the antigenic peptide for 2 h on ice. Peptides were diluted in lysis buffer to their elution concentration (PI4KIIα antigenic peptide, 200 μM; 3X FLAG peptide, 340 μM). Samples were resolved by SDS–PAGE electrophoresis, followed by either silver stain or immunoblotting to detect individual proteins. For SILAC analysis, 18 immunoaffinity chromatography and 18 control reactions were conducted. Eluates from these reactions were combined and trichloroacetic acid precipitated to concentrate the samples for SDS–PAGE and mass spectrometry analysis. Samples were analyzed for SILAC protein identification by MS Bioworks (Ann Arbor, MI) and the Emory CND Proteomics Facility (Atlanta, GA). SILAC-labeled samples were separated on a 4–12% Bis-Tris Novex minigel (Invitrogen) using the MOPS buffer system and stained with Coomassie, and the lane was excised into 20 equal segments using a grid. Gel pieces were processed using a robot (ProGest; Digilab, Marlborough, MA) as described (Gokhale et al., 2012a; Perez-Cornejo et al., 2012).
Transient protein knockdown and immunoblot analysis
For shRNA-mediated protein knockdown, constructs in the pLKO.1 vector were obtained from Open Biosystems (Huntsville, AL). Clone IDs for the targeting constructs are as follows: muted (Bloc1s5), TRCN0000129117; pallidin (Bloc1s6), TRCN0000122781; strumpellin, TRCN0000128018; and RhoGEF1, TRCN0000033567. A scramble control vector was obtained from Addgene (Cambridge, MA; Vector 1864). HEK293T cells seeded in six-well plates were transfected with 1 μg of shRNA construct DNA overnight by Lipofectamine 2000 (Life Technologies) according to the manufacturer's instructions. MNT1 and SHSY-5Y cells were seeded in six-well plates and treated with 1–2 μl of high-titer lentiviral particles containing the appropriate lentiviral construct. The Emory NINDS Viral Vector Core prepared all high-titer lentiviruses. Two days after delivery of shRNA construct, transformed cells were selected by treatment with 2 μg/ml puromycin (Invitrogen) in the appropriate growth media as described. The total length of treatment was 5–10 d to allow for efficient knockdown of the proteins of interest. Small interfering RNA (siRNA) oligonucleotide sequences against PI4KIIα and controls, as well as procedures, were described previously (Craige et al., 2008; Salazar et al., 2009).
For immunoblot analysis of protein levels, cells were grown to confluence in six-well plates. Cells were treated with lentiviruses for 7 d. Plates were rapidly moved to an ice bath and immediately washed two times in ice-cold PBS. Cells were then lifted in buffer A and spun at 16,100 × g for 1 min to pellet. MNT1 cells were incubated for 10 min in buffer A before lifting to facilitate release from the tissue culture plate. Cell pellets were resuspended in 100 μl of buffer A supplemented with Complete antiprotease. Cells were then lysed by sonication (three rounds of five 1-s bursts). Lysates were diluted to 1 mg/ml and resolved by SDS–PAGE and immunoblotting. From 5 to 15 μg of cell lysate was loaded per well.
Immunofluorescence microscopy
HEK293T cells were transfected in six-well plates with 1 μg of the DNA construct by Lipofectamine 2000 for 4 h and then switched into growth medium. After 24 h, cells were seeded on glass coverslips coated with Matrigel. The next day, coverslips were moved to ice, washed two times in ice-cold PBS, and then fixed with 4% paraformaldehyde in PBS for 20 min. For cells stained with the A488 MitoTracker dye (Invitrogen), cells were incubated with 500 nM MitoTracker dye diluted in growth medium for 30 min. After this incubation, cells were immediately fixed at 37°C in 4% paraformaldehyde. After fixation, all coverslips were washed two times in PBS and then blocked and permeabilized for 30 min at room temperature in a solution of 0.02% saponin (Sigma-Aldrich, St. Louis, MO), 15% horse serum (HyClone), 2% bovine serum albumin (Roche), and 1% fish skin gelatin (Sigma-Aldrich) in PBS. Primary antibodies (Supplemental Table S3) were diluted in blocking solution and applied for 30 min at 37°C. After primary antibody incubation, coverslips were washed three times in blocking solution. Coverslips were incubated for 30 min at 37°C with fluorophore-conjugated secondary antibodies (Supplemental Table S3) diluted in blocking solution. Coverslips were washed two times in blocking solution, one time in PBS, and one time in ultrapure water and then mounted on glass slides in Gelvatol mounting medium (Sigma-Aldrich).
For fixed-cell deconvolution microscopy, images were collected on a Zeiss Axiovert 200M inverted microscope (Carl Zeiss, Jena, Germany) with a Sedat filter set (Chroma Technology, Brattleboro, VT). Slidebook 4.0 OSX software was used to run a multiwavelength, wide-field, three-dimensional microscopy system (Intelligent Imaging Innovations, Denver, CO). Images were visualized with a 100× oil differential interference contrast objective with numerical aperture 1.4 and captured using a scientific-grade cooled, charge-coupled Cool-Snap HQ camera (Photometrics, Tucson, AZ) with an ORCA-ER chip (Hamamatsu Photonics, Hamamatsu, Japan). Images were captured with 200 nm between z-planes. A nearest-neighbor, constrained iterative deconvolution algorithm with Gaussian smoothing was used to remove out-of-focus light (Swedlow et al., 1997, 2002). Images were exported from the Slidebook 4.0 software as 8-bit TIF series. For colocalization analysis, in-focus image planes were background subtracted with a 10-pixel rolling-ball algorithm in Fiji (ImageJ; National Institutes of Health, Bethesda, MD). A region of interest immediately surrounding individual cells was selected, and the Pearson coefficient was calculated by the Coloc 2 plug-in. Three-dimensional reconstructions were created from deconvolved 8-bit TIF series images imported into Imaris 6.3.1 software using manual thresholding and a minimal volume limit of 4 voxels (Bitplane Scientific Software, Zurich, Switzerland).
For live-cell imaging of EGFP-tagged PI4KIIα and mCherry-tagged LifeAct peptide, HEK293T cells expressing fluorescently tagged protein constructs were seeded in Matrigel-coated glass-bottom culture dishes (Matek Corporation, Ashland, MA). Before imaging, cells were maintained in HEK293T growth media as described at 37ºC and 10% CO2. Immediately before imaging, an imaging medium consisting of Hank's balanced salt solution minus phenol red and NaHCO2 (Sigma-Aldrich) supplemented with 10% FBS (HyClone) was applied. Cells were imaged using a Nikon TE2000 inverted microscope (Nikon Instruments, Melville, NY) equipped with a hybrid scanner, Perfect Focus, and an environmental chamber for temperature regulation to 37ºC and 5% CO2. Images were captured with a 100×, 1.49 numerical aperture oil objective and 500–550/570–620 filter sets using NIS-Elements AR 3.1 (Nikon) software. Cells were alternately excited with 488- and 568-nm-wavelength lasers every 4.25 s, and a single confocal plane image was captured. Images were exported as an 8-bit TIF series in the NIS-Elements software, and figures were compiled in Photoshop (Adobe, San Jose, CA).
For live-cell imaging of PI4KIIα with labeled functionally labeled endosomes, fluorescent labels were from expressed protein constructs (GFP and dsRed) or anti-FLAG M1 antibody conjugated to Alexa 647 dye. Cells were grown on coverslips and maintained in 10% FBS (Life Technologies), high-glucose DMEM (HyClone) and imaged in Opti-MEME (Life Technologies) with 10% FBS and buffered to pH 7.4 with 40 mM HEPES. Live-cell confocal images were collected on a Revolution XD Spinning Disk system (Andor, Belfast, United Kingdom) with a Nikon Eclipse Ti inverted microscope. Objective used for capture was a 100×/1.49 numerical aperture total internal reflection fluorescence objective from Nikon. Both microscope and objectives were housed in a temperature control case maintained at 37°C. Light sources were 488-, 561-, and 647-nm solid-state lasers, and images were captured on an iXON + 897 electron-multiplying charge-coupled device camera using Andor iQ. Image analysis was performed on raw images in ImageJ. Images were not modified in any way other than color and brightness/contrast assignment for multichannel images. For tubule scoring, cells were blindly selected in the β2-adregenergic receptor channel. We considered a cell to have the tubuled phenotype if it contained more than one object that was approximately four times longer than wide in the size range of endosomes. Cells were set in focus using only the β2-dregenergic receptor channel before agonist to avoid bias selecting for cells with the phenotype. All file names were randomized for blind assessment.
Cell fractionation
Confluent HEK293T cells were washed two times in ice-cold PBS and then lifted in intracellular buffer (38 mM potassium aspartate, 38 mM potassium glutamate, 38 mM potassium gluconate, 20 mM MOPS-KOH, pH 7.2, 5 mM reduced glutathione, 5 mM sodium carbonate, 2.5 mM magnesium sulfate, 2 mM EGTA). Cells were spun at 800 × g to pellet and then resuspended in a minimal volume of intracellular buffer supplemented with 5× concentrated Complete antiprotease. Cells were homogenized using a cell cracker with a 12-μm clearance (Clift-O'Grady et al., 1998; Lichtenstein et al., 1998). Between 200 and 250 μl of homogenate (450–800 μg) was layered on 10–45% sucrose gradients buffered with 20 mM MOPS (pH 7.2), which were prepared using the Gradient Master (Biocomp Instruments, Fredericton, Canada). Sucrose gradients were centrifuged for 1 h at 210,000 × g at 4°C in a Beckman SW55Ti rotor and fractions analyzed by immunoblot. Sucrose refractive indices for each fraction were measured by refractometer (Leica, Wetzlar, Germany).
Surface labeling and streptavidin pull-downs
After 7 d treatment with scramble or strumpellin-targeted lentiviruses, plates of confluent MNT1 cells were moved to an ice bath and washed two times with ice-cold PBS-Ca-Mg (0.1 mM CaCl2 and 1 mM MgCl2 in PBS). All solutions used were ice-cold when applied to cells, and cells were maintained on an ice bath throughout. After washes, cells were incubated in a 1 mM sodium periodate (Sigma-Aldrich) solution prepared in PBS-Ca-Mg for 30 min, after which the periodate solution was removed and a quenching solution of 1 mM glycerol in PBS-Ca-Mg was added (Zeng et al., 2009). Cells were then washed two times with PBS-Ca-Mg, and a biotin ligation solution of 100 μM aminooxy biotin (Biotium, Hayward, CA) and 10 mM aniline (Sigma-Aldrich) was applied for 90 min. After biotin ligation, cells were washed two times in PBS-Ca-Mg and lysed for 30 min in buffer A with 0.5% Triton X-100 supplemented with Complete antiprotease. This biotinylation procedure is based on a periodate-generated aldehyde on sialic acids. This process is catalyzed by aniline-dependent ligation with an appropriate tag. Aniline catalysis accelerates ligation, facilitating the reaction with low concentrations of aminooxy-biotin at neutral pH to label the most cell-surface sialylated glycoproteins (Zeng et al., 2009). After lysis, cell debris were scraped from the plates, and lysates were spun at 16,100 × g for 15 min. The supernatant was recovered and diluted to 0.2 mg/ml. A small volume (50 μl) of NeutrAvidin-coated agarose beads (Thermo Scientific, Waltham, MA) was prewashed two times in buffer A with 0.1% Triton X-100, and 100 μg of cell lysate was incubated with the beads for 2 h with end-over-end rotation at 4°C. Beads were then washed five times in buffer A with 0.1% Triton X-100 for 5 min with end-over-end rotation at 4°C. Proteins were eluted from the beads by boiling in Laemmli sample buffer at 75°C for 5 min, and samples were analyzed by SDS–PAGE followed by silver stain or immunoblot.
Computational and statistical analysis
Functional gene annotation for the set of PI4KIIα-interacting proteins (Supplemental Table S2) was performed using the Database for Annotation, Visualization, and Integrated Discovery (DAVID, version 6.7) and gene ontology (GO) terms (Dennis et al., 2003; Huang da et al., 2007). The Cytoscape plug-in Enrichment Map was used to display overlap between gene ontology terms within Cytoscape (version 2.8.3; Merico et al., 2010). Node size was mapped to number of genes within a GO category, and node color was mapped to the p value for enrichment of a given category. For network analysis, a network interaction map was built using GeneGo Metacore (version 6.11, build 41105) for PI4KIIα-interacting proteins (Supplemental Table S1) and BLOC-1 subunits. The network was visualized in Cytoscape, and node color was mapped to proteins of interest.
Experimental conditions were compared using KaleidaGraph, version 4.1.3 (Synergy Software, Reading, PA), or StatPlus Mac Built5.6.0pre/Universal (AnalystSoft, Vancouver, Canada). Data are represented as box plots displaying the four quartiles of the data, with the “box” comprising the two middle quartiles, separated by the median. The upper and lower quartiles are represented by the single lines extending from the box.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institutes of Health (GM077569 and NS42599) and CHOA Children's Center for Neuroscience to V.F. P.V.R. was supported by National Research Service Award Fellowship F31NS0765. This research project was supported in part by the Emory University Integrated Cellular Imaging Microscopy Core and Viral Cores of the Emory Neuroscience National Institute of Neurological Disorders and Stroke Core Facilities Grant P30NS055077. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Abbreviations used:
- ATP7A
ATPase Cu++ transporting alpha polypeptide
- BLOC
biogenesis of liposome-related organelles complex
- DSP
dithiobis(succin-imidylpropionate)
- GFP
green fluorescent protein
- HA
hemagglutinin
- HPLC
high-performance liquid chromatography
- HPS
Hermansky-Pudlak syndrome
- HSP
hereditary spastic paraplegia
- MS
mass spectrometry
- MS/MS
tandem mass spectrometry
- PI4KIIα
phosphatidylinositol-4-kinase type IIα
- PI4P
phosphatidylinositol 4-phosphate
- PBS
phosphate-buffered saline
- SILAC
stable isotope labeling with amino acids in cell culture
- siRNA
small interfering
- RNA
shRNA short hairpin RNA
- TrfR
transferrin receptor
- VAMP7
vesicle-associated membrane protein 7
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-02-0088) on May 15, 2013.
REFERENCES
- Aittaleb M, Boguth CA, Tesmer JJ. Structure and function of heterotrimeric G protein-regulated Rho guanine nucleotide exchange factors. Mol Pharmacol. 2010;77:111–125. doi: 10.1124/mol.109.061234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anitei M, Hoflack B. Bridging membrane and cytoskeleton dynamics in the secretory and endocytic pathways. Nat Cell Biol. 2012;14:11–19. doi: 10.1038/ncb2409. [DOI] [PubMed] [Google Scholar]
- Anitei M, Stange C, Parshina I, Baust T, Schenck A, Raposo G, Kirchhausen T, Hoflack B. Protein complexes containing CYFIP/Sra/PIR121 coordinate Arf1 and Rac1 signalling during clathrin-AP-1-coated carrier biogenesis at the TGN. Nat Cell Biol. 2010;12:330–340. doi: 10.1038/ncb2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balla A, Tuymetova G, Barshishat M, Geiszt M, Balla T. Characterization of type II phosphatidylinositol 4-kinase isoforms reveals association of the enzymes with endosomal vesicular compartments. J Biol Chem. 2002;277:20041–20050. doi: 10.1074/jbc.M111807200. [DOI] [PubMed] [Google Scholar]
- Barylko B, Wlodarski P, Binns DD, Gerber SH, Earnest S, Sudhof TC, Grichine N, Albanesi JP. Analysis of the catalytic domain of phosphatidylinositol 4-kinase type II. J Biol Chem. 2002;277:44366–44375. doi: 10.1074/jbc.M203241200. [DOI] [PubMed] [Google Scholar]
- Baust T, Anitei M, Czupalla C, Parshyna I, Bourel L, Thiele C, Krause E, Hoflack B. Protein networks supporting AP-3 function in targeting lysosomal membrane proteins. Mol Biol Cell. 2008;19:1942–1951. doi: 10.1091/mbc.E08-02-0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bear JE. Sorting out endosomes in the WASH. Dev Cell. 2009;17:583–584. doi: 10.1016/j.devcel.2009.10.019. [DOI] [PubMed] [Google Scholar]
- Birkenfeld J, Nalbant P, Yoon SH, Bokoch GM. Cellular functions of GEF-H1, a microtubule-regulated Rho-GEF: is altered GEF-H1 activity a crucial determinant of disease pathogenesis. Trends Cell Biol. 2008;18:210–219. doi: 10.1016/j.tcb.2008.02.006. [DOI] [PubMed] [Google Scholar]
- Blackstone C. Cellular pathways of hereditary spastic paraplegia. Annu Rev Neurosci. 2012;35:25–47. doi: 10.1146/annurev-neuro-062111-150400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasius AL, Brandl K, Crozat K, Xia Y, Khovananth K, Krebs P, Smart NG, Zampolli A, Ruggeri ZM, Beutler BA. Mice with mutations of Dock7 have generalized hypopigmentation and white-spotting but show normal neurological function. Proc Natl Acad Sci USA. 2009;106:2706–2711. doi: 10.1073/pnas.0813208106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino JS, Glick BS. The mechanisms of vesicle budding and fusion. Cell. 2004;116:153–166. doi: 10.1016/s0092-8674(03)01079-1. [DOI] [PubMed] [Google Scholar]
- Brocker C, Engelbrecht-Vandre S, Ungermann C. Multisubunit tethering complexes and their role in membrane fusion. Curr Biol. 2010;20:R943–R952. doi: 10.1016/j.cub.2010.09.015. [DOI] [PubMed] [Google Scholar]
- Bultema JJ, Di Pietro SM. Cell type-specific Rab32 and Rab38 cooperate with the ubiquitous lysosome biogenesis machinery to synthesize specialized lysosome-related organelles. Small GTPases. 2013;4:16–21. doi: 10.4161/sgtp.22349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campellone KG, Welch MD. A nucleator arms race: cellular control of actin assembly. Nat Rev. Mol Cell Biol. 2010;11:237–251. doi: 10.1038/nrm2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnell M, Zech T, Calaminus SD, Ura S, Hagedorn M, Johnston SA, May RC, Soldati T, Machesky LM, Insall RH. Actin polymerization driven by WASH causes V-ATPase retrieval and vesicle neutralization before exocytosis. J Cell Biol. 2011;193:831–839. doi: 10.1083/jcb.201009119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemen CS, et al. Strumpellin is a novel valosin-containing protein binding partner linking hereditary spastic paraplegia to protein aggregation diseases. Brain. 2010;133:2920–2941. doi: 10.1093/brain/awq222. [DOI] [PubMed] [Google Scholar]
- Clift-O'Grady L, Desnos C, Lichtenstein Y, Faundez V, Horng JT, Kelly RB. Reconstitution of synaptic vesicle biogenesis from PC12 cell membranes. Methods. 1998;16:150–159. doi: 10.1006/meth.1998.0662. [DOI] [PubMed] [Google Scholar]
- Conner SD, Schmid SL. Regulated portals of entry into the cell. Nature. 2003;422:37–44. doi: 10.1038/nature01451. [DOI] [PubMed] [Google Scholar]
- Craige B, Salazar G, Faundez V. Phosphatidylinositol-4-hinase type II alpha contains an AP-3 sorting motif and a kinase domain that are both required for endosome traffic. Mol Biol Cell. 2008;19:1415–1426. doi: 10.1091/mbc.E07-12-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delevoye C, et al. AP-1 and KIF13A coordinate endosomal sorting and positioning during melanosome biogenesis. J Cell Biol. 2009;187:247–264. doi: 10.1083/jcb.200907122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell'Angelica EC. AP-3-dependent trafficking and disease: the first decade. Curr Opin Cell Biol. 2009;21:552–559. doi: 10.1016/j.ceb.2009.04.014. [DOI] [PubMed] [Google Scholar]
- Dell'Angelica EC, Shotelersuk V, Aguilar RC, Gahl WA, Bonifacino JS. Altered trafficking of lysosomal proteins in Hermansky-Pudlak syndrome due to mutations in the beta 3A subunit of the AP-3 adaptor. Mol Cell. 1999;3:11–21. doi: 10.1016/s1097-2765(00)80170-7. [DOI] [PubMed] [Google Scholar]
- Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- Derivery E, Sousa C, Gautier JJ, Lombard B, Loew D, Gautreau A. The Arp2/3 activator WASH controls the fission of endosomes through a large multiprotein complex. Dev Cell. 2009;17:712–723. doi: 10.1016/j.devcel.2009.09.010. [DOI] [PubMed] [Google Scholar]
- Di Pietro SM, Falcon-Perez JM, Tenza D, Setty SR, Marks MS, Raposo G, Dell'Angelica EC. BLOC-1 interacts with BLOC-2 and the AP-3 complex to facilitate protein trafficking on endosomes. Mol Biol Cell. 2006;17:4027–4038. doi: 10.1091/mbc.E06-05-0379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duleh SN, Welch MD. WASH and the Arp2/3 complex regulate endosome shape and trafficking. Cytoskeleton (Hoboken) 2010;67:193–206. doi: 10.1002/cm.20437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duleh SN, Welch MD. Regulation of integrin trafficking, cell adhesion, and cell migration by WASH and the Arp2/3 complex. Cytoskeleton (Hoboken) 2012;69:1047–1058. doi: 10.1002/cm.21069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson SM, De Camilli P. Dynamin, a membrane-remodelling GTPase. Nat Rev Mol Cell Biol. 2012;13:75–88. doi: 10.1038/nrm3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finsterer J, Loscher W, Quasthoff S, Wanschitz J, Auer-Grumbach M, Stevanin G. Hereditary spastic paraplegias with autosomal dominant, recessive, X-linked, or maternal trait of inheritance. J Neurol Sci. 2012;318:1–18. doi: 10.1016/j.jns.2012.03.025. [DOI] [PubMed] [Google Scholar]
- Freeman C, Seaman MN, Reid E. The hereditary spastic paraplegia protein strumpellin: characterisation in neurons and of the effect of disease mutations on WASH complex assembly and function. Biochim Biophys Acta. 2013;1832:160–173. doi: 10.1016/j.bbadis.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Zorman S, Gundersen G, Xi Z, Ma L, Sirinakis G, Rothman JE, Zhang Y. Single reconstituted neuronal SNARE complexes zipper in three distinct stages. Science. 2012;337:1340–1343. doi: 10.1126/science.1224492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokhale A, Larimore J, Werner E, So L, Moreno-De-Luca A, Lese-Martin C, Lupashin VV, Smith Y, Faundez V. Quantitative proteomic and genetic analyses of the schizophrenia susceptibility factor dysbindin identify novel roles of the biogenesis of lysosome-related organelles complex 1. J Neurosci. 2012a;32:3697–3711. doi: 10.1523/JNEUROSCI.5640-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokhale A, Perez-Cornejo P, Duran C, Hartzell HC, Faundez V. A comprehensive strategy to identify stoichiometric membrane protein interactomes. Cell Logist. 2012b;2:1–8. doi: 10.4161/cl.22717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez TS, Billadeau DD. A FAM21-containing WASH complex regulates retromer-dependent sorting. Dev Cell. 2009;17:699–711. doi: 10.1016/j.devcel.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez TS, Gorman JA, de Narvajas AA, Koenig AO, Billadeau DD. Trafficking defects in WASH-knockout fibroblasts originate from collapsed endosomal and lysosomal networks. Mol Biol Cell. 2012;23:3215–3228. doi: 10.1091/mbc.E12-02-0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Wenk MR, Pellegrini L, Onofri F, Benfenati F, De Camilli P. Phosphatidylinositol 4-kinase type IIalpha is responsible for the phosphatidylinositol 4-kinase activity associated with synaptic vesicles. Proc Natl Acad Sci USA. 2003;100:3995–4000. doi: 10.1073/pnas.0230488100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao YH, Doyle JM, Ramanathan S, Gomez TS, Jia D, Xu M, Chen ZJ, Billadeau DD, Rosen MK, Potts PR. Regulation of WASH-dependent actin polymerization and protein trafficking by ubiquitination. Cell. 2013;152:1051–1064. doi: 10.1016/j.cell.2013.01.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbour ME, Breusegem SY, Antrobus R, Freeman C, Reid E, Seaman MN. The cargo-selective retromer complex is a recruiting hub for protein complexes that regulate endosomal tubule dynamics. J Cell Sci. 2010;123:3703–3717. doi: 10.1242/jcs.071472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbour ME, Breusegem SY, Seaman MN. Recruitment of the endosomal WASH complex is mediated by the extended “tail” of Fam21 binding to the retromer protein Vps35. Biochem J. 2012;442:209–220. doi: 10.1042/BJ20111761. [DOI] [PubMed] [Google Scholar]
- Helfer E, Harbour ME, Henriot V, Lakisic G, Sousa-Blin C, Volceanov L, Seaman MN, Gautreau A. Endosomal recruitment of the WASH complex: active sequences and mutations impairing interaction with the retromer. Biol Cell. 2013;105:191–207. doi: 10.1111/boc.201200038. [DOI] [PubMed] [Google Scholar]
- Huang da W, et al. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007;35:W169–W175. doi: 10.1093/nar/gkm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huizing M, Helip-Wooley A, Westbroek W, Gunay-Aygun M, Gahl WA. Disorders of lysosome-related organelle biogenesis: clinical and molecular genetics. Annu Rev Genomics Hum Genet. 2008;9:359–386. doi: 10.1146/annurev.genom.9.081307.164303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia D, Gomez TS, Billadeau DD, Rosen MK. Multiple repeat elements within the FAM21 tail link the WASH actin regulatory complex to the retromer. Mol Biol Cell. 2012;23:2352–2361. doi: 10.1091/mbc.E11-12-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia D, Gomez TS, Metlagel Z, Umetani J, Otwinowski Z, Rosen MK, Billadeau DD. WASH and WAVE actin regulators of the Wiskott-Aldrich syndrome protein (WASP) family are controlled by analogous structurally related complexes. Proc Natl Acad Sci USA. 2010;107:10442–10447. doi: 10.1073/pnas.0913293107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaksonen M, Peng HB, Rauvala H. Association of cortactin with dynamic actin in lamellipodia and on endosomal vesicles. J Cell Sci. 2000;113:4421–4426. doi: 10.1242/jcs.113.24.4421. [DOI] [PubMed] [Google Scholar]
- Kaksonen M, Toret CP, Drubin DG. A modular design for the clathrin- and actin-mediated endocytosis machinery. Cell. 2005;123:305–320. doi: 10.1016/j.cell.2005.09.024. [DOI] [PubMed] [Google Scholar]
- Kaksonen M, Toret CP, Drubin DG. Harnessing actin dynamics for clathrin-mediated endocytosis. Nat Rev Mol Cell Biol. 2006;7:404–414. doi: 10.1038/nrm1940. [DOI] [PubMed] [Google Scholar]
- Kantheti P, Diaz ME, Peden AE, Seong EE, Dolan DF, Robinson MS, Noebels JL, Burmeister ML. Genetic and phenotypic analysis of the mouse mutant mh(2J), an Ap3d allele caused by IAP element insertion. Mamm Genome. 2003;14:157–167. doi: 10.1007/s00335-002-2238-8. [DOI] [PubMed] [Google Scholar]
- Kennerson ML, et al. Missense mutations in the copper transporter gene ATP7A cause X-linked distal hereditary motor neuropathy. Am J Hum Genet. 2010;86:343–352. doi: 10.1016/j.ajhg.2010.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larimore J, et al. The schizophrenia susceptibility factor dysbindin and its associated complex sort cargoes from cell bodies to the synapse. Mol Biol Cell. 2011;22:4854–4867. doi: 10.1091/mbc.E11-07-0592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, et al. Hermansky-Pudlak syndrome type 7 (HPS-7) results from mutant dysbindin, a member of the biogenesis of lysosome-related organelles complex 1 (BLOC-1) Nat Genet. 2003;35:84–89. doi: 10.1038/ng1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenstein Y, Desnos C, Faundez V, Kelly RB, Clift-O'Grady L. Vesiculation and sorting from PC12-derived endosomes in vitro. Proc Natl Acad Sci USA. 1998;95:11223–11228. doi: 10.1073/pnas.95.19.11223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R, Abreu-Blanco MT, Barry KC, Linardopoulou EV, Osborn GE, Parkhurst SM. Wash functions downstream of Rho and links linear and branched actin nucleation factors. Development. 2009;136:2849–2860. doi: 10.1242/dev.035246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomant AJ, Fairbanks G. Chemical probes of extended biological structures: synthesis and properties of the cleavable protein cross-linking reagent [35S]dithiobis(succinimidyl propionate) J Mol Biol. 1976;104:243–261. doi: 10.1016/0022-2836(76)90011-5. [DOI] [PubMed] [Google Scholar]
- Mann M. Functional and quantitative proteomics using SILAC. Nat Rev. Mol Cell Biol. 2006;7:952–958. doi: 10.1038/nrm2067. [DOI] [PubMed] [Google Scholar]
- McMahon HT, Boucrot E. Molecular mechanism and physiological functions of clathrin-mediated endocytosis. Nat Rev. Mol Cell Biol. 2011;12:517–533. doi: 10.1038/nrm3151. [DOI] [PubMed] [Google Scholar]
- Merico D, Isserlin R, Stueker O, Emili A, Bader GD. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PloS One. 2010;5:e13984. doi: 10.1371/journal.pone.0013984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minogue S, Waugh MG, De Matteis MA, Stephens DJ, Berditchevski F, Hsuan JJ. Phosphatidylinositol 4-kinase is required for endosomal trafficking and degradation of the EGF receptor. J Cell Sci. 2006;119:571–581. doi: 10.1242/jcs.02752. [DOI] [PubMed] [Google Scholar]
- Monfregola J, Napolitano G, D'Urso M, Lappalainen P, Ursini MV. Functional characterization of Wiskott-Aldrich syndrome protein and scar homolog (WASH), a bi-modular nucleation-promoting factor able to interact with biogenesis of lysosome-related organelle subunit 2 (BLOS2) and gamma-tubulin. J Biol Chem. 2010;285:16951–16957. doi: 10.1074/jbc.M109.078501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooren OL, Galletta BJ, Cooper JA. Roles for actin assembly in endocytosis. Annu Rev Biochem. 2012;81:661–686. doi: 10.1146/annurev-biochem-060910-094416. [DOI] [PubMed] [Google Scholar]
- Mossinger J, Wieffer M, Krause E, Freund C, Gerth F, Krauss M, Haucke V. Phosphatidylinositol 4-kinase IIalpha function at endosomes is regulated by the ubiquitin ligase Itch. EMBO Rep. 2012;13:1087–1094. doi: 10.1038/embor.2012.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Setou M, Seog D, Ogasawara K, Dohmae N, Takio K, Hirokawa N. A novel motor, KIF13A, transports mannose-6-phosphate receptor to plasma membrane through direct interaction with AP-1 complex. Cell. 2000;103:569–581. doi: 10.1016/s0092-8674(00)00161-6. [DOI] [PubMed] [Google Scholar]
- Newell-Litwa K, Chintala S, Jenkins S, Pare JF, McGaha L, Smith Y, Faundez V. Hermansky-Pudlak protein complexes, AP-3 and BLOC-1, differentially regulate presynaptic composition in the striatum and hippocampus. J Neurosci. 2010;30:820–831. doi: 10.1523/JNEUROSCI.3400-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newell-Litwa K, Salazar G, Smith Y, Faundez V. Roles of BLOC-1 and AP-3 complexes in cargo sorting to synaptic vesicles. Mol Biol Cell. 2009;20:1441–1453. doi: 10.1091/mbc.E08-05-0456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newell-Litwa K, Seong E, Burmeister M, Faundez V. Neuronal and non-neuronal functions of the AP-3 sorting machinery. J Cell Sci. 2007;120:531–541. doi: 10.1242/jcs.03365. [DOI] [PubMed] [Google Scholar]
- Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteom. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- Ong SE, Mann M. A practical recipe for stable isotope labeling by amino acids in cell culture (SILAC) Nat Protoc. 2006;1:2650–2660. doi: 10.1038/nprot.2006.427. [DOI] [PubMed] [Google Scholar]
- Peden AA, Rudge RE, Lui WW, Robinson MS. Assembly and function of AP-3 complexes in cells expressing mutant subunits. J Cell Biol. 2002;156:327–336. doi: 10.1083/jcb.200107140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Cornejo P, Gokhale A, Duran C, Cui Y, Xiao Q, Hartzell HC, Faundez V. Anoctamin 1 (Tmem16A) Ca2+-activated chloride channel stoichiometrically interacts with an ezrin-radixin-moesin network. Proc Natl Acad Sci USA. 2012;109:10376–10381. doi: 10.1073/pnas.1200174109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piotrowski JT, Gomez TS, Schoon RA, Mangalam AK, Billadeau DD. WASH knockout T cells demonstrate defective receptor trafficking, proliferation, and effector function. Mol Cell Biol. 2013;33:958–973. doi: 10.1128/MCB.01288-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puthenveedu MA, Lauffer B, Temkin P, Vistein R, Carlton P, Thorn K, Taunton J, Weiner OD, Parton RG, von Zastrow M. Sequence-dependent sorting of recycling proteins by actin-stabilized endosomal microdomains. Cell. 2010;143:761–773. doi: 10.1016/j.cell.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qualmann B, Kessels MM, Kelly RB. Molecular links between endocytosis and the actin cytoskeleton. J Cell Biol. 2000;150:F111–F116. doi: 10.1083/jcb.150.5.f111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raposo G, Marks MS. Melanosomes—dark organelles enlighten endosomal membrane transport. Nat Rev. Mol Cell Biol. 2007;8:786–797. doi: 10.1038/nrm2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raposo G, Marks MS, Cutler DF. Lysosome-related organelles: driving post-Golgi compartments into specialisation. Curr Opin Cell Biol. 2007;19:394–401. doi: 10.1016/j.ceb.2007.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedl J, et al. Lifeact: a versatile marker to visualize F-actin. Nat Methods. 2008;5:605–607. doi: 10.1038/nmeth.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossman KL, Der CJ, Sondek J. GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nat Rev Mol Cell Biol. 2005;6:167–180. doi: 10.1038/nrm1587. [DOI] [PubMed] [Google Scholar]
- Rottner K, Hanisch J, Campellone KG. WASH, WHAMM and JMY: regulation of Arp2/3 complex and beyond. Trends Cell Biol. 2010;20:650–661. doi: 10.1016/j.tcb.2010.08.014. [DOI] [PubMed] [Google Scholar]
- Rotty JD, Wu C, Bear JE. New insights into the regulation and cellular functions of the ARP2/3 complex. Nat Rev Mol Cell Biol. 2012;14:7–12. doi: 10.1038/nrm3492. [DOI] [PubMed] [Google Scholar]
- Salazar G, et al. BLOC-1 complex deficiency alters the targeting of adaptor protein complex-3 cargoes. Mol Biol Cell. 2006;17:4014–4026. doi: 10.1091/mbc.E06-02-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar G, Zlatic S, Craige B, Peden AA, Pohl J, Faundez V. Hermansky-Pudlak syndrome protein complexes associate with phosphatidylinositol 4-kinase type II alpha in neuronal and non-neuronal cells. J Biol Chem. 2009;284:1790–1802. doi: 10.1074/jbc.M805991200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid EM, McMahon HT. Integrating molecular and network biology to decode endocytosis. Nature. 2007;448:883–888. doi: 10.1038/nature06031. [DOI] [PubMed] [Google Scholar]
- Schmid SL, Frolov VA. Dynamin: functional design of a membrane fission catalyst. Annu Rev Cell Dev Biol. 2011;27:79–105. doi: 10.1146/annurev-cellbio-100109-104016. [DOI] [PubMed] [Google Scholar]
- Seaman MN. The retromer complex—endosomal protein recycling and beyond. J Cell Sci. 2012;125:4693–4702. doi: 10.1242/jcs.103440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setty SR, Tenza D, Sviderskaya EV, Bennett DC, Raposo G, Marks MS. Cell-specific ATP7A transport sustains copper-dependent tyrosinase activity in melanosomes. Nature. 2008;454:1142–1146. doi: 10.1038/nature07163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setty SR, et al. BLOC-1 is required for cargo-specific sorting from vacuolar early endosomes toward lysosome-related organelles. Mol Biol Cell. 2007;18:768–780. doi: 10.1091/mbc.E06-12-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons JP, et al. Loss of phosphatidylinositol 4-kinase 2alpha activity causes late onset degeneration of spinal cord axons. Proc Natl Acad Sci USA. 2009;106:11535–11539. doi: 10.1073/pnas.0903011106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starcevic M, Dell'Angelica EC. Identification of snapin and three novel proteins (BLOS1, BLOS2, and BLOS3/reduced pigmentation) as subunits of biogenesis of lysosome-related organelles complex-1 (BLOC-1) J Biol Chem. 2004;279:28393–28401. doi: 10.1074/jbc.M402513200. [DOI] [PubMed] [Google Scholar]
- Styers ML, Salazar G, Love R, Peden AA, Kowalczyk AP, Faundez V. The endo-lysosomal sorting machinery interacts with the intermediate filament cytoskeleton. Mol Biol Cell. 2004;15:5369–5382. doi: 10.1091/mbc.E04-03-0272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swedlow JR, Hu K, Andrews PD, Roos DS, Murray JM. Measuring tubulin content in Toxoplasma gondii: a comparison of laser-scanning confocal and wide-field fluorescence microscopy. Proc Natl Acad Sci USA. 2002;99:2014–2019. doi: 10.1073/pnas.022554999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swedlow JR, Sedat JW, Agard DA. Deconvolution in optical microscopy. In: Jansson PA, editor. Deconvolution of Images and Spectra. San Diego, CA: Academic Press; 1997. pp. 284–307. [Google Scholar]
- Taylor MJ, Perrais D, Merrifield CJ. A high precision survey of the molecular dynamics of mammalian clathrin-mediated endocytosis. PLoS Biol. 2011;9:e1000604. doi: 10.1371/journal.pbio.1000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinkle-Mulcahy L, et al. Identifying specific protein interaction partners using quantitative mass spectrometry and bead proteomes. J Cell Biol. 2008;183:223–239. doi: 10.1083/jcb.200805092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdmanis PN, et al. Mutations in the KIAA0196 gene at the SPG8 locus cause hereditary spastic paraplegia. Am J Hum Genet. 2007;80:152–161. doi: 10.1086/510782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veltman DM, Insall RH. WASP family proteins: their evolution and its physiological implications. Mol Biol Cell. 2010;21:2880–2893. doi: 10.1091/mbc.E10-04-0372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei AH, Li W. Hermansky-Pudlak syndrome: pigmentary and non-pigmentary defects and their pathogenesis. Pigment Cell Melanoma Res. 2013;26:176–192. doi: 10.1111/pcmr.12051. [DOI] [PubMed] [Google Scholar]
- Wickner W, Schekman R. Membrane fusion. Nat Struct Mol Biol. 2008;15:658–664. doi: 10.1038/nsmb.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi L, Donsante A, Kennerson ML, Mercer JF, Garbern JY, Kaler SG. Altered intracellular localization and valosin-containing protein (p97 VCP) interaction underlie ATP7A-related distal motor neuropathy. Human Mol Genet. 2012;21:1794–1807. doi: 10.1093/hmg/ddr612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zech T, Calaminus SD, Caswell P, Spence HJ, Carnell M, Insall RH, Norman J, Machesky LM. The Arp2/3 activator WASH regulates alpha5beta1-integrin-mediated invasive migration. J Cell Sci. 2011;124:3753–3759. doi: 10.1242/jcs.080986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, Ramya TN, Dirksen A, Dawson PE, Paulson JC. High-efficiency labeling of sialylated glycoproteins on living cells. Nat Methods. 2009;6:207–209. doi: 10.1038/nmeth.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlatic SA, Ryder PV, Salazar G, Faundez V. Isolation of labile multi-protein complexes by in vivo controlled cellular cross-linking and immuno-magnetic affinity chromatography. J Vis Exp. 2010;pii:1855. doi: 10.3791/1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.