Abstract
The membrane protein FlhB is a highly conserved component of the flagellar secretion system, and it plays an active role in the regulation of protein export. In this study conserved properties of FlhB that are important for its function were investigated. Replacing the flhB gene (or part of the gene) in Salmonella typhimurium with the flhB gene of the distantly related bacterium Aquifex aeolicus greatly reduces motility. However, motility can be restored to some extent by spontaneous mutations in the part of flhB gene coding for the cytoplasmic domain of Aquifex FlhB. Structural analysis suggests that these mutations destabilize the structure. The secondary structure and stability of the mutated cytoplasmic fragments of FlhB have been studied by circular dichroism spectroscopy. The results suggest that conformational flexibility could be important for FlhB function. An extragenic suppressor mutation in the fliS gene, which decreases the affinity of FliS to FliC, partially restores motility of the FlhB substitution mutants.
Introduction
The bacterial flagellum is a large, complex molecular machine made up of more than 30 different proteins. It contains three major substructures: the basal body, the hook and the filament. Most of the flagellar proteins are localized outside of the cell and are exported across the cytoplasmic membrane by the flagellum-specific secretion apparatus. This apparatus is evolutionarily related to the type III secretion system that is used by many pathogenic bacteria for secretion of virulence factors into the host eukaryote cell cytoplasm [1], [2].
In the case of Salmonella enterica serovar Typhimurium (S. typhimurium), the flagellar secretion system consists of six integral membrane proteins: FlhA, FlhB, FliO, FliP, FliQ and FliR; and three cytoplasmic proteins: FliH, FliI, and FliJ [3]. Protein export by the flagellar type III secretion system is highly regulated. When the hook reaches an appropriate length (55.0±5.9 nm in Salmonella [4]), the secretion system switches substrate specificity from rod/hook-type export to filament-type export [3], [5]. Two proteins, the membrane protein FlhB and the hook-length control protein FliK, are critical for the substrate switching.
FlhB consists of two domains: a hydrophobic N-terminal part (FlhBTM) that is predicted to contain four transmembrane helices, and a C-terminal cytoplasmic domain (FlhBC) [6]. The two domains are connected by a flexible linker. The linker is important for FlhB function: its deletion or point mutations altering its flexibility have a significant effect on substrate secretion [7], [8]. The cytoplasmic domain of wild-type Salmonella FlhB undergoes autocatalytic cleavage between amino-acid residues Asn269 and Pro270 within a highly conserved Asn-Pro-Thr-His sequence [8], [9]. This auto-cleavage is essential for the switching process [7], [10]. Mutation of Asn269 to Ala prevents cleavage and locks the export apparatus in the rod/hook-type specificity state.
To switch substrate specificity, FlhB receives a signal from FliK [11], [12]. In the case of a deleted fliK gene the substrate switching does not occur and this results in a very long hook, termed “polyhook”, without any filament attached [4]. Several extragenic suppressor mutations, which allow the switching even in the absence of FliK, have been isolated and mapped to the part of flhB gene coding for FlhBC [11], [13].
In the current work, to reveal functionally important properties of FlhB, we replaced the flhB gene of Salmonella with the flhB gene of A. aeolicus, or with a fusion gene encoding a chimera FlhB composed of FlhBTM of S. typhimurium and FlhBC of A. aeolicus. All substitution mutants were substantially less motile than wild-type cells. However, several spontaneous mutations were found altering the C-terminal part of FlhB and resulting in enhanced motility. The sites of these mutations were mapped onto the structure of Aquifex FlhBC that was recently solved by X-ray crystallography [14], [15], [16]. Structural analysis suggested that the suppressor mutations destabilize the structure of FlhBC. The secondary structure and the stability of the mutated Aquifex FlhBC protein were studied by circular dichroism spectroscopy. We conclude that conformational flexibility of the cytoplasmic part of FlhB could be important for its function. Additionally, an extragenic bypass mutation in the fliS gene that partially restores motility of FlhB substitution mutants has been found. This mutation affects affinity of FliS to FliC but not to FlhB.
Results
Design of flhB Fusion Genes Encoding FlhB Chimeras
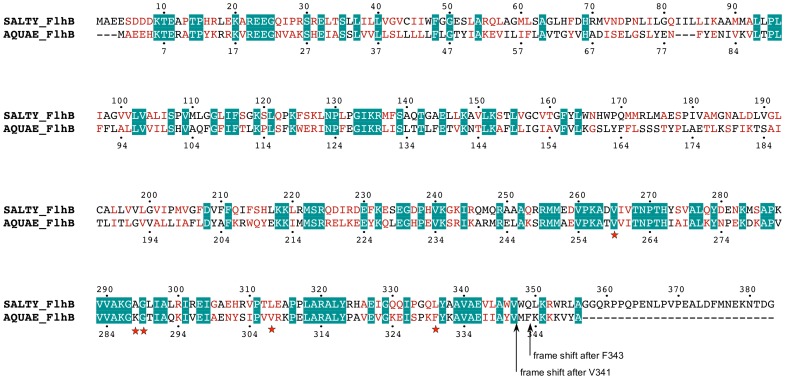
Aquifex FlhB shares a 32% sequence identity with FlhB of Salmonella (Figure 1). To investigate the ability of the cytoplasmic domain from Aquifex FlhB to function within the Salmonella flagellar export apparatus, we constructed three genes to produce chimeric FlhB proteins where the N-terminal transmembrane region of Salmonella FlhB was fused to the C-terminal cytoplasmic domain of Aquifex FlhB. All of these chimeric FlhB proteins differed in their C-terminal sequences and have different levels of sequence homology to wild-type Salmonella FlhB (Figure 2).
Figure 1. Amino acid sequence alignment of FlhB from S. typhimurium (SALTY_FlhB) and A. aeolicus (AQUAE_FlhB).
Identical residues are shown with a teal background; similar residues are colored red. Suppressor mutations found in this study are marked by red stars (for point mutations) and black arrows (for frame shift mutations).
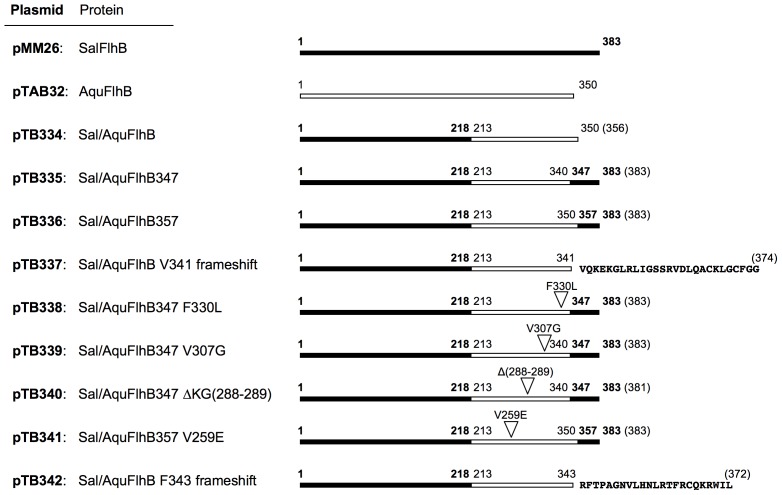
Figure 2. Schematic representations of FlhB products encoded by the plasmids used for the motility assay.
Black and white bars indicate the Salmonella and Aquifex parts of FlhB, respectively. Triangles mark the positions of point and deletion suppressor mutations. Numbers in parentheses show the total number of amino acids in each protein.
The first chimera, Sal/AquFlhB, consisted of Salmonella FlhBTM (residues 1–218) fused to Aquifex FlhBC (residues 213–350). The C-terminus of Aquifex FlhB is shorter in comparison to Salmonella FlhB by 33 residues. Therefore, we also produced chimeric FlhB constructs with C-termini having the extra 33 residues just like in the case of wild-type Salmonella FlhB. One of these, Sal/AquFlhB357, consisted of Salmonella FlhBTM fused to Aquifex FlhBC followed by C-terminal residues 357–383 of Salmonella FlhB. Another chimera, Sal/AquFlhB347, consisted of Salmonella FlhBTM fused to truncated Aquifex FlhBC (residues 213–340) followed by C-terminal residues 347–383 from Salmonella FlhB.
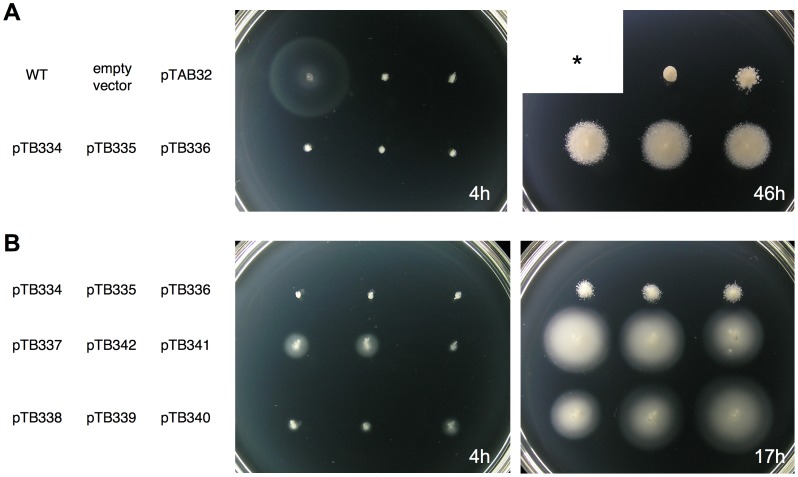
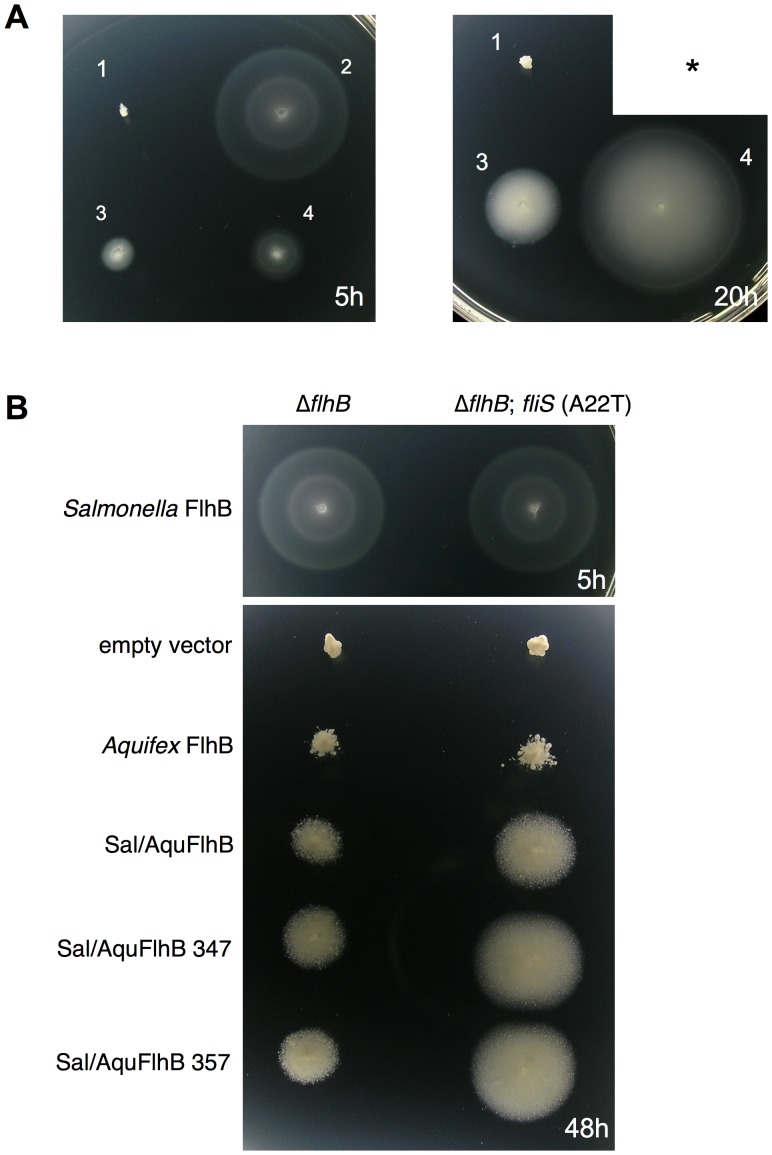
pTrc99A-FF4-based plasmids [17] expressing the different variants of FlhB proteins were transformed into ΔflhB Salmonella cells and tested for their ability to restore motility. Transformants were inoculated into tryptone soft agar plates. We found that all transformants were substantially less motile than wild-type cells (Figure 3A).
Figure 3. Swimming motility assay.
(A) Motility of ΔflhB Salmonella strain transformed with plasmids coding for different variants of FlhB. The asterisk “*” marks the previous emplacement for the wild type strain that was removed because it would have overgrown after 46 hours. (B) Rescue of motility by mutations in the cytoplasmic domain of FlhB. The plates were incubated at 30°C for the indicated time. Indicated plasmids carry genes of the following proteins: wild-type SalFlhB (WT); AquFlhB (pTAB32); Sal/AquFlhB (pTB334); Sal/AquFlhB347 (pTB335); Sal/AquFlhB357 (pTB336); Sal/AquFlhB V341 frame-shift (pTB337); Sal/AquFlhB F343 frame-shift (pTB342); Sal/AquFlhB357 V259E (pTB341); Sal/AquFlhB347 F330L (pTB338); Sal/AquFlhB347 V307G (pTB339); Sal/AquFlhB347 ΔKG(288–289) (pTB340).
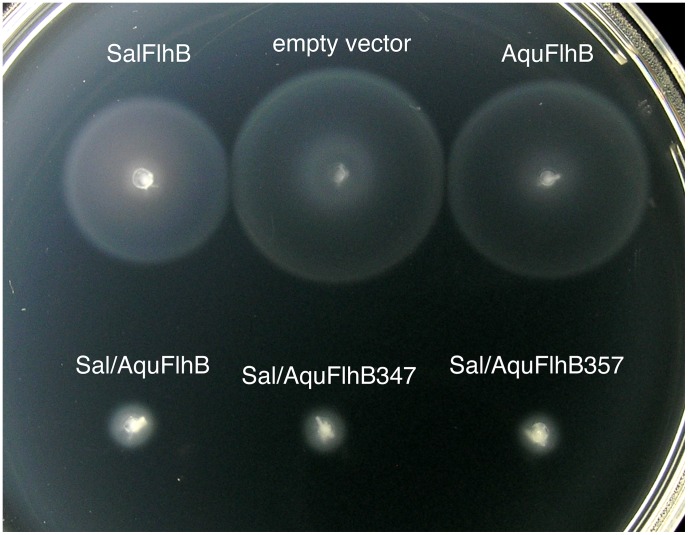
To examine negative dominant effects on the motility of the wild-type cells, we tested the swimming of Salmonella wild-type strain SJW1103 transformed with the pTrc99A-based plasmids (Figure 4). All chimera proteins (Sal/AquFlhB, Sal/AquFlhB347, and Sal/AquFlhB357) inhibited motility of the wild-type cells. This finding suggests that these proteins could be incorporated into the Salmonella export apparatus. In contrast, motility of SJW1103 cells producing wild-type Aquifex FlhB was the same as that of the cells with the vector control, demonstrating that Aquifex FlhB cannot efficiently compete with the Salmonella protein. These results indicate the importance of the transmembrane region for FlhB to be inserted into secretion system.
Figure 4. Dominance effect on motility of wild-type Salmonella strain SJW1103 transformed with the plasmids encoding different FlhB proteins.
Soft agar plate containing 0.1 mM IPTG was incubated at 30°C for 5 hours.
Isolation of Suppressor Mutants from ΔflhB Strains Producing Chimeric FlhB Proteins
After extended incubation, cells expressing chimeric flhB genes gave rise to suppressor mutants with enhanced motility, although motility was less than of the wild-type strain (Figure 3B).
To determine the positions of the suppressor mutations, the plasmids carrying the flhB genes that were initially transformed in ΔflhB Salmonella cells, were isolated and sequenced. Most of the suppressor mutations were located at the 3′-end of chimeric flhB gene, which encodes the C-terminal cytoplasmic part of FlhB. These mutations lead to single amino acid substitutions (V259E, V307G, and F330L), frame shifts (after V341 and F343), and an amino acid deletion (ΔKG 288–289) (Figure 2). Mutation ΔKG was encountered several times.
To confirm the effects of the found mutations on motility, isolated plasmids with the mutated flhB genes were re-transformed in ΔflhB Salmonella cells. All re-transformants, except one (see below), showed the same motility phenotype as originally obtained pseudorevertants (data not shown).
Effect of the Suppressor Mutations on FlhB Autocleavage
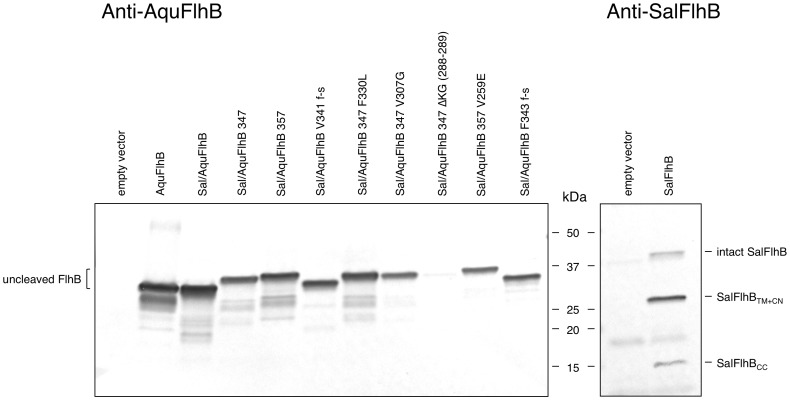
The cytoplasmic domain of FlhB undergoes autocatalytic cleavage that is important for its function [10]. To examine whether the suppressor mutations affect FlhB cleavage, we performed affinity western blotting analysis with anti-SalFlhB and anti-AquFlhB polyclonal antibodies (Figure 5). In the case of wild-type Salmonella FlhB three bands were detected that correspond to uncleaved protein and the cleavage products. For Aquifex FlhB, the FlhB chimeras, and the suppressor mutants only uncleaved proteins could be identified (Figure 5). This does not mean that autocleavage of AquFlhBC does not occur at all. All the Aquifex FlhBC proteins we purified were fully cleaved (data not shown). Furthermore, the Salmonella cells carrying the chimera genes were motile and this also suggests cleavage occurred, since without autocleavage of FlhB switching of substrate specificity (and therefore motility) is not possible [7]. This indicates that the cytoplasmic domain of Aquifex FlhB was more stable within Salmonella cells, which might be because Salmonella is a mesophilic bacterium and has a lower growth temperature than the hyperthermophile Aquifex.
Figure 5. Autocleavage properties of FlhB.
Cells carrying the plasmids encoding different FlhB proteins were grown at 37°C until OD600 = 0.5–0.6. Protein expression was induced by 0.1 mM IPTG. After 2 hours cells were harvested by centrifugation and analyzed by immunoblotting using polyclonal antibodies against full-length Aquifex and Salmonella FlhB.
In the case of ΔKG (288–289) mutant, the protein was detected at very low level that could be explained by its instability or reduced affinity of polyclonal anti-Aquifex FlhB antibodies to this mutant.
Mapping of the Suppressor Mutations into the Structure of FlhBC
The structures of Salmonella and Aquifex FlhBC are available in the Protein Data Bank (PDB) with the accession numbers 3B0Z and 3B1S, respectively [16].
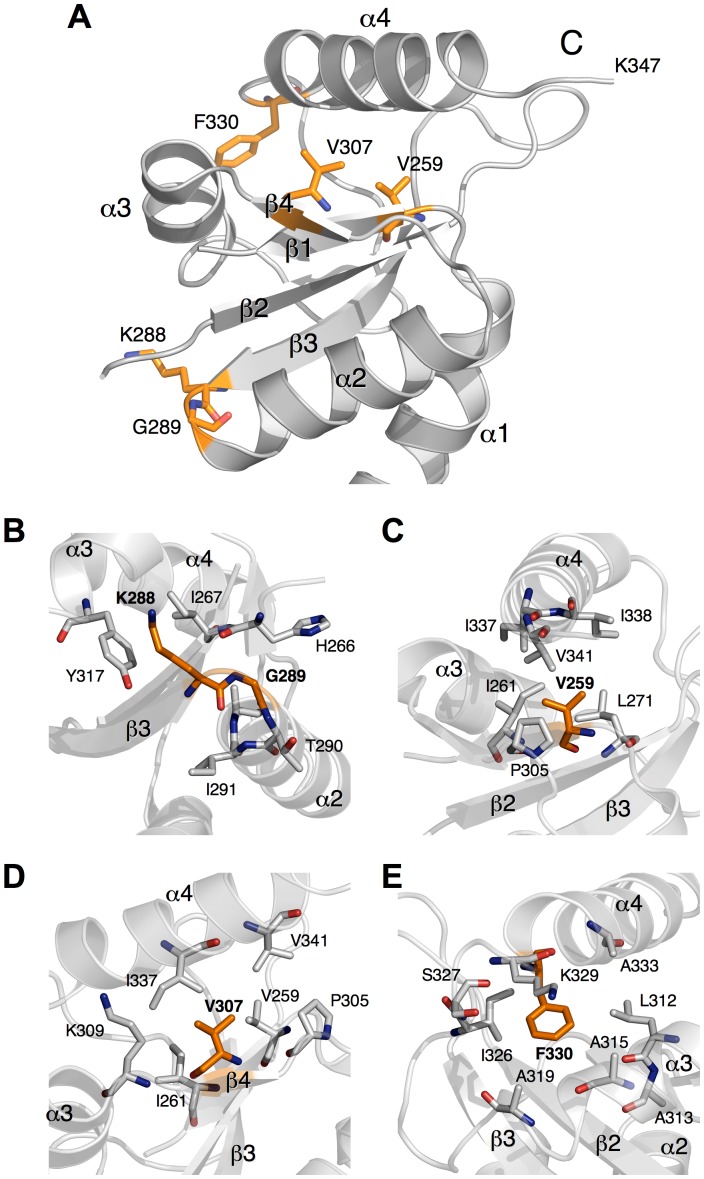
The positions of the suppressor mutations were mapped onto the structure of AquFlhBC (Figure 6). Two frame shift mutations, V341 and F343, are positioned very close to the C-terminus of the molecule. It is difficult to predict how they could affect the whole FlhBC structure. Residues V259 and V307 are located close to each other in a hydrophobic core between helix α4 and the β-sheet. Mutations in these positions, especially V259E with a change from a hydrophobic to a charged side-chain, are likely to destabilize the hydrophobic core and disrupt packing of the helix against the β-sheet. Residues K288 and G289 are located between strand β3 and helix α2. Deletion of these residues would disrupt helix α2 and interfere with the hydrophobic core formed by the β-sheet and helices α1 and α2. Mutation F330L may affect the hydrophobic surface between helices α3 and α4. Thus, most of the suppressor mutations (except the frame shift mutations) are located in or close to a hydrophobic core of the protein, and are likely to result in destabilization of its structure.
Figure 6. Mapping of the suppressor mutations in Aquifex FlhBC (PDB accession code: 3B1S).
(A) Ribbon diagram of Aquifex FlhBC with the suppressor mutations shown as sticks. Panels (B) to (E) show close-up views of the mutated residues and their surroundings.
Circular Dichroism and Stability Studies
Structural analysis suggests that the suppressor mutations could affect the stability of FlhB. To test this idea, Aquifex FlhBC mutant proteins were purified and their stability was checked. First, we measured thermal stability of the proteins using a fluorescence-based thermal shift assay [18]–[20]. The melting temperatures were 63.7°C±0.5 for Salmonella FlhBC; 78.9°C±0.2 for Aquifex FlhBC V259E; and 54.5°C±0.3 for Aquifex FlhBC ΔKG (288–289). Thermal denaturation was not observed for the native Aquifex FlhBC and for the mutants V307G, F330L, and frameshift V341, most likely because of the high thermal stability of these proteins (higher than 100°C - the temperature limit of the thermal shift assay).
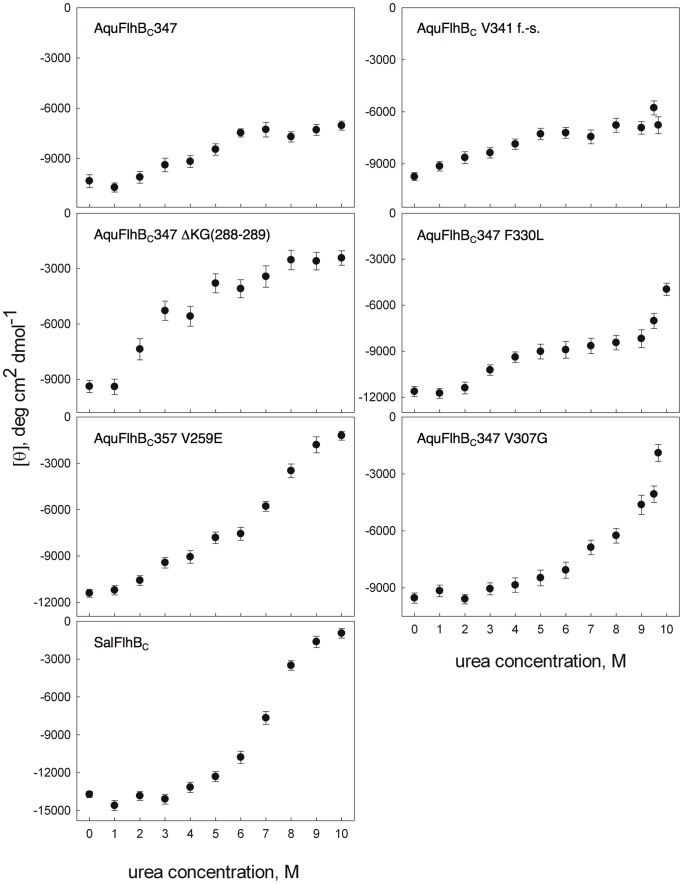
The secondary structures of wild-type FlhBC and mutants were calculated from their circular dichroism (CD) spectra. To perform analysis of the stability of the mutated AquFlhBC, we used chemical denaturation in urea. Monitoring of unfolding was done by circular dichroism spectroscopy. The dependence of mean residue ellipticity on urea concentration for wild-type and mutant Aquifex FlhBC is shown in Figure 7.
Figure 7. Urea denaturation of Salmonella and Aquifex FlhBC and various Aquifex FlhBC mutants.
Protein unfolding was monitored by ellipticity at 222 nm.
The shapes of the titration curves of Aquifex FlhBC, wild-type and mutants, suggest that there are two transition states. There might be at least two cooperatively unfolding domains in AquFlhBC with the midpoint of the first transition corresponds to 4 M urea, whereas the second domain stays folded in wild-type protein even at 10 M urea. These domains might correspond to the two fragments generated by the auto-cleavage of FlhB at the conserved NPTH sequence.
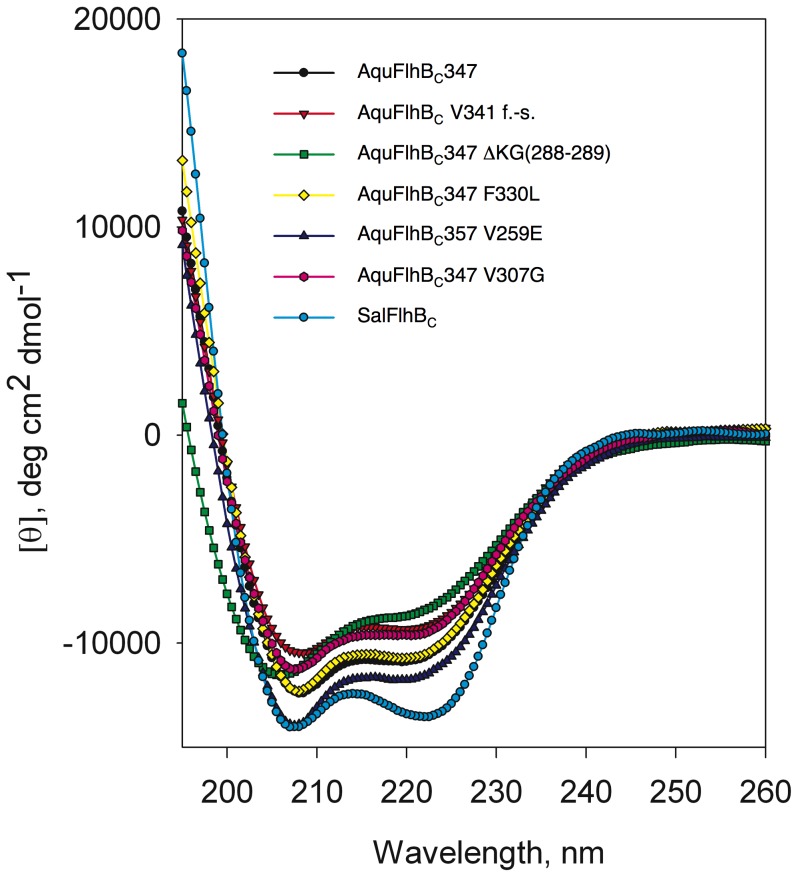
Of all the suppressor mutations, only the frame shift V341 had no effect on the secondary structure and stability: there was no difference in the CD spectra (Figure 8), and the unfolding curves (Figure 7) and the calculated secondary structure content (Table 1) between Aquifex FlhBC and this mutant are similar.
Figure 8. Circular dichroism measurements of Aquifex FlhBC, Aquifex FlhBC mutants, and Salmonella FlhBC.
Table 1. Secondary structure content of various FlhBC. Secondary structure content was calculated from the CD spectra using a multilinear regression [36].
| Protein | α-helix, % | β-structure, % | β-turn, % | random coil, % |
| AquFlhBC347 | 28.5±0.1 | 28.7±0.1 | 5.2±2.9 | 37±1.8 |
| AquFlhBC V341 f.-s. | 31.3±1.5 | 25.3±0.7 | 6.1±0.5 | 37.4±2.8 |
| AquFlhBC347 ΔKG(288–289) | 17.3±1.3 | 30.4±0.3 | 6.1±0.01 | 46.4±1.1 |
| AquFlhBC347 F330L | 32±3.1 | 26.1±1.3 | 4.45±1.1 | 37.5±0.01 |
| AquFlhBC357 V259E | 27.3±1.3 | 25.8±1.6 | 6.5±0.7 | 40.4±0.4 |
| AquFlhBC347 V307G | 28.4±1.8 | 27.7±1.0 | 4.6±0.3 | 39.4±0.4 |
| SalFlhBC | 40.8±0.7 | 14.4±0.6 | 9.9±1.5 | 34.9±0.2 |
Mutations V307G and F330L had no measured effect on the secondary structure. Mutation V259E caused a slight decrease of β-structure content and a corresponding increase of random coil content (Table 1). However, the effect of these three mutations on the stability of AquFlhBC differed drastically. F330L and V259E decreased stability of the more stable second domain of AquFlhBC, V259E having the greater effect. For the V259E mutation, the transition midpoint is 8 M urea, and for F330L, 10 M urea. The V307G mutation increased stability of the first domain but decreased stability of the second domain to intermediate between the stabilities of F330L and V259E.
The ΔKG (288–289) deletion had the biggest effect on structure and stability: an almost 2-fold decrease in helical content with a corresponding increase in random coil content (Table 1). Stability of both putative domains substantially decreased, and with these mutations AquFlhBC became completely unfolded at 8 M urea, the midpoint of transition is ∼3.5 M urea (Figure 7). This effect may be explained by the disruption of helix α2 as well as by the destabilization of an adjacent part of helix α1 when these two residues are deleted. This supports the conclusions based on the crystal structure analysis.
The range of effects seen for the suppressor mutations is consistent with the structural analysis. V307G and F330L, being on the protein surface, would be predicted to have less effect on stability. In contrast, the introduction of a charged Glu residue instead of Val (V259E) inside the hydrophobic core, or deletion of part of the structurally important helix α2 (ΔKG(288–289)) would be expected to have larger effects on stability.
Extragenic Suppressor Mutation in fliS
We found that two suppressor mutants carried an identical mutation of the fusion flhB gene, frame shift F343. However motility of these strains was substantially different (Figure 9A). Therefore we concluded that one of the strains, CB351, must bear an additional extragenic mutation. Genomic DNA of this strain was isolated and sequenced. A single point mutation, which was responsible for the suppression effect, localized to the fliS gene and encoded a FliS A22T mutant. To further investigate the effect of this mutation on motility, Salmonella strain ΔflhB fliS(A22T) was constructed from strain MKM50. We found that the A22T mutation of FliS enhanced motility of Salmonella cells expressing Aquifex FlhB or different FlhB chimera (Figure 9B), although its suppression ability was much weaker than that of the intragenic FlhB suppressor mutations identified earlier. The FliS A22T mutant did not affect the motility with wild type Salmonella FlhB.
Figure 9. Effect of fliS mutation on motility of Salmonella cells.
Cells were transformed with pTrc99A-based plasmids encoding various FlhB proteins. Soft agar plates were incubated at 30°C for the indicated time. (A) Motility of ΔflhB Salmonella strains producing Sal/AquFlhB F343 frame shift. One of the strains contains additional mutation in fliS. 1 - empty vector, 2 - wild type Salmonella FlhB, 3 - Sal/AquFlhB F343 frame shift, 4 - Sal/AquFlhB F343 frame shift plus FliS (A22T). The asterisk “*” marks the previous emplacement for the wild type strain that was removed because it would have overgrown after 20 hours. (B) Comparison of the motility of ΔflhB and ΔflhB fliS(A22T) Salmonella strains producing different FlhB variants.
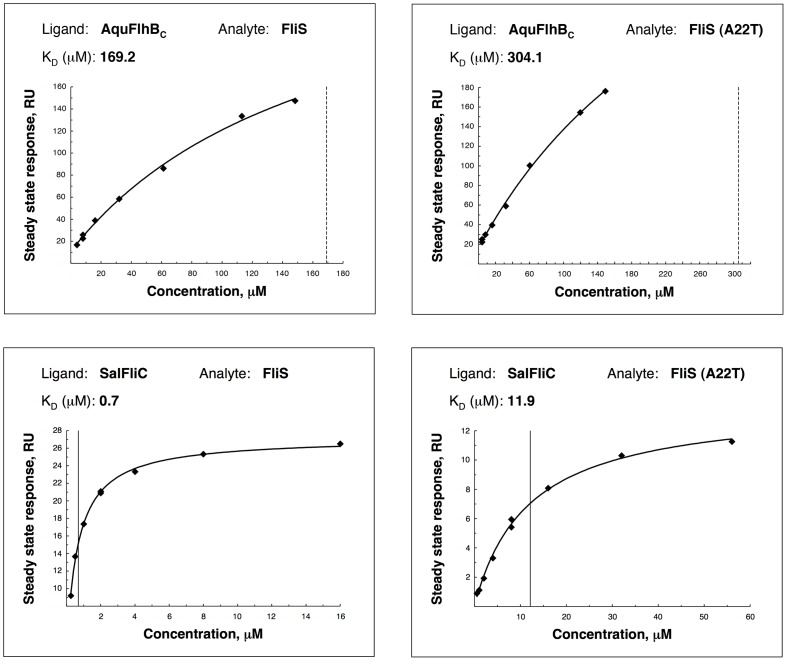
The mutation of FliS that enhances motility of FlhB mutant cells suggests interaction between these two proteins. One of the possible effects of this mutation could be a change in the affinity of FliS to FlhBC. On the other hand, Ala22 lies in the region that directly contacts with FliC [21], the natural binding partner of FliS in Salmonella cells [22]. Obviously, mutation of this amino acid could affect FliS-FliC interaction. To check both possibilities, interactions between Salmonella FliS and Salmonella FliC or Aquifex FlhBC were analyzed using surface plasmon resonance technique (Biacore, GE Healthcare). AquFlhBC, or Salmonella FliC, was amine coupled to a sensor chip and allowed to bind to flowing FliS and FliS A22T. A dissociation constant KD was calculated using steady state analysis with 1∶1 binding model (Figure 10). There were some differences between affinities of AquFlhBC to FliS and FliS A22T (KD is 169.2 µM for wild-type FliS, and 304.1 µM for FliS A22T), however we cannot make any conclusions since the affinity for both cases was very low and as a result the estimation was not very confident. The affinity of FliS mutant A22T to FliC was greatly reduced in comparison with wild type protein (0.7 µM for wild-type FliS and 11.9 µM for FliS A22T). Thus mutation of FliS affects interaction between FliS and FliC, but not between FliS and FlhB.
Figure 10. Steady-state surface plasmon resonance analysis of FliS binding to Aquifex FlhBC and FliC.
FliS (or FliS A22T) of various concentrations was flowed over the sensor surface with immobilized ligand (AquFlhBC or FliC) in Biacore buffer (10 mM HEPES pH 7.4, 150 mM NaCl, 0.05% (v/v) Surfactant P20). Response at steady state was plotted against analyte concentration. KD is measured as the protein concentration that gives response equal to 50% saturation.
Discussion
FlhB is a key protein in the regulation of secretion by the type III secretion system. Homologues of FlhB were found in all type III secretion systems including Aquifex, one of the deepest-branching lineages within the bacteria having flagella. In this work we applied a new genetic approach, which allowed us to reveal functionally important properties of FlhB. Complementation properties of FlhB from two very distantly related organisms: S. typhimurium and A. aeolicus, have been investigated. We showed that Sal/AquFlhB chimeras (or the whole Aquifex FlhB) could not replace native FlhB in Salmonella cells. But functionality of the substituted protein could be restored to some degree by mutations that destabilize the protein. It is well known that proteins from hyperthermophilic organisms (like Aquifex) possess rigid structure at moderate temperature. For this reason thermophilic enzymes are not functional at normal temperature since they do not have enough flexibility to perform their function [23]–[25]. FlhB has been suggested to exist in two conformational states corresponding to two substrate specificity modes: rod/hook mode and filament mode [9], . The flexibility might be essential for transition from one state to another. The necessary degree of conformational freedom, in particular, might result from the autocleavage within the highly conserved NPTH sequence. In our experiments we could not detect any improvement in autocleavage for the suppressor mutations that improve the functionality of FlhB. However, while we cannot completely exclude possible effect of these mutations on FlhB autocleavage, we think that there could be other factors important for the protein function. One of such factors could be flexibility of FlhB molecule. It is also possible that the suppressor mutations in AquFlhBC affect the binding of FlhB to other proteins of the type III secretion system. Investigation of this possibility is a subject for future research.
The conformation changes in wild-type Salmonella FlhB are catalyzed through interaction between FlhBC and FliK. However, a fliK gene was not found in the Aquifex genome [26]. It could be that at extremely high temperatures flexibility of FlhBC is enough to switch even without help of FliK. It is not clear whether Aquifex FlhB can recognize FliK the same way as Salmonella FlhB. Such recognition could be important for the function of Aquifex FlhB in Salmonella cells.
Proteins of the FlhB family exhibit significant variation in length (mainly because of differences at the C-terminus). Salmonella FlhB is longer than Aquifex by 33 residues. To check importance of these residues for function of the protein, we created chimera FlhB having C-termini identical to wild-type Salmonella FlhB. We found that addition of extra residues to C-terminus of chimera FlhB almost has no effect on motility of Salmonella cells. Moreover, one of the frame shift suppressor mutation (F343 frame shift) occurred in Sal/AquFlhB357. This chimera has a C-terminus native to Salmonella FlhB, however the C-terminus is apparently not functional and must be modified to restore protein activity. Surprisingly, the C-terminus created by the frame-shift mutation was identical to that found in several frame-shift mutants of Salmonella FlhB that suppress the phenotype of the fliK deletion [11], [13]. Currently existing ideas suggest that ΔfliK suppressor mutations of FlhB could change conformational flexibility of FlhB or its affinity to other proteins [8], [11], [27]. However, there is no clear understanding how frame shift mutations of FlhB could affect protein function. Apparently, the C-terminal part of Salmonella FlhB is involved in the secretion regulation specific for Salmonella. We strongly believe that there is something common between ΔfliK suppressor mutations of Salmonella FlhB mutations and the mutations we describe in this paper. Definitely, investigation of this relationship deserves more detailed study in the future.
One extragenic suppressor mutation we identified was found in the fliS gene. FliS is a chaperone, which specifically binds to flagellin (FliC), the major component of flagellar filament, and prevents its polymerization [21]. Presumably FliC-FliS complex is delivered to the export apparatus, where it must dissociate, since only FliC is translocated through the cell membrane [28], [29]. However details of FliS-FliC dissociation are not known. We found that the mutation of FliS (A22T) improves the motility of Salmonella cells expressing chimeric flhB genes while it decreases the affinity of FliS to FliC. These data suggests that FlhB might be involved in the process of FliS-FliC dissociation. Although we could not detect stable complexes between Salmonella FlhB and FliS-FliC or isolated FliS and FliC proteins in vitro (data are not shown), such interactions can be possible in vivo where FlhB works together with other proteins of type III secretion system. The cytoplasmic domain of FlhA, another membrane protein of type III secretion system, is known to interact strongly with FliS-FliC complex [30]. Whether FlhB also participates in this process releasing FliC prior to secretion is a question for future work.
Materials and Methods
Bacterial Strains, Plasmids and Growth Conditions
The bacterial strains and plasmids used in this study are listed in Tables 2 and 3, respectively. Bacteria were routinely cultured in Luria-Bertani Broth medium with continuous shaking at 37°C. For Salmonella strains, ampicillin was used in media at 100 µg ml−1, and for E. coli strains it was used at 50 µg ml−1. Tetracycline was used at 15 µg ml−1 where appropriate.
Table 2. Strains used in this study.
| Strain | Genotype | Source/ref. | ||
| Escherichia coli | ||||
| NovaBlue | Recipient for cloning experiments | Novagen | ||
| BW25113 | Plasmid pKD46 (AmpR) | CGSC7739a | ||
| BL21(DE3) | Host for overexpression from T7 promoter | Novagen | ||
| Rosetta™ | Host for overexpression from T7 promoter | Novagen | ||
| Salmonella enterica serovar Typhimurium | ||||
| JR501 | R−m+ for converting plasmids to Salmonella compatibility | FBSb [39] | ||
| TT13206 | LT7 phoN51::Tn10-11(TetR) | SGSC3718c | ||
| SJW1103 | Wild-type for motility and chemotaxis | FBSb [40] | ||
| MKM50 | ΔflhB null mutant | FBSb [7] | ||
| CB351 | ΔflhB fliS22264(A22T) carrying plasmid pTB342 (AmpR) | This study | ||
| VM001 | ΔflhB fliS22264(A22T) derived from MKM50 (ΔflhB) | This study | ||
CGSC, Escherichia coli Genetic Stock Center, Yale University, USA.
FBS, School of Frontier Biosciences, Osaka University, Japan.
SGSC, Salmonella Genetic Stock Centre, University of Calgary, Canada.
Table 3. Plasmids used in this study.
| Plasmid | Relevant characteristic | Ref. |
| pKD46 | λ-Red genetic engineering plasmid; temperature-sensitive ori (30°C); AmpR | CGSC |
| pTrc99A-FF4 | Modified pTrc99A expression vector; AmpR | [17] |
| pMM26 | pTrc99A-FF4 derivative encoding wild-type S. typhimurium FlhB (1–383) | [9] |
| pTAB32 | pTrc99A-FF4 derivative encoding wild-type A. aeolicus FlhB (1–350) | [32] |
| pTB334 | pTrc99A-FF4 derivative encoding a FlhB chimera: | This study |
| S. typhimurium FlhB1–218-A. aeolicus FlhB213–350 | ||
| pTB335 | pTrc99A-FF4 derivative encoding a FlhB chimera: | This study |
| S. typhimurium FlhB1–218-A. aeolicus FlhB213–340-S. typhimurium FlhB347–383 | ||
| pTB336 | pTrc99A-FF4 derivative encoding a FlhB chimera: | This study |
| S. typhimurium FlhB1–218-A. aeolicus FlhB213–350-S. typhimurium FlhB357–383 | ||
| pTB337 | pTrc99A-FF4 derivative encoding a FlhB chimera: | This study |
| S. typhimurium FlhB1–218-A. aeolicus FlhB213–341-VQKEKGLRLIGSSRVDLQACKLGCFGG | ||
| pTB338 | pTrc99A-FF4 derivative encoding a FlhB chimera: | This study |
| S. typhimurium FlhB1–218-A. aeolicus FlhB213–340/F330L-S. typhimurium FlhB347–383 | ||
| pTB339 | pTrc99A-FF4 derivative encoding a FlhB chimera: | This study |
| S. typhimurium FlhB1–218-A. aeolicus FlhB213–340/V307G-S. typhimurium FlhB347–383 | ||
| pTB340 | pTrc99A-FF4 derivative encoding a FlhB chimera: | This study |
| S. typhimurium FlhB1–218-A. aeolicus FlhB213–340/Δ(288–289)-S. typhimurium FlhB347–383 | ||
| pTB341 | pTrc99A-FF4 derivative encoding a FlhB chimera: | This study |
| S. typhimurium FlhB1–218-A. aeolicus FlhB213–350/V259E-S. typhimurium FlhB357–383 | ||
| pTB342 | pTrc99A-FF4 derivative encoding a FlhB chimera: | This study |
| S. typhimurium FlhB1–218-A. aeolicus FlhB213–343-RFTPAGNVLHNLRTFRCQKRWIL |
CGSC, Escherichia coli Genetic Stock Center, Yale University, USA.
Plasmid and Strain Constructions
The oligonucleotides used in construction of plasmids and strains are listed in Table 4. The flhB chimera gene encoded by plasmid pTB334 was derived from S. typhimurium and A. aeolicus genomic DNA by PCR-driven overlap extension, similar to previously described [31], [32]. For the swapping of domains within genes carried on plasmids, the plasmid was first made linear at the desired location using PCR with a thermophilic DNA polymerase lacking strand-displacing activity. The DNA fragment to be inserted was also derived by PCR, and contained ends homologous to the linear plasmid. The two DNA fragments were then fused to recreate circular plasmid DNA, using an In-fusion cloning kit (Clontech Laboratories, Inc., USA).
Table 4. Oligonucleotides used in the strain and plasmid constructions.
| Primer name | Sequence (5′ to 3′)a |
| 5′-Sal-flhB | CATATGGCAGAAGAGAGCGACGAC |
| 3′-Sal-flhB-1-218 | GACATCATTATCTTTTTCAGGTGGCTAAAGATCTGG |
| 5′-Aqu-flhB-213-350 | CTTTAGCCACCTGAAAAAGATAATGATGTCGAGAAGGGAATTG |
| 3′-Aqu-flhB | CTAGAGGATCCTATTAGGCGTAAACC |
| 5′-pMM26-Linear-347 | GTCTGGCAGCTTAAACGCTGG |
| 3′-pMM26-Linear-218 | TTTCAGGTGGCTAAAGATCTGG |
| 5′-Aqu-flhB-213 | TTTAGCCACCTGAAAAAGATAATGATGTCGAGAAGGG |
| 3′-Aqu-flhB-340 | TTTAAGCTGCCAGACGTAGGCTATTATTTCCGCTACGG |
| 5′-pMM26-Linear-357 | GGCGGGCAACGTCCTCCAC |
| 3′-Aqu-flhB-350 | AGGACGTTGCCCGCCGGCGTAAACCTTTTTCTTTTTGAAC |
Restriction sites are underlined.
To genetically engineer strains, bacteriophage λ-Red-based recombination was used [33]. Genomic DNA was first prepared from E. coli strain TT13206, which contains a Tn10-11 element on the chromosome, conferring tetracycline-resistance [34]. Tetracycline-resistance (tetRA) cassettes, flanked by ends homologous to the intended chromosomal target site were obtained by PCR using TT13206 DNA as template. These cassettes were then inserted into the chromosome of the desired strain by homologous recombination. Following this, it was possible to counter-select against tetracycline-resistance on medium containing fusaric acid, and replace the tetRA-cassette with a PCR product containing ends homologous to the target site.
Soft Tryptone Agar Motility Assays and Isolation of Motile Suppressor Mutants
Soft tryptone agar contained 0.35% (w/v) agar, and was used in motility assays at 30°C [35]. Freshly transformed cells were inoculated as colonies directly into soft tryptone agar. To test for dominant effects, 0.1 mM IPTG was included in the agar plates. To isolate motile suppressor mutants, 10 µl of an overnight culture were inoculated as a streak into soft tryptone agar. After incubation, suppressor mutants were purified from the edges of outgrowths generated by motile cells.
Protein Purification
Purification of Aquifex FlhBC, Salmonella FlhBC and the mutated AquFlhBC proteins was done as described previously [14]. Purification of the F343 frame shift mutant was not successful; therefore this mutant was not used for further experiments.
Thermal Shift Assay
TSA experiments were carried out in triplicates using a 7500 Real-Time PCR System (Applied Biosystems, Inc.). The reaction mixture contained 0.2 mg ml−1 purified protein, 5× SYPRO Orange dye (Molecular Probes, Invitrogen) in 10 mM K-phosphate pH 6.1, 50 mM NaCl. Reactions were heated from 20 to 95°C with a rate of 1°C per minute. Protein unfolding was monitored by detection of changes in fluorescence of SYPRO Orange dye. Fluorescence intensities were plotted as a function of temperature, and the midpoint of the unfolding transition was taken as an estimation of the melting temperature.
Circular Dichroism Spectroscopy and Chemical Denaturation
Far-UV CD spectra were measured using an Aviv model 400 spectropolarimeter (Lakewood, NJ) in 1 mm cuvettes in 10 mM K-phosphate pH 6.1, 50 mM NaCl. Changes in helical content during urea denaturation were monitored at 222 nm. Secondary structure content was calculated from the CD spectra using a multilinear regression [36]. The errors represent standard deviations for the secondary structure content calculated from the CD spectra of different preparations of the same proteins. The changes that we describe as significant ones based on the calculations of the secondary structure are confirmed by the shifts of the spectra intersection with the X-axis to lower wavelengths, which is characteristic for the increase of random coil content.
Biacore Analysis
All analyses were carried out on a Biacore T100 (T200 Sensitivity Enhanced) (GE Healthcare). Ligand AquFlhBC or FliC was immobilized on a CM5 chip by amine cross-linking. Analyte FliS or FliS (A22T) of various concentrations in binding buffer (10 mM HEPES pH 7.4, 150 mM NaCl, 0.05% (v/v) Surfactant P20) was passed over the sensor surface at a flow rate of 30 µl min−1 for 12 min. Dissociation of the sample was monitored in binding buffer for 12 min. Surface was regenerated using 10 mM Glycine pH 2.0. All measurements were performed at 25°C. Data were analyzed by Biacore T200 Evaluation software using a 1∶1 binding model.
Sequence Alignment
Acknowledgments
We thank I. Meshcheryakova for helping in the experiments. We thank H. Matsunami and Y.-H. Yoon for useful discussions on the results. We are grateful to R. Huber (University of Regensberg) for providing us with genomic DNA of A. aeolicus and to School of Frontier Biosciences (Osaka University) for Salmonella strains. We thank E. Martz for critical reading of the manuscript.
Funding Statement
This work was supported by direct funding provided by OIST. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Aizawa S-I (2001) Bacterial flagella and type III secretion systems. FEMS Microbiol Lett 202: 157–164. [DOI] [PubMed] [Google Scholar]
- 2. Blocker A, Komoriya K, Aizawa S-I (2003) Type III secretion systems and bacterial flagella: Insights into their function from structural similarities. Proc Natl Acad Sci USA 100: 3027–3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Minamino T, Macnab RM (1999) Components of the Salmonella flagellar export apparatus and classification of export substrates. J Bacteriol 181: 1388–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hirano T, Yamaguchi S, Oosawa K, Aizawa S-I (1994) Roles of FliK and FlhB in determination of flagellar hook length in Salmonella typhimurium . J Bacteriol 176: 5439–5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hirano T, Minamino T, Namba K, Macnab RM (2003) Substrate specificity classes and the recognition signal for Salmonella type III flagellar export. J Bacteriol 185: 2485–2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Minamino T, Iino T, Kutsukake K (1994) Molecular characterization of the Salmonella typhimurium flhB operon and its protein products. J Bacteriol 176: 7630–7637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fraser GM, Hirano T, Ferris HU, Devgan LL, Kihara M, et al. (2003) Substrate specificity of type III flagellar protein export in Salmonella is controlled by subdomain interactions in FlhB. Mol Microbiol 48: 1043–1057. [DOI] [PubMed] [Google Scholar]
- 8. Zarivach R, Deng W, Vuckovic M, Felise HB, Nguyen HV, et al. (2008) Structural analysis of the essential self-cleaving type III secretion proteins EscU and SpaS. Nature 453: 124–127. [DOI] [PubMed] [Google Scholar]
- 9. Minamino T, Macnab RM (2000) Domain structure of Salmonella FlhB, a flagellar export component responsible for substrate specificity switching. J Bacteriol 182: 4906–4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ferris HU, Furukawa Y, Minamino T, Kroetz MB, Kihara M, et al. (2005) FlhB regulates ordered export of flagellar components via autocleavage mechanism. J Biol Chem 280: 41236–41242. [DOI] [PubMed] [Google Scholar]
- 11. Williams A, Yamaguchi S, Togashi F, Aizawa S-I, Kawagishi I, et al. (1996) Mutations in fliK and flhB affecting flagellar hook and filament assembly in Salmonella typhimurium . J Bacteriol 178: 2960–2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Minamino T, Ferris HU, Moriya N, Kihara M, Namba K (2006) Two parts of the T3S4 domain of the hook-length control protein FliK are essential for the substrate specificity switching of the flagellar type III export apparatus. J Mol Biol 362: 1148–1158. [DOI] [PubMed] [Google Scholar]
- 13. Kutsukake K, Minamino T, Yokoseki T (1994) Isolation and characterization of FliK-independent flagellation mutants from Salmonella typhimurium . J Bacteriol 176: 7625–7629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Meshcheryakov VA, Yoon Y-H, Samatey FA (2011) Purification, crystallization and preliminary X-ray crystallographic analysis of the C-terminal cytoplasmic domain of FlhB from Aquifex aeolicus . Acta Crystallogr F 67: 280–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Meshcheryakov VA, Samatey FA (2011) Purification, crystallization and preliminary X-ray crystallographic analysis of the C-terminal cytoplasmic domain of FlhB from Salmonella typhimurium . Acta Crystallogr F 67: 808–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Meshcheryakov VA, Kitao A, Matsunami H, Samatey FA (2013) Inhibition of a type III secretion system by the deletion of a short loop in one of its membrane proteins. Acta Crystallogr D 69: 812–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ohnishi K, Fan F, Schoenhals GJ, Kihara M, Macnab RM (1997) The FliO, FliP, FliQ, and FliR proteins of Salmonella typhimurium: putative components for flagellar assembly. J Bacteriol 179: 6092–6099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pantoliano MW, Petrella EC, Kwasnoski JD, Lobanov VS, Myslik J, et al. (2001) High-density miniaturized thermal shift assays as a general strategy for drug discovery. J Biomol Screen 6: 429–440. [DOI] [PubMed] [Google Scholar]
- 19. Vedadi M, Niesen FH, Allali-Hassani A, Fedorov OY, Finerty PJ Jr, et al. (2006) Chemical screening methods to identify ligands that promote protein stability, protein crystallization, and structure determination. Proc Natl Acad Sci USA 103: 15835–15840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Niesen FH, Berglund H, Vedadi M (2007) The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat Protoc 2: 2212–2221. [DOI] [PubMed] [Google Scholar]
- 21. Evdokimov AG, Phan J, Tropea JE, Routzahn KM, Peters HK, et al. (2003) Similar modes of polypeptide recognition by export chaperones in flagellar biosynthesis and type III secretion. Nat Struct Biol 10: 789–793. [DOI] [PubMed] [Google Scholar]
- 22. Auvray F, Thomas J, Fraser GM, Hughes C (2001) Flagellin polymerisation control by a cytosolic export chaperone. J Mol Biol 308: 221–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Varley PG, Pain RH (1991) Relation between stability, dynamics and enzyme activity in 3-phosphoglycerate kinases from yeast and Thermus thermophilus . J Mol Biol 220: 531–538. [DOI] [PubMed] [Google Scholar]
- 24. Zavodszky P, Kardos J, Svingor A, Petsko GA (1998) Adjustment of conformational flexibility is a key event in the thermal adaptation of proteins. Proc Natl Acad Sci USA 95: 7406–7411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Merz A, Yee MC, Szadkowski H, Pappenberger G, Crameri A, et al. (2000) Improving the catalytic activity of a thermophilic enzyme at low temperatures. Biochemistry 39: 880–889. [DOI] [PubMed] [Google Scholar]
- 26. Deckert G, Warren PV, Gaasterland T, Young WG, Lenox AL, et al. (1998) The complete genome of the hyperthermophilic bacterium Aquifex aeolicus . Nature 392: 353–358. [DOI] [PubMed] [Google Scholar]
- 27. Deane JE, Graham SC, Mitchell EP, Flot D, Johnson S, et al. (2008) Crystal structure of Spa40, the specificity switch for the Shigella flexneri type III secretion system. Mol Microbiol 69: 267–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Thomas J, Stafford GP, Hughes C (2004) Docking of cytosolic chaperone-substrate complexes at the membrane ATPase during flagellar type III protein export. Proc Natl Acad Sci USA 101: 3945–3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Evans DBL, Stafford GP, Ahmed S, Fraser GM, Hughes C (2006) An escort mechanism for cycling of export chaperones during flagellar assembly. Proc Natl Acad Sci USA 103: 17474–17479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bange G, Kummerer N, Engel C, Bozkurt G, Wild K, et al. (2010) FlhA provides the adaptor for coordinated delivery of late flagella building blocks to the type III secretion system. Proc Natl Acad Sci USA 107: 11295–11300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Heckman KL, Pease LR (2007) Gene splicing and mutagenesis by PCR-driven overlap extension. Nat Protoc 2: 924–932. [DOI] [PubMed] [Google Scholar]
- 32. Barker CS, Samatey FA (2012) Cross-complementation study of the flagellar type III export apparatus membrane protein FlhB. PLoS One 7: e44030 doi:10.1371/journal.pone.0044030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Karlinsey JE (2007) λ-Red genetic engineering in Salmonella enterica serovar Typhimurium. Methods Enzymol 421: 199–209. [DOI] [PubMed] [Google Scholar]
- 34. Jiang W, Metcalf WW, Lee KS, Wanner BL (1995) Molecular cloning, mapping, and regulation of Pho regulon genes for phosphonate breakdown by the phosphonatase pathway of Salmonella typhimurium LT2. J Bacteriol 177: 6411–6421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Toker AS, Kihara M, Macnab RM (1996) Deletion analysis of the FliM flagellar switch protein of Salmonella typhimurium . J Bacteriol 178: 7069–7079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Greenfield NJ (2006) Using circular dichroism spectra to estimate protein secondary structure. Nat Protoc 1(6): 2876–2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948. [DOI] [PubMed] [Google Scholar]
- 38. Goujon M, McWilliam H, Li W, Valentin F, Squizzato S, et al. (2010) A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res 38: W695–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ryu J, Hartin RJ (1990) Quick transformation in Salmonella typhimurium LT2. Biotechniques 8: 43–44. [PubMed] [Google Scholar]
- 40. Yamaguchi S, Fujita H, Sugata K, Taira T, Iino T (1984) Genetic analysis of H2, the structural gene for phase-2 flagellin in Salmonella . J Gen Microbiol 130: 255–265. [DOI] [PubMed] [Google Scholar]