Abstract
Mutations in the park2 gene, encoding the RING-inBetweenRING-RING E3 ubiquitin ligase parkin, cause 50% of autosomal recessive juvenile Parkinsonism cases. More than 70 known pathogenic mutations occur throughout parkin, many of which cluster in the inhibitory amino-terminal ubiquitin-like domain, and the carboxy-terminal RING2 domain that is indispensable for ubiquitin transfer. A structural rationale showing how autosomal recessive juvenile Parkinsonism mutations alter parkin function is still lacking. Here we show that the structure of parkin RING2 is distinct from canonical RING E3 ligases and lacks key elements required for E2-conjugating enzyme recruitment. Several pathogenic mutations in RING2 alter the environment of a single surface-exposed catalytic cysteine to inhibit ubiquitination. Native parkin adopts a globular inhibited conformation in solution facilitated by the association of the ubiquitin-like domain with the RING-inBetweenRING-RING C-terminus. Autosomal recessive juvenile Parkinsonism mutations disrupt this conformation. Finally, parkin autoubiquitinates only in cis, providing a molecular explanation for the recessive nature of autosomal recessive juvenile Parkinsonism.
 Mutations in the E3 ubiquitin ligase parkin are associated with juvenile Parkinson’s disease. Here the authors report the solution structure of the Parkin RING2 domain, revealing how disease-associated mutations affect its function and providing a molecular explanation for the recessive nature of the disease.
Mutations in the E3 ubiquitin ligase parkin are associated with juvenile Parkinson’s disease. Here the authors report the solution structure of the Parkin RING2 domain, revealing how disease-associated mutations affect its function and providing a molecular explanation for the recessive nature of the disease.
Parkinson’s disease (PD) is a neurodegenerative disorder that arises from the loss of dopaminergic neurons in the substantia nigra of the midbrain. Typical symptoms include bradykinesia and loss of motor control. In addition to sporadic PD, mutations in six different genes lead to heritable forms of the disease. In particular, homozygous and compound heterozygous mutations in the gene encoding parkin (PARK2) cause 50% of autosomal recessive juvenile Parkinsonism (ARJP) cases resulting in early-onset PD1.
Parkin is a RING-inBetweenRING-RING (RBR) E3 ubiquitin ligase involved in the ubiquitination of substrate proteins2,3 and mitochondrial mitophagy4,5. The hallmark of RBR ligases is the presence of two RING (Really Interesting New Gene) domains (RING1 and RING2), separated by an inBetweenRING (IBR) domain (Fig. 1a). The family includes the human homologue of the Drosophila Ariadne (HHARI/ARI1), androgen-receptor-associated protein 54, haem-oxidized IRP2 ubiquitin ligase 1 (HOIL-1), HOIL-1-interacting protein (HOIP) and Dorfin2,3. Parkin also contains an inhibitory N-terminal ubiquitin-like domain (Ubl) (ref. 6). Pathogenic mutations leading to ARJP include numerous deletions, truncations and point mutations that occur throughout parkin, but cluster in the Ubl and RING2 domains7,8.
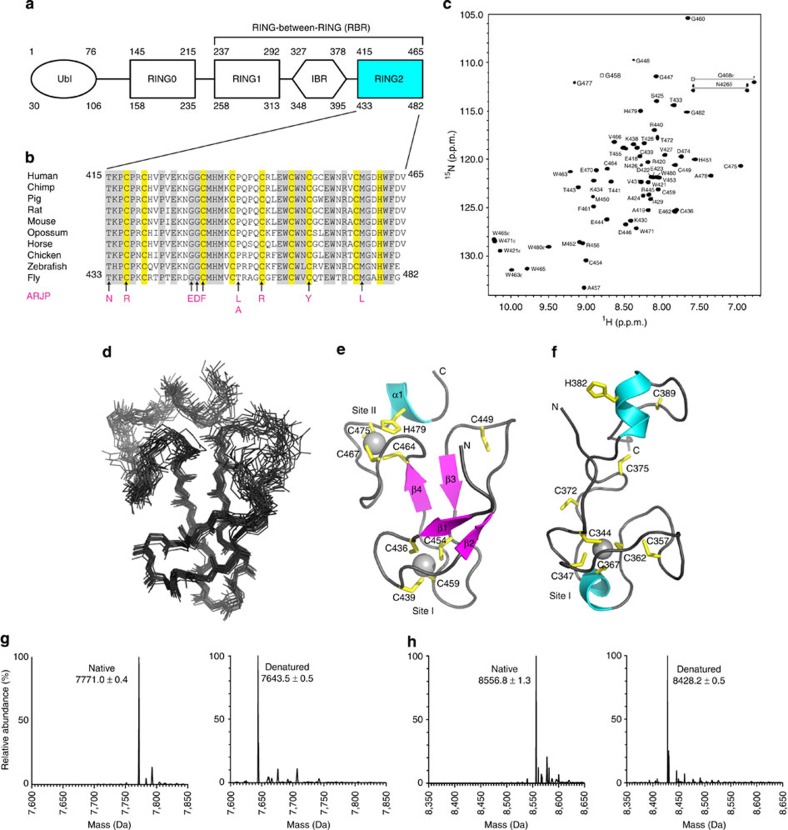
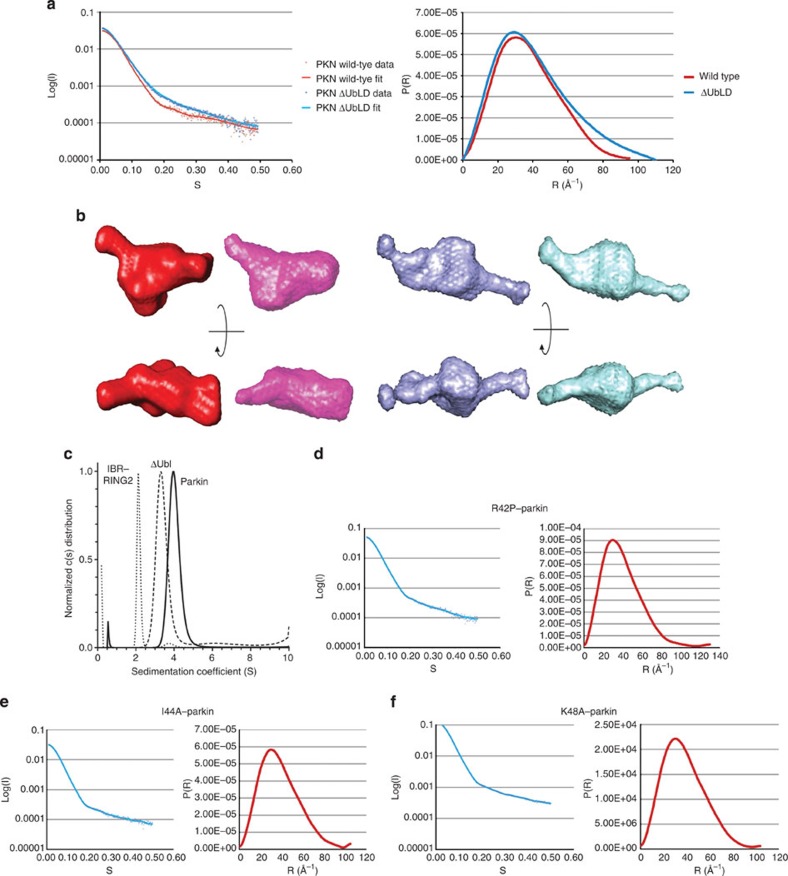
Figure 1. Sequence alignment, structure and metal analysis of parkin RING2 domain.
(a) Domain structure of parkin showing the C-terminal RING1, IBR and RING2 domains found in all RBR E3 ligase proteins. The ubiquitin-like (Ubl) and RING0 domains are specific to the parkin E3 ligase. Residue numbering is shown for both the human (top) and D. melanogaster (bottom) parkin sequences. (b) Multiple sequence alignment of the RING2 domain for parkin orthologues. Sequence numbers are indicated for the human and D. melanogaster species only. Conserved (grey) and cysteine/histidine (yellow) residues are highlighted. Substitutions that contribute to ARJP in the parkin RING2 domain are shown below the sequences (magenta). (c) Assigned 600 MHz 1H–15N HSQC spectrum of 13C,15N-labelled parkin RING2 (500 μM in 20 mM Tris–HCl, 120 mM NaCl, 1 mM DTT, pH 7.25), labelled using the one-letter amino acid code and residue number according to the D. melanogaster parkin sequence. (d) Superposition of the 20 lowest energy solution structures of parkin RING2 (residues 430–482, backbone r.m.s.d. 0.82±0.17 Å). (e) Ribbon structure of parkin RING2 showing β-strands β1 (T433–P435), β2 (P442–E444), β3 (H451–V453) and β4 (E462–C464), and helix α1 (M476–W480). Side chains for Zn2+-coordinating residues are shown in yellow. (f) Structure of the HHARI RING2 (ref. 35) (PDB accession code 1WD2) showing different zinc occupancy and fold compared with parkin (e). Deconvoluted mass spectra for native and denatured parkin RING2 (g), and native and denatured HHARI RING2 (h). The mass differences of 127.5 Da (g) and 128.4 Da (h) indicate the presence of two bound Zn2+ ions in the native proteins. Raw data are found in Supplementary Figure S2. Structures were visualized using Pymol (PyMOL Molecular Graphics System, Version 1.5.0.4 Schrödinger, LLC).
RING E3 ligases function by recruiting both a ubiquitin-loaded E2 enzyme and a substrate to catalyse the transfer of ubiquitin. Parkin was originally thought to be a subclass of the canonical RING E3 enzymes, as early studies showed that the RING1 and RING2 domains could each recruit E2-conjugating enzymes, including UbcH7 (UBE2L3), UbcH8 (UBE2L6) and UbcH13/Uev1a9,10,11,12,13, and lead to autoubiquitination10,11. Numerous studies show that the RBR domains of parkin, HOIP and HHARI are sufficient for ubiquitination14,15,16. However, the ubiquitination activity of parkin is modulated by its Ubl domain (Fig. 1a), proposed to render parkin in an autoinhibited state6,17. ARJP mutations in the Ubl domain result in constitutively active parkin that is capable of autoubiquitination6. Such mutants are unstable in cells, often only detectable in the presence of proteasome inhibitors6,18. In contrast most C-terminal mutations, particularly those found in RING2, abolish ligase activity11,18,19,20,21,22,23,24,25,26.
Studies with HOIP and HHARI suggest that RBR ligases form a thiolester intermediate between a cysteine in the RING2 domain and ubiquitin14,15,27. By this mechanism, the RING1 module recruits the E2~ubiquitin complex, whereas RING2 acts as a true catalytic domain performing the transthiolation reaction14,15,27 by transferring ubiquitin to a substrate, in a manner analogous to the homologous to E6AP C terminus (HECT)-type ligases28. Both parkin and HOIP can catalyse ubiquitin conjugation in the absence of an E2 enzyme14,19. The loss of the RING1 domain of parkin, thought to be essential for E2 recruitment, does not result in a loss of ligase activity19,25. Parkin RING2 mutants are catalytically dead in the absence or presence of E2s, whereas mutations outside of RING2 retain both E2-dependent and -independent catalytic activity24. These insights suggest that the RING2 of RBR ligases is a catalytic entity that can function in the absence of an E2 enzyme.
The mechanism of RBR ligase-catalysed ubiquitin transfer is poorly understood, partly because of a lack of biophysical and structural data on these enzymes. Structures of Ubl domains from parkin and HOIL-1, and the IBR of parkin and HOIP (PDB 2CT7), are available29,30,31,32,33,34. However, little structural data exist for any RBR RING2 domain that would support a catalytic role, identify potential catalytic residues needed to catalyse transthiolation, or identify how this domain might differ from conventional RING E3 ligases. The only available structure of the HHARI RING2 domain shows a loosely organized structure containing a single zinc ion35 compared with typical RING proteins that possess two metal ions. Although potentially interesting, the HHARI RING2 structure cannot be reconciled with available functional data. Further, the structure sheds little insight on the function of the parkin RING2 domain and how ARJP-causing mutations contribute to the loss of E3 ligase activity. The opposing effects of different mutations in the Ubl and RING2 domains on parkin activity presents an enigma for understanding the loss-of-function phenotype associated with recessive PD.
To understand how inhibitory and catalytic domains function in both wild-type parkin and in the disease context, we carried out an extensive biophysical and biochemical characterization of parkin and mutants in solution. We determined structures of the parkin RING2 and IBR–RING2 domains, revealing that the RING2 is neither a canonical RING E3 ligase nor an E2-recruiting module. We find that RING2 is catalytic, identify catalytic residues and show that pathogenic mutations in RING2 perturb the catalytic environment. We show that the tertiary structure of native parkin is dependent on the interaction of the Ubl domain with the RBR region and is disrupted by pathogenic mutations within the N-terminal domain. Finally, we find that constitutively active mutant parkin autoubiquitinates in cis, and not in trans, thereby providing a molecular rationale for the homozygous nature of ARJP.
Results
Parkin RING2 is distinct from canonical RING E3 ligases
In addition to the characteristic RING1, RING2 and IBR domains, parkin contains N-terminal Ubl and RING0 (ref. 36) domains (Fig. 1a). We used the RING2 region from Drosophila melanogaster (A417–G482; Fig. 1b), because it resulted in improved solubility compared with the human protein (residues A398–V465). Numerous studies show that loss of D. melanogaster parkin recapitulates characteristics of PD37,38. In addition, ARJP variants are conserved between species with the exception of T455 (P437 in human). Initial characterization of fly parkin RING2 by 1H–15N heteronuclear single quantum coherence (HSQC) spectroscopy (Fig. 1c) and analytical ultracentrifugation (Supplementary Fig. S1) shows the domain is monomeric and well-folded, but with a disordered N terminus (A417–I429). The solution structure of parkin RING2 shows a well-defined globular fold (root mean squared deviation (r.m.s.d.) 0.82 Å) comprising two pairs of antiparallel β-strands (β1, T433–P435; β2, P442–E444; β3, H451–V453; and β4, E462–C464) and a one-turn C-terminal α-helix (M476–W480) (Fig. 1d and Supplementary Table S1). Two Zn2+-binding sites are present comprising residues in the β1–β2 and β3–β4 loops (Site I: C436, C439, C454 and C459) and the loop after β4 and α1 (Site II: C464, C467, C475 and H479). A single cysteine residue (C449; C431 in human) is exposed on the β2–β3 loop, which, according to chemical-shift analysis, was not oxidized or involved in Zn2+ coordination (Supplementary Table S2). This residue is conserved in all parkin orthologues (Fig. 1b) and other RBR sequences.
The parkin RING2 structure clearly reveals two Zn2+ ions coordinated in a linear arrangement consistent with observations that full-length parkin can accommodate eight Zn2+ ions (that is, two Zn2+ ions per RING0, RING1, IBR and RING2 domains)36. Disagreement exists for the Zn2+-coordinating ability of RBR proteins, as the structure of HHARI RING2 shows a single bound Zn2+ and several non-coordinating cysteine residues35 (Fig. 1f). Substitutions of cysteine residues in parkin render the protein inactive23,24. Given the importance for the Zn2+ ion coordination in canonical E3 ligases and cysteine residues in parkin on its function, we used native-state mass spectrometry to identify the metal–ion stoichiometry of the RING2 domains for both parkin and HHARI. Our analyses reproducibly showed that the mass differences between native and denatured states of parkin and HHARI RING2 (Fig. 1g and Supplementary Fig. S2) corresponded to those expected for two Zn2+ ions, indicating both RING2 domains bind two Zn2+ ions in their native states.
We next used nuclear magnetic resonance (NMR) spectroscopy to reconcile the zinc-binding differences between our data and the previously solved structure of HHARI RING2 (ref. 35). The well-dispersed nature of the 1H–15N HSQC spectrum of HHARI RING2 (Supplementary Fig. S3) indicates the protein is well-folded. Analysis of Cα and Cβ chemical shifts for HHARI RING2 reveal that seven cysteines are involved in Zn2+ coordination (Supplementary Table S2), including C372, C375 and C389 that appear unstructured in HHARI (Fig. 1f) but form a well-defined metal centre (Site II) in parkin (Fig. 1e). The NMR and mass spectral data clearly show that parkin, HHARI and, probably, all related RBR RING2 domains require the coordination of two Zn2+ ions for correct folding.
The RING2 domain lacks features required for E2 recruitment
Comparison of parkin RING2 with canonical RING E3s (Fig. 2) reveals little similarity in protein fold. Parkin RING2 displays a sequential Zn2+ ion coordination (Fig. 2a) where the first and second cysteine pairs and the third and fourth cysteine/histidine pairs act as metal–ion ligands. In contrast, canonical RING E3 ligases, such as TRAF6 or BRCA1, adopt a cross-brace coordination motif39,40 (Fig. 2c), using the first and third pairs of cysteines and the second and fourth pair of cysteine/histidine residues. This difference in coordination leads to a more elongated structure for the parkin RING2 domain compared with the more compact fold of canonical RING domains. These structural differences indicate that the RING2 function will not parallel that of canonical RING E3 ligases.
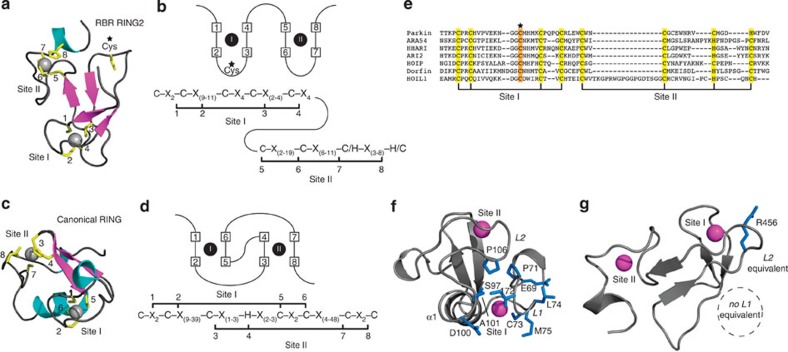
Figure 2. RING2 domain of parkin and other RBR E3 ligases do not recruit E2 enzymes.
Parkin RING2 defines a novel structure common to RBR ubiquitin ligases that are unable to recruit E2 enzymes. The structures and schematic representations of (a,b) RBR RING2 (parkin) and (c,d) RING (TRAF6, PDB 3HCS) domains showing differences in Zn2+ coordination and consensus sequences. (e) Alignment of the RING2 domains of RBR ligases showing residues predicted to coordinate Zn2+ (yellow) based on the parkin RING2 structure and the proposed catalytic cysteine conserved in all sequences (orange). (f) Key residues and elements used for E2 recruitment by the canonical RING E3 ligase TRAF6 are found in two loops (L1, L2) formed through Zn2+ coordination that are absent in (g) parkin RING2. The structures are oriented using the loops involved in Zn2+ coordination for TRAF6 site II and parkin RING2 site I, which exhibit the closest structural similarity.
In contrast to the structural dissimilarity with RING E3 ligases, parkin RING2 possesses several features in common with the IBR domains from parkin32 (PDB accession code 2JMO) and HOIP (PDB accession code 2CT7). With identities below 20%, the sequence of the RING2 domain does not suggest an IBR-fold. However, parkin RING2 superposed well with both IBR structures (backbone r.m.s.d. 2.07 Å and 2.28 Å, respectively) and shares similar Zn2+-coordination topology32. The surface-exposed cysteine residue (C449) observed in the parkin RING2 structure (Fig. 2a) is notably absent in the sequences of other IBR domains32. Alignment of the parkin RING2 sequence with other RBR E3s (Fig. 2e) shows that all proteins contain conserved cysteine and histidine residues required to coordinate two Zn2+ ions as observed in the parkin RING2 structure (Fig. 1e). This indicates that the architecture and Zn2+ coordination observed for parkin RING2 is common to all RBR family members.
Several structures exist for complexes between RING E3 ligases and E2-conjugating enzymes that allow regions of interaction and function to be identified. For example, the E2:E3 complexes of Ubc13/TRAF6 (ref. 41), UbcH5b/cIAP2 (ref. 42) and UbcH7/c-Cbl (ref. 43) show that two loop regions (L1 and L2) of the RING are integral for E2 recruitment (Fig. 2f). Substitutions in these regions compromise the interaction and ubiquitination activity, indicating E2 interaction is necessary for ubiquitin transfer41,42,44. As parkin RING2 is essential for ubiquitination, we reasoned that examining the E2-interacting regions observed in canonical RING E3 ligases might reveal how the RING2 domain in an RBR E3 ligase is involved in a ubiquitination reaction. Using TRAF6 as a template, we find that the RING2 domain lacks the surface and residues typically used by a RING domain to recruit an E2 enzyme as a requisite for ubiquitin transfer. The closest similarity between parkin RING2 and TRAF6 RING domains exists with L2 (Fig. 2f, r.m.s.d. 1.2 Å for RING2 T434–E444, T455–G458, and TRAF6 V81–K91 and C105–D108). In TRAF6, hydrophobic residues (P71, I72, L74, M75 and P106) within the adjacent L1 and L2 loop regions are used to recruit Ubc13 (ref. 41). In particular, a conserved proline (P106 in TRAF6), central to E2 binding, is replaced by a polar side chain group (R456) in parkin RING2. Further, this region lying between the third and fourth chelating cysteine residues from Site I is amongst the poorest conserved in the RBR RING2 sequences (Fig. 2e). The most significant difference between the RBR RING2 and canonical RING domain structures occurs for the L1 loop. In parkin RING2, the different Zn2+-binding topology places its L1 equivalent on the opposite side of the domain distal from the L2 loop (Fig. 2g). These observations indicate that parkin RING2 should be incapable of E2 recruitment using the same binding mode as a canonical E3 RING ligase. To test this hypothesis, we examined the interaction between parkin RING2 and the E2s UbcH7 and UbcH8, which can both facilitate autoubiquitination with full-length parkin6,10,11. As predicted from our structure, we were unable to identify any direct interaction between the IBR or RING2 domains and the E2 enzymes using NMR and isothermal titration calorimetry experiments, even at high concentrations (Supplementary Fig. S4). Our observations demonstrate that parkin RING2, and, by corollary, other RBR RING2 domains are not an E2 recruitment module.
Pathogenic RING2 mutations disrupt catalysis
The parkin RING2 structure positions a single cysteine (C449 in fly; C431 in human), required for transthiolation and proposed to be catalytic, on the surface of the domain (Fig. 3a). To confirm whether this residue is absolutely required for catalysis, we assayed for the formation of a reducible ubiquitin thiolester. We find that active parkin forms a reducible band consistent with the formation of a thiolester (Fig. 3b, lanes 7 and 8). In contrast, a C431S mutant forms a non-reducible band that is base labile, consistent with an ester adduct (Fig. 3b lanes 9–11). C431A-parkin does not form either band, indicating that C431 is required for activity and forms a thiolester (Fig. 3b), as seen for HHARI and HOIP15,27. Several ARJP mutations in RING2 (T433N, C436R, G447E, G448D, C449F and C459R in fly; T415N, C418R, G429E, G430D, C431F and C441R in human), which lead to loss of parkin’s E3 ligase activity, have been reported11,18,19,20,21,22,23,24,25,26. There are two possible explanations for the loss of ligase activity in these mutants—(i) the substitutions lead to the loss of the integrity of the fold of RING2; or (ii) the substitutions alter the chemical environment of the catalytic cysteine, leading to a loss of catalytic competency. To test these hypotheses, ARJP RING2 substitutions were examined to identify how they might alter the structure of parkin. As expected, C436R- and C459R-substituted proteins were insoluble, consistent with their role in Zn2+ coordination. All other RING2 proteins were soluble and well-folded as determined by SDS–polyacrylamide gel electrophoresis and NMR analysis (Supplementary Figs S5 and S6), including C449F, the site of the proposed catalytic cysteine. NMR experiments show that most of the substitutions alter the chemical shifts of residues near the unligated cysteine C449 (Fig. 3a and Supplementary Fig. S6). Our observations show that several ARJP substitutions in the RING2 domain likely modify the chemical environment of C449 and interfere with ubiquitin transthiolation. To test this hypothesis, we assayed each of the pathogenic mutants for autoubiquitination activity and observed that all of the mutants exhibited reduced or no activity (Fig. 3c).
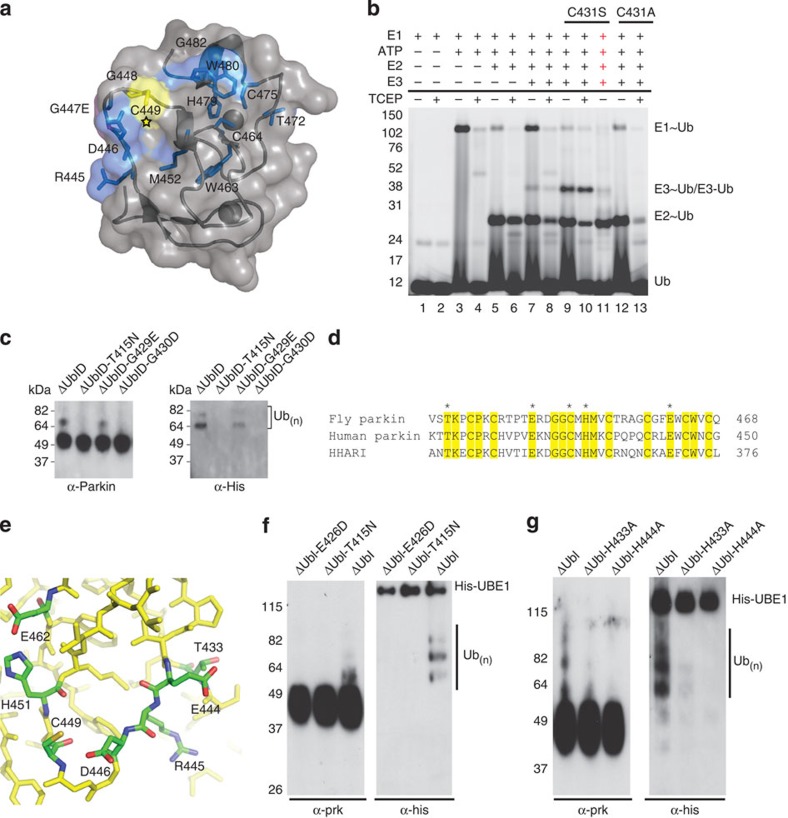
Figure 3. Pathogenic mutations in parkin RING2.
The structure of parkin predicts catalytic residues conserved between parkin and HHARI. (a) The structure of parkin with the catalytic cysteine highlighted (star) and chemical-shift perturbation experiments show affected residues (blue) in parkin RING2 by the ARJP substitution G447E (magenta). (b) The formation of a reducible parkin~Ub thiolester was monitored using His-SUMO-IBR-R2-parkin. Lanes 1–2 contain E1, Mg2+ and Cy5–ubiquitin incubated for 10 min at 37 oC. Lanes 3–4 have ATP added for 10 min. Lanes 5–6 have E2 added followed by a further 10-min incubation. Lanes 7–8 have wild-type parkin added. Lanes 9–11 have C431S–parkin added, with NaOH added to lane 11, indicated by red crosses. Lanes 12–13 have C431A–parkin added; ‘−’ and ‘+’ below the line indicate the absence or presence of TCEP. (c) An autoubiquitination assay of folded pathogenic mutants. Parkin–ubiquitin conjugates are detected by western blotting using parkin (left) and His-ubiquitin (right) antibodies. (d) An alignment of the RING2 domains of fly and human parkin, and HHARI. Conserved residues are shaded yellow and the potential catalytic residues are marked with an asterisk. (e) Close-up view of the surface of parkin RING2 with the potential catalytic residues indicated. (f) Autoubiquitination assay of active parkin (ΔUbl) and parkin ΔUbl-E426D. (g) Autoubiquitination assays of active parkin (ΔUbl) and parkin ΔUbl-H433A and ΔUbl-H444A. In f and g, parkin–ubiquitin conjugates are detected by western blotting using parkin (left) and His-ubiquitin (right) antibodies.
One substitution (T433N) had little effect on the environment of C449; hence, a rationale for the loss of activity is less obvious. In human parkin, the analogous T415N mutation consistently results in an inactive form of parkin regardless of experimental setup11,18,19,24,45. This threonine is conserved in all parkin orthologues and HHARI (Figs 1b and 3d), and rests on the surface of RING2 ~14 Å from the proposed catalytic cysteine. Analysis of the structure revealed a potential catalytic surface hydrogen-bonding network between T433 and a neighbouring glutamate residue (E444; E426 in human) located 3.5 Å from T433, (Fig. 3e), which is also conserved in parkin and HHARI. Consistent with this observation, a large chemical shift occurs for E444 in the 1H–15N HSQC spectrum of the T433N-parkin RING2 (Supplementary Fig. S6a). This led us to hypothesize that this glutamate could be acting in conjunction with the threonine as a catalytic residue to support the transthiolation reaction by C449. To test this hypothesis, we generated a conservative E426D mutation (human numbering; E444D in fly parkin) in an active form of parkin lacking the N-terminal Ubl domain6 (ΔUbl, Fig. 3f). We find that in contrast to ΔUbl–parkin, ΔUbl–E426D is not active for autoubiquitination, further supporting the notion that the RING2 domain is a catalytic entity. In addition to the catalytic cysteine, H451 exhibits a conformation and protonation state consistent with a catalytic role. To test whether H451 (human H433) could act as the proton shuttle between C449 and E462 (human E444), we conducted autoubiquitination assays using H433A and E444A substitutions. Both of these substitutions are unable to support ubiquitination (Fig. 3f). Taken together, the structure and subsequent assays reveal the existence of a hydrogen-bonding network involving T433, E444, H451, E462 and semi-conserved R445 and D446 (Fig. 3e) that supports the catalytic cysteine required for ubiquitination.
IBR and RING2 facilitate interaction with RING0 and RING1
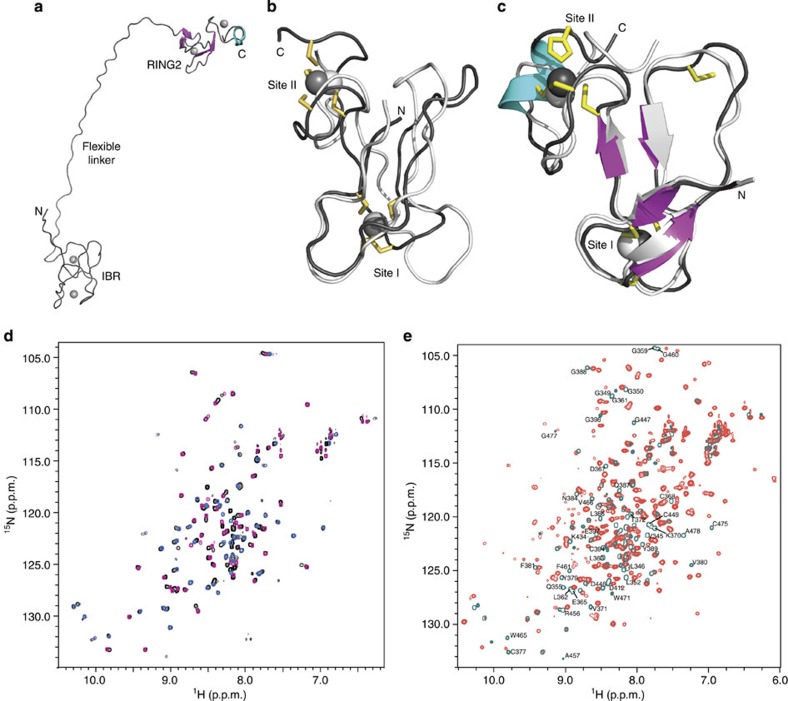
To determine the structural relationship between the IBR domain and the catalytic RING2 domain, we analysed both human and fly IBR–RING2 fragments by NMR spectroscopy and mass spectrometry, and determined the solution structure of fly IBR–RING2. Both human and fly IBR–RING2 structures bind a total of four Zn2+ ions consistent with structures of the isolated IBR and RING2 domains. The IBR–RING2 structure shows that IBR and RING2 domains are each well-folded but separated by a 26-residue flexible linker between the domains (Fig. 4a and Supplementary Table S3). The lack of interaction between the two domains is supported by the absence of interdomain nuclear overhauser effects (NOEs), near identical peak positions when individual domain 1H–15N HSQC spectra are superimposed with that of IBR–RING2 (Fig. 4d), and no observable chemical-shift changes in NMR titrations between RING2 and 15N-labelled IBR domains (Supplementary Fig. S7). The structure also reveals that the IBR and RING2 domains in IBR–RING2 have similar folds to their independent structures (Fig. 4b). Analysis of chemical-shift data indicates that fly and human IBR–RING2 utilize analogous cysteine and histidine residues to coordinate two Zn2+ ions in each domain (Supplementary Table S4). Together, these results indicate that in the context of native parkin the IBR and RING2 domains are not juxtaposed in the structure.
Figure 4. The structure of fly IBR–RING2 shows the domains are remote in parkin but interact with RING0 and RING1.
(a) Representative ribbon structure of parkin IBR–RING2 showing the flexible linker between the IBR and RING2 domains. (b) The superposition of parkin IBR domains from human (white, PBD accession code 2JMO) and fly (black). (c) The superposition of RING2 domains from fly parkin RING2417–482 (white) and fly parkin IBR–RING2342–482 (coloured). The two domains adopt similar folds, although the IBR domains do not contain formal β-strands. (d) Superposition of 1H–15N HSQC spectra for the fly IBR–RING2 (black contours) with spectra for the individual fly parkin IBR342–402 (pink) and RING2417–482 (blue) domains. (e) Superposition of 1H–15N HSQC spectra for parkin RING0–RING1–IBR–RING2 (red contours) and IBR–RING2342–482 (teal contours). The large number of chemical-shift changes indicates that RING0 and RING1 interact with IBR–RING2. Residues in both IBR and RING2 that undergo the largest changes in chemical shift are indicated near their positions from the IBR–RING2 assignment.
To identify how the IBR and RING2 domains might be arranged with respect to the RING0 and RING1 domains in native parkin, we acquired 1H–15N HSQC spectra of fly RING0–RING1–IBR–RING2 (Fig. 4e) and compared the data with spectra of IBR–RING2. Unlike the near-perfect superposition of spectra exhibited for the IBR and RING2 domains with the IBR–RING2 fragment, many chemical-shift changes were observed in each of the IBR (that is, D367, V380, V381 and N384) and RING2 (that is, G447, A457, W465 and C475) domains in RING0–RING1–IBR–RING2. This indicates that the environments of these residues in IBR and RING2 are changed by the presence of the RING0 and RING1 domains. The most logical explanation is that the RING0 and/or RING1 domains are interacting with the IBR–RING2 structure forming a more compact arrangement in the protein.
Parkin tertiary structure is maintained by the Ubl domain
To understand the parkin structure in the context of the native protein, we obtained a molecular envelope of human parkin in solution using small-angle X-ray scattering (SAXS) (Fig. 5a). The analysis of the SAXS data, generated from a monodispersed pure sample, indicates that full-length parkin adopts a globular conformation with a radius of gyration of 29.0 Å (Dmax 95 Å). Removal of the Ubl domain leads to an extended conformation of parkin (Fig. 5b), with a larger radius of gyration (32.0 Å; Dmax 110 Å) than the 8-kDa larger full-length protein. These observations were validated using sedimentation velocity analysis (Fig. 5c) that showed sedimentation coefficients of 4.1 S (parkin) and 3.5 S (ΔUblD–parkin), reflecting the higher mass for parkin. However, the frictional ratio (f/f0) for ΔUblD–parkin (1.53) was larger than that observed for parkin (1.38). This indicates that ΔUblD–parkin has a more extended conformation, and that in the presence of the Ubl domain parkin adopts a more compact conformation. Activating point mutations in the Ubl domain, R42P, I44A and K48A6 were also subjected to SAXS analysis and have radii of gyration that were all larger than native parkin (31.8, 30.3 and 29.8 Å, Dmax 130, 105 and 104 Å, respectively). Despite high-quality data from non-aggregated samples (Fig. 5d–f), ab initio modelling cannot be used to interpret the data due to inherent conformational flexibility. Taken together, these data suggest the Ubl associates with the RBR domains in the full-length protein.
Figure 5. The tertiary structure of parkin is maintained by the Ubl.
(a) Superposition of the scattering data (left) and distance distribution (right) plots from purified human parkin samples, wild-type (red) and ΔUblD–parkin (blue). The plots show the quality of the data and the radius of gyration. (b) Representative and averaged ab initio models of wild-type parkin (red/pink) and ΔUbl–parkin (blue/cyan). Two views for each protein are shown rotated 90o about the x axis. (c) Sedimentation velocity experiments of parkin, ΔUblD–parkin, and IBR–RING2. All data were analysed using the Lamm equation and fit to a c(s) distribution. Sedimentation coefficients, corrected to 20 °C and in H2O, were determined to be 4.1 S for full-length parkin, 3.5 S for ΔUblD–parkin and 2.2 S for IBR–RING2. Fitted frictional ratios (f/f0) were calculated to be 1.38 for full-length parkin, 1.53 for ΔUbl–parkin and 1.31 for IBR–RING2. Sedimentation velocity experiments were performed at 20 °C using 10–16 μM protein in 25 mM Tris-HCl, 50 mM NaCl, 0.5 mM TCEP, pH 8.0. (d) Scattering data (left) and distance distribution (right) plots for R42P–parkin, (e) I44A–parkin, and (f) K48A–parkin.
Parkin is not a substrate of parkin in trans
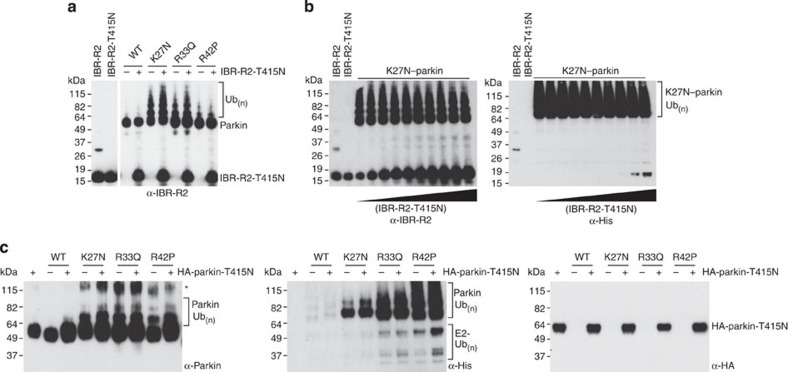
Our structural data indicate that parkin and parkin mutants form monomers in solution, even at the very high concentrations required for SAXS analysis (up to 0.5 mM). As parkin is reported to be a substrate of parkin, these insights led us to ask whether parkin self-ubiquitination occurs in cis or in trans. To address this question, we first probed the targets of mutant human parkin autoubiquitination by mass spectrometry and identified a conserved site at K349, located within the IBR domain (Supplementary Fig. S8). The IBR–RING2 is sufficient for autoubiquitination activity of parkin19,25, suggesting that IBR–RING2 is a plausible candidate substrate of full-length mutant parkin. Furthermore, at 17-kDa, IBR–RING2 is easily distinguishable from full-length parkin (52 kDa), or parkin lacking the Ubl domain (46 kDa). The pathogenic mutation T415N in RING2 results in well-folded but inactive protein (Fig. 3f and Supplementary Fig. S6) and, therefore, we used IBR–RING2–T415N as a model substrate for parkin to distinguish between cis and trans autoubiquitination. In contrast to the mutants themselves, IBR–RING2–T415N is not ubiquitinated by any of the active parkin mutants (Fig. 6a), even in the presence of a large excess of ‘substrate’ (Fig. 6b). To rule out that IBR–RING2–T415N simply does not contain a recognition site for parkin, we expressed and purified full-length haemagglutinin (HA)-tagged T415N–parkin (Fig. 6c). Again, even at high concentrations full-length parkin is not a substrate of active parkin. Taken together, these data show that parkin autoubiquitinates in cis, and not in trans.
Figure 6. Parkin is not a substrate of parkin in trans.
(a) Ubiquitination assay using IBR–RING2–T415N (IBR–R2–T415N) as a substrate. Each parkin species point mutation is assayed in the absence (−) or presence (+) of substrate. The first lane shows wild-type active IBR–RING2, the second lane shows the inactive mutant. Parkin–ubiquitin conjugates are detected by western blotting using a parkin antibody. (b) K27N–parkin is assayed for activity towards IBR-R2-T415N with increasing substrate concentration. Parkin–ubiquitin conjugates are detected by western blotting using parkin (left) and His-ubiquitin (right) antibodies. (c) Ubiquitination assay using full-length HA–parkin–T415N as a substrate. Parkin–ubiquitin conjugates are detected by western blotting using parkin and His-ubiquitin antibodies. Unmodified HA-parkin-T415N is detected using HA antibodies. The asterisk denotes a non-specific band.
Discussion
There are several classes of E3 ligases, including the RING, U-box and HECT E3s. HECT E3 ligases form a catalytic intermediate via a thiolester with the C terminus of ubiquitin before transferring it to a substrate protein28,46. In contrast, the RING and U-box E3 ligases function primarily as scaffolds by orienting the E2~ubiquitin thiolester complex and target protein for ubiquitin transfer47. Recently, the RBR ligases were proposed to function as a hybrid between the HECT and RING ligases that can also form a catalytic thiolester intermediate14,15,27.
In this study we set out to understand the RBR ligase parkin and its domains both in detail, and in the broader context of the whole protein. We determined the structure of parkin RING2 to decipher how this domain might account for the unique RING/HECT hybrid mechanism. We show that parkin RING2 adopts a different fold and Zn2+-coordination topology than its RING namesake that likely exists for all RBR ligases. RING2 lacks key residues required for E2/RING interactions, which, in combination with our isothermal titration calorimetry experiments showing no interaction with either UbcH7 or UbcH8, reinforces that RING2 is not an E2 recruitment domain. Likewise, recent observations that parkin IBR–RING2 is capable of E2-independent ubiquitination19 supports the notion of this RING2 domain having different structural features and activity than canonical RINGs. A key observation from the RING2 structure is the location of a solvent-exposed cysteine that is absolutely conserved in all RBR-containing E3 ligases. This cysteine is perfectly poised to accept an ubiquitin molecule, be it from an E1 or an E2 enzyme, and perform a transthiolation reaction. Clearly, RING2 is the true catalytic domain for the RBR ligases that is not dependent on interacting with an E2 enzyme. Consistent with this, there are numerous ARJP mutations that affect the RING2 domain, not only point mutations but also truncations and deletions that lead to the loss of the RING2 fold16 (http://www.molgen.ua.ac.be/PDmutDB).
Understanding the regulation of E3 ubiquitin ligases has become increasingly important in the ubiquitin field. A prime example is the autoinhibited state of the Cullin-RING ligases that can be alleviated by the conjugation of the ubiquitin-like protein NEDD8 to the extreme C terminus of the Cullin, thus increasing the mobility of the Rbx1/ROC1 RING E3 protein for guiding an E2~ubiquitin complex to successfully transfer its ubiquitin cargo to a substrate48. Interestingly, several RBR ligases have been recently shown to use autoinhibition to regulate their ubiquitination activity6,14,15, a regulatory mechanism also employed by HECT E3s49.
There are many aspects of parkin function that are poorly understood, including the structural basis of parkin regulation by a variety of post-translational modifications, and how different E2s impact on parkin activity10,11,17. However, our findings that an affected copy of parkin does not ubiquitinate an unaffected copy suggest a unifying ‘loss-of-function’ rationale for parkin mutations, whereby mutations in the catalytic domain lead to a catalytically inactive parkin, whereas ‘activating’ mutations in the inhibitory domain lead to ubiquitination and subsequent degradation. Our structural analyses in solution combined with biochemical data suggest that in a heterozygous scenario, only the mutant copy of parkin is affected. If the mutation is inactivating, only the mutant copy loses function, and if the mutation is activating only the mutant copy is destroyed. Wild-type parkin is not preyed upon by mutant parkin and remains functional. Our data also provide a molecular rationale for the existence of non-homozygous ARJP sufferers who have compound heterozygous mutations.
Methods
Protein expression and purification
All domains from D. melanogaster PARK2 were cloned into a modified pGEX-6P-2 vector having an N-terminal GST tag and TEV cleavage site (ENLYFQ). Site-directed mutagenesis was used to create all substitutions in parkin RING2. The full length (residues 1–465), ΔUbl (residues 77–465), IBR–RING2 (residues 321–465) domains and substituted proteins from human PARK2 were cloned, expressed and purified as described previously6. The RING2 domain of HHARI (residues 325–396) was synthesized by DNA 2.0 (Menlo Park, CA, USA) and cloned into the pGEX-TEV vector.
GST-tagged constructs were transformed into Escherichia coli BL21(DE3)-RIL (Stratagene) and grown in Luria broth supplemented with 500 μM ZnCl2 at 37 °C until OD600 reached 0.8. Expression was induced with 1 mM isopropylthiogalactoside at 16 °C for 20 h. For 13C/15N-labelled proteins, cells were grown similarly in M9 media supplemented with 15NH4Cl (1 g l−1), 13C6-glucose (2 g l−1) and 500 μM ZnCl2. Purification used GSTrap FF columns (GE Healthcare) before and following TEV cleavage.
Protein solubility experiments
Isopropylthiogalactoside-induced overnight cultures (10 ml) were collected and resuspended in 20 mM Tris-HCl, 120 mM NaCl, 5 mM dithiothreitol (DTT), pH 7.4 (1 ml), sonicated for 20 s and centrifuged at 16,000g for 5 min. The cleared supernatant was separated and the pellet was resuspended in 2% SDS. The preinduction, postinduction, supernatant and pellet samples were then heated in SDS-loading buffer with 5 mM DTT, run on a 16.5% SDS–PAGE gel and stained with Coomassie Brilliant Blue R-250 dye.
Metal-binding experiments using mass spectrometry
Zinc ion content was assessed by mass spectrometry as previously described36. Samples were dialysed twice against 120 mM ammonium acetate pH 7.4, with a final exchange against 10 mM ammonium acetate to remove sodium adducts. Mass spectrometry was performed in positive-ion mode on a Q-TOF Micro mass spectrometer (Waters) with Z-spray source. Denatured proteins were prepared by mixing the native samples with acetonitrile and formic acid (5%) in a 50:50 ratio. Data showed the following masses; fly parkin RING2 C449S (MWcalc 7643.6 Da, MWobs (native) 7771.99 Da, MWobs (denatured) 7643.47 Da), human HHARI RING2 C357S (MWcalc 8428.2 Da, MWobs (native) 8556.56 Da, MWobs (denatured) 8428.15 Da), human parkin IBR–RING2 (MWcalc 16095.34 Da, MWobs (native) 16346.39 Da, MWobs (denatured) 16093.80 Da), fly parkin IBR–RING2 (MWcalc 15589.6 Da, MWobs (native) 15842.06 Da, MWobs (denatured) 15588.65 Da), human parkin ΔUbl (MWcalc 42834.53 Da, MWobs (native) 43342.07 Da, MWobs (denatured) 42836.07 Da) and fly parkin ΔUbl (MWcalc 42470.8 Da, MWobs (native) 42974.25 Da, MWobs (denatured) 42472.0 Da).
Isothermal titration calorimetry experiments
The interaction of human or D. melanogaster parkin IBR, RING2 or IBR–RING2 with either UbcH7 or UbcH8 was probed by isothermal titration calorimetry using an iTC200 Microcalorimeter (GE Healthcare). Sample cells contained 30–50 μM of the appropriate parkin domain, and 10 × sample cell concentration in the injection syringe of either UbcH7 or UbcH8. All samples were buffered using 50 mM HEPES, 200 mM NaCl and 250 μM TCEP (tris-(2-carboxyethyl)phosphine) at pH 8.0. Injections of 1.5 μl were dispensed with a 5-s addition time and a spacing of 250 s, with a total of 20 injections at 18 °C. The stirrer speed was 1,000 r.p.m.
NMR spectroscopy and structure calculations
NMR data were collected using 15N- or 13C/15N-labelled parkin domains (100–500 μM) and HHARI RING2 (400 μM) at 25 °C using a Varian Inova 600 MHz NMR spectrometer equipped with a triple resonance probe and z-field gradients. Data were processed using NMRPipe50 and were analysed using NMRViewJ51. Structures were calculated using manual and automatic NOE assignments and dihedral angular restraints (ϕ,Ψ) from TALOS+ (ref. 52) incorporated into the programme CYANA53. The standard CYANA protocol was used with default settings, involving eight cycles of structure generation and refinement (100 structures per round). Cysteine residues involved in Zn2+ coordination were identified using Cα and Cβ chemical shifts54. The protonation state of histidine residues was determined from Cδ chemical shifts55. Initial structures were calculated in the absence of any Zn2+-ion restraints so as to not bias the fold of the domain. Following this, zinc atoms were added using virtual linkers. Restraints between the atom pairs Zn–Sγ, Sγ–Cβ, (His Nε2)–Sγ and Sγ–Sγ were imposed to maintain tetrahedral geometry around the zinc ion as previously described32. The final 50 structures were water refined using a modified force field in Xplor-NIH56. The 20 structures with the lowest NOE energies were chosen as representative of the calculation.
Mass spectrometric identity of autoubiquitination sites
Ubiquitination experiments were carried out in 50 mM HEPES pH 8.0, 2 mM DTT and 5 mM MgCl2 containing 5 mM ATP, 200 nM UBE1, 4 μM UbcH7, 16 μM His6-ubiquitin and 2 μM of either R42P, K48A or ΔUblD. Reactions were incubated at 37 °C for 60 min before addition of LDS sample buffer and β-ME. Boiled samples were resolved by gel electrophoresis and gel bands corresponding to ubiquitinated parkin were subjected to further analysis by LC–MS/MS.
In vitro ubiquitination assays
Reactions were carried out in a total volume of 25 μl in 50 mM Tris pH 7.5, 2 mM DTT and 5 mM MgCl2. His mouse UBE1 virus (6 × ) was a kind gift from Kazuhiro Iwai and was purified as a His fusion using Ni-NTA resin (QIAGEN) affinity chromatography followed by size-exclusion liquid chromatography. UbcH7 was purified as previously described6. Ubiquitination components were added as follows: 4 mM ATP, 15 nM UBE1, 1.1 μM UbcH7 and 5 μM His6-ubiquitin. Untagged parkin (0.77–1 μM) was added with or without the stated parkin substrate. IBR–R2–T415N (1 μM) or HA–parkin-T415N (0.77 μM) were added in the respective assays unless otherwise specified. Incubation at 37 °C for 60 min preceded addition of SDS buffer and denaturation by boiling. Samples were resolved by gel electrophoresis and analysed by western blot. Antibodies used for visualization were as follows: anti-parkin (1/5,000, 1A1; IBL), anti-6 × His (1/2,000; GE Healthcare), anti-HA (1/1,000; Mono-HA.11, Covance) and anti-IBR-R2 raised against recombinant IBR-R2 (Pettingill Technology Ltd). Full blots and gel scans are included in Supplementary Fig. S9 where appropriate.
Parkin thiolester in vitro ubiquitination assays
Cy5-labelled His6-ubiquitin was generated as described15. A cysteine residue was inserted between the 6xHis tag and the first methionine by site-directed mutagenesis. Purification was carried out initially on a HiLoad 16/10 Q-Sepharose High Performance column (GE Healthcare) equilibrated in 25 mM Tris, 1 mM EDTA, at pH 8. His6-ubiquitin eluted in the flowthrough was then concentrated and purified further using size-exclusion chromatography. Protein was stored in 50 mM HEPES pH 7.5. Purified protein (1 mg) was incubated at room temperature with one vial of Cy5 Maleimide Mono-Reactive Dye (GE Healthcare) in a 550 μl total reaction volume for 90 min with gentle agitation. Excess dye was removed on a 5 ml HiTrap Desalting column (GE Healthcare) equilibrated in 50 mM HEPES pH 7.5 and 150 mM NaCl. Labelled protein was further purified by size-exclusion chromatography.
Reactions were buffered in 50 mM Tris pH 7.5 and contained 2 mM DTT and 5 mM MgCl2. Cy5-His6-ubiquitin (1 μM), 2 μM UBE1 and 1 mM ATP were mixed and incubated at 37 °C for 10 min before addition of 10 μM UbcH7, and a further 10 min incubation at 37 °C. 20 μM His-SUMO-IBR-R2, His-SUMO-IBR-R2-C431S or His-SUMO-IBR-R2-C431A-parkin was then added. The native parkin was incubated for only 15 s, C431S was incubated at 37 °C for 1 h, and a sample incubated with 0.15 M NaOH. Reactions were stopped using NuPAGE LDS Sample Buffer (Invitrogen) and were reduced by the addition of TCEP and boiling where indicated. Gels were visualized using an ImageQuant LAS 4000 imaging system.
Analytical ultracentrifugation
Sedimentation velocity experiments were performed using a Beckman Coulter XL-A analytical ultracentrifuge equipped with an An-60 Ti rotor. Samples (400 μl) were prepared in 25 mM Tris–HCl, 50 mM NaCl and 0.5 mM TCEP pH 8.0, and centrifuged in a double sector cell (1.2 cm) equipped with sapphire windows at 45,000 r.p.m. at 20 °C. Samples were scanned at equal intervals (60 scans total) and the absorbance at 280 nm measured with three replicates averaged per scan. Data were analysed using the programme Sedfit using non-linear regression to determine the frictional ratio (f/f0) for each species as well as the sedimentation coefficient corrected to 20 °C. All data were fit to an r.m.s.d. equal or less than 0.008.
SAXS data collection and processing
SAXS data were collected at Synchrotron Soleil; beamline SWING. Samples ranging in concentration from 5 to 25 mg ml−1 were directly passed into the beam from HPLC gel filtration using an Agilent BioSec-3 300 column. Data were collected from the peak(s) comprising the protein of interest, avoiding any aggregates with data for the buffer blank collected before the elution of protein from the column.
Initial data processing was undertaken automatically using the FOXTROT software at the beamline. Further processing, using the ATSAS suite (version 2.5.0— http://www.embl-hamburg.de/biosaxs/software.html), was undertaken using PRIMUS57 (for initial Rg and Kratky plot analysis), GNOM58 (generation of distance distribution plots) and DAMMIF59 (generation of ab initio models). Models were converted to Situs format using PDB2VOL (part of the SITUS suite, http://situs.biomachina.org/). Figures were produced using CHIMERA60 followed by superimposition of envelopes.
Author contributions
D.E.S. completed NMR experiments with parkin and HHARI RING2, analysed the structures, prepared and characterized all mutant protein and wrote the manuscript. R.J.M. expressed and purified human parkin species and performed isothermal titration calorimetry experiments. SAXS data collection and analysis was carried out by R.J.M. and A.P. Y.J.N. cloned and expressed fly IBR–RING2, collected and analysed NMR data. P.M. calculated the structures of RING2 and IBR–RING2 and analysed the structures. N.M. cloned and expressed fly parkin RING2, collected NMR data and assigned NMR spectra. L.B. performed ubiquitination and thiolester formation assays. J.D.A. performed sedimentation velocity and NMR experiments for several parkin proteins. K.R.B. expressed human IBR–RING2, collected and analysed NMR data. H.W. designed experiments and wrote the manuscript. G.S.S. designed, analysed experiments and wrote the manuscript.
Additional information
Accession codes: Atomic coordinates and structural restraints have been deposited in the RCSB Protein Data Bank under PDB accession codes 2LWR and 2M48. NMR assignments have been deposited to the Biological Magnetic Resonance Data bank under accession codes 18642 and 18990.
How to cite this article: Spratt, D. E. et al. A molecular explanation for the recessive nature of parkin-linked Parkinson’s disease. Nat. Commun. 4:1983 doi: 10.1038/ncomms2983 (2013).
Supplementary Material
Supplementary Figures S1-S9 and Supplementary Tables S1-S4
Acknowledgments
We thank Paula Pittock (Biological Mass Spectrometry Laboratory, University of Western Ontario) for mass spectroscopy data collection and analysis, and Lee-Ann Briere and Sue Safadi (Biological Interactions and Conformations Facility, University of Western Ontario) for help with analytical ultracentrifugation data collection and analysis. We thank Viduth Chaugule and Mark Skehel for mass spectrometry (London Research Institute). We are grateful to Pierre Roblin and Javier Perez for help and advice with the SAXS data collection, which received funding from the European Community’s Seventh Framework Programme (FP7/2007-2013) under BioStruct-X (grant agreement number 283570). This work was supported by a grant from the Canadian Institutes of Health Research (MOP-14606), the Canada Research Chairs Program (G.S.S.) and fellowships (D.E.S.) from the Canadian Institutes of Health Research and a Natural Sciences and Engineering Research Council of Canada. This work was also supported by Cancer Research UK, and an EMBO Young Investigator award (H.W.). The work was presented at the International Conference on NMR in Biological Systems, August 19–24, 2012, Lyon, France.
References
- Kitada T. et al. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature 392, 605–608 (1998). [DOI] [PubMed] [Google Scholar]
- Lucas J. I., Arnau V. & Marin I. Comparative genomics and protein domain graph analyses link ubiquitination and RNA metabolism. J. Mol. Biol. 357, 9–17 (2006). [DOI] [PubMed] [Google Scholar]
- Marin I., Lucas J. I., Gradilla A. C. & Ferrus A. Parkin and relatives: the RBR family of ubiquitin ligases. Physiol. Genomics 17, 253–263 (2004). [DOI] [PubMed] [Google Scholar]
- Narendra D., Kane L. A., Hauser D. N., Fearnley I. M. & Youle R. J. p62/SQSTM1 is required for Parkin-induced mitochondrial clustering but not mitophagy; VDAC1 is dispensable for both. Autophagy 6, 1090–1106 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziviani E., Tao R. N. & Whitworth A. J. Drosophila parkin requires PINK1 for mitochondrial translocation and ubiquitinates mitofusin. Proc. Natl Acad. Sci. USA 107, 5018–5023 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaugule V. K. et al. Autoregulation of Parkin activity through its ubiquitin-like domain. EMBO J. 30, 2853–2867 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuytemans K., Theuns J., Cruts M. & Van Broeckhoven C. Genetic etiology of Parkinson disease associated with mutations in the SNCA, PARK2, PINK1, PARK7, and LRRK2 genes: a mutation update. Hum. Mutat. 31, 763–780 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S., Zhang Z., Kavitha G., Tan E. K. & Ng S. K. MDPD: an integrated genetic information resource for Parkinson’s disease. Nucleic. Acids Res. 37, D858–D862 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai Y., Soda M. & Takahashi R. Parkin suppresses unfolded protein stress-induced cell death through its E3 ubiquitin-protein ligase activity. J. Biol. Chem. 275, 35661–35664 (2000). [DOI] [PubMed] [Google Scholar]
- Shimura H. et al. Familial Parkinson disease gene product, parkin, is a ubiquitin-protein ligase. Nat. Genet. 25, 302–305 (2000). [DOI] [PubMed] [Google Scholar]
- Zhang Y. et al. Parkin functions as an E2-dependent ubiquitin- protein ligase and promotes the degradation of the synaptic vesicle-associated protein, CDCrel-1. Proc. Natl Acad. Sci. USA 97, 13354–13359 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doss-Pepe E. W., Chen L. & Madura K. Alpha-synuclein and parkin contribute to the assembly of ubiquitin lysine 63-linked multiubiquitin chains. J. Biol. Chem. 280, 16619–16624 (2005). [DOI] [PubMed] [Google Scholar]
- Moynihan T. P. et al. The ubiquitin-conjugating enzymes UbcH7 and UbcH8 interact with RING finger/IBR motif-containing domains of HHARI and H7-AP1. J. Biol. Chem. 274, 30963–30968 (1999). [DOI] [PubMed] [Google Scholar]
- Smit J. J. et al. The E3 ligase HOIP specifies linear ubiquitin chain assembly through its RING-IBR-RING domain and the unique LDD extension. EMBO J. 31, 3833–3844 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stieglitz B., Morris-Davies A. C., Koliopoulos M. G., Christodoulou E. & Rittinger K. LUBAC synthesizes linear ubiquitin chains via a thioester intermediate. EMBO Rep. 13, 840–846 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardley H. C., Tan N. G., Rose S. A., Markham A. F. & Robinson P. A. Features of the parkin/ariadne-like ubiquitin ligase, HHARI, that regulate its interaction with the ubiquitin-conjugating enzyme, Ubch7. J. Biol. Chem. 276, 19640–19647 (2001). [DOI] [PubMed] [Google Scholar]
- Walden H. & Martinez-Torres R. J. Regulation of Parkin E3 ubiquitin ligase activity. Cell. Mol. Life Sci. 69, 3053–3067 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sriram S. R. et al. Familial-associated mutations differentially disrupt the solubility, localization, binding and ubiquitination properties of parkin. Hum. Mol. Genet. 14, 2571–2586 (2005). [DOI] [PubMed] [Google Scholar]
- Chew K. C. et al. Parkin mediates apparent E2-independent monoubiquitination in vitro and contains an intrinsic activity that catalyzes polyubiquitination. PLoS ONE 6, e19720 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darios F. et al. Parkin prevents mitochondrial swelling and cytochrome c release in mitochondria-dependent cell death. Hum. Mol. Genet. 12, 517–526 (2003). [DOI] [PubMed] [Google Scholar]
- Fallon L. et al. A regulated interaction with the UIM protein Eps15 implicates parkin in EGF receptor trafficking and PI(3)K-Akt signalling. Nat. Cell Biol. 8, 834–842 (2006). [DOI] [PubMed] [Google Scholar]
- Fournier M. et al. Parkin deficiency delays motor decline and disease manifestation in a mouse model of synucleinopathy. PLoS ONE 4, e6629 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W. J. et al. The C289G and C418R missense mutations cause rapid sequestration of human Parkin into insoluble aggregates. Neurobiol. Dis. 14, 357–364 (2003). [DOI] [PubMed] [Google Scholar]
- Hampe C., Ardila-Osorio H., Fournier M., Brice A. & Corti O. Biochemical analysis of Parkinson’s disease-causing variants of Parkin, an E3 ubiquitin-protein ligase with monoubiquitylation capacity. Hum. Mol. Genet. 15, 2059–2075 (2006). [DOI] [PubMed] [Google Scholar]
- Matsuda N., Kitami T., Suzuki T., Mizuno Y., Hattori N. & Tanaka K. Diverse effects of pathogenic mutations of Parkin that catalyze multiple monoubiquitylation in vitro. J. Biol. Chem. 281, 3204–3209 (2006). [DOI] [PubMed] [Google Scholar]
- Tsai Y. C., Fishman P. S., Thakor N. V. & Oyler G. A. Parkin facilitates the elimination of expanded polyglutamine proteins and leads to preservation of proteasome function. J. Biol. Chem. 278, 22044–22055 (2003). [DOI] [PubMed] [Google Scholar]
- Wenzel D. M., Lissounov A., Brzovic P. S. & Klevit R. E. UBCH7 reactivity profile reveals parkin and HHARI to be RING/HECT hybrids. Nature 474, 105–108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L. et al. Structure of an E6AP-UbcH7 complex: insights into ubiquitination by the E2-E3 enzyme cascade. Science 286, 1321–1326 (1999). [DOI] [PubMed] [Google Scholar]
- Sakata E. et al. Parkin binds the Rpn10 subunit of 26S proteasomes through its ubiquitin-like domain. EMBO Rep. 4, 301–306 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tashiro M. et al. NMR structure of ubiquitin-like domain in PARKIN: gene product of familial Parkinson’s disease. J. Biomol. NMR 25, 153–156 (2003). [DOI] [PubMed] [Google Scholar]
- Tomoo K. et al. Crystal structure and molecular dynamics simulation of ubiquitin-like domain of murine parkin. Biochim. Biophys. Acta 1784, 1059–1067 (2008). [DOI] [PubMed] [Google Scholar]
- Beasley S. A., Hristova V. A. & Shaw G. S. Structure of the Parkin in-between-ring domain provides insights for E3-ligase dysfunction in autosomal recessive Parkinson’s disease. Proc. Natl Acad. Sci. USA 104, 3095–3100 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beasley S. A., Safadi S. S., Barber K. R. & Shaw G. S. Solution structure of the E3 ligase HOIL-1 Ubl domain. Protein Sci. 21, 1085–1092 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi H. et al. A non-canonical UBA-UBL interaction forms the linear-ubiquitin-chain assembly complex. EMBO Rep. 13, 462–468 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capili A. D., Edghill E. L., Wu K. & Borden K. L. Structure of the C-terminal RING finger from a RING-IBR-RING/TRIAD motif reveals a novel zinc-binding domain distinct from a RING. J. Mol. Biol. 340, 1117–1129 (2004). [DOI] [PubMed] [Google Scholar]
- Hristova V. A., Beasley S. A., Rylett R. J. & Shaw G. S. Identification of a novel Zn2+-binding domain in the autosomal recessive juvenile Parkinson-related E3 ligase parkin. J. Biol. Chem. 284, 14978–14986 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J. et al. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature 441, 1157–1161 (2006). [DOI] [PubMed] [Google Scholar]
- Greene J. C., Whitworth A. J., Andrews L. A., Parker T. J. & Pallanck L. J. Genetic and genomic studies of Drosophila parkin mutants implicate oxidative stress and innate immune responses in pathogenesis. Hum. Mol. Genet. 14, 799–811 (2005). [DOI] [PubMed] [Google Scholar]
- Borden K. L. RING fingers and B-boxes: zinc-binding protein-protein interaction domains. Biochem. Cell. Biol. 76, 351–358 (1998). [DOI] [PubMed] [Google Scholar]
- Budhidarmo R., Nakatani Y. & Day C. L. RINGs hold the key to ubiquitin transfer. Trends Biochem. Sci. 37, 58–65 (2012). [DOI] [PubMed] [Google Scholar]
- Yin Q. et al. E2 interaction and dimerization in the crystal structure of TRAF6. Nat. Struct. Mol. Biol. 16, 658–666 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mace P. D. et al. Structures of the cIAP2 RING domain reveal conformational changes associated with ubiquitin-conjugating enzyme (E2) recruitment. J. Biol. Chem. 283, 31633–31640 (2008). [DOI] [PubMed] [Google Scholar]
- Zheng N., Wang P., Jeffrey P. D. & Pavletich N. P. Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell 102, 533–539 (2000). [DOI] [PubMed] [Google Scholar]
- Spratt D. E., Wu K., Kovacev J., Pan Z. Q. & Shaw G. S. Selective recruitment of an E2~ubiquitin complex by an E3 ubiquitin ligase. J. Biol. Chem. 287, 17374–17385 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. Y., Nagano Y., Taylor J. P., Lim K. L. & Yao T. P. Disease-causing mutations in parkin impair mitochondrial ubiquitination, aggregation, and HDAC6-dependent mitophagy. J. Cell Biol. 189, 671–679 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee Y. & Huibregtse J. M. Regulation of catalytic activities of HECT ubiquitin ligases. Biochem. Biophys. Res. Commun. 354, 329–333 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies R. J. & Joazeiro C. A. RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 78, 399–434 (2009). [DOI] [PubMed] [Google Scholar]
- Yamoah K., Oashi T., Sarikas A., Gazdoiu S., Osman R. & Pan Z. Q. Autoinhibitory regulation of SCF-mediated ubiquitination by human cullin 1's C-terminal tail. Proc. Natl Acad. Sci. USA 105, 12230–12235 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesner S. et al. Autoinhibition of the HECT-type ubiquitin ligase Smurf2 through its C2 domain. Cell 130, 651–662 (2007). [DOI] [PubMed] [Google Scholar]
- Deleglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J. & Bax A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 (1995). [DOI] [PubMed] [Google Scholar]
- Johnson B. A. & Belvins R. A. NMRView: a computer program for the visualization and analysis of NMR data. J. Biomol. NMR 4, 603–614 (1994). [DOI] [PubMed] [Google Scholar]
- Shen Y., Delaglio F., Cornilescu G. & Bax A. TALOS+: A hybrid method for predicting backbone torsion angles from NMR chemical shifts. J. Biomol. NMR 44, 213–223 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guntert P., Mumenthaler C. & Wuthrich K. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol. 273, 283–298 (1997). [DOI] [PubMed] [Google Scholar]
- Kornhaber G. J., Snyder D., Moseley H. N. & Montelione G. T. Identification of zinc-ligated cysteine residues based on 13Calpha and 13Cbeta chemical shift data. J. Biomol. NMR 34, 259–269 (2006). [DOI] [PubMed] [Google Scholar]
- Sudmeier J. L. et al. Identification of histidine tautomers in proteins by 2D 1H/13C(delta2) one-bond correlated NMR. J. Am. Chem. Soc. 125, 8430–8431 (2003). [DOI] [PubMed] [Google Scholar]
- Schwieters C. D., Kuszewski J. J. & Clore G. M. Using Xplor-NIH for NMR molecular structure determination. Progr. NMR Spectroscopy 48, 47–62 (2006). [Google Scholar]
- Konarev P. V., Volkov V. V., Sokolova A. V., Koch M. H. J. & Svergun D. I. PRIMUS—a Windows-PC based system for small-angle scattering data analysis. J. Appl. Crystallogr. 36, 1277–1282 (2003). [Google Scholar]
- Svergun D. I. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 25, 495–503 (1992). [Google Scholar]
- Franke D. & DAMMIF Svergun D. I. a program for rapid ab-initio shape determination in small-angle scattering. J. Appl. Crystallogr. 42, 342–346 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E. F. et al. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures S1-S9 and Supplementary Tables S1-S4