Abstract
Eukaryotic nucleus is functionally as well as spatially compartmentalized and maintains dynamic organization of sub-nuclear bodies. This organization is supported by a non-chromatin nuclear structure called the nuclear matrix. Although the precise molecular composition and ultra-structure of the nuclear matrix is not known, proteins and RNA molecules are its major components and several nuclear matrix proteins have been identified. However, the nature of its RNA component is unknown. Here we show that in Drosophila melanogaster, transcripts from AAGAG repeats of several hundred nucleotide in length are critical constituents of the nuclear matrix. While both the strands of this repeat are transcribed and are nuclear matrix associated, the polypurine strand is predominantly detected in situ. We also show that AAGAG RNA is essential for viability. Our results reveal the molecular identity of a critical RNA component of the nuclear architecture and point to one of the utilities of the repetitive part of the genome that has accumulated in higher eukaryotes.
Keywords: nuclear matrix, nuclear architecture, non-coding RNA, AAGAG repeat, Drosophila
Introduction
Nucleus, the most prominent organelle in eukaryotic cells, houses the genetic material and is separated from the cytoplasm by a double membrane layer, the nuclear envelope. The inner nuclear membrane is supported by the nuclear lamina, which is made of intermediate filament family of proteins, the lamins. Within the nucleus, there is considerable spatial organization, with chromosomes occupying non-random positions that are often cell type-specific.1 A prominent example of chromosome organization observed in nearly all eukaryotic cells is the nucleolus where the ribosomal genes and multiple tRNA genes associate together to generate a hub for rDNA transcription and ribosome assembly. Similarly, many loci converge to form heterochromatin foci in many organisms, including S. cervisiae, S. pombe and Drosophila.2,3 In addition, most nuclear processes, including replication, repair, transcription and splicing show spatial sequestration within the nucleus. This organization is clearly important for genome maintenance and control of gene expression during growth and development.
While it is clear that nucleus is both structurally and functionally compartmentalized, it is not at all clear what the structural basis of this compartmentalization is.4-8 Biochemical analysis earlier showed existence of nuclear skeleton or nuclear matrix (NuMat) but its direct involvement in organizing sub-nuclear compartments has not been demonstrated. It is possible that the nuclear compartmentalization is established and maintained through the attachment of these components with the NuMat. The underlying NuMat structure is made of a fibrillar network that runs across the nucleus and consists of protein, RNA and some DNA. The DNA that remains associated with the matrix after nuclease treatment is called the matrix attachment region (MAR) or scaffold attachment region (SAR). NuMat is thought to serve as a base for the attachment of chromatin loops via the MAR sequences and, thereby, help in the assembly of many of the nuclear compartments.7,9,10 Recent studies have shown that origin of replication sequences are enriched in MARs.11 Significance of nuclear organization in multiple process is also reflected by the finding that disruption in this structure is often correlated with disease states such as the loss of subnuclear promyelocytic leukemia bodies in acute promyelocytic leukemia, loss of lamina in Hutchinson-Progeria and altered profile of nuclear matrix (NuMat) proteins in many cancers.12-14
Early studies on the ultrastructure of the eukaryotic nucleus have shown that RNA and protein components are critical for the integrity of this structure.15,16 While the structural and functional compartmentalization of the nucleus is well known,4-8 much of the molecular composition of the structural framework, which are likely to be the components of the NuMat, is unknown.17-19 Proteins like nuclear lamins are one of the prominent protein constituents of the internal fibrils that form the NuMat and provide strength to the nucleus. Nuclei assembled in the absence of lamins are fragile.20-22 In addition, several more protein components of the NuMat have been identified.23-27
NuMat is sensitive to RNase, indicating that RNA is its important component;28-30 however, the molecular identity of RNA component(s) is not known. To understand the nature and function of this RNA, we have isolated and characterized the nuclear matrix_RNA (NuMat_RNA) from Drosophila melanogaster. We report here that an essential component of NuMat_RNA in fly is transcribed from the pentameric micro satellite repeat AAGAG. We also demonstrate that this repetitive NuMat_RNA is essential for normal development and is a critical component of nuclear architecture.
Results
Transcripts from AAGAG repeat are component of NuMat
NuMat was prepared from 0–16 h Drosophila embryos and Schneider cell-line (S2 cells) as described earlier31 and used for isolation of RNA. We found that 30% of nuclear RNA remains associated with NuMat. The NuMat_RNA (120–550 bp) was used for making a cDNA library, and several clones from the library were sequenced. We found many clones corresponding to single-copy mRNA sequences, rRNA sequences and most intriguingly non-coding repeat sequences. The presence of rRNA and nascent mRNA in NuMat was on expected lines as splicing machinery and nucleolar components are retained in NuMat preparations.15,32,33 Interestingly, RNA corresponding to pentameric satellite repeat, AAGAG, was identified as one of the components of NuMat_RNA. Of ~450 clones sequenced, almost ~70% had AAGAG repeats, suggesting that AAGAG repeats were abundant in the nuclear matrix. In order to explore the significance of long non-coding repeat RNA in nuclear architecture, we performed a detailed study on AAGAG repeat RNA in NuMat.
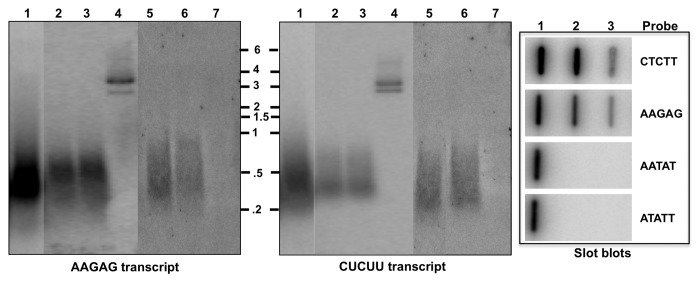
AAGAG repeats are present at the pericentromeric region of all the chromosomes in Drosophila melanogaster.34-36 Since the fly genome does not have long AAGAG repeats elsewhere, it is likely that NuMat_RNA components are transcribed from this locus. To determine the abundance and size range of the transcripts, nuclear RNA and NuMat_RNA from embryos were analyzed by northern hybridization with strand-specific probe. The transcripts ranged in size from 0.5–3 Kb and smaller transcripts were enriched in the nucleus as well as NuMat (Fig. 1). Strand-specific northern blots showed that AAGAG satellite repeats were transcribed from both strands and transcripts corresponding to AAGAG as well as CTCTT strand were retained in the NuMat. We then investigated whether transcription and NuMat association of satellite repeats is common to other satellite sequences or specific to AAGAG. For this, the transcription and distribution of RNA corresponding to the second most abundant micro satellite repeat in D. melanogaster, AATAT, was tested in the nuclear/NuMat preparations. Slot-blot northern hybridization shows that transcription and association of AAGAG/CUCUU transcripts is unique and another major satellite repeat sequence in not transcribed. To further confirm that these molecules are RNA (and not contaminating DNA), we repeated the northern after treating the NuMat_RNA samples with RNase-free DNase I. The northern blot (lane 3) shows that the signal is resistant to DNase I treatment. To reveal the molecular nature of these RNAs, systematic treatment with RNases H and RNase I (not RNase A, known not to digest purine rich RNA preferentially) was performed (Lanes 6, 7). The AAGAG/CUCUU RNAs were completely digested by RNase I (single strand-specific ribonuclease), indicating that these RNA molecules were predominantly single stranded. We also detected insensitivity of both the transcripts to RNase H (ribonuclease that digests RNA when RNA-DNA hybrid is the substrate).
Figure 1. AAGAG/CUCUU repetitive RNAs are components of NuMat. AAGAG and CUCUU transcripts in nuclear and NuMat_RNA were compared for size and abundance. Plasmid DNA with AAGAG insert and 1 kb RNA ladder were loaded as size marker. Northern hybridization with strand-specific probes revealed signal pertaining to either of the strands as specified at bottom of the panels. Lane 1, Nuclear RNA; 2, NuMat_RNA; 3, NuMat_RNA + DNaseI; 4, Plasmid DNA with AAGAG insert; 5, NuMat_RNA; 6, NuMat + RNaseH; 7, NuMat_RNA + RNaseI. Slot blot hybridization shows that no transcripts corresponding to AATAT satellite repeat were present in nuclear or NuMat_RNA. Slot 1, Genomic DNA; 2, Emb nuclear RNA; 3, Emb NuMat_RNA.
Sub-cellular localization of AAGAG and CUCUU RNA
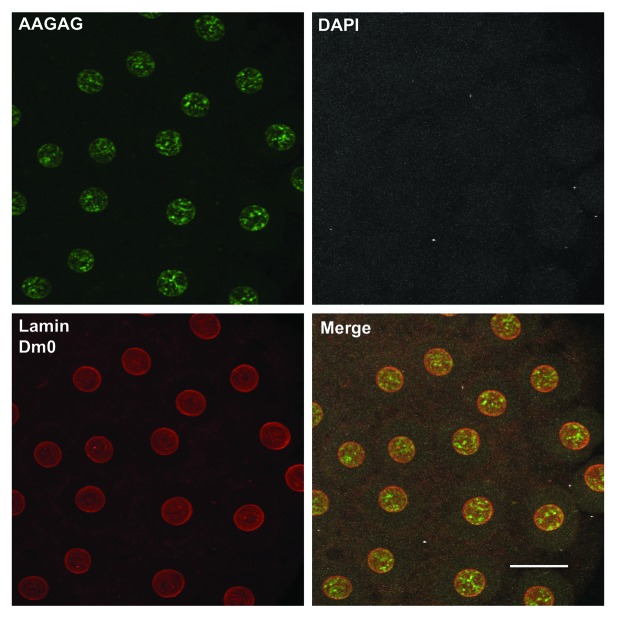
We determined the distribution of the AAGAG and CUCUU transcripts by RNA-FISH in early Drosophila embryos using fluorescently labeled strand-specific probes (Fig. 2) that under non-denaturing conditions detects only single-stranded RNA molecules. In situ NuMat was prepared using early syncytial embryos where nuclei had not yet cellularized. Using RNA-FISH, we found AAGAG transcript predominantly in the NuMat of interphase nuclei. Under the non-denaturing conditions used for FISH, we could not see any CUCUU transcript in NuMat although our northern blot hybridization had indicated that both the transcripts associate with NuMat. To look for possible reasons, we scanned many more embryos and found that both the transcripts associate with dividing chromosomal scaffold (Fig. S1). Thus, AAGAG/CUCUU transcripts appear to be have differential association with interphase NuMat/mitotic chromosome scaffold. We swapped the fluorescent labels on the probe to rule out any artifact that may result due to artificial sticking of probe to NuMat (Biotin-AAGAG/DIG-CTCTT and DIG-AAGAG/Biotin-CTCTT). The results were the same with both the combinations used (data not shown). Further to confirm that the nuclear sub-structure AAGAG transcripts associate with is NuMat indeed, we performed an immunoFISH where NuMat was demarcated by immunostaining of lamin Dm0 (its most prominent protein component). The immunoFISH shows that the transcripts co-localize with lamin Dm0 and, hence, are confirmed as a component of NuMat. To find out whether these transcripts are a part of NuMat of other tissues, we performed RNA_FISH on S2 cells and salivary gland nuclei. We find that both the transcripts are a component of NuMat in these nuclei (Fig. S2).
Figure 2. Distribution of AAGAG transcripts in vivo. RNA-FISH shows that AAGAG transcripts were readily detected in NuMat of early embryo. FITC (green), AAGAG RNA; Cy3 (red), Lamin Dm0. DNA was stained with DAPI (gray). NuMat preparation was judged by the absence of DAPI signal that indicates complete digestion and removal of chromatin. Scale Bar, 10 μm.
These results confirm that both the strands of AAGAG satellite are transcribed giving rise to a novel class of long non-coding RNA (lncRNA). Both of the transcripts associate with NuMat, but probably exist as single strands. Of the two transcripts, the poly-purine AAGAG RNA is more abundant as seen in the NuMat of interphase nuclei by non-denaturing in situ hybridizations. However, transcripts corresponding to both the strands are associated with the mitotic chromosome scaffold. They are also present in the NuMat of differentiated tissue like salivary gland and in the NuMat of S2 cells.
AAGAG NuMat_RNA is essential
To investigate the functional relevance of the AAGAG and CUCUU repeat RNAs, we generated RNAi lines using the UAS-Gal4 system in flies.37 Induction of RNAi by ubiquitous drivers like tubulin-Gal4 and actin-Gal4, a significant reduction in the levels of AAGAG/CUCUU transcripts was seen that leads to complete late larval/early pupal lethality. While wild-type larvae secrete pupal coat, transform to mature pupae and eventually hatch, the knockdown larvae initiate pupation but fail to progress to transformation for several days, which eventually leads to death and degradation (Fig. 3). These results indicate that while maternal contribution and residual levels of this NuMat_RNA can take development up to third instar larval stage, possibly due to massive reprogramming and reorganization that is needed during pupation, the NuMat_RNA components become more critical and a reduction in these, as seen by northern analysis is not viable. We also noticed abnormal growth at the late larval stages that results in bigger early pupae, it remains, however, to be investigated how reduction in NuMat_RNA affects growth regulation. Taken together, these results establish that the transcripts from the AAGAG satellite sequences are essential and play a critical role in regulation of development in Drosophila.
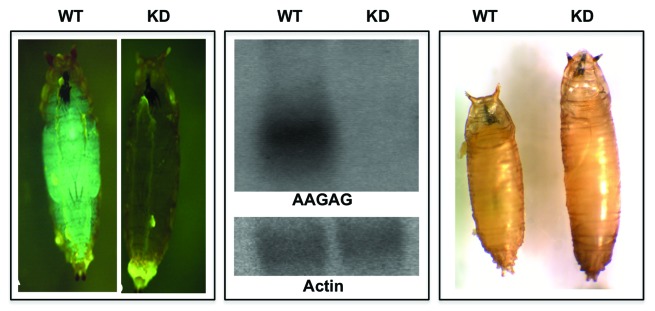
Figure 3. Knockdown of AAGAG/CUCUU transcripts is lethal. Transgenic line (pSympAAGAG/CyOGFP) was crossed with actinGal4/CyOGFP. GFP positive (CyOGFP/actin-Gal4 or CyOGFP/pSympAAGAG) embryo and larvae were wild-type (WT) and GFP negative (pSympAAGAG/actin-Gal4) were knockdown (KD). Northern analysis with RNA from WT and KD larvae shows a clear knockdown of AAGAG/CUCUU transcript levels in the RNAi lines when compared with the WT. Loading control is shown by actin hybridization. WT larvae pupate and shrink in length to become mature pupae and hatch to produce flies. KD larvae begin to pupate but stop at early pupal stage, stay at this stage for several days and eventually die.
NuMat_RNA maintains sub-nuclear compartments
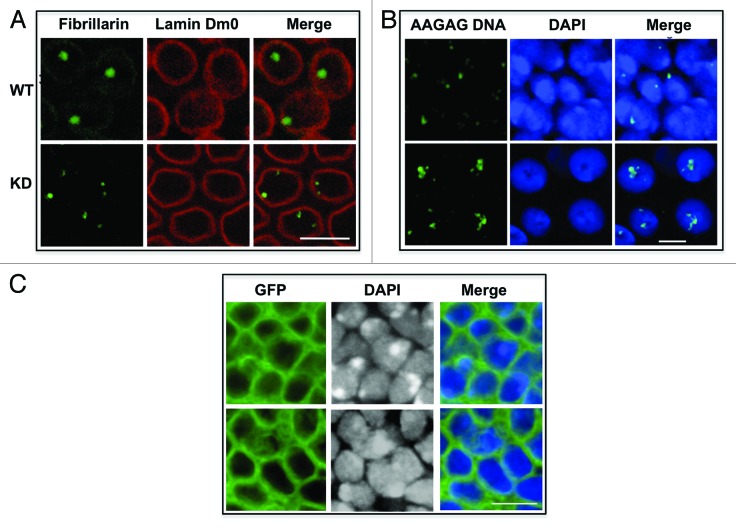
Several studies have demonstrated the importance of the RNA in maintaining nuclear architecture.38-40 We hypothesized that if AAGAG satellite transcripts are a critical component of NuMat_RNA, their knockdown might disturb the organization of the nuclear compartments. We investigated this possibility by staining for heterochromatin components and nucleolus in wild-type and knockdown nuclei. Immunostaining with fibrillarin, an abundant nucleolar protein, showed a prominent single nucleolus in the wild-type nuclei whereas in the knockdown embryos the nucleolus showed fragmentation in all nuclei (two to eight spots, with 89% of nuclei showing more than one spot) (Fig. 4A). The nuclear lamina retained its integrity in both the sets of embryos. These data show that reduction in the levels of AAGAG/CUCUU transcripts leads to perturbation of nucleolar organization.
Figure 4. AAGAG/CUCUU RNA knockdown lead to nuclear defects. (A) AAGAG/CUCUU KD embryos show multiple nucleoli. WT and KD embryos were immunostained for fibrillarin and lamin Dm0. WT nuclei show single nucleolus, whereas nuclei in KD embryos show multiple nucleoli. No change was detected in lamin Dm0 staining. Scale bar, 10 μm. (B) Localization of satellite DNA is disturbed in nuclei of wing imaginal discs of AAGAG-knockdown larvae. DNA-FISH with WT and KD nuclei was done using DIG-labeled AAGAG probe. DNA was visualized by DAPI. In WT nuclei, satellite DNA was seen as one or two condensed spots close to the nuclear periphery, while in KD it moves toward interior of the nuclei and was split into many foci. Scale bar, 5 μm. (C) AAGAG-knockdown nuclei show defective chromatin packaging. In this experiment, RNAi was induced in localized areas of wing imaginal disc using omb-Gal4 driver. GFP positive cells mark the area where RNAi is executed. KD wing imaginal discs show disturbed nuclear DAPI staining where heterochromatin was spread out and diffused. WT discs showed condensed heterochromatic spots in equivalent positions. Scale bar, 5 μm.
Organization of heterochromatin was the next nuclear feature that was investigated by DNA-FISH of pericentromeric AAGAG satellite. In normal wing imaginal disc cells, AAGAG satellite DNA stains as a single spot, depicting the convergence of pericentromeric satellite sequences from multiple chromosomes at the chromocenter, whereas in the knockdown nuclei a diffused staining was observed along with more number of AAGAG satellite foci (Fig. 4B). While about 24% of wild-type nuclei appeared to have split/multiple AAGAG signal, over 90% of the knockdown nuclei had more than one AAGAG satellite signal per nuclei. This dispersed staining could be due to improper heterochromatin condensation in the knockdown tissues. Alternatively, greater number of foci in knockdown tissues may reflect failure of regions from different chromosomes containing these repeats34,35 to converge due to weakened nuclear architecture. Similar results after RNase treatment were reported in mouse cells immunostained for HP1.41 These results point to the importance of RNA component in the maintenance of heterochromatic organization.
As ubiquitous knockdown of AAGAG RNA is lethal, we reasoned that a restricted knockdown could be more informative. Tissue-specific drivers like tsh-Gal4, omb-Gal4 and neuralized-Gal4 were used to knockdown AAGAG transcripts in specific sets of cells. Surprisingly, we found that RNAi with these drivers also caused larval/pupal lethality. However, as we could obtain knockdown larvae, we observed a section of wing imaginal disc where the omb-Gal4 driver is active (Fig. 4C). The wild-type nuclei showed condensed regions of heterochromatic DNA, stained deeply with DAPI, while rest of the nuclear space showed light and diffused euchromatic staining. The knockdown imaginal discs at the equivalent positions showed larger regions of intense DAPI staining with no prominent condensed heterochromatin. These data are consistent with the earlier conclusion that nuclear organization requires normal levels of AAGAG transcripts and knockdown triggers the nuclear organization defects. Taken together, our data suggest that AAGAG transcripts are needed for the integrity of sub-nuclear compartments and facilitate long range interactions needed for formation of chromocenter or nucleolus.
Discussion
Several studies have implicated lncRNAs in essential cellular functions that are modulated in a cell- and developmental stage-specific manner.42-48 Similarly, lncRNA are shown to be involved in organization of heterochromatin,38,41 paraspeckles,39,49 nuclear stress bodies and multiple chromatin features.50-53 In several other instances, lncRNAs act as guides for chromatin-modifying machinery and mediate the formation of a defined active/inactive chromatin compartment. Recently, GAA-repeat containing RNA was found in the nuclear matrix of mammalian cells.54 Although functional significance of this RNA was not investigated, it was shown to interact with multiple nuclear matrix proteins and associate with the midbody during cell division. NuMat has been shown earlier to be dependent on RNA component but molecular nature of these RNAs has not been reported.
Earlier studies have identified lncRNA transcribed from repeat regions38,55-57 that function in cis close to their site of transcription. In this work, we show that a novel class of repeat-containing lncRNA from the AAGAG satellite DNA repeat of Drosophila is a component of the nuclear architecture. Genetic studies reveal the essential nature of this component in normal development, concomitant with loss of the sub-nuclear structural integrity. Our studies suggest that AAGAG/CUCUU RNA are critical structural components of the core nuclear architecture that contributes to anchor different nuclear compartments. Reduction in this RNA, therefore, causes disruption of nucleoskeleton and, consequently, the assembly and stability of the chromosome compartments are disturbed.
These studies point to regulatory as well as structural, and potentially epigenetic maintenance role of NuMat constituents including NuMat_RNA. It will be of interest to identify other NuMat_RNAs and understand how the AAGAG/CUCUU RNA and other structural NuMat_RNA integrate with the NuMat structural features. It would also be of interest to find out whether there are proteins that interact with such RNAs and, if so, whether they are also part of the structural framework.
Materials and Methods
Drosophila S2 cells and embryo collection
S2 cells were grown on Schneider’s medium (Himedia Laboratory, Mumbai, India) with 10% fetal bovine serum and penicillin and streptomycin antibiotics. Embryos (0–16 h or 0–2 h) were collected from a laboratory population of Drosophila melanogaster (Canton-S) maintained at 25°C.
NuMat preparation from Drosophila embryos and S2 cells
All the chemicals used for NuMat isolation and other methods were obtained from Sigma-Aldrich unless mentioned otherwise. Nuclei were isolated from Drosophila S2 cells and Drosophila embryos. NuMat was prepared according to published procedure.31 Briefly, the nuclear pellet was removed of chromatin by digesting with digestion buffer [DB: 20 mM Tris pH 7.4, 20 mM KCl, 70 mM NaCl, 10 mM MgCl2, 0.125 mM spermidine 1 mM PMSF, 0.5% Triton-X 100, 10 U/ml RNasin (Promega Corporation) and 200 µg/ml DNase I (Sigma)] at 4°C for 1 h. Digestion was followed by extraction with 0.4 M NaCl for 5 min in extraction buffer [EB: 10 mM Hepes pH 7.5, 4 mM EDTA, 0.25 mM spermidine, 0.1 mM PMSF, 0.5% (v/v) Triton X-100] and further 5 min with 2 M NaCl in EB. The final pellet after extraction was washed twice with wash buffer (WB: 5 mM Tris pH 7.4, 20 mM KCl, 1 mM EDTA, 0.25 mM spermidine, 0.1 mm PMSF). After washing, TRIzol (Invitrogen, Life Technologies) was added immediately to the NuMat pellet and stored at -70°C until it was used for RNA isolation.
Isolation and characterization of RNA from NuMat
RNA was isolated from NuMat pellet stored in TRIzol according to manufacturer’s protocol. NuMat_RNA was in the size range of 125–500 bp. The NuMat_RNA was treated with RNase free, DNase I (NEB), reverse transcribed with Superscript II, (Invitrogen) and made double stranded using random primers. Double-stranded cDNA was treated with T4 DNA polymerase (NEB) to polish the ends and ligated to pMOS blunt end vector (GE Healthcare) according to the manufacturer’s instructions. Transformed colonies were screened on blue-white selection plates and checked for inserts by restriction enzyme digestions. Inserts were sequenced by cycle sequencing method using Big Dye terminator kit (Applied Biosystems, Life Technologies) on an ABI Prism 310 Automated DNA sequencer (Applied Biosystems) with M13F and T7 primers.
Northern blotting
Equal amount (10 μg) of nuclear and NuMat_RNA were resolved on a 1.5% denaturing formaldehyde agarose gel and transferred to nylon membrane by capillary transfer method in 20X SSC. NuMat_RNA treated with RNase free, DNase I (100 U/ml), RNase H (100 U/ml) or RNase I (200 U/ml) was also resolved on separate gels and transferred similarly. These enzymes were obtained from NEB. The blots were UV cross-linked and processed for northern hybridization. Strand-specific AAGAG/CTCTT and AATAT/ATATT probes were made by phosphorylating oligonucleotides using gamma 32P-ATP. For control hybridization, actin probe was random primer labeled using random primer labeling kit (NEB). Probes were used at a concentration of 105 cpm/ml of hybridization solution. The blots were hybridized at 55°C for AAGAG/CTCTT and actin, and at 37°C for AATAT/ATATT. After stringent washing, the blots were exposed to a Phosphor-imaging screen to obtain the images on a Fujifilm Fluorescent Image Analyzer FLA 3000 (Fujifilm Corporation).
In situ NuMat preparation in early Drosophila embryo
Drosophila embryos (0–2 h old) were collected, de-chorionated and washed thoroughly with running water to remove sodium hypochlorite. Embryos (0.1 g) were then rinsed in distilled water and pre-treated in 4:1 heptane:PBS for 2 min. Fixation was performed in 0.8 ml heptane, 0.1 ml 10X PBS with 4% formaldehyde for 20 min at room temperature with continuous mixing. After fixation, heptane and aqueous phase was removed. A mixture of 1:1 (v/v) ice cold methanol: heptane was added to devitellinize the embryos. The tube was shaken vigorously and the embryos were allowed to settle to the bottom. This step is repeated several times. These fixed embryos were brought to aqueous medium by several washes in PBS + 0.1% Triton-X-100 (PBT) and used directly for RNA-FISH as control embryos. To prepare NuMat, the fixed embryos were brought to aqueous medium by several washes in DB without DNase I. Digestion was performed with DNase I (200 ug/ml) in DB for 30 min at room temperature along with 10 U/ml RNasin. Embryos were then washed two times in EB without NaCl by suspending and then allowing to settle down in the tube due to gravity. Extraction was performed in EB with 2 M NaCl for 20 min. The samples were finally washed several times in PBT. These embryos/salivary glands with in situ nuclear matrices were used for RNA-FISH or immunostaining.
Probes for RNA-FISH
Dual color RNA-FISH was performed using differentially labeled probes generated by single-stranded PCR. For preparation of AAGAG probe, the pMOS plasmid containing the AAGAG insert was linearized using an appropriate restriction enzyme so that the PCR product is terminated by run-off. M13 forward primer and biotin-labeled dATP or DIG-labeled dUTP (Roche) was used in the PCR reaction mix to obtain the biotin/DIG-labeled AAGAG probe. Similarly for CUCUU probe, appropriately linearized plasmid was used for PCR with T7 primer and biotin-labeled dATP or DIG-labeled dUTP (Roche). The T7 primer synthesizes the CUCUU strand, whereas the M13 forward primer synthesizes AAGAG strand. PCR products were gel eluted and checked for labeling. These probes were used for RNA in situ hybridizations.
RNA fluorescence in situ hybridization (RNA-FISH)
Control embryos and in situ nuclear matrices prepared from them, were incubated in RNA hybridization buffer (HB: 40% formamide, 2X SSC, 10 mM EDTA, 0.1% Tween-20, 20 µg/ml of yeast tRNA) for 2 h at 37°C. After pre-incubation, the single-stranded probes in HB were added sequentially one after the other to prevent self-hybridization of the probes to each other. First, biotin/DIG-labeled AAGAG probe was diluted (1:50) in HB and added to the samples. Hybridization was performed at 37°C for 8 h after which the solution was removed and the samples were washed three times with the HB. Then diluted (1:50), biotin/DIG-labeled CUCUU probe was added to the samples. After another 8 h incubation, the samples were washed three times for 5 min each (2X SSC, 40% formamide, 0.1% tween 20) and brought finally to PBS. Anti-DIG FITC and anti-biotin Cy3 (1:500) (Jackson ImmunoResearch) were added and the samples were incubated at room temperature for an hour. Finally, after washing with PBS, the samples were mounted in Vectasheild with DAPI (Vector Laboratories). In case RNA-FISH was followed by antibody detection, after hybridization the samples were brought into PBS and incubated with 1:100 anti-lamin Dm0 (DSHB, University of Iowa). Secondary antibody (mouse Cy3, Jackson ImmunoResearch) was included along with the anti-DIG and anti-biotin secondary antibodies in the subsequent step.
DNA fluorescence in situ hybridization (DNA-FISH)
As DNA-FISH was performed to visualize the satellite DNA repeats, a double-stranded DIG-dUTP-labeled probe obtained by PCR was used. The probe corresponds to both the strands of satellite DNA repeat. DNA-FISH was performed on fixed embryos (as detailed above) in hybridization buffer (40% formamide, 2X SSC, 0.1% SDS, 20 μg/ml yeast tRNA) at 60°C overnight. Embryos and dsAAGAG probe were denatured 70°C and 95°C respectively for 5 min. Probe was chilled immediately and added to the samples. After hybridization, washings, detection of DIG and mounting was done as described above.
Immunostaining
Drosophila embryos
Control or knockdown embryos were de-chorionated, devitellenized and fixed as mentioned above and were processed for immunostaining. Salivary glands dissected out from control or knockdown third instar larvae were also fixed and processed in a similar manner. For antibody detection, anti-lamin Dm0 (mouse) and anti-fibrillarin (rabbit) primary antibodies at 1:100 dilutions in PBT were used. Incubation was performed for 1 h at 37°C or at 4°C overnight. Primary antibody was followed by secondary antibody treatment (anti-mouse Cy3 or FITC in 1:300 dilutions) for another hour. Samples were then washed and mounted in Vectasheild with DAPI.
Imaginal discs
Third instar crawling larvae were separated as wild-type and knockdown based on GFP expression. They were made inside-out with the help of needle exposing the inner organs. Inside out larvae were fixed in 4% paraformaldehyde and immunostained as mentioned above. After immunostaining, imaginal discs were separated and mounted.
Microscopy
Confocal laser scanning was performed on a Zeiss LSM 510 META (Carl Zeiss Inc.) with excitation at 488 nm, 543 nm, 633 nm (Ar-ion and HeNe lasers) and 760 nm at a pinhole of 1 AU. The scanning was done in the multi-track mode using a 10X, 63X and 100X 1.4NA objective. The emission of FITC was acquired using a 500–530 BP filter, that of Cy3 with a 565–615 BP filter and that of Cy5 with 650–710 BP filter set. Optical sections were taken 0.35 μm intervals. Individual optical sections were projected to give information in 3D using the Zeiss LSM software version 3.2 SP2. Later the images were assembled using Adobe Photoshop 6.0.
Generation and analysis of the transgenic lines
The AAGAG sequence (125 bp) in pMOS vector was excised using EcoRI-HindIII and inserted into the EcoRI-HindIII sites of pBluescript (Agilent Technologies, Inc.). From pBleuscript the AAGAG sequence were excised with EcoRI-NotI and inserted at EcoRI-NotI site in between the UAS elements of pSymp vector.
The pSympAAGAG construct (0.5 mg/ml DNA) was introduced into the Drosophila germ line of w-y-Δ2–3kstb embryos following standard protocol.58,59 Transformants were identified by the presence of the mini-white selectable marker. Multiple transformed lines were generated for the construct, and several independent lines were made. Chromosomal locations of transgene in the different lines was determined by crossing the transformed flies with balanced fly stock: w-;Pin/CyO;TM2/Tb. Further, to follow the chromosome at embryonic and larval stages, the balanced flies were crossed with different Gal4 driver lines.37 All the fly stocks were obtained from Bloomington Drosophila Stock Center (Indiana University).
Supplementary Material
Acknowledgments
We thank Durgadas Kasbekar and Jerry Workman for reading the manuscript and suggestions. A.M. thanks Department of Science and Technology (DST) for young scientist fellowship, R.P.K. thanks Council of Scientific and Industrial Research (CSIR) for senior research fellowship. We also thank Alka Dwivedi and Parul Varma for help during early stages of this work. Work in R.K.M’s lab has been supported by CSIR, young investigator grant of Human Frontier Science Program (HFSP) and Indo-French Centre for the Promotion of Advanced Research (CEFIPRA). Authors declare that there is no conflict of interest.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Author Contributions
A.M. execution (NuMat library preparation and sequencing), R.U.P. execution (staining and northern analysis), N.R. execution (microscopy), R.P.K. execution (generation of transgenic flies), D.V. execution (genetic analysis of flies), K.M. interpretation of the data and R.K.M. conception and design.
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/24326
References
- 1.Meldi L, Brickner JH. Compartmentalization of the nucleus. Trends Cell Biol. 2011;21:701–8. doi: 10.1016/j.tcb.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akhtar A, Gasser SM. The nuclear envelope and transcriptional control. Nat Rev Genet. 2007;8:507–17. doi: 10.1038/nrg2122. [DOI] [PubMed] [Google Scholar]
- 3.Bank EM, Gruenbaum Y. The nuclear lamina and heterochromatin: a complex relationship. Biochem Soc Trans. 2011;39:1705–9. doi: 10.1042/BST20110603. [DOI] [PubMed] [Google Scholar]
- 4.Meaburn KJ, Misteli T. Cell biology: chromosome territories. Nature. 2007;445:379–781. doi: 10.1038/445379a. [DOI] [PubMed] [Google Scholar]
- 5.Spector DL. Macromolecular domains within the cell nucleus. Annu Rev Cell Biol. 1993;9:265–315. doi: 10.1146/annurev.cb.09.110193.001405. [DOI] [PubMed] [Google Scholar]
- 6.Jackson DA, Cook PR. The structural basis of nuclear function. Int Rev Cytol. 1995;162A:125–49. doi: 10.1016/s0074-7696(08)61230-9. [DOI] [PubMed] [Google Scholar]
- 7.Razin SV, Gromova II, Iarovaia OV. Specificity and functional significance of DNA interaction with the nuclear matrix: new approaches to clarify the old questions. Int Rev Cytol. 1995;162B:405–48. doi: 10.1016/s0074-7696(08)62623-6. [DOI] [PubMed] [Google Scholar]
- 8.Cremer T, Kreth G, Koester H, Fink RH, Heintzmann R, Cremer M, et al. Chromosome territories, interchromatin domain compartment, and nuclear matrix: an integrated view of the functional nuclear architecture. Crit Rev Eukaryot Gene Expr. 2000;10:179–212. doi: 10.1615/CritRevEukarGeneExpr.v10.i2.60. [DOI] [PubMed] [Google Scholar]
- 9.Ma H, Siegel AJ, Berezney R. Association of chromosome territories with the nuclear matrix. Disruption of human chromosome territories correlates with the release of a subset of nuclear matrix proteins. J Cell Biol. 1999;146:531–42. doi: 10.1083/jcb.146.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Galande S, Purbey PK, Notani D, Kumar PP. The third dimension of gene regulation: organization of dynamic chromatin loopscape by SATB1. Curr Opin Genet Dev. 2007;17:408–14. doi: 10.1016/j.gde.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 11.Rivera-Mulia JC, Hernández-Muñoz R, Martínez F, Aranda-Anzaldo A. DNA moves sequentially towards the nuclear matrix during DNA replication in vivo. BMC Cell Biol. 2011;12:3. doi: 10.1186/1471-2121-12-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weis K, Rambaud S, Lavau C, Jansen J, Carvalho T, Carmo-Fonseca M, et al. Retinoic acid regulates aberrant nuclear localization of PML-RAR alpha in acute promyelocytic leukemia cells. Cell. 1994;76:345–56. doi: 10.1016/0092-8674(94)90341-7. [DOI] [PubMed] [Google Scholar]
- 13.Ellis JA, Shackleton S. Nuclear envelope disease and chromatin organization. Biochem Soc Trans. 2011;39:1683–6. doi: 10.1042/BST20110744. [DOI] [PubMed] [Google Scholar]
- 14.Gadji M, Vallente R, Klewes L, Righolt C, Wark L, Kongruttanachok N, et al. Nuclear remodeling as a mechanism for genomic instability in cancer. Adv Cancer Res. 2011;112:77–126. doi: 10.1016/B978-0-12-387688-1.00004-1. [DOI] [PubMed] [Google Scholar]
- 15.Nickerson J. Experimental observations of a nuclear matrix. J Cell Sci. 2001;114:463–74. doi: 10.1242/jcs.114.3.463. [DOI] [PubMed] [Google Scholar]
- 16.Nickerson JA, Blencowe BJ, Penman S. The architectural organization of nuclear metabolism. Int Rev Cytol. 1995;162A:67–123. doi: 10.1016/s0074-7696(08)61229-2. [DOI] [PubMed] [Google Scholar]
- 17.Fey EG, Krochmalnic G, Penman S. The nonchromatin substructures of the nucleus: the ribonucleoprotein (RNP)-containing and RNP-depleted matrices analyzed by sequential fractionation and resinless section electron microscopy. J Cell Biol. 1986;102:1654–65. doi: 10.1083/jcb.102.5.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hancock R. A new look at the nuclear matrix. Chromosoma. 2000;109:219–25. doi: 10.1007/s004120000077. [DOI] [PubMed] [Google Scholar]
- 19.Pederson T. Half a century of “the nuclear matrix”. Mol Biol Cell. 2000;11:799–805. doi: 10.1091/mbc.11.3.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lammerding J, Schulze PC, Takahashi T, Kozlov S, Sullivan T, Kamm RD, et al. Lamin A/C deficiency causes defective nuclear mechanics and mechanotransduction. J Clin Invest. 2004;113:370–8. doi: 10.1172/JCI19670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Newport JW, Wilson KL, Dunphy WG. A lamin-independent pathway for nuclear envelope assembly. J Cell Biol. 1990;111:2247–59. doi: 10.1083/jcb.111.6.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lenz-Böhme B, Wismar J, Fuchs S, Reifegerste R, Buchner E, Betz H, et al. Insertional mutation of the Drosophila nuclear lamin Dm0 gene results in defective nuclear envelopes, clustering of nuclear pore complexes, and accumulation of annulate lamellae. J Cell Biol. 1997;137:1001–16. doi: 10.1083/jcb.137.5.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Albrethsen J, Knol JC, Jimenez CR. Unravelling the nuclear matrix proteome. J Proteomics. 2009;72:71–81. doi: 10.1016/j.jprot.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 24.Calikowski TT, Meulia T, Meier I. A proteomic study of the arabidopsis nuclear matrix. J Cell Biochem. 2003;90:361–78. doi: 10.1002/jcb.10624. [DOI] [PubMed] [Google Scholar]
- 25.Nakayasu H, Berezney R. Nuclear matrins: identification of the major nuclear matrix proteins. Proc Natl Acad Sci USA. 1991;88:10312–6. doi: 10.1073/pnas.88.22.10312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kallappagoudar S, Varma P, Pathak RU, Senthilkumar R, Mishra RK. Nuclear matrix proteome analysis of Drosophila melanogaster. Mol Cell Proteomics. 2010;9:2005–18. doi: 10.1074/mcp.M110.001362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Varma P, Mishra RK. Dynamics of nuclear matrix proteome during embryonic development in Drosophila melanogaster. J Biosci. 2011;36:439–59. doi: 10.1007/s12038-011-9081-6. [DOI] [PubMed] [Google Scholar]
- 28.Barboro P, D’Arrigo C, Mormino M, Coradeghini R, Parodi S, Patrone E, et al. An intranuclear frame for chromatin compartmentalization and higher-order folding. J Cell Biochem. 2003;88:113–20. doi: 10.1002/jcb.10378. [DOI] [PubMed] [Google Scholar]
- 29.Nickerson JA, Krochmalnic G, Wan KM, Penman S. Chromatin architecture and nuclear RNA. Proc Natl Acad Sci USA. 1989;86:177–81. doi: 10.1073/pnas.86.1.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fey EG, Ornelles DA, Penman S. Association of RNA with the cytoskeleton and the nuclear matrix. J Cell Sci Suppl. 1986;5:99–119. doi: 10.1242/jcs.1986.supplement_5.6. [DOI] [PubMed] [Google Scholar]
- 31.Pathak RU, Rangaraj N, Kallappagoudar S, Mishra K, Mishra RK. Boundary element-associated factor 32B connects chromatin domains to the nuclear matrix. Mol Cell Biol. 2007;27:4796–806. doi: 10.1128/MCB.00305-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hozák P, Sasseville AM, Raymond Y, Cook PR. Lamin proteins form an internal nucleoskeleton as well as a peripheral lamina in human cells. J Cell Sci. 1995;108:635–44. doi: 10.1242/jcs.108.2.635. [DOI] [PubMed] [Google Scholar]
- 33.Wan KM, Nickerson JA, Krockmalnic G, Penman S. The nuclear matrix prepared by amine modification. Proc Natl Acad Sci USA. 1999;96:933–8. doi: 10.1073/pnas.96.3.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lohe AR, Brutlag DL. Multiplicity of satellite DNA sequences in Drosophila melanogaster. Proc Natl Acad Sci USA. 1986;83:696–700. doi: 10.1073/pnas.83.3.696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lohe AR, Hilliker AJ, Roberts PA. Mapping simple repeated DNA sequences in heterochromatin of Drosophila melanogaster. Genetics. 1993;134:1149–74. doi: 10.1093/genetics/134.4.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith CD, Shu S, Mungall CJ, Karpen GH. The Release 5.1 annotation of Drosophila melanogaster heterochromatin. Science. 2007;316:1586–91. doi: 10.1126/science.1139815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–15. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 38.Grewal SI, Elgin SC. Transcription and RNA interference in the formation of heterochromatin. Nature. 2007;447:399–406. doi: 10.1038/nature05914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, et al. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell. 2009;33:717–26. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sunwoo H, Dinger ME, Wilusz JE, Amaral PP, Mattick JS, Spector DL. MEN epsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. 2009;19:347–59. doi: 10.1101/gr.087775.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maison C, Bailly D, Peters AH, Quivy JP, Roche D, Taddei A, et al. Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nat Genet. 2002;30:329–34. doi: 10.1038/ng843. [DOI] [PubMed] [Google Scholar]
- 42.Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS. Specific expression of long noncoding RNAs in the mouse brain. Proc Natl Acad Sci USA. 2008;105:716–21. doi: 10.1073/pnas.0706729105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dinger ME, Pang KC, Mercer TR, Crowe ML, Grimmond SM, Mattick JS. NRED: a database of long noncoding RNA expression. Nucleic Acids Res. 2009;37(Database issue):D122–6. doi: 10.1093/nar/gkn617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–7. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–14. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mallik M, Lakhotia SC. Pleiotropic consequences of misexpression of the developmentally active and stress-inducible non-coding hsrω gene in Drosophila. J Biosci. 2011;36:265–80. doi: 10.1007/s12038-011-9061-x. [DOI] [PubMed] [Google Scholar]
- 47.Singh AK, Lakhotia SC. The hnRNP A1 homolog Hrp36 is essential for normal development, female fecundity, omega speckle formation and stress tolerance in Drosophila melanogaster. J Biosci. 2012;37:659–78. doi: 10.1007/s12038-012-9239-x. [DOI] [PubMed] [Google Scholar]
- 48.Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39:925–38. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sasaki YT, Ideue T, Sano M, Mituyama T, Hirose T. MENepsilon/beta noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc Natl Acad Sci USA. 2009;106:2525–30. doi: 10.1073/pnas.0807899106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Onorati MC, Lazzaro S, Mallik M, Ingrassia AM, Carreca AP, Singh AK, et al. The ISWI chromatin remodeler organizes the hsrω ncRNA-containing omega speckle nuclear compartments. PLoS Genet. 2011;7:e1002096. doi: 10.1371/journal.pgen.1002096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lakhotia SC. Long non-coding RNAs coordinate cellular responses to stress. Wiley Interdiscip Rev RNA 2012. [DOI] [PubMed] [Google Scholar]
- 52.Lakhotia SC, Mallik M, Singh AK, Ray M. The large noncoding hsrω-n transcripts are essential for thermotolerance and remobilization of hnRNPs, HP1 and RNA polymerase II during recovery from heat shock in Drosophila. Chromosoma. 2012;121:49–70. doi: 10.1007/s00412-011-0341-x. [DOI] [PubMed] [Google Scholar]
- 53.Valgardsdottir R, Chiodi I, Giordano M, Cobianchi F, Riva S, Biamonti G. Structural and functional characterization of noncoding repetitive RNAs transcribed in stressed human cells. Mol Biol Cell. 2005;16:2597–604. doi: 10.1091/mbc.E04-12-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zheng R, Shen Z, Tripathi V, Xuan Z, Freier SM, Bennett CF, et al. Polypurine-repeat-containing RNAs: a novel class of long non-coding RNA in mammalian cells. J Cell Sci. 2010;123:3734–44. doi: 10.1242/jcs.070466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Topp CN, Zhong CX, Dawe RK. Centromere-encoded RNAs are integral components of the maize kinetochore. Proc Natl Acad Sci USA. 2004;101:15986–91. doi: 10.1073/pnas.0407154101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bouzinba-Segard H, Guais A, Francastel C. Accumulation of small murine minor satellite transcripts leads to impaired centromeric architecture and function. Proc Natl Acad Sci USA. 2006;103:8709–14. doi: 10.1073/pnas.0508006103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Valgardsdottir R, Chiodi I, Giordano M, Rossi A, Bazzini S, Ghigna C, et al. Transcription of Satellite III non-coding RNAs is a general stress response in human cells. Nucleic Acids Res. 2008;36:423–34. doi: 10.1093/nar/gkm1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rubin GM, Spradling AC. Genetic transformation of Drosophila with transposable element vectors. Science. 1982;218:348–53. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- 59.Spradling AC, Rubin GM. Transposition of cloned P elements into Drosophila germ line chromosomes. Science. 1982;218:341–7. doi: 10.1126/science.6289435. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.