Abstract
In bacteria, trans-translation rescues stalled ribosomes by the combined action of tmRNA (transfer-mRNA) and its associated protein SmpB. The tmRNA 5′ and 3′ ends fold into a tRNA-like domain (TLD), which shares structural and functional similarities with tRNAs. As in tRNAs, the UUC sequence of the T-arm of the TLD is post-transcriptionally modified to m5UψC. In tRNAs of gram-negative bacteria, formation of m5U is catalyzed by the SAM-dependent methyltransferase TrmA, while formation of m5U at two different positions in rRNA is catalyzed by distinct site-specific methyltransferases RlmC and RlmD. Here, we show that m5U formation in tmRNAs is exclusively due to TrmA and should be considered as a dual-specific enzyme. The evidence comes from the lack of m5U in purified tmRNA or TLD variants recovered from an Escherichia coli mutant strain deleted of the trmA gene. Detection of m5U in RNA was performed by NMR analysis.
Keywords: methylation, methyltransferase, TrmA, NMR, post-transcriptional modification, t-RNA like domain (TLD), tmRNA
Introduction
Defective mRNAs can cause stalling of translating ribosomes. In bacteria, ribosomes are mainly rescued by the trans-translation process, the essential components of which are tmRNA (transfer-mRNA, formerly called 10Sa RNA), a large ubiquitous RNA of about 360 nt with both mRNA and tRNA activities, and a partner protein SmpB. tmRNA is alanylated prior to entering the ribosome (for recent reviews. see refs. 1 and 2). Alanine is transferred to the nascent peptide whose synthesis has been stopped. Translation then resumes on the open reading frame internal to tmRNA (mRNA-like domain or MLD) and ends normally at an internal stop codon, allowing ribosome recovery. The tmRNA-encoded C-terminal fusion peptide targets the incomplete protein for degradation.
The biosynthesis of tmRNA shares many features with that of tRNAs. The precursor tmRNA is processed at the 5′ end by RNase P and at the 3′ end by several enzymes, including RNase PH and RNase T. Both ends then fold into a tRNA-like domain (TLD) containing an amino acid acceptor stem and a T-arm closed by a TΨC-loop.3-6 The 3′-CCA sequence of the TLD is either encoded in the gene or added post-transcriptionally and is charged in vivo by the Ala-RNA synthetase (AlaRS).3,7 There is no D-stem in tmRNAs. However, their D-loop contains the consensus GG doublet found in all canonical tRNAs, which is known to form two base pairs with T-loop residues (G18.Ψ55 and G19.C56). An NMR study of the D/T loop interactions stabilizing the Escherichia coli and Aquifex aeolicus TLDs and two crystallographic structures of the TLDs of A. aeolicus and Thermus thermophilus in complex with the partner protein SmpB, confirm that the fold of the TΨC-loop and the existence of the two base pairs between doublets of the T-loop are very similar to what is observed in all cytoplasmic tRNAs.8-10
Methylation occurs in most non-coding RNAs and accounts for more than half of the post-transcriptional modifications.11 As in regular tRNAs of all three phylogenetic domains, the UUC sequence of the tmRNA T-arm loop is usually post-transcriptionally modified into TψC.6 Because of obvious structural similarity between the T-arms of tmRNA and tRNA, it has been thought that formation of ribo-T (m5U), as well as ψ in both types of nucleic acids, would be catalyzed by the same set of modification enzymes. However, in the case of tmRNA, no experimental evidence has yet been provided.
In E. coli, three m5U-RNA methyltransferases exist: TrmA (formerly called RumT) catalyzing the formation of m5U at position 54 of tRNA, RlmD catalyzing the formation of m5U at position 1939 of 23S rRNA and RlmC that methylates U747 in 23S rRNA.12,13
Here, we aimed at identifying which of the three rRNA-m5U methyltransferase paralogs in E. coli is responsible for the methylation of uridine of the T-arm of tmRNA. For this purpose, we used three RNAs recapitulating the functional 5′ and 3′ tmRNA ends (Fig. 1). One of them is the TLD fragment of A. aeolicus (TLDaa, 60 nt in length) whose structure was formerly investigated by NMR.8 The other two RNAs are longer: an A. aeolicus TLD-H2 derivative of 143 nucleotides (TLD-Haa) and the full-length processed E. coli tmRNA itself (ectmRNA, 363 nt). All RNAs were overproduced in three E. coli strains, wild-type or deleted either of the trmA gene alone or of the set of the three trmA, rumB and rumC genes. The triple deletion was used as negative control. After recovery and purification of the expressed RNAs, the presence or the absence of methylation of the first uridine of the T-loop UUC sequence, was investigated by NMR. One goal of our work was also to expand the application of the NMR technique to test the presence or absence of a naturally occurring methyl group in a long cellular RNA molecule. At the same time, this approach gives conformational and dynamic information around the site of methylation, which would not be easily obtained by other techniques, such as in vitro tests with recombinant enzymes. While tedious for sample preparation, this technique is nevertheless particularly suited to analyze the TLD portion of tmRNA because it can rapidly and unambiguously search for the presence or absence of methylation, determining the position of the methylated nucleotide.
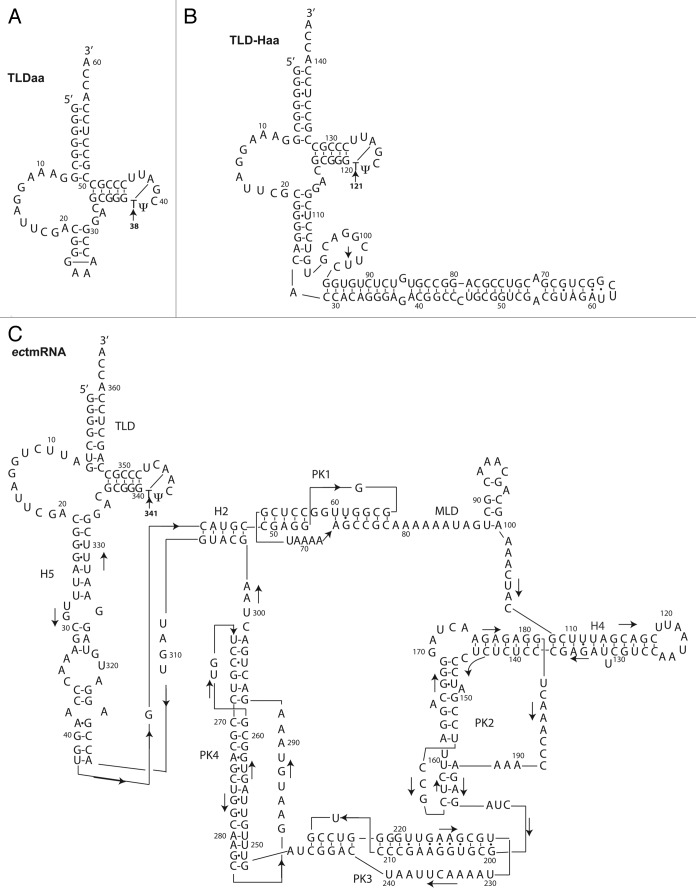
Figure 1. Secondary structure models of (A) TLDaa, (B) TLD-Haa and (C) full-length ectmRNA. The TLD parts of the A. aeolicus and E. coli models are very similar in sequence. An arrow stresses the position of the ribothymidine m5U (labeled T for simplification) (i.e. 38 in TLDaa, 121 in TLD-Haa and 341 in ectmRNA). The second methylated uridine observed in TLD-Haa, as explained in text, is most probably U97.
Results and Discussion
Investigations were performed exclusively with E. coli, a Gram-negative bacterium. Three RNAs were used as templates to probe methylation of the uridines in vivo (Fig. 1). The shorter fragment (TLDaa) recapitulates the TLD part of the A. aeolicus tmRNA and is closed by an artificial GAAA loop. The m5U was expected at position 38. The second RNA was originally designed so as to extend the natural part of the TLDaa by an extended bulged helical segment mimicking the tmRNA H2 connecting stem. Unfortunately, this construct proved to be toxic for the bacteria. However, the bacteria evolved a mutation, in which one adenine in the D-loop is deleted and that bears an extra nine-residue bulge whose sequence (CUUCGGACG) resembles that of the TLDaa T-loop. The corresponding RNA is called hereafter TLD-Haa. According to MFOLD, a secondary structure prediction program, deletion of the adenine does not break the L-shaped fold of the TLD part.14 This prediction is confirmed by the overproduction of an intact RNA, that attests the formation of a protecting tRNA-like scaffold. In this TLD-Haa, uridine methylation was expected at position 121. The third RNA is the full-length ectmRNA itself. The T-arms of the three RNAs are identical. The T-loops only differ by the fourth and sixth residues, which however remain in all cases respectively a purine and a pyrimidine. The position of the m5U in the naturally occurring full-length E. coli tmRNA is 341.
The wild-type and the mutant E. coli strains lacking either tRNA:m5U methyltransferase trmA, or trmA and both rRNA:m5U methyltransferases rlmC and rlmD were transformed with each of the three plasmids. Bacterial growth of the mutant strains was not totally abolished by the deletion of one, or all three RNA:m5U-methyltransferase(s). We were able to recover enough material (i.e. between 1–5 mg) from the wild-type and ΔtrmA strains to perform NMR studies. For simplification, the names of the RNAs produced in the wild-type strain will be preceded by “wt-,” and those produced from the ΔtrmA and ΔtrmA-rlmC-rlmD mutant strains, respectively by “Δ1-” and “Δ3-.”
For each sample, the 1D and NOESY spectra disclose the characteristic resonance(s) of a methyl proton (sharp peak between 1.0–1.5 ppm) when the RNAs are overproduced in the wt-BW25113 strain, but not when produced in the mutant strains (Figs. 2–4).
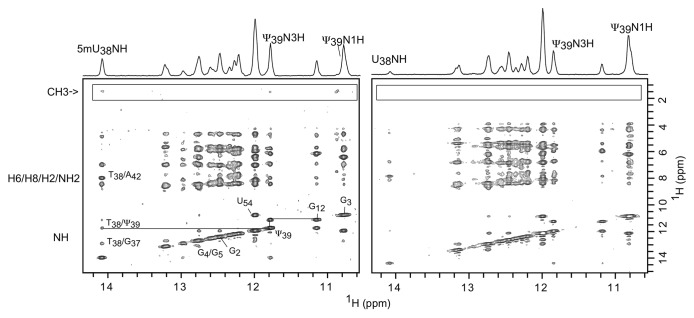
Figure 2. Fingerprint regions of two NOESY spectra recorded at 150 ms mixing times and 288 K of the wt-TLDaa (left) and Δ1-TLDaa (right). The spectra are almost identical, except for the boxed cross-peaks arising from m5U38 that are absent when the RNA is overproduced in the ΔtrmA strain.
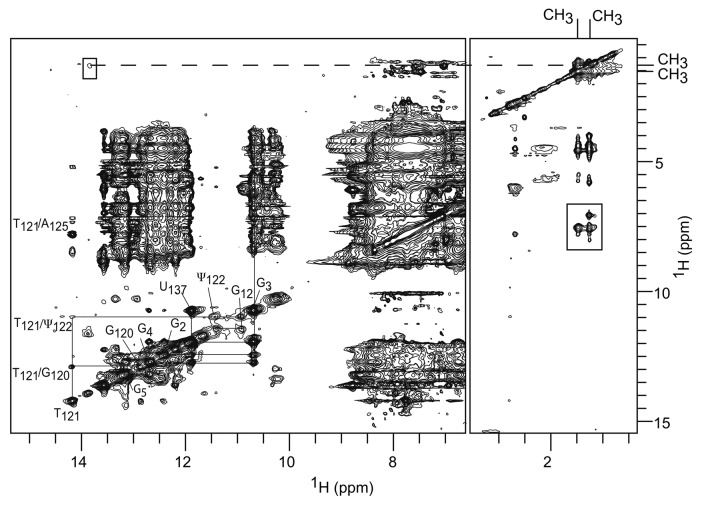
Figure 4. Superposition of TLD-Haa jrNOESY spectra recorded at 298 K at 100 and 150 ms mixing times: imino proton region (left) and methyl proton region (right). The pattern of the TLDaa cross-peaks and its assignment is transposable to these spectra, showing that the TLD parts of both RNAs fold similarly. The spectra disclose two methyl proton resonances (boxed cross-peaks), one arising from the expected m5U121 and the other probably from m5U97.
In a previous study, we investigated the TLDaa by NMR and proved that it adopts a tRNA-like tri-dimensional fold. In this former study, the TLDaa was overproduced in JM101 strains and we were able to unambiguously assign the resonances of both the imino and the methyl protons of m5U38 (or T38), the equivalent position of m5U55 in tRNAs.8 The spectra recorded here for the TLDaa produced in BW25113 E. coli cells are identical to our previous spectra, confirming: (1) that the RNA is correctly folded, and (2) that the position of methylation is independent of the nature of the E. coli strains. TLDaa 1D and NOESY spectra recorded in both strains also display one set of the two imino proton resonances of the Ψ39, emphasizing the quality of our RNAs. The 1D spectrum of wt-TLDaa exhibits an extra sharp resonance at 1.19 ppm compared with that of Δ1-TLDaa (Fig. S1). As expected, the NOESY spectrum of the wt-TLDaa discloses a cross-peak pattern similar to that of our previously studied TLD, which allowed the resonance at 1.19 ppm to be assigned to m5U38 (T38) methyl protons (Fig. 2, left panel) and therefore confirming the site of methylation.8 This set of cross-peaks is totally absent from the equivalent spectra recorded for Δ1-TLDaa (Fig. 2, right panel) and Δ3-TLDaa (not shown).
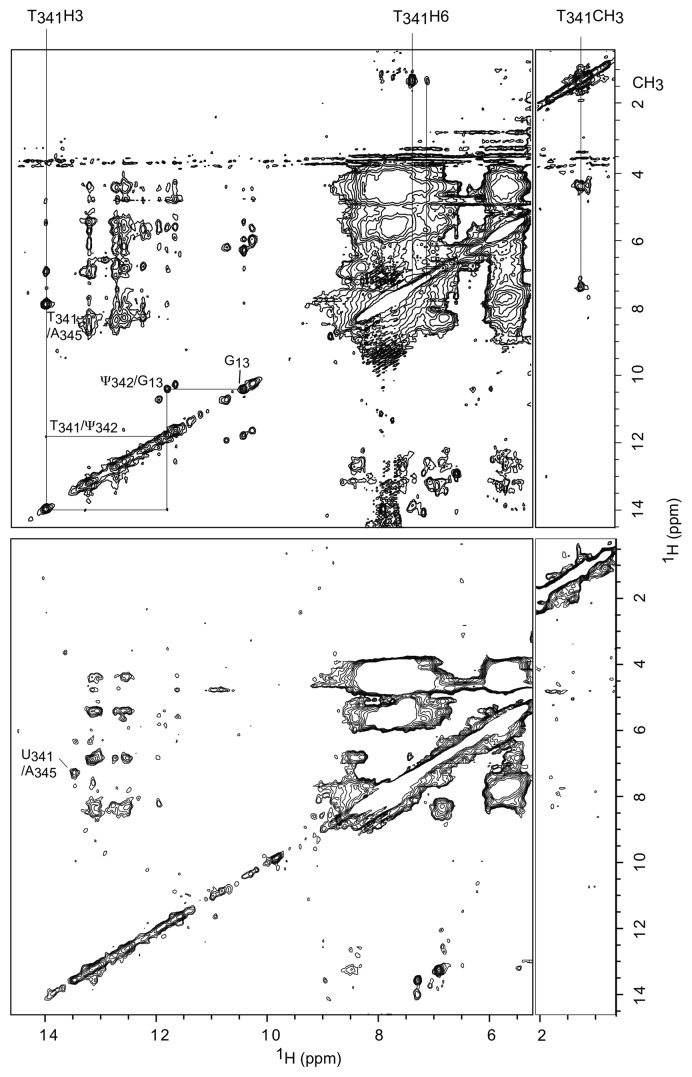
The NOESY spectrum of the full-length wt-ectmRNA presents very broad resonances, which is not surprising for a RNA of 117.4 kDa (Fig. 3). Contrary to the cases of the shorter RNA derivatives, the spectral resolution is thus much too low to allow direct identification of the methylation position. However, the wt-ectmRNA spectrum discloses two well-resolved cross-peaks, between a methyl group and two aromatic protons demonstrating the presence of a ribothymidine (m5U). The stronger cross-peak arises from the intra-residual CH3-H6 NOE. It is connected to the resonance at 14.0 ppm whose connection pathway to G13 is reminiscent of that of wt-TLDaa T38H3 to G12 (Fig. 2) and that of T39H3 to G13 in the spectrum we recorded for the RNA recapitulating the E. coli tmRNA TLD.8 As only one uridine is post-transcriptionally methylated, we could infer that it indeed stems from m5U341 in the T-loop and that the imino proton resonance that is at 14.0 ppm is that of m5U341H3 (or T341).6 As expected, this resonance is also connected to the N3H imino resonance of the neighboring Ψ342 at 11.85 ppm, which is, in turn, connected to G13 N3H at 10.2 ppm.
Figure 3. wgNOESY spectra recorded at 150 ms mixing times and 293 K of wt-ectmRNA 0.2 mM (upper spectrum) and in Δ1-ectmRNA 50 μM (lower spectrum). Upper: a strong cross-peak connects a methyl group and an H6 proton. T341H3 connectivity pathway to G13 is similar to that linking wt-TLDaa T38H3 to G12H3. Lower: the methyl resonance and the linked cross-peaks are absent.
The overproduction of each of the three RNAs was a little less efficient in the deleted strains than in the wild-type strain (not shown). This is not surprising since the lack of m5U54 in tRNA was reported to only marginally reduce (4%) the growth rate (reviewed in ref. 15). Consequently, we can rule out any saturation of the methylation enzymes to explain the absence of observable methylation on each of the RNAs produced in the ΔtrmA and ΔtrmA-rlmC-rlmD strains. Our data thus definitively prove that under in vivo experimental conditions, only tRNA methyltransferase TrmA of E. coli is able to catalyze the formation of m5U in both tRNA and tmRNA, while neither rRNA methyltransferases RlmD nor RlmC was able to catalyze even trace amounts of m5U in any of the tmRNA-TLD derivatives tested.
Unexpectedly, the spectrum of TLD-Haa also discloses cross-peaks connected to a second methyl group (Fig. 4). The presence of this second spin system connected to a methyl group might result from the formation of two alternative conformations in slow equilibrium on the NMR time scale or from two methylation sites within the same conformation. We ruled out the first possible explanation because if two conformations of the TLD were in equilibrium, then two sets of proton resonances, one for each conformation, should be visible. This is not the case. The imino spectrum, assigned by analogy with the wt-TLDaa equivalent spectrum, discloses only one characteristic imino pathway (Fig. 4). Moreover, the intensities of the two methyl resonances remain identical whatever the experimental conditions (concentration, temperature, thermal denaturation of the sample followed by fast freezing). Therefore, the observed second methyl resonance results from the existence of a second cryptic methylation site in the molecule, identified as m5U97. Indeed, the additional bulge whose sequence resembles that of the TLD T-loop and fits the consensus descriptors, i.e. the triplet UUC next to a stack of purines, makes its first uridine a target for TrmA.16 This second methylation is absent when TLD-Haa is produced in the ΔtrmA strain attesting that TrmA also catalyzes the methylation of this second uridine target (Fig. S2). Nevertheless, beyond the scope of this report, it would be interesting to assess the methylation by TrmA of an oligonucleotide recapitulating this bulge.
The tRNA mimicry of the 5′ and 3′ ends of bacterial tmRNAs was postulated before the discovery of its structure and the exact chemical nature of the post-transcriptional modifications. We have now demonstrated that the E. coli SAM-dependent U-methyltransferase acting on the T-loop of tRNA, also acts on the T-loop of the tRNA-like domain of tmRNA and should therefore be considered as a dual-specific enzyme, as the pseudouridine synthase RluA acting on 23S rRNA (position 746) and tRNA (position 32) or the SAM-dependent methyltransferase RlmN acting on 23S rRNA (position 2503) and tRNA (position 37).17,18 In the same line, we expect TruB, the Psi-synthase catalyzing formation of Ψ55 within the same T-loop in tRNAs in E. coli, to also be a dual-specific enzyme responsible for pseudouridylation of the T-loop in both the tmRNA and tRNA.
In the Gram-positive B. subtilis, only one enzyme (RlmCD) catalyzes the formation of m5U at both positions 747 and 1939 in 23S RNA, instead of two enzymes (RlmC and RlmD) in the Gram-negative E. coli. Moreover, in B. subtilis, m5U at position 54 in tRNA is catalyzed by TrmFO, a methylene-tetrahydrofolate-dependent enzyme, while in E. coli it is the SAM-dependent TrmA enzyme that catalyzes the formation of the same m5U-54 in tRNA.19-21 Very recently, Yamagami and collaborators have demonstrated that TrmFO, like TrmA, recognizes the T-arm structure of T. thermophilus tRNAs.22 We can thus infer from this and our results that TrmFO most probably methylates the TLD T-loop in tmRNAs of Gram-positive bacteria (another dual-specific enzyme).
Materials and Methods
E. coli strains and media
E. coli simple mutant strains are derivatives of BW 25113 [F-, Δ(araD-araB)567, ΔlacZ4787(::rrnB-3), λ−, rph-1, Δ(rhaD-rhaB)568, hsdR514] and were obtained from the Coli Genetic Stock Center. The BW 25113 single ΔtrmA and triple ΔtrmA/ΔrlmC/ΔrlmD strains were initially constructed by Damien Brégeon (University of Orsay) and previously described in Desmolaize et al.19 Electrocompetent cells were transformed with the pBSTNAV vectors and selected on LB plates containing ampicillin (100 μg/ml).23
Preparation and purification of the RNAs
The genes of TLDaa, TLD-Haa and ectmRNA were cloned in the pBSTNAV vector (AmpR) between EcoRI and PstI according to the procedure described by Ponchon et al.24,25 The expression of the recombinant RNAs was monitored by carrying out extractions of total soluble RNAs from small-scale overnight cultures in 2XTY growth medium. Large-scale expression was performed overnight at 37°C in 2 L of 2XTY medium containing 100 μg/ml ampicillin. The RNAs were recovered and purified as described in Gaudin et al. and in Ponchon et al.8,25 Pure fractions of monomeric ectmRNA were separated from aggregates on a Superdex-200 prep grade gel filtration column (GE-Healthcare) equilibrated with 50 mM NaCl, 20 mM potassium phosphate pH 6.5 and 1 mM EDTA water solution. The purity of all samples was checked by 12% PAGE containing 7 M urea. The gels were visualized by UV shadowing. The final yields were about 10 mg/L for the production in wild-type strains and 2 mg/L in ΔtrmA and ΔtrmA/ΔrlmC/ΔrlmD strains.
NMR sample preparation and analysis
The purified samples were deacylated in Tris buffer pH 8 at 37°C for 1 h and dialyzed several times against 1 mM EDTA water solutions and finally extensively against water. They were then freeze-dried before re-suspension in 95% H2O-5% D2O. The pH was adjusted to 6.5 by addition of small aliquots of NaOH 0.1 N under stirring. The sample strand concentrations were between 0.3 mM and 50 μM. TLDaa and TLD-Haa samples were heated at 95°C and snap-cooled in ice water to ensure monomeric species before NMR experiments. The spectra were recorded in 5 mm Shigemi tubes at 600 MHz on a Bruker Avance DRX600 spectrometer equipped with a cryoprobe and Z-axis gradient. 1D and 2D NOESY experiments were recorded at 288 K and 293 K using the Jump and Returm procedure (jrNOESY) to avoid excitation of the water resonance, or the Watergate pulse sequence (wgNOESY) for solvent suppression.26,27 The data were processed using TOPSIN 3.1 (Bruker).
Supplementary Material
Acknowledgments
We are really grateful to Dr Damien Brégeon (University Paris VI) for the kind gift of the BW 25113 mutant strains and to Dr Anne-Lise Haenni (University Paris VII) for careful editing of the manuscript.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by grant (ANR-09-MIEN-030-01) from the “Agence Nationale pour la Recherche” to S.N.-L. and F.D., as well as from the CNRS and from the French Department of Research and Education. H.G. is an emeritus scientist at the Center of Molecular Genetics of the CNRS in Gif-sur-Yvette.
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/24327
References
- 1.Barends S, Kraal B, van Wezel GP. The tmRNA-tagging mechanism and the control of gene expression: a review. Wiley Interdiscip Rev RNA. 2011;2:233–46. doi: 10.1002/wrna.48. [DOI] [PubMed] [Google Scholar]
- 2.Felden B, Gillet R. SmpB as the handyman of tmRNA during trans-translation. RNA Biol. 2011;8:440–9. doi: 10.4161/rna.8.3.15387. [DOI] [PubMed] [Google Scholar]
- 3.Komine Y, Kitabatake M, Yokogawa T, Nishikawa K, Inokuchi H. A tRNA-like structure is present in 10Sa RNA, a small stable RNA from Escherichia coli. Proc Natl Acad Sci USA. 1994;91:9223–7. doi: 10.1073/pnas.91.20.9223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li Z, Pandit S, Deutscher MP. 3′ exoribonucleolytic trimming is a common feature of the maturation of small, stable RNAs in Escherichia coli. Proc Natl Acad Sci USA. 1998;95:2856–61. doi: 10.1073/pnas.95.6.2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Felden B, Himeno H, Muto A, McCutcheon JP, Atkins JF, Gesteland RF. Probing the structure of the Escherichia coli 10Sa RNA (tmRNA) RNA. 1997;3:89–103. [PMC free article] [PubMed] [Google Scholar]
- 6.Felden B, Hanawa K, Atkins JF, Himeno H, Muto A, Gesteland RF, et al. Presence and location of modified nucleotides in Escherichia coli tmRNA: structural mimicry with tRNA acceptor branches. EMBO J. 1998;17:3188–96. doi: 10.1093/emboj/17.11.3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ushida C, Himeno H, Watanabe T, Muto A. tRNA-like structures in 10Sa RNAs of Mycoplasma capricolum and Bacillus subtilis. Nucleic Acids Res. 1994;22:3392–6. doi: 10.1093/nar/22.16.3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gaudin C, Nonin-Lecomte S, Tisné C, Corvaisier S, Bordeau V, Dardel F, et al. The tRNA-like domains of E coli and A.aeolicus transfer-messenger RNA: structural and functional studies. J Mol Biol. 2003;331:457–71. doi: 10.1016/S0022-2836(03)00760-5. [DOI] [PubMed] [Google Scholar]
- 9.Gutmann S, Haebel PW, Metzinger L, Sutter M, Felden B, Ban N. Crystal structure of the transfer-RNA domain of transfer-messenger RNA in complex with SmpB. Nature. 2003;424:699–703. doi: 10.1038/nature01831. [DOI] [PubMed] [Google Scholar]
- 10.Bessho Y, Shibata R, Sekine S, Murayama K, Higashijima K, Hori-Takemoto C, et al. Structural basis for functional mimicry of long-variable-arm tRNA by transfer-messenger RNA. Proc Natl Acad Sci USA. 2007;104:8293–8. doi: 10.1073/pnas.0700402104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Motorin Y, Helm M. RNA nucleotide methylation. Wiley Interdiscip Rev RNA. 2011;2:611–31. doi: 10.1002/wrna.79. [DOI] [PubMed] [Google Scholar]
- 12.Ny T, Björk GR. Cloning and restriction mapping of the trmA gene coding for transfer ribonucleic acid (5-methyluridine)-methyltransferase in Escherichia coli K-12. J Bacteriol. 1980;142:371–9. doi: 10.1128/jb.142.2.371-379.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Madsen CT, Mengel-Jørgensen J, Kirpekar F, Douthwaite S. Identifying the methyltransferases for m(5)U747 and m(5)U1939 in 23S rRNA using MALDI mass spectrometry. Nucleic Acids Res. 2003;31:4738–46. doi: 10.1093/nar/gkg657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–15. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Björk GR. The role of modified nucleosides in tRNA interactions. In Hatfield DL, Lee BJ and Pirtle RM, ed. Transfer RNA in Protein Synthesis. CRC Press, Boca Raton, FL, 1992:23-85. [Google Scholar]
- 16.Alian A, Lee TT, Griner SL, Stroud RM, Finer-Moore J. Structure of a TrmA-RNA complex: A consensus RNA fold contributes to substrate selectivity and catalysis in m5U methyltransferases. Proc Natl Acad Sci USA. 2008;105:6876–81. doi: 10.1073/pnas.0802247105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wrzesinski J, Nurse K, Bakin A, Lane BG, Ofengand J. A dual-specificity pseudouridine synthase: an Escherichia coli synthase purified and cloned on the basis of its specificity for psi 746 in 23S RNA is also specific for psi 32 in tRNA(phe) RNA. 1995;1:437–48. [PMC free article] [PubMed] [Google Scholar]
- 18.Benítez-Páez A, Villarroya M, Armengod ME. The Escherichia coli RlmN methyltransferase is a dual-specificity enzyme that modifies both rRNA and tRNA and controls translational accuracy. RNA. 2012;18:1783–95. doi: 10.1261/rna.033266.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Desmolaize B, Fabret C, Brégeon D, Rose S, Grosjean H, Douthwaite S. A single methyltransferase YefA (RlmCD) catalyses both m5U747 and m5U1939 modifications in Bacillus subtilis 23S rRNA. Nucleic Acids Res. 2011;39:9368–75. doi: 10.1093/nar/gkr626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Urbonavicius J, Skouloubris S, Myllykallio H, Grosjean H. Identification of a novel gene encoding a flavin-dependent tRNA:m5U methyltransferase in bacteria--evolutionary implications. Nucleic Acids Res. 2005;33:3955–64. doi: 10.1093/nar/gki703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamagami R, Yamashita K, Nishimasu H, Tomikawa C, Ochi A, Iwashita C, et al. The tRNA recognition mechanism of folate/FAD-dependent tRNA methyltransferase (TrmFO) J Biol Chem. 2012;287:42480–94. doi: 10.1074/jbc.M112.390112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nishimasu H, Ishitani R, Yamashita K, Iwashita C, Hirata A, Hori H, et al. Atomic structure of a folate/FAD-dependent tRNA T54 methyltransferase. Proc Natl Acad Sci USA. 2009;106:8180–5. doi: 10.1073/pnas.0901330106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meinnel T, Mechulam Y, Fayat G. Fast purification of a functional elongator tRNAmet expressed from a synthetic gene in vivo. Nucleic Acids Res. 1988;16:8095–6. doi: 10.1093/nar/16.16.8095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ponchon L, Dardel F. Recombinant RNA technology: the tRNA scaffold. Nat Methods. 2007;4:571–6. doi: 10.1038/nmeth1058. [DOI] [PubMed] [Google Scholar]
- 25.Ponchon L, Beauvais G, Nonin-Lecomte S, Dardel F. A generic protocol for the expression and purification of recombinant RNA in Escherichia coli using a tRNA scaffold. Nat Protoc. 2009;4:947–59. doi: 10.1038/nprot.2009.67. [DOI] [PubMed] [Google Scholar]
- 26.Guéron M, Plateau P, Descorps M. Solvent signal suppression in NMR. Prog Nucl Magn Reson Spectrosc. 1991;23:135–209. doi: 10.1016/0079-6565(91)80007-O. [DOI] [Google Scholar]
- 27.Piotto M, Saudek V, Sklenár V. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J Biomol NMR. 1992;2:661–5. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.