Abstract
Discovered in eukaryotes as a modification essential for mRNA function, polyadenylation was then identified as a means used by all cells to destabilize RNA. In Escherichia coli, most accessible 3′ RNA extremities are believed to be potential targets of poly(A) polymerase I. However, some RNAs might be preferentially adenylated. After a short statement of the current knowledge of poly(A) metabolism, we discuss how Hfq could affect recognition and polyadenylation of RNA terminated by Rho-independent terminators. Comparison of RNA terminus leads to the proposal that RNAs harboring 3′ terminal features required for Hfq binding are not polyadenylated, whereas those lacking these structural elements can gain the oligo(A) tails that initiate exonucleolytic degradation. We also speculate that Hfq stimulates the synthesis of longer tails that could be used as Hfq-binding sites involved in non-characterized functions of Hfq-dependent sRNAs.
Keywords: RNA degradation, Rho-independent terminators, exoribonucleases, oligo(A) tails, poly(A) polymerase, polyadenylation, protein Hfq
Introduction
Polyadenylation is a universal post-transcriptional modification, which profoundly affects the activity and fate of RNA. First discovered in eukaryotes, where it contributes to export of RNA to the cytoplasm and promotes mRNA stability and translation, polyadenylation was then demonstrated to have an RNA destabilizing function that is conserved in bacteria, organelles and nuclei.1-12 In prokaryotes, it is widely accepted that oligo(A) tails primarily expedite degradation of short-structured mRNA decay intermediates and control the turnover of several non-coding RNAs that regulate plasmid replication and maintenance, viral lysogeny and translation efficiency.13-20 In addition, poly(A) assisted decay also has been shown to be involved in the quality control of precursors of a defective and a wild-type tRNAs, to affect gene expression and to compensate for a deficiency in the main pathway of RNA decay orchestrated by RNase E.21-25
In Escherichia coli, all types of RNA are polyadenylated by poly(A) polymerase I (PAP I) encoded by the pcnB gene;26-35 these RNAs include mRNAs, mature and precursor tRNAs and rRNAs, decay intermediates and by-products of processing as well as small regulatory non-coding RNAs (sRNAs) and viral RNAs. Nearly all mRNAs were shown to be adenylated at the 3′-extremities, which can result from transcription termination, processing and/or exonucleolytic nibbling.28,29,34,36 Besides this set of data leading to the widely accepted idea that any accessible 3′ end of an RNA can be polyadenylated, there are clues that PAP I is able to discriminate between RNA substrates. Indeed, global analysis of polyadenylated mRNAs showed that stimulation of polyadenylation of individual mRNAs ranges from 2‒50 times when PAP I is overexpressed.29 Moreover, the structures of the 5′ and 3′ extremities of the RNA were reported to affect its efficiency. In particular, 3′ ends resulting from rho-dependent transcription termination were reported to be preferentially polyadenylated by PAP I.29,35,37-39 By analogy with the situation in eukaryotic cells, where the activity of PAP I depends on a complex machinery that includes a poly(A) binding protein,40 we have demonstrated that the Hfq protein, which exhibits a very high affinity for A-rich sequences, stimulates the polyadenylation activity in E. coli.36,41-44 In contrast, another poly(A) binding protein, ribosomal protein S1, has no effect on this reaction.45 At that time, Hfq was known as a cellular factor involved in the replication of a viral RNA.46 Its function in the control of gene expression was just beginning to be investigated47,48 and neither its structure nor its mode of action were elucidated. We scrutinize here how the properties of Hfq, which are now well-characterized, may account for the current understanding of poly(A) metabolism and functions.
Our knowledge of poly(A) metabolism and poly(A) dependent degradation of RNA in E. coli was gained from the study of actors of poly(A) metabolism as well as by a limited set of transcripts such as rpsO, rpsT, ompA, lpp, RNA I of ColE1 and few other regulatory RNAs where the abundance or stability was dependent on polyadenylation.2,3,9,13-16,18,49-53 We present below a rough outline of the poly(A) dependent mechanism of RNA degradation as it is known to date.
Poly(A) synthesis
The current model postulates that the poly(A) tails detected in bacteria result from the equilibrium between the activity of PAP I and the activity of 3′‒5′ exoribonucleases, which attack RNA 3′ extremities (see ref. 3 for a review). The fraction of oligoadenylated molecules range from 0.011‒40% depending on the RNA, and poly(A) tails ranging from 1‒50 nucleotides have been described.39 Ten percent of rpsO transcripts harbor short oligo(A) tails of 1–5 nucleotides in a wild-type strain.36 The slow lenghtening of poly(A) tails in vivo, which most probably results from the low intracellular concentration of PAP I, explains at least in part the limited size of oligo(A) tails.29,54
In spite of the fact that all the 3′ extremities may be recognized and elongated by PAP I, different studies reported that poly(A) tails were often detected downstream of secondary structures of Rho-independent transcription terminators, which stabilize RNAs. This is the case of RNA I, which controls ColE1 plasmid replication and of rpsO, ompA and lpp mRNAs.8,13,29,34,39 The presence of a few unpaired nucleotides downstream of the hairpin (at least two) is required for the RNA to be polyadenylated.38,55 PAP I is a distributive enzyme.56 It only polymerizes the addition of A residues in vivo but it can also incorporate C residues in vitro but with a lower efficiency. GTP and UTP are not PAP I substrates.44
Exoribonucleases that degrade RNA from the 3′ end recognize single-stranded extremities
It was demonstrated both in vivo and in vitro that oligo(A) tails are efficiently recognized and degraded by the major exoribonucleases, which are implicated in RNA catabolism; these include RNase II, PNPase and RNase R.49,57-61 Oligo(A) tails are longer in cells when PNPase and RNase II are inactive.8,57,62-65 Moreover, efficient degradation of RNA by these three exoribonucleases can only occur if 3′ terminal stable secondary structures are followed by single-stranded sequences, which are used as recognition sites. PNPase degrades transcripts harboring a Rho-independent transcription terminator provided it exhibits 10-12 3′ terminal unpaired nucleotides.66 Secondary structures impeding PNPase progression are more efficiently degraded when the ribonuclease is associated with the RhlB helicase within the degradosome complex.59,67 In contrast, RNase II, which is the most active ribonuclease in the cell as evaluated by its ability to degrade oligo(A)68 is unable to go through secondary structures.66,69,70 It nibbles single-stranded stretches longer than 9–10 nucleotides.54,66 Finally, RNase R, the less abundant of these three exoribonucleases, can degrade structured RNA harboring a 3′ single-stranded extension with a minimum of seven nucleotides without being slowed down by annealed nucleotides.71,72 These properties allow us to understand on the one hand why oligo(A) tail addition by PAP I downstream of terminator hairpins allow PNPase and RNase R to degrade folded transcripts and on the other hand why RNase II, which shortens poly(A) tails, protects RNAs against the attack of both PNPase and RNase R.16,64,73,74 We also have observed that primary transcripts harboring Rho-independent terminators may be nibbled by RNase II, which reduces the number of unpaired uridine residues downstream of the 3′ hairpin of the rpsO mRNA.36,54,75 These extremities may then be readenylated by PAP I. Exoribonucleases and PAP I can act distributively or processively depending on the length of the oligo(A) extension. The distributive synthesis of short tails, namely the dissociation of the enzyme from oligo(A) after the addition of each nucleotide, implies that the adenyl residues that have just been polymerized can be immediately removed by exoribonucleases that are able to bind to the accessible 3′ RNA extremities. In addition, in vitro experiments reveal that longer oligo(A) tails (more than 10 As downstream of the terminal Us) are very rapidly degraded processively by exoribonucleases while short oligo(A) tails (1‒5 As) are degraded slowly and through a distributive reaction.54,75 As a consequence, short oligo tails (1–5 As downstream of the Us) result from the dynamic equilibrium mediated by distributive enzymes while longer oligo(A) extensions are hydrolysed processively and more rapidly. We also have demonstrated that oligo(A) tail synthesis also becomes progressively processive in the presence of Hfq.
Hfq stimulates poly(A) synthesis
Oligo(A) tails are shorter and less abundant in cells deficient for Hfq.36,39,43 Consistently, Hfq stimulates poly(A) synthesis by PAP I in vitro.39,43,76 Hfq affects the synthesis rate when tails reach about 20 nucleotides in length. The elongation is slightly quicker at first becoming strongly stimulated when tails reach about 30–35 nucleotides. The reaction becomes processive in the presence of Hfq; processivity can be detected as soon as tails reach about 5–10 As. Tails of several hundred As are rapidly synthesized processively in vitro in the presence of Hfq. Preferential binding of Hfq to 3′ oligo(A) extensions presumably accounts for the speed and the processivity of the reaction.43,44 The correlation between the low affinity for poly(C) and the failure to stimulate PAP I-mediated poly(C) synthesis reinforces the idea that Hfq must bind at the 3′ end of RNA in order to stimulate elongation by PAP I.44 Moreover, the fact that Hfq does not affect ADP polymerization by PNPase implies that it does not impair access to the 3′OH extremity of oligo(A) tails. The many poly(A) Hfq complexes detected when protein concentration increases suggest that Hfq sequentially binds every 14 residues to form complexes looking like perls on a string.42,75 Although, Hfq was reported to bind preferentially to pre-existing oligo(A)-Hfq complexes,77 the cooperativity of Hfq binding remains controversial.42,75 Besides the evidence presented above that Hfq stimulates poly(A) synthesis, there are some clues that its binding at the 3′ end of non-adenylated RNAs terminated by Rho-independent terminators impairs polyadenylation.
Hfq recognizes the 3′ ends of Rho-independent terminators
Hfq was indeed reported to bind Rho-independent terminators harboring a 3′ stretch of Us; Hfq affinity decreases when the stretch of Us downstream of the terminal hairpin is shortened.39 Moreover, locations of polyadenylation sites in vivo suggests that 3′ ends of the rpsO mRNA are more efficently adenylated when Hfq is inactive thereby indicating that Hfq inhibits poly(A) synthesis downstream of the hairpin of the Rho-independent terminators.36 In the case of the rpsO transcript, oligo(A) tails appended 3′ to the terminator hairpin increase Hfq binding, which is 15 times more efficient when 18 As are added downsteam of the U6C-OH of the terminator.44 These observations suggest that Hfq can bind both the 3′ terminal oligo(A) of polyadenylated RNAs and the stretch of Us following the terminal hairpin of Rho-independent terminators. Intriguingly, they also indicate that the consequences of Hfq binding are different: polyadenylation is stimulated in the first case while elongation of the RNA is inhibited in the latter one. It must be pointed out here that enhanced polyadenylation due to a 5′ terminal mono-P extremity37,44 does not result from Hfq binding, which is not affected by the phosphorylation status of the 5′ RNA extremity.44
Hfq has various effects on RNA stability
These investigations suggest that Hfq could either destabilize RNA when it stimulates synthesis of poly(A) tails or protect RNA from exoribonucleases when it binds at their 3′ end.78 It was in fact observed that RNAs whose stability depends upon polyadenylation are stabilized when Hfq is inactive; this is the case of the rpsO, rpsT, ompA and lpp transcripts.39,43 However, this stabilizing effect is pretty weak, and it has been attributed to translation activation as respect to the rpsO and rpsT mRNAs.79 In addition, the stabilizing effect was not observed in the case of RNA I of ColE1 plasmids, whose decay is dramatically dependent on polyadenylation.13 While poly(A) tail lengths decrease slighly, RNA I stability is not modified in the Hfq mutant and the copy number of pBR322 is not affected (Hajnsdorf unpublished data). In contrast, many sRNAs, also terminated by a Rho-independent transcription terminators, are stabilized by Hfq, which prevents ribonuclease attacks. Several sRNAs that were efficiently degraded exonucleolytically by PNPase in the absence of Hfq become resistant to this nuclease when Hfq is present.80 For example, the MicA sRNA, whose degradation by PNPase is facilitated by poly(A), is stabilized by Hfq, which, in this case, protects the RNAs against ribonucleases instead of stimulating synthesis of destabilizing tails.18,80,81 Similarly, the SraL sRNA (also named RyjA), whose degradation is poly(A) dependent, is stabilized by Hfq.18 Paradoxically, SraL does not co-immunoprecipitate with Hfq,82 which contrasts with the majority of sRNAs.82-86 One of them, SgrS, forms a stable complex with Hfq, which presumably prevents exonucelolytic degradation.87 Hfq also protects the oligo(A) tails of rpsO transcript from degradation by PNPase and RNase II.75 Moreover, Hfq also impairs endonucleolytic cleavages. Its binding just upstream of the 3′ terminal hairpins protects the rpsO and cspA transcripts from cleavage by RNase E.75,88 In the case of rps0, this cleavage is rate-limiting for decay.89 Hfq also protects several sRNAs from RNase E.18,90 In contrast, polyadenylation does not affect the stability of the CsrC sRNA, which is mostly degraded by endonucleases.18 CsrC does not form a stable complex with Hfq,85 which does not affect its stability.18 This series of observations highlight the different roles played by Hfq in RNA metabolism. It forms very strong complexes with most sRNAs and some mRNAs while some fail to be co-immunoprecipitated with Hfq.83-85 In respect to RNA stability, it can either destabilize RNA fragments and some mRNAs probably through stimulation of poly(A) synthesis,3 or have a stabilizing effect that presumably results from the formation of complexes, which may reduce the access of ribonucleases to the RNA.90 In contrast, Hfq also facilitates the coordinated degradation of sRNAs and of their associated mRNA targets.90-92 In an effort to rationalize these observations, we postulate below that the accessibility of RNA to the exonucleolytic degradation machinery depends, at least in part, on the interactions of Hfq with the structural features of Rho-independent terminators, which have been recently demonstrated to play a major part in Hfq-dependent sRNA-mediated regulation of gene expression.
Molecular and functional interactions of Hfq with the 3′ end of RNAs
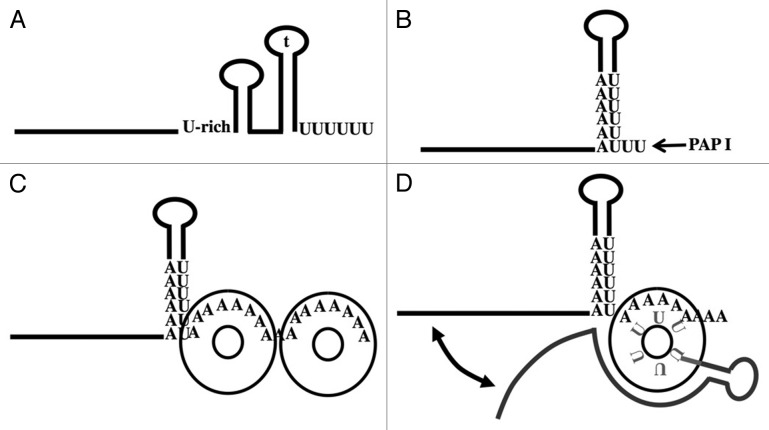
Recent data showed that the binding of Hfq to the 3′ terminal structural features resulting from Rho-independent termination is required for the regulatory function of sRNA78,87 (Fig. 1A). In addition to the 3′ terminal U-stretch and the stable hairpin of the terminator (t on Fig. 1A), formation of a stable Hfq-RNA complex requires a second single-stranded U-rich sequence located upstream of the terminator and, in several cases, a second hairpin between this U-rich sequence and the terminator. Moreover, current models based on the solved crystal structure of Hfq-RNA complexes and RNA-binding properties of Hfq mutants postulate that Hfq exhibits three different RNA-binding surfaces that can interact with the 3′ ends of polyadenylated and non-adenylated Rho-independent terminators.77,93-95 The proximal domain, located on the internal rim of the ring formed by the six identical protomers, establishes strong interactions with the 3′ terminal U-stretch of Rho-independent terminators. Importantly, the 3′-OH terminus is masked in this complex.95 The distal site, located at the other face of the ring, recognizes repetitions of A-R-N triplets (for Adenine, Purine and A, U, G or C), and it strongly binds a A15 oligoribonucleotide.93,94 Finally, the lateral site located on the outer rim of the ring is involved in the binding of U-rich sequences and base-paired elements of sRNAs.77
Figure 1. Structural features and potential interactions at the 3′ end of adenylated RNAs terminated by Rho-independent terminators. (A) The four structural elements involved in Hfq binding87 are described in the text. (B) RNAs that are polyadenylated by PAP I contain an oligo(A) or an A-rich sequence located upstream of the hairpin of the terminator which can hybridize with the 3′ terminal single standed U-stretch recognized by Hfq. (C) Processive elongation of oligo(A) tails in the presence of Hfq presumably generates stable oligo(A)-Hfq complexes. Several Hfq probably bind side by side on the oligo(A) tail. The number of As interacting with the distal RNA-binding surface of Hfq on the scheme is arbitrary. See the text for the description of this hypothetical complex. (D) The 3′ end of polyadenylated RNA can in principle also bind an Hfq-sRNA complex. The terminal U-stretch of the sRNA and the oligo(A) tail can bind simultaneously to the proximal and the distal-binding surfaces located on the opposite faces of the hexamer. The U-stretch hidden by the protein is in light gray. The sRNA shown in dark gray also interacts with the lateral surface of Hfq. There are indications that a sRNA and a mRNA can compete for binding at the same site.114 We propose in the text that such complexes may favor the interaction of the sRNA with potential target sites located in 3′ UTR close to the terminator.
PAP I selectively adenylates RNA extremities that are not recognized by Hfq
It was noticed that sRNAs that do not co-immunoprecipitate with Hfq lack the 3′ terminal Hfq-binding site depicted above.95 The same holds true for the 3′ regions of sRNAs and mRNAs, which were shown to be polyadenylated in vivo. In effect, the polyadenylated SraG and SraL sRNAs, which were not isolated as stable Hfq-RNA complexes, miss the U-rich sequence upstream of the Rho-independent terminator hairpin (Table 1).19,85,95 In addition, their 3′ terminal U-stretch is presumably masked because it anneals with the stretch of As laying immediately upstream of the terminator hairpin.95 Similarly, other sRNAs (RNA I, Oop, CsrC, RNA OUT, Sok, CopA)15,16,19,73,96,97 and mRNAs, such as rpsO, lpp, rpsT and ompA39,62,98,99 that are terminated by Rho-independent terminators, can be polyadenylated in vivo probably because they lack accessible structural features required for binding of Hfq at the 3′ end (Table 1). The strong affinity of Hfq for polyadenylated rpsO mRNA compared with the non-adenytated transcript75 indeed confirms that the 3′ terminal U-stretch is weakly bound by Hfq. The cspA mRNA whose 3′ extremity can both be polyadenylated and form a stable Hfq-RNA complex confirms that Hfq binding and polyadenylation are not exclusive.76 Interestingly, internal base-pairing masks the U-rich singled-standed stretches of nucleotides located in the 3′ UTR of the cspA transcripts (Table 1).88 The 3′ terminal Us could either form a stable complex with Hfq, implying also an upstream U-rich region when they are both single-stranded, or be accessible and polyadenylated when the 3′ UTR is folded. Similarly, melting of the A-U base pairs that mask the terminal U-stretch of the rpsO Rho-independent terminator may facilitate the binding of Hfq at the 3′end of the mRNA (Table 1). This could explain why polyadenylation of the rpsO mRNA is affected by Hfq in vivo.36 The 3′ region of some mRNAs, e.g., flgL of Salmonella typhimurium that exhibits a typical potential Hfq binding site, co-immunoprecipitate with Hfq (Table1).85 In the case of the dapB gene of Salmonella typhimurium co-immunoprecipitation of the 3′ region of the mRNA with Hfq reflects the synthesis of the DapK sRNA from a promoter located at the end of the dapB coding sequence.84 A similar origin was proposed for the RyeF sRNA in E. coli and S. typhimurium.83,84 These data indicate that Hfq interacts in the same way with the 3′ ends of mRNAs and sRNAs. Indeed, the 3′ end of the cspA mRNA forms a complex with the proximal RNA-binding surface of Hfq, which also interacts with the 3′ terminal U-stretch of the RybB sARN.76,95 Consistently, a large part (35%) of the predicted Rho-independent terminators of Salmonella co-immunoprecipitate with Hfq.84 The RybD sRNA characterized in E. coli and S. typhimurium presumably belongs to this category.83,84 The observations above suggest that the 3′ends of RNAs resulting from Rho-independent transcription termination affect the fate of RNA that can either form a stable functional complex with Hfq or be attacked by the oligo(A)-dependent exonucleolytic degradation machinery. A speculative model taking in account the molecular events occurring at the 3′ ends of these transcripts is presented below.
Table 1. The 3′ends of polyadenylated RNAs.
| Polyadenylated mRNAs |
|---|
| rpsO—UAAUUUCUUGCGAGUUUCAGAAAAGGGGCCUGAGUGGCCCCUUUUUUC |
| cspA—UAAUCUCUGCUUAAAAGCACAGAAUCUAAGAUCCCUGCCAUUUGGCGGGGAUUUUUUU |
| lpp—UAAUAGUACCUGUGAAGUGAAAAAUGGCGCACAUUGUGCGCCAUUUUUUUU |
| ompA—UAAGUUCUCGUCUGGUAGAAAAACCCCGCUGCUGCGGGGUUUUUUUU |
| rpsT—UCAACAAACUGGCUUAAUCGCCAAUUUGCUGAAGCUUUGUGAAAAAGCCCGCG |
| CAAGCGGGUUUUUUU |
| Polyadenylated sRNAs |
|---|
| RNA 1 pAUUUGGUAUCUGCGCUCUGCUGAAGCCAGUUACCUUCGGAAAAAGAGUUGGUAG |
| CUCUUGAUCCGGCAAACAAACCACCGCUGGUAGCGGUGGUUUUUUUGUU |
| CopA pUUUAAGUGGGCCCCGGUAAUCUUUUCGUACUCGCCAAAGUUGAAGAAGAUUAUC |
| GGGGUUUUUGCUU |
| Oop pppGUUGAUAGAUCCAGUAAUGACCUCAGAACUCCAUCUGGAUUUGUUC |
| AGAACGCUCGGUUGCCGCCGGGCGUUUUUUA |
| Sok pppGACUAGACAUAGGGAUGCCUCGUGGUGGUUAAUGAAAAUUAACUUACUACGG |
| GGCUAUUUCCUU |
| RNA OUT |
|---|
| pppUCGCACAUCUUGUUGUCUGAUUAUUGAUUUUUCGCGAAACCAUUUGAUCAUA |
| UGACAAGAUGUGUAUCC |
| SraL—GAUAGAGAGAAAGACAAAGACCGGAAAACAAACUAAAGCGCCCUUGUGGCGCUUUAGUUU |
| SraG— AUUAGUUUCCAGUGAUUGCUGCCGUCAGCUUGAAAAAAGGGGCCACUCAGGCCCCCUUUUCU |
| CsrC— CCCGUUAAGGGUUAAGAGUCAGGAAAAAAGGCGACAGAGUAAUCUGUCGCCUUUUUUCUU |
| GlmY—GCUUAUUCCAUAACAAAGCCGGGUAAUUCCCGGCUUUGUU |
| Hfq complexes |
|---|
| SgrS—GUAUUGGUGUAAAAUCACCCGCCAGCAGAUUAUACCUGCUGGUUUUUUUU |
| flgL—UAACGCCUCUUUUUGAAACAUAUCACGAAACUGGAUAUGUUUUGUCUGCCCGCGCC |
| AUCCACCCCGGCGCGGGCAUUUUUUUA |
Sequence data are from references quoted in the text and from databases. SgrS and flgL are taken as examples of an sRNA and mRNA-forming stable complexes with Hfq.85 The 3′ terminal oligo (U) sequences and the complementary stretches of As are underlined. Secondary structures are shaded un gray (light gray for the terminator and dark gray for the upstream hairpin). Termination codons of mRNAs are in bold, ppp and p at the 5′ ends means that the full-length or a processed sRNA are shown, respectively.
The role of Hfq in RNA polyadenylation: A mechanistic speculation
RNAs terminated by Rho-independent terminators harboring the structural motifs required for Hfq binding probably rapidly establish strong interactions with the proximal and lateral sites of Hfq.77 In the case of sRNA, these complexes probably facilitate annealing of the seeding sequence with the complementarity sequence of the target mRNA, which may be made accessible through an association with the distal sites on the other face of Hfq.100,101 Similarly, mRNAs resulting from Rho-independent termination, whose 3′ ends associate with the proximal site of Hfq, are presumably not accessible to exoribonucleases and PAP I.77 Some of these stabilized 3′ terminal mRNA fragments are probably regulatory RNAs as proposed for RybD.84 In contrast, 3′ fragments of mRNAs generated by endonucleases that cannot be bound by Hfq are presumably marked for degradation. This is the case of rpsO, rpsT, lpp, ompA mRNAs and of the SraL,18 SraG,19 RNA I,13 Oop,15 Sok,16 RNA OUT73 and CopA14 sRNAs that are all polyadenylated and mostly degraded by a oligo(A)-dependent exonucleolytic process (see above). It is interesting to point out that most polyadenylated sRNA mentioned above (RNA I, RNA OUT, Oop, CopA and Sok) are cis-acting regulators that do not require Hfq for annealing the complementary target transcribed from the opposite DNA strand. Polyadenylation is one of the parameters that control their activity. Biological activity of the GlmY sRNA also depends on polyadenylation, which controls its stability.17 Consistent with the hypothesis above, GlmY miss a strong putative 3′ terminal Hfq-binding site (Table 1).95 However, in this case, polyadenylation takes place at an endonucleolytic processing site just upstream of the terminator.
It is reasonable to assume that oligo(A) tails less than 10 As in length, which are too short to form stable complexes with Hfq,42,77 are nevertheless long enough to be used as “toe-holds” by exoribonucleases able to carry out the degradation of structured RNAs (see above). Because PNPase dissociates when it encounters secondary structures the current model postulates that the oligo(A) “toe-holds” are repetitively degraded and resynthesized until the base-paired nucleotides are removed.7,52,66 RNase R is also acting in the oligo(A)-dependent degradation of the rpsO mRNA and several structured RNA fragments.57,58 In contrast, RNase II activity, which overpasses PNPase and RNase R activities in the cell,68 efficiently prevents oligo(A) tail extension.54,66 Thereby, it presumably moderates the amount of RNA that has to be degraded by the poly(A) dependent machinery of degradation and prevents the formation of oligo(A) tails long enough for Hfq binding.52,61,64 However, the growth-rate could stimulate their appearance; it has been reported that PAP I expression, as well as Oop RNA and lpp mRNA polyadenylation, increase when the growth rate slows down.102
The impact of these long tails on RNA metabolism remains mysterious. One can imagine that Hfq, which stimulates poly(A) synthesis and strongly binds tails that it protects against ribonucleases, plays a major part in poly(A) metabolism. Oligo(A) tails longer than 14 As most likely form stable complexes with Hfq.42,44,75,77,93,94 The interaction would involve the distal face of Hfq that is capable of interacting with consecutive “A-R-N” triplets. Several Hfq molecules presumably bind side by side on long poly(A) (Fig. 1B) so that four protomers of each Hfq interacts with 12 A residues. The observation that oligo(A) tails as long as 5–10 As are sufficient to transform PAP I into a processive enzyme suggests that the interaction of two triplets of the oligo(A) tail with two Hfq protomers is sufficient to impact polyadenylation activity.43 That polymerization progressively becomes processive upon oligo(A) elongation supports the idea that the fixation of several Hfq molecules facilitate PAP I access to the 3′ RNA extremity. How Hfq stimulates PAP I activity is unknown. An interaction between PAP I and Hfq has been reported,39 suggesting that Hfq may act by increasing the local concentration of PAP I. Such a complex could also account for the processivity of the reaction. Polynucleotide phosphorylase could associate with this complex and as a consequence coordinate degradation and RNA elongation.39 It is worth remembering here that PNPase was proposed to synthesize long heterogeneous tails at the 3′ ends of transcripts.103 In contrast to short tails used as “toe-holds” by exoribonucleases, long oligo(A) tails are strong Hfq-binding sites that likely protect RNAs against exonucleolytic degradation.75 In the case of rpsO mRNAs, Hfq could bind simultaneously to the oligo(A) tail via its distal site and the U-rich sequence recognized by RNase E just upstream of the terminator hairpin through its lateral surface.75,77 Such an interaction could explain why Hfq protects the transcripts from RNase E processing in vitro. Two Hfq hexamers bound independently on both sides of the hairpin would also protect the RNA from endo- and exoribonuclease attacks.75 It is worth noting here that simultaneous interaction of Hfq with the 3′ extremity and an internal segment of the Qß RNA has been reported.104,105 It is also appealing to speculate that Hfq bound to oligo(A) tails still has accessible proximal and lateral RNA-binding surfaces that can be occupied by sRNAs (Fig. 1D). Such oligo(A) tail-Hfq-sRNA complexes may favor sRNA annealing with complementary sequences located on the oligoadenylated mRNA.101,106 The locations of facilitating oligo(A) sequences in the vicinity of sRNA target sites on the mRNA suggests that oligo(A) tails may contribute to the recognition of hypothetical sRNA target sites located at the 3′ end of the mRNA. Moreover, as demonstrated for sRNA-mRNA, interactions occurring upstream or in coding sequences formation of such Hfq-sRNA-mRNA complexes may end up in RNase E cleavage.91,92,107 However, there is no indications that the RNase E cleavage rate limiting for decay of the rpsO mRNA, occurring just upstream of the Rho-independent terminator, is activated by Hfq in vivo. In contrast, the fact that poly(A) dependent decay is only operational when RNase E is inactive suggests that a direct, or sRNA-mediated, RNase E-Hfq complex interacting with the 3′ extremity of the mRNA could switch off poly(A) dependent decay of the rpsO mRNA and account for the coordinated activities of the RNase E and poly(A) dependent decay pathways.24,108,109 One can also imagine that oligo(A) tails would also compete with the repetition of “A-R-N” motifs for binding to the distal RNA-binding surface of Hfq.87,100,106 Indeed, it is now recognized that the intracellular concentration of Hfq is limiting in respect to the concentration of its potential partners. An exchange mechanism based on the progressive binding of competing RNAs to the multiple sites of the RNA-binding surfaces of Hfq has been proposed to explain the paradoxical rapid dissociation of strong Hfq-RNA complexes in the presence of a competing RNA.110-113 Hfq-sRNA complexes could interact with oligo(A) tails in the absence of target mRNAs. This might be a means used by the cell to store sRNAs that could then be rapidly delivered to their cognate mRNA. Finally, one could also speculate that long tails bound by Hfq are synthesized when free Hfq or unpartnered Hfq-sRNA complexes, harboring an unoccupied distal site that is able to bind oligo(A) sequences are present in the cell. Under these conditions, short tails contributing to the degradation of tightly folded 3′ RNA extremities might be processively elongated and generate Hfq docking sites that are degraded when Hfq becomes limiting.
Acknowledgments
This work was supported by the Centre National de la Recherche scientifique (UPR9073), University Paris-Diderot and Agence Nationale de la Recherche: asSUPYCO (ANR-12-BSV6-0007-03). Kyle Tanner is acknowledged for reading the manuscript.
Glossary
- Abbrevations: PAP I
Poly(A) polymerase
- PNPase
Polynucleotide phosphorylase
- RNase II
Ribonuclease II
- RNase R
Ribonuclease R
- sRNA
small regulatory RNA
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/23664
References
- 1.Edmonds M. A history of poly A sequences: from formation to factors to function. Prog Nucleic Acid Res Mol Biol. 2002;71:285–389. doi: 10.1016/S0079-6603(02)71046-5. [DOI] [PubMed] [Google Scholar]
- 2.Sarkar N. Polyadenylation of mRNA in prokaryotes. Annu Rev Biochem. 1997;66:173–97. doi: 10.1146/annurev.biochem.66.1.173. [DOI] [PubMed] [Google Scholar]
- 3.Régnier P, Hajnsdorf E. Poly(A)-assisted RNA decay and modulators of RNA stability. Prog Mol Biol Transl Sci. 2009;85:137–85. doi: 10.1016/S0079-6603(08)00804-0. [DOI] [PubMed] [Google Scholar]
- 4.Schuster G, Stern D. RNA polyadenylation and decay in mitochondria and chloroplasts. Prog Mol Biol Transl Sci. 2009;85:393–422. doi: 10.1016/S0079-6603(08)00810-6. [DOI] [PubMed] [Google Scholar]
- 5.Wyers F, Rougemaille M, Badis G, Rousselle JC, Dufour ME, Boulay J, et al. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell. 2005;121:725–37. doi: 10.1016/j.cell.2005.04.030. [DOI] [PubMed] [Google Scholar]
- 6.Kadaba S, Krueger A, Trice T, Krecic AM, Hinnebusch AG, Anderson J. Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes Dev. 2004;18:1227–40. doi: 10.1101/gad.1183804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dreyfus M, Régnier P. The poly(A) tail of mRNAs: bodyguard in eukaryotes, scavenger in bacteria. Cell. 2002;111:611–3. doi: 10.1016/S0092-8674(02)01137-6. [DOI] [PubMed] [Google Scholar]
- 8.Hajnsdorf E, Braun F, Haugel-Nielsen J, Régnier P. Polyadenylylation destabilizes the rpsO mRNA of Escherichia coli. Proc Natl Acad Sci USA. 1995;92:3973–7. doi: 10.1073/pnas.92.9.3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carpousis AJ, Vanzo NF, Raynal LC. mRNA degradation. A tale of poly(A) and multiprotein machines. Trends Genet. 1999;15:24–8. doi: 10.1016/S0168-9525(98)01627-8. [DOI] [PubMed] [Google Scholar]
- 10.Lange H, Sement FM, Canaday J, Gagliardi D. Polyadenylation-assisted RNA degradation processes in plants. Trends Plant Sci. 2009;14:497–504. doi: 10.1016/j.tplants.2009.06.007. [DOI] [PubMed] [Google Scholar]
- 11.Doma MK, Parker R. RNA quality control in eukaryotes. Cell. 2007;131:660–8. doi: 10.1016/j.cell.2007.10.041. [DOI] [PubMed] [Google Scholar]
- 12.Houseley J, Tollervey D. The nuclear RNA surveillance machinery: the link between ncRNAs and genome structure in budding yeast? Biochim Biophys Acta. 2008;1779:239–46. doi: 10.1016/j.bbagrm.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 13.Xu F, Lin-Chao S, Cohen SN. The Escherichia coli pcnB gene promotes adenylylation of antisense RNAI of ColE1-type plasmids in vivo and degradation of RNAI decay intermediates. Proc Natl Acad Sci USA. 1993;90:6756–60. doi: 10.1073/pnas.90.14.6756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Söderbom F, Binnie U, Masters M, Wagner EGH. Regulation of plasmid R1 replication: PcnB and RNase E expedite the decay of the antisense RNA, CopA. Mol Microbiol. 1997;26:493–504. doi: 10.1046/j.1365-2958.1997.5871953.x. [DOI] [PubMed] [Google Scholar]
- 15.Szalewska-Pałasz A, Wróbel B, Wegrzyn G. Rapid degradation of polyadenylated oop RNA. FEBS Lett. 1998;432:70–2. doi: 10.1016/S0014-5793(98)00834-5. [DOI] [PubMed] [Google Scholar]
- 16.Dam Mikkelsen N, Gerdes K. Sok antisense RNA from plasmid R1 is functionally inactivated by RNase E and polyadenylated by poly(A) polymerase I. Mol Microbiol. 1997;26:311–20. doi: 10.1046/j.1365-2958.1997.5751936.x. [DOI] [PubMed] [Google Scholar]
- 17.Reichenbach B, Maes A, Kalamorz F, Hajnsdorf E, Görke B. The small RNA GlmY acts upstream of the sRNA GlmZ in the activation of glmS expression and is subject to regulation by polyadenylation in Escherichia coli. Nucleic Acids Res. 2008;36:2570–80. doi: 10.1093/nar/gkn091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Viegas SC, Pfeiffer V, Sittka A, Silva IJ, Vogel J, Arraiano CM. Characterization of the role of ribonucleases in Salmonella small RNA decay. Nucleic Acids Res. 2007;35:7651–64. doi: 10.1093/nar/gkm916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Argaman L, Hershberg R, Vogel J, Bejerano G, Wagner EGH, Margalit H, et al. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr Biol. 2001;11:941–50. doi: 10.1016/S0960-9822(01)00270-6. [DOI] [PubMed] [Google Scholar]
- 20.Urban JH, Vogel J. Two seemingly homologous noncoding RNAs act hierarchically to activate glmS mRNA translation. PLoS Biol. 2008;6:e64. doi: 10.1371/journal.pbio.0060064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Z, Reimers S, Pandit S, Deutscher MP. RNA quality control: degradation of defective transfer RNA. EMBO J. 2002;21:1132–8. doi: 10.1093/emboj/21.5.1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Joanny G, Le Derout J, Bréchemier-Baey D, Labas V, Vinh J, Régnier P, et al. Polyadenylation of a functional mRNA controls gene expression in Escherichia coli. Nucleic Acids Res. 2007;35:2494–502. doi: 10.1093/nar/gkm120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hajnsdorf E, Braun F, Haugel-Nielsen J, Le Derout J, Régnier P. Multiple degradation pathways of the rpsO mRNA of Escherichia coli. RNase E interacts with the 5′ and 3′ extremities of the primary transcript. Biochimie. 1996;78:416–24. doi: 10.1016/0300-9084(96)84748-1. [DOI] [PubMed] [Google Scholar]
- 24.Marujo PE, Braun F, Haugel-Nielsen J, Le Derout J, Arraiano CM, Régnier P. Inactivation of the decay pathway initiated at an internal site by RNase E promotes poly(A)-dependent degradation of the rpsO mRNA in Escherichia coli. Mol Microbiol. 2003;50:1283–94. doi: 10.1046/j.1365-2958.2003.03753.x. [DOI] [PubMed] [Google Scholar]
- 25.Maes A, Gracia C, Hajnsdorf E, Régnier P. Search for poly(A) polymerase targets in E. coli reveals its implication in surveillance of Glu tRNA processing and degradation of stable RNAs. Mol Microbiol. 2012;83:436–51. doi: 10.1111/j.1365-2958.2011.07943.x. [DOI] [PubMed] [Google Scholar]
- 26.Cao G-J, Sarkar N. Poly(A) RNA in Escherichia coli: nucleotide sequence at the junction of the lpp transcript and the polyadenylate moiety. Proc Natl Acad Sci USA. 1992;89:7546–50. doi: 10.1073/pnas.89.16.7546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson MD, Popowski J, Cao G-J, Shen P, Sarkar N. Bacteriophage T7 mRNA is polyadenylated. Mol Microbiol. 1998;27:23–30. doi: 10.1046/j.1365-2958.1998.00649.x. [DOI] [PubMed] [Google Scholar]
- 28.Li Z, Pandit S, Deutscher MP. Polyadenylation of stable RNA precursors in vivo. Proc Natl Acad Sci USA. 1998;95:12158–62. doi: 10.1073/pnas.95.21.12158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mohanty BK, Kushner SR. The majority of Escherichia coli mRNAs undergo post-transcriptional modification in exponentially growing cells. Nucleic Acids Res. 2006;34:5695–704. doi: 10.1093/nar/gkl684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Briani F, Del Vecchio E, Migliorini D, Hajnsdorf E, Régnier P, Ghisotti D, et al. RNase E and polyadenyl polymerase I are involved in maturation of CI RNA, the P4 phage immunity factor. J Mol Biol. 2002;318:321–31. doi: 10.1016/S0022-2836(02)00085-2. [DOI] [PubMed] [Google Scholar]
- 31.van Meerten D, Zelwer M, Régnier P, Duin J. In vivo oligo(A) insertions in phage MS2: role of Escherichia coli poly(A) polymerase. Nucleic Acids Res. 1999;27:3891–8. doi: 10.1093/nar/27.19.3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Z, Pandit S, Deutscher MP. Maturation of 23S ribosomal RNA requires the exoribonuclease RNase T. RNA. 1999;5:139–46. doi: 10.1017/S1355838299981669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taljanidisz J, Karnik P, Sarkar N. Messenger ribonucleic acid for the lipoprotein of the Escherichia coli outer membrane is polyadenylated. J Mol Biol. 1987;193:507–15. doi: 10.1016/0022-2836(87)90263-4. [DOI] [PubMed] [Google Scholar]
- 34.Haugel-Nielsen J, Hajnsdorf E, Regnier P. The rpsO mRNA of Escherichia coli is polyadenylated at multiple sites resulting from endonucleolytic processing and exonucleolytic degradation. EMBO J. 1996;15:3144–52. [PMC free article] [PubMed] [Google Scholar]
- 35.Hajnsdorf E, Régnier P. E. coli RpsO mRNA decay: RNase E processing at the beginning of the coding sequence stimulates poly(A)-dependent degradation of the mRNA. J Mol Biol. 1999;286:1033–43. doi: 10.1006/jmbi.1999.2547. [DOI] [PubMed] [Google Scholar]
- 36.Le Derout J, Folichon M, Briani F, Dehò G, Régnier P, Hajnsdorf E. Hfq affects the length and the frequency of short oligo(A) tails at the 3′ end of Escherichia coli rpsO mRNAs. Nucleic Acids Res. 2003;31:4017–23. doi: 10.1093/nar/gkg456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Feng Y, Cohen SN. Unpaired terminal nucleotides and 5′ monophosphorylation govern 3′ polyadenylation by Escherichia coli poly(A) polymerase I. Proc Natl Acad Sci USA. 2000;97:6415–20. doi: 10.1073/pnas.120173797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yehudai-Resheff S, Schuster G. Characterization of the E.coli poly(A) polymerase: nucleotide specificity, RNA-binding affinities and RNA structure dependence. Nucleic Acids Res. 2000;28:1139–44. doi: 10.1093/nar/28.5.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mohanty BK, Maples VF, Kushner SR. The Sm-like protein Hfq regulates polyadenylation dependent mRNA decay in Escherichia coli. Mol Microbiol. 2004;54:905–20. doi: 10.1111/j.1365-2958.2004.04337.x. [DOI] [PubMed] [Google Scholar]
- 40.Wahle E, Keller W. The biochemistry of polyadenylation. Trends Biochem Sci. 1996;21:247–50. [PubMed] [Google Scholar]
- 41.Carmichael GG, Weber K, Niveleau A, Wahba AJ. The host factor required for RNA phage Qbeta RNA replication in vitro. Intracellular location, quantitation, and purification by polyadenylate-cellulose chromatography. J Biol Chem. 1975;250:3607–12. [PubMed] [Google Scholar]
- 42.de Haseth PL, Uhlenbeck OC. Interaction of Escherichia coli host factor protein with oligoriboadenylates. Biochemistry. 1980;19:6138–46. doi: 10.1021/bi00567a029. [DOI] [PubMed] [Google Scholar]
- 43.Hajnsdorf E, Régnier P. Host factor Hfq of Escherichia coli stimulates elongation of poly(A) tails by poly(A) polymerase I. Proc Natl Acad Sci USA. 2000;97:1501–5. doi: 10.1073/pnas.040549897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Folichon M, Allemand F, Régnier P, Hajnsdorf E. Stimulation of poly(A) synthesis by Escherichia coli poly(A)polymerase I is correlated with Hfq binding to poly(A) tails. FEBS J. 2005;272:454–63. doi: 10.1111/j.1742-4658.2004.04485.x. [DOI] [PubMed] [Google Scholar]
- 45.Hajnsdorf E, Boni IV. Multiple activities of RNA-binding proteins S1 and Hfq. Biochimie. 2012;94:1544–53. doi: 10.1016/j.biochi.2012.02.010. [DOI] [PubMed] [Google Scholar]
- 46.Kamen R, Kondo M, Römer W, Weissmann C. Reconstitution of Q replicase lacking subunit with protein-synthesis-interference factor i. Eur J Biochem. 1972;31:44–51. doi: 10.1111/j.1432-1033.1972.tb02498.x. [DOI] [PubMed] [Google Scholar]
- 47.Tsui H-C, Leung HC, Winkler ME. Characterization of broadly pleiotropic phenotypes caused by an hfq insertion mutation in Escherichia coli K-12. Mol Microbiol. 1994;13:35–49. doi: 10.1111/j.1365-2958.1994.tb00400.x. [DOI] [PubMed] [Google Scholar]
- 48.Muffler A, Fischer D, Hengge-Aronis R. The RNA-binding protein HF-I, known as a host factor for phage Qbeta RNA replication, is essential for rpoS translation in Escherichia coli. Genes Dev. 1996;10:1143–51. doi: 10.1101/gad.10.9.1143. [DOI] [PubMed] [Google Scholar]
- 49.Régnier P, Arraiano CM. Degradation of mRNA in bacteria: emergence of ubiquitous features. Bioessays. 2000;22:235–44. doi: 10.1002/(SICI)1521-1878(200003)22:3<235::AID-BIES5>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 50.Arraiano CM, Andrade JM, Domingues S, Guinote IB, Malecki M, Matos RG, et al. The critical role of RNA processing and degradation in the control of gene expression. FEMS Microbiol Rev. 2010;34:883–923. doi: 10.1111/j.1574-6976.2010.00242.x. [DOI] [PubMed] [Google Scholar]
- 51.Mohanty BK, Kushner SR. Bacterial/archaeal/organellar polyadenylation. Wiley Interdiscip Rev RNA. 2011;2:256–76. doi: 10.1002/wrna.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Coburn GA, Mackie GA. Reconstitution of the degradation of the mRNA for ribosomal protein S20 with purified enzymes. J Mol Biol. 1998;279:1061–74. doi: 10.1006/jmbi.1998.1842. [DOI] [PubMed] [Google Scholar]
- 53.Coburn GA, Mackie GA. Degradation of mRNA in Escherichia coli: an old problem with some new twists. Prog Nucleic Acid Res Mol Biol. 1999;62:55–108. doi: 10.1016/S0079-6603(08)60505-X. [DOI] [PubMed] [Google Scholar]
- 54.Folichon M, Marujo PE, Arluison V, Le Derout J, Pellegrini O, Hajnsdorf E, et al. Fate of mRNA extremities generated by intrinsic termination: detailed analysis of reactions catalyzed by ribonuclease II and poly(A) polymerase. Biochimie. 2005;87:819–26. doi: 10.1016/j.biochi.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 55.Lisitsky I, Schuster G. Preferential degradation of polyadenylated and polyuridinylated RNAs by the bacterial exoribonuclease polynucleotide phosphorylase. Eur J Biochem. 1999;261:468–74. doi: 10.1046/j.1432-1327.1999.00285.x. [DOI] [PubMed] [Google Scholar]
- 56.Sano H, Feix G. Terminal riboadenylate transferase from Escherichia coli. Characterization and application. Eur J Biochem. 1976;71:577–83. doi: 10.1111/j.1432-1033.1976.tb11148.x. [DOI] [PubMed] [Google Scholar]
- 57.Andrade JM, Hajnsdorf E, Régnier P, Arraiano CM. The poly(A)-dependent degradation pathway of rpsO mRNA is primarily mediated by RNase R. RNA. 2009;15:316–26. doi: 10.1261/rna.1197309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cheng Z-F, Deutscher MP. An important role for RNase R in mRNA decay. Mol Cell. 2005;17:313–8. doi: 10.1016/j.molcel.2004.11.048. [DOI] [PubMed] [Google Scholar]
- 59.Blum E, Carpousis AJ, Higgins CF. Polyadenylation promotes degradation of 3′-structured RNA by the Escherichia coli mRNA degradosome in vitro. J Biol Chem. 1999;274:4009–16. doi: 10.1074/jbc.274.7.4009. [DOI] [PubMed] [Google Scholar]
- 60.Xu F, Cohen SN. RNA degradation in Escherichia coli regulated by 3′ adenylation and 5′ phosphorylation. Nature. 1995;374:180–3. doi: 10.1038/374180a0. [DOI] [PubMed] [Google Scholar]
- 61.Mohanty BK, Kushner SR. Polynucleotide phosphorylase, RNase II and RNase E play different roles in the in vivo modulation of polyadenylation in Escherichia coli. Mol Microbiol. 2000;36:982–94. doi: 10.1046/j.1365-2958.2000.01921.x. [DOI] [PubMed] [Google Scholar]
- 62.O’Hara EB, Chekanova JA, Ingle CA, Kushner ZR, Peters E, Kushner SR. Polyadenylylation helps regulate mRNA decay in Escherichia coli. Proc Natl Acad Sci USA. 1995;92:1807–11. doi: 10.1073/pnas.92.6.1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hajnsdorf E, Steier O, Coscoy L, Teysset L, Régnier P. Roles of RNase E, RNase II and PNPase in the degradation of the rpsO transcripts of Escherichia coli: stabilizing function of RNase II and evidence for efficient degradation in an ams pnp rnb mutant. EMBO J. 1994;13:3368–77. doi: 10.1002/j.1460-2075.1994.tb06639.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Marujo PE, Hajnsdorf E, Le Derout J, Andrade R, Arraiano CM, Régnier P. RNase II removes the oligo(A) tails that destabilize the rpsO mRNA of Escherichia coli. RNA. 2000;6:1185–93. doi: 10.1017/S135583820000073X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cao GJ, Kalapos MP, Sarkar N. Polyadenylated mRNA in Escherichia coli: modulation of poly(A) RNA levels by polynucleotide phosphorylase and ribonuclease II. Biochimie. 1997;79:211–20. doi: 10.1016/S0300-9084(97)83508-0. [DOI] [PubMed] [Google Scholar]
- 66.Spickler C, Mackie GA. Action of RNase II and polynucleotide phosphorylase against RNAs containing stem-loops of defined structure. J Bacteriol. 2000;182:2422–7. doi: 10.1128/JB.182.9.2422-2427.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Py B, Higgins CF, Krisch HM, Carpousis AJ. A DEAD-box RNA helicase in the Escherichia coli RNA degradosome. Nature. 1996;381:169–72. doi: 10.1038/381169a0. [DOI] [PubMed] [Google Scholar]
- 68.Deutscher MP, Reuven NB. Enzymatic basis for hydrolytic versus phosphorolytic mRNA degradation in Escherichia coli and Bacillus subtilis. Proc Natl Acad Sci USA. 1991;88:3277–80. doi: 10.1073/pnas.88.8.3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cannistraro VJ, Kennell D. The reaction mechanism of ribonuclease II and its interaction with nucleic acid secondary structures. Biochim Biophys Acta. 1999;1433:170–87. doi: 10.1016/S0167-4838(99)00136-3. [DOI] [PubMed] [Google Scholar]
- 70.Arraiano CM, Matos RG, Barbas A. RNase II: the finer details of the Modus operandi of a molecular killer. RNA Biol. 2010;7:276–81. doi: 10.4161/rna.7.3.11490. [DOI] [PubMed] [Google Scholar]
- 71.Vincent HA, Deutscher MP. Substrate recognition and catalysis by the exoribonuclease RNase R. J Biol Chem. 2006;281:29769–75. doi: 10.1074/jbc.M606744200. [DOI] [PubMed] [Google Scholar]
- 72.Cheng Z-F, Deutscher MP. Purification and characterization of the Escherichia coli exoribonuclease RNase R. Comparison with RNase II. J Biol Chem. 2002;277:21624–9. doi: 10.1074/jbc.M202942200. [DOI] [PubMed] [Google Scholar]
- 73.Pepe CM, Maslesa-Galić S, Simons RW. Decay of the IS10 antisense RNA by 3′ exoribonucleases: evidence that RNase II stabilizes RNA-OUT against PNPase attack. Mol Microbiol. 1994;13:1133–42. doi: 10.1111/j.1365-2958.1994.tb00504.x. [DOI] [PubMed] [Google Scholar]
- 74.Mohanty BK, Kushner SR. Genomic analysis in Escherichia coli demonstrates differential roles for polynucleotide phosphorylase and RNase II in mRNA abundance and decay. Mol Microbiol. 2003;50:645–58. doi: 10.1046/j.1365-2958.2003.03724.x. [DOI] [PubMed] [Google Scholar]
- 75.Folichon M, Arluison V, Pellegrini O, Huntzinger E, Régnier P, Hajnsdorf E. The poly(A) binding protein Hfq protects RNA from RNase E and exoribonucleolytic degradation. Nucleic Acids Res. 2003;31:7302–10. doi: 10.1093/nar/gkg915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hankins JS, Denroche H, Mackie GA. Interactions of the RNA-binding protein Hfq with cspA mRNA, encoding the major cold shock protein. J Bacteriol. 2010;192:2482–90. doi: 10.1128/JB.01619-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sauer E, Schmidt S, Weichenrieder O. Small RNA binding to the lateral surface of Hfq hexamers and structural rearrangements upon mRNA target recognition. Proc Natl Acad Sci USA. 2012;109:9396–401. doi: 10.1073/pnas.1202521109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Otaka H, Ishikawa H, Morita T, Aiba H. PolyU tail of rho-independent terminator of bacterial small RNAs is essential for Hfq action. Proc Natl Acad Sci USA. 2011;108:13059–64. doi: 10.1073/pnas.1107050108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Le Derout J, Boni IV, Régnier P, Hajnsdorf E. Hfq affects mRNA levels independently of degradation. BMC Mol Biol. 2010;11:17. doi: 10.1186/1471-2199-11-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Andrade JM, Pobre V, Matos AM, Arraiano CM. The crucial role of PNPase in the degradation of small RNAs that are not associated with Hfq. RNA. 2012;18:844–55. doi: 10.1261/rna.029413.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Andrade JM, Arraiano CM. PNPase is a key player in the regulation of small RNAs that control the expression of outer membrane proteins. RNA. 2008;14:543–51. doi: 10.1261/rna.683308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wassarman KM, Repoila F, Rosenow C, Storz G, Gottesman S. Identification of novel small RNAs using comparative genomics and microarrays. Genes Dev. 2001;15:1637–51. doi: 10.1101/gad.901001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang A, Wassarman KM, Rosenow C, Tjaden BC, Storz G, Gottesman S. Global analysis of small RNA and mRNA targets of Hfq. Mol Microbiol. 2003;50:1111–24. doi: 10.1046/j.1365-2958.2003.03734.x. [DOI] [PubMed] [Google Scholar]
- 84.Chao Y, Papenfort K, Reinhardt R, Sharma CM, Vogel J. An atlas of Hfq-bound transcripts reveals 3′ UTRs as a genomic reservoir of regulatory small RNAs. EMBO J. 2012;31:4005–19. doi: 10.1038/emboj.2012.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sittka A, Lucchini S, Papenfort K, Sharma CM, Rolle K, Binnewies TT, et al. Deep sequencing analysis of small noncoding RNA and mRNA targets of the global post-transcriptional regulator, Hfq. PLoS Genet. 2008;4:e1000163. doi: 10.1371/journal.pgen.1000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Vogel J, Luisi BF. Hfq and its constellation of RNA. Nat Rev Microbiol. 2011;9:578–89. doi: 10.1038/nrmicro2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ishikawa H, Otaka H, Maki K, Morita T, Aiba H. The functional Hfq-binding module of bacterial sRNAs consists of a double or single hairpin preceded by a U-rich sequence and followed by a 3′ poly(U) tail. RNA. 2012;18:1062–74. doi: 10.1261/rna.031575.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hankins JS, Zappavigna C, Prud’homme-Généreux A, Mackie GA. Role of RNA structure and susceptibility to RNase E in regulation of a cold shock mRNA, cspA mRNA. J Bacteriol. 2007;189:4353–8. doi: 10.1128/JB.00193-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Régnier P, Hajnsdorf E. Decay of mRNA encoding ribosomal protein S15 of Escherichia coli is initiated by an RNase E-dependent endonucleolytic cleavage that removes the 3′ stabilizing stem and loop structure. J Mol Biol. 1991;217:283–92. doi: 10.1016/0022-2836(91)90542-E. [DOI] [PubMed] [Google Scholar]
- 90.Massé E, Escorcia FE, Gottesman S. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev. 2003;17:2374–83. doi: 10.1101/gad.1127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Morita T, Maki K, Aiba H. RNase E-based ribonucleoprotein complexes: mechanical basis of mRNA destabilization mediated by bacterial noncoding RNAs. Genes Dev. 2005;19:2176–86. doi: 10.1101/gad.1330405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bandyra KJ, Said N, Pfeiffer V, Górna MW, Vogel J, Luisi BF. The seed region of a small RNA drives the controlled destruction of the target mRNA by the endoribonuclease RNase E. Mol Cell. 2012;47:943–53. doi: 10.1016/j.molcel.2012.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Link TM, Valentin-Hansen P, Brennan RG. Structure of Escherichia coli Hfq bound to polyriboadenylate RNA. Proc Natl Acad Sci USA. 2009;106:19292–7. doi: 10.1073/pnas.0908744106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mikulecky PJ, Kaw MK, Brescia CC, Takach JC, Sledjeski DD, Feig AL. Escherichia coli Hfq has distinct interaction surfaces for DsrA, rpoS and poly(A) RNAs. Nat Struct Mol Biol. 2004;11:1206–14. doi: 10.1038/nsmb858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sauer E, Weichenrieder O. Structural basis for RNA 3′-end recognition by Hfq. Proc Natl Acad Sci USA. 2011;108:13065–70. doi: 10.1073/pnas.1103420108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cohen SN. Surprises at the 3′ end of prokaryotic RNA. Cell. 1995;80:829–32. doi: 10.1016/0092-8674(95)90284-8. [DOI] [PubMed] [Google Scholar]
- 97.Söderbom F, Wagner EGH. Degradation pathway of CopA, the antisense RNA that controls replication of plasmid R1. Microbiology. 1998;144:1907–17. doi: 10.1099/00221287-144-7-1907. [DOI] [PubMed] [Google Scholar]
- 98.Hajnsdorf E, Carpousis AJ, Régnier P. Nucleolytic inactivation and degradation of the RNase III processed pnp message encoding polynucleotide phosphorylase of Escherichia coli. J Mol Biol. 1994;239:439–54. doi: 10.1006/jmbi.1994.1387. [DOI] [PubMed] [Google Scholar]
- 99.Mohanty BK, Kushner SR. Analysis of the function of Escherichia coli poly(A) polymerase I in RNA metabolism. Mol Microbiol. 1999;34:1094–108. doi: 10.1046/j.1365-2958.1999.01673.x. [DOI] [PubMed] [Google Scholar]
- 100.Beisel CL, Updegrove TB, Janson BJ, Storz G. Multiple factors dictate target selection by Hfq-binding small RNAs. EMBO J. 2012;31:1961–74. doi: 10.1038/emboj.2012.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Panja S, Woodson SA. Hfq proximity and orientation controls RNA annealing. Nucleic Acids Res. 2012;40:8690–7. doi: 10.1093/nar/gks618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jasiecki J, Wegrzyn G. Growth-rate dependent RNA polyadenylation in Escherichia coli. EMBO Rep. 2003;4:172–7. doi: 10.1038/sj.embor.embor733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mohanty BK, Kushner SR. Polynucleotide phosphorylase functions both as a 3′ right-arrow 5′ exonuclease and a poly(A) polymerase in Escherichia coli. Proc Natl Acad Sci USA. 2000;97:11966–71. doi: 10.1073/pnas.220295997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Miranda G, Schuppli D, Barrera I, Hausherr C, Sogo JM, Weber H. Recognition of bacteriophage Qbeta plus strand RNA as a template by Qbeta replicase: role of RNA interactions mediated by ribosomal proteins S1 and host factor. J Mol Biol. 1997;267:1089–103. doi: 10.1006/jmbi.1997.0939. [DOI] [PubMed] [Google Scholar]
- 105.Klovins J, Berzins V, van Duin J. A long-range interaction in Qbeta RNA that bridges the thousand nucleotides between the M-site and the 3′ end is required for replication. RNA. 1998;4:948–57. doi: 10.1017/S1355838298980177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Soper T, Mandin P, Majdalani N, Gottesman S, Woodson SA. Positive regulation by small RNAs and the role of Hfq. Proc Natl Acad Sci USA. 2010;107:9602–7. doi: 10.1073/pnas.1004435107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Prévost K, Desnoyers G, Jacques JF, Lavoie F, Massé E. Small RNA-induced mRNA degradation achieved through both translation block and activated cleavage. Genes Dev. 2011;25:385–96. doi: 10.1101/gad.2001711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Worrall JA, Górna M, Crump NT, Phillips LG, Tuck AC, Price AJ, et al. Reconstitution and analysis of the multienzyme Escherichia coli RNA degradosome. J Mol Biol. 2008;382:870–83. doi: 10.1016/j.jmb.2008.07.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ikeda Y, Yagi M, Morita T, Aiba H. Hfq binding at RhlB-recognition region of RNase E is crucial for the rapid degradation of target mRNAs mediated by sRNAs in Escherichia coli. Mol Microbiol. 2011;79:419–32. doi: 10.1111/j.1365-2958.2010.07454.x. [DOI] [PubMed] [Google Scholar]
- 110.Olejniczak M. Despite similar binding to the Hfq protein regulatory RNAs widely differ in their competition performance. Biochemistry. 2011;50:4427–40. doi: 10.1021/bi102043f. [DOI] [PubMed] [Google Scholar]
- 111.Moon K, Gottesman S. Competition among Hfq-binding small RNAs in Escherichia coli. Mol Microbiol. 2011;82:1545–62. doi: 10.1111/j.1365-2958.2011.07907.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hussein R, Lim HN. Disruption of small RNA signaling caused by competition for Hfq. Proc Natl Acad Sci USA. 2011;108:1110–5. doi: 10.1073/pnas.1010082108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Fender A, Elf J, Hampel K, Zimmermann B, Wagner EG. RNAs actively cycle on the Sm-like protein Hfq. Genes Dev. 2010;24:2621–6. doi: 10.1101/gad.591310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hwang W, Arluison V, Hohng S. Dynamic competition of DsrA and rpoS fragments for the proximal binding site of Hfq as a means for efficient annealing. Nucleic Acids Res. 2011;39:5131–9. doi: 10.1093/nar/gkr075. [DOI] [PMC free article] [PubMed] [Google Scholar]