Abstract
Over the past years, small non-coding RNAs (sRNAs) emerged as important modulators of gene expression in bacteria. Guided by partial sequence complementarity, these sRNAs interact with target mRNAs and eventually affect transcript stability and translation. The physiological function of sRNAs depends on the protein Hfq, which binds sRNAs in the cell and promotes the interaction with their mRNA targets. This important physiological function of Hfq as a central hub of sRNA-mediated regulation made it one of the most intensely studied proteins in bacteria. Recently, a new model for sRNA binding by Hfq has been proposed that involves the direct recognition of the sRNA 3′ end and interactions of the sRNA body with the lateral RNA-binding surface of Hfq. This review summarizes the current understanding of the RNA binding properties of Hfq and its (s)RNA complexes. Moreover, the implications of the new binding model for sRNA-mediated regulation are discussed.
Keywords: 3′ end recognition, LSm ring, RNA chaperone, RNA degradation, crystal structure, gene regulation, non-coding RNAs, prokaryotes
Introduction
Post-transcriptional regulation of gene expression by non-coding RNAs (ncRNAs) is a crucial mechanism for the cell to impose complex and rapid control over its proteome and, hence, its physiological state. In bacteria, a large variety of ncRNAs regulate genes that are responsible for the specific adaptation to constantly changing metabolite and environmental conditions (for review, see refs. 1 and 2). A special group of bacterial ncRNAs are the so-called Hfq-binding small RNAs (sRNAs) that specifically interact with mRNA targets based on partial sequence complementarity. Ultimately, sRNAs affect bacterial gene expression by regulating the stability and translation of the respective transcripts (for review, see refs. 3 and 4). The physiological function of these sRNAs depends on the homohexameric (L)Sm protein Hfq, which is now established as a central mediator of sRNA-based gene regulation in bacteria. Hfq specifically recognizes the structurally diverse sRNAs and facilitates the interaction with their target mRNAs (for review, see ref. 5). The specificity of Hfq for sRNAs, however, could not be explained by previously described RNA-binding modes of this protein.6-8 Therefore, one of the central aims in the field is to understand how Hfq recognizes and binds RNA in general and sRNAs in particular. Recently, several studies provided new insights into the RNA binding properties of Hfq, ultimately suggesting a new model for sRNA recognition.9-12 This review therefore summarizes the current knowledge of the atomic structure of Hfq and its interactions with RNA. Furthermore, the new sRNA binding model and its implications for sRNA-mediated regulation are discussed in the context of current and previously published results. A detailed discussion of the (L)Sm protein superfamily as well as a revision of the role of Hfq in the context of sRNA-mediated regulation and mRNA degradation is provided in the accompanying articles within this Special Focus.
Hfq - The bacterial (L)Sm protein
Discovery and cellular functions
Hfq is the only bacterial LSm homolog and one of the first characterized RNA binding proteins. The first known and also eponymous function of Hfq coincided with its original identification as a host factor for the replication of the Qβ-phage in Escherichia coli.13 In this context, Hfq was shown to bind at the cytosine-rich 3′ end of the plus-strand viral RNA and Hfq binding was required for the initiation of minus-strand RNA synthesis by the Qβ-replicase.14-16 Moreover, Hfq has been implicated in various processes of bacterial RNA metabolism and several studies suggested a function of Hfq in polyadenylation-mediated mRNA degradation.17-20 It was shown that Hfq binds oligo-adenosine stretches of degradation intermediates and that stoichiometric Hfq binding stimulates the processive polyadenylation by poly-(A) polymerase I (PAPI).21,22 The resulting poly-(A)n termini represent a bacterial degradation signal and ultimately promote RNA decay by the degradosome and/or serve as a toehold for exonucleases.23,24
Inactivation of the hfq gene in Escherichia coli and Salmonella typhimurium resulted in severe phenotypes especially under adaptive growth conditions. The observed pleiotropic effects included changes in cell viability and morphology accompanied by decreased growth rates and virulence.25,26 The bacteria showed general defects in transcription and/or translation: in Salmonella, for example, transcriptomic profiling and deep-sequencing of Hfq-associated RNAs revealed that Hfq, as a global regulator, (in)directly affects approximately one-fifth of the Salmonella genome.27 Furthermore, the cells were strongly impaired in their adaptability to stress conditions like stationary growth, oxidation or UV light.28-30 The strong influence of Hfq on bacterial gene expression is now attributed to its central role in sRNA-mediated regulation, where Hfq was shown to (1) bind and stabilize sRNAs in the cell, (2) facilitate base pairing between sRNAs and their targets and (3) trigger subsequent steps like translational repression and decay (for review, see ref. 5).
The (L)Sm fold and oligomerization
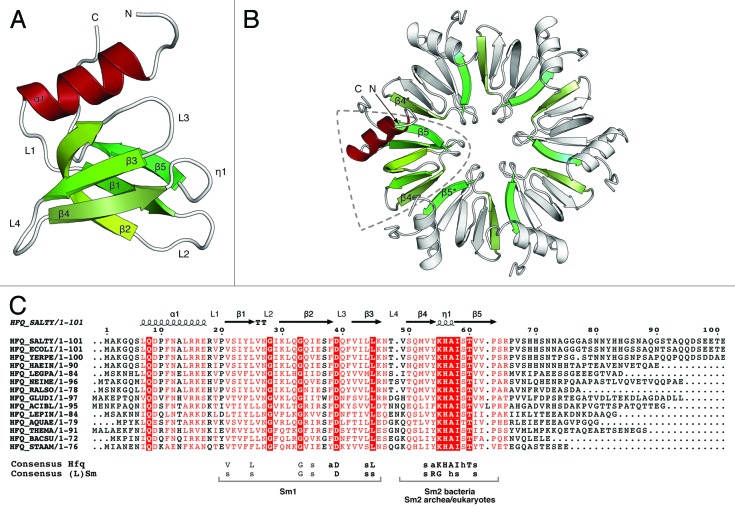
Hfq is a member of the (L)Sm protein superfamily. The founding members of this large group are the classical Sm and like-Sm (LSm) proteins, which are conjointly referred to as (L)Sm proteins (for review, see refs. 31 and 32, as well as the reviews by C. and J. Wilusz and C. Mura et al. in this Special Focus). (L)Sm proteins are found in all three domains of life and are characterized by the presence of a conserved protein fold, the so-called LSm-domain.33 Topologically, the LSm fold consists of an N-terminal α-helix (α1) followed by five β-strands (β1-5) and the secondary structure elements are separated by five loops (L1-5) of variable length (Fig. 1A). The β-strands form an antiparallel, strongly bent β-sheet with the α-helix stacked on top of the open barrel. On the primary sequence level, the LSm fold is characterized by two conserved sequence signatures Sm1 and Sm2, where residues from Sm1 reside in the first three β-strands and residues from Sm2 reside in strands β4 and β5 (Fig. 1C). The Sm1 signature can be identified in all (L)Sm proteins, while Sm2 is divergent in the bacterial Hfq proteins (Fig. 2). Although, the sequence conservation between prokaryotic,6,34 archeal35,36 and eukaryotic37-40 ring-forming (L)Sm proteins is low, the characteristic LSm fold is preserved.
Figure 1. Fold and oligomerization of the LSm domain. (A) Cartoon representation of the LSm domain of Salmonella typhimurium Hfq (PDB-ID: 2YLB9). Secondary structure elements (α-helix 1, red; β-sheets 1–5, green; loops 1–4, white) as well as the N- and C-termini are indicated. The five β-strands form a half-open barrel with the N-terminal α-helix stacked on top. (B) Cartoon representation of the Salmonella typhimurium Hfq6 ring. A single LSm domain is highlighted and colored as in (A); secondary structure elements involved in intersubunit interactions are colored green. Six LSm domains assemble into a homohexameric ring resulting in an extended β-sheet spanning the entire hexamer. Intersubunit contacts are provided by backbone interactions between strands β4 and β5 to strands β5* and β4* in the neighboring (indicated by a *) monomers, respectively. All (L)Sm rings assemble in a polar way with the N-terminal α-helices located on the same side of the oligomer. (C) Multiple sequence alignment of bacterial Hfq proteins. The secondary structure of Salmonella typhimurium Hfq (PDB-ID: 2YLB9) is superimposed on the primary sequence. The Sm consensus sequences are shown below the alignment (the nature of the amino acid side-chains is: s = small hydrophobic, I, L, V; h = hydrophilic, S, T; a = aromatic, Y, F). Highly conserved residues are red (> 70% conservation) or white in red boxes (100% conservation). While the Sm1 signature is conserved in all domains of life, the Sm2 is divergent in bacteria. The species abbreviations and UniProt-IDs are: γ-Proteobacteria: SALTY, Salmonella typhimurium (P0A1R0); ECOLI Escherichia coli (P0A6X3); YERPE, Yersinia pestis (A4TRN9); HAEIN, Hemophilus influenza, (P44437); LEGPA Legionella pneumophila (Q5X982). β-Proteobacteria: NEIME, Neisseiriameningitides (B9VV05); RALSO, Ralstonia solanacearum (Q8Y025). α-Proteobacteria: GLUDI, Gluconacetobacter diazotrophicus (Q8RMG6); Acidobacteria: ACIBL, Acidobacteria bacterium (Q1IIF9). Spirochaetales: LEPIN, Leptospira interogans (Q8F5Z7). Aquafecales: AQUAE Aquifex aeolicus, (O66512). Thermotogales: THEMA, Thermotoga maritime (Q9WYZ6). Fermicutes: BACSU, Bacillus subtilis (O31796); STAAM, Staphylococcus aureus (Q99UG9).
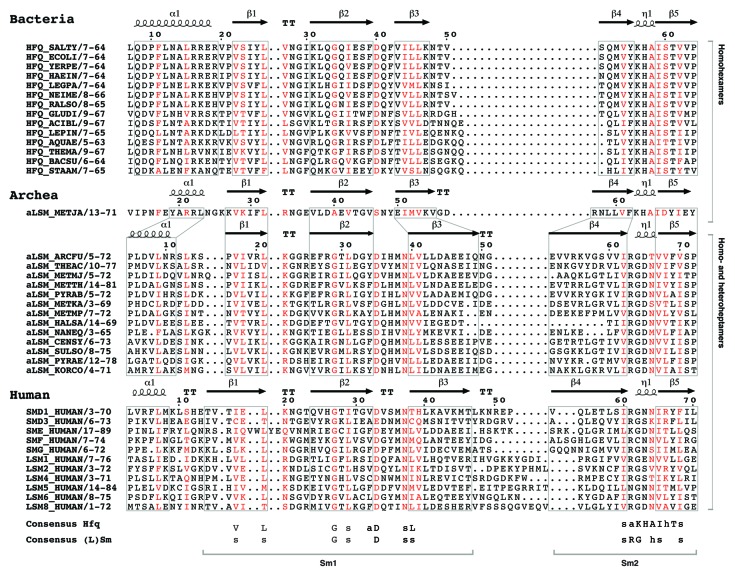
Figure 2. The LSm fold is conserved in all domains of life. Multiple sequence alignment of the LSm domain found in bacterial (Hfq), archeal (aLSM) and human (L)Sm proteins. Residues with > 70% sequence conservation are shown in red. The secondary structure of always the top species of each subgroup is indicated on top of the primary sequence. Although the sequence conservation of the different LSm domains is low, the LSm fold is conserved. The main differences are the length of the N-terminal α-helix, the strands β3 and β4 as well as loop L1 and L4. Interestingly, the only known archeal homohexameric Hfq protein (aLSM_METJA) comprises an Sm2 signature very similar to its homohexameric bacterial homologs, while the Sm2 motif of homoheptameric archeal homologs is more related to the human (L)Sm proteins. Species abbreviations and UniProt accession numbers for the selected bacterial sequences are given in Figure 1. Species abbreviations and UniProt accession numbers of archeal LSm proteins are: Methanococci: METJA, Methanococcus jannaschii (Q58830); Archeoglobi: ARCFU, Archeoglobus fulgidus, (O29386); Thermoplasmata: THEAC, Thermoplasma acidophilum, (P57670); Methanomicrobia: METMJ, Methanoculleus marisnigri, (A3CS14); Methanobacteria: METTH, Methanobacterium thermoautotrophicum, (O26745); Thermococci: PYRAB, Pyrococcus abyssi, (Q9V0Y8); Methanopyri: METKA, Methanopyrus kandleri, (Q8TYS2); Methanococci: METMP, Methanococcus maripaludis, (Q6LY45); Halobacteria: HALSA, Halobacterium salinarum, (Q9HPS2); Nanoarcheoata NANEQ, Nanoarcheum equitans, (Q74N54); Thaumarcheoata CENSY, Cenarcheum symbiosum, (A0RZA4); Crenarcheoata SULSO, Sulfolubus solfataricus, (Q97ZQ0); Crenarcheoata PYRAE, Pyrobacculum aerophilum, (Q8ZYG5); Koracheoata CORCO, Korarcheum cryptofilum, (B1L734). The accession numbers for the HUMAN (Homo sapiens) Sm proteins are: SmD1, (P62314); SmD3, (P62318); SmE, (P62304); SmF, (P62306); SmG, (P62308). LSm proteins: LSm1, (O15116); LSm2, (Q9Y333); LSm4, (Q9Y4Z0); LSm5, (Q9Y4Y9); LSm6, (P62312); LSm8, (O95777).
All classical eukaryotic Sm (SmB/B’, SmD1, SmD2, SmD3, SmE, SmF, SmG) and LSm (LSm1-8 and LSm10-11) proteins assemble into ring-shaped heteroheptamers, which represent the functional biological entity (for review, see ref. 32). The interface between two subunits is made up from residues in the β4 and β5* strands (* indicates a neighboring subunit) and adjacent subunits interact in an oriented way, resulting in an oligomerization where all N-terminal α-helices are located on the same face of the ring (Fig. 1B).
In bacteria, there is usually only one (L)Sm homolog, the Hfq protein, which assembles into homohexamers.3,46 Interestingly, enterobacterial Hfq proteins comprise unusually long C-terminal extensions (Fig. 1C). The sequence of this unstructured region is not conserved and its biological function is controversial.41-45 It has been shown that Hfq proteins, which lack the C-terminal extension, can functionally replace the full-length protein27,46and, recently, it has been suggested that the C-terminal tails may be involved in additional RNA interactions.47
Although several crystal structures of ring-forming (L)Sm proteins have been determined, their oligomerization state is still a matter of debate. One reason complicating the interpretation of experimental data is the intrinsic propensity of the wedge-shaped LSm domain to assemble into oligomers and consequently pentameric,48,49 hexameric,6,34 heptameric36,40,50 and octameric51 rings have been described for (L)Sm proteins of bacterial, archeal and eukaryotic origin. For bacterial Hfq, a homopentameric form has once been suggested,48 but no further experimental evidence supporting this hypothesis could be provided in the following years. Furthermore, the published crystal structures of bacterial Hfq proteins6,9,34,42,52-54 as well as electron microscopy55 and small-angle X-ray scattering43 data have established the hexamer as the functional state of Hfq. In contrast to eukaryotic Sm protein heteroheptamers,37-39,56,57 the assembly of archeal LSm and bacterial Hfq oligomers is less well understood. Recent in vitro evidence however, suggests that E. coli Hfq exists in a monomer-hexamer equilibrium in solution with the hexamer being most active in RNA binding and annealing.58
The RNA Binding Properties of Hfq and the Initial sRNA Binding Model
Hfq achieves its different cellular tasks by the differential use of distinct RNA binding sites and already very early experiments indicated a preference of Hfq for single-stranded adenosine- and uridine-rich sequences.59,60 Subsequently, two independent RNA binding sites—distal and proximal—with respective specificities, could be characterized on opposite surfaces of the Hfq hexamer.8
The distal RNA binding site
The so-called distal RNA binding site shows specificity for purine-rich sequences and is thought to be relevant for the interaction of Hfq with internal adenosine-rich sequences of mRNAs47,61,62 and with poly-(A)n tracts found at the 3′ end of RNA degradation intermediates21,22. In 2008, Soper et al. originally showed that an adenosine-rich motif (5′-AAYAA-3′; A, adenosine; Y, pyrimidine) in the leader sequence of the rpoS mRNA is important for the stimulation of rpoS translation by the Hfq-dependent sRNA DsrA, ultimately resulting in the expression of the rpoS encoded transcription factor σS.61 Similar Hfq-binding sequence motifs could also be identified in the leader sequences of flhA62 and glmS47 mRNAs and by genomic SELEX (5′-AAYAAYAA-3′),63 indicating that adenosine-rich sequences represent important Hfq binding sites in target mRNAs. The structural basis for RNA binding to the distal surface was provided by several crystal structures of Hfq hexamers bound to short, purine-rich oligonucleotides.7,64,65 In 2009, the complex structure of Escherichia coli Hfq bound to poly-(A)15 RNA revealed a tripartite RNA binding motif on each Hfq monomer (A-R-E motif: A site, adenosine specificity site; R site, purine selectivity site; E site, non-discriminatory entrance/exit site) resulting in specific binding of up to six poly-(ARN)n repeats (A, adenosine; R, purine; N, any nucleotide) per hexamer.7 In contrast, the recent complex structures of Hfq homologs from the gram-positive bacteria Staphylococcus aureus and Bacillus subtilis in complex with poly-(A)4 64 and (AG)3A65 RNA respectively, revealed a bipartite RNA-binding motif composed of a purine nucleotide-specificity site and a sequence-independent linker site. These data indicate that RNA binding to the distal surface can differ between bacterial species: Hfq proteins from gram-negative bacteria comprise a tripartite binding motif for poly-(ARN)n repeats, while Hfq homologs from gram-positive species are likely to interact with poly-(AN)n sequences via bipartite binding sites.64
The distal RNA binding site is thought to be important for the function of Hfq as an RNA interaction platform and it has been suggested that Hfq promotes sRNA/mRNA base pairing by simultaneous interactions with poly-(ARN)n sequences of mRNAs via its distal and with sRNA via its other RNA binding surface (for review, see ref. 5). Consistently, it has been demonstrated by Mikulecky et al. that the distal site is indeed independent and available for simultaneous RNA binding.8
The proximal RNA binding site
The second, so-called proximal RNA binding site of Hfq was shown to preferably bind uridine-rich sequences and has been implicated in sRNA recognition. In analogy to the eukaryotic Sm heteroheptamer, where the RNA threads through the central pore of the ring, the proximal site of Hfq has been assumed to bind internal A/U-rich sequences and Staphylococcus aureus Hfq was hence co-crystallized with a A(U)5G RNA substrate.6 In the complex, the first six nucleotides were bound in specific binding pockets around the central pore and the 3′ terminal guanosine was exposed. Additionally, probing experiments of Hfq/sRNA complexes indicated a preference of Hfq for single-stranded A/U-rich sequences in vicinity to secondary structure elements.66,67 Combined, these observations led to a widely accepted binding model where Hfq interacts with internal A/U-rich sequences of sRNAs via its proximal surface.6,7,23,66,67 This binding model however could not explain (1) the general protection of sRNAs by Hfq beyond few nucleotides and (2) the apparent specificity of Hfq for the structurally very diverse sRNAs.
Hfq and its Interaction with (s)RNAs—A Changing Perspective
The prevailing sRNA binding model has been challenged by the discovery that Hfq specifically binds 3′ terminal uridine-rich sequences and that this binding specificity is utilized to recognize a common feature in bacterial sRNA transcripts, namely the ρ-independent transcription terminator. The molecular details of this new binding mode were examined using a U6 RNA substrate and in vitro binding experiments demonstrated that a free 3′ hydroxyl group is crucial for the high-affinity interaction.9 The crystal structure of Salmonella typhimurium Hfq in complex with the U6 oligonucleotide revealed that the specificity and affinity of Hfq for RNA 3′ ends relies on a special mode of direct recognition of the 3′ hydroxyl group in the context of a constricted RNA backbone conformation.
In parallel, the importance of sRNA 3′ end binding by Hfq was demonstrated in vivo. Using the model sRNA SgrS, Otaka and colleagues found that shortening of the 3′ uridine-rich sequence eliminates the interaction of the sRNA with Hfq and abolishes target mRNA regulation.10 Given that ρ-independent terminators are found at the 3′ end of most Hfq binding sRNAs,27 it is, therefore, very likely that 3′ end binding contributes significantly to the selective recognition of sRNAs by Hfq.
A new RNA binding surface
Importantly, the interaction of the sRNA 3′ end with the proximal RNA binding site of Hfq was found to be not the only determinant for sRNA binding. A subsequent analysis of Hfq RNA binding mutants identified an additional binding surface for the sRNA body on the lateral surface of the hexamer.12 This so-called lateral RNA binding surface consists of six patches (one per monomer) of conserved polar residues (R16, R17, R19 and K47), and a mutational analysis of the model sRNA RybB demonstrated that especially single-stranded, internal uridine-rich sequences interact with these sites.
The new sRNA binding model
Combined, the results of the recent studies have changed the view of how sRNAs are thought to interact with Hfq.9-12 Ultimately, the data suggest a new model for sRNAs recognition where, the specific interaction of Hfq with the sRNA 3′ end anchors the sRNA on the proximal face of the hexamer, whereas internal uridine-rich sRNA sequences contribute additively to complex stability by interacting with several of the six lateral RNA binding sites of Hfq (Fig. 3). Although binding experiments with RybB mutants indicated that also the 3′ terminator structure contributes to sRNA binding, its detailed binding mode remains unclear. Given the binding specificities of the proximal, distal and lateral RNA-binding sites of Hfq and its low affinity for double-stranded sequences,68 it seems unlikely that the terminator stem-loop interacts with one these surfaces.
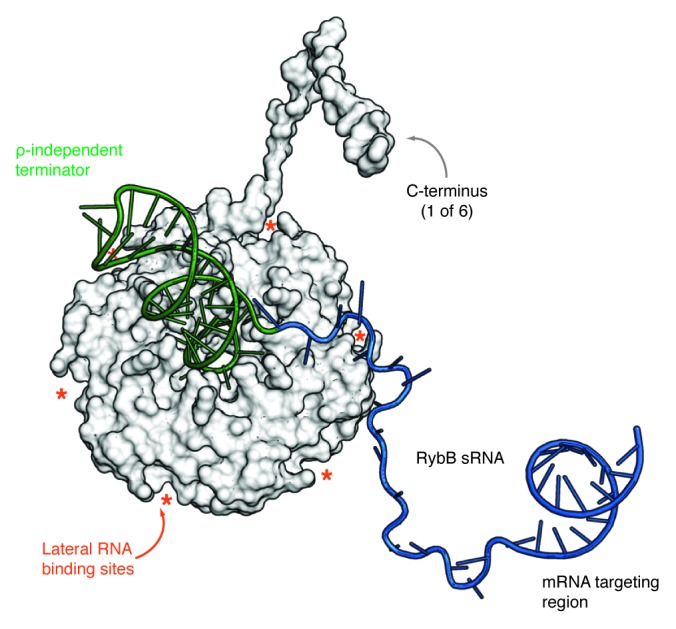
Figure 3. Model of an Hfq/sRNA complex. Proximal side view of a model of the Hfq/RybB complex. The Hfq hexamer (PDB-ID: 2YLC9) is shown as a surface representation with a superimposed model of only one of the six Hfq C-termini for clarity. RybB sRNA is depicted in cartoon representation with the ρ-independent terminator colored in green, the single-stranded sequence is blue and the location of the seed region is indicated. The asterisks mark the location of the six lateral RNA binding sites of Hfq. The model was assembled using COOT98 considering the biochemical and structural evidence summarized in this review. The depicted structure of the C terminus was modeled using HHpred.99 The model shows how Hfq might interact with RybB sRNA and also gives an impression of the proportions of the sRNA body with respect to the size of the terminator stem-loop and the Hfq protein.
Therefore, recognition of the sRNA 3′ end on the proximal surface of Hfq should rather position the terminator stem-loop above the proximal side of the ring. Possible interaction sites for the terminator stem-loop could be the C-terminal extensions of Hfq, which are very flexible43,47 and contain several polar and aromatic residues that could interact with the grooves of the RNA helix.
Hfq—The RNA interaction platform
The partial sequence complementarity between sRNAs and their mRNA targets determines the specificity of sRNA-mediated regulation.3,69 Consequently, the putative complex formation between the Hfq/sRNA complex and an mRNA target has been addressed by several studies.61,70-74 The application of high-resolution chromatography techniques combined with optimized RNA and protein constructs recently provided a more detailed understanding of multipart Hfq/(s)RNA complexes and confirmed that the three RNA binding sites of Hfq are independent and can be used simultaneously in any combination.12 The experimental data showed that in a binary complex composed of the model sRNA RybB and Hfq, the sRNA is presented in a hybridization competent state on Hfq and readily forms a duplex with its mRNA target. In the resulting sRNA-mediated ternary complex, the sRNA 3′ end remained anchored to the proximal site of Hfq, while the sRNA body was released from the lateral surface to base pair with the mRNA target sequence. In the case of a natural, full-length mRNA, however, it has to be considered that remote (ARN)n-repeats in the mRNA sequence could simultaneously interact with the distal surface of Hfq and influence complex formation and stability.62,70 The physiological relevance of these in vitro observations is strongly supported by recent evidence, which indicates that the ternary complex of Hfq, sRNA and mRNA guides the endoribonuclease RNase E to initiate degradation of the target mRNA.75
sRNA Recognition by Hfq—Implications and Future Directions
sRNA 3′ end binding by Hfq—The key to selectivity?
The apparent selectivity of Hfq for bacterial sRNAs is still an unresolved question in the field of sRNA research. The discovery that Hfq directly recognizes sRNA 3′ ends therefore suggests an elegant explanation for the selectivity of Hfq for many sRNAs despite their structural diversity. The high sequence conservation of the proximal RNA binding site (KHAI motif, Fig. 1C) in bacteria implies that the recognition of uridine-rich sRNA 3′ ends is a general property of Hfq and likely represents the predominant function of the proximal surface. Clearly, this hypothesis has to be verified experimentally in the future, as to date, direct 3′ end recognition by Hfq has only been demonstrated for a few sRNAs (RybB,9 SgrS and RyhB10,11).
Although a uridine-rich 3′ end is the only recurrent similarity of Hfq-binding sRNAs, the ρ-independent transcription terminator is not a unique feature. Actually, the transcription of a majority of bacterial mRNAs is terminated using this mechanism resulting in uridine-rich 3′ ends prone to bind Hfq.27,76-78 An immediate question raised by this fact is how Hfq discriminates between these different RNA substrates in the cell. An important factor for efficient proximal site binding seems to be the accessibility of the single-stranded uridine-rich 3′ sequence and a first quantitative picture of Hfq-bound RNAs was provided by deep sequencing experiments.27 The data showed that sRNAs that display uridine-rich tails are highly enriched on Hfq. In contrast, Hfq-independent RNAs either had processed 3′ ends or base-paired terminal uridines. In the future, a comprehensive analysis of RNA sequences that are directly bound by Hfq in the cell could reveal the in vivo relevance of 3′ end recognition and, furthermore, show which additional sRNA sequences are involved in Hfq binding.
The observation that the sRNA/mRNA duplex remains associated with Hfq via the sRNA 3′ end12 further emphasizes the importance of this binding mode for sRNA regulation. Apparently, this high affinity interaction is sufficient to tether the sRNA to Hfq and may also explain why no additional general Hfq binding sites have been conserved among sRNAs. It is tempting to hypothesize that only those Hfq/sRNA complexes are functional in sRNA-mediated regulation, whose sRNA 3′ end is bound to the proximal site and allow for ternary complex formation with the mRNA followed by RNaseE recruitment.75,79 An interesting (in vivo) experiment to prove such a model would be to test the regulatory potential of an sRNA, which on the one hand is stably bound to Hfq via the lateral site, but where 3′ binding is prevented for example by a 2′-3′ cyclic phosphate.
The Lateral RNA Binding Surface of Hfq—The Missing Link?
In the past, probing experiments of several Hfq/sRNA complexes have shown that internal A/U-rich sequences of sRNAs are protected by Hfq.61,66,72,80 However, the protection of multiple sites was difficult to reconcile with sRNA binding to the single proximal surface of Hfq. The identification of the six lateral sites as a third RNA binding surface furthermore provide a rational explanation for the protection of a much larger portion of the sRNA body.
In principle, every sRNA could occupy a different number of lateral sites depending on the length and spatial distance of its A/U-rich sequences. In an extreme scenario, it is even possible that every individual sRNA takes a different path on the lateral surface of Hfq resulting in heterogeneity of Hfq/sRNA complexes, which, in turn, could have complicated the interpretation of probing data.
Several single point mutants of lateral site residues have been described previously, however, only a partial reduction in RNA binding affinity was observed indicating that each residue additively contributes to (s)RNA binding.8,71,81 The sequence alignment of bacterial Hfq homologs shows that the lateral surface is conserved and, therefore, likely a general property of Hfq proteins (Fig. 1C). Arginine 16 shows the highest sequence conservation, while in position 17 and 19, positively charged (Arginine or Lysine) and aromatic (Histidine) residues are found. The side chains of these amino acids are often involved in RNA binding interactions, as they can interact with RNA by stacking or hydrogen bonding.82,83 Moreover, single-stranded RNA sequences are more accessible for this type of intermolecular interactions and, therefore, the lateral site may represent an ideal binding surface. Also in Staphylococcus aureus, where Hfq function is currently under debate (for review, see ref. 84), the sequence conservation suggests that the key biochemical properties of Hfq should be conserved (Fig. 1C): the proximal RNA binding site (KHAI motif) is identical with enterobacterial Hfq proteins and several residues on the lateral surface (K10, K41) could be engaged in RNA interactions.
Implications for sRNA competition
In 2010, Fender and colleagues demonstrated that sRNAs can displace each other on Hfq and similar observations were later made in vivo.85,86 The suggested model for sRNA competition consequently implied (1) a transient association of the competitor sRNA with existing sRNA/Hfq complexes, followed by (2) an exchange of RNA binding sites and (3) the eventual dissociation of one of the sRNAs. Because the A/U content and the localization of single-stranded sequences in the sRNA body differ between individual sRNAs, binding of the sRNA body to lateral surface could be an important factor in the context of sRNA competition for Hfq. In contrast to the proximal site, which is likely to be occupied by only one sRNA 3′ end, the six lateral sites could serve as individual entry points for a competitor sRNA and allow for a gradual displacement of a bound sRNA. Clearly, this hypothesis has to be tested in the future e.g., by competition experiments using selective Hfq lateral site mutants.12 Moreover, the weak and transient interaction of the sRNA body with Hfq suggests that the lateral sites might represent the “chaperoning” surface of Hfq87-90 (for review, see ref. 91). Evidently, structural information is required to understand the detailed interaction of RNA with the lateral surface and the associated binding parameters have to be determined. Nevertheless, the interaction of the sRNA body with the lateral surface of Hfq emerges as an additional element in the sRNA/Hfq interaction and has to be further investigated.
(s)RNA Binding to the Distal Surface of Hfq
Although the lateral and proximal RNA binding sites of Hfq seem to be sufficient for sRNA binding, this was only shown on the example of the model sRNA RybB, which does not contain poly-(ARN)n repeats.12 In principle, other sRNAs that comprise poly-(ARN)n sequences could (also) bind to the distal site of Hfq or additional Hfq rings could associate with an Hfq/sRNA complex via the distal surface. The underlying biochemical property of the Hfq distal RNA binding site is the cooperative, length-dependent assembly of Hfq rings on poly-(A)n RNA, suggesting that for a stable interaction with the distal surface, at least four Hfq monomers have to be occupied by (four) consecutive (ARN)-repeats that are rarely found in sRNAs associated with Hfq.7,12 Importantly, the cooperative effect could be due to the homo-polymeric sequence and, hence, the high local concentration of binding sites.12 It is therefore normal to expect that cooperative binding is rather limited to the interaction of Hfq with poly-(A)n sequences. Thus, in sRNA-mediated regulation, RNA binding to the distal surface of Hfq may rather be relevant for the simultaneous interaction of the Hfq/sRNA complex with an mRNA target and may contribute to the stabilization of the sRNA/mRNA duplex.47,61,62 Again, this hypothesis could be tested in vivo by analyzing the functionality of selective Hfq distal site mutants12 in sRNA-mediated regulation.
Hfq/sRNA/mRNA Ternary Complex Formation—Implications for sRNA Function
RNaseE was shown to directly interact with Hfq and to initiate the degradation of the mRNA target as well as the base paired sRNA by endonucleolytic cleavage.92-94 A very recent study furthermore suggested an active role for the sRNA/mRNA/Hfq complex in stimulation of RNase E cleavage.75 Consequently, it might be interesting to investigate whether the observed structural rearrangement of Hfq/sRNA complexes upon target mRNA binding12,68,70,95 is recognized by downstream factors like RNaseE. Because RNaseE cleavage preferably occurs at single stranded A/U-rich sites,66,94,96 the release of the sRNA body from the lateral surface of Hfq would expose putative RNaseE sites both in the sRNA and the mRNA target, which could initiate their degradation. Furthermore, it is possible that the freed surfaces of Hfq serve as an interaction site for the direct recruitment of RNaseE to the Hfq/sRNA/mRNA particle.
Conclusions
The systematic discovery of bacterial sRNAs and the growing understanding of their mechanism of action opened an exciting field of RNA research. Furthermore, the observation that the biological function of sRNAs depends on the Hfq protein, did finally explain the pleiotropic effects of Hfq on bacterial gene expression. The specific interaction of Hfq with sRNAs turned out to be rather complex and is still not fully understood. However, the accumulated knowledge of the RNA binding properties of Hfq and the characterization of its (s)RNA complexes, has now set the starting point for the design of new experiments toward an ever more detailed molecular analysis of Hfq function in the cell and the mechanisms of sRNA-mediated regulation.
Cleary, atomic structures of Hfq/sRNA complexes represent one of the most important but also challenging aims in the field and a first glimpse was provided by a study using small-angle X-ray scattering to analyze the overall shape of Vibrio cholerae Hfq in complex with Qrr sRNA.97 The determination of high-resolution structures is complicated by the inherent flexibility and heterogeneity of Hfq/sRNA complexes, which interferes with their crystallization. The design and utilization of minimal sRNAs and Hfq mutants in combination with elaborated purification protocols (e.g., high-resolution chromatography) and stabilization (e.g., by cross-linking) techniques should identify the most promising candidate complexes for structure determination in near future.
Most importantly, the biochemical properties of Hfq and its (s)RNA complexes have to be reconciled with the in vivo situation. Therefore, a global analysis of sRNA sequences that are directly bound by wild-type Hfq (and/or its RNA binding mutants) could allow for a critical evaluation of the proposed sRNA binding model. Furthermore, the inclusion of selective Hfq RNA binding mutants in in vivo experiments should facilitate a further differentiation between the various cellular functions of Hfq and, thereby, contribute to an advanced understanding of sRNA-mediated regulation in bacteria.
Acknowledgments
The author is very grateful to Oliver Weichenrieder, Steffen Schmidt and many other colleagues at the Max Planck Institute for Developmental Biology and the members of the German Research Foundation Priority Program SPP1258 “Sensory and Regulatory RNAs in Prokaryotes” for stimulating discussions about the biochemistry of Hfq and its RNA binding properties. During her work on the Hfq protein, the author was supported by a grant from the German Research Foundation Priority Program SPP1258 “Sensory and Regulatory RNAs in Prokaryotes” and a fellowship by the International PhD Program Tübingen.
Glossary
Abbreviations:
- ncRNA
non-coding RNA
- sRNA
small RNA
- Hfq
host factor for Qβ replication
- (L)Sm
like-Sm
- poly-(A)
poly-adenosine
- mRNA
messenger RNA
- PAPI
poly-(A) polymerase I
- aLSM protein
archeal (L)Sm protein
- A
adenosine
- U
uridine
- Y
pyrimidine
- R
purine
- SELEX
systematic evolution of ligands by exponential enrichment
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/24201
References
- 1.Waters LS, Storz G. Regulatory RNAs in bacteria. Cell. 2009;136:615–28. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gottesman S, Storz G. Bacterial small RNA regulators: versatile roles and rapidly evolving variations. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a003798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Papenfort K, Vogel J. Multiple target regulation by small noncoding RNAs rewires gene expression at the post-transcriptional level. Res Microbiol. 2009;160:278–87. doi: 10.1016/j.resmic.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 4.Storz G, Vogel J, Wassarman KM. Regulation by small RNAs in bacteria: expanding frontiers. Mol Cell. 2011;43:880–91. doi: 10.1016/j.molcel.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vogel J, Luisi BF. Hfq and its constellation of RNA. Nat Rev Microbiol. 2011;9:578–89. doi: 10.1038/nrmicro2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schumacher MA, Pearson RF, Møller T, Valentin-Hansen P, Brennan RG. Structures of the pleiotropic translational regulator Hfq and an Hfq-RNA complex: a bacterial Sm-like protein. EMBO J. 2002;21:3546–56. doi: 10.1093/emboj/cdf322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Link TM, Valentin-Hansen P, Brennan RG. Structure of Escherichia coli Hfq bound to polyriboadenylate RNA. Proc Natl Acad Sci USA. 2009;106:19292–7. doi: 10.1073/pnas.0908744106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mikulecky PJ, Kaw MK, Brescia CC, Takach JC, Sledjeski DD, Feig AL. Escherichia coli Hfq has distinct interaction surfaces for DsrA, rpoS and poly(A) RNAs. Nat Struct Mol Biol. 2004;11:1206–14. doi: 10.1038/nsmb858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sauer E, Weichenrieder O. Structural basis for RNA 3′-end recognition by Hfq. Proc Natl Acad Sci USA. 2011;108:13065–70. doi: 10.1073/pnas.1103420108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Otaka H, Ishikawa H, Morita T, Aiba H. PolyU tail of rho-independent terminator of bacterial small RNAs is essential for Hfq action. Proc Natl Acad Sci USA. 2011;108:13059–64. doi: 10.1073/pnas.1107050108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ishikawa H, Otaka H, Maki K, Morita T, Aiba H. The functional Hfq-binding module of bacterial sRNAs consists of a double or single hairpin preceded by a U-rich sequence and followed by a 3′ poly(U) tail. RNA. 2012;18:1062–74. doi: 10.1261/rna.031575.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sauer E, Schmidt S, Weichenrieder O. Small RNA binding to the lateral surface of Hfq hexamers and structural rearrangements upon mRNA target recognition. Proc Natl Acad Sci USA. 2012;109:9396–401. doi: 10.1073/pnas.1202521109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Franze de Fernandez MT, Eoyang L, August JT. Factor fraction required for the synthesis of bacteriophage Qbeta-RNA. Nature. 1968;219:588–90. doi: 10.1038/219588a0. [DOI] [PubMed] [Google Scholar]
- 14.Franze de Fernandez MT, Hayward WS, August JT. Bacterial proteins required for replication of phage Q ribonucleic acid. Pruification and properties of host factor I, a ribonucleic acid-binding protein. J Biol Chem. 1972;247:824–31. [PubMed] [Google Scholar]
- 15.Barrera I, Schuppli D, Sogo JM, Weber H. Different mechanisms of recognition of bacteriophage Q beta plus and minus strand RNAs by Q beta replicase. J Mol Biol. 1993;232:512–21. doi: 10.1006/jmbi.1993.1407. [DOI] [PubMed] [Google Scholar]
- 16.Miranda G, Schuppli D, Barrera I, Hausherr C, Sogo JM, Weber H. Recognition of bacteriophage Qbeta plus strand RNA as a template by Qbeta replicase: role of RNA interactions mediated by ribosomal proteins S1 and host factor. J Mol Biol. 1997;267:1089–103. doi: 10.1006/jmbi.1997.0939. [DOI] [PubMed] [Google Scholar]
- 17.Folichon M, Arluison V, Pellegrini O, Huntzinger E, Régnier P, Hajnsdorf E. The poly(A) binding protein Hfq protects RNA from RNase E and exoribonucleolytic degradation. Nucleic Acids Res. 2003;31:7302–10. doi: 10.1093/nar/gkg915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Folichon M, Allemand F, Régnier P, Hajnsdorf E. Stimulation of poly(A) synthesis by Escherichia coli poly(A)polymerase I is correlated with Hfq binding to poly(A) tails. FEBS J. 2005;272:454–63. doi: 10.1111/j.1742-4658.2004.04485.x. [DOI] [PubMed] [Google Scholar]
- 19.Mohanty BK, Maples VF, Kushner SR. The Sm-like protein Hfq regulates polyadenylation dependent mRNA decay in Escherichia coli. Mol Microbiol. 2004;54:905–20. doi: 10.1111/j.1365-2958.2004.04337.x. [DOI] [PubMed] [Google Scholar]
- 20.Le Derout J, Folichon M, Briani F, Dehò G, Régnier P, Hajnsdorf E. Hfq affects the length and the frequency of short oligo(A) tails at the 3′ end of Escherichia coli rpsO mRNAs. Nucleic Acids Res. 2003;31:4017–23. doi: 10.1093/nar/gkg456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hajnsdorf E, Régnier PE. E. coli RpsO mRNA decay: RNase E processing at the beginning of the coding sequence stimulates poly(A)-dependent degradation of the mRNA. J Mol Biol. 1999;286:1033–43. doi: 10.1006/jmbi.1999.2547. [DOI] [PubMed] [Google Scholar]
- 22.Hajnsdorf E, Régnier P. Host factor Hfq of Escherichia coli stimulates elongation of poly(A) tails by poly(A) polymerase I. Proc Natl Acad Sci USA. 2000;97:1501–5. doi: 10.1073/pnas.040549897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brennan RG, Link TM. Hfq structure, function and ligand binding. Curr Opin Microbiol. 2007;10:125–33. doi: 10.1016/j.mib.2007.03.015. [DOI] [PubMed] [Google Scholar]
- 24.Worrall JA, Górna M, Crump NT, Phillips LG, Tuck AC, Price AJ, et al. Reconstitution and analysis of the multienzyme Escherichia coli RNA degradosome. J Mol Biol. 2008;382:870–83. doi: 10.1016/j.jmb.2008.07.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsui HC, Leung HC, Winkler ME. Characterization of broadly pleiotropic phenotypes caused by an hfq insertion mutation in Escherichia coli K-12. Mol Microbiol. 1994;13:35–49. doi: 10.1111/j.1365-2958.1994.tb00400.x. [DOI] [PubMed] [Google Scholar]
- 26.Sittka A, Pfeiffer V, Tedin K, Vogel J. The RNA chaperone Hfq is essential for the virulence of Salmonella typhimurium. Mol Microbiol. 2007;63:193–217. doi: 10.1111/j.1365-2958.2006.05489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sittka A, Lucchini S, Papenfort K, Sharma CM, Rolle K, Binnewies TT, et al. Deep sequencing analysis of small noncoding RNA and mRNA targets of the global post-transcriptional regulator, Hfq. PLoS Genet. 2008;4:e1000163. doi: 10.1371/journal.pgen.1000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muffler A, Traulsen DD, Fischer D, Lange R, Hengge-Aronis R. The RNA-binding protein HF-I plays a global regulatory role which is largely, but not exclusively, due to its role in expression of the sigmaS subunit of RNA polymerase in Escherichia coli. J Bacteriol. 1997;179:297–300. doi: 10.1128/jb.179.1.297-300.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsui HC, Feng G, Winkler ME. Negative regulation of mutS and mutH repair gene expression by the Hfq and RpoS global regulators of Escherichia coli K-12. J Bacteriol. 1997;179:7476–87. doi: 10.1128/jb.179.23.7476-7487.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vytvytska O, Moll I, Kaberdin VR, von Gabain A, Bläsi U. Hfq (HF1) stimulates ompA mRNA decay by interfering with ribosome binding. Genes Dev. 2000;14:1109–18. [PMC free article] [PubMed] [Google Scholar]
- 31.Khusial P, Plaag R, Zieve GW. LSm proteins form heptameric rings that bind to RNA via repeating motifs. Trends Biochem Sci. 2005;30:522–8. doi: 10.1016/j.tibs.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 32.Wilusz CJ, Wilusz J. Eukaryotic Lsm proteins: lessons from bacteria. Nat Struct Mol Biol. 2005;12:1031–6. doi: 10.1038/nsmb1037. [DOI] [PubMed] [Google Scholar]
- 33.Achsel T, Stark H, Lührmann R. The Sm domain is an ancient RNA-binding motif with oligo(U) specificity. Proc Natl Acad Sci USA. 2001;98:3685–9. doi: 10.1073/pnas.071033998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sauter C, Basquin J, Suck D. Sm-like proteins in Eubacteria: the crystal structure of the Hfq protein from Escherichia coli. Nucleic Acids Res. 2003;31:4091–8. doi: 10.1093/nar/gkg480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mura C, Cascio D, Sawaya MR, Eisenberg DS. The crystal structure of a heptameric archaeal Sm protein: Implications for the eukaryotic snRNP core. Proc Natl Acad Sci USA. 2001;98:5532–7. doi: 10.1073/pnas.091102298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nielsen JS, Bøggild A, Andersen CB, Nielsen G, Boysen A, Brodersen DE, et al. An Hfq-like protein in archaea: crystal structure and functional characterization of the Sm protein from Methanococcus jannaschii. RNA. 2007;13:2213–23. doi: 10.1261/rna.689007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kambach C, Walke S, Young R, Avis JM, de la Fortelle E, Raker VA, et al. Crystal structures of two Sm protein complexes and their implications for the assembly of the spliceosomal snRNPs. Cell. 1999;96:375–87. doi: 10.1016/S0092-8674(00)80550-4. [DOI] [PubMed] [Google Scholar]
- 38.Pomeranz Krummel DA, Oubridge C, Leung AK, Li J, Nagai K. Crystal structure of human spliceosomal U1 snRNP at 5.5 A resolution. Nature. 2009;458:475–80. doi: 10.1038/nature07851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leung AK, Nagai K, Li J. Structure of the spliceosomal U4 snRNP core domain and its implication for snRNP biogenesis. Nature. 2011;473:536–9. doi: 10.1038/nature09956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Törö I, Thore S, Mayer C, Basquin J, Séraphin B, Suck D. RNA binding in an Sm core domain: X-ray structure and functional analysis of an archaeal Sm protein complex. EMBO J. 2001;20:2293–303. doi: 10.1093/emboj/20.9.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arluison V, Folichon M, Marco S, Derreumaux P, Pellegrini O, Seguin J, et al. The C-terminal domain of Escherichia coli Hfq increases the stability of the hexamer. Eur J Biochem. 2004;271:1258–65. doi: 10.1111/j.1432-1033.2004.04026.x. [DOI] [PubMed] [Google Scholar]
- 42.Beich-Frandsen M, Večerek B, Sjöblom B, Bläsi U, Djinović-Carugo K. Structural analysis of full-length Hfq from Escherichia coli. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2011;67:536–40. doi: 10.1107/S174430911100786X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beich-Frandsen M, Vecerek B, Konarev PV, Sjöblom B, Kloiber K, Hämmerle H, et al. Structural insights into the dynamics and function of the C-terminus of the E. coli RNA chaperone Hfq. Nucleic Acids Res. 2011;39:4900–15. doi: 10.1093/nar/gkq1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Olsen AS, Møller-Jensen J, Brennan RG, Valentin-Hansen P. C-terminally truncated derivatives of Escherichia coli Hfq are proficient in riboregulation. J Mol Biol. 2010;404:173–82. doi: 10.1016/j.jmb.2010.09.038. [DOI] [PubMed] [Google Scholar]
- 45.Vecerek B, Rajkowitsch L, Sonnleitner E, Schroeder R, Bläsi U. The C-terminal domain of Escherichia coli Hfq is required for regulation. Nucleic Acids Res. 2008;36:133–43. doi: 10.1093/nar/gkm985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sonnleitner E, Moll I, Bläsi U. Functional replacement of the Escherichia coli hfq gene by the homologue of Pseudomonas aeruginosa. Microbiology. 2002;148:883–91. doi: 10.1099/00221287-148-3-883. [DOI] [PubMed] [Google Scholar]
- 47.Salim NN, Faner MA, Philip JA, Feig AL. Requirement of upstream Hfq-binding (ARN)x elements in glmS and the Hfq C-terminal region for GlmS upregulation by sRNAs GlmZ and GlmY. Nucleic Acids Res. 2012;40:8021–32. doi: 10.1093/nar/gks392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kajitani M, Kato A, Wada A, Inokuchi Y, Ishihama A. Regulation of the Escherichia coli hfq gene encoding the host factor for phage Q beta. J Bacteriol. 1994;176:531–4. doi: 10.1128/jb.176.2.531-534.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Das D, Kozbial P, Axelrod HL, Miller MD, McMullan D, Krishna SS, et al. Crystal structure of a novel Sm-like protein of putative cyanophage origin at 2.60 A resolution. Proteins. 2009;75:296–307. doi: 10.1002/prot.22360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Collins BM, Harrop SJ, Kornfeld GD, Dawes IW, Curmi PM, Mabbutt BC. Crystal structure of a heptameric Sm-like protein complex from archaea: implications for the structure and evolution of snRNPs. J Mol Biol. 2001;309:915–23. doi: 10.1006/jmbi.2001.4693. [DOI] [PubMed] [Google Scholar]
- 51.Naidoo N, Harrop SJ, Sobti M, Haynes PA, Szymczyna BR, Williamson JR, et al. Crystal structure of Lsm3 octamer from Saccharomyces cerevisiae: implications for Lsm ring organisation and recruitment. J Mol Biol. 2008;377:1357–71. doi: 10.1016/j.jmb.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 52.Nikulin A, Stolboushkina E, Perederina A, Vassilieva I, Blaesi U, Moll I, et al. Structure of Pseudomonas aeruginosa Hfq protein. Acta Crystallogr D Biol Crystallogr. 2005;61:141–6. doi: 10.1107/S0907444904030008. [DOI] [PubMed] [Google Scholar]
- 53.Bøggild A, Overgaard M, Valentin-Hansen P, Brodersen DE. Cyanobacteria contain a structural homologue of the Hfq protein with altered RNA-binding properties. FEBS J. 2009;276:3904–15. doi: 10.1111/j.1742-4658.2009.07104.x. [DOI] [PubMed] [Google Scholar]
- 54.Baba S, Someya T, Kawai G, Nakamura K, Kumasaka T. Expression, crystallization and preliminary crystallographic analysis of RNA-binding protein Hfq (YmaH) from Bacillus subtilis in complex with an RNA aptamer. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2010;66:563–6. doi: 10.1107/S1744309110009942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Arluison V, Mura C, Guzmán MR, Liquier J, Pellegrini O, Gingery M, et al. Three-dimensional structures of fibrillar Sm proteins: Hfq and other Sm-like proteins. J Mol Biol. 2006;356:86–96. doi: 10.1016/j.jmb.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 56.Weber G, Trowitzsch S, Kastner B, Lührmann R, Wahl MC. Functional organization of the Sm core in the crystal structure of human U1 snRNP. EMBO J. 2010;29:4172–84. doi: 10.1038/emboj.2010.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang R, So BR, Li P, Yong J, Glisovic T, Wan L, et al. Structure of a key intermediate of the SMN complex reveals Gemin2’s crucial function in snRNP assembly. Cell. 2011;146:384–95. doi: 10.1016/j.cell.2011.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Panja S, Woodson SA. Hexamer to monomer equilibrium of E. coli Hfq in solution and its impact on RNA annealing. J Mol Biol. 2012;417:406–12. doi: 10.1016/j.jmb.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hori K, Yanazaki Y. Nucleotide sequence specific interaction of host factor I with bacteriophage Q beta RNA. FEBS Lett. 1974;43:20–2. doi: 10.1016/0014-5793(74)81095-1. [DOI] [PubMed] [Google Scholar]
- 60.Carmichael GG. Isolation of bacterial and phage proteins by homopolymer RNA-cellulose chromatography. J Biol Chem. 1975;250:6160–7. [PubMed] [Google Scholar]
- 61.Soper TJ, Woodson SA. The rpoS mRNA leader recruits Hfq to facilitate annealing with DsrA sRNA. RNA. 2008;14:1907–17. doi: 10.1261/rna.1110608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Salim NN, Feig AL. An upstream Hfq binding site in the fhlA mRNA leader region facilitates the OxyS-fhlA interaction. PLoS One. 2010;5 doi: 10.1371/journal.pone.0013028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lorenz C, Gesell T, Zimmermann B, Schoeberl U, Bilusic I, Rajkowitsch L, et al. Genomic SELEX for Hfq-binding RNAs identifies genomic aptamers predominantly in antisense transcripts. Nucleic Acids Res. 2010;38:3794–808. doi: 10.1093/nar/gkq032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Horstmann N, Orans J, Valentin-Hansen P, Shelburne SA, 3rd, Brennan RG. Structural mechanism of Staphylococcus aureus Hfq binding to an RNA A-tract. Nucleic Acids Res. 2012;40:11023–35. doi: 10.1093/nar/gks809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Someya T, Baba S, Fujimoto M, Kawai G, Kumasaka T, Nakamura K. Crystal structure of Hfq from Bacillus subtilis in complex with SELEX-derived RNA aptamer: insight into RNA-binding properties of bacterial Hfq. Nucleic Acids Res. 2012;40:1856–67. doi: 10.1093/nar/gkr892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Moll I, Afonyushkin T, Vytvytska O, Kaberdin VR, Bläsi U. Coincident Hfq binding and RNase E cleavage sites on mRNA and small regulatory RNAs. RNA. 2003;9:1308–14. doi: 10.1261/rna.5850703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Udekwu KI, Darfeuille F, Vogel J, Reimegård J, Holmqvist E, Wagner EG. Hfq-dependent regulation of OmpA synthesis is mediated by an antisense RNA. Genes Dev. 2005;19:2355–66. doi: 10.1101/gad.354405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hopkins JF, Panja S, Woodson SA. Rapid binding and release of Hfq from ternary complexes during RNA annealing. Nucleic Acids Res. 2011;39:5193–202. doi: 10.1093/nar/gkr062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vogel J. A rough guide to the non-coding RNA world of Salmonella. Mol Microbiol. 2009;71:1–11. doi: 10.1111/j.1365-2958.2008.06505.x. [DOI] [PubMed] [Google Scholar]
- 70.Lease RA, Woodson SA. Cycling of the Sm-like protein Hfq on the DsrA small regulatory RNA. J Mol Biol. 2004;344:1211–23. doi: 10.1016/j.jmb.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 71.Sun X, Wartell RM. Escherichia coli Hfq binds A18 and DsrA domain II with similar 2:1 Hfq6/RNA stoichiometry using different surface sites. Biochemistry. 2006;45:4875–87. doi: 10.1021/bi0523613. [DOI] [PubMed] [Google Scholar]
- 72.Sledjeski DD, Whitman C, Zhang A. Hfq is necessary for regulation by the untranslated RNA DsrA. J Bacteriol. 2001;183:1997–2005. doi: 10.1128/JB.183.6.1997-2005.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Updegrove TB, Correia JJ, Chen Y, Terry C, Wartell RM. The stoichiometry of the Escherichia coli Hfq protein bound to RNA. RNA. 2011;17:489–500. doi: 10.1261/rna.2452111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Papenfort K, Bouvier M, Mika F, Sharma CM, Vogel J. Evidence for an autonomous 5′ target recognition domain in an Hfq-associated small RNA. Proc Natl Acad Sci USA. 2010;107:20435–40. doi: 10.1073/pnas.1009784107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bandyra KJ, Said N, Pfeiffer V, Górna MW, Vogel J, Luisi BF. The seed region of a small RNA drives the controlled destruction of the target mRNA by the endoribonuclease RNase E. Mol Cell. 2012;47:943–53. doi: 10.1016/j.molcel.2012.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kingsford CL, Ayanbule K, Salzberg SL. Rapid, accurate, computational discovery of Rho-independent transcription terminators illuminates their relationship to DNA uptake. Genome Biol. 2007;8:R22. doi: 10.1186/gb-2007-8-2-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wilson KS, von Hippel PH. Transcription termination at intrinsic terminators: the role of the RNA hairpin. Proc Natl Acad Sci USA. 1995;92:8793–7. doi: 10.1073/pnas.92.19.8793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Santangelo TJ, Artsimovitch I. Termination and antitermination: RNA polymerase runs a stop sign. Nat Rev Microbiol. 2011;9:319–29. doi: 10.1038/nrmicro2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Morita T, Aiba H. RNase E action at a distance: degradation of target mRNAs mediated by an Hfq-binding small RNA in bacteria. Genes Dev. 2011;25:294–8. doi: 10.1101/gad.2030311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Brescia CC, Mikulecky PJ, Feig AL, Sledjeski DD. Identification of the Hfq-binding site on DsrA RNA: Hfq binds without altering DsrA secondary structure. RNA. 2003;9:33–43. doi: 10.1261/rna.2570803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Updegrove TB, Wartell RM. The influence of Escherichia coli Hfq mutations on RNA binding and sRNA•mRNA duplex formation in rpoS riboregulation. Biochim Biophys Acta. 2011;1809:532–40. doi: 10.1016/j.bbagrm.2011.08.006. [DOI] [PubMed] [Google Scholar]
- 82.Bayer TS, Booth LN, Knudsen SM, Ellington AD. Arginine-rich motifs present multiple interfaces for specific binding by RNA. RNA. 2005;11:1848–57. doi: 10.1261/rna.2167605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Weiss MA, Narayana N. RNA recognition by arginine-rich peptide motifs. Biopolymers. 1998;48:167–80. doi: 10.1002/(SICI)1097-0282(1998)48:2<167::AID-BIP6>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 84.Romilly C, Caldelari I, Parmentier D, Lioliou E, Romby P, Fechter P. Current knowledge on regulatory RNAs and their machineries in Staphylococcus aureus. RNA Biol. 2012;9:402–13. doi: 10.4161/rna.20103. [DOI] [PubMed] [Google Scholar]
- 85.Fender A, Elf J, Hampel K, Zimmermann B, Wagner EG. RNAs actively cycle on the Sm-like protein Hfq. Genes Dev. 2010;24:2621–6. doi: 10.1101/gad.591310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Moon K, Gottesman S. Competition among Hfq-binding small RNAs in Escherichia coli. Mol Microbiol. 2011;82:1545–62. doi: 10.1111/j.1365-2958.2011.07907.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hopkins JF, Panja S, McNeil SA, Woodson SA. Effect of salt and RNA structure on annealing and strand displacement by Hfq. Nucleic Acids Res. 2009;37:6205–13. doi: 10.1093/nar/gkp646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Soper TJ, Doxzen K, Woodson SA. Major role for mRNA binding and restructuring in sRNA recruitment by Hfq. RNA. 2011;17:1544–50. doi: 10.1261/rna.2767211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hopkins JF, Panja S, Woodson SA. Rapid binding and release of Hfq from ternary complexes during RNA annealing. Nucleic Acids Res. 2011;39:5193–202. doi: 10.1093/nar/gkr062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Panja S, Woodson SA. Hfq proximity and orientation controls RNA annealing. Nucleic Acids Res. 2012;40:8690–7. doi: 10.1093/nar/gks618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rajkowitsch L, Chen D, Stampfl S, Semrad K, Waldsich C, Mayer O, et al. RNA chaperones, RNA annealers and RNA helicases. RNA Biol. 2007;4:118–30. doi: 10.4161/rna.4.3.5445. [DOI] [PubMed] [Google Scholar]
- 92.Massé E, Majdalani N, Gottesman S. Regulatory roles for small RNAs in bacteria. Curr Opin Microbiol. 2003;6:120–4. doi: 10.1016/S1369-5274(03)00027-4. [DOI] [PubMed] [Google Scholar]
- 93.Morita T, Maki K, Aiba H. RNase E-based ribonucleoprotein complexes: mechanical basis of mRNA destabilization mediated by bacterial noncoding RNAs. Genes Dev. 2005;19:2176–86. doi: 10.1101/gad.1330405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Prévost K, Desnoyers G, Jacques JF, Lavoie F, Massé E. Small RNA-induced mRNA degradation achieved through both translation block and activated cleavage. Genes Dev. 2011;25:385–96. doi: 10.1101/gad.2001711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hopkins JF, Panja S, McNeil SA, Woodson SA. Effect of salt and RNA structure on annealing and strand displacement by Hfq. Nucleic Acids Res. 2009;37:6205–13. doi: 10.1093/nar/gkp646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pfeiffer V, Papenfort K, Lucchini S, Hinton JC, Vogel J. Coding sequence targeting by MicC RNA reveals bacterial mRNA silencing downstream of translational initiation. Nat Struct Mol Biol. 2009;16:840–6. doi: 10.1038/nsmb.1631. [DOI] [PubMed] [Google Scholar]
- 97.Vincent HA, Henderson CA, Stone CM, Cary PD, Gowers DM, Sobott F, et al. The low-resolution solution structure of Vibrio cholerae Hfq in complex with Qrr1 sRNA. Nucleic Acids Res. 2012;40:8698–710. doi: 10.1093/nar/gks582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 99.Biegert A, Mayer C, Remmert M, Söding J, Lupas AN. The MPI Bioinformatics Toolkit for protein sequence analysis. Nucleic Acids Res. 2006;34(Web Server issue):W335-9. doi: 10.1093/nar/gkl217. [DOI] [PMC free article] [PubMed] [Google Scholar]