Abstract
We previously found that azido-containing β-diketo acid derivatives (DKAs) are potent inhibitors of human immunodeficiency virus type 1 (HIV-1) integrase (IN) (X. Zhang et al., Bioorg. Med. Chem. Lett., 13:1215-1219, 2003). To characterize the intracellular mechanisms of action of DKAs, we analyzed the antiviral activities of two potent azido-containing DKAs with either a monosubstitution or a disubstitution of azido groups, using single- and multiple-replication-cycle assays. Both azido-containing DKAs significantly inhibited HIV-1 infection in 293T, CEM-SS, and H9 cells (50% inhibitory concentration = 2 to 13 μM) and exhibited low cytotoxicity (50% cytotoxic concentration = 60 to 600 μM). Inhibition of HIV-1 IN in vivo was demonstrated by the observation that previously described L-708,906 resistance mutations in HIV-1 IN (T66I and T66I/S153Y) also conferred resistance to the azido-group-containing DKAs. In vitro assays and in vivo analysis indicated that the DKAs did not significantly inhibit the 3′ processing and selectively inhibited the strand transfer reaction. In addition, quantitative PCR indicated that two-long-terminal-repeat (2-LTR) circles were elevated in the presence of the azido-containing DKAs, confirming that HIV-1 IN was the intracellular target of viral inhibition. To gain insight into the mechanism by which the DKAs increased 2-LTR-circle formation of 3′-processed viral DNAs, we performed extensive DNA sequencing analysis of 2-LTR-circle junctions. The results indicated that the frequency of deletions at the circle junctions was elevated from 19% for the untreated controls to 32 to 41% in the presence of monosubstituted (but not disubstituted) DKAs. These results indicate that the structure of the DKAs can influence the extent of degradation of viral DNA ends by host nucleases and the frequency of deletions at the 2-LTR-circle junctions. Thus, sequencing analysis of 2-LTR-circle junctions can elucidate the intracellular mechanisms of action of HIV-1 IN inhibitors.
All antiretroviral drugs that are currently being used for the treatment of human immunodeficiency virus type 1 (HIV-1), with the exception of a recently approved peptide inhibitor of HIV-1 entry, target the viral pol-encoded enzymes reverse transcriptase (RT) and protease (PR) (13, 51). Combination therapy with RT and PR inhibitors has provided significant clinical benefits to HIV-1-infected individuals by suppressing viral replication and reducing mortality associated with AIDS (2, 48, 65). However, the efficacy of these drugs is limited by rapid selection of viral variants resistant to the drugs, toxicity, lack of adherence, and high cost (27, 59). Therefore, new inhibitors of HIV-1 replication that can be used as antiviral drugs are needed. HIV-1 integrase (IN), another enzyme encoded by the pol gene of HIV-1, is an attractive target for drug development because it is necessary and specific for viral replication (41). Similarities between HIV-1 IN and cellular enzymes such as RAG1/2 have suggested that some IN inhibitors interfere with B- and T-cell development (49). Nevertheless, because integration of viral DNA into the host chromosome is an enzymatic reaction that is not needed for host cell replication, inhibitors of HIV-1 IN have the potential to exhibit high specificity and low cytotoxicity.
HIV-1 IN is produced by proteolytic processing of the Gag-Pol polyprotein and consists of N-terminal, core, and C-terminal domains (10, 17). The crystal and/or nuclear magnetic resonance structures of the three domains have been determined in various combinations, but the structure of the entire HIV-1 IN protein has not been determined (8, 15, 16, 25, 43, 66). The N-terminal domain, which contains a zinc binding motif, is believed to be important for multimerization of IN (71); the core domain contains the catalytic site residues that constitute the highly conserved D,D,35-E motif (18); the C-terminal domain is believed to be involved in DNA binding (19, 23, 44). The interactions of HIV-1 IN with the substrate DNA complex have not been fully determined.
The first catalytic step in integration, 3′ processing, involves recognition of specific sequences at the ends of the long terminal repeats (LTRs) and removal of GT dinucleotides from the 3′ ends (10, 57). After 3′ processing, which occurs in the cytoplasm, the viral DNA/IN complex is transported to the nucleus, where the second catalytic step, strand transfer, is carried out. During strand transfer, IN cleaves the host genomic DNA to generate staggered ends as it ligates the 3′ OH ends of viral DNA to the 5′ phosphate ends of host DNA at the site of cleavage. Host cell enzymes are implicated in filling the gaps at the site of integration and removing unpaired dinucleotides from the viral DNA ends to complete the integration reaction (4, 11, 12, 68).
In addition to the linear form of viral DNA that serves as a substrate for integration, two other circular forms containing either one or two LTRs are observed in infected cells (7, 32, 38). The 2-LTR-circle form is generated by ligation of the ends of the viral DNA, and the 1-LTR-circle form may arise during reverse transcription or by recombination between the LTRs of a 2-LTR circle (36, 62). It has been previously shown that the 2-LTR-circle forms are increased in the presence of anti-IN inhibitors (6, 31). The mechanism by which the 2-LTR-circle forms are elevated is unknown; one possibility is that the viral DNA ends become more accessible to the cellular nucleases and ligases when the integration reaction is inhibited.
Potential inhibitors of HIV-1 IN have been identified by in vitro assays using purified IN and substrate oligonucleotides (5, 20, 21, 29, 30, 33, 35, 42). Potential IN inhibitors identified in such assays interfere with either the formation of a complex between IN and substrate DNA, 3′ processing, or the strand transfer reaction (56). Many potential inhibitors identified through these assays do not exhibit antiviral activity in cell culture assays or exhibit high levels of cytotoxicity (53, 56). In addition, when potential IN inhibitors exhibit antiviral activity, the molecular target of viral inhibition may not be the integration reaction. For example, G-tetrads and l-chicoric acid, two compounds that inhibit HIV-1 IN in vitro and exhibit antiviral activity, were recently shown to inhibit viral infection, at least in part, by binding to the viral envelope protein and inhibiting entry (9, 34, 40, 42, 55, 58). These studies underscore the need for reliable cell-based assays to analyze the antiviral activities and intracellular mechanisms of action of potential IN inhibitors.
β-Diketo acid derivatives (DKAs) are potent inhibitors of HIV-1 integration in vitro and in vivo (3, 31, 45, 53, 70). High-throughput screening assays were performed to identify DKAs such as L-708,906 that are selective and potent inhibitors of the strand transfer reaction catalyzed by HIV-1 IN (31). Another DKA, 4-(5-chloroindol-3-yl)-2,4-dioxobutanoic acid (5-CITEP), was also identified as a structural lead on the basis of in vitro anti-HIV-1 IN activity (22, 63). Crystal structures of the HIV-1 IN core domain with 5-CITEP indicate that this DKA is bound to the enzyme between the catalytic triad residues, supporting the hypothesis that the inhibitor competes with the substrate DNA for binding to IN (25).
DKAs L-731,988 and L-708,906 selectively inhibit the strand transfer reaction at nanomolar concentrations but require significantly higher concentrations for inhibition of the 3′-processing reaction (31, 46, 52, 69). Both of these DKAs have been shown to inhibit HIV-1 in single- as well as multiple-replication-cycle assays. In addition, HIV-1 IN was confirmed as the molecular target of viral inhibition by identification of mutations in HIV-1 IN that confer resistance to these DKAs (31). Moreover, it was shown that the 2-LTR-circle forms of viral DNA were elevated in the presence of the DKAs, further confirming that the integration reaction was inhibited in vivo (6, 31).
Recently, we described azido-group-containing DKAs that potently inhibit the strand transfer reaction catalyzed by HIV-1 IN (45, 70). In the present study, we have further evaluated the antiviral activity of these compounds and analyzed their intracellular mechanism of action. The results indicate that in the presence of azido-containing DKAs, the spread of replication-competent HIV-1 is inhibited and 2-LTR-circle formation is increased. In addition, mutations previously shown to confer resistance to the DKA L-708,906 also conferred resistance to the azido-containing DKAs, confirming that HIV-1 IN is the molecular target of viral inhibition. Furthermore, we demonstrated that the DKAs did not affect the 3′-processing reaction during HIV-1 infection. Interestingly, the structures of the azido-containing DKAs affected the frequency of deletions at the 2-LTR-circle junctions, providing evidence that the monosubstituted DKAs increased the extent to which viral DNA ends are degraded by host nucleases.
MATERIALS AND METHODS
Plasmids and site-directed mutagenesis.
Plasmids pHCMV-G, pHDV-EGFP (kindly provided by Derya Unutmaz), and pNL4-3 were described previously (1, 64, 67). Plasmid pNLuc was a kind gift from Eric Freed (39). Plasmids pD64E, pT66I, and pT66I/S153Y were constructed using the QuikChange site-directed mutagenesis kit (Stratagene). An ApaI-SalI restriction fragment of pNL4-3 was subcloned into pBS-KS+ to generate plasmid pES-HIVApa-Sal, which was used for site-directed mutagenesis (Stratagene) (1). The mutagenic primers that were used are available upon request. After mutagenesis, an AgeI-PflMI fragment from the mutated pES-HIVApa-Sal plasmids was sequenced to verify the presence of the desired mutation and absence of undesired mutations. The AgeI-PflMI fragment from the mutated pES-HIVApa-Sal plasmids was used to replace the same restriction fragment in pNLuc.
To quantify the 2-LTR-circle forms by real-time PCR analysis, a control plasmid, pRT-2LTRK, was generated by deletion of the BssHII-KpnI fragment (8,294 bp) of pNL4-3. The structure of this plasmid is similar to that of 2-LTR-circle forms, except that the LTRs are separated by 146 nucleotides (nt).
Cells, transfection, and virus production.
293T cells (American Type Culture Collection [ATCC]) were maintained in the presence of Dulbecco's modified Eagle's medium (Cellgro), 10% fetal calf serum (HyClone Laboratories), penicillin (50 U/ml; Gibco), and streptomycin (50 μg/ml; Gibco). H9 T-lymphoid cells (ATCC) were maintained in the presence of RPMI 1640 medium (Cellgro), 20% fetal calf serum, penicillin (50 U/ml), and streptomycin (50 μg/ml). CEM-SS cells (ATCC) were maintained in the presence of RPMI 1640 medium, 10% fetal calf serum, penicillin (50 U/ml), and streptomycin (50 μg/ml).
To produce concentrated pNLuc or pNL4-3 virus, 293T cells were plated at a density of 5 × 106 cells per 100-mm-diameter dish and transfected with pNLuc or pNL4-3 DNA by using the modified bovine serum mammalian transfection kit (Stratagene). Cell supernatants were harvested 48 h after transfection and filtered using 0.45-μm-pore-size membranes (Nalgene). pNLuc virus was concentrated 10-fold by centrifugation at 25,000 rpm (Surespin; Sorvall) for 90 min at 4°C and stored at −80°C.
Single-replication-cycle assays.
293T cells were plated at a density of 105 cells per 35-mm-diameter dish and used as targets of infection. Concentrated pNLuc virus was diluted 100-fold and used to infect the 293T cells for 1 h as previously described (26). The DKAs were dissolved in 100% dimethyl sulfoxide. The 293T target cells were incubated with medium containing DKAs for 4 h prior to infection, 1 h during infection, and 24 h after infection. To minimize cytotoxicity from dimethyl sulfoxide, the final concentration in the cell culture medium was maintained at less than 0.5%. To determine luciferase activities, 293T cells were washed with phosphate-buffered saline 72 h after infection and lysed in 400 μl of reporter lysis buffer (Promega). The cell lysates were subjected to one freeze-thaw cycle, and cell membranes were removed by centrifugation for 5 min at maximum speed in a microcentrifuge. After the addition of 100 μl of substrate to 20 μl of cell lysate, luciferase activity was determined using a TD20/20 luminometer (Promega).
H9 or CEM-SS target cells were plated in 96-well plates at a density of 5 × 104 cells per well in 100 μl of medium. The DKAs were added to the target cells to achieve the desired concentrations. The cells were incubated with the DKAs for 4 h and then infected with 10 μl of concentrated virus per well. Forty-eight hours later, luciferase activities in H9 and CEM-SS infected cells were determined by addition of 100 μl of reconstituted reagent (SteadyGlow kit; Promega), incubation for at least 5 min, and measurement of chemiluminescence by using a 96-well luminometer (Lumistar Galaxy; BMG Labtechnologies).
Multiple-replication-cycle assays.
Replication-competent pNL4-3 virus was used to infect H9 or CEM-SS target cells. H9 target cells were plated at a density of 5 × 105 cells per 35-mm-diameter dish and infected using culture supernatants containing 1 μg of HIV-1 p24 antigen in 2 ml of medium. After 4 h, the cells were washed, resuspended in 2 ml of cell culture medium containing a DΚΑ, and maintained in 35-mm-diameter dishes. The culture medium was replenished every 3 days by replacing 1 ml of cell culture suspension with an equivalent amount of fresh medium containing the appropriate DΚΑ. After each 3-day interval, the harvested supernatants were clarified by filtration and assayed for p24 antigen, using an enzyme-linked immunosorbent assay (p24 core profile kit; DuPont).
Infections of CEM-SS target cells with replication-competent pNL43 virus were performed as follows: 2.5 × 105 cells in 100 μl were mixed with 100 μl of virus-containing supernatant from 293T transfected cells, incubated for 2 h, and then diluted with 5 ml of medium containing an appropriate concentration of a DKA. The cells were placed in a 25-ml flask, and the culture medium was replaced every day with fresh medium. Virus replication was monitored by enzyme-linked immunosorbent assay for HIV-1 p24 antigen.
Cell proliferation inhibition assay.
The effects of DKAs on cell toxicity were determined by using the Roche cell proliferation kit II (sodium 3′-[1-(phenylaminocarbonyl)-3,4-tetrazolium]-bis(4-methoxy-6-nitro) benzene sulfonic acid hydrate [XTT] assay; Roche Molecular Biochemicals). 293T cells were plated at a density of 103 cells per well in a 96-well plate. Twenty-four hours later, the cells were incubated with various concentrations of a DKA for 36 h. Subsequently, the cells were incubated for an additional 48 h in the absence of the inhibitor and stained with 50 μl of the XTT labeling mix (Roche Molecular Biochemicals) per well. After 4 h, spectrophotometric absorbance of the samples was measured at a wavelength of 450 nm with a reference point at 700 nm in a 96-well plate reader (V-max Kinetic microplate reader; Molecular Devices). Proliferation of H9 or CEM-SS cells was measured in a similar manner by plating 5 × 103 cells per well and incubating the cells with various concentrations of the DKAs for 48 h.
Viral DNA quantification by real-time PCR.
Real-time PCR assays were performed using the TaqMan Universal PCR Master Mix kit (Roche Molecular Biochemicals), and the products were quantified using the ABI Prism 7700 sequence detection system.
To quantify viral DNA products by real-time PCR, 293T target cells were plated at a density of 105 cells per 60-mm-diameter dish. Twenty-four hours later, the cells were incubated in medium containing a 25 μM concentration of an azido-containing DKA with either a monosubstitution (MA-DKA) or a disubstitution (DA-DKA) of azido groups, or L-708,906 for 4 h. Control experiments were performed in the absence of any drug or in the presence of 1 μM 3-azido-3-deoxythymidine. Concentrated pNLuc virus was thawed, treated with 30 U of RNase-free DNase I (Roche Molecular Biochemicals) per ml of virus for 20 min in the presence of 2 mM MgCl2, and diluted 10-fold. The 293T cells were infected with pNLuc virus for 2 h in the presence of the DKA of interest. The virus was removed, and the cells were further incubated with medium containing the DKA for up to 48 h. Twelve, 24, and 48 h after infection, the cells were harvested and total cellular DNA was extracted (QIAamp DNA blood mini kit; Qiagen). DNA from approximately 2 ×103 to 5 ×103 cells was used for each real-time PCR assay. The following previously described primer-probe sets were used: for the 2-LTR-circle forms, the forward primer was MH535 (5′-AACTAGGGAACCCACTGCTTAAG-3′), the reverse primer was MH536 (5′-TCCACAGATCAAGGATATCTTGTC-3′), and the probe was 2LTR-P {5′-(carboxyfluorescein [FAM])-ACACTACTTTGAGCACTCAA GGCAAGCTTT-(5-carboxytetramethylrhodamine [TAMRA])-3′} (6). Total viral DNA was quantified by using the previously described U5-Ψ primer-probe set that detected late RT products (6). The late RT primer sequences were ES531 (5′-TGTGTGCCCGTCTGTTGTGT-3′) and ES532 (5′-GAGTCCTGCGTCGAGAGATC-3′), and the probe was LRT-P [5′-(FAM)-CAGTGGCGCCCGAACAGGGA-(TAMRA)-3′]. Plasmid pRT-2LTRK was used to generate standard curves for the 2-LTR and late RT primer-probe sets. A primer-probe set designed to quantify the copy numbers of the cellular gene porphobilinogen deaminase (PBGD; kindly provided by Michael Piatak, Science Applications International Corp., Frederick, Md.) was used to normalize the amount of DNA in each PCR assay. The PBGD primer-probe set consisted of the forward primer DF3 (5′-AGGGCAGGAACCAGGGATTATG-3′) the reverse primer DR1 (5′-GGGCACCACACTCTCCTATCTTT-3′), and the probe PBGD 01 (5′-ATGTCCACCACAGGGGACAAGATTC-3′). A plasmid containing a PBGD gene fragment was used to generate a standard curve for the PBGD primer-probe set.
Sequencing analysis of 2-LTR-circle junctions.
293T cells (2 × 105 cells per 60-mm-diameter dish) were infected in the absence or presence of DKA (25 μM) for 2 h. Twenty-four hours later, cells were harvested and total cellular DNA was isolated. The total DNA was used to amplify 2-LTR-circle junctions by PCR using the primers FOR-2LTR-EcoR (5′-GAGCTCTGAATTCAACTAGGGAACCCACTGCTTAAG-3′) and REV-2LTR-Xba (5′-GTGTCTAGATCCACAGATCAAGGATATCTTGTC-3′). PCR primers contained EcoRI and XbaI restriction sites that were used to clone the amplified PCR products into pBS-KS+. The PCR products were purified by a PCR purification kit (Qiagen), digested with EcoRI and XbaI restriction enzymes (Roche Molecular Biochemicals) for 3 h, and ligated with pBS-KS+ EcoRI/XbaI vector for 12 h at 16°C. The ligated products were transformed into DH5α competent cells (Gene Choice). Plasmid DNA was isolated from individual bacterial colonies by using a BioRobot 9600 (Qiagen). The EcoRI-XbaI inserted fragments were sequenced using the M13 sequencing primer 5′-GTAAAACGACGGCCAGT-3′ (Laboratory of Molecular Technology, Science Applications International Corp.).
In vitro assays for HIV-1 IN 3′-processing and strand transfer activities.
The in vitro HIV-1 IN assays were performed as previously described (46). Briefly, wild-type HIV-1 enzyme was purified, IN-DNA complexes were preformed, and the integration reactions were carried out for 1 h at 37°C in the presence of MgCl2 (45). Radioactively labeled products of the integration reaction were separated by gel electrophoresis, and the intensities of bands representing the 3′-processing and strand transfer reactions were quantified by using a phosphorimager and densitometric analysis.
In vivo analysis of HIV-1 IN 3′-processing reaction.
A cell clone of 293T cells that stably expresses pHDV-EGFP was transfected with pHCMV-G; 24 h later, virus was harvested, concentrated fivefold, and used to infect 293T cells (5 × 106 cells per 100-mm-diameter dish) in the absence or presence of 25 μM DKAs. Cytoplasmic DNA was extracted 6 h after infection as described previously (50), digested with HindIII for 4 h, treated with RNase A, and purified by phenol-chloroform extraction and ethanol precipitation. DNA pellets were resuspended in 7 μl of water at 68°C for 30 min; 7 μl of loading buffer (95% formamide, 20 mM EDTA, 0.05% bromophenol blue, 0.05% xylene cyanol) was subsequently added, and the samples were incubated at 68°C for 30 min and at 95°C for 10 min. The samples were resolved by electrophoresis through a 10% polyacrylamide gel containing 8 M urea in 0.5× Tris-borate-EDTA (TBE) buffer and then transferred to a Duralon-UV membrane (Stratagene) for 30 min at 200 mA in 0.5× TBE (Pharmacia Biotech). The membrane was cross-linked with 2 × 105 μJ of UV (Stratalinker) and incubated with UltraHyb (Ambion) for 20 h at 40°C. DNAs were detected by hybridization to 32P-labeled RNA probes (107 cpm/ml in 12 ml of UltraHyb [Ambion]) for 20 h at 39°C. To generate RNA probe, a PCR was performed using the pHDV-EGFP plasmid and the following primers: Ribo-F, 5′-AAAAAAGCTTGCCTTGAGTGCTCAA-3′; T7-Ribo-R, 5′-TAATACGACTCACTATAGGACTGCTAGAGATTTTCCACACTGAC-3′. The resulting PCR product was gel purified and utilized as a template for in vitro transcription (MAXIscript; Ambion). The hybridized membrane was washed twice for 10 min in Northern MAX low-stringency wash solution (Ambion) and for 10 min in Northern MAX high-stringency wash solution (Ambion). The radioactive signals were detected by using a Molecular Imager FX (Bio-Rad) and quantified using Quantity One (Bio-Rad). To generate molecular weight control markers for processed and unprocessed bands, two oligonucleotides (103 and 105 nt) were synthesized. The sequence of the 103-nt oligonucleotide was identical to the plus strand of 3′-processed product: 5′-AGCTTGCCTTGAGTGCTCAAAGTAGTGTGTGCCCGTCTGTTGTGTGACTCTGGTAACTAGAGATCCCTCAGACCCTTTTAGTCAGTGTGGAAAATCTCTAGCA-3′. The sequence of the 105-nt oligonucleotide was identical to the plus-strand product that did not undergo 3′ processing: 5′-AGCTTGCCTTGAGTGCTCAAAGTAGTGTGTGCCCGTCTGTTGTGTGACTCTGGTAACTAGAGATCCCTCAGACCCTTTTAGTCAGTGTGGAAAATCTCTAGCAGT-3′. These markers were mixed at a 1:1 molar ratio and loaded in polyacrylamide gels in adjacent lanes, transferred to the membrane, and detected by hybridization to the RNA probe.
RESULTS
Structures of azido-containing DKAs that inhibit HIV-1 IN.
We recently synthesized a series of DKA derivatives and determined their abilities to inhibit HIV-1 IN in an in vitro assay (53, 70). In these studies, DKAs that contained one or two azido moieties exhibited potent and selective inhibition of the strand transfer reaction catalyzed by HIV-1 IN (46, 70). The structures of the previously reported DKA L-708,906 (31), another structurally related benzyloxy-containing DKA (MB-DKA), and two azido-containing DKAs (DA-DKA and MA-DKA) are shown in Fig. 1A. These DKAs contain a diketone that separates a carboxylic acid and a benzene ring moiety. A disubstitution of the benzene ring with benzyloxy groups forms L-708,906. Removal of one of the benzyloxy groups results in the MB-DKA. The structure of the DA-DKA is similar to L-708,906 and is disubstituted with azido groups instead of benzyloxy groups. The structure of the MA-DKA is similar to MB-DKA and is monosubstituted with an azido group.
FIG. 1.
Structures of azido-containing DKAs and single-replication-cycle assay for screening inhibitors of HIV-1 replication. (A) Structures of DKAs shown are disubstituted azido-containing DKA (DA-DKA), disubstituted benzyloxy-containing DKA (L-708,906), monosubstituted azido-containing DKA (MA-DKA), and monosubstituted benzyloxy-containing DKA (MB-DKA). (B) Schematic representation of pNLuc vector and single-replication-cycle assay for testing antiviral activities of compounds. The pNLuc vectorcontains HIV-1 LTRs, all other cis-acting elements, and genes encoding all HIV-1 viral proteins (Gag, Pro, Pol, Vif, Vpr, Tat, Vpu, and Rev) except envelope (Env) and Nef. The vector also expresses the firefly luciferase gene (luc). Plasmid pHCMV-G encodes the G glycoprotein of the vesicular stomatitis virus envelope, which was expressed from the cytomegalovirus promoter. The shaded area represents treatment of target cells with DKAs. (C) Luciferase activity in 293T cells infected with pNLuc virus. Luciferase activity of infected cells in the absence of any drug (set to 100%) or in the presence of L-708,906 (25 μM) is shown. Luciferase activity in cells infected with pNLuc virus containing the D64E mutation in HIV-1 IN is also shown. The mean of three independent infections and standard error of the mean (error bars) are indicated. In these experiments, the average luminometer signal for wild-type (WT) virus infection in the absence of drug was 1,092 relative light units (RLU), whereas the negative control (uninfected cells) resulted in luminometer signals of <1 RLU.
Development of a sensitive single-replication-cycle assay for determination of antiviral activities of potential inhibitors.
To further investigate antiviral activities of the azido-containing DKAs, we developed a single-replication-cycle assay for testing IN inhibitors in vivo (Fig. 1B). The HIV-1 retroviral vector pNLuc expresses the firefly luciferase reporter gene and all HIV-1 proteins except for the Env and Nef proteins. pNLuc was cotransfected into 293T cells with pHCMV-G, which expresses the G glycoprotein of vesicular stomatitis virus (67). Virus harvested from the transfected cells was used to infect target cells in the presence or absence of the DKAs tested. The DKAs were added to target cell cultures at least 4 h prior to infection and were present for the next 48 to 72 h, at which point the ability of the DKAs to inhibit viral replication was measured by determining the amount of luciferase activity in the infected cells.
To evaluate whether the luciferase activity was dependent on integration of viral DNA into the host cell DNA, we substituted an aspartic acid at position 64 of HIV-1 IN with a glutamic acid (D64E). Because D64 is a component of the highly conserved D,D,35E catalytic site motif in HIV-1 IN, the mutation was expected to abolish the HIV-1 IN activity (14, 18, 24). When pNLuc containing the D64E mutation was used in the single-replication-cycle assay, the luciferase activity was decreased to less than 1% of the activity present in cells infected with wild-type pNLuc (Fig. 1C). This result indicated that expression of the luciferase gene was dependent on integration of viral DNA mediated by HIV-1 IN.
To verify that inhibition of HIV-1 IN with DKAs could be detected in the single-replication-cycle assay, we performed infection with pNLuc virus in the presence of 25 μM L-708,906 (Fig. 1C). The luciferase activity was decreased to 7% of the activity present in cells infected with pNLuc in the absence of an inhibitor, indicating that inhibition of HIV-1 IN with a DKA derivative resulted in a substantial reduction in luciferase expression.
Quantification of antiviral and cytotoxic activities of azido-containing DKAs.
To quantify the antiviral activities of DKA derivatives, we determined the concentrations of drugs needed to inhibit viral replication by 50% (IC50) in 293T cells as well as two human T-cell lines (CEM-SS and H9). First, to determine the IC50 values using 293T target cells, we performed the single-replication-cycle assay in the presence of a series of concentrations of the DKAs (0.05, 0.5, 5, and 50 μM). The results indicated that the IC50 values for the MA-DKA and DA-DKA were 2.1 and 5.0 μM, respectively (Table 1). These IC50 values were similar to the IC50 value that we observed for L-708,906 (1.9 μM), which was in good agreement with the previously reported results (2.5 μM) (31).
TABLE 1.
IC50 and CC50 values for L-708,906, MA-DKA, and DA-DKA in 293T, CEM-SS, and H9 cells by using single- and multiple-replication-cycle assays
| Assay and DKA analyzed | 293T
|
CEM-SS
|
H9
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| IC50a (μM) | CC50b (μM) | TIc | IC50a (μM) | CC50b (μM) | TIc | IC50a (μM) | CC50b (μM) | TIc | |
| Single-replication-cycle assay | |||||||||
| L-708,906 | 1.9 | >50 | >26 | 0.43 | 65 | 151 | 1.8 | 90 | 50 |
| MA-DKA | 2.1 | >50 | >24 | 7.0 | 600 | 86 | 6.3 | 400 | 63 |
| DA-DKA | 5.0 | >50 | >10 | 4.2 | 180 | 43 | 7.0 | 60 | 9 |
| HIV-1 replication-competent assay | |||||||||
| L-708,906 | 0.86 | 65 | 76 | 1.4 | 90 | 64 | |||
| MA-DKA | 11 | 600 | 55 | 7 | 400 | 57 | |||
The IC50 was determined as an average of two or three independent experiments. For single-replication-cycle assays, luciferase activity of infected cells was determined 72 h after infection. For multiple-replication-cycle assays, the extent of virus replication was determined by quantifying p24 antigen present in culture supernatants 72 h after infection.
The CC50 was determined as an average of three independent experiments by using the XTT assay.
TI was calculated as the ratio of the corresponding CC50 and IC50 values.
We also quantified the cytotoxicities of the azido-containing DKAs by using 293T cells and an XTT assay (Table 1). The MA-DKA and DA-DKA displayed a 50% cytotoxic concentration (CC50) that was greater than 50 μM. Therefore, the reduction in luciferase activity in the presence of azido-containing DKAs was indeed due to inhibition of HIV-1 infection and not due to cytotoxicity.
The IC50 and CC50 values for MA-DKA, DA-DKA, and L-708,906 were also determined using CEM-SS and H9 cells as targets of infection. The results indicated that most of the IC50 values for the azido-containing DKAs were similar to those obtained in 293T cells (within fourfold). The CC50 values for the CEM-SS and H9 cells were comparable to each other. The therapeutic index (TI) for L-708,906 was similar to that of MA-DKA in H9 cells (50 versus 63) and approximately twofold higher in CEM-SS cells (151 versus 86). The MA-DKA was significantly less cytotoxic to both CEM-SS and H9 cells than L-708,906 and DA-DKA. The TI of MA-DKA was higher than that of DA-DKA because of its lower cytotoxicity.
MA-DKA and DA-DKA inhibit replication-competent HIV-1.
To further establish the efficacy of azido-containing DKAs in inhibiting HIV-1 replication, we performed a multiple-replication-cycle assay. H9 cells were infected with replication-competent pNL4-3 virus, and the cultures were maintained in media containing 0.6 to 40 μM MA-DKA or DA-DKA. The extent of virus replication was monitored by determining the concentration of p24 CA in the culture medium every 3 to 4 days over a 2-week period. The results indicated that both DKAs were able to inhibit virus replication in a dose-dependent manner (Fig. 2). At the highest concentration tested (40 μM), the p24 CA amounts in the culture medium were reduced to approximately 12% for MA-DKA-treated cultures and 4% for DA-DKA-treated cultures on day 14, relative to the amounts present in HIV-1-infected cultures in the absence of DKAs. These results indicated that azido-containing DKA derivatives strongly inhibited the spread of replication-competent HIV-1 in cell culture.
FIG. 2.
Dose-dependent inhibition of replication-competent HIV-1 virus by MA-DKA and DA-DKA in H9 cells. Inhibition of virus replication was evaluated by measuring the amounts of p24 capsid antigen at days 3 to 14. MA-DKA and DA-DKA were tested at concentrations ranging between 0.6 and 40 μM as indicated.
To quantify the ability of MA-DKA to inhibit replication-competent HIV-1, we determined the IC50 values for MA-DKA and L-708,906 by using pNL4-3 virus and CEM-SS as well as H9 target cells. The results indicated that the IC50 values for L-708,906 were significantly lower than those for MA-DKA (Table 1). However, the L-708,906 compound was also significantly more toxic to CEM-SS and H9 cells than MA-DKA; as a result, the TI values in CEM-SS and H9 target cells for MA-DKA (55 and 57, respectively) were similar to the TIs for L-708,906 (76 and 64, respectively). These results indicated that MA-DKA has antiviral activity similar to that of L-708,906.
Mutations in HIV-1 IN confer resistance to azido-containing DKAs.
To dissect the mechanism of HIV-1 inhibition by azido-containing DKAs, we introduced mutations into HIV-1 IN that were previously shown to confer resistance to DKAs (31). We generated T66I and T66I/S153Y mutations in the IN gene of pNLuc and performed single-replication-cycle assays. The T66I mutant and T66I/S153Y mutants displayed 10- and 1,000-fold reductions in viral titers, respectively, as estimated by the reduction in luciferase activity in comparison to that of the wild-type HIV-1 IN. In addition, we performed single-replication-cycle assays in the absence or presence of 0.05, 0.5, 5, and 50 μM MA-DKA, DA-DKA, or L-708,906. The results indicated that T66I and T66I/S153Y mutations in HIV-1 IN conferred resistance to L-708,906 DKA as well as MA-DKA and DA-DKA, indicating that HIV-1 IN is the target of viral inhibition for azido-containing DKAs in vivo (Fig. 3). Furthermore, the results suggested that the mechanism of inhibition of HIV-1 IN by azido-containing DKAs is similar to that of L-708,906.
FIG. 3.
Resistance to DKAs conferred by T66I and T66I/S153Y mutations in HIV-1 IN. Inhibition of integration was determined in the single-replication-cycle assay in the presence of 50 μM DKAs. Luciferase activities for control infections (No drug) were set at 100% for each experiment. The mean of three independent experiments and standard error of the mean (error bars) are indicated. To achieve similar luciferase activity in infected target cells, infections were performed at different dilutions for wild-type (WT) and mutant viruses (WT virus was diluted 100-fold; T66I virus was diluted 10-fold; T66I/S153Y mutant virus was diluted 2-fold). In these experiments, the average luminometer signal for WT virus infection in the absence of drug was 21,654 relative light units (RLU); the average luminometer signal for T66I virus infection in the absence of drug was 84,791 RLU; the average luminometer signal for T66I/S153Y virus infection in the absence of drug was 12,719 RLU. A negative control (uninfected cells) resulted in luminometer signals lower than 100 RLU.
Interestingly, the luciferase activity in target cells infected with the T66I/S153Y mutant was elevated in the presence of the DKAs, suggesting that the replication of this mutant was more efficient in the presence of the DKAs.
Quantification of viral DNA products in the presence of IN inhibitors.
To investigate the effect of azido-containing DKAs on viral DNAs produced during infection, we performed quantitative real-time PCR assays for detection of full-length linear and 2-LTR-circle forms of HIV-1 DNA. To quantify the viral DNA products, we infected 293T cells with pNLuc virus in the presence or absence of a DKA. Total cellular DNA was extracted 12, 24, and 48 h after infection. The full-length viral DNA products were measured by using a primer-probe set that was designed to amplify reverse transcription products that underwent plus-strand strong-stop DNA transfer (Fig. 4A, late RT-P). To quantify 2-LTR circles in HIV-1-infected cells, we used a primer-probe set that was designed to amplify and detect 2-LTR-circle junctions (Fig. 4A, 2LTR-P). The amounts of full-length as well as 2-LTR-circle forms of viral DNAs were normalized to the cell number equivalents in each sample, which were determined by real-time PCR using a primer-probe set that was used to quantify the cellular PBGD gene copy number.
FIG. 4.
Quantitative real-time PCR analysis of late RT product and 2-LTR-circle DNAs in the presence of DKAs. (A) Schematic representation of primer-probe sets that were used for real-time PCR. Primers ES531 and ES532 and probe LateRT-P were used to amplify late RT products (full-length viral DNA). Primers MH535 and MH536 and probe 2LTR-P were used to amplify the 2-LTR-circle form of viral DNA. (B) Full-length viral DNA copies detected by using primers andprobe for detecting late RT product at 12, 24, and 48 h postinfection in the absence or presence of DKAs (L-708,906, MA-DKA, and DA-DKA). Cell numbers were determined by quantification of the PBGD gene, which was present in a single copy in chromosomal DNA. (C) Copies of 2-LTR circles detected per cell at 12, 24, and 48 h postinfection in the absence or presence of DKAs and the cell numbers were determined by PBGD quantification. (D) 2-LTR circles presented as a percentage of full-length viral DNA transcripts.
The quantification of full-length viral DNAs showed that the late viral DNA products ranged from 6 to 13 copies per cell for 12, 24, and 48 h in the absence of IN inhibitors (Fig. 4B). Similar results were obtained when the cells were infected in the presence of the DKAs (approximately 10 copies of late viral DNA products per cell at 12, 24, and 48 h). These results indicated that viral DNA synthesis was not affected by the presence of azido-containing DKAs (MA-DKA and DA-DKA) or the benzyloxy-containing DKA L-708,906.
The quantification of 2-LTR-circle DNA forms in the absence and presence of DKAs is shown in Fig. 4C. The results indicated that in the absence of the DKAs, there were approximately one or two copies of 2-LTR circles per cell. In the presence of the DKAs, the 2-LTR-circle forms were significantly increased 24 and 48 h after infection. The amounts of 2-LTR circles were elevated to a greater extent in the presence of L-708,906 than in the presence of the azido-containing DKAs.
We also normalized the numbers of 2-LTR circles to the numbers of the late viral DNA products in order to calculate the percentage of viral DNAs that underwent 2-LTR-circle formation in the absence or presence of IN inhibitors (Fig. 4D). In the presence of 25 μM L-708,906, 80 to 100% of HIV-1 DNAs formed 2-LTR circles. Although the fraction of 2-LTR-circle viral DNA in cells infected in the presence of azido-containing DKAs was not as high as in the presence of L-798,906, it was approximately fourfold higher than in cells infected in the absence of any IN inhibitor (40 versus 10% at 24 h after infection). This result provided additional evidence that azido-containing DKAs inhibited HIV-1 infection at the level of integration and indicated that the azido-containing DKAs increased the frequency of 2-LTR-circle formation.
In vitro inhibition of 3′-processing and strand transfer reactions for monosubstituted or disubstituted DKAs.
To further dissect which step of the integration reaction is inhibited by the azido-group-containing DKAs, we performed a previously described in vitro assay in the presence of MgCl2 to determine the IC50 values for the strand transfer and 3′-processing reactions (31, 45, 46). As shown in Table 2, all four of the DKAs tested were potent and selective inhibitors of the strand transfer reaction. The IC50 values for the benzyloxy-containing L-708,906 and MB-DKA were 0.06 and 0.09 μM for the strand transfer reaction, respectively. Similarly, the IC50 values for the DA-DKA and MA-DKA were 0.46 and 0.33 μM for the strand transfer reaction, respectively. Inhibition of the 3′-processing reaction required significantly higher concentrations than those needed to inhibit the strand transfer reaction. The ratios of the IC50 values for the 3′-processing and strand transfer reactions (selectivity index) ranged from 42 to 212.
TABLE 2.
In vitro analysis of inhibition of 3′ processing and strand transfer reactions for monosubstituted or disubstituted DKAs
| DKA analyzed | 3′ processing IC50 (μM)a | Strand transfer IC50 (μM)a | Selectivity index for strand transfer (n-fold)b |
|---|---|---|---|
| Benzyloxy-DKAs | |||
| L-708,906 (disubstituted) | 2.5 | 0.06 | 42 |
| MB-DKA (monosubstituted) | 7.0 | 0.09 | 78 |
| Azido-DKAs | |||
| DA-DKA (disubstituted) | 24.5 | 0.46 | 53 |
| MA-DKA (monosubstituted) | 70 | 0.33 | 212 |
In vitro IC50 values for 3′-processing and strand transfer reactions were determined as an average of two or three independent experiments.
Selectivity index was calculated as the ratio of the IC50 value for the 3′-processing reaction and corresponding IC50 value for the strand transfer reaction.
In vivo efficiency of the 3′-processing reaction in the presence of DKAs.
To determine whether monosubstituted and disubstituted DKAs exhibited any differences in inhibition of 3′-processing reactions in vivo, we performed HIV-1 infection in the absence or presence of 25 μM concentrations of the DKAs. Cytoplasmic DNA was isolated 6 h after infection and digested with HindIII restriction enzyme. HindIII digestion of HIV-1 unintegrated viral DNA generated three different fragments that were derived from the 3′ end of viral DNA (Fig. 5A), which were separated on a denaturing polyacrylamide gel and visualized by using a probe that specifically hybridized to the plus-strand DNA. The fragment labeled +SSS (122 nt in length) was generated from the plus-strand strong-stop DNA and included 18 nt of the primer binding site sequence generated from copying the primer tRNA at the 3′ end of the LTR. The fragment labeled un (105 nt) was generated from the viral DNA end that resulted from completed synthesis of the 3′ LTR; this fragment did not undergo the 3′-processing reaction. The fragment labeled proc (103 nt) was generated from the viral DNA end that resulted from completed synthesis of the 3′ LTR; this fragment underwent the 3′-processing reaction. Interestingly, we also observed a 106-nt fragment that was most likely derived from completed synthesis of the 3′ LTR which did not undergo the 3′-processing reaction. This 106-nt fragment was equally present in the presence or absence of the DKAs, indicating that this fragment was not generated due to DKA treatment. We hypothesize that this fragment was either generated as a result of alternative cleavage of the tRNA primer by the RNase H domain of RT or by the addition of a nontemplated nucleotide at the 3′ end of the plus-strand DNA (54, 61).
FIG. 5.
In vivo analysis of 3′ processing in the absence or presence of DKAs. (A) Reverse transcription products that were detected by the RNA probe that hybridizes to the plus-strand of HIV-1 DNA (thick line) after HindIII digestion. The fragment of plus-strand strong-stop DNA that was detected by the probe was 122 nt in length (labeled +SSS). Unprocessed DNA generated a 105- or a 106-nt fragment (see text). The DNA ends that underwent the 3′-processing reaction carried out by HIV-1 IN generated a 103-nt product. RNA primers (PPT and tRNA) are shown as a dashed line. Minus-strand DNA is shown as a thin line. (B) Representative electrophoretic analysis of DNA products during HIV-1 infection. The lanes are labeled according to the drug treatment during infection. The sizes of the processed and unprocessed bands were determined by the migration of oligonucleotide markers of 103 and 105 nt length as described in Materials and Methods (results not shown). The band derived from plus-strand strong-stop DNA is labeled as +SSS, the unprocessed band is labeled as un, and the 3′-processed band is labeled as proc.
We quantified the efficiency of the 3′-processing reaction in the absence or presence of DKAs from two independent experiments by determining the percentage of total completed DNA ends that underwent 3′ processing (Fig. 5B). In the absence of DKAs, 51 to 57% of viral DNA ends underwent the 3′-processing reaction. No significant differences were observed in the in vivo efficiency of 3′ processing in the presence of the DKAs. The percentages of 3′-processed viral DNA ends were as follows: 55 to 60% for MA-DKA, 44 to 52% for MB-DKA, 57 to 63% for DA-DKA, and 46 to 56% for L-708,906. These results indicated that the monosubstituted and disubstituted DKAs diminish viral replication by inhibition of viral replication after the 3′-processing reaction.
Sequencing analysis of 2-LTR-circle junctions.
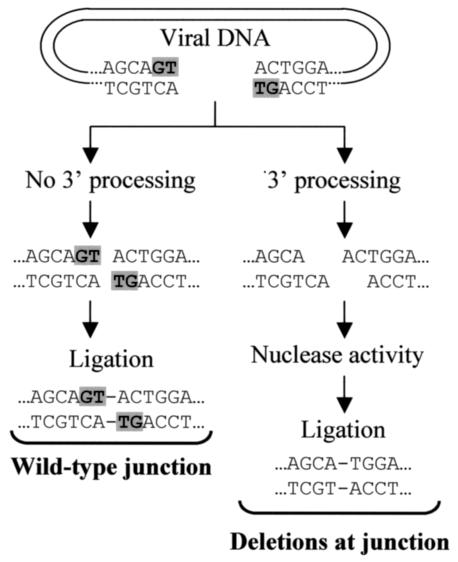
We sought to gain insight into the mechanism of IN inhibition and 2-LTR-circle formation by analyzing the structure of the 2-LTR-circle junctions. As outlined in Fig. 6, formation of viral DNA through reverse transcription is expected to generate blunt ends. In the absence of 3′ processing, viral DNA can potentially undergo blunt-end ligation mediated by host enzymes to create 2-LTR circles containing “wild-type” junctions. These wild-type junctions will contain the GT dinucleotide at each 3′ end of the junction. In the presence of a DKA that effectively inhibits 3′ processing, wild-type 2-LTR-circle junctions should be generated more frequently. On the other hand, in the presence of IN inhibitors that selectively inhibit the strand transfer reaction but have no effect on the 3′-processing reaction, the GT dinucleotides are expected to be removed, resulting in staggered nonpalindromic ends that cannot be directly ligated by host enzymes. We hypothesized that these 3′-processed DNA ends are processed further by host cell nucleases so that they can be ligated into 2-LTR circles; thus, we hypothesized that increased accessibility of viral DNA ends to the cellular nucleases would result in a higher frequency of deletions at the 2-LTR-circle junctions.
FIG. 6.
Deletions at 2-LTR-circle junctions that result from the host nuclease activity. A schematic representation of viral DNA is shown; the shaded GT dinucleotides were removed by HIV-1 IN during the 3′-processing reaction. Removal of the GT dinucleotide by HIV-1 IN through the 3′-processing reaction resulted in the generation of nonpalindromic ends. Deletion of additional sequences by host cell nuclease activity and ligation by host enzymes resulted in the formation of deletions at the 2-LTR-circle junctions. Absence of 3′ processing results in the generation of viral DNA with blunt ends, and ligation of the blunt-ended viral DNA with host enzymes results in the formation of wild-type circle junctions.
To analyze the effect of IN inhibitors on the structure of 2-LTR-circle junctions, we first performed sequencing analysis of 2-LTR circles formed in the absence of inhibitors. The results revealed that 60% of a total of 96 2-LTR-circle junctions were wild type and contained sequences that retained the GT dinucleotide, 19% contained deletions, and 18% contained insertions (Table 3; Fig. 7). Most of the circle junction sequences that were different from wild type were also different from each other, suggesting that each sequence represented an independent infection event. The majority of the insertions were derived from viral sequences such as polypurine tract (PPT) and the primer-binding site (PBS), as well as sequences adjacent to the PPT and/or PBS, confirming previous observations that these products were generated as a result of aberrant cleavage by RNase H during reverse transcription and were unable to serve as substrates for the integration reaction (37). Finally, very few of the 2-LTR-circle junctions isolated from the control infections contained substitution mutations (2 of 96) in the GT dinucleotide at the end of the LTR.
TABLE 3.
Frequencies of wild-type sequences, deletions, insertions, and mutations present at 2-LTR-circle junctionsa
| Nature of circle junction | Frequency (mean % ± SE)c in the presence of:
|
|||||
|---|---|---|---|---|---|---|
| No drugb | D64Eb | Monosubstituted DKAs
|
Disubstituted DKAs
|
|||
| MA-DKAb | MB-DKAb | DA-DKAb | L-708,906b | |||
| WT 2-LTR | 60 ± 5 | 88 ± 4 | 56 ± 8 | 64 ± 6 | 71 ± 13 | 75 ± 7 |
| Deletions | 19 ± 4 | 5 ± 3 | 41 ± 6 | 32 ± 5 | 19 ± 10 | 20 ± 5 |
| Insertions | 18 ± 3 | 5 ± 2 | 3 ± 2 | 2 ± 2 | 2 ± 2 | 5 ± 5 |
| Mutations | 3 ± 2 | 1 ± 1 | 0 | 3 ± 3 | 5 ± 3 | 0 |
The numbers of each type of 2-LTR-circle junction are shown in Fig. 6.
The 2-LTR-circle junctions were isolated from 3 to 12 independent experiments.
SE represents the standard error of the mean. The likelihood chi-square decomposition test was performed to determine if the frequencies of wild-type (WT) circle junctions and circle junctions with deletions were statistically different for no drug and DKA-treated experiments. It was found that frequency of WT junctions was not different for monosubstituted DKA-treated infections versus no-drug control infections (P = 0.83), whereas the frequency of WT junctions was higher for disubstituted DKA-treated infections than for no-drug control infections (P = 0.014). We also found that the frequency of junctions containing deletions was not different for disubstituted DKA-treated infections versus no-drug control infections (P = 0.241), whereas the frequency of junctions with deletions was higher for monosubstituted DKA-treated infections than for no-drug control infections (P = 0.010).
FIG.7.
Sequencing analysis of 2-LTR-circle junctions. The normal 2-LTR-circle junction is indicated as wild type (WT). GT and AC terminal nucleotides that were removed by IN during the 3′-processing reaction are shown in bold. Dashes represent deletions. All sequences containinga deletion or mutation are aligned to the WT sequence. All sequences containing an insertion are aligned to each other. PPT and PBS sequences are underlined. Insertions containing PBS sequences are adjacent to the 3′ LTR. Insertions containing PPT sequences are adjacent to the 5′ LTR. Large deletions or insertions are indicated by − or + followed by the numbers of total nucleotides deleted or inserted, respectively. The total number of each detected sequence is shown for wild-type virus in the absence of drug treatment (No drug), for D64E mutant of HIV-1 IN in the absence of drug treatment, and for wild-type virus in the presence of monosubstituted and disubstituted DKAs. The total number of sequences that were analyzed is also indicated in the bottom of the table.
To verify the hypothesis that the frequency of wild-type circle junctions will increase with inhibition of 3′ processing, we analyzed the sequences of 2-LTR-circle junctions that were formed during infection with a virus that contained a D64E mutation at the active site of HIV-1 IN. The D64E mutation completely abolished IN function; consequently, no 3′ processing of the viral DNA ends was expected to occur and the frequency of wild-type junctions was expected to increase. Consistent with this expectation, the frequency of the wild-type junctions was significantly increased from 60% for the control infections with wild-type virus to 88% for the D64E mutant virus, confirming that inhibition of 3′ processing leads to an increase in the frequency of 2-LTR circles containing wild-type junctions (Table 3; Fig. 7).
In the presence of the monosubstituted or disubstituted DKAs as well as the D64E mutation in HIV-1 IN, the frequencies of 2-LTR circles containing insertions were very low (2 to 5%) (Table 3; Fig. 7). It was previously reported that inhibition of HIV-1 RNase H activity results in an increased frequency of PPT and/or PBS insertions at the 2-LTR-circle junctions (37). It has also been reported that some DKAs inhibit HIV-1 RNase H activity in vitro (60). The low frequencies of insertions at the circle junctions indicated that DKAs did not affect the cleavage specificity and/or activity of HIV-1 RNase H.
To examine the mechanisms by which DKAs inhibit integration and increase 2-LTR-circle formation, we performed extensive DNA sequencing analysis of 2-LTR-circle junctions generated in the presence of monosubstituted (MA-DKA and MB-DKA) or disubstituted (DA-DKA and L-708,906) DKAs (Table 3; Fig. 7). The results indicated that for the monosubstituted DKAs, the frequency of deletions at the 2-LTR-circle junctions was elevated from 19% for the control infections to 32 to 41%. However, the frequency of deletions at the 2-LTR-circle junctions was not elevated for the disubstituted DKAs (19 to 20%). The frequency of the wild-type junctions was elevated in the presence of the disubstituted DKA L-708,906 in comparison to the control infections (75%). These results suggested that the monosubstituted DKAs, but not disubstituted DKAs, increase the accessibility of viral DNA ends to host cell nucleases, resulting in a higher frequency of deletions at the 2-LTR-circle junctions.
DISCUSSION
The results of these studies confirm and extend our previous findings that azido-containing DKAs are potent inhibitors of HIV-1 IN (45, 53, 70). The azido-containing DKAs inhibited replication-competent virus in a dose-dependent manner. The MA-DKA exhibited a lower cytotoxicity and a similar TI relative to L-708,906 in H9 cells. The similar TI resulted from a lower antiviral activity coupled with a lower cytotoxicity, suggesting that substitution of the benzyloxy groups with the azido groups resulted in a reduction in cytotoxicity. Recently, 8-hydroxy-(1,6)naphthyridines were shown to inhibit HIV-1 IN with a substantially higher potency than L-708,906 and the azido-containing DKAs described here (28, 72). In addition, another DKA compound related to 5-CITEP (S-1360) is in clinical trial (3). Although the azido-containing DKA derivatives described here exhibit lower antiviral activities than the newly described HIV-1 IN inhibitors, our characterization of the intracellular mechanisms of action of the azido-containing DKAs could be useful in further development of potent antiviral agents that target HIV-1 IN.
Mutations that were previously shown to confer resistance to L-708,906 (31) also conferred resistance to the azido-containing DKAs, suggesting that the benzyloxy group-containing- and azido-containing DKAs bind to HIV-1 IN similarly. Modeling of the azido-containing and benzyloxy-containing DKAs indicates that they are essentially superimposable, but the benzyloxy groups occupy more space (70). Interestingly, recent modeling studies have indicated that the azido-containing DKAs might bind to the active site of HIV-1 IN in two different modes: in addition to binding to HIV-1 IN in a manner similar to that of L-708,906, another mode of binding could take place in which the azido moiety, rather than the diketo moiety, participates in metal chelation (45). The observation that the T66I/S153Y mutation confers resistance to azido-containing DKAs as well as L-708,906 suggests that these mutations interfere with both of the proposed binding modes.
Interestingly, the T66I/S153Y mutants replicated more efficiently in the presence of the azido-containing DKAs and also the benzyloxy-containing DKAs, suggesting that in the presence of the DKAs these mutants exhibited higher levels of integration activity. It is conceivable that because the T66I/S153Y mutations of HIV-1 IN were selected in the presence of a DKA, the geometry of the active site of the drug-resistant enzyme is optimal for catalysis only when a DKA is bound to HIV-1 IN. The phenomenon of drug-dependent viral replication has been previously described (47); these studies also suggest that viruses containing drug resistance mutations evolved to enhance viral replication in the presence of the drug. The azido-containing DKAs could serve as useful tools for understanding the mechanisms by which these drug-resistant variants replicate more efficiently in the presence of the DKAs, the mechanisms by which DKAs inhibit HIV-1 integration, and the mechanisms of viral drug resistance.
In the presence of the azido-containing DKAs, the frequency of 2-LTR-circle formation was increased without affecting total viral cDNA synthesis. This result indicated that the DKAs did not significantly inhibit viral DNA synthesis. The in vivo analysis of 3′ processing also supported this view and indicated that there was no significant effect on reverse transcription at the ends of the viral DNAs, because the ratio of the plus-strand strong-stop DNA product (Fig. 6, +SSS) to the 3′-processed and unprocessed products (103 plus 105 plus 106 nt) was not altered in the presence of the DKAs. Our observation that the unprocessed DNA products were variable in length (105 and 106 nt) is consistent with the previously observed heterogeneity of the unprocessed bands (50). The observed 2- to 10-fold increase in 2-LTR-circle formation was in good agreement with previous studies indicating that HIV-1 IN is the molecular target of viral inhibition (6, 31). Furthermore, the results of these studies showed that analysis of the sequences at the 2-LTR-circle junctions can provide insight into the metabolism of unintegrated viral DNA in the presence of IN inhibitors. Our analysis of the 2-LTR-circle junctions in the absence of any drug treatment indicated that the frequencies of wild-type junctions (60%), deletions (19%), and insertions (18%) were in close agreement with those previously published (37). The structures of the DKAs clearly affected the frequency of deletions at circle junctions, indicating that there is a relationship between the DKA structure and the nature of the 2-LTR-circle junction. The in vivo analysis indicated that both monosubstituted and disubstituted DKAs do not inhibit the 3′-processing reaction. We hypothesize that the monosubstituted DKAs alter the structure of the preintegration complex to make the viral DNA ends more accessible to host nucleases, which results in an increase in the frequency of deletions.
The results of this study also showed that the DKAs did not affect the frequency of insertions at the 2-LTR-circle junctions, indicating that these DKAs do not have a significant influence on the in vivo RNase H activity. It is important to note that some DKAs have been shown to inhibit HIV-1 RNase H activity in vitro (60), highlighting the need to analyze the effects of potential DKA inhibitors of HIV-1 IN on the HIV-1 RNase H activity in vivo. Thus, analysis of 2-LTR-circle junctions can provide insights into the intracellular mechanisms of action of potential inhibitors with respect to the 3′-processing and strand transfer reactions catalyzed by HIV-1 IN, the nuclease activity catalyzed by HIV-1 RNase H, and the accessibility of unintegrated viral DNA ends to host nucleases.
Acknowledgments
We especially thank Wei-Shau Hu and John Coffin for valuable intellectual input and discussions throughout this project, Michael D. Miller (Merck Laboratories) for valuable advice and the protocol for in vivo analysis of 3′ processing, Doug Powell (Data Management Services—Frederick) for help with statistical analysis, and Anne Arthur for her editorial expertise and revisions. We also thank Jean Mbisa, Galina Nikolenko, and David Thomas for critical reading of the manuscript and useful discussions.
This work was supported by the HIV Drug Resistance Program, Laboratory of Medicinal Chemistry, and the Laboratory of Molecular Pharmacology, National Cancer Institute.
REFERENCES
- 1.Adachi, A., H. E. Gendelman, S. Koenig, T. Folks, R. Willey, A. Rabson, and M. A. Martin. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beale, K. K., and W. E. Robinson, Jr. 2000. Combinations of reverse transcriptase, protease, and integrase inhibitors can be synergistic in vitro against drug-sensitive and RT inhibitor-resistant molecular clones of HIV-1. Antivir. Res. 46:223-232. [DOI] [PubMed] [Google Scholar]
- 3.Billich, A. 2003. S-1360 Shionogi-GlaxoSmithKline. Curr. Opin. Investig. Drugs 4:206-209. [PubMed] [Google Scholar]
- 4.Brin, E., J. Yi, A. M. Skalka, and J. Leis. 2000. Modeling the late steps in HIV-1 retroviral integrase-catalyzed DNA integration. J. Biol. Chem. 275:39287-39295. [DOI] [PubMed] [Google Scholar]
- 5.Burke, T. R., Jr., M. R. Fesen, A. Mazumder, J. Wang, A. M. Carothers, D. Grunberger, J. Driscoll, K. Kohn, and Y. Pommier. 1995. Hydroxylated aromatic inhibitors of HIV-1 integrase. J. Med. Chem. 38:4171-4178. [DOI] [PubMed] [Google Scholar]
- 6.Butler, S. L., M. S. Hansen, and F. D. Bushman. 2001. A quantitative assay for HIV DNA integration in vivo. Nat. Med. 7:631-634. [DOI] [PubMed] [Google Scholar]
- 7.Butler, S. L., E. P. Johnson, and F. D. Bushman. 2002. Human immunodeficiency virus cDNA metabolism: notable stability of two-long terminal repeat circles. J. Virol. 76:3739-3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, J. C., J. Krucinski, L. J. Miercke, J. S. Finer-Moore, A. H. Tang, A. D. Leavitt, and R. M. Stroud. 2000. Crystal structure of the HIV-1 integrase catalytic core and C-terminal domains: a model for viral DNA binding. Proc. Natl. Acad. Sci. USA 97:8233-8238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cherepanov, P., J. A. Este, R. F. Rando, J. O. Ojwang, G. Reekmans, R. Steinfeld, G. David, E. De Clercq, and Z. Debyser. 1997. Mode of interaction of G-quartets with the integrase of human immunodeficiency virus type 1. Mol. Pharmacol. 52:771-780. [DOI] [PubMed] [Google Scholar]
- 10.Coffin, J. M., S. H. Hughes, and H. E. Varmus. 1997. Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 11.Daniel, R., R. A. Katz, G. Merkel, J. C. Hittle, T. J. Yen, and A. M. Skalka. 2001. Wortmannin potentiates integrase-mediated killing of lymphocytes and reduces the efficiency of stable transduction by retroviruses. Mol. Cell. Biol. 21:1164-1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daniel, R., R. A. Katz, and A. M. Skalka. 1999. A role for DNA-PK in retroviral DNA integration. Science 284:644-647. [DOI] [PubMed] [Google Scholar]
- 13.De Clercq, E. 2001. New developments in anti-HIV chemotherapy. Curr. Med. Chem. 8:1543-1572. [DOI] [PubMed] [Google Scholar]
- 14.Drelich, M., R. Wilhelm, and J. Mous. 1992. Identification of amino-acid-residues critical for endonuclease and integration activities of HIV-1 in protein in vitro. Virology 188:459-468. [DOI] [PubMed] [Google Scholar]
- 15.Dyda, F., A. B. Hickman, T. M. Jenkins, A. Engelman, R. Craigie, and D. R. Davies. 1994. Crystal structure of the catalytic domain of HIV-1 integrase—similarity to other polynucleotidyl transferases. Science 266:1981-1986. [DOI] [PubMed] [Google Scholar]
- 16.Eijkelenboom, A., R. Sprangers, K. Hard, R. A. P. Lutzke, R. H. A. Plasterk, R. Boelens, and R. Kaptein. 1999. Refined solution structure of the C-terminal DNA-binding domain of human immunovirus-1 integrase. Proteins Struct. Funct. Genet. 36:556-564. [DOI] [PubMed] [Google Scholar]
- 17.Engelman, A., F. D. Bushman, and R. Craigie. 1993. Identification of discrete functional domains of HIV-1 integrase and their organization within an active multimeric complex. EMBO J. 12:3269-3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Engelman, A., and R. Craigie. 1992. Identification of conserved amino acid residues critical for human immunodeficiency virus type-1 integrase function in vitro. J. Virol. 66:6361-6369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Esposito, D., and R. Craigie. 1999. HIV integrase structure and function. Adv. Virus Res. 52:319-333. [DOI] [PubMed] [Google Scholar]
- 20.Farnet, C. M., B. Wang, J. R. Lipford, and F. D. Bushman. 1996. Differential inhibition of HIV-1 preintegration complexes and purified integrase protein by small molecules. Proc. Natl. Acad. Sci. USA 93:9742-9747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fesen, M. R., K. W. Kohn, F. Leteurtre, and Y. Pommier. 1993. Inhibitors of human immunodeficiency virus integrase. Proc. Natl. Acad. Sci. USA 90:2399-2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fujishita, T., T. Yoshinaga, and A. Sato. 2000. Preparation of aromatic heterocycle compounds having HIV integrase inhibiting activities. Shionogi & Co., Ltd. patent PCT int. appl. WO-00039086.
- 23.Gao, K., S. L. Butler, and F. Bushman. 2001. Human immunodeficiency virus type 1 integrase: arrangement of protein domains in active cDNA complexes. EMBO J. 20:3565-3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gerton, J. L., S. Ohgi, M. Olsen, J. De Risi, and P. O. Brown. 1998. Effects of mutations in residues near the active site of human immunodeficiency virus type 1 integrase on specific enzyme-substrate interactions. J. Virol. 72:5046-5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goldgur, Y., R. Craigie, G. H. Cohen, T. Fujiwara, T. Yoshinaga, T. Fujishita, H. Sugimoto, T. Endo, H. Murai, and D. R. Davies. 1999. Structure of the HIV-1 integrase catalytic domain complexed with an inhibitor: a platform for antiviral drug design. Proc. Natl. Acad. Sci. USA 96:13040-13043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Halvas, E. K., E. S. Svarovskaia, E. O. Freed, and V. K. Pathak. 2000. Wild-type and YMDD mutant murine leukemia virus reverse transcriptases are resistant to 2′,3′-dideoxy-3′-thiacytidine. J. Virol. 74:6669-6674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Havlir, D. V. 2002. Structured intermittent treatment for HIV disease: necessary concession or premature compromise? Proc. Natl. Acad. Sci. USA 99:4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hazuda, D. 2002. A novel HIV-1 integrase inhibitor mediates sustained suppression of viral replication and CD4 depletion in a SHIV rhesus macaque model of infection. Antivir. Ther. 7:S3. [Google Scholar]
- 29.Hazuda, D., C. U. Blau, P. Felock, J. Hastings, B. Pramanik, A. Wolfe, F. Bushman, C. Farnet, M. Goetz, M. Williams, K. Silverman, R. Lingham, and S. Singh. 1999. Isolation and characterization of novel human immunodeficiency virus integrase inhibitors from fungal metabolites. Antivir. Chem. Chemother. 10:63-70. [DOI] [PubMed] [Google Scholar]
- 30.Hazuda, D., P. Felock, J. Hastings, B. Pramanik, A. Wolfe, G. Goodarzi, A. Vora, K. Brackmann, and D. Grandgenett. 1997. Equivalent inhibition of half-site and full-site retroviral strand transfer reactions by structurally diverse compounds. J. Virol. 71:807-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hazuda, D. J., P. Felock, M. Witmer, A. Wolfe, K. Stillmock, J. A. Grobler, A. Espeseth, L. Gabryelski, W. Schleif, C. Blau, and M. D. Miller. 2000. Inhibitors of strand transfer that prevent integration and inhibit HIV-1 replication in cells. Science 287:646-650. [DOI] [PubMed] [Google Scholar]
- 32.Hong, T., K. Drlica, A. Pinter, and E. Murphy. 1991. Circular DNA of human immunodeficiency virus: analysis of circle junction nucleotide sequences. J. Virol. 65:551-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hwang, Y., D. Rhodes, and F. Bushman. 2000. Rapid microtiter assays for poxvirus topoisomerase, mammalian type IB topoisomerase and HIV-1 integrase: application to inhibitor isolation. Nucleic Acids Res. 28:4884-4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jing, N., C. Marchand, Y. Guan, J. Liu, L. Pallansch, C. Lackman-Smith, E. De Clercq, and Y. Pommier. 2001. Structure-activity of inhibition of HIV-1 integrase and virus replication by G-quartet oligonucleotides. DNA Cell Biol. 20:499-508. [DOI] [PubMed] [Google Scholar]
- 35.Johnson, A. A., C. Marchand, and Y. Pommier. HIV-1 integrase inhibitors: a decade of research and two drugs in clinical trial. Curr. Top. Med. Chem., in press. [DOI] [PubMed]
- 36.Ju, G., and A. M. Skalka. 1980. Nucleotide sequence analysis of the long terminal repeat (LTR) of avian retroviruses: structural similarities with transposable elements. Cell 22:379-386. [DOI] [PubMed] [Google Scholar]
- 37.Julias, J. G., M. J. McWilliams, S. G. Sarafianos, E. Arnold, and S. H. Hughes. 2002. Mutations in the RNase H domain of HIV-1 reverse transcriptase affect the initiation of DNA synthesis and the specificity of RNase H cleavage in vivo. Proc. Natl. Acad. Sci. USA 99:9515-9520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jurriaans, S., A. Deronde, J. Dekker, J. Goudsmit, and M. Cornelissen. 1992. Analysis of human immunodeficiency virus type-1 LTR-LTR junctions in peripheral blood mononuclear cells of infected individuals. J. Gen. Virol. 73:1537-1541. [DOI] [PubMed] [Google Scholar]
- 39.Kiernan, R. E., A. Ono, G. Englund, and E. O. Freed. 1998. Role of matrix in an early postentry step in the human immunodeficiency virus type 1 life cycle. J. Virol. 72:4116-4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.King, P. J., D. J. Lee, R. A. Reinke, J. G. Victoria, K. Beale, J. Robinson, and W. Edward. 2003. Human immunodeficiency virus type-1 integrase containing a glycine to serine mutation at position 140 is attenuated for catalysis and resistant to integrase inhibitors. Virology 306:147-161. [DOI] [PubMed] [Google Scholar]
- 41.Lafemina, R. L., C. L. Schneider, H. L. Robbins, P. L. Callahan, K. Legrow, E. Roth, W. A. Schleif, and E. A. Emini. 1992. Requirement of active human immunodeficiency virus type-1integrase enzyme for productive infection of human T-lymphoid cells. J. Virol. 66:7414-7419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin, Z., N. Neamati, H. Zhao, Y. Kiryu, J. A. Turpin, C. Aberham, K. Strebel, K. Kohn, M. Witvrouw, C. Pannecouque, Z. Debyser, E. De Clercq, W. G. Rice, Y. Pommier, and T. R. Burke, Jr. 1999. Chicoric acid analogues as HIV-1 integrase inhibitors. J. Med. Chem. 42:1401-1414. [DOI] [PubMed] [Google Scholar]
- 43.Lodi, P. J., J. A. Ernst, J. Kuszewski, A. B. Hickman, A. Engelman, R. Craigie, G. M. Clore, and A. M. Gronenborn. 1995. Solution structure of the DNA-binding domain of HIV-1 integrase. Biochemistry 34:9826-9833. [DOI] [PubMed] [Google Scholar]
- 44.Lutzke, R. A., and R. H. Plasterk. 1998. Structure-based mutational analysis of the C-terminal DNA-binding domain of human immunodeficiency virus type 1 integrase: critical residues for protein oligomerization and DNA binding. J. Virol. 72:4841-4848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marchand, C., A. Johnson, R. G. Karki, G. C. G. Pais, X. Zhang, K. Cowansage, T. A. Patel, M. C. Nicklaus, T. R. Burke, Jr., and Y. Pommier. Metal-dependent inhibition of HIV-1 integrase by beta-diketo acids and resistance of the soluble double-mutant (F185K/C280S). Mol. Pharmacol. 64:600-609. [DOI] [PubMed]
- 46.Marchand, C., X. Zhang, G. C. Pais, K. Cowansage, N. Neamati, T. R. Burke, Jr., and Y. Pommier. 2002. Structural determinants for HIV-1 integrase inhibition by beta-diketo acids. J. Biol. Chem. 277:12596-12603. [DOI] [PubMed] [Google Scholar]
- 47.Matsuoka-Aizawa, S., H. Sato, A. Hachiya, K. Tsuchiya, Y. Takebe, H. Gatanaga, S. Kimura, and S. Oka. 2003. Isolation and molecular characterization of a nelfinavir (NFV)-resistant human immunodeficiency virus type 1 that exhibits NFV-dependent enhancement of replication. J. Virol. 77:318-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matsushita, S. 2000. Current status and future issues in the treatment of HIV-1 infection. Int. J. Hematol. 72:20-27. [PubMed] [Google Scholar]
- 49.Melek, M., J. M. Jones, M. H. O'Dea, G. Pais, T. R. Burke, Jr., Y. Pommier, N. Neamati, and M. Gellert. 2002. Effect of HIV integrase inhibitors on the RAG1/2 recombinase. Proc. Natl. Acad. Sci. USA 99:134-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miller, M., C. Farnet, and F. Bushman. 1997. Human immunodeficiency virus type 1 preintegration complexes: studies of organization and composition. J. Virol. 71:5382-5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.O'Hara, B. M., and W. C. Olson. 2002. HIV entry inhibitors in clinical development. Curr. Opin. Pharmacol. 2:523-528. [DOI] [PubMed] [Google Scholar]
- 52.Pais, G. C. G., and T. R. Burke. 2002. Novel aryl diketo-containing inhibitors of HIV-1 integrase. Drugs Future 27:1101-1111. [Google Scholar]
- 53.Pais, G. C. G., X. C. Zhang, C. Marchand, N. Neamati, K. Cowansage, E. S. Svarovskaia, V. K. Pathak, Y. Tang, M. Nicklaus, Y. Pommier, and T. R. Burke. 2002. Structure activity of 3-aryl-1,3-diketo-containing compounds as HIV-1 integrase inhibitors. J. Med. Chem. 45:3184-3194. [DOI] [PubMed] [Google Scholar]
- 54.Patel, P., and B. Preston. 1994. Marked infidelity of human immunodeficiency virus type 1 reverse transcriptase at RNA and DNA template ends. Proc. Natl. Acad. Sci. USA 91:549-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pluymers, W., N. Neamati, C. Pannecouque, V. Fikkert, C. Marchand, T. R. Burke, Jr., Y. Pommier, D. Schols, E. De Clercq, Z. Debyser, and M. Witvrouw. 2000. Viral entry as the primary target for the anti-HIV activity of chicoric acid and its tetra-acetyl esters. Mol. Pharmacol. 58:641-648. [DOI] [PubMed] [Google Scholar]
- 56.Pommier, Y., and N. Neamati. 1999. Inhibitors of human immunodeficiency virus integrase. Adv. Virus Res. 52:427-458. [DOI] [PubMed] [Google Scholar]
- 57.Pommier, Y., A. A. Pilon, K. Bajaj, A. Mazumder, and N. Neamati. 1997. HIV-1 integrase as a target for antiviral drugs. Antivir. Chem. Chemother. 8:463-483. [Google Scholar]
- 58.Reinke, R. A., P. J. King, J. G. Victoria, B. R. McDougall, G. X. Ma, Y. Q. Mao, M. G. Reinecke, and W. E. Robinson. 2002. Dicaffeoyltartaric acid analogues inhibit human immunodeficiency virus type 1 (HIV-1) integrase and HIV-1 replication at nontoxic concentrations. J. Med. Chem. 45:3669-3683. [DOI] [PubMed] [Google Scholar]
- 59.Richman, D. D. 2001. HIV chemotherapy. Nature 410:995-1001. [DOI] [PubMed] [Google Scholar]
- 60.Shaw-Reid, C. A., V. Munshi, P. Graham, A. Wolfe, M. Witmer, R. Danzeisen, D. B. Olsen, S. S. Carroll, M. Embrey, J. S. Wai, M. D. Miller, J. L. Cole, and D. J. Hazuda. 2003. Inhibition of HIV-1 ribonuclease H by a novel diketo acid, 4-[5-(benzoylamino)thien-2-yl]-2,4-dioxobutanoic acid. J. Biol. Chem. 278:2777-2780. [DOI] [PubMed] [Google Scholar]
- 61.Smith, J., and M. Roth. 1992. Specificity of human immunodeficiency virus-1 reverse transcriptase-associated ribonuclease H in removal of the minus-strand primer, tRNALys3. J. Biol. Chem. 267:15071-15079. [PubMed] [Google Scholar]
- 62.Swanstrom, R., W. J. Delorbe, J. M. Bishop, and H. E. Varmus. 1981. Nucleotide sequence of cloned unintegrated avian sarcoma virus DNA-viral DNA contains direct and inverted repeats similar to those in transposable elements. Proc. Natl. Acad. Sci. USA 78:124-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Uenaka, M., K. Kawata, M. Nagai, and T. Endoh. 2000. Novel processes for the preparation of substituted propenone derivatives. Shionogi & Co., Ltd. patent PCT int. appl. WO-00075122.
- 64.Unutmaz, D., V. N. KewalRamani, S. Marmon, and D. R. Littman. 1999. Cytokine signals are sufficient for HIV-1 infection of resting human T lymphocytes. J. Exp. Med. 189:1735-1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vittinghoff, E., S. Scheer, P. O'Malley, G. Colfax, S. D. Holmberg, and S. P. Buchbinder. 1999. Combination antiretroviral therapy and recent declines in AIDS incidence and mortality. J. Infect. Dis. 179:717-720. [DOI] [PubMed] [Google Scholar]
- 66.Wang, J. Y., H. Ling, W. Yang, and R. Craigie. 2001. Structure of a two-domain fragment of HIV-1 integrase: implications for domain organization in the intact protein. EMBO J. 20:7333-7343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yee, J. K., A. Miyanohara, P. Laporte, K. Bouic, J. C. Burns, and T. Friedmann. 1994. A general method for the generation of high-titer, pantropic retroviral vectors: highly efficient infection of primary hepatocytes. Proc. Natl. Acad. Sci. USA 91:9564-9568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yoder, K. E., and F. D. Bushman. 2000. Repair of gaps in retroviral DNA integration intermediates. J. Virol. 74:11191-11200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Young, S. D. 2001. Inhibition of HIV-1 integrase by small molecules: the potential for a new class of AIDS chemotherapeutics. Curr. Opin. Drug Discov. Dev. 4:402-410. [PubMed] [Google Scholar]
- 70.Zhang, X., G. C. G. Pais, E. S. Svarovskaia, C. Marchand, A. A. Johnson, R. G. Karki, M. C. Nicklaus, V. K. Pathak, Y. Pommier, J. Burke, and R. Terrence. 2003. Azido-containing aryl β-diketo acid HIV-1 integrase inhibitors. Bioorg. Med. Chem. Lett. 13:1215-1219. [DOI] [PubMed] [Google Scholar]
- 71.Zheng, R., T. M. Jenkins, and R. Craigie. 1996. Zinc folds the N-terminal domain of HIV-1 integrase, promotes multimerization, and enhances catalytic activity. Proc. Natl. Acad. Sci. USA 93:13659-13664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhuang, L. C., J. S. Wai, M. W. Embrey, T. E. Fisher, M. S. Egbertson, L. S. Payne, J. P. Guare, J. P. Vacca, D. J. Hazuda, P. J. Felock, A. L. Wolfe, K. A. Stillmock, M. V. Witmer, G. Moyer, W. A. Schleif, L. J. Gabryelski, Y. M. Leonard, J. J. Lynch, S. R. Michelson, and S. D. Young. 2003. Design and synthesis of 8-hydroxy-1,6 naphthyridines as novel inhibitors of HIV-1 integrase in vitro and in infected cells. J. Med. Chem. 46:453-456. [DOI] [PubMed] [Google Scholar]