Abstract
Accurate replication in the presence of DNA damage is essential to genome stability and viability in all cells. In Escherichia coli, DNA replication forks blocked by UV-induced damage undergo a partial resection and RecF-catalyzed regression before synthesis resumes. These processing events generate distinct structural intermediates on the DNA that can be visualized in vivo using 2D agarose gels. However, the fate and behavior of the stalled replisome remains a central uncharacterized question. Here, we use thermosensitive mutants to show that the replisome’s polymerases uncouple and transiently dissociate from the DNA in vivo. Inactivation of α, β, or τ subunits within the replisome is sufficient to signal and induce the RecF-mediated processing events observed following UV damage. By contrast, the helicase–primase complex (DnaB and DnaG) remains critically associated with the fork, leading to a loss of fork integrity, degradation, and aberrant intermediates when disrupted. The results reveal a dynamic replisome, capable of partial disassembly to allow access to the obstruction, while retaining subunits that maintain fork licensing and direct reassembly to the appropriate location after processing has occurred.
Keywords: replication fork processing, RecF pathway
The replisome consists of several, multisubunit protein complexes and is responsible for duplicating the genome. In Escherichia coli, it is comprised of three DNA polymerase complexes tethered to the DNA template by dimeric processivity factors, a τ complex that couples leading and lagging strand synthesis, and a helicase–primase complex that separates the duplex DNA and primes lagging strand synthesis (1–3).
When the replisome encounters DNA damage that blocks its progression, the potential for mutagenesis, rearrangements, and lethality increases significantly. Replication in the presence of DNA damage can generate mutations if the wrong base is incorporated, rearrangements if it resumes from the wrong site, or lethality if the obstructing lesion cannot be overcome. Following the arrest of replication at UV-induced damage, the nascent lagging strand is partially resected by the combined action of the RecQ helicase and RecJ nuclease (4, 5). RecF-O-R, along with RecA, limit this degradation and promote a transient regression of the DNA branch point, which is thought to be important for restoring the damaged region to a form that can be acted on by repair enzymes or translesion DNA polymerases (4–10). These processing events generate distinct structural intermediates on the DNA that can be readily visualized using 2D agarose gel analysis, a technique that allows one to identify the shape and structure of DNA molecules (5, 11).
Although the processing that occurs on the DNA is well characterized, little is known about the behavior or composition of the replisome itself during these events. If the replisome remains bound to the arresting lesion, it may sterically obstruct repair or bypass from occurring. Conversely, complete dissociation of the replisome would likely abolish the licensing for the replication fork and expose DNA ends that have the potential to recombine, generating deletions, duplications, or rearrangements on the chromosome. Recent studies in vitro have suggested that dynamic interactions between replisome components may play a role in allowing the machinery to overcome specific challenges such as collisions with the transcription apparatus or DNA-bound proteins (1, 12, 13). In this study, we used thermosensitive replication mutants to characterize how the composition of the replisome changes following encounters with UV-induced photoproducts, a biologically relevant lesion that is known to block the progression of the replisome when located in the leading strand template (6, 14–16). The results demonstrate that the DNA polymerases can dissociate from the replisome in a modular manner without compromising the integrity of the replication fork. Dissociation of the DNA polymerase from the replisome is sufficient and can serve to initiate the processing of the replication fork DNA via the RecF pathway, similar to that seen when replication is arrested by UV-induced damage. By comparison, the helicase complex remains associated with the replication fork throughout the recovery process. If the helicase is disrupted, aberrant intermediates, degradation, and loss of fork integrity ensue. We propose that the retention of the helicase is needed to maintain licensing for the replication fork and direct reassembly to the appropriate location after processing has occurred.
Results
Dissociation of the Polymerase from the Replisome Is Sufficient to Induce Processing Events Similar to Those Observed After UV-Induced Arrest.
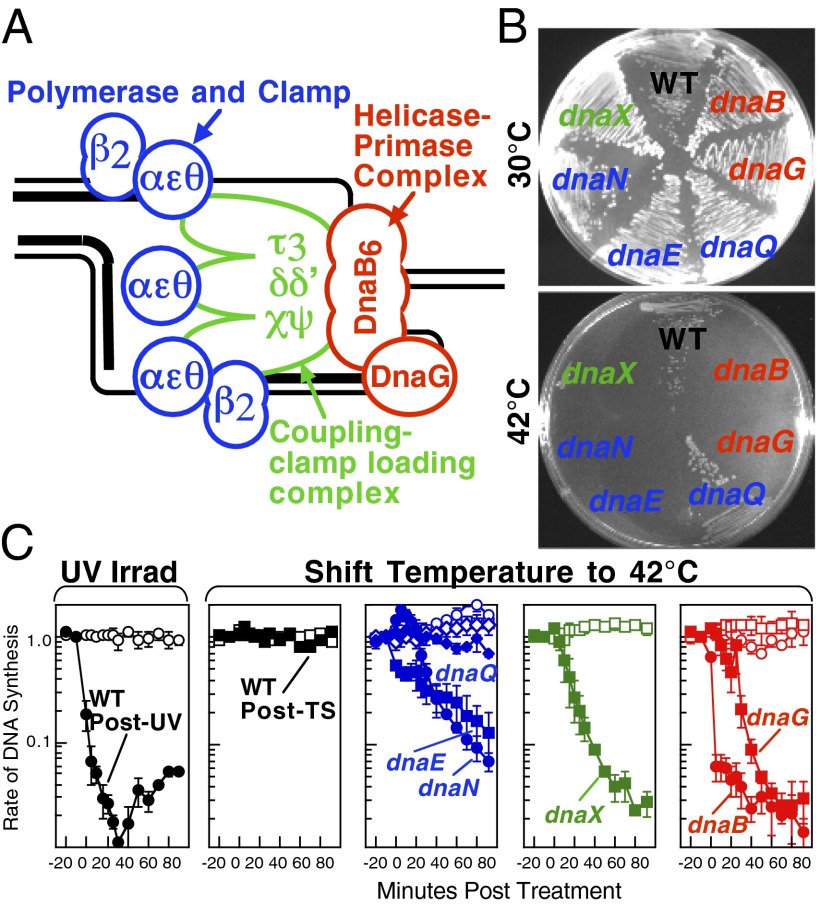
A schematic of each of the components of the replisome tested in this study and their function is presented in Fig. 1A. Temperature-sensitive mutants exist in subunits from each of replisome’s complexes for which viability or functionality is supported at 30 °C, but not at 42 °C (Fig. 1B). Although replication proceeds normally at the permissive temperature, it rapidly decreases following inactivation of the thermosensitive protein at the restrictive temperature, similar to that seen after UV irradiation (Fig. 1C). The exception to this is in the proofreading subunit ε, encoded by dnaQts, which is mutagenic at the restrictive temperature, but is not essential for viability or replication (17).
Fig. 1.
Replication is disrupted by UV-induced damage or following inactivation of the DNA polymerase, τ complex, or helicase–primase complex. (A) A diagram of the replisome, indicating the subunits of each protein complex. (B) Thermosensitive mutants that inactivate the polymerase core, τ complex, or helicase complex are viable at 30 °C but fail to grow at the restrictive temperature of 42 °C following overnight incubation. (C) The rate of DNA synthesis is inhibited following UV-induced damage or inactivation of the replisome’s essential subunits. Wild-type or mutant cultures, grown at 30 °C were pulse-labeled with 1 µCi per 10 µg/mL [3H]thymidine for 2 min at the indicated times following mock treatment (open symbols), 50 J/m2 UV irradiation (filled symbols), or a shift to 42 °C (filled symbols). The amount of radioactivity incorporated into the DNA, relative to pretreated cultures is plotted. Error bars represent SE of two experiments.
To determine how the replisome behaves or is modified following encounters with DNA damage in vivo, these thermosensitive mutants were used to deliberately disrupt the specific components of the replisome. We then compared the effect that the loss of that component had on replication processing to that seen when replication encounters DNA damage. Replication and processing intermediates were visualized in vivo on replicating fragments of the plasmid pBR322 (Fig. 2). This plasmid replicates using the host’s replication machinery, it can be linearized at its single origin of replication, and it maintains a moderate copy number, making it useful to detect rare events such as replication through a defined fragment (18). In this type of analysis, actively replicating molecules appear as “Y”-shaped structures that migrate in an arc that extends out from the prominent spot of nonreplicating linear molecules. Following the arrest of replication in irradiated wild-type cultures, replication forks undergo a transient regression and other processing events that generate intermediates having two branch points (18). These intermediates, form double-Y or “X”-shaped molecules that migrate more slowly due to their nonlinear shape, forming a cone region above the arc of replicating molecules that can be clearly distinguished from the normal replicating molecules (Fig. 2). Importantly, no processing intermediates or aberrant DNA structures are observed in wild-type cultures shifted to 42 °C.
Fig. 2.
Inactivation of the DNA polymerase or τ complex, but not the helicase complex, induces replication fork processing intermediates similar to UV-induced damage. (A) Diagram depicting the migration pattern of replicating DNA fragments and intermediates associated with processing forks disrupted by UV-induced damage in 2D agarose gels. Strains containing plasmid pBR322 were UV-irradiated with 50 J/m2 or filtered and placed in prewarmed media at 42 °C. Genomic and plasmid DNA was then purified, digested with PvuII, and analyzed by 2D agarose gel analysis at 15 min following UV-irradiation and 90 min following temperature shift. (B) In wild-type cultures, replication fork processing intermediates are observed after disruption by UV-irradiation but not after a shift to 42 °C. (C) Disruption of the DNA polymerase core (dnaEts), processivity factor (dnaNts) or τ (dnaXts) is sufficient to induce replication fork processing similar to that seen after UV-induced damage. (D) Abnormal intermediates, distinct from any of those associated with processing UV-induced damage, arise after disruption of the helicase (dnaBts) or primase (dnaGts). (E) No atypical intermediates are observed following disruption of the nonessential proofreading subunit of the DNA polymerase (dnaQts).
To characterize how the composition of the replisome changes or is modified during these processing events, cultures of thermosensitive replication mutants were grown at the permissive temperature, split in half, and then either UV-irradiated with 50 J/m2 or shifted to the restrictive temperature at 42 °C. Aliquots of each culture were taken at various times and the DNA was prepared and examined by 2D agarose gel analysis. In all mutants, only normal Y-shaped replication intermediates were observed before treatment, and normal damage-induced processing of the replication fork occurred following UV-induced damage, as evidenced by their resistance to UV, and the timely appearance of cone region intermediates in each mutant (Fig. 2 and Figs. S1 and S2).
Following inactivation of either the catalytic subunit of the DNA polymerase, α (DnaE), or the processivity factor that tethers the DNA polymerase to the DNA template, β (DnaN), processing events were induced at the replication fork DNA that appeared similar to those seen after UV irradiation. Both α and β are required to maintain DNA polymerase binding to the DNA template (19, 20). The processing intermediates were induced specifically by the disruption of the DNA polymerase III subunits, because no processing intermediates were induced in wild-type cells at the restrictive temperature or following inactivation of the nonessential proofreading subunit, ε (DnaQ) (Fig. 2 and Fig. S1).
Similar to the DNA polymerase subunits, UV-like processing intermediates were also induced following inactivation of τ (DnaX), which is responsible for coupling the polymerases to the replisome, coordinating leading and lagging strand synthesis, and repetitive cycling of the processivity factor onto the lagging strand template (21–25). The observed induction of UV-like intermediates following inactivation of the DNA polymerase or coupling factor indicates that the UV-induced processing of replication forks is likely to involve the transient dissociation of the polymerase from the DNA.
The helicase–primase complex interacts with τ and the holoenzyme and tracks along the lagging strand template, serving to unwind and prime it during replication (26–28). In contrast to α, β, or τ, inactivation of either the helicase (DnaB) or primase (DnaG) led to the production of aberrant structural intermediates that were unlike any of those that are observed during the processing of UV-induced damage (Fig. 2 and Fig. S1), arguing that disruption of the helicase complex does not normally occur during the recovery of replication after UV-induced arrest.
Similar to UV, the Integrity of the Replication Fork DNA Is Maintained After Polymerase Dissociation, and Replication Fork Processing Is Catalyzed by RecF.
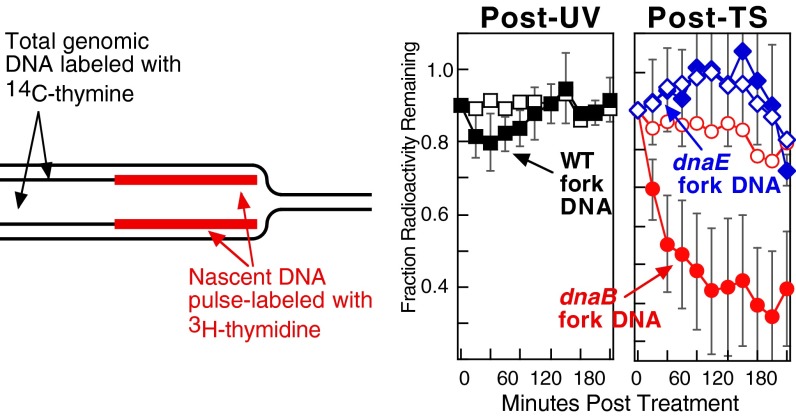
The above results are consistent with the idea that the DNA polymerases dissociate from the replisome upon encounters with UV-induced damage whereas the helicase complex remains associated with the DNA. Following UV irradiation, the integrity of the arrested replication fork is normally maintained and the nascent DNA undergoes only limited degradation before the resumption of replication (5). To further characterize how the dissociation of the DNA polymerase or helicase affect replication fork integrity, we monitored the fate of the nascent DNA after disruption of these components and compared it to that at UV-arrested replication forks. To this end, wild-type, dnaEts, and dnaBts cultures, grown at 30 °C in the presence of [14C]thymine, were pulse-labeled with [3H]thymidine for 20 s and immediately placed in fresh nonradiactive media before either UV-irradiating or shifting the culture to 42 °C. In wild-type cells after UV irradiation, no degradation was detected in the [14C]-labeled genomic DNA and only ∼10% of the [3H]-labeled nascent DNA at the arrested fork was degraded following DNA damage (Fig. 3). Similarly, following inactivation of the DNA polymerase, both the genomic and nascent DNA remained protected and was maintained to a similar extent as that seen after UV-induced arrest. The results strongly suggest that DNA polymerase dissociation can occur without compromising the integrity of the replication fork. By contrast and distinct from UV-arrested replication forks, extensive degradation of the nascent DNA was observed following disruption of the helicase, DnaB (Fig. 3), suggesting that the aberrant structural intermediates are associated with the collapse and degradation of the replication fork. The observed degradation was targeted specifically to the DNA at the replication fork because no degradation was detected in the overall genomic DNA. Neither the abnormal intermediates nor the extensive degradation of the fork are observed during the recovery of replication after UV-induced damage, arguing that the helicase complex normally remains associated with the DNA during the recovery process and is needed to maintain the integrity of the replication fork.
Fig. 3.
Similar to after UV, the integrity of the replication fork DNA is maintained after DNA polymerase dissociation. The integrity of the replication fork is maintained following arrest by UV irradiation or polymerase dissociation, but collapses after disruption of the helicase. [14C]thymine-prelabeled cultures were pulse-labeled with [3H]thymidine for 20 s before being filtered and placed in nonradioactive media and either UV-irradiated with 50 J/m2 or shifted to 42 °C as indicated. The amount of radioactivity remaining in the total genomic DNA (open symbols) and nascent DNA at the replication fork (filled symbols) is plotted over time. Error bars represent SE of two independent experiments.
Consistent with these interpretations, the replication fork processing induced by either polymerase inactivation or UV-induced damage remains unchanged when the second form of challenge is administered (Fig. S3). By contrast, disruption of the helicase by thermal inactivation prevents the UV-induced processing from occurring and destroys any processing intermediates that exist at the time of inactivation.
We have previously shown that the formation of the UV-induced processing intermediates is catalyzed by the RecF pathway gene products, depends on active replication, and resolves at a time that correlates with when the lesions are repaired and replication resumes (5). In the absence of RecF, the transient regression and processing intermediates fail to form at the replication fork after UV-induced damage (Fig. 4). To determine if the DNA processing events induced by DNA polymerase dissociation are identical to those occurring after UV-induced damage, we examined whether the intermediates formed by DNA polymerase dissociation also depend on RecF. Wild-type, dnaEts, and dnaBts cultures containing an intact recF gene or a recF deletion were grown at 30 °C, UV irradiated or shifted to the nonpermissive temperature, and then examined by 2D agarose gel analysis, as before. Similar to UV treatment, the formation of the replication fork intermediates induced following temperature shift in dnaE mutants were dependent on RecF, strongly arguing that the processing intermediates observed after polymerase dissociation are identical to those occurring following the arrest of replication at UV-induced damage (Fig. 4). By contrast, the aberrant intermediates observed following disruption of the helicase complex appeared irrespective of whether RecF was present or absent, further supporting the idea that these intermediates and processing events are distinct from those that occur during the recovery of replication at UV damage.
Fig. 4.

RecF mediates the formation of replication fork intermediates following DNA polymerase dissociation. The formation and processing of the replication intermediates after DNA polymerase dissociation are mediated by RecF, similar to UV. The formation of the abnormal intermediates that arise following helicase disruption is not dependent on RecF. Isogenic wild-type, dnaBts, and dnaEts cultures containing a normal or mutated copy of the recF gene were UV-irradiated or temperature shifted and analyzed by 2D agarose gel analysis as in Fig. 2.
Discussion
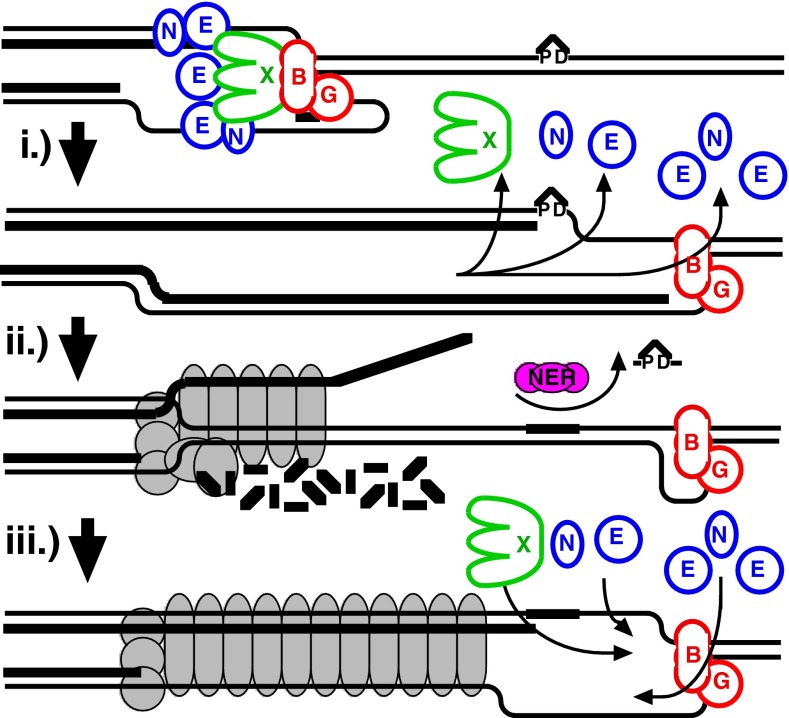
Following encounters with DNA damage that blocks the progression of replication, the DNA at the arrest site undergoes a partial resection and transient regression to allow the lesion to be repaired or bypassed before DNA synthesis resumes. How the replisome allows repair enzymes to access the lesion without compromising the replication fork’s integrity or license to resume replication has remained a challenging question. These data indicate that, in vivo, dissociation of the replisome’s polymerases from the DNA is sufficient and can serve as a signal to initiate the RecF-mediated processing that occurs following arrest by a UV-induced adduct. The DNA polymerase dissociation occurs without compromising the integrity of the fork DNA, which remains protected even after the polymerase dissociates or uncouples from the replisome. By contrast, the helicase complex remains associated with the DNA, and is necessary to maintain the integrity of the replication fork. We propose that uncoupling of the polymerase from the replisome may be required to allow repair enzymes or translesion DNA polymerases to gain access to and effect repair (Fig. 5). Retention of the helicase complex at the arrest site maintains licensing for the replication fork and may serve to signal and recruit replisome reassembly to the correct site after the lesion has been processed.
Fig. 5.
Model of replisome at UV-induced damage. Upon encountering an arresting lesion (PD, pyrimidine dimer) (i), DNA synthesis becomes uncoupled and the polymerases transiently dissociate. (ii) This serves as a signal to initiate the replication fork DNA processing by the RecF-pathway gene products (gray circles) allowing repair enzymes (NER) or translesion polymerases to access the lesion. (iii) The helicase–primase complex remains bound to the template DNA and serves to maintain the licensing and integrity of the replication fork, directing replisome reassembly to the correct location once the lesion has been processed.
dnaE486 contains a S885P mutation that falls within the domains necessary for interactions with β (29, 30). dnaX2016 contains a G118D mutation that disrupts the protein’s ATPase domain (31). dnaN159 contains two amino acid substitutions, G66E and G174A, which impair its interactions with α (32). Considering that α, β, and τ are all necessary to maintain polymerase binding to the DNA and replisome, and that all three temperature-sensitive mutants induce a similar phenotype, it seems reasonable to suggest that the temperature shift is likely to result in polymerase dissociation, rather than having it remain bound but inactive within the replisome. dnaQ49 contains a V96G mutation that decreases the ability of ε to interact with α and θ subunits (33, 34). We confirmed that our strain did not contain spq-2, a suppressor mutation (Fig. S4), which can frequently arise in dnaQ mutants (35).
dnaG3 contains a G247D mutation within the catalytic core domain (36). dnaB266 is an amber mutation that requires coexpression of suppressor tRNAs (38, 39). It has been characterized to be phenotypically similar to dnaB8, an A130V mutation in the hinge region that affects ATPase activity, primer synthesis, and helicase activity at 42 °C (40, 41). The accessibility of the fork DNA to nucleases and its loss of structural integrity suggests that thermal inactivation in this case is likely to result in helicase dissociation in vivo.
We would emphasize that the assays involving thermal inactivation are likely to reflect the events that occur upon the initial arrest of replication. Because thermal inactivation is presumably irreversible, this type of analysis does not permit inferences to be made about the mechanism by which replication resumes, which is almost certain to involve all of the proteins of the replisome.
The idea that the DNA polymerase can function as a modular unit within the replisome and dissociate without disrupting the integrity of the replication fork is consistent with a number of observations from previous studies. Functional replisomes can be reconstituted in vitro that contain one, two, or three polymerases (2, 42), arguing that the presence of all three polymerases is not essential to maintain replication fork integrity (12). Increasing the concentrations of DNA polymerase II or DNA polymerase IV can reduce the speed of the replisome and its helicase in vitro and reduces the rate of DNA synthesis in vivo (43). These observations imply that the DNA polymerases have the capability to swap into a functional replisome, replacing DNA polymerase III during synthesis. A similar form of polymerase dynamics is also observed in phage T4 and T7, in which the polymerases transiently release and rebind the DNA template without losing contact or compromising replisome integrity (44, 45). In eukaryotes, two subunits of the replicative lagging strand polymerase, δ, are also components of translesion DNA polymerase ζ and are required for translesion synthesis in vivo (46–48). Although it remains unclear whether translesion DNA synthesis is occurring within the active replisome or at DNA gaps following replication, the observations are suggestive that eukaryotic DNA polymerases can also function in a modular fashion in some instances.
Replication is differentially affected by lesions in the leading strand and lagging strand template. Both in vitro and in vivo approaches suggest that lesions in the leading strand arrest the overall progression of the fork, whereas the priming activity associated with lagging strand synthesis allows replication to continue past lesions in this template, leaving a gap in the DNA (15, 16, 28, 49). A number of studies have shown that, when replication encounters a leading strand lesion in vivo, nucleotide excision repair plays a predominant role in allowing DNA synthesis to resume following arrest (7, 50). Translesion DNA synthesis affects the timing and kinetics of the recovery only when nascent strand processing or repair cannot occur (7, 50, 51). The observed dissociation of the DNA polymerase seems likely to be required to allow repair enzymes to gain access to and effect repair of the lesions at the arrest site. A similar displacement of RNA polymerase is necessary before nucleotide excision repair can act when transcription is arrested by UV-induced damage (52–54).
In vivo, we observed that RecF-mediated processing was triggered following dissociation of the DNA polymerase from the replisome. Consistent with this observation, in vitro approaches have shown that RecF pathway proteins can displace a DNA polymerase arrested at a leading strand block (28). Although it remains unclear from these studies whether the dissociation of the DNA polymerase in vivo requires RecF or occurs spontaneously and serves to trigger the RecF-mediated processing.
Biochemical studies have led to different models for how the helicase may behave when replication is blocked. In vitro, purified DnaB was shown to be capable of loading onto synthetic fork substrates when combinations of the primosomal proteins, PriA, -B, and -C, or Rep are present, leading some to speculate that the helicase may dissociate and reassemble as a mechanism to bypass lesions or other obstructions on the DNA (55). However, other in vitro studies have shown that DnaB remains stably associated with the DNA when replication encounters various impediments or is arrested by a leading strand block, suggesting the helicase remains associated until the obstruction can be processed (15, 56). The observations reported in this study show that disruption of the helicase in vivo leads to aberrant replication intermediates and a loss of fork integrity. Considering that these events are not observed following encounters with UV-induced damage, the observations imply that the helicase complex is normally retained at the site of arrest and is likely to be necessary to maintain the integrity and licensing of the replication forks.
Both prokaryotes and eukaryotes tightly regulate helicase loading onto DNA as a mechanism to control where replication initiates and license regions of DNA for replication to prevent rereplication (57, 58). In eukaryotes, origins are prelicensed for initiation in G1 by loading the DNA helicase, and no new helicase loading is thought to occur once S phase has begun (59–61). Following activation, the remaining components of the replisome are recruited to the helicase and replication begins (61). Thus, in eukaryotic systems, it is generally thought that disruption of the helicase abolishes the ability of the fork to resume replication. A similar mechanism of helicase loading and activation regulates replisome assembly and initiation at origins in prokaryotes (58, 62). The observed retention of the helicase at UV-arrested replication forks suggests a mechanism by which the replication fork may retain licensing to resume replication. In addition, the presence of the helicase at the replication fork may serve as a signal to recruit the replisome components to reassemble at the correct site once the lesion has been processed, similar to the process that occurs at origins.
The mechanisms of replication and origin initiation are highly conserved among both prokaryotes and eukaryotes organisms, making it likely that the observed replisome modularity and importance of helicase retention will extend broadly among evolutionarily diverse organisms.
Materials and Methods
Strains and Plasmids.
Strains used in this study are presented in Table S1. All recF strains were made isogenic with their parents for this study. Mutants were constructed using standard P1 transduction and verified according to their temperature- and UV-sensitive phenotypes, as appropriate. The strains containing temperature sensitive alleles, in many cases, are related and have been used comparatively in the literature, but are not strictly isogenic. Thus, in each case, comparisons of the temperature-shift response to the UV response were made within each strain, rather than between strains. In this way, each strain effectively served as its own parental control for the analysis. In addition, temperature-sensitive alleles were verified to have a normal response to UV irradiation as measured by survival, replication recovery, and UV-induced processing intermediates.
Rate of DNA Synthesis.
Overnight cultures were diluted 1:100 and grown in Davis medium supplemented with 0.4% glucose, 0.2% casamino acid, and 10 μg/mL thymine (DGCthy) to an OD600 of 0.4 in a 30 °C shaking water bath. For UV, cultures were split in half and either UV irradiated with 50 J/m2 (Sylvania 15-W germicidal lamp, 254 nm, 0.9 J/m2 per s incident dose) or mock irradiated. For temperature shift, cultures were filtered and resuspended in fresh prewarmed media at either 30 °C or 42 °C. At the indicated times, duplicate 0.5-mL aliquots of the culture were pulse-labeled with [3H]thymidine (1.0 μCi/10 μg/mL) for 2 min before cells were lysed, and DNA was precipitated by the addition of 5 mL of ice-cold 5% (wt/wt) trichloroacetic acid. The precipitate was collected and the amount of radioactivity was determined as described (63).
Two-Dimensional Agarose Gel Analysis.
Overnight cultures containing the plasmid pBR322 grown in 100 μg/mL ampicillin were pelleted, diluted 1:100 in DGCthy without antibiotic, and grown to an OD600 of 0.4 in a 30 °C shaking water bath. Cultures were split in half and either UV irradiated with 50 J/m2 or filtered, and resuspended in prewarmed 42 °C media. At the indicated times, DNA was prepared from 0.75-mL aliquots of the cultures and then analyzed by 2D agarose gel electrophoresis as described (63).
Nascent DNA Degradation and Fork Integrity.
Overnight cultures were diluted 1:100 and grown in DGCthy and [14C]thymine (0.1 μCi/10 μg/mL) to an OD600 of 0.4 in a 30 °C shaking water bath. Cultures were then pulse labeled with [3H]thymidine (1 μCi/10 μg/mL) for 20 s, filtered on Whatman 0.4-μm membrane filters, and washed twice with 3 mL of cold NET buffer (100 mM NaCl/10 mM EDTA, pH 8.0/10 mM Tris, pH 8.0). For UV experiments, the filter was then resuspended in prewarmed nonradioactive DGCthy media, UV irradiated with 50 J/m2, and incubated at 30 °C. For temperature-shift experiments, the filter was instead immediately resuspended in fresh media that had been prewarmed to 42 °C. At the times indicated, duplicate 200-μL aliquots of cells in culture were lysed, the DNA was precipitated, and the amount of radioactivity was determined as described (63).
Supplementary Material
Acknowledgments
This work was funded by the National Institutes of Health/National Institute of Environmental Health Research Grants R21ES018940 and R15ES021594.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1300624110/-/DCSupplemental.
References
- 1.O’Donnell M. Replisome architecture and dynamics in Escherichia coli. J Biol Chem. 2006;281(16):10653–10656. doi: 10.1074/jbc.R500028200. [DOI] [PubMed] [Google Scholar]
- 2.McInerney P, Johnson A, Katz F, O’Donnell M. Characterization of a triple DNA polymerase replisome. Mol Cell. 2007;27(4):527–538. doi: 10.1016/j.molcel.2007.06.019. [DOI] [PubMed] [Google Scholar]
- 3.Reyes-Lamothe R, Sherratt DJ, Leake MC. Stoichiometry and architecture of active DNA replication machinery in Escherichia coli. Science. 2010;328(5977):498–501. doi: 10.1126/science.1185757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Courcelle J, Hanawalt PC. RecQ and RecJ process blocked replication forks prior to the resumption of replication in UV-irradiated Escherichia coli. Mol Gen Genet. 1999;262(3):543–551. doi: 10.1007/s004380051116. [DOI] [PubMed] [Google Scholar]
- 5.Courcelle J, Donaldson JR, Chow KH, Courcelle CT. DNA damage-induced replication fork regression and processing in Escherichia coli. Science. 2003;299(5609):1064–1067. doi: 10.1126/science.1081328. [DOI] [PubMed] [Google Scholar]
- 6.Courcelle J, Carswell-Crumpton C, Hanawalt PC. recF and recR are required for the resumption of replication at DNA replication forks in Escherichia coli. Proc Natl Acad Sci USA. 1997;94(8):3714–3719. doi: 10.1073/pnas.94.8.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Courcelle CT, Chow KH, Casey A, Courcelle J. Nascent DNA processing by RecJ favors lesion repair over translesion synthesis at arrested replication forks in Escherichia coli. Proc Natl Acad Sci USA. 2006;103(24):9154–9159. doi: 10.1073/pnas.0600785103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Webb BL, Cox MM, Inman RB. Recombinational DNA repair: The RecF and RecR proteins limit the extension of RecA filaments beyond single-strand DNA gaps. Cell. 1997;91(3):347–356. doi: 10.1016/s0092-8674(00)80418-3. [DOI] [PubMed] [Google Scholar]
- 9.Bork JM, Cox MM, Inman RB. The RecOR proteins modulate RecA protein function at 5′ ends of single-stranded DNA. EMBO J. 2001;20(24):7313–7322. doi: 10.1093/emboj/20.24.7313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robu ME, Inman RB, Cox MM. RecA protein promotes the regression of stalled replication forks in vitro. Proc Natl Acad Sci USA. 2001;98(15):8211–8218. doi: 10.1073/pnas.131022698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Friedman KL, Brewer BJ. Analysis of replication intermediates by two-dimensional agarose gel electrophoresis. Methods Enzymol. 1995;262:613–627. doi: 10.1016/0076-6879(95)62048-6. [DOI] [PubMed] [Google Scholar]
- 12.Furukohri A, Goodman MF, Maki H. A dynamic polymerase exchange with Escherichia coli DNA polymerase IV replacing DNA polymerase III on the sliding clamp. J Biol Chem. 2008;283(17):11260–11269. doi: 10.1074/jbc.M709689200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Langston LD, Indiani C, O’Donnell M. Whither the replisome: Emerging perspectives on the dynamic nature of the DNA replication machinery. Cell Cycle. 2009;8(17):2686–2691. doi: 10.4161/cc.8.17.9390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Setlow RB, Swenson PA, Carrier WL. Thymine dimers and inhibition of DNA synthesis by ultraviolet irradiation of cells. Science. 1963;142(3598):1464–1466. doi: 10.1126/science.142.3598.1464. [DOI] [PubMed] [Google Scholar]
- 15.McInerney P, O’Donnell M. Replisome fate upon encountering a leading strand block and clearance from DNA by recombination proteins. J Biol Chem. 2007;282(35):25903–25916. doi: 10.1074/jbc.M703777200. [DOI] [PubMed] [Google Scholar]
- 16.Higuchi K, et al. Fate of DNA replication fork encountering a single DNA lesion during oriC plasmid DNA replication in vitro. Genes Cells. 2003;8(5):437–449. doi: 10.1046/j.1365-2443.2003.00646.x. [DOI] [PubMed] [Google Scholar]
- 17.Scheuermann R, Tam S, Burgers PM, Lu C, Echols H. Identification of the epsilon-subunit of Escherichia coli DNA polymerase III holoenzyme as the dnaQ gene product: A fidelity subunit for DNA replication. Proc Natl Acad Sci USA. 1983;80(23):7085–7089. doi: 10.1073/pnas.80.23.7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martín-Parras L, Hernández P, Martínez-Robles ML, Schvartzman JB. Unidirectional replication as visualized by two-dimensional agarose gel electrophoresis. J Mol Biol. 1991;220(4):843–853. doi: 10.1016/0022-2836(91)90357-c. [DOI] [PubMed] [Google Scholar]
- 19.Stukenberg PT, Studwell-Vaughan PS, O’Donnell M. Mechanism of the sliding beta-clamp of DNA polymerase III holoenzyme. J Biol Chem. 1991;266(17):11328–11334. [PubMed] [Google Scholar]
- 20.Marians KJ, Hiasa H, Kim DR, McHenry CS. Role of the core DNA polymerase III subunits at the replication fork. Alpha is the only subunit required for processive replication. J Biol Chem. 1998;273(4):2452–2457. doi: 10.1074/jbc.273.4.2452. [DOI] [PubMed] [Google Scholar]
- 21.Kim S, Dallmann HG, McHenry CS, Marians KJ. Tau protects beta in the leading-strand polymerase complex at the replication fork. J Biol Chem. 1996;271(8):4315–4318. doi: 10.1074/jbc.271.8.4315. [DOI] [PubMed] [Google Scholar]
- 22.Kim S, Dallmann HG, McHenry CS, Marians KJ. tau couples the leading- and lagging-strand polymerases at the Escherichia coli DNA replication fork. J Biol Chem. 1996;271(35):21406–21412. doi: 10.1074/jbc.271.35.21406. [DOI] [PubMed] [Google Scholar]
- 23.Leu FP, Georgescu R, O’Donnell M. Mechanism of the E. coli tau processivity switch during lagging-strand synthesis. Mol Cell. 2003;11(2):315–327. doi: 10.1016/s1097-2765(03)00042-x. [DOI] [PubMed] [Google Scholar]
- 24.Xiao H, Naktinis V, O’Donnell M. Assembly of a chromosomal replication machine: Two DNA polymerases, a clamp loader, and sliding clamps in one holoenzyme particle. IV. ATP-binding site mutants identify the clamp loader. J Biol Chem. 1995;270(22):13378–13383. doi: 10.1074/jbc.270.22.13378. [DOI] [PubMed] [Google Scholar]
- 25.Studwell-Vaughan PS, O’Donnell M. Constitution of the twin polymerase of DNA polymerase III holoenzyme. J Biol Chem. 1991;266(29):19833–19841. [PubMed] [Google Scholar]
- 26.LeBowitz JH, McMacken R. The Escherichia coli dnaB replication protein is a DNA helicase. J Biol Chem. 1986;261(10):4738–4748. [PubMed] [Google Scholar]
- 27.Gao D, McHenry CS. tau binds and organizes Escherichia coli replication proteins through distinct domains. Domain IV, located within the unique C terminus of tau, binds the replication fork, helicase, DnaB. J Biol Chem. 2001;276(6):4441–4446. doi: 10.1074/jbc.M009830200. [DOI] [PubMed] [Google Scholar]
- 28.McInerney P, O’Donnell M. Functional uncoupling of twin polymerases: Mechanism of polymerase dissociation from a lagging-strand block. J Biol Chem. 2004;279(20):21543–21551. doi: 10.1074/jbc.M401649200. [DOI] [PubMed] [Google Scholar]
- 29.Kim DR, McHenry CS. Identification of the beta-binding domain of the alpha subunit of Escherichia coli polymerase III holoenzyme. J Biol Chem. 1996;271(34):20699–20704. doi: 10.1074/jbc.271.34.20699. [DOI] [PubMed] [Google Scholar]
- 30.Vandewiele D, Fernández de Henestrosa AR, Timms AR, Bridges BA, Woodgate R. Sequence analysis and phenotypes of five temperature sensitive mutator alleles of dnaE, encoding modified alpha-catalytic subunits of Escherichia coli DNA polymerase III holoenzyme. Mutat Res. 2002;499(1):85–95. doi: 10.1016/s0027-5107(01)00268-8. [DOI] [PubMed] [Google Scholar]
- 31.Blinkova A, et al. The Escherichia coli DNA polymerase III holoenzyme contains both products of the dnaX gene, tau and gamma, but only tau is essential. J Bacteriol. 1993;175(18):6018–6027. doi: 10.1128/jb.175.18.6018-6027.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sutton MD. The Escherichia coli dnaN159 mutant displays altered DNA polymerase usage and chronic SOS induction. J Bacteriol. 2004;186(20):6738–6748. doi: 10.1128/JB.186.20.6738-6748.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jonczyk P, Nowicka A, Fijałkowska IJ, Schaaper RM, Cieśla Z. In vivo protein interactions within the Escherichia coli DNA polymerase III core. J Bacteriol. 1998;180(6):1563–1566. doi: 10.1128/jb.180.6.1563-1566.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takano K, Nakabeppu Y, Maki H, Horiuchi T, Sekiguchi M. Structure and function of dnaQ and mutD mutators of Escherichia coli. Mol Gen Genet. 1986;205(1):9–13. doi: 10.1007/BF02428026. [DOI] [PubMed] [Google Scholar]
- 35.Slater SC, Lifsics MR, O’Donnell M, Maurer R. holE, the gene coding for the theta subunit of DNA polymerase III of Escherichia coli: Characterization of a holE mutant and comparison with a dnaQ (epsilon-subunit) mutant. J Bacteriol. 1994;176(3):815–821. doi: 10.1128/jb.176.3.815-821.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grompe M, Versalovic J, Koeuth T, Lupski JR. Mutations in the Escherichia coli dnaG gene suggest coupling between DNA replication and chromosome partitioning. J Bacteriol. 1991;173(3):1268–1278. doi: 10.1128/jb.173.3.1268-1278.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.D’Ari R, Jaffé-Brachet A, Touati-Schwartz D, Yarmolinsky MB. A dnaB analog specified by bacteriophage P1. J Mol Biol. 1975;94(3):341–366. doi: 10.1016/0022-2836(75)90207-7. [DOI] [PubMed] [Google Scholar]
- 39.Ogawa T. Analysis of dnaB function of Escherichia coli K12 and the dnaB-like function of P1 prophage. J Mol Biol. 1975;94(3):327–340. doi: 10.1016/0022-2836(75)90206-5. [DOI] [PubMed] [Google Scholar]
- 40.Saluja D, Godson GN. Biochemical characterization of Escherichia coli temperature-sensitive dnaB mutants dnaB8, dnaB252, dnaB70, dnaB43, and dnaB454. J Bacteriol. 1995;177(4):1104–1111. doi: 10.1128/jb.177.4.1104-1111.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Belle JJ, Casey A, Courcelle CT, Courcelle J. Inactivation of the DnaB helicase leads to the collapse and degradation of the replication fork: A comparison to UV-induced arrest. J Bacteriol. 2007;189(15):5452–5462. doi: 10.1128/JB.00408-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Georgescu RE, Kurth I, O’Donnell ME. Single-molecule studies reveal the function of a third polymerase in the replisome. Nat Struct Mol Biol. 2012;19(1):113–116. doi: 10.1038/nsmb.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Indiani C, Langston LD, Yurieva O, Goodman MF, O’Donnell M. Translesion DNA polymerases remodel the replisome and alter the speed of the replicative helicase. Proc Natl Acad Sci USA. 2009;106(15):6031–6038. doi: 10.1073/pnas.0901403106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hamdan SM, et al. Dynamic DNA helicase-DNA polymerase interactions assure processive replication fork movement. Mol Cell. 2007;27(4):539–549. doi: 10.1016/j.molcel.2007.06.020. [DOI] [PubMed] [Google Scholar]
- 45.Yang J, Zhuang Z, Roccasecca RM, Trakselis MA, Benkovic SJ. The dynamic processivity of the T4 DNA polymerase during replication. Proc Natl Acad Sci USA. 2004;101(22):8289–8294. doi: 10.1073/pnas.0402625101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baranovskiy AG, et al. DNA polymerase δ and ζ switch by sharing accessory subunits of DNA polymerase δ. J Biol Chem. 2012;287(21):17281–17287. doi: 10.1074/jbc.M112.351122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Johnson RE, Prakash L, Prakash S. Pol31 and Pol32 subunits of yeast DNA polymerase δ are also essential subunits of DNA polymerase ζ. Proc Natl Acad Sci USA. 2012;109(31):12455–12460. doi: 10.1073/pnas.1206052109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hanna M, Ball LG, Tong AH, Boone C, Xiao W. Pol32 is required for Pol zeta-dependent translesion synthesis and prevents double-strand breaks at the replication fork. Mutat Res. 2007;625(1-2):164–176. doi: 10.1016/j.mrfmmm.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 49.Svoboda DL, Vos JM. Differential replication of a single, UV-induced lesion in the leading or lagging strand by a human cell extract: Fork uncoupling or gap formation. Proc Natl Acad Sci USA. 1995;92(26):11975–11979. doi: 10.1073/pnas.92.26.11975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bichara M, Pinet I, Lambert IB, Fuchs RP. RecA-mediated excision repair: A novel mechanism for repairing DNA lesions at sites of arrested DNA synthesis. Mol Microbiol. 2007;65(1):218–229. doi: 10.1111/j.1365-2958.2007.05790.x. [DOI] [PubMed] [Google Scholar]
- 51.Courcelle J, Crowley DJ, Hanawalt PC. Recovery of DNA replication in UV-irradiated Escherichia coli requires both excision repair and recF protein function. J Bacteriol. 1999;181(3):916–922. doi: 10.1128/jb.181.3.916-922.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Selby CP, Sancar A. Molecular mechanism of transcription-repair coupling. Science. 1993;260(5104):53–58. doi: 10.1126/science.8465200. [DOI] [PubMed] [Google Scholar]
- 53.Mellon I, Spivak G, Hanawalt PC. Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell. 1987;51(2):241–249. doi: 10.1016/0092-8674(87)90151-6. [DOI] [PubMed] [Google Scholar]
- 54.Mellon I, Hanawalt PC. Induction of the Escherichia coli lactose operon selectively increases repair of its transcribed DNA strand. Nature. 1989;342(6245):95–98. doi: 10.1038/342095a0. [DOI] [PubMed] [Google Scholar]
- 55.Heller RC, Marians KJ. Replication fork reactivation downstream of a blocked nascent leading strand. Nature. 2006;439(7076):557–562. doi: 10.1038/nature04329. [DOI] [PubMed] [Google Scholar]
- 56.Georgescu RE, Yao NY, O’Donnell M. Single-molecule analysis of the Escherichia coli replisome and use of clamps to bypass replication barriers. FEBS Lett. 2010;584(12):2596–2605. doi: 10.1016/j.febslet.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sclafani RA, Holzen TM. Cell cycle regulation of DNA replication. Annu Rev Genet. 2007;41:237–280. doi: 10.1146/annurev.genet.41.110306.130308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Remus D, Diffley JF. Eukaryotic DNA replication control: Lock and load, then fire. Curr Opin Cell Biol. 2009;21(6):771–777. doi: 10.1016/j.ceb.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 59.Ishimi Y, Komamura-Kohno Y, You Z, Omori A, Kitagawa M. Inhibition of Mcm4,6,7 helicase activity by phosphorylation with cyclin A/Cdk2. J Biol Chem. 2000;275(21):16235–16241. doi: 10.1074/jbc.M909040199. [DOI] [PubMed] [Google Scholar]
- 60.Maiorano D, Moreau J, Méchali M. XCDT1 is required for the assembly of pre-replicative complexes in Xenopus laevis. Nature. 2000;404(6778):622–625. doi: 10.1038/35007104. [DOI] [PubMed] [Google Scholar]
- 61.Nishitani H, Lygerou Z. Control of DNA replication licensing in a cell cycle. Genes Cells. 2002;7(6):523–534. doi: 10.1046/j.1365-2443.2002.00544.x. [DOI] [PubMed] [Google Scholar]
- 62.Makowska-Grzyska M, Kaguni JM. Primase directs the release of DnaC from DnaB. Mol Cell. 2010;37(1):90–101. doi: 10.1016/j.molcel.2009.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Courcelle CT, Courcelle J. Monitoring DNA replication following UV-induced damage in Escherichia coli. Methods Enzymol. 2006;409:425–441. doi: 10.1016/S0076-6879(05)09025-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.