Significance
We report the use of the magnetic tweezers approach to monitor the dynamics of double-stranded DNA break resection at single-molecule resolution. In this work, we unveil the inner workings of the Bacillus subtilis AddAB complex by showing that the domain responsible for the recognition of the recombination hotspot sequence Crossover hotspot instigator (Chi) antagonizes the translocation activity, causing the complex to transiently stall at Chi and Chi-like sequences. We propose a simple model for failed and successful Chi recognition in which the battle between the translocation and sequence-specific binding activities acts as a selectivity filter for bona fide Chi sequences.
Keywords: protein motor, single molecule biophysics, DNA-end processing, real-time measurements, protein–DNA interactions
Abstract
Double-stranded DNA break repair by homologous recombination is initiated by resection of free DNA ends to produce a 3′-ssDNA overhang. In bacteria, this reaction is catalyzed by helicase–nuclease complexes such as AddAB in a manner regulated by specific recombination hotspot sequences called Crossover hotspot instigator (Chi). We have used magnetic tweezers to investigate the dynamics of AddAB translocation and hotspot scanning during double-stranded DNA break resection. AddAB was prone to stochastic pausing due to transient recognition of Chi-like sequences, unveiling an antagonistic relationship between DNA translocation and sequence-specific DNA recognition. Pauses at bona fide Chi sequences were longer, were nonexponentially distributed, and resulted in an altered velocity upon restart of translocation downstream of Chi. We propose a model for the recognition of Chi sequences to explain the origin of pausing during failed and successful hotspot recognition.
Double-stranded DNA breaks (DSBs) are formed frequently, both as a result of exogenous and endogenous DNA damaging agents and as intermediates in programmed DNA rearrangements. Failure to properly repair DSBs results in loss of chromosome structural integrity and genomic instability and is associated with developmental defects, deficiencies of the immune system, and cancer predisposition. There are multiple mechanisms for DSB repair, but faithful repair generally requires the homologous recombination pathway, which can occur only if a suitable donor molecule such as the sister chromatid is available (1). Recombinational repair of DSBs is initiated by the long-range resection of the DNA end to form a 3′-terminated ssDNA overhang that acts as a substrate for the RecA/Rad51 recombinase. In all domains of life, this reaction is catalyzed by an array of helicases and nucleases but is best characterized in bacteria where either an AddAB- or a RecBCD-type helicase–nuclease complex recognizes the DNA end structure and then promotes its processive unwinding and concomitant resection (2, 3). A third class of helicase–nuclease named AdnAB is found in the mycobacterial niche and it shares limited structural similarity with AddAB enzymes (4). An apparently unique feature of the DNA break processing in bacteria is its control by cis-acting DNA sequences called Crossover hotspot instigator (Chi) sequences. In the absence of hotspot sequences, AddAB or RecBCD complexes processively and rapidly degrade DNA in an ATP-dependent fashion, a mode of action that probably acts to degrade foreign DNA or in the restart of regressed replication forks (5). Recognition of Chi sequences by the translocating enzymes has many different effects on AddAB and/or RecBCD complexes, all of which serve to promote downstream recombination (6). These include the attenuation of the nuclease activity downstream of Chi on the 3′ strand (7), the promotion of DNA unwinding (8), and the loading of RecA protein (9). Together, these factors can help to ensure the formation of a recombinogenic RecA-ssDNA filament as the product of the reaction. For this reason, Chi sequences, which vary both in length and in sequence in different bacteria (10), act as recombination hotspots and have played an important role in shaping bacterial genomes.
In comparison with other sequence-specific DNA-binding events, the recognition of Chi is unusual in several respects. The sequence is recognized as single-stranded DNA during break resection and unwinding (11) but must then remain bound within the moving enzyme complex beyond Chi to prevent cleavage of the 3′ strand (12). This is an inefficient process, even under optimized in vitro conditions, with estimates of about 0.2–0.4 for the probability of recognition of a single Chi sequence (7, 8, 13). This may relate in part to the mechanism for target site location, which does not involve facilitated diffusion, but rather a 1D unidirectional scanning of the DNA driven by an ATP-dependent helicase motor. Importantly, although many different consequences of Chi recognition have been documented and rationalized for AddAB or RecBCD complexes, the underlying mechanism is poorly understood not least because a crystal structure of either an AddAB-Chi or a RecBCD-Chi complex remains elusive. However, structures of these enzymes bound to DNA ends have provided limited structural insight into the process (14–17). Residues important for Chi recognition are located in a catalytically inactivated helicase domain that forms a “Chi-scanning module” positioned immediately behind the 3′–5′ ssDNA motor (Fig. 1A). Interestingly, the structural organization of these two activities may be superficially regarded as antagonistic. On the one hand, the motor pumps DNA through the scanning module extremely rapidly (at rates of up to 2,000 bp⋅s−1). This presumably results in dwell times (<1 ms) of the Chi sequence at the binding locus, within which the binding event has a single chance to occur. On the other hand, stable binding of Chi behind the motor would inhibit translocation of ssDNA through the 3′ channel, presumably stalling the motor until ssDNA can be extruded through a proposed alternative exit channel in the complex (details in Fig. 1A). To gain further insight into Chi recognition and how it affects DNA translocation, we have investigated the dynamics of AddAB on single DNA molecules, using magnetic tweezers. Our work shows that the Chi scanning module is indeed inhibitory to DNA translocation and causes the moving enzyme complex to make short stochastic pauses at the many sites on DNA that closely resemble Chi sequences. The enzyme pauses efficiently at locations containing multiple bona fide Chi sites, but the durations of these pauses are not exponentially distributed, consistent with a multistep process associated with successful hotspot recognition.
Fig. 1.
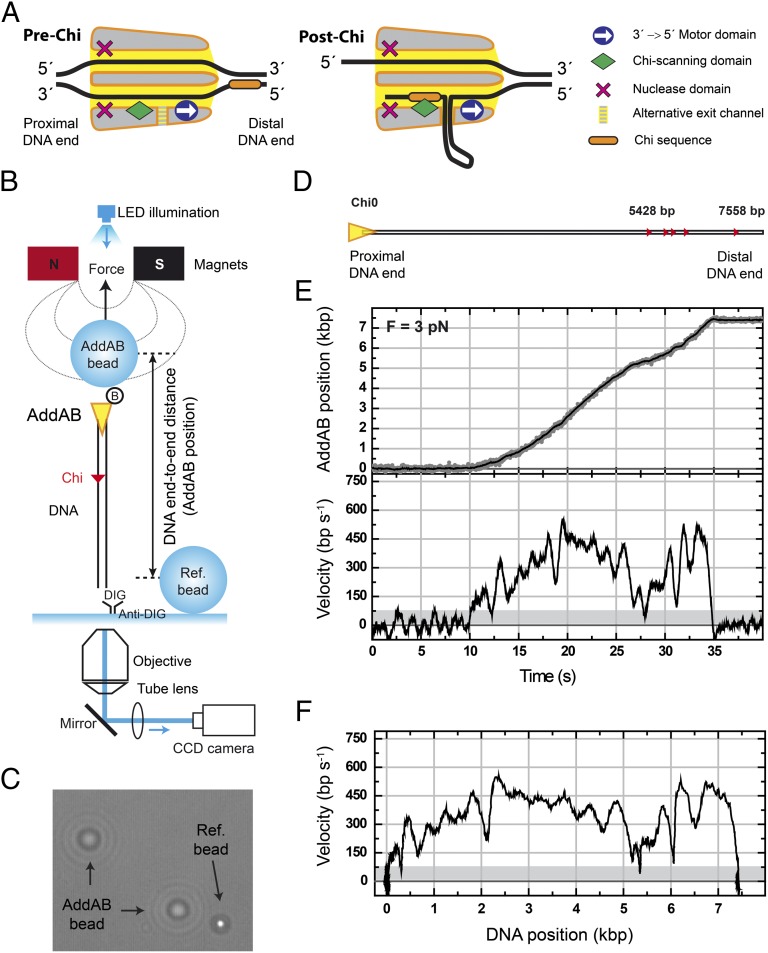
Real-time measurements of AddAB translocation using magnetic tweezers. (A) Cartoon of AddAB showing the two single-stranded DNA channels and the main domains of the heterodimer. The 3′→5′ motor domain and the Chi-scanning domain are situated along the 3′ channel. Between these domains a hypothetical alternative DNA exit channel is depicted. Nuclease domains are shown for completeness. The cartoon highlights the antagonistic role of specific sequence-recognition and translocation functions of AddAB. (B) Scheme of the experiment used to measure real-time dynamics of AddAB at the single-molecule level. (C) Optical image showing the different appearance of a reference bead (in focus) and AddAB beads (out of focus). Diffraction rings are used to determine the height difference between beads (AddAB position). (D) Schematic representation of Chi0 DNA substrate. Triangles represent recombination hotspot sequences within the parental vector. Five Chi sequences appear at the distal end of the DNA at positions approximately in scale with the length of the substrate. (E) Representative time trace of a complete AddAB translocation event. DNA end-to-end distance measured in nanometers is translated to base pairs, using the worm-like chain model for a given force. (Upper) This sets the position of AddAB on the DNA track. Gray points are raw data taken at 60 Hz; black line is the data averaged over ∼0.33 s. (Lower) AddAB velocity time trace calculated using a running window of 0.33 s. Gray area determines the threshold to separate pauses and translocation data. (F) AddAB velocity as a function of its position along the track. Experiments were performed at 3 pN.
Results
ATP-Dependent Single-Molecule Translocation of AddAB Helicase–Nuclease.
In previous work we monitored the DNA unwinding (i.e., strand separation) kinetics of a synchronized population of AddAB molecules in real time (18) and imaged the products of AddAB–DNA reactions at the single-molecule level (8). That work showed that Chi recognition greatly stimulates productive strand separation beyond Chi, but ruled out the possibility of large effects of Chi recognition on DNA translocation kinetics. In this work, to study DNA translocation by single AddAB molecules at high resolution we used a magnetic tweezers (MT) apparatus (19, 20), which consists of a pair of magnets positioned over a flow cell on an inverted optical microscope (Fig. 1B). Magnetic beads are used in the flow cell as probes that are manipulated by an external force that pulls them toward the magnets. Because AddAB starts translocation from a DNA end, we used biotinylated AddAB to bridge one end of a DNA molecule to 1-μm superparamagnetic beads coated with streptavidin (AddAB-bead). The other end of the DNA was labeled with digoxigenin and attached to a glass surface covered with antidigoxigenin. Note that this strategy naturally selects for AddAB–DNA assemblies with the correct orientation as complexes with AddAB bound to the digoxigenin end or to both DNA ends are unable to interact with the glass surface. The glass surface was used as a zero-height reference determined with fixed beads that were not affected by the magnetic field (Ref-bead). The difference in height between both AddAB- and Ref-beads was readily distinguishable by their diffraction rings (Fig. 1C). Comparison of these rings with a calibration profile provided the DNA end-to-end distance as a function of time. Experiments were performed in a chamber that allows exchange of buffers, and injection of ATP quickly triggered unidirectional translocation of AddAB along the DNA, dragging the micrometer bead toward the surface. Because AddAB always begins translocation from the nonlabeled end, the position of the translocating bead can be directly correlated with the position of AddAB along the DNA track. An illustrative example of a typical experiment is included in Movie S1.
Our standard substrate (Chi0) consists of a ∼7,500-bp DNA with a proximal Chi-free section of 5,400 bp and five recombination hotspots (small red triangles, Fig. 1D) located at the distal end of the DNA within the parental vector DNA (details in SI Materials and Methods). Following the introduction of 1 mM ATP into the fluid chamber at 20 °C, the height of the AddAB-bead decreased monotonically. Backsliding events of tens of nanometers were observed in less than 1% of traces. A representative example of a single AddAB translocation trace is shown in Fig. 1E, Upper and the first derivative of the filtered position signal yielded the instantaneous velocity curve (Fig. 1E, Lower). AddAB translocated the entire substrate in about 20 s, providing a mean translocation rate for this molecule of ∼375 bp⋅s−1. To ensure that we were observing a bona fide ATP-dependent translocation signal, experiments were carried out with varying [ATP]. The translocation rate changed with [ATP] in accordance with Michaelis–Menten kinetics, yielding a maximum translocation rate of Vmax = 344 ± 7 bp⋅s−1 and Km = 31 ± 3 µM (Fig. S1A). The measurements shown below were made at 1 mM ATP and this corresponds to saturating conditions. The translocation rate is in good agreement with values from bulk triplex displacement translocation assays at the same temperature (Fig. S1B), demonstrating that the immobilization of the AddAB on the magnetic bead did not perturb the activity. This view is also supported by experiments in which AddAB was conjugated to the bead, using a longer linker that produced no significant difference in the observed activity (details in Materials and Methods). Visual inspection of the data identified stalling or pausing events that, for quantification purposes, were defined as instantaneous translocation velocity values below 75–100 bp⋅s−1, a threshold set as three times the SD of the instantaneous velocity at zero ATP (i.e., the noise, shaded region in Fig. 1E, Lower; details in SI Materials and Methods). A particularly informative representation of the data is shown in Fig. 1F, where the instantaneous velocity is plotted as a function of the position of AddAB on the track. In the example shown, we observed two short pauses, at 300 bp and 5,400 bp, and varying translocation velocities ranging from 300 to 600 bp⋅s−1. This assay allows direct measurement of AddAB instantaneous velocities and their correlation with positions along the DNA track.
AddAB Pauses at Chi Sequences.
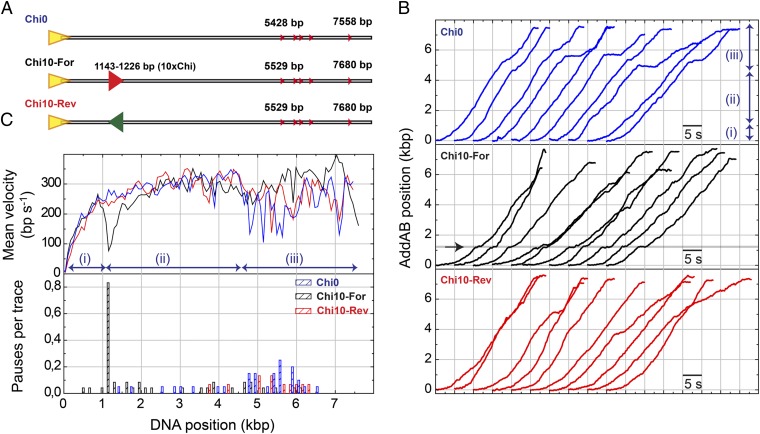
We first characterized the behavior of AddAB on Chi0 DNA (Fig. 2A). To help visualize these data, a representative set of instantaneous-velocity traces is displayed with 5-s offsets in Fig. 2B, Top (blue data). Individual AddAB enzymes displayed large variability in their velocity distributions (static disorder, Fig. S2A) and three distinct regimes within a single trace: (i) a rapid increase in velocity over the first 1,000 bp, (ii) a region with approximately constant velocity, and (iii) a region containing pauses and a significant reduction of velocity. These three regimes are clearly distinguished in a mean velocity curve (Fig. 2C, Upper, blue data). We attribute the initial acceleration to the arrival of ATP in a concentration gradient that stabilizes in ∼5 s (Materials and Methods). From individual translocation activities the positions of pauses were recorded and represented as a pause frequency per trace histogram (Fig. 2C, Lower, blue bars). Note that most of the pauses for Chi0 (74%) are located in the distal region of the substrate, beyond about 4,500 bp, which contains several individual Chi sequences. Importantly, however, most of these pauses do not correlate with the position of the five individual Chi sequences and we return to this point later.
Fig. 2.
AddAB pauses at Chi sequences. (A) The standard DNA substrate Chi0 and DNA substrates containing 10 closely spaced Chi sequences (10×Chi) either correctly oriented for AddAB recognition (Chi10-For) or in opposite orientation (Chi10-Rev). The 10×Chi locus is located ∼1.1 kbp from the entry point. (B) Representative sets of wild-type AddAB translocation time traces for Chi0, Chi10-For, and Chi10-Rev. In the standard substrate (blue data) three regimes were identified: (i) arrival of ATP, (ii) constant AddAB velocity, and (iii) variable rates and pauses. A clear pause was found at the correctly oriented Chi locus (arrow in Chi10-For data). A control experiment using Chi10-Rev did not show a pause at Chi. (C) Mean AddAB velocity as a function of DNA position (Upper) and pauses per trace distribution for different DNA substrates (Lower) were calculated as described in SI Materials and Methods. About 80% of traces presented a clear pause of AddAB at the Chi locus.
Previous studies showed that Chi recognition is an inefficient process, but that the number of successful Chi recognition events increases with the number of consecutive Chi sequences present in DNA substrates (8). Therefore, to promote efficient Chi recognition at a defined position in our substrates, we fabricated a DNA molecule (Chi10-For) with 10 closely spaced Chi sequences located approximately ∼1.1 kbp from the entry point but that is otherwise identical to Chi0. A control substrate (Chi10-Rev) with the same 10 Chi sequences in the opposite orientation was also purified as a control (Fig. 2A). The 10×Chi loci were recognized efficiently and provoked the expected down-regulation of nuclease activity in conventional bulk Chi-recognition assays (Fig. S2B). A set of 10 representative single-molecule translocation curves for Chi10-For is shown in Fig. 2B, Middle (black data). In most of the traces a pause is detected precisely at the position of the 10×Chi region (arrow in Fig. 2B, Middle). Downstream of the 10×Chi locus, AddAB moved at variable translocation rates between 150 and 450 bp⋅s−1, punctuated by short stochastic pauses. As observed previously, these pauses were mainly located at the end of the trace but were reduced in frequency compared with Chi0 substrates. Multiple individual translocation traces were processed to produce mean velocity curves and the pause distribution as shown previously (Fig. 2C, black data). Approximately 80% of all traces exhibited a well-defined pause at the 10×Chi locus, and this was reflected by a downward spike in the mean translocation curve. A control experiment with Chi10-Rev did not show any pause at the inverted 10×Chi locus (Fig. 2 B and C, red data). An additional control used a mutant enzyme (AddABF210A) with a point mutation in the Chi recognition domain of AddB. We have shown that AddABF210A displays wild-type ATPase and DSB resection activities when processing Chi-free DNA (Fig. S2C and ref. 15). This mutant is specifically defective in generating Chi-specific DNA fragments on Chi-containing substrates (Fig. S2B and ref. 15). The AddABF210A showed no pausing at the 10×Chi locus and only three pauses in the entire molecule (Fig. S3, blue data, and Table S1). The observed pausing was therefore specific to Chi recognition (Fig. 2C and Fig. S3).
In current models for Chi recognition, AddAB (or RecBCD) remains bound to the Chi sequence during translocation beyond Chi, resulting in the formation of a ssDNA loop and the inability of the Chi-modified form of the enzyme to recognize additional Chi sequences (12, 15). To test this, we performed conditional Chi recognition experiments involving substrates with two 10×Chi loci in either correct (“For”) or incorrect (“Rev”) orientation for recognition (Fig. 3A). In agreement with the model, these experiments showed that the presence of a correctly oriented 10×Chi sequence in the proximal region of the substrate strongly inhibited pausing at an equivalent 10×Chi locus in the distal region of the substrate (compare For-For and Rev-For substrates in Fig. 3 C and F, Lower). Interestingly, about 15% of AddAB enzymes paused at both Chi loci in Chi10-For-For substrates (Table S1). In the context of our current structural understanding of AddAB–Chi interactions, this observation can be accommodated only if (i) the AddAB–Chi complex can decay during translocation to the second locus or (ii) a proportion of the pauses that are observed at the first locus actually represent failed recognition events. Interestingly, the pause duration at the first locus (for traces containing pauses at both loci) was relatively short, which is consistent with the latter possibility in the context of a model for Chi recognition that is presented later (see below). However, the low number of observed events prevents us from reaching a statistically rigorous conclusion.
Fig. 3.
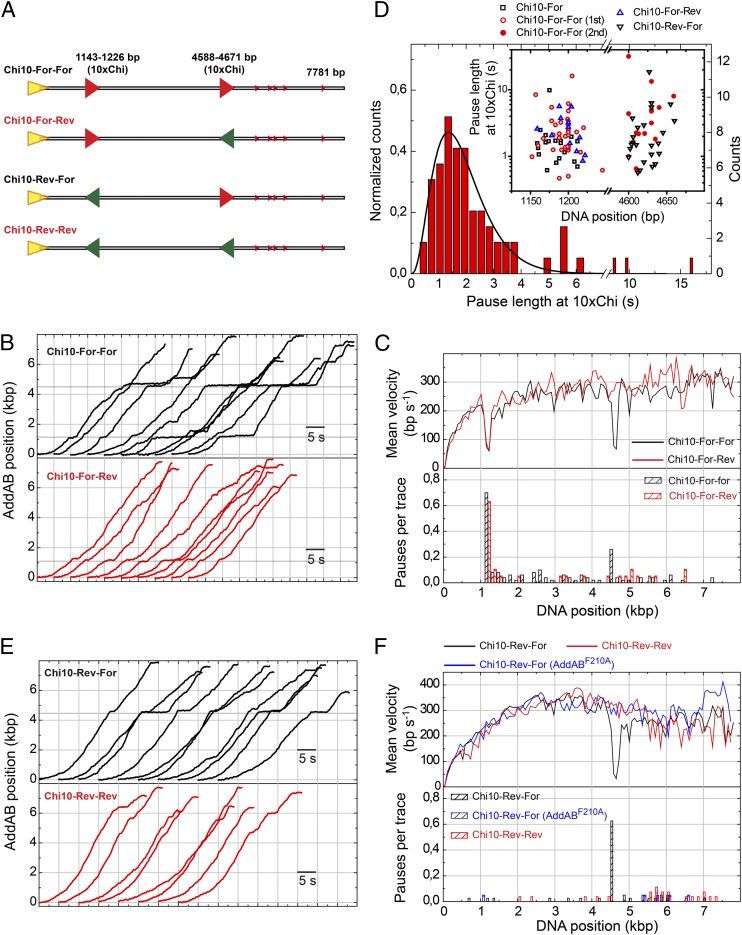
Conditional Chi recognition experiments and pause duration at Chi. (A) Cartoon representation of DNA substrates containing two Chi loci in all possible orientations (For-For, For-Rev, Rev-For, and Rev-Rev). (B) Representative sets of WTAddAB translocation time traces for the Chi10-For-For and Chi10-For-Rev substrates. Gray regions represent 10×Chi loci appropriately oriented for AddAB recognition. (C) Mean velocity graph and pause distribution histogram for wild-type AddAB translocation on Chi10-For-For and Chi10-For-Rev substrates, showing Chi-dependent pausing. (D) Normalized pause length distribution for all pauses at the first 10×Chi locus. Data were fitted to a normalized gamma function (SI Materials and Methods), giving N = 3.9 ± 0.3 and k = 2.1 ± 0.3 s−1. This distribution suggests a process made of multiple stochastic steps. (Inset) Pause position and duration at the first and second 10×Chi loci. No significant differences were found between pauses at the first and second loci in terms of their duration. (E) Representative sets of WTAddAB translocation time traces for the Chi10-Rev-For and Chi10-Rev-Rev substrates. (F) Mean velocity graph and pause distribution histogram for wild-type AddAB and AddABF210A translocation on Chi10-Rev-For and Chi10-Rev-Rev substrates. Data show that AddAB paused precisely at the Chi loci and implies that the positional accuracy of our measurements is ∼75 bp.
These experiments also confirmed the pausing of AddAB at correctly oriented 10×Chi loci and allowed the measurement of the pause duration at the first 10×Chi loci and its representation in a histogram (Fig. 3D). Data showed a nonexponential distribution that was fitted with a gamma function providing a maximum at 1.4 s, with N = 3.9 ± 0.3 and a decay rate of K = 2.1 ± 0.3 s−1. The pauses at the first and second Chi loci were identified with ∼75 bp precision and were of similar duration (Fig. 3D, Inset). The fact that the Chi-induced pause distribution is not exponential indicates that several kinetic steps must be overcome to exit the pause state at a 10×Chi locus. However, we cannot exclude the possibility that the pauses observed at the 10×Chi locus may represent the sum of both failed and successful Chi recognition events, because the spacing of the Chi loci is of a similar magnitude to the spatial resolution of the apparatus.
Chi Recognition Decreases the AddAB Translocation Velocity.
Previous work using the related Escherichia coli RecBCD observed pausing at recombination hotspots and a reduction of translocation rate after Chi (21, 22). Given the fundamentally different structural organization of AddAB-type helicase–nuclease complexes (3), we were interested in understanding how Chi recognition by AddAB may affect its movement on DNA. Our pause-free velocity histograms hinted at a general slowing effect on the translocation rate of AddAB, above and beyond the stalling effect at the position of the Chi sequences (Fig. S4A). However, those distributions were broad due to static and dynamic disorder. To address this question more directly, we compared the translocation rates of many individual AddAB complexes immediately pre- and post-Chi. To avoid any issues arising from the arrival of ATP, we compared data from substrates in which the 10×Chi locus was located at ∼4.5 kbp in either the correct or the incorrect orientation for recognition (Fig. 3A, Rev-For and Rev-Rev substrates). As expected, traces for these substrates showed a high-frequency pause that correlated perfectly with the position of the 10×Chi region, but only if it was appropriately oriented for recognition (Fig. 3 E and F). The duration of the pauses at the second Chi locus followed a distribution described by a gamma function with N = 4.2 ± 0.4 and a decay rate of K = 3.1 ± 0.3 s−1, similar values to those obtained previously at the first Chi locus (Table S1).
Focusing on individual traces, we calculated the mean velocity over the 1 kbp immediately before and after the pause at Chi (Fig. 4A) and plotted these values against one another (Fig. 4B). As a control we made equivalent measurements at the second Chi locus of the Chi10-Rev-Rev substrate that did not display pausing because of its inappropriate orientation. In this analysis, enzymes that do not change velocity at the Chi locus produce a data point on the diagonal shown in Fig. 4B. As might be expected, the control experiment provided values clustered closely along the diagonal (correlation coefficient = 0.88) with a slight tendency to decrease velocity for higher rates (above 500 bp/s). In clear contrast, enzymes that paused at the 10×Chi locus showed greater dispersion of velocities from the diagonal line (correlation coefficient = 0.04) and an average decrease of 16% in the translocation rate compared with the control. Interestingly, the population of enzymes that did not detectably pause in Chi10-Rev-For substrates also showed a tendency to decrease velocity after Chi (12/14 molecules decreased velocity after Chi). This either could mean that the effect of Chi on the velocity of AddAB is independent of pausing or might simply reflect our inability to detect pauses that are shorter than the time resolution of our apparatus.
Fig. 4.
Chi recognition decreases the AddAB translocation velocity. (A) Detail of pauses at the 10×Chi locus in Chi10-Rev-For substrates. (B) Scatter plot of velocities before and after Chi for Chi10-Rev-For and Chi10-Rev-Rev time traces. The velocity was calculated by fitting a straight line over 1,000 bp before and after the pause. Enzymes translocating on Chi10-Rev-For substrates change velocity at the Chi locus and also showed a slight tendency to slow down after Chi.
Pauses That Do Not Occur at Chi Are Short, Stochastic, and Caused by Interactions with Chi-Like Sequences.
We now return to the origin of pausing that does not apparently occur at Chi, for example as was found at the distal end of the DNA in Chi0 substrates (Fig. 2, blue data). This pausing was largely eliminated by placing 10×Chi loci in the proximal region of the substrate (Fig. 2, black data). This implies that the pausing is in some way related to the process of Chi recognition, because we know that successful Chi recognition inhibits downstream recognition of additional hotspots (Fig. 3). To test this idea further, we used a substrate (Chi3-For) with fewer intervening Chi sequences at ∼1,140 bp from the AddAB-binding end (Fig. 5A). Individual translocation traces, the mean velocity curve, and the pause distribution histogram revealed pausing at the distal end of the substrate similar to that observed for Chi0 substrates (compare blue data in Fig. 2 B and C and black data in Fig. 5 B and C). The lack of any clear feature in the translocation curve at the triple-Chi locus shows that recognition of a triple-Chi locus is relatively inefficient, in agreement with bulk Chi recognition assays performed at room temperature (Fig. S2B). This also suggests that the increased pause frequency at the distal end of the substrates was unlikely to be accounted for by the five single Chi sequences common to both of them. Indeed, as discussed above and given the positional accuracy of our assay (details in Materials and Methods), we can conclude that the majority of the stalling events observed in both Chi0 and Chi3-For substrates do not occur at the location of the Chi sequences.
Fig. 5.
Pauses that do not occur at Chi were caused by interactions with Chi-like sequences. (A) Substrates used in this study. The substrate Chi3-For contains a triple-chi locus located at ∼1 kbp from the AddAB-binding end and otherwise is identical to Chi0 with five recombination sequences within the parental plasmid at the distal end (small triangles) and multiple Chi-like sequences in this region (Fig. S5D). Chi3-For-Inverted is equivalent to Chi3-For but is attached to the glass surface at the opposite end of the substrate, presenting an inverted sequence to AddAB. (B) Representative sets of time traces for Chi3-For and Chi3-For-Inverted substrates. Traces for Chi3-For were virtually identical to those obtained with Chi0. (C) Mean wild-type AddAB velocity as a function of DNA position (Upper) and pauses per trace distribution. In contrast with data from Chi3-For, Chi3-For-Inverted showed pauses in the region closer to the AddAB entry point, a region containing multiple Chi-like sequences (red data). (D) Single-molecule time traces of AddABF210A on Chi3-For and Chi3-For-Inverted substrates. (E) Mean AddABF210A velocity as a function of DNA position (Upper) and pauses per trace distribution. Note that the mutant protein generated far smoother traces with virtually no pauses occurring along the substrate.
To determine whether the distal pausing behavior of AddAB was an artifact associated with the enzyme nearing the surface of the flow cell, we fabricated the Chi3-For-Inverted substrate with identical base pair content but the reverse orientation with respect to Chi3-For (Fig. 5A). In distinct contrast to Chi3-For, the individual translocation traces for Chi3-For-Inverted substrates displayed slow translocation and pausing in the proximal region with faster translocation in the distal region (compare red and black data in Fig. 5 B and C). Regardless of the orientation of the substrate, the overall frequency of pausing, calculated as the average number of pauses per trace divided by the mean translocation time without pauses, was kin = 0.06 s−1 (Fig. S4C), indicating that, on average, AddAB makes a pause every 6.3 kbp. Pauses were short and their duration followed an exponential distribution (Fig. S5A). This indicates that the transition from a paused state to a translocating state is stochastic and occurs with a rate constant of kout = 2.5 ± 0.3 s−1, almost two orders of magnitude faster than the average rate for entry into the pause. The overall picture is that AddAB is a processive motor protein that pauses at Chi, but also makes other short and infrequent pauses along the track. Pausing is not caused by the approach to the surface, so we considered other aspects of the DNA substrates that might influence translocation, including the location of Chi-like sequences and their GC content.
We define a Chi-like sequence as a set of 5 nt with one mismatch from the consensus Bacillus subtilis Chi: 5′-AGCGG. A histogram with the occurrences of Chi-like sequences for Chi3-For and Chi3-For-Inverted substrates is shown in Fig. S5B. Intriguingly, this correlates well with the distribution of pauses observed in our experiments (Fig. S5C). Motivated by the fact that the energy needed to separate a GC base pair is larger than for AT (23) and that this is known to affect the movement of some DNA helicases (24–26), we also analyzed the GC content of our substrates. Interestingly, the regions of the substrates associated with elevated pausing clearly also coincided with areas of high GC content (Fig. S5D). To distinguish between the possible role of Chi-like sequences and high-GC content in AddAB pausing, we repeated our experiments with the mutant enzyme AddABF210A. Experiments using the Chi3-For and Chi3-For-Inverted substrates generated far smoother single-molecule translocation traces (Fig. 5D) with an almost complete absence of pauses in all regions of the substrate (Fig. 5E). The frequency of pausing for AddABF210A was sixfold lower than for wild-type AddAB (kin, F210A = 0.01 s−1, Fig. S4C). The F210 residue is located in the Chi-scanning domain of the AddB subunit. Therefore, we conclude that virtually all pausing in the wild-type enzyme is caused by interactions of the Chi-recognition domain of AddAB with the DNA during translocation. Because these pauses also correlate with the location of Chi-like sequences, it is highly likely that they represent failed Chi recognition attempts that stall the progression of the translocating motor. Representative differences in the overall shape of the mean translocation curves were still maintained (Fig. 5E, Upper); AddABF210A translocated at a slightly slower velocity in the first half and clearly faster in the second half of Chi3-For-Inverted compared with data obtained on Chi3-For. This may mean that GC content also has subtle effects on AddAB translocation, with the enzyme moving slightly faster in AT-rich regions compared with GC-rich areas, or might alternatively reflect residual Chi recognition (possibly with a shorter stalling duration) in the mutant protein. Histograms of AddAB instantaneous velocity (which exclude pauses as defined above, but would include pausing on a timescale too short to be detected in our instrument) showed that the mutant protein was faster than the wild type, consistent with the general idea that Chi (or Chi-like) sequence recognition is inhibitory to translocation (Fig. S4B). However, these distributions were broad due to the large variability of velocities within one trace (dynamic disorder) and between different independent traces (static disorder, Fig. S2A) (27, 28). Such variability between traces has been previously described in single-molecule experiments (29, 30).
Discussion
The crystal structure of the heterodimeric AddAB complex showed that the AddA helicase motor and the AddB inactivated helicase domain responsible for Chi recognition are situated along a narrow channel that accommodates the moving single-stranded DNA chain (15). This structure presents a paradox for AddAB translocation and Chi recognition functions, both of which are important for its biological role. Continuous translocation would prevent interaction with recombination hotspots and conversely, blockage of the channel by the strong AddAB–Chi interaction would prevent translocation (Fig. 1A). A model for the post-Chi translocation of AddAB based on the crystal structure resolves this paradox, by proposing that the translocated ssDNA strand exits the enzyme complex in the form of a loop via a new exit channel between the AddA motor and the AddB Chi recognition domain (Fig. 1A). A similar exit channel is proposed to form in RecBCD upon Chi recognition (31), and in both systems the opening of such a channel may be controlled by an “ionic latch” that is released upon Chi binding (15–17). However, detailed insight into this process, especially the manner in which the initial interactions with Chi would affect translocation and promote the opening of the alternative exit channel, is lacking.
Previous AddAB DNA unwinding measurements have uncovered static disorder and pausing during translocation, but the effect of Chi sequences has never been directly investigated (32, 33). In this work, we explored the effect of Chi recognition on AddAB translocation at the single-molecule level, using magnetic tweezers. Our assay allows parallel tracking of many individual AddAB enzymes operating under identical conditions. AddAB translocation was highly processive because release of the magnetic bead into solution occurred only at the lower surface of the flow cell following complete processing of the ∼7-kbp substrate.
Movement along DNA was characterized by continuous changes in velocity punctuated by short and infrequent pauses, with minimal backsliding, unlike that observed for RecBCD using high-resolution optical trapping (34). The location of most of the pauses suggests that they could not have been caused by bona fide Chi sequences, but did correlate with regions of the substrate DNA that were rich in Chi-like sequences. The use of an AddAB complex with a point mutation (F210A) in the Chi-scanning domain that severely reduces its ability to recognize hotspots was particularly informative with respect to the origin of this pausing. The mutant protein paused extremely rarely on all DNA substrates studied, strongly suggesting that the transient (and presumably failed) recognition of Chi-like sequences momentarily stalls the complex on DNA. Pausing of DNA-unwinding enzymes has also been attributed to the unfolding of kinetic barriers dependent on sequence (25, 26, 35). However, the experiment with the mutant AddAB complex excludes the possibility that the pauses are caused by high GC content, as this would be expected to directly affect the AddA helicase motor as opposed to the Chi-scanning module. The same would be true for any damage that may be present in our DNA substrates and that might contribute to the observed pausing. In fact, we have deliberately introduced nicks and UV-induced damage into our substrates and these do cause stalling of AddAB, a finding that led us to take special care in the preparation of DNA substrates for these studies (Materials and Methods). We envisage that the pause of AddAB at Chi-like sequences reflects the formation of an unstable complex with a nonideal recombination hotspot that decays stochastically. The duration of these short pauses is close to the limit of our temporal resolution, and so it is quite probable that we have underestimated the extent of these transient interactions with Chi-like sequences. Indeed, the observation that the instantaneous velocity histograms for the F210A mutant reveal it to be generally faster than wild-type AddAB would be consistent with this idea.
We also found that the presence of several bona fide Chi sequences in the substrate led to a pause precisely at the location of the recombination hotspots, and translocation then typically resumed at a different and (on average) lower rate. We propose that this nonexponential pausing at Chi is the consequence of a set of steps associated with conformational changes required to open the alternative exit channel, thereby beginning the process of single-stranded DNA loop extrusion that will eventually produce the substrate for RecA-mediated recombination. The “resetting” of the translocation rate following a pause may indicate a resampling of the disorder that is inherent in the enzyme that is triggered by Chi recognition and/or pausing. However, we have also found that pauses induced by nicks in the DNA track do not provoke this rate change phenomenon. Alternatively, or additionally, it may indicate that the passage of Chi through the Chi-scanning domain of AddAB alters the translocation mode of AddAB, perhaps due to the inhibitory effects of a growing ssDNA loop on the moving complex. However, we found no evidence for a continued decrease of translocation rate with the distance traveled beyond Chi. An altered translocation mode could also relate to the conformational changes that would be required to accommodate the extrusion of a ssDNA loop (Fig. 1A). These changes most frequently result in a lower translocation velocity post-Chi, but events with substantially higher velocities after the hotspot were also observed.
Pausing and translocation rate changes at recombination hotspots were first observed in the related E. coli RecBCD enzyme that contains dual motor domains (17, 21, 22, 36). Two alternative models were initially proposed to explain the origin of the pause observed in RecBCD and the subsequent decrease in velocity. In the first model, the faster motor, RecD stops upon Chi recognition and the pause accounts for the time it takes the slower motor, RecB, to catch up. An alternative model attributed the pause to the time required for a Chi-induced conformational change. In both models, the change in translocation rate observed after Chi could be simply related to a change in the identity of the lead motor subunit. AddAB-type helicase–nuclease complexes have a simpler organization compared with their RecBCD cousins and contain only a single-motor subunit (3). Therefore, the first model for pausing can be excluded, and the change in translocation velocity after Chi must have an alternative explanation. In contrast to the results presented here, shorter stochastic pauses at Chi-like sequences have never been observed for wild-type RecBCD, and so it might be argued that these are not a general feature of recombination hotspot scanning. Although this could simply reflect a lower time resolution of previous studies, it is critical to note that such pauses would not be expected to be detectable in the RecBCD system. It is the slower motor (RecB) that delivers the Chi sequence to RecC for recognition and consequently, any short-lived stalling events at Chi-like sequences would be buffered by a ssDNA loop that is always present ahead of RecB.
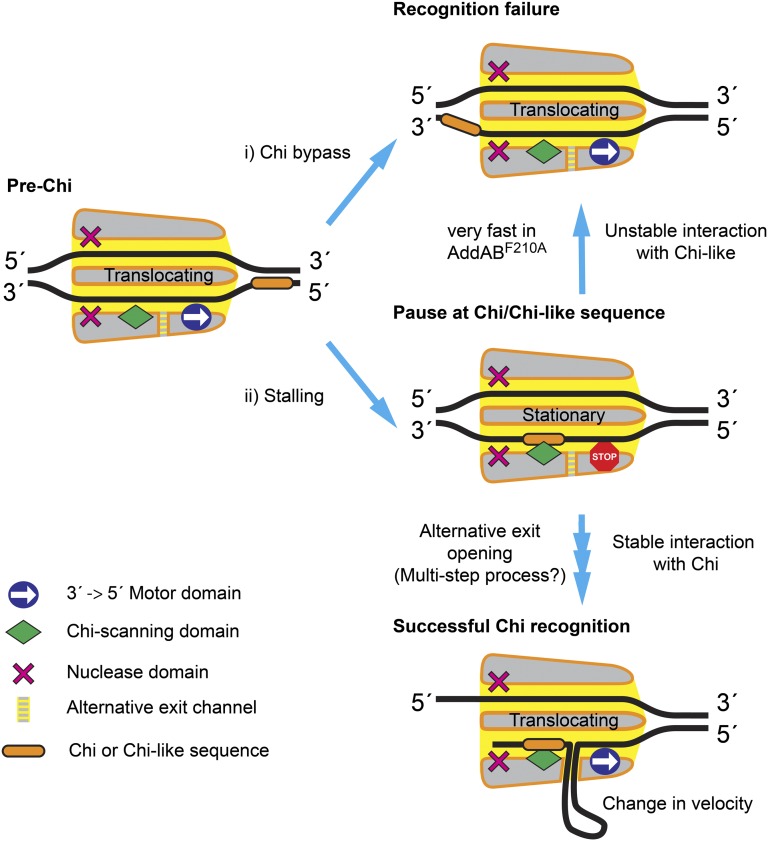
A unified model to explain the origin of pausing at both Chi-like and bona fide Chi sequences and why this may lead to either successful or unsuccessful Chi recognition (i.e., down-regulation of nuclease activity via the formation of a ssDNA loop) is presented in Fig. 6. We propose that the “battle” between the DNA translocation and Chi-binding domains acts as a selectivity filter for correct Chi sequences, as only these form sufficiently stable, long-lived intermediate complexes to resist the motor domain, simply forcing the Chi sequence out of the scanning locus, thereby allowing the opening of an alternative exit channel and escape of the ssDNA in the form of a loop (Fig. 1A). In previous work, we have imaged intermediates of DNA break resection that are consistent with the formation of a ssDNA loop as a consequence of Chi recognition (8). This is thought to exit the Chi-modified form of AddAB at some point between the AddA motor subunit and the inactivated helicase AddB domain, but an in-depth understanding of this exit channel and how it is controlled by Chi binding will await further work. Nevertheless, our detection of Chi-induced stalling intermediates in this work now provides a method for characterizing mutations that may interfere with the opening of the alternative exit channel, including, for example, mutations designed to unlock an ionic latch that is proposed to mediate conformational changes in response to Chi binding.
Fig. 6.
A model for Chi recognition by AddAB. (i) During DNA translocation a fraction of AddAB enzymes simply bypass hotspot sequences, resulting in Chi recognition failure. (ii) The initial binding of Chi or Chi-like sequences to AddAB antagonizes translocation and causes the enzyme to briefly stall. The frequency of pausing (i.e., the average rate of entry into the pause) is 0.06 s−1 for wild-type AddAB. Short pauses are attributed to failed recognition events at Chi-like sequences. These form transient complexes with the wild-type enzyme that decay at a rate of 2.5 s−1, and this results in recognition failure. The AddABF210A complex forms highly unstable complexes with Chi-like sequences that are essentially undetectable at the time resolution of our assay (i.e., much greater than 3 s−1). The longer, nonexponentially distributed pauses of the wild-type enzyme at bona fide Chi loci imply that successful Chi recognition involves several kinetic steps. These would include conformational changes in the protein that allow continued DNA translocation beyond Chi while remaining bound at Chi: for example, the opening of an alternative exit channel that facilitates formation of a single-stranded DNA loop pumped out of the enzyme complex by the motor domain. We postulate that only interactions between wild-type AddAB and bona fide Chi sequences are sufficiently stable to allow these changes to occur in the enzyme. The altered structure of Chi-modified enzyme is reflected by a change in the translocation rate, which is typically slower than the pre-Chi rate.
Materials and Methods
Proteins and DNA Substrates.
Biotinylated AddAB and AddABF210A were purified as described previously (32). DNA molecules for single-molecule experiments were produced by PCR, using one digoxigenin (DIG)-labeled primer (distal end), one unlabeled primer (proximal end), and a high-fidelity DNA polymerase. See SI Materials and Methods and Tables S2 and S3 for detailed methods.
Magnetic Tweezers.
The magnetic tweezers was set up as described in ref. 37 and is similar to that used in refs. 38 and 39. Force values were calculated using the Brownian motion method applied to a DNA-tethered bead (37). Measurements of AddAB mean translocation rate as a function of the applied force showed that it remained constant over our working range of 0.5–4 pN. Because the Brownian motion of the bead is inversely proportional to the applied force, we set 3 pN as our standard force and, after filtering the signals to 3 Hz, our system provides 5–10 nm positional precision in all three dimensions in the absence of flow. Experiments were performed under continuous flow and with enzyme activity that adds experimental noise to the measurement. The estimated resolution of our approach was around 75–100 bp based on the noise of the measurement in the absence of ATP but in the presence of flow. Tethered bead positions were recorded relative to fixed beads to cancel out thermal drift effects. Experiments used streptavidin-coated beads of 1 µm (Dynabeads, MyOne streptavidin; Invitrogen). Alignment of magnets and instrument performance were checked by obtaining a force-extension curve of Chi0 DNA molecules containing a single biotin and digoxigenin. Data were well fitted to the worm-like chain model with a contour length of 2.7 µm and persistence length of 50 nm. In AddAB translocation experiments, beads were selected for analysis showing the maximum possible height (i.e., with the AddAB–DNA connection at the lowest point of the DNA bead). ATP solution flow was set to 65 µL/min. Injecting the ATP faster compromised the bead tracking but reduced the time taken to reach saturating ATP conditions (regime 1, Fig. 2B). Duration of regime 1 did not change either with ATP concentration or with temperature. Therefore, arrival of ATP accounts for regime 1 and lasts for ∼5 s at standard flow conditions.
Analysis of Translocation Data.
Experimental bead positions were recorded at 60 Hz. These data were smoothed by a running window of 20 points. The derivative of the data (translocation rate) was calculated by fitting a linear function to the smoothed data over a running window of 20 data points. The measured end-to-end distance of translocated DNA is always less than the contour length and is dependent on the applied force. Quoted distances in base pairs were corrected using the value given by the worm-like chain model of rise per base pair of DNA at a given force.
Supplementary Material
Acknowledgments
We are grateful to Dr. M. D. Szczelkun, B. Gollnick, Dr. S. A. Khan, and Dr. S. H. Leuba for their comments on the manuscript, and to Dr. N. H. Dekker, Dr. C. Dekker, and Dr. R. Seidel for their generous help in setting up our magnetic tweezers machine and fruitful discussions. This work was supported by a starting grant from the European Research Council (206117) (to F.M.-H., M.S.D., and C.C.), by a grant from the Spanish Ministry of Economy and Competitiveness (FIS2011-24638) (to F.M.-H.), by a Royal Society University Research Fellowship (to M.S.D.), by a Biotechnology and Biological Sciences Research Council Studentship (to N.S.G.), by a Juan de la Cierva contract [JCI-2011-10277, Spanish Ministry of Science and Innovation (MICINN)] (to C.C.), and by Spanish National Research Council Grant I-LINK0331 (to F.M.-H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1303035110/-/DCSupplemental.
References
- 1.Wyman C, Kanaar R. DNA double-strand break repair: All’s well that ends well. Annu Rev Genet. 2006;40:363–383. doi: 10.1146/annurev.genet.40.110405.090451. [DOI] [PubMed] [Google Scholar]
- 2.Mimitou EP, Symington LS. Nucleases and helicases take center stage in homologous recombination. Trends Biochem Sci. 2009;34(5):264–272. doi: 10.1016/j.tibs.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 3.Yeeles JT, Dillingham MS. The processing of double-stranded DNA breaks for recombinational repair by helicase-nuclease complexes. DNA Repair. 2010;9(3):276–285. doi: 10.1016/j.dnarep.2009.12.016. [DOI] [PubMed] [Google Scholar]
- 4.Unciuleac MC, Shuman S. Characterization of the mycobacterial AdnAB DNA motor provides insights into the evolution of bacterial motor-nuclease machines. J Biol Chem. 2010;285(4):2632–2641. doi: 10.1074/jbc.M109.076133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dillingham MS, Kowalczykowski SC. RecBCD enzyme and the repair of double-stranded DNA breaks. Microbiol Mol Biol Rev. 2008;72(4):642–671. doi: 10.1128/MMBR.00020-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wigley DB. Bacterial DNA repair: Recent insights into the mechanism of RecBCD, AddAB and AdnAB. Nat Rev Microbiol. 2013;11(1):9–13. doi: 10.1038/nrmicro2917. [DOI] [PubMed] [Google Scholar]
- 7.Dixon DA, Kowalczykowski SC. The recombination hotspot chi is a regulatory sequence that acts by attenuating the nuclease activity of the E. coli RecBCD enzyme. Cell. 1993;73(1):87–96. doi: 10.1016/0092-8674(93)90162-j. [DOI] [PubMed] [Google Scholar]
- 8.Yeeles JT, van Aelst K, Dillingham MS, Moreno-Herrero F. Recombination hotspots and single-stranded DNA binding proteins couple DNA translocation to DNA unwinding by the AddAB helicase-nuclease. Mol Cell. 2011;42(6):806–816. doi: 10.1016/j.molcel.2011.04.012. [DOI] [PubMed] [Google Scholar]
- 9.Anderson DG, Kowalczykowski SC. The translocating RecBCD enzyme stimulates recombination by directing RecA protein onto ssDNA in a chi-regulated manner. Cell. 1997;90(1):77–86. doi: 10.1016/s0092-8674(00)80315-3. [DOI] [PubMed] [Google Scholar]
- 10.Halpern D, et al. Identification of DNA motifs implicated in maintenance of bacterial core genomes by predictive modeling. PLoS Genet. 2007;3(9):1614–1621. doi: 10.1371/journal.pgen.0030153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bianco PR, Kowalczykowski SC. The recombination hotspot Chi is recognized by the translocating RecBCD enzyme as the single strand of DNA containing the sequence 5′-GCTGGTGG-3′. Proc Natl Acad Sci USA. 1997;94(13):6706–6711. doi: 10.1073/pnas.94.13.6706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chédin F, Handa N, Dillingham MS, Kowalczykowski SC. The AddAB helicase/nuclease forms a stable complex with its cognate chi sequence during translocation. J Biol Chem. 2006;281(27):18610–18617. doi: 10.1074/jbc.M600882200. [DOI] [PubMed] [Google Scholar]
- 13.Taylor AF, Smith GR. RecBCD enzyme is altered upon cutting DNA at a chi recombination hotspot. Proc Natl Acad Sci USA. 1992;89(12):5226–5230. doi: 10.1073/pnas.89.12.5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Singleton MR, Dillingham MS, Gaudier M, Kowalczykowski SC, Wigley DB. Crystal structure of RecBCD enzyme reveals a machine for processing DNA breaks. Nature. 2004;432(7014):187–193. doi: 10.1038/nature02988. [DOI] [PubMed] [Google Scholar]
- 15.Saikrishnan K, et al. Insights into Chi recognition from the structure of an AddAB-type helicase-nuclease complex. EMBO J. 2012;31(6):1568–1578. doi: 10.1038/emboj.2012.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Handa N, et al. Molecular determinants responsible for recognition of the single-stranded DNA regulatory sequence, χ, by RecBCD enzyme. Proc Natl Acad Sci USA. 2012;109(23):8901–8906. doi: 10.1073/pnas.1206076109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang L, et al. Alteration of χ recognition by RecBCD reveals a regulated molecular latch and suggests a channel-bypass mechanism for biological control. Proc Natl Acad Sci USA. 2012;109(23):8907–8912. doi: 10.1073/pnas.1206081109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yeeles JT, Gwynn EJ, Webb MR, Dillingham MS. The AddAB helicase-nuclease catalyses rapid and processive DNA unwinding using a single Superfamily 1A motor domain. Nucleic Acids Res. 2011;39(6):2271–2285. doi: 10.1093/nar/gkq1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Strick TR, Allemand JF, Bensimon D, Bensimon A, Croquette V. The elasticity of a single supercoiled DNA molecule. Science. 1996;271(5257):1835–1837. doi: 10.1126/science.271.5257.1835. [DOI] [PubMed] [Google Scholar]
- 20.Gosse C, Croquette V. Magnetic tweezers: Micromanipulation and force measurement at the molecular level. Biophys J. 2002;82(6):3314–3329. doi: 10.1016/S0006-3495(02)75672-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spies M, et al. A molecular throttle: The recombination hotspot chi controls DNA translocation by the RecBCD helicase. Cell. 2003;114(5):647–654. doi: 10.1016/s0092-8674(03)00681-0. [DOI] [PubMed] [Google Scholar]
- 22.Spies M, Amitani I, Baskin RJ, Kowalczykowski SC. RecBCD enzyme switches lead motor subunits in response to chi recognition. Cell. 2007;131(4):694–705. doi: 10.1016/j.cell.2007.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.SantaLucia J., Jr A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc Natl Acad Sci USA. 1998;95(4):1460–1465. doi: 10.1073/pnas.95.4.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Betterton MD, Jülicher F. Velocity and processivity of helicase unwinding of double-stranded nucleic acids. J Phys Condens Matter. 2005;17(47):S3851–S3869. doi: 10.1088/0953-8984/17/47/015. [DOI] [PubMed] [Google Scholar]
- 25.Manosas M, Xi XG, Bensimon D, Croquette V. Active and passive mechanisms of helicases. Nucleic Acids Res. 2010;38(16):5518–5526. doi: 10.1093/nar/gkq273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morin JA, et al. Active DNA unwinding dynamics during processive DNA replication. Proc Natl Acad Sci USA. 2012;109(21):8115–8120. doi: 10.1073/pnas.1204759109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zwanzig R. Dynamical disorder: Passage through a fluctuating bottleneck. J Chem Phys. 1992;97:3587. [Google Scholar]
- 28.Zwanzig R. Rate processes with dynamical disorder. Acc Chem Res. 1990;23(5):148–152. [Google Scholar]
- 29.Park J, et al. PcrA helicase dismantles RecA filaments by reeling in DNA in uniform steps. Cell. 2010;142(4):544–555. doi: 10.1016/j.cell.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuo TL, et al. Probing static disorder in Arrhenius kinetics by single-molecule force spectroscopy. Proc Natl Acad Sci USA. 2010;107(25):11336–11340. doi: 10.1073/pnas.1006517107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wong CJ, Rice RL, Baker NA, Ju T, Lohman TM. Probing 3′-ssDNA loop formation in E. coli RecBCD/RecBC-DNA complexes using non-natural DNA: A model for “Chi” recognition complexes. J Mol Biol. 2006;362(1):26–43. doi: 10.1016/j.jmb.2006.07.016. [DOI] [PubMed] [Google Scholar]
- 32.Fili N, et al. Visualizing helicases unwinding DNA at the single molecule level. Nucleic Acids Res. 2010;38(13):4448–4457. doi: 10.1093/nar/gkq173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reuter M, Parry F, Dryden DT, Blakely GW. Single-molecule imaging of Bacteroides fragilis AddAB reveals the highly processive translocation of a single motor helicase. Nucleic Acids Res. 2010;38(11):3721–3731. doi: 10.1093/nar/gkq100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perkins TT, Li HW, Dalal RV, Gelles J, Block SM. Forward and reverse motion of single RecBCD molecules on DNA. Biophys J. 2004;86(3):1640–1648. doi: 10.1016/S0006-3495(04)74232-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheng W, Dumont S, Tinoco I, Jr, Bustamante C. NS3 helicase actively separates RNA strands and senses sequence barriers ahead of the opening fork. Proc Natl Acad Sci USA. 2007;104(35):13954–13959. doi: 10.1073/pnas.0702315104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fan HF, Li HW. Studying RecBCD helicase translocation along Chi-DNA using tethered particle motion with a stretching force. Biophys J. 2009;96(5):1875–1883. doi: 10.1016/j.bpj.2008.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Strick TR, Allemand JF, Bensimon D, Croquette V. Behavior of supercoiled DNA. Biophys J. 1998;74(4):2016–2028. doi: 10.1016/S0006-3495(98)77908-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seidel R, et al. Real-time observation of DNA translocation by the type I restriction modification enzyme EcoR124I. Nat Struct Mol Biol. 2004;11(9):838–843. doi: 10.1038/nsmb816. [DOI] [PubMed] [Google Scholar]
- 39.van der Heijden T, et al. Homologous recombination in real time: DNA strand exchange by RecA. Mol Cell. 2008;30(4):530–538. doi: 10.1016/j.molcel.2008.03.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.