Abstract
Protein ectodomain shedding by ADAM17 (a disintegrin and metalloprotease 17), a principal regulator of EGF-receptor signaling and TNFα release, is rapidly and posttranslationally activated by a variety of signaling pathways, and yet little is known about the underlying mechanism. Here, we report that inactive rhomboid protein 2 (iRhom2), recently identified as essential for the maturation of ADAM17 in hematopoietic cells, is crucial for the rapid activation of the shedding of some, but not all substrates of ADAM17. Mature ADAM17 is present in mouse embryonic fibroblasts (mEFs) lacking iRhom2, and yet ADAM17 is unable to support stimulated shedding of several of its substrates, including heparin-binding EGF and Kit ligand 2 in this context. Stimulated shedding of other ADAM17 substrates, such as TGFα, is not affected in iRhom2−/− mEFs but can be strongly reduced by treating iRhom2−/− mEFs with siRNA against iRhom1. Activation of heparin-binding EGF or Kit ligand 2 shedding by ADAM17 in iRhom2−/− mEFs can be rescued by wild-type iRhom2 but not by iRhom2 lacking its N-terminal cytoplasmic domain. The requirement for the cytoplasmic domain of iRhom2 for stimulated shedding by ADAM17 may help explain why the cytoplasmic domain of ADAM17 is not required for stimulated shedding. The functional relevance of iRhom2 in regulating shedding of EGF receptor (EGFR) ligands is established by a lack of lysophasphatidic acid/ADAM17/EGFR-dependent crosstalk with ERK1/2 in iRhom2−/− mEFs, and a significant reduction of FGF7/ADAM17/EGFR-stimulated migration of iRhom2−/− keratinocytes. Taken together, these findings uncover functions for iRhom2 in the regulation of EGFR signaling and in controlling the activation and substrate selectivity of ADAM17-dependent shedding events.
Keywords: ADAMs, Rhbdf1/2
ADAM17 (a disintegrin and metalloprotease17) (also referred to TNFα convertase) is a membrane-anchored metalloproteinase that controls the release of tumor necrosis factor (TNF)α and ligands of the epidermal growth factor receptor (EGFR) from cells (1–5). Inactivation of ADAM17 in myeloid cells protects from endotoxin shock and from inflammatory arthritis, both of which depend on the release of soluble TNFα from myeloid cells (6, 7). Moreover, mice lacking ADAM17 resemble mice lacking the EGFR in that they have open eyes at birth, skin barrier defects, enlarged heart valves, abnormal mammary ductal morphogenesis, and lung defects (3, 8–11). ADAM17 is also responsible for the proteolytic release, or “ectodomain shedding” of a variety of other membrane proteins from the cell surface, such as Kit ligand (KitL) 1 and 2 (12), the vascular endothelial growth factor receptor 2 and many others (13, 14). However, most phenotypes caused by inactivation of ADAM17 can be explained through loss of shedding of TNFα or of EGFR ligands (3, 6–11), so these can be considered the functionally dominant substrates of ADAM17.
The sheddase activity of ADAM17 in cells can be rapidly and posttranslationally activated by intracellular signaling pathways through a mechanism that requires the transmembrane domain of ADAM17, but not its cytoplasmic domain (15–18). These findings raise the possibility that other transmembrane proteins that interact with ADAM17 could have a role in activation of ADAM17-dependent shedding. Recently, the inactive Rhomboid iRhom2, a catalytically inactive member of the Rhomboid family of intramembrane proteinases (19–22), was identified as a crucial regulator of the maturation of ADAM17 in hematopoietic cells. Interestingly, mature ADAM17 is present in several tissues and cell types of iRhom2−/− mice, including mouse embryonic fibroblasts (mEFs) and keratinocytes (7, 23). This raises questions about whether iRhom2 affects the function of ADAM17 in cells outside of immune cells. Here, we show that iRhom2 is not only required for the maturation of ADAM17 in hematopoietic cells but also that it has essential functions in controlling the activation and substrate selectivity of ADAM17-dependent shedding events.
Results and Discussion
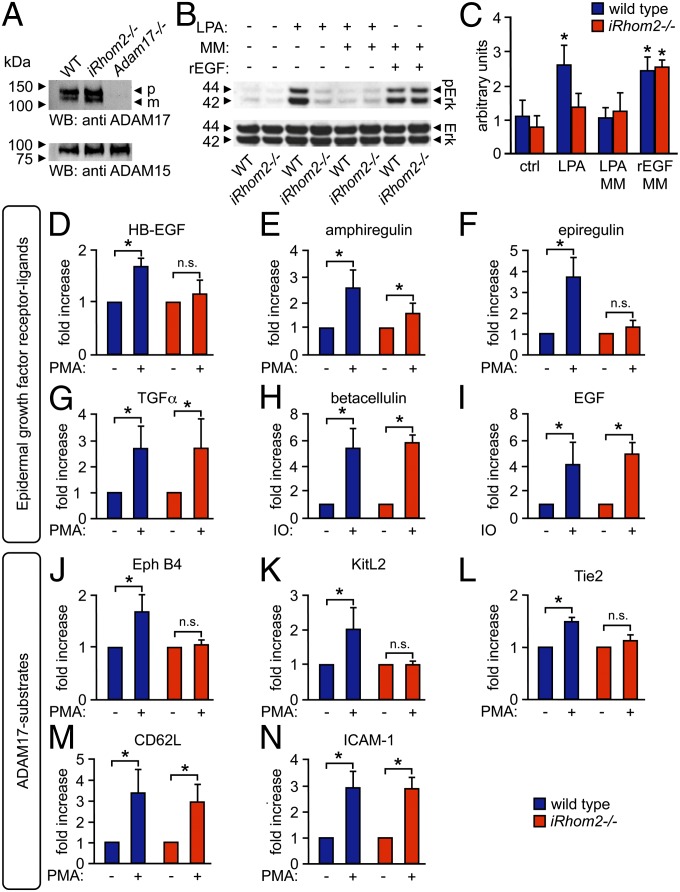
Previous studies have demonstrated that ADAM17 is essential for crosstalk between G protein coupled receptors and EGFR (24–26). To gain a better understanding of the role of iRhom2 in regulating ADAM17-dependent shedding events, we tested whether iRhom2−/− mEFs, which have similar levels of mature ADAM17 as wild-type (WT) cells [Fig.1A; Adam17−/− mEFs shown as control (7)], might have a defect in the metalloproteinase-dependent activation of ERK1/2 following stimulation of these cells with lysophasphatidic acid (LPA). We found that LPA-stimulated phosphorylation of ERK1/2 was significantly reduced in iRhom2−/− mEFs compared with WT controls (Fig.1 B and C), just like in WT cells treated with the metalloproteinase inhibitor marimastat (MM). Because LPA is known to stimulate ADAM17 (16), these findings suggested that iRhom2−/− mEFs could have a defect in activation of ADAM17-dependent crosstalk with ERK1/2. iRhom2 was not directly required for activation of ERK1/2, as recombinant (r) EGF promoted ERK1/2 phosphorylation similarly in iRhom2−/− and WT mEFs (Fig.1 B and C). We also found that phorbol-12-myristate-13-acetate (PMA)-stimulated shedding of heparin-binding (HB)-EGF, an ADAM17 substrate and EGFR ligand (4, 10) that is crucial for G protein-coupled receptor-mediated crosstalk with the EGFR/ERK1/2 signaling axis (25, 26), was almost completely abrogated in iRhom2−/− mEFs compared with WT controls, despite the presence of mature ADAM17 (experiments with immortalized mEFs shown in Fig.1D, experiments with 3 separately isolated cultures of primary mEFs shown in Fig. S1A). Taken together, these results suggest a function of iRhom2 in controlling the rapid activation of ADAM17-mediated cleavage of HB-EGF (or other EGFR ligands), leading to activation of the EGFR and phosphorylation of ERK1/2.
Fig. 1.
iRhom2 is required for metalloproteinase-dependent crosstalk between the LPA receptor and ERK1/2 and for the substrate selectivity of stimulated ADAM17-dependent shedding events. (A) Western blot analysis shows similar levels of mature ADAM17 in mEFs from iRhom2−/− mice and littermate controls (Adam17−/− mEFs are shown as negative control; ADAM15 Western blot is shown as loading control; n = 3). (B) WT and iRhom2−/− mEFs, starved for 24 h, were treated for 15 min with 10 µM LPA, 20 ng/mL recombinant EGF (rEGF), or medium alone. Where indicated, cells were pretreated for 20 min with 20 µM the hydroxamate metalloproteinase inhibitor MM. The Western blot was probed for phospho-ERK1/2, with total ERK1/2 as loading control. (C) Densitometric quantification of ERK1/2 phosphorylation relative to total ERK (n = 3). (D–N) WT and iRhom2−/− mEFs were transfected with the AP-tagged EGFR ligands HB-EGF (D), amphiregulin (E), epiregulin (F), TGFα (G), BTC (H), or EGF (I) or the ADAM17 substrates EphB4 (J), KitL2 (K), Tie2 (L), CD62L (M), or ICAM-1 (N). Shedding of all ADAM17 substrates was activated by treatment with 25 ng/mL PMA for 30 min (D–G and J–N), whereas shedding of the ADAM10 substrates BTC and EGF was activated by 2.5 µM ionomycin (IO) (H and I). *P ≤ 0.05; ±SEM (n = 3).
We next tested whether iRhom2 also controls activated shedding of other EGFR ligands (3–5, 27). The PMA-stimulated shedding of the ADAM17 substrates and EGFR ligands amphiregulin and epiregulin (4, 5) was also strongly reduced in iRhom2−/− mEFs compared with WT controls (Fig.1 E and F). However, the PMA-stimulated shedding of the ADAM17 substrate TGFα (3, 4) was not affected (Fig.1G and Fig. S1B). Moreover, the ionomycin-stimulated shedding of the ADAM10 substrates and EGFR ligands betacellulin (BTC) and EGF (4, 15, 27) was normal in iRhom2−/− mEFs (Fig.1 H and I), arguing against a role of iRhom2 in ADAM10-dependent protein ectodomain shedding.
We then tested how lack of iRhom2 affects shedding of additional ADAM17 substrates unrelated to EGFR signaling. We found that PMA-stimulated shedding of the Eph receptor B4 (EphB4), KitL2, and Tie2 (12, 14) was strongly reduced in iRhom2−/− mEFs compared with WT control mEFs (Fig.1 J–L; KitL2 shedding in primary mEFs is shown in Fig. S1C), whereas the PMA-stimulated shedding of the ADAM17 substrates CD62 ligand [cluster of differentiation 62L (CD62L)] and intercellular adhesion molecule-1 (ICAM-1) (14, 28) was not affected (Fig.1 M and N). The absence of iRhom2 did not significantly influence the constitutive shedding of any of the ADAM17 substrates tested here over a 4-h period, although these experiments cannot rule out a minor difference in constitutive shedding (Fig. S2; cells treated with the metalloproteinase inhibitor MM served to establish the contribution of metalloproteinases to constitutive shedding of these substrates). Taken together, these results demonstrate that iRhom2 controls the substrate selectivity of ADAM17 under stimulated conditions; it is required for the stimulated shedding of some substrates of ADAM17, including HB-EGF, but not for the stimulated shedding of other substrates, such as TGFα. In this context, it is interesting to note that Herrlich and coworkers have described differences in the processing of HB-EGF and TGFα in response to distinct stimuli (29, 30), which could be a reflection of differential regulation of the shedding of these EGFR ligands by iRhom2.
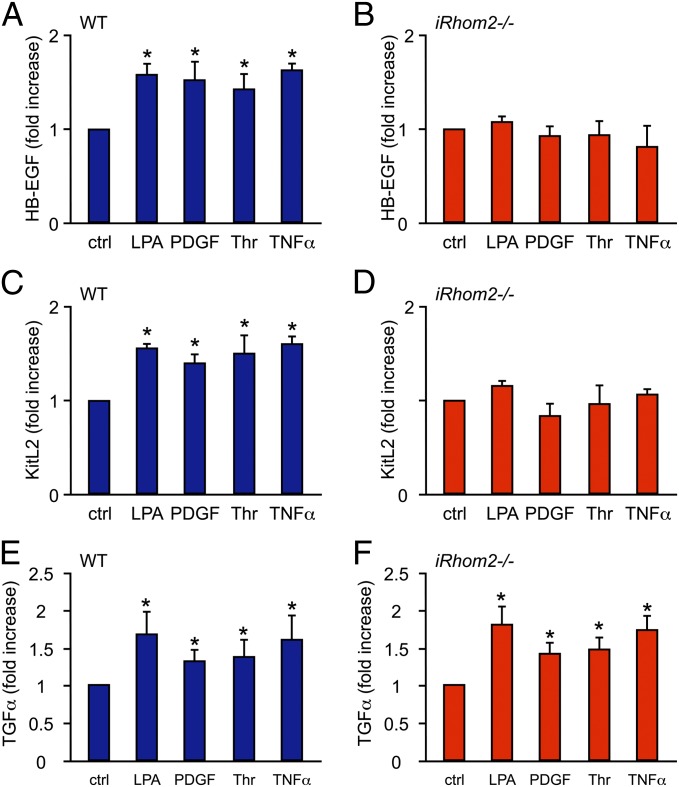
Because PMA is a pleiotropic activator of cellular signaling pathways, we also tested whether iRhom2 controls the stimulation of ADAM17-dependent shedding by more specific physiological signaling pathways. Consistent with the results obtained with PMA-stimulated cells, we found that the stimulated release of HB-EGF and KitL2 in response to the physiological stimuli LPA, PDGFβ (PDGF), Thrombin (Thr), or TNFα (16, 31) was strongly reduced in iRhom2−/− cells compared with WT controls (Fig. 2 A–D), whereas the activation of TGFα shedding by these stimuli in iRhom2−/− cells remained normal (Fig.2 E and F). Moreover, we found that both LPA and TNFα stimulated the shedding of amphiregulin, epiregulin, EphB4, and Tie2 in WT mEFs, but not in iRhom2−/− mEFs, whereas the activation of CD62L or ICAM-1 shedding in response to these stimuli was comparable in iRhom2−/− mEFs and WT controls (Fig. S3). The reduction in LPA-stimulated shedding of amphiregulin and epiregulin in iRhom2−/− mEFs raises the possibility that, in addition to HB-EGF, these EGFR ligands could also contribute to the LPA/ERK1/2 crosstalk in mEFs shown in Fig. 1B.
Fig. 2.
iRhom2 is required for the stimulated shedding of HB-EGF and KitL2 in response to physiological stimuli but not of TGFα. (A–F) WT and iRhom2−/− mEFs were transfected with the AP-tagged ADAM17 substrates HB-EGF (A and B), KitL2 (C and D), or TGFα (E and F) and then activated with different stimuli for 30 min, as indicated [LPA (10 µM), PDGF (50 ng/mL), Thr (2 units/mL), or TNFα (10 ng/mL)]. All stimuli tested here activated ADAM17-mediated shedding, as evidenced by the significantly increased cleavage of HB-EGF (A), KitL2 (C), or TGFα (E) in WT mEFs. Identical experiments were performed with iRhom2−/− mEFs, in which stimulation for 30 min with LPA, Thr, PDGF, or TNFα did not increase the shedding of HB-EGF (B) or KitL2 (D) but activated the release of TGFα (F). *P ≤ 0.05; ±SEM (n = 3 for A–F). ctrl, control.
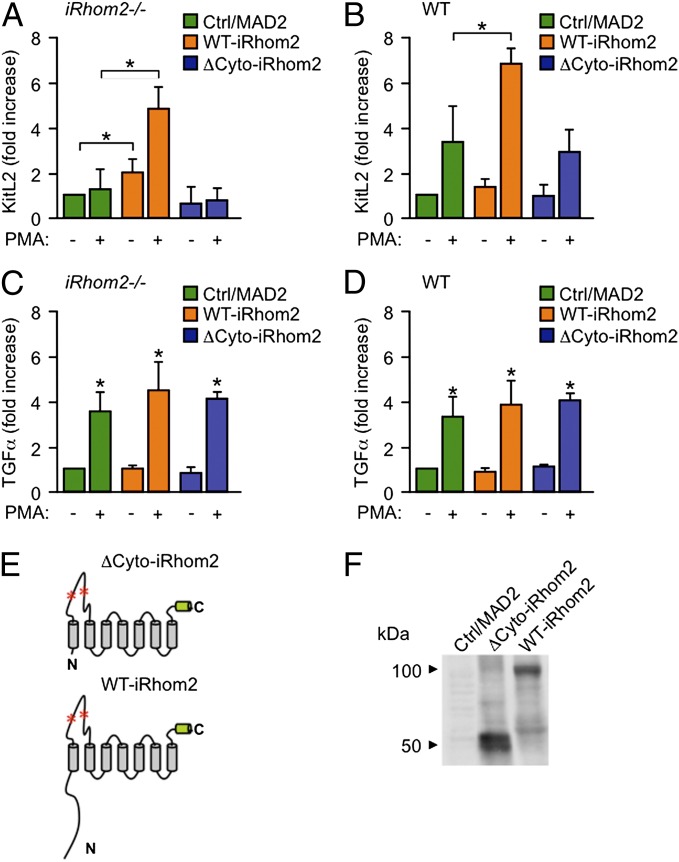
Previous studies have shown that the cytoplasmic domain of ADAM17 is not required for rapid activation of the shedding of its substrates by PMA or other signaling pathways (15–18). Because the stimulated shedding of KitL2 was abolished in iRhom2−/− mEFs, this provided an opportunity to test whether the cytoplasmic domain of iRhom2 is required for the stimulation of ADAM17-dependent shedding of KitL2, used here as a model substrate. We found that the defect in PMA-stimulated KitL2 shedding in iRhom2−/− mEFs could be rescued with WT iRhom2 but not with a mutant form of iRhom2 lacking its N-terminal cytoplasmic domain (Fig. 3A). These findings provide evidence that the cytoplasmic domain of iRhom2 could be involved in controlling the rapid and posttranslational activation of ADAM17-dependent shedding events. These results also support the notion that mutations in the cytoplasmic domain of iRhom2 that have been linked to the development of tylosis esophageal cancer (32) could act, at least in part, by promoting ADAM17-dependent shedding. We also found that overexpression of iRhom2 enhanced stimulated KitL2 shedding from WT cells, further corroborating that iRhom2 regulates the activation of shedding by ADAM17 (Fig. 3B). However, overexpression of iRhom2 did not affect the constitutive or stimulated shedding of TGFα from iRhom2−/− or WT cells (Fig. 3 C and D; a diagram of the membrane topology of ΔCyto-iRhom2 and WT-iRhom2 is shown in Fig. 3E, and a representative Western blot of the overexpressed ΔCyto-iRhom2 and WT-iRhom2 is shown in Fig. 3F).
Fig. 3.
Analysis of the requirement for the N-terminal cytoplasmic domain of iRhom2 in the rapid and posttranslational activation of ADAM17-dependent shedding of KitL2. (A–D) iRhom2−/− (A and C) or WT mEFs (B and D) were cotransfected with KitL2-AP (A and B) or TGFα-AP together with full-length WT-iRhom2, ΔCyto-iRhom2 (lacking the N-terminal cytoplasmic domain), or MAD2 as a control and either left untreated or stimulated with PMA (25 ng/mL; 30 min). (E) The membrane topology and domain organization of ΔCyto-iRhom2 and WT-iRhom2 is shown, with red asterisks indicating potential N-linked glycosylation sites in the large extracellular loop. C indicates C terminus; N indicates N terminus. The green cylinder at the C terminus marks the position of the T7 tag. (F) Representative Western blot of ΔCyto-iRhom2 or WT-iRhom2 expressed in iRhom2−/− mEFs and detected with antibodies against a C-terminal T7-tag. *P ≤ 0.05; ±SEM (n = 3 for A–F).
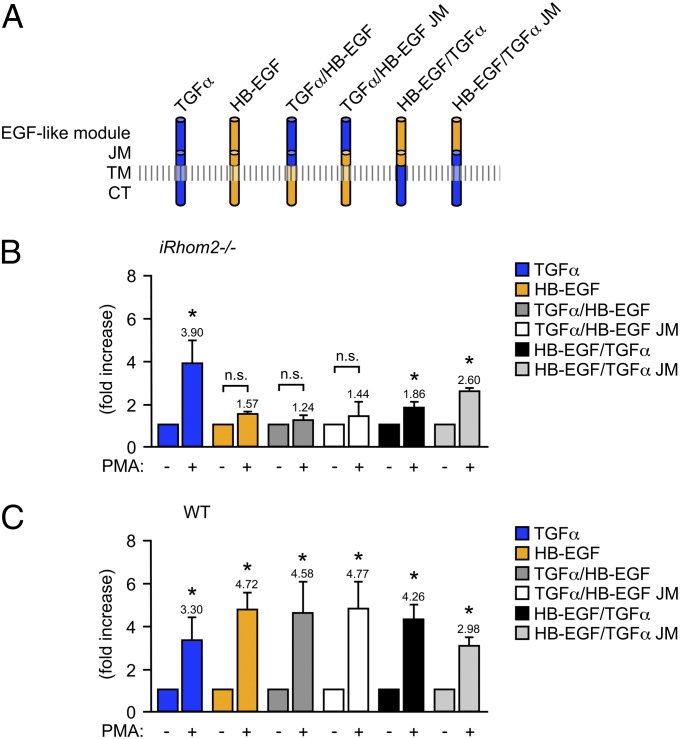
To understand which substrate domains confer selectivity to stimulated shedding by iRhom2/ADAM17, we generated TGFα/HB-EGF chimeras (see Fig.4A for a diagram of the HB-EGF/TGFα chimeras used here). We found that neither the replacement of the entire extracellular domain of HB-EGF with that of TGFα [the EGF-like module and the juxtamembrane domain (JM) including the membrane proximal cleavage site, TGFα/HB-EGF] nor a swap of the EGF module alone (TGFα/HB-EGF-JM) was sufficient to allow stimulated shedding in iRhom2−/− mEFs (Fig. 4B), even though the stimulated shedding of both chimeras from WT mEFs was comparable to that of HB-EGF (Fig.4C). Shedding of a chimera with the transmembrane and cytoplasmic domain (TM-CT) of TGFα and the EGF module and JM of HB-EGF (HB-EGF/TGFα) was slightly, but significantly increased upon PMA-stimulation in iRhom2−/− mEFs, although the response was much weaker than in WT control mEFs. However, the PMA-stimulated shedding of a chimera containing the JM and TM-CT from TGFα and only the EGF-like module from HB-EGF (HB-EGF/TGFα JM) was similar in iRhom2−/− mEFs compared with WT control mEFs (for this chimera, stimulated shedding was slightly reduced in WT controls compared with the shedding of TGFα or HB-EGF or the other chimera). Taken together, these results suggest that the main determinants for PMA-stimulated shedding of TGFα in iRhom2−/− mEFs most likely reside in its JM and TM-CT domains and probably not in the EGF-like module, raising the possibility that the JM and TM-CT of HB-EGF (and likely other substrates as well) could be important for its selective stimulated shedding by iRhom2/ADAM17.
Fig. 4.
Analysis of the contribution of the extracellular or cytoplasmic domain of TGFα and HB-EGF to their stimulated shedding in iRhom2−/− mEFs. (A) Full-length TGFα-AP or HB-EGF-AP or chimeras of these EGFR ligands containing the extracellular EGF module and JM with the cleavage site of TGFα but the TM-CT of HB-EGF (TGFα/HB-EGF) or the EGF module of TGFα with the JM and TM-CT of HB-EGF (TGFα/HB-EGF JM) or the EGF module and JM of HB-EGF attached to the TM-CT of TGFα (HB-EGF/TGFα) or the EGF module of HB-EGF with the JM and TM-CT of TGFα (HB-EGF/TGFα-JM). The constructs depicted in A were transfected into iRhom2−/− (B) or WT (C) mEFs, and their constitutive and PMA-stimulated shedding into the supernatant was determined. *P ≤ 0.05, ±SEM (n = 3 for B and C).
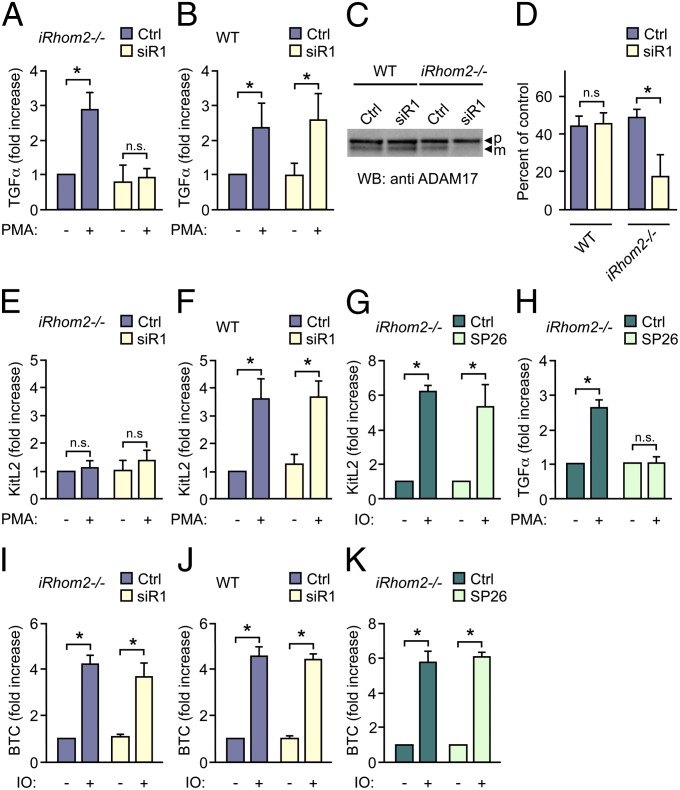
These observations, and our previous work (7), raised questions about the potential role of the related iRhom1 in the stimulated shedding of TGFα in iRhom2−/− mEFs. When iRhom2−/− mEFs were treated with small interfering (si) RNA against iRhom1 (siR1), this abolished the PMA-stimulated shedding of TGFα (Fig. 5A). However, treatment of WT mEFs with iRhom1 siRNA had no effect on the stimulated release of TGFα (Fig. 5B), so iRhom1 and iRhom2 presumably have redundant or compensatory functions in stimulated TGFα shedding. Western blots showed normal mature ADAM17 levels in siR1-treated WT mEFs but strongly reduced mature ADAM17 in siR1-treated iRhom2−/− mEFs (Fig. 5 C and D and ref. 7). Treatment of iRhom2−/− mEFs with iRhom1 siRNA did not further reduce the constitutive or stimulated shedding of KitL2 (Fig. 5E), and iRhom1 siRNA also did not affect KitL2 shedding from WT mEFs (Fig. 5F).
Fig. 5.
Effect of inactivation of iRhom1 or iRhom2 or both iRhom1 and -2 on ADAM17- and ADAM10-dependent shedding. iRhom2−/− mEFs (A, E, G, H, I, and K) or WT controls (B, F, and J) were transfected with the AP-tagged ADAM17 substrates TGFα (A, B, and H), KitL2 (E–G), or the AP-tagged ADAM10 substrate BTC (I–K) and stimulated with 25 ng/mL PMA to activate ADAM17-dependent shedding (A, B, E, F, and H) or 2.5 µM ionomycin (IO) to activate ADAM10-dependent shedding (G and I–K) with or without siRNA against iRhom1 (siR1) (10 nM) (A, B, E, F, I, and J) or the ADAM17-specific inhibitor SP26 (G, H, and K). (C) ADAM17 Western blot shows a decrease of mature ADAM17 only in iRhom2−/− mEFs treated with iRhom1 siRNA but not in WT controls treated with iRhom1 siRNA. (D) Densitometric quantification of the percentage of mature ADAM17 relative to the proform in blots from three separate experiments like the one shown in C. *P ≤ 0.05; ±SEM (n = 3 for A–K).
ADAM10-dependent–stimulated processing of ADAM17 substrates can be observed when Adam17−/− mEFs are treated with ionomycin (28), allowing us to test whether iRhom2 affects the ability of KitL2 to be shed by other ADAMs. Stimulation of iRhom2−/− mEFs with ionomycin activated shedding of KitL2 to the same extent as in WT cells and was not inhibited by the ADAM17-selective inhibitor SP26 (28) (Fig. 5G). SP26 blocks PMA-stimulated ADAM17-dependent shedding of TGFα from iRhom2−/− cells (Fig. 5H), confirming the selectivity of SP26 toward ADAM17 over ADAM10 under the conditions used here. Furthermore, the observation that shedding of KitL2 by ADAM10 is not affected in iRhom2−/− cells argues against a general defect in trafficking and shedding of KitL2 in the absence of iRhom2. The constitutive and ionomycin-stimulated shedding of the ADAM10 substrate BTC was similarly unaffected in iRhom2−/− or WT mEFs in the presence or absence of iRhom1 siRNA (Fig. 5 I and J) and insensitive to the ADAM17 inhibitor SP26 (Fig. 5K), providing additional evidence that substrate trafficking, and the function of the related ADAM10, is normal in iRhom2−/− mEFs and in iRhom2−/− mEFs treated with iRhom1 siRNA (7).
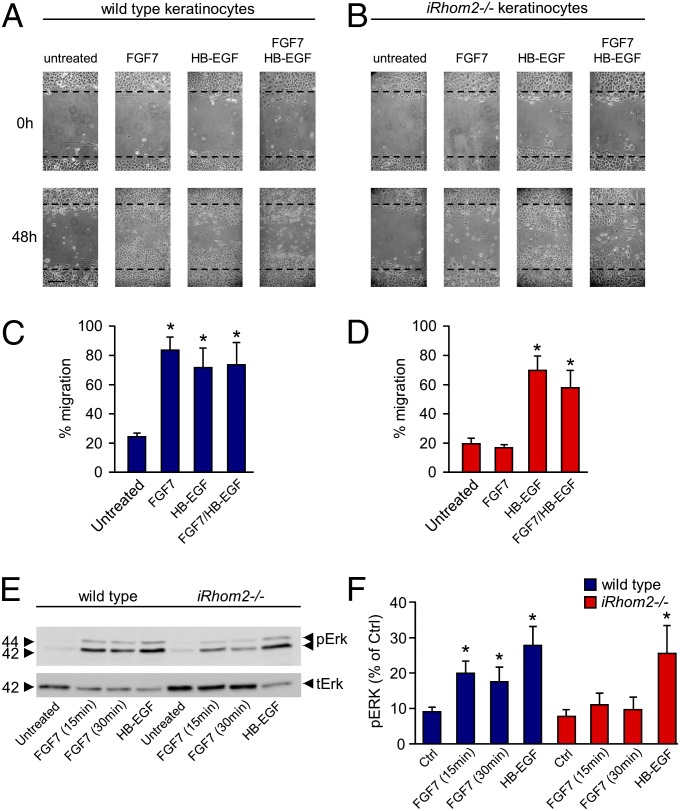
The functional relevance of inactivation of iRhom2 in nonhematopoietic cells was assessed in a keratinocyte-migration assay that depends on the release of HB-EGF by ADAM17 (26). Treatment of WT keratinocytes with fibroblast growth factor (FGF)7 stimulated cell migration, whereas stimulation of iRhom2−/− keratinocytes did not (Fig. 6 A and B; quantification is shown in Fig. 6 C and D). In WT and iRhom2−/− keratinocytes, addition of HB-EGF alone stimulated scratch wound healing, and HB-EGF also rescued the defect in FGF7-stimulated cell migration in iRhom2−/− keratinocytes. Moreover, crosstalk between FGF7/FGFR2 and ERK1/2, which depends on ADAM17 and the release of HB-EGF (26), was reduced in iRhom2−/− keratinocytes compared with WT controls, whereas the ERK1/2 stimulation by exogenous soluble HB-EGF was comparable (Fig. 6E; quantification is shown in Fig. 6F).
Fig. 6.
iRhom2 controls ADAM17-dependent keratinocyte migration. (A and B) Primary WT (A) or iRhom2−/− (B) keratinocytes from 12-wk-old animals were cultured to confluence, and then a scratch wound was introduced, and the cultures treated with or without FGF7 (50 ng/mL) or HB-EGF (50 ng/mL), as indicated. Micrographs were taken at 0 and 48 h after scratch wounding. (Scale bar: 100 μm.) (C and D) Quantification of the results obtained with WT keratinocytes (C) or iRhom2−/− keratinocytes (D) (n = 3). (E) Western blot of ERK1/2 phosphorylation in primary WT or iRhom2−/− keratinocytes incubated with or without FGF7 (20 ng/mL) or HB-EGF (50 ng/mL) (ERK1/2 was loading control in E). (F) Densitometric quantification of the levels of pERK1/2 of three experiments like the one shown in E. *P ≤ 0.05; ±SEM.
Taken together, these results uncover a crucial role for iRhom2 in controlling the substrate selectivity and stimulation of ADAM17-dependent shedding events. Maturation of ADAM17 is not affected in iRhom2−/− mEFs, at least as determined by Western blot analysis, but the available mature ADAM17 is nevertheless unable to promote stimulated shedding of HB-EGF, KitL2, and several other substrates, whereas the release of TGFα, ICAM, and CD62L is normal (Table 1). The lack of iRhom2 does not affect the function of the related ADAM10, nor does it appear to block maturation of substrates of ADAM17, because these can still be cleaved by ADAM10 in ionomycin-stimulated iRhom2−/− mEFs. This study extends our understanding of the functions of iRhom2 by demonstrating that it is not only required for the maturation of ADAM17 but is also an essential component of the mechanism that controls substrate selectivity and the rapid posttranslational activation of ADAM17-dependent shedding events.
Table 1.
iRhom2 substrate selectivity in mEFs
| iRhom2-dependent substrates | iRhom2-independent substrates |
| HB-EGF | TGFα |
| Amphiregulin | ICAM-1 |
| Epiregulin | L-selectin |
| EphB4 | |
| KitL2 | |
| Tie2 |
Materials and Methods
Primary Cells and Cell Lines.
iRhom2−/− mice [C57BL/6; described previously (20)] were crossed twice onto a 129Sv/C57BL/6 mixed background. Primary mouse keratinocytes were isolated and cultured in Keratinocyte Growth Medium KGM-2 supplemented with 8 ng/mL cholera toxin and 20 ng/mL murine EGF (Sigma). The offspring of matings between heterozygous iRhom2+/− mice were used for isolation of primary keratinocytes from WT or iRhom2−/− littermates as follows: 12-wk-old mice were euthanized, and their tails were removed and incubated for 5 min in Triadine (Triad Disposables). After washing in 70% (vol/vol) ethanol, the skin was removed with sterile instruments and placed epidermis side down on sterile Whatman paper and incubated in serum-free DMEM with 1% trypsin for no more than 2 h at 37 °C. The epidermis was peeled off from the dermis and transferred to 10% (vol/vol) FCS in PBS. This suspension was passed through a 40-μm cell filter and centrifuged for 5 min at 200 × g. The sedimented cells were resuspended in KGM-2, seeded in collagen-coated 6- or 12-well plates (Becton-Dickinson) and used at a confluence of 80–90%. All animal experiments were approved by the Internal Animal Use and Care Committee of the Hospital for Special Surgery. mEFs were grown in DMEM supplemented with antibiotics and 10% (vol/vol) FCS.
Growth Factors and Inhibitors.
Recombinant human HB-EGF, murine EGF, and murine FGF7 (also referred to as keratinocyte growth factor) were from R&D Systems. PMA and the calcium ionophore, ionomycin, were from Sigma. The metalloproteinase inhibitor marimastat (BB2516) was from Tocris Biosciences. The ADAM17-selective inhibitor SP26 was provided by Daniel Lundell and Xioada Niu (Schering Plough Research Institute, Kenilworth, NJ).
Antibodies.
Rabbit anti-phospho ERK1/2 was from Cell Signaling, rabbit anti-ERK2 was from Santa Cruz Biotechnology, anti-T7 tag mouse monoclonal antibodies were from Novagen. Rabbit polyclonal anti-ADAM17 cytotail and anti-ADAM15 antibodies have been described previously (33).
Expression Vectors.
The expression vectors for iRhom2, mitotic arrest deficient 2 (MAD2), and plasmids encoding alkaline phosphatase (AP)-tagged EGFR ligands, CD62L, ICAM-1, EphB4, and KitL2, have been described previously (4, 12, 14, 20, 28). The chimeras between TGFα and HB-EGF were generated by overlap extension PCR (34) using human TGFα-AP and HB-EGF-AP cDNAs as a template and cloned into pRc/CMV (Invitrogen) (see Fig. S4 for details). The cytoplasmic domain-deleted mutant of iRhom2 was generated by PCR and cloned into pcDNA 3.1 (+) (see Fig. S5 for details).
Transfection and Ectodomain-Shedding Assay.
mEFs were transfected with Lipofectamine 2000 (Invitrogen). For shedding experiments (performed 1 or 3 d after transfection, respectively), cells were washed with OptiMEM (without added serum or growth factors) (Gibco), which was replaced after 1 h with fresh OptiMEM with or without the indicated inhibitors or stimuli, and then incubated for 30 min to assess stimulated shedding or for 4 h to monitor constitutive shedding. The AP activity in the supernatant and cell lysates was measured at an absorbance of A405 after incubation with the AP substrate 4-nitrophenyl phosphate. No AP activity was present in conditioned media of nontransfected cells. Three identical wells were prepared, and the ratio between the AP activity in the supernatant and the cell lysate plus supernatant was calculated. For constitutive shedding, ratios of AP activity in supernatant vs. cell lysate plus supernatant are reported. For stimulated shedding, the fold increase in the ratio of AP activity obtained after stimulation is shown (relative to ratio of AP activity in control wells without stimulation). Each experiment was repeated at least three times.
siRNA Transfection.
For silencing of iRhom1, mEFs were grown to 40–50% confluence and transfected with 10 nM of Stealth siRNA duplex [rhomboid family member 1 (Rhbdf1)-MSS203813, Rhbdf1-MSS203814, Rhbdf1-MSS203815; Invitrogen] using Lipofectamine 2000. Random Stealth siRNA duplexes coding for nonfunctional RNAs served as controls. After 72 h of incubation at 37 °C, the cells were used in shedding assays. Afterward, the mEFs were processed for Western blot analysis to analyze the effect of knocking down iRhom1 on the maturation of ADAM17.
Western Blot Analysis.
Cells were lysed on ice in Tris-buffered saline (TBS) Triton-X100 (1%), EDTA (1 mM), 1,10-phenanthroline (10 mM), protease inhibitor mixture and phosphatase inhibitor (Roche Applied Science). Comparable amounts of protein were separated on 10% (wt/vol) SDS/polyacrylamide gels and transferred onto poly(vinylidene difluoride) membranes (BioTrace; Pall Corporation). These were blocked with 3% (wt/vol) skim milk in TBS and then incubated with primary antibodies and washed in 0.1% Tween-TBS, and bound primary antibodies were detected with peroxidase-conjugated goat anti-mouse or goat anti-rabbit antibodies (Promega) using the ECL detection system (Amersham Biosciences) and a Chemdoc image analyzer (Bio-Rad). To generate the control blots, membranes were incubated in stripping reagent [100 mM 2-mercaptoethanol, 2% (wt/vol) SDS, 62.5 mM Tris⋅HCl (pH 6.7)] at 55 °C for 30 min and then reprobed with anti ERK1/2 antibodies.
In Vitro Scratch Wound-Healing Assays.
Primary mouse keratinocytes used for in vitro scratch wound-healing assays were seeded in 12-well plates and cultured until they reached confluence (26). A scratch wound was introduced with a 200-μL pipette tip. After washing with PBS, the cells were incubated with or without the indicated stimuli. After different periods of time, cells at the same positions along the scratch wound (marked with an indelible marker) were photographed using an inverted phase-contrast microscope (Nikon; Eclipse TS100), and National Institutes of Health Image J software was used for quantification of scratch wound assays.
Statistical Analysis.
All values are expressed as means ± SEM. An unpaired two-tailed Student t test was used for all statistical tests.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grant R01-GM64750 (to C.P.B., T.M., and P.D.A.I.), Alexander von Humboldt Foundation Grant Sofia Kovalevskaya Award (SKA2010) (to P.A.L.), and German Research Council Grant LA2558/3-1 (to P.A.L.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1302553110/-/DCSupplemental.
References
- 1.Black RA, et al. A metalloproteinase disintegrin that releases tumour-necrosis factor-alpha from cells. Nature. 1997;385(6618):729–733. doi: 10.1038/385729a0. [DOI] [PubMed] [Google Scholar]
- 2.Moss ML, et al. Cloning of a disintegrin metalloproteinase that processes precursor tumour-necrosis factor-α. Nature. 1997;385(6618):733–736. doi: 10.1038/385733a0. [DOI] [PubMed] [Google Scholar]
- 3.Peschon JJ, et al. An essential role for ectodomain shedding in mammalian development. Science. 1998;282(5392):1281–1284. doi: 10.1126/science.282.5392.1281. [DOI] [PubMed] [Google Scholar]
- 4.Sahin U, et al. Distinct roles for ADAM10 and ADAM17 in ectodomain shedding of six EGFR ligands. J Cell Biol. 2004;164(5):769–779. doi: 10.1083/jcb.200307137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sunnarborg SW, et al. Tumor necrosis factor-alpha converting enzyme (TACE) regulates epidermal growth factor receptor ligand availability. J Biol Chem. 2002;277(15):12838–12845. doi: 10.1074/jbc.M112050200. [DOI] [PubMed] [Google Scholar]
- 6.Horiuchi K, et al. Cutting edge: TNF-alpha-converting enzyme (TACE/ADAM17) inactivation in mouse myeloid cells prevents lethality from endotoxin shock. J Immunol. 2007;179(5):2686–2689. doi: 10.4049/jimmunol.179.5.2686. [DOI] [PubMed] [Google Scholar]
- 7.Issuree PD, et al. iRHOM2 is a critical pathogenic mediator of inflammatory arthritis. J Clin Invest. 2013;123(2):928–932. doi: 10.1172/JCI66168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chalaris A, et al. Critical role of the disintegrin metalloprotease ADAM17 for intestinal inflammation and regeneration in mice. J Exp Med. 2010;207:1617–1624. doi: 10.1084/jem.20092366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Franzke CW, et al. Epidermal ADAM17 maintains the skin barrier by regulating EGFR ligand-dependent terminal keratinocyte differentiation. J Exp Med. 2012;209(6):1105–1119. doi: 10.1084/jem.20112258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jackson LF, et al. Defective valvulogenesis in HB-EGF and TACE-null mice is associated with aberrant BMP signaling. EMBO J. 2003;22(11):2704–2716. doi: 10.1093/emboj/cdg264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sternlicht MD, et al. Mammary ductal morphogenesis requires paracrine activation of stromal EGFR via ADAM17-dependent shedding of epithelial amphiregulin. Development. 2005;132(17):3923–3933. doi: 10.1242/dev.01966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kawaguchi N, et al. Different ADAMs have distinct influences on Kit ligand processing: Phorbol-ester-stimulated ectodomain shedding of Kitl1 by ADAM17 is reduced by ADAM19. J Cell Sci. 2007;120(Pt 6):943–952. doi: 10.1242/jcs.03403. [DOI] [PubMed] [Google Scholar]
- 13.Swendeman S, et al. VEGF-A stimulates ADAM17-dependent shedding of VEGFR2 and crosstalk between VEGFR2 and ERK signaling. Circ Res. 2008;103(9):916–918. doi: 10.1161/CIRCRESAHA.108.184416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weskamp G, et al. Pathological neovascularization is reduced by inactivation of ADAM17 in endothelial cells but not in pericytes. Circ Res. 2010;106(5):932–940. doi: 10.1161/CIRCRESAHA.109.207415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horiuchi K, et al. Substrate selectivity of EGF-receptor ligand sheddases and their regulation by phorbol esters and calcium influx. Mol Biol Cell. 2007;18:176–188. doi: 10.1091/mbc.E06-01-0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Le Gall SM, et al. ADAM17 is regulated by a rapid and reversible mechanism that controls access to its catalytic site. J Cell Sci. 2010;123(Pt 22):3913–3922. doi: 10.1242/jcs.069997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reddy P, et al. Functional analysis of the domain structure of tumor necrosis factor-alpha converting enzyme. J Biol Chem. 2000;275(19):14608–14614. doi: 10.1074/jbc.275.19.14608. [DOI] [PubMed] [Google Scholar]
- 18.Hall KC, Blobel CP. Interleukin-1 stimulates ADAM17 through a mechanism independent of its cytoplasmic domain or phosphorylation at threonine 735. PLoS ONE. 2012;7(2):e31600. doi: 10.1371/journal.pone.0031600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adrain C, Zettl M, Christova Y, Taylor N, Freeman M. Tumor necrosis factor signaling requires iRhom2 to promote trafficking and activation of TACE. Science. 2012;335(6065):225–228. doi: 10.1126/science.1214400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McIlwain DR, et al. iRhom2 regulation of TACE controls TNF-mediated protection against Listeria and responses to LPS. Science. 2012;335(6065):229–232. doi: 10.1126/science.1214448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adrain C, Freeman M. New lives for old: Evolution of pseudoenzyme function illustrated by iRhoms. Nat Rev Mol Cell Biol. 2012;13(8):489–498. doi: 10.1038/nrm3392. [DOI] [PubMed] [Google Scholar]
- 22.Siggs OM, et al. iRhom2 is required for the secretion of mouse TNFα. Blood. 2012;119(24):5769–5771. doi: 10.1182/blood-2012-03-417949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lichtenthaler SF. iRHOM2 takes control of rheumatoid arthritis. J Clin Invest. 2013;123(2):560–562. doi: 10.1172/JCI67548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gschwind A, Hart S, Fischer OM, Ullrich A. TACE cleavage of proamphiregulin regulates GPCR-induced proliferation and motility of cancer cells. EMBO J. 2003;22(10):2411–2421. doi: 10.1093/emboj/cdg231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prenzel N, et al. EGF receptor transactivation by G-protein-coupled receptors requires metalloproteinase cleavage of proHB-EGF. Nature. 1999;402(6764):884–888. doi: 10.1038/47260. [DOI] [PubMed] [Google Scholar]
- 26.Maretzky T, et al. Migration of growth factor-stimulated epithelial and endothelial cells depends on EGFR transactivation by ADAM17. Nat Commun. 2011;2:229. doi: 10.1038/ncomms1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanderson MP, et al. ADAM10 mediates ectodomain shedding of the betacellulin precursor activated by p-aminophenylmercuric acetate and extracellular calcium influx. J Biol Chem. 2005;280(3):1826–1837. doi: 10.1074/jbc.M408804200. [DOI] [PubMed] [Google Scholar]
- 28.Le Gall S, et al. ADAMs 10 and 17 represent differentially regulated components of a general shedding machinery for membrane proteins such as TGFα, L-Selectin and TNFα. Mol Biol Cell. 2009;20:1785–1794. doi: 10.1091/mbc.E08-11-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Herrlich A, Klinman E, Fu J, Sadegh C, Lodish H. Ectodomain cleavage of the EGF ligands HB-EGF, neuregulin1-beta, and TGF-alpha is specifically triggered by different stimuli and involves different PKC isoenzymes. FASEB J. 2008;22(12):4281–4295. doi: 10.1096/fj.08-113852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hartmann M, Herrlich A, Herrlich P. Who decides when to cleave an ectodomain? Trends Biochem Sci. 2013;38(3):111–120. doi: 10.1016/j.tibs.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 31.Mendelson K, Swendeman S, Saftig P, Blobel CP. Stimulation of platelet-derived growth factor receptor beta (PDGFRbeta) activates ADAM17 and promotes metalloproteinase-dependent crosstalk between the PDGFRbeta and epidermal growth factor receptor (EGFR) signaling pathways. J Biol Chem. 2010;285(32):25024–25032. doi: 10.1074/jbc.M110.102566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blaydon DC, et al. RHBDF2 mutations are associated with tylosis, a familial esophageal cancer syndrome. Am J Hum Genet. 2012;90(2):340–346. doi: 10.1016/j.ajhg.2011.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schlöndorff J, Becherer JD, Blobel CP. Intracellular maturation and localization of the tumour necrosis factor alpha convertase (TACE) Biochem J. 2000;347(Pt 1):131–138. [PMC free article] [PubMed] [Google Scholar]
- 34.Nelson MD, Fitch DH. Overlap extension PCR: An efficient method for transgene construction. Methods Mol Biol. 2011;772:459–470. doi: 10.1007/978-1-61779-228-1_27. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.