Abstract
Gram-negative bacteria acquire iron with TonB-dependent uptake systems. The TonB–ExbBD inner membrane complex is hypothesized to transfer energy to outer membrane (OM) iron transporters. Fluorescence microscopic characterization of green fluorescent protein (GFP)-TonB hybrid proteins revealed an unexpected, restricted localization of TonB in the cell envelope. Fluorescence polarization measurements demonstrated motion of TonB in living cells, which likely was rotation. By determining the anisotropy of GFP-TonB in the absence and presence of inhibitors, we saw the dependence of its motion on electrochemical force and on the actions of ExbBD. We observed higher anisotropy for GFP-TonB in energy-depleted cells and lower values in bacteria lacking ExbBD. However, the metabolic inhibitors did not change the anisotropy of GFP-TonB in ΔexbBD cells. These findings demonstrate that TonB undergoes energized motion in the bacterial cell envelope and that ExbBD couples this activity to the electrochemical gradient. The results portray TonB as an energized entity in a regular array underlying the OM bilayer, which promotes metal uptake through OM transporters by a rotational mechanism.
Keywords: bioenergetics, membrane transport, FepA, iron transport
From its importance in aerobic metabolism, iron is essential to most pro- and eukaryotes and therefore is a determinant of bacterial disease. Its sequestration by transferrin, lactoferrin, ferritin, heme compounds, and lipocalins defends animal cells, fluids, and tissues by “nutritional immunity” (1). However, efficient pathogens overcome this barrier and capture Fe3+ either by producing siderophores (2) or by directly removing the metal from eukaryotic proteins (3). The trilaminar cell envelope of Gram-negative bacteria, composed of inner membrane (IM), outer membrane (OM), and the periplasm between them, contains protein components that confer the uptake of metabolic solutes, including sugars, amino acids, nucleotides, vitamins, and metals such as iron (4, 5). Enigmatic OM active transporters acquire metal complexes (ferric siderophores, heme, vitamin B12) from the environment (6). The OM protein ferric enterobactin permease A (FepA), for example, internalizes the siderophore ferric enterobactin (FeEnt) (7). It is typical of many homologous metal transporters in commensal and pathogenic organisms. These uptake reactions also require TonB (8), a cell envelope protein that long ago was proposed to transduce energy (9–12). However, many questions exist about TonB’s mediation of iron uptake, including its physical mechanism and its relationship to bioenergetics. Proton motive force (PMF) may drive OM active transport (6–8), but the mode of energy transmission to the OM and TonB’s potential role in it are unknown. We addressed these topics by characterizing the localization of TonB in the cell membranes, by monitoring its physical motion, and by determining the dependence of its movements on metabolic energy and the additional IM proteins ExbBD.
Iron chelates bind to their OM transporters on the cell surface (13). The subnanomolar affinities of these receptor ferric siderophores (14) impart efficiency and specificity to the transport process. These proteins, also called ligand-gated porins (LGP) (13) or TonB-dependent transporters (15), contain a C-terminal porin (16) channel (C-domain) that surrounds an N-terminal globule (N-domain) within the pore (17–22). When LGP bind ligands, structural changes expose an N-terminal polypeptide [the TonB-box (20–22)] in the periplasm, transmitting a signal of receptor occupancy to the internal surface of the OM bilayer. The N-domain somehow regulates the subsequent stage of energy-dependent ligand transport through the transmembrane channel, which also requires the actions of TonB. Ultimately, periplasmic binding proteins (23) adsorb the transported metal complexes and transfer them to ABC-type IM permeases (24, 25).
Bioinformatic, biochemical, and biophysical data suggest that TonB comprises three parts in the cell envelope: a hydrophobic N-terminal sequence in the IM (26–28), a central rigid section in the periplasm (29–31), and a C-terminal ββαβ domain that may transiently associate with the TonB-box of LGP in the OM (32–35). By spanning the periplasm, TonB may link the IM and OM in a manner that facilitates energy transmission to the metal transporters (32, 36). However, its participation in energy metabolism remains hypothetical: TonB is not known to generate, use, or transfer bioenergetic force. Despite this gap, most theories postulate that TonB transduces energy (11, 31, 32, 37, 38). Furthermore, the ExbBD proteins that associate with TonB in the IM show homology to MotAB, the presumed “stator” element of the bacterial flagellar motor (39). Both LGP and the flagellar motor require an electrochemical gradient for activity. These realizations led to the theory (32) or implication (40) that TonB functions by rotation. Using GFP fusion proteins, we conducted experiments to observe the disposition and motion of TonB in vivo. The results suggest that it undergoes constant motion, driven by electrochemical force.
Results
Localization of TonB in the Cell Envelope.
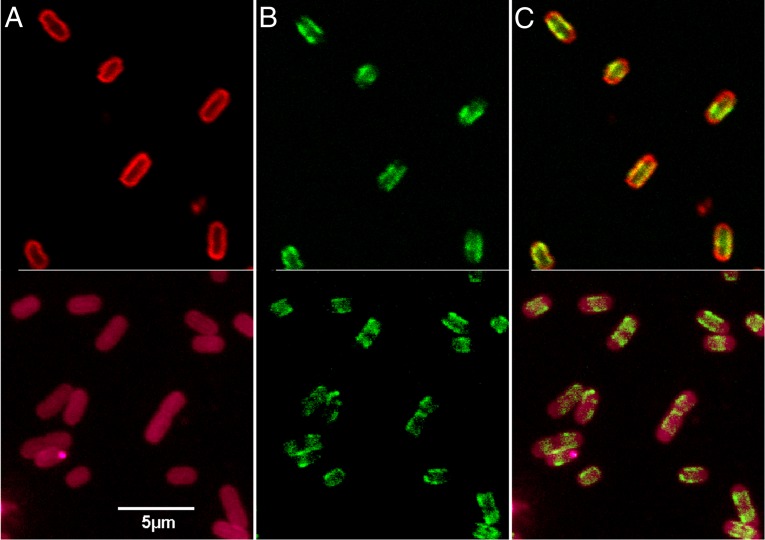
We microscopically characterized hybrid proteins (36) that encode either cytoplasmic GFP (pTpG) or membrane-localized GFP-TonB (pGT). The latter plasmid introduced GFP upstream of and in frame with wild-type TonB (GFP-TonB); it produced a fluorescent hybrid protein with wild-type TonB activity (36). Fluorescence microscopic observations of the chimera originally showed its association with the IM, in contrast to GFP alone, which localized in the cytoplasm (36, 41). Higher-resolution confocal images of GFP-TonB revealed more detail about the distribution of TonB in the cell (Fig. 1). Unlike other fluorescently labeled OM (FepA-AM; Fig. 1) and IM (GFP-LacY; Fig. S1) proteins, which were distributed uniformly throughout the bacterial cell envelope, GFP-TonB localized to the central regions of the cells and usually was absent from the poles. The microscopic images suggested a defined organization of GFP-TonB underlying the cell surface. Its OM partner, FepA, fully encircled the perimeter of bacterial cells, but TonB did not inhabit the poles, where roughly one-third of the total FepA localized (Fig. 1). The absence of the accessory proteins ExbBD did not change the distribution of TonB in the IM (Fig. S1): TonB localization was autonomous from the presence or absence of ExbBD. This restriction of TonB to the central regions of the bacterial cell is germane to proposed mechanisms of TonB action. At any instant, all the FepA proteins in the OM are active in that they all bind FeEnt and within seconds transport it into the periplasm (8). Hence, the images raise questions about the physical relationship between TonB and OM proteins such as FepA during metal transport reactions through the OM (31–35, 37, 38).
Fig. 1.
Fluorescence microscopic localization of FepA and TonB in the E. coli cell envelope. E. coli strain OKN3 harboring pFepAS271C (7) and pGT (36) was grown in MOPS minimal medium to late log, which derepressed the synthesis of FepAS271C and GFP-TonB, respectively. We pelleted the bacteria by centrifugation and exposed them to 5 μM A680M (Upper) or A555M (Lower) in PBS at pH 6.5, which restricts the labeling reaction to the genetically engineered Cys sulfhydryl in FepA (46, 48). (A) FepA. The bacteria were illuminated with 680 nm light and observed at 700 nm (Upper) or illuminated with 553 nm light and observed at 570 nm (Lower), which visualized FepAS271C-A680M and FepAS271C-A555M, respectively, from pFepAS271C. The images with different fluorophores and depths of focus show uniform distribution of the OM transporter around the entire cell surface. (B) GFP-TonB. The bacteria were illuminated with 488 nm light and observed at 520 nm, which visualized GFP-TonB from pGT, showing the membrane localization of the fusion protein, its centralized distribution, and general absence from the poles of the cell. (C) Covisualization of FepA and TonB. Superposition of the images in A and B reiterated the presence of FepA and absence of TonB at the poles of the cell.
Measurements of TonB Motion.
Using the same constructs, we monitored the directional dependence of GFP emissions, either free in the cytoplasm or linked to TonB. Steady-state fluorescence anisotropy examined the motion of the GFP protein. We measured its depolarization in terms of the anisotropy value (R) according to
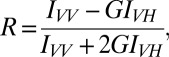 |
where IVV and IVH are fluorescence intensity parallel and perpendicular to the excitation polarization, respectively, and G is an instrument correction factor for its uneven response to s and p polarized fluorescence. In practice, R ranges from 0 to 0.4; a smaller R value signifies a more mobile fluorophore.
The fluorescence anisotropy measurements are sensitive only to molecular motions on the nanosecond timescale, during the excitation/emission interval. When randomly oriented fluorophores are excited by polarized light, the excited molecules are oriented within a range of angles to the applied polarization. If the fluorophore reorients before light emission occurs, then the extent of polarization will decrease. The anisotropy values strongly depend on the fraction of GFP-TonB molecules undergoing rapid motion during the nanosecond observation window and on the frequency of the motion. These observations record changes in the relative magnitude of the s and p polarized fluorescence, as will arise from molecular rotation. Discernible changes in R require a large fraction of molecules in a population displaying asynchronized motion, and the time frame of the measurements excludes potential effects from translational motion in the membrane bilayer, which occurs much more slowly (41).
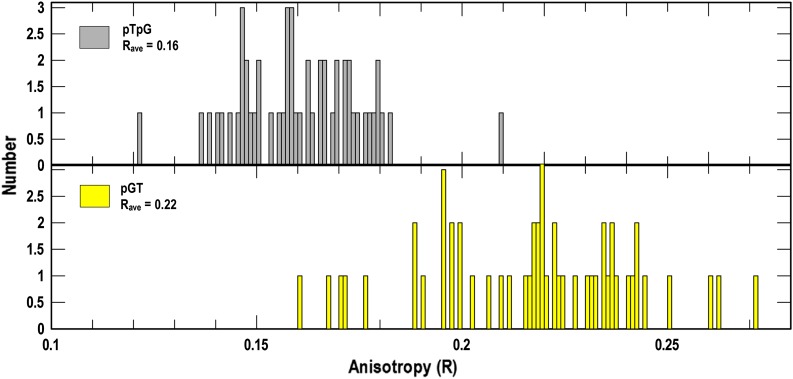
We expected free GFP to show lower R values than membrane-anchored GFP, and we determined the parameter for Escherichia coli BN1071/pTpG (native, cytoplasmic GFP) and BN1071/pGT (GFP anchored to the TonB N-terminus at the cytoplasmic side of the IM) (36). The same ferric uptake regulator (Fur)-regulated TonB promoter controlled expression of both molecules. Histograms comparing R from GFP with that from GFP-TonB (Fig. 2) revealed statistically significant (P < 0.01) differences in mean anisotropy between the cytoplasmic (Rave = 0.160; n = 51) and membrane-anchored (Rave = 0.22; n = 53) fluorophore. These data came from measurements of individual bacteria in each population expressing cytoplasmic GFP or membrane-localized GFP-TonB. The results confirmed that covalent attachment of GFP to TonB in the IM restricted the motion of the fluorescent protein relative to its tumbling when free in the cytoplasm.
Fig. 2.
Fluorescence anisotropy measurements of bacteria expressing GFP. E. coli BN1071/pTpG (Upper) or /pGT (Lower) (36), which produce cytoplasmic GFP or membrane-associated GFP-TonB, respectively, was grown in MOPS medium and subjected to fluorescence microscopy. We recorded anisotropy (R) of GFP in the two cells for ∼50 measurements from each construct. The resulting histograms revealed a higher R-value, reflecting less rapid motion, for GFP fused to the N-terminus of TonB, resident in the IM bilayer.
Effects of Metabolic Inhibitors on TonB Motion.
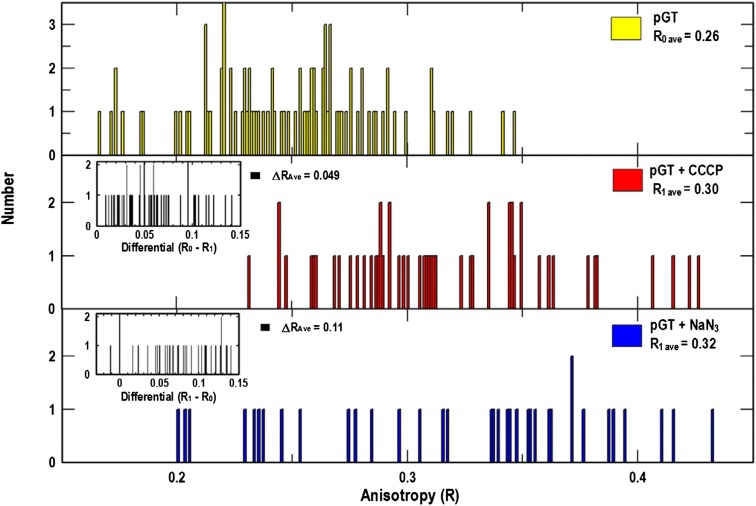
Models of TonB action and its postulated participation in cell envelope bioenergetics raised the possibility that the anisotropy of GFP-TonB may change in response to inhibition of energy metabolism. To evaluate this idea, we observed the effects of the proton ionophore carbonyl cyanide m-chlorophenyl hydrazone (CCCP) on the anisotropy of the GFP constructs. In such studies, we measured the fluorescence of a single cell before and after the addition of the inhibitor. The microscope itself remained fixed during this procedure: the field and focus were unchanged, allowing determinations of the difference in anisotropy (ΔR) before and after CCCP addition (Fig. 3 and Table 1). Statistical analyses of ∼50 measurements showed a positive change in the anisotropy of GFP-TonB after CCCP addition (ΔR = +0.047; P < 0.01) at the stringent 99% confidence level. The absolute values of R varied on different days, but observations of an individual cell before and after CCCP addition consistently showed that anisotropy increased. On the other hand, the much smaller change in anisotropy (ΔR = −0.015) seen for cytoplasmic GFP after CCCP addition was statistically insignificant at a relaxed confidence level of 90%. These data indicate that CCCP-induced depletion of the electrochemical gradient reduced the mobility of TonB in the IM. An electron transport blocker (azide) and dinitrophenol (DNP), another protonophore that collapses PMF, similarly increased the anisotropy of GFP-TonB (Table 1 and Fig. S2). Thus in wild-type bacteria, depletion of the electrochemical gradient always increased the polarization of GFP tethered to TonB, indicating a reduction in the motion of fusion protein in the IM. Identical experiments involving the addition, binding, and transport of FeEnt did not comparably alter the anisotropy of either GFP or GFP-TonB: the magnitude of its effects was smaller (ΔR = +0.008), and the change was not significant at the 99% confidence level (Table 1 and Fig. S2).
Fig. 3.
Effect of CCCP and azide on anisotropy of GFP-TonB. E. coli strain BN1071 harboring pGT was grown in MOPS minimal medium and subjected to fluorescence microscopy in 1-mL cuvettes. After the initial observation and anisotropy measurement (R0) of GFP-TonB in single cells (Top), either CCCP (Middle) or sodium azide (Bottom) was added at 1 or 10 mM, respectively, and incubated for 30 min before a second anisotropy determination (R1). The microscope was not focused or adjusted during exposure to the inhibitors. In this experiment, CCCP increased anisotropy in each of the 51 samples (Middle Inset: ΔRave = 0.049); azide increased anisotropy in 35 of 36 samples (Bottom Inset: ΔRave = 0.11). Hence, both dissipation of the PMF and inhibition of electron transport increased the anisotropy of GFP-TonB.
Table 1.
Effect of metabolic inhibitors on the anisotropy of cytoplasmic GFP and membrane-localized GFP-TonB
| Construct | Agent | Sample size | ΔRave | SD | z | 90%* | 95%† | 99%‡ |
| BN1071/pGT | CCCP | 52 | +0.047 | 0.0302 | +11.3172 | Pass | Pass | Pass |
| BN1071/pTpG | CCCP | 23 | −0.015 | 0.0431 | −1.6627 | Fail | Fail | Fail |
| ΔexbBD/pGT | CCCP | 30 | −0.005 | 0.0177 | +0.052 | Fail | Fail | Fail |
| BN1071/pGT | NaN3 | 58 | +0.060 | 0.0371 | +12.3404 | Pass | Pass | Pass |
| BN1071/pTpG | NaN3 | 40 | +0.019 | 0.0674 | +1.7975 | Fail | Fail | Fail |
| BN1071/pGT | DNP | 53 | +0.056 | 0.0512 | +7.9198 | Pass | Pass | Pass |
| BN1071/pTpG | DNP | 44 | −0.012 | 0.0717 | −1.0806 | Fail | Fail | Fail |
| BN1071/pGT | FeEnt | 51 | 0.008 | 0.0240 | +2.3815 | Pass | Pass | Fail |
BN1071 harboring pTpG or pGT, and BN1071 ΔexbBD/pGT were grown in MOPS minimal medium, suspended in PBS, and subjected to fluorescence microscopy. We measured anisotropy (R) in individual cells adhered to the coverslip before and after the addition of the noted agents to the cuvette. In each case, we conducted z-tests for paired dependent samples to determine the statistical significance of the different mean values.
The observation is deemed statistically significant if |z| > 1.960.
The observation is deemed statistically significant if |z| > 2.241.
The observation is deemed statistically significant if |z| > 2.807.
The anisotropy measurements in the presence of inhibitors revealed that depletion of the electrochemical gradient decreased GFP-TonB motion in the IM. These results demonstrated an energy-dependent biochemical activity of TonB in vivo: depletion of electrochemical force decreased the motion of TonB. The absence of ExbBD also affected the anisotropy of GFP-TonB. However, whereas inhibition of energy metabolism increased the polarization of GFP-TonB, ΔexbBD decreased it (ΔRave = −0.184, P < 0.01) relative to that of the same construct in wild-type cells (Table 2). Lastly, we determined the effects of PMF depletion on TonB motion in the ΔexbBD strain and found that CCCP did not change the anisotropy of GFP-TonB in bacteria lacking ExbBD (Fig. S2 and Table 1).
Table 2.
Measurement of GFP-TonB anisotropy in exbBD± bacteria
| BN1071/pGT |
BN1071ΔexbBD /pGT |
|||||||
| Study | RAvg | SD | n | RAvg | SD | n | t test result | Null hypothesis probability |
| 1 | 0.547 | 0.100 | 12 | 0.298 | 0.179 | 24 | 4.92 | <0.0001 |
| 2 | 0.493 | 0.142 | 36 | 0.311 | 0.080 | 24 | 5.72 | <0.0001 |
| 3 | 0.414 | 0.166 | 26 | 0.306 | 0.053 | 31 | 3.42 | 0.0012 |
| 4 | 0.377 | 0.160 | 29 | 0.173 | 0.096 | 37 | 6.55 | <0.0001 |
| 5 | 0.459 | 0.198 | 50 | 0.281 | 0.153 | 49 | 4.99 | <0.0001 |
On five occasions, E. coli strains BN1071/pGT and BN1071 ΔexbBD/pGT were grown in MOPS minimal media, suspended in PBS, and subjected to fluorescence microscopy. We measured anisotropy (R) in individual cells adhered to the coverslip. After n measurements, we calculated mean anisotropy (RAvg) and SD of the mean, and performed Student t tests to determine the statistical significance of the different mean values. In each case, the null hypothesis probability was <0.01, indicating that GFP-TonB was more mobile in ΔexbBD bacteria.
Discussion
The first conclusion of our study is that unlike FepA, TonB does not uniformly inhabit the Gram-negative bacterial IM, but tends away from the poles of the cell. This result was unexpected because all FepA proteins in the OM are functionally active (8), and iron uptake requires direct, protein–protein interactions between the TonB C-terminus and LGP N-termini (33, 35). The observation of different FepA/TonB localization in the cell envelope does not readily reconcile with these findings. If TonB is their physical partner in OM active transport, then how do polar-localized LGP internalize solutes? In addition, the bacterial IM is fluid at 37 °C, so why does TonB not diffuse to the poles? Fusion of GFP to the TonB N-terminus was not responsible for its restricted distribution, because GFP-LacY hybrids uniformly inhabited the IM. Peptidoglycan (PG) beneath the OM potentially hinders the lateral motion of a periplasm-spanning protein such as TonB (42), and TonB associates with PG (36). Murein is less metabolically active near the poles (43, 44), resulting in an inert form that restricts OM protein mobility (43). These or other phenomena may underlie the localization of TonB in the central regions of the cell envelope.
It was intuitive to understand the decreased motion that resulted from tethering GFP to a membrane protein (TonB) (Fig. S3). The further decreases in motion from reagents that collapse the electrochemical gradient led to a second conclusion, that PMF drives GFP-TonB motion. These movements likely are rotational, because the anisotropy experiments are sensitive only to motions on the nanosecond timescale, which excludes the observation of much slower, translational motions (41). TonB rotation and FeEnt uptake are functionally linked in that all the metabolic inhibitors we tested had the same dual effect: they all increased the anisotropy (decreased the motion) of TonB, and they all blocked FeEnt transport by FepA (7, 8). These findings consistently supported the conclusion that energized movements of TonB facilitate OM metal transport (32, 36, 42). They constitute experimental evidence for a physical mechanism of TonB action. If the TonB/ExbBD complex transfers energy from the IM to the OM by rotational motion, then it resembles an electric motor in which the rotor (TonB) moves within a stator (ExbBD) in response to current flow (PMF) (Fig. S3 and Movie S1). Sequence homology in the proposed transmembrane helices of ExbBD and MotAB (39) supports this notion, suggesting a relationship to rotatory flagellar motion (40). However, the MotAB stators encircle a large flagellum; the proposed ExbBD-TonB machine is much smaller and may function by a different process to achieve its different result. At the same time, TonB rotation appears consistent with the vertical scaffold model of PG architecture (45), it provides a mechanism to move TonB through the PG matrix, and it may supply a force that promotes conformational change in OM transporters, compelling bound metal complexes through their transmembrane channels. It is noteworthy that this hypothesis is unsubstantiated by any structural information on the proposed TonB-ExbBD complex: notwithstanding biochemical estimates (38, 40), no structural data exist to describe the nature of their physical associations or their component stoichiometry. Furthermore, despite the correspondence between the biological and mechanical systems, the electric motor analogy has limitations. Energized TonB motion must occur in the context of biochemical associations between the TonB C-terminus and the TonB-box of iron transporters (33), in response to their cell surface binding of ligands. The turnover number of TonB-facilitated FeEnt uptake by FepA is quite low [on the order of 10−1 s−1 (8, 15)]; at present, we have no estimate of a rate of motion for TonB.
To rationalize the effect of ΔexbBD, we distinguish between random motion and energy-driven, biochemically relevant motion. Removal of ExbBD from a protein complex [GFP-TonB2(ExbBD)n (6, 26, 32, 38)] decreases its aggregate mass and thereby increases the rate of random motion of the remaining component (GFP-TonB), as we observed. Next, ΔexbBD disconnected GFP-TonB anisotropy from the actions of proton ionophores and other inhibitors. The deletion compromised the connection between the electrochemical gradient and GFP-TonB motion, suggesting that ExbBD physically links proton movement to TonB rotation. Hence, whereas the overall effect of ΔexbBD was more rapid random movements of GFP-TonB, that motion was nonproductive in promoting OM transport. According to this explanation, depletion of PMF and deletion of ExbBD blocked OM transport in different ways. In the former case, CCCP, azide, etc. dissipated the bioenergetic force that drives transport, but the TonB–ExbBD membrane complex remained intact. In the latter case, the membrane complex lost protein components (ExbBD), which removed its mechanical interface to the electrochemical gradient.
Overall, the results suggest that in wild-type bacteria, ExbBD engage TonB to PMF, creating controlled motion in response to the electrochemical gradient. This model implies that deflation of PMF will retard TonB motion, as we observed. It further predicts that ΔexbBD disconnects TonB from PMF, resulting in insensitivity of GFP-TonB anisotropy to inhibitors, as we also observed. Hence, the results reconcile with the expectations of the electric motor analogy. TonB is moving in the IM bilayer, energized by the electrochemical gradient, which determines the frequency of the motion. This summarizes our understanding of the data, but other models that we cannot yet envision also may explain the results. TonB motion may allow surveillance of the OM for receptors with bound metals (36), and during interactions with these proteins (33, 35), rotation may transfer energy that triggers ligand transport.
Methods
Bacterial Strains and Plasmids.
OKN3 (ΔfepA) (46), OKN13 (ΔfepA, ΔtonB) (46), and their parent, BN1071 (F-, entA, pro, trp, B1) (47), were the hosts for pTpG and pGT (36). These plasmids are derivatives of the low-copy vector pHSG575 that express cytoplasmic GFP and GFP-TonB, respectively. The latter hybrid protein has normal TonB activity, including ferric siderophore uptake and colicin susceptibility (36). We also transformed OKN3/pGT and OKN13/pGT with pFepAS271C, a pUC18 derivative that carries fepAS271C under control of its natural promoter (7).
Fluorescence Microscopy.
Bacteria were grown overnight in LB broth with appropriate antibiotics, subcultured into 3-(N-morpholino)propanesulfonic acid (MOPS) minimal medium with the same antibiotics, and grown for 5.5 h to late exponential phase (46, 48). The cells were washed with Tris-buffered saline (TBS), adsorbed to slides coated with poly-l-lysine hydrobromide (8.33 mg/mL) for 15 min, and observed by a Nikon confocal microscope. In experiments with BN1071/pGT/pFepAS271C, after growth the cells were washed with 50 mM NaHPO4, pH 6.5, and subjected to Alexa Fluor 555 maleimide (A555M) or A680M at 5 μM in the same buffer for 15 min at 37 °C, which specifically modifies FepA residue S271C with the Alexa Fluors (48).
Analysis of Anisotropy.
Sample cuvettes were made from a hollow plastic tube with a glass coverslip glued to the bottom. The coverslip was coated with 300 μL of poly-l-lysine hydrobromide (8.33 mg/mL) for 15 min. Bacteria were grown in LB and then in MOPS minimal medium to induce their Fur-regulated promoters, and then analyzed by confocal fluorescence microscopy in TBS. One hundred microliters of cell suspension was added and adsorbed to the poly-l-lysine–coated surface for 15 min. Unadsorbed cells were removed, and the cuvette was rinsed twice with 500 μL of TBS, pH 7.0. Five hundred microliters of TBS plus 0.4% glucose was added to the sample cuvette, which was transferred to a confocal microscope for anisotropy measurement.
Variations in the setup and calibration of the microscope resulted in day-to-day differences in the absolute values of R, even in the same strain, so we evaluated samples under comparison on the same day, with minimal adjustment of microscopic parameters between the different groups. These variations arise from several factors that affected the individual experiments, including cell shape, pixel selection, sample size, detector alignment, reproducibility of focus, laser intensity, the concentration of fluorescein used to obtain the G-factor, and temperature. We attempted to normalize and minimize the effects of these factors during the study. In a typical experiment, cells were imaged in the field and focused. The entire field was scanned; background values in areas without cells were less than 10. In the regions of membranes illuminated by the laser, we collected data from representative single pixels near the center of a cell, which represents an area of 0.01 μM2. We collected a stream of data from a single pixel for 5–15 s, recording both IVV and IVH, which produced more than 200 simultaneous individual measurements of IVV and IVH, which we averaged for calculations of R. The microscopic measurements were not particular noisy in that SD for IVV and IVH was around 6%. In two representative experiments, raw IVV (SD):IVH (SD) values were 771.3 (46.9):321.6 (20.5) and 902.4 (53.0):249.6 (16.2). In all the experiments, the background intensity was below 10.
Effects of Metabolic Inhibitors.
After focusing the microscope on a single cell that was immobilized on the glass surface and determining its anisotropy value, we diluted the metabolic inhibitors 100-fold into the 500-μL cuvette [the final concentrations of CCCP, NaN3, and DNP were 1, 10, and 2 mM (7), respectively], equilibrated the sample for 10 min, and remeasured the anisotropy of the same cell. Because the agent affected all cells within the cuvette, this protocol produced only one measurement per sample, but it produced an accurate record of anisotropy changes in response to extrinsic inhibitors.
GFP emission intensity is sharply pH dependent (49), and because ExbBD/TonB may transfer protons from one side of the membrane to the other (Discussion), its mechanism may decrease pH in the vicinity of GFP, potentially changing its emission intensity. However, fluorescence anisotropy is relatively immune to intensity fluctuations because it simultaneously measures IVV and IVH . As long as IVV and IVH are affected to the same extent (e.g., both drop by 20% because of altered pH), then R will not be skewed, because the 20% decrease occurs in both the numerator and denominator of the anisotropy equation for R and therefore cancels. Thus, the anisotropy measurement is insensitive to pH effects on fluorescence emissions, unless intensity drops so low that poor signal-to-noise ratios degrade data quality. We did not see much drop in fluorescence intensity upon addition of different chemicals to the sample, which led to any concerns about data quality.
Supplementary Material
Acknowledgments
The research reported herein was supported by National Institutes of Health Grant GM53836 and National Science Foundation Grant MCB09522999 (to P.E.K., S.M.N., and K.R.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1304243110/-/DCSupplemental.
References
- 1.Kochan I, Kvach JT, Wiles TI. Virulence-associated acquisition of iron in mammalian serum by Escherichia coli. J Infect Dis. 1977;135(4):623–632. doi: 10.1093/infdis/135.4.623. [DOI] [PubMed] [Google Scholar]
- 2.Neilands JB. Siderophores: Structure and function of microbial iron transport compounds. J Biol Chem. 1995;270(45):26723–26726. doi: 10.1074/jbc.270.45.26723. [DOI] [PubMed] [Google Scholar]
- 3.Cornelissen CN, Sparling PF. Iron piracy: Acquisition of transferrin-bound iron by bacterial pathogens. Mol Microbiol. 1994;14(5):843–850. doi: 10.1111/j.1365-2958.1994.tb01320.x. [DOI] [PubMed] [Google Scholar]
- 4.Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev. 2003;67(4):593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nikaido H, Vaara M. Molecular basis of bacterial outer membrane permeability. Microbiol Rev. 1985;49(1):1–32. doi: 10.1128/mr.49.1.1-32.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Noinaj N, Guillier M, Barnard TJ, Buchanan SK. TonB-dependent transporters: Regulation, structure, and function. Annu Rev Microbiol. 2010;64:43–60. doi: 10.1146/annurev.micro.112408.134247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao Z, Warfel P, Newton SM, Klebba PE. Spectroscopic observations of ferric enterobactin transport. J Biol Chem. 2003;278(2):1022–1028. doi: 10.1074/jbc.M210360200. [DOI] [PubMed] [Google Scholar]
- 8.Newton SM, Trinh V, Pi H, Klebba PE. Direct measurements of the outer membrane stage of ferric enterobactin transport: postuptake binding. J Biol Chem. 2010;285(23):17488–17497. doi: 10.1074/jbc.M109.100206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang CC, Newton A. Iron transport in Escherichia coli: Roles of energy-dependent uptake and 2,3-dihydroxybenzoylserine. J Bacteriol. 1969;98(3):1142–1150. doi: 10.1128/jb.98.3.1142-1150.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang CC, Newton A. An additional step in the transport of iron defined by the tonB locus of Escherichia coli. J Biol Chem. 1971;246(7):2147–2151. [PubMed] [Google Scholar]
- 11.Konisky J. Specific Transport Systems and Receptors for Colicins and Phages. New York: Wiley; 1979. pp. 319–359. [Google Scholar]
- 12.Bradbeer C. The proton motive force drives the outer membrane transport of cobalamin in Escherichia coli. J Bacteriol. 1993;175(10):3146–3150. doi: 10.1128/jb.175.10.3146-3150.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rutz JM, et al. Formation of a gated channel by a ligand-specific transport protein in the bacterial outer membrane. Science. 1992;258(5081):471–475. doi: 10.1126/science.1411544. [DOI] [PubMed] [Google Scholar]
- 14.Newton SM, Igo JD, Scott DC, Klebba PE. Effect of loop deletions on the binding and transport of ferric enterobactin by FepA. Mol Microbiol. 1999;32(6):1153–1165. doi: 10.1046/j.1365-2958.1999.01424.x. [DOI] [PubMed] [Google Scholar]
- 15.Schauer K, Rodionov DA, de Reuse H. New substrates for TonB-dependent transport: Do we only see the ‘tip of the iceberg’? Trends Biochem Sci. 2008;33(7):330–338. doi: 10.1016/j.tibs.2008.04.012. [DOI] [PubMed] [Google Scholar]
- 16.Yen MR, et al. Protein-translocating outer membrane porins of Gram-negative bacteria. Biochim Biophys Acta. 2002;1562(1-2):6–31. doi: 10.1016/s0005-2736(02)00359-0. [DOI] [PubMed] [Google Scholar]
- 17.Buchanan SK, et al. Structure of colicin I receptor bound to the R-domain of colicin Ia: Implications for protein import. EMBO J. 2007;26(10):2594–2604. doi: 10.1038/sj.emboj.7601693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buchanan SK, et al. Crystal structure of the outer membrane active transporter FepA from Escherichia coli. Nat Struct Biol. 1999;6(1):56–63. doi: 10.1038/4931. [DOI] [PubMed] [Google Scholar]
- 19.Ferguson AD, et al. Structural basis of gating by the outer membrane transporter FecA. Science. 2002;295(5560):1715–1719. doi: 10.1126/science.1067313. [DOI] [PubMed] [Google Scholar]
- 20.Ferguson AD, Hofmann E, Coulton JW, Diederichs K, Welte W. Siderophore-mediated iron transport: Crystal structure of FhuA with bound lipopolysaccharide. Science. 1998;282(5397):2215–2220. doi: 10.1126/science.282.5397.2215. [DOI] [PubMed] [Google Scholar]
- 21.Locher KP, et al. Transmembrane signaling across the ligand-gated FhuA receptor: Crystal structures of free and ferrichrome-bound states reveal allosteric changes. Cell. 1998;95(6):771–778. doi: 10.1016/s0092-8674(00)81700-6. [DOI] [PubMed] [Google Scholar]
- 22.Chimento DP, Mohanty AK, Kadner RJ, Wiener MC. Substrate-induced transmembrane signaling in the cobalamin transporter BtuB. Nat Struct Biol. 2003;10(5):394–401. doi: 10.1038/nsb914. [DOI] [PubMed] [Google Scholar]
- 23.Sprencel C, et al. Binding of ferric enterobactin by the Escherichia coli periplasmic protein FepB. J Bacteriol. 2000;182(19):5359–5364. doi: 10.1128/jb.182.19.5359-5364.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Borths EL, Locher KP, Lee AT, Rees DC. The structure of Escherichia coli BtuF and binding to its cognate ATP binding cassette transporter. Proc Natl Acad Sci USA. 2002;99(26):16642–16647. doi: 10.1073/pnas.262659699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cui J, Davidson AL. ABC solute importers in bacteria. Essays Biochem. 2011;50(1):85–99. doi: 10.1042/bse0500085. [DOI] [PubMed] [Google Scholar]
- 26.Chu BC, Peacock RS, Vogel HJ. Bioinformatic analysis of the TonB protein family. Biometals. 2007;20(3-4):467–483. doi: 10.1007/s10534-006-9049-4. [DOI] [PubMed] [Google Scholar]
- 27.Klebba PE, Rutz JM, Liu J, Murphy CK. Mechanisms of TonB-catalyzed iron transport through the enteric bacterial cell envelope. J Bioenerg Biomembr. 1993;25(6):603–611. doi: 10.1007/BF00770247. [DOI] [PubMed] [Google Scholar]
- 28.Roof SK, Allard JD, Bertrand KP, Postle K. Analysis of Escherichia coli TonB membrane topology by use of PhoA fusions. J Bacteriol. 1991;173(17):5554–5557. doi: 10.1128/jb.173.17.5554-5557.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brewer S, et al. Structure and function of X-Pro dipeptide repeats in the TonB proteins of Salmonella typhimurium and Escherichia coli. J Mol Biol. 1990;216(4):883–895. doi: 10.1016/S0022-2836(99)80008-4. [DOI] [PubMed] [Google Scholar]
- 30.Evans JS, Levine BA, Trayer IP, Dorman CJ, Higgins CF. Sequence-imposed structural constraints in the TonB protein of E. coli. FEBS Lett. 1986;208(2):211–216. doi: 10.1016/0014-5793(86)81020-1. [DOI] [PubMed] [Google Scholar]
- 31.Krewulak KD, Vogel HJ. TonB or not TonB: Is that the question? Biochem Cell Biol. 2011;89(2):87–97. doi: 10.1139/o10-141. [DOI] [PubMed] [Google Scholar]
- 32.Chang C, Mooser A, Plückthun A, Wlodawer A. Crystal structure of the dimeric C-terminal domain of TonB reveals a novel fold. J Biol Chem. 2001;276(29):27535–27540. doi: 10.1074/jbc.M102778200. [DOI] [PubMed] [Google Scholar]
- 33.Pawelek PD, et al. Structure of TonB in complex with FhuA, E. coli outer membrane receptor. Science. 2006;312(5778):1399–1402. doi: 10.1126/science.1128057. [DOI] [PubMed] [Google Scholar]
- 34.Peacock SR, Weljie AM, Peter Howard S, Price FD, Vogel HJ. The solution structure of the C-terminal domain of TonB and interaction studies with TonB box peptides. J Mol Biol. 2005;345(5):1185–1197. doi: 10.1016/j.jmb.2004.11.026. [DOI] [PubMed] [Google Scholar]
- 35.Shultis DD, Purdy MD, Banchs CN, Wiener MC. Outer membrane active transport: Structure of the BtuB:TonB complex. Science. 2006;312(5778):1396–1399. doi: 10.1126/science.1127694. [DOI] [PubMed] [Google Scholar]
- 36.Kaserer WA, et al. Insight from TonB hybrid proteins into the mechanism of iron transport through the outer membrane. J Bacteriol. 2008;190(11):4001–4016. doi: 10.1128/JB.00135-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kadner RJ. Vitamin B12 transport in Escherichia coli: Energy coupling between membranes. Mol Microbiol. 1990;4(12):2027–2033. doi: 10.1111/j.1365-2958.1990.tb00562.x. [DOI] [PubMed] [Google Scholar]
- 38.Postle K, Larsen RA. TonB-dependent energy transduction between outer and cytoplasmic membranes. Biometals. 2007;20(3-4):453–465. doi: 10.1007/s10534-006-9071-6. [DOI] [PubMed] [Google Scholar]
- 39.Kojima S, Blair DF. Conformational change in the stator of the bacterial flagellar motor. Biochemistry. 2001;40(43):13041–13050. doi: 10.1021/bi011263o. [DOI] [PubMed] [Google Scholar]
- 40.Zhai YF, Heijne W, Saier MH., Jr Molecular modeling of the bacterial outer membrane receptor energizer, ExbBD/TonB, based on homology with the flagellar motor, MotAB. Biochim Biophys Acta. 2003;1614(2):201–210. doi: 10.1016/s0005-2736(03)00176-7. [DOI] [PubMed] [Google Scholar]
- 41.Lill Y, et al. Single-molecule study of molecular mobility in the cytoplasm of Escherichia coli. Phys Rev E Stat Nonlin Soft Matter Phys. 2012;86(2 Pt 1):021907. doi: 10.1103/PhysRevE.86.021907. [DOI] [PubMed] [Google Scholar]
- 42.Klebba PE. Three paradoxes of ferric enterobactin uptake. Front Biosci. 2003;8:s1422–s1436. doi: 10.2741/1233. [DOI] [PubMed] [Google Scholar]
- 43.de Pedro MA, Grünfelder CG, Schwarz H. Restricted mobility of cell surface proteins in the polar regions of Escherichia coli. J Bacteriol. 2004;186(9):2594–2602. doi: 10.1128/JB.186.9.2594-2602.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lybarger SR, Maddock JR. Polarity in action: Asymmetric protein localization in bacteria. J Bacteriol. 2001;183(11):3261–3267. doi: 10.1128/JB.183.11.3261-3267.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meroueh SO, et al. Three-dimensional structure of the bacterial cell wall peptidoglycan. Proc Natl Acad Sci USA. 2006;103(12):4404–4409. doi: 10.1073/pnas.0510182103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ma L, et al. Evidence of ball-and-chain transport of ferric enterobactin through FepA. J Biol Chem. 2007;282(1):397–406. doi: 10.1074/jbc.M605333200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Klebba PE, McIntosh MA, Neilands JB. Kinetics of biosynthesis of iron-regulated membrane proteins in Escherichia coli. J Bacteriol. 1982;149(3):880–888. doi: 10.1128/jb.149.3.880-888.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smallwood CR, et al. Fluoresceination of FepA during colicin B killing: Effects of temperature, toxin and TonB. Mol Microbiol. 2009;72(5):1171–1180. doi: 10.1111/j.1365-2958.2009.06715.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Haupts U, Maiti S, Schwille P, Webb WW. Dynamics of fluorescence fluctuations in green fluorescent protein observed by fluorescence correlation spectroscopy. Proc Natl Acad Sci USA. 1998;95(23):13573–13578. doi: 10.1073/pnas.95.23.13573. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.