Abstract
The E7 oncoprotein of high-risk human papillomaviruses (HPVs) binds to and alters the action of cell cycle regulatory proteins such as members of the retinoblastoma (Rb) family of proteins as well as the histone deacetylases (HDACs). To examine the significance of the binding of E7 to HDACs in the viral life cycle, a mutational analysis of the E7 open reading frame was performed in the context of the complete HPV type 31 (HPV-31) genome. Human foreskin keratinocytes were transfected with wild-type HPV-31 genomes or HPV-31 genomes containing mutations in HDAC binding sequences as well as in the C-terminal zinc finger-like domain, and stable cell lines were isolated. All mutant genomes, except those with E7 mutations in the HDAC binding site, were found to be stably maintained extrachromosomally at an early passage following transfection. Upon further passage in culture, genomes containing mutations to the Rb binding domain as well as the zinc finger-like region quickly lost the ability to maintain episomal genomes. Genomes containing mutations abolishing E7 binding to HDACs or to Rb or mutations to the zinc finger-like motifs failed to extend the life span of transfected keratinocytes and caused cells to arrest at the same time as the untransfected keratinocytes. When induced to differentiate by suspension in methylcellulose, cells maintaining genomes with mutations in the Rb binding domain or the zinc finger-like motifs were impaired in their abilities to activate late viral functions. This study demonstrates that the interaction of E7 with HDACs and the integrity of the zinc finger-like motifs are essential for extending the life span of keratinocytes and for stable maintenance of viral genomes.
Human papillomaviruses (HPVs) are small, double-stranded DNA viruses which infect epithelial cells. Approximately one-third of all HPV types specifically target the genital epithelia. These viruses can be divided into the high-risk types (including HPV-16, -18, -31, and -33) that are associated with the development of anogenital malignancies and the low-risk viruses (HPV-6 and -11) that induce hyperproliferative lesions but rarely are associated into malignancies. Following infection of cells in the basal layer of the epithelium, viral genomes are established in the nucleus as episomes at approximately 50 to 100 copies per cell. As infected basal cells divide, viral DNA is partitioned to daughter cells, one of which migrates from the basal layer and begins differentiation (25, 37). In highly differentiated suprabasal cells, viral genomes are amplified to thousands of copies per cell, late genes are expressed, and production of mature virions is induced (21, 26).
HPV genomes contain six to eight open reading frames carried on one strand of DNA. The early proteins, designated E1, E2, E6, and E7, are expressed prior to productive replication (Fig. 1). The late proteins include L1 and L2, as well as E1^E4 and E5, and are expressed upon productive replication in differentiated suprabasal cells. Among the first viral genes expressed following infection are the replication proteins, E1 and E2, which have been shown to form a complex and bind to the viral origin sequences (11, 30, 41). In addition, recent studies have shown that stable replication of the HPV-31 genome requires the expression of E6 and E7, which also function as the two viral transforming proteins or oncoproteins (40). The functions of the late viral proteins E1^E4 and the E5 proteins are not yet fully understood but may be important for activation of late viral functions (9, 13). The L1 and L2 genes encode the capsid proteins (16, 21).
FIG. 1.
Diagram of the HPV-31 genome and the series of mutations introduced into the E7 open reading frame. The three conserved regions of E7 are indicated as follows: CR1 (light gray); CR2 (horizontal bars); zinc finger-like motif (dark gray). The individual mutations and their approximate locations in the E7 open reading frame are also indicated. The names of the corresponding plasmids containing the mutated HPV-31 genomes are indicated to the right of each mutant E7 protein. Locations of early and late HPV-31 promoters are shown in the top line.
The high-risk E6 and E7 proteins have been shown to be efficient immortalizing agents in tissue culture models (18, 31, 35). The HPV E7 proteins act by binding to members of the retinoblastoma (Rb) family of proteins, which allows cells to rapidly progress into S phase (3, 8). The unphosphorylated form of the Rb protein binds to E2F/DP1 heterodimers and recruits histone deacetylase (HDAC) complexes to repress transcription from promoters containing E2F binding sites (27, 42, 43). As many as 11 different HDACs have been identified, and the most extensively studied are human HDAC1 and HDAC2. HDACs 1 and 2 usually associate with cellular DNA binding proteins that recruit them to genomic regions as well as modulate their deacetylase activities (19, 24). HDACs repress transcription from promoters through deacetylation of lysine residues present in the N-terminal tails of core histone proteins. This deacetylation results in the unmasking of a positive charge on the lysine residue, resulting in a tight interaction between the histone proteins and the DNA. The tight binding then leads to heterochromatin formation and repression of transcription. The HPV-16 E7 protein has been shown to bind to HDACs, and this interaction occurs through an intermediary protein called Mi2β (2). Mi2β is a member of the nucleosome remodeling and histone deacetylation (NURD) complex which has the ability to modify chromatin structure through both deacetylation of histones and ATP-dependent nucleosome repositioning (44, 45). The binding of HDACs to E7 is independent of binding to Rb (2). Mutation of HPV-16 E7 at amino acid 67 (L67R), as well as mutation of cysteine residues in the C terminus, has been shown to abolish binding to HDAC1 and results in a loss of E7's ability to efficiently transform rodent fibroblasts (2).
All E7 proteins contain conserved Cys-X-X-Cys motifs or zinc finger-like domains at their C termini. Mutation to one or both of the cysteines in the second Cys-X-X-Cys motif in E7 abolished HPV-16's ability to extend the life span of human foreskin keratinocytes (HFKs) (23). As mentioned previously, this region has been implicated in binding to HDACs but the zinc finger-like motifs also play a role in mediating protein stability as well as dimerization of E7 (15, 29).
In this study, we investigated the significance of the binding of E7 to HDACs as well as the integrity of the zinc finger-like motifs on the ability of HPV genomes to be stably maintained as episomes. We found that mutants of E7 unable to bind to HDAC1 and HDAC2 as well as mutants with alterations in the zinc finger-like motifs are severely impaired in episomal maintenance capability and ability to extend cellular life span.
MATERIALS AND METHODS
Cell culture.
HFKs were isolated from neonatal human foreskin epithelium and maintained in E medium as described previously (17, 28). Prior to harvesting of RNA, DNA, or protein from keratinocytes, fibroblast feeders were removed by treatment with phosphate-buffered saline (PBS) containing 0.1% 0.5 M EDTA for 2 min, followed by three washes with PBS. COS cells are a monkey kidney cell line and were maintained in Dulbecco's modified Eagle medium (Gibco Invitrogen) containing 10% fetal bovine serum.
Plasmids.
The pBR322 min plasmid contains the complete HPV-31 genome (22), and all of the E7 mutations were generated in this plasmid by using a QuikChange XL site-directed mutagenesis kit (Stratagene, La Jolla, Calif.). The specific mutations made and the corresponding plasmids generated are as follows: pΔLHCYE contains an in-frame deletion of the Rb binding domain, pC61A contains a point mutation to the second cysteine of the first zinc finger-like motif in E7, pL67R contains a point mutation converting a leucine to an arginine, pCVQ68-70AAA contains alanine substitutions at residues 68 through 70, pS71C contains a point mutation converting a serine to a cysteine, pΔLQELL contains a deletion of the residues numbered 79 through 83, pLL82/83RR contains two point mutations converting two leucines to two arginines, pC91G contains a point mutation to the first cysteine of the second zinc finger-like motif in E7, and pC94A contains a point mutation to the second cysteine present in the second zinc finger-like motif of E7. All of these plasmids were sequenced to confirm the presence of the mutation. To generate glutathione S-transferase (GST) fusions, the wild-type and mutant E7 open reading frames were each cloned into the pGEX4T3 vector (Amersham, Piscataway, N.J.) at the BamHI and XhoI restriction endonuclease sites and the resulting plasmids were sequenced. Hemagglutinin (HA)-tagged E7 proteins were derived from an expression vector called pGWIHA18E6 (provided by Ron Javier, Baylor University). The wild-type and mutant E7 open reading frames were each cloned into the HindIII and EcoRI sites present in the multiple cloning site of the vector and verified by sequencing.
GST pull-down assays.
GST fusion protein synthesis and purification were performed according to the manufacturer's instructions (Amersham). GST proteins were quantitated using a commercial protein assay (Bio-Rad, Hercules, Calif.), and levels were confirmed by visualization following sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis. Cell extracts were harvested from monolayer HFKs by using NP-40 lysis buffer (50 mM Tris [pH 7.4], 0.5% NP-40, 150 mM NaCl, 200 μM sodium orthovanadate, 100 mM sodium fluoride, 5 mM EDTA [pH 8.0], 0.5 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, and 1 Complete mini protease inhibitor cocktail tablet [Roche Diagnostics, Mannheim, Germany]/10 ml of solution). After preclearing of cell lysates, purified GST fusions were incubated with cell extracts. GST pull-down assays and analysis of bound proteins were then performed as described previously (5).
Immunoblot analysis.
Whole-cell extracts were prepared with NP-40 lysis buffer and quantitated using the Bio-Rad protein assay. Unless otherwise indicated, 100 μg of protein per sample was electrophoresed on an SDS-polyacrylamide gel and transferred onto a polyvinylidene difluoride membrane (Immobilon-P; Millipore, Bedford, Mass.). Following the transfer, the membrane was blocked for 1 h with 50 ml of PBST solution (PBS plus 0.1% Tween; J.T. Baker, Phillipsburg, N.J.) containing 5% nonfat dry milk. After incubation with the primary antibody in PBST-milk solution for 1 h or overnight according to the manufacturer's protocol, the blot was washed, exposed to ECL Western blotting detection reagents (Amersham), and visualized by chemiluminescence. Primary antibodies used included anti-HDAC1 (Cell Signaling Technologies, Beverly, Mass.), anti-HDAC-2 (Calbiochem, San Diego, Calif.), anti-GST (Cell Signaling Technologies), anti-Rb (Cell Signaling Technologies), and anti-HA clone 12CA5 (Roche Diagnostics). Secondary antibodies used included horseradish peroxidase-linked anti-rabbit (Cell Signaling Technologies) and horseradish peroxidase-linked anti-mouse (Amersham).
Stable transfection of HFKs.
Stable transfections of HFKs were performed as previously described (40). Briefly, HPV-31 genomes were released from the pBR322 min plasmid DNA by restriction enzyme digest, followed by heat inactivation of enzymes and unimolecular religation with freshly added T4 DNA ligase (New England Biolabs, Beverly, Mass.). The genomes were then precipitated in 35% isopropyl alcohol and 10% NaCl overnight at −20°C and resuspended in TE (10 mm Tris-1 mM EDTA [pH 7.5]). HFKs grown to 30% confluence in 10-cm-diameter tissue culture dishes were transfected with 1 μg of HPV-31 DNA along with 1 μg of pSV2neo by using Fugene (Roche Diagnostics) according to the manufacturer's protocol. After 24 h, the transfected cells were trypsinized and replated onto a 10-cm-diameter tissue culture dish containing 10 ml of E medium with epidermal growth factor and J2 fibroblast feeder cells. Selection was done as previously described (40). After selection was completed, pooled populations were expanded for further analyses.
Stable replication assay.
Stably transfected HFKs were treated with 1 ml of 0.5 M EDTA in 1 liter of PBS to remove the J2 fibroblast feeders, trypsinized, and washed once with PBS. Keratinocytes were then isolated and centrifuged, and the pellet was resuspended in 3 ml of DNA lysis buffer (400 mM sodium chloride, 10 mM Tris hydrochloride [pH 7.4], 10 mM EDTA). RNase A was added to a final concentration of 50 μg/ml, and SDS was added to a final concentration of 0.2%. This solution was incubated overnight at 37°C and was then passed through an 18-gauge needle 10 times to shear the DNA. DNA was extracted by centrifugation three times with a 25:24:1 solution of phenol-chloroform-isoamyl ethanol, followed by two washes in a 24:1 chloroform-isoamyl ethanol solution. The DNA was then ethanol precipitated overnight at −20°C and resuspended in TE, after which the DNA samples were analyzed by Southern analyses as previously described (40).
Determination of cell growth rates.
Approximately 2 × 105 cells that had been transfected with HPV-31 wild-type or mutant constructs and selected for drug resistance were plated onto J2 fibroblast feeders in 5-cm-diameter tissue culture dishes. At 2-day intervals from the day of plating, for a total of 6 days, fibroblast feeders were removed and keratinocytes from two plates were trypsinized and counted using a hemacytometer.
Episomal copy amplification assay.
Cells were treated with 1 ml of 0.5 M EDTA in 1 liter of PBS to remove fibroblast feeders, trypsinized, and replated onto a 10-cm-diameter petri dish containing 25 ml of 1.5% methylcellulose in E medium to induce differentiation. The methylcellulose solution was prepared as previously described (9). The cells in methylcellulose were incubated at 37°C for 24 or 48 h and were then transferred into four 50-ml conical tubes into which 40 ml of cold PBS was added. Following centrifugation and washing, the DNA was harvested for Southern blot analysis as described above.
HA-tagged E7 protein expression.
Two micrograms of the wild-type and mutant HA-tagged HPV-31 E7 vectors were transfected into COS cells by using Fugene according to the manufacturer's instruction at a confluence of approximately 30% in a 10-cm-diameter tissue culture dish. After 72 h, the cells were trypsinized and protein was harvested with NP-40 lysis buffer. Protein levels were determined by immunoblot analysis.
RESULTS
To investigate the importance of E7's interaction with HDACs, as well as the zinc finger-like motifs, in the productive life cycle of HPV-31, a genetic analysis was initiated (Fig. 1). We chose to analyze the effects on extension of cellular life span and episomal maintenance due to changes at amino acids 82 and 83 (LL82/83RR), 91 (C91G), and 67 (L67R), all of which have previously been shown in HPV-16 E7 to abolish binding to HDAC1 (2). In addition, the role of the zinc finger-like motifs was investigated through changes in the cysteine residues at amino acids 61 (C61A), 91 (C91G), and 94 (C94A). We also sought to compare the effects of Rb binding and chose to examine a deletion of the Rb binding domain, ΔLHCYE, as well as two other mutations that were previously shown to be Rb binding competent but transformation defective, CVQ68-70AAA and ΔLQELL (20). Finally, we also investigated effects due to an S71C mutation, which has been shown in HPV-16 E7 to act like wild type (5, 20).
The L67R mutation is defective for binding to HDAC1 and HDAC2.
It was important first to confirm that the mutations in HPV-16 E7 that were previously reported to be defective for HDAC binding exhibited a similar phenotype in HPV-31 E7. The wild-type HPV-31 E7, along with the nine different E7 mutants described above, were used to generate GST fusion proteins to determine whether they retained binding to HDAC1 and/or HDAC2. Equal amounts of GST fusion proteins were incubated with primary HFK cell lysates, complexes were precipitated with glutathione, and the ability to bind to HDAC1 and HDAC2 was examined by Western blot analysis. It was observed that all of the mutations in E7 except for the L67R mutation resulted in the retention of some ability to bind to both HDAC1 and HDAC2 (Fig. 2). The L67R mutation in HPV-31 E7 abolished binding to both HDAC1 and HDAC2 (Fig. 2). In HPV-16 E7, two other mutations, LL82/83RR and C91G, have been shown to be deficient in HDAC binding. For HPV-31, the LL82/83RR mutant retained binding ability comparable to that of wild-type E7 (Fig. 2) while the C91G mutant exhibited significantly reduced binding to HDAC1 and low level binding to HDAC2. The remaining mutant E7 proteins, including the ΔLHCYE, CVQ68-70, ΔLQELL, and S71C mutants, as well as the remaining zinc finger mutants, all bound HDAC1 and HDAC2 at levels similar to that of wild-type. We also confirmed the ability of E7 to bind to Rb through similar GST pull-down assays followed by Western blot analysis with a monoclonal antibody to the Rb protein (Fig. 2). All of the mutant proteins were found to bind Rb except for the ΔLHCYE E7 protein, which was expected as it is missing the entire Rb binding domain. The binding levels of GST-E7 protein to HDAC1 and HDAC2 are comparable to those seen with wild-type E7 binding to Rb as judged by the amount of total input bound in Fig. 2. This indicates that the binding of HPV-31 E7 to HDACs occurs at physiologically significant levels.
FIG. 2.
Binding of HDAC1, HDAC2, and Rb to wild-type and mutant E7 proteins in GST pull-down assays. GST fusions of wild-type and mutant E7 proteins were incubated with HFK cell extracts, and bound proteins were isolated through the use of glutathione beads and visualized by Western analysis. Each quadrant represents a single GST pull-down experiment with Western analysis for HDAC1, HDAC2, and Rb. Equal amounts of GST fusion proteins were examined, and equal loading was confirmed by Western analysis for GST (data not shown). At the right end of each panel is a lane containing 5% of the total cell extract.
The HDAC binding mutation to E7 in the HPV-31 genome abolishes the ability to maintain episomal copy numbers at an early passage.
We next constructed HPV-31 genomes that contained the nine mutations in E7 described above. The wild-type and mutant HPV-31 genomes were transfected into HFKs along with a neomycin resistance plasmid as previously described, and HPV-31-positive cells were selected through G418 drug selection (40). Pooled populations of drug-resistant cells were expanded, and DNA was extracted following cessation of drug selection (passage 4) for use in Southern analysis. The L67R mutation, which results in an E7 protein unable to bind HDAC1 or HDAC2 (Fig. 2), completely abolished episomal maintenance (Fig. 3, lower panel). In contrast, all three of the zinc finger-like motif mutations (C94A, C91G, and C61A) resulted in genomes that were significantly reduced in episomal copy number in comparison to wild-type levels (Fig. 3). Similar significant reductions in copy number were observed with these mutant genomes in three separate experiments, each using different host keratinocytes. In addition, the Rb binding site deletion (ΔLHCYE) as well as the ΔLQELL and LL82/83RR mutations resulted in a moderate reduction in the level of episomal copy numbers compared to what was seen with wild-type HPV-31 (Fig. 3). Finally, the S71C and CVQ68-70AAA mutations had little or no effect on the ability of genomes to be maintained as episomes, as the genomes containing these mutations exhibited copy numbers comparable to wild type (Fig. 3, upper panel). Identical results were obtained following transfection of these mutant genomes into two other primary HFK isolates. These results indicate that the HDAC binding site in the E7 protein is essential for stable maintenance of episomes in host keratinocytes. The zinc finger-like motifs of E7 are also very important for the ability of the HPV-31 genome to replicate at early passages after transfection.
FIG. 3.
Southern analysis of HPV-31 DNA in cell lines transfected with wild-type and mutant HPV-31 genomes at passage 2 after selection. Plasmids used for transfection are indicated above each lane. Lanes designated with the letter “U” include total DNA sheared but digested only with DPN1. Lanes designated by “C” were restricted with DPN1 and XbaI, which cuts the HPV-31 genome once. In several of the “C” lanes, there was incomplete digestion of viral DNA. Copy number standards are indicated to the left of each gel. The lower panel of gels was exposed approximately four times longer than was the upper panel of gels to allow for visualization of low copy numbers of episomes for several HPV-31 mutant genomes. The four forms of HPV-31 DNA found in these analyses are present: supercoiled (I), relaxed circle (II), integrated/multimers (III), and liner (IV). Integrated copies of the L67R genomes are seen more clearly at longer exposures of this gel (data not shown).
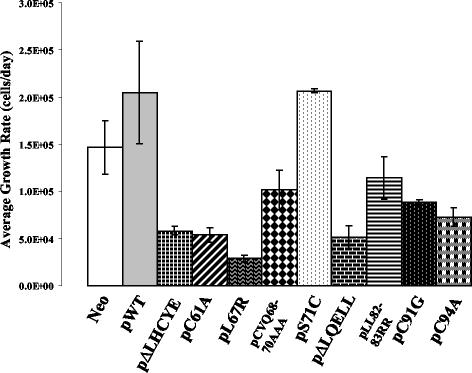
HFKs transfected with the HDAC binding mutant genome grow more slowly than cells transfected with all of the other E7 mutant genomes.
Upon passaging of the cell lines containing wild-type and mutant HPV-31 genomes, we noted consistent differences in growth characteristics. Similar effects were seen in two additional experiments using two different sets of transfected cells. The average growth rates for the various cell lines are shown in Fig. 4. Cells transfected with most of the mutant genomes were reduced in their average growth rate compared with cells maintaining wild-type genomes (Fig. 4). Only the cells transfected with the S71C mutant genome continued to grow at a rate similar to that of wild-type transfected cells. Interestingly, the cells transfected with the mutant genome defective for HDAC binding (pL67R) exhibited the lowest growth rate of all of the cell lines tested.
FIG. 4.
Average growth rates of keratinocytes transfected with wild-type and mutant HPV-31 genomes after completion of drug selection. The average growth rates were determined at passage 3 following completion of the transfection and drug selection protocol as described in Materials and Methods. The plasmid constructs used in the transfections are indicated below each column. Data shown are the averages of three experiments in two different transfected cell lines.
The HDAC binding site and zinc finger-like motifs of E7 are essential for extended life span of HFKs.
The cell lines stably transfected with E7 mutant genomes were passaged continuously until they either senesced or reached passage 9 after selection, at which time DNA was collected and Southern analyses were again performed to look for episomal maintenance (Fig. 5). HFKs transfected with neomycin alone senesced at approximately passage 4 after selection and similar effects were seen in cells isolated from three different hosts. Cells transfected with five of the mutant genomes consistently underwent senescence at passage numbers comparable to that of HFKs transfected with neomycin vectors alone (Table 1). These included cells transfected with mutant genomes containing the HDAC binding mutation L67R, the zinc finger-like motif mutations C61A, C91G, and C94A, and the Rb binding site deletion mutation. Consistent with the results seen at early passage, cells transfected with genomes containing mutated HDAC binding sites failed to maintain HPV episomes at a later passage, prior to senescence. Genomes containing zinc finger-like motif mutations in E7 or a deletion of the Rb binding domain exhibited significantly reduced levels of episomes prior to senescence. These results confirm that in the context of the complete HPV-31 genome the ability of the E7 protein to bind Rb is essential for extending the life span of HFKs and identify the HDAC binding sequence and the zinc finger-like motifs of the E7 protein as being equally important for extension of life span.
FIG. 5.
Southern analysis of cell lines transfected with HPV-31 mutant genomes at passage 9 following selection. Data are from a single exposure of the same gel, and copy number standards are shown at the left. Plasmids used for transfection are indicated above each lane. Lanes designated with “U” indicate that total DNA was sheared but digested only with DPN1, which does not cut the genome. Lanes designated with “C” were restricted with DPN1 and XbaI, which cuts the HPV-31 genome once. In several of the “C” lanes, there was incomplete digestion of viral DNA.
TABLE 1.
Ability to extend life span of transfected cells
| E7 mutant | Extended life spana |
|---|---|
| pWT | Yes |
| pΔLHCYE | No |
| pC61A | No |
| pL67R | No |
| pCVQ68-70AAA | Yes |
| pS71C | Yes |
| pLL82/83RR | Yes |
| pΔLQELL | Yes |
| pC91G | No |
| pC94A | No |
Able to extend life span past passage 9 after selection (HFK controls senesced at passage 4 after selection).
Cells transfected with genomes containing the four remaining E7 mutations (S71C, ΔLQELL, LL82/83RR, and CVQ68-70AAA) were all able to extend the life span of keratinocytes until at least passage 9 after selection (Table 1). We suspect that these cells are immortal, but further passaging in culture would be required before this conclusion can be made definitively.
Mutations to E7 result in impaired late viral functions.
In our experiments, all of the cells transfected with E7 mutant genomes except L67R initially maintained these DNAs extrachromosomally, although some were maintained at low copy number. We next investigated whether the cells that retained episomes could induce late viral functions upon differentiation. Previous studies have demonstrated that genomes need to be maintained as episomes for the induction of late functions upon differentiation (12). In addition, E7 has been implicated in blocking cell cycle exit upon differentiation, at least in part, through the binding of Rb family members (4, 10). For these studies, stably transfected cells at passage 5 were trypsinized and suspended in 1.5% methylcellulose for either 24 or 48 h. Cells were then harvested by centrifugation, DNA was isolated, and Southern analysis was performed to look at episomal amplification. We observed that the wild-type genome and the S71C genome were amplified at 24 and 48 h to similar levels. In contrast, all of the other mutant genomes were deficient in their amplification (Fig. 6). Since cells transfected with the HDAC binding mutant genome contained only integrated HPV DNAs (Fig. 6), no amplification was observed. Table 2 summarizes the average levels of amplification for each of the mutant genomes from two experiments. RNA samples were also taken from the methylcellulose-treated cell lines and used in Northern analysis for late transcript production. As expected, all of the mutant E7 genomes tested, except for the S71C genome, were significantly impaired in late transcript production when compared to the wild-type transfected cells (data not shown).
FIG. 6.
Amplification of wild-type and mutant HPV-31 genomes following suspension in methylcellulose. Cells transfected with wild-type and mutant genomes at passage 3 following completion of drug selection were incubated in methylcellulose for 24 or 48 h, DNA harvested, and examined by Southern analysis for the levels of viral episomes. The fastest migrating band is supercoiled viral DNA.
TABLE 2.
Copy number amplification in methylcellulose-treated cells
| E7 mutant | Avg amplification (n-fold) after methylcellulose treatment forb:
|
|
|---|---|---|
| 24 h | 48 h | |
| WT | 3.8 | 4.2 |
| ΔLHCYE | 1.25 | 1.72 |
| C61A | 1.6 | 2.1 |
| L67R | NAa | NA |
| CVQ68-70AAA | 1.4 | 2.24 |
| S71C | 3.7 | 5.2 |
| ΔLQELL | 1.2 | 1.3 |
| LL82/83RR | 1.3 | 1.6 |
| C91G | 1 | 1.4 |
| C94A | 1.2 | 1.8 |
NA, not applicable.
Levels are in comparison to copy numbers present at 0 h in methylcellulose treatment.
All of the HPV-31 E7 mutations, except the mutations to the zinc finger-like motifs, are expressed stably in mammalian cells. Finally, it was important to confirm that the E7 mutants that were used in this study were stably expressed in vivo. No effective antibody to HPV-31 E7 is available and we therefore constructed fusions of wild-type and mutant E7 proteins with an HA tag to facilitate protein identification. Each of the E7 mutants was cloned into a cytomegalovirus expression vector that also fused an HA tag at the 5′ end of the coding sequences. These vectors were transiently transfected into COS cells, cell lysates were harvested 72 h after transfection, and Western analysis was performed using an antibody to the HA tag. All of the mutant E7 proteins were found to be expressed in the COS cells; however, the mutations to the zinc finger-like motifs (C91G, C61A, and C94A) resulted in proteins that were present at levels significantly lower than those seen in the wild-type E7 protein (Fig. 7). The L67R mutant protein was expressed at a slightly reduced level compared to the wild-type protein. These observations indicate that mutations to the zinc finger-like motifs of the HPV-31 E7 protein destabilize the protein, resulting in lower levels of protein. It has been reported that the E7 protein is degraded through ubiquitin-mediated proteolysis, which most likely is mediated through sequences at the N terminus (36). Any effects due to the insertion of a tag at the N terminus would, however, have a similar effect on all of our constructs and so should not alter our conclusions.
FIG. 7.
Stability of wild-type and mutant E7 proteins. Expression vectors for HA-tagged wild-type and mutant E7 proteins were transfected into Cos cells and total cell extracts were harvested after 48 h. (A) Western analysis was performed using an antibody to HA. Two experiments are shown in the left and right panels. Each experiment included a mock transfection and wild-type E7 control. The Western blots were stripped and reprobed with an antibody to GAPDH to confirm equal loading. (B) Quantitation of average stabilities of wild-type and mutant E7 proteins. The average E7 protein levels seen at 72 h after transfection, obtained from two experiments, are shown. E7 protein quantitation was performed by scanning of autoradiographs of gels.
DISCUSSION
In this study we demonstrate that the binding of E7 to HDAC1 and HDAC2 is important for the maintenance of HPV episomes in undifferentiated keratinocytes as well as for extension of cellular life span. The binding of HDACs to E7 was found to be as important as binding to Rb family members in mediating E7's role in the viral life cycle, since mutation of either interaction sequence resulted in a rapid loss of the ability to maintain episomes. The E7 protein is unlikely to act directly to modulate viral replication, but it is more likely to function indirectly by altering cellular regulatory mechanisms to allow for the sustained presence of extrachromosomal DNAs.
The binding of E7 to HDACs may act in several ways to facilitate episomal maintenance and extend the life span of cells. HDACs modulate gene transcription indirectly through deacetylation of histones and act directly to deacetylate cell cycle regulatory proteins such as p53 and E2F to modulate their function (reviewed in reference 39). Among the genes negatively regulated by HDACs are those involved in cell cycle progression, and E7 could act to sequester HDACs away from these promoters. It has been shown that HPV-16 E7 activates cdc25A expression in an HDAC-dependent manner, and similar effects have been suggested for interferon regulatory factor 1 expression (33, 34). In a similar manner, the adenovirus E1A gene has recently been shown to sequester HDACs away from the cyclin A and cdc6 promoters in quiescent mouse fibroblasts, resulting in activation of expression (14). Our observation that binding of HDACs to an HPV protein is essential for stable replication could indicate that HDACs are directly involved in transcriptional regulation of early viral genes. Alternatively, E7 could act through an indirect mechanism in which HDACs are recruited by the E7 protein to promoters of cell cycle regulatory factors that act to repress viral replication as well as block the cell cycle program in differentiating cells. For instance, HDACs could be recruited by E7 to the E2F4 and E2F5 promoters in order to repress their transcription, leading to the expression of “activator” E2Fs (E2F1, E2F2, and E2F3) as well as cdc25A and cyclins A and E. These latter genes have been shown to be repressed by binding of the E2F4 protein to their promoters (38). Consistent with this hypothesis, our preliminary evidence suggests that during differentiation, E7 downregulates expression of E2F4, while E2F2 is upregulated (M. S. Longworth and L. A. Laimins, unpublished data).
In our study, E7-HDAC interactions were also found to be important in modulating cellular proliferation contributing to the extension of cell life span. Keratinocytes that maintain HPV genomes deficient in HDAC binding exhibited reduced growth rates compared to those seen with cells maintaining wild-type HPV-31 genomes, and this correlated with an inability to extend the life span of cells. This reduction could be due in part to the sequestering of Rb but not HDACs, leading to alteration of S-phase progression. Elucidation of the cellular targets of E7-HDAC action should provide insights into this mechanism. Our study also demonstrates that HDAC binding to HPV-31 E7 is mediated through a different domain than that used for binding Rb family members, and this is consistent with the results of previous studies with HPV-16 (2). No residual binding of HDACs was observed with the HDAC binding-deficient E7 L67R protein, despite the ability to bind Rb. In contrast, high levels of HDAC binding were observed in mutant E7 proteins that were unable to bind Rb. This suggests either that E7 binds only Rb family members that are not associated with HDACs or that binding of Rb by E7 induces HDACs to dissociate, which has been demonstrated previously in vitro (1). It is also still possible that other factors bind E7 at amino acid 67 and that these are vital for its role in the viral life cycle.
Our study confirms that the binding of Rb family members to E7 is also important for maintenance of episomes and extension of life span. We observed that mutation of the Rb binding site in HPV-31 E7 resulted in a reduction of episomal copy numbers compared to those seen with cells maintaining wild-type HPV-31 as well as in a failure to extend life span. A previous study reported that there was a complete loss of episomal maintenance when the Rb binding site was mutated in the context of the complete HPV-31 genome (40). Our present study represents a more extensive analysis and we believe that it more accurately reflects the effects due to loss of Rb binding ability. While we detected a low level of episomes with cells transfected with Rb deletion genomes, an immediate loss of all episomes was detected in cells with genomes that contained mutated HDAC binding sites. We believe that this represents a modest difference in phenotypes, and overall we believe that binding to Rb and binding to HDACs are equally important for episomal maintenance and extended life span. It has previously been reported for HPV-16 that the mutation of the Rb binding domain and zinc finger-like regions in the context of the complete viral genome does not alter the ability to immortalize keratinocytes (23). We are unsure whether the differences between these results and those of our study are due to different viral types or to differences in biological assay protocols.
In this study examining E7 mutant genomes, we observed a correlation between low copy numbers of episomes and the ability to extend life span. In other studies, we have generated a series of immortalized HPV-31 cell lines that contain mutations in other viral genes or regulatory sequences in which only low copy numbers of integrated genomes are found (22). Therefore, we do not feel that low copy numbers of episomes strictly correlate with a failure to extend the life span of cells. Rb and p130 repress transcription by binding directly to E2F proteins bound to promoter sequences as well as through the recruitment of HDAC complexes to deacetylate histones (27, 42, 43). E7 binding to Rb and p130 acts to overcome this repression and to push cells into S phase (7, 8, 32). p107 is expressed only upon entry into S phase, and E7 binding may or may not induce different activities (reviewed in reference 6). Rb family members have also been implicated in controlling cell cycle exit upon epithelial differentiation, and E7 has been shown to block this process (4, 10). Since we observed that cells transfected with genomes containing mutated Rb binding sites maintained low levels of episomes, it was possible to investigate whether loss of Rb binding influenced differentiation-dependent amplification following suspension in methylcellulose. In agreement with studies from the Lambert laboratory, we observed a significant impairment in the differentiation-dependent viral activities when E7's ability to bind to Rb was abolished (10). This is likely the result of an impaired ability of cells to remain active in the cell cycle following differentiation due to the repressive activities of Rb family members. We were unable to investigate whether genomes containing mutations of the HDAC binding residue could undergo amplification following suspension in methylcellulose due to the rapid loss of episomes following transfection. Interestingly, mutations within E7 that have been shown previously to be impaired in transformation, such as ΔLQELL and several mutations in the zinc-finger cysteines, also failed to induce wild-type levels of amplification upon differentiation. Only genomes containing the S71C mutation amplified viral DNAs to wild-type levels. This indicates that several activities of E7 along with binding of Rb family members are important for activating HPV late functions following differentiation.
The third motif in E7 found to be necessary for maintenance of viral episomes consists of the cysteines present in the zinc finger-like motifs at the C terminus. Zinc finger motifs are commonly found in DNA binding proteins, but for E7 they have been shown to facilitate dimerization (29). Two of the cysteine mutations (C94A and C91G) allowed for low-level maintenance of episomes at early passage, in contrast to the total absence of episomal forms with the L67R mutant genomes. We further observed that E7 proteins with mutations in the cysteines in the zinc finger regions (C61A, C91G, and C94A) exhibited decreased protein half-lives, while mutations that abolished Rb or HDAC binding had minimal effect on protein stability. The C94A and C91G mutations also impaired the binding of HDAC1 and HDAC2 to E7. However, since the half-lives of these proteins were significantly altered from wild type, we cannot conclude that the loss of these mutants' ability to extend life span was specifically due to impaired HDAC binding. Instead, we believe that the deficiencies we observed in episomal maintenance and in extension of life span with genomes containing these cysteine mutations were the result of reduced levels of E7 protein.
In summary, our study has identified activities of E7 in addition to the binding of Rb family members that are important for the viral life cycle. The binding of E7 to HDACs 1 and 2 was found to be as important as Rb association in mediating episomal maintenance and in extending the life span of keratinocytes. Elucidation of the mechanisms by which E7 complex formation with HDACs facilitates these activities will depend on the identification of the cellular targets of these complexes.
Acknowledgments
We thank the members of the Laimins laboratory for advice and comments on protocols. We also thank Ashok Aiyar (Northwestern University) for the pGEX4T-3 vector and Ron Javier (Baylor University) for the use of the GW1-CMV polylinker mammalian expression vector.
M.S.L. was supported by National Cancer Institute Carcinogenesis Training Grant no. T32CA09560 and by a Gramm Travel Fellowship Grant. This work was supported by a Sexually Transmitted Diseases Cooperative Research Center grant funded through the National Institute of Allergy and Infectious Diseases.
REFERENCES
- 1.Brehm, A., E. A. Miska, D. J. McCance, J. L. Reid, A. J. Bannister, and T. Kouzarides. 1998. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature 391:597-601. [DOI] [PubMed] [Google Scholar]
- 2.Brehm, A., S. J. Nielsen, E. A. Miska, D. J. McCance, J. L. Reid, A. J. Bannister, and T. Kouzarides. 1999. The E7 oncoprotein associates with Mi2 and histone deacetylase activity to promote cell growth. EMBO J. 18:2449-2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chellappan, S., V. B. Kraus, B. Kroger, K. Munger, P. M. Howley, W. C. Phelps, and J. R. Nevins. 1992. Adenovirus E1A, simian virus 40 tumor antigen, and human papillomavirus E7 protein share the capacity to disrupt the interaction between transcription factor E2F and the retinoblastoma gene product. Proc. Natl. Acad. Sci. USA 89:4549-4553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, J., P. Jackson, M. Kirschner, and A. Dutta. 1995. Separate domains of p21 involved in the inhibition of cdk kinase and PCNA. Nature 374:386-388. [DOI] [PubMed] [Google Scholar]
- 5.Ciccolini, F., G. Di Pasquale, F. Carlotti, L. Crawford, and M. Tommasino. 1994. Functional studies of E7 proteins from different HPV types. Oncogene 9:2633-2638. [PubMed] [Google Scholar]
- 6.Classon, M., and E. Harlow. 2002. The retinoblastoma tumour suppressor in development and cancer. Nat. Rev. Cancer 2:910-917. [DOI] [PubMed] [Google Scholar]
- 7.Dyson, N., P. Guida, K. Munger, and E. Harlow. 1992. Homologous sequences in adenovirus E1A and human papillomavirus E7 proteins mediate interaction with the same set of cellular proteins. J. Virol. 66:6893-6902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dyson, N., P. M. Howley, K. Munger, and E. Harlow. 1989. The human papilloma virus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science 243:934-937. [DOI] [PubMed] [Google Scholar]
- 9.Fehrmann, F., D. J. Klumpp, and L. A. Laimins. 2003. Human papillomavirus type 31 E5 protein supports cell cycle progression and activates late viral functions upon epithelial differentiation. J. Virol. 77:2819-2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flores, E. R., B. L. Allen-Hoffman, D. Lee, and P. F. Lambert. 2000. The human papillomavirus type 16 E7 oncogene is required for the productive stage of the life cycle. J. Virol. 74:6622-6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frattini, M. G., and L. A. Laimins. 1994. Binding of the human papillomavirus E1 origin-recognition protein is regulated through complex formation with the E2 enhancer-binding protein. Proc. Natl. Acad. Sci. USA 91:12398-12402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frattini, M. G., H. B. Lim, J. Doorbar, and L. A. Laimins. 1997. Induction of human papillomavirus type 18 late gene expression and genomic amplification in organotypic cultures from transfected DNA templates. J. Virol. 71:7068-7072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Genther, S. M., S. Sterling, S. Duensing, K. Munger, C. Sattler, and P. F. Lambert. 2003. Quantitative role of the human papillomavirus type 16 E5 gene during the productive stage of the viral life cycle. J. Virol. 77:2832-2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghosh, M. K., and M. L. Harter. 2003. A viral mechanism for remodeling chromatin structure in G0 cells. Mol. Cell 12:255-260. [DOI] [PubMed] [Google Scholar]
- 15.Grossman, S. R., R. Mora, and L. A. Laimins. 1989. Intracellular localization and DNA-binding properties of human papillomavirus type 18 E6 protein expressed with a baculovirus vector. J. Virol. 63:366-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hagensee, M. E., N. Yaegashi, and D. A. Galloway. 1993. Self-assembly of human papillomavirus type 1 capsids by expression of the L1 protein alone or by coexpression of the L1 and L2 capsid proteins. J. Virol. 67:315-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Halbert, C. L., G. W. Demers, and D. A. Galloway. 1992. The E6 and E7 genes of human papillomavirus type 6 have weak immortalizing activity in human epithelial cells. J. Virol. 66:2125-2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Halbert, C. L., G. W. Demers, and D. A. Galloway. 1991. The E7 gene of human papillomavirus type 16 is sufficient for immortalization of human epithelial cells. J. Virol. 65:473-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hassig, C. A., T. C. Fleischer, A. N. Billin, S. L. Schreiber, and D. E. Ayer. 1997. Histone deacetylase activity is required for full transcriptional repression by mSin3A. Cell 89:341-347. [DOI] [PubMed] [Google Scholar]
- 20.Helt, A. M., and D. A. Galloway. 2001. Destabilization of the retinoblastoma tumor suppressor by human papillomavirus type 16 E7 is not sufficient to overcome cell cycle arrest in human keratinocytes. J. Virol. 75:6737-6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Howley, P. M. 1996. Papillomaviridae: the viruses and their replication, p. 947-978. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fundamental virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 22.Hubert, W., and L. A. Laimins. 2002. HPV 31 replication modes during early phases of viral life cycle depend upon transcriptional and posttranscriptional regulation of E1 and E2 expression. J. Virol. 76:2263-2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jewers, R. J., P. Hildebrandt, J. W. Ludlow, B. Kell, and D. J. McCance. 1992. Regions of human papillomavirus type 16 E7 oncoprotein required for immortalization of human keratinocytes. J. Virol. 66:1329-1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laherty, C. D., W. M. Yang, J. M. Sun, J. R. Davie, E. Seto, and R. N. Eisenman. 1997. Histone deacetylases associated with the mSin3 corepressor mediate Mad transcriptional repression. Cell 89:349-356. [DOI] [PubMed] [Google Scholar]
- 25.Laimins, L. A. 1998. Regulation of transcription and replication by human papillomaviruses, p. 201-223. In D. J. McCance (ed.), Human tumor viruses. American Society for Microbiology, Washington, D.C.
- 26.Lambert, P. F. 1991. Papillomavirus DNA replication. J. Virol. 65:3417-3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo, R. X., A. A. Postigo, and D. C. Dean. 1998. Rb interacts with histone deacetylase to repress transcription. Cell 92:463-473. [DOI] [PubMed] [Google Scholar]
- 28.McCance, D., R. Kopan, E. Fuchs, and L. Laimins. 1988. HPV 16 alters human epithelial cell differentiation in vitro. Proc. Natl. Acad. Sci. USA 85:7169-7173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McIntyre, M. C., M. G. Frattini, S. R. Grossman, and L. A. Laimins. 1993. Human papillomavirus type 18 E7 protein requires intact Cys-X-X-Cys motifs for zinc binding, dimerization, and transformation but not for Rb binding. J. Virol. 67:3142-3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mohr, I. J., R. Clark, S. Sun, E. J. Androphy, P. MacPherson, and M. R. Botchan. 1990. Targeting the E1 replication protein to the papillomavirus origin of replication by complex formation with the E2 transactivator. Science 250:1694-1699. [DOI] [PubMed] [Google Scholar]
- 31.Munger, K., W. C. Phelps, V. Bubb, P. M. Howley, and R. Schlegel. 1989. The E6 and E7 genes of the human papillomavirus type 16 together are necessary and sufficient for transformation of primary human keratinocytes. J. Virol. 63:4417-4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Munger, K., B. Werness, N. Dyson, W. Phelps, E. Harlow, and P. Howley. 1989. Complex formation of human papillomavirus E7 proteins with the retinoblastoma tumor suppressor gene product. EMBO J. 8:4099-4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nguyen, D. X., T. F. Westbrook, and D. J. McCance. 2002. Human papillomavirus type 16 E7 maintains elevated levels of the cdc25A tyrosine phosphatase during deregulation of cell cycle arrest. J. Virol. 76:619-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park, J. S., E. J. Kim, H. J. Kwon, E. S. Hwang, S. E. Namkoong, and S. J. Um. 2000. Inactivation of interferon regulatory factor-1 tumor suppressor protein by HPV E7 oncoprotein. Implication for the E7-mediated immune evasion mechanism in cervical carcinogenesis. J. Biol. Chem. 275:6764-6769. [DOI] [PubMed] [Google Scholar]
- 35.Phelps, W. C., C. L. Yee, K. Munger, and P. M. Howley. 1988. The human papillomavirus type 16 E7 gene encodes transactivation and transformation functions similar to those of adenovirus E1A. Cell 53:539-547. [DOI] [PubMed] [Google Scholar]
- 36.Reinstein, E., M. Scheffner, M. Oren, A. Ciechanover, and A. Schwartz. 2000. Degradation of the E7 human papillomavirus oncoprotein by the ubiquitin-proteasome system: targeting via ubiquitination of the N-terminal residue. Oncogene 19:5944-5950. [DOI] [PubMed] [Google Scholar]
- 37.Stubenrauch, F., and L. A. Laimins. 1999. Human papillomavirus life cycle: active and latent phases. Semin. Cancer Biol. 9:379-386. [DOI] [PubMed] [Google Scholar]
- 38.Takahashi, Y., J. B. Rayman, and B. D. Dynlacht. 2000. Analysis of promoter binding by the E2F and pRB families in vivo: distinct E2F proteins mediate activation and repression. Genes Dev. 14:804-816. [PMC free article] [PubMed] [Google Scholar]
- 39.Thiagalingam, S., K. H. Cheng, H. J. Lee, N. Mineva, A. Thiagalingam, and J. F. Ponte. 2003. Histone deacetylases: unique players in shaping the epigenetic histone code. Ann. N. Y. Acad. Sci. 983:84-100. [DOI] [PubMed] [Google Scholar]
- 40.Thomas, J. T., W. G. Hubert, M. N. Ruesch, and L. A. Laimins. 1999. Human papillomavirus type 31 oncoproteins E6 and E7 are required for the maintenance of episomes during the viral life cycle in normal human keratinocytes. Proc. Natl. Acad. Sci. USA 96:8449-8454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ustav, M., and A. Stenlund. 1991. Transient replication of BPV-1 requires two viral polypeptides encoded by the E1 and E2 open reading frames. EMBO J. 10:449-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weintraub, S. J., K. N. Chow, R. X. Luo, S. H. Zhang, S. He, and D. C. Dean. 1995. Mechanism of active transcriptional repression by the retinoblastoma protein. Nature 375:812-815. [DOI] [PubMed] [Google Scholar]
- 43.Weintraub, S. J., C. A. Prater, and D. C. Dean. 1992. Retinoblastoma protein switches the E2F site from positive to negative element. Nature 358:259-261. [DOI] [PubMed] [Google Scholar]
- 44.Xue, Y., J. Wong, G. T. Moreno, M. K. Young, J. Cote, and W. Wang. 1998. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol. Cell 2:851-861. [DOI] [PubMed] [Google Scholar]
- 45.Zhang, Y., G. LeRoy, H. P. Seelig, W. S. Lane, and D. Reinberg. 1998. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell 95:279-289. [DOI] [PubMed] [Google Scholar]