Abstract
Solid tumors with disorganized, insufficient blood supply contain hypoxic cells that are resistant to radiotherapy and chemotherapy. Drug resistance, an obstacle to curative treatment of solid tumors, can occur via suppression of apoptosis, a process controlled by pro- and antiapoptotic members of the Bcl-2 protein family. Oxygen deprivation of human colon cancer cells in vitro provoked decreased mRNA and protein levels of proapoptotic Bid and Bad. Hypoxia-inducible factor 1 (HIF-1) was dispensable for the down-regulation of Bad but required for that of Bid, consistent with the binding of HIF-1α to a hypoxia-responsive element (positions −8484 to −8475) in the bid promoter. Oxygen deprivation resulted in proteosome-independent decreased expression of Bax in vitro, consistent with a reduction in global translation efficiency. The physiological relevance of Bid and Bax down-regulation was confirmed in tumors in vivo. Oxygen deprivation resulted in decreased drug-induced apoptosis and clonogenic resistance to agents with different mechanisms of action. The contribution of Bid and/or Bax down-regulation to drug responsiveness was demonstrated by the relative resistance of normoxic cells that had no or reduced expression of Bid and/or Bax and by the finding that forced expression of Bid in hypoxic cells resulted in increased sensitivity to the topoisomerase II inhibitor etoposide.
Solid tumors contain subpopulations of hypoxic cells that are refractory to radiotherapy and some forms of chemotherapy (45). While the mechanism of hypoxic cell radioresistance is well characterized, the underlying mechanisms contributing to hypoxia-mediated chemoresistance are poorly understood. Precedents exist for hypoxia being a strong physiological stimulus for genome instability, whereby cells acquire drug resistance (45). In addition, there is direct evidence that hypoxia in tumors selects for cells with decreased potential for apoptosis through the loss of p53 or overexpression of Bcl-2 (15).
Cellular responses to hypoxia include processes leading to enhanced oxygen delivery, increased glucose transport, increased glycolytic metabolism, and a switch from oxidative phosphorylation to anaerobic glycolysis. These responses occur via changes in gene expression mediated by hypoxia-inducible factor 1 (HIF-1). HIF-1 is comprised of two subunits, HIF-1α and HIF-1β, both of which are required for DNA binding and transactivation of target genes (49). HIF-1 binds to specific sequences within target genes with the consensus 5′-RCGTG-3′ regions, called hypoxia response elements (HREs) (38), and targets include vascular endothelial growth factor, lactate dehydrogenase (LDH) A, phosphoglycerate kinase 1 (PGK-1), and carbonic anhydrase IX (CA-IX) (52, 56).
A number of studies highlight a positive role for HIF-1 in tumorigenesis (16, 18, 19, 27, 33, 34, 42, 54), and the influence of hypoxia and HIF-1 on the regulation of apoptosis as a component of tumor development has been considered (39). However, there remains controversy regarding the potential of HIF-1 in altering apoptosis in the tumor microenvironment. Studies using tumors derived from HIF-1-deficient embryonic stem cells have yielded conflicting data (8, 33). Whereas the slower growth of HIF-1α-deficient embryonic stem cell tumors was attributable in part to an increased rate of apoptosis (33), the opposite effect on hypoxia-induced apoptosis and tumor growth was observed in the same tumor type by Carmeliet and colleagues (8). The fact that constitutive expression of HIF-1α rendered pancreatic cells resistant to hypoxia-induced apoptosis in vitro (2) is perhaps more consistent with HIF-1-mediated suppression of apoptotic signaling. In keeping with the findings of the majority of studies, there is also a growing body of evidence that HIF-1 is deregulated in many human tumors (5, 60, 61), and the resulting constitutively high level of HIF-1 expression is associated with more aggressive tumors and treatment failure (4).
The Bcl-2 family of pro- and antiapoptotic proteins plays a central role in establishing the threshold for apoptosis (1), and it is thought that there is a degree of redundancy within the family in this role. The relationship between the major Bcl-2 family proteins, HIF-1, and the chemosensitivity of tumors has not been assessed and is the subject of our investigation.
MATERIALS AND METHODS
Cell culture.
HCT116, HT29, and SW480 human colon carcinoma cells were grown in McCoy's, RPMI, and Dulbecco's modified Eagle medium (DMEM), respectively, all supplemented with 10% fetal bovine serum and antibiotics (from GIBCO BRL). Peter Ratcliffe kindly provided CHO and HEPA-1 cell lines (Institute of Molecular Medicine, Oxford, United Kingdom). The HIF-1β-deficient cell line (HEPA-1 c4) was derived from the murine hepatoma cell line Hepa1c1c7 (HEPA-1 wild type [wt]) and lacks a HIF-1-mediated transcription response to hypoxia (27). A mutant revertant cell line (Rc4) was used as a control. HEPA-1 cells were cultured in DMEM supplemented with 10% fetal calf serum and 2 mM glutamine. The HIF-1α-deficient cell line, Ka13.5, was derived from a Chinese hamster ovary strain (C4.5) stably transfected with HRE-regulated surface marker expression constructs following drug-induced mutagenesis, as previously described (55). The C15 cell line is a pooled population of CHO HIF-1 wt cells that have been through the same mutagenic procedure used to generate the Ka13.5 clone from the parental C4.5 strain. This cell line has been shown in vivo to behave in the same way as the wt cells (54). V79 Chinese hamster lung fibroblasts were cultured in DMEM exactly as described previously by Smith and colleagues (40). HCT116 Bax null cells were a kind gift from Bert Vogelstein (Johns Hopkins, Baltimore, Md.) and were cultured as previously described (59). The SV40-transformed wt and Bid knockout (KO) mouse embryonic fibroblasts (MEFs) were a kind gift from Stanley Korsmeyer (Harvard Medical School, Boston, Mass.) and were cultured as previously described (47). All cells were routinely cultured at 37°C in 95% air-5% CO2.
Oxygen deprivation.
Cells were maintained under anoxic (<0.1% O2) conditions at 37°C within an anaerobic chamber (Bactron 2; Sheldon Scientific) or under hypoxic (1 to 2% O2) conditions at 37°C within a modular incubator chamber filled with 5% CO2 and 1 to 2% O2 balanced with N2 (phiTEC).
Measurement of cell death.
Cell death was assessed by trypan blue uptake. Apoptosis was detected by staining cells with Hoechst 33342 (0.1 mg/ml; Molecular Probes) and identifying those cells with condensed and fragmented nuclei.
Flow cytometry.
Cell cycle analysis was performed by using flow cytometry and propidium iodide as previously described (3). Cells expressing green fluorescent protein (GFP) were isolated with a fluorescence-activated cell sorter (FACS) Vantage flow cytometer set to excite at 488 nm, and cells exhibiting fluorescence at 520 ± 30 nm were gated for sorting.
Immunoblotting.
Immunoblotting was performed as previously described (10). Immunoreactive bands were developed with an enhanced chemiluminescence kit (Amersham) and analyzed on a Fuji LAS-1000plus imaging system using AIDA software. The primary antibodies used were Bcl-2 (Dako), Bcl-xL, human HIF-1α (Transduction Labs), murine and human HIF-1α (Novus Biologicals), Bax (N20; Santa Cruz), Bcl-w (Calbiochem), Bad and Bid (C20; Santa Cruz or R&D), actin (Act40; Sigma), and porin (Santa Cruz). Antibodies to Nip3 and CA-IX were kindly provided by Adrian Harris (Institute of Molecular Medicine).
Growth and analysis of HCT116 cells as xenograft tumors in nude mice.
HCT116 cells were grown as subcutaneous xenografts in 8-week-old female CD-1 nude mice following intradermal injection of 5 × 106 cells in 0.1 ml of media 1 cm from the tail base on the midline. Mice were housed in an individually ventilated caging system on a 12 -h light-12-h dark environment maintained at constant temperature and humidity. The mice were fed a standard irradiated diet and allowed water ad libitum. The hypoxic marker pimonidazole (Hypoxyprobe-1; Chemicon International Inc.) was administered at 100 mg per kg of body weight in phosphate-buffered saline (PBS) to mice bearing tumors of 600 to 800 mm3 2 h before sacrifice. All procedures were carried out in accordance with the Scientific Procedures Act of 1986 and in line with the UKCCCR guidelines of 1999 and by approved protocols (Home Office Project License number 40-1770).
Immunohistochemistry.
The protocol of Wykoff and colleagues (56) was used to assess the levels of Bid, Bax, and CA-IX in HT29 and SW480 cell pellets. Xenograft tumors were formalin fixed and paraffin embedded and 5 μM sections cut onto 3-aminopropyltriethoxysilane-coated slides (Maldini Diagnostics). Slides were dewaxed in xylene, washed in 96% vol/vol ethanol, and incubated for 30 min in 0.3% hydrogen peroxide solution before being washed in PBS. Slides were then placed in citrate buffer and microwaved at 900 W for 20 min. Slides were washed, dried, and blocked for 1 h with the appropriate serum (Vector Laboratories, Peterborough, United Kingdom) in a humidified chamber. Primary antibody was added overnight at 4°C. Hypoxic tumor cells were identified by using anti-pimonidazole antibody as previously described (31). Bax was analyzed by using a rabbit polyclonal antibody raised against a synthetic peptide corresponding to amino acids 43 to 61 of human Bax (Pharmingen) diluted 1 in 4,000. Bid was analyzed by using a 1 in 2,000 dilution of a polyclonal goat antibody that was a gift from X. Wang (University of Texas). The sections were incubated with secondary antibodies for 30 min and then processed by using the Vectastain Elite ABC kit per the manufacturer's instructions (Vector Laboratories) prior to digital photography on a Nikon Eclipse E600 microscope with a Spot RT slider camera and imaging software (supplied by Imsol Imaging Solutions).
RNA isolation and quantitative reverse transcriptase (RT)-PCR analysis.
Cells were harvested following incubation under normoxic or anoxic conditions. RNA was extracted by using RNAzol (Biogenesis) according to the manufacturer's instructions. To remove traces of genomic DNA, the RNA was digested with an RQ1 RNase-free DNase kit (Promega) per the manufacturer's instructions. cDNA strand synthesis was performed by using a Moloney murine leukemia virus cDNA synthesis kit (GIBCO BRL). cDNA samples were diluted 1:20, and 10 μl was used as a template for the Taqman real-time PCR technique to quantify mRNA expression (11) by using a qPCR core kit (Eurogentec) and an ABI Prism 7700 sequence detection system (PE Applied Biosystems). Taqman PCR primers were designed for each gene based on the mRNA sequence by using Primer Express software (Perkin-Elmer) supplied by Sigma Genosys. Their sequences were as follows: for actin, ACCATGGATGATGATATCGCC and GCCTTGCACATGCCGG; for Bid, GCTGTATAGCTGCTTCCAGTGTA and GCTATCTTCCAGCCTGTCTTCTC; for Bad, GCACAGCAACGCAGATGC and AAGTTCCGATCCCACCAGG; for Bax, CTGCAGAGGATGATTGCCG and TGCCACTCGGAAAAAGACCT; and for GAPDH (glyceraldehyde-3-phosphate dehydrogenase), ACACTCAGACCCCCACCACA and CATAGGCCCCTCCCCTCTT.
Assessment of HIF-1 proficiency.
Cells were transiently transfected with firefly luciferase reporter constructs containing either no HRE sequences (pGL3-Con; Promega) or HRE sequences from either PGK or LDH (pGL3-Prom; Promega). The assay was performed by using a dual luciferase reporter assay system (Promega) according to the manufacturer's instructions and a dual injector MicroLumat LB 96 luminometer (EG & G Berthold).
Transient transfection of cells.
All cells were transfected by using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. In some of the studies described below, high concentrations of etoposide were necessary in order to achieve appropriate levels of apoptosis in a short time frame. HT29 cells were transfected with pSilencer1 vector (Ambion) (43), containing a HIF-1α-specific targeting sequence (5′-GTCTCGAGATGCAGCCAGA-3′). This sequence was checked against the database to confirm specificity. As a negative control, a scrambled sequence was used (5′-TCAGCACGGTGACTGAGAC-3′). The cells harvested 48 h after transfection had experienced oxygen deprivation in the last 16 h. The same RNA interference (RNAi) sequences were successfully used to transfect transformed MEFs; this sequence exhibits a 2-bp difference with respect to the analogous human sequence but does not interfere with the expression of other mouse genes.
HCT116 Bax null cells were transfected with RNAi specifically targeted to Bid (a kind gift from Olivier Geneste, Institute Servier, Paris, France). Cells were treated with 1 mM etoposide 32 h posttransfection. The drug was then removed and cells counted 2 days later when Bid levels were still reduced (determined by Western blotting) (see Fig. 8B).
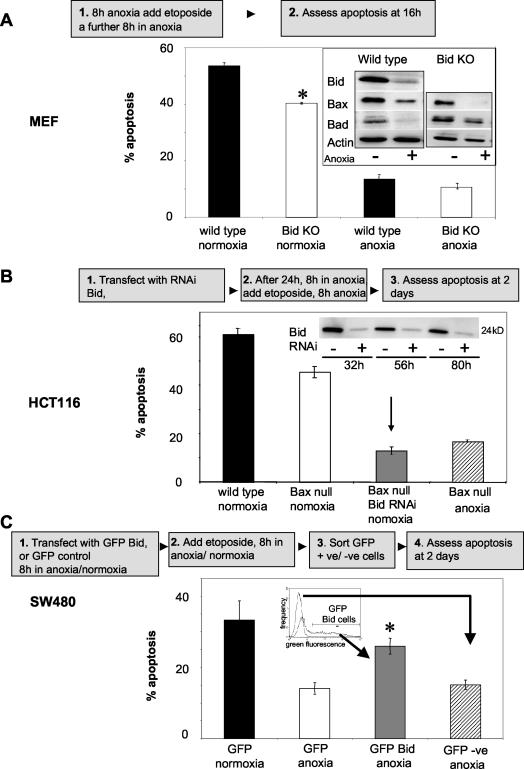
FIG. 8.
Impact of decreased Bid and/or Bax expression on response to etoposide. (A) SV40-transformed wt and Bid KO cells were incubated for 8 h under normoxic or anoxic conditions prior to 8 h of treatment with 50 μM etoposide (with continued normoxic or anoxic incubation), and apoptosis was assessed. Inset: Cells were incubated for 16 h under normoxic (−) or anoxic (+) conditions, and lysates were immunoblotted for Bid, Bax, Bad, and actin (as a loading control). Statistical analysis using Student's two-tailed t test showed significance (P ≤ 0.05) when comparing normoxic wt and Bid KO cells. (B) Normoxic HCT116 wt (black bar) and Bax null (white bar) cells were treated for 8 h with 1 mM etoposide, and apoptosis was assessed 2 days later. HCT116 Bax null cells were transiently transfected with Bid RNAi (grey bar) and then left for 24 h prior to 8-h treatment with etoposide, and apoptosis was assessed 2 days later. Inset: Western blot analysis to confirm that Bid was down-regulated during the course of the experiment. The apoptotic response was compared to that in HCT116 Bax null cells that were oxygen deprived 8 h prior to and after treatment with etoposide (hatched bar). For Western blots, 40 μg of protein per sample was loaded, and equal loading was confirmed by using actin as a control. (C) SW480 cells were transfected with GFP empty vector and immediately incubated for 8 h under normoxic (black bar) or anoxic (white bar) conditions prior to treatment for 8 h with 500 μM etoposide with continued incubation. GFP-expressing cells were isolated by FACS analysis and replated, and apoptosis was assessed 2 days later. SW480 cells transfected with GFP Bid were immediately incubated for 8 h under anoxia prior to 8-h etoposide treatment with continued anoxic incubation. GFP Bid-positive (grey bar) and -negative (hatched bar) cells were sorted by FACS analysis and replated, and apoptosis was assessed 2 days later. Statistical analysis using Student's two-tailed t test showed significance (P ≤ 0.05) when comparing GFP Bid-positive (grey bar) to GFP Bid-negative (hatched bar) cells, as well as to GFP vector anoxic cells (white bar). All data are plotted as means ± SEs (n ≥ 3). +ve, positive; −ve, negative.
SW480 cells were transfected with pdEGFP-Bid (BD Biosciences). Cells were immediately subjected to oxygen deprivation and drug treatment posttransfection. Cells were then sorted by FACS analysis and replated at 2 × 105 cells per well on a 24-well plate, and apoptosis was assessed 2 days later. As a control, SW480 cells were transfected with a cytomegalovirus-driven GFP empty vector control that was a kind gift from Simon Scott (Karmanos Cancer Institute, Detroit, Mich.).
Demonstration of HIF-1α binding to an HRE on the bid promoter by using the EMSA.
The double-stranded DNA probes used in the electrophoresis mobility shift assay (EMSA) experiments contained the following sequences: 5′-ATCTGTGTTGTAGCGTGTGTCAATTGTATG-3′ for the wt HIF-1α binding site and 5′-ATCTGTGTTGTAGCACGTGTCAATTGTATG-3′ for the mutant HIF-1α binding site. An unrelated double-stranded oligonucleotide was used (ISRE [insulin receptor responsive element]) as a nonspecific competitor. The oligonucleotides were end-labeled with T4 polynucleotide kinase. SW480 cells incubated under anoxic or normoxic conditions were harvested and lysed in extraction buffer (20 mM HEPES [pH 7.9], 1 mM EDTA, 400 mM NaCl [25%], 0.1% NP-40, 1× protease inhibitors cocktail, 0.5 mM phenylmethylsulfonyl fluoride, 2.5 mM dithiothreitol). An equal amount of protein (1 μg) from the nuclear extract was used for binding reactions with the radiolabeled wt or mutant bid probe for 20 min at room temperature in binding buffer (8 mM HEPES [pH 7.4], 80 mM KCl, 0.8 mM EDTA, 1 mM dithiothreitol) at a 20-μl final volume. Immediately after, binding reaction samples were loaded on a 4% EMSA gel under nondenaturing conditions. For supershift experiments, 1 μg of monoclonal antibody against HIF-1 (NB 100-105K3; Novus Biologicals) was added to the reaction mixture before the addition of labeled oligonucleotides. Equivalent amounts of radiolabeled probe were used for all samples. For the binding competition experiment, unlabeled oligonucleotides were added into the reaction mixture in a 50-fold excess. DNA-protein complexes were analyzed in a 4% polyacrylamide gel with 0.5× Tris-borate-EDTA at 200 V. The gel was vacuum dried and exposed for autoradiography.
Effect of proteosome inhibitors on anoxia-mediated down-regulation of Bax and Bad.
Cells were incubated under normoxic or anoxic conditions with 10 μM Z-Leu-Leu-Norvalinal (MG115; Sigma) or Z-Leu-Leu-Leu-Al (MG132; Sigma) or with 0.001% dimethyl sulfoxide (as a control) for 16 h.
Measurement of translation efficiency by polysome fractionation and analysis.
Following exposure to hypoxia or normoxia conditions, cells were incubated for 5 min with 0.1 mg of cycloheximide (CHX) per ml at 37°C and then placed on ice. Cells were rinsed twice with PBS containing 0.1 mg of CHX/ml and scraped in lysis buffer (1% Triton X-100, 300 mM NaCl, 15 mM MgCl2, 15 mM Tris-HCl [pH 7.4], 0.1 mg of CHX/ml, 0.33 U of Superase-In [Ambion]/μl). Nuclei were pelleted at 2,000 × g and 200 μg of heparin/ml was added to the supernatant. Debris was pelleted at 10,000 × g, and the lysates were loaded on 20 to 50% sucrose gradients and centrifuged at 39,000 rpm for 90 min in a Beckman SW41Ti rotor. The absorbance at 254 nm as a function of depth in the gradient was subsequently measured by using a Bio-Rad UV monitor with a flowthrough cuvette. Simultaneously, 0.5-ml fractions were collected in TRI reagent (Sigma). The area under the curve in the polysomal and nonpolysomal fractions was approximated by using an in-house written algorithm. The background signal was subtracted and the area under the curve normalized to 1. The fraction of rRNA in polysomes was calculated as the area under the curve representing two or more ribosomes divided by the total area under the curve. Polysome fractions were spiked with an in vitro transcribed bacterial mRNA (pGIBS-TRP, ATCC 87485) for normalization purposes. RNA was isolated from the fractions according to the manufacturer's protocol and subjected to a Moloney murine leukemia virus (Sigma) reverse transcriptase reaction. Eurogentec's qPCRMastermix for Sybr Green I was used for real-time PCR amplification in a Taqman (ABI 7000; Applied Biosystems) machine. The primers for bid, bad, bax, and actin were the same as described above. Primers for CA-IX were CATCCTAGCCCTGGTTTTTGG and GCTCACACCCCCTTTGGTT; primers for the bacterial mRNA were ATATTGCGGCATACGGTCACT and CGGAGATACTTTTCGGTAGCTTTC.
Drug treatment under normoxic or anoxic conditions.
HCT116, HT29, SW480, and MEF cells were incubated for 8 h under normoxic or anoxic conditions and then treated with increasing concentrations of etoposide (Sigma) or oxaliplatin (Alexis Biochemicals) for 8 h with continuous normoxic or anoxic incubation. Cells were reseeded at a subconfluent density for short-term viability assays or into 6-well plates for clonogenic assays as previously described (54). Cells were left for 3 days for the short-term assay and counted and reseeded at a subconfluent density, and viable cell numbers were counted 4 days later. V79 cells were oxygen deprived for 16 h and then incubated with 5-FU for a further 4 h under anoxic conditions.
RESULTS
Effects of oxygen deprivation on colon cancer cell survival.
The effects of oxygen deprivation (<0.1% O2 [anoxia] and/or 1 to 2% O2 [hypoxia]) for 16 h on the viability of three human colon carcinoma cell lines, HCT116, HT29, and SW480, were examined. Anoxia did not promote apoptosis above basal levels (<4%) in any of the cell lines. There was no difference in the S phase traverse of cells cultured for 16 h in normoxia compared to those in anoxia or in the total cell number at 16 h. Additionally, there was no impact of oxygen deprivation on the subsequent clonogenicity of any of the three cell lines.
Effects of oxygen deprivation on the levels of Bcl-2 family proteins.
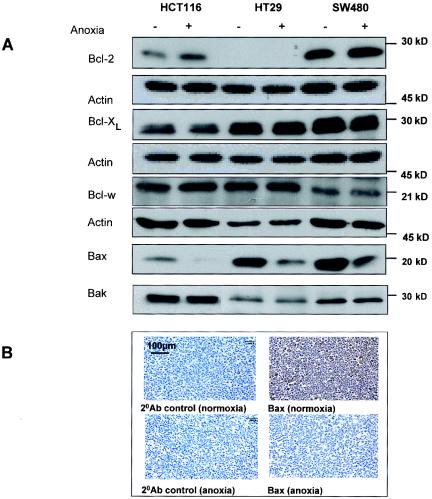
When the human colon carcinoma cells were cultured for up to 24 h under anoxia (or hypoxia), there were no major changes in the levels of the antiapoptotic proteins Bcl-2, Bcl-xL, and Bcl-w. There was, however, a slight but reproducible increase in Bcl-2 in HCT116 cells, confirming a previous report (Fig. 1A) (22). Bax and Bak were assessed. Studies of Bax−/−/Bak−/− double-KO MEFs have demonstrated that one or the other of these proapoptotic proteins is required for hypoxia-induced apoptosis (28). Bak levels were not down-regulated under anoxic conditions (Fig. 1A). In contrast, Bax levels were decreased in all cell lines, an effect that was first observed at 8 h (data not shown) and maintained at 16 h (Fig. 1A). The anoxia-mediated decrease in Bax was confirmed in SW480 cells by using immunohistochemistry (Fig. 1B).
FIG. 1.
Effect of anoxia on Bcl-2 family protein levels. (A) HCT116, HT29, and SW480 cells were incubated for 16 h under normoxic (−) or anoxic (+) conditions, and lysates were immunoblotted for Bcl-2 family proteins. Forty micrograms of protein per sample was loaded, and equal loading was confirmed by using actin as a control. (B) Immunohistochemical analysis of Bax in SW480 cells that were pelleted, fixed, embedded, and stained after incubation of cells in normoxic and anoxic conditions as described above. Results are representative of at least three independent repeat experiments.
BH-3-only proapoptotic proteins of the Bcl-2 family, Bid and Bad, were also decreased by oxygen deprivation.
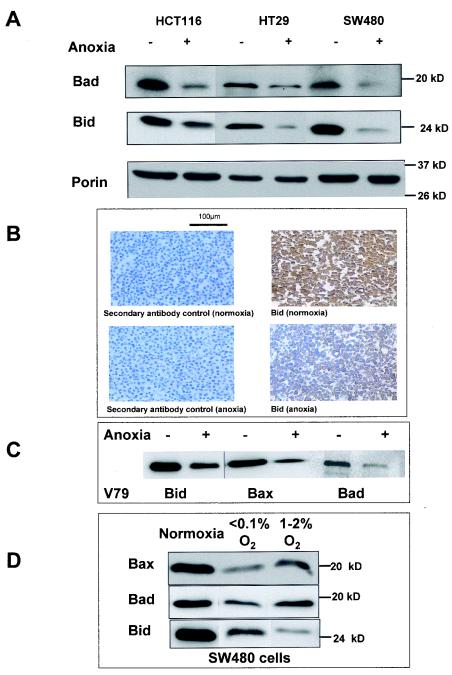
The function of Bax as a proapoptotic protein is regulated directly or indirectly by BH-3-only Bcl-2 family members (1, 9). For example, Bax can be activated via the binding of truncated Bid (50), and protection against Bax-mediated apoptosis is countered by the binding of Bid or Bad to Bcl-2 or Bcl-xL (9, 58). Figure 2A shows that in all three colon carcinoma cell lines, Bid and Bad protein levels were decreased after 16 h of oxygen deprivation. This effect was first observed at 8 h for Bid and Bad (data not shown). The down-regulation of Bid was confirmed in SW480 cells by using immunohistochemistry (Fig. 2B).
FIG. 2.
Effect of anoxia on protein levels of BH-3-only members of the Bcl-2 family. (A) HCT116, HT29, and SW480 cells were incubated for 16 h under normoxic (−) or anoxic (+) conditions, and lysates were immunoblotted for Bad, Bid, and porin (as a mitochondrial protein control). (B) Immunohistochemical analysis of Bid in SW480 cells that were pelleted, fixed, embedded, and stained after incubation under the same conditions of normoxia and anoxia. (C) V79 Chinese hamster lung fibroblasts cells were incubated for 16 h under normoxic or anoxic conditions. Lysates were prepared and immunoblotted for Bid, Bax, and Bad. (D) SW480 cells were incubated for 16 h under normoxic, anoxic (<0.1% O2), and hypoxic (1 to 2% O2) conditions, and lysates were immunoblotted for Bax, Bad, and Bid. For Western blotting, 40 μg of protein per sample was loaded (equal loading was confirmed by using actin as a control [data not shown]). Results are representative of at least three independent repeat experiments.
Oxygen deprivation-mediated reductions in Bid, Bad, and Bax were also observed in V79 Chinese hamster lung fibroblasts (Fig. 2C), in HEPA-1 murine hepatoma cells (see Fig. 5C), and in SV40-transformed MEFs (see Fig. 8A, inset), confirming that this effect is not restricted to the colon cancer cells.
FIG. 5.
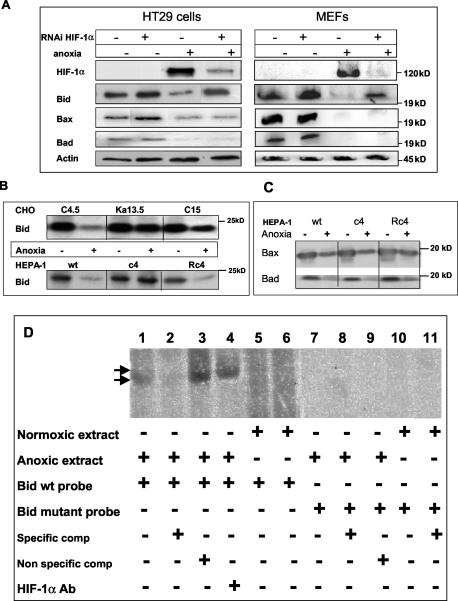
HIF-1-dependent down-regulation of Bid by oxygen deprivation. (A) HT29 cells and MEFs were transiently transfected with either a vector containing a HIF-1α scrambled sequence (−) or a vector containing a HIF-1α targeting sequence (+) 32 h prior to incubation under normoxic (−) or anoxic conditions (+) for 16 h. Lysates were then prepared and immunoblotted for Bid, Bax, Bad, and HIF-1α. (B) CHO wt (C4.5), HIF-1α-deficient (Ka13.5), and C15 (a pooled population control [see Materials and Methods]) cells and HEPA-1 wt, HIF-1β-deficient (c4), and mutant revertant (Rc4) cells were incubated for 16 h under normoxic or anoxic conditions, and the levels of Bid were assessed. (C) HEPA-1 wt, HIF-1β-deficient (c4), and mutant revertant (Rc4) cells were incubated for 16 h under normoxic or anoxic conditions and assessed for Bax and Bad levels. For all Western blots, 20 to 40 μg of protein per sample was loaded, and equal loading was confirmed by using actin as a control. (D) SW480 cells were incubated for 16 h under normoxic or anoxic conditions. Nuclear extracts were prepared and incubated with radiolabeled wt or mutant bid probes in the presence and absence of HIF-1α antibody and subjected to EMSAs (see Materials and Methods). Unlabeled oligonucleotides were used in competitive binding experiments to determine the specificity of the HIF-1α bid promoter complex. Arrows indicate the shift in complex size caused by the addition of the HIF-1α antibody (Ab). All results are representative of at least three independent repeat experiments.
Effects of oxygen concentration on proapoptotic Bcl-2 family proteins.
In vivo tumor cells experience a range of oxygen concentrations, thus, in order to assess the dose responses of proapoptotic Bcl-2 family proteins to oxygen, cells were also incubated for 16 h under hypoxic conditions (1 to 2% O2). Figure 2D shows that in SW480 cells, Bax, Bad, and Bid are all decreased under hypoxic conditions. For Bax and Bad, the magnitude of the decrease was positively correlated with the degree of oxygen deprivation. In contrast, for Bid, the decrease in the protein level observed for hypoxia was larger than that for anoxia (Fig. 2D).
Proapoptotic Bcl-2 family proteins were also down-regulated by oxygen deprivation in vivo.
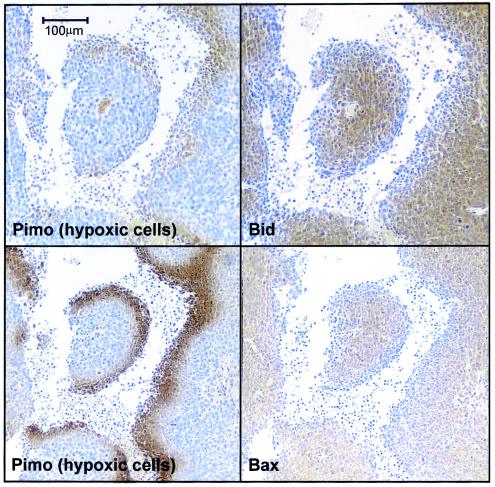
In order to assess the physiological relevance of our observations, HCT116 cells were grown as tumor xenografts in nude mice. Prior to sacrifice and excision of tumors, the mice were administered pimonidazole, a compound that binds irreversibly to hypoxic cells (31). Serial sections of the tumors were subsequently stained for pimonidazole and for Bid or Bax. An inverse correlation between the hypoxic cell marker and Bid or Bax was observed at the hypoxic rim around necrotic regions of the tumor, a location typically associated with chronic hypoxia (Fig. 3). These data have important ramifications when considering therapeutic strategies to induce apoptosis in tumors in vivo.
FIG. 3.
Down-regulation of Bax and Bid in HCT116 tumors in vivo. HCT116 cells were grown as xenograft tumors in CD1 nude mice, and tumors were excised, fixed in formalin, and paraffin embedded, and 5-μm serial sections were prepared. Data show a typical tumor chord with hypoxic penumbra marked by pimonidazole (Pimo) immunostaining with reciprocal immunostaining for Bid or Bax.
Down-regulation of Bid and Bad mRNA by oxygen deprivation.
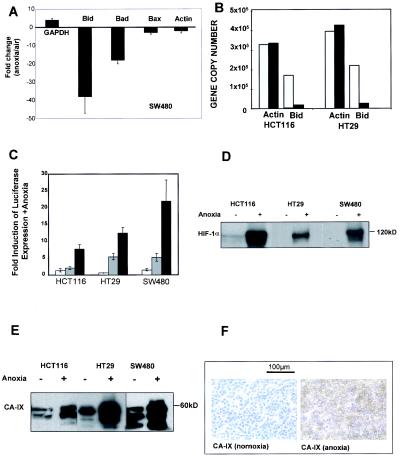
Quantitative real-time RT-PCR (Taqman) was used to investigate the mechanism by which the proapoptotic proteins of the Bcl-2 family were down-regulated by oxygen deprivation. Figure 4A shows that in SW480 cells that had been incubated under anoxic conditions for 16 h, the mRNA levels were decreased for Bid (38-fold ± 9-fold, mean ± standard error [SE], n = 3) and for Bad (18-fold ± 2-fold, mean ± SE, n = 3) compared to those for control cells incubated under normoxia. The severalfold difference between Bax mRNA in normoxia and anoxia SW480 cells was not significantly different than those observed for actin.
FIG. 4.
Effect of anoxia on mRNA levels of proapoptotic Bcl-2 family members and assessment of HIF-1 proficiency in HCT116, HT29, and SW480 cell lines. (A) cDNA was synthesized from total RNA extracted from 16-h incubations of SW480 cells under normoxic conditions (white bars) or anoxic conditions (black bars). Changes in levels of actin, GAPDH, Bid, Bad, and Bax copy numbers were assessed by quantitative RT-PCR (Taqman) analysis. The mean (± SE) changes in gene copy number in the SW480 cell line (normoxia/anoxia, n = 3) are represented. (B) Data representative of at least three experimental repeats for the change in Bid mRNA levels in HCT116 and HT29 cell lines with actin serving as a housekeeping gene control. (C) Cells were transiently transfected with vectors containing HRE sequences from either control (empty vector) (white bars), PGK (grey bars), or LDH (black bars) cloned upstream of a luciferase transcriptional unit and exposed to anoxia or normoxia (see Materials and Methods). Luciferase activity was normalized to the measured activity of controls under normoxia. Data shown are means ± SEs (n = 3). (D) Lysates were prepared from HCT116, HT29, and SW480 cells incubated for 16 h under normoxic or anoxic conditions and immunoblotted for HIF-1α. (E) Lysates were prepared from HCT116, HT29, and SW480 cells incubated for 16 h under normoxic or anoxic conditions and immunoblotted for CA-IX. (F) HT29 cells were incubated for 16 h under normoxic or anoxic conditions and then pelleted and subjected to immunohistochemical analysis of CA-IX. For Western blotting, 40 to 80 μg of protein per sample was loaded (equal loading was confirmed by using actin as a control [data not shown]). Results are representative of at least three independent repeat experiments.
GAPDH was analyzed as a positive control for the experiment, as it is known to increase under hypoxic conditions (62). In SW480 cells, GAPDH showed a 4-fold ± 1-fold increase under anoxia compared to that for normoxia (mean ± SE, n = 3), consistent with its role in the glycolytic pathway that is stimulated by oxygen deprivation. Bid mRNA levels were also decreased in HCT116 and HT29 cells incubated under anoxia by 9-fold ± 1-fold (for both, mean ± SE, n = 3) (Fig. 4B) compared to those for normoxia; there was no significant change in the mRNA levels of actin that was included as a housekeeping gene control. These data provoked an investigation into the possibility of oxygen deprivation-mediated transcriptional repression of proapoptotic members of the Bcl-2 family and the role of the HIF-1 transcription factor in their down-regulation.
All three colon carcinoma cell lines express a functional HIF-1 pathway.
The function of the HIF-1 pathway was investigated in all three colon carcinoma cells by using a transient reporter assay in which endogenous HIF-1 binds HREs cloned upstream of a luciferase transcriptional reporter. Figure 4C shows that endogenous HIF-1 in anoxic HCT116, HT29, and SW480 cells can bind to exogenous HREs of PGK and LDH. Consistent with our findings for a range of tumor cell types, the amplitude of response driven by the LDH HRE exceeds that of the PGK HRE (S. Robinson, K. J. Williams, and I. J. Stratford, personal communication). Oxygen deprivation resulted in increased levels of HIF-1α in all three colon cell lines (Fig. 4D). Additionally, the protein levels of CA-IX (another transcriptional target of HIF-1) were increased in each cell line in response to anoxia, as measured by Western blotting (Fig. 4E) and by immunohistochemistry for SW480 cells (Fig. 4F). Lastly, each cell line synthesized and secreted vascular endothelial growth factor, another HIF-1 transcriptional target in response to anoxia (data not shown). Of the three cell lines, SW480 cells displayed the most robust HIF-1-mediated responses (Fig. 4C), consistent with the magnitude of the decrease in Bid mRNA (see Fig. 4A).
Oxygen deprivation-mediated down-regulation of Bid occurs via a HIF-1-dependent pathway, whereas Bax and Bad down-regulation occurs independently of HIF-1.
In order to explore the relationship between HIF-1 function and the hypoxia-mediated down-regulation of Bcl-2 proapoptotic proteins, HT29 cells and MEFs were transfected with a vector containing a HIF-1α-specific targeting sequence 32 h prior to the exposure of cells to oxygen deprivation for 16 h. Figure 5A shows a substantial reduction in HIF-1α protein in oxygen-deprived HT29 cells and MEF cells expressing HIF-1α small interfering RNA that is correlated with the inhibition of Bid down-regulation. Conversely, Bax and Bad remain down-regulated in oxygen-deprived cells regardless of the small interfering RNA-mediated reduced expressed of HIF-1α (Fig. 5A). These data strongly suggest that Bid down-regulation in anoxic cells is dependent on HIF-1 function.
To determine the generality of this functional relationship between HIF-1 and Bid, we exploited two further cell types in which HIF-1 function was compromised. Firstly, Chinese hamster ovary (CHO, Ka13.5) cells with defective HIF-1α (55) were compared to CHO cells with wt HIF-1 function for their response to anoxia with respect to proapoptotic Bcl-2 family protein levels (Fig. 5B). Secondly, we examined the effects of anoxia on a panel of murine hepatoma cell lines, including a subline with HIF-1β deficiency (27) (Fig. 5B). In both cell types, the loss of HIF-1 function prevented the anoxia-mediated down-regulation of Bid but had no effect on the down-regulation of Bax or Bad (e.g., Fig. 5C, HEPA-1 cells). These differences between HIF-1α- and HIF-1β-deficient cells and their wt counterparts are unlikely to be selection artifacts, since anoxia-induced down-regulation of Bid was also observed in CHO C15 control cells and in the mutant revertant HEPA-1 clone Rc4 (Fig. 5C). Taken together, the data demonstrate that the down-regulation of Bid is a HIF-1-dependent event in several cell types. In contrast, the down-regulation of Bax and Bad occurred via a HIF-1-independent mechanism(s).
HIF-1α binding sites in bid.
The selection and design of an oligonucleotide corresponding to a HIF-1α binding site on the promoter region of the human bid gene was carried out by identifying the human genomic sequence in the NCBI database and analyzing the upstream region. The regulatory sequence analysis tools found at http://rsat.ulb.ac.be/rsat/ were used to identify 10,000-bp regions upstream from the translation start site of the bid gene in the human genome. In order to identify the most likely HIF-1α binding site in the upstream region of the human bid gene, the following information was considered: (i) the formation of a heterodimeric complex of HIF-1α with arlhydrocarbon receptor nuclear translocator (ARNT; HIF-1β) is required for efficient DNA binding where the minimal motif is [A or G]CGTG, (ii) ARNT heterodimers bind two half-sites of a motif CANNTG, (iii) one part of the dimer binds CAN, the other binds NTG, (iv) the third nucleotide downstream of the minimal GCGTG motif can affect binding of ARNT heterodimers to DNA and if an A or a T is present, binding is enhanced, whereas C has never been observed in this position, (v) the ARNT homodimer consensus binding sequence is [A or G]TCACGTGA[C or T], and (vi) if a heterodimer was formed with HIF-1α, a putative consensus binding site is TN[A or G]CGTGNN[A or T]. By combining all of the above information (44), it was possible to suggest that the HIF-1α binding site is located −8484 bp from the translation start site of the human bid gene. The location of the putative HIF-1α binding site is 1,860 bp downstream of the reported p53 binding site on the human bid promoter (35).
Confirmation of HIF-1α binding to the bid promoter.
The EMSA, incorporating an oligonucleotide that contained the putative HRE consensus in the bid promoter identified by bioinformatics (positions −8484 to −8475), was employed to investigate whether HIF-1α binds to the bid promoter. Figure 5D shows that the putative HRE is active, binding specifically to HIF-1α in cellular extract prepared from oxygen-deprived SW480 cells. The binding between HIF-1α and the bid HRE was prevented by 50-fold excess unlabeled oligonucleotides, indicating the specificity of the interaction (compare lane 1 with lane 2). The complex seen in lane 4 on the bid HRE is shifted by exposure to a HIF-1α-specific antibody (compare lanes 1 and 3 with lane 4), suggesting that the complex indeed contains HIF-1α. Finally, in the absence of HIF-1α in normoxic cell extracts, there is no complex formation on the bid HRE (lanes 6 and 7). To further test the specificity of the HIF-1 Bid HRE complex, the bid HRE consensus site was mutated (two point mutations) and no longer supported complex formation with HIF-1α (lanes 7 to 11). Taken together, the data in Fig. 5, panels A to E show that oxygen deprivation-mediated decrease of Bid expression occurs at the transcriptional level and is mediated by HIF-1.
HIF-1-independent down-regulation of Bax and Bad.
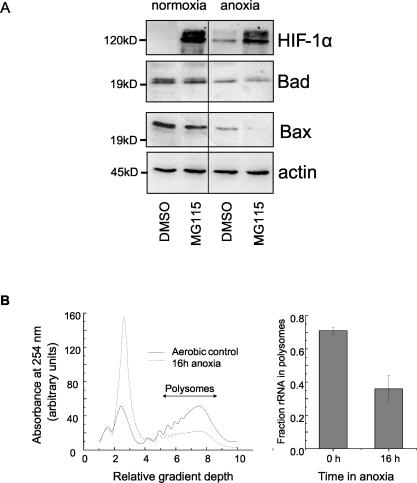
To examine whether the decreased levels of Bad and Bax protein expression under anoxia was due to increased proteasome-mediated degradation, SW480 cells treated with the proteasome inhibitor MG115 (10 μM) were incubated under anoxic conditions (Fig. 6A). The efficacy of MG115 treatment was demonstrated by the increased HIF-1α expression in normoxia (as expected of a protein known to be degraded by the proteosome [39]). However, MG115 did not prevent the reduction of Bad and Bax expression mediated by oxygen deprivation. Similar data were obtained with the proteosome inhibitor MG132. These data suggest that the decrease in Bax and Bad protein levels in anoxic colon cancer cells is not a function of enhanced proteosome-mediated protein degradation.
FIG. 6.
Oxygen deprivation-induced, HIF-1-independent decreased expression of Bax and Bad. (A) SW480 cells were treated with the proteosome inhibitor MG115 (10 μM) or dimethyl sulfoxide (DMSO) as a solvent control and immediately incubated for 16 h under normoxic or anoxic conditions prior to analysis of HIF-1α, Bad, and Bax by Western blotting. Twenty to 40 μg of protein per sample was loaded, and equal loading was confirmed by using actin as a control. Data are representative of three independent repeat experiments. (B) SW480 cells were incubated for 16 h under aerobic or anoxic conditions, and the amount of rRNA actively involved in translation (associated with polysomes) was measured (see Materials and Methods). The data show a representative polysome profile from normoxic and anoxic cells, and the average amounts of rRNA in the polysomal fraction are depicted in the bar chart (n = 3).
To determine whether oxygen deprivation altered translational efficiency, the fractional amount of rRNA actively involved in translation (associated with polysomes) was measured. Figure 6B shows an example of the polysome profile from normoxic and anoxic SW480 cells and indicates a strong, oxygen deprivation-mediated inhibition of translational efficiency. In normoxic samples, 75% of RNA was in polysomes, and this dropped to 30% under anoxic conditions. This reduction in global translation by analysis of polysomes was confirmed for bax and bad individual mRNAs by measuring their levels in polysomal fractions (data not shown). We are currently investigating the relative magnitude and importance of translational control of proapoptotic Bcl-2 family members in drug resistance in tumor cells deprived of oxygen.
Oxygen deprivation results in drug resistance in vitro.
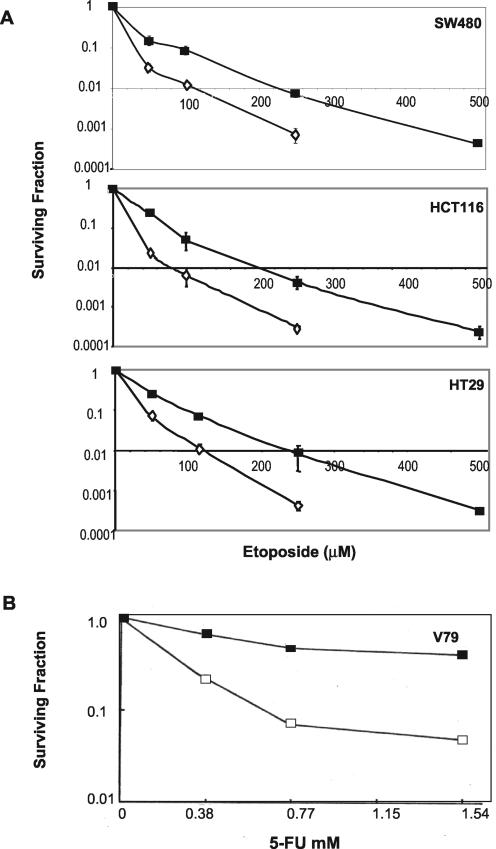
The down-regulation of Bax, Bid, and Bad in the absence of up-regulation of antiapoptotic proteins Bcl-2, Bcl-w, and Bcl-xL may be predicted to raise the threshold for drug-induced apoptosis. To investigate this possibility, HCT116, HT29, and SW480 cells were subjected to a standard clonogenic assay and to viability assay over days 3 to 7 following treatment with etoposide under normoxic and anoxic conditions. The clonogenic survival data show significant resistance to etoposide in all three cell lines (Fig. 7A). Anoxic exposure resulted in a twofold or greater resistance to etoposide; for example, under anoxic conditions, 2 logs of cell kill required 2.7-, 2.6-, and 2.3-fold higher concentrations of etoposide for the SW480, HCT116, and HT29 cells, respectively, compared to that required under normoxic conditions. Anoxic SW480 cells were also 2.7-fold more resistant to oxaliplatin (which is undergoing clinical trial for the treatment of colon cancer) in three repeat clonogenic assays (data not shown).
FIG. 7.
Effect of oxygen deprivation on drug response. (A) Colon cancer cells were incubated for 8 h under normoxic (open diamonds) or anoxic conditions (black squares) prior to 8 h of treatment with a range of concentrations of etoposide (with continued normoxic or anoxic incubation). (B) V79 fibroblasts were deprived of oxygen for 16 h prior to treatment with 5-FU for 4 h. Colonies were counted after 10 to 14 days. Data shown are means ± SEs of three independent repeat experiments.
Given the ongoing debate about the advantages and limitations of clonogenic assays for studies of drug-induced apoptosis (6, 37), the suppression of etoposide-induced cell death by oxygen deprivation was also assessed and demonstrated in a short-term viability assay. Although the kinetics of cell death were different for each cell line, the percentage of apoptotic cells that had been oxygen deprived prior to and after etoposide addition was consistently lower; for example, in three repeat experiments, on days 3 and 11 there was 10 to 19% less apoptosis and on day 7 there was 24% less apoptosis in cultures that had been oxygen deprived prior to drug treatment than in those maintained under normoxic conditions by using a drug concentration corresponding to a 2-log kill in the clonogenic assays. This difference could not be attributed to the altered kinetics of S phase traverse in anoxic versus normoxic SW480 cells as assessed by using the bromodeoxyuridine labeling index; 33% ± 2% and 30% ± 2% of cells moved through S phase within a 2-h period for anoxic and normoxic cells, respectively (mean ± SE, n = 3). Moreover, there was no difference in the kinetics of G2/M arrest after drug treatment (Table 1). Similar results were obtained with HCT116 cells (data not shown). Furthermore, oxygen deprivation did not alter the levels of the drug target topoisomerase II or of the multidrug resistance efflux pump P-gp (data not shown). Anoxic SW480 cells treated with a concentration of oxaliplatin corresponding to a 3-log kill in clonogenic assays showed 18 and 36% less apoptosis at 3 and 7 days after drug treatment, respectively (mean, n = 3).
TABLE 1.
Effect of oxygen deprivation on etoposide-induced G2/M arrest in SW480 colon cancer cells
| Oxygen status and drug treatment | Cell cycle phase distributiona
|
||
|---|---|---|---|
| % of cells in G1 phase | % of cells in S phase | % of cells in G2/M phase | |
| Normoxia | |||
| Control | 52.6 ± 4.5 | 31.5 ± 0.7 | 15.9 ± 3.9 |
| Etoposide | 39.1 ± 0.8 | 28.3 ± 1.7 | 32.5 ± 2.2 |
| Anoxia | |||
| Control | 48.1 ± 4.8 | 31.9 ± 1.3 | 20.1 ± 3.5 |
| Etoposide | 40.6 ± 1.2 | 27.4 ± 1.3 | 32.1 ± 2.2 |
Mean ± SE for the percentage of cells in each cell cycle phase as determined by flow cytometric analysis of propidium-bound DNA.
In the above experiments, cells were oxygen deprived for 8 h prior to drug addition and for 8 h after drug addition, i.e., changes in gene expression due to oxygen deprivation, including the down-regulation of proapoptotic Bcl-2 family proteins, will have occurred before drug-target interactions. The 8-h preexposure to conditions of low oxygen was required for drug resistance, as normoxic cells that were oxygen deprived simultaneously with etoposide or oxaliplatin addition showed a loss of viability similar to those cells that never experienced low oxygen in both short-term and long-term (clonogenic) cell viability assays (data not shown).
Bid, Bax, and Bad were also down-regulated in anoxic V79 fibroblasts (Fig. 2C). Oxygen deprivation-mediated resistance to adriamycin in these V79 cells could not be explained by differences in drug uptake and efflux (40). Moreover, oxygen deprivation also rendered these cells resistant to 5-FU (Fig. 7B), to etoposide (21), and to amsacrine (53). Taken together, the data suggest that the down-regulation of proapoptotic proteins of the Bcl-2 family by oxygen deprivation is not specific to a single cell type and that it is correlated with resistance to a wide range of anticancer drugs with different mechanisms of action.
Down-regulation of Bid and/or Bax contributes to resistance to etoposide.
We took several approaches to assess the functional relevance of the hypoxia-mediated down-regulation of Bcl-2 proapoptotic proteins Bax and Bid in drug responsiveness. Firstly, we assessed the response to etoposide of normoxic cells in which either Bid or Bax was not expressed or in which we had down-regulated Bid in a Bax null background in order to mimic the scenario observed in hypoxic cells. To more robustly test the role in drug resistance of Bid down-regulation in hypoxic cells, we expressed an exogenous Bid that could not be down-regulated by HIF-1 and assessed the response to etoposide in hypoxic cells.
Sixteen hours of oxygen deprivation resulted in the down-regulation of Bid, Bax, and Bad in wt MEFs and of Bax and Bad in Bid KO MEFs but did not affect cell viability in either case (Fig. 8A, inset). While Bid KO MEFs exhibited a decreased apoptotic response in normoxia to 8 h of exposure to etoposide compared to that of their wt counterparts, this differential response was lost when MEFs experienced oxygen deprivation, suggesting that the loss of Bid contributes to etoposide resistance in anoxia (Fig. 8A).
Similarly, normoxic Bax null HCT116 cells were more resistant to etoposide than were wt HCT116 cells (Fig. 8B), consistent with previous observations of decreased drug responsiveness in these cells (46, 59). Etoposide resistance was further increased in the Bax null HCT116 cells by the down-regulation of Bid with RNAi (Fig. 8B and inset). The etoposide response in these cells was not significantly different than that observed in anoxic wt HCT116 cells in which Bid and Bax were down-regulated by the oxygen deprivation. These data suggest that Bid and Bax act together to suppress etoposide-induced apoptosis and predict that their down-regulation in hypoxic subpopulations of cells in solid tumors may have a negative impact on drug response. Finally, when the expression of GFP Bid was maintained in anoxic SW480 cells (Fig. 8C), the percentage of apoptosis was significantly increased compared to that seen in nonexpressing GFP Bid transfected anoxic cells or compared to vector GFP transfected cells. However, when Bid expression was maintained under anoxia, it did not fully restore the apoptotic response to etoposide observed in vector GFP control normoxic cells (Fig. 8C). These data demonstrate that Bid plays a role in etoposide-induced apoptosis in SW480 cells and that its down-regulation by oxygen deprivation is an important contributor to the observed drug resistance.
DISCUSSION
In solid tumors, a hostile cellular microenvironment due to the limited delivery of oxygen and nutrients drives selection for those tumor cells that can adapt to survive these insults. Pertinent to our study, hypoxia selects for cells with increased capacities to survive (15, 39), and this increased survival potential not only facilitates tumor development but may also be expected to result in resistance to drug-induced apoptosis (12, 20), a prediction borne out in solid tumors in vivo (45).
The Bcl-2 family members are considered to be integrators of both survival and damage signals and play a critical role in setting the threshold at which a cell in a specific context will commit to apoptosis (26). We therefore conducted a survey to assess the impact of oxygen deprivation on multiple key pro- and antiapoptotic proteins of the Bcl-2 family in a range of cell types.
There have been sporadic reports that hypoxia can provoke changes in Bcl-2 family proteins in cancer cell types. Oxygen and serum deprivation of HepG2 hepatoma cells that also express wt p53 resulted in Bax down-regulation (3). Hypoxia provoked an increase in Bcl-2 and Bcl-xL levels in human lung carcinoma cells (30) and an increase in Bcl-2 levels in HCT116 cells (22). The results presented here show that Bax, Bid, and Bad are down-regulated in colon cancer cells, hepatomas, ovarian epithelial cells, and both lung and embryonic fibroblasts subjected to periods of oxygen deprivation that have no detrimental effect on cell growth. This occurred without a compensatory rise in antiapoptotic family members and in both p53 wt (HCT116) and p53 mutant-containing cells (SW480 and HT29) (48).
The BH-3-only proapoptotic proteins are considered to be sentinels of damage signals and function to inactivate the antiapoptotic proteins of the family (9). Bid and Bad may also bind to and activate Bax and Bak (14, 25, 50). Studies of Bax/Bak double-KO fibroblasts demonstrate the requirement of either Bak or Bax to mount an apoptotic response to damage signals (51). Bax expression is lost in a proportion of human colon cancers with defective mismatch repair (32). Of the proapoptotic Bcl-2 family proteins studied here, Bak was the least responsive to oxygen deprivation, suggesting that many human colon cancer cells may rely heavily on Bak to affect drug-induced apoptosis.
In regard to cellular survival, the down-regulation of Bid under anoxic and hypoxic conditions is important for several reasons. Firstly, this member of the family serves to link the mitochondrial and death receptor pathways for apoptosis. This is particularly relevant in so-called type II cells (the vast majority) that require Bid function for death receptor-mediated apoptosis (36). In this pathway, Bid is cleaved by caspase 8 and translocates to the mitochondrial surface to activate Bax and/or Bak. Hence, the down-regulation of Bid in hypoxic type II tumor cells may protect against a range of apoptotic stimuli. Secondly, further interplay has been reported between the pathways that lead to death receptor- and drug-induced apoptosis in which Bax and Bid are key components (24). Thirdly, we recently demonstrated that Bid normally functions as a lipid transfer protein that shuttles lipid between the endoplasmic reticulum and mitochondria. We suggested that upon receipt of apoptotic stimuli, Bid-mediated lipid transport processes are deregulated and serve to destabilize mitochondrial membranes (10). The down-regulation of Bad by oxygen deprivation in addition to its dephosphorylation due to HIF-1-mediated amplification of PI-3K signaling (39) may make for a robust negation of its proapoptotic function.
Bad and Bid were down-regulated at the mRNA level in colon cancer cell lines, and we discovered that the bid promoter contained putative HRE sequences. It was therefore interesting that in contrast to Bad, Bid protein was down-regulated to a greater extent under hypoxic conditions than it was under anoxic conditions, and it is the oxygen concentration of 1 to 2% (that we term hypoxia) at which HIF-1 is most active (13). Consistent with this observation, we showed that the prominent down-regulation of Bid mRNA was dependent on HIF-1 transcriptional activity in four different cell types and that HIF-1α binds specifically to an HRE in the bid promoter. In addition to this discovery that HIF-1 causes the transcriptional repression of bid, at least one precedent exists for HIF-1-mediated transcriptional repression. The peroxisome proliferator-activated receptor alpha is negatively regulated by HIF-1 (29), and emerging DNA microarray data show the down-regulation of several genes by HIF-1 (57).
The murine promoter of the proapoptotic BH-3-only homolog Nip3 contains an HRE (7), and Nip3 mRNA and protein have been shown to be up-regulated by oxygen deprivation in several cell types, including LS174T human colon cancer cells (17, 41). However, we found this response of Nip3 to decreased oxygen to be cell type specific, occurring in an HIF-1-dependent fashion in both HEPA-1 and CHO cells but not in HCT116, HT29, or SW480 colon cancer cells where, paradoxically, the protein level of Nip3 was decreased (data not shown). Further studies are required to identify the cell-type-specific determinants of the Nip3 response to hypoxia and their impact on cell fate.
The down-regulation of Bax under conditions of oxygen deprivation was not associated with decreased levels of Bax mRNA, and the observed reduction in Bax protein did not reflect increased proteosomal degradation. However, we observed a global decrease in translation efficiency that would be expected to contribute to the reduced expression of many proteins, including Bax and Bad. Ongoing studies show the decrease in translation of Bax and Bad to be greater than that for actin, and further studies are now required to validate and expand this analysis. It has recently been shown that hypoxia results in the repression of protein synthesis via phosphorylation of the translational initiation factor eIF2α (23). Studies to investigate the relevance of this mechanism to the observed HIF-1-independent, hypoxia-mediated down-regulation of Bax and Bad are also under way.
Given the mechanistic data regarding how these proapoptotic proteins function (1), we considered that the down-regulation of Bax, Bid, and Bad by oxygen deprivation would render cells resistant to a variety of apoptotic stimuli. Bax and Bid participate in oxygen deprivation-induced cell death (28), implying that their role in this pathway must be abolished in cells that adapt to survive hypoxia in solid tumors. Drug resistance of human solid tumors is a major obstacle for successful treatment. Our data now show that significant resistance to etoposide and oxaliplatin is engendered when colon cancer cells are subjected to a period of oxygen deprivation before and during drug treatment. We show that this drug resistance is most unlikely to be mediated solely by upstream determinants, i.e., those that modulate drug-target interaction. Oxygen deprivation of V79 fibroblasts resulted in resistance to a range of anticancer drugs with differing mechanisms of action, again arguing that the modulation of events downstream of drug-target interactions are the most likely causes for the reduced response.
Several approaches were undertaken to test the importance of Bid and Bax down-regulation in the response of oxygen-deprived cells to etoposide, bearing in mind that lack of oxygen drives a multitude of adaptive cellular response. Lack of Bid expression in anoxic cells resulted in the loss in the differential in drug response seen in normoxic cells. RNAi-mediated Bid down-regulation in normoxic Bax null cells produced the equivalent pronounced resistance to etoposide seen in anoxic Bax null cells, and the maintained expression of Bid in anoxic cells significantly increased their drug sensitivity. Taken together, the data demonstrate that the down-regulation of Bid and Bax by oxygen deprivation contributes to the drug resistance of anoxic cells.
Critically, the down-regulation of Bax and Bid in human colon cancer xenograft tumors demonstrates the physiological relevance of our observations in vitro. The area of tumor in which reciprocity of hypoxic markers and Bid or Bax was seen, i.e., the penumbra around necrotic zones, is suggestive of the fact that the effect is associated with chronic hypoxia. It is open to question whether acutely hypoxic cells would rapidly restore their proapoptotic protein complement and whether reoxygenation could promote apoptosis.
Considering that the Bcl-2 family of proteins work together to determine the threshold for drug-induced apoptosis, our data predict that novel therapeutic strategies to combat solid tumors must take into account a hypoxia-mediated, increased threshold for apoptosis that is Bcl-2 family controlled. New anticancer drugs targeted to survival signaling pathway components may not be entirely effective if the proapoptotic proteins of the Bcl-2 family acting downstream are down-regulated to an extent that they cannot couple the apoptotic stimuli to the release of mitochondrial factors that activate the caspase cascade. We suggest that in chronically hypoxic cells, the down-regulation of Bax simultaneously with that of Bid and of Bad will provide a significant survival advantage, as Bid activates Bak and Bax, and Bad is an inactivator of Bcl-2 and Bcl-xL (25).
In summary, the data presented here provide a mechanistic explanation for the enhanced survival of hypoxic tumor cells via oxygen deprivation-mediated, coordinated down-regulation of three proapoptotic proteins of the Bcl-2 family. The HIF-1-dependent down-regulation of Bid in hypoxic tumor cells offers an additional impetus to antitumor strategies that target HIF-1 (39), as one favorable consequence of such a strategy will be to restore the levels of a potent proapoptotic protein that couples extrinsic and intrinsic pathways to the machinery of apoptosis.
Acknowledgments
The studies described here were funded via program grants from Cancer Research UK (C.D.), the Medical Research Council (I.J.S.), and the EORTC (grant TRF 01/03 to the Screening Pharmacology Group) (I.J.S.). We acknowledge the BBSRC and Aventis for their sponsorship of J.E. via a Case Ph.D. studentship.
We thank Mike Jackson, Tania Nolan, and Ged Brady for technical assistance with flow cytometry and Taqman. We thank Brian Telfer for his technical assistance and Mauro Degli Esposti for his advice regarding the function(s) of Bid.
REFERENCES
- 1.Adams, J. M., and S. Cory. 1998. The Bcl-2 protein family: arbiters of cell survival. Science 281:1322-1326. [DOI] [PubMed] [Google Scholar]
- 2.Akakura, N., M. Kobayashi, I. Horiuchi, A. Suzuki, J. Wang, J. Chen, H. Niizeki, K. Kawamura, M. Hosokawa, and M. Asaka. 2001. Constitutive expression of hypoxia-inducible factor-1α renders pancreatic cancer cells resistant to apoptosis induced by hypoxia and nutrient deprivation. Cancer Res. 61:6548-6554. [PubMed] [Google Scholar]
- 3.Baek, J. H., J. E. Jang, C. M. Kang, H. Y. Chung, N. D. Kim, and K. W. Kim. 2000. Hypoxia-induced VEGF enhances tumor survivability via suppression of serum deprivation-induced apoptosis. Oncogene 19:4621-4631. [DOI] [PubMed] [Google Scholar]
- 4.Birner, P., M. Schindl, A. Obermair, G. Breitenecker, and G. Oberhuber. 2001. Expression of hypoxia-inducible factor 1α in epithelial ovarian tumors: its impact on prognosis and response to chemotherapy. Clin. Cancer Res. 7:1661-1668. [PubMed] [Google Scholar]
- 5.Bos, R., H. Zhong, C. F. Hanrahan, E. C. Mommers, G. L. Semenza, H. M. Pinedo, M. D. Abeloff, J. W. Simons, P. J. van Diest, and E. van der Wall. 2001. Levels of hypoxia-inducible factor-1 alpha during breast carcinogenesis. J. Natl. Cancer Inst. 93:309-314. [DOI] [PubMed] [Google Scholar]
- 6.Brown, J. M., and B. Wouters. 2001. Apoptosis: mediator or mode of cell killing by anticancer drugs? Drug Resist. Updates 4:135-136. [DOI] [PubMed] [Google Scholar]
- 7.Bruick, R. K. 2000. Expression of the gene encoding the proapoptotic Nip3 protein is induced by hypoxia. Proc. Natl. Acad. Sci. USA 97:9082-9087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carmeliet, P., Y. Dor, J. M. Herbert, D. Fukumura, K. Brusselmans, M. Dewerchin, M. Neeman, F. Bono, R. Abramovitch, P. Maxwell, C. J. Koch, P. Ratcliffe, L. Moons, R. K. Jain, D. Collen, E. Keshert, and E. Keshet. 1998. Role of HIF-1α in hypoxia-mediated apoptosis, cell proliferation and tumour angiogenesis. Nature 394:485-490. [DOI] [PubMed] [Google Scholar]
- 9.Cheng, E. H., M. C. Wei, S. Weiler, R. A. Flavell, T. W. Mak, T. Lindsten, and S. J. Korsmeyer. 2001. BCL-2, BCL-X(L) sequester BH3 domain-only molecules preventing BAX- and BAK-mediated mitochondrial apoptosis. Mol. Cell 8:705-711. [DOI] [PubMed] [Google Scholar]
- 10.Degli Esposti, M., J. T. Erler, J. A. Hickman, and C. Dive. 2001. Bid, a widely expressed proapoptotic protein of the Bcl-2 family, displays lipid transfer activity. Mol. Cell. Biol. 21:7268-7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DePreter, K., F. Speleman, V. Combaret, J. Lunec, G. Lautreys, B. H. Eussen, N. Francotte, J. Board, A. D. Pearson, A. De Paepe, N. Van Roy, and J. Vandesompele. 2002. Quantification of MYCN, DDX1 and NAG gene copy number in neuroblastoma using real time quantitative PCR assay. Mod. Pathol. 15:59-66. [DOI] [PubMed] [Google Scholar]
- 12.Dive, C., and J. A. Hickman. 1991. Drug target interactions: only the first step in the commitment of a cell to a programmed cell death. Br. J. Cancer 64:192-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dong, Z., M. A. Venkatachalam, J. Wang, Y. Patel, P. Saikumar, G. L. Semenza, T. Force, and J. Nishiyana. 2001. Up-regulation of apoptosis inhibitory protein IAP-2 by hypoxia. HIF-1-independent mechanisms. J. Biol. Chem. 276:18702-18709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eskes, R., S. Desagher, B. Antonsson, and J.-C. Martinou. 2000. Bid induces the oligomerization and insertion of Bax into the outer mitochondrial membrane. Mol. Cell. Biol. 20:929-935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Graeber, T. G., C. Osmanian, T. Jacks, D. E. Housman, S. J. Koch, S. W. Lowe, and A. J. Giaccia. 1996. Hypoxia-mediated selection of cells with diminished apoptotic potential in solid tumours. Nature 379:88-91. [DOI] [PubMed] [Google Scholar]
- 16.Griffiths, J. R., P. M. McSheehy, S. P. Robinson, H. Troy, Y. L. Chung, R. D. Leek, K. J. Williams, I. J Stratford, A. L. Harris, and M. Stubbs. 2002. Metabolic changes detected by in vivo magnetic resonance studies of HEPA-1 wild-type tumors and tumors deficient in hypoxia-inducible factor-1beta (HIF-1beta): evidence of an anabolic role for the HIF-1 pathway. Cancer 62:688-695. [PubMed] [Google Scholar]
- 17.Guo, K., G. Searfossm, D. Krolikowskim, M. Pagnoni, C. Granksm, K. Clark, K. T. Yu, M. Jaye, and Y. Ivashchenko. 2001. Hypoxia induces the expression of the pro-apoptotic gene BNIP3. Cell Death Differ. 8:367-376. [DOI] [PubMed] [Google Scholar]
- 18.Hopfl, G., R. H. Wenger, U. Ziegler, T. Stallmach, O. Gardelle, R. Achermann, M. Wergin, B. Kaser-Hotz, H. M. Saunders, K. Williams, I. Stratford, M. Gassman, and I. Desbaillets. 2002. Rescue of hypoxia-inducible factor-1alpha-deficient tumor growth by wild-type cells is independent of vascular endothelial growth factor. Cancer Res. 62:2962-2970. [PubMed] [Google Scholar]
- 19.Jiang, B. H., F. Agani, A. Passaniti, and G. L. Semenza. 1997. V-Src induces expression of hypoxia-inducible factor 1 (HIF-1) and transcription of genes encoding vascular endothelial growth factor and enolase 1: involvement of HIF-1 in tumour progression. Cancer Res. 57:5328-5335. [PubMed] [Google Scholar]
- 20.Johnstone, R. W., A. A. Ruefi, and S. W. Lowe. 2002. Apoptosis: a link between cancer genetics and chemotherapy. Cell 108:153-164. [DOI] [PubMed] [Google Scholar]
- 21.Kalra, R., A.-M. Jones, J. Kirk, G. E. Adams, and I. J. Stratford. 1993. The effect of hypoxia on acquired drug resistance and response to epidermal growth factor in Chinese hamster lung fibroblasts and human breast cancer cells in vitro. Int. J. Cancer 54:650-655. [DOI] [PubMed] [Google Scholar]
- 22.Kinoshita, M., D. L. Johnson, C. H. Shatney, Y. L. Lee, and H. Mochizuki. 2001. Cancer cells surviving hypoxia obtain hypoxia resistance and maintain anti-apoptotic potential under reoxygenation. Int. J. Cancer 91:322-326. [DOI] [PubMed] [Google Scholar]
- 23.Koumenis, C., C. Naczki, M. Koritzinsky, S. Rastani, A. Diehl, N. Sonenberg, A. Koromilas, and B. G. Wouters. 2002. Regulation of protein synthesis by hypoxia via activation of the endoplasmic reticulum kinase PERK and phosphorylation of the translation initiation factor eIF2α. Mol. Cell. Biol. 22:7405-7416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.LeBlanc, H., D. Lawrence, E. Varfolomeev, K. Totpal, J. Morlan, P. Schow, S. Fong, R. Schwall, D. Sinicropi, and A. Ashkenazi. 2002. Tumor-cell resistance to death receptor-induced apoptosis through mutational inactivation of the proapoptotic Bcl-2 homolog Bax. Nat. Med. 8:274-281. [DOI] [PubMed] [Google Scholar]
- 25.Letai, A., M. C. Bassik, L. D. Walensky, M. D. Sorcinelli, S. Weiler, and J. Korsmeyer. 2002. Distinct BH3 domains either sensitize or activate mitochondrial apoptosis, serving as prototype cancer therapeutics. Cancer Cell 2:183-192. [DOI] [PubMed] [Google Scholar]
- 26.Makin, G., and C. Dive. 2001. Apoptosis and cancer chemotherapy. Trends Cell Biol. 11:S22-S26. [DOI] [PubMed] [Google Scholar]
- 27.Maxwell, P. H., G. U. Dachs, J. M. Gleadle, L. G. Nicholls, A. L. Harris, I. J. Stratford, O. Hankinson, C. W. Pugh, and P. Ratcliffe. 1997. Hypoxia-inducible factor-1 modulates gene expression in solid tumours and influences both angiogenesis and tumour growth. Proc. Natl. Acad. Sci. USA 94:8104-8109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McClintock, D. S., M. T. Santore, V. Y. Lee, J. Brunelle, G. R. Scott Bidinger, W.-X. Zong, C. B. Thompson, N. Hay, and N. S. Chandel. 2002. Bcl-2 family members and functional electron transport chain regulate oxygen deprivation-induced cell death. Mol. Cell. Biol. 22:94-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Narravula, S., and S. P. Colgan. 2001. Hypoxia-inducible factor 1-mediated inhibition of peroxisome proliferator-activated receptor α expression during hypoxia. J. Immunol. 166:7543-7548. [DOI] [PubMed] [Google Scholar]
- 30.Park, S. Y., T. R. Billiar, and D. W. Seol. 2002. Hypoxia inhibition of apoptosis induced by tumor necrosis factor-related apoptosis-inducing ligand (TRAIL). Biochem. Biophys. Res. Commun. 291:150-153. [DOI] [PubMed] [Google Scholar]
- 31.Raleigh, J. A., S. C. Chou, G. E. Arteel, and M. R. Horsman. 1999. Comparisons among pimonidazole binding, oxygen electrode measurements, and radiation response in C3H mouse tumors. Radiat. Res. 151:580-589. [PubMed] [Google Scholar]
- 32.Rampino, N., H. Yamamoto, Y. Ionov, Y. Li, H. Sawai, J. C. Reed, and M. Perucho. 1997. Somatic frameshift mutations in the BAX gene in colon cancers of the microsatellite mutator phenotype. Science 275:967-969. [DOI] [PubMed] [Google Scholar]
- 33.Ryan, H. E., J. Lo, and R. S. Johnson. 1998. HIF-1α is required for solid tumor formation and embryonic vascularization. EMBO J. 17:3005-3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ryan, H. E., M. Poloni, W. McNutty, D. Elson, M. Grassmann, J. M. Arbeit, and R. S. Johnson. 2000. Hypoxia-inducible factor-1alpha is a positive factor in solid tumor growth. Cancer Res. 60:4010-4015. [PubMed] [Google Scholar]
- 35.Sax, J. K., P. Fei, M. E. Murphy, E. Bernhard, S. J. Korsmeyer, and W. S. El-Deiry. 2002. BID regulation by p53 contributes to chemosensitivity. Nat. Cell Biol. 4:842-849. [DOI] [PubMed] [Google Scholar]
- 36.Scaffidi, C., I. Schmitz, J. Zha, S. J. Korsmeyer, P. H. Krammer, and M. E. Peter. 1999. Differential modulation of apoptosis sensitivity in CD95 type I and type II cells. J. Biol. Chem. 6:22532-22538. [DOI] [PubMed] [Google Scholar]
- 37.Schmitt, C. A., and S. W. Lowe. 2001. Apoptosis is critical for drug response in vivo. Drug Resist. Updates 4:132-134. [DOI] [PubMed] [Google Scholar]
- 38.Semenza, G. L., B. H. Jiang, S. W. Leung, R. Passantino, J. P. Concordet, P. Maire, and A. Giallongo. 1996. Hypoxia response elements in the aldolase A, enolase 1, and lactate dehydrogenase A gene promoters contain essential binding sites for hypoxia-inducible factor 1. J. Biol. Chem. 271:32529-32537. [DOI] [PubMed] [Google Scholar]
- 39.Semenza, G. L. 2002. HIF-1 and tumour progression: pathophysiology and therapeutics. Trends Mol. Med. 8:S62-S67. [DOI] [PubMed] [Google Scholar]
- 40.Smith, E., I. J. Stratford, and G. E. Adams. 1980. Cytotoxicity of adriamycin on aerobic and hypoxic Chinese hamster V79 cells in vitro. Br. J. Cancer 41:568-573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sowter, H. S., P. J. Ratcliffe, P. Watson, A. H. Greenberg, and A. L. Harris. 2001. HIF-1-dependent regulation of hypoxic induction of cell death factors BNIP3 and NIX in human tumors. Cancer Res. 61:6669-6673. [PubMed] [Google Scholar]
- 42.Stratford, I. J., A. V. Patterson, G. U. Dachs, B. Telfer, and K. Williams. 1999. Hypoxia-mediated gene expression, p. 107-113. In P. Vaupel and D. K. Kelleher (ed.), Tumour hypoxia: pathophysiology, clinical significance and therapeutic perspectives. Wissenchaftliche Verlagsgeselschaft GmbH, Stuttgart, Germany.
- 43.Sui, G., C. Soohoo, E. B. Affar, F. Gay, Y. Shi, W. C. Forrester, and Y. Shi. 2002. A DNA vector-based RNAi technology to suppress gene expression in mammalian cells. Proc. Natl. Acad. Sci. USA 99:5515-5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Swanson, H. I. 2002. DNA binding and protein interactions of the AHR/ARNT heterodimer that facilitate gene activation. Chem.-Biol. Interact. 141:63-76. [DOI] [PubMed] [Google Scholar]
- 45.Teicher, B. A. 1994. Hypoxia and drug resistance. Cancer Metastasis Rev. 13:139-168. [DOI] [PubMed] [Google Scholar]
- 46.Theodorakis, P., E. Lomonosova, and D. Chinnadurai. 2002. Critical requirement of BAX for manifestation of apoptosis induced by multiple stimuli in human epithelial cancer cells. Cancer Res. 62:3373-3376. [PubMed] [Google Scholar]
- 47.Thomas, D. A., L. Scorrano, G. V. Putcha, S. J. Korsmeyer, and T. J. Ley. 2001. Granzyme B can cause mitochondrial depolarisation and cell death in the absence of BID, BAX and BAK. Proc. Natl. Acad. Sci. USA 98:14985-14990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Violette, S., L. Poulain, E. Dussaulx, D. Pepin, A. M. Faussat, J. Chambaz, J. M. Lacorte, C. Staedel, and T. Lesuffleur. 2002. Resistance of colon cancer cells to long-term 5-fluorouracil exposure is correlated to the relative level of Bcl-2, Bcl-xl in addition to BAX and p53 status. Int. J. Cancer 98:498-504. [DOI] [PubMed] [Google Scholar]
- 49.Wang, G. 1995. Hypoxia-inducible factor 1 is a basic helix-loop-helix-PAS heterodimer regulated by cellular oxygen tension. Proc. Natl. Acad. Sci. USA 92:5510-5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wei, M. C., T. Lindsten, V. K. Mootha, S. Weiler, A. Gross, M. Ashiya, C. B. Thompson, and S. J. Korsmeyer. 2000. tBid, a membrane-targeted death ligand, oligomerizes BAK to release cytochrome c. Genes Dev. 14:2060-2071. [PMC free article] [PubMed] [Google Scholar]
- 51.Wei, M. C., W. X. Zong, E. H. Cheng, T. Lindsten, V. Panoutsakopoulou, A. J. Ross, K. A. Roth, G. R. MacGregor, C. B. Thompson, and S. J. Korsmeyer. 2001. Pro-apoptotic Bax and Bak: a requisite gateway to mitochondrial dysfunction and death. Science 292:727-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wenger, R. H., and M. Gassman. 1997. Oxygen(es) and the hypoxia-inducible factor-1. Biol. Chem. 378:609-616. [PubMed] [Google Scholar]
- 53.West, C., I. J. Stratford, N. Barrass, and E. Smith. 1981. A comparison of adriamycin and mAMSA in vitro: cell lethality and sister chromatid exchange studies. Br. J. Cancer 44:798-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Williams, K. J., B. A. Telfer, R. E. Airley, H. P. Peters, M. R. Sheridan, A. J. van der Kogel, A. L. Harris, and I. J. Stratford. 2002. A protective role for HIF-1 in response to redox manipulation and glucose deprivation: implications for tumorigenesis. Oncogene 21:282-290. [DOI] [PubMed] [Google Scholar]
- 55.Wood, S. M., M. S. Wiesener, K. M. Yeates, N. Okada, C. W. Pugh, P. H. Maxwell, and P. J. Ratcliffe. 1998. Selection and analysis of a mutant cell line defective in the hypoxia-inducible factor-1α-subunit (HIF-1α). J. Biol. Chem. 273:8360-8368. [DOI] [PubMed] [Google Scholar]
- 56.Wykoff, C. C., N. J. Beasley, P. H. Watson, K. J. Turner, J. Pastorek, A. Sibtain, G. D. Wilson, H. Turley, K. L. Talks, P. H. Maxwell, C. W. Pugh, P. J. Ratcliffe, and A. L. Harris. 2000. Hypoxia-inducible expression of tumor-associated carbonic anhydrases. Cancer Res. 60:7075-7083. [PubMed] [Google Scholar]
- 57.Wykoff, C. C., C. W. Pugh, A. L. Harris, P. H. Maxwell, and P. J. Ratcliffe. 2001. The HIF pathway: implications for patterns of gene expression in cancer. Novartis Found. Symp. 240:212-225. [DOI] [PubMed] [Google Scholar]
- 58.Yang, E., J. Zha, J. Jockel, L. H. Boise, C. B. Thompson, and S. J. Korsmeyer. 1995. Bad, a heterodimeric partner for Bcl-xL and Bcl-2, displaces Bax and promotes cell death. Cell 80:285-291. [DOI] [PubMed] [Google Scholar]
- 59.Zhang, L., J. Yu, B. H. Park, K. W. Kinzler, and B. Vogelstein. 2000. Role of BAX in the apoptotic response to anticancer agents. Science 290:989-992. [DOI] [PubMed] [Google Scholar]
- 60.Zhong, H., F. Agani, A. A. Baccala, E. Laughner, N. Rioseco-Camacho, W. B. Isaacs, J. W. Simons, and G. L. Semenza. 1998. Increased expression of hypoxia-inducible factor-1α in rat and human prostate cancer. Cancer Res. 58:5280-5284. [PubMed] [Google Scholar]
- 61.Zhong, H., A. M. De Marzom, E. Laughnerm, M. Lim, D. A. Hilton, D. Zagzag, P. Buechler, W. B. Isaacs, G. L. Semenza, and J. W. Simons. 1999. Overexpression of hypoxia-inducible factor 1alpha in common human cancers and their metastases. Cancer Res. 59:5830-5835. [PubMed] [Google Scholar]
- 62.Zhong, H., and J. W. Simons. 1999. Direct comparison of GAPDH, beta-actin, cyclophilin, and 28S rRNA as internal standards for quantifying RNA levels under hypoxia. Biochem. Biophys. Res. Commun. 259:523-526. [DOI] [PubMed] [Google Scholar]