Abstract
Human tumors result from an evolutionary process operating on somatic cells within tissues, whereby natural selection operates on the phenotypic variability generated by the accumulation of genetic, genomic and epigenetic alterations. This somatic evolution leads to adaptations such as increased proliferative, angiogenic, and invasive phenotypes. In this review we outline how cancer genomes are beginning to be investigated from an evolutionary perspective. We describe recent progress in the cataloging of somatic genetic and genomic alterations, and investigate the contributions of germline as well as epigenetic factors to cancer genome evolution. Finally, we outline the challenges facing researchers who investigate the processes driving the evolution of the cancer genome.
Organismal and somatic evolution
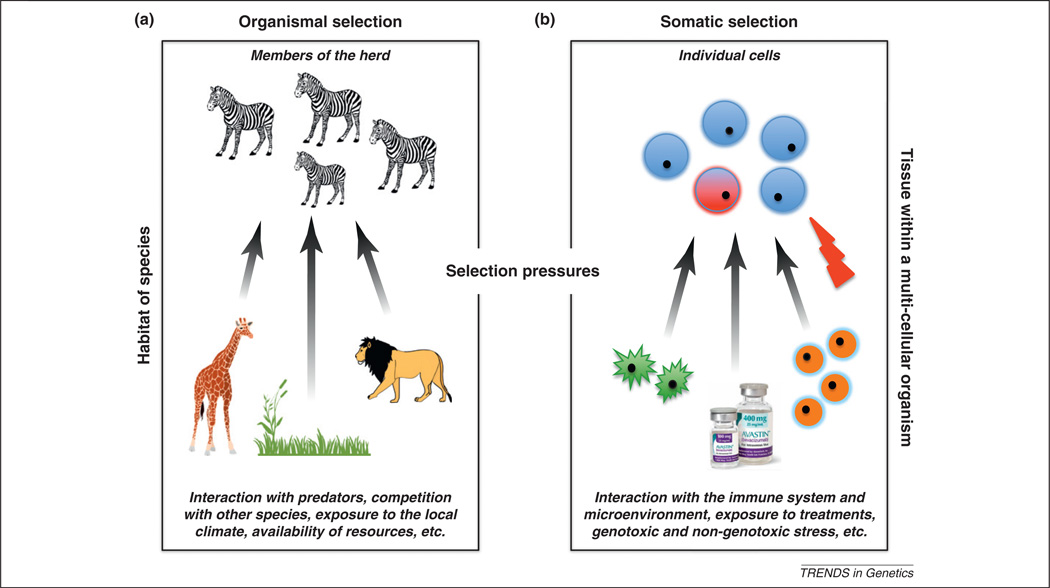
Evolution by means of natural selection is most commonly discussed in conjunction with organismal change over generations, thereby explaining phenomena such as speciation, adaptation, and invasion of new habitats [1]. The same evolutionary framework can be applied at the cellular level within an organism to understand processes driven by somatic evolution, such as tumorigenesis. Precisely as predation and competition impose selective pressures on organisms in their habitats, the human immune system and the surrounding microenvironment exert selective pressures on cells within a tissue (Figure 1). A tissue compartment with in a multicellular organism represents a complex environment with diverse mechanisms to constrain cells from abnormal proliferation, death, selfrenewal, recruitment of blood supply, and dissemination [2]. On occasion, however, changes in the DNA sequence, genome structure or epigenetic regulation may arise and enable an individual cell to overcome these constraints, thereby gaining a selective advantage over its normal counterparts. Such alterations can ‘sweep’ through the cell population and cause clone expansion [3]. The pinnacle of the somatic adaptation process is represented by tumor cells that acquire the ability to proliferate autonomously and evade selective pressures exerted by the immune system, microenvironmental interactions within tissues, exposures to genotoxic and non-genotoxic external stress, and clinical interventions and other factors (Figure 1), enabling them to invade surrounding tissues and metastasize to distant organs.
Figure 1.
Natural selection acting at the organismal and somatic levels. Multiple selective pressures are present at both the organismal (a) and somatic (b) levels. In the organismal realm, competition with other species, predation, resource limitation and others dictate the habitat boundaries and frequencies of organisms. In the somatic realm, constant immune system surveillance for abnormality, targeted therapy to inhibit division of particular cells, the constraints of the cell cycle program, the blood supply for nutrients, and many other factors determine the microenvironment within which each cell exists. Any cell that gains the capability to overcome these physiological constraints and to avoid immune system attack gains a reproductive advantage over its neighbors, thus leading to clonal expansion. An analogy in the organismal world to such clonal expansion is a growing herd of zebras that out-competes other grazers and successfully evades predators. An important distinction between organismal and somatic evolution is that adaptive traits are inherited via sexual and asexual reproduction, respectively.
But how do cancer cells evolve the properties enabling them to survive and proliferate despite the mechanisms that have emerged to constrain them? What are the genomic bases for cancer initiation and progression? And are cancer-associated phenotypes gained via gradual accumulation of alterations or through punctuated, catastrophic events? Although the notion that cancer progression is an evolutionary process has been recognized for decades [4–6], this framework of thinking has recently started to gain new appreciation among cancer biologists and clinicians. Understanding the forces that govern cancer initiation and lead to the acquisition of resistance to anti-cancer therapy represent very pressing issues in medical oncology. Here, we review recent progress in elucidating the evolution of cancer cells on the genetic, genomic, and epigenetic levels, and provide new perspectives on the experimental and theoretical challenges of cancer genome research.
Somatic genetic evolution of cancer
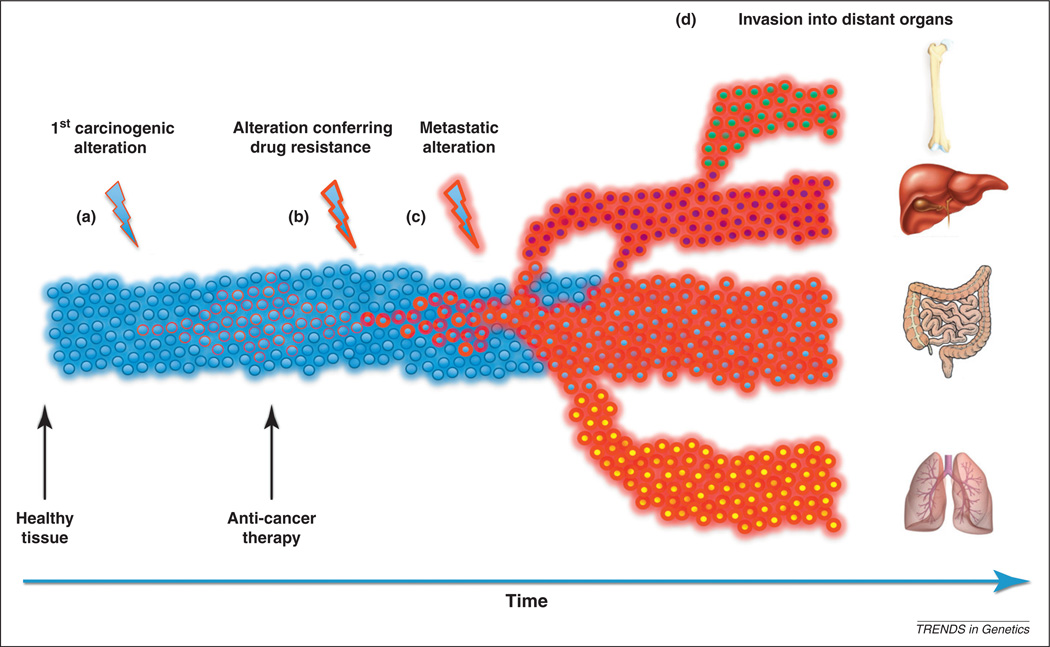
Somatic genomic alterations are DNA sequence aberrations that have accumulated in the genome of a cell during the lifespan of the patient; these include point mutations, deletions, gene fusions, gene amplifications and chromosomal rearrangements. In some cases these mutations are benign, leading to somatic mosaicism in healthy individuals [7], whereas in other cases they lead to diseases such as cancer. In 1914, chromosomal abnormalities were first suggested to be causally related to tumorigenesis [8]. This notion was confirmed half a century later with the arrival of sophisticated cytogenetic techniques, which facilitated the identification of the first specific translocation driving human neoplasia: the t(9;22)(q24;q11) translocation responsible for generating the Philadelphia chromosome [9], resulting in the BCR–ABL fusion oncoprotein causing leukemiagenesis [9,10]. Since these groundbreaking findings, somatic alterations of nearly 500 genes [11] have been linked to cancer initiation and progression. Traditionally, these alterations have been considered to arise sequentially [12] and to give rise to the progressively more aggressive and invasive phenotypes observed during tumorigenesis (Figure 2).
Figure 2.
Cancer progression. The progression to cancer begins with the emergence of the first genetic, epigenetic or genomic alteration in normal cells (blue circles) and usually ends with a large population of malignant cells invading multiple tissues (a). This process involves the evolution of multiple ‘novel’ cellular traits. Most somatic alterations in epithelial cells lining the colon, for instance, are not advantageous and will disappear with the death of a cell. Occasionally, an alteration that increases the proliferation rate of a cell arises, allowing this cell to increase in number. This population of ‘rogue’ cells can decline with the onset of anti-cancer therapy; however, the arrival of an alteration conferring drug resistance reverses the effects of treatment and allows new growth (b). In some cases, resistance to an anti-cancer drug may already be present in a small subset of tumor cells. In such a scenario, the population of sensitive cancer cells will decline and eventually be replaced by drug-resistant cells. Further alterations may be necessary to enable tumor cells to metastasize (c) and spread to other tissues (d); these changes might arise before diagnosis and treatment or, as shown in this example, thereafter.
The advent of next-generation sequencing technologies has stimulated rapid cataloging of all alterations in cancer genomes and has enabled researchers to look at large-scale genome events such as chromosomal lesions and copy-number variations as well as small-scale aberrations represented by point mutations, small insertions and deletions. The analysis of glioblastoma (GBM) and ovarian cancer samples by the Cancer Genome Atlas project, for example, revealed interesting differences between these two cancers [12,13]. At the genome level, ovarian cancer and glioblastoma stand in marked contrast regarding somatic copynumber alterations (SCNAs). High-grade ovarian cancers have a larger degree of both chromosome-arm and focal SCNAs when compared to GBMs. At the gene level, both cancers show tumor protein 53 (TP53) mutations in a majority of the samples; the retinoblastoma (RB) and RTK/ RAS/pi(3)K pathways are the most frequently altered signaling pathways in ovarian cancer and GBM, respectively. Many other whole-genome analyses of cancer cells have been completed to date, and the collective picture of the mutation spectra provides important insights into the dynamics of cancer genomes. Several trends are emerging:
The number and patterns of somatic alterations vary dramatically across cancer types. At one extreme, childhood medulloblastomas can harbor fewer than ten genomic alterations [14], whereas over 50 000 somatic changes have been observed in primary lung adenocarcinoma samples [15]. These diverging patterns may be accounted for by a combination of factors such as the total number of cell divisions a clone has undergone, the cell-intrinsic rate at which alterations are accumulated, the selective environment of a cell, and the presence and effects of exogenous mutagens. For instance, the total number of cell divisions undergone by adult lung cancer cells is expected to be larger than that generating a childhood medullo-blastoma cell. The intrinsic mutation rate of a cell depends on the status of its telomeres and DNA repair enzymes; for instance, colorectal or pancreatic cancer cells with mutations in mutL homolog (MLH1), a gene involved in DNA repair, suffer increased frequencies of nucleotide changes and small insertions and deletions [16]. The neighborhood of copy-number alteration boundaries often have increased single-nucleotide variations, probably due to the deployment of error-prone DNA polymerases for trans-lesion synthesis [17]. Eroded telomeres, on the other hand, contribute to chromosomal rearrangements and instability [18]. Additionally, exposure to cell-extrinsic mutagens such as tobacco smoke or ultraviolet (UV) radiation further escalates the rate of alterations, leaving behind distinctive signatures of DNA damage [19].
The mutation rate may not be constant over time. Recent evidence suggests that the notion of cancer cells gradually accumulating genomic alterations in a stepwise manner may be too simplistic. Investigations of cancer cell karyotypes have led researchers to suggest a genome-based stochastic evolutionary model of cancer, observing that cancer cells switch between specific punctuated and stepwise phases during the course of their genome evolution [20,21]. The punctuated phase is characterized by large-scale genome alterations and tremendous heterogeneity among cells, driven by genomic instability. The stepwise phase, by contrast, is marked by relatively low-level genome alterations such as gene mutations or small rearrangements and is comparatively more stable, generating more homogeneous clones. The dynamics of switching from the punctuated to the stepwise phase of genome evolution was hypothesized to reflect complex phenomena such as the establishment of immortality, drug resistance, or overcoming of particular selective pressures. Later on, detailed studies of chromosomal rearrangements in chronic lymphocytic leukemia and osteosarcoma samples led to the hypothesis that cancer cells may undergo ‘catastrophic’ events, termed chromothripsis, wherein the genome acquires a large number of rearrangements within a single breakage–fusion event [22]. The fact that cells can tolerate this magnitude of genomic reshuffling implies a level of selective advantage for such events. These rapid changes in cancer genomes are more reminiscent of punctual rather than gradual evolution, as proposed in several recent studies [22,23]. In organismal evolution, the punctuated equilibrium hypothesis relates to a controversial claim made about the pattern of morphological change in the fossil record [24]. According to this idea, most phenotypic traits change relatively little over long periods of time, but when phenotypic change does occur, it tends to result in a radical and rapid transformation [25,26]. The hypothesis of a punctuated equilibrium does not exclude intermediate phenotypic states, but rather advocates a rapid transition from one equilibrium state to another, and hence the two competing models are not necessarily mutually exclusive. The question remains concerning the extent to which this idea also applies to cancer genome evolution. Future investigations will elucidate the mechanisms, frequency and general applicability of these concepts to cancer in general.
There is a relatively limited number of recurrent alterations within and across cancer types. With several prominent exceptions, such as frequent rearrangements of the erythroblastosis virus E26 oncogene homolog (ERG) in prostate cancer [27], characteristic TP53 mutations in high-grade ovarian cancers [13] and universal fusions in the breakpoint cluster region c-Abl1 oncogene (BCR-ABL1) in chronic myeloid leukemia [28], there is a relatively limited number of recurrent alterations within and across cancer types. For instance, alterations in TP53, Kirsten rat sarcoma viral oncogene (KRAS), murine sarcoma viral oncogene homolog B1 (BRAF) and phosphoinositide-3-kinase catalytic α polypeptide (PIK3CA) have been found across many cancer types, but their frequency within each type is relatively small [11,29]. A case in point is PIK3CA, the most frequently altered gene in breast cancer, which is, however, altered in only 26% of all breast cancer samples (n > 5000) [29]. Similarly, serine/threonine kinase 11 (STK11) is the most frequently mutated gene in cervical cancer but is found in only 14% of samples (n > 200) [29]. It is thus more meaningful to cluster altered genes based on gene family membership or affected signaling pathways. This approach has identified several gene families prominently altered across cancer types and subtypes, most notably the protein kinase gene family and the RAS-regulated signaling pathways controlling apoptosis, cell differentiation, migration, and others [12]. These findings highlight the variability among cancer cells within the same sample and across patients, and support the necessity of single-cell studies, as discussed below.
Identifying causal genetic events
Given that cancer genomes often harbor hundreds or thousands of genetic alterations, how many of these play a causal role in tumorigenesis? Genetic lesions accumulating in the genome of a cell create heritable phenotypic variation, which is subject to natural selection. Distinguishing between genomic alterations that are positively selected during tumorigenesis (‘drivers’) and those that tag along (‘passengers’) is a primary focus of cancer genomics research. Owing to the lack of recombination in mitotically dividing cancer cells and the limited ability to analyze the genomes of single cells, traditional population genetics approaches [30] are not appropriate to search for drivers. Instead, methods designed to uncover driver alterations aim to identify genes or regions in which changes occur across independent cancer samples and more frequently than expected by chance [31–33]. To aid in the identification of genes that are mutated in cancer cells more frequently than expected in the absence of selection, a method was devised where the probability for each gene to be mutated was calculated given the local background mutation level [34], while correcting for the number of nucleotides sequenced, the effects of nucleotide composition, and the number of tumor samples analyzed. The resulting cancer mutation prevalence (CaMP) score yielded 122 and 69 candidate driver genes in breast and colorectal cancers, respectively. This method has instigated extensive discussion about such approaches [35–37]. The dN/dS ratio test, commonly used in molecular evolution studies, has also been implemented in cancer genome analyses to identify the relative proportions of driver and passenger alterations [38,39]. This test relies on the assumption that natural selection will skew the frequency of non-synonymous mutations in cancer cells as a result of their potential effects on protein function. A high dN/dS ratio suggests the presence of positive selection and thus driver mutations. A recent study [39] reported >1000 base substitutions in protein kinase genes across various cancer types, with a dN/dS ratio of 1.66. This analysis suggests that, indeed, the protein kinome (the full complement of human protein kinases) harbors many drivers in cancer samples, with 20% of all substitutions representing driver mutations. Similarly, the GISTIC methodology (genomic identification of significant targets in cancer [40]) aims to identify significant copy-number alteration events. This method was recently applied to data of > 3000 cancer samples from 26 tissue types, identifying 158 genomic hotspots that are prone to focal somatic copy-number alterations and are enriched for several gene families, including the B-cell leukemia/lymphoma 2 (BCL2) family of apoptosis regulators [32]. Another approach [41] utilized information from sequence alignments of a large number of homologs to calculate a ‘functional impact score’, which was used to categorize somatic DNA mutations based on the extent of its impact upon protein function. Using this approach, a ranked list of the top ~1000 genes was assembled with mutations affecting protein function relevant to cancer [41]. A variety of additional approaches have been implemented to uncover drivers by, for example, integrating copy-number variation and gene expression [31] and phylogenetic analysis [23,42].
Germline predisposition to cancer
The task of identifying causative changes in cancer genomes is further complicated by the intricate interplay of somatic mutations and inherited alterations. Three recent studies [43–45] have made progress towards identifying germline susceptibility alleles, showing that Janus kinase 2 V617F (JAK2V617F) mutations in myeloproliferative neoplasms (MPNs) are preferentially found within a particular inherited JAK2 haplotype. This haplotype block is tagged by a single-nucleotide polymorphism (SNP) close to the JAK2V617F mutation, and heterozygotes for this haplotype were demonstrated to be significantly more likely to acquire the JAK2 mutation in cis to the predisposition allele as compared to the other chromosome. According to these studies, this single haplotype accounts for about half of the MPN risk attributed to inherited factors. The effect size is larger than that of most susceptibility alleles identified in genome-wide association studies, and is comparable in effect size to that of germline ataxia telangiectasia mutated (ATM) mutations in breast cancer [46]. Possible mechanisms explaining this interplay between germline and somatic mutations remain to be identified. So far, two hypotheses have been suggested. First, the haplotype could confer a hypermutator property to the JAK2 locus and could thus enable the rapid accumulation of JAK2V617F mutations in cells of individuals carrying this susceptibility allele. This hypothesis is supported by the observation that three patients harbored clones with JAK2V617F mutations on different alleles [45], suggesting that the mutation arose at least twice in these individuals. The biological basis for this hypermutator phenotype remains to be identified. Second, the haplotype could lead to a different fitness effect of cells carrying the JAK2V617F mutation once it emerges, thus leading to a larger chance that the clone harboring this mutation survives stochastic fluctuations and leads to clinically manifest disease. The hypothesis of a functional difference is supported by the finding that the numbers of granulocyte–macrophage progenitors in normal individuals carrying the risk allele are smaller than in those without the allele. Again, the biological underpinnings of how such differences would lead to fitness effects remain to be elucidated. These and many more genome-wide association studies of melanoma [47], gastric [48], breast [49], ovarian [50]and other cancers suggest that the generation of somatic alterations in cancer genomes might have germline underpinnings, thus complicating the search for novel driver alleles for therapeutic targeting.
Epigenetic determinants of cancer genome evolution
DNA methylation and histone modifications play a crucial role in gene regulation and the spatial architecture of the genome [51]. It has been known for decades that DNA methylation patterns change dramatically in cancer cells as compared to their normal counterparts [52], and can transcriptionally silence specific tumor suppressor genes [53]. With the recent development of the Chip-seq [54] and high-throughput bisulfate sequencing [55] technologies, whole genomes can now be interrogated at single-nucleotide resolution to gain insights about histone modifications and DNA methylation, respectively. Growing evidence suggests that epigenetic changes – which involve extensive alterations of chromatin and lead to altered expression profiles of particular genes without affecting the primary DNA sequence – also play a key role in cancer initiation and progression [51].
In a classic study, genome-wide loss of DNA methylation was observed to be one of the earliest epigenetic alterations in cancer [52]. Since then, many reports have documented that synthetically induced epigenetic changes can trigger tumorigenesis. Embryonic stem cells from DNA methyltransferase 1 (Dnmt1) knockout mice, for instance, display an increased rate of accumulating genomic alterations [56]. Furthermore, genetically engineered mice carrying hypomorphic Dnmt1, which leads to a global erosion of DNA methylation, spontaneously develop tumors [57]. These findings highlight the fact that epigenetic abnormalities may have the potential to increase the risk of tumorigenesis.
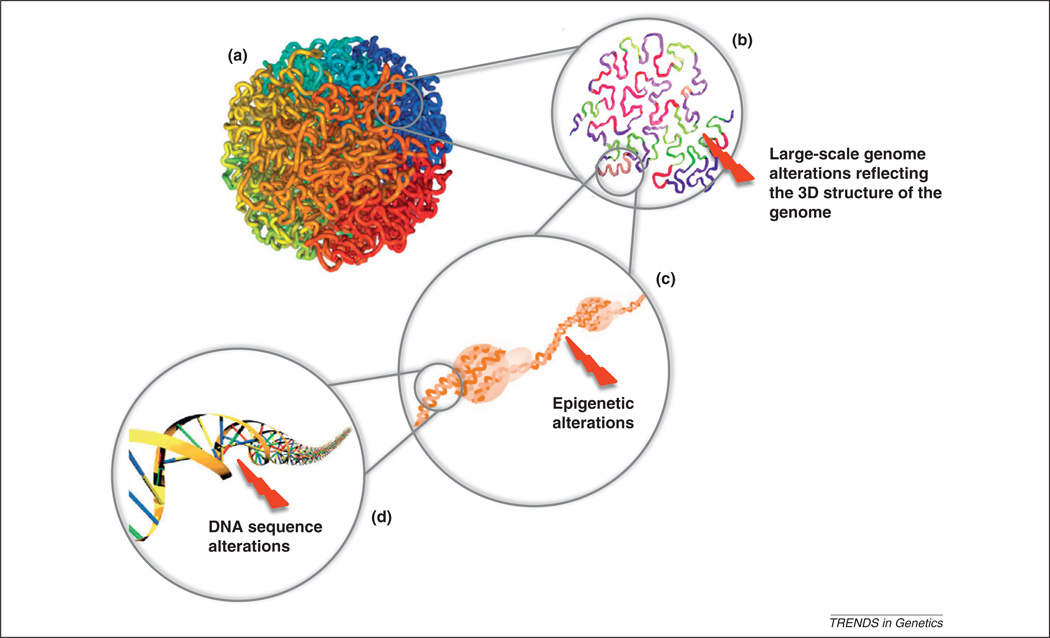
Epigenetic abnormalities commonly arise in healthy cells of somatic tissues; in fact, the frequency of epigenetic changes in mice is one to two orders of magnitude larger than the rate of somatic DNA sequence alterations [58], and the extent of such abnormalities typically increases with age [59]. Epigenetic abnormalities can predispose cells to genomic instability and an increased risk of accumulating the genomic alterations leading to cancer. Indeed, several of the frequently mutated cancer genes [e.g. Dnmt1 and enhancer of zeste homolog 2 (Ezh2)] are epigenetic modifier enzymes [29]. An aberrant epigenome can introduce genomic alterations via multiple mechanisms (Figure 3):
Epimutation of DNA repair genes. Several pieces of evidence suggest that epigenetic alterations of DNA repair genes contribute to tumorigenesis. For instance, epigenetic silencing of the MGMT gene, which encodes the O6-methylguanine DNA methyltransferase, inhibits repair of DNA guanosine adducts, leading to an increased ratio of G:C to A:T mutations [60]. Similarly, epigenetic silencing of another DNA repair gene, breast cancer 1 (BRCA1), is associated with a poor prognosis in breast cancer patients [61], whereas de novo constitutional MLH1 epimutations confer early-onset colorectal cancer [62].
Epimutation of genes located in signaling pathways. Emerging evidence also supports the notion that epigenetic alterations of particular signaling pathways influence tumorigenesis. For instance, methylation of the estrogen receptor CpG island is linked to the development of colorectal cancer in humans [63]. Furthermore, epigenetic silencing of secreted frizzled-related protein (SFRP) leads to an activation of the Wnt pathway, thus promoting cell proliferation during tumorigenesis [64]. Such epigenetic changes may lead to early oncogenic pathway addiction [65].
Facilitation of DNA secondary structure-mediated mutagenesis. Many DNA secondary structures, such as the G-quadruplex structure, have mutagenic potential, and epigenetic modifications can affect the stability of such structures, thus leading to an increased risk of genomic breaks at the sites of such structures [66].
Control of replication timing and nuclear architecture. DNA replication timing and higher-order chromatin structures are determined, at least partly, by epigenetic states of genomic regions, which in turn can affect the patterns of alterations in cancer genomes [67,68].
Non-coding RNA-dependent regulation of crucial pathways. It was recently demonstrated that competing endogenous mRNAs regulate the expression of phosphatase and tensin homolog (PTEN), a known tumor-suppressor gene [69]. Additionally, a micro-RNA-mediated network was found to regulate established oncogenic pathways in glioblastoma [70] and T-cell lymphoblastic leukemia [71]. These findings support the hypothesis that hitherto little-studied factors, such as non-coding RNAs, are causally implicated in tumorigenesis
Epigenetic regulation of repeat elements. Changes in the methylation patterns of long and short interspersed elements (LINEs and SINEs) are frequent in cancer cells. Interestingly, aberrant expression of satellite repeats in pancreatic and other cancers was found to be correlated with overexpression of LINE-1 elements [72], which can lead to a drastic remodeling of the cancer genome. Applying high-throughput methods to identify LINE-1 mutations, a recent study discovered 7,743 somatic LINE-1 insertions in three healthy adult human brain samples [73]. Furthermore, hypomethylation of intragenic LINE-1 can repress transcription of genes in cancer cells through argonaute 2 (AGO2) [74].
Epigenetic promotion of metastasis. Recent findings suggest that the long non-coding RNA HOTAIR reprograms the chromatin state of cancer cells, thereby promoting metastasis [75].
Figure 3.
Genetic and epigenetic determinants of cancer genome evolution. The human genome is organized into highly complex structures with multiple levels of organization. The highest level comprises chromosome packaging into the cell nucleus (a). DNA strands in close spatial proximity are more likely to interact during replication and transcription, leading to chromosomal rearrangements and gene fusions (b). Aberrant methylation and acetylation of histone tails can result in gene expression and splicing variation (c). DNA sequence alterations may modulate gene expression and change protein amino acid composition (d). Aberrations at all of these levels may influence the mutational landscape of cancer genomes.
These findings, although fascinating, represent only the beginning of the investigation into epigenetic determinants of cancer evolution.
Epigenetic changes have several interesting attributes, such as reversibility and the ability to affect the penetrance of somatic mutations. Although cancer genomes accumulate a large number of genetic alterations, some of the disease-causing genetic variants can be masked by epigenetic modifications [76]. Furthermore, epigenetic states are reversible, thereby allowing tumor cells to exploit or suppress various genetic alterations under different circumstances [51,76]. Moreover, epigenetic heterogeneity in a clonal population of cancer cells allows selection to operate dynamically on the same mutation pool. As a result, promoter methylation patterns of specific genes are often associated with sensitivity to chemotherapeutic drugs [77].
Taken together, it has become increasingly evident that epigenetic abnormalities are not only widespread in many cancer types, but that some of them, either independently or in conjunction with genetic alterations, play a crucial role in establishing some of the hallmarks of neoplasms [65,78]. These findings also contribute to the ongoing debate about the epigenetic origin of cancer [78].
Intra-tumor heterogeneity
The studies outlined above suggest that each tumor may contain a large number of individual clones, each defined by different landscapes of genetic and epigenetic alterations [79–83]. This heterogeneity is also obvious pathologically, because different cross-sections of tumor tissue often reveal diverse cell morphologies [84,85]. A series of landmark papers reported that some breast tumors consist of clones with different copy-number profiles [81,82]. Similarly, it has been observed that breast tumors are highly heterogeneous at both the genetic and cell-differentiation levels [80]. In this study, immunofluorescence in situ hybridization (immunoFISH) was used to analyze the degree of variability in individual breast tumors, finding that the extent of heterogeneity was related to patient outcome. Similar associations were also observed in a pre-malignant condition, Barrett’s esophagus, in which a larger extent of intra-sample heterogeneity was correlated with an increased risk of progression to esophageal adenocarcinoma [86]. Following up on these findings, it was recently demonstrated that breast cancers are either monoclonal, with the majority of tumor cells showing the same stable chromosome structure, or polyclonal [79]. Both the mono- and polyclonal types appeared to be equally common. Similar observations have been made in lymphoblastic leukemia, in which copy-number alteration data showed that polyclonal evolution is common in leukemiagenesis [12,13]. Furthermore, an analysis of somatic mutations in primary pancreatic tumors and their matched metastases uncovered multiple differences between the primary and the metastatic samples [83]; the authors concluded that metastatic capacity is obtained late in pancreatic cancer evolution. In glioblastoma, primary tumors consisting of clones with mutually exclusive receptor tyrosine kinase amplifications have been reported [17]. In this study, mutational data showed that these clones were derived from common precursors instead of representing independent primary tumors. These findings underscore the widespread heterogeneity within cancer samples and highlight the need to design cost-effective ways to analyze tumors at the single-cell level.
An important question in oncology is whether tumors become resistant to chemotherapy because of de novo somatic mutations or because of resistant clones already present at the time of diagnosis or surgery. From a Darwinian point of view, such resistant clones might not have a fitness advantage when untreated and might not be detectable in the tumor bulk [79,80,87,88]. A timely detection of resistant clones would, however, be essential for a reliable prediction of treatment responses and the choice of therapy. Recent technological breakthroughs have made possible the identification of single-cell heterogeneity and are likely to provide answers to such questions [23]. With current high-throughput methods, it is now possible to obtain the copy-number status of 200 cells in a single sequencing run, and this number is likely to increase dramatically in the next years [89]. Using such techniques, the landscape of copy-number alterations from a polyclonal breast tumor was inferred [79], as well as from a monoclonal breast tumor and its liver metastasis. Their analysis allowed the authors to identify as well as approximately date clonal expansion events, leading to the conclusion that tumor evolution might be driven by a very small number of such clonal expansions. Furthermore, single-cell analysis has the potential to confirm or reject the self-seeding hypothesis [90], which states that tumor cells from a metastatic site could return to the primary tumor.
Although whole-genome sequencing of single cells is currently not possible [89], promising progress has been made in single-cell transcriptomics [91,92]. Measurements of the tumor genome, transcriptome, proteome and epigenome at the single-cell level at high resolution will become a cornerstone of oncology in the future and will allow the timely detection of driver events of tumor evolution and therapeutic resistance
But what are the underlying mechanisms creating these patterns of genetic and phenotypic diversity in tumors? The answer to this question is important because the molecular mechanisms that generate tumor heterogeneity are directly linked to cancer initiation and progression. From a clinical perspective, tumor heterogeneity can dictate the response of a patient to anti-cancer therapy and increase the likelihood of drug resistance. For example, stable coexistence of genetically distinct clones has been documented in malignant brain tumors [93],in which three different tyrosine kinases were amplified in different cells in a mutually exclusive manner, bearing important clinical implications for tumor resistance to targeted therapies. Two models of tumor progression have been proposed – the cancer stem cell and the clonal evolution models, each making qualitative predictions about tumor heterogeneity. The cancer stem-cell model postulates that a tumor originates from a single (or a few) cancer stem cell, giving rise to differentiated cells with limited proliferation capability [86,94]. In this scenario, cells arising from a cancer stem cell would be genetically nearly identical, resulting in a homogeneous tumor. Several analyses of leukemia [95], breast [96] and colon cancer [97] samples support the cancer stem-cell model, showing that only a small fraction of cells are capable of inducing a tumor in mice upon transplantation. The clonal evolution model, on the other hand, states that each cancer cell gives rise to descendants with unlimited proliferation ability and that further genetic and epigenetic alterations generate phenotypic variation. Natural selection then sifts through this variation, driving expansions or contractions of particular clones based on their relative fitness [4]. These theories have recently been shown not to be mutually exclusive [12,13]. In fact, these models serve as an umbrella for more specific monoclonal and polyclonal models, all of which share the above stated assumptions but differ in the number of distinct clones that ultimately dominate the tumor cell population ([98] for more details).
Concluding remarks
We have outlined recent advances elucidating the mutational landscapes of cancer genomes at the genomic, genetic and epigenetic scales. The level of complexity, however, does not stop here. The recently proposed fractal organization of the genome is starting to reveal intriguing relationships among phenomena such as the timing of genome replication and locations of DNA breakpoints in cancer genomes [66]. These findings provide fertile ground for further investigations. Experimentally, challenges remain to obtain single-cell genomic and epigenetic profiling data, which are both rapidly obtainable and cost-effective. The ultimate dataset will contain information on DNA sequence, copy number, DNA methylation, histone modifications, gene and protein expression as well as on chromatin architecture for the same sample, obtained from single cells over a time-course of several years, both before and after the administration of therapy. Computational and mathematical modeling tools can then be applied to address a flurry of questions. For instance, the relationship between genomic, epigenomic, and higher-order chromatin structure needs to be investigated, potentially leading to insights into the regulation of genomic stability and gene expression by the epigenome. Furthermore, insights into the evolutionary dynamics of a tumor can be gained by analyzing single-cell-based information, leading to better diagnostic and prognostic tools as well as novel drug targets.
Unique opportunities to gain new insights into the evolution of cancer genome can also arise from unlikely sources, such as from the study of organisms that successfully evade cancer altogether. Compared to mammals of similar body size, naked mole rats have an exceptionally long lifespan, slow senescence, and are resistant to spontaneous and experimentally induced tumorigenesis [99]. Recent sequencing of the naked mole rat genome is starting to shed light onto the molecular basis of these extraordinary traits and might provide a deeper understanding of cancer resistance mechanisms [100]. In particular, distinct modifications of proteins involved in telomerase lengthening, DNA repair and replication, cell cycle, and DNA torsion control during transcription were identified. These discoveries mark the beginning of efforts to decipher how these insights can be implemented in cancer treatments. The future might hold investigations of similar exceptions to the rule of cancer as a general fate of long-lived multicellular organisms, and will open up the opportunity for further evolutionary inquiries into tumorigenesis.
References
- 1.Futuyma DJ. Evolutionary Biology. Sinauer Associates; 1997. [Google Scholar]
- 2.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 3.Campbell PJ, et al. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature. 2010;467:1109–1113. doi: 10.1038/nature09460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 5.Cairns J. Mutation selection and the natural history of cancer. Nature. 1975;255:197–200. doi: 10.1038/255197a0. [DOI] [PubMed] [Google Scholar]
- 6.Fisher JC. Multiple-mutation theory of carcinogenesis. Nature. 1958;181:651–652. doi: 10.1038/181651b0. [DOI] [PubMed] [Google Scholar]
- 7.De S. Somatic mosaicism in healthy human tissues. Trends Genet. 2011;27:217–223. doi: 10.1016/j.tig.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 8.Boveri T. Zur Frage der Entstehung Maligner Tumoren. Gustav Fisher; 1914. [Google Scholar]
- 9.Rowley JD. Letter: a new consistent chromosomal abnormality in chronic myelogenous leukaemia identified by quinacrine fluorescence and Giemsa staining. Nature. 1973;243:290–293. doi: 10.1038/243290a0. [DOI] [PubMed] [Google Scholar]
- 10.Deininger MW, et al. The molecular biology of chronic myeloid leukemia. Blood. 2000;96:3343–3356. [PubMed] [Google Scholar]
- 11.Forbes SA, et al. The Catalogue of Somatic Mutations in Cancer (COSMIC) Curr. Protoc. Hum. Genet. 2008 doi: 10.1002/0471142905.hg1011s57. Chapter 10 Unit 10.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parsons DW, et al. The genetic landscape of the childhood cancer medulloblastoma. Science. 2011;331:435–439. doi: 10.1126/science.1198056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee W, et al. The mutation spectrum revealed by paired genome sequences from a lung cancer patient. Nature. 2010;465:473–477. doi: 10.1038/nature09004. [DOI] [PubMed] [Google Scholar]
- 16.Lengauer C, et al. Genetic instabilities in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 17.De S, Babu MM. A time-invariant principle of genome evolution. Natl Acad Sci U. S. A. 2010;107:13004–13009. doi: 10.1073/pnas.0914454107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sahin E, Depinho RA. Linking functional decline of telomeres, mitochondria and stem cells during ageing. Nature. 2010;464:520–528. doi: 10.1038/nature08982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olivier M, et al. TP53 mutation spectra and load: a tool for generating hypotheses on the etiology of cancer. IARC Sci. Publ. 2004:247–270. [PubMed] [Google Scholar]
- 20.Heng HH, et al. Evolutionary mechanisms and diversity in cancer. Adv. Cancer Res. 2011;112:217–253. doi: 10.1016/B978-0-12-387688-1.00008-9. [DOI] [PubMed] [Google Scholar]
- 21.Heng HH, et al. Stochastic cancer progression driven by nonclonal chromosome aberrations. J. Cell. Physiol. 2006;208:461–472. doi: 10.1002/jcp.20685. [DOI] [PubMed] [Google Scholar]
- 22.Stephens PJ, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Navin N, et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472:90–94. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eldredge N, Gould SJ. Punctuated equilibria: an alternative to phyletic gradualism. In: Schopf TJM, editor. Models in Paleobiology. Freeman Cooper; 1972. pp. 82–115. [Google Scholar]
- 25.Gould SJ, Eldredge N. Punctuated equilibria: the tempo and mode of evolution reconsidered. Paleobiology. 1977:115–151. [Google Scholar]
- 26.Gould SJ, Eldredge N. Punctuated equilibrium comes of age. Nature. 1993:223–227. doi: 10.1038/366223a0. [DOI] [PubMed] [Google Scholar]
- 27.Scheble VJ, et al. ERG rearrangement is specific to prostate cancer and does not occur in any other common tumor. Mod. Pathol. 2010;23:1061–1067. doi: 10.1038/modpathol.2010.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shtivelman E, et al. Fused transcript of abl and bcr genes in chronic myelogenous leukaemia. Nature. 1985;315:550–554. doi: 10.1038/315550a0. [DOI] [PubMed] [Google Scholar]
- 29.Forbes SA, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39:D945–D950. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Charlesworth B, Charlesworth D. Elements of Evolutionary Genetics. Roberts and Company Publishers; 2010. [Google Scholar]
- 31.Akavia UD, et al. An integrated approach to uncover drivers of cancer. Cell. 2010;143:1005–1017. doi: 10.1016/j.cell.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beroukhim R, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bignell GR, et al. Signatures of mutation and selection in the cancer genome. Nature. 2010;463:893–898. doi: 10.1038/nature08768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sjoblom T, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 35.Forrest WF, Cavet G. Comment on ‘The consensus coding sequences of human breast and colorectal cancers’. Science. 2007;317:1500. doi: 10.1126/science.1138179. [DOI] [PubMed] [Google Scholar]
- 36.Getz G, et al. Comment on ‘The consensus coding sequences of human breast and colorectal cancers’. Science. 2007;317:1500. doi: 10.1126/science.1138764. [DOI] [PubMed] [Google Scholar]
- 37.Rubin AF, Green P. Comment on ‘The consensus coding sequences of human breast and colorectal cancers’. Science. 2007;317:1500. doi: 10.1126/science.1138956. [DOI] [PubMed] [Google Scholar]
- 38.Goldman N, Yang Z. A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol. Biol. Evol. 1994;11:725–736. doi: 10.1093/oxfordjournals.molbev.a040153. [DOI] [PubMed] [Google Scholar]
- 39.Greenman C, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beroukhim R, et al. Assessing the significance of chromosomal aberrations in cancer: methodology and application to glioma. Proc. Natl. Acad. Sci. U.S.A. 2007;104:20007–20012. doi: 10.1073/pnas.0710052104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reva B, et al. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 2011;39:e118. doi: 10.1093/nar/gkr407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Y, et al. Molecular evolutionary analysis of cancer cell lines. Mol. Cancer Ther. 2010;9:279–291. doi: 10.1158/1535-7163.MCT-09-0508. [DOI] [PubMed] [Google Scholar]
- 43.Jones AV, et al. JAK2 haplotype is a major risk factor for the development of myeloproliferative neoplasms. Nat. Genet. 2009;41:446–449. doi: 10.1038/ng.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kilpivaara O, et al. A germline JAK2 SNP is associated with predisposition to the development of JAK2(V617F)-positive myeloproliferative neoplasms. Nat. Genet. 2009;41:455–459. doi: 10.1038/ng.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Olcaydu D, et al. A common JAK2 haplotype confers susceptibility to myeloproliferative neoplasms. Nat. Genet. 2009;41:450–454. doi: 10.1038/ng.341. [DOI] [PubMed] [Google Scholar]
- 46.Stratton MR, Rahman N. The emerging landscape of breast cancer susceptibility. Nat. Genet. 2008;40:17–22. doi: 10.1038/ng.2007.53. [DOI] [PubMed] [Google Scholar]
- 47.Macgregor S, et al. Genome-wide association study identifies a new melanoma susceptibility locus at 1q21.3. Nat. Genet. 2011;43:1114–1118. doi: 10.1038/ng.958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shi Y, et al. A genome-wide association study identifies new susceptibility loci for non-cardia gastric cancer at 3q13.31 and 5p13.1. Nat. Genet. 2011;43:1215–1218. doi: 10.1038/ng.978. [DOI] [PubMed] [Google Scholar]
- 49.Haiman CA, et al. A common variant at the TERT–CLPTM1L locus is associated with estrogen receptor-negative breast cancer. Nat. Genet. 2011;43:1210–1214. doi: 10.1038/ng.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rafnar T, et al. Mutations in BRIP1 confer high risk of ovarian cancer. Nat. Genet. 2011;43:1104–1107. doi: 10.1038/ng.955. [DOI] [PubMed] [Google Scholar]
- 51.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Feinberg AP, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983;301:89–92. doi: 10.1038/301089a0. [DOI] [PubMed] [Google Scholar]
- 53.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat. Rev. Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 54.Johnson DS, et al. Genome-wide mapping of in vivo protein–DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 55.Meissner A, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen RZ, et al. DNA hypomethylation leads to elevated mutation rates. Nature. 1998;395:89–93. doi: 10.1038/25779. [DOI] [PubMed] [Google Scholar]
- 57.Gaudet F, et al. Induction of tumors in mice by genomic hypomethylation. Science. 2003;300:489–492. doi: 10.1126/science.1083558. [DOI] [PubMed] [Google Scholar]
- 58.Bennett-Baker PE, et al. Age-associated activation of epigenetically repressed genes in the mouse. Genetics. 2003;165:2055–2062. doi: 10.1093/genetics/165.4.2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bocker MT, et al. Genome-wide promoter DNA methylation dynamics of human hematopoietic progenitor cells during differentiation and aging. Blood. 2011;117:e182–e189. doi: 10.1182/blood-2011-01-331926. [DOI] [PubMed] [Google Scholar]
- 60.Esteller M, et al. Promoter hypermethylation of the DNA repair gene O(6)-methylguanine-DNA methyltransferase is associated with the presence of G:C to A:T transition mutations in p53 in human colorectal tumorigenesis. Cancer Res. 2001;61:4689–4692. [PubMed] [Google Scholar]
- 61.Chiang JW, et al. BRCA1 promoter methylation predicts adverse ovarian cancer prognosis. Gynecol. Oncol. 2006;101:403–410. doi: 10.1016/j.ygyno.2005.10.034. [DOI] [PubMed] [Google Scholar]
- 62.Goel A, et al. De novo constitutional MLH1 epimutations confer early-onset colorectal cancer in two new sporadic Lynch syndrome cases, with derivation of the epimutation on the paternal allele in one. Int. J. Cancer. 2011;128:869–878. doi: 10.1002/ijc.25422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Issa JP, et al. Methylation of the oestrogen receptor CpG island links ageing and neoplasia in human colon. Nat. Genet. 1994;7:536–540. doi: 10.1038/ng0894-536. [DOI] [PubMed] [Google Scholar]
- 64.Suzuki H, et al. Epigenetic inactivation of SFRP genes allows constitutive WNT signaling in colorectal cancer. Nat. Genet. 2004;36:417–422. doi: 10.1038/ng1330. [DOI] [PubMed] [Google Scholar]
- 65.Baylin SB, Ohm JE. Epigenetic gene silencing in cancer – a mechanism for early oncogenic pathway addiction? Nat. Rev. Cancer. 2006;6:107–116. doi: 10.1038/nrc1799. [DOI] [PubMed] [Google Scholar]
- 66.De S, Michor F. DNA secondary structures and epigenetic determinants of cancer genome evolution. Nat. Struct. Mol. Biol. 2011;18:950–955. doi: 10.1038/nsmb.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.De S, Michor F. DNA replication timing and long-range DNA interactions predict mutational landscapes of cancer genomes. Nat. Biotechnol. 2011;29:1103–1108. doi: 10.1038/nbt.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fudenberg G, et al. High order chromatin architecture shapes the landscape of chromosomal alterations in cancer. Nat. Biotechnol. 2011;29:1109–1113. doi: 10.1038/nbt.2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tay Y, et al. Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell. 2011;147:344–357. doi: 10.1016/j.cell.2011.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sumazin P, et al. An extensive microRNA-mediated network of RNA-RNA interactions regulates established oncogenic pathways in glioblastoma. Cell. 2011;147:370–381. doi: 10.1016/j.cell.2011.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mavrakis KJ, et al. Genome-wide RNA-mediated interference screen identifies miR-19 targets in Notch-induced T-cell acute lymphoblastic leukaemia. Nat. Cell Biol. 2010;12:372–379. doi: 10.1038/ncb2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ting DT, et al. Aberrant overexpression of satellite repeats in pancreatic and other epithelial cancers. Science. 2011;331:593–596. doi: 10.1126/science.1200801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Baillie JK, et al. Somatic retrotransposition alters the genetic landscape of the human brain. Nature. 2011;479:534–537. doi: 10.1038/nature10531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Aporntewan C, et al. Hypomethylation of intragenic LINE-1 represses transcription in cancer cells through AGO2. PLoS ONE. 2011;6:e17934. doi: 10.1371/journal.pone.0017934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gupta RA, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bjornsson HT, et al. An integrated epigenetic and genetic approach to common human disease. Trends Genet. 2004;20:350–358. doi: 10.1016/j.tig.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 77.Toyota M, et al. Cancer epigenomics: implications of DNA methylation in personalized cancer therapy. Cancer Sci. 2009;100:787–791. doi: 10.1111/j.1349-7006.2009.01095.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Feinberg AP, et al. The epigenetic progenitor origin of human cancer. Nat. Rev. Genet. 2006;7:21–33. doi: 10.1038/nrg1748. [DOI] [PubMed] [Google Scholar]
- 79.Navin N, et al. Inferring tumor progression from genomic heterogeneity. Genome Res. 2010;20:68–80. doi: 10.1101/gr.099622.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Park SY, et al. Cellular and genetic diversity in the progression of in situ human breast carcinomas to an invasive phenotype. J. Clin. Invest. 2010;120:636–644. doi: 10.1172/JCI40724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Teixeira MR, et al. Clonal heterogeneity in breast cancer: karyotypic comparisons of multiple intra- and extra-tumorous samples from 3 patients. Int. J. Cancer. 1995;63:63–68. doi: 10.1002/ijc.2910630113. [DOI] [PubMed] [Google Scholar]
- 82.Torres L, et al. Intratumor genomic heterogeneity in breast cancer with clonal divergence between primary carcinomas and lymph node metastases. Breast Cancer Res. Treat. 2007;102:143–155. doi: 10.1007/s10549-006-9317-6. [DOI] [PubMed] [Google Scholar]
- 83.Yachida S, et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467:1114–1117. doi: 10.1038/nature09515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fitzgerald PJ. Homogeneity and heterogeneity in pancreas cancer: presence of predominant and minor morphological types and implications. Int. J. Pancreatol. 1986;1:91–94. doi: 10.1007/BF02788442. [DOI] [PubMed] [Google Scholar]
- 85.Komaki K, et al. Problems in histological grading of malignancy and its clinical significance in patients with operable breast cancer. Breast Cancer. 2006;13:249–253. doi: 10.2325/jbcs.13.249. [DOI] [PubMed] [Google Scholar]
- 86.Maley CC, et al. Genetic clonal diversity predicts progression to esophageal adenocarcinoma. Nat. Genet. 2006;38:468–473. doi: 10.1038/ng1768. [DOI] [PubMed] [Google Scholar]
- 87.Khalique L, et al. Genetic intra-tumour heterogeneity in epithelial ovarian cancer and its implications for molecular diagnosis of tumours. J. Pathol. 2007;211:286–295. doi: 10.1002/path.2112. [DOI] [PubMed] [Google Scholar]
- 88.Bachtiary B, et al. Gene expression profiling in cervical cancer: an exploration of intratumor heterogeneity. Clin. Cancer Res. 2006;12:5632–5640. doi: 10.1158/1078-0432.CCR-06-0357. [DOI] [PubMed] [Google Scholar]
- 89.Navin N, Hicks J. Future medical applications of single-cell sequencing in cancer. Genome Med. 2011;3:31. doi: 10.1186/gm247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Norton L, Massague J. Is cancer a disease of self-seeding? Nat. Med. 2006;12:875–878. doi: 10.1038/nm0806-875. [DOI] [PubMed] [Google Scholar]
- 91.Tan RZ, van Oudenaarden A. Transcript counting in single cells reveals dynamics of rDNA transcription. Mol. Syst. Biol. 2010;6:358. doi: 10.1038/msb.2010.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tang F, et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat. Methods. 2009;6:377–382. doi: 10.1038/nmeth.1315. [DOI] [PubMed] [Google Scholar]
- 93.Snuderl M, et al. Mosaic amplification of multiple receptor tyrosine kinase genes in glioblastoma. Cancer Cell. 2011;20:810–817. doi: 10.1016/j.ccr.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 94.Reya T, et al. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 95.Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat. Med. 1997;3:730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- 96.Al-Hajj M, et al. Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. U.S.A. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Dalerba P, et al. Phenotypic characterization of human colorectal cancer stem cells. Proc. Natl. Acad. Sci. U.S.A. 2007;104:10158–10163. doi: 10.1073/pnas.0703478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Navin NE, Hicks J. Tracing the tumor lineage. Mol. Oncol. 2010;4:267–283. doi: 10.1016/j.molonc.2010.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Liang S, et al. Resistance to experimental tumorigenesis in cells of a long-lived mammal, the naked mole-rat (Heterocephalus glaber) Aging Cell. 2010;9:626–635. doi: 10.1111/j.1474-9726.2010.00588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kim EB, et al. Genome sequencing reveals insights into physiology and longevity of the naked mole rat. Nature. 2011;479:223–227. doi: 10.1038/nature10533. [DOI] [PMC free article] [PubMed] [Google Scholar]