Abstract
DNA methylation is one of the best-characterized epigenetic modifications and has been implicated in numerous biological processes, including transposable element silencing, genomic imprinting and X chromosome inactivation. Compared with other epigenetic modifications, DNA methylation is thought to be relatively stable. Despite its role in long-term silencing, DNA methylation is more dynamic than originally thought as active DNA demethylation has been observed during specific stages of development. In the past decade, many enzymes have been proposed to carry out active DNA demethylation and growing evidence suggests that, depending on the context, this process may be achieved by multiple mechanisms. Insight into how DNA methylation is dynamically regulated will broaden our understanding of epigenetic regulation and have great implications in somatic cell reprogramming and regenerative medicine.
Eukaryotic chromatin contains a wealth of information required for the growth and development of a multicellular organism. This information is not only stored genetically in the DNA sequence itself but also epigenetically through DNA methylation and post-translational modifications of histone proteins1,2. Although every nucleotide in the genome has the potential to be transcribed3, the presence or absence of specific epigenetic marks influences gene expression, resulting in a transcriptional programme that specifies for a particular cell type. For example, in embryonic stem (ES) cells, active gene expression marks are found at pluripotent genes and repressive marks are found at lineage-specific genes. Thus, different cell types can be defined by their epigenetic and gene expression profiles.
During development, these transcriptional programmes undergo dynamic changes that ultimately lead to the production of distinct cell types and tissues that make up an organism. Accommodating such a transcriptional programme requires an epigenome that is both dynamic and flexible. Furthermore, the diversity of genetic material to be regulated necessitates the use of marks corresponding to short-term and long-term epigenetic memory, depending on the transcriptional requirements of the cell (as well as those of future generations). Developmental genes that are needed during the later stages of development are transiently held in a repressed state during early development. This is achieved through short-term epigenetic marks such as histone modifications, which can be removed before or within a few cell divisions.
By contrast, other regions of the genome are marked with epigenetic information that is stably maintained and heritable after many cell divisions. For example, imprinted genes, transposons and the inactive X chromosome require long-term silencing that is sustained throughout the development and lifespan of an organism. This is generally achieved by DNA methylation, an epigenetic mark that refers to the addition of a methyl group to the fifth carbon of base C. Because DNA methylation provides heritable, long-term silencing that is crucial for an organism, aberrant DNA methylation has been associated with cancer, imprinting-related diseases and psychiatric disorders4-7.
In mammals, DNA methylation occurs predominantly in the context of CpG (C followed by G) dinucleotides, whereas DNA methylation in plants can occur at C bases in diverse sequence contexts8. The enzymes responsible for this modification, DNA methyltransferases (DNMTs), are well characterized and conserved in mammals and plants8. DNMTs fall under two categories: de novo and maintenance9. Patterns of DNA methylation are initially established by the de novo DNA methyltransferases DNMT3A and DNMT3B during the blastocyst stage of embryonic development10,11 (FIG. 1). These methyl marks are then faithfully maintained during cell divisions through the action of the maintenance methyltransferase, DNMT1, which has a preference for hemi-methylated DNA12-14. Both the establishment and maintenance of DNA methylation patterns are crucial for development as mice deficient in DNMT3B or DNMT1 are embryonic lethal11,15 and DNMT3A-null mice die by 4 weeks of age11.
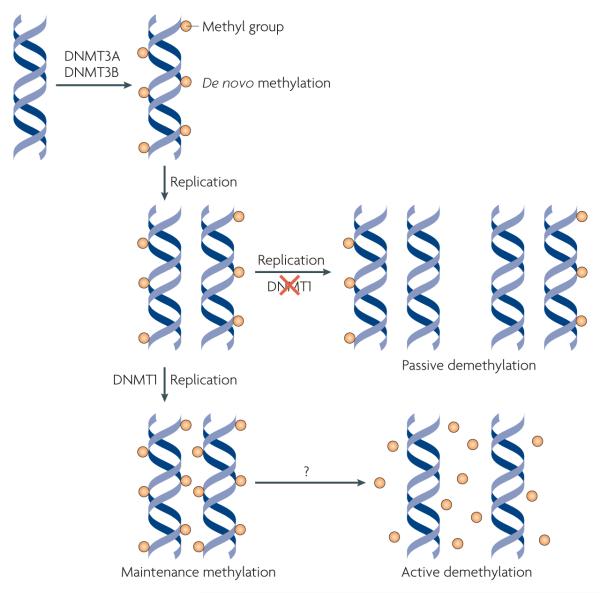
Figure 1. Mechanisms of DNa methylation and demethylation.
During early development, methylation patterns are initially established by the de novo DNA methyltransferases DNMT3A and DNMT3B. When DNA replication and cell division occur, these methyl marks are maintained in daughter cells by the maintenance methyltransferase, DNMT1, which has a preference for hemi-methylated DNA. If DNMT1 is inhibited or absent when the cell divides, the newly synthesized strand of DNA will not be methylated and successive rounds of cell division will result in passive demethylation. By contrast, active demethylation can occur through the enzymatic replacement of 5-methylcytosine (5meC) with C.
Although DNA methylation has been viewed as a stable epigenetic mark, studies in the past decade have revealed that this modification is not as static as once thought. In fact, loss of DNA methylation, or DNA demethylation, has been observed in specific contexts (see below) and can occur through active or passive mechanisms (FIG. 1). Active DNA demethylation is the enzymatic process that results in the removal of the methyl group from 5-methylcytosine (5meC) by breaking a carbon-carbon bond. By contrast, passive DNA demethylation refers to the loss of the methyl group from 5meC when DNMT1 is inhibited or absent during successive rounds of DNA replication. whereas passive DNA demethylation is generally understood and accepted, the subject of active DNA demethylation has been controversial16.
In this Review, we explore what is known about active DNA demethylation and the disputes that are embedded in this field. First, we describe the contexts in which DNA demethylation has been observed and discuss the evidence that supports an active mechanism. we then present the many enzymes that have been proposed to carry out active DNA demethylation. we conclude by discussing emerging themes and highlighting the remaining questions in this exciting field.
Evidence for active DNA demethylation
Even though DNA methylation contributes to stable, long-term and heritable silencing, it has become apparent that in some instances DNA methylation levels can rapidly change by mechanisms involving active DNA demethylation. Genome-wide and gene-specific demethylation events have both been observed, but current evidence suggests that the former only occurs at specific times during early development, whereas the latter occurs in somatic cells responding to specific signals.
Genome-wide DNA demethylation of paternal pronuclei
Prior to fertilization, mammalian gametes are at different stages of the cell cycle and their genomes are organized differently. The egg is meiotically arrested at metaphase II, resulting in a diploid genome that is packaged with histones. Mature sperm, however, have completed meiosis, but their haploid genomes are packaged with protamines instead of histones. when a sperm penetrates the zona pellucida to fertilize the egg, both gametes undergo rapid changes. The egg completes its second meiosis resulting in the extrusion of one copy of the genome as the polar body; the sperm reorganizes its genomic DNA by replacing protamines with histone proteins.
Shortly after the protamine-histone exchange, the sperm-derived paternal pronucleus undergoes genome-wide DNA demethylation17,18, an event that occurs quite rapidly within 4-8 hours post-fertilization (FIG. 2a). Although there are some disputes regarding the timing and synchrony of DNA replication in the zygote19-25, loss of DNA methylation is seen before the completion of the first cell division. Thus, it is unlikely that a passive demethylation mechanism is the cause for this observation. Furthermore, when zygotes were treated with the replication inhibitor aphidicolin, paternal genome demethylation was still detected17,26, further supporting an active demethylation mechanism.
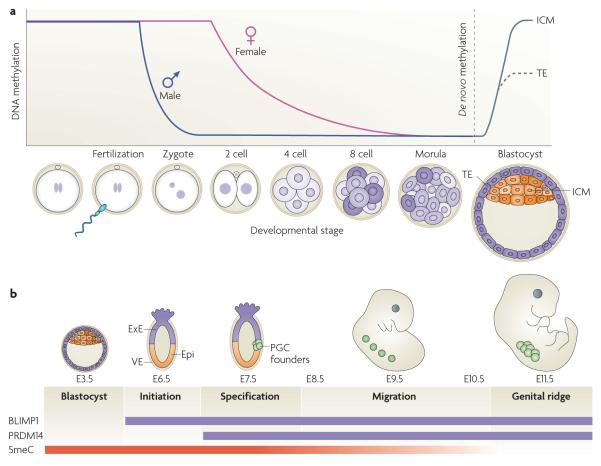
Figure 2. Dynamics of DNa methylation during development.
a | Active demethylation in the zygotic paternal genome. Shortly after a sperm fertilizes an egg, the paternal genome rapidly undergoes genome-wide active DNA demethylation and remains demethylated following multiple rounds of cell division. During this time, the maternal genome experiences gradual, passive demethylation. De novo methylation patterns are established by the DNA methyltransferases DNMT3A and DNMT3B during the development of the blastocyst. b | Active demethylation in primordial germ cells (PGCs). After implantation of the blastocyst at embryonic day 7.5 (E7.5), the extraembryonic ectoderm (ExE) and visceral endoderm (VE) produce signals that specify a subset of epiblast cells (Epi) to become PGCs. This process requires two key transcription factors, BLIMP1 (also known as PR domain zinc finger protein 1 (PRDM1)) and PDRM14, which are expressed during this stage of development. Following specification, PGC founder cells divide in the presence of the DNA methyltransferase DNMT1 and migrate towards the genital ridge. During this migration and on arrival at the genital ridge, 5-methylcytosine (5meC) is erased through an active mechanism. ICM, inner cell mass; TE, trophectoderm.
Paternal genome demethylation has been observed in many mammalian organisms, including human, mouse, rat, bovine and pig17,18,27,28, but seems to be absent from others, such as sheep29. when sheep sperm are injected into mouse oocytes, demethylation is seen in the sheep-derived paternal genome30, suggesting that the demethylating factor or factors are contributed by the oocyte. However, sheep oocytes injected with mouse sperm also resulted in demethylation of the mouse-derived paternal genome30. Although this occurs to a lesser extent compared to mouse oocytes, it is likely that factors present in the sperm or features unique to the paternal genome also contribute to demethylation. Consistent with this notion, mouse oocytes can demethylate multiple male pronuclei31, but are incapable of demethylating the additional maternal genome in parthenogenetic, gynogenetic and digynic triploid zygotes32.
Although immunostaining studies suggest that demethylation occurs globally, bisulphite sequencing indicates that some genomic regions are resistant to such a wave of demethylation. These genomic regions include imprinting control regions33, intracisternal A-particle (IAP) retrotransposons34 and centric and pericentric heterochromatin31,35. It is not clear why these genomic regions are resistant to this wave of DNA demethylation, but one possibility is that methylation of these regions may be required to ensure transcriptional repression and chromosomal stability. Additionally, the maternal genome remains methylated during this time even though it is exposed to the same cytoplasmic factors. Insight into how some regions in the paternal genome are targeted for DNA demethylation whereas other regions are resistant may also provide clues as to how the maternal genome is protected from active demethylation (BOX 1).
Box 1. Protection of the maternal genome from demethylation.
Whereas the paternal genome undergoes extensive demethylation, the maternal genome remains methylated even though it is exposed to the same cytoplasmic factors. This may be due to a mechanism that protects the maternal genome from this wave of demethylation or to a putative DNA demethylase that is specifically recruited to the paternal genome.
Sperm DNA is packaged with protamines, which are exchanged for canonical and noncanonical histones on fertilization. Interestingly, deposition of the histone variant H3.3 occurs asymmetrically, with a strong preference for the paternal pronucleus158,159. This raises the possibility that asymmetric H3.3 deposition may trigger the paternal genome-specific demethylation process. Asymmetric patterns of histone modifications have also been seen in the maternal and paternal pronuclei and may also contribute to the asymmetric demethylation process. For example, methylation, dimethylation and trimethylation at H3 Lys27 (H3K27me1, H3K27me2 and H3K27me3, respectively) and at Lys9 (H3K9me2 and H3K9me3) are clearly seen in the maternal pronucleus of zygotes, but are virtually undetectable in the paternal pronucleus159-164. Thus, the maternal genome may use a protective mechanism against demethylation by carrying specific histone variants or modifications.
Alternatively, a recent study has suggested that non-histone factors present in the oocyte might protect the maternal genome from demethylation165. Zygotes lacking stella (also known as DPPA3 and PGC7), a maternal effect gene required for early development166, exhibited demethylation of both pronuclei. Although stella can directly bind DNA in vitro, it seems to lack specificity for methylated DNA165. Therefore, how stella protects the maternal genome from demethylation remains to be determined.
The significance of zygotic paternal genome DNA demethylation is unclear at present. Genome-wide demethylation may facilitate transcriptional activation of the paternal genome36, which has been reported to occur before transcriptional activation of the maternal genome in some species37. Although some transposable elements and repeat sequences have been identified to be resistant to DNA demethylation, it is likely that others are still targets of DNA demethylation, given that these types of sequences account for half of the genome. whether demethylation of transposable elements and repeat sequences results in their reactivation and, if so, what the significance of their reactivation is remains to be determined.
Genome-wide DNA demethylation of primordial germ cells
After fertilization, the one-cell zygote undergoes several cell divisions that ultimately lead to formation of the blastocyst. During this developmental period, the maternal genome undergoes passive DNA demethylation (FIG. 2a) — a gradual loss of DNA methylation occurs with each cell division38 in a replication-dependent manner39. Consistent with this, maternally contributed DNMT1 (also known as DNMT1o) is excluded from the nucleus40. Although passive DNA demethylation seems to affect a large part of the genome, imprinted genes still retain their methylation marks. Recent genetic studies indicate that maternal and zygotic DNMT1 (ReF. 41) and the zinc finger protein ZFP57 (ReF. 42) are required to maintain the DNA methylation imprints during pre-implantation development.
At embryonic day 7.5 (E7.5), signals originating from the extraembryonic ectoderm and the visceral endoderm instruct a subset of posterior epiblast cells to become primordial germ cells (PGCs). Specification of PGCs involves the BMP4 and BMP8 signalling pathway and activation of transcription factors BLIMP1 (also known as PR domain zinc finger protein 1 (PRDM1)) and PRDM14 (REFs 43,44). These founder cells of the germ line begin to migrate at E8.5 and arrive at the genital ridge at E11.5. At the beginning of their specification and migration, PGCs are thought to have the same epigenetic marks as other epiblast cells. However, by the time they arrive at the genital ridge, many of these marks including DNA methylation have been erased45-47 (FIG. 2b). Given that PGCs have undergone several cell cycles in the presence of DNMT1, this demethylation event is considered to be active. It is thought that global demethylation, including that of imprinted genes, takes place so that new DNA methylation patterns can be re-established, although experimental evidence supporting this remains to be shown.
Loci-specific active demethylation in somatic cells
Active DNA demethylation has also been reported in somatic cells, but only at specific genomic loci in response to certain signals. For example, within 20 minutes of stimulation, activated T lymphocytes undergo active demethylation at the interleukin-2 promoter-enhancer region in the absence of DNA replication48. Locus-specific demethylation has also been reported to occur at the promoter of brain-derived neurotrophic factor (BDNF)49, the protein product of which is important for adult neural plasticity (FIG. 3a). In unstimulated neurons, the BDNF promoter is methylated, allowing for the recruitment of the repressive meC-binding protein, MeCP2. when depolarized with KCl, BDNF is upregulated, coinciding with the release of MeCP2 and demethylation of the promoter49. Because this event takes place in post-mitotic neurons, active demethylation is thought to be the underlying mechanism. In addition to T cells and neurons, active DNA demethylation has been reported to take place during nuclear hormone-regulated gene activation (FIG. 3b). For example, the pS2 (also known as TFF1) promoter exhibits periodic methylation and demethylation that coincides with cyclical binding of oestrogen receptor-α (Erα) and expression of pS2 (REFs 50,51). Similarly, active DNA demethylation occurs at the cytochrome p450, subfamily 27B, polypeptide 1 (CYP27B1) promoter in response to parathyroid hormone (PTH)52. These studies suggest that DNA methylation may not function solely as a long-term silencing mark, but could also function in the dynamic regulation of genes that require rapid responses to specific stimuli.
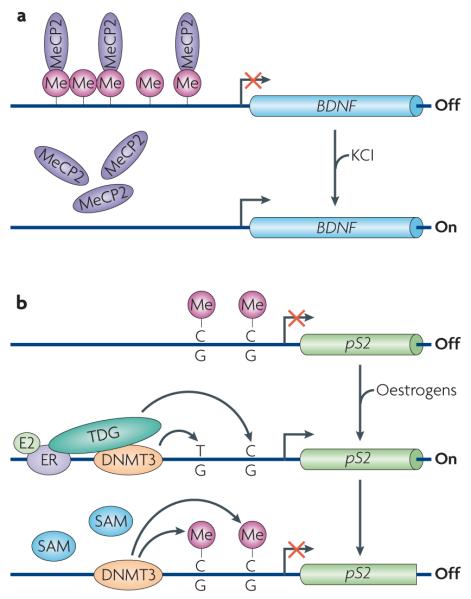
Figure 3. Locus-specific active DNa demethylation in somatic cells.
a | Active demethylation at the brain-derived neurotrophic factor (BDNF) promoter. In neurons, BDNF is maintained in a repressed state through DNA methylation and binding of the repressive methylcytosine (meC)-binding protein MeCP2. On depolarization with KCl, DNA methylation and MeCP2 binding are lost, concomitant with increased BDNF expression. This demethylation event is considered to be active because it occurs in post-mitotic neurons. b | Active demethylation at nuclear receptor target promoters. The promoter of the oestrogen receptor (ER) target gene pS2 (also known as TFF1) undergoes cyclical rounds of methylation and demethylation that correspond to the repression and expression of the gene, respectively. Transcriptional activation of pS2 occurs in the presence of oestrogens (E2) and coincides with demethylation of the promoter. This is achieved by deamination of 5meC by DNA methyltransferase 3 (DNMT3) followed by base excision repair (BER) of the T•G mismatch by T DNA glycosylase (TDG). To revert to repression, DNMT3 re-methylates the promoter. Although DNMT3 is involved in both methylation and demethylation, it is important to note that DNMT3 can only carry out the deamination step in the absence or at low concentrations of the methyl donor S-adenosylmethionine (SAM).
Mechanisms of active DNA demethylation
The importance of DNA methylation in diverse biological processes coupled with the observations of active DNA demethylation in embryonic development and somatic cells have led to extensive efforts in identifying DNA demethylases. DNA demethylase activity was first reported in murine erythroleukaemic nuclear extracts53. Although it was determined that 5meC was ultimately replaced by C in a replication-independent manner, this activity has not been further characterized. A DNA demethylase activity was also seen in rat myoblasts54. However, its sensitivities towards RNase and protease treatments were conflicting55 and this activity was not pursued further.
Since then, several studies have led to the proposal of various mechanisms by which active DNA demethylation can occur. These include: enzymatic removal of the methyl group of 5meC, base excision repair (BER) through direct excision of 5meC, deamination of 5meC to T followed by BER of the T•G mismatch, nucleotide excision repair (NER), oxidative demethylation and radical S-adenosylmethionine (SAM)-based demethylation.
Enzymatic removal of the methyl group of 5meC
The simplest way to achieve DNA demethylation is through enzymatic removal of the methyl group of 5meC. This requires an enzyme with enormous catalytic power because of the strength of the carbon-carbon bond that needs to be broken. Methyl-CpG-binding domain protein 2 (MBD2) was the first reported protein to carry out this reaction. It did not require any specific cofactors, and was proposed to release methanol56. This thermodynamically unfavourable mechanism was hotly contested for several reasons. First, previous studies had shown that MBD2 can stably bind methylated DNA57,58, making it unclear how binding could occur if MBD2 was so efficient at removing the methyl group. Further concerns were raised when MBD2-null mice were not only viable, but also exhibited normal methylation patterns59. Importantly, the paternal pronucleus of MBD2-null zygotes still exhibit normal demethylation31. These observations, coupled with the fact that no other laboratories could reproduce the reported enzymatic activity, have raised serious doubts on the capacity of MBD2 to serve as a DNA demethylase. Regardless of the controversy surrounding MBD2, it is still conceivable that a bona fide DNA demethylation mechanism exists. In fact, numerous histone demethylases that can break a carbon-nitrogen bond have recently been discovered60,61. Although carbon-carbon bonds are inherently more difficult to break than carbon-nitrogen bonds, enzymes that have the capacity to do so have been reported in the thymidine salvage pathway62 and the cholesterol synthesis pathway63.
BER through direct excision of 5meC
It has been proposed for some time that DNA demethylation can be achieved through the BEr DNA repair pathway (FIG. 4a). This type of repair involves a DNA glycosylase that removes the target base resulting in an abasic (apurinic and apyrimidinic (AP)) site. The DNA backbone is subsequently nicked by an AP lyase activity to generate a 5′ phosphomonoester and a 3′ sugar phosphate residue. An AP endonuclease then removes the 3′ sugar group leaving a single nucleotide gap that is ultimately filled in by DNA repair polymerases and ligases64.
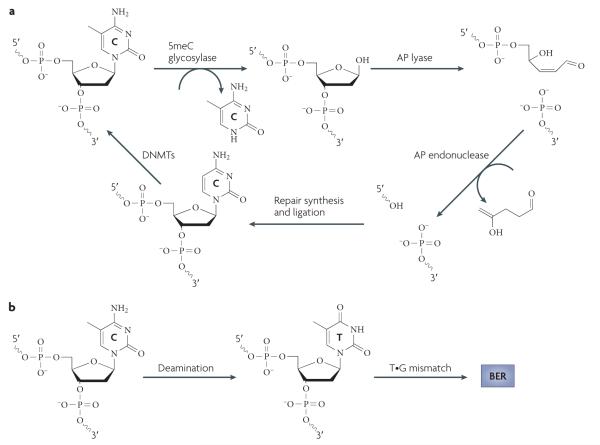
Figure 4. Base excision repair-based mechanisms for DNa demethylation.
a | Base excision repair (BER) through direct excision of 5-methylcytosine (5meC). Initiation of the BER pathway can be carried out by a glycosylase that directly excises 5meC to generate an abasic (apurinic and apyrimidinic (AP)) site. The DNA backbone is nicked by an AP lyase (or by the glycosylase itself if it is bifunctional). The 3′ sugar group is then cleaved by an AP endonuclease and the resulting single nucleotide gap is filled in with an unmethylated C by an unknown polymerase and ligase. It has been well established in plants that the demeter (Dme; also known as repressor of silencing 1 (ROS1)) family of enzymes can carry out the 5meC glycosylase reaction, but to date no mammalian enzymes have been reported to be capable of carrying out this step efficiently. b | Deamination of 5meC followed by BER. In contrast to direct excision of 5meC, deamination of 5meC produces T, which can be repaired by BER by a T•G mismatch glycosylase such as T DNA glycosylase (TDG) or methyl-CpG-binding domain protein 4 (MBD4) to regenerate an unmethylated C. DNMT, DNA methyltransferase.
Active DNA demethylation can be accomplished by a DNA glycosylase that directly excises 5meC to initiate BER (FIG. 4a). Strong genetic and biochemical evidence supports the use of this mechanism in plants65. In Arabidopsis thaliana, DNA demethylation is mediated by the Demeter (Dme) family of DNA glycosylases, which consists of four members: DME, repressor of silencing 1 (ROS1; also known as DML1), DML2 and DML3 (REF. 65). The discovery that these DNA glycosylases suppress DNA methylation initially came from forward-genetic screens in A. thaliana. whereas DME was discovered owing to the loss of expression of the imprinted gene MEDEA in a loss-of-function DME mutant66, ROS1 was recovered in a genetic screen for mutants that confer promoter hypermethylation and transgene silencing defects67.
DME, ROS1, DML2 and DML3 possess glycosylase activity against oligonucleotides containing 5meC67-71. In addition, all members of the Dme family possess AP lyase activity and are thus considered bifunctional glycosylases69-71. Besides CpG, DNA methylation in plants can occur in the context of CpNpG (where N is A, T or C) and CpNpN. All members of the Dme family have the capacity to recognize and remove meC bases from double-stranded DNA (dsDNA) oligonucleotides, irrespective of their sequence context in vitro71. However, attempts to determine the substrate specificity of these enzymes have resulted in conflicting reports owing to the use of different substrates and reaction conditions68-71. In vivo studies indicate that mutation of each of these genes results in hypermethylation in all sequence contexts but at distinct genomic loci69,71-73, indicating that each of these enzymes has a unique in vivo function.
Although it is clear that plants use BER to achieve DNA demethylation, evidence supporting a similar mechanism in mammals has been less compelling. Despite the lack of an obvious mammalian orthologue of the ROS1 family, the first indication that a repair mechanism could contribute to DNA demethylation came from early studies in chicken embryo extracts74, revealing 5meC glycosylase activity against hemi-methylated DNA75. Subsequent purification of this activity showed that it has three components: RNA, an RNA helicase related to the human p68 DEAD-box protein and a homologue of human T DNA glycosylase (TDG)76-78. Thus, 5meC glycosylase activity initially detected in chicken embryo extracts was attributed to TDG. However, its excision activity against 5meC was 30-40-fold lower compared with that against T78. Although TDG can flip C and C analogues into its active site, it does not possess the catalytic power to break the N-glycosidic bond79. It should be noted that the excision activity of TDG against 5meC is stimulated by the presence of both RNA and the RNA helicase78. Similarly, both DNMT3A and DNMT3B have been reported to interact with and stimulate the enzymatic activity of TDG80,81. Future work should determine whether these interactions have an effect on substrate preference in vitro and whether loss of function of TDG has an effect on DNA methylation status in vivo.
In addition to TDG, the methyl-CpG-binding protein MBD4 has glycosylase activity against 5meC, but again this activity is 30-40-fold lower than its T•G mismatch glycosylase activity82. Not surprisingly, MBD4-null zygotes exhibit normal demethylation of the zygotic paternal pronucleus83, and MBD4-null mice have an increased number of C to T mutations regardless of whether the C is methylated or not84. Despite its unfavourable biochemical properties, MBD4 was reported to carry out active DNA demethylation of the CYP27B1 promoter in response to PTH52. Interestingly, phosphorylation by protein kinase C enhanced MBD4 glycosylase activity against 5meC52, which may partially explain earlier enzymatic studies showing MBD4’s preference for C over 5meC85.
Deamination of 5meC to T followed by BER
DNA demethylation can also be achieved by deamination of 5meC to produce T, followed by BER to replace the mismatched T with unmethylated C (FIG. 4b). Both cytidine deaminases and DNMTs have been proposed to carry out the first step of this mechanism. on deamination of 5meC, T glycosylases such as TDG and MBD4 (see above) may function by repairing the mismatch.
Cytidine deaminases are important players in diverse biological processes such as the generation of antibody diversity, RNA editing and retroviral defence86. These processes require the production of mutations in DNA and RNA, which is achieved, in part, through the deamination of cytidine to uridine by the activation-induced deaminase (AID) and apolipoprotein B mRNA editing enzyme, catalytic polypeptide (APOBEC) family of proteins. APOBEC1, the founding member of this family, is involved in editing apolipoprotein B pre-mRNA87,88. The related deaminase AID was discovered to be essential for somatic hypermutation and class switch recombination of immunoglobulin genes in B cells89,90. Consistent with its role in the diversification of antibodies, AID-deficient mice are viable and fertile and significant phenotypic abnormalities are seen only in B cells89,90.
Despite the lack of developmental defects in AID-knockout mice, both AID and APOBEC1 have been shown in vitro and in an E. coli assay to have the capacity to deaminate 5meC to T in the context of single-stranded DNA91. AID and APOBEC1 are also expressed in mouse oocytes, ES cells and PGCs, which may be a consequence of their genomic location in a cluster of pluripotency genes that include nanog and stella (also known as DPPA3 and PGC7)91. Nevertheless, expression of AID in PGCs and the early embryo points to a possible role in global DNA demethylation. Indeed, a recent large-scale bisulphite sequencing study indicates that DNA methylation levels of male and female PGCs derived from AID-null embryos increased about 4% (from 18% to 22%) and 13% (from 7% to 20%), respectively, when compared to their wildtype counterpart92, suggesting that AID may contribute to PGC demethylation. However, because the DNA methylation levels in AID-null PGCs (~20%) are still relatively low compared with ES or somatic cells (70-80%), considerable demethylation still occurs in the absence of AID, indicating that other factors responsible for PGC demethylation remain to be identified.
Nevertheless, studies in zebrafish embryos have suggested that Aid, Mbd4 and the DNA repair protein Gadd45a (growth arrest and DNA-damage-inducible 45α) can cooperate in demethylating a methylated DNA duplex93. In this study, when a methylated linear dsDNA of ~740 bp was injected into a zebrafish embryo, demethylation of the injected DNA was seen when Aid and Mbd4 were co-expressed. The authors postulated that Aid de aminated 5meC, allowing Mbd4 to excise the T•G mismatch. Indeed, the T•G mismatch was detected using a PCR strategy, but only when Aid was expressed with a catalytic mutant of Mbd4 because the wild-type version excised the mismatch too quickly for it to be detected. Furthermore, when Aid and Mbd4 were titrated to levels that did not cause demethylation, the inclusion of Gadd45a elicited demethylation, indicating that these three proteins act cooperatively93.
Although the above studies have provided some evidence that AID may contribute to mammalian DNA demethylation, decisive biochemical and genetic evidence supporting a major role in this process is still lacking. Biochemically, AID can act on 5meC in the context of single-stranded DNA but not dsDNA91. Genetically, AID-knockout mice exhibit the expected B cell and immunological defects89,90, but no gross developmental or reproductive defects. Similarly, APOBEC1-knockout mice are also viable and fertile94,95. Although genetic redundancy may be a possible cause of the lack of expected developmental and reproductive phenotypes, such explanation needs to be confirmed by generating combinational knockouts. Furthermore, because DNA methylation occurs symmetrically, deamination of both strands would give rise to a TG•GT double mismatch. There is no evidence indicating that either TDG or MBD4 can use a double mismatch as a substrate. Furthermore, processing of a double mismatch by the AP endonuclease would generate a DNA double-strand break. This would put tremendous pressure on the repair machinery if such a mechanism were used for global demethylation. However, for locus-specific DNA demethylation, such a mechanism would not present a big problem.
In addition to AID and APOBEC, DNMTs have recently been implicated in 5meC deamination, even though they are commonly known for their ability to catalyse DNA methylation. Evidence indicating their involvement in the deamination process initially came from studies in bacteria where the methyltransferases M. HpaII96-98 and M. EcorII99,100 were shown to possess deaminase activities. Consistent with bacterial studies, the mammalian counterparts, DNMT3A and DNMT3B, have also been shown to possess deaminase activity in vitro51. As discussed above, the promoters of oestrogen-responsive genes undergo cyclical rounds of methylation and demethylation. Thus, the participation of DNMT3A and DNMT3B in both methylation and demethylation would facilitate rapid transcriptional cycling (FIG. 3b). Interestingly, ERα associates with and stimulates the activity of TDG101,102, allowing for the repair of the T•G mismatch. DNMT3A and DNMT3B also associate with TDG and this interaction stimulates glycosylase activity80,81. Indeed, DNA demethylation was found to coincide with the recruitment of TDG and other BER enzymes51.
However, it is surprising that DNMTs possess two opposing enzymatic activities. Although the methyltransferase activity of DNMT3A is inhibited by TDG81, the 5meC deamination reaction can only occur under conditions where SAM concentrations are very low or nonexistent51. In order for DNMT3A to carry out efficient methylation and demethylation during transcriptional cycling, levels of SAM must fluctuate rapidly. Given that SAM is crucial for many essential biochemical and biological processes, it is difficult to imagine how this could be achieved without serious biological consequences.
Nucleotide excision repair
Another DNA repair pathway, NER, has also been proposed to carry out DNA demethylation. This type of repair is generally used to repair DNA containing bulky lesions, which form after exposure to chemicals or radiation. After damaged DNA is recognized, dual incisions flanking the lesion are made and a 24-32 nucleotide oligomer is released. The resulting gap is then filled in by repair polymerases and sealed by a ligase64.
In an assay aimed at identifying proteins required for activation of a reporter that is silenced by DNA methylation, Niehrs and colleagues uncovered a novel function for GADD45A103, which is encoded by a p53- and breast cancer type 1 susceptibility protein (BRCA1)-inducible gene and participates in diverse biological processes, including DNA damage response, cell cycle progression, apoptosis and NER104. Overexpression of GADD45A in mammalian cell lines leads to loci-specific and global demethylation, whereas knockdown results in DNA hypermethylation103. Because GADD45A had previously been implicated in NER105,106, Barretto et al. explored the role of NER in DNA demethylation and found that loss of DNA methylation is accompanied by DNA synthesis and requires the NER endonuclease xeroderma pigmentosum group G-complementing protein (XPG), which interacts with GADD45A103. The recruitment of GADD45A and other components of the NEr repair machinery to ribosomal RNA (rRNA) genes is facilitated by TBP-associated factor 12 (TAF12) and leads to DNA demethylation and rRNA gene activation107. However, it is not clear how the demethylation process is initiated and whether GADD45A is directly involved. More importantly, two independent studies have raised doubt on the role of GADD45A in the active DNA demethylation process. In the first study, the Pfeifer group carried out a series of experiments that were similar to those carried out by the Niehrs group, but obtained no evidence indicating that GADD45A had any effect on DNA methylation108. In the second study, analysis of the GADD45A-null mice indicated that loss of GADD45A function had neither loci-specific nor global effects on DNA methylation levels109.
GADD45B, another member of the GADD45 family, has also been implicated in active demethylation of genes that are crucial for adult neurogenesis110. Loss of GADD45B results in defects in neural progenitor proliferation and dendritic growth. This was attributed to promoter hypermethylation and the repression of BDNF and fibroblast growth factor 1 (FGF1), two genes crucial for neurogenesis110. However, GADD45B is not involved in zygotic DNA demethylation as GADD45B-null zygotes undergo normal paternal genome demethylation111. Because GADD45B has not been biochemically characterized, it is unknown whether it is directly involved in the active demethylation of neurogenesis genes.
Oxidative demethylation
Another possible mechanism by which DNA demethylation can be carried out is through oxidative demethylation. The E. coli enzyme AlkB is a member of the 2-oxoglutarate (2OG)-dependent dioxygenases and is involved in the bacterial response to alkylation damage to DNA. using oxygen, iron and 2OG as cofactors, AlkB is able to carry out oxidative demethylation of 1-methyladenine and 3meC by releasing the methyl group as formaldehyde112,113. The same mechanism is used by the JmjC family of enzymes to demethylate histon e substrates60,114.
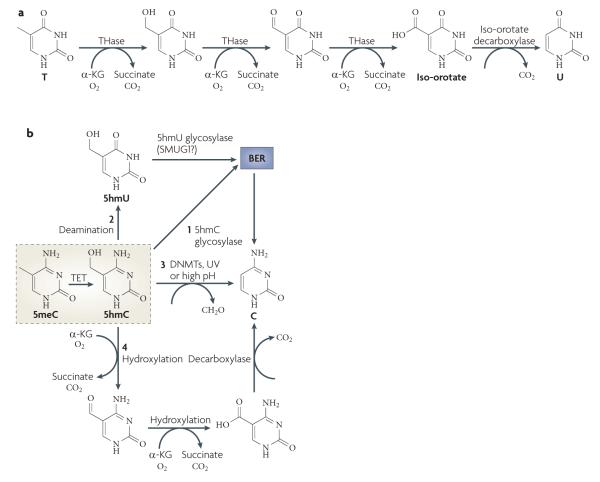
Although breakage of a carbon-carbon bond is energetically difficult, enzymes that catalyse such reactions do exist. As shown in FIG. 5a, thymine 7-hydroxylase can catalyse the conversion of T to iso-orotate through three consecutive oxidation reactions using oxygen, iron and 2OG as cofactors115. Iso-orotate can be further converted to C through a decarboxylation reaction. Although thymine 7-hydroxylase and iso-orotate decarboxylase have been isolated from fungi, such as Rhodotorula glutinis, Neurospora crassa and Aspergillus nidulans62, no homologue of thymine 7-hydroxylase has been found in mammals. Interestingly, the trypanosome base J-binding proteins, JBP1 and JBP2, have properties similar to that of thymine 7-hydroxylase 116,117, prompting the Rao group to search for mammalian homologues with similarity to the dioxygenase domains of the JBP proteins. This effort led to the identification of the ten-eleven translocation (TET) family of proteins118. we have also independently characterized the mouse TET family119.
Figure 5. Oxidative demethylation by TET proteins.
a | Part of the thymidine salvage pathway. Direct removal of the methyl group of 5-methylcytosine (5meC) involves breaking a carbon-carbon bond, which requires an enzyme with great catalytic power. Such an enzyme exists in the thymidine salvage pathway. Starting with T, thymine-7-hydroxylase (THase) carries out three consecutive hydroxylation reactions to produce iso-orotate, which is processed by a decarboxylase to produce U. A similar mechanism may be used in active DNA demethylation, particularly by the ten-eleven translocation (TET) family of proteins. b | The fate of 5-hydroxymethylcytosine (5hmC). The TET family of proteins catalyses the conversion of 5meC to 5hmC, which may be an intermediate that can be further processed by one of the following mechanisms. BER may be initiated by a 5hmC glycosylase (1); 5hmC may undergo deamination to produce 5hmU (2), which is repaired by BER through a 5hmU glycosylase such as SMUG1 (single-strand-selective monofunctional U DNA glycosylase 1); 5hmC may directly be converted to C by DNA methyltransferases (DNMTs), ultraviolet (UV) exposure or high pH (3); or consecutive hydroxylation reactions followed by a decarboxylation reaction similar to the thymidine salvage pathway may be used to ultimately replace 5hmC with C (4). Alternatively, 5hmC itself may be a functional modification. α-KG, α-ketoglutarate.
TET1, the founding member of the TET family, was initially discovered in acute myeloid leukaemia (AMl) as a fusion partner of the histone H3 Lys4 methyltransferase MLL120,121. Subsequent studies in vitro and in cultured cells showed that human TET1 is capable of hydrolysing 5meC to produce 5-hydroxymethylcytosine (5hmC) in DNA118. Similarly, all three members of the mouse TET family possess this enzymatic activity119. Consistent with the presence of a dioxygenase domain in the proteins and the predicted reaction mechanism, the putative iron-binding sites are required for their enzymatic activities118,119. Furthermore, TET1 is capable of acting on both fully methylated and hemi-methylated DNA118.
Although 5hmC has previously been reported to exist in animal DNA122, this modified base is not found in some cell types and tissues118,123, thus raising the question of whether 5hmC is present in mammalian DNA at physiologically relevant levels. This issue was directly addressed in two cell types. In Purkinje neurons, 5hmC is ~40% as abundant as 5meC124, whereas the frequency of 5hmC in ES cells was estimated to be approximately 1 in every 3,000 nucleotides118. Thus, it is evident that 5hmC constitutes a large fraction of mammalian DNA in some cell types.
The consequences of 5hmC in genomic DNA are currently unclear. Because 5hmC seems to be stable, it may function like other modifications by altering local chromatin structure or contributing to the recruitment or exclusion of other factors that influence transcription. For example, the transcriptional repressors MeCP2, MBD1, MBD2 and MBD4 bind to methylated DNA, but do not recognize 5hmC125,126. It is also possible that the TET proteins may facilitate passive demethylation in dividing cells such as ES cells as 5hmC is not recognized by DNMT1 (REF. 127); thus, newly replicated DNA would not maintain patterns of methylation. Alternatively, 5hmC may be an intermediate in an active demethylation pathway that ultimately leads to the replacement of 5meC with C (FIG. 5b). This could be achieved by several ways that include: BER by a 5hmC-specific DNA glycosylase (as such activity has been previously reported to exist in calf thymus extracts128), deamination of 5hmC to generate 5hmu followed by BER initiated by a 5hmu-specific glycosylase such as single-strand-selective monofunctional u DNA glycosylase 1 (SMUG1)129, conversion of 5hmC to C through loss of formaldehyde on ultraviolet light exposure130 or high pH131 (or possibly carried out by DNMTs)132, and two consecutive oxidation steps followed by decarboxylation similar to that used by the thymidine salvage pathway (FIG. 5a). It is not clear why TET proteins cannot catalyse consecutive reactions such as that of thymine 7-hydroxylase. Because all in vitro assays carried out so far used recombinant TET proteins alone, it is possible that association of TET proteins with their in vivo partners is necessary to confer such a capability. In this case, a decarboxylase may eventually remove the carboxyl group to complete the demethylation process.
Consistent with the relative enrichment of 5hmC in ES cells, recent studies have shed light on the role of TET1 in ES cell biology. During ES cell differentiation, TET1 mRNA levels decline, coinciding with a decrease in 5hmC levels118, which suggests that TET1 may be important for ES cell identity. Indeed, knockdown of TET1, but not TET2 or TET3, in mouse ES cells results in impairment of ES cell self-renewal and maintenance119. Analysis of the differentiated TET1-knockdown ES cells revealed a bias towards the trophoectoderm and primitive endoderm lineages. Furthermore, knockdown of TET1 at two-cell stage embryos followed by cell lineage tracing revealed that the knockdown cells are biased towards the trophoectoderm119, indicating that TET1 is required for inner cell mass cell specification. Consistent with its role in ES cell self-renewal and maintenance, knockout of TET1 resulted in embryonic lethality (K. Hong and Y.Z., unpublished observations), making the evaluation of the role of TET1 on global demethylation of the paterna l genome difficult.
With regard to the mechanism underlying the role of TET1 in ES cells, TET1 maintains nanog expression in ES cells by directly binding to the nanog promoter and protecting it from becoming hypermethylated, as knockdown of TET1 in ES cells resulted in downregulation of nanog expression concomitant with increased nanog promoter methylation119. Nanog seems to be one of the main TET1 targets as the phenotypes associated with TET1 knockdown can largely be rescued by ectopic expression of nanog119.
Although TET2 is also expressed in ES cells, it seems that TET2 does not play a significant part in ES cell biology as knockdown of TET2 does not confer any obvious phenotype 119. However, a flurry of recent studies have uncovered that dysfunction of human TET2 may be a key event in leukaemogenesis as human TET2 is mutated in a range of human myeloid malignancies, including myelodysplastic syndromes (MDSs), myeloproliferative disorders (MPDs) and acute myeloid leukaemias (AMls)133-140. Currently, TET2 is the most frequently mutated gene that has been identified in patients with MDS and these mutations have been suggested to occur early during the pathogenesis of the disease137. Consistent with a role for TET2 in regulating DNA demethylation, aberrant DNA methylation is frequently found in patients with MDS141. Indeed, mutations of TET2 that mimic mutations identified in patients with MDS abolished the enzymatic activity of TET2 (A. C. D’Alessio and Y.Z., unpublished observations). Furthermore, the DNA methyltransferase inhibitor 5-azacytidine (5-azaC) has been shown to be an effective treatment for patients with high-risk MDS and secondary AML142,143, indicating that aberrant DNA methylation plays a crucial part in MDS development and progression. The participation of TET2 in DNA demethylation may provide a molecular basis for the effectiveness of using methyltransferase inhibitors in the treatment of patients with MDS, thus setting the stage for understanding the molecular mechanism underlying the pathogenesis of leukaemias.
Radical SAM mechanism
Although many proteins have been proposed to carry out active DNA demethylation, none of the proteins discussed above have been shown to have a role in paternal genome demethylation in zygotes. To identify proteins involved in paternal genome demethylation, our laboratory used a candidate gene knockdown approach coupled with live-cell imaging. To facilitate a screen of candidate proteins, we developed a probe that consists of the Cys-X-X-Cys domain of MLL fused to enhanced green fluorescent protein (EGFP). Because the Cys-X-X-Cys domain has high affinity for unmethylated CpG144, injection of mRNA encoding the probe into zygotes results in the accumulation of the probe at the demethylated paternal pronucleus111, allowing live-cell imaging of the paternal genome demethylation process. Using this imaging system, we screened several candidate proteins by injecting small interfering RNAs (siRNAs) against each of the candidates into eggs before carrying out intracytoplasmic sperm injection (ICSI), and monitored the effect of the siRNA on the accumulation of the probe at the paternal pronucleus. This screen uncovered a role for elongator complex protein 3 (ELP3) in paternal genome demethylation. ELP3 knockdown prevented the accumulation of the probe in the paternal pronucleus at pronuclear stages 4 and 5 (REF. 111). In addition, immunostaining and bisulphite sequencing of selected retrotransposon elements further support a role for ELP3 in paternal genome demethylation111.
ELP3 is a member of the core elongator complex (ELP1-ELP3), which combines with another subcomplex (ELP4-ELP6) to form the holo-elongator complex145,146. Because knockdown of the ELP1 and ELP4 components also impaired paternal genome demethylation, it is likely that the entire elongator complex may be involved in the demethylation process111. Interestingly, the Fe-S radical SAM domain of ELP3, but not the histone acetyltransferase (HAT) domain, is required for paternal genome demethylation111. Although this may provide a clue regarding the enzymatic mechanism of ELP3, recent studies in yeast suggest that the Cys-rich domain of Elp3 is required for the integrity of the elongator complex147,148, raising the possibility that the Fe-S radical SAM motif may have a structural rather than an enzymatic role. Thus, direct biochemical evidence of the enzymatic activity of the elongator complex and genetic evidence using ELP3-null oocytes remain to be shown.
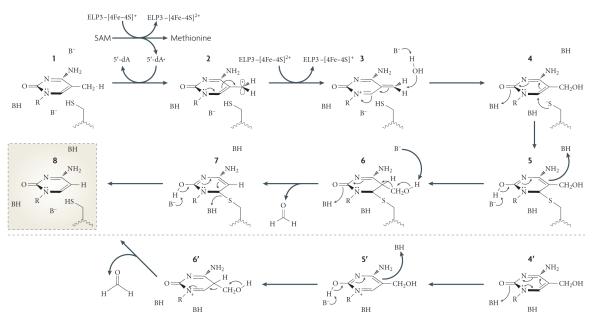
Interestingly, a recent study confirmed the presence of an Fe-S cluster in the bacteria Methanocaldococcus jannaschii Elp3 protein149. The assumption that mammalian ELP3 is a radical SAM protein has led to a potential mechanism for ELP3-catalysed DNA demethylation as outlined in FIG. 6 (S. J. Booker, personal communication). like every radical SAM enzyme, the reaction is initiated by the generation of a powerful oxidizing agent, the 5′-deoxyadenosyl (5′-dA) radical, from SAM. The 5′-dA radical could extract a hydrogen atom from the 5-methyl group to generate a 5meC radical. In the next step, an electron is returned back to the Fe-S cluster to generate a third intermediate, which can be converted to the relatively stable 5hmC by the addition of a water molecule. In order to break the carbon-carbon bond, the next step requires the generation of an intermediate, the resulting carbanion of which would be stabilized. This can probably be achieved by a thymidylate synthase or methyltransferase type of mechanism, whereby a Cys residue carries out a nucleophilic attack at carbon 6, leading to the release of formaldehyde. In the absence of an external nucleophile, an alternative pathway leading to the release of formaldehyde can also take place. Finally, an elimination at the formaldehyde release step results in the final product of C.
Figure 6. Proposed mechanism for elP3-mediated DNa demethylation.
Mammalian elongator complex protein 3 (ELP3) contains an Fe-S radical S-adenosylmethionine (SAM) domain that is important for active DNA demethylation of the zygotic paternal genome. If ELP3 is indeed a functional radical SAM protein, it may directly carry out DNA demethylation through the following mechanism. First, ELP3 uses SAM to generate a 5′-deoxyadenosyl radical, which could extract a hydrogen atom from the 5-methyl group of 5-methylcytosine (5meC; 1) to form a 5meC radical (2). After an electron is donated back to the Fe -S to create the third intermediate (3), a water molecule would promote the formation of 5-hydroxymethylcytosine (5hmC) (4). A nucleophilic attack at carbon 6 can result in the carbon-carbon bond breaking to release formaldehyde (5-7). In the absence of an external nucleophile, an alternative pathway (4′-6′) that leads to the release of formaldehyde can also take place. Finally, an elimination step would produce an end product of C (8).
Although future studies are required to validate or refute this proposed mechanism, we note that this work is not trivial for three reasons. First, the identities of mammalian ELP5 and ELP6 still need to be determined as an apparent orthologue of yeast Elp5 and Elp6 cannot be identified by sequence homology searches. Second, the radical SAM reaction occurs under anaerobic conditions and reconstitution of the elongator complex under anaerobic conditions is challenging. Finally, given that zygotic DNA demethylation occurs only on the paternal genome, some unique features of the paternal genome may be required in order for it to serve as a substrate. Despite these challenges, identification of the elongator complex as an important factor for paternal genome demethylation in zygotes allows for further studies towards understanding the molecular mechanisms of active DNA demethylation.
Concluding remarks
Observations of active DNA demethylation during embryonic development and in somatic cells have opened the door for many questions to be answered. In particular, how DNA demethylation is achieved in mammalian cells remains debatable as no single enzyme or mechanism has gained decisive biochemical and genetic support (see Supplementary information S1 (table)). It is possible that multiple mechanisms exist to carry out DNA demethylation and that the use of each one is dictated by the specific biological context.
Although repair-based mechanisms, particularly deamination of 5meC followed by BER, have offered an attractive mechanism for active DNA demethylation, genetic evidence is still lacking. Furthermore, the involvement of a repair-based mechanism in global DNA demethylation would put tremendous pressure on the repair machinery when considering that paternal pronucleus demethylation is completed within 4 hours17,18.
Although AID seems to contribute to active demethylation in PGCs, it is only responsible for a small part of it as considerable demethylation still takes place in the AID-null PGCs92. Nevertheless, this mechanism does provide a reasonable explanation for loci-specific demethylation in response to gene-activation signals. Although AID deficiency has some effect on PGC demethylation, there is no evidence that it affects paternal DNA demethylation in zygotes. Similarly, MBD4-null zygotes still experience paternal genome demethylation83. It seems that although repair-based mechanisms may be responsible for loci-specific DNA demethylation and partial demethylation in PGCs, their role in zygotic paternal genome demethylation is less likely. To date, the only factor shown to have a role in zygotic paternal genome demethylation is the elongator complex, although it is unclear whether its role is direct or indirect111. Future work should focus on gaining additional genetic evidence using elongator-null zygotes and elucidating its enzymatic activity. The recent demonstration that the TET family proteins are capable of catalysing the conversion of 5meC to 5hmC has raised the possibility that these proteins may have a role in active DNA demethylation118,119. we anticipate that work evaluating their role in demethylation of the zygotic paternal genome and PGCs is forthcoming. Furthermore, analysis of the fate and function of 5hmC will also attract a lot of attention.
In addition to determining the mechanism of active demethylation, one open question that remains is to what extent the paternal genome and PGCs are demethylated. Although this event is considered to be global, as determined by 5meC immunostaining, it is evident that some regions of the paternal genome are protected from this wave of demethylation. The advent of high-throughput analyses including chromatin immunoprecipitation-on-chip (ChIP-chip), ChIP sequencing (ChIP-Seq) and bisulphite sequencing (BS-Seq; bisulphite treatment followed by high-throughput sequencing) has allowed for genome-wide profiling of epigenetic marks such as DNA methylation92,150-153. using single-molecule, realtime sequencing, a recent study showed the feasibility of direct detection of modified nucleotides in DNA, including N6-meA, 5mC and 5hmC154. Future studies using these tools will undoubtedly determine precisely which genomic regions are demethylated and which regions are protected. However, improvements in the sensitivity of these technologies will be necessary for such experiments, given that paternal genomic DNA would need to be obtained from individual zygotes.
As well as being fundamental to our knowledge in epigenetics, a better understanding of how DNA demethylation occurs will allow for the development of techniques and approaches that will improve somatic cell reprogramming (BOX 2) and cancer treatment. Tumour suppressor gene silencing by promoter DNA methylation is thought to play an important part in cancer development155. Consistently, inhibitors of DNMTs have been used in the treatment of certain cancers156. Owing to the reversible nature of epigenetic modifications, developing drugs that target epigenetic factors is becoming one of the top priorities for many biotechnology and pharmaceutical companies157. It is anticipated that targeted demethylation of tumour suppressor genes may reactivate the silenced tumour suppressor genes, which can lead to cellular differentiation or halt uncontrolled cell proliferation.
The mechanism underlying the regulation of DNA methylation is a question that has elicited much attention and controversy over the past decade. Although recent studies have proposed numerous ideas as to how active DNA demethylation is carried out, many aspects are still contentious and a consensus has yet to be achieved. with the development of new technology and the studies described above, our continued and collective efforts in this field will hopefully provide clearer answers in the coming decade.
Supplementary Material
Box 2. Implications of active DNA demethylation in reprogramming.
Induced pluripotent stem (iPS) cells can be generated by introducing four transcription factors — octamer-binding protein 3 (OCT3; also known as OCT4 and POU5F1), SRY box-containing factor 2 (SOX2), krüppel-like factor 4 (KLF4) and MYC — into somatic cells167,168. Successful reprogramming requires the activation of endogenous OCT4 and nanog genes, which are known to be silenced by DNA methylation in somatic cells169-172. Demethylation of the OCT4 and nanog promoters is thus an integral event in iPS cell generation173. In fact, inefficient DNA demethylation is thought to be one of the causes of the low efficiency in iPS cell generation because the use of the DNA methylation inhibitor 5-azacytidine can increase the efficiency of iPS cell generation by converting partially reprogrammed cells to fully reprogrammed iPS cells173.
Transcription factor-based iPS cell generation is a slow process compared to somatic cell nuclear transfer (SCNT) and cell fusion174,175. One possible explanation for this difference may be the mechanisms used to reactivate endogenous OCT4 and nanog. Epigenetic reprogramming of somatic cells to pluripotent iPS cells may necessitate several cell divisions176 owing to the absence of the DNA demethylase or demethylases required for demethylation of the OCT4 and nanog promoters. By contrast, reactivation of OCT4 and nanog can occur quickly during SCNT and cell fusion because the DNA demethylase or demethylases may already be present in eggs and embryonic stem (ES) cells. Consistent with this notion, reprogramming by cell fusion requires activation-induced deaminase (AID)-dependent demethylation and reactivation of OCT4 and nanog177. Surprisingly, although AID was present at the OCT4 and nanog promoters in fibroblasts, these promoters are methylated, suggesting that other factors or regulatory events are required for demethylation. Given that genetic evidence does not support an important role for AID in ES cells (see main text), it is unclear whether AID directly participates in promoter demethylation of these genes during somatic cell reprogramming. Regardless, it is evident that activation of pluripotent genes through DNA demethylation is an important step during the somatic cell reprogramming process. Identification and characterization of the enzymes involved should improve protocols of somatic cell reprogramming.
Acknowledgements
We thank S. J. Booker for discussions regarding the radical SAM mechanism, and K. Hong and A. D’Alessio for critical comments on the manuscript. We apologize to colleagues whose work cannot be cited owing to space constraints. Work in the Zhang laboratory is supported by the National Institutes of Health (GM68804) and the Howard Hughes Medical Institute, of which Y.Z. is an investigator.
Glossary
- Imprinted gene
A gene that is expressed in a parent-of-origin-specific manner.
- Inactive X chromosome
The copy of X chromosome that is silenced in female chromosomes in order to equalize the expression of genes located in the X chromosome in males and females.
- DNA methyltransferase
An enzyme that catalyses the addition of a methyl group to C or A.
- Hemi-methylated DNA
Duplex DNA in which only one of the two strands is methylated.
- Zona pellucida
The glycoprotein coat that surrounds the oocytes and the early embryos of mammals.
- Polar body
The structure that is extruded from the oocyte during meiosis and contains one haploid set of chromosomes
- Parthenogenesis
The production of a diploid offspring from two sets of haploid maternal gametes and no paternal contribution
- Gynogenesis
Parthenogenesis in which the embryo contains only maternal chromosomes owing to the failure of the sperm to fuse with the egg nucleus
- Digynic triploid
An embryo that contains two maternal genomes and one paternal genome.
- Bisulphite sequencing
A technique in which the treatment of DNA with bisulphite, which converts C to U but does not modify meC, is used to determine the DNA methylation pattern.
- Blastocyst
An embryonic stage that is characterized by the formation of the first definitive lineages.
- Primordial germ cell
One of a population of embryonic cells from which germ cells are formed.
- RNA editing
The post-transcriptional modification of RNA primary sequence by the insertion and/or deletion of specific bases, or the chemical modification of adenosine to inosine or cytidine to uridine.
- Somatic hypermutation
The mutation of the immunoglobulin variable region in mature B cells during an immune response. It results in affinity maturation of the antibody response. Like class switch recombination, it requires activation-induced cytidine deaminase.
- Class switch recombination
A mechanism that changes the class or isotype of antibody produced by an activated B cell. This does not change the affinity towards an antigen, but instead allows for interaction with different effector molecules.
- JmjC
(Jumonji C). An evolutionarily conserved motif. Proteins containing this domain are predicted to be protein hydroxylases or histone demethylases.
- Base J-binding protein
A protein that binds to base J (β-D-glucosylhydroxymethyl-U), a modified T produced by hydroxylation and glucosylation of the methyl group of T.
- Elongator complex
A protein complex originally identified in budding yeast to be associated with the elongating and hyperphosphorylated RNA polymerase II. It has also been implicated in tRNA modification, exocytosis and neuronal maturation.
- SAM domain
A protein domain containing an Fe-s cluster that uses s-adenosylmethionine (sAM) to catalyse various radical reactions.
Footnotes
Competing interests statement The authors declare no competing financial interests.
DATABASES Entrez Gene: http://www.ncbi.nlm.nih.gov/gene BDNF | CYP27B1 | FGF1 | MEDEA | nanog | pS2 | stella UniProtKB: http://www.uniprot.org AID | AlkB | APOBEC1 | BLIMP1 | DME | DML2 | DML3 | DNMT1 |DNMT3A | DNMT3B | ELP3 | ERα | Gadd45a | GADD45B | JBP1 | JBP2 | MBD2 | MBD4 | MeCP2 | M.EcoRII | M.HpaII | MLL |PRDM1 | PRDM14 | PTH | ROS1 | SMUG1 | TAF12 | TDG | TET1 |XPG | ZFP57
FURTHER INFORMATION Yi Zhang’s homepage: http://www.med.unc.edu/~zhangyi/lab.htm
SUPPLEMENTARY INFORMATION See online article: S1 (table) ALL LINKS ARE ACTIVE IN THE ONLINE PDF
References
- 1.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nature Genet. 2003;33:245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 2.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 3.Birney E, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nature Rev. Genet. 2007;8:286–298. doi: 10.1038/nrg2005. [DOI] [PubMed] [Google Scholar]
- 5.Feinberg AP, Tycko B. The history of cancer epigenetics. Nature Rev. Cancer. 2004;4:143–153. doi: 10.1038/nrc1279. [DOI] [PubMed] [Google Scholar]
- 6.Pogribny IP, Beland FA. DNA hypomethylation in the origin and pathogenesis of human diseases. Cell. Mol. Life Sci. 2009;66:2249–2261. doi: 10.1007/s00018-009-0015-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Santos-Reboucas CB, Pimentel MM. Implication of abnormal epigenetic patterns for human diseases. Eur. J. Hum. Genet. 2007;15:10–17. doi: 10.1038/sj.ejhg.5201727. [DOI] [PubMed] [Google Scholar]
- 8.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nature Rev. Genet. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- 10.Okano M, Xie S, Li E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nature Genet. 1998;19:219–220. doi: 10.1038/890. [DOI] [PubMed] [Google Scholar]
- 11.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 12.Bestor TH, Ingram VM. Two DNA methyltransferases from murine erythroleukemia cells: purification, sequence specificity, and mode of interaction with DNA. Proc. Natl Acad. Sci. USA. 1983;80:5559–5563. doi: 10.1073/pnas.80.18.5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bestor T, Laudano A, Mattaliano R, Ingram V. Cloning and sequencing of a cDNA encoding DNA methyltransferase of mouse cells. The carboxyl-terminal domain of the mammalian enzymes is related to bacterial restriction methyltransferases. J. Mol. Biol. 1988;203:971–983. doi: 10.1016/0022-2836(88)90122-2. [DOI] [PubMed] [Google Scholar]
- 14.Hermann A, Goyal R, Jeltsch A. The Dnmt1 DNA-(cytosine-C5)-methyltransferase methylates DNA processively with high preference for hemimethylated target sites. J. Biol. Chem. 2004;279:48350–48359. doi: 10.1074/jbc.M403427200. [DOI] [PubMed] [Google Scholar]
- 15.Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 16.Ooi SK, Bestor TH. The colorful history of active DNA demethylation. Cell. 2008;133:1145–1148. doi: 10.1016/j.cell.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 17.Mayer W, Niveleau A, Walter J, Fundele R, Haaf T. Demethylation of the zygotic paternal genome. Nature. 2000;403:501–502. doi: 10.1038/35000656. [DOI] [PubMed] [Google Scholar]
- 18.Oswald J, et al. Active demethylation of the paternal genome in the mouse zygote. Curr. Biol. 2000;10:475–478. doi: 10.1016/s0960-9822(00)00448-6. [DOI] [PubMed] [Google Scholar]
- 19.Ajduk A, Yamauchi Y, Ward MA. Sperm chromatin remodeling after intracytoplasmic sperm injection differs from that of in vitro fertilization. Biol. Reprod. 2006;75:442–51. doi: 10.1095/biolreprod.106.053223. [DOI] [PubMed] [Google Scholar]
- 20.Aoki E, Schultz RM. DNA replication in the 1-cell mouse embryo: stimulatory effect of histone acetylation. Zygote. 1999;7:165–172. doi: 10.1017/s0967199499000532. [DOI] [PubMed] [Google Scholar]
- 21.Bouniol-Baly C, Nguyen E, Besombes D, Debey P. Dynamic organization of DNA replication in one-cell mouse embryos: relationship to transcriptional activation. Exp. Cell Res. 1997;236:201–211. doi: 10.1006/excr.1997.3708. [DOI] [PubMed] [Google Scholar]
- 22.Ferreira J, Carmo-Fonseca M. Genome replication in early mouse embryos follows a defined temporal and spatial order. J. Cell Sci. 1997;110:889–897. doi: 10.1242/jcs.110.7.889. [DOI] [PubMed] [Google Scholar]
- 23.Howlett SK, Bolton VN. Sequence and regulation of morphological and molecular events during the first cell cycle of mouse embryogenesis. J. Embryol. Exp. Morphol. 1985;87:175–206. [PubMed] [Google Scholar]
- 24.Luthardt FW, Donahue RP. Pronuclear DNA synthesis in mouse eggs. An autoradiographic study. Exp. Cell Res. 1973;82:143–151. doi: 10.1016/0014-4827(73)90256-5. [DOI] [PubMed] [Google Scholar]
- 25.Yamauchi Y, Ward MA, Ward WS. Asynchronous DNA replication and origin licensing in the mouse one-cell embryo. J. Cell. Biochem. 2009;107:214–223. doi: 10.1002/jcb.22117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kishigami S, et al. Epigenetic abnormalities of the mouse paternal zygotic genome associated with microinsemination of round spermatids. Dev. Biol. 2006;289:195–205. doi: 10.1016/j.ydbio.2005.10.026. [DOI] [PubMed] [Google Scholar]
- 27.Dean W, et al. Conservation of methylation reprogramming in mammalian development: aberrant reprogramming in cloned embryos. Proc. Natl Acad. Sci. USA. 2001;98:13734–13738. doi: 10.1073/pnas.241522698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fulka H, Mrazek M, Tepla O, Fulka J., Jr. DNA methylation pattern in human zygotes and developing embryos. Reproduction. 2004;128:703–708. doi: 10.1530/rep.1.00217. [DOI] [PubMed] [Google Scholar]
- 29.Beaujean N, et al. Non-conservation of mammalian preimplantation methylation dynamics. Curr. Biol. 2004;14:R266–R267. doi: 10.1016/j.cub.2004.03.019. [DOI] [PubMed] [Google Scholar]
- 30.Beaujean N, et al. The effect of interspecific oocytes on demethylation of sperm DNA. Proc. Natl Acad. Sci. USA. 2004;101:7636–7640. doi: 10.1073/pnas.0400730101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Santos F, Hendrich B, Reik W, Dean W. Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev. Biol. 2002;241:172–182. doi: 10.1006/dbio.2001.0501. [DOI] [PubMed] [Google Scholar]
- 32.Barton SC, et al. Genome-wide methylation patterns in normal and uniparental early mouse embryos. Hum. Mol. Genet. 2001;10:2983–2987. doi: 10.1093/hmg/10.26.2983. [DOI] [PubMed] [Google Scholar]
- 33.Olek A, Walter J. The pre-implantation ontogeny of the H19 methylation imprint. Nature Genet. 1997;17:275–276. doi: 10.1038/ng1197-275. [DOI] [PubMed] [Google Scholar]
- 34.Lane N, et al. Resistance of IAPs to methylation reprogramming may provide a mechanism for epigenetic inheritance in the mouse. Genesis. 2003;35:88–93. doi: 10.1002/gene.10168. [DOI] [PubMed] [Google Scholar]
- 35.Rougier N, et al. Chromosome methylation patterns during mammalian preimplantation development. Genes Dev. 1998;12:2108–2113. doi: 10.1101/gad.12.14.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dean W, Santos F, Reik W. Epigenetic reprogramming in early mammalian development and following somatic nuclear transfer. Semin. Cell Dev. Biol. 2003;14:93–100. doi: 10.1016/s1084-9521(02)00141-6. [DOI] [PubMed] [Google Scholar]
- 37.Aoki F, Worrad DM, Schultz RM. Regulation of transcriptional activity during the first and second cell cycles in the preimplantation mouse embryo. Dev. Biol. 1997;181:296–307. doi: 10.1006/dbio.1996.8466. [DOI] [PubMed] [Google Scholar]
- 38.Monk M, Boubelik M, Lehnert S. Temporal and regional changes in DNA methylation in the embryonic, extraembryonic and germ cell lineages during mouse embryo development. Development. 1987;99:371–382. doi: 10.1242/dev.99.3.371. [DOI] [PubMed] [Google Scholar]
- 39.Howlett SK, Reik W. Methylation levels of maternal and paternal genomes during preimplantation development. Development. 1991;113:119–127. doi: 10.1242/dev.113.1.119. [DOI] [PubMed] [Google Scholar]
- 40.Carlson LL, Page AW, Bestor TH. Properties and localization of DNA methyltransferase in preimplantation mouse embryos: implications for genomic imprinting. Genes Dev. 1992;6:2536–2541. doi: 10.1101/gad.6.12b.2536. [DOI] [PubMed] [Google Scholar]
- 41.Hirasawa R, et al. Maternal and zygotic Dnmt1 are necessary and sufficient for the maintenance of DNA methylation imprints during preimplantation development. Genes Dev. 2008;22:1607–1616. doi: 10.1101/gad.1667008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li X, et al. A maternal-zygotic effect gene, Zfp57, maintains both maternal and paternal imprints. Dev. Cell. 2008;15:547–557. doi: 10.1016/j.devcel.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ohinata Y, et al. A signaling principle for the specification of the germ cell lineage in mice. Cell. 2009;137:571–584. doi: 10.1016/j.cell.2009.03.014. [DOI] [PubMed] [Google Scholar]
- 44.Saitou M. Germ cell specification in mice. Curr. Opin. Genet. Dev. 2009;19:386–395. doi: 10.1016/j.gde.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 45.Hajkova P, et al. Epigenetic reprogramming in mouse primordial germ cells. Mech. Dev. 2002;117:15–23. doi: 10.1016/s0925-4773(02)00181-8. [DOI] [PubMed] [Google Scholar]
- 46.Lee J, et al. Erasing genomic imprinting memory in mouse clone embryos produced from day 11.5 primordial germ cells. Development. 2002;129:1807–1817. doi: 10.1242/dev.129.8.1807. [DOI] [PubMed] [Google Scholar]
- 47.Yamazaki Y, et al. Reprogramming of primordial germ cells begins before migration into the genital ridge, making these cells inadequate donors for reproductive cloning. Proc. Natl Acad. Sci. USA. 2003;100:12207–12212. doi: 10.1073/pnas.2035119100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bruniquel D, Schwartz RH. Selective, stable demethylation of the interleukin-2 gene enhances transcription by an active process. Nature Immunol. 2003;4:235–240. doi: 10.1038/ni887. [DOI] [PubMed] [Google Scholar]
- 49.Martinowich K, et al. DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science. 2003;302:890–893. doi: 10.1126/science.1090842. [DOI] [PubMed] [Google Scholar]
- 50.Kangaspeska S, et al. Transient cyclical methylation of promoter DNA. Nature. 2008;452:112–115. doi: 10.1038/nature06640. [DOI] [PubMed] [Google Scholar]
- 51.Metivier R, et al. Cyclical DNA methylation of a transcriptionally active promoter. Nature. 2008;452:45–50. doi: 10.1038/nature06544. [DOI] [PubMed] [Google Scholar]
- 52.Kim MS, et al. DNA demethylation in hormoneinduced transcriptional derepression. Nature. 2009;461:1007–1012. doi: 10.1038/nature08456. [DOI] [PubMed] [Google Scholar]
- 53.Gjerset RA, Martin DW., Jr. Presence of a DNA demethylating activity in the nucleus of murine erythroleukemic cells. J. Biol. Chem. 1982;257:8581–8583. [PubMed] [Google Scholar]
- 54.Weiss A, Keshet I, Razin A, Cedar H. DNA demethylation in vitro: involvement of RNA. Cell. 1996;86:709–718. doi: 10.1016/s0092-8674(00)80146-4. [DOI] [PubMed] [Google Scholar]
- 55.Swisher JF, Rand E, Cedar H, Marie Pyle A. Analysis of putative RNase sensitivity and protease insensitivity of demethylation activity in extracts from rat myoblasts. Nucleic Acids Res. 1998;26:5573–5580. doi: 10.1093/nar/26.24.5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bhattacharya SK, Ramchandani S, Cervoni N, Szyf M. A mammalian protein with specific demethylase activity for mCpG DNA. Nature. 1999;397:579–583. doi: 10.1038/17533. [DOI] [PubMed] [Google Scholar]
- 57.Ng HH, et al. MBD2 is a transcriptional repressor belonging to the MeCP1 histone deacetylase complex. Nature Genet. 1999;23:58–61. doi: 10.1038/12659. [DOI] [PubMed] [Google Scholar]
- 58.Hendrich B, Bird A. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol. Cell. Biol. 1998;18:6538–6547. doi: 10.1128/mcb.18.11.6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hendrich B, Guy J, Ramsahoye B, Wilson VA, Bird A. Closely related proteins MBD2 and MBD3 play distinctive but interacting roles in mouse development. Genes Dev. 2001;15:710–723. doi: 10.1101/gad.194101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Klose RJ, Kallin EM, Zhang Y. JmjC-domain-containing proteins and histone demethylation. Nature Rev. Genet. 2006;7:715–727. doi: 10.1038/nrg1945. [DOI] [PubMed] [Google Scholar]
- 61.Klose RJ, Zhang Y. Regulation of histone methylation by demethylimination and demethylation. Nature Rev. Mol. Cell Biol. 2007;8:307–318. doi: 10.1038/nrm2143. [DOI] [PubMed] [Google Scholar]
- 62.Smiley JA, Kundracik M, Landfried DA, Barnes VR, Sr, Axhemi AA. Genes of the thymidine salvage pathway: thymine-7-hydroxylase from a Rhodotorula glutinis cDNA library and iso-orotate decarboxylase from Neurospora crassa. Biochim. Biophys. Acta. 2005;1723:256–264. doi: 10.1016/j.bbagen.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 63.Lepesheva GI, Waterman MR. Sterol 14α-demethylase cytochrome P450 (CYP51), a P450 in all biological kingdoms. Biochim. Biophys. Acta. 2007;1770:467–477. doi: 10.1016/j.bbagen.2006.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 65.Zhu JK. Active DNA demethylation mediated by DNA glycosylases. Annu. Rev. Genet. 2009;43:143–166. doi: 10.1146/annurev-genet-102108-134205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Choi Y, et al. DEMETER, a DNA glycosylase domain protein, is required for endosperm gene imprinting and seed viability in Arabidopsis. Cell. 2002;110:33–42. doi: 10.1016/s0092-8674(02)00807-3. [DOI] [PubMed] [Google Scholar]
- 67.Gong Z, et al. ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell. 2002;111:803–814. doi: 10.1016/s0092-8674(02)01133-9. [DOI] [PubMed] [Google Scholar]
- 68.Agius F, Kapoor A, Zhu JK. Role of the Arabidopsis DNA glycosylase/lyase ROS1 in active DNA demethylation. Proc. Natl Acad. Sci. USA. 2006;103:11796–11801. doi: 10.1073/pnas.0603563103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gehring M, et al. DEMETER DNA glycosylase establishes MEDEA polycomb gene self-imprinting by allele-specific demethylation. Cell. 2006;124:495–506. doi: 10.1016/j.cell.2005.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Morales-Ruiz T, et al. DEMETER and REPRESSOR OF SILENCING 1 encode 5-methylcytosine DNA glycosylases. Proc. Natl Acad. Sci. USA. 2006;103:6853–6858. doi: 10.1073/pnas.0601109103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Penterman J, et al. DNA demethylation in the Arabidopsis genome. Proc. Natl Acad. Sci. USA. 2007;104:6752–6757. doi: 10.1073/pnas.0701861104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gehring M, Bubb KL, Henikoff S. Extensive demethylation of repetitive elements during seed development underlies gene imprinting. Science. 2009;324:1447–1451. doi: 10.1126/science.1171609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ortega-Galisteo AP, Morales-Ruiz T, Ariza RR, Roldan-Arjona T. Arabidopsis DEMETER-LIKE proteins DML2 and DML3 are required for appropriate distribution of DNA methylation marks. Plant Mol. Biol. 2008;67:671–681. doi: 10.1007/s11103-008-9346-0. [DOI] [PubMed] [Google Scholar]
- 74.Jost JP. Nuclear extracts of chicken embryos promote an active demethylation of DNA by excision repair of 5-methyldeoxycytidine. Proc. Natl Acad. Sci. USA. 1993;90:4684–4688. doi: 10.1073/pnas.90.10.4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jost JP, Siegmann M, Sun L, Leung R. Mechanisms of DNA demethylation in chicken embryos. Purification and properties of a 5-methylcytosine-DNA glycosylase. J. Biol. Chem. 1995;270:9734–9739. doi: 10.1074/jbc.270.17.9734. [DOI] [PubMed] [Google Scholar]
- 76.Fremont M, et al. Demethylation of DNA by purified chick embryo 5-methylcytosine-DNA glycosylase requires both protein and RNA. Nucleic Acids Res. 1997;25:2375–2380. doi: 10.1093/nar/25.12.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jost JP, et al. A chicken embryo protein related to the mammalian DEAD box protein p68 is tightly associated with the highly purified protein-RNA complex of 5-MeC-DNA glycosylase. Nucleic Acids Res. 1999;27:3245–3252. doi: 10.1093/nar/27.16.3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhu B, et al. 5-methylcytosine-DNA glycosylase activity is present in a cloned G/T mismatch DNA glycosylase associated with the chicken embryo DNA demethylation complex. Proc. Natl Acad. Sci. USA. 2000;97:5135–5139. doi: 10.1073/pnas.100107597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bennett MT, et al. Specificity of human thymine DNA glycosylase depends on N-glycosidic bond stability. J. Am. Chem. Soc. 2006;128:12510–12519. doi: 10.1021/ja0634829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Boland MJ, Christman JK. Characterization of Dnmt3b:thymine-DNA glycosylase interaction and stimulation of thymine glycosylase-mediated repair by DNA methyltransferase(s) and RNA. J. Mol. Biol. 2008;379:492–504. doi: 10.1016/j.jmb.2008.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li YQ, Zhou PZ, Zheng XD, Walsh CP, Xu GL. Association of Dnmt3a and thymine DNA glycosylase links DNA methylation with base-excision repair. Nucleic Acids Res. 2007;35:390–400. doi: 10.1093/nar/gkl1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhu B, et al. 5-Methylcytosine DNA glycosylase activity is also present in the human MBD4 (G/T mismatch glycosylase) and in a related avian sequence. Nucleic Acids Res. 2000;28:4157–4165. doi: 10.1093/nar/28.21.4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Santos F, Dean W. Epigenetic reprogramming during early development in mammals. Reproduction. 2004;127:643–651. doi: 10.1530/rep.1.00221. [DOI] [PubMed] [Google Scholar]
- 84.Millar CB, et al. Enhanced CpG mutability and tumorigenesis in MBD4-deficient mice. Science. 2002;297:403–405. doi: 10.1126/science.1073354. [DOI] [PubMed] [Google Scholar]
- 85.Hendrich B, Hardeland U, Ng HH, Jiricny J, Bird A. The thymine glycosylase MBD4 can bind to the product of deamination at methylated CpG sites. Nature. 1999;401:301–304. doi: 10.1038/45843. [DOI] [PubMed] [Google Scholar]
- 86.Conticello SG. The AID/APOBEC family of nucleic acid mutators. Genome Biol. 2008;9:229. doi: 10.1186/gb-2008-9-6-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Navaratnam N, et al. The p27 catalytic subunit of the apolipoprotein B mRNA editing enzyme is a cytidine deaminase. J. Biol. Chem. 1993;268:20709–20712. [PubMed] [Google Scholar]
- 88.Teng B, Burant CF, Davidson NO. Molecular cloning of an apolipoprotein B messenger RNA editing protein. Science. 1993;260:1816–1819. doi: 10.1126/science.8511591. [DOI] [PubMed] [Google Scholar]
- 89.Muramatsu M, et al. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 90.Revy P, et al. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2) Cell. 2000;102:565–575. doi: 10.1016/s0092-8674(00)00079-9. [DOI] [PubMed] [Google Scholar]
- 91.Morgan HD, Dean W, Coker HA, Reik W, Petersen-Mahrt SK. Activation-induced cytidine deaminase deaminates 5-methylcytosine in DNA and is expressed in pluripotent tissues: implications for epigenetic reprogramming. J. Biol. Chem. 2004;279:52353–52360. doi: 10.1074/jbc.M407695200. [DOI] [PubMed] [Google Scholar]
- 92.Popp C, et al. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature. 2010;463:1101–1105. doi: 10.1038/nature08829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rai K, et al. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and GADD45. Cell. 2008;135:1201–1212. doi: 10.1016/j.cell.2008.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hirano K, et al. Targeted disruption of the mouse apobec-1 gene abolishes apolipoprotein B mRNA editing and eliminates apolipoprotein B48. J. Biol. Chem. 1996;271:9887–9890. doi: 10.1074/jbc.271.17.9887. [DOI] [PubMed] [Google Scholar]
- 95.Morrison JR, et al. Apolipoprotein B RNA editing enzyme-deficient mice are viable despite alterations in lipoprotein metabolism. Proc. Natl Acad. Sci. USA. 1996;93:7154–7159. doi: 10.1073/pnas.93.14.7154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bandaru B, Wyszynski M, Bhagwat AS. HpaII methyltransferase is mutagenic in Escherichia coli. J. Bacteriol. 1995;177:2950–2952. doi: 10.1128/jb.177.10.2950-2952.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Shen JC, Rideout WM, 3rd, Jones PA. High frequency mutagenesis by a DNA methyltransferase. Cell. 1992;71:1073–1080. doi: 10.1016/s0092-8674(05)80057-1. [DOI] [PubMed] [Google Scholar]
- 98.Zingg JM, Shen JC, Yang AS, Rapoport H, Jones PA. Methylation inhibitors can increase the rate of cytosine deamination by (cytosine-5)-DNA methyltransferase. Nucleic Acids Res. 1996;24:3267–3275. doi: 10.1093/nar/24.16.3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wyszynski M, Gabbara S, Bhagwat AS. Cytosine deaminations catalyzed by DNA cytosine methyltransferases are unlikely to be the major cause of mutational hot spots at sites of cytosine methylation in Escherichia coli. Proc. Natl Acad. Sci. USA. 1994;91:1574–1578. doi: 10.1073/pnas.91.4.1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yebra MJ, Bhagwat AS. A cytosine methyltransferase converts 5-methylcytosine in DNA to thymine. Biochemistry. 1995;34:14752–14757. doi: 10.1021/bi00045a016. [DOI] [PubMed] [Google Scholar]
- 101.Chen D, et al. T:G mismatch-specific thymine-DNA glycosylase potentiates transcription of estrogen-regulated genes through direct interaction with estrogen receptor α. J. Biol. Chem. 2003;278:38586–38592. doi: 10.1074/jbc.M304286200. [DOI] [PubMed] [Google Scholar]
- 102.Jost JP, Thiry S, Siegmann M. Estradiol receptor potentiates, in vitro, the activity of 5-methylcytosine DNA glycosylase. FEBS Lett. 2002;527:63–66. doi: 10.1016/s0014-5793(02)03166-6. [DOI] [PubMed] [Google Scholar]
- 103.Barreto G, et al. Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature. 2007;445:671–675. doi: 10.1038/nature05515. [DOI] [PubMed] [Google Scholar]
- 104.Zhan Q. Gadd45a, a p53- and BRCA1-regulated stress protein, in cellular response to DNA damage. Mutat. Res. 2005;569:133–143. doi: 10.1016/j.mrfmmm.2004.06.055. [DOI] [PubMed] [Google Scholar]
- 105.Smith ML, et al. Interaction of the p53-regulated protein Gadd45 with proliferating cell nuclear antigen. Science. 1994;266:1376–1380. doi: 10.1126/science.7973727. [DOI] [PubMed] [Google Scholar]
- 106.Smith ML, et al. Antisense GADD45 expression results in decreased DNA repair and sensitizes cells to UV-irradiation or cisplatin. Oncogene. 1996;13:2255–2263. [PubMed] [Google Scholar]
- 107.Schmitz KM, et al. TAF12 recruits Gadd45a and the nucleotide excision repair complex to the promoter of rRNA genes leading to active DNA demethylation. Mol. Cell. 2009;33:344–353. doi: 10.1016/j.molcel.2009.01.015. [DOI] [PubMed] [Google Scholar]
- 108.Jin SG, Guo C, Pfeifer GP. GADD45A does not promote DNA demethylation. PLoS Genet. 2008;4:e1000013. doi: 10.1371/journal.pgen.1000013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Engel N, et al. Conserved DNA methylation in Gadd45a−/− mice. Epigenetics. 2009;4:98–9. doi: 10.4161/epi.4.2.7858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ma DK, et al. Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science. 2009;323:1074–1077. doi: 10.1126/science.1166859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Okada Y, Yamagata K, Hong K, Wakayama T, Zhang Y. A role for the elongator complex in zygotic paternal genome demethylation. Nature. 2010;463:554–558. doi: 10.1038/nature08732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Falnes PO, Johansen RF, Seeberg E. AlkB-mediated oxidative demethylation reverses DNA damage in Escherichia coli. Nature. 2002;419:178–182. doi: 10.1038/nature01048. [DOI] [PubMed] [Google Scholar]
- 113.Trewick SC, Henshaw TF, Hausinger RP, Lindahl T, Sedgwick B. Oxidative demethylation by Escherichia coli AlkB directly reverts DNA base damage. Nature. 2002;419:174–178. doi: 10.1038/nature00908. [DOI] [PubMed] [Google Scholar]
- 114.Tsukada Y, et al. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–816. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- 115.Warn-Cramer BJ, Macrander LA, Abbott MT. Markedly different ascorbate dependencies of the sequential α-ketoglutarate dioxygenase reactions catalyzed by an essentially homogeneous thymine 7-hydroxylase from Rhodotorula glutinis. J. Biol. Chem. 1983;258:10551–10557. [PubMed] [Google Scholar]
- 116.Cliffe LJ, et al. JBP1 and JBP2 are two distinct thymidine hydroxylases involved in J biosynthesis in genomic DNA of African trypanosomes. Nucleic Acids Res. 2009;37:1452–1462. doi: 10.1093/nar/gkn1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Yu Z, et al. The protein that binds to DNA base J in trypanosomatids has features of a thymidine hydroxylase. Nucleic Acids Res. 2007;35:2107–2115. doi: 10.1093/nar/gkm049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Tahiliani M, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ito S, et al. Role of Tet proteins in 5mC to 5hmC conversion, ES cell self-renewal, and ICM specification. Nature. 2010 Jul 18; doi: 10.1038/nature09303. (doi:10.1038/nature09303) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lorsbach RB, et al. TET1, a member of a novel protein family, is fused to MLL in acute myeloid leukemia containing the t(10;11)(q22;q23) Leukemia. 2003;17:637–641. doi: 10.1038/sj.leu.2402834. [DOI] [PubMed] [Google Scholar]
- 121.Ono R, et al. LCX, leukemia-associated protein with a CXXC domain, is fused to MLL in acute myeloid leukemia with trilineage dysplasia having t(10;11) (q22;q23) Cancer Res. 2002;62:4075–4080. [PubMed] [Google Scholar]
- 122.Penn NW, Suwalski R, O’Riley C, Bojanowski K, Yura R. The presence of 5-hydroxymethylcytosine in animal deoxyribonucleic acid. Biochem. J. 1972;126:781–790. doi: 10.1042/bj1260781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kothari RM, Shankar V. 5-Methylcytosine content in the vertebrate deoxyribonucleic acids: species specificity. J. Mol. Evol. 1976;7:325–329. doi: 10.1007/BF01743628. [DOI] [PubMed] [Google Scholar]
- 124.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Jin SG, Kadam S, Pfeifer GP. Examination of the specificity of DNA methylation profiling techniques towards 5-methylcytosine and 5-hydroxymethylcytosine. Nucleic Acids Res. 2010;38:e125. doi: 10.1093/nar/gkq223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Valinluck V, et al. Oxidative damage to methyl-CpG sequences inhibits the binding of the methyl-CpG binding domain (MBD) of methyl-CpG binding protein 2 (MeCP2) Nucleic Acids Res. 2004;32:4100–4108. doi: 10.1093/nar/gkh739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Valinluck V, Sowers LC. Endogenous cytosine damage products alter the site selectivity of human DNA maintenance methyltransferase DNMT1. Cancer Res. 2007;67:946–950. doi: 10.1158/0008-5472.CAN-06-3123. [DOI] [PubMed] [Google Scholar]
- 128.Cannon SV, Cummings A, Teebor GW. 5-Hydroxymethylcytosine DNA glycosylase activity in mammalian tissue. Biochem. Biophys. Res. Commun. 1988;151:1173–1179. doi: 10.1016/s0006-291x(88)80489-3. [DOI] [PubMed] [Google Scholar]
- 129.Boorstein RJ, et al. Definitive identification of mammalian 5-hydroxymethyluracil DNA N-glycosylase activity as SMUG1. J. Biol. Chem. 2001;276:41991–41997. doi: 10.1074/jbc.M106953200. [DOI] [PubMed] [Google Scholar]
- 130.Privat E, Sowers LC. Photochemical deamination and demethylation of 5-methylcytosine. Chem. Res. Toxicol. 1996;9:745–750. doi: 10.1021/tx950182o. [DOI] [PubMed] [Google Scholar]
- 131.Alegria AH. Hydroxymethylation of pyrimidine mononucleotides with formaldehyde. Biochim. Biophys. Acta. 1967;149:317–324. doi: 10.1016/0005-2787(67)90159-1. [DOI] [PubMed] [Google Scholar]
- 132.Liutkeviciute Z, Lukinavicius G, Masevicius V, Daujotyte D, Klimasauskas S. Cytosine-5-methyltransferases add aldehydes to DNA. Nature Chem. Biol. 2009;5:400–402. doi: 10.1038/nchembio.172. [DOI] [PubMed] [Google Scholar]
- 133.Abdel-Wahab O, et al. Genetic characterization of TET1, TET2, and TET3 alterations in myeloid malignancies. Blood. 2009;114:144–147. doi: 10.1182/blood-2009-03-210039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Delhommeau F, et al. Mutation in TET2 in myeloid cancers. N. Engl. J. Med. 2009;360:2289–2301. doi: 10.1056/NEJMoa0810069. [DOI] [PubMed] [Google Scholar]
- 135.Jankowska AM, et al. Loss of heterozygosity 4q24 and TET2 mutations associated with myelodysplastic/myeloproliferative neoplasms. Blood. 2009;113:6403–6410. doi: 10.1182/blood-2009-02-205690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kosmider O, et al. TET2 mutation is an independent favorable prognostic factor in myelodysplastic syndromes (MDSs) Blood. 2009;114:3285–3291. doi: 10.1182/blood-2009-04-215814. [DOI] [PubMed] [Google Scholar]
- 137.Langemeijer SM, et al. Acquired mutations in TET2 are common in myelodysplastic syndromes. Nature Genet. 2009;41:838–842. doi: 10.1038/ng.391. [DOI] [PubMed] [Google Scholar]
- 138.Mohamedali AM, et al. Novel TET2 mutations associated with UPD4q24 in myelodysplastic syndrome. J. Clin. Oncol. 2009;27:4002–4006. doi: 10.1200/JCO.2009.22.6985. [DOI] [PubMed] [Google Scholar]
- 139.Saint-Martin C, et al. Analysis of the ten-eleven translocation 2 (TET2) gene in familial myeloproliferative neoplasms. Blood. 2009;114:1628–1632. doi: 10.1182/blood-2009-01-197525. [DOI] [PubMed] [Google Scholar]
- 140.Tefferi A, et al. Detection of mutant TET2 in myeloid malignancies other than myeloproliferative neoplasms: CMML, MDS, MDS/MPN and AML. Leukemia. 2009;23:1343–1345. doi: 10.1038/leu.2009.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Nolte F, Hofmann WK. Myelodysplastic syndromes: molecular pathogenesis and genomic changes. Ann. Hematol. 2008;87:777–795. doi: 10.1007/s00277-008-0502-z. [DOI] [PubMed] [Google Scholar]
- 142.Daskalakis M, et al. Demethylation of a hypermethylated P15/INK4B gene in patients with myelodysplastic syndrome by 5-Aza-2′-deoxycytidine (decitabine) treatment. Blood. 2002;100:2957–64. doi: 10.1182/blood.V100.8.2957. [DOI] [PubMed] [Google Scholar]
- 143.Silverman LR, et al. Randomized controlled trial of azacitidine in patients with the myelodysplastic syndrome: a study of the cancer and leukemia group B. J. Clin. Oncol. 2002;20:2429–2440. doi: 10.1200/JCO.2002.04.117. [DOI] [PubMed] [Google Scholar]
- 144.Allen MD, et al. Solution structure of the nonmethyl-CpG-binding CXXC domain of the leukaemia-associated MLL histone methyltransferase. EMBO J. 2006;25:4503–4512. doi: 10.1038/sj.emboj.7601340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Hawkes NA, et al. Purification and characterization of the human elongator complex. J. Biol. Chem. 2002;277:3047–3052. doi: 10.1074/jbc.M110445200. [DOI] [PubMed] [Google Scholar]
- 146.Kim JH, Lane WS, Reinberg D. Human elongator facilitates RNA polymerase II transcription through chromatin. Proc. Natl Acad. Sci. USA. 2002;99:1241–1246. doi: 10.1073/pnas.251672198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Greenwood C, Selth LA, Dirac-Svejstrup AB, Svejstrup JQ. An iron-sulfur cluster domain in Elp3 important for the structural integrity of elongator. J. Biol. Chem. 2009;284:141–149. doi: 10.1074/jbc.M805312200. [DOI] [PubMed] [Google Scholar]
- 148.Li Q, et al. The elongator complex interacts with PCNA and modulates transcriptional silencing and sensitivity to DNA damage agents. PLoS Genet. 2009;5:e1000684. doi: 10.1371/journal.pgen.1000684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Paraskevopoulou C, Fairhurst SA, Lowe DJ, Brick P, Onesti S. The elongator subunit Elp3 contains a Fe4S4 cluster and binds S-adenosylmethionine. Mol. Microbiol. 2006;59:795–806. doi: 10.1111/j.1365-2958.2005.04989.x. [DOI] [PubMed] [Google Scholar]
- 150.Cokus SJ, et al. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215–219. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Li M, et al. Sensitive digital quantification of DNA methylation in clinical samples. Nature Biotechnol. 2009;27:858–863. doi: 10.1038/nbt.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Zilberman D, Gehring M, Tran RK, Ballinger T, Henikoff S. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nature Genet. 2007;39:61–69. doi: 10.1038/ng1929. [DOI] [PubMed] [Google Scholar]
- 153.Lister R, et al. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Flusberg BA, et al. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nature Methods. 2010;7:461–465. doi: 10.1038/nmeth.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nature Rev. Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 156.Gal-Yam EN, Saito Y, Egger G, Jones PA. Cancer epigenetics: modifications, screening, and therapy. Annu. Rev. Med. 2008;59:267–280. doi: 10.1146/annurev.med.59.061606.095816. [DOI] [PubMed] [Google Scholar]
- 157.Karberg S. Switching on epigenetic therapy. Cell. 2009;139:1029–1031. doi: 10.1016/j.cell.2009.11.038. [DOI] [PubMed] [Google Scholar]
- 158.Torres-Padilla ME, Bannister AJ, Hurd PJ, Kouzarides T, Zernicka-Goetz M. Dynamic distribution of the replacement histone variant H3.3 in the mouse oocyte and preimplantation embryos. Int. J. Dev. Biol. 2006;50:455–461. doi: 10.1387/ijdb.052073mt. [DOI] [PubMed] [Google Scholar]
- 159.van der Heijden GW, et al. Asymmetry in histone H3 variants and lysine methylation between paternal and maternal chromatin of the early mouse zygote. Mech. Dev. 2005;122:1008–1022. doi: 10.1016/j.mod.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 160.Arney KL, Bao S, Bannister AJ, Kouzarides T, Surani MA. Histone methylation defines epigenetic asymmetry in the mouse zygote. Int. J. Dev. Biol. 2002;46:317–320. [PubMed] [Google Scholar]
- 161.Cowell IG, et al. Heterochromatin, HP1 and methylation at lysine 9 of histone H3 in animals. Chromosoma. 2002;111:22–36. doi: 10.1007/s00412-002-0182-8. [DOI] [PubMed] [Google Scholar]
- 162.Liu H, Kim JM, Aoki F. Regulation of histone H3 lysine 9 methylation in oocytes and early preimplantation embryos. Development. 2004;131:2269–2280. doi: 10.1242/dev.01116. [DOI] [PubMed] [Google Scholar]
- 163.Santos F, Peters AH, Otte AP, Reik W, Dean W. Dynamic chromatin modifications characterise the first cell cycle in mouse embryos. Dev. Biol. 2005;280:225–236. doi: 10.1016/j.ydbio.2005.01.025. [DOI] [PubMed] [Google Scholar]
- 164.Erhardt S, et al. Consequences of the depletion of zygotic and embryonic enhancer of zeste 2 during preimplantation mouse development. Development. 2003;130:4235–4248. doi: 10.1242/dev.00625. [DOI] [PubMed] [Google Scholar]
- 165.Nakamura T, et al. PGC7/Stella protects against DNA demethylation in early embryogenesis. Nature Cell Biol. 2007;9:64–71. doi: 10.1038/ncb1519. [DOI] [PubMed] [Google Scholar]
- 166.Payer B, et al. Stella is a maternal effect gene required for normal early development in mice. Curr. Biol. 2003;13:2110–2117. doi: 10.1016/j.cub.2003.11.026. [DOI] [PubMed] [Google Scholar]
- 167.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 168.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 169.Gidekel S, Bergman Y. A unique developmental pattern of Oct-3/4 DNA methylation is controlled by a cis-demodification element. J. Biol. Chem. 2002;277:34521–34530. doi: 10.1074/jbc.M203338200. [DOI] [PubMed] [Google Scholar]
- 170.Hattori N, et al. Epigenetic regulation of Nanog gene in embryonic stem and trophoblast stem cells. Genes Cells. 2007;12:387–396. doi: 10.1111/j.1365-2443.2007.01058.x. [DOI] [PubMed] [Google Scholar]
- 171.Hattori N, et al. Epigenetic control of mouse Oct-4 gene expression in embryonic stem cells and trophoblast stem cells. J. Biol. Chem. 2004;279:17063–17069. doi: 10.1074/jbc.M309002200. [DOI] [PubMed] [Google Scholar]
- 172.Li JY, et al. Synergistic function of DNA methyltransferases Dnmt3a and Dnmt3b in the methylation of Oct4 and Nanog. Mol. Cell Biol. 2007;27:8748–8759. doi: 10.1128/MCB.01380-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Mikkelsen TS, et al. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Han DW, et al. Pluripotential reprogramming of the somatic genome in hybrid cells occurs with the first cell cycle. Stem Cells. 2008;26:445–454. doi: 10.1634/stemcells.2007-0553. [DOI] [PubMed] [Google Scholar]
- 175.Tada M, Takahama Y, Abe K, Nakatsuji N, Tada T. Nuclear reprogramming of somatic cells by in vitro hybridization with ES cells. Curr. Biol. 2001;11:1553–1558. doi: 10.1016/s0960-9822(01)00459-6. [DOI] [PubMed] [Google Scholar]
- 176.Hanna J, et al. Direct cell reprogramming is a stochastic process amenable to acceleration. Nature. 2009;462:595–601. doi: 10.1038/nature08592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Bhutani N, et al. Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature. 2010;463:1042–1047. doi: 10.1038/nature08752. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.