Abstract
Upon antigenic challenge, B cells enter the dark-zone (DZ) of germinal-centers (GC) to proliferate and hypermutate their immunoglobulin genes. Mutants with increased affinity are positively selected in the light-zone (LZ) to either differentiate into plasma and memory cells, or re-enter the DZ. The molecular circuits governing GC positive selection are not known. We show that the GC reaction requires the biphasic regulation of c-MYC expression, involving its transient induction during early GC commitment, its repression by BCL6 in DZ B cells, and its re-induction in B cells selected for DZ re-entry. Inhibition of MYC in vivo leads to GC collapse, indicating an essential role in GCs. These results have implications for the mechanism of GC selection and the role of MYC in lymphomagenesis.
Germinal centers (GC) are transient structures that form within secondary lymphoid organs. Within these structures, B cells are selected based on their ability to produce high-affinity antibodies1-3. The GC reaction is triggered by T cell-dependent antigens, in response to which B cells initiate vigorous proliferation coupled with somatic hypermutation (SHM) of their Immunoglobulin (Ig) genes. These events take place in the dark zone (DZ) of GCs4, and produce B cell populations expressing cell surface B cell receptors (BCR) with a range of affinities for the initiating antigen. DZ B cells quickly transit to the light zone (LZ)5, 6, where they exit the cell cycle and are selected, based on the affinity of their mutated B cell receptors (BCRs), to differentiate into memory B cells or plasma cells1-3.
Upon selection, B cells can also re-enter the DZ for additional cycles of SHM and division, in an iterative process known as ‘cyclic re-entry’ 6, 7. Thus, GC development requires coordinated signals dictating the induction of proliferation, cell cycle exit, cyclic re-entry, and differentiation, as well as the elimination of non-selected B cells by apoptosis. These signals, and their corresponding nuclear effectors, are only partially understood, in part due to the fact that the GC reaction cannot be reproduced in vitro. Nevertheless, their elucidation is important not only for the understanding of normal GC physiology, but also to explain the pathogenesis of B cell lymphomas, most of which derive from GC B cells and are thought to represent their deregulated phenotypes.
Gene expression profile analysis of purified GC B cells (originally identified as centroblasts and centrocytes) has provided a rather descriptive and schematic picture of the phases of GC development by identifying genes specifically expressed in these populations8, 9. Some of these genes have been shown to have essential roles in GC formation by genetic ablation experiments in mice10-12. In particular, the analysis of the genes targeted by the BCL6 proto-oncogene, which encodes a transcriptional repressor required for GC formation and involved in lymphomagenesis, has shed light on several unique features of GC B cells13. These include BCL6-mediated suppression of cell cycle arrest and plasma cell differentiation, modulation of the DNA damage response, and silencing of anti-apoptotic molecules such as BCL213, 14. However, these studies were performed on bulk GC purified B cells, CD77+ or CD77- GC B cell subpopulations, all of which represent heterogeneous mixtures of GC cells. More recently, the identification of novel markers for the isolation of mouse and human DZ and LZ GC B cells has led to a more precise definition of their respective phenotypes, which notably, appear to differ more in terms of functional status than differentiation stage6.
One surprising finding common to several of the above studies is that the expression of the proto-oncogene c-MYC (MYC) appears to be extremely low8 or virtually absent9 in bulk GC B cells. MYC encodes a transcription factor thought to be ubiquitously expressed in proliferating cells, where it controls cellular processes necessary for normal cell growth and proliferation via transcriptional and non-transcriptional mechanisms including ribosome biogenesis, metabolism, cell cycle and DNA replication, and telomere maintenance15-17. Hence MYC plays a fundamental role in all tissues undergoing rapid cellular expansion and renewal15-17. Thus, the apparent absence of MYC RNA and protein from most GC B cells, along with the observation that the MYC promoter is bound by BCL6, a known transcriptional repressor14, 18, 19, appeared inconsistent with the rapid-cycling, proliferative phenotype of GC B cells. This paradox, together with the growing notion that GC B cell populations may be more complex and heterogeneous than previously envisioned, prompted us to examine MYC expression in detail during GC development, and to determine its role in GC formation and maintenance.
Results
MYC expression is restricted to a subset of LZ B cells
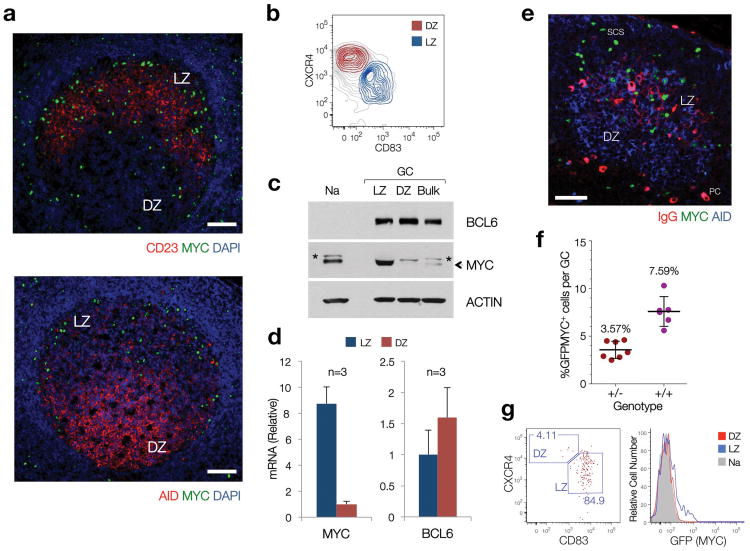
The issue of MYC expression in GC B cells remains a matter of debate. While previous studies have reported the virtual absence of MYC protein and/or mRNA in total populations of purified GC B cells9, recent work provided molecular evidence suggestive of MYC activation in the LZ of murine and human GC4, 6. We therefore investigated whether MYC expression could be detected in specific GC subpopulations or at specific times during GC formation. Immunofluorescence analysis of human reactive lymph nodes and tonsils revealed the presence of a small number of GC B cells expressing MYC, located preferentially within the GC LZ (Fig. 1a). Consistent with this observation, MYC mRNA and protein were found in B cells purified from the LZ of human tonsillar GCs (CXCR4lo, CD83hi), but not in their DZ counterparts (CXCR4hi, CD83lo)4, 6 (Fig. 1b-d; Supplementary Fig. 1). MYC protein levels in LZ B cells were several fold higher than those observed in Naive tonsillar B cells. Similarly, immunofluorescence analysis of murine lymphoid tissues after immunization identified a small number of MYC+ B cells within the LZ of GC (Fig. 1e). This topographical polarity was confirmed by flow cytometry of lymphoid tissues from a ‘reporter’ mouse strain in which one copy of the endogenous MYC allele was modified to encode a GFP-MYC fusion protein (GFPMYC mice20, Fig. 1f,g, Supplementary Fig. 1). Using this approach, we estimate that MYC+ GC B cells account for ∼8% of all GC B cells in a ‘mature’ murine GC (a 10-12 day steady-state GC, polarized into LZ and DZ2, 3, 21) (range 5-10%, Fig. 1f). Thus, MYC expression can be reproducibly detected in a small subpopulation of B cells in the LZ of both human and mouse GCs.
Figure 1. A subset of LZ GC B cells express MYC under physiologic conditions.
(a) Immunofluorescence staining, paraffin-embedded sections of reactive human lymph nodes. CD23, expressed in Follicular Dendritic cells50 (FDC), highlights the boundaries of the LZ. AID is used as a DZ marker, as previously reported4, 51. Scale Bars = 200μm. (b) Sorting profiles of LZ and DZ GC B cell subpopulations in human tonsils (See Supplementary Fig. 1) (c) Immunoblot of populations shown in (b). (Na), Naive B cells. (Bulk), bulk CD77+ GC B cells, isolated as in9. The asterisk denotes a non-specific band. Actin is used as loading control. (d) Quantitative RT-PCR for MYC and BCL6 mRNA levels in LZ and DZ B cell pools. Average of 3 independent cell pools per population (n=3). Error bars, standard deviation (SD). (e) Immunofluorescence staining, paraffin-embedded section, mouse lymph node (12 days after SRBC immunization). AID highlights the GC (DZ). IgG highlights the FDC network (i.e. LZ). Scale bar = 50μm. PC, plasma cell. SCS, subcapsular sinus (f) Number of MYC+ B cells, as assessed by flow cytometry (GFP=MYC) in LZ and DZ GC B cell subsets from GFPMYC mice (12 days post-immunization). (g) Distribution of GFPMYC+ GC B cells among LZ and DZ subsets. Left panel, dot plot analysis. Right panel, histogram shows the relative GFPMYC fluorescence intensities within these cell subsets. Shadowed histogram, Naive B cells (Na), used as a reference.
Kinetics of MYC expression during GC formation
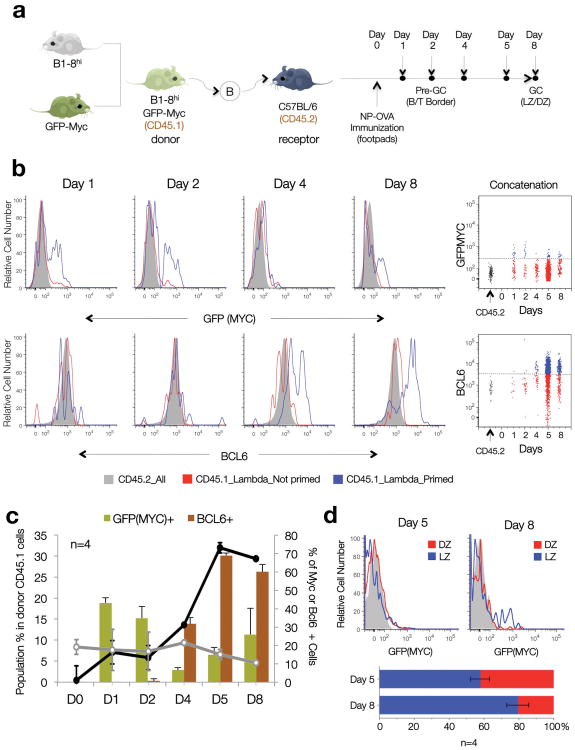
B cells commit to the GC fate in response to T-dependent antigens shortly after being primed by T cells in the interfollicular areas and at the ‘B-T border’ of secondary lymphoid tissues22, 23. Division of the GC into LZ and DZ takes place only later in the response1-3, and thus the presence of MYC+ GC B cells in the LZ suggested that their appearance could be a late event in the GC reaction. To address this issue, we analyzed the time of appearance of MYC+ GC B cells in the course of GC maturation (Fig. 2a). GFPMYC mice were crossed to B1-8hi mice, which bear a gene-targeted BCR that binds with high affinity to the hapten 4-hydroxy-3- nitrophenyl-acetyl (NP) when paired to an endogenous Igλ light chain24. Naive B cells from allotypically marked (CD45.1+) GFPMYC × B1-8hi mice were subsequently transferred into wildtype (WT) hosts, and their behavior was monitored by flow cytometry for GFPMYC expression at different time points after immunization with NP-conjugated ovalbumin (NP-OVA6). This protocol ensured the specific recruitment of B1-8hi B cells to GC at the expense of the host B cell pool25. In order to follow the kinetics of GC development, we also monitored transferred cells for BCL6 expression, which is induced relatively early upon GC commitment (3-4 days post-immunization) and is maintained throughout the GC reaction (>15 days)11, 26.
Figure 2. Alternating peaks of MYC and BCL6 expression during T cell-dependent antigen responses and GC formation.
(a) Diagrammatic representation of the experimental approach. (b) Representative histogram plots depict the distribution of GFPMYC (top) and BCL6 (bottom) protein expression in primed, Igλ, CD45.1+ (blue profiles); non-primed, Igλ, CD45.1+ B1-8hi B cells (red profiles), and CD45.2 host B cells (shadowed histograms) at different time points after NP-OVA immunization (see also Supplementary Fig. 2). The bidimensional dotplots shown on the right panels summarize the data by concatenating all samples (n=4). Dashed lines mark the background levels for GFPMYC and BCL6 fluorescent signals. (c) Bar and line graphs summarize the temporal evolution in cell numbers (black and grey lines, left axis) and GFPMYC and BCL6 protein expression (right axis), in the antigen-primed B1-8hi B cell population during GC formation. Error bars=SD (n=4). (d) Topographic distribution of GFPMYC positive, primed B cells, within each GC compartment (LZ, DZ) at Day 5 and 8 post-immunization (flow cytometry). The average number of GFPMYC positive cells in each compartment is summarized in the graph below (n=4). Error bars=SD.
As previously reported22, 23, 26, immunization with NP-OVA triggered a T-dependent antigen response that began at the B-T border and interfollicular areas and progressed to mature GCs by day 5-8 post-immunization (Supplementary Fig. 2). Coinciding with the initial expansion of the CD45.1+ donor cell population, an early peak of GFPMYC expression occurred 1-2 days after immunization in small clusters of B cells located in the interfollicular areas and, less frequently, at the B-T border (Fig. 2b, Supplementary Fig. 2). By day 4, the percentage of MYC-expressing cells rapidly decreased in coincidence with the progressive acquisition of BCL6 expression, a marker of GC fate commitment (Fig. 2b, Supplementary Fig. 2). At this time, most MYC+ cells were preferentially located in small-sized BCL6+ clusters within B cell follicles, corresponding to early GCs (Supplementary Fig. 2c-e; Day 4). From day 4 to day 8, the number of cells within each developing GC increased exponentially (Fig. 2c, Supplementary Fig. 2; equivalent to ∼10-12 population doublings contributing to a final ∼70% of the total CD45.1+ Igλ B cell pool) as the total number of MYC+ antigen specific B cells within these GC clusters decreased progressively, prior to increasing again in mature GCs (Day 8; Fig. 2c, Supplementary Fig. 2f). The topographic distribution of MYC+ cells within the GC also changed, shifting from the sparse distribution in early GC (Day 4-5, Fig. 2d, Supplementary Fig. 2e) to the preferential location in the LZ of mature GC (Day 8; Fig. 2d).
These observations indicate that, during T cell dependent immune responses MYC expression is induced early during B cell priming, is progressively suppressed during early GC expansion and DZ formation, and is then re-induced in a subpopulation of LZ B cells at the time when LZ and DZs segregate and GC selection begins.
BCL6 directly represses MYC transcription in DZ B cells
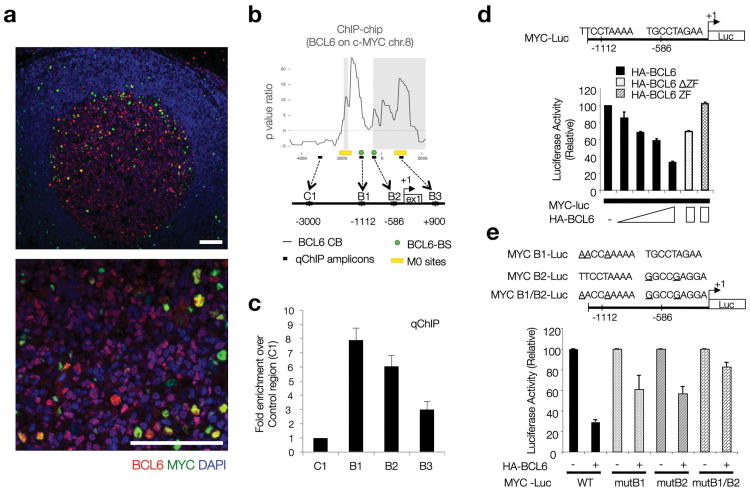
The progressive reduction in MYC+ cells at the onset of the GC reaction suggested that MYC expression might be actively regulated in this compartment. BCL6 is a candidate MYC repressor in this setting because it binds to the MYC promoter region in pre-B and transformed B cells14, 18, 19 and its expression appears to coincide with a decrease of MYC expression during the early stages of GC formation (Fig. 2, Supplementary Fig. 2). Consistent with this hypothesis, and analogous to the pattern in mouse B cells, immunofluorescence analysis of human secondary lymphoid tissues confirmed that expression of MYC and BCL6 in mature GCs was inversely correlated in the majority of GC B cells, with most GC B cells expressing either MYC or BCL6 (∼91% of all GC B cells; Fig. 3a, Supplementary Fig. 3), and only a small fraction (∼8%) of GC B cells showing coexpression of both proteins (Supplementary Fig. 3b,c). Accordingly, BCL6 mRNA and protein levels (as measured by mean fluorescence intensity upon specific immunostaining) were significantly lower in MYC+ GC B cells (Supplementary Fig. 3d).
Figure 3. BCL6 represses MYC protein expression in DZ GC B cells.
(a) Immunofluorescence co-staining for MYC and BCL6 in human reactive lymph nodes. BCL6 (red) highlights the boundaries of the GC. DAPI (blue), nuclear counterstain. Scale bars= 200μm. (b) BCL6 chromatin binding profile (ChIP-chip) at the MYC locus in human CD77+ GC B cells. The diagram shows the organization of this locus in the region around Exon1 and the transcription start site (+1). B6BS and M0 potential BCL6 binding sites are based on previously defined consensus14. The qChIP amplicons referred to in panel (c) are shown here as black boxes (B1-3, C1). Raw data corresponding to the ChIP-chip analysis at the MYC locus is provided in Supplementary Table 1, available online. (c) Quantitative PCR on DNA isolated from an independent BCL6 chromatin immunoprecipitation assay in CD77+ GC B cells. Fold enrichment refers to the ‘normalized’ binding at each region, relative to C1 (arbitrarily set to 1). ‘Normalized binding’ measures the relative enrichment in BCL6 immunoprecipitates over background (i.e. species or isotype matched irrelevant antibody). Average of 3 technical replicates (Error bar=SD). (d) Dual-Luciferase reporter assay, HEK293T cells. Effects of WT BCL6 or two defective truncations (as indicated) on the 1.2 Kb upstream MYC regulatory region depicted in the diagram, which includes two BCL6 potential binding sites. Average of 2 technical replicates from a representative experiment (Error bar=SD). (e) Same assay as in panel (d), but using Luciferase reporter constructs where point mutations on each putative BCL6 consensus site were introduced, as indicated. Average of 2 technical replicates from a representative experiment (Error bar=SD).
ChIP-on-chip analysis of human GC B cells isolated from reactive tonsils confirmed physical binding of BCL6 to the MYC gene promoter, within a ∼2Kb region 5′ to the Transcription Start Site (TSS, +1 in Fig. 3b, Supplementary Table 1). Quantitative PCR on ChIP-DNA confirmed the existence of two adjacent BCL6 binding sites that, upon sequence analysis, were found to include two clusters of BCL6 consensus binding sites (B6BS, M0)14(Fig. 3b,c). BCL6 binding to the MYC promoter appeared to occur equally in LZ and DZ GC B cells (Supplementary Fig. 3e,f). To verify the regulatory role of these binding sites, we performed transient transfection assays using a reporter construct in which the ∼2Kb MYC promoter region drives the transcription of a Luciferase gene (Fig. 3d). These assays showed a dose-dependent repression of Luciferase expression by BCL6 in HEK293T cells (Fig. 3d) and B cell lines (Supplementary Fig. 3g-j), which was dependent on the DNA-binding and transrepressor domains of BCL6 (ΔZF, ZF mutants, Fig. 3d), and largely on the presence of BCL6 consensus binding sites (Fig. 3e). The residual repression activity (<20%) observed after mutation of these two major consensus binding sites (last two bars in Fig. 3e) may be due the presence of lower affinity BCL6 binding motifs such as M0 sites (Fig. 3b). Together, these results indicate that BCL6 directly represses MYC transcription in the majority of GC B cells, and thus may be responsible for the loss of MYC expression in DZ and most LZ GC B cells.
Immune activation and high affinity BCRs in MYC+ GC cells
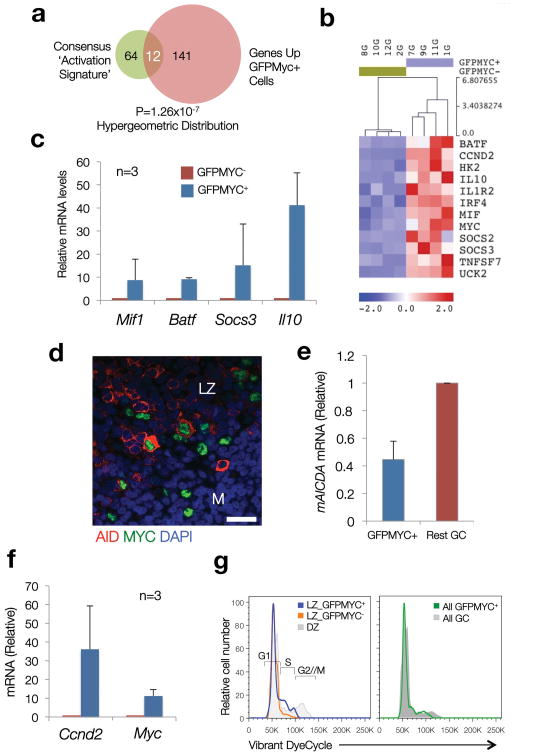
To gain insight into the factors controlling MYC expression in LZ B cells, we examined the phenotype of MYC+ GC B cells by gene expression profiling (GEP). To this end, we sorted GFP+ and GFP- GC B cells from spleens of GFPMYC reporter mice, 12 days after immunization with sheep red blood cells (SRBC). A signature of 425 genes reproducibly defined the GEP of GFPMYC positive vs. negative B cells by supervised analysis (Supplementary Fig. 4, Supplementary Table 2). This gene signature was highly enriched in LZ GC B cells, as predicted from the immunofluorescence and flow cytometry data (Supplementary Fig. 4c), and included: i) the expected upregulation of MYC target genes, with significant representation of genes involved in metabolism, a finding consistent with the significant enrichment in MYC target gene signatures detected in the GFPMYC positive population by GeneSet Enrichment Analysis27 (Table 1, Supplementary Table 3, Supplementary Fig. 4); ii) and the upregulation of genes encoding cell surface and adhesion molecules and cytokine receptors (i.e. Cd70 [Tnfsf7], Cd59a, Cd47 or Vcam1). This analysis also revealed the coordinated upregulation of early response genes (Egr2, Socs3, Irf4, Nfkbi, Il10, Batf) and genes involved in cell-cycle entry (Ccnd2, Cdk4 and Cdk6), which coincided with the enrichment for gene signatures related to immune activation (i.e. by BCR, CD40 and IL-2 signaling8, 28, 29) and the expression of AID (Aicda) in a substantial fraction of MYC+ cells (Fig. 4a-e, Table 1, Supplementary Table 3 and 4). Consistent with this activated phenotype, GFPMYC-positive GC B cells were found to be actively cycling (G0-G1 to S), in clear contrast to the quiescent nature of the remaining LZ GC B cells (Fig. 4f,g).
Table 1. Summary of gene signatures found enriched in GFPMYC+ GC B cells.
Gene Sets were scored and grouped in clusters based on similarities. Only gene sets significantly enriched (FDR<25%, P value ≤0.05) were included. See Methods and Supplementary Figure 4, Supplementary Tables 2 to 4 online for further details.
| MSigDB Compendium | Signature Cluster | Number of Gene Sets enriched | Average NES (St Dev) | Average P value (St Dev) | Average FDR (St Dev) |
|---|---|---|---|---|---|
| C2.CGP | MYC Gene Signatures | 10 | 2.30 (0.51) | 0.0004 (0.001) | 0.021 (0.052) |
| Embryonic Stem Cell and Hematopoietic Progenitor Signatures | 8 | 2.28 (0.55) | 0.0000 (0) | 0.018 (0.037) | |
| Immune Activation Signatures | 8 | 1.97 (0.42) | 0.0009 (0.001) | 0.044 (0.058) | |
| Metabolism-related Signatures | 7 | 2.77 (0.41) | 0.0000 (0) | 0.000 (0.001) | |
| Hypoxia pathways (HIF1α, HIF2α) | 7 | 2.05 (0.52) | 0.0044 (0.121) | 0.019 (0.028) | |
| C3.TFT | MYC motifs (Ebox) | 3 | 1.82 (0.42) | 0.0000 (0) | 0.032 (0.028) |
| C5.GO_BP | Mitochondrial Biogenesis and Metabolism | 5 | 2.02 (0.23) | 0.0010 (0.002) | 0.012 (0.010) |
| Biosynthesis | 6 | 1.73 (0.20) | 0.0027 (0.003) | 0.063 (0.054) | |
| RNA Metabolism | 7 | 2.19 (0.25) | 0.0008 (0.002) | 0.006 (0.015) | |
| Cell Cycle Entry & Progression | 6 | 1.53 (0.10) | 0.0160 (0.019) | 0.128 (0.050) | |
| Cell Survival (Negative Regulation of Apoptosis) | 3 | 1.60 (0.04) | 0.0012 (0.002) | 0.086 (0.020) |
Abbreviations: MsigDB, Molecular Signatures Database, Broad Institute; NES, Normalized Enrichment Score; FDR: False Discovery Rate; StDev, Standard Deviation.
Figure 4. Coordinated upregulation of immune activation signatures and cell cycle entry genes in GFPMYC+ GC B cells.
(a) A consensus ‘Activation Signature’, integrated by genes common to two or more published ‘immune activation’ signatures, was built (76 genes, details in Supplementary Table 4). The overlap between this signature and that of GFPMYC+ GC B cells (Venn diagram) was determined using a hypergeometric distribution (=12 genes). (Normalized Enrichment Score (NES)=2.34 (P val<0.00001; FDR=0%) using the GSEA algorithm). (b) Expression profile of the 12 consensus ‘immune activation’ genes enriched in GFPMYC+ cells. (c) Relative mRNA levels of selected ‘immune activation’ genes in GFPMYC+ cells, average of 3 independent cell pools (2 mice per pool). Error bars=SD. (d) Co-expression of AID (red) and MYC (green) in murine GC B cells (immunofluorescence, murine lymph node; DAPI (blue), nuclear counterstain). Scale bar, 20 μm. (e) Relative Aicda (AID) mRNA levels in GC B cell subsets isolated from GFPMYC mice, 12 days after SRBC immunization. Average of 2 independent experiments. Error bars, SD. (f) Relative Ccnd2 mRNA levels in GFPMYC+ and GFPMYC- populations (qRT-PCR). Average of 3 independent cell pools (2 mice per pool). (g) Cell cycle profile analysis in LZ and DZ GC B cells. Surface CXCR4-CD86 define the LZ and DZ subpopulations6. These histograms correspond to one mouse, representative of 3. Cell cycle phases are defined based on DNA content, adjusted to a Watson-Pragmatic model. Right panel, cell cycle distribution in bulk GFPMYC+ GC B cells, as compared to whole GC B cells.
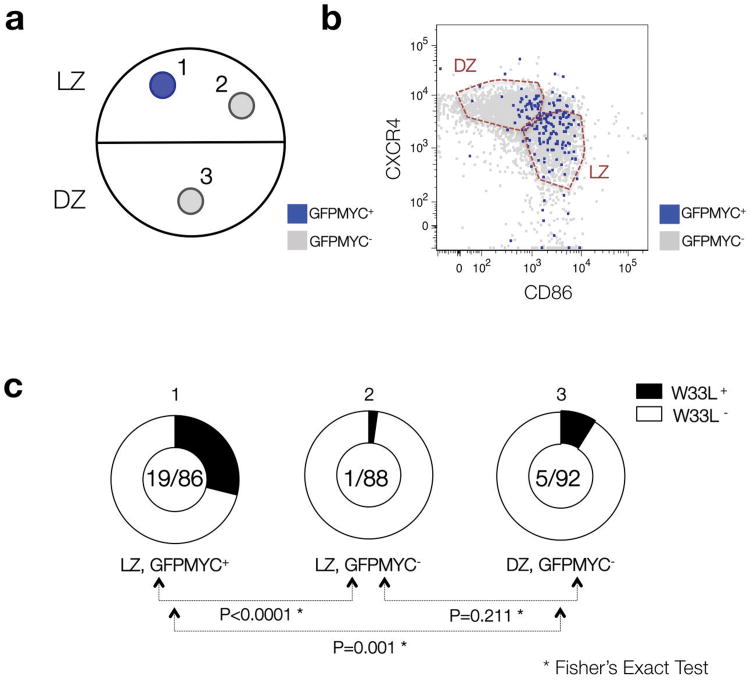
Since only positively-selected GC B cells are expected to be activated and re-enter the cell cycle in the LZ6, we hypothesized that MYC+ cells in the LZ might bear higher-affinity B cell receptors, which would lead to their positive selection. To address this possibility, we analyzed the antibody repertoire of MYC+ B cells sorted from GCs of GFPMYC mice immunized with NP-conjugated keyhole limpet hemocyanin (KLH). In NP-immunized C57BL6 mice, a Trp33 to Leu (W33L) mutation within the CDR1 region of the V186.2 Vh segment is associated with high affinity for the NP hapten21, 30, 31. On day 9 after immunization, GFP+ cells in the GC LZ (CXCR4loCD83hi population) were highly enriched in W33L+ sequences when compared to GFP- LZ cells (∼22% vs. ∼1%, respectively, P<0.0001) (Fig. 5; Supplementary Table 5). GC cells sorted from the DZ (CXCR4hiCD83lo) showed an intermediate proportion of W33L+ cells (∼5%), likely reflecting the absence of selection in this compartment. Together, these results strongly suggest that the MYC+ B cells in the GC LZ represent cells that are being actively selected for their high-affinity BCRs.
Figure 5. The B cell receptor (BCR) repertoire of GFPMYC positive GC B cells is enriched in high affinity variants.
(a) Diagrammatic representation of the 3 different populations purified from GC B cell pools of GFPMYC mice, 9 days after immunization with NP-KLH. (b) Distribution of GFPMYC+ and GFPMYC- subpopulations within these compartments. The detailed gating strategy to isolate the GC cell subsets is described in Supplementary Figure 1. (c) Doughnut charts show the fraction of sequenced Vh186.2 segments (CDR1 region) with a W33L mutation among all sequenced segments (W33L mutants/Total segments). See Supplementary Table 5 for additional details. The results correspond to the pooled data of two independent experiments, 2-3 mice analyzed per experiment. P value calculations are based on a Fisher's exact test.
MYC is induced in LZ B cells by T cell selection
LZ GC B cells with high-affinity receptors have increased access to T helper cells, which leads to their positive selection6, 22, 32. GFPMYC+ B cells expressed Cd70 (Tnfsf7, CD27L, see Fig. 4b), which encodes for a TNF family surface protein transiently induced in B cells by T cells, and known to modulate B cell proliferation through activation of the PI(3)K pathway (reviewed in33, 34). The robust upregulation of Cd70 mRNA in GFPMYC+ GC B cells correlated with its surface expression, which was restricted and specific to this GC population (Supplementary Fig. 5). This finding suggested that MYC+ GC B cells could be involved in productive interactions with T cells, an idea also supported by the observation that MYC+ GC B cells are preferentially located in the vicinity of LZ CD3+ T cells (as revealed by immunofluorescence analyses, Supplementary Fig. 5c).
To test the hypothesis that access to T cell help and GC positive selection involve induction of MYC expression, we mimicked the events taking place in the GC during affinity-based selection using DEC-205-mediated antigen delivery. In this system, a T cell antigen (OVA) fused to a chimeric antibody specific to the surface lectin DEC-205 (CD205, Ly75) was used to deliver antigen to DEC-205 expressing, NP-specific B cells within an otherwise DEC-205 deficient GC6. Targeted antigen delivery induces an increase in peptide-MHC presentation on DEC-205+ B cells, leading to their efficient interaction with GC T cells and subsequent selection for cyclic re-entry and differentiation into plasma cells6 (scheme in Fig. 6a, and Methods online).
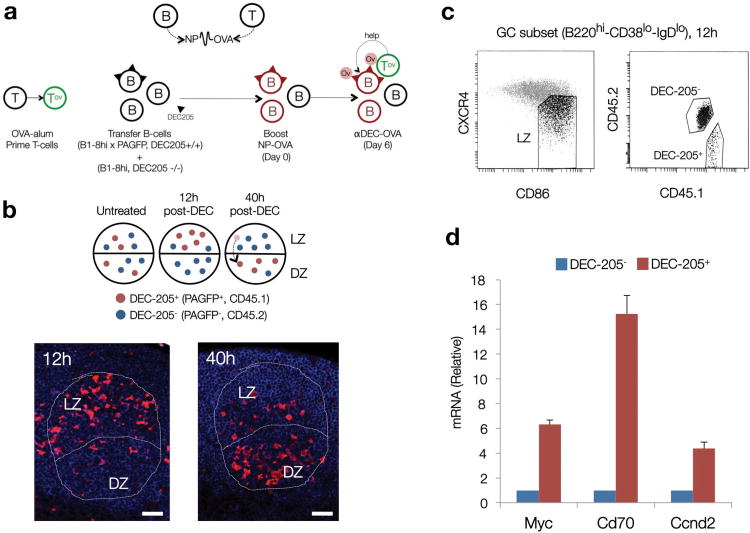
Figure 6. Access to T cell help triggers MYC expression prior to DZ re-entry.
(a) Diagrammatic representation of the experimental strategy. NP haptens are detected by B cells. Ovalbumin peptide conjugates (OVA), by T cells. DEC-205+ (CD205+, Ly75) cells are represented with triangular segments on their surface. Red cells, NP primed. OV labeled circles, OVA. (b) Top panel, expected dynamics and topographical distribution of DEC-205+ (PAGFP+) cells before (untreated) and after injection of OVA conjugated anti-DEC-205 antibodies (αDEC-OVA). Red cells, DEC-205+. Blue cells, DEC-205-. Bottom panels, immunofluorescence analysis on representative paraffin-embedded sections of popliteal lymph nodes after αDEC-OVA injection, at the indicated time points, to detect PAGFP+ (DEC-205+) B cells within B220+ GC compartments (outlined). LZ (Light Zone), DZ (Dark Zone). Scale bars = 50 μm. (c) Gating strategy used to isolate DEC-205+ and DEC-205- GC B cells from the LZ of the experimental mouse cohort 12 hours after αDEC-OVA injection. (d) Quantitative analysis for Myc, Cd70 and Ccnd2 mRNA levels in DEC-205+ and DEC-205- populations, isolated as shown in panel (c). Shown is one representative experiment (n=2). Average of 3 technical replicates +/- SD). See also Supplementary Fig. 5.
As previously reported6, injection of anti-DEC-205-OVA resulted in retention of DEC-205+ (B1-8hi, PAGFP+) GC B cells in the LZ at 12 hours after treatment, followed by their accumulation and expansion in the DZ at 40 hours (Fig. 6b). When in the LZ (12hr) these cells displayed robust upregulation of Myc, Cd70 and Ccnd2 mRNAs when compared to DEC-205- B cells in the same compartment (Fig. 6c,d). We thus conclude that increased access to T cell help triggers MYC expression in LZ GC B cells, establishing MYC expression as a bona fide marker for positive selection in the GC.
MYC is required for GC maintenance
According to the cyclic re-entry model, affinity-based selection of B cells in the LZ results in initiation of the cell cycle and return to the DZ. This selective flow of cells feeds the proliferating pool in the DZ, and is required to maintain the GC3, 6, 7, 35.
Having established a direct correlation between MYC expression, high BCR affinity and T cell-mediated selection (Figs. 4 and 5), we examined whether MYC was in fact required for GC maintenance through cyclic re-entry. To block the biological activity of MYC, we took advantage of a previously published combined Tet-On mouse model in which Omomyc, a selective MYC antagonist36, is placed under the control of a tetracycline-inducible element, responsive to a rtTA transgene driven by a CMV enhancer- β-actin promoter (TRE-Omomyc × actin-rtTA mouse37, 38). Omomyc causes severe perturbation of MYC transcriptional programs by abrogating its ability to bind canonical target genes, leading to cell-cycle arrest and apoptosis in a MYC-dependent manner37, 39. Since MYC expression within the GC is restricted to B cells (B220+) and specifically absent in follicular T-helper cells (CD4+CXCR5+PD1+) or non-lymphoid GC cell types (B220-, CD4-) (Supplementary Fig. 6), we reasoned that induction of Omomyc expression by doxycycline treatment would have cell-autonomous effects only in B cells.
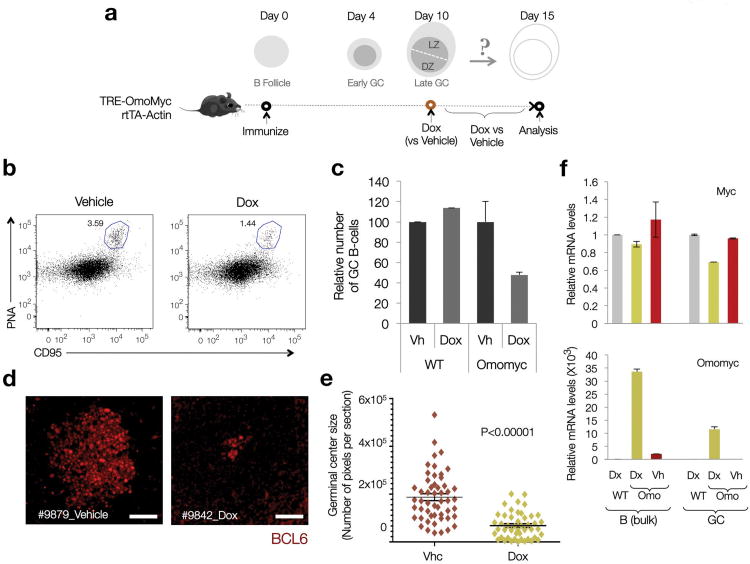
GCs were allowed to develop for 10 days after immunization with SRBC before induction of Omomyc expression for an additional 5 days (Fig. 7a). This treatment led to a ∼50% reduction in the percentage of GC B cells (B220+PNAhiCD95hi cells) (Fig. 7b,c) and a marked reduction in the average size of GCs, with a sizable fraction of the remaining GCs being composed of clusters of no more than 5-10 BCL6+ B cells (Fig. 7d,e; p<0.0001). No significant effects in GC numbers were observed in WT mice exposed to Doxycycline (Fig. 7c). In the few GCs that displayed a normal size, Omomyc expression was significantly lower than in the non-GC B cell population (Fig. 7f), suggesting suboptimal Omomyc induction, by either limited Doxycycline delivery or variability in the induction or response to the rtTA transgene40. Taken together this data indicates that MYC is required for GC maintenance.
Figure 7. MYC biological activity is required for normal GC maintenance.
(a) Diagrammatic representation of the experimental approach, as described in the main text, and the expected dynamics in T-dependent antigen responses and GC formation (Dox=Doxycycline) (SRBC= Sheep Red Blood Cells) (b) Representative Dot Plots on B cell splenic pools from Control and Dox-induced TRE-Omomyc rtTA-actin mice. The CD95hiPNAhi gate defines the GC population (blue outline). The number indicates the percentage of GC B cells. 2 mice per condition were analyzed. (c) Bar graphs show the average percentage of GC B cells in each group of samples (n=2 per condition). Error bars=SD. (d) Representative images of GC in paraffin-embedded sections of spleens from control (Vehicle) and Dox-induced (Dox) mice. BCL6 stain highlights GC B cells. Scale bars= 50 μm.(e) GC size distribution (surface) in control (Vhc) and Dox-induced (Dox) Omomyc mice (2 mice per condition pooled). Each marker corresponds to a single GC, its surface represented in number of pixels (∼50-55 GCs per mouse, see Methods online). Horizontal lines highlight average values. Error bars=SE. P value calculated using a two-tailed Student's T-test, unequal variance. (f) Quantitative analysis of Myc and Omomyc mRNA levels in bulk B cell pools and GC fractions. mRNA obtained from SRBC immunized wild-type (WT) mice was used as a reference control. Data analysis was performed by the ΔΔCt method, upon normalization to the geometric mean of Ct values for 3 different housekeeping genes (Gapdh, Actb and Hprt1). Bar graphs correspond to the average of 2 mice (Error bars=SD).
Discussion
Our results show that MYC expression is restricted to specific phases of GC development, namely during the initial expansion of naive B cells upon antigenic priming and at stages immediately preceding the LZ to DZ transition in mature GCs. MYC expression is repressed in the GC DZ. This specific pattern of expression clarifies and corrects previous observations on the absence of MYC expression in GC B cells8, 9, 41, 42. Notably, MYC expression is concurrent with important positive selection checkpoints promoting survival and proliferative expansion of affinity-selected B cells during the GC reaction. These findings have direct implications for our understanding of GC physiology, GC and pre-GC selection, and of the role of chromosomal translocations deregulating MYC expression in B cell lymphomas.
MYC is transiently expressed in the few B cells interacting with the antigen and accessing T cell help during the early steps of GC commitment, prior to the induction of BCL6 expression. Subsequent suppression of MYC expression is at least in part caused by active transcriptional repression by BCL6. This conclusion is based on kinetic data showing the disappearance of MYC expression at the time of BCL6 induction; on the binding of BCL6 to the MYC promoter14, 18, 19; and on our evidence that BCL6 directly represses MYC transcription. The signals that are thought to initiate the GC reaction – namely engagement of the BCR, CD40 and cytokine receptors (i.e. the receptor for the T cell-derived cytokine IL-2) – are known to negatively regulate BCL6 activity13, 43. Such signals would allow MYC to escape BCL6 repression during the first rounds of cell division that will give rise to the bulk of GC B cells. The transient nature of these signals, and the decreasing number of positive selection events found in GC as they mature21, would explain why MYC is not detectable in the majority of ‘mature’ GC B cells8, 9. Although not directly addressed by our results, MYC is required for the initiation of the GC reaction, as demonstrated by the lack of GCs in mice in which MYC is ablated early during GC induction (Calado et al., accompanying paper).
The finding that MYC transcription is suppressed by BCL6 in DZ B cells explains previous reports regarding the absence of MYC in GC B cells, and more specifically in DZ cells6, 8, 9. In fact, the virtual absence of MYC expression in DZ cells (which represent over two-thirds of all GC cells in mouse and human4, 6); and in the majority (>90%) of LZ cells, interferes with its detection in the small MYC+ subset of LZ cells when bulk GC populations are analyzed. We show that this can be overcome by use of assays with single-cell sensitivity (such as histology and flow cytometry) or by the separate analysis of fractionated LZ and DZ populations4, 6.
One question that remains unanswered is why MYC expression needs to be actively repressed in dividing DZ B cells, an observation that is at odds with the widely accepted notion of MYC as a proliferation marker in most tissues15-17. One possibility is that MYC repression is required to limit the rounds of cell division in the DZ, consistent with the idea that only a single round of cell-division may be required prior to each round of affinity-based selection3, 7. In addition, active repression of MYC expression could be important in order to allow the normal progression of SHM, since MYC, by abolishing transcription pausing of RNA polymerase II44 (an activity that is critical for AID-mediated SHM activity45) would impair the affinity maturation process in DZ B cells.
Our results show that, in mature GCs, MYC expression is detectable in a subset of LZ B cells with an activated phenotype that are re-entering cell cycle, suggesting that MYC+ cells are undergoing positive selection. Consistent with this idea, MYC-expressing LZ B cells are more likely to bear hypermutated Ig genes encoding high-affinity B cell receptors than their MYC- counterparts. Most notably, triggering positive selection by forcing B cell–T cell interactions using DEC-205 targeting leads to robust induction of MYC in the DEC-205+ population. Analogous to the early steps of the GC reaction, induction of MYC expression at this stage depends on BCR affinity and on the ability of B cells to recruit T cell help. Signals delivered through the BCR and/or by T cell help may thus block BCL6 activity, allowing for the transient induction of MYC transcription and the apparent co-expression of BCL6 and MYC in the LZ of ‘mature’ GCs (∼8% of all GC B cells) similar to what is observed in early GCs. Post-translational mechanisms (e.g. acetylation), which can rapidly inactivate the repressive activity of BCL6 in cells still expressing the protein46, may contribute to this phenomenon.
T cell-mediated selection induces the re-entry of selected LZ cells into the DZ for further rounds of proliferation and hypermutation, but also triggers their differentiation into plasmablasts6, 47. This raises the question of whether MYC expression is required for cyclic re-entry (and therefore for GC maintenance) or is instead upregulated only in cells leaving the GC as plasmablasts, which express the plasma cell marker and transcriptional repressor Blimp-1 (Prdm1), and are therefore in the process of downregulating BCL613. Our data is consistent with the first possibility, given that blockade of MYC activity in mature GC B cells leads to the collapse of the GC, which would not occur if MYC+ cells were simply destined to exit the GC. Aside from the transcription factor IRF4, which can also be induced early in response to immune activation signals8, 28, the gene expression profile of isolated MYC-expressing LZ GC B cells did not reveal signatures of plasma cell commitment (e.g. Blimp1 expression8, 47). Thus, we speculate that MYC induction may specify DZ re-entry as opposed to plasma cell differentiation.
The specific pattern of MYC expression during GC development has implications for our understanding of MYC-driven lymphomagenesis. Chromosomal translocations involving Ig loci and the c-MYC gene occur in all Burkitt Lymphomas (BL) and a fraction (∼20%) of Diffuse Large B cell Lymphomas48, 49, probably as a result of abnormal AID activity on an actively transcribed MYC locus49. The co-expression of MYC and AID proteins in a fraction of MYC+ GC B cells suggests that these events occur in this specific GC B cell subpopulation. Lymphoma-associated translocations, which juxtapose Ig transcription control elements to the c-MYC promoter and in some cases remove the BCL6 binding sites in the c-MYC 5′ region, may override BCL6-mediated repression due to strong Ig enhancer activity. These mechanisms can thus prevent the suppression of MYC transcription by BCL6 in the DZ and notably, uncouple MYC expression from positive selection. Consistent with this idea, BL display gene expression signatures that represent an admixture of DZ B cells and MYC-dependent signatures that are otherwise confined to the LZ4. Therefore, by overriding the physiologic control of MYC expression at critical checkpoints in the GC reaction (GC entry and DZ re-entry), Ig-MYC translocations may allow B cells to bypass affinity-based selection signals and impose a continuous re-entry phenotype that, by perturbing normal GC dynamics, may contribute to lymphomagenesis.
Accession numbers for gene expression data
Gene expression profile data has been deposited in the Gene Expression Omnibus (GEO) database, record number GSE38304.
Methods
Human tissue samples
Tonsils were obtained from routine tonsillectomies performed at the Children's Hospital of Columbia-Presbyterian Medical Center. Reactive lymph node sections were obtained from archived tissue. All samples were exempt from informed consent for being anonymized and de-identified residual material obtained after diagnosis, in compliance with the HHS Regulatory Guideline 45 CFR 46.101 (b)(4) for Exempt Human Research Subjects. All procedures were approved by the Institutional Ethics Committees.
Mouse strains
C57BL/6 mice were obtained from The Jackson Laboratory (USA). GFPMYC mice20, were a kind gift of Dr Barry Sleckman (Washington University). TRE-Omomyc/rtTA-Actin mice37, 38, were a kind gift of Drs Gerard Evan (University of Cambridge, UK) and Laura Soucek (VHIO, Barcelona). rtTA-Actin transgenic mice were generated in Steve Artandi's laboratory (Stanford University). B1-8hi 25 and DEC-205- (Ly75tm1Mnz) mice52 have been previously described. B1-8hi-GFPMYC compound mice were generated breeding B1-8hi and GFPMYC mice (back-crossed to C57BL/6 for >5 generations). Animals were housed in specific pathogen–free environments. All experiments were conformed to ethical principles and guidelines, revised and approved by the Institutional Animal Care and Use Committee (IACUC) in our institutions.
Immunization protocols and treatments
To generate GC responses, mice were immunized with either sheep red blood cells (SRBC; Cocalico Biologicals, Inc.) or 4-hydroxy-3-nitrophenyl-acetyl hapten (NP), conjugated to either Keyhole Limpet Hemocyanin (KLH) or ovalbumin (OVA) (BioSearch). SRBC immunization was performed by intraperitoneal injection (1×109 SRBC) and analysis at day 10-12. To generate larger amounts of GC B cells, we performed two sequential SRBC injections (Day 0, 1-2×108; Day 5, 1×109), and collected cells at Day 12. This protocol yielded ∼3 fold more GC B cells (∼7-9% of the B cell pool).
To analyze the dynamics of GC formation (Fig. 2), 5×106 naive B cells isolated from B1-8hi-GFPMyc donors, and 2×105 dsRed+ OT-II cells, isolated from dsRed-transgenic mice (Jackson Laboratory) (CD4+ T cell isolation kit, Miltenyi), were transferred to each C57BL/6 host mice (2 mice per time point, 3 experiments). Mice were subcutaneously immunized 24hr later in multiple sites (totaling 50 μg of NP3-OVA precipitated in alum; Imject Alum, ThermoScientific). Skin-draining lymph nodes were dissected for cell isolation and histology.
The experiment shown in Fig. 6 has been reported in detail 6. Briefly, host C57BL/6 mice were primed with an intraperitoneal injection of 50 μg of ovalbumin (OVA) precipitated in alum (T cell priming). 2-4 weeks after priming, a mixture of naive B cells isolated from CD45.1+-B1-8hi-DEC-205+ mice (15%) and CD45.2-B1-8hi-DEC-205- mice (85%) were transferred to each host mouse (∼5-10×106 total cells). Mice were boosted 24 hours later with 25 μg of NP-OVA (=Day 0) in the footpad. 6-7 days after NP-OVA immunization, mice received 3 μg of anti-DEC-OVA per footpad6, 53 prior to isolation of draining lymph nodes (12, 40 hours later).
In the experiment shown in Fig. 7, mice were immunized with SRBC (i.p.), and GCs were allowed to develop for 10 days. At day 10, 2 mg Doxycycline (Sigma, #9891) in 250 μl of PBS or PBS alone was administered (i.p.) to induce expression of the TRE-Omomyc transgene, which was further maintained by adding Doxycycline (2mg/mL) to the drinking water (+5% sucrose). Water was replaced every 48 hours for a 5-day period. Mice were sacrificed at Day 15, and tissues processed for flow cytometry, cell sorting and histology analyses.
Mononuclear and B cell isolation
Mononuclear cells were isolated from spleens of immunized mice by tissue homogenization, as described in 4. For gene expression analyses and DNA purification, ‘untouched’ total B cell fractions were isolated using magnetic beads (Mouse B cell Isolation Kit, Miltenyi; or EasySep Mouse B Cell Enrichment Kit, StemCell Technologies). Isolation of mononuclear cells from human tonsils was performed as previously described9.
Cell staining, flow cytometry analysis and cell sorting
Mononuclear cell pools or enriched B cell fractions were stained on ice in PBS-BSA (or PBS-FCS) with specific antibodies, washed and analyzed or sorted using BD LSRII or BD FACSAria flow cytometers. Biotin-conjugated antibodies were incubated separately first in a separate step (Supplementary Table 7 online for antibody details). Marker-based isolation of mouse GC B cell fractions was performed as shown in Supplementary Fig. 1 online.
For the experiments shown in Fig. 2 and Supplementary Fig. 2, mononuclear cells suspensions were stained and gated on the B220hi-CD45.1+ pool. IgD-Igλ distinguished primed and non-primed B cells. CD95, BCL6 (GC markers) allowed us to track GC commitment. To co-detect Bcl6 and GFP(MYC), cells were fixed for 10 minutes in 1% paraformaldehyde after surface staining, post-fixed and permeabilized using Cytofix-Cytoperm and Cytoperm plus buffers (BD Biosciences) as per manufacturer's instructions. BCL6 was detected using a PE-conjugated anti-BCL6 antibody (45-60 min at RT).
To generate cell cycle profiles, we used the Vibrant DyeCycle Violet Stain (Invitrogen-Life Technologies) in RPMI+2%FCS (5 μM) following manufacturer's instructions. Flow data was collected at 400 events per second.
All flow cytometry analyses and cell sorting procedures were performed at the HICCC Flow Cytometry Facility (Columbia University), or in the Nussenzweig laboratory. FlowJo software (v 8.8.6; TreeStar Inc.) was used for data analyses and plot rendering.
Immunofluorescence analysis on paraffin-embedded lymphoid tissues
Immunofluorescence was performed on 3 µm-thick sections of formalin-fixed, paraffin-embedded tissues, as previously described51. Heat-induced epitope retrieval was performed in citrate buffer (pH 6.0). Endogenous peroxidase was blocked in PBS+3% H2O2; endogenous biotin, using the Avidin-Biotin Blocking Kit (Vector). All slides were blocked in PBS-0.1%Tween-3%BSA plus 5% goat serum (Jackson Immunoresearch), prior to 4°C overnight incubation with specific primary antibodies. When using mouse primary antibodies on mouse tissues, an extra block with ‘M.O.M. Ig Block reagent’ (Vector) was performed. After repeated washes in PBS-0.1%Tween, tissue sections were incubated with fluorochrome-, HRP- or biotin-conjugated species and isotype-specific secondary antibodies (1hr, RT), washed and mounted (ProLong Gold Anti-Fade Reagent, Invitrogen). For Myc immunodetection, a polymer-enhanced HRP-conjugated secondary antibody (EnVision+ system, Dako) was used, and immunocomplexes detected using Tyramide-FITC amplification (Perkin-Elmer; 1:1000 for 3 min). For biotin-conjugated secondary antibodies (i.e., BCL6 immunodetection in human tissues), streptavidin-fluorochrome was added as a final step. Antibody details are provided in Supplementary Table 7 online. Technical validation of the anti-MYC antibody (Epitomics) was previously reported54 and confirmed in B cells (Supplementary Fig. 7).
Immunoblot analysis
LZ and DZ human tonsillar GC fractions sorted based on their surface markers (see Supplementary Fig. 1) were lysed in 1% SDS lysis buffer (50 mM Tris 8.0, 1 mM EDTA, 100 mM NaCl, 5 mM DTT, 1% SDS), and sonicated. Protein lysates were resolved in 4-12% Tris-Glycine gels (Novex, Life Technologies). Primary antibody incubations were performed overnight at 4°C. HRP-conjugated secondary antibodies and ECL reagent (Thermo) were used for detection.
Chromatin immunoprecipitation (ChIP)
BCL6 ChIP-on-chip profiles at the human MYC locus in CD77+ GC B cells were extracted from previous results14. For qChIP validation, an independent pool of CD77+ tonsillar GC B cells isolated as in9, 14 was used. Briefly, 40×106 cells (106/mL) were crosslinked in IMDM+1% formalin for 10 minutes, quenched in 0.1M Glycine, washed twice in PBS. Lysis procedures, antibodies and wash buffers have been previously described in detail14. Bound DNA was isolated as previously14, and PCR amplification of selected regions was achieved using primers listed in Supplementary Table 6 online.
Luciferase reporter assays
HEK293T cells were maintained in DMEM (Life Technologies) plus 10% FBS and 100 μg/mL Penicillin-Streptomycin (5% CO2, 37°C). A region corresponding to the ∼1.2Kb 5′ proximal segment of the human MYC gene (-1128/+63, GenBank NT_008046.16: 42020689-42021880) was PCR amplified and subcloned into pGL3-basic (Promega). This construct and a control reporter plasmid (pRL-SV40, Promega) were co-transfected in HEK293T cells using calcium-phosphate precipitation, together with plasmids encoding HA-tagged human wild-type BCL6 (pMT2T-HABcl6), or selective truncations deleting the entire Zinc Finger domain (Delta-ZF) or the N-terminal transrepression domain (ZF)46, 55. Luciferase expression was assessed 48 hours later (Dual-Luciferase reporter kit, Promega). Selected point mutations aimed at Bcl6 consensus sites were introduced by Site-Directed Mutagenesis. All constructs were further verified by Sanger sequencing.
IgH-V gene analysis
For the experiment shown in Fig. 5, 2 independent pools of 3 GFPMYC+ mice were immunized by single intraperitoneal injections of 100 μg of NP-KLH precipitated in complete Freund's adjuvant (Sigma). Genomic DNA from selected GC B cell subpopulations (∼2×105 cells per pool) was extracted (QIAamp DNA Micro Kit, QIAGEN), and 60-90 ng used as template for PCR amplification of Vh186.2-JH2 segments, predominant in GC responses to NP immunization21 (see Supplementary Table 6 online for primer sequences). PCR products were generated with high-fidelity DNA polymerase (36 cycles; Pfu TurboCx, Agilent-Stratagene), subcloned in the pSC-B vector (Strataclone Blunt PCR cloning kit, Stratagene) and sequenced (∼45-50 clones per sample, 2 independent experiments). IgH-V gene sequences were analyzed using the IMGT V-Quest tool (www.imgt.org), matched and compared to mouse germline Vh186.2 sequences, and clones with W33L mutations (CDR1) identified. All sequencing results are summarized in Supplementary Table 5. Differences between groups were determined using a Fisher's Exact Test (GraphPad Prism software).
DNA, RNA extraction, cDNA synthesis and quantitative RT-PCR
When isolating cells for gene expression profiling, cell pools were sorted in GFP+ and GFP- GC (PNAhi-CD95hi) B cell fractions (∼10,000-50,000 and 100,000 cells, respectively) and collected in Trizol Reagent (Life Technologies). Total RNA from sorted fractions was isolated using the Nucleospin RNA XS RNA isolation kit (Macherey-Nagel). RNA integrity was assessed using a BioAnalyzer 2100 (Agilent), and samples with RIN>9 were processed for cDNA synthesis (10-25 ng of total RNA) using the Ovation RNA Amplification System (NuGEN), as per manufacturer's instructions. This yielded ∼4-5 μg of cDNA (size range, 0.2 to 2 Kb).
Quantitative RT-PCR analyses (20-30 ng cDNA per reaction) were performed using a SYBR-green based PCR mix (Absolute Blue SYBR Green ROX mix, Thermo Scientific) and gene specific primers spanning at least one intron in the target gene (Supplementary Table 6, available online). All reactions were performed in triplicates using a 7300 Real Time PCR System (Applied Biosystems). Relative mRNA levels were calculated using the 2(-ΔΔCt) method, with Actb, Gapdh and Hprt1 as housekeeping reference genes.
Gene expression profile analysis, GSEA and statistical analyses
3.75 μg of total cDNA, obtained as described above, labeled and fragmented using the Encore Biotin Module (NuGEN), was hybridized on Mouse M430.2 microarrays (Affymetrix). Hybridization images were obtained using a GeneChip Scanner 3000 7G, connected to Command Console software (Affymetrix), and row analysis performed using the Affymetrix Expression Console Software. Gene expression data was extracted and normalized using the MAS5.0 algorithm. Four paired samples per condition (GFPMYC+, GFPMYC-), isolated from 4 independent pools of 2 mice, were analyzed. Unsupervised and Supervised Gene Expression analyses were performed using the SPLASH pattern discovery algorithm, part of the Bluegenes and Genes@Work software packages56, 57. Supervised analysis was performed using a Delta=2% (maximum deviation in normalized expression units) and full support (all samples in each phenotype satisfying the Delta condition). Under these parameters, we found 637 probesets differentially expressed between both groups. A list of these probesets is provided in Supplementary Table 2 online.
For the identification of enriched gene signatures, we used the Gene Set Enrichment Analysis tool27 (GSEA) available at the Broad Institute public server (http://www.broadinstitute.org/gsea/index.jsp). We screened the collection of signatures under categories C2 (curated gene sets), C3 (motif gene sets) and C5 (GO gene sets) to look for significant enrichments. Data was collapsed to gene names (median of probes). A total of 452 (C2.CGP), 38 (C3.TFT) and 74 (C5.GO_BP) signatures were significantly enriched in this comparison. A representative list of signatures (FDR<0.25, p≤0.05), grouped by categories, is shown in Table 1 and Supplementary Table 3. Given the number of samples (4×4), enrichment was assessed by using gene set permutations (×1000).
All statistical methods used to estimate p-values for differences in expression levels (qRT-PCR), numbers of events (cells, GC size) or other associations (IgH-V genes) were performed using GraphPad Prism software, and are detailed in each figure legend.
Omomyc GC image analysis
To analyze GCs size distribution (Fig. 7), we acquired high power view pictures of each individual GC present in a 12-mm2 spleen section (∼50-55 per mouse group). The size of each GC was recorded as number of pixels within the GC outline (highlighted by anti-Bcl6 staining), using the Analysis tool in Adobe Photoshop CS3 Software.
Supplementary Material
Acknowledgments
We are indebted to B.P. Sleckman, S. Artandi, L. Soucek and G. Evan for sharing mouse models; L. Reavie, I. Aifantis (NYU), M. Chesi and L. Bergsagel (Mayo Clinic) for providing access to these mice; and F.W. Alt for advice on immunization protocols. We thank R. Bosch for experimental help, and K. Basso, A.B. Holmes, L. Pasqualucci, M. Bansal, C. Lefebvre, P. Sumazin, and A. Califano for advice on data analysis; T. Mo for mouse husbandry, H. Tang, Q. Shen, V. Miljkovic (HICCC Genomics Facility), K. Gordon, C. Liu, and S. Tetteh (HICCC Flow Cytometry Facility) for technical assistance. We thank S. Serrano (Parc de Salut Mar, Barcelona) for providing human tissue samples; M. Jara-Acevedo (CSIC-USAL) for technical advice and U. Klein for insights and critically reading this manuscript.
Work supported by NIH grants PO1-CA092625 and RO1-CA37295 (to R.D-F.), RO1-AI037526 (to M.C.N.), and a Specialized Center of Research grant (Leukemia and Lymphoma Society) to R.D-F. D.D.S. was supported by NIH-NCI award K99/R00-CA151827. G.D.V is a Whitehead Fellow. M.C.N. is an HHMI investigator.
Footnotes
Competing Financial Interests: The authors declare no competing financial interests.
Author Contributions: D.D.S and G.D.V designed the study, performed the experiments, analyzed the data and wrote the manuscript. D.D.S performed the computational analyses. C.Y.Y. performed experiments. C.Y.Y., R.T.P. and M.S identified and characterized the BCL6-MYC transcriptional interaction. R.D.F., and M.C.N. supervised the project, provided direction in the study design and wrote the manuscript. All authors approved the final manuscript.
References
- 1.Allen CD, Okada T, Cyster JG. Germinal-center organization and cellular dynamics. Immunity. 2007;27:190–202. doi: 10.1016/j.immuni.2007.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.MacLennan IC. Germinal centers. Annu Rev Immunol. 1994;12:117–139. doi: 10.1146/annurev.iy.12.040194.001001. [DOI] [PubMed] [Google Scholar]
- 3.Victora GD, Nussenzweig MC. Germinal centers. Annu Rev Immunol. 2012;30:429–457. doi: 10.1146/annurev-immunol-020711-075032. [DOI] [PubMed] [Google Scholar]
- 4.Victora GD, et al. Identification of human germinal center light and dark zone cells and their relationship to human B cell lymphomas. Blood. 2012 doi: 10.1182/blood-2012-03-415380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allen CD, Okada T, Tang HL, Cyster JG. Imaging of germinal center selection events during affinity maturation. Science. 2007;315:528–531. doi: 10.1126/science.1136736. [DOI] [PubMed] [Google Scholar]
- 6.Victora GD, et al. Germinal center dynamics revealed by multiphoton microscopy with a photoactivatable fluorescent reporter. Cell. 2010;143:592–605. doi: 10.1016/j.cell.2010.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oprea M, Perelson AS. Somatic mutation leads to efficient affinity maturation when centrocytes recycle back to centroblasts. J Immunol. 1997;158:5155–5162. [PubMed] [Google Scholar]
- 8.Shaffer AL, et al. Signatures of the immune response. Immunity. 2001;15:375–385. doi: 10.1016/s1074-7613(01)00194-7. [DOI] [PubMed] [Google Scholar]
- 9.Klein U, et al. Transcriptional analysis of the B cell germinal center reaction. Proc Natl Acad Sci U S A. 2003;100:2639–2644. doi: 10.1073/pnas.0437996100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dent AL, Shaffer AL, Yu X, Allman D, Staudt LM. Control of inflammation, cytokine expression, and germinal center formation by BCL-6. Science. 1997;276:589–592. doi: 10.1126/science.276.5312.589. [DOI] [PubMed] [Google Scholar]
- 11.Ye BH, et al. The BCL-6 proto-oncogene controls germinal-centre formation and Th2-type inflammation. Nat Genet. 1997;16:161–170. doi: 10.1038/ng0697-161. [DOI] [PubMed] [Google Scholar]
- 12.Schubart DB, Rolink A, Kosco-Vilbois MH, Botteri F, Matthias P. B-cell-specific coactivator OBF-1/OCA-B/Bob1 required for immune response and germinal centre formation. Nature. 1996;383:538–542. doi: 10.1038/383538a0. [DOI] [PubMed] [Google Scholar]
- 13.Basso K, Dalla-Favera R. Roles of BCL6 in normal and transformed germinal center B cells. Immunol Rev. 2012;247:11. doi: 10.1111/j.1600-065X.2012.01112.x. [DOI] [PubMed] [Google Scholar]
- 14.Basso K, et al. Integrated biochemical and computational approach identifies BCL6 direct target genes controlling multiple pathways in normal germinal center B cells. Blood. 2010;115:975–984. doi: 10.1182/blood-2009-06-227017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dang CV. MYC on the Path to Cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eilers M, Eisenman RN. Myc's broad reach. Genes Dev. 2008;22:2755–2766. doi: 10.1101/gad.1712408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grandori C, Cowley SM, James LP, Eisenman RN. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu Rev Cell Dev Biol. 2000;16:653–699. doi: 10.1146/annurev.cellbio.16.1.653. [DOI] [PubMed] [Google Scholar]
- 18.Ci W, et al. The BCL6 transcriptional program features repression of multiple oncogenes in primary B cells and is deregulated in DLBCL. Blood. 2009;113:5536–5548. doi: 10.1182/blood-2008-12-193037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nahar R, et al. Pre-B cell receptor-mediated activation of BCL6 induces pre-B cell quiescence through transcriptional repression of MYC. Blood. 2011;118:4174–4178. doi: 10.1182/blood-2011-01-331181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang CY, Bredemeyer AL, Walker LM, Bassing CH, Sleckman BP. Dynamic regulation of c-Myc proto-oncogene expression during lymphocyte development revealed by a GFP-c-Myc knock-in mouse. Eur J Immunol. 2008;38:342–349. doi: 10.1002/eji.200737972. [DOI] [PubMed] [Google Scholar]
- 21.Jacob J, Kelsoe G. In situ studies of the primary immune response to (4-hydroxy-3-nitrophenyl)acetyl. II. A common clonal origin for periarteriolar lymphoid sheath-associated foci and germinal centers. J Exp Med. 1992;176:679–687. doi: 10.1084/jem.176.3.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schwickert TA, et al. A dynamic T cell-limited checkpoint regulates affinity-dependent B cell entry into the germinal center. J Exp Med. 2011;208:1243–1252. doi: 10.1084/jem.20102477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garside P, et al. Visualization of specific B and T lymphocyte interactions in the lymph node. Science. 1998;281:96–99. doi: 10.1126/science.281.5373.96. [DOI] [PubMed] [Google Scholar]
- 24.Shih TA, Roederer M, Nussenzweig MC. Role of antigen receptor affinity in T cell-independent antibody responses in vivo. Nat Immunol. 2002;3:399–406. doi: 10.1038/ni776. [DOI] [PubMed] [Google Scholar]
- 25.Shih TA, Meffre E, Roederer M, Nussenzweig MC. Role of BCR affinity in T cell dependent antibody responses in vivo. Nat Immunol. 2002;3:570–575. doi: 10.1038/ni803. [DOI] [PubMed] [Google Scholar]
- 26.Kitano M, et al. Bcl6 protein expression shapes pre-germinal center B cell dynamics and follicular helper T cell heterogeneity. Immunity. 2011;34:961–972. doi: 10.1016/j.immuni.2011.03.025. [DOI] [PubMed] [Google Scholar]
- 27.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Glynne R, et al. How self-tolerance and the immunosuppressive drug FK506 prevent B-cell mitogenesis. Nature. 2000;403:672–676. doi: 10.1038/35001102. [DOI] [PubMed] [Google Scholar]
- 29.Klaus SJ, Parker DC. Inducible cell contact signals regulate early activation gene expression during B-T lymphocyte collaboration. J Immunol. 1992;149:1867–1875. [PubMed] [Google Scholar]
- 30.Cumano A, Rajewsky K. Structure of primary anti-(4-hydroxy-3-nitrophenyl)acetyl (NP) antibodies in normal and idiotypically suppressed C57BL/6 mice. Eur J Immunol. 1985;15:512–520. doi: 10.1002/eji.1830150517. [DOI] [PubMed] [Google Scholar]
- 31.Rajewsky K, Forster I, Cumano A. Evolutionary and somatic selection of the antibody repertoire in the mouse. Science. 1987;238:1088–1094. doi: 10.1126/science.3317826. [DOI] [PubMed] [Google Scholar]
- 32.Batista FD, Iber D, Neuberger MS. B cells acquire antigen from target cells after synapse formation. Nature. 2001;411:489–494. doi: 10.1038/35078099. [DOI] [PubMed] [Google Scholar]
- 33.Nolte MA, van Olffen RW, van Gisbergen KP, van Lier RA. Timing and tuning of CD27-CD70 interactions: the impact of signal strength in setting the balance between adaptive responses and immunopathology. Immunol Rev. 2009;229:216–231. doi: 10.1111/j.1600-065X.2009.00774.x. [DOI] [PubMed] [Google Scholar]
- 34.Borst J, Hendriks J, Xiao Y. CD27 and CD70 in T cell and B cell activation. Curr Opin Immunol. 2005;17:275–281. doi: 10.1016/j.coi.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 35.Hauser AE, Shlomchik MJ, Haberman AM. In vivo imaging studies shed light on germinal-centre development. Nat Rev Immunol. 2007;7:499–504. doi: 10.1038/nri2120. [DOI] [PubMed] [Google Scholar]
- 36.Soucek L, et al. Design and properties of a Myc derivative that efficiently homodimerizes. Oncogene. 1998;17:2463–2472. doi: 10.1038/sj.onc.1202199. [DOI] [PubMed] [Google Scholar]
- 37.Soucek L, et al. Modelling Myc inhibition as a cancer therapy. Nature. 2008;455:679–683. doi: 10.1038/nature07260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sarin KY, et al. Conditional telomerase induction causes proliferation of hair follicle stem cells. Nature. 2005;436:1048–1052. doi: 10.1038/nature03836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Savino M, et al. The action mechanism of the Myc inhibitor termed Omomyc may give clues on how to target Myc for cancer therapy. PLoS One. 2011;6:e22284. doi: 10.1371/journal.pone.0022284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Premsrirut PK, et al. A rapid and scalable system for studying gene function in mice using conditional RNA interference. Cell. 2011;145:145–158. doi: 10.1016/j.cell.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cutrona G, et al. c-myc proto-oncogene expression by germinal center B cells isolated from human tonsils. Ann N Y Acad Sci. 1997;815:436–439. doi: 10.1111/j.1749-6632.1997.tb52096.x. [DOI] [PubMed] [Google Scholar]
- 42.Martinez-Valdez H, et al. Human germinal center B cells express the apoptosis-inducing genes Fas, c-myc, P53, and Bax but not the survival gene bcl-2. J Exp Med. 1996;183:971–977. doi: 10.1084/jem.183.3.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oestreich KJ, Mohn SE, Weinmann AS. Molecular mechanisms that control the expression and activity of Bcl-6 in T(H)1 cells to regulate flexibility with a T(FH)-like gene profile. Nat Immunol. 2012;13:405–411. doi: 10.1038/ni.2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rahl PB, et al. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pavri R, et al. Activation-induced cytidine deaminase targets DNA at sites of RNA polymerase II stalling by interaction with Spt5. Cell. 2010;143:122–133. doi: 10.1016/j.cell.2010.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bereshchenko OR, Gu W, Dalla-Favera R. Acetylation inactivates the transcriptional repressor BCL6. Nat Genet. 2002;32:606–613. doi: 10.1038/ng1018. [DOI] [PubMed] [Google Scholar]
- 47.Phan TG, et al. High affinity germinal center B cells are actively selected into the plasma cell compartment. J Exp Med. 2006;203:2419–2424. doi: 10.1084/jem.20061254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kuppers R. Mechanisms of B-cell lymphoma pathogenesis. Nat Rev Cancer. 2005;5:251–262. doi: 10.1038/nrc1589. [DOI] [PubMed] [Google Scholar]
- 49.Nussenzweig A, Nussenzweig MC. Origin of chromosomal translocations in lymphoid cancer. Cell. 2010;141:27–38. doi: 10.1016/j.cell.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hardie DL, Johnson GD, Khan M, MacLennan IC. Quantitative analysis of molecules which distinguish functional compartments within germinal centers. Eur J Immunol. 1993;23:997–1004. doi: 10.1002/eji.1830230502. [DOI] [PubMed] [Google Scholar]
- 51.Cattoretti G, et al. Nuclear and cytoplasmic AID in extrafollicular and germinal center B cells. Blood. 2006;107:3967–3975. doi: 10.1182/blood-2005-10-4170. [DOI] [PubMed] [Google Scholar]
- 52.Guo M, et al. A monoclonal antibody to the DEC-205 endocytosis receptor on human dendritic cells. Hum Immunol. 2000;61:729–738. doi: 10.1016/s0198-8859(00)00144-0. [DOI] [PubMed] [Google Scholar]
- 53.Jiang W, et al. The receptor DEC-205 expressed by dendritic cells and thymic epithelial cells is involved in antigen processing. Nature. 1995;375:151–155. doi: 10.1038/375151a0. [DOI] [PubMed] [Google Scholar]
- 54.Gurel B, et al. Nuclear MYC protein overexpression is an early alteration in human prostate carcinogenesis. Mod Pathol. 2008;21:1156–1167. doi: 10.1038/modpathol.2008.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Phan RT, Dalla-Favera R. The BCL6 proto-oncogene suppresses p53 expression in germinal-centre B cells. Nature. 2004;432:635–639. doi: 10.1038/nature03147. [DOI] [PubMed] [Google Scholar]
- 56.Califano A, Stolovitzky G, Tu Y. Analysis of gene expression microarrays for phenotype classification. Proceedings/… International Conference on Intelligent Systems for Molecular Biology; ISMB. International Conference on Intelligent Systems for Molecular Biology. 2000;8:75–85. [PubMed] [Google Scholar]
- 57.Klein U, et al. Gene expression profiling of B cell chronic lymphocytic leukemia reveals a homogeneous phenotype related to memory B cells. J Exp Med. 2001;194:1625–1638. doi: 10.1084/jem.194.11.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.