Abstract
Many plant and animal bacterial pathogens assemble a needle-like nanomachine, the type III secretion system (T3SS), to inject virulence proteins directly into eukaryotic cells to initiate infection. The ability of bacteria to inject effectors into host cells is essential for infection, survival, and pathogenesis for many Gram-negative bacteria, including Salmonella, Escherichia, Shigella, Yersinia, Pseudomonas, and Chlamydia spp. These pathogens are responsible for a wide variety of diseases, such as typhoid fever, large-scale food-borne illnesses, dysentery, bubonic plague, secondary hospital infections, and sexually transmitted diseases. The T3SS consists of structural and nonstructural proteins. The structural proteins assemble the needle apparatus, which consists of a membrane-embedded basal structure, an external needle that protrudes from the bacterial surface, and a tip complex that caps the needle. Upon host cell contact, a translocon is assembled between the needle tip complex and the host cell, serving as a gateway for translocation of effector proteins by creating a pore in the host cell membrane. Following delivery into the host cytoplasm, effectors initiate and maintain infection by manipulating host cell biology, such as cell signaling, secretory trafficking, cytoskeletal dynamics, and the inflammatory response. Finally, chaperones serve as regulators of secretion by sequestering effectors and some structural proteins within the bacterial cytoplasm. This review will focus on the latest developments and future challenges concerning the structure and biophysics of the needle apparatus.
OVERVIEW OF THE NEEDLE APPARATUS
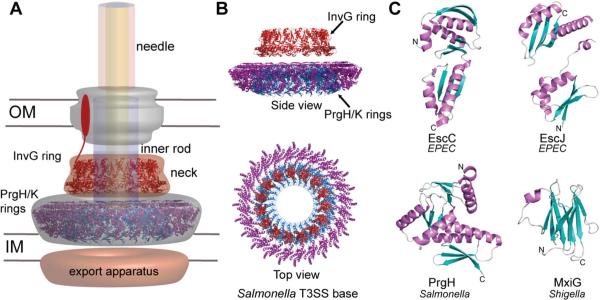
More than a dozen different structural proteins assemble to form the type III secretion system (T3SS) needle apparatus. The needle complex, which consists of a basal structure and an external needle, was first visualized by electron microscopy of osmotically shocked Salmonella typhimurium.1 Similar structures were also observed in other bacteria, such as Shigella flexneri,2Yersinia pestis,3Escherichia coli,4 and Pseudomonas aeruginosa.5 Following years of extensive genetic, biochemical, and biophysical research, the core components of the complex were identified. The ~3.5 MDa needle complex contains a basal structure spanning the periplasm that is embedded within the inner and outer bacterial membranes followed by a needle-like structure that protrudes into the extracellular space (Figure 1). In Salmonella, the basal structure is assembled from oligomeric rings of InvG, PrgK, and PrgH,6 and an inner rod structure formed by PrgJ.7 The basal structure supports the entire needle complex by anchoring the needle on the bacterial membranes. The needle itself is composed of protomers of PrgI arranged in a spiral symmetry around a central ~25 Å diameter channel to allow the passage of effector proteins.8 At the needle tip, multiple copies of SipD form a platform for the translocon.9 Upon contact with the host cell, the assembly of the entire needle apparatus is completed by the formation of the translocon, which is formed by the transmembrane proteins SipB and SipC.9 Structural homologues of each protein are present in the T3SSs of other Gram-negative bacteria (Table 1). The assembly of the needle apparatus is a highly regulated process involving specific protein–protein, protein–membrane, and protein–small molecule interactions. Likewise, the order of protein secretion is also regulated;10 for example, needle proteins pass through the nascent apparatus prior to the tip and the translocon proteins. Each protein plays a critical role in the proper assembly of the needle apparatus, as null mutations of any of the structural components render bacteria incapable of assembling a functional needle apparatus.11 The following sections will focus on the structure, biophysics, and protein–protein interactions of the core components of the T3SS needle apparatus.
Figure 1.
Cartoon of the type III secretion system showing the needle apparatus in contact with the host cell.
Table 1.
Structural Proteins of the T3SS Needle Apparatus
| role | Salmonella | Shigella | Yersinia | EPEC | Pseudomonas |
|---|---|---|---|---|---|
| translocon | SipB | IpaB | YopB | EspB | PopB |
| translocon | SipC | IpaC | YopD | EspD | PopD |
| needle tip | SipD | IpaD | LcrV | EspA | PcrV |
| needle | PrgI | MxiH | YscF | EscF | PscF |
| OM ring | InvG | MxiD | YscC | EscC | PscC |
| inner rod | PrgJ | MxiI | YscI | EscI | PscI |
| IM ring | PrgK | MxiJ | YscJ | EscJ | PscJ |
| PrgH | MxiG | YscD | EscD | PscD | |
| export apparatus | SpaP | Spa24 | YscR | EscR | PscR |
| SpaQ | Spa9 | YscS | EscS | PscS | |
| SpaR | Spa29 | YscT | EscT | PscT | |
| SpaS | Spa40 | YscU | EscU | PscU | |
| InvA | MxiA | YscV | EscV | PerD | |
| ATPase | InvC | Spa47 | YscN | EscN | PscN |
BASAL STRUCTURE
The basal structure anchors the needle to the bacterial membranes by traversing the inner membrane (IM), the periplasm, and the outer membrane (OM).1 The base measures approximately 250 Å × 300 Å and is comparable in size to the flagellar basal body.6 PrgH, PrgK, and InvG assemble into ringlike structures at the IM and OM in Salmonella.6 The IM and OM rings enclose a tubelike inner rod structure formed by PrgJ7 in Salmonella, MxiI12 in Shigella, and YscI13 in Yersinia. The atomic structure of the assembled inner rod is currently unknown; however, NMR and circular dichroism spectroscopy indicate that PrgJ in the monomeric form is a partially folded protein.14 Further, yeast two-hybrid and GST pull-down experiments showed a direct binding interaction between the Yersinia inner rod protein YscI and the needle protomer YscF.15
In Salmonella, the IM ring structure is formed by PrgH and PrgK, which assemble into two concentric rings, with PrgK forming the smaller ring and PrgH forming the outer ring6,12,16 (Figure 2A,B). PrgK belongs to the highly conserved YscJ/EscJ family of periplasmic lipoproteins, and NMR results for the enteropathogenic E. coli (EPEC) EscJ revealed a two-domain structure connected by a flexible linker (Figure 2C).17 These proteins are localized to the outer leaflet of the IM with their lipidated N-terminal domain and contain N-terminal signal sequences that are cleaved upon insertion in the membrane.12,17,18 X-ray crystallography showed that EscJ packs into a 24-mer ring structure18 with overall dimensions similar to those of the IM rings.19 Homology modeling based on the EscJ template generated the atomic model of Salmonella PrgK, which was then fit into the EM density map of the Salmonella base.20 The outer IM ring consists of 24 monomers of PrgH, a bitopic membrane protein with an N-terminal cytoplasmic domain and a large C-terminal periplasmic domain.20,21 The crystal structure of PrgH (residues 170–362) showed a distinctive “boot-shaped” organization of its modular domains (Figure 2C).21 The NMR structure of the N-terminal domain of Shigella MxiG (Figure 2C) (the homologue of Salmonella PrgH) supported the 24-subunit structure of the Shigella IM ring.22
Figure 2.
(A) Salmonella T3SS basal structure that spans the outer membrane (OM), the periplasm, and the inner membrane (IM) and is formed by (B) rings of InvG (red), PrgH (purple), and PrgK (blue).20 (C) Structures of proteins that form the T3SS basal body: EPEC EscC,21 EPEC EscJ,18Salmonella PrgH,21 and Shigella MxiG.22
The OM ring is formed by the secretin family of integral membrane proteins that includes Salmonella InvG,20Yersinia YscC,23Shigella MxiD,24 and EPEC EscC.21 A neck region spanning the periplasm connects the OM ring to the IM rings. The N-terminal domain of InvG forms the neck region of the base and is associated with the PrgH IM ring,20 whereas the C-terminal domain forms the rest of the OM ring.21 Electron microscopy of the Shigella base at 21–25 Å resolution showed an OM ring with a 12-fold symmetry.24 Recently, the refined structure of the Salmonella base at 10 Å resolution showed a 15-fold symmetry for the OM ring (Figure 2B).20 This higher-resolution structure of the base was achieved by selectively disassembling the basal components into stable IM and OM rings by pH treatment.20 The structural model of the Salmonella InvG based on the crystal structure of EPEC EscC (Figure 2C) was fit into the EM density map of the Salmonella basal structure to generate an atomic model of the Salmonella OM ring.21
Secretins are transported to the OM with the help of pilotins, which are small lipoproteins that assist in the assembly of the OM ring.25 The Shigella pilotin protein MxiM forms a pseudo-β-barrel with a hydrophobic pocket for binding lipids.26 MxiM interacts with the secretin protein MxiD, and the binding interaction blocks the lipid-binding pocket in MxiM.25 The pilotins also contain a characteristic signal peptide leader sequence with a conserved cysteine residue can be lipidated.25 These features are thought to help in the membrane targeting of the secretin–pilotin complex.25
Secretion through the base is controlled by an export apparatus,27 which serves as a platform for the assembly of the basal structure and controls substrate specificity.27–29 The export apparatus is a complex of highly conserved integral membrane proteins: YscR, YscS, YscT, YscU, and YscV in Yersinia28,29 and SpaP, SpaQ, SpaR, SpaS, and InvA in Salmonella.27 In Yersinia, the assembly of the base and export apparatus is initiated independently at the outer and inner membranes.28,29 In Yersinia, the OM ring of the base is formed by the secretin protein YscC (and assisted by the YscW pilotin), which then recruits the outer IM ring protein, YscD.28 Simultaneously, the export apparatus proteins YscR, YscS, and YscT assemble at the inner membrane, which promote the polymerization of YscV.29 These two pathways can independently recruit the inner IM ring protein, YscJ, which is thus thought to link these two substructures.29 However, in Salmonella, the prior assembly of the export apparatus proteins (SpaP, SpaQ, and SpaR) at the IM is required for the assembly of the IM ring.27
The hierarchy of secretion of structural components and virulence effectors through the base is regulated in part by the export apparatus protein YscU in Yersinia,30 SpaS in Salmonella,31 Spa40 in Shigella,32 and EscU in EPEC.31 The overall structure of these proteins consists of an N-terminal transmembrane domain that is connected to a C-terminal globular cytoplasmic domain by a flexible linker.30,31 The cytoplasmic domain shows a conserved fold composed of a central five-stranded mixed β-sheet surrounded by four α-helices.30–32 The cytoplasmic domain undergoes an autocatalytic cleavage at a highly conserved NPTH motif, resulting in an altered surface feature, which is important for substrate specificity switching.32 The largest component of the export apparatus, the YscV29 family of proteins (including Salmonella InvA33 and Shigella MxiA34), forms a ring directly below the base. The atomic structures of the cytosolic domains of InvA,33 MxiA,34 and the flagellar homologue FlhA35 show a conserved fold composed of four subdomains. The cytosolic domain of Shigella MxiA formed a nonameric ring, and residues lining the internal surface of this ring are important for secretion of T3SS substrates.34
An ATPase complex associated with the bacterial IM36 is responsible for energizing the export of substrates through the needle apparatus. The ATPase complex couples ATP hydrolysis to the unfolding and release of effector proteins from their cognate chaperones prior to secretion through the needle apparatus.34,37 The major component of the ATPase complex is a homohexamer formed by the EscN protein family (with homologues listed in Table 1), which is related to the α and β subunits of the F1 ATP synthase.38
STRUCTURE OF THE NEEDLE
The needle is assembled from multiple copies of a single protein, the needle protomer. Needle protomers are small polar proteins of <90 residues.12 Early experiments with recombinant forms of these proteins impeded structure determination because of their tendency to aggregate in solution.39 However, deletion of the last five residues at the C-terminus allowed for determination of the atomic structure of the Shigella needle protomer MxiH by crystallography,40 and Burkholderia BsaL41 and Salmonella PrgI42 needle protomers by NMR spectroscopy (Figure 3A). Unfortunately, this deletion also inhibited polymerization of the needle and abolished host cell invasion.43 Shortly after, the crystal and NMR structures of a soluble and functional V65A/V67A double mutant of full-length PrgI were determined (Figure 3A).43 The monomeric forms of needle protomers adopt α-helical hairpin structures, essentially two α-helices joined by a four-residue PXXP motif (where X is any amino acid), containing a turn in the central region of the protein (Figure 3A). The Pseudomonas PscF, however, lacks a PXXP motif and instead contains an AXXP motif.44 Complete structures of the Yersinia YscF and Pseudomonas PscF needle protomers are currently unknown; however, their partial structures in complex with their chaperones (YscE–YscG45 and PscE–PscG,44 respectively) are known. The structured regions of YscF (Figure 3A) and PscF in these complexes also form α-helices. Although YscF and PscF have chaperones, no chaperones have been reported for BsaL, MxiH, or PrgI.
Figure 3.
(A) Structures of needle protomers Salmonella PrgICΔ5 (CΔ5, deletion of five C-terminal residues),42 PrgI* (a functional V65A/V67A double mutant),43 PrgI polymerized into needles,48Shigella MxiHCΔ5,40 MxiH polymerized into needles,47 and Burkholderia BsaLCΔ5.41 Regions that adopted β-strands upon needle assembly are colored green. (B) Atomic model of the Salmonella needle derived by solid-state NMR showing that the N-terminus of PrgI faces the exterior.48
Multiple copies of needle protomers assemble to form the T3SS needle. Electron microscopy and X-ray fiber diffraction of Shigella needles allowed the atomic scale modeling of a T3SS needle at 16 Å resolution.46 This model revealed a structure measuring 500 Å in length and 70 Å in width enclosing an inner channel with a diameter of ~25 Å.46 Subsequently, when the atomic structures of needle protomers became available, they were used to fit the electron density maps of the Shigella40 and Salmonella8 needles. The N-terminal region of MxiH lacked electron density in the crystal structure.40 Therefore, it was modeled in the Shigella needle as an α-helix facing the lumen, based on the predicted α-helical propensities of residues within the region.40 Similarly, in the Salmonella needle model, approximately half of the length of PrgI containing the flexible N- and C-termini was absent because of the conformational heterogeneity of the PrgI protomer in the NMR structure.8 The Shigella needle was shown to contain ~5.6 subunits per turn with a helical pitch of 24 Å,40 while the Salmonella needle contained ~6.3 subunits per turn with a helical pitch of 26 Å.8
Recently, higher-resolution atomic models of the Shigella47 and Salmonella48 needles were reported. These models represent our current understanding of the atomic structures of the T3SS needles. Fujii et al. used cryo-electron microscopy (cryo-EM) to determine the structure of purified Shigella needles formed by the overexpression of recombinant full-length MxiH.47 The atomic model of the Shigella needle was obtained by fitting the crystal structure of MxiH residues 26–51 (the “head” structure) into the 7.7 Å resolution electron density map.47 In this model, the first 38 residues of MxiH form a straight α-helix facing the lumen and the exterior surface of the needle is formed by the C-terminus of MxiH, which contains a short α-helix (residues 44–50), a β-hairpin (residues 51–64), and a kinked helix (residues 65–83).47 Intersubunit interactions are present between the β-hairpin and the PXXP motif, as well as the β-hairpin and the C-terminus of adjacent subunits. The needle is further stabilized by helix–helix interactions between adjacent subunits.
Loquet et al.,48 on the other hand, used solid-state NMR spectroscopy (ssNMR) to determine the structure of Salmonella needles polymerized from full-length recombinant PrgI (Figure 3B). The use of [1-13C]glucose or [2-13C]glucose instead of the traditional uniform labeling with 13C-labeled glucose49 as the carbon source for bacteria expressing recombinant PrgI reduced the NMR signal complexity of the megadalton needle. Coupled with 15N labeling, ssNMR allowed the identification of intra- and intersubunit 15N–13C distance restraints,48 which were used in combination with Rosetta modeling50 to generate the atomic structure of the Salmonella needle48 (Figure 3B). According to the ssNMR model, the Salmonella needle is composed of PrgI protomers with an α-helical hairpin head structure similar to those that have been described previously.42,43 However, unlike previously determined crystal and NMR structures, ssNMR secondary structure analysis excluded conformational heterogeneity at the N- and C-termini, except for Met1 and Ala2.48 The Salmonella needle has an 80 Å outer diameter with a 25 Å lumen, similar to values previously described.8,51 Unlike previous reconstructions,8 however, the needle was shown to contain ~5.7 subunits per turn with a helical pitch of 24 Å.48 The needle is held together by multiple intra- and intersubunit contacts. The intrasubunit contacts are primarily hydrophobic interactions between the two helices of the hairpin, and the intersubunit interactions are present on the lateral and axial surfaces of each subunit.48
There are two major differences between the ssNMR model48 of the Salmonella needle and previous needle models.8,40,47 The first difference is that in the ssNMR Salmonella needle model, the N-terminus of PrgI faces the exterior of the needle while the C-terminal tail faces the interior48 (Figure 3B). Immunoelectron microscopy against the N-terminus of PrgI confirmed that nonconserved residues in the N-terminal region face the exterior of the needle, while highly conserved residues in the C-terminus are contained within the lumen.48 In contrast, the EM models have reported that the N-terminus of the needle protomer faces the lumen while the C-terminus faces the exterior.8,40,47 This was supported by secretion experiments in which the N-terminus of MxiH tolerated more substantial modifications than the C-terminus.47 Another difference is that throughout the Salmonella needle protomer, no β-hairpin was detected, in contrast to the structure of the Shigella needle protomer.47 Poyraz et al.,43 however, reported that residues I71–I76 of PrgI V65A/V67A undergo a backbone α-helix–β-strand conversion upon polymerization. In the Shigella needle, the β-hairpin is hypothesized to regulate assembly and the positioning of the subunits within the needle.47 ssNMR data of Shigella needles should clarify these differences.
NEEDLE TIP
On the distal end of the needle sits a tip protein complex that functions to sense the presence of eukaryotic cells, to serve as a platform for the assembly of the translocon, and to regulate the secretion of effector proteins into the host cell.9,52 The T3SS tip proteins can be grouped into two classes based on their structures and properties. One class is formed by Salmonella SipD, Shigella IpaD, and Burkholderia BipD tip proteins, which have sequences that are 30–50% identical and are structural homologues with a Cα root-mean-square deviation of <1.5 Å.53–56 Members of the SipD family of tip proteins share a more distant resemblance to another class of tip proteins, the Yersinia LcrV and the Pseudomonas PcrV tip proteins, with sequences that are ~37% identical.
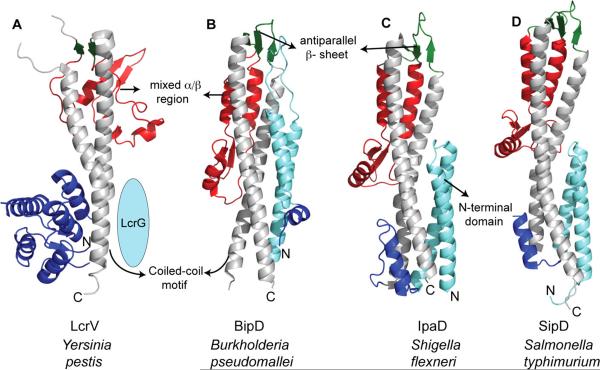
The SipD, IpaD, and BipD53–56 tip proteins contain eight α-helices and five β-strands, which are arranged into three distinct structural elements: an N-terminal α-helical hairpin, a long central coiled-coil, and a C-terminal region of mixed α-helices and β-strands (Figure 4A–D). The central coiled-coil is a distinct structural feature of all tip proteins and is responsible for their overall oblong shape. The coiled-coil motif, especially the C-terminal residues, is important for the interaction of the tip protein with the needle, and the needle protomer is expected to bind at multiple sites on the coiled-coil motif.52,55–58 A three-stranded antiparallel β-sheet connects the central coiled-coil to the mixed α/β region.53–55 The first 30–40 residues of the tip proteins are disordered in the crystal structures of SipD, IpaD, and BipD.52 The N-terminal α-hairpin folds independently and functions as a self-chaperone by preventing the self-oligomerization of SipD and IpaD53,56,59 and hinders the interaction between the needle protomer PrgI and the tip protein SipD.56 Upon assembly of SipD at the needle tip, the α-hairpin is expected to be displaced by the needle protomer, which interacts with the coiled-coil domain.53,56,58
Figure 4.
Structure of tip proteins from (A) Y. pestis,60 (B) Burkholderia pseudomallei,54 (C) S. flexneri,53 and (D) S. typhimurium.55
The 2.2 Å crystal structure of LcrV60 (Figure 4) represents what is currently known about the atomic structure of the Yersinia/Pseudomonas LcrV/PcrV family of tip proteins. Similar to the SipD, the structure of LcrV also contains a long central coiled-coil, which is flanked by globular domains on the N- and C-termini, giving an overall “dumbbell shape” to LcrV.60 LcrV, however, lacks the self-chaperoning α-helical hairpin found in SipD. Instead, a small cytoplasmic protein, LcrG, functions as the chaperone for LcrV.11 Likewise, the LcrG homologue in Pseudomonas, PcrG, functions as a chaperone for the PcrV tip protein. In addition to binding their cognate tip proteins, LcrG and PcrG negatively regulate secretion of effectors61,62 in Yersinia and Pseudomonas. Deletion of lcrG decreases the level of LcrV secretion,63 and host cell contact with Yersinia upregulates the expression of LcrV, which then forms a tight binding complex with LcrG; this interaction relieves the negative block on effector secretion by titrating away LcrG.61,62,64 The N-terminal regions of LcrG and PcrG are required for binding to their cognate tip proteins,61,65,66 and the coiled-coil region of LcrV is involved in the interaction with LcrG.65,67 The absence of an intramolecular chaperoning domain in LcrV and PcrV hints at divergent assembly processes at the needle tip between the Salmonella/Shigella and Yersinia/Pseudomonas species.
A major challenge in this field is the determination of atomic-resolution structures of the tip protein docked at the needle tip. Currently, low-resolution electron micrographs of the Yersinia LcrV tip68 and the Shigella IpaD tip69,70 as well as models made by docking the crystal structures of tip proteins on the needle11,40 are available. Electron micrographs of the Yersinia needle tip show a well-defined tip complex that was estimated to be formed by an LcrV pentameric ring.68,71 The Yersinia LcrV tip complex showed a distinct head, neck, and a base, and Broz et al.71 concluded that the LcrV N-terminal globular domain forms the base, the C-terminal globular domain forms the head, and the coiled-coil region forms the neck. The base was also shown to be responsible for the correct insertion of the Yersinia translocon protein YopB into the host cell membrane.71
Results from electron microscopy of the Shigella tip complex also estimated a stoichiometry of a pentameric IpaD at the needle tip.70 However, the Shigella tip complex did not show the head, neck, and base morphology that was seen in the Yersinia needle tip. This difference was partly ascribed to the difference in the structure of IpaD and LcrV where the domains flanking the IpaD coiled-coil were more elongated in shape compared to the more globular domains flanking the LcrV coiled-coil. In the Shigella tip complex, the IpaD coiled-coil is aligned with the vertical axis of the tip complex with a 20° tilt and the tip complex forms an internal channel diameter from 40 Å at the middle to 22 Å at the top.70 The most distinctive feature of this structure is that the IpaD mixed α/β domain undergoes a 165° rotation about the horizontal axis, forming a scepterlike structure at the top of the tip complex.70 In support of this drastic rotation, tip complexes made of IpaD lacking the mixed α/β domain (residues 192–267) were shorter by ~37 Å and lacked electron density for the scepterlike structure.70
Prior to contact with the host cell membrane, the tip complex is present at the needle tip while the translocon is yet to be assembled.9,69,72 The tip complex therefore controls the secretion of the translocon proteins, and this process is not well understood. It has been proposed that the pentameric tip complex forms a closed conformation that prevents protein secretion11 and, upon receiving an extracellular signal in the presence of the host cell,72,73 changes into an open conformation that allows protein secretion to complete the assembly of the needle apparatus. Bile salts, levels of which are elevated in the intestines, have been identified as extracellular signals that can affect type III secretion in the enteric pathogens Shigella74–76 and Salmonella.77,78 Bile salts bind to IpaD72,73,75,76 and SipD;56,76,79 however, despite the structural homology of IpaD and SipD, bile salts bind these proteins at different sites and affect their respective T3SSs in opposite ways. Bile salts increase the level of type III secretion and invasiveness of Shigella74–76 but repress the transcription of T3SS proteins and decrease the invasiveness of Salmonella.77,78 The mechanism of how extracellular signals affect the structure and conformation of the tip complex remains to be elucidated.
TRANSLOCON
The final stage in the assembly of the needle apparatus is the formation of the translocon, which forms a 20–30 Å pore on the host cell membrane to allow the passage of effectors into the host cell.2,80–82 The translocon is assembled from two integral membrane proteins named SipB and SipC in Salmonella,83 IpaB and IpaC in Shigella,2,84 YopB and YopD in Yersinia,82 and PopB and PopD in Pseudomonas.85 The translocon proteins are important for the entry of bacteria into eukaryotic cells as demonstrated by the internalization of latex beads coated with IpaB and IpaC into HeLa cells.86 While inside the bacterial cytosol, the translocon proteins are bound to their chaperones to maintain their intracellular stability. SipB and SipC are chaperoned by SicA in Salmonella.87 The chaperones for the translocon proteins in other species are the Shigella IpgC,88Yersinia SycD (also named LcrH),89 and Pseudomonas PcrH90 proteins.
The major translocon proteins from Shigella and Salmonella (IpaB and SipB) are 62 kDa α-helical proteins that can bind to cell membranes.91 The Yersinia YopB and Pseudomonas PopB share a higher degree of conservation and are ~40 kDa in size.92 The N-terminal region of IpaB and SipB is responsible for their self-oligomerization.91 Two transmembrane helices facilitate intimate attachment to lipid membranes.91,93 The extreme C-terminus is predicted to form an amphipathic helix and is important for invasion of the host cell and regulation of effector secretion.91,94,95 The crystal structures of the protease resistant fragments in the N-terminal regions of IpaB (residues 74–224) and SipB (residues 82–226) revealed a trimeric coiled-coil domain formed by three antiparallel α-helices96 (Figure 5A).
Figure 5.
(A) Structures of the N-terminal domains of the Shigella IpaB and Salmonella SipB translocon proteins.96 (B) Cocrystal structures of IpgC,115 SycD,113 and PcrH90 chaperones bound to peptides derived from their cognate translocon proteins.
The Shigella IpaC,52Salmonella SipC,9Yersinia YopD,97 and Pseudomonas PopD98 are the minor translocon proteins and form complexes with the major translocon proteins, and this interaction is necessary for the attachment and entry into epithelial cells.9 Incubation of Shigella with recombinant IpaC led to an increased level of invasion of epithelial cells,99,100 and recombinant SipC can interact with actin.101 Similar to other T3SS proteins, the N-terminus of these molecules is required for their secretion,9,100,102,103 and a central hydrophobic region allows binding to lipid membranes.104 The C-terminus, which is predicted to be helical, has been implicated in actin modulation105–107 and hemolytic activity.108 In addition, the conserved C-terminus is also involved in the homotypic oligomerization of IpaC.100
The chaperones of the translocon proteins belong to class II of T3SS chaperones and are responsible for partitioning the intracellular pools of the major and minor translocon proteins, thereby preventing their premature degradation.87,109,110 Chaperones bind at the N-terminal regions of translocon proteins.90,111–114 Cocrystal structures of chaperones bound to peptides derived from the chaperone-binding regions of the translocon proteins revealed how chaperones recognize their cognate translocon proteins (Figure 5B).90,113,115 The chaperones form a homodimer of tetratricopeptide motifs composed of antiparallel α-helical repeats. In the cocrystal structure of the Shigella IpgC–IpaB peptide complex,115 IpgC forms a dimer with each IpgC molecule forming a concave cleft ~10 Å in width to accommodate the IpaB peptide, and the IpgC–IpaB protein–protein contacts are mediated by conserved residues on both proteins. The 32 N-terminal residues of IpgC are responsible for its homodimerization, and this dimerization pattern is conserved in the Salmonella SicA115 and Yersinia SycD116 chaperones. Amino acid substitutions in the IpgC N-terminal region decreased the level of secretion of effectors and bacterial invasion.115
The major translocon proteins IpaB and SipB interact with lipids such as cholesterol and sphingomyelin,117,118 and cholesterol is required for effector translocation in Shigella, Salmonella, and EPEC.117,118 Further, the interaction of sphingomyelin and cholesterol with IpaB is proposed to induce the recruitment of IpaC to assemble the translocon.118 In addition to their structural role in the assembly of the translocon, the translocon proteins are also delivered into the host cell where they interact with a number of cellular factors.119 IpaB binds to caspase-1, and this leads to apoptosis of macrophages.120,121 Furthermore, the C-terminus of IpaC induces actin polymerization and formation of filopodia and lamellipodia in fibroblast cells.105,106 Cytoskeletal rearrangements induced by IpaC were dependent on the function of Cdc42 and Rac GTPases.105,106 The C-terminus of SipC (within residues 200–409) could also induce actin polymerization in HeLa cells.105,106
CONCLUSION
Since the visualization of parts of the needle apparatus at low resolution 15 years ago, much progress has been achieved in determining atomic-resolution structures of components there-of from electron microscopy, crystallography, and NMR spectroscopy. However, much remains to be elucidated in understanding this macromolecular nanoinjector device. Among them are the atomic structures of the translocon and the tip complex. Because the needle apparatus plays a critical role in pathogenesis, understanding its assembly will contribute to the development of novel anti-infectives.
Acknowledgments
Funding This work was supported by National Institutes of Health Grants T32-GM008359 (A.C.M.) and R01AI074856 (R.N.D.G.).
ABBREVIATIONS
- T3SS
type III secretion system
- OM
outer membrane
- IM
inner membrane
- EPEC
enteropathogenic E. coli
- NMR
nuclear magnetic resonance
- ssNMR
solid-state NMR
- EM
electron microscopy
Footnotes
Author Contributions S. Chatterjee and S. Chaudhury contributed equally to this work.
The authors declare no competing financial interest.
REFERENCES
- (1).Kubori T, Matsushima Y, Nakamura D, Uralil J, Lara-Tejero M, Sukhan A, Galan JE, Aizawa SI. Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science. 1998;280:602–605. doi: 10.1126/science.280.5363.602. [DOI] [PubMed] [Google Scholar]
- (2).Blocker A, Gounon P, Larquet E, Niebuhr K, Cabiaux V, Parsot C, Sansonetti P. The tripartite type III secreton of Shigella flexneri inserts IpaB and IpaC into host membranes. J. Cell Biol. 1999;147:683–693. doi: 10.1083/jcb.147.3.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Cornelis GR. Yersinia type III secretion: Send in the effectors. J. Cell Biol. 2002;158:401–408. doi: 10.1083/jcb.200205077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Sekiya K, Ohishi M, Ogino T, Tamano K, Sasakawa C, Abe A. Supermolecular structure of the enteropathogenic Escherichia coli type III secretion system and its direct interaction with the EspA-sheath-like structure. Proc. Natl. Acad. Sci. U.S.A. 2001;98:11638–11643. doi: 10.1073/pnas.191378598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Pastor A, Chabert J, Louwagie M, Garin J, Attree I. PscF is a major component of the Pseudomonas aeruginosa type III secretion needle. FEMS Microbiol. Lett. 2005;253:95–101. doi: 10.1016/j.femsle.2005.09.028. [DOI] [PubMed] [Google Scholar]
- (6).Marlovits TC, Kubori T, Sukhan A, Thomas DR, Galan JE, Unger VM. Structural insights into the assembly of the type III secretion needle complex. Science. 2004;306:1040–1042. doi: 10.1126/science.1102610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Marlovits TC, Kubori T, Lara-Tejero M, Thomas D, Unger VM, Galan JE. Assembly of the inner rod determines needle length in the type III secretion injectisome. Nature. 2006;441:637–640. doi: 10.1038/nature04822. [DOI] [PubMed] [Google Scholar]
- (8).Galkin VE, Schmied WH, Schraidt O, Marlovits TC, Egelman EH. The structure of the Salmonella typhimurium type III secretion system needle shows divergence from the flagellar system. J. Mol. Biol. 2010;396:1392–1397. doi: 10.1016/j.jmb.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Lara-Tejero M, Galan JE. Salmonella enterica serovar typhimurium pathogenicity island 1-encoded type III secretion system translocases mediate intimate attachment to nonphagocytic cells. Infect. Immun. 2009;77:2635–2642. doi: 10.1128/IAI.00077-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Lara-Tejero M, Kato J, Wagner S, Liu X, Galan JE. A sorting platform determines the order of protein secretion in bacterial type III systems. Science. 2011;331:1188–1191. doi: 10.1126/science.1201476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Blocker AJ, Deane JE, Veenendaal AK, Roversi P, Hodgkinson JL, Johnson S, Lea SM. What's the point of the type III secretion system needle? Proc. Natl. Acad. Sci. U.S.A. 2008;105:6507–6513. doi: 10.1073/pnas.0708344105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Blocker A, Jouihri N, Larquet E, Gounon P, Ebel F, Parsot C, Sansonetti P, Allaoui A. Structure and composition of the Shigella flexneri “needle complex”, a part of its type III secretion. Mol. Microbiol. 2001;39:652–663. doi: 10.1046/j.1365-2958.2001.02200.x. [DOI] [PubMed] [Google Scholar]
- (13).Wood SE, Jin J, Lloyd SA. YscP and YscU switch the substrate specificity of the Yersinia type III secretion system by regulating export of the inner rod protein YscI. J. Bacteriol. 2008;190:4252–4262. doi: 10.1128/JB.00328-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Zhong D, Lefebre M, Kaur K, McDowell MA, Gdowski C, Jo S, Wang Y, Benedict SH, Lea SM, Galan JE, De Guzman RN. The Salmonella Type III Secretion System Inner Rod Protein PrgJ Is Partially Folded. J. Biol. Chem. 2012;287:25303–25311. doi: 10.1074/jbc.M112.381574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Yang H, Tan Y, Zhang T, Tang L, Wang J, Ke Y, Guo Z, Yang X, Yang R, Du Z. Identification of Novel Protein-Protein Interactions of Yersinia pestis Type III Secretion System by Yeast Two Hybrid System. PLoS One. 2013;8:e54121. doi: 10.1371/journal.pone.0054121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Schraidt O, Lefebre MD, Brunner MJ, Schmied WH, Schmidt A, Radics J, Mechtler K, Galan JE, Marlovits TC. Topology and organization of the Salmonella typhimurium type III secretion needle complex components. PLoS Pathog. 2010;6:e1000824. doi: 10.1371/journal.ppat.1000824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Crepin VF, Prasannan S, Shaw RK, Wilson RK, Creasey E, Abe CM, Knutton S, Frankel G, Matthews S. Structural and functional studies of the enteropathogenic Escherichia coli type III needle complex protein EscJ. Mol. Microbiol. 2005;55:1658–1670. doi: 10.1111/j.1365-2958.2005.04508.x. [DOI] [PubMed] [Google Scholar]
- (18).Yip CK, Kimbrough TG, Felise HB, Vuckovic M, Thomas NA, Pfuetzner RA, Frey EA, Finlay BB, Miller SI, Strynadka NC. Structural characterization of the molecular platform for type III secretion system assembly. Nature. 2005;435:702–707. doi: 10.1038/nature03554. [DOI] [PubMed] [Google Scholar]
- (19).Moraes TF, Spreter T, Strynadka NC. Piecing together the type III injectisome of bacterial pathogens. Curr. Opin. Struct. Biol. 2008;18:258–266. doi: 10.1016/j.sbi.2007.12.011. [DOI] [PubMed] [Google Scholar]
- (20).Schraidt O, Marlovits TC. Three-dimensional model of Salmonella's needle complex at subnanometer resolution. Science. 2011;331:1192–1195. doi: 10.1126/science.1199358. [DOI] [PubMed] [Google Scholar]
- (21).Spreter T, Yip CK, Sanowar S, Andre I, Kimbrough TG, Vuckovic M, Pfuetzner RA, Deng W, Yu AC, Finlay BB, Baker D, Miller SI, Strynadka NC. A conserved structural motif mediates formation of the periplasmic rings in the type III secretion system. Nat. Struct. Mol. Biol. 2009;16:468–476. doi: 10.1038/nsmb.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).McDowell MA, Johnson S, Deane JE, Cheung M, Roehrich AD, Blocker AJ, McDonnell JM, Lea SM. Structural and functional studies on the N-terminal domain of the Shigella type III secretion protein MxiG. J. Biol. Chem. 2011;286:30606–30614. doi: 10.1074/jbc.M111.243865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Burghout P, van Boxtel R, Van Gelder P, Ringler P, Muller SA, Tommassen J, Koster M. Structure and electrophysiological properties of the YscC secretin from the type III secretion system of Yersinia enterocolitica. J. Bacteriol. 2004;186:4645–4654. doi: 10.1128/JB.186.14.4645-4654.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Hodgkinson JL, Horsley A, Stabat D, Simon M, Johnson S, da Fonseca PC, Morris EP, Wall JS, Lea SM, Blocker AJ. Three-dimensional reconstruction of the Shigella T3SS transmembrane regions reveals 12-fold symmetry and novel features throughout. Nat. Struct. Mol. Biol. 2009;16:477–485. doi: 10.1038/nsmb.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Okon M, Moraes TF, Lario PI, Creagh AL, Haynes CA, Strynadka NC, McIntosh LP. Structural characterization of the type-III pilot-secretin complex from Shigella flexneri. Structure. 2008;16:1544–1554. doi: 10.1016/j.str.2008.08.006. [DOI] [PubMed] [Google Scholar]
- (26).Lario PI, Pfuetzner RA, Frey EA, Creagh L, Haynes C, Maurelli AT, Strynadka NC. Structure and biochemical analysis of a secretin pilot protein. EMBO J. 2005;24:1111–1121. doi: 10.1038/sj.emboj.7600610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Wagner S, Konigsmaier L, Lara-Tejero M, Lefebre M, Marlovits TC, Galan JE. Organization and coordinated assembly of the type III secretion export apparatus. Proc. Natl. Acad. Sci. U.S.A. 2010;107:17745–17750. doi: 10.1073/pnas.1008053107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Diepold A, Amstutz M, Abel S, Sorg I, Jenal U, Cornelis GR. Deciphering the assembly of the Yersinia type III secretion injectisome. EMBO J. 2010;29:1928–1940. doi: 10.1038/emboj.2010.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Diepold A, Wiesand U, Cornelis GR. The assembly of the export apparatus (YscR,S,T,U,V) of the Yersinia type III secretion apparatus occurs independently of other structural components and involves the formation of an YscV oligomer. Mol. Microbiol. 2011;82:502–514. doi: 10.1111/j.1365-2958.2011.07830.x. [DOI] [PubMed] [Google Scholar]
- (30).Wiesand U, Sorg I, Amstutz M, Wagner S, van den Heuvel J, Luhrs T, Cornelis GR, Heinz DW. Structure of the type III secretion recognition protein YscU from Yersinia enterocolitica. J. Mol. Biol. 2009;385:854–866. doi: 10.1016/j.jmb.2008.10.034. [DOI] [PubMed] [Google Scholar]
- (31).Zarivach R, Deng W, Vuckovic M, Felise HB, Nguyen HV, Miller SI, Finlay BB, Strynadka NC. Structural analysis of the essential self-cleaving type III secretion proteins EscU and SpaS. Nature. 2008;453:124–127. doi: 10.1038/nature06832. [DOI] [PubMed] [Google Scholar]
- (32).Deane JE, Graham SC, Mitchell EP, Flot D, Johnson S, Lea SM. Crystal structure of Spa40, the specificity switch for the Shigella flexneri type III secretion system. Mol. Microbiol. 2008;69:267–276. doi: 10.1111/j.1365-2958.2008.06293.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Worrall LJ, Vuckovic M, Strynadka NC. Crystal structure of the C-terminal domain of the Salmonella type III secretion system export apparatus protein InvA. Protein Sci. 2010;19:1091–1096. doi: 10.1002/pro.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Abrusci P, Vergara-Irigaray M, Johnson S, Beeby MD, Hendrixson DR, Roversi P, Friede ME, Deane JE, Jensen GJ, Tang CM, Lea SM. Architecture of the major component of the type III secretion system export apparatus. Nat. Struct. Mol. Biol. 2013;20:99–104. doi: 10.1038/nsmb.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Saijo-Hamano Y, Imada K, Minamino T, Kihara M, Shimada M, Kitao A, Namba K. Structure of the cytoplasmic domain of FlhA and implication for flagellar type III protein export. Mol. Microbiol. 2010;76:260–268. doi: 10.1111/j.1365-2958.2010.07097.x. [DOI] [PubMed] [Google Scholar]
- (36).Pozidis C, Chalkiadaki A, Gomez-Serrano A, Stahlberg H, Brown I, Tampakaki AP, Lustig A, Sianidis G, Politou AS, Engel A, Panopoulos NJ, Mansfield J, Pugsley AP, Karamanou S, Economou A. Type III protein translocase: HrcN is a peripheral ATPase that is activated by oligomerization. J. Biol. Chem. 2003;278:25816–25824. doi: 10.1074/jbc.M301903200. [DOI] [PubMed] [Google Scholar]
- (37).Akeda Y, Galan JE. Chaperone release and unfolding of substrates in type III secretion. Nature. 2005;437:911–915. doi: 10.1038/nature03992. [DOI] [PubMed] [Google Scholar]
- (38).Zarivach R, Vuckovic M, Deng W, Finlay BB, Strynadka NC. Structural analysis of a prototypical ATPase from the type III secretion system. Nat. Struct. Mol. Biol. 2007;14:131–137. doi: 10.1038/nsmb1196. [DOI] [PubMed] [Google Scholar]
- (39).Kenjale R, Wilson J, Zenk SF, Saurya S, Picking WL, Picking WD, Blocker A. The needle component of the type III secreton of Shigella regulates the activity of the secretion apparatus. J. Biol. Chem. 2005;280:42929–42937. doi: 10.1074/jbc.M508377200. [DOI] [PubMed] [Google Scholar]
- (40).Deane JE, Roversi P, Cordes FS, Johnson S, Kenjale R, Daniell S, Booy F, Picking WD, Picking WL, Blocker AJ, Lea SM. Molecular model of a type III secretion system needle: Implications for host-cell sensing. Proc. Natl. Acad. Sci. U.S.A. 2006;103:12529–12533. doi: 10.1073/pnas.0602689103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Zhang L, Wang Y, Picking WL, Picking WD, De Guzman RN. Solution structure of monomeric BsaL, the type III secretion needle protein of Burkholderia pseudomallei. J. Mol. Biol. 2006;359:322–330. doi: 10.1016/j.jmb.2006.03.028. [DOI] [PubMed] [Google Scholar]
- (42).Wang Y, Ouellette AN, Egan CE, Rathinavelan T, Im W, De Guzman RN. Differences in the electrostatic surfaces of the type III secretion needle proteins PrgI, BsaL, and MxiH. J. Mol. Biol. 2007;371:1304–1314. doi: 10.1016/j.jmb.2007.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Poyraz O, Schmidt H, Seidel K, Delissen F, Ader C, Tenenboim H, Goosmann C, Laube B, Thunemann AF, Zychlinsky A, Baldus M, Lange A, Griesinger C, Kolbe M. Protein refolding is required for assembly of the type three secretion needle. Nat. Struct. Mol. Biol. 2010;17:788–792. doi: 10.1038/nsmb.1822. [DOI] [PubMed] [Google Scholar]
- (44).Quinaud M, Ple S, Job V, Contreras-Martel C, Simorre JP, Attree I, Dessen A. Structure of the heterotrimeric complex that regulates type III secretion needle formation. Proc. Natl. Acad. Sci. U.S.A. 2007;104:7803–7808. doi: 10.1073/pnas.0610098104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Sun P, Tropea JE, Austin BP, Cherry S, Waugh DS. Structural Characterization of the Yersinia pestis Type III Secretion System Needle Protein YscF in Complex with Its Heterodimeric Chaperone YscE/YscG. J. Mol. Biol. 2008;377:819–830. doi: 10.1016/j.jmb.2007.12.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Cordes FS, Komoriya K, Larquet E, Yang S, Egelman EH, Blocker A, Lea SM. Helical structure of the needle of the type III secretion system of Shigella flexneri. J. Biol. Chem. 2003;278:17103–17107. doi: 10.1074/jbc.M300091200. [DOI] [PubMed] [Google Scholar]
- (47).Fujii T, Cheung M, Blanco A, Kato T, Blocker AJ, Namba K. Structure of a type III secretion needle at 7-Å resolution provides insights into its assembly and signaling mechanisms. Proc. Natl. Acad. Sci. U.S.A. 2012;109:4461–4466. doi: 10.1073/pnas.1116126109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Loquet A, Sgourakis NG, Gupta R, Giller K, Riedel D, Goosmann C, Griesinger C, Kolbe M, Baker D, Becker S, Lange A. Atomic model of the type III secretion system needle. Nature. 2012;486:276–279. doi: 10.1038/nature11079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Loquet A, Lv G, Giller K, Becker S, Lange A. 13C spin dilution for simplified and complete solid-state NMR resonance assignment of insoluble biological assemblies. J. Am. Chem. Soc. 2011;133:4722–4725. doi: 10.1021/ja200066s. [DOI] [PubMed] [Google Scholar]
- (50).Das R, Andre I, Shen Y, Wu Y, Lemak A, Bansal S, Arrowsmith CH, Szyperski T, Baker D. Simultaneous prediction of protein folding and docking at high resolution. Proc. Natl. Acad. Sci. U.S.A. 2009;106:18978–18983. doi: 10.1073/pnas.0904407106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Kubori T, Sukhan A, Aizawa SI, Galan JE. Molecular characterization and assembly of the needle complex of the Salmonella typhimurium type III protein secretion system. Proc. Natl. Acad. Sci. U.S.A. 2000;97:10225–10230. doi: 10.1073/pnas.170128997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Picking WL, Nishioka H, Hearn PD, Baxter MA, Harrington AT, Blocker A, Picking WD. IpaD of Shigella flexneri is independently required for regulation of Ipa protein secretion and efficient insertion of IpaB and IpaC into host membranes. Infect. Immun. 2005;73:1432–1440. doi: 10.1128/IAI.73.3.1432-1440.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Johnson S, Roversi P, Espina M, Olive A, Deane JE, Birket S, Field T, Picking WD, Blocker AJ, Galyov EE, Picking WL, Lea SM. Self-chaperoning of the type III secretion system needle tip proteins IpaD and BipD. J. Biol. Chem. 2007;282:4035–4044. doi: 10.1074/jbc.M607945200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Erskine PT, Knight MJ, Ruaux A, Mikolajek H, Wong Fat Sang N, Withers J, Gill R, Wood SP, Wood M, Fox GC, Cooper JB. High Resolution Structure of BipD: An Invasion Protein Associated with the Type III Secretion System of Burkholderia pseudomallei. J. Mol. Biol. 2006;363:125–136. doi: 10.1016/j.jmb.2006.07.069. [DOI] [PubMed] [Google Scholar]
- (55).Chatterjee S, Zhong D, Nordhues BA, Battaile KP, Lovell SW, De Guzman RN. The Crystal Structure of the Salmonella Type III Secretion System Tip Protein SipD in Complex with Deoxycholate and Chenodeoxycholate. Protein Sci. 2011;20:75–86. doi: 10.1002/pro.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Lunelli M, Hurwitz R, Lambers J, Kolbe M. Crystal structure of PrgI-SipD: Insight into a secretion competent state of the type three secretion system needle tip and its interaction with host ligands. PLoS Pathog. 2011;7:e1002163. doi: 10.1371/journal.ppat.1002163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Zhang L, Wang Y, Olive AJ, Smith ND, Picking WD, De Guzman RN, Picking WL. Identification of the MxiH needle protein residues responsible for anchoring invasion plasmid antigen D to the type III secretion needle tip. J. Biol. Chem. 2007;282:32144–32151. doi: 10.1074/jbc.M703403200. [DOI] [PubMed] [Google Scholar]
- (58).Rathinavelan T, Tang C, De Guzman RN. Characterization of the Interaction between the Salmonella Type III Secretion System Tip Protein SipD and the Needle Protein PrgI by Paramagnetic Relaxation Enhancement. J. Biol. Chem. 2011;286:4922–4930. doi: 10.1074/jbc.M110.159434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Espina M, Ausar SF, Middaugh CR, Picking WD, Picking WL. Spectroscopic and calorimetric analyses of invasion plasmid antigen D (IpaD) from Shigella flexneri reveal the presence of two structural domains. Biochemistry. 2006;45:9219–9227. doi: 10.1021/bi060625v. [DOI] [PubMed] [Google Scholar]
- (60).Derewenda U, Mateja A, Devedjiev Y, Routzahn KM, Evdokimov AG, Derewenda ZS, Waugh DS. The structure of Yersinia pestis V-antigen, an essential virulence factor and mediator of immunity against plague. Structure. 2004;12:301–306. doi: 10.1016/j.str.2004.01.010. [DOI] [PubMed] [Google Scholar]
- (61).Lee PC, Stopford CM, Svenson AG, Rietsch A. Control of effector export by the Pseudomonas aeruginosa type III secretion proteins PcrG and PcrV. Mol. Microbiol. 2010;75:924–941. doi: 10.1111/j.1365-2958.2009.07027.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Hamad MA, Nilles ML. Roles of YopN, LcrG and LcrV in controlling Yops secretion by Yersinia pestis. Adv. Exp. Med. Biol. 2007;603:225–234. doi: 10.1007/978-0-387-72124-8_20. [DOI] [PubMed] [Google Scholar]
- (63).Fields KA, Nilles ML, Cowan C, Straley SC. Virulence role of V antigen of Yersinia pestis at the bacterial surface. Infect. Immun. 1999;67:5395–5408. doi: 10.1128/iai.67.10.5395-5408.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Matson JS, Nilles ML. LcrG-LcrV interaction is required for control of Yops secretion in Yersinia pestis. J. Bacteriol. 2001;183:5082–5091. doi: 10.1128/JB.183.17.5082-5091.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Lawton DG, Longstaff C, Wallace BA, Hill J, Leary SE, Titball RW, Brown KA. Interactions of the type III secretion pathway proteins LcrV and LcrG from Yersinia pestis are mediated by coiled-coil domains. J. Biol. Chem. 2002;277:38714–38722. doi: 10.1074/jbc.M203632200. [DOI] [PubMed] [Google Scholar]
- (66).Matson JS, Nilles ML. Interaction of the Yersinia pestis type III regulatory proteins LcrG and LcrV occurs at a hydrophobic interface. BMC Microbiol. 2002;2:16. doi: 10.1186/1471-2180-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Hamad MA, Nilles ML. Structure-function analysis of the C-terminal domain of LcrV from Yersinia pestis. J. Bacteriol. 2007;189:6734–6739. doi: 10.1128/JB.00539-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Mueller CA, Broz P, Muller SA, Ringler P, Erne-Brand F, Sorg I, Kuhn M, Engel A, Cornelis GR. The V-antigen of Yersinia forms a distinct structure at the tip of injectisome needles. Science. 2005;310:674–676. doi: 10.1126/science.1118476. [DOI] [PubMed] [Google Scholar]
- (69).Espina M, Olive AJ, Kenjale R, Moore DS, Ausar SF, Kaminski RW, Oaks EV, Middaugh CR, Picking WD, Picking WL. IpaD localizes to the tip of the type III secretion system needle of Shigella flexneri. Infect. Immun. 2006;74:4391–4400. doi: 10.1128/IAI.00440-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Epler CR, Dickenson NE, Bullitt E, Picking WL. Ultrastructural Analysis of IpaD at the Tip of the Nascent MxiH Type III Secretion Apparatus of Shigella flexneri. J. Mol. Biol. 2012;420:29–39. doi: 10.1016/j.jmb.2012.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Broz P, Mueller CA, Muller SA, Philippsen A, Sorg I, Engel A, Cornelis GR. Function and molecular architecture of the Yersinia injectisome tip complex. Mol. Microbiol. 2007;65:1311–1320. doi: 10.1111/j.1365-2958.2007.05871.x. [DOI] [PubMed] [Google Scholar]
- (72).Dickenson NE, Zhang L, Epler CR, Adam PR, Picking WL, Picking WD. Conformational Changes in IpaD from Shigella flexneri upon Binding Bile Salts Provide Insight into the Second Step of Type III Secretion. Biochemistry. 2011;50:172–180. doi: 10.1021/bi101365f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Barta ML, Guragain M, Adam P, Dickenson NE, Patil M, Geisbrecht BV, Picking WL, Picking WD. Identification of the bile salt binding site on IpaD from Shigella flexneri and the influence of ligand binding on IpaD structure. Proteins. 2012;80:935–945. doi: 10.1002/prot.23251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Pope LM, Reed KE, Payne SM. Increased protein secretion and adherence to HeLa cells by Shigella spp. following growth in the presence of bile salts. Infect. Immun. 1995;63:3642–3648. doi: 10.1128/iai.63.9.3642-3648.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75).Olive AJ, Kenjale R, Espina M, Moore DS, Picking WL, Picking WD. Bile salts stimulate recruitment of IpaB to the Shigella flexneri surface, where it colocalizes with IpaD at the tip of the type III secretion needle. Infect. Immun. 2007;75:2626–2629. doi: 10.1128/IAI.01599-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (76).Stensrud KF, Adam PR, La Mar CD, Olive AJ, Lushington GH, Sudharsan R, Shelton NL, Givens RS, Picking WL, Picking WD. Deoxycholate interacts with IpaD of Shigella flexneri in inducing the recruitment of IpaB to the type III secretion apparatus needle tip. J. Biol. Chem. 2008;283:18646–18654. doi: 10.1074/jbc.M802799200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (77).Prouty AM, Brodsky IE, Manos J, Belas R, Falkow S, Gunn JS. Transcriptional regulation of Salmonella enterica serovar Typhimurium genes by bile. FEMS Immunol. Med. Microbiol. 2004;41:177–185. doi: 10.1016/j.femsim.2004.03.002. [DOI] [PubMed] [Google Scholar]
- (78).Prouty AM, Gunn JS. Salmonella enterica serovar typhimurium invasion is repressed in the presence of bile. Infect. Immun. 2000;68:6763–6769. doi: 10.1128/iai.68.12.6763-6769.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79).Wang Y, Nordhues BA, Zhong D, De Guzman RN. NMR Characterization of the Interaction of the Salmonella Type III Secretion System Protein SipD and Bile Salts. Biochemistry. 2010;49:4220–4226. doi: 10.1021/bi100335u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (80).Miki T, Okada N, Shimada Y, Danbara H. Characterization of Salmonella pathogenicity island 1 type III secretion-dependent hemolytic activity in Salmonella enterica serovar Typhimurium. Microb. Pathog. 2004;37:65–72. doi: 10.1016/j.micpath.2004.04.006. [DOI] [PubMed] [Google Scholar]
- (81).Faudry E, Vernier G, Neumann E, Forge V, Attree I. Synergistic pore formation by type III toxin translocators of Pseudomonas aeruginosa. Biochemistry. 2006;45:8117–8123. doi: 10.1021/bi060452+. [DOI] [PubMed] [Google Scholar]
- (82).Neyt C, Cornelis GR. Insertion of a Yop translocation pore into the macrophage plasma membrane by Yersinia enterocolitica: Requirement for translocators YopB and YopD, but not LcrG. Mol. Microbiol. 1999;33:971–981. doi: 10.1046/j.1365-2958.1999.01537.x. [DOI] [PubMed] [Google Scholar]
- (83).Kaniga K, Tucker S, Trollinger D, Galan JE. Homologs of the Shigella IpaB and IpaC invasins are required for Salmonella typhimurium entry into cultured epithelial cells. J. Bacteriol. 1995;177:3965–3971. doi: 10.1128/jb.177.14.3965-3971.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (84).Menard R, Sansonetti PJ, Parsot C. Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J. Bacteriol. 1993;175:5899–5906. doi: 10.1128/jb.175.18.5899-5906.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (85).Frithz-Lindsten E, Holmstrom A, Jacobsson L, Soltani M, Olsson J, Rosqvist R, Forsberg A. Functional conservation of the effector protein translocators PopB/YopB and PopD/YopD of Pseudomonas aeruginosa and Yersinia pseudotuberculosis. Mol. Microbiol. 1998;29:1155–1165. doi: 10.1046/j.1365-2958.1998.00994.x. [DOI] [PubMed] [Google Scholar]
- (86).Menard R, Prevost MC, Gounon P, Sansonetti P, Dehio C. The secreted Ipa complex of Shigella flexneri promotes entry into mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 1996;93:1254–1258. doi: 10.1073/pnas.93.3.1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (87).Tucker SC, Galan JE. Complex function for SicA, a Salmonella enterica serovar typhimurium type III secretion-associated chaperone. J. Bacteriol. 2000;182:2262–2268. doi: 10.1128/jb.182.8.2262-2268.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (88).Page AL, Ohayon H, Sansonetti PJ, Parsot C. The secreted IpaB and IpaC invasins and their cytoplasmic chaperone IpgC are required for intercellular dissemination of Shigella flexneri. Cell. Microbiol. 1999;1:183–193. doi: 10.1046/j.1462-5822.1999.00019.x. [DOI] [PubMed] [Google Scholar]
- (89).Neyt C, Cornelis GR. Role of SycD, the chaperone of the Yersinia Yop translocators YopB and YopD. Mol. Microbiol. 1999;31:143–156. doi: 10.1046/j.1365-2958.1999.01154.x. [DOI] [PubMed] [Google Scholar]
- (90).Job V, Mattei PJ, Lemaire D, Attree I, Dessen A. Structural basis of chaperone recognition of type III secretion system minor translocator proteins. J. Biol. Chem. 2010;285:23224–23232. doi: 10.1074/jbc.M110.111278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (91).Hume PJ, McGhie EJ, Hayward RD, Koronakis V. The purified Shigella IpaB and Salmonella SipB translocators share biochemical properties and membrane topology. Mol. Microbiol. 2003;49:425–439. doi: 10.1046/j.1365-2958.2003.03559.x. [DOI] [PubMed] [Google Scholar]
- (92).Broms JE, Forslund AL, Forsberg A, Francis MS. Dissection of homologous translocon operons reveals a distinct role for YopD in type III secretion by Yersinia pseudotuberculosis. Microbiology. 2003;149:2615–2626. doi: 10.1099/mic.0.26322-0. [DOI] [PubMed] [Google Scholar]
- (93).Ryndak MB, Chung H, London E, Bliska JB. Role of predicted transmembrane domains for type III translocation, pore formation, and signaling by the Yersinia pseudotuberculosis YopB protein. Infect. Immun. 2005;73:2433–2443. doi: 10.1128/IAI.73.4.2433-2443.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (94).Shen DK, Saurya S, Wagner C, Nishioka H, Blocker AJ. Domains of the Shigella flexneri type III secretion system IpaB protein involved in secretion regulation. Infect. Immun. 2010;78:4999–5010. doi: 10.1128/IAI.00470-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (95).Roehrich AD, Martinez-Argudo I, Johnson S, Blocker AJ, Veenendaal AK. The extreme C terminus of Shigella flexneri IpaB is required for regulation of type III secretion, needle tip composition, and binding. Infect. Immun. 2010;78:1682–1691. doi: 10.1128/IAI.00645-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (96).Barta ML, Dickenson NE, Patil M, Keightley A, Wyckoff GJ, Picking WD, Picking WL, Geisbrecht BV. The structures of coiled-coil domains from type III secretion system translocators reveal homology to pore-forming toxins. J. Mol. Biol. 2012;417:395–405. doi: 10.1016/j.jmb.2012.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (97).Montagner C, Arquint C, Cornelis GR. Translocators YopB and YopD from Yersinia enterocolitica form a multimeric integral membrane complex in eukaryotic cell membranes. J. Bacteriol. 2011;193:6923–6928. doi: 10.1128/JB.05555-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (98).Goure J, Pastor A, Faudry E, Chabert J, Dessen A, Attree I. The V antigen of Pseudomonas aeruginosa is required for assembly of the functional PopB/PopD translocation pore in host cell membranes. Infect. Immun. 2004;72:4741–4750. doi: 10.1128/IAI.72.8.4741-4750.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (99).Marquart ME, Picking WL, Picking WD. Soluble invasion plasmid antigen C (IpaC) from Shigella flexneri elicits epithelial cell responses related to pathogen invasion. Infect. Immun. 1996;64:4182–4187. doi: 10.1128/iai.64.10.4182-4187.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (100).Picking WL, Coye L, Osiecki JC, Barnoski SA, Schaper E, Picking WD. Identification of functional regions within invasion plasmid antigen C (IpaC) of Shigella flexneri. Mol. Microbiol. 2001;39:100–111. doi: 10.1046/j.1365-2958.2001.02210.x. [DOI] [PubMed] [Google Scholar]
- (101).Hayward RD, Koronakis V. Direct nucleation and bundling of actin by the SipC protein of invasive Salmonella. EMBO J. 1999;18:4926–4934. doi: 10.1093/emboj/18.18.4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (102).Amer AA, Ahlund MK, Broms JE, Forsberg A, Francis MS. Impact of the N-terminal secretor domain on YopD translocator function in Yersinia pseudotuberculosis type III secretion. J. Bacteriol. 2011;193:6683–6700. doi: 10.1128/JB.00210-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (103).Tomalka AG, Stopford CM, Lee PC, Rietsch A. A translocator-specific export signal establishes the translocator-effector secretion hierarchy that is important for type III secretion system function. Mol. Microbiol. 2012;86:1464–1481. doi: 10.1111/mmi.12069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (104).De Geyter C, Vogt B, Benjelloun-Touimi Z, Sansonetti PJ, Ruysschaert JM, Parsot C, Cabiaux V. Purification of IpaC, a protein involved in entry of Shigella flexneri into epithelial cells and characterization of its interaction with lipid membranes. FEBS Lett. 1997;400:149–154. doi: 10.1016/s0014-5793(96)01379-8. [DOI] [PubMed] [Google Scholar]
- (105).Tran Van Nhieu G, Caron E, Hall A, Sansonetti PJ. IpaC induces actin polymerization and filopodia formation during Shigella entry into epithelial cells. EMBO J. 1999;18:3249–3262. doi: 10.1093/emboj/18.12.3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (106).Myeni SK, Zhou D. The C terminus of SipC binds and bundles F-actin to promote Salmonella invasion. J. Biol. Chem. 2010;285:13357–13363. doi: 10.1074/jbc.M109.094045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (107).Kueltzo LA, Osiecki J, Barker J, Picking WL, Ersoy B, Picking WD, Middaugh CR. Structure-function analysis of invasion plasmid antigen C (IpaC) from Shigella flexneri. J. Biol. Chem. 2003;278:2792–2798. doi: 10.1074/jbc.M208383200. [DOI] [PubMed] [Google Scholar]
- (108).Terry CM, Picking WL, Birket SE, Flentie K, Hoffman BM, Barker JR, Picking WD. The C-terminus of IpaC is required for effector activities related to Shigella invasion of host cells. Microb. Pathog. 2008;45:282–289. doi: 10.1016/j.micpath.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (109).Cornelis GR. The type III secretion injectisome. Nat. Rev. Microbiol. 2006;4:811–825. doi: 10.1038/nrmicro1526. [DOI] [PubMed] [Google Scholar]
- (110).Menard R, Sansonetti P, Parsot C, Vasselon T. Extracellular association and cytoplasmic partitioning of the IpaB and IpaC invasins of S. flexneri. Cell. 1994;79:515–525. doi: 10.1016/0092-8674(94)90260-7. [DOI] [PubMed] [Google Scholar]
- (111).Kim BH, Kim HG, Kim JS, Jang JI, Park YK. Analysis of functional domains present in the N-terminus of the SipB protein. Microbiology. 2007;153:2998–3008. doi: 10.1099/mic.0.2007/007872-0. [DOI] [PubMed] [Google Scholar]
- (112).Lokareddy RK, Lunelli M, Eilers B, Wolter V, Kolbe M. Combination of two separate binding domains defines stoichiometry between type III secretion system chaperone IpgC and translocator protein IpaB. J. Biol. Chem. 2010;285:39965–39975. doi: 10.1074/jbc.M110.135616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (113).Schreiner M, Niemann HH. Crystal structure of the Yersinia enterocolitica type III secretion chaperone SycD in complex with a peptide of the minor translocator YopD. BMC Struct. Biol. 2012;12:13. doi: 10.1186/1472-6807-12-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (114).Adam PR, Patil MK, Dickenson NE, Choudhari S, Barta M, Geisbrecht BV, Picking WL, Picking WD. Binding affects the tertiary and quaternary structures of the Shigella translocator protein IpaB and its chaperone IpgC. Biochemistry. 2012;51:4062–4071. doi: 10.1021/bi300243z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (115).Lunelli M, Lokareddy RK, Zychlinsky A, Kolbe M. IpaB-IpgC interaction defines binding motif for type III secretion translocator. Proc. Natl. Acad. Sci. U.S.A. 2009;106:9661–9666. doi: 10.1073/pnas.0812900106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (116).Buttner CR, Sorg I, Cornelis GR, Heinz DW, Niemann HH. Structure of the Yersinia enterocolitica Type III Secretion Translocator Chaperone SycD. J. Mol. Biol. 2008;375:997–1012. doi: 10.1016/j.jmb.2007.11.009. [DOI] [PubMed] [Google Scholar]
- (117).Hayward RD, Cain RJ, McGhie EJ, Phillips N, Garner MJ, Koronakis V. Cholesterol binding by the bacterial type III translocon is essential for virulence effector delivery into mammalian cells. Mol. Microbiol. 2005;56:590–603. doi: 10.1111/j.1365-2958.2005.04568.x. [DOI] [PubMed] [Google Scholar]
- (118).Epler CR, Dickenson NE, Olive AJ, Picking WL, Picking WD. Liposomes recruit IpaC to the Shigella flexneri type III secretion apparatus needle as a final step in secretion induction. Infect. Immun. 2009;77:2754–2761. doi: 10.1128/IAI.00190-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (119).Collazo CM, Galan JE. The invasion-associated type III system of Salmonella typhimurium directs the translocation of Sip proteins into the host cell. Mol. Microbiol. 1997;24:747–756. doi: 10.1046/j.1365-2958.1997.3781740.x. [DOI] [PubMed] [Google Scholar]
- (120).Thirumalai K, Kim KS, Zychlinsky A. IpaB, a Shigella flexneri invasin, colocalizes with interleukin-1β-converting enzyme in the cytoplasm of macrophages. Infect. Immun. 1997;65:787–793. doi: 10.1128/iai.65.2.787-793.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (121).Obregon C, Dreher D, Kok M, Cochand L, Kiama GS, Nicod LP. Human alveolar macrophages infected by virulent bacteria expressing SipB are a major source of active interleukin-18. Infect. Immun. 2003;71:4382–4388. doi: 10.1128/IAI.71.8.4382-4388.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]